The integral membrane protein stomatin inhibits the gating of acid-sensing ion channels by an unknown mechanism. Klipp et al. suggest that stomatin binds to the C-terminus of ASIC3 and stabilizes the desensitized state via an interaction with the channels’ first transmembrane domain.
Abstract
Stomatin (STOM) is a monotopic integral membrane protein found in all classes of life that has been shown to regulate members of the acid-sensing ion channel (ASIC) family. However, the mechanism by which STOM alters ASIC function is not known. Using chimeric channels, we combined patch-clamp electrophysiology and FRET to search for regions of ASIC3 critical for binding to and regulation by STOM. With this approach, we found that regulation requires two distinct sites on ASIC3: the distal C-terminus and the first transmembrane domain (TM1). The C-terminal site is critical for formation of the STOM–ASIC3 complex, while TM1 is required only for the regulatory effect. We then looked at the mechanism of STOM-dependent regulation of ASIC3 and found that STOM does not alter surface expression of ASIC3 or shift the pH dependence of channel activation. However, a point mutation (Q269G) that prevents channel desensitization also prevents STOM regulation, suggesting that STOM may alter ASIC3 currents by stabilizing the desensitized state of the channel. Based on these findings, we propose a model whereby STOM is anchored to the channel via a site on the distal C-terminus and stabilizes the desensitized state of the channel via an interaction with TM1.
Introduction
Acid-sensing ion channels (ASICs) are members of the degenerin/epithelial Na+ channel family of ion channels. ASICS are Na+-selective, voltage-insensitive channels that are activated by extracellular protons. There are five ASIC isoforms that give rise to at least seven distinct channel subunits, which can form both heteromeric or homomeric trimers (Hesselager et al., 2004; Jasti et al., 2007). ASIC1a, ASIC1b, ASIC2a, and ASIC3 all form functional pH-sensitive channels as homotrimers, while ASIC2b and ASIC4 homotrimers are not gated by protons (Akopian et al., 2000; Gründer et al., 2000; Lingueglia et al., 1997). ASICs are expressed throughout the central and peripheral nervous systems, where they are thought to play a role in a range of physiological and pathophysiological functions, including nociception, fear conditioning, neuronal death following ischemia, baroreception and autonomic control of circulation, and sensing myocardial ischemia (Benson et al., 1999; Jones et al., 2004; Lu et al., 2009; Ugawa et al., 2002; Wemmie et al., 2002; Xiong et al., 2004). Like many of the ion channels of the sensory system, ASICs are multimodal receptors; in addition to activation by protons, ASICs are regulated by lipids, phosphorylation, numerous extracellular ligands, and accessory proteins (for a review, see Boscardin et al., 2016).
A number of high-resolution structures of ASIC1a from chicken have been solved (Baconguis et al., 2014; Baconguis and Gouaux, 2012; Dawson et al., 2012; Jasti et al., 2007; Yoder et al., 2018). These structures have provided hypotheses for how protons and toxins derived from animal venoms might act on the extracellular domain of the channel and lead to opening and closing of the gate. However, in each structure, the intracellular domains are either missing from the protein or not resolved in the structure. Consequently, little is known about how the intracellular termini contribute to channel function and how proteins that interact in this region might impact channel function.
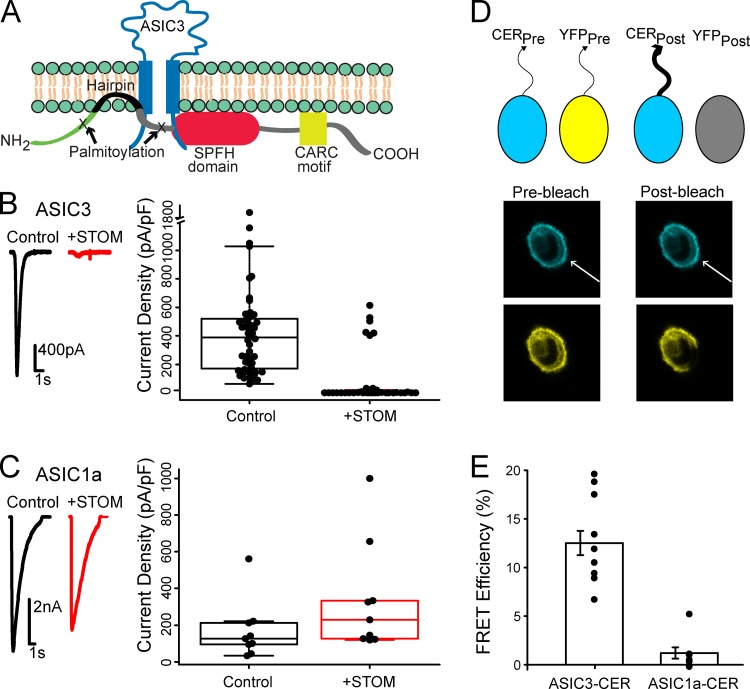
Stomatin (STOM) is a 31.5-kD monotopic integral membrane protein ubiquitously found throughout the central and peripheral nervous system. STOM is associated with the cytoplasmic face of the plasma membrane via a short hydrophobic hairpin region and a number of palmitoylation sites (Snyers et al., 1999; Fig. 1 A). In addition, STOM contains a STOM, Prohibitin, Flotillin, and HflK/C (SPFH) domain and at least one cholesterol-binding site and is frequently localized to cholesterol-rich lipid rafts. In humans, there are four proteins closely related to STOM: STOM-like (STOML) proteins 1–3 (STOML1, STOML2, and STOML3) and a protein important for proper filtration in the kidney called Podocin (for a review, see Browman et al., 2007). In addition, STOM is part of a superfamily of proteins that contain a conserved SPFH domain. STOM has previously been shown to regulate several membrane proteins, including the glucose transporter GLUT-1, the anion exchanger AE-1, and ASICs (Brand et al., 2012; Genetet et al., 2017; Moshourab et al., 2013; Price et al., 2004; Zhang et al., 2001).
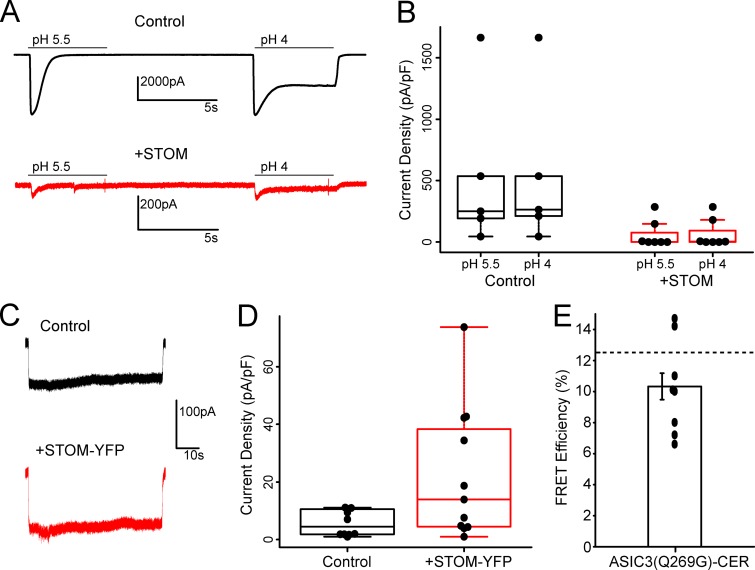
Figure 1.
STOM inhibits ASIC3 via a direct interaction. (A) Topological cartoon of STOM interaction with ASIC3. Shown are several of STOM's topological features that influence its localization and interaction with membrane proteins including: palmitoylation sites, a hydrophobic hairpin, an SPFH domain, and a cholesterol recognition motif (CARC). (B) Left: Representative pH 5.5–evoked ASIC3 currents alone (black) and cotransfected with STOM (red). Right: Plot shows every measurement made for the two conditions plotted as the current density (peak current amplitude/cell capacitance) superimposed onto a boxplot that summarizes the data. Average current densities of the control and +STOM conditions were 364.0 ± 32.6 pA/pF (n = 45) and 2.0 ± 0.4 pA/pF (n = 27), respectively. (C) Identical experiments as in B performed for ASIC1a. Average current densities of ASIC1a in the absence (black) and presence (red) of STOM were 122.5 ± 22.9 pA/pF (n = 8) and 200.3 ± 33.7 pA/pF (n = 7), respectively. (D) Cartoon showing FRET photobleaching assay. Bottom panel shows a representative cell where the YFP was bleached, which led to a corresponding increase in CER intensity. The displayed images are 36 x 36 μm. (E) FRET efficiency measured between ASIC3-CER and STOM-YFP and ASIC1a-CER and STOM-YFP. FRET efficiencies for ASIC3-CER and ASIC1a-CER were 12.5 ± 1.2% (n = 11) and 1.2 ± 0.6% (n = 8), respectively. All data are given as mean ± SEM.
Previous work showed that recombinant expression of STOM into mammalian cells decreased ASIC3 current magnitude and increased the speed of ASIC2a desensitization but had no effect on ASIC1a (Price et al., 2004). STOML proteins have also been shown to regulate ASICs in an isoform-dependent manner. A STOM homologue, MEC-2, is essential for the function of the mechanosensitive ASIC homologue MEC-4–MEC-10 complex in Caenorhabditis elegans (Brown et al., 2008; Goodman et al., 2002; Huang et al., 1995). Despite the ubiquity of this family of proteins, little is known about the mechanisms through which STOM and its family members regulate ion channels.
Here, we paired patch-clamp electrophysiology with FRET to localize the binding site for STOM on ASIC3. Making chimeric channels between ASIC3, which is regulated by STOM, and ASIC1a, which is not regulated, allowed us to localize two sites on ASIC3 that are critical for STOM-dependent regulation. First, we found that the distal C-terminus of the channel was necessary for both complex formation and regulation of the channel. Second, the first transmembrane domain (TM1) of ASIC3, while not sufficient for binding, was necessary for STOM-dependent regulation. In addition, we examined the mechanism of STOM-dependent regulation of ASIC3. We used fluorescence imaging, a surface biotinylation assay, and patch-clamp recording to examine three potential hypotheses for how ASIC3 currents are reduced by STOM. These data suggest that surface expression and activation of the channel are not impacted by STOM. However, a point mutation in the extracellular domain that nearly eliminates desensitization also prevented STOM-dependent regulation of ASIC3. Taken together, this led us to a model whereby STOM is anchored to ASIC3 via an interaction site on the distal C-terminus and potentially acts to stabilize the desensitized state through a second interaction with TM1 of the channel. These results extend our understanding of the STOM–ASIC3 complex and may shed light on how the SPFH domain proteins regulate ASICs more generally.
Materials and methods
Mutagenesis
Plasmids for ASIC1a, ASIC2a, and ASIC3 each from rat were gifts from David Julius (University of California, San Francisco, San Francisco, CA) and subcloned into our pcDNA3.1 vector. STOM from mouse was purchased from Dharmacon/Horizon Discovery in a mammalian expression pCMV vector. The mouse and rat STOM share 96% sequence identity and mouse and rat ASIC3 share 97% identity. This combination (rat ASIC3 and mouse STOM) has been used previously (Brand et al., 2012). Chimeric channels were created using Gibson Assembly. Sanger sequencing was used to verify correct sequences for all constructs in this study (AGCT). For our fluorescently labeled ASIC variants, a short proline-rich linker was used to join our fluorophore to the C-terminus of the indicated ASIC isoform. We tried multiple naturally occurring linkers found in the SynLinker database (synlinker.syncti.org) and found that a short linker from the α subunit of DNA polymerase worked well (ILPLPYPNSPV; Liu et al., 2015). Importantly, we found that STOM was unable to regulate ASIC3 when a fluorescent protein was attached directly to the C-terminus of ASIC3. Fluorescently labeled STOM was constructed by joining the fluorophore directly to STOM’s C-terminus. Fluorophores used are indicated throughout and include mCerulean3 (CER), EGFP (GFP), EYFP (YFP), mTurquoise (TUR), and TagRFP (RFP).
Cell lines and transfection
CHO-K1 cells (ATCC) were cultured in Ham’s F12 media with 10% FBS at 37°C in 5% CO2. Cells at ∼70% confluency were transfected via electroporation with a Lonza 4D Nucleofector unit following the manufacturer’s protocol. Plasmid DNA (1 µg) encoding for our WT and mutant rat ASIC3, ASIC2a, or ASIC1a proteins in the presence or absence of plasmid DNA (3 µg) encoding for mouse STOM was used for transfection. Nonfluorescent ASICs were also transfected with free Citrine plasmid DNA (0.1 µg) to identify cells containing transfected DNA. Following transfection, cells were plated on 12-mm glass coverslips coated in poly-L-lysine.
Biotinylation assay and Western blotting
Biotinylation of plasma membrane proteins was performed using a slightly modified protocol from a commercially available cell surface protein isolation kit (BioVision). CHO-K1 cells were first transfected with ASIC3-TUR or ASIC1a-CER with and without unlabeled STOM. Approximately 18 h after transfection, cells were quick-washed in ice-cold PBS followed by incubation with Sulfo-NHS-SS-Biotin at 4°C with gentle shaking. After 1 h of biotin labeling, reaction was quenched, and cells were scraped and collected followed by centrifugation at 1,000 ×g for 5 min. Cells were washed twice in Tris-buffered saline (TBS) buffer followed by centrifugation at 1,000 ×g for 5 min. Pellet was collected, and cells were resuspended in RIPA buffer for 1 hr at 4°C with end-over-end mixing (in mM): 150 NaCl, 50 Tris, 1% NP-40, 0.5% sodium deoxycholate, and 1X protease inhibitor added before lysis (Thermo Fisher Scientific, Pierce). Lysed cells were centrifuged at 10,000 ×g for 7 min, and supernatant was transferred onto packed streptavidin beads and incubated with end-over-end mixing for 1 h at room temperature. A portion of each sample was also collected before loading onto beads to quantify total protein concentration. Beads were centrifuged at 800 ×g for 60 s, and supernatant was collected as the non–biotin-bound (intracellular) fraction. Beads were washed three times in TBS followed by centrifugation at 800 ×g for 60 s. Biotin-labeled protein bound to streptavidin beads was eluted by incubating beads with 100 mM DTT for 1 h at room temperature. Sample was centrifuged at 800 ×g for 60 s, and supernatant was collected as biotinylated (plasma membrane) protein.
Samples that were collected just after lysis were measured for the total protein concentration using a Bicinchoninic Acid (BCA) assay kit (Thermo Fisher Scientific, Pierce). Both the biotinylated and intracellular fractions were normalized to total protein and loaded on a 4–12% Bis-Tris precast gel (Thermo Fisher Scientific, Invitrogen) and run at 200 V for 30 min. Protein was transferred from gel to a PVDF membrane at 100 V for 1 hr. Membrane was incubated in blocking buffer for 1 h followed by overnight incubation in primary antibody (purified rabbit anti-GFP; Torrey Pines Biolabs) at 4°C. Membrane was then washed six times with TBS-T (Tris-buffered saline + Tween20) followed by 1-h incubation with secondary antibody (goat anti-rabbit IgG; KPL). Membrane was washed another six times in TBS-T and then developed in the dark for 5 min with chemiluminescent reagent (Immobilon Forte; Millipore).
Electrophysiological recordings
All experiments were performed in the whole-cell patch-clamp configuration 16–48 h after transfection as described earlier. Borosilicate glass pipettes (Harvard Apparatus) pulled to a resistance of 2–6 MΩ (P-1000; Sutter Instrument) and filled with an internal solution containing (in mM): 20 EGTA, 10 HEPES, 50 CsCl, 10 NaCl, and 60 CsF, pH 7.2. Extracellular solution contained (in mM): 110 NaCl, 5 KCl, 40 NMDG, 10 MES, 10 HEPES, 5 glucose, 10 Trizma base, 2 CaCl2, and 1 MgCl2, and pH was adjusted as desired with HCl or NaOH. An Axopatch 200B amplifier and pCLAMP 10.6 (Axon Instruments) were used to record whole-cell currents. Recordings were performed at a holding potential of −80 mV with a 5-kHz low-pass filter and sampling at 10 kHz. Channel activation was performed via a rapid change in solution from a resting pH 8.0 to pH 5.5 (unless indicated otherwise) for 5 s. Following activation of the channel by pH 5.5 solution, pH was returned to the resting pH (8.0) for 9 s, and protocol was repeated for a total of six activations. Rapid perfusion was achieved using a SF-77B Fast-Step perfusion system (Warner Instruments). Fluorescence was visualized on an Olympus IX73 microscope (Olympus) with a CoolLED pE-4000 illumination system (CoolLED).
FRET
Cells expressing fluorescent ASIC3 and STOM were examined 16–40 h after transfection using the confocal laser scanning microscope LSM 710 (Zeiss). The same external solutions used for our patch-clamp recordings were used for imaging. An area of 500–2,500 µm2 was selected from the overall field of view. Images were taken through a 40× oil objective with a numerical aperture of 1.3. CER and YFP were excited with separate sweeps of the 458- and 514-nm laser lines of an argon laser directed at the cell with a 458/514-nm dual dichroic mirror. Relative to full power, the excitation power for the imaging sweeps was attenuated to 1% for CER and 0.5% for YFP. Bleaching was performed by using multiple (20–60) sweeps of the YFP laser at full power. Bleaching was usually complete within 30–90 s. Emitted light was collected between 460 and 496 nm for CER and 526 and 579 nm for YFP. With this setup, there was no contamination of the relevant CER signal from the YFP. For each experiment, the photomultiplier tube gain was adjusted to ensure that the maximum pixel intensity was not >70% saturated. Fluorescence intensity was then measured by drawing regions of interest (ROIs) around the cell in ImageJ (Schneider et al., 2012). Masks were used to eliminate bright fluorescent puncta within the cell. This was a rare occurrence in the CER signal. We also made measurements with ROIs that, to the best of our ability, only surrounded the plasma membrane. This approach did not change the results. Percent FRET (E) was calculated as
where ICERpost is the CER intensity after bleaching and ICERpreis the CER intensity before bleaching.
Confocal imaging
ASIC3-TUR and a protein serving as a membrane marker were coexpressed in CHO-K1 cells with and without STOM-YFP and examined using a Zeiss LSM 710 confocal microscope. The membrane marker protein used was a TagRFP-labeled portion of the L-type calcium channel, consisting of the I-II loop of Cav1.2, joined to the N-terminus of Cav1.1 that has previously been shown to associate with the plasma membrane (Kaur et al., 2015; Polster et al., 2018). Excitation and emission for the fluorescent proteins were TUR (excitation 458 nm, emission 460–496 nm), YFP (excitation 514 nm, emission 530–565 nm), and TagRFP (excitation 543 nm, emission 582–754 nm). Relative to full power output, the excitation was attenuated to ∼2.5–5% (458 nm), ∼5% (514 nm), and ∼5–8% (543 nm). Images were obtained with a 40× (1.3 numerical aperture) oil-immersion objective as a single, midlevel optical slice that was halfway between the lower and upper cell surface for CHO-K1 cells.
Data analysis and statistics
Whole-cell patch-clamp current recordings were analyzed with Clampfit 10.6 software (Axon Instruments). Desensitization rates for most currents were fitted well with a single exponential using a homemade MATLAB script (MathWorks). These data are reported in Table S1. Currents were normalized to cell capacitance, and the raw current densities and box plot were plotted for each condition using R software (R Core Team, 2017). All data points collected were plotted, and those points that are >1.5 times the boxplot interquartile range (IQR) were plotted as outliers. Data reported throughout are calculated as the mean ± the standard error (SE) excluding the outliers. Means were also calculated with outliers in supplement Table S1, and statistical significance for both conditions were calculated using the Mann–Whitney–Wilcoxon test.
Results
STOM binds to ASIC3 and inhibits current magnitude
To begin to understand how STOM regulates ASIC3 currents, we sought to localize the regions on ASIC3 that are critical for both binding and the regulatory effect. To do this, we combined patch-clamp electrophysiology to measure the functional effect of STOM on ASIC3 with FRET to measure binding of STOM to the channel.
Previous reports have suggested that STOM bound to ASIC3 and dramatically reduced channel currents (Price et al., 2004). We first sought to confirm these original findings. Whole-cell patch-clamp recordings on Chinese hamster ovary (CHO) K1 cells expressing ASICs were performed by rapidly switching between solutions at pH 8.0 and pH 5.5 using a stepper motor-driven solution exchange system. Representative current traces show that control ASIC3 acid evoked currents (black) are drastically reduced in the presence of STOM (red; Fig. 1 B). Boxplot summary of the data show that STOM reduced ASIC3 mean current density from 364.0 ± 32.6 pA/pF (n = 45) to 1.96 ± 0.44 pA/pF (n = 27; Fig. 1 B). Performing the same experiment for cells expressing ASIC1a with and without STOM also confirmed previous findings that ASIC1a was not functionally regulated by STOM (Fig. 1 C). In 6 out of the 35 recordings, ASIC3 displayed control-like current densities even in the presence of STOM. This is likely an artifact of our transient transfection system where STOM may be either absent or weakly expressing in these cells. Given these outliers, we calculated the mean current density for every experiment in this study in two ways. First, we averaged all the data collected. Second, we calculated the adjusted mean, excluding outliers that were in excess of 1.5*IQR, where IQR is the interquartile range of the data. We will use this adjusted mean to discuss the data, but both calculations appear in Table S1. Additionally, all data, including outliers, are shown throughout in the boxplots.
We next wanted to develop an approach that would allow us to measure direct interaction between ASIC3 and STOM. To do this, we used acceptor photobleaching FRET. We tagged ASIC3 with a C-terminal Cerulean (CER) and STOM with a C-terminal YFP and recorded two cyan (in response to weak 458-nm excitation) and two yellow (in response to weak 514-nm excitation) images. Subsequently, the cell was subjected to repeated illuminations with high-intensity 514-nm light, which bleached YFP but had no effect on CER. Finally, two more images were measured in each color using the same conditions as before bleaching the YFP. Because of the near-total bleaching of YFP, if the two fluorophores are within ∼70 nm of each other, then the postbleaching CER signal will be larger due to loss of resonant energy being donated to the YFP. A cartoon of the approach and a representative set of images illustrating this method, one before and one after bleaching, can be seen in Fig. 1 D. Using this approach, we measured an average FRET efficiency between ASIC3-CER and STOM-YFP of 12.5 ± 1.2% (n = 11; Fig. 1 E), suggesting that these two proteins were interacting. Previous reports suggested that STOM could bind ASIC1a as well, but our FRET assay did not detect interaction between the two molecules. Cells coexpressing only free CER and YFP showed no change in CER intensity between the pre- and postbleaching images (data not shown). To ensure bleaching of the CER signal was minimal during the measurement, we used very low laser power to excite the CER. Overall, the average decrease in CER intensity from the first to the final image in cells where only ASIC3-CER was expressed was 0.8 ± 0.3% (n = 7). This suggested to us that bleaching made only a minimal contribution to the results. With these data, we could now pair our functional measurement with our FRET measurement to attempt to localize sites on ASIC3 that are critical for binding STOM and sites that are critical for the functional regulation of ASIC3 by STOM.
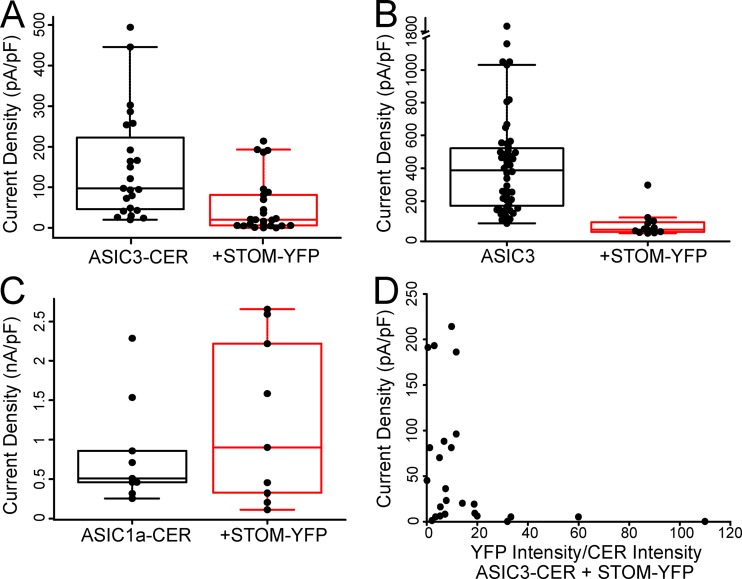
To confirm that our fluorescent labels did not prevent STOM inhibition, we also performed functional assays identical to those in Fig. 1 using our fluorescently labeled proteins. Although inhibition of ASIC3 was maintained, we did observe that the magnitude of STOM inhibition of ASIC3 was diminished in the presence of the fluorophores (Fig. S1 A). Compared with the control, mean current density decreased approximately threefold when ASIC3-CER was expressed with STOM-YFP. We believe this reduced STOM effect occurs due to the YFP tag on STOM, because tagged ASIC3 was fully regulated by an untagged STOM (see Fig. 6 D) and STOM-YFP also showed a reduced inhibition of untagged ASIC3 (Fig. S1 B). ASIC1a-CER currents were not affected by STOM (Fig. S1 C). The YFP on the C-terminus of STOM could reduce the effect on ASIC3 in several ways. First, the STOM-YFP may show lower expression in our transient transfection system. Second, the presence of the fluorophore on the C-terminus of the channel allowed for us to select the brightest cells in each of the control and +STOM-YFP cases. This may have caused us to select cells where there was not enough STOM-YFP to fully regulate all of the ASIC3 in the cells. Consistent with each of these ideas, it appears from our data that cells with higher levels of STOM-YFP can inhibit ASIC3-CER as well as in the untagged case (Fig. S1 D). Lastly, the YFP may simply interfere with the ability of STOM to regulate ASIC3. Even with this possibility, STOM-YFP clearly interacted with ASIC3-CER and significantly reduced currents.
Figure S1.
Data examining the effect of fluorescent tags on the regulation of ASIC3 by STOM. (A) Boxplots showing the current density measurements for ASIC3-CER with and without STOM-YFP. Average current densities of ASIC3-CER in the absence (black) and presence (red) of STOM-YFP were 137.0 ± 23.5 pA/pF (n = 22) and 47.1 ± 12.4 pA/pF (n = 24), respectively. (B) Current densities of the untagged ASIC3 in the absence (black; replotted from Fig. 1 B) and presence of STOM-YFP. Average current densities were 364.0 ± 32.6 pA/pF (n = 45) and 34.3 ± 9.1 pA/pF (n = 11), respectively. (C) Boxplots showing average current densities of ASIC1a in the absence (black) and presence (red) of STOM-YFP, which were 509.7 ± 74.2 pA/pF (n = 7) and 1,227.5 ± 329.8 pA/pF (n = 9), respectively. (D) Current densities of ASIC3-CER with STOM-YFP from Fig. 3 C plotted as a function of the ratio of fluorescence intensity of YFP/CER. These data show that after about a 15:1 ratio of YFP/CER, the magnitude of the regulation of ASIC3-CER was as large as in the untagged STOM case. Data are given as mean ± SEM.
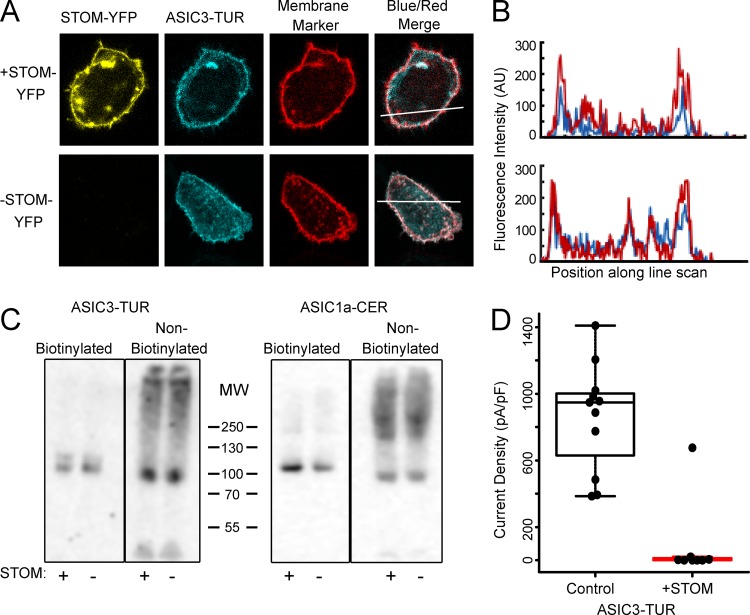
Figure 6.
STOM does not alter ASIC3 expression on the plasma membrane. (A) Confocal images of cells expressed with ASIC3-TUR, membrane marker (TagRFP-labeled I-II loop of Cav1.2 with Cav1.1 N-terminus), with (top) and without STOM-YFP (bottom). The displayed images are 30 x 36 μm. (B) Line scan of the blue and red signals from A (indicated by the bar) shows that ASIC3-TUR peak intensity overlaps with membrane marker on the plasma membrane in the presence and absence of STOM-YFP. This experiment was repeated for an n = 10. (C) Western blot against the fluorescent tag of ASIC3-TUR and ASIC1a-CER with and without STOM after performance of a cell surface biotinylation assay. (D) Boxplot showing the current density measured from pH 5.5–evoked currents from ASIC3-TUR with and without STOM. Current density for control and +STOM were 859.2 ± 94.1 pA/pF (n = 11) and 4.6 ± 2.7 pA/pF (n = 7), respectively. Data are given as mean ± SEM.
STOM inhibition of ASIC3 requires the C-terminus
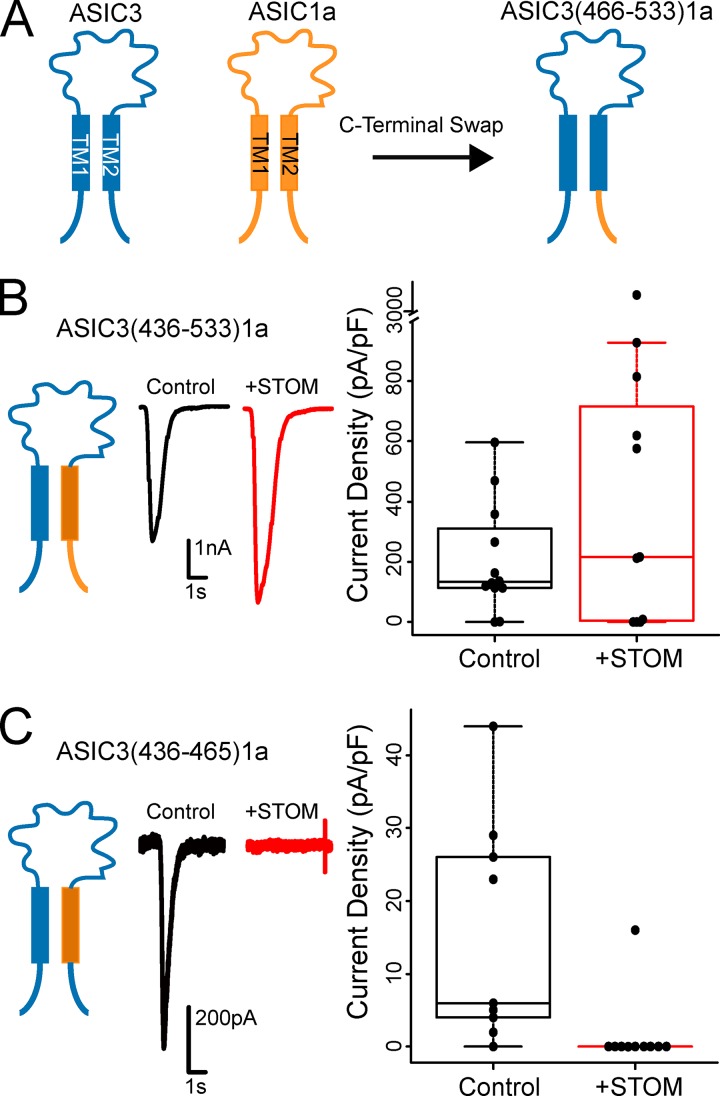
Having measured a direct interaction between ASIC3 and STOM, we then sought to identify the specific sites on ASIC3 that are critical for STOM regulation. To do this, we systematically created chimeras where portions of ASIC1a replaced the corresponding residues of ASIC3. For naming of the chimeras used in this study, we first indicate the isoform backbone followed by the backbone’s residue numbers being swapped for the corresponding residues of the other ASIC isoform (Fig. 2 A). The protein alignment and domains are given in Fig. S2. With these chimeric channels, we measured both the functional effect of coexpressing STOM as well as FRET between STOM and ASIC3. This allowed us to look for sites on ASIC3 that when mutated altered binding, regulation, or both. For these first experiments, we used the untagged versions of both the channel and STOM because of the large effect and then used the tagged versions for the FRET measurements. Given the topology of STOM (Fig. 1 A), we reasoned that the binding site for STOM on ASIC3 must involve the transmembrane domains and/or the intracellular termini.
Figure 2.
STOM inhibition of ASIC3 involves the C-terminus, but not TM2. (A) Cartoon showing swap of ASIC3's C-terminus (residues 466–533) with ASIC1a's C-terminus (residues 459–526) is named ASIC3(466–533)1a. (B and C) Left: Cartoon depicting the chimera being examined. Middle: Representative pH 5.5–evoked currents of the indicated chimeric channel alone (black) and cotransfected with STOM (red). Right: Boxplots of the current density measurements for the chimeric channel with and without STOM. Average current densities for ASIC3(436–533)1a in the absence (black) and presence (red) of STOM were 205.3 ± 50.6 pA/pF (n = 12) and 337.1 ± 109.0 pA/pF (n = 10), respectively. Average current densities for ASIC3(436–465)1a in the absence (black) and presence (red) of STOM were 15.4 ± 4.9 pA/pF (n = 9) and STOM mean 0 ± 0 pA/pF (n = 9), respectively. Data are given as mean ± SEM.
Figure S2.
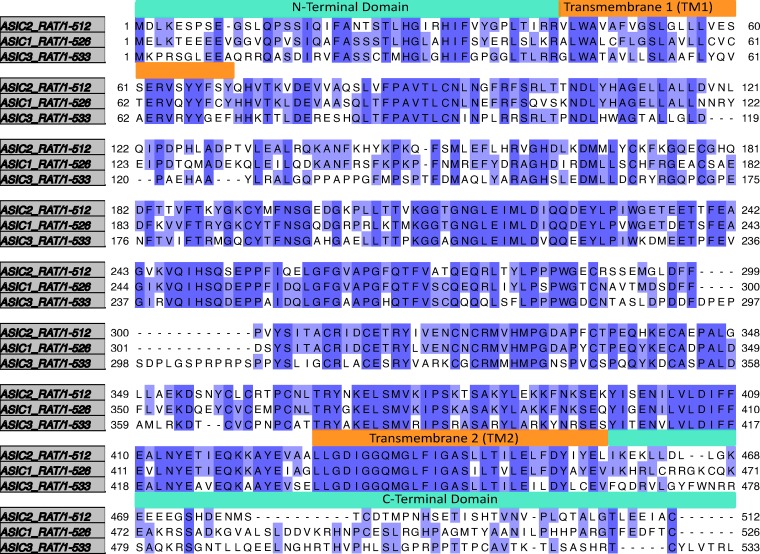
Sequence alignment of rat ASIC3, rat ASIC1a, and rat ASIC2a. Sequence alignment generated in Jalview v.2.10.5 using Mafft under default settings. Dark purple regions show residues that are identical between the three proteins. Light purple shows residues that are identical in two of the proteins. Orange boxes are present above the residues that make up the transmembrane domains. Cyan boxes are present above the residues that comprise the N- and C-terminus.
We first investigated the C-terminus and second transmembrane domain (TM2) of ASIC3. Swap of TM2 and the C-terminus of ASIC3 with the corresponding residues of ASIC1a, ASIC3(436–533)1a, eliminated STOM’s functional inhibition, suggesting that this region is necessary for regulation (Fig. 2 B). Replacement of TM2 alone, ASIC3(436–465)1a, resulted in a chimeric channel that expressed poorly. However, when we coexpressed STOM, there was a clear and dramatic reduction in current, which suggested that TM2 likely does not play a role in STOM-dependent regulation of ASIC3 (Fig. 2 C).
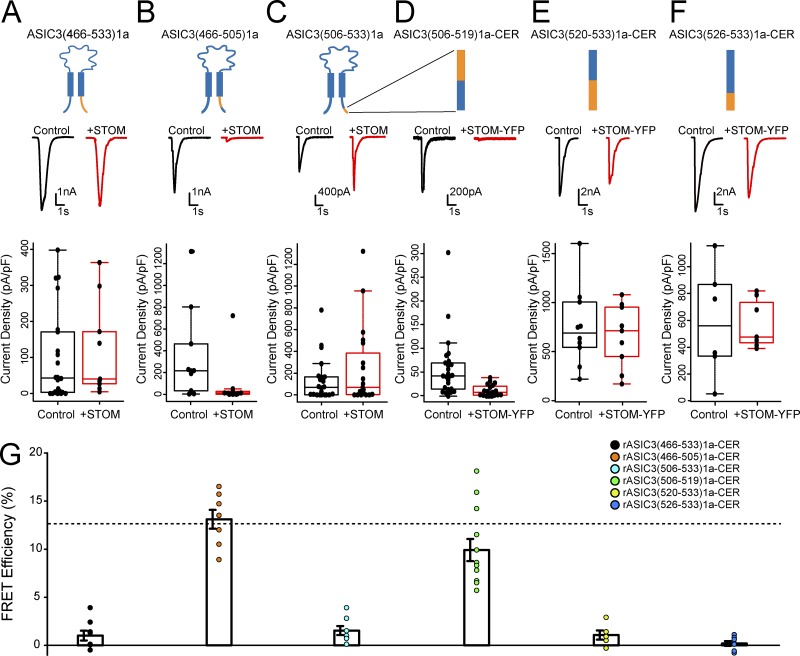
Together, these data suggested that the C-terminus is a critical determinant of the STOM-dependent regulation of ASIC3. To test this, we broke down the C-terminus of ASIC3 further. Replacing the entire ASIC3 C-terminus with that of ASIC1a, ASIC3(466–533)1a, eliminated STOM’s functional inhibition of ASIC3 (Fig. 3 A). FRET measurements between ASIC3(466–533)1a-CER and STOM-YFP indicated that this loss of functional interaction was associated with loss of binding (Fig. 3 G). Further breakdown of this region indicated that replacement of the first 40 residues of the C-terminus of ASIC3, ASIC3(466–505)1a, did not disrupt STOM inhibition, and this chimera still showed a robust FRET with STOM-YFP (Fig. 3, B and G). However, swap of the final 28 residues of the C-terminus, ASIC3(506–533)1a, eliminated STOM inhibition as well as STOM binding to ASIC3 (Fig. 3, C and G). These data point to the distal C-terminus as a critical determinant for the function and interaction of the STOM–ASIC3 complex.
Figure 3.
STOM binding at the distal C-terminus of ASIC3 is necessary for inhibition. (A–F) Each column shows the cartoon of the chimera being tested. In the case of D–F, the cartoon shows a blow up of the distal C-terminus. In addition, each column has a representative set of pH 5.5–evoked currents of the indicated chimeric channel alone (black) and cotransfected with STOM (red). Finally, each column shows a boxplot of the current density of the channel with and without STOM coexpression. (A) Average current densities for ASIC3(466–533)1a in the absence (black) and presence (red) of STOM were 99.5 ± 26.2 pA/pF (n = 22) and 121.4 ± 41.7 pA/pF (n = 9), respectively. (B) Average current densities for ASIC3(466–505)1a in the absence (black) and presence (red) of STOM were 239.9 ± 80.3 pA/pF (n = 9) and 13.4 ± 5.1 pA/pF (n = 9), respectively. (C) Average current densities for ASIC3(506–533)1a in the absence (black) and presence (red) of STOM were 69.6 ± 18.7 pA/pF (n = 19) and 193.1 ± 61.8 pA/pF (n = 18), respectively. (D) Average current densities of ASIC3(506–519)1a-CER in the absence (black) and presence (red) of STOM-YFP were 42.6 ± 6.3 pA/pF (n = 23) and 12.0 ± 2.7 pA/pF (n = 19), respectively. (E) Average current densities of ASIC3(520–533)1a-CER in the absence (black) and presence (red) of STOM-YFP were 750.1 ± 118.7 pA/pF (n = 10) and 661.9 ± 101.2 pA/pF (n = 9), respectively. (F) Average current densities of ASIC3(526–533)1a-CER in the absence (black) and presence (red) of STOM-YFP were 586.3 ± 152.7 pA/pF (n = 6) STOM-YFP mean 572.9 ± 64.2 pA/pF (n = 7), respectively. (G) Plot showing the FRET efficiency for each chimeric construct. Average FRET efficiency for ASIC3(466–533)1a-CER + STOM-YFP = 1.0 ± 0.5% (n = 8). Average FRET efficiency for ASIC3(466–505)1a-CER and STOM-YFP = 13.1 ± 1.0% (n = 7). Average FRET efficiency for ASIC3(506–533)-CER + STOM-YFP = 1.5 ± 0.5% (n = 7). Average FRET efficiency for ASIC3(506–519)1a-CER + STOM-YFP = 9.9 ± 1.1% (n = 12). Average FRET efficiency for ASIC3(520–533)1a-CER + STOM-YFP = 1.1 ± 0.5% (n = 5). Average FRET efficiency for ASIC3(526–533)1a-CER + STOM-YFP = 0.2 ± 0.3% (n = 7). Dotted line in FRET plot corresponds to control WT ASIC3-CER + STOM-YFP FRET signal replotted from Fig. 1 E for comparison. Data are given as mean ± SEM.
We then systematically replaced smaller segments of the ASIC3 C-terminus with the corresponding residues from ASIC1a to further narrow down the residues important for the interaction. The rectangular cartoons above the data in Fig. 3, D–F, provide visual representations of the chimeric channels that we made between residues 506 and 533. Several of these chimeras exhibited poor transfection efficiency, creating difficulties in selecting cells that consistently exhibited pH-evoked currents. In these cases, we elected to use our fluorescently labeled ASIC3 and STOM for both the functional measurements and the FRET measurements. First, we split the final 28 residues into two chimeras, ASIC3(506–519)1a and ASIC3(520–533)1a. The more proximal chimera, ASIC3(506–519)1a-CER, was still regulated by STOM-YFP and was also still able to bind STOM-YFP (Fig. 3, D and G). However, the more distal chimera, ASIC3(520–533)1a-CER, was not functionally regulated and failed to show any appreciable FRET with STOM-YFP (Fig. 3, E and G). Finally, we made a chimera where the final 8 amino acids of the channel were mutated to their counterparts in ASIC1a, ASIC3(526–533)1a-CER, and both STOM-YFP regulation and binding were lost (Fig. 3, F and G). This approach was able to localize a critical binding site on ASIC3 for STOM to the final eight residues of the channel. Previous work has shown that ASIC3 has a PDZ-binding motif in this region that is critical for binding of a number of other proteins, including Lin-7B, CIPP, and PSD-95 (Anzai et al., 2002; Eshcol et al., 2008; Hruska-Hageman et al., 2004). Lin-7B and PSD-95 appear to alter ASIC3 surface expression, while CIPP shifts the pH dependence of the channel in the basic direction. Interestingly, STOM appears to bind to this same region but lacks a PDZ domain.
STOM requires TM1 to fully regulate ASIC3
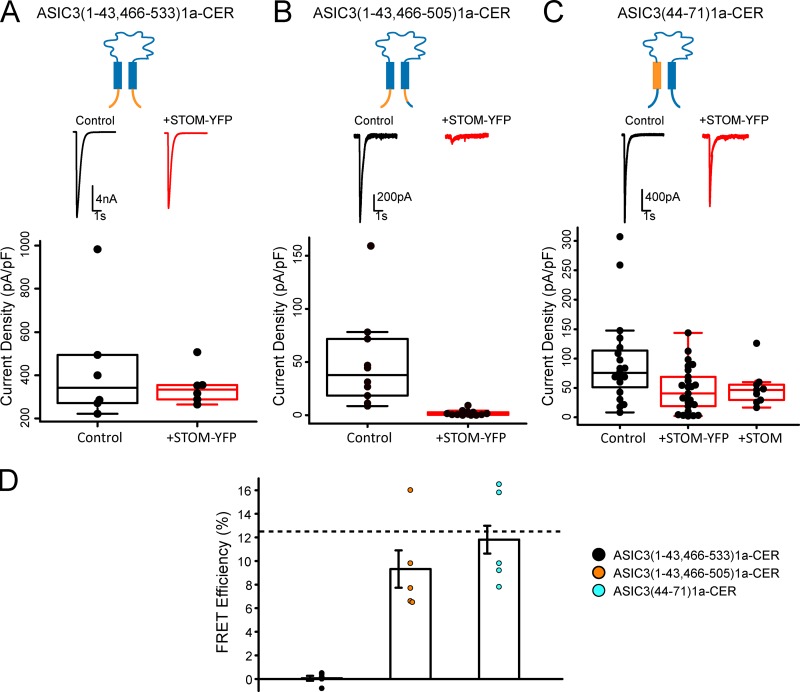
Although swap of distal C-terminus of ASIC3 with the corresponding residues of ASIC1a is sufficient to eliminate STOM functional inhibition, it did not exclude the possibility that STOM interaction could also involve the N-terminus or TM1. To test this, we attempted to make a number of chimeric channels on the N-terminal side of the channel. Insertion of N-terminus of ASIC1a into ASIC3 (residues 1–43) or the N-terminus and a portion of TM1 (residues 1–55) both resulted in nonfunctional channels, consistent with previous reports (Salinas et al., 2009).
However, this same report showed that simultaneous swap of both the N- and C-termini resulted in functional channels (Salinas et al., 2009). Therefore, to investigate the role, if any, that the N-terminus of ASIC3 plays in STOM inhibition, we created chimeras that simultaneously swapped out the N- and C-termini of ASIC3. Replacing the full N- and C-terminus of ASIC3 with the termini of ASIC1a, ASIC3(1–43,466–533)1a-CER, resulted in a functional channel that was not inhibited by STOM-YFP (Fig. 4 A). FRET measurements also showed no signs of interaction between the two proteins (Fig. 4 D). These data were expected because this chimera is missing the critical binding site on the distal C-terminus. Since the distal C-terminus of ASIC3 was sufficient for STOM-dependent regulation of ASIC3, we inserted this portion of ASIC3 back into the N-C-terminal double chimera. This chimera, ASIC3(1–43,466–505)1a-CER, was also functional, and STOM-YFP successfully inhibited and bound to this chimeric channel (Fig. 4, B and D). Together, these data show that the N-terminus of ASIC3 is not a critical region for inhibition by STOM.
Figure 4.
TM1 of ASIC3, but not the N-terminus, is important for STOM regulation. (A–C) Each column shows the cartoon of the chimera being tested. Each column has a representative set of pH 5.5–evoked currents of the indicated chimeric channel alone (black) and cotransfected with STOM (red) and shows a boxplot of the current density of the channel with and without STOM coexpression. (A) Average current densities for ASIC3(1–43,466-533)1a-CER in the absence (black) and presence (red) of STOM-YFP were 334.2 ± 44.2 pA/pF (n = 5) and 314.0 ± 15.9 pA/pF (n = 5), respectively. (B) Average current densities for ASIC3(1–43,466-505)1a-CER in the absence (black) and presence (red) of STOM-YFP were 37.6 ± 7.9 pA/pF (n = 9) and 1.4 ± 0.4 pA/pF (n = 11), respectively. (C) Average current densities for ASIC3(44–71)1a-CER in the absence (black) and presence (red) of STOM-YFP or untagged STOM (red) were 72.8 ± 9.0 pA/pF (n = 18), 47.3 ± 7.5 pA/pF (n = 25), and 40.2 ± 5.0 pA/pF (n = 8), respectively. (D) Plot of the FRET efficiency between each chimera and STOM-YFP. Average FRET efficiency of ASIC3(1–43,466-533)1a-CER + STOM-YFP = 0.1 ± 0.2% (n = 5). Average FRET efficiency of ASIC3(1–43,466-505)1a-CER + STOM-YFP = 9.3 ± 1.6% (n = 5). Average FRET efficiency of ASIC3(44–71)1a-CER + STOM-YFP = 11.8 ± 1.2% (n = 8). Dotted line in FRET plots corresponds to control WT ASIC3-CER/STOM-YFP FRET signal replotted from Fig. 1 E for comparison. Data are given as mean ± SEM.
To look at the possible role of TM1, we wanted to make a chimera that swapped the TM1 of ASIC3 with the TM1 of ASIC1a. Salinas and colleagues made a similar chimeric construct where they replaced residues 44–69 of ASIC3 with residues 44–65 of ASIC1a and found the channel to be nonfunctional (Salinas et al., 2009). However, we found that replacing residues 44–71 of ASIC3 with the corresponding residues 44–71 of ASIC1a resulted in functional channels. This mutant channel, ASIC3(44–71)1a-CER, while functional, was only modestly affected by STOM-YFP coexpression (Fig. 4 C). Since we showed that the YFP on STOM reduces the overall regulatory effect, we also measured ASIC3(44–71)1a-CER currents in the presence of untagged STOM. Again, the currents for this chimera were only slightly reduced (approximately twofold; Fig. 4 C), as opposed to the nearly 200-fold reduction seen when tagged ASIC3 channels were coexpressed with untagged STOM (see Fig. 6 D). Interestingly, although this construct did not seem to be regulated by STOM, FRET measurements indicated that STOM-YFP still bound to the channel (Fig. 4 D). These data suggest that STOM regulation of ASIC3 is governed by an interaction site on TM1 in addition to the site on the distal C-terminus.
ASIC2a does not directly interact with STOM
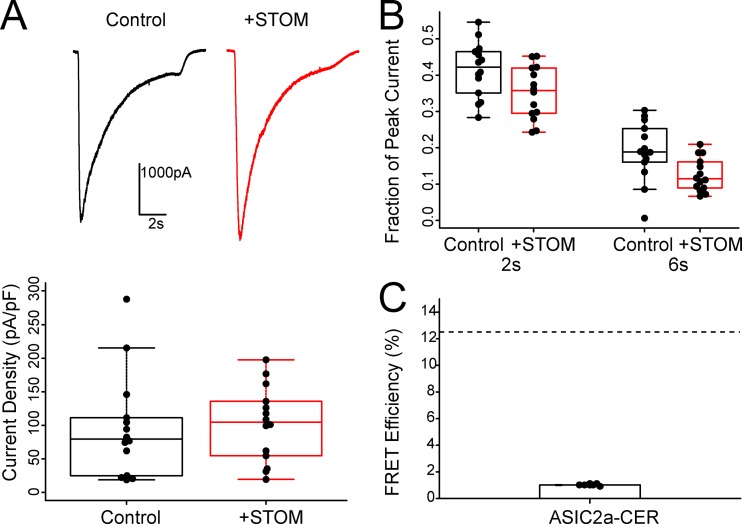
Previous work has suggested that STOM can also bind to and regulate ASIC2a, leading to a speeding of the desensitization rate of the channel. Our data have identified a crucial binding site on the distal C-terminus of ASIC3 that is required for complex formation and channel regulation. ASIC2a does not have this critical sequence (Fig. S2). Thus, we sought to test for interaction and regulation between STOM and ASIC2a using our patch-clamp and FRET assays. Fig. 5 A shows representative ASIC2a traces with and without coexpressed STOM elicited by a rapid switch to pH 4.0. We confirmed that there was no change in ASIC2a current density in the presence of STOM, as found previously (Fig. 5 A, lower panel; Price et al., 2004). To look at the desensitization rate of ASIC2a, we plotted the ratio of the remaining current to the peak current at two time points (2 and 6 s; Fig. 5 B). The results show a trend toward a modest speeding of desensitization. At 2 s, the fraction current remaining goes from 0.42 ± 0.02 to 0.35 ± 0.2 (n = 14, P = 0.04) while at 6 s, the fraction current remaining goes from 0.19 ± 0.02 to 0.13 ± 0.1 (n = 14, P = 0.01). Lastly, we were not able to detect any complex formation between ASIC2a-CER and STOM-YFP using our FRET approach (Fig. 5 D). This suggests that ASIC2a does not directly interact with STOM. We postulate that the small effect on the currents might be the result of an indirect effect on the channel (see Discussion). In addition, these data are consistent with the hypothesis that the distal eight amino acids of ASIC3 are required for STOM binding.
Figure 5.
ASIC2a does not directly interact with STOM. (A) Top: Representative current traces of ASIC2a elicited by a switch to pH 4.0 with and without coexpression of STOM. Bottom: Average current densities for ASIC2a in the absence (black) and presence (red) of STOM were 95.9 ± 20.7 pA/pF (n = 14) and 101.9 ± 14.9 pA/pF (n = 14), respectively. (B) Plot showing the fraction of ASIC2a current remaining at 2 and 6 s. (C) Plot of the FRET efficiency between ASIC2a-CER and STOM-YFP. Average FRET efficiency of ASIC2a-CER + STOM-YFP = 1.0 ± 0.02% (n = 8). Dotted line in FRET plots corresponds to control WT ASIC3-CER/STOM-YFP FRET signal replotted from Fig. 1 E for comparison. Data are given as mean ± SEM.
STOM does not alter ASIC3 surface expression
We next sought to try and determine the mechanism for the STOM-dependent reduction in ASIC3 current. A change in channel current density must occur either via a change in channel gating or a change in surface expression. We first examined membrane expression of ASIC3 in two ways. First, we used confocal microscopy to look at the localization of fluorescently tagged ASIC3 to determine if STOM coexpression dramatically altered the distribution of channels in the cell. To do this, we coexpressed ASIC3 with a C-terminal Turquoise tag (ASIC3-TUR) and STOM with a C-terminal YFP tag (STOM-YFP). To help ensure that we could identify the plasma membrane, we used a TagRFP-labeled portion of the L-type calcium channel, consisting of the I-II loop of Cav1.2, joined to the N-terminus of Cav1.1. This protein, which we will designate “membrane marker” in the figure, has previously been shown to associate with the plasma membrane (Kaur et al., 2015; Polster et al., 2018). Fig. 6 A shows representative images with and without coexpressed STOM-YFP. In each case, ASIC3-TUR was strongly associated with the cell surface. Fig. 6 B shows line scans of the RFP and TUR signals both with and without STOM-YFP. Peaks in fluorescence intensity for ASIC3-TUR coincided with peaks from our RFP membrane marker, suggesting that ASIC3-TUR was on the plasma membrane. In 10 cells for each condition, we saw no substantial change in membrane localization that would explain an ∼200-fold decrease in current.
To more quantitatively examine this question, we employed a cell-surface biotinylation assay where we expressed the same ASIC3-TUR with and without untagged STOM. 1 d after transfection of our proteins into CHO-K1 cells, we isolated plasma membrane–localized proteins by labeling them with Sulfo-NHS-SS-Biotin and pulling them down with NeutrAvidin resin. We then performed Western blots looking at the surface membrane and intracellular fractions of ASIC3. To ensure equal loading of protein in our Western blots, we quantified total protein concentration in our lysate using a BCA assay and adjusted our gel loading to ensure that we loaded equal amounts of total protein into each lane. One representative blot is shown in Fig. 6 C. These blots show all the protein from 35 kD to the top of the gel. There was no immunoreactive protein below this point on the gel. These data show that coexpression of STOM did not dramatically reduce the total amount of ASIC3-TUR on the membrane. In this example, there was no change in the intensity of the band in the with- and without-STOM lanes. We found, on average, a 37 ± 31.8% increase in ASIC3 surface expression when STOM was present (n = 3). Overall, we did not see a change in total ASIC3 expression as measured by looking at the intensity of the ASIC3 band in the nonbiotinylated fraction. These data are consistent with previous observations that this change in ASIC3 current does not occur due to a change in cell surface expression (Price et al., 2004). To ensure these results were not impacted by the presence of the fluorescent protein on the C-terminus of ASIC3, we measured whole-cell acid-evoked currents of ASIC3-TUR with and without STOM. We found that STOM was able to dramatically inhibit ASIC3-TUR to the same extent as the untagged channel (Fig. 6 D). ASIC1a surface expression also appeared to be modestly increased by coexpression with STOM (Fig. 6 C). These data are consistent with our functional measurements that showed a trend toward larger currents when STOM was present in the case of ASIC1a (Fig. 1 C). On average, we saw a modest increase in surface expression of both ASIC1a and ASIC3 when STOM was coexpressed, which may or may not be significant, but the data show clearly that the nearly 200-fold decrease in ASIC3 current when STOM was present cannot be explained by a change in cell surface expression.
STOM binding stabilizes ASIC3 in the desensitized state
Since STOM did not impact ASIC3 surface expression, the effect is likely via a change in channel gating. There are a number of ways this could occur, but we chose to look at two simple hypotheses first. The first hypothesis is that STOM stabilizes the closed state of the channel. This would manifest as a shift in the pH dependence of channel opening toward more acidic pH. To test this, we attempted to open the channel by perfusing a significantly more acidic solution (pH 4.0) onto the cells. Fig. 7 A shows that currents elicited from a pH pulse to 5.5 followed by a recovery at pH 8.0 and a second pH pulse to 4.0. With ASIC3 alone, there is no significant change in the peak current between pH 5.5 and pH 4.0. However, as previously reported, pH 4.0 elicits a slowly activating steady-state current (Babinski et al., 1999; Bassilana et al., 1997; de Weille et al., 1998). Currents measured in cells coexpressing both ASIC3 and STOM showed the typical small currents at both pH 5.5 and pH 4.0 (Fig. 7 B). This suggested to us that either an acidic shift in the pH dependence was not the mechanism of STOM-dependent regulation or that the shift is more severe than is experimentally tractable.
Figure 7.
Potential mechanism for STOM-dependent regulation of ASIC3. (A) Currents from ASIC3 with and without STOM coexpression. Currents were measured first with a switch to pH 5.5 for 5 s followed by a 9-s recovery period at pH 8.0. Then currents were measured with a switch to pH 4.0 for 5 s. (B) Boxplot showing the current density measured from pH 5.5– and pH 4.0–evoked currents from ASIC3 with and without STOM. Current densities for control at pH 5.5 and pH 4.0 were 538.0 ± 261.7 pA/pF (n = 5) and 544.5 ± 260.2 pA/pF (n = 5), respectively. Current densities for the +STOM case at pH 5.5 and pH 4.0 were 63.4 ± 39.4 pA/pF (n = 7) and 68.1 ± 41.0 pA/pF (n = 7), respectively. (C) Representative pH 5.0–evoked currents from the ASIC3(Q269G)-CER mutant alone (black) and cotransfected with STOM (red). (D) Boxplot of the current density of the ASIC3(Q269G)-CER channel with and without STOM coexpression. Average current densities for ASIC3(Q269G)-CER in the absence (black) and presence (red) of STOM were 5.7 ± 1.3 pA/pF (n = 10) and 22.5 ± 6.6 pA/pF (n = 11), respectively. (E) Plot of the FRET efficiency between ASIC3(Q269G)-CER and STOM-YFP. Average FRET efficiency of ASIC3(Q269G)-CER + STOM-YFP = 10.3 ± 0.85% (n = 10). Dotted line in FRET plots corresponds to control WT ASIC3-CER/STOM-YFP FRET signal replotted from Fig. 1 E for comparison. Data are given as mean ± SEM.
The second hypothesis for explaining how STOM regulates ASIC3 is that STOM binding could stabilize the desensitized state of the channel. To test this, we used a recently reported point mutation in the extracellular domain that nearly eliminates desensitization (Wu et al., 2019). We reasoned that by preventing the transition to the desensitized state, we would prevent STOM from being able to stabilize the channel in that state. Fig. 7 C shows representative currents from the ASIC3(Q269G)-CER mutation. The mutant currents were small, but addition of STOM did not further reduce current density (Fig. 7 D). It is possible that the Q269 mutation could disrupt STOM binding. This is unlikely given that the mutation is on the extracellular side of the channel. We tested this possibility using our FRET assay and found a robust FRET signal between ASIC3(Q269)-CER and STOM-YFP (Fig. 7 E). This result is consistent with a model where STOM acts by stabilizing the desensitized state, and in a mutant that prevents this transition, STOM cannot regulate the channel.
Discussion
STOM has been shown to dramatically reduce ASIC3 currents (Brand et al., 2012; Price et al., 2004). In addition, the closely related STOML proteins have been shown to regulate ASICs in an isoform-dependent manner, with STOML3 also inhibiting ASIC3 currents (Kozlenkov et al., 2014; Lapatsina et al., 2012; Moshourab et al., 2013; Wetzel et al., 2007). The mechanisms for this inhibition and the binding site for this family of proteins on ASIC3 are not known. Using chimeric ASICs, we found two critical regions for ASIC3 regulation by STOM. A site on the distal C-terminus that involves the final eight amino acids of ASIC3 was critical for binding of STOM to the channel. Mutation of that site eliminated binding of STOM to the channel, which in turn prevented regulation as well. A second site on TM1 was found to be critical for the regulatory effect, as mutation greatly reduced the effect of STOM on ASIC3 but was not critical for binding. The TM1-mutated ASIC3 still showed robust FRET with STOM.
Our FRET assay showed that neither ASIC1a nor ASIC2a bound STOM. We found ASIC1a currents were not impacted by STOM coexpression, and ASIC2a currents showed a slight speeding of the desensitization rate. Members of the STOM and SPFH families have been suggested to change the curvature of the membrane (Brand et al., 2012; Frick et al., 2007). In addition, STOML3 has been shown to change membrane stiffness, and STOM localizes to cholesterol-rich lipid rafts. Given the small effect of STOM on ASIC2a and the lack of a direct interaction, we hypothesize that STOM may alter ASIC2a desensitization rate by changing the character of the membrane. If true, this would suggest that ASIC2a is sensitive to this change in the membrane in a way that ASIC1a and ASIC3 are not.
A previous study suggested that both ASIC1a and ASIC2a were capable of binding STOM by using a coimmunoprecipitation assay (Price et al., 2004). Since we do not detect FRET between STOM and these isoforms, and we do not measure a functional effect on ASIC1a and only a very slight effect on ASIC2a, we believe that easiest interpretation of our results is that if STOM cannot bind then it cannot regulate the channel. However, FRET requires that the fluorophores be within ∼70 Å of each other. It is possible that STOM bound to ASIC1a or ASIC2a, but the conformation of the complex is such that the fluorophores are outside the usable range for FRET and that this alternate bound conformation does not result in a regulated channel complex.
There has been very little work examining the binding site between ASIC3 and STOM. However, in one study, the authors posited that a hydrophobic dipeptide (L488 and L489) at the proximal portion of the C-terminus was critical for STOM regulation of the channel (Brand et al., 2012). When these two leucine residues were mutated to aspartate (LL/DD), the effect of STOM was greatly reduced, but not eliminated, while the binding was unaffected. Our data agree that this region is not critical for the interaction between the two proteins but do not support this region as critical for STOM-dependent regulation. One possible explanation for the discrepancy in these conclusions stems from the observation that effect of STOM on ASIC3 depends on the expression levels of each protein. In the previous study, the LL/DD mutant increased the ASIC3 currents by ∼5- to 10-fold, suggesting the possibility that the STOM expression level was not enough to fully inhibit this mutant channel. The current density plot for this mutant looks to have a large population of cells with very small currents that appear regulated by STOM and a second population with larger potentially unregulated currents.
Interestingly, the binding site on the distal C-terminus of ASIC3 includes a well-characterized PDZ-binding motif. This site has been shown to interact with several other PDZ domain–containing regulatory proteins, including PSD-95, CIPP, and Lin-7b (Anzai et al., 2002; Eshcol et al., 2008; Hruska-Hageman et al., 2004). However, STOM does not contain a PDZ domain, and how this interaction occurs is a mystery. Little is known about the targets for SPFH domains, but it is possible that this domain on STOM could be recognizing residues in the distal C-terminus. Further work will be needed to find which domains on STOM are critical for binding to ASIC3. In addition, our data suggest a trend toward decreased ASIC3 currents with just the C-terminal binding site intact (Fig. 4 C), which suggests that binding of STOM to the C-terminus might have a modest impact on ASIC3 gating alone.
However, the bulk of the regulatory effect of STOM appears to be mediated through TM1 of the channel. Given the fact that STOM contains a small 29–amino acid membrane hairpin, we hypothesize that this region interacts with and alters TM1 of ASIC3. There are a number of residues that are different in this region between ASIC1a and ASIC3. How interaction with this region of the channel could dramatically inhibit gating is unknown. It is possible that the interaction alters the selectivity filter, which is thought to be on the intracellular side of TM2, or alters the ability of the transmembrane domains to move during gating (Baconguis et al., 2014; Lynagh et al., 2017; Yoder et al., 2018).
In addition to examining the binding site for STOM on ASIC3, we considered three major hypotheses for the regulatory mechanism of ASIC3 by STOM. First, STOM could act by reducing the surface expression of ASIC3. Previous work has suggested that this is not the case and we confirmed that here (Price et al., 2004). Second, STOM could stabilize the closed state of the channel, which would result in an acidic shift of the pH curve. We attempted to test this by using more acidic solutions (pH 4.0) to try and open the channel but found that this was not sufficient to open the channel. These data suggest that stabilization of the closed state does not explain the effect, but it is possible that the acidic shift is more severe and that the channel would require a pH that is not experimentally achievable. Finally, we considered the possibility that STOM could be stabilizing the desensitized state of the channel. To look at this question, we used a point mutation (Q269G) in the extracellular domain of the channel that prevents ASIC3 from undergoing desensitization (Wu et al., 2019). We reasoned that if the channel cannot enter the desensitized state, it cannot be stabilized in that state. We found that this mutant was not regulated by STOM, suggesting the possibility that STOM acts by stabilizing the desensitized state of ASIC3. It is worth noting that the Q269 mutation reduced current density dramatically but that STOM did not further decrease it. These data lead to an interesting hypothesis for how STOM and its relatives regulate ASICs, but more work is needed to confirm this hypothesis.
The physiological role of the ASIC3–STOM complex has not been uncovered to date. However, the closely related STOML3 (60.5% identical, 74.9% similar) has been shown to impact nociceptor mechanosensitivity and be critical for touch sensation (Moshourab et al., 2013; Wetzel et al., 2007). In addition, these studies showed that pH-sensitive currents were larger in DRG neurons of STOML3 knockout mice and that the loss of the STOM–ASIC complex can have profound effects on mechanosensitivity in some neurons innervating the skin. We focused this study on the ASIC3–STOM complex, in part, due to the large effect of STOM on ASIC3, but we believe that the results here likely shed light on the regulation of ASIC3 by STOML3 as well. However, this mechanism of regulation may not be ubiquitous across all STOM family members. The homologous complex in C. elegans (MEC-2–MEC-4) has been shown to be transient and low affinity (Chen et al., 2015). Moreover, MEC-2, the STOM homologue, appears to bind to the N-terminus of MEC-4, the ASIC homologue, and increase instead of decrease current (Goodman et al., 2002; Zhang et al., 2004). We see no evidence that the critical C-terminal binding site we found in ASIC3 is present anywhere in the MEC-4 sequence. However, there are a number of hydrophobic residues in the lower portion of TM1 that are shared between ASIC3 and MEC-4, so it is possible that the interaction site varies but the site of regulation may be similar. It is also possible that the mechanism of STOM-dependent regulation of ASICs is not shared by MEC-2.
It is worth wondering about how a complex that renders the channels nonfunctional might be important for neuronal physiology. We imagine a possible scenario where dynamic regulation of this complex could act to rapidly increase or decrease the amount of functional ASIC3 present on the membrane. There is no direct evidence to date for how this complex might be regulated, but there are palmitoylation sites on STOM that are required for proper membrane targeting of the protein that could be a site of dynamic regulation (Rungaldier et al., 2017). MEC-2, the C. elegans homologue of STOM, requires the palmitoylation of a conserved cysteine in order to regulate the ASIC homologue MEC-4/10 (Brown et al., 2008). In addition, the final 30 amino acids of ASIC3 contain six threonines and three serines, including one threonine in the STOM-binding site, raising the possibility of a number of potential phosphorylation sites in this region that could lead to dynamic regulation of this complex. Previous work has shown that residue S523 on ASIC3 can be phosphorylated by PKC and that this phosphorylation, which is very near the critical STOM-binding site, increases the association of a regulatory protein called NHERF-1 with ASIC3 (Deval et al., 2006, 2004). A great deal more work needs to be done to uncover the role this complex plays in neurons.
The data presented here show that STOM can inhibit ASIC3 currents by nearly 200-fold. We found two critical sites on ASIC3 for this regulatory effect. The first is a site on the distal C-terminus of the channel that is necessary for complex formation. Disruption of this site eliminates both binding of STOM to ASIC3 as well as the regulatory effect. The second critical site on ASIC3 is TM1, which appears to be necessary for regulation of the channel by STOM. Mutation of this region greatly reduces the effect of STOM without impacting binding of STOM and ASIC3. In addition, we showed that a mutation that cannot desensitize prevented STOM-dependent regulation of the channel. Taken together, we propose a model whereby STOM binds to the distal C-terminus of ASIC3, which leads to a modest reduction in current and anchors the complex together. Then, ASIC3 currents are dramatically reduced via a second interaction between TM1 of ASIC3 and the membrane-imbedded hairpin of STOM. We hypothesize that this dramatic reduction is a result of a stabilization of the desensitized state of the channel. To fully understand this complex, it will be important to understand which parts of STOM are critical for this interaction as well. It will also be interesting to see if the mechanisms that we show here for STOM binding to and regulation of ASIC3 can shed light on the broader question of how this family of proteins regulates such a wide variety of ion channels and transporters.
Online supplemental material
Table S1 provides summary of all current density data reported throughout the paper in addition to statistical analysis. In addition, the table shows the desensitization rates of a number of the chimeric channels used. Fig. S1 shows that presence of YFP on STOM reduces inhibition of ASIC3 and that this reduction may result from a decreased expression of STOM-YFP compared with untagged STOM. Fig. S2 provides the sequence alignment for the ASIC isoforms used in this study.
Supplementary Material
provides a summary of data.
Acknowledgments
Merritt C. Maduke served as editor.
The authors would like to thank Prafulla Aryal for helpful discussions. In addition, we would like to thank Kurt Beam (University of Colorado Anschutz Medical Campus, School of Medicine, Aurora, CO) for providing the TagRFP-CaV1.2 I-II loop-CaV1.1 N-terminal construct as well as for the use of his Zeiss LSM710 confocal microscope.
Research reported in this publication was supported by the National Eye Institute of the National Institutes of Health under award number R00EY024267 (to J.R. Bankston), the National Institute of Dental and Craniofacial Research under award number F31DE028739 (to M.M. Cullinan), and the National Heart, Lung, and Blood Institute under award 2T32HL007822 (to R.C. Klipp).
The authors declare no competing financial interests.
Author contributions: All authors participated in the experimental design and conceptualization. J. R. Bankston, R.C. Klipp, and M.M. Cullinan participated in data collection. J.R. Bankston and R.C. Klipp analyzed the data. J.R. Bankston and R.C. Klipp wrote the paper. All authors approved the final version of the manuscript.
References
- Akopian A.N., Chen C.C., Ding Y., Cesare P., and Wood J.N.. 2000. A new member of the acid-sensing ion channel family. Neuroreport. 11:2217–2222. 10.1097/00001756-200007140-00031 [DOI] [PubMed] [Google Scholar]
- Anzai N., Deval E., Schaefer L., Friend V., Lazdunski M., and Lingueglia E.. 2002. The multivalent PDZ domain-containing protein CIPP is a partner of acid-sensing ion channel 3 in sensory neurons. J. Biol. Chem. 277:16655–16661. 10.1074/jbc.M201087200 [DOI] [PubMed] [Google Scholar]
- Babinski K., Lê K.-T., and Séguéla P.. 1999. Molecular cloning and regional distribution of a human proton receptor subunit with biphasic functional properties. J. Neurochem. 72:51–57. 10.1046/j.1471-4159.1999.0720051.x [DOI] [PubMed] [Google Scholar]
- Baconguis I., and Gouaux E.. 2012. Structural plasticity and dynamic selectivity of acid-sensing ion channel-spider toxin complexes. Nature. 489:400–405. 10.1038/nature11375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baconguis I., Bohlen C.J., Goehring A., Julius D., and Gouaux E.. 2014. X-ray structure of acid-sensing ion channel 1-snake toxin complex reveals open state of a Na(+)-selective channel. Cell. 156:717–729. 10.1016/j.cell.2014.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassilana F., Champigny G., Waldmann R., de Weille J.R., Heurteaux C., and Lazdunski M.. 1997. The acid-sensitive ionic channel subunit ASIC and the mammalian degenerin MDEG form a heteromultimeric H+-gated Na+ channel with novel properties. J. Biol. Chem. 272:28819–28822. 10.1074/jbc.272.46.28819 [DOI] [PubMed] [Google Scholar]
- Benson C.J., Eckert S.P., and McCleskey E.W.. 1999. Acid-evoked currents in cardiac sensory neurons: A possible mediator of myocardial ischemic sensation. Circ. Res. 84:921–928. 10.1161/01.RES.84.8.921 [DOI] [PubMed] [Google Scholar]
- Boscardin E., Alijevic O., Hummler E., Frateschi S., and Kellenberger S.. 2016. The function and regulation of acid-sensing ion channels (ASICs) and the epithelial Na(+) channel (ENaC): IUPHAR Review 19. Br. J. Pharmacol. 173:2671–2701. 10.1111/bph.13533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand J., Smith E.S.J., Schwefel D., Lapatsina L., Poole K., Omerbašić D., Kozlenkov A., Behlke J., Lewin G.R., and Daumke O.. 2012. A stomatin dimer modulates the activity of acid-sensing ion channels. EMBO J. 31:3635–3646. 10.1038/emboj.2012.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browman D.T., Hoegg M.B., and Robbins S.M.. 2007. The SPFH domain-containing proteins: more than lipid raft markers. Trends Cell Biol. 17:394–402. 10.1016/j.tcb.2007.06.005 [DOI] [PubMed] [Google Scholar]
- Brown A.L., Liao Z., and Goodman M.B.. 2008. MEC-2 and MEC-6 in the Caenorhabditis elegans sensory mechanotransduction complex: auxiliary subunits that enable channel activity. J. Gen. Physiol. 131:605–616. 10.1085/jgp.200709910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Bharill S., Isacoff E.Y., and Chalfie M.. 2015. Subunit composition of a DEG/ENaC mechanosensory channel of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 112:11690–11695. 10.1073/pnas.1515968112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson R.J.P., Benz J., Stohler P., Tetaz T., Joseph C., Huber S., Schmid G., Hügin D., Pflimlin P., Trube G., et al. 2012. Structure of the acid-sensing ion channel 1 in complex with the gating modifier Psalmotoxin 1. Nat. Commun. 3:936 10.1038/ncomms1917 [DOI] [PubMed] [Google Scholar]
- de Weille J.R., Bassilana F., Lazdunski M., and Waldmann R.. 1998. Identification, functional expression and chromosomal localisation of a sustained human proton-gated cation channel. FEBS Lett. 433:257–260. 10.1016/S0014-5793(98)00916-8 [DOI] [PubMed] [Google Scholar]
- Deval E., Salinas M., Baron A., Lingueglia E., and Lazdunski M.. 2004. ASIC2b-dependent regulation of ASIC3, an essential acid-sensing ion channel subunit in sensory neurons via the partner protein PICK-1. J. Biol. Chem. 279:19531–19539. 10.1074/jbc.M313078200 [DOI] [PubMed] [Google Scholar]
- Deval E., Friend V., Thirant C., Salinas M., Jodar M., Lazdunski M., and Lingueglia E.. 2006. Regulation of sensory neuron-specific acid-sensing ion channel 3 by the adaptor protein Na+/H+ exchanger regulatory factor-1. J. Biol. Chem. 281:1796–1807. 10.1074/jbc.M509669200 [DOI] [PubMed] [Google Scholar]
- Eshcol J.O., Harding A.M.S., Hattori T., Costa V., Welsh M.J., and Benson C.J.. 2008. Acid-sensing ion channel 3 (ASIC3) cell surface expression is modulated by PSD-95 within lipid rafts. Am. J. Physiol. Cell Physiol. 295:C732–C739. 10.1152/ajpcell.00514.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick M., Bright N.A., Riento K., Bray A., Merrified C., and Nichols B.J.. 2007. Coassembly of flotillins induces formation of membrane microdomains, membrane curvature, and vesicle budding. Curr. Biol. 17:1151–1156. 10.1016/j.cub.2007.05.078 [DOI] [PubMed] [Google Scholar]
- Genetet S., Desrames A., Chouali Y., Ripoche P., Lopez C., and Mouro-Chanteloup I.. 2017. Stomatin modulates the activity of the Anion Exchanger 1 (AE1, SLC4A1). Sci. Rep. 7:46170 10.1038/srep46170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M.B., Ernstrom G.G., Chelur D.S., O’Hagan R., Yao C.A., and Chalfie M.. 2002. MEC-2 regulates C. elegans DEG/ENaC channels needed for mechanosensation. Nature. 415:1039–1042. 10.1038/4151039a [DOI] [PubMed] [Google Scholar]
- Gründer S., Geissler H.S., Bässler E.L., and Ruppersberg J.P.. 2000. A new member of acid-sensing ion channels from pituitary gland. Neuroreport. 11:1607–1611. 10.1097/00001756-200006050-00003 [DOI] [PubMed] [Google Scholar]
- Hesselager M., Timmermann D.B., and Ahring P.K.. 2004. pH Dependency and desensitization kinetics of heterologously expressed combinations of acid-sensing ion channel subunits. J. Biol. Chem. 279:11006–11015. 10.1074/jbc.M313507200 [DOI] [PubMed] [Google Scholar]
- Hruska-Hageman A.M., Benson C.J., Leonard A.S., Price M.P., and Welsh M.J.. 2004. PSD-95 and Lin-7b interact with acid-sensing ion channel-3 and have opposite effects on H+- gated current. J. Biol. Chem. 279:46962–46968. 10.1074/jbc.M405874200 [DOI] [PubMed] [Google Scholar]
- Huang M., Gu G., Ferguson E.L., and Chalfie M.. 1995. A stomatin-like protein necessary for mechanosensation in C. elegans. Nature. 378:292–295. 10.1038/378292a0 [DOI] [PubMed] [Google Scholar]
- Jasti J., Furukawa H., Gonzales E.B., and Gouaux E.. 2007. Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature. 449:316–323. 10.1038/nature06163 [DOI] [PubMed] [Google Scholar]
- Jones N.G., Slater R., Cadiou H., McNaughton P., and McMahon S.B.. 2004. Acid-induced pain and its modulation in humans. J. Neurosci. 24:10974–10979. 10.1523/JNEUROSCI.2619-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur G., Pinggera A., Ortner N.J., Lieb A., Sinnegger-Brauns M.J., Yarov-Yarovoy V., Obermair G.J., Flucher B.E., and Striessnig J.. 2015. A Polybasic Plasma Membrane Binding Motif in the I-II Linker Stabilizes Voltage-gated CaV1.2 Calcium Channel Function. J. Biol. Chem. 290:21086–21100. 10.1074/jbc.M115.645671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlenkov A., Lapatsina L., Lewin G.R., and Smith E.S.J.. 2014. Subunit-specific inhibition of acid sensing ion channels by stomatin-like protein 1. J. Physiol. 592:557–569. 10.1113/jphysiol.2013.258657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapatsina L., Jira J.A., Smith E.S.J., Poole K., Kozlenkov A., Bilbao D., Lewin G.R., and Heppenstall P.A.. 2012. Regulation of ASIC channels by a stomatin/STOML3 complex located in a mobile vesicle pool in sensory neurons. Open Biol. 2:120096 10.1098/rsob.120096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingueglia E., de Weille J.R., Bassilana F., Heurteaux C., Sakai H., Waldmann R., and Lazdunski M.. 1997. A modulatory subunit of acid sensing ion channels in brain and dorsal root ganglion cells. J. Biol. Chem. 272:29778–29783. 10.1074/jbc.272.47.29778 [DOI] [PubMed] [Google Scholar]
- Liu C., Chin J.X., and Lee D.Y.. 2015. SynLinker: an integrated system for designing linkers and synthetic fusion proteins. Bioinformatics. 31:3700–3702. 10.1093/bioinformatics/btv447 [DOI] [PubMed] [Google Scholar]
- Lu Y., Ma X., Sabharwal R., Snitsarev V., Morgan D., Rahmouni K., Drummond H.A., Whiteis C.A., Costa V., Price M., et al. 2009. The ion channel ASIC2 is required for baroreceptor and autonomic control of the circulation. Neuron. 64:885–897. 10.1016/j.neuron.2009.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynagh T., Flood E., Boiteux C., Wulf M., Komnatnyy V.V., Colding J.M., Allen T.W., and Pless S.A.. 2017. A selectivity filter at the intracellular end of the acid-sensing ion channel pore. eLife. 6:e24630 10.7554/eLife.24630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshourab R.A., Wetzel C., Martinez-Salgado C., and Lewin G.R.. 2013. Stomatin-domain protein interactions with acid-sensing ion channels modulate nociceptor mechanosensitivity. J. Physiol. 591:5555–5574. 10.1113/jphysiol.2013.261180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polster A., Nelson B.R., Papadopoulos S., Olson E.N., and Beam K.G.. 2018. Stac proteins associate with the critical domain for excitation-contraction coupling in the II-III loop of CaV1.1. J. Gen. Physiol. 150:613–624. 10.1085/jgp.201711917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M.P., Thompson R.J., Eshcol J.O., Wemmie J.A., and Benson C.J.. 2004. Stomatin modulates gating of acid-sensing ion channels. J. Biol. Chem. 279:53886–53891. 10.1074/jbc.M407708200 [DOI] [PubMed] [Google Scholar]
- R Core Team 2017. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. Available at: http://www.R-project.org/
- Rungaldier S., Umlauf E., Mairhofer M., Salzer U., Thiele C., and Prohaska R.. 2017. Structure-function analysis of human stomatin: A mutation study. PLoS One. 12:e0178646 10.1371/journal.pone.0178646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas M., Lazdunski M., and Lingueglia E.. 2009. Structural elements for the generation of sustained currents by the acid pain sensor ASIC3. J. Biol. Chem. 284:31851–31859. 10.1074/jbc.M109.043984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyers L., Umlauf E., and Prohaska R.. 1999. Cysteine 29 is the major palmitoylation site on stomatin. FEBS Lett. 449:101–104. 10.1016/S0014-5793(99)00417-2 [DOI] [PubMed] [Google Scholar]
- Ugawa S., Ueda T., Ishida Y., Nishigaki M., Shibata Y., and Shimada S.. 2002. Amiloride-blockable acid-sensing ion channels are leading acid sensors expressed in human nociceptors. J. Clin. Invest. 110:1185–1190. 10.1172/JCI0215709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemmie J.A., Chen J., Askwith C.C., Hruska-Hageman A.M., Price M.P., Nolan B.C., Yoder P.G., Lamani E., Hoshi T., Freeman J.H. Jr., and Welsh M.J.. 2002. The acid-activated ion channel ASIC contributes to synaptic plasticity, learning, and memory. Neuron. 34:463–477. 10.1016/S0896-6273(02)00661-X [DOI] [PubMed] [Google Scholar]
- Wetzel C., Hu J., Riethmacher D., Benckendorff A., Harder L., Eilers A., Moshourab R., Kozlenkov A., Labuz D., Caspani O., et al. 2007. A stomatin-domain protein essential for touch sensation in the mouse. Nature. 445:206–209. 10.1038/nature05394 [DOI] [PubMed] [Google Scholar]
- Wu Y., Chen Z., and Canessa C.M.. 2019. A valve-like mechanism controls desensitization of functional mammalian isoforms of acid-sensing ion channels. eLife. 8:e45851 10.7554/eLife.45851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z.-G., Zhu X.-M., Chu X.-P., Minami M., Hey J., Wei W.-L., MacDonald J.F., Wemmie J.A., Price M.P., Welsh M.J., and Simon R.P.. 2004. Neuroprotection in ischemia: blocking calcium-permeable acid-sensing ion channels. Cell. 118:687–698. 10.1016/j.cell.2004.08.026 [DOI] [PubMed] [Google Scholar]
- Yoder N., Yoshioka C., and Gouaux E.. 2018. Gating mechanisms of acid-sensing ion channels. Nature. 555:397–401. 10.1038/nature25782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.-Z., Abbud W., Prohaska R., and Ismail-Beigi F.. 2001. Overexpression of stomatin depresses GLUT-1 glucose transporter activity. Am. J. Physiol. Cell Physiol. 280:C1277–C1283. 10.1152/ajpcell.2001.280.5.C1277 [DOI] [PubMed] [Google Scholar]
- Zhang S., Arnadottir J., Keller C., Caldwell G.A., Yao C.A., and Chalfie M.. 2004. MEC-2 is recruited to the putative mechanosensory complex in C. elegans touch receptor neurons through its stomatin-like domain. Curr. Biol. 14:1888–1896. 10.1016/j.cub.2004.10.030 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
provides a summary of data.