Characterization of two loci influencing flowering initiation and reproductive development, LATE3 and LATE4, reveals an important role for the deeply conserved Mediator complex.
Abstract
Control of flowering time has been a major focus of comparative genetic analyses in plant development. This study reports on a forward genetic approach to define previously uncharacterized components of flowering control pathways in the long-day legume, pea (Pisum sativum). We isolated two complementation groups of late-flowering mutants in pea that define two uncharacterized loci, LATE BLOOMER3 (LATE3) and LATE4, and describe their diverse effects on vegetative and reproductive development. A map-based comparative approach was employed to identify the underlying genes for both loci, revealing that that LATE3 and LATE4 are orthologs of CYCLIN DEPENDENT KINASE8 (CDK8) and CYCLIN C1 (CYCC1), components of the CDK8 kinase module of the Mediator complex, which is a deeply conserved regulator of transcription in eukaryotes. We confirm the genetic and physical interaction of LATE3 and LATE4 and show that they contribute to the transcriptional regulation of key flowering genes, including the induction of the florigen gene FTa1 and repression of the floral repressor LF. Our results establish the conserved importance of the CDK8 module in plants and provide evidence for the function of CYCLIN C1 orthologs in the promotion of flowering and the maintenance of normal reproductive development.
The initiation of flowering is one of the key developmental changes in the plant life cycle and is regulated by different environmental factors and endogenous cues. Evidence from Arabidopsis (Arabidopsis thaliana) indicates that it is a highly complex process, regulated by hundreds of genes through transcriptional, posttranscriptional, and epigenetic pathways (Bratzel and Turck, 2015; Song et al., 2015; Whittaker and Dean, 2017). One well-known control point is the FT gene, which encodes a small protein that is formed in leaf vasculature and moves through the phloem to the shoot apical meristem, where it interacts with the basic leucine zipper (bZIP) domain transcription factor FD. This complex then activates transcription of floral meristem identity genes such as MADS box genes LEAFY (LFY) and APETALA1 (AP1) via SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), leading to initiation of flowering (Wigge et al., 2005; Andrés and Coupland, 2012).
There is growing appreciation of the importance of regulatory mechanisms at the FT locus. The effects of many different environmental and endogenous factors on flowering are integrated through effects on FT expression (Andrés and Coupland, 2012; Song et al., 2015; Cho et al., 2017), and numerous proteins have been reported to associate with the FT promoter and other regulatory regions in or near the FT gene, including generalist transcription factors, transcriptional coregulators, and histone-modifying proteins (Bratzel and Turck, 2015; Luo et al., 2018). However, relatively little detail is known about specific mechanisms and interactions by which these factors regulate FT transcription.
Transcriptional regulation is also critical at many other different points in the flowering time network. For example, in addition to direct regulation of and by FT itself, pathways upstream and downstream also feature transcriptional control. Examples include the circadian and light control of the FT activator CONSTANS (Shim et al., 2017), repression of the FT repressor FLC in response to cold (Whittaker and Dean, 2017), and the mutually repressive interactions that establish organ identity and govern the patterning of inflorescences and flowers (Wagner, 2017). In addition to these largely flowering-specific factors, many general transcriptional and epigenetic regulators have also been identified from their effects on flowering time and reproductive development or have been shown to participate in these processes. These include NUCLEAR FACTOR-Y, the TOPLESS corepressor, and polycomb repressive complex2 (Causier et al., 2012; Eom et al., 2018).
In comparison to Arabidopsis, less is known about flowering time control in other plant groups. Loci controlling natural variation for flowering time have been identified across many major crop species (Fjellheim et al., 2014; Blümel et al., 2015; Brambilla et al., 2017; Cao et al., 2017; Higuchi, 2018), and such studies have highlighted aspects of regulation that are deeply conserved but others that may be confined to specific groups. As in Arabidopsis, flowering time control involves both specific pathways and general transcriptional and chromatin regulators (Shi et al., 2015; Brambilla et al., 2017). However, our understanding of the mechanisms controlling flowering time is still relatively limited in many species.
Legumes are a major plant group that includes many crop plants that display wide, agriculturally relevant variation in flowering time (Weller and Ortega, 2015). They include both short-day (SD) and long-day (LD) responsive species for which soybean (Glycine max) and pea (Pisum sativum) have been prominent examples. Characterization of induced mutants and natural variation in these and other species have been useful in defining flowering-associated loci (Weller and Ortega, 2015; Cao et al., 2017), and reverse genetics is increasingly employed for defining specific gene functions (e.g. Laurie et al., 2011; Berbel et al., 2012; Cai et al., 2018). A forward genetic strategy in pea has previously identified a number of loci that control flowering time through primary roles in light perception and signaling, circadian clock function, and regulation and function of florigen genes (e.g. Sussmilch et al., 2015; Weller and Ortega, 2015; Ridge et al., 2016). In this study, we have characterized two additional loci, LATE BLOOMER3 (LATE3) and LATE4, that have extremely late-flowering mutant phenotypes and other pleiotropic effects on vegetative and reproductive development. We identify these genes as likely components of the Mediator transcriptional coregulator complex and present evidence that their effects on flowering may in part result from effects on the transcription of FT and TFL1 homologs.
RESULTS
LATE3 and LATE4 Promote Flowering and Impair Responsiveness to Photoperiod
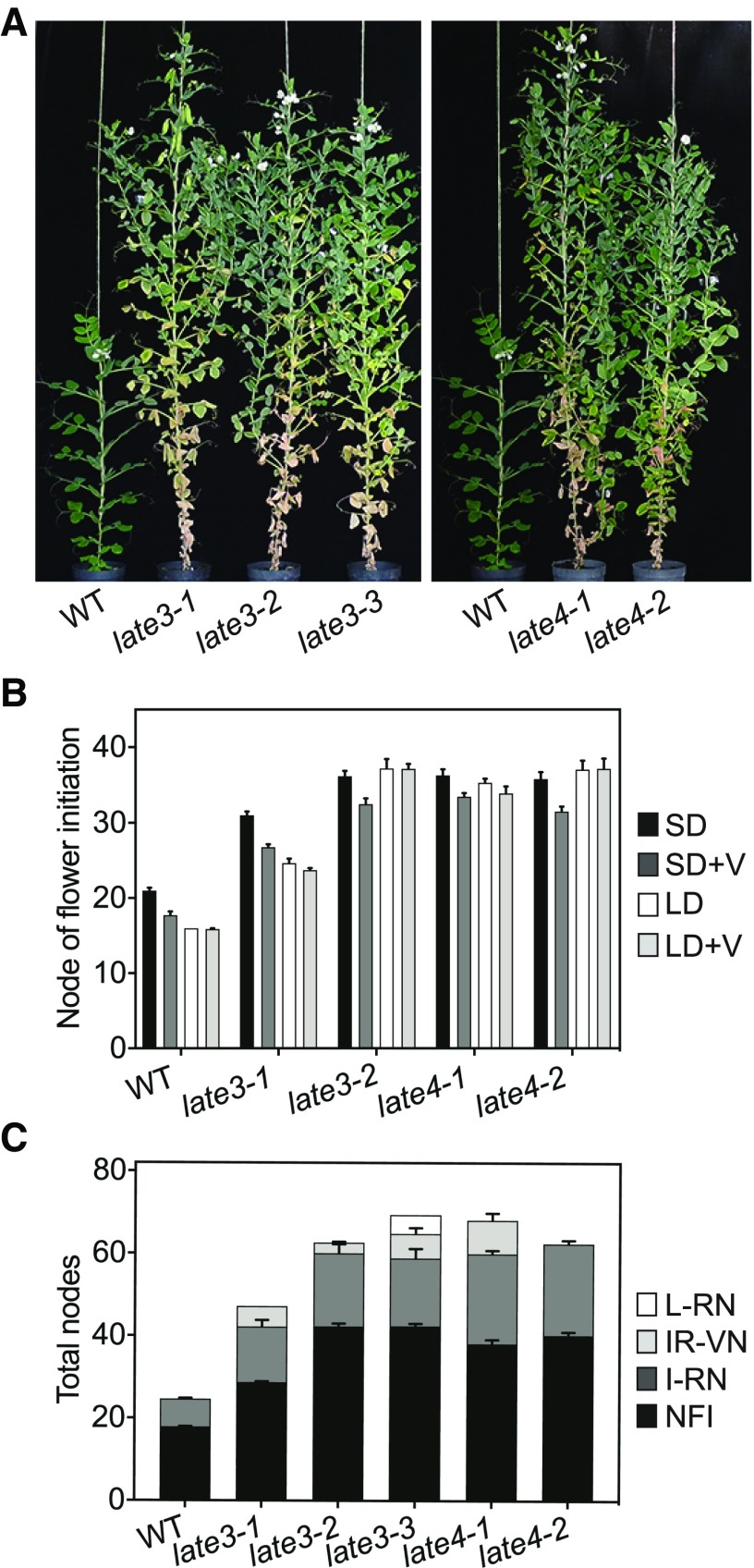
Among a number of flowering-time mutants generated through ethylmethanesulfonate mutagenesis (Hecht et al., 2007), we identified five similar fully recessive mutants that showed a substantial delay in flowering and maturity under LD conditions. These mutants defined two genetic loci: LATE BLOOMER3 (LATE3), with three mutant alleles, and LATE4, with two alleles (Fig. 1A). Four of the five mutants flowered equivalently late at around node 35 in comparison to wild-type line NGB5839, which flowered at around node 16 under LD (Fig. 1B). Only the late3-1 mutant was notably earlier in flowering than the other mutants. Overall, the late3 and late4 late-flowering phenotype was notably more severe than that of other previously described late-flowering mutants at the PHYTOCHROME A (PHYA), LATE1, and LATE2 loci (Ridge et al., 2016). We also examined the response of late3 and late4 mutants to photoperiod and vernalization. Whereas wild type and late3-1 flowered earlier under LD than under SD, other mutants at both loci showed no effect of photoperiod on flowering initiation (Fig. 1B). By contrast, all mutant lines showed a small but significant response to vernalization under SD (P < 0.05 in all cases), similar to wild type (Fig. 1B).
Figure 1.
Mutations at LATE3 and LATE4 loci delay flowering and prolong the reproductive phase. A, Representative wild-type (WT; NGB5839), late3, and late4 plants grown under 16-h long-day conditions. To account for the disparity in flowering time, this image compares WT and mutants at 62 and 130 d after sowing, respectively. B and C, Effect of photoperiod and vernalization on flowering initiation in WT, late3, and late4, mutant plants. Data represent mean ± se for n = 6 to 8 plants. +V, vernalization; NFI, node of flower initiation; I-RN, initial reproductive nodes; IR-VN, inflorescence reverted-vegetative nodes; l-RN, later reproductive nodes.
LATE3 and LATE4 Have Pleiotropic Effects throughout Reproductive Development
The late3 and late4 mutants also shared a number of additional defects that differed from those seen in previously described flowering-time mutants in pea. The most conspicuous of these was an extreme delay in maturity and senescence illustrated by the substantial increase in the number of reproductive nodes relative to wild type (initial reproductive nodes; P < 0.05 for all comparisons; Fig. 1C). This was accompanied by, and probably in part enhanced by, numerous defects in other aspects of reproductive development, including flower and inflorescence formation, flower fertility, pod formation, and seed content.
For example, whereas most secondary inflorescences in late3 and late4 mutants had a normal structure and developed to produce open flowers, a substantial minority showed growth defects in which they remained arrested or aborted at an early growth stage (Supplemental Fig. S1, A–C). Other secondary inflorescences displayed defects in identity, failing to suppress leaflet and bract formation and/or exhibiting reduced determinacy (Supplemental Fig. S1, A–C).
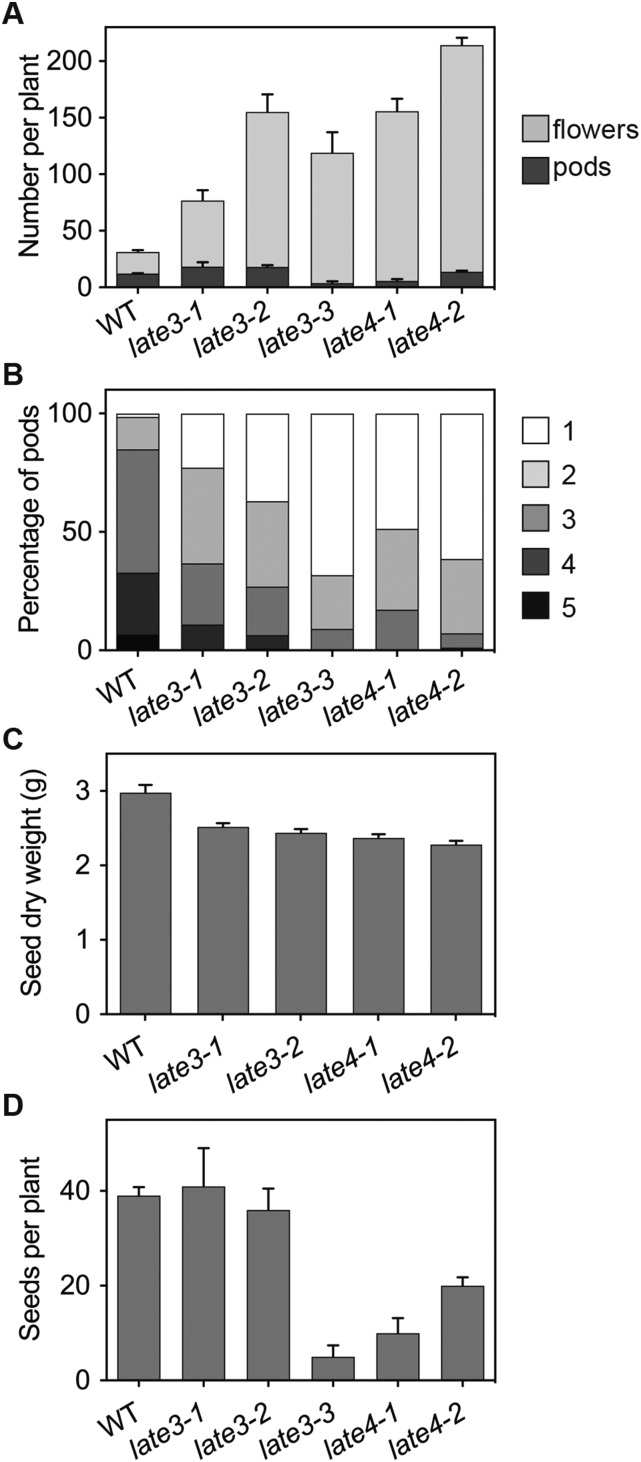
Where flowers did develop fully and open, other defects were evident. Some had abnormal organ morphology and number, and in the more severe mutants, pollen abundance was low and most flowers were sterile, with pods forming on only a small proportion of flowers (Fig. 2A). During the process of genetic analysis, it became apparent that the success rate of crosses made with wild-type pollen onto late3 and late4 mutants was also markedly reduced, suggesting that the reduced fertility derived from both paternal and maternal defects. When pods were formed, they often arrested in a partially developed state, and where they developed fully, they were generally shorter and contained fewer, smaller seeds (Fig. 2, B–D; Supplemental Fig. S2).
Figure 2.
Mutations at LATE3 and LATE4 loci affect varied aspects of reproductive development. A, Total number of flowers and pods at maturity. B, Seed content per pod (from one to five seeds) expressed as a proportion of the total number of seed-bearing pods. C, 10-seed dry weight and (D) total number of seeds per plant. Data were collected at the time of harvest and represented as mean ± se for n = 6 (A, B, and D) or n = 3 (C).
Finally, all mutants showed variable expression of these defects across the reproductive phase, with zones of more advanced development followed by stages of greater impairment. In addition, after forming 12 to 20 reproductive nodes, the mutants reverted to production of vegetative axillary buds, and in some cases subsequently reinitiated reproductive development at later nodes (Fig. 1C; Supplemental Fig. S1, D–E). Overall, provided they remained disease free, late3 and late4 mutant plants under glasshouse LD continued to grow for more than 6 months without signs of terminal senescence, in contrast to wild type, which generally reached maturity within 90 d.
LATE3 and LATE4 Also Affect Aspects of Vegetative Development
Initial observations in segregating progenies suggested late3 and late4 mutants could also be distinguished from wild type early in development, on the basis of a number of vegetative growth traits. Both late3 and late4 mutants showed significant reduction in leaflet area, petiole and proximal rachis length (Supplemental Fig. S3, A–C; both P < 0.0001), and stem diameter compared to wild type (Supplemental Fig. S3E; P < 0.0001). By 4 weeks after sowing, mutants also exhibited lower shoot and root dry weight in comparison to wild type (Supplemental Fig. S3, F and G; both P < 0.05). The late3 and late4 mutants also showed a substantial increase in shoot branching relative to wild type, when assessed as the ratio of total branch length to total height at maturity. (Supplemental Fig. S3H; P < 0.0001). For individual late3 and late4 plants, branching started at around node 11 or 12 and continued for a few nodes, followed by a gap, with branches reappearing just below the node of flower initiation (Supplemental Figs. S1, D and E, and S4). Whereas late3 mutants showed only aerial branching, we observed both basal and aerial branching for late4 mutants. Previously, various photoperiod-sensitive and nonsensitive late-flowering mutants were shown to exhibit only basal and aerial branching, respectively (Hecht et al., 2007; Berbel et al., 2012; Sussmilch et al., 2015). Also, whereas wild-type plants typically show an increase in the length of internodes immediately below the node of first open flower (e.g. Weller et al., 1997), this was not seen in the stronger late3 and late4 mutants (Supplemental Fig. S3I). Finally, the mutants also showed a delay in the normal progression of compound leaf morphology from one to two pairs of leaflets (Supplemental Fig. S3J) and never progressed to the three-pair stage.
LATE3 and LATE4 Are Putative Components of the Mediator Complex
Analysis of the F2 progeny of a cross between cv Térèse and late3-1 (n = 255) located LATE3 in a 0.3-cM interval on pea linkage group III (LG III) between markers BTB1 and SPS1 (Supplemental Fig. S5A; Supplemental Table S1). The corresponding interval on Medicago truncatula chromosome 3 (version 4.0) is 0.9 Mb in length and includes 62 annotated genes (Supplemental Table S1). A similar mapping population (cv Térèse × late4-1 F2; n = 189) was used initially to define the position of the LATE4 locus within a 3.2-cM interval on linkage group V, flanked by markers MCO1 and BZIP1 (Supplemental Fig. S5B). Genotyping of further markers within this region in relevant recombinant individuals refined this position to an interval corresponding to a 0.5-Mb region of M. truncatula chromosome 7 containing 54 annotated genes (Supplemental Fig. S5C; Supplemental Table S2).
The very close phenotypic similarity between late3 and late4 mutants suggested the possibility that the LATE3 and LATE4 genes might encode proteins with complementary functions, potentially acting within the same pathway or protein complex. Therefore, the two regions were scanned for pairs of genes that might be closely related in function. These analyses revealed the presence in both intervals of genes encoding to components of the Mediator transcriptional regulator complex, CYCLIN DEPENDENT KINASE8 (CDK8, Medtr3g096960) and CYCLIN C1 (CYCC1, Medtr7g055650; Supplemental Figs. S6 and S7). This complex is deeply conserved from baker's yeast (Saccharomyces cerevisiae) to humans (Homo sapiens) and flowering plants and consists of 28 to 34 component proteins that form four distinct modules (Allen and Taatjes, 2015; Jeronimo and Robert, 2017). The CDK8 and CYCC1 proteins associate with two other proteins, MED12 and MED13, to form the so-called CDK8 module (Dolan and Chapple, 2017; Jeronimo and Robert, 2017).
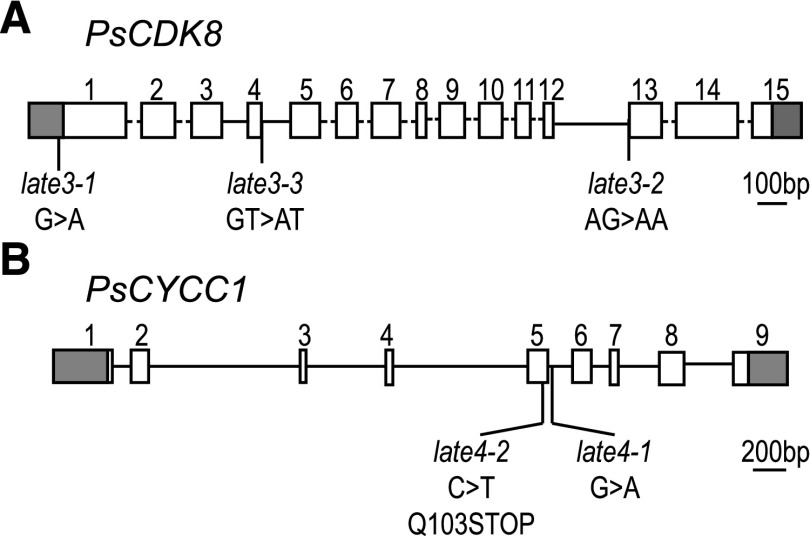
In parallel, RNA sequencing was used to screen transcripts from genes inferred to be within the LATE3 mapping interval for polymorphisms between late3-1 and LATE3 genotypes. Whereas only partial coverage of the transcripts within the region was achieved (Supplemental Table S3), this analysis nevertheless identified a G-to-A mutation typical of ethylmethanesulfonate exposure at position −17 in the 5′ untranslated region (UTR) of the CDK8 ortholog in late3-1, which was verified by Sanger sequencing. This mutation potentially introduces an alternative start codon (GTG/ATG), defining a short (25 amino acid) open reading frame (ORF) out of frame with the CDK8 coding sequence (Fig. 3A; Supplemental Figs. S8 and S9). Perfect cosegregation of the PsCDK8 genotype with the late3 phenotype in the mapping population confirmed the presence of this gene within the defined genetic interval (Supplemental Fig. S4A). Sequencing of PsCDK8 complementary DNA (cDNA) and genomic DNA (gDNA) in late3-2 and late3-3 subsequently revealed splice site mutations in both mutants. In late3-2, a mutation in the −1 position of the 3′ splice site of intron 12 (AG/AA) resulted in skipping of exon 13 (Fig. 3A; Supplemental Figs. S8 and S9), whereas in late3-3, a mutation in the +1 position of the 5′ splice site of intron 4 (GT/AT) resulted in retention of 7 bp from intron 4 in the cDNA, consistent with the activation of an alternative splice site (Fig. 3A; Supplemental Figs. S8 and S9). Both splicing defects were verified by PCR from cDNA using primers specific for either wild type or mutant transcript and would be predicted to result in frameshift and a truncated protein (Supplemental Fig. S9).
Figure 3.
Mutant alleles at LATE3 and LATE4 loci carry mutations in genes encoding Mediator complex components CYCLIN-DEPENDENT KINASE8 and CYCLIN C1, respectively. Diagrams showing (A) gene structure of PsCDK8 and the nature and location of mutations in late3 alleles and (B) gene structure of PsCYCC1 and the nature and location of mutations in late4 alleles. Exons are represented by numbered boxes, with gray shading designating 5′ and 3′ UTRs. Dashed lines in A represent introns not fully characterized. Sequence details of mutations and splice variants are shown in Supplemental Figs. S9, S12, and S13.
In view of these results, the pea CYCC1 gene (corresponding to transcript PsCam050605) was sequenced from the late4-1 and late4-2 mutants. This revealed a nonsense mutation in exon 5, introducing a premature stop codon (Q103X) in late4-2, and in late4-1, a G-to-A mutation at the +5 position of the 5′ splice site in intron 5 (GTAAGC/GTAAAC; Fig. 3C; Supplemental Figs. S10 and S11). As in the analysis of CDK8/LATE3, mapping of PsCYCC1 confirmed its presence within the defined LATE4 interval and demonstrated the absence of recombination with the late4 phenotype in the original mapping population (Supplemental Fig. S4B).
Splicing Defects in late4 Mutants
Amplification of cDNA from late4 mutants indicated the presence of multiple bands suggestive of possible splicing variants (Supplemental Fig. S11A). This was confirmed by sequencing of cloned fragments, which identified multiple distinct transcripts with variations around the site of the mutations (Supplemental Fig. S11, B and C). Whereas the majority of transcripts in the late4-2 mutant were wild type in structure (22 out of 27 clones sequenced), instances of skipping, partial deletion, and partial intron retention involving exon 5 were detected (Fig. 3D; Supplemental Figs. S11 and S12). However, all transcripts would be expected to be nonfunctional, in view of the presence of the late4-2 nonsense mutation and/or frameshift. In the case of late4-1, a small proportion of wild-type transcripts were also detected (3 out of 17), but the majority of transcripts displayed splicing defects around exon 5 (Fig. 3D; Supplemental Figs. S11 and S13), indicative of selection of alternative/cryptic splice sites in preference to the standard site affected by the mutation. All proteins hypothetically encoded by the aberrant transcripts would show significant disruption of the major functional domain of the PsCYCC1 protein, the cyclin N domain, and would therefore likely be inactive.
LATE3 and LATE4 Interact Genetically and Physically
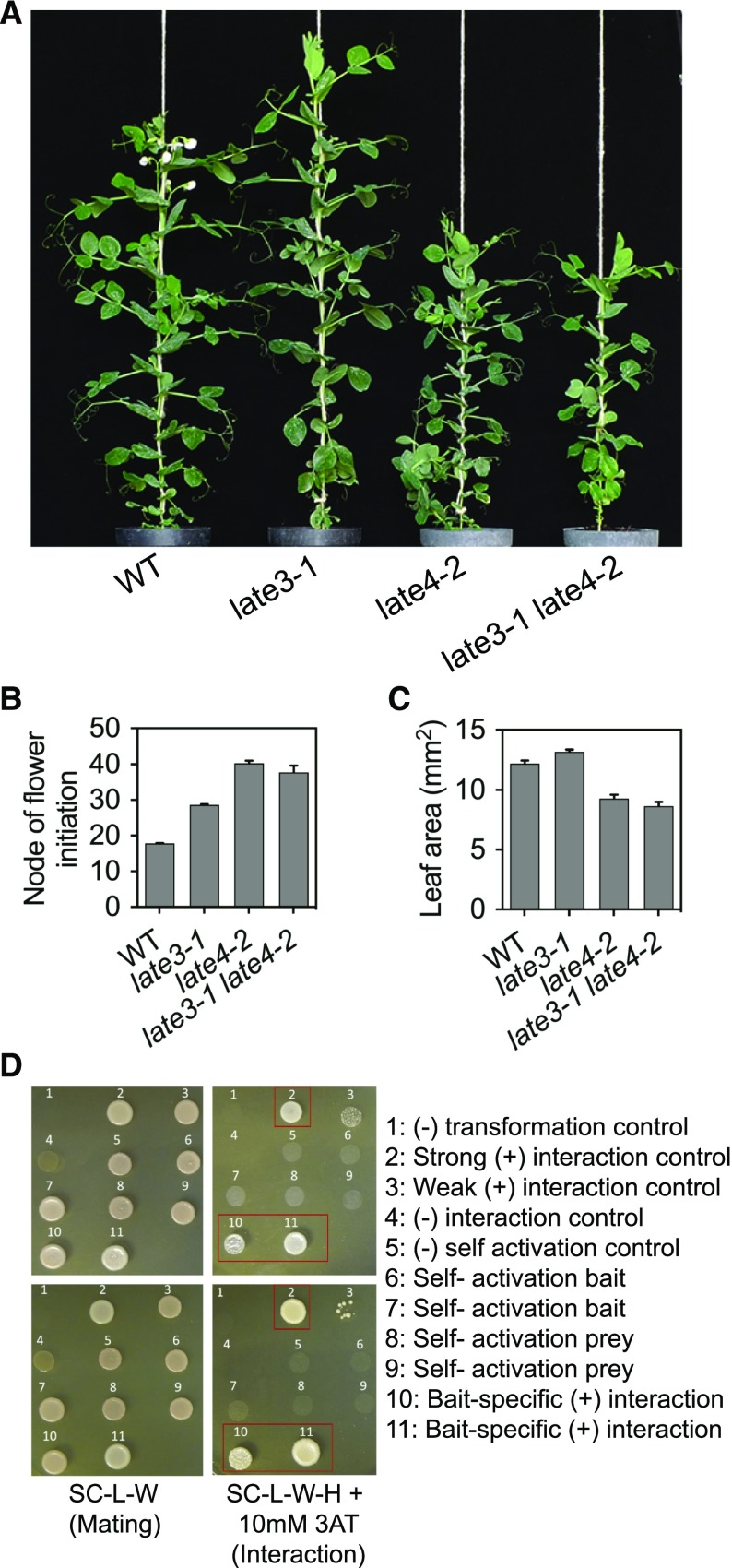
The molecular identities of LATE3 and LATE4 and the similarity of their mutant phenotypes implied their likely genetic and physical interaction. To examine their genetic interaction, we selected late3-1 late4-2 double mutants. The results in Figure 4, A–C, showed that the double mutant did not differ from the stronger of the two single mutants with respect to either node of flower initiation or leaflet area, indicating that LATE3 and LATE4 act in the same genetic pathway. The potential direct physical interaction between LATE3 and LATE4 was then examined using the yeast two-hybrid assay. We found that diploid yeast colonies carrying PsCDK8 and PsCYCC1 bait and prey plasmids displayed growth similar to a strong positive interaction control on selective medium (SC-L-W-H+10 mm 3-amino-1,2,4-triazole; Fig. 4D), whereas all negative controls showed no growth. These results indicate that LATE3 and LATE4 also show a strong physical interaction, consistent with their molecular identity as components of the same deeply conserved protein complex.
Figure 4.
LATE3 and LATE4 show genetic and physical interaction. A to C, Comparison of wild type, late3-1, and late4-2 single mutants and the late3-1 late4-2 double mutant grown under LD conditions. A, Representative 75-d-old plants. B, Node of flower initiation. C, Representative leaflet area (single leaflet from leaf 10). Data represent mean ± se for n = 6 to 10 plants. D, Yeast two-hybrid analysis for interaction between PsCDK8 and PsCYCC1 proteins from wild-type (NGB5839) genotype. The image shows diploid yeast colonies derived via mating of haploid yeast strains PJ694 alpha and PJ694 A carrying different bait and prey plasmids for experimental and control interactions (as indicated). For each interaction tested, two colonies derived from independent matings (top, colony 1; bottom, colony 2) were used grown in selective interaction-specific (SC-L-W-H +10 mm 3A-amino-1,2,4-triazole [3AT], right) and selective mating-specific (SC-L-W, left) medium and incubated at 30°C for 4 d. Key interactions are highlighted in red.
Genetic Interactions between LATE3 and LATE4 and Other Flowering Genes
Previous genetic analyses in pea have outlined a genetic pathway for flowering time control (Hecht et al., 2011; Sussmilch et al., 2015; Weller and Ortega, 2015). In an attempt to locate LATE3 and LATE4 within this model, we examined the genetic interaction of late3 and late4 with two early-flowering mutants, namely sn and lf. SN has primary role as a component of the circadian clock evening complex, which acts to repress flowering and FT expression (Liew et al., 2014; Rubenach et al., 2017). LF is one of three pea TFL1 co-orthologs (Foucher et al., 2003), and appears to act downstream of the FT gene FTa1 to repress expression of inflorescence identity genes (Hecht et al., 2011; Sussmilch et al., 2015).
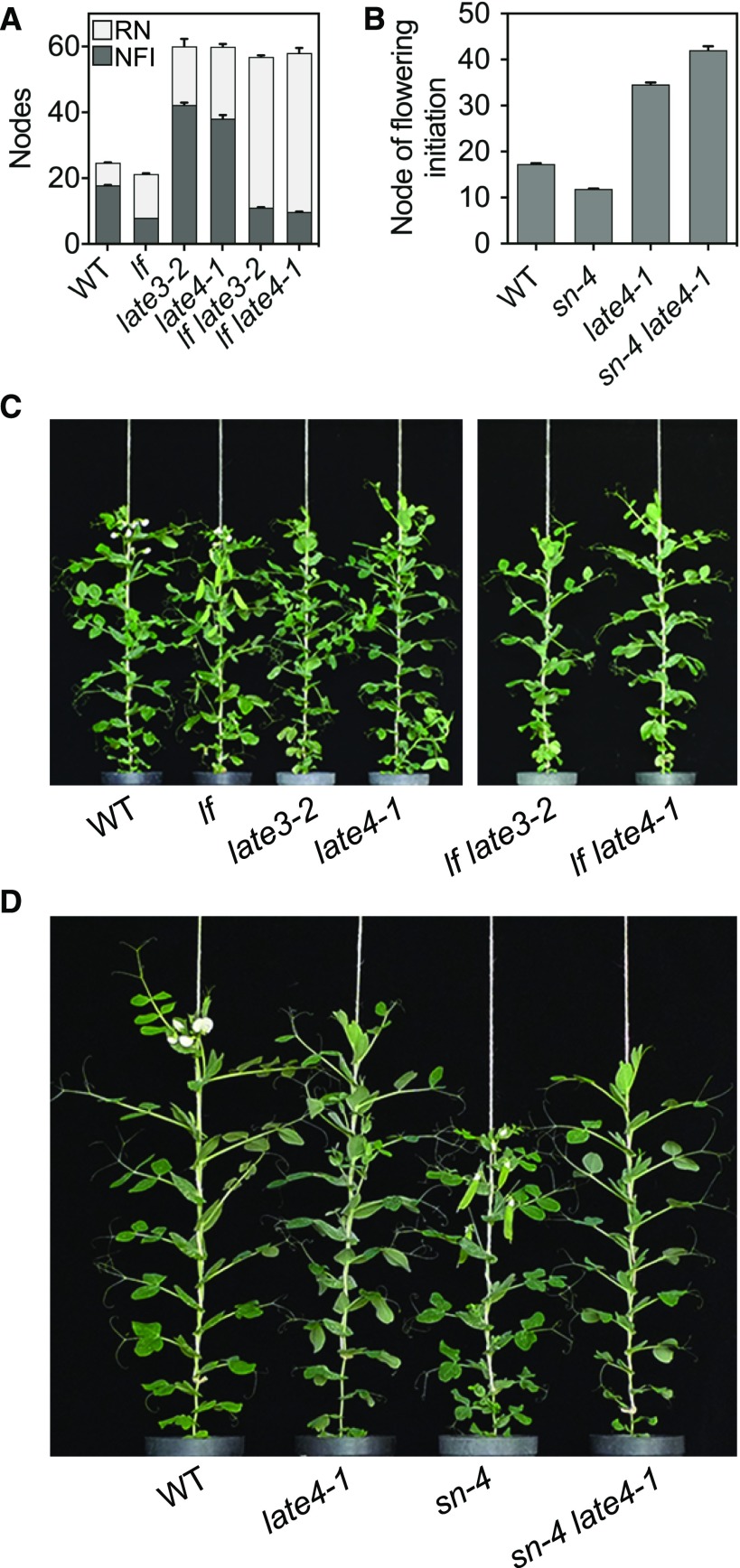
Fig. 5A shows that both lf late3 and lf late4 mutants initiated flowering very early and in this respect were much more similar to lf single mutants than to late3 or late4. This indicates that the effects of late3 and late4 mutations on flower initiation largely depend on the repressive effects of LF, although a small but significant increase in flowering node in the double mutants relative to the lf single mutant (P < 0.01 for both comparisons) indicates that LATE3 and LATE4 can also influence the initiation of flowering independently of LF to a small extent. In other respects, the double-mutant phenotypes were more similar to late3 and late4, with a massively extended reproductive phase and delayed maturity (Supplemental Fig. S14). In the case of SN, we identified plants with sn late4 genotype as late-flowering segregants in F3 progeny derived from sn individuals in the F2 of a cross between the sn-4 and late4-1 mutants. Figure 5B shows that in the presence of late4, the sn mutation was unable to promote flowering, and sn late4 plants in fact initiated flowering even later than late4 single-mutant controls. This suggests that LATE4 acts downstream of the changes to FT expression that are assumed to be the primary cause of the sn early-flowering phenotype (Liew et al., 2014; Rubenach et al., 2017).
Figure 5.
Genetic interactions of late3 and late4 mutants with early-flowering mutants lf and sn. A, Node of flower initiation (NFI) and number of reproductive nodes (RN) in wild-type, lf, late3-2, late4-1, lf late3-2, and lf late4-1 genotypes. B, Node of flower initiation in wild-type, sn-4, late4-1, and sn-4 late4-1 genotypes. C and D, Representative plants at 75 (C) and 63 d (D) after sowing. All plants were grown in long days. Data in A and B represent mean ± se for n = 6 to 10 plants.
LATE3 and LATE4 Regulate Expression of FT Genes and Inflorescence-Identity Genes
We next sought to understand how LATE3 and LATE4 might regulate the initiation of flowering by examining the effect of late3 and late4 mutations on expression of several key flowering-time genes. This analysis focused on the late3-1 and late4-1 mutants, which were the only two alleles for which sufficient quantities of seeds were available. Under LD conditions, wild-type and late3-1 visible flower buds were first detected in dissected apices of wild-type and late3-1 plants by 42 and 80 d after sowing respectively, whereas late4-1 mutants did not flower before termination of the experiment. In Arabidopsis, CDK8 module genes influence the flowering pathway in several different ways, including partially independent effects on FLC, FT, and SOC1/FUL expression (Imura et al., 2012). As FLC-like genes are absent from the genomes of pea and related legumes (Hecht et al., 2005), we focused on an analysis of FT genes and inflorescence-identity genes.
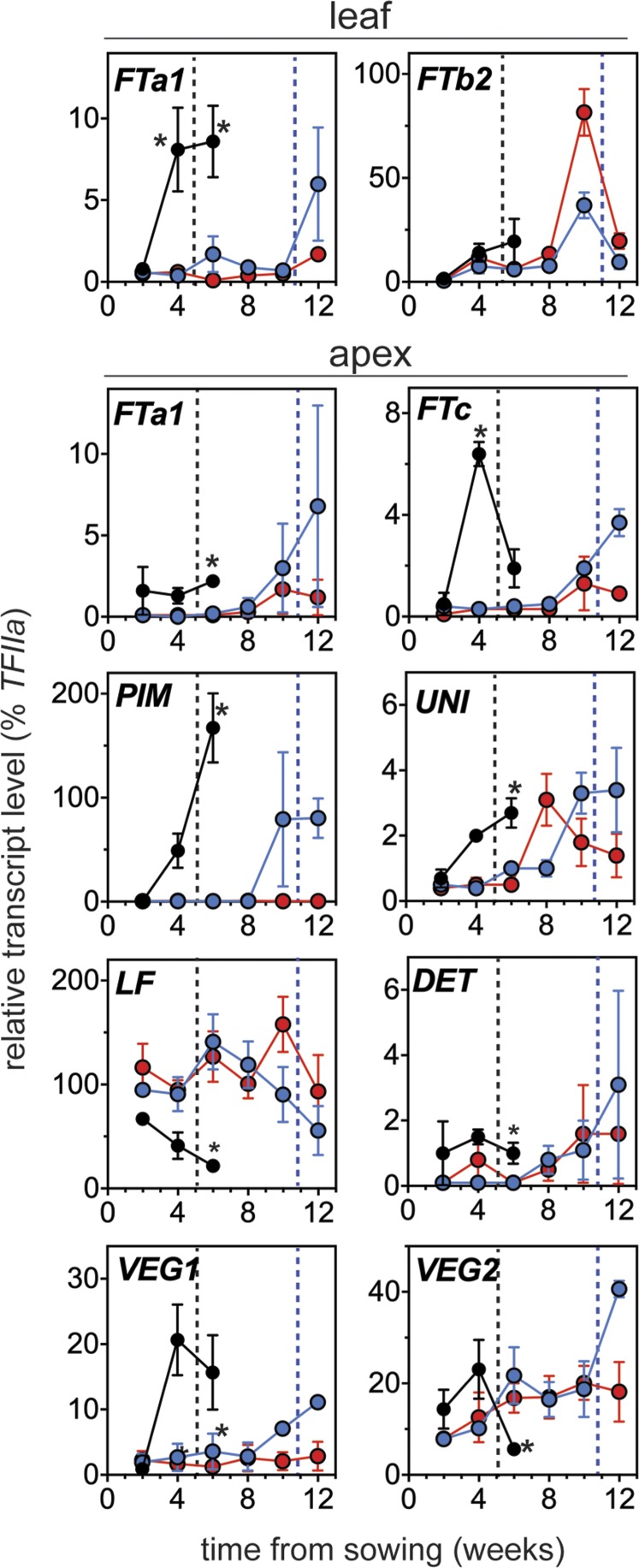
Previous studies have shown that two of the six pea FT genes, FTa1 and FTb2, are induced in leaves under LD, whereas a third gene (FTc) is induced at the shoot apex in parallel with inflorescence-identity genes VEG1 and PIM (Hecht et al., 2011; Sussmilch et al., 2015). Figure 6 shows that the expression of the FTa1 gene in leaves was significantly induced above background by 28 d after sowing in wild-type plants but remained low in late3 and late4 mutants. By contrast, the induction of FTb2 at this same time point was apparently unaffected.
Figure 6.
LATE3 and LATE4 loci affect expression of several florigen family and inflorescence-identity genes. Developmental time courses for expression of key flowering genes in leaf and shoot apex material from wild type NGB5839 (black), late3-1 mutant (blue), and late4-2 mutant (red) grown under long-day conditions. Data have been normalized to the reference gene TFIIa and represent mean ± se for n = 3 biological replicates, each consisting of material pooled from two different plants. Black and blue dashed lines indicate the time that flower buds first became visible in dissected shoot apices of wild type and late3-1 mutants, respectively. Flower initiation did not occur in late4-1 mutants for the duration of the experiment. Time points at which expression in wild type was significantly different from both mutants (P ≤ 0.05) are indicated with an asterisk.
Similar to FTa1 in leaves, expression of the inflorescence-identity genes VEG1 and PIM in shoot apices was induced by 28 d after sowing in wild-type plants but remained at background levels in late3 and late4 mutants, only rising in late3 at around the time of flower initiation, 10 weeks after sowing. A similar pattern of expression in the shoot apex was also shown by the LFY ortholog UNI and the apex-specific FTc gene. Effects on expression of the FTa1, DET/TFL1a, and VEG2/FD genes were not clear, but LF/TFL1c was expressed at a higher level in late3 and late4 than in wild type.
DISCUSSION
LATE3 and LATE4 Are Mediator Components
Mapping and sequencing from multiple independent mutant alleles have established the identity of pea flowering-time loci LATE3 and LATE4 as the pea orthologs of CDK8 and CYCC1; two genes that encode physically interacting components of the Mediator transcriptional regulation complex. This conclusion is further supported by the very similar pleiotropic phenotypes of late3 and late4 mutants and by their genetic and physical interactions.
Mediator is a large and dynamically variable multiprotein complex with diverse and deeply conserved roles in regulation of gene expression from yeast to animals and plants (Allen and Taatjes, 2015; Dolan and Chapple, 2017; Jeronimo and Robert, 2017). It comprises four different modules, of which three (head, tail, and middle) form the so-called “core” Mediator, which has a positive role in general regulation of transcription. The core Mediator forms a preinitiation complex with transcription factors at promoters of target genes, in which it acts to convey signals from gene-specific transcription and to enable the continuous reinitiation of transcription by RNA polymerase II (PolII; Knuesel et al., 2009a). LATE3 and LATE4 are orthologous to components of the fourth, cyclin-dependent kinase module (CKM), which has been shown to bind reversibly with the core Mediator to modify its transcriptional activity (Knuesel et al., 2009a; Allen and Taatjes, 2015).
In plants, a number of different Mediator subunits have been functionally characterized. The first to be described was Arabidopsis PFT1/MED25 (Cerdán and Chory, 2003), which is a component of the middle module of the core Mediator complex. More recently, functional studies have been reported on a number of other core Mediator components, which have been shown to participate in distinctive ways in diverse processes related to root, shoot, and reproductive development and responses to disease and abiotic stress (Dolan and Chapple, 2017; Kumar et al., 2018; Zhang et al., 2018).
Three of the four subunits of the kinase module (CDK8/CDKE1/HEN3, MED12/CRP/CCT, and MED13/MAB2/GCT) have also been directly functionally characterized in Arabidopsis by analysis of loss-of-function, gain-of-function, and overexpression phenotypes. Mutants have been isolated from several different screens and show diverse defects in embryonic and floral patterning (Wang and Chen, 2004; Gillmor et al., 2010; Ito et al., 2011), developmental phase transitions (Imura et al., 2012; Gillmor et al., 2014), stress and defense responses (Zhu et al., 2014), and hormone signaling (Ito et al., 2011, 2016). The phenotypic effects of the fourth subunit, CYCC1, have not previously been clearly defined (Dolan and Chapple, 2017). This reflects the fact that in Arabidopsis, CYCC1 is present as a recently duplicated tandem pair (Supplemental Fig. S6B), making generation of double mutants by recombination challenging. Zhu et al. (2014) isolated T-DNA-insertion mutants specific for each of these genes but reported no relevant phenotypes, although a mutant with a T-DNA insertion between the two genes showed reduced expression of both genes and some similarities to the defense-related phenotypes of the cdk8 mutant. Our description of LATE4 as the single pea ortholog of CYCC1 and characterization of the late4-2 nonsense mutant therefore describes the direct consequence of loss of CYCC1 in plants. We detected no clear phenotypic differences between strong late3 and late4 mutants, suggesting that their function is intimately related through essential and complementary roles in the CDK8 module.
Unusual Mutations in late3 and late4 Alleles
Among the five mutant alleles characterized in this study, only one, the late4-2 nonsense mutation, directly affected the coding region. Two others were typical splicing mutations affecting a consensus +1 donor (late3-3) or −1 acceptor site (late3-2). However, the two remaining mutations were notable for being somewhat less often described.
The late3-1 mutant carried a single G > A transition at position −17 in the 5′ UTR of CDK8, introducing a novel upstream potential ATG start codon (uATG) and an ORF overlapping and out-of-frame with the canonical CDK8 coding sequence. The most straightforward interpretation is that this uATG might provide an alternative translation initiation site and reduce to some extent the efficiency of translation from the normal AUG (Kozak, 1987). Interference of this nature is known to be greatest when the uORF extends into the major ORF (Kozak, 1987), as seen for late3-1. However, relative to the other late3 and late4 alleles, the late3-1 mutant was distinctly less severe (Fig. 1), implying that the wild-type CDK8 ORF is still translated to some extent in late3-1. This might in part reflect a weaker context of the upstream initiator codon introduced by the late3-1 mutation (AACAAAAUGA), which retains the conserved A in position −3 but not the G in position +4, whereas both are present in the CDK8 translation initiation site (GCAACCAUGG). However, in a recent yeast example, targeted introduction of diverse uAUG revealed effects on both transcription and translation of the associated major ORF that were independent of immediate sequence context (Yun et al., 2012), suggesting the possibility of a broader influence of uAUG beyond simply providing a competing site for initiation of transcription.
The second unusual mutant, late 4-1, carried only a single G > A transition at position +5 of the donor site of CYCC1 intron 5, which interferes with normal processing of the CYCC1 transcript (Fig. 3). Although +5 G is highly conserved (>75%) in U2 type GT-AG introns in yeast and animals, genome-wide analyses in Arabidopsis indicate a weaker consensus of around 50% (Sheth et al., 2006; Buratti et al., 2011), and some degree of tolerance for +5 A is therefore likely to explain the presence of normally spliced transcript in late4-1. However, the fact that the late4-1 phenotype appears equivalently severe to that of the late4-2 nonsense allele (Fig. 1) may therefore suggest that a threshold level of expression is required for CYCC1 function that exceeds that seen in late4-1. Although less common by far than mutations affecting the highly conserved +1 and +2 positions, +5 mutations affecting splicing have been described for several human disease genes (e.g. Tran et al., 2005; Fiorentino et al., 2018).
LATE3 and LATE4 Influence Multiple Steps in Flowering Time Control and Reproductive Development
The dramatic effects of LATE3 and LATE4 mutations point to a key role for the Mediator kinase module in promotion of flowering and maintenance of diverse aspects of reproductive development. In Arabidopsis, effects of CDK8 and CYCC on flowering have not been examined in detail. However, single mutants for the other two CKM components MED12 and MED13 show similar, relatively strong LD-specific late-flowering phenotypes, again consistent with a close functional relationship (Imura et al., 2012). A weaker late-flowering phenotype has also been reported for an RNA-null CDK8 insertion mutant (Zhu et al., 2014), but this effect has not been further characterized. Thus, based on this relatively limited evidence, it appears that there may be some difference in the relative effects of CKM components on flowering time in Arabidopsis. This is supported by observations from other systems indicating that CKM components, in addition to their co-operative functions, may also function independently to some extent (e.g. Loncle et al., 2007). It also points to a potential difference between pea and Arabidopsis with respect to CDK8 function in flowering time control.
Mutants for LATE3 and LATE4 have similar effects on flowering gene expression, with reduced expression of GIGAS/FTa1 in leaves and multiple inflorescence-identity genes in the shoot apex. One interpretation of this is that FTa1 might be the primary target of CKM regulation and that effects on other genes might be a downstream consequence of FTa1 misregulation, as most are known to be regulated by FTa1 (Hecht et al., 2011). However, the epistasis of late4 over sn, a mutant in which photoperiod-insensitive early flowering is due to elevated expression of FTa1 and other FT genes in leaves (Liew et al., 2014; Rubenach et al., 2017), suggests that the impaired flowering may not be primarily due to altered FT gene expression but because of effects on genes downstream. It is also notable that expression of the FTb2 gene, which is qualitatively induced by LD in wild-type pea and not detectable in late-flowering photoperiod response mutants (Hecht et al., 2011; Ridge et al., 2016), does not appear to be affected in the late3 and late4 mutants (Fig. 6) despite their clear insensitivity to photoperiod for induction of flowering (Fig. 1). This again suggests a primary requirement for CKM in regulation of signaling from FT genes, rather than in their regulation.
This interpretation is also consistent with observations that expression of LF, a TFL1 paralog, was elevated in late3 and late4 mutants (Fig. 6), and that for initiation of flowering, an lf null mutant was epistatic to both late3 and late4 (Fig. 5). Formally, this suggests that LF is required for expression of the late3/4 late-flowering phenotype and that the promotion of flowering by the CKM may at least in part involve the transcriptional repression of LF. However, the effect of late3 and late4 on other aspects of reproductive development in lf mutant plants clearly indicates the existence of LF-independent effects of CKM action.
No direct information about molecular effects of CDK8 and CYCC1 on flowering time is available from Arabidopsis, but characterization of med12 and med13 mutants revealed increased expression of FLC and decreased expression of FT, LFY, and MADS-domain genes SOC1, FUL, and AP1 (Imura et al., 2012). In the same study, analysis in flc and ft mutant backgrounds further established that MED12 and MED13 act at multiple steps, with control of FT expression partly independent of FLC and control of SOC1 and FUL expression at least partly independent of both FT and FLC. This has been interpreted as a potential feed-forward mechanism that may confer robustness of the flowering transition and is interesting because it supports the idea that the CKM, in addition to its more well-established repressive role, may also activate expression in specific contexts (Nemet et al., 2014). Despite the fact that FLC is not present in pea and related legumes, it is still probable that the CKM acts at multiple steps of the flowering and inflorescence-development pathway, including in FT-independent effects on MADS domain genes and other targets.
LATE3 and LATE4 Have Diverse Pleiotropic Effects
Phenotypic effects of late3 and late4 mutants beyond flowering and reproductive development indicate that the CDK8 module in pea has pervasive effects throughout development (Figs. 1 and 2; Supplemental Figs. S1–S4). These include effects on stem thickness, leaflet size and shape, seed size and shape, and the timing of changes in compound leaf structure, which has been implicated as a possible marker of vegetative phase change in pea (Wiltshire et al., 1994). These effects are generally similar to those described for Arabidopsis CKM mutants (e.g. Imura et al., 2012; Gillmor et al., 2014; Chhun et al., 2016). This suggests that despite the taxonomic distance between pea and Arabidopsis and despite the fundamental role of the CKM in regulation of gene expression, its preferential involvement in certain aspects of growth and development may be conserved to some extent. This is likely to reflect conservation in the interactions of the CKM with specific transcription factors and corepressors.
In yeast and animal systems, CKM may act by interfering with the positive transcriptional role of the core Mediator complex by blocking its association with PolII, or by directly regulating PolII activity through phosphorylation (Nemet et al., 2014). However, there is also evidence that the CKM can have bidirectional effects on transcription through phosphorylation of transcription factors and may also act independently of core Mediator by direct and indirect modification of histones and regulation of chromatin (Knuesel et al., 2009b; Tsutsui et al., 2013; Allen and Taatjes, 2015). Recent transcriptome analyses have revealed broad effects of Arabidopsis CDK8 on expression of genes involved in processes such as growth regulation, photosynthesis, and light, hormone, defense, and stress responses (Zhu et al., 2014; Mao et al., 2019), but there are few examples in plants where the mechanisms of CKM action have been examined in detail. One recent report has demonstrated the importance of the CDK8 kinase function for some but not all effects on defense-related gene expression (Zhu et al., 2014). Another has linked the CKM to auxin-dependent gene expression through its role in relaying repressive signals from ARF/IAA proteins in association with the TOPLESS corepressor (Ito et al., 2016). Future genomic-scale studies will help define the global effects of the pea CDK8 module and the extent to which they may be shared with Arabidopsis. Such studies should also help clarify the effects of pea CDK8/CYCC1 on flowering-time pathways and shed light on other developmental mechanisms responsible for other aspects of the late3/late4 phenotype. Finally, in view of reports that the Arabidopsis CKM is important for defense and abiotic stress tolerance, it may be of interest to examine the effects of late3 and late4 on these traits.
MATERIALS AND METHODS
Plant Materials
The origins of late3, late4, lf, det, and sn-4 mutants in pea (Pisum sativum) have been described previously by Foucher et al. (2003) and Hecht et al. (2007). Plants for phenotypic characterizations and genetic analysis (Figs. 1, 2, 4, and 5) were grown in a glasshouse or phytotron under extended natural daylight, whereas plants for gene expression analysis (Fig. 6) were grown in growth chambers. Growth media, light sources, and growth conditions have been described previously by Hecht et al. (2007). Vernalization treatment was given by subjecting imbibed seeds to 4°C for 4 weeks.
Mapping
Mapping of LATE3 and LATE4 utilized a combination of previously described by Aubert et al. (2006) and new gene-based markers, developed from transcript sequences obtained from the pea gene atlas (http://bios.dijon.inra.fr/FATAL/cgi/pscam.cgi) based on sequence comparisons with orthologous genes within syntenic regions of the Medicago truncatula genome (Mt4.0v1, https://phytozome.jgi.doe.gov/pz/portal.html#). Marker details are provided in Supplemental Table S3. Linkage analysis was performed using JoinMAP 4 (Kyazma) software.
RNA Sequencing and Data Analysis
RNA sequencing from isogenic late3-1 and LATE3 genotypes was performed on RNA pooled from entire embryos isolated from seeds 2 d after imbibition and leaves and shoot apices from 4-week-old plants. Samples were harvested from three plants in two independent replications, and one replication was used for cDNA library construction. Samples from the three different tissues were used for RNA extraction according to SV total RNA isolation (Promega). One microgram of total RNA from each of the three tissues was pooled for preparation and indexing cDNA library using the TruSeq Stranded Total RNA library preparation kit with Ribozero Plant (Illumina). Pools of indexed cDNA libraries of about 260 bp diluted to 6 pM were then used for sequencing in a Miseq next-generation sequencing machine using Miseq Reagent v3 150 cycles kit (Illumina). Quality of the reads generated was assessed in FASTQC in galaxy (Giardine et al., 2005). Paired-end reads were aligned to pea transcript sequences located within the defined interval of PsLGIII (Supplemental Table S3) in Geneious 8.0.4 software.
Other Molecular Analyses
PCR fragments were purified using Promega Wizard SV gel and PCR clean-up system (Promega) and cloned using pGEM-T easy vector (Promega) by following manufacturer’s protocol. Sequencing was performed at Macrogen. For gene expression assays, both leaflets from the second youngest fully expanded leaf and a dissected apical bud containing the shoot apex (∼2 mm in length) were harvested from two plants per replicate. These samples were frozen in liquid nitrogen and processed for RNA extraction, reverse transcription, and reverse transcription quantitative PCR, according to procedures described previously by Sussmilch et al. (2015). Two technical and three biological replicates were used for each sample point. Details of primers are given in Supplemental Table S3.
Phylogenetic Analysis
Phylogenetic analysis was performed by identification of genes through BLAST searches of the M. truncatula genome (Mt4.0v1) and pea gene atlas with reciprocal BLAST searches against the Arabidopsis genome at TAIR (www.arabidopsis.org) to confirm gene identity. Full-length amino acid sequences were aligned using ClustalX (Thompson et al., 1997), adjusted manually, and analyzed using distance based methods in PAUP* (Supplemental Figs. S6, S7, and S10).
Yeast Two-Hybrid Assay
Full-length coding sequences of PsCDK8 and PsCYCC1 were amplified from wild-type (NGB5839) cDNA, cloned into yeast two-hybrid destination vectors, and tested for interactions following methods described previously by Ridge et al. (2016). Empty vector controls were performed to test, and controls for strong and negative interactions provided as part of the ProQuest two-hybrid system were performed according to the manufacturer’s instructions. Relevant details of primers and constructs are listed in Supplemental Tables S4 and S5.
Statistical Analysis
For statistical analysis of data presented in Figures 1, 2, and 4, Welch t test (two tailed) with 95% confidence interval was performed, whereas for Figure 6, a one-way ANOVA followed by Dunnett test was used. Analyses were conducted in GraphPad Prism (v7, GraphPad Software).
Accession Numbers
Sequences referred to in this article can be found in the P. sativum v1a genome database (urgi.versailles.inra.fr) under loci Psat5g058480/PsCam048317 (PsCDK8/LATE3) and Psat3g149520/PsCam050605 (PsCYCC1/LATE4).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Inflorescence development and reversion in late3 and late4 mutants.
Supplemental Figure S2. Pod and seed morphology in late3 and late4 mutants.
Supplemental Figure S3. Effect of and late3 and late4 mutations on vegetative growth traits.
Supplemental Figure S4. Diagram illustrating branching pattern in late3 and late4 mutants.
Supplemental Figure S5. Genetic mapping of LATE3 and LATE4 loci.
Supplemental Figure S6. Phylogenetic trees showing identity and relationships of PsCDK8 and PsCYCC1 protein sequences.
Supplemental Figure S7. Alignment of CDK8 protein sequences.
Supplemental Figure S8. Alternative splicing and putative ORF generation for wild-type and late3 mutant alleles of PsCDK8.
Supplemental Figure S9. Sites and consequences of mutations in PsCDK8 in late3 mutant alleles.
Supplemental Figure S10. Alignment of CYCC1 protein sequences.
Supplemental Figure S11. Alternative splicing and putative ORF generation for wild-type and late4 mutant alleles of PsCYCC1.
Supplemental Figure S12. Sites of mutations in PsCYCC1 in late4 mutant alleles and consequences for splicing in late4-2.
Supplemental Figure S13. Consequences of the late4-1 mutation for splicing of PsCYCC1.
Supplemental Figure S14. Genetic interaction of late3-2 and late4-1 mutants with lf-22 and det mutants.
Supplemental Table S1. Details of gene-based markers used for mapping of LATE3 and their Medicago orthologs.
Supplemental Table S2. Details of gene-based markers used for mapping of LATE4 and their Medicago orthologs.
Supplemental Table S3. Comparison of RNA sequencing data analysis for pea transcriptome sequences inferred by mapping and/or synteny with Medicago to derive from genes located in the mapping interval for LATE3 (BTB1-SPS1 in PsLGIII).
Supplemental Table S4. Primer details.
Supplemental Table S5. Details of bait and prey plasmids used in yeast two hybrid assays for testing interactions between pea CDK8 and CYCC1 proteins.
Acknowledgments
We thank Ian Cummings, Michelle Lang, and Tracey Winterbottom for help with plant husbandry.
Footnotes
This project was funded in part by the Australian Research Council (grants DP120101241 and DP160100793 to J.L.W.).
Articles can be viewed without a subscription.
References
- Allen BL, Taatjes DJ (2015) The Mediator complex: A central integrator of transcription. Nat Rev Mol Cell Biol 16: 155–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639 [DOI] [PubMed] [Google Scholar]
- Aubert G, Morin J, Jacquin F, Loridon K, Quillet MC, Petit A, Rameau C, Lejeune-Hénaut I, Huguet T, Burstin J (2006) Functional mapping in pea, as an aid to the candidate gene selection and for investigating synteny with the model legume Medicago truncatula. Theor Appl Genet 112: 1024–1041 [DOI] [PubMed] [Google Scholar]
- Berbel A, Ferrándiz C, Hecht V, Dalmais M, Lund OS, Sussmilch FC, Taylor SA, Bendahmane A, Ellis TH, Beltrán JP, et al. (2012) VEGETATIVE1 is essential for development of the compound inflorescence in pea. Nat Commun 3: 797. [DOI] [PubMed] [Google Scholar]
- Blümel M, Dally N, Jung C (2015) Flowering time regulation in crops—what did we learn from Arabidopsis? Curr Opin Biotechnol 32: 121–129 [DOI] [PubMed] [Google Scholar]
- Brambilla V, Martignago D, Goretti D, Cerise M, Somssich M, de Rosa M, Galbiati F, Shrestha R, Lazzaro F, Simon R, et al. (2017) Antagonistic transcription factor complexes modulate the floral transition in rice. Plant Cell 29: 2801–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratzel F, Turck F (2015) Molecular memories in the regulation of seasonal flowering: From competence to cessation. Genome Biol 16: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Chivers M, Hwang G, Vorechovsky I (2011) DBASS3 and DBASS5: Databases of aberrant 3′- and 5′-splice sites. Nucleic Acids Res 39: D86–D91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Chen L, Liu X, Guo C, Sun S, Wu C, Jiang B, Han T, Hou W (2018) CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Plant Biotechnol J 16: 176–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Takeshima R, Zhao C, Liu B, Jun A, Kong F (2017) Molecular mechanisms of flowering under long days and stem growth habit in soybean. J Exp Bot 68: 1873–1884 [DOI] [PubMed] [Google Scholar]
- Causier B, Ashworth M, Guo W, Davies B (2012) The TOPLESS interactome: A framework for gene repression in Arabidopsis. Plant Physiol 158: 423–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdán PD, Chory J (2003) Regulation of flowering time by light quality. Nature 423: 881–885 [DOI] [PubMed] [Google Scholar]
- Chhun T, Chong SY, Park BS, Wong ECC, Yin JL, Kim M, Chua NH (2016) HSI2 repressor recruits MED13 and HDA6 to down-regulate seed maturation gene expression directly during Arabidopsis early seedling growth. Plant Cell Physiol 57: 1689–1706 [DOI] [PubMed] [Google Scholar]
- Cho LH, Yoon J, An G (2017) The control of flowering time by environmental factors. Plant J 90: 708–719 [DOI] [PubMed] [Google Scholar]
- Dolan WL, Chapple C (2017) Conservation and divergence of mediator structure and function: Insights from plants. Plant Cell Physiol 58: 4–21 [DOI] [PubMed] [Google Scholar]
- Eom H, Park SJ, Kim MK, Kim H, Kang H, Lee I (2018) TAF15b, involved in the autonomous pathway for flowering, represses transcription of FLOWERING LOCUS C. Plant J 93: 79–91 [DOI] [PubMed] [Google Scholar]
- Fiorentino A, Yu J, Arno G, Pontikos N, Halford S, Broadgate S, Michaelides M, Carss KJ, Raymond FL, Cheetham ME, et al. ; NIHR-BioResource Rare Diseases Consortium; U.K. Inherited Retinal Dystrophy Consortium (2018) Novel homozygous splicing mutations in ARL2BP cause autosomal recessive retinitis pigmentosa. Mol Vis 24: 603–612 [PMC free article] [PubMed] [Google Scholar]
- Fjellheim S, Boden S, Trevaskis B (2014) The role of seasonal flowering responses in adaptation of grasses to temperate climates. Front Plant Sci 5: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foucher F, Morin J, Courtiade J, Cadioux S, Ellis N, Banfield MJ, Rameau C (2003) DETERMINATE and LATE FLOWERING are two TERMINAL FLOWER1/CENTRORADIALIS homologs that control two distinct phases of flowering initiation and development in pea. Plant Cell 15: 2742–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. (2005) Galaxy: A platform for interactive large-scale genome analysis. Genome Res 15: 1451–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillmor CS, Park MY, Smith MR, Pepitone R, Kerstetter RA, Poethig RS (2010) The MED12-MED13 module of Mediator regulates the timing of embryo patterning in Arabidopsis. Development 137: 113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillmor CS, Silva-Ortega CO, Willmann MR, Buendía-Monreal M, Poethig RS (2014) The Arabidopsis Mediator CDK8 module genes CCT (MED12) and GCT (MED13) are global regulators of developmental phase transitions. Development 141: 4580–4589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V, Foucher F, Ferrándiz C, Macknight R, Navarro C, Morin J, Vardy ME, Ellis N, Beltrán JP, Rameau C, et al. (2005) Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol 137: 1420–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V, Knowles CL, Vander Schoor JK, Liew LC, Jones SE, Lambert MJ, Weller JL (2007) Pea LATE BLOOMER1 is a GIGANTEA ortholog with roles in photoperiodic flowering, deetiolation, and transcriptional regulation of circadian clock gene homologs. Plant Physiol 144: 648–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V, Laurie RE, Vander Schoor JK, Ridge S, Knowles CL, Liew LC, Sussmilch FC, Murfet IC, Macknight RC, Weller JL (2011) The pea GIGAS gene is a FLOWERING LOCUS T homolog necessary for graft-transmissible specification of flowering but not for responsiveness to photoperiod. Plant Cell 23: 147–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi Y. (2018) Florigen and anti-florigen: Flowering regulation in horticultural crops. Breed Sci 68: 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imura Y, Kobayashi Y, Yamamoto S, Furutani M, Tasaka M, Abe M, Araki T (2012) CRYPTIC PRECOCIOUS/MED12 is a novel flowering regulator with multiple target steps in Arabidopsis. Plant Cell Physiol 53: 287–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J, Fukaki H, Onoda M, Li L, Li C, Tasaka M, Furutani M (2016) Auxin-dependent compositional change in Mediator in ARF7- and ARF19-mediated transcription. Proc Natl Acad Sci USA 113: 6562–6567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J, Sono T, Tasaka M, Furutani M (2011) MACCHI-BOU 2 is required for early embryo patterning and cotyledon organogenesis in Arabidopsis. Plant Cell Physiol 52: 539–552 [DOI] [PubMed] [Google Scholar]
- Jeronimo C, Robert F (2017) The mediator complex: At the nexus of RNA polymerase II transcription. Trends Cell Biol 27: 765–783 [DOI] [PubMed] [Google Scholar]
- Knuesel MT, Meyer KD, Bernecky C, Taatjes DJ (2009a) The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev 23: 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuesel MT, Meyer KD, Donner AJ, Espinosa JM, Taatjes DJ (2009b) The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of mediator. Mol Cell Biol 29: 650–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1987) An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 15: 8125–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar KRR, Blomberg J, Björklund S (2018) The MED7 subunit paralogs of Mediator function redundantly in development of etiolated seedlings in Arabidopsis. Plant J 96: 578–594 [DOI] [PubMed] [Google Scholar]
- Laurie RE, Diwadkar P, Jaudal M, Zhang L, Hecht V, Wen J, Tadege M, Mysore KS, Putterill J, Weller JL, et al. (2011) The Medicago FLOWERING LOCUS T homolog, MtFTa1, is a key regulator of flowering time. Plant Physiol 156: 2207–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew LC, Hecht V, Sussmilch FC, Weller JL (2014) the pea photoperiod response gene STERILE NODES is an ortholog of LUX ARRHYTHMO. Plant Physiol 165: 648–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncle N, Boube M, Joulia L, Boschiero C, Werner M, Cribbs DL, Bourbon HM (2007) Distinct roles for Mediator Cdk8 module subunits in Drosophila development. EMBO J 26: 1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Gao Z, Wang Y, Chen Z, Zhang W, Huang J, Yu H, He Y (2018) The NUCLEAR FACTOR-CONSTANS complex antagonizes Polycomb repression to de-repress FLOWERING LOCUS T expression in response to inductive long days in Arabidopsis. Plant J 95: 17–29 [DOI] [PubMed] [Google Scholar]
- Mao X, Kim JI, Wheeler MT, Heintzelman AK, Weake VM, Chapple C (2019) Mutation of Mediator subunit CDK8 counteracts the stunted growth and salicylic acid hyperaccumulation phenotypes of an Arabidopsis MED5 mutant. New Phytol 223: 233–245 [DOI] [PubMed] [Google Scholar]
- Nemet J, Jelicic B, Rubelj I, Sopta M (2014) The two faces of Cdk8, a positive/negative regulator of transcription. Biochimie 97: 22–27 [DOI] [PubMed] [Google Scholar]
- Ridge S, Sussmilch FC, Hecht V, Vander Schoor JK, Lee R, Aubert G, Burstin J, Macknight RC, Weller JL (2016) Identification of LATE BLOOMER2 as a CYCLING DOF FACTOR homolog reveals conserved and divergent features of the flowering response to photoperiod in pea. Plant Cell 28: 2545–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenach AJ, Hecht V, Vander Schoor JK, Liew LC, Aubert G, Burstin J, Weller JL (2017) EARLY FLOWERING3 redundancy fine-tunes photoperiod sensitivity. Plant Physiol 173: 2253–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth N, Roca X, Hastings ML, Roeder T, Krainer AR, Sachidanandam R (2006) Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res 34: 3955–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Dong A, Shen WH (2015) Epigenetic regulation of rice flowering and reproduction. Front Plant Sci 5: 803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim JS, Kubota A, Imaizumi T (2017) Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol 173: 5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Shim JS, Kinmonth-Schultz HA, Imaizumi T (2015) Photoperiodic flowering: time measurement mechanisms in leaves. Annu Rev Plant Biol 66: 441–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussmilch FC, Berbel A, Hecht V, Vander Schoor JK, Ferrándiz C, Madueño F, Weller JL (2015) Pea VEGETATIVE2 is an FD homolog that is essential for flowering and compound inflorescence development. Plant Cell 27: 1046–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran VK, Zhang Z, Yagi M, Nishiyama A, Habara Y, Takeshima Y, Matsuo M (2005) A novel cryptic exon identified in the 3′ region of intron 2 of the human dystrophin gene. J Hum Genet 50: 425–433 [DOI] [PubMed] [Google Scholar]
- Tsutsui T, Fukasawa R, Shinmyouzu K, Nakagawa R, Tobe K, Tanaka A, Ohkuma Y (2013) Mediator complex recruits epigenetic regulators via its two cyclin-dependent kinase subunits to repress transcription of immune response genes. J Biol Chem 288: 20955–20965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D. (2017) Key developmental transitions during flower morphogenesis and their regulation. Curr Opin Genet Dev 45: 44–50 [DOI] [PubMed] [Google Scholar]
- Wang W, Chen X (2004) HUA ENHANCER3 reveals a role for a cyclin-dependent protein kinase in the specification of floral organ identity in Arabidopsis. Development 131: 3147–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Ortega R (2015) Genetic control of flowering time in legumes. Front Plant Sci 6: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Reid JB, Taylor SA, Murfet IC (1997) The genetic control of flowering in pea. Trends Plant Sci 2: 412–418 [Google Scholar]
- Whittaker C, Dean C (2017) The FLC locus: A platform for discoveries in epigenetics and adaptation. Annu Rev Cell Dev Biol 33: 555–575 [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Wiltshire RJE, Murfet IC, Reid JB (1994) The genetic control of heterochrony: Evidence from developmental mutants of Pisum sativum L. J Evol Biol 7: 447–465 [Google Scholar]
- Yun Y, Adesanya TM, Mitra RD (2012) A systematic study of gene expression variation at single-nucleotide resolution reveals widespread regulatory roles for uAUGs. Genome Res 22: 1089–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhou W, Chen Q, Fang M, Zheng S, Scheres B, Li C (2018) Mediator subunit MED31 is required for radial patterning of Arabidopsis roots. Proc Natl Acad Sci USA 115: E5624–E5633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Schluttenhoffer CM, Wang P, Fu F, Thimmapuram J, Zhu JK, Lee SY, Yun DJ, Mengiste T (2014) CYCLIN-DEPENDENT KINASE8 differentially regulates plant immunity to fungal pathogens through kinase-dependent and -independent functions in Arabidopsis. Plant Cell 26: 4149–4170 [DOI] [PMC free article] [PubMed] [Google Scholar]