MYB proteins regulate carbon flow into the phenylpropanoid pathway for lignin biosynthesis.
Abstract
Lignin is a phenylpropanoid-derived polymer that functions as a major component of cell walls in plant vascular tissues. Biosynthesis of the aromatic amino acid Phe provides precursors for many secondary metabolites, including lignins and flavonoids. Here, we discovered that MYB transcription factors MYB20, MYB42, MYB43, and MYB85 are transcriptional regulators that directly activate lignin biosynthesis genes and Phe biosynthesis genes during secondary wall formation in Arabidopsis (Arabidopsis thaliana). Disruption of MYB20, MYB42, MYB43, and MYB85 resulted in growth development defects and substantial reductions in lignin biosynthesis. In addition, our data showed that these MYB proteins directly activated transcriptional repressors that specifically inhibit flavonoid biosynthesis, which competes with lignin biosynthesis for Phe precursors. Together, our results provide important insights into the molecular framework for the lignin biosynthesis pathway.
Plant cell walls are commonly classified into primary and secondary based on their composition and cellular location. In general, all plant cells form primary cell walls during cell division, while secondary cell walls are produced after cessation of cell division and expansion only in specific types of cells, such as vessel elements and fibers (Cosgrove and Jarvis, 2012). Secondary cell walls play important roles in providing long-distance water transport, mechanical support, and plant defense (Zhao and Dixon, 2014; Hoson and Wakabayashi, 2015).
The main components of typical secondary cell walls are cellulose, hemicellulose, and lignin. Lignins are phenylpropanoid-derived polymers resulting from oxidative polymerization of three major hydroxycinnamyl alcohols (Bonawitz and Chapple, 2010; Zhao, 2016). Natural lignin polymers are only composed of p-hydroxyphenyl, guaiacyl, and syringyl as basic units; however, occasionally some unconventional units, such as coniferaldehyde, sinapaldehyde, catechyl, and 5-hydroxyl guaiacyl units, are also incorporated into lignin monomers (Weng et al., 2010; Chen et al., 2012, 2013; Zhao et al., 2013).
It is widely accepted that lignin biosynthesis is controlled by a regulatory hierarchy involving NAC (no apical meristem [NAM], Arabidopsis transcription activation factor [ATAF1/2], and cup-shaped cotyledon [CUC2]) and myeloblastosis (MYB) transcription factors (Zhao, 2016; Ohtani and Demura, 2019). In this network, NAC proteins, including VASCULAR-RELATED NAC DOMAIN1 (VND1) to VND7 and NAC SECONDARY WALL THICKNING PROMOTING FACTOR1 (NST1) to NST3 were identified in Arabidopsis (Arabidopsis thaliana) to serve as a first layer of transcription factors (Kubo et al., 2005; Mitsuda et al., 2007). These NACs are master regulators in various cell types. NST3, also called SND1, together with NST1 redundantly regulates secondary cell wall formation in fibers (Mitsuda et al., 2007), and NST1 and NST2 activate the same process in the endothecium of anthers (Mitsuda et al., 2005). MYB46 and MYB83 in Arabidopsis, the downstream targets of the NAC proteins, are a second layer of the network, redundantly regulating secondary cell wall formation (McCarthy et al., 2009; Ko et al., 2014). Recently, two types of cis-element sequences were reported by two different groups, secondary wall MYB-responsive element, ACC(A/T)A(A/C)(T/C), and MYB46-responsive cis-regulatory element, (T/C)ACC(A/T)A(A/C)(T/C) (Kim et al., 2012; Zhong and Ye, 2012). Comprehensive survey of these cis-elements and DNA protein-binding assays identified the direct targets of MYB46/83, including MYB genes implicated in the regulation of lignin biosynthesis (e.g. MYB58, MYB63, MYB103, and KNAT7), secondary cell wall biosynthesis genes such as cellulose synthase genes (CesA4, CesA7, and CesA8), xylan biosynthesis genes (IRX7, IRX8, and IRX9), and lignin biosynthesis genes (PAL1, C4H, 4CL1, CCoAOMT, HCT, CCR1, and F5H1; Kim et al., 2012, 2013a, 2013b, 2014; Wang et al., 2020a).
It was previously revealed that the majority of promoters of lignin biosynthesis genes contain so-called AC elements, including AC-I (ACCTACC), AC-II (ACCAACC), and AC-III (ACCTAAC), that are recognized by transcription factors (Raes et al., 2003). MYB58 and MYB63 were shown to activate lignin biosynthesis genes by directly targeting the AC elements in Arabidopsis (Zhou et al., 2009). Members from subfamily 4 of the R2R3-MYB family can also target the AC elements to function as transcriptional repressors of the lignin biosynthesis pathway. AmMYB308 and AmMYB330 in Antirrhinum majus, EgMYB1 and EgMYB2 in Eucalyptus gunnii, PtMYB1, PtMYB4, PtMYB8, and PtMYB14 in Pinus teada, PgMYB1, PgMYB8, PgMYB14, and PgMYB15 in Picea glauca, ZmMYB31 and ZmMYB42 in maize (Zea mays), and Arabidopsis MYB4, MYB32, and MYB75 were all reported to directly regulate lignin biosynthesis genes (Tamagnone et al., 1998; Jin et al., 2000; Patzlaff et al., 2003; Preston et al., 2004; Goicoechea et al., 2005; Bhargava et al., 2010; Fornalé et al., 2010; Legay et al., 2010; Bomal et al., 2014).
The synthesis of lignin monomers involves the phenylpropanoid pathway, which is also shared by many other metabolites such as suberin, flavonoids, tannins, and lignans (Fraser and Chapple, 2011). The aromatic amino acid Phe, the end product of the shikimate pathway, serves as the precursor of downstream phenylpropanoids (Maeda and Dudareva, 2012; Tohge et al., 2013). Lignin biosynthesis is a high-energy-consuming and irreversible process; therefore, it is critical that lignin deposition is tightly controlled to avoid overconsumption of Phe. Transcription factors have been identified that coordinately regulate the expression of Phe biosynthesis and the downstream metabolism of Phe. Ectopic expression of the petunia (Petunia hybrida) transcription factor ODORANT1 (ODO1) in tomato (Solanum lycopersicum) can induce shikimate biosynthesis genes and phenylpropanoid-associated genes (Dal Cin et al., 2011). Green tea (Camellia sinensis) MYB4 negatively regulates phenylpropanoid and shikimate biosynthesis by directly targeting the AC elements of promoters of the genes involved in the two pathways (Li et al., 2017).
l-Phe is an aromatic amino acid required not only for protein biosynthesis but also for many secondary metabolites, such as flavonoids, condensed tannins, lignans, and lignins (Maeda and Dudareva, 2012). In plants, Phe is produced by two alternative pathways. Most Phe is produced via the chloroplast-localized arogenate pathway (Maeda et al., 2010). In addition, a cytosolic postchorismate Phe biosynthesis pathway has also been functionally characterized (Yoo et al., 2013; Qian et al., 2019). It was proposed that the alternative cytosolic pathway may contribute to produce specialized Phe-derived metabolites in response to biotic and abiotic stress in plants (Qian et al., 2019).
MYB20, MYB42, MYB43, and MYB85 were previously identified as secondary cell wall-related transcription factors based on a genome-wide transcriptome analysis of NST1 down-regulated mutants in Arabidopsis (Zhong et al., 2008). Dominant repression of MYB85, which is a transcriptional activator, substantially reduced secondary wall thickening in fiber cells, and overexpression of MYB85 caused ectopic deposition of lignin in epidermal and cortical cells in stems. However, how these MYBs are involved in secondary cell wall formation remains elusive. In this study, we report that MYB20, MYB42, MYB43, and MYB85 redundantly activate both Phe and lignin biosynthesis. In addition, MYB20, MYB42, MYB43, and MYB85 activate the transcription factor MYB4, which represses flavonoid biosynthesis to optimize Phe flux to the lignin biosynthesis pathway. This discovery uncovers another level of complexity in the regulatory network of plant lignin biosynthesis.
RESULTS
MYB20, MYB42, MYB43, and MYB85 Genes Are Regulated by NST3 and MYB46
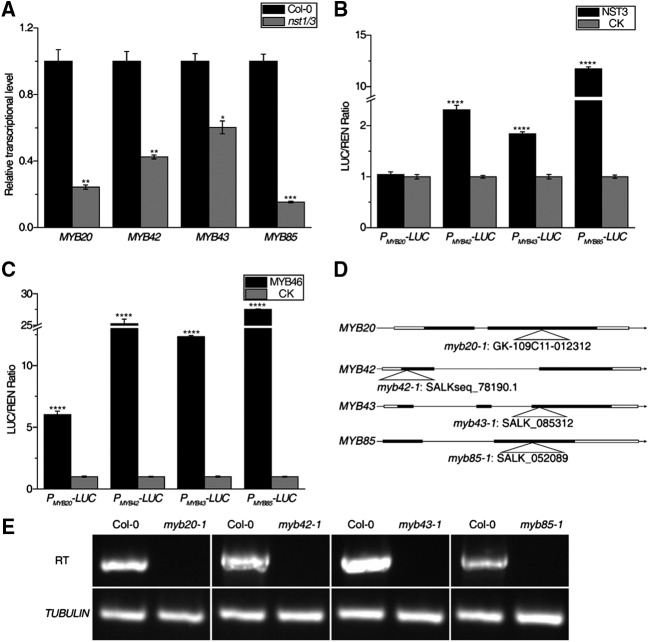
It was previously revealed that several unknown transcription factors are under the control of SND1 and NST1, including two NACs (SND2 and SND3) and eight MYB genes (MYB20, MYB42, MYB43, MYB52, MYB54, MYB69, MYB85, and MYB103; Zhong et al., 2008). Phylogenetic analysis demonstrated that MYB42 and MYB85 are closely related to each other, while MYB20 and MYB43 are likely more closely related to one another (Supplemental Fig. S1). Furthermore, the two subgroups are also closely related, but they are remotely related to the previously reported lignin transcriptional activators MYB58 and MYB63 (Supplemental Fig. S1). Therefore, we selected MYB20, MYB42, MYB43, and MYB85 to functionally characterize their roles in secondary cell wall biosynthesis. Consistent with the fact that MYB20, MYB42, MYB43, and MYB85 are regulated by NST3, the transcript levels of all four MYB genes were reduced in the double mutant nst1/3 (Fig. 1A). MYB46, which is a direct target of NST3, is also a master regulator of secondary cell wall biosynthesis, potentially controlling the expression of MYB20, MYB42, MYB43, and MYB85. To determine whether MYB46 and NST3 can activate MYB20, MYB42, MYB43, and MYB85 transcription, we performed transient expression assays in Nicotiana benthamiana leaves. The promoters of MYB20, MYB42, MYB43, and MYB85 were fused with Luciferase (LUC) to generate PMYB20-LUC, PMYB42-LUC, PMYB43-LUC, and PMYB85-LUC, respectively. MYB46 and NST3 were driven by the 35S promoter to act as effectors. Coexpression of MYB46 or NST3 can activate PMYB20-LUC, PMYB42-LUC, PMYB43-LUC, and PMYB85-LUC activity (Fig. 1, B and C). However, neither NST1 to NST3 nor MYB46 and MYB83 were able to bind these promoters in yeast one-hybrid assays (Supplemental Fig. S2).
Figure 1.
Description of myb20, myb42, myb43, and myb85 mutants. A, Relative transcript levels of MYB20, MYB42, MYB43, and MYB85 in the inflorescence stems of 33-d-old wild-type (Columbia-0 [Col-0]) and nst1/3 mutant plants were determined by reverse transcription quantitative PCR (RT-qPCR). The expression level in Col-0 was set to 1. Tubulin was used as the internal control. B and C, Transient expression assays showed that the promoters of MYB20, MYB42, MYB43, and MYB85 can be activated by NST3 (B) and MYB46 (C). The PMYB20-LUC, PMYB42-LUC, PMYB43-LUC, and PMYB85-LUC reporters were cotransformed with the indicated constructs in N. benthamiana leaves. The LUC/REN ratio represents the LUC activity relative to the REN activity. CK was the vector of GUS-pEarleyGate 101, used as a control, and was set to 1. Data are means ± sd of three biological replicates. Asterisks indicate significant differences by Student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001). D, Schematic diagrams of the structures of MYB20, MYB42, MYB43, MYB85, and T-DNA insertions. Solid boxes represent exons, black lines represent introns, and hollow boxes represent untranslated regions. E, RT-PCR analysis of MYB20, MYB42, MYB43, and MYB85 transcripts in the inflorescence stems of 33-d-old plants. Tubulin was used as a positive control.
Simultaneous Disruption of MYB20, MYB42, MYB43, and MYB85 Causes Plant Growth Defects
To further investigate the functions of these genes, the T-DNA insertion lines of myb20-1 (GK-109C11-012312), myb42-1 (SALKseq_78190.1), myb43-1 (SALK_085312), and myb85-1 (SALK_052089) were characterized and the insertion positions were verified by genomic sequencing (Fig. 1D). Reverse transcription PCR showed that these T-DNA insertion lines were null mutants (Fig. 1E). The double mutants myb20/43 and myb42/85 exhibited a normal growth phenotype, similar to the wild type (Fig. 2A). However, the quadruple mutant myb20/42/43/85 showed a semidwarfism phenotype compared with myb20/43 and myb42/85, indicating genetic redundancy among these genes (Fig. 2A). In addition, the quadruple mutant also showed shorter siliques with compromised fertility (Fig. 2, B–D).
Figure 2.
Disruption of MYB20/42/43/85 causes growth and fertility defects. A, Five-week-old plants of Col-0, the double mutants myb20/43 and myb42/85, and the quadruple mutant myb20/42/43/85. B, The inflorescence stems of 6-week-old plants of Col-0 and myb20/42/43/85. C, The upper inflorescence stems as in B. D, The middle inflorescence stems as in B.
MYB20, MYB42, MYB43, and MYB85 Coordinately Regulate Phe and Lignin Biosynthesis
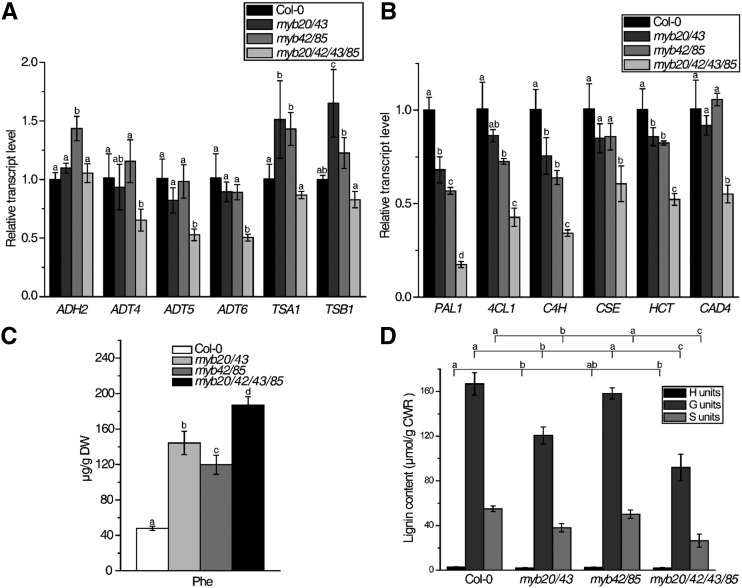
To determine the possible effects on gene expression of disrupting MYB20, MYB42, MYB43, and MYB85, we used high-throughput mRNA sequencing to identify differentially expressed genes in the quadruple mutant myb20/42/43/85 compared with wild-type plants. In the quadruple mutant myb20/42/43/85, a total of 2,596 genes were expressed at levels significantly different from the wild type (Supplemental Data Sets S1 and S2). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis showed that biosynthesis of secondary metabolites was changed greatly (Supplemental Fig. S3) and genes involved in lignin and Phe biosynthesis showed reduced expression in the quadruple mutant. To further verify the RNA sequencing data, we performed RT-qPCR to compare gene expression among the double mutants myb20/43 and myb42/85, the quadruple mutant myb20/42/43/85, and the wild type. myb20/42/43/85 plants exhibited reduced transcript levels of lignin biosynthesis genes such as PAL1, 4CL1, C4H, CSE, HCT, and CAD4, while the reduction was less severe in both double mutants (Fig. 3B). The transcript levels of ADT4, ADT5, and ADT6, which are specifically involved in Phe biosynthesis, were greatly reduced in myb20/42/43/85 plants but were not significantly changed in the myb20/43 and myb42/85 mutants, indicating genetic redundancy among the MYB genes (Fig. 3A). On the other hand, the expression of ADH2, TSA1, and TSB1, involved in Tyr and Trp biosynthesis, was not significantly changed in myb20/42/43/85 mutants even though these genes are also related to the shikimate biosynthesis pathway (Fig. 3A).
Figure 3.
MYB20, MYB42, MYB43, and MYB85 redundantly regulate Phe and lignin biosynthesis. A and B, Relative transcript levels of aromatic amino acid (A) and lignin (B) biosynthesis genes in the inflorescence stems of 33-d-old Arabidopsis wild-type, myb20/43, myb42/85, and myb20/42/43/85 mutant plants as determined by RT-qPCR. C, Phe level in the inflorescence stems of plants with the same genotypes and period as in A. DW, Dry weight. D, Lignin composition analysis of the inflorescence stems of plants with the same genotypes and period as in A. Gas chromatography (GC) quantification is shown for p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) lignin units by thioacidolysis. CWR, Cell wall residue. Data are means ± sd of three biological replicates. Based on Tukey’s test, columns with the same letters are not significantly different and columns with different letters are significantly different (P < 0.05; one-way ANOVA).
The thioacidolysis method was used to determine the lignin composition of double mutants myb20/43 and myb42/85 and the quadruple mutant myb20/42/43/85. As shown in Figure 3D, the G and S subunits were significantly reduced in myb20/42/43/85 plants, while the reduction was less severe in the two double mutants, which is consistent with the growth phenotypes and gene expression analysis (Figs. 2A and 3B). To further confirm the impact on lignin biosynthesis of disrupting MYB20, MYB42, MYB43, and MYB85, the total lignin content of mutants and control plants was quantified by Klason analysis. As shown in Supplemental Figure S4, myb20/42/43/85 plants produced significantly reduced lignin, while double mutants myb20/43 and myb42/85 contained similar lignin content to wild-type plants.
Phe content was also quantified using the same mature stem tissues used for lignin measurements. Interestingly, both myb20/43 and myb42/85 accumulated greatly elevated levels of Phe, and myb20/42/43/85 plants contained even more Phe than the double mutants (Fig. 3C). These results were not consistent with the fact that ADT genes were down-regulated in the mutants. However, the increase of the free form of Phe may be due to the reduction of downstream Phe-derived lignin biosynthesis.
MYB20, MYB42, MYB43, and MYB85 Activate Phe and Lignin Biosynthesis Gene Expression
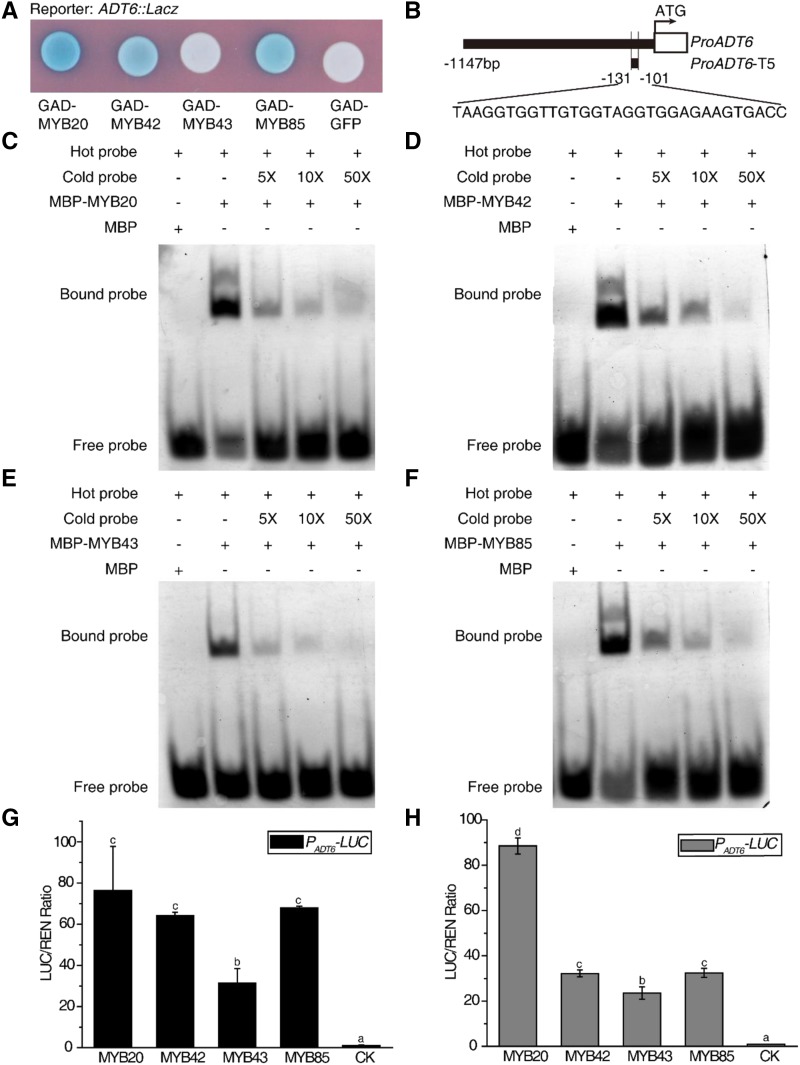
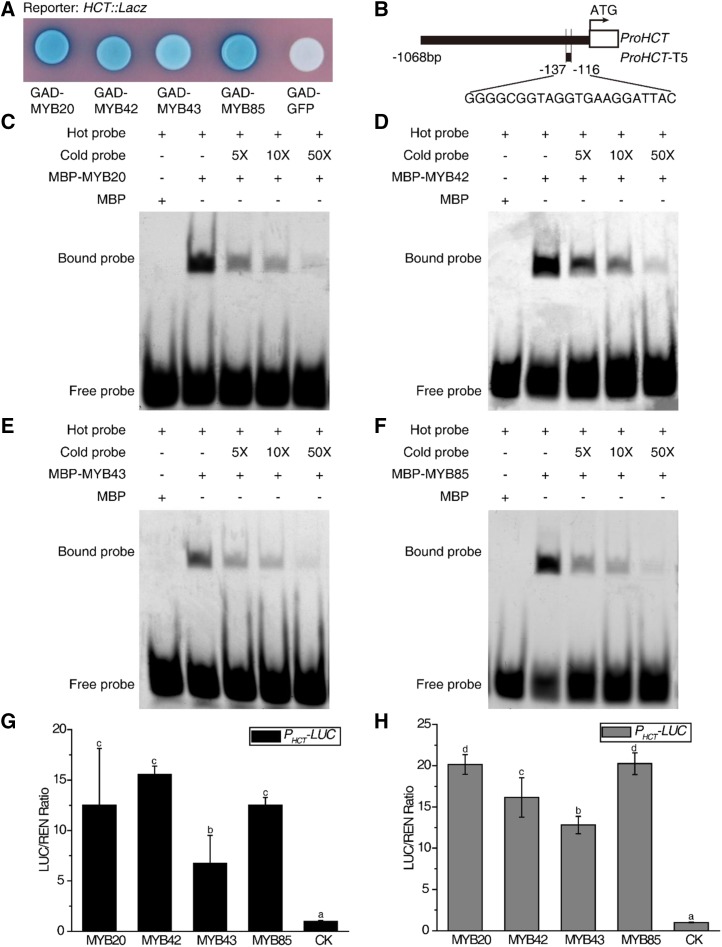
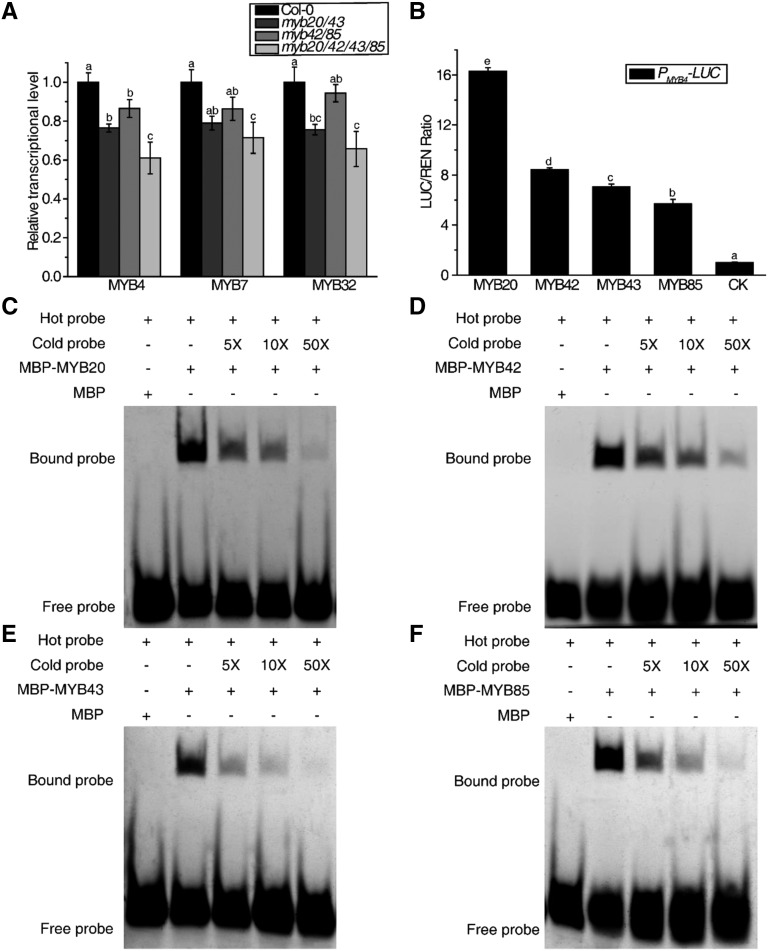
To investigate the molecular mechanism underlying the roles of the MYB transcription factors in lignin and Phe biosynthesis pathways, a yeast one-hybrid assay was performed. We found that full-length MYB proteins had very low expression levels in the EGY48 yeast strain, but truncated MYB proteins could be easily expressed (Supplemental Fig. S5). Therefore, we used truncated proteins in the yeast one-hybrid assay. ADT6 and HCT were selected as representative Phe and lignin biosynthesis genes, respectively. MYB20, MYB42, and MYB85 were able to activate the LacZ reporter genes driven by the ADT6 and HCT promoters, while MYB43 could only activate the LacZ reporter gene driven by the HCT promoter (Figs. 4A and 5A). We utilized the electrophoretic mobility shift assay (EMSA) to further verify the direct binding to the ADT6 and HCT promoters. All four recombinant MBP-MYB fusion proteins caused an upshift of the labeled ADT6 promoter or HCT promoter. By contrast, MBP alone did not generate the same mobility shift (Figs. 4, C–F–F, and 5, C–F–F). The schematic diagrams of the promoters in yeast one-hybrid assays and EMSA are shown in Figures 4B and 5B.
Figure 4.
MYBs activate Phe biosynthesis genes. A, GAD-MYBs activate the expression of the LacZ reporter gene driven by the ADT6 promoter in yeast. GFP was used as the negative control. B, The top schematic diagram and the bottom one show the promoter regions of ADT6 in A and in C to F separately. C to F, EMSA showed that MYB20 (C), MYB42 (D), MYB43 (E), and MYB85 (F) bind to the promoter of ADT6. The cold probes used as competitors were with the same sequence as the TAMRA-labeled probes. Truncated MYB20 (Δ166–282 amino acids), MYB42 (Δ138–286 amino acids), MYB43 (Δ201–327 amino acids), and MYB85 (Δ168–266 amino acids) were used for yeast one-hybrid assay (A) and EMSA (C–F). G and H, Transient expression assays showed that the ADT6 promoter can be activated by MYB20/42/43/85 in Arabidopsis Col-0 protoplasts (G) and in Arabidopsis myb20/42/43/85 mutant protoplasts (H). CK was the empty vector used as a control and was set to 1. Data are means ± sd of three biological replicates. Based on Tukey’s test, columns with the same letters are not significantly different and columns with different letters are significantly different (P < 0.05; one-way ANOVA).
Figure 5.
MYBs activate lignin biosynthesis genes. A, GAD-MYBs activate the expression of the LacZ reporter gene driven by the HCT promoter in yeast. GFP was used as the negative control. B, The top schematic diagram and the bottom one show the promoter regions of HCT in A and in C to F separately. C to F, EMSA showed that MYB20 (C), MYB42 (D), MYB43 (E), and MYB85 (F) bind to the promoter of HCT. The cold probes used as competitors were with the same sequence as the FAM-labeled probes. Truncated MYB20 (Δ166–282 amino acids), MYB42 (Δ138–286 amino acids), MYB43 (Δ201–327 amino acids), and MYB85 (Δ168–266 amino acids) were used for yeast one-hybrid assay (A) and EMSA (C–F). G and H, Transient expression assays showed that the HCT promoter can be activated by MYB20/42/43/85 in Arabidopsis Col-0 protoplasts (G) and in Arabidopsis myb20/42/43/85 mutant protoplasts (H). CK was the empty vector used as a control and was set to 1. Data are means ± sd of three biological replicates. Based on Tukey’s test, columns with the same letters are not significantly different and columns with different letters are significantly different (P < 0.05; one-way ANOVA).
To further confirm that MYB20, MYB42, MYB43, and MYB85 were able to activate the expression of ADT6 and HCT, we performed a transient expression assay in Arabidopsis protoplasts. Coexpression of MYB20, MYB42, MYB43, or MYB85 dramatically activated PADT6-LUC and PHCT-LUC activity in Arabidopsis Col-0 protoplasts (Figs. 4G and 5G). To further examine whether each MYB is able to activate the genes independently, a similar transient expression assay was carried out in Arabidopsis myb20/42/43/85 mutant protoplasts. It turned out that each MYB was able to activate downstream targets alone (Figs. 4H and 5H), suggesting that these four MYBs are not interdependent.
MYB20, MYB42, MYB43, and MYB85 Optimize Phe Distribution in the Phenylpropanoid Biosynthesis Pathway
Our RNA sequencing data indicated that the previously identified phenylpropanoid pathway transcription factor MYB7 was down-regulated in myb20/42/43/85 compared with the wild type (Supplemental Data Set S2). Through RT-qPCR analysis, we showed that not only MYB7 but also two homologous genes, MYB4 and MYB32, showed lower transcript levels in the myb20/42/43/85 mutant compared with the wild type (Fig. 6A). Transient expression assays in N. benthamiana leaves indicated that MYB20, MYB42, MYB43, and MYB85 activate the promoters of MYB4 (Fig. 6B). EMSA was performed to investigate whether MYB4 was directly targeted, as described previously (Zhao et al., 2007). EMSA showed that all four MBP-MYB proteins could bind the MYB4 promoter but not the MBP tag (Fig. 6, C–F).
Figure 6.
MYBs activate MYB4/7/32 genes directly. A, MYB4, MYB7, and MYB32 transcript levels in the inflorescence stems of 33-d-old wild-type, myb20/43, myb42/85, and myb20/42/43/85 mutant plants as determined by RT-qPCR. B, Transient expression assays showed that the MYB4 promoter can be activated by MYB20, MYB42, MYB43, and MYB85 in N. benthamiana leaves. CK was the vector of GUS-pEarleyGate 101, used as a control, and was set to 1. C to F, EMSA showed that MYB20 (C), MYB42 (D), MYB43 (E), and MYB85 (F) bind to the promoter of MYB4. The cold probes used as competitors were with the same sequence as the FAM-labeled probes. Truncated MYB20 (Δ166–282 amino acids), MYB42 (Δ138–286 amino acids), MYB43 (Δ201–327 amino acids), and MYB85 (Δ168–266 amino acids) were used for EMSA (C–F). Data are means ± sd of three biological replicates. Based on Tukey’s test, columns with the same letters are not significantly different and columns with different letters are significantly different (P < 0.05; one-way ANOVA).
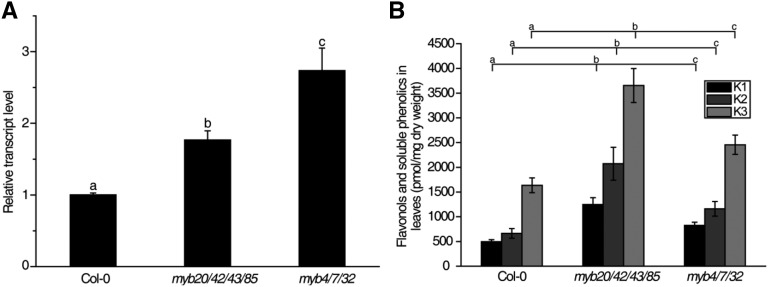
It has been reported that MYB4 can repress the expression of Chalcone Synthase (CHS), encoding the enzyme that catalyzes the first step of flavonoid biosynthesis, which competes with lignin biosynthesis for Phe as a precursor (Jin et al., 2000). Consistent with this finding, the transcript level of CHS was significantly enhanced in myb20/42/43/85 compared with the wild type and was increased even more in the myb4/7/32 mutant (Fig. 7A). These results suggest that MYB20, MYB42, MYB43, and MYB85 activate MYB4, MYB7, and MYB32, resulting in the down-regulation of CHS expression. To investigate the possible effects on the phenylpropanoid biosynthesis pathway of disrupting MYB20, MYB42, MYB43, and MYB85, we compared flavonoid accumulation among myb20/42/43/85, myb4/7/32, and wild-type plants. As shown in Figure 7B, kaempferol 3-O-[6′′-O-(rhamnosyl)glucoside] 7-O-rhamnoside, kaempferol 3-O-glucoside 7-O-rhamnoside, and kaempferol 3-O-rhamnoside 7-O-rhamnoside accumulated at significantly higher amounts in myb20/42/43/85 and myb4/7/32. In addition, myb20/43, myb42/85, and myb20/42/43/85 also accumulated enhanced levels of anthocyanin (Supplemental Fig. S6). These data suggest that down-regulation of the transcription repressors MYB4, MYB7, and MYB32 in myb20/42/43/85 leads to enhanced flavonoid biosynthesis due to elevated expression of CHS.
Figure 7.
Disruption of MYB20/42/43/85 promotes soluble flavonoid accumulation. A, RT-qPCR analysis of CHS in the inflorescence stems of 5-week-old wild-type, myb20/42/43/85, and myb4/7/32 mutant plants. B, Quantification of the three major flavonols in the inflorescence stems of the same plants as in A. K1, Kaempferol 3-O-[6′′-O-(rhamnosyl)glucoside] 7-O-rhamnoside; K2, kaempferol 3-O-glucoside 7-O-rhamnoside; K3, kaempferol 3-O-rhamnoside 7-O-rhamnoside. Data are means ± sd of three biological replicates. Based on Tukey’s test, columns with the same letters are not significantly different and columns with different letters are significantly different (P < 0.05; one-way ANOVA).
DISCUSSION
During the last decade, the transcriptional regulation of secondary cell wall formation has been extensively studied. Lignin biosynthesis has been shown to be under the control of the NAC-MYB-based transcriptional regulatory network (Ohtani and Demura, 2019). NAC proteins as master switches coordinately regulate the entirety of secondary cell wall formation, including cellulose, hemicellulose, and lignin (Zhong et al., 2010). Lignin biosynthesis is derived from the aromatic amino acid Phe, and the deposition process is believed to be irreversible. Therefore, lignin biosynthesis is not only coordinately regulated along with other secondary cell wall components but also interacts with primary metabolic pathways related to Phe biosynthesis and other branches of the phenylpropanoid pathway (Li et al., 2016; Ohtani et al., 2016).
MYB58 and MYB63 were first reported in Arabidopsis as lignin-specific transcriptional activators (Zhou et al., 2009). These MYBs are able to directly activate lignin biosynthesis genes, including early genes such as PAL, C4H, and 4CL that are shared by other branches of the phenylpropanoid pathway such as flavonoid biosynthesis. MYB4 was shown to directly target lignin biosynthesis genes as repressors (Shen et al., 2012; Li et al., 2017). Currently, there is no evidence that MYB58/63 and MYB4 dynamically regulate lignin biosynthesis. MYB103, an NST1-regulated transcription factor, was recently suggested to be involved in syringyl lignin biosynthesis (Öhman et al., 2013). However, MYB103 did not directly target F5H, which is required for syringyl lignin formation (Öhman et al., 2013).
Phe derived from the shikimate pathway, which also gives rise to the other two aromatic amino acids Tyr and Trp, serves as a precursor for many secondary metabolites such as lignins, flavonoids, coumarins, and lignans (Maeda and Dudareva, 2012). Since lignin deposition is a very energy-consuming and nonreversible process, it is critical that Phe distribution among the phenylpropanoid pathway and protein synthesis is tightly controlled. AtMYB12 was previously shown to not only increase flavonoid biosynthesis but also to enhance the supply of precursors and carbon from primary metabolism (Zhang et al., 2015). It was also reported that a MYB transcription factor ODO1 from petunia was able to activate genes involved in the shikimate pathway and the biosynthesis of Phe-derived volatile compounds in tomato (Dal Cin et al., 2011). However, it remained unclear how plants coordinately regulate Phe formation in response to lignin biosynthesis.
It was previously shown that MYB85 was able to activate the expression of a lignin biosynthesis gene, 4CL1, in a transient assay in Arabidopsis protoplasts (Zhong et al., 2008). In this study, we provided evidence that MYB85 and MYB42, along with MYB20 and MYB43, function as regulators of the lignin and shikimate biosynthesis pathways. MYB20, MYB42, MYB43, MYB85, MYB58, and MYB63 belong to a monophyletic group in the tree of MYB transcription factors, indicating that they have highly similar protein sequences (Supplemental Fig. S1). Among them, AtMYB20 and AtMYB43 have the highest similarity with AtMYB42 and AtMYB85 in Arabidopsis, forming a monophyletic clade with their putative orthologs in Amborella trichopoda and rice (Oryza sativa; Supplemental Fig. S1).
Disruption of MYB20, MYB42, MYB43, and MYB85 significantly reduced the expression of genes involved in lignin biosynthesis (Fig. 3B). Consistent with gene reduction, lignin content was significantly reduced in myb20/42/43/85 plants, indicating that MYB20, MYB42, MYB43, and MYB85 are regulators of lignin biosynthesis (Fig. 3D; Supplemental Fig. S4). Interestingly, ADT genes that are specifically involved in Phe biosynthesis were also down-regulated in myb20/42/43/85 plants (Fig. 3A). However, free Phe content was increased despite the fact that ADT genes were down-regulated (Fig. 3C). This may be a consequence of down-regulated flux toward lignin biosynthesis. Even though Phe must be shunted toward enhanced flavonoid biosynthesis, it seems that lignin biosynthesis is down-regulated to a higher extent. Our data suggested that MYB20, MYB42, and MYB85 could directly target both lignin biosynthesis genes and ADT genes (Figs. 4A and 5A). However, MYB43 was only able to bind to the promoter of lignin biosynthesis genes, and not ADT6, in our yeast one-hybrid assay. EMSA data indicated that all four recombinant MBP-MYB fusion proteins caused an upshift of the labeled ADT6 or HCT promoters. Interestingly, each MYB protein could activate ADT or lignin biosynthesis genes independently (Figs. 4G and 5G).
Like lignin, flavonoids are derived from Phe. Studies in Arabidopsis, as well as in maize, have pointed out that R2R3-MYBs have dual activation/repression effects on genes from flavonoid and lignin pathways that compete for carbon. For example, MYB32 and MYB4 in Arabidopsis were shown to influence the composition of the pollen wall by changing the flux through phenylpropanoid pathways (Preston et al., 2004). Similarly, Arabidopsis MYB75, a positive regulator of anthocyanin biosynthesis, was shown to regulate lignin accumulation negatively in secondary cell walls, which reflects carbon flux redistribution within the branches of phenylpropanoid metabolism (Bhargava et al., 2010). Overexpression of ZmMYB31 from maize down-regulated several genes involved in monolignol biosynthesis and up-regulated flavonoid biosynthesis in Arabidopsis (Fornalé et al., 2010). However, it remains unclear how these MYBs fine-tune the biosynthesis of lignins and flavonoids. The channeling of carbon toward lignin and flavonoid pathways is thought to be highly coordinated by cross talk between these pathways. It was reported that AtMYB75 can interact with AtKNAT7, which functions as a negative regulator of secondary cell wall formation in interfascicular fibers (Bhargava et al., 2010; Wang et al., 2020a). A recent study further demonstrated that AtMYB4, a transcriptional repressor of lignin biosynthesis, along with its homologs MYB7 and MYB32, can attenuate the transcriptional function of MYB75-TT8-TTG1 complexes in flavonoid metabolism by interacting with the TT8 transcription factor (Wang et al., 2020b). The coordinated action of MYB repressors and activators has been proposed as a fine-tuning mechanism for the regulation of plant secondary metabolism (Bomal et al., 2014). In this study, we found that MYB20, MYB42, MYB43, and MYB85 redundantly activate MYB4, which was reported to repress the expression of CHS, which encodes the enzyme that catalyzes the first step of flavonoid biosynthesis. Consistent with the finding that the transcript level of CHS is enhanced in myb20/42/43/85, flavonoid biosynthesis is up-regulated in the mutant. In addition, MYB20 was shown to positively regulate the CHS promoter (Öhman et al., 2013), indicating that multiple levels of transcriptional control by MYB regulators can allow fine-tuned regulation of carbon flux along the phenylpropanoid pathway. In conclusion, our results showed that MYB20, MYB42, MYB43, and MYB85, in addition to positively regulating lignin biosynthesis by directly activating lignin and Phe biosynthesis genes, repress flavonoid accumulation through MYB4-mediated regulation of flavonoid biosynthesis. This suggests that the transcription factors MYB20, MYB42, MYB43, and MYB85 control carbon flow into the phenylpropanoid pathway by activating Phe biosynthesis to ensure precursor supply while at the same time repressing other competing metabolic pathways to optimize lignin biosynthesis. This discovery uncovers another level of complexity in the regulatory network of the plant lignin biosynthesis pathway.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) mutants myb20-1 (GK-109C11-012312), myb42-1 (SALKseq_78190.1), myb43-1 (SALK_085312), myb85-1 (SALK_052089), nst1 (SALK_120377), and nst3 (SALK_149909) were obtained from the Arabidopsis Biological Resource Center at Ohio State University. Seeds were sterilized with 3% (v/v) sodium hypochlorite, sown on one-half-strength Murashige and Skoog (MS) plates, stored at 4°C, and then transferred into a chamber under long-day conditions at 22°C (day/night cycle of 16/8 h). The primers used for genotyping are listed in Supplemental Table S1.
Phylogenetic Tree Analysis
Arabidopsis MYB20 was used as query to perform a BLASTP search against all genes annotated on 19 published genomes covering all main archaeplastida clades (Supplemental Table S2). Then, similar sequences with E < 1e-3 and an annotation term MYB transcription factors assigned by InterProScan (v5.24) were selected (Jones et al., 2014). Multiple protein sequence alignments of MYB transcription factors were performed using MAFFT (v7), and positions with more than 50% gaps were removed using the Phyutility program (v2.2.6; Smith and Dunn, 2008; Katoh and Standley, 2013). Phylogenetic analyses were performed with a maximum likelihood method by FastTree (v2.1) with optimal model of amino acid substitution (LG+G) chosen based on the results of ProtTest (v3.4.2), and a bootstrap analysis with 500 replicates was performed (Price et al., 2010; Darriba et al., 2011). Sequences belonging to the monophyletic group of Arabidopsis MYB20, MYB43, MYB42, MYB85, MYB58, and MYB63 were identified and selected for another round of phylogenetic analyses as described above. Only sequences annotated from Arabidopsis, rice (Oryza sativa), Amborella trichopoda, Salvinia cucullata, Azolla filiculoides, Sphagnum fallax, and Physcomitrella patens genomes were displayed on the final tree. Sequences with very atypical sizes and extremely long braches were manually removed.
Anthocyanin Measurement
Seedlings were grown on one-half strength MS medium containing 100 μm norflurazon. The seedlings were then grown for 4 d in a growth chamber. Photographs of the seedlings were taken by a microscope. To analyze anthocyanin content, the seeds were grown on plates with one-half strength MS medium containing 1% (w/v) Suc for 14 d and then transferred to one-half strength MS medium containing 12% (w/v) Suc for 3 d (Yonekura-Sakakibara et al., 2012). Plants were harvested and frozen with liquid nitrogen immediately. Anthocyanins were extracted with methanol containing 0.2% (v/v) formic acid and 30% (v/v) water and then placed at −20°C overnight to remove chlorophyll. The determination of anthocyanin content was carried out by spectrophotometry at 530 nm. The anthocyanin level of wild-type Col-0 (1.13 mg cyanidin 3-O-glucoside equivalence g–1) was set to 100.
RNA Analyses
Total RNA was extracted from the inflorescence stems of 33-d-old Arabidopsis wild-type Col-0 and myb20/42/43/85 mutants using the RNeasy Plant Mini Kit (Qiagen). Three biological replicates of each sample were sent to BGI Genomics (The Beijing Genomics Institute) for the construction of libraries and RNA sequencing. Data were analyzed on the Dr. Tom system from BGI.
The double mutants, myb20/43 and myb42/85, were planted and harvested at the same time as wild-type Col-0 and the myb20/42/43/85 mutant. Total RNA was reverse transcribed by Superscript Reverse Transcriptase III (Invitrogen). RT-qPCR was performed with SYBR Premix Ex Taq (Takara). The average levels of transcripts were from three biological replicates, and each biological replicate was from triplicate cDNA. The Arabidopsis Tubulin gene was used as a reference. The primers are listed in Supplemental Table S1.
KEGG Pathway Analysis
KEGG pathway enrichment analysis was performed using the Dr. Tom system from BGI. According to KEGG pathway annotation, phyper function in the R project was used to calculate P values and false discovery rates (FDRs). Q value was the correction of the P value. Terms with Q ≤ 0.05 were considered as enriched.
Dual-Luciferase Assay
To test transcriptional activation, the dual-luciferase assay was performed in Nicotiana benthamiana leaves. About 1 kb of the promoters of MYB20, MYB42, MYB43, MYB85, and MYB4 was cloned into the pGreenII 0800-LUC vector to generate P-LUC reporter constructs (Hellens et al., 2005). Each promoter in pGreenII 0800-LUC was transformed into Agrobacterium tumefaciens strain GV3101 containing the helper plasmid pSoup-P19 (Hellens et al., 2005). The full-length coding sequences (without stop codons) of NST3, MYB46, MYB20, MYB42, MYB43, MYB85, and GUS were cloned into the pEarleyGate 101 vector. These six constructs were transformed into A. tumefaciens strain GV3101. Overnight cultures of A. tumefaciens strains were resuspended in infiltration buffer (10 mm MgCl2, 10 mm MES, and 0.1 mm acetosyringone) to OD600 ∼ 0.6 and incubated at room temperature for 2 h. The reporter:effector ratio was 2:8. Different combinations of A. tumefaciens suspension were infiltrated onto N. benthamiana leaves. After 48 h, leaf samples were collected and ground in liquid nitrogen for the dual-luciferase assay. Luciferase activities were detected using the Dual-Luciferase Reporter Assay System (Promega). Each sample contained three biological repeats.
To examine whether each MYB was able to activate the genes independently, a transcriptional activation assay was conducted in Arabidopsis Col-0 and myb20/42/43/85 protoplasts. About 1 kb of promoters of ADT6 and HTC was cloned into pGreenII 0800-LUC. The coding sequences of MYB20, MYB42, MYB43, and MYB85 were inserted into p2GW7 under the control of the 35S promoter. After the protoplast preparation and subsequent transfection were completed (Yoo et al., 2007), samples were collected. The test of luciferase activities was described above. The raw data are given in Supplemental Table S3.
Yeast One-Hybrid Assays
GAD fusion constructs were cotransformed with the LexAop::LacZ (Clontech) reporter gene plasmid into the EGY48 yeast strain. Transformants were grown on appropriate dropout plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside for blue color test.
EMSA
The DNA fragments of MYBs were cloned into pMAL-c2x vector and expressed in the Escherichia coli BL21 (DE3) cell line, which were cultured in Luria-Bertani culture at 37°C to an OD600 between 0.6 and 0.8. To induce protein expression, a final concentration of 0.5 mm isopropyl β-d-1-thiogalactopyranoside was added to the cultures, after which the cells were incubated for 8 h at 28°C. Cells were harvested and resuspended in buffer containing 20 mm Tris-HCl, pH 7.4, 200 mm NaCl, and 1 mm EDTA, pH 8. After sonication and centrifugation, the MBP-TF proteins were purified using amylose resin from the supernatant.
The oligonucleotide probes were labeled with TAMRA or FAM and synthesized from Sangon Biotech. Then, the oligonucleotides were diluted to 10 μm, heated to 95°C for 5 min, and slowly cooled to room temperature. Briefly, 1 pmol of labeled probe was incubated with 0.1 μg of the indicated protein in 20 μL of reaction buffer (25 mm HEPES, pH 8, 40 mm KCl, 5 mm MgCl2, 1 mm DTT, 1 mm EDTA, 8% [v/v] glycerol, and 1 μg of poly[dI-dC]) for 20 min at room temperature. Then, 5 μL of 5× loading buffer (12.5% [w/v] Ficoll-400, 0.2% [w/v] bromophenol blue, and 0.2% [w/v] xylene cyanol FF) was added to the reaction mixture and loaded onto a 4.5% polyacrylamide gel in 0.5× Tris-borate/EDTA. The labeled DNAs were detected using a Typhoon FLA 9500 Instrument (General Electric).
Amino Acid Extraction and Analysis
Amino acids were extracted from 50 mg (dry weight) of 5-week-old Arabidopsis stem samples with 1 mL of 0.5 m HCl solution. The supernatants were subsequently filtered using a 0.22-μm nylon membrane, and 250 μL of the extraction supernatant was transferred into autosamper vials. Finally, the mixture was diluted to 1 mL with 20% (v/v) water in acetonitrile prior to further analysis. Ultra-performance liquid chromatography (UPLC)-electrospray ionization-tandem mass spectrometry analysis was carried out on an Agilent 1290 LC system coupled to an Ultivo Triple Quadrupole mass spectrometer by means of an electrospray ionization probe. Eluent A was 20 mm ammonium formate in water (pH 3), while eluent B was 20 mm aqueous ammonium formate (pH 3) in 9:1 acetonitrile:water. The separation gradient was as follows: 100% B at 0 min, 70% B at 5 min, and 100% B at 8 min. The column temperature was maintained at 25°C. The injection volume of each sample was 1 μL. The multiple reaction monitoring mass spectrometry method was employed for the quantitation of amino acids. Data for Phe were acquired in positive ion mode under the following operating conditions: desolvation temperature, 330°C; desolvation gas (N2) rate, 13 L min−1; collision energy, 13 V for quantitative ion (mass-to-charge ratio [m/z] 120.1) and 29 V for qualitative ion (m/z 103).
Determination of Lignin Composition
Thioacidolysis analysis was performed to determine lignin composition. Briefly, 30-mg samples were reacted with 3 mL of 0.2 m BF3-etherate in an 8.75:1 dioxane:ethanethiol mixture at 100°C for 4 h. After adding 4 mL of water, the mixture was extracted using 4 mL of dichloromethane (three times). The organic layer was separated and dried under a stream of nitrogen. After derivatization with N,O-bis-(trimethylsilyl)trifluoroacetamide + 1% (w/v) trimethylchlorosilane (Sigma-Aldrich), lignin-derived monomers were identified by GC-mass spectrometry and quantified by GC as their trimethylsilyl derivatives. GC-mass spectrometry was performed on an Agilent Intuvo 9000 GC system with a 5977B series mass selective detector (column, HP-5MS; 30 m × 0.25 mm; 0.25-μm film thickness), and mass spectra were recorded in electron impact mode (70 eV) with a 50 to 650 m/z scanning range.
Quantification of Flavonoids
Samples of three replicates of Arabidopsis stems were collected and ground into a fine powder with a mortar and pestle under liquid nitrogen. Twenty-microgram dried samples were extracted with 1 mL of 80% (v/v) methanol (containing 100 μm naringenin as an internal reference), and then ultrasonic processing was conducted for 30 min at room temperature. After that, the samples were centrifuged at 14,000g for 10 min, and the supernatant was transferred to a sterile 1.5-mL Eppendorf tube for another centrifugation as above. A total of 300 μL of sterile Milli-Q water was added to each 1-mL supernatant solution and stored at −20°C overnight to precipitate chlorophyll. The following day, 5 μL of the supernatant solution after another centrifugation was injected for UPLC analysis. The identification of soluble flavonoid compounds was described previously (Yonekura-Sakakibara et al., 2008), and quantification of detected compounds was based on the standard curves of a kaempferol standard detected in the same UPLC runs.
Statistical Analyses
The SPSS 20.0 statistical package (IBM SPSS Statistics for Windows, Version 20.0; IBM Corp) was used for the statistical data analysis. Statistical analysis was done with univariate analysis and Scheffe posthoc testing at the 5% level. Samples included in the analysis were arranged based on at least three biological replicates.
Data Availability
RNA sequencing data have been uploaded to the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) with the accession number PRJNA578015.
Accession Numbers
The Arabidopsis Information Resource accession numbers of genes mentioned are as follows: AT2G46770 (NST1), AT3G61910 (NST2), AT1G32770 (NST3), AT5G12870 (MYB46), AT1G66230 (MYB20), AT4G12350 (MYB42), AT5G16600 (MYB43), AT4G22680 (MYB85), AT4G38620 (MYB4), AT2G16720 (MYB7), AT4G34990 (MYB32), AT2G37040 (PAL1), AT2G30490 (C4H), AT1G15710 (ADH2), AT3G44720 (ADT4), AT5G22630 (ADT5), AT1G08250 (ADT6), AT1G51680 (4CL1), AT3G19450 (CAD4), AT1G52760 (CSE), AT5G48930 (HCT), AT3G54640 (TSA1), AT5G54810 (TSB1), and AT5G13930 (CHS).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogenetic tree analysis of MYBs in 19 published genomes in main archaeplastida clades.
Supplemental Figure S2. NST1/2/3 and MYB46/83 could not bind the promoters of MYB20/42/43/85 directly.
Supplemental Figure S3. KEGG pathway enrichment analysis.
Supplemental Figure S4. Lignin content Klason analysis of the wild type (Col-0), myb20/43, myb42/85, and myb20/42/43/85.
Supplemental Figure S5. Truncated MYB proteins have higher expression in yeast.
Supplemental Figure S6. Disruption of MYB20/42/43/85 promotes anthocyanin accumulation.
Supplemental Table S1. Primers used for vector construction and gene expression analysis.
Supplemental Table S2. Resources of 19 representative genomes used in phylogenetic tree analysis.
Supplemental Table S3. Source data of the firefly and Renilla luciferase signal in the dual-luciferase assay.
Supplemental Data Set S1. A full list of the genes that are up-regulated in myb20/42/43/85 (FDR < 0.05, log2 fold change > 1).
Supplemental Data Set S2. A full list of the genes that are down-regulated in myb20/42/43/85 (FDR < 0.05, log2 fold change < −1).
Footnotes
This work was supported by the Youth Thousand Scholar Program of China (grant no. 31522008), the National Natural Science Foundation of China (grant no. 31570296), and the School of Life Sciences, Tsinghua University.
Articles can be viewed without a subscription.
References
- Bhargava A, Mansfield SD, Hall HC, Douglas CJ, Ellis BE (2010) MYB75 functions in regulation of secondary cell wall formation in the Arabidopsis inflorescence stem. Plant Physiol 154: 1428–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomal C, Duval I, Giguère I, Fortin É, Caron S, Stewart D, Boyle B, Séguin A, MacKay JJ (2014) Opposite action of R2R3-MYBs from different subgroups on key genes of the shikimate and monolignol pathways in spruce. J Exp Bot 65: 495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz ND, Chapple C (2010) The genetics of lignin biosynthesis: Connecting genotype to phenotype. Annu Rev Genet 44: 337–363 [DOI] [PubMed] [Google Scholar]
- Chen F, Tobimatsu Y, Havkin-Frenkel D, Dixon RA, Ralph J (2012) A polymer of caffeyl alcohol in plant seeds. Proc Natl Acad Sci USA 109: 1772–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tobimatsu Y, Jackson L, Nakashima J, Ralph J, Dixon RA (2013) Novel seed coat lignins in the Cactaceae: Structure, distribution and implications for the evolution of lignin diversity. Plant J 73: 201–211 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ, Jarvis MC (2012) Comparative structure and biomechanics of plant primary and secondary cell walls. Front Plant Sci 3: 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Cin V, Tieman DM, Tohge T, McQuinn R, de Vos RC, Osorio S, Schmelz EA, Taylor MG, Smits-Kroon MT, Schuurink RC, et al. (2011) Identification of genes in the phenylalanine metabolic pathway by ectopic expression of a MYB transcription factor in tomato fruit. Plant Cell 23: 2738–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D (2011) ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 27: 1164–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornalé S, Shi X, Chai C, Encina A, Irar S, Capellades M, Fuguet E, Torres JL, Rovira P, Puigdomènech P, et al. (2010) ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. Plant J 64: 633–644 [DOI] [PubMed] [Google Scholar]
- Fraser CM, Chapple C (2011) The phenylpropanoid pathway in Arabidopsis. The Arabidopsis Book 9: e0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goicoechea M, Lacombe E, Legay S, Mihaljevic S, Rech P, Jauneau A, Lapierre C, Pollet B, Verhaegen D, Chaubet-Gigot N, et al. (2005) EgMYB2, a new transcriptional activator from Eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. Plant J 43: 553–567 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoson T, Wakabayashi K (2015) Role of the plant cell wall in gravity resistance. Phytochemistry 112: 84–90 [DOI] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19: 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. (2014) InterProScan 5: Genome-scale protein function classification. Bioinformatics 30: 1236–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol 30: 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WC, Kim JY, Ko JH, Kang H, Han KH (2014) Identification of direct targets of transcription factor MYB46 provides insights into the transcriptional regulation of secondary wall biosynthesis. Plant Mol Biol 85: 589–599 [DOI] [PubMed] [Google Scholar]
- Kim WC, Kim JY, Ko JH, Kim J, Han KH (2013a) Transcription factor MYB46 is an obligate component of the transcriptional regulatory complex for functional expression of secondary wall-associated cellulose synthases in Arabidopsis thaliana. J Plant Physiol 170: 1374–1378 [DOI] [PubMed] [Google Scholar]
- Kim WC, Ko JH, Han KH (2012) Identification of a cis-acting regulatory motif recognized by MYB46, a master transcriptional regulator of secondary wall biosynthesis. Plant Mol Biol 78: 489–501 [DOI] [PubMed] [Google Scholar]
- Kim WC, Ko JH, Kim JY, Kim J, Bae HJ, Han KH (2013b) MYB46 directly regulates the gene expression of secondary wall-associated cellulose synthases in Arabidopsis. Plant J 73: 26–36 [DOI] [PubMed] [Google Scholar]
- Ko JH, Jeon HW, Kim WC, Kim JY, Han KH (2014) The MYB46/MYB83-mediated transcriptional regulatory programme is a gatekeeper of secondary wall biosynthesis. Ann Bot 114: 1099–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 19: 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legay S, Sivadon P, Blervacq AS, Pavy N, Baghdady A, Tremblay L, Levasseur C, Ladouce N, Lapierre C, Séguin A, et al. (2010) EgMYB1, an R2R3 MYB transcription factor from eucalyptus negatively regulates secondary cell wall formation in Arabidopsis and poplar. New Phytol 188: 774–786 [DOI] [PubMed] [Google Scholar]
- Li M, Li Y, Guo L, Gong N, Pang Y, Jiang W, Liu Y, Jiang X, Zhao L, Wang Y, et al. (2017) Functional characterization of tea (Camellia sinensis) MYB4a transcription factor using an integrative approach. Front Plant Sci 8: 943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Omranian N, Neumetzler L, Wang T, Herter T, Usadel B, Demura T, Giavalisco P, Nikoloski Z, Persson S (2016) A transcriptional and metabolic framework for secondary wall formation in Arabidopsis. Plant Physiol 172: 1334–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Dudareva N (2012) The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu Rev Plant Biol 63: 73–105 [DOI] [PubMed] [Google Scholar]
- Maeda H, Shasany AK, Schnepp J, Orlova I, Taguchi G, Cooper BR, Rhodes D, Pichersky E, Dudareva N (2010) RNAi suppression of Arogenate Dehydratase1 reveals that phenylalanine is synthesized predominantly via the arogenate pathway in petunia petals. Plant Cell 22: 832–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy RL, Zhong R, Ye ZH (2009) MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol 50: 1950–1964 [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M (2007) NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19: 270–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M (2005) The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17: 2993–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öhman D, Demedts B, Kumar M, Gerber L, Gorzsás A, Goeminne G, Hedenström M, Ellis B, Boerjan W, Sundberg B (2013) MYB103 is required for FERULATE-5-HYDROXYLASE expression and syringyl lignin biosynthesis in Arabidopsis stems. Plant J 73: 63–76 [DOI] [PubMed] [Google Scholar]
- Ohtani M, Demura T (2019) The quest for transcriptional hubs of lignin biosynthesis: Beyond the NAC-MYB-gene regulatory network model. Curr Opin Biotechnol 56: 82–87 [DOI] [PubMed] [Google Scholar]
- Ohtani M, Morisaki K, Sawada Y, Sano R, Uy ALT, Yamamoto A, Kurata T, Nakano Y, Suzuki S, Matsuda M, et al. (2016) Primary metabolism during biosynthesis of secondary wall polymers of protoxylem vessel elements. Plant Physiol 172: 1612–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzlaff A, McInnis S, Courtenay A, Surman C, Newman LJ, Smith C, Bevan MW, Mansfield S, Whetten RW, Sederoff RR, et al. (2003) Characterisation of a pine MYB that regulates lignification. Plant J 36: 743–754 [DOI] [PubMed] [Google Scholar]
- Preston J, Wheeler J, Heazlewood J, Li SF, Parish RW (2004) AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J 40: 979–995 [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP (2010) FastTree 2: Approximately maximum-likelihood trees for large alignments. PLoS ONE 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Lynch JH, Guo L, Rhodes D, Morgan JA, Dudareva N (2019) Completion of the cytosolic post-chorismate phenylalanine biosynthetic pathway in plants. Nat Commun 10: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W (2003) Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol 133: 1051–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, He X, Poovaiah CR, Wuddineh WA, Ma J, Mann DG, Wang H, Jackson L, Tang Y, Stewart CN Jr., et al. (2012) Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytol 193: 121–136 [DOI] [PubMed] [Google Scholar]
- Smith SA, Dunn CW (2008) Phyutility: A phyloinformatics tool for trees, alignments and molecular data. Bioinformatics 24: 715–716 [DOI] [PubMed] [Google Scholar]
- Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, Roberts K, Martin C (1998) The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10: 135–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, Watanabe M, Hoefgen R, Fernie AR (2013) Shikimate and phenylalanine biosynthesis in the green lineage. Front Plant Sci 4: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Yamaguchi M, Grienenberger E, Martone PT, Samuels AL, Mansfield SD (2020a) The class II KNOX genes KNAT3 and KNAT7 work cooperatively to influence deposition of secondary cell walls that provide mechanical support to Arabidopsis stems. Plant J 101: 293–309 [DOI] [PubMed] [Google Scholar]
- Wang XC, Wu J, Guan ML, Zhao CH, Geng P, Zhao Q (2020b) Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis. Plant J 101: 637–652 [DOI] [PubMed] [Google Scholar]
- Weng JK, Mo H, Chapple C (2010) Over-expression of F5H in COMT-deficient Arabidopsis leads to enrichment of an unusual lignin and disruption of pollen wall formation. Plant J 64: 898–911 [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Fukushima A, Nakabayashi R, Hanada K, Matsuda F, Sugawara S, Inoue E, Kuromori T, Ito T, Shinozaki K, et al. (2012) Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana. Plant J 69: 154–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tohge T, Matsuda F, Nakabayashi R, Takayama H, Niida R, Watanabe-Takahashi A, Inoue E, Saito K (2008) Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell 20: 2160–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H, Widhalm JR, Qian Y, Maeda H, Cooper BR, Jannasch AS, Gonda I, Lewinsohn E, Rhodes D, Dudareva N (2013) An alternative pathway contributes to phenylalanine biosynthesis in plants via a cytosolic tyrosine:phenylpyruvate aminotransferase. Nat Commun 4: 2833. [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Butelli E, Alseekh S, Tohge T, Rallapalli G, Luo J, Kawar PG, Hill L, Santino A, Fernie AR, et al. (2015) Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat Commun 6: 8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Zhang W, Zhao Y, Gong X, Guo L, Zhu G, Wang X, Gong Z, Schumaker KS, Guo Y (2007) SAD2, an importin-like protein, is required for UV-B response in Arabidopsis by mediating MYB4 nuclear trafficking. Plant Cell 19: 3805–3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q. (2016) Lignification: Flexibility, biosynthesis and regulation. Trends Plant Sci 21: 713–721 [DOI] [PubMed] [Google Scholar]
- Zhao Q, Dixon RA (2014) Altering the cell wall and its impact on plant disease: From forage to bioenergy. Annu Rev Phytopathol 52: 69–91 [DOI] [PubMed] [Google Scholar]
- Zhao Q, Tobimatsu Y, Zhou R, Pattathil S, Gallego-Giraldo L, Fu C, Jackson LA, Hahn MG, Kim H, Chen F, et al. (2013) Loss of function of cinnamyl alcohol dehydrogenase 1 leads to unconventional lignin and a temperature-sensitive growth defect in Medicago truncatula. Proc Natl Acad Sci USA 110: 13660–13665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Ye ZH (2010) Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol Plant 3: 1087–1103 [DOI] [PubMed] [Google Scholar]
- Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH (2008) A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20: 2763–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH (2012) MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant Cell Physiol 53: 368–380 [DOI] [PubMed] [Google Scholar]
- Zhou J, Lee C, Zhong R, Ye ZH (2009) MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21: 248–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNA sequencing data have been uploaded to the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) with the accession number PRJNA578015.