JGP study suggests that stomatin may trap the acid-sensing ASIC3 channel in its desensitized state.
Abstract
JGP study suggests that stomatin may trap the acid-sensing ASIC3 channel in its desensitized state.
Acid-sensing ion channels (ASICs) are trimeric, Na+-conducting, voltage-insensitive channels that are expressed throughout the central and peripheral nervous systems and are activated by low extracellular pH (1). The channels have been implicated in a range of physiological functions, including learning and memory, as well as fear responses. Blocking ASICs, meanwhile, can inhibit pain and reduce neuronal damage caused by ischemic stroke. In this issue of JGP, Klipp et al. provide new insights into how the integral monotopic membrane protein stomatin (STOM) suppresses the activity of one family member, ASIC3 (2).
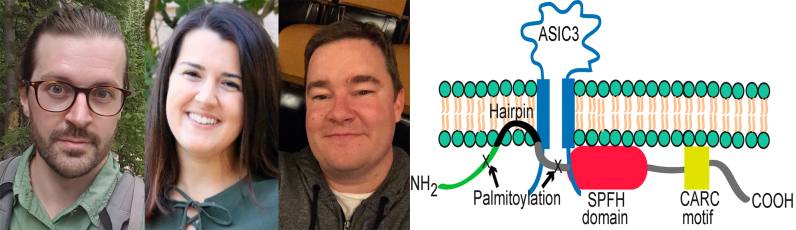
Robert Klipp (left), Megan Cullinan (center), and John Bankston (right) present new details of how the integral monotopic membrane protein STOM inhibits the acid-sensing ion channel ASIC3. The researchers find that STOM interacts with the distal C-terminus of ASIC3 and suppresses the channel’s activity through an interaction with its first transmembrane domain. This interaction may stabilize ASIC3 in its desensitized state because STOM-dependent regulation is abolished by a point mutation that prevents channel desensitization.
STOM associates with the cytoplasmic face of the plasma membrane via a short, hydrophobic hairpin and several palmitoylation sites (3). Along with a set of closely related STOM-like proteins, STOM has been shown to regulate ASICs in an isoform-dependent manner. STOM itself, for example, can almost completely suppress the activity of ASIC3 when the two proteins are coexpressed in mammalian cells, but has no effect on ASIC1a (4). “However, little is known about how STOM and STOM-like proteins discriminate between different ASIC family members and how they exert their effects,” says John Bankston, an assistant professor at the University of Colorado Anschutz Medical Campus.
Bankston, together with postdoctoral fellow Robert Klipp and graduate student Megan Cullinan, first set out to identify how STOM interacts with ASIC3. The researchers determined that STOM does not interact with ASIC1a, so they created a series of chimeric proteins in which portions of this channel replaced the corresponding regions of ASIC3. Using a FRET-based assay in cells, the team found that STOM binds to the final eight amino acids in ASIC3’s intracellular C-terminus. “Strangely, these eight amino acids bind to PDZ domains in other proteins, but STOM does not contain a PDZ domain,” Bankston notes.
Patch-clamp electrophysiology confirmed that this C-terminal region of ASIC3 is required for STOM to suppress channel currents. However, the researchers found that STOM-dependent regulation of ASIC3 also requires the channel’s first transmembrane region (TM1), a key region involved in channel gating (5). A chimera in which this region was replaced by the TM1 of ASIC1a was barely suppressed by STOM, even though STOM was still able to bind to the channel’s C-terminus.
“We speculate that, after binding to the distal C-terminus of ASIC3, STOM’s hydrophobic hairpin can interact with ASIC3’s TM1 and prevent it from undergoing the conformational changes associated with channel gating,” Bankston says.
This interaction could potentially stabilize the closed state of ASIC3, but Bankston and colleagues found that STOM does not alter the pH dependence of channel activation. Nor does STOM inhibit ASIC3 by reducing the channel’s expression at the cell surface.
Instead, the researchers suggest, STOM may stabilize ASIC3 in its desensitized state: a point mutation in ASIC3 that prevents channel desensitization abolished STOM-dependent regulation. Bankston and colleagues now plan to investigate this potential mechanism in more detail.
Another outstanding question is whether the interaction between ASIC3 and STOM can be dynamically regulated, allowing neurons to rapidly alter channel functionality at the plasma membrane. Bankston notes that there are potential phosphorylation sites for PKC and other protein kinases near the C-terminal region of ASIC3 that binds to STOM, while the availability of STOM itself could be controlled by reversible palmitoylation.
References
- 1.Boscardin E., et al. Br. J. Pharmacol. 2016 doi: 10.1111/bph.13533. [DOI] [Google Scholar]
- 2.Klipp R.C., et al. J. Gen. Physiol. 2020 doi: 10.1085/jgp.201912471. [DOI] [Google Scholar]
- 3.Snyers L., et al. FEBS Lett. 1999 doi: 10.1016/S0014-5793(99)00417-2. [DOI] [PubMed] [Google Scholar]
- 4.Price M.P., et al. J. Biol. Chem. 2004 doi: 10.1074/jbc.M407708200. [DOI] [Google Scholar]
- 5.Yoder N., et al. Nature. 2018 doi: 10.1038/nature25782. [DOI] [Google Scholar]