Resupply of free zinc ions via autophagic degradation suppresses photosynthesis-related Fenton-like reaction-induced chlorosis under zinc starvation in Arabidopsis thaliana.
Abstract
Zinc (Zn) is an essential micronutrient for plant growth. Accordingly, Zn deficiency (−Zn) in agricultural fields is a serious problem, especially in developing regions. Autophagy, a major intracellular degradation system in eukaryotes, plays important roles in nutrient recycling under nitrogen and carbon starvation. However, the relationship between autophagy and deficiencies of other essential elements remains poorly understood, especially in plants. In this study, we focused on Zn due to the property that within cells most Zn is tightly bound to proteins, which can be targets of autophagy. We found that autophagy plays a critical role during −Zn in Arabidopsis (Arabidopsis thaliana). Autophagy-defective plants (atg mutants) failed to grow and developed accelerated chlorosis under −Zn. As expected, −Zn induced autophagy in wild-type plants, whereas in atg mutants, various organelle proteins accumulated to high levels. Additionally, the amount of free Zn2+ was lower in atg mutants than in control plants. Interestingly, −Zn symptoms in atg mutants recovered under low-light, iron-limited conditions. The levels of hydroxyl radicals in chloroplasts were elevated, and the levels of superoxide were reduced in −Zn atg mutants. These results imply that the photosynthesis-mediated Fenton-like reaction, which is responsible for the chlorotic symptom of −Zn, is accelerated in atg mutants. Together, our data indicate that autophagic degradation plays important functions in maintaining Zn pools to increase Zn bioavailability and maintain reactive oxygen species homeostasis under −Zn in plants.
Plant essential nutrients, defined as those elements indispensable for optimal plant growth, are classified as macronutrients or micronutrients according to the amounts required. Thus, the macronutrients are carbon, hydrogen, oxygen, nitrogen, phosphorus, potassium, sulfur, calcium, and magnesium, whereas the micronutrients are iron (Fe), manganese, zinc (Zn), copper (Cu), nickel, molybdenum, chlorine, and boron. Carbon and oxygen can be obtained from air, and hydrogen from water, but the other nutrients must be absorbed from the soil through the roots.
In this study, we focused on Zn, a metallic element that is essential for all living organisms. Most cellular Zn is tightly bound to proteins; thus levels of free Zn ions are quite low within cells. Zn serves as a catalytic or structural cofactor in a large number of enzymes including alcohol dehydrogenase, superoxide dismutase (SOD), and regulatory proteins such as transcription factors containing Zn-finger domains (Vallee and Auld, 1990; Maret, 2009). Therefore, Zn deficiency (−Zn) disturbs cellular homeostasis. In the context of agriculture, −Zn is a serious problem because it dramatically decreases the quality and quantity of crop commodities, especially in developing regions. Previous research on −Zn in plants has focused primarily on uptake of Zn by transporters (Grotz et al., 1998) and gene regulation by transcription factors that function under −Zn (Assunção et al., 2010). By contrast, relatively few studies have focused on the redistribution of intracellular Zn (Eguchi et al., 2017) and the detailed mechanisms of the onset of −Zn symptoms remains unclear.
Autophagy is a major intracellular degradation mechanism that is conserved throughout the eukaryotes. During autophagy, degradation targets are surrounded by an isolation membrane and encapsulated in an autophagosome (AP). The outer membrane of the AP fuses with the vacuolar membrane, and the inner membrane of the AP and its contents (i.e. degradation targets) are released into the vacuolar lumen. This single membrane-bound vesicle inside the vacuole is called the autophagic body (AB). The AB is rapidly degraded by vacuolar lipases and proteases, and the contents are recycled for use as nutrients.
Autophagic processes are driven by a number of autophagy-related (ATG) proteins (Mizushima et al., 2011). The ATG genes were first discovered in yeast (Saccharomyces cerevisiae), and many ATG genes are highly conserved in plants (Hanaoka et al., 2002; Yoshimoto, 2012). In Arabidopsis (Arabidopsis thaliana), transfer DNA insertions that disrupted ATG genes were identified and the mutants were shown to be defective in autophagy (Doelling et al., 2002; Hanaoka et al., 2002; Yoshimoto et al., 2004; Thompson et al., 2005). These mutants, referred to as atg (e.g. atg2 and atg5), exhibit accelerated senescence under normal conditions, but complete the normal life cycle and form seeds for the next generation.
The physiological significance of autophagy in plants has been gradually clarified by analyses of atg mutants. For example, it has become clear that autophagy suppresses salicylic acid (SA) signaling. When NahG, a SA hydroxylase, is overexpressed in an atg5 plant, the level of endogenous SA is reduced and SA signaling is inhibited, resulting in suppression of senescence and immunity-related programmed cell death (PCD). Additionally, a knockout mutant of SALICYLIC ACID INDUCTION DEFICIENT2, a gene for a SA biosynthetic enzyme, suppresses senescence and PCD in the atg2 mutant. These data suggest that excessive SA signaling causes accelerated PCD during senescence and immunity in atg mutants (Yoshimoto et al., 2009).
Nitrogen or carbon starvation induces autophagy in Arabidopsis (Thompson et al., 2005; Izumi et al., 2010; Merkulova et al., 2014), as in yeast and mammals. However, the relationship between autophagy and deficiencies in many other essential elements remains poorly understood, especially in plants. In yeast, −Zn induces autophagy and plays important roles in adaptation to −Zn. The transcription factor Zap1, the master regulator of the −Zn response in yeast, does not directly control −Zn-induced autophagy. Zn is thought to be supplied by bulk degradation of cytoplasm via nonselective autophagy (Kawamata et al., 2017). The association between autophagy and Zn has also been examined in cultured mammalian cells (Liuzzi et al., 2014). These studies have revealed, for example, that n,n,n′,n′-tetrakis(2-pyridylmethyl)ethylenediamine, a Zn-specific–chelating agent that can cross biological membranes, inhibits autophagy, whereas the addition of Zn promotes autophagy (Liuzzi and Yoo, 2013).
In this study, we sought to characterize the relationship between −Zn and autophagy in plants. For this purpose, we established an experimental system for inducing micronutrient deficiency in hydroponic culture, and grew atg mutants under these conditions. Using various cell biological, biochemical, histochemical, and physiological methods, we analyzed the phenotypes of atg mutants under −Zn. Our results indicated that −Zn-induced autophagy improves Zn bioavailability in plants, as it does in yeast. In addition, we found that autophagy suppresses the symptoms of −Zn in a photosynthesis-dependent manner. Our findings regarding the mechanism by which plants respond to −Zn should contribute to methods aimed at preventing −Zn in crops and help to solve important agricultural problems.
RESULTS
atg Mutants Exhibit Accelerated −Zn Symptoms
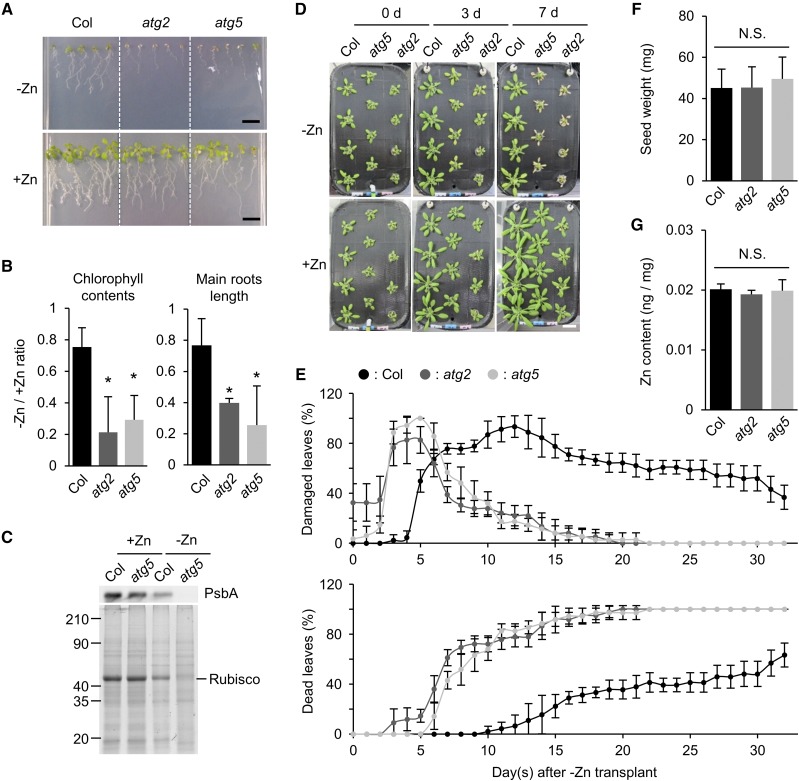
To evaluate the relationship between −Zn and autophagy in plants, we grew wild-type Arabidopsis Columbia (Col) and two autophagy-defective mutants, atg2 and atg5, under −Zn conditions. Col was used as a control for normal autophagic activity. First, we sowed seeds on solid media and grew the seedlings for 14 d. In the presence of sufficient Zn (+Zn), the atg mutants grew as rapidly as Col (Fig. 1A, lower). On the other hand, as shown in the upper representations of Figure 1A, the atg mutants exhibited severe symptoms under −Zn: Chlorophyll contents and main root length were significantly lower in the atg mutants than in Col (Fig. 1B), and the levels of chloroplastic proteins, such as D1 protein of PSII (PsbA) and Rubisco, were much lower in the atg5 mutant than in Col 10 d after sowing on −Zn media (Fig. 1C). These results suggested that autophagy has an important function in adaptation to −Zn.
Figure 1.
Autophagy-defective plants exhibit accelerated −Zn symptoms. A, Phenotypes of Col, atg2, and atg5 plants on solid media, grown vertically for 14 d after sowing under long-day conditions. −Zn: Zn deficiency (0 μm); +Zn: normal media containing a sufficient amount of Zn (1 μm). Scale bars = 1 cm. B, Values of −Zn/+Zn ratios of chlorophyll concentration (left) and main root length (right) shown in (A). n = 3, *P < 0.05 Student’s t test, error bars show sd. C, Protein levels in Col and atg5 plants under +Zn and −Zn conditions. PsbA was detected by western blot. Total proteins were separated by SDS-PAGE and stained with Coomassie Brilliant Blue (CBB). Approximate molecular mass (kilodalton) is displayed on the left. Equal amount of total protein was loaded in each lane. D, Phenotypes of Col, atg5, and atg2 plants in hydroponic transplant experiments. Twenty-one days after sowing on +Zn media, plants were transplanted to −Zn or +Zn media. Images were acquired at 0, 3, and 7 DAT. Scale bar = 2 cm. E, Quantified data of (D). Percentages of damaged (chlorotic) leaves (upper) and dead (completely dried) leaves (lower) in a time-course manner. n = 4. Error bars show sd. F, Seed weight of Col, atg2, and atg5 plants. Seeds were harvested from soil-grown plants and 500 seeds were counted and weighted. Individual seed weight was calculated by dividing the total weight by 500. n = 4. Error bars show sd. No significant difference was detected among the three samples in Student’s t test (P > 0.05). G, Zn contents per 1 mg of seeds of Col, atg2, and atg5 plants. Quantification was performed by ICP-MS. n = 4. Error bars show sd. No significant difference was detected among the three samples in Student’s t test (P > 0.05). N.S., not significant.
However, these phenotypes could be explained by lower Zn contents in atg mutant seeds. If autophagy is involved in remobilization of Zn ions from leaves to seeds, the level of Zn could have been lower in the seeds of atg mutants than in Col seeds, leading to a growth defect on media lacking Zn. To exclude this possibility, we next conducted hydroponic transplant experiments. Plants were first grown in +Zn hydroponic media for 21 d. After the roots were washed well in −Zn media, the plants were transferred to −Zn and then grown for another 32 d. The results revealed that atg mutants had −Zn phenotypes even in transplant experiments. Under −Zn conditions, the leaves of atg mutants started to yellow at 3 d after transplanting (DAT); and at 7 DAT, many of the mutant leaves were dead (Fig. 1D). We quantified the changes in leaf phenotypes under −Zn conditions. The peak percentage of damaged (yellow) leaves occurred at 12 DAT in Col versus 5 DAT in the atg mutants (Fig. 1E, top). In addition, the percentage of completely dead leaves was elevated earlier in the atg mutants, and the rate of the increase was also more rapid (Fig. 1E, bottom). These data suggested that a difference in seed Zn level is not responsible for the −Zn-sensitive phenotype of atg mutants. Indeed, neither seed weight (Fig. 1F) nor seed Zn content (Fig. 1G) differed between atg mutants and Col.
Excessive SA Signaling in atg Mutant Plants Is Not a Main Cause of −Zn-Induced Chlorosis
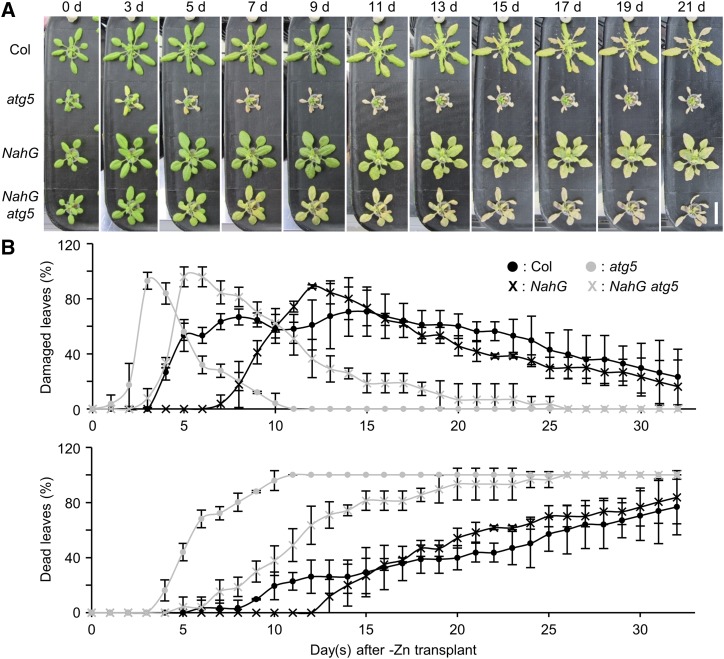
SA is a phytohormone involved in defense against pathogens and senescence, and autophagy negatively regulates SA signaling. We previously showed that atg mutants exhibit accelerated PCD during senescence and immunity. When NahG is overexpressed, leading to suppression of SA signaling, the early PCD phenotype in atg mutants is suppressed (Yoshimoto et al., 2009). To determine whether excessive SA signaling is the cause of −Zn-induced chlorosis in atg mutants, we compared NahG and NahG atg5 plants. In hydroponic transplant experiments, NahG atg5 plants exhibited more severe −Zn symptoms (chlorosis) than NahG plants. In NahG atg5 plants, chlorosis occurred at 5 DAT, and the progression of symptoms was also much faster than in NahG plants (Fig. 2). This suggests that the defect in autophagy is a major factor in −Zn-induced chlorosis, independent of SA signaling. Because the severe chlorosis phenotype of atg mutants under −Zn causes difficulties for sampling enough materials, we used NahG plants as a control and NahG atg5 plants as an autophagy defective plant for further experiments.
Figure 2.
Phenotypes of NahG and NahG atg5 plants under −Zn. A, Phenotypes of Col, atg5, NahG, and NahG atg5 plants in hydroponic transplant experiments. Twenty-one days after sowing on +Zn media, plants were transplanted to −Zn media. Images were acquired at 0, 3, 5, 7, 9, 11, 13, 15, 17, 19, and 21 DAT. Scale bar = 2 cm. B, Quantified data of (A). Temporal changes in the percentages of damaged leaves and dead leaves were shown in the same manner as in Fig. 1E. n = 3. Error bars show sd.
−Zn Induces Bulk Autophagy
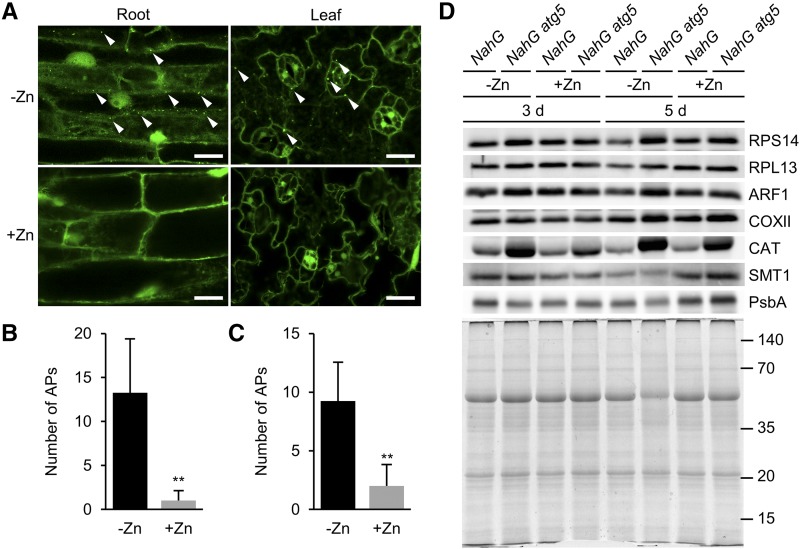
Starvation of nitrogen or carbon induces autophagy in Arabidopsis. To determine whether −Zn also induces autophagy in plants, we observed APs using plants expressing GFP-fused AUTOPHAGY-RELATED PROTEIN8a (GFP-ATG8a). ATG8 is a lipid-conjugated protein localized to ATG membrane structures such as the AP (Kirisako et al., 1999; Kabeya et al., 2000; Yoshimoto et al., 2004). GFP-ATG8a–expressing plants grown on +Zn solid media for 7 d were transferred to −Zn or +Zn media, and then their cotyledons and roots were observed by confocal laser scanning microscopy. At 18 h after transplanting, plants under −Zn exhibited more dots labeled with GFP-ATG8a in both cotyledon and root cells than plants under +Zn (Fig. 3, A–C). Temporal changes in the number of APs in roots under +/−Zn are shown in Supplemental Figure S1.
Figure 3.
Zinc starvation induces autophagic degradation of many targets in plants. A, GFP-ATG8a–labeled dots indicate APs. Images of main root cells (left) and cotyledon cells (right) were acquired at 18 h after transplanting to +/−Zn conditions. Under +Zn, APs were barely observed (lower), but under −Zn, many APs were observed in both roots and cotyledons (upper). Representative APs are indicated by white arrowheads. Scale bars = 20 μm. B and C, Quantified data of (A). Number of APs per image in roots (B) and cotyledons (C) at 18 h after transplant to +/−Zn media. n = 4, **P < 0.01, Student’s t test, error bars show sd. D, Various organelle proteins were compared by western blot under +/−Zn. Proteins were extracted from leaves of NahG and NahG atg5 plants at 3 and 5 DAT under +/−Zn. Antibodies used in this experiment are indicated on the right. The lower representation shows a CBB-stained gel confirming equal protein loading. Approximate molecular mass (kilodaltons) is displayed on the right.
To further confirm the autophagy induction by −Zn, we checked autophagy flux using a V-ATPase inhibiter, concanamycin A (ConA). When roots are treated with ConA, vacuolar acidification is inhibited, leading to accumulation of ABs inside vacuolar lumens if autophagy is activated. Under −Zn, AB numbers were significantly increased compared to under +Zn (Supplemental Fig. S2, A and B). In addition, we quantified autophagic degradation activity by a GFP-ATG8 processing assay. In GFP-ATG8 plants, amounts of free GFP were increased inside the vacuole along with the activation of autophagy. Therefore, the activity of autophagy can be estimated by detecting the ratio of free GFP against GFP-ATG8 by western blot analysis using anti-GFP antibody. As expected, under −Zn, the GFP/GFP-ATG8 ratio was increased compared to +Zn conditions (Supplemental Fig. S3). These data indicate that autophagy activity was upregulated by −Zn.
Furthermore, we checked autophagy activity in roots using a split-root assay. After splitting a root of a GFP-ATG8 plant, two roots were transplanted to a +Zn- and −Zn-separated media plate (−Zn/+Zn plate) as shown in Supplemental Figure S4A and incubated for 18 h under continuous light conditions. Then, we quantified the number of APs using a confocal microscope. The result showed that the −Zn side root had significantly higher autophagy activity than that of the control (+Zn/+Zn plate). On the other hand, the +Zn side of the −Zn/+Zn plate did not show an increase of APs (Supplemental Fig. S4B), implying that autophagy is induced locally at the −Zn site, such as at organ or tissue levels.
In yeast, it has been suggested that induction of autophagy by −Zn is not a selective process (Kawamata et al., 2017). To investigate the degradation targets of plant autophagy under −Zn, we conducted western blot analyses. Using antibodies against representative organelle marker proteins, we compared the amount of each organelle protein in NahG and NahG atg5 leaves under −Zn and +Zn at 3 and 5 DAT. We reasoned that if −Zn conditions induced selective autophagy, specific organelles fated for degradation would accumulate to higher levels in NahG atg5 than in NahG plants under −Zn. ADP-ribosylation factor1 (ARF1), cytochrome oxidase subunit II (COXII), catalase (CAT), sterol methyltransferase1 (SMT1), and PsbA were used as markers for Golgi apparatus, mitochondrion, peroxisome, endoplasmic reticulum, and chloroplast, respectively. Ribosomes are complexes of proteins and RNAs, and several ribosomal proteins, such as RPL37, are Zn-binding proteins (Klinge et al., 2011). In addition, because ribosomes can be degraded by ribophagy, a form of selective autophagy (Kraft et al., 2008; Floyd et al., 2015; Bassham and MacIntosh, 2017), we performed western blot using antibodies against 40S ribosomal protein S14-1 (RPS14) and 60S ribosomal protein L13-1 (RPL13), along with the other organelle markers. Under +Zn, the levels of each protein were almost equal in NahG atg5 and NahG plants, with the exception of CAT. Higher accumulation of CAT is consistent with our previous report that peroxisomes constantly accumulate in atg mutant leaves (Yoshimoto et al., 2014). However, under −Zn, many organelle markers accumulated to higher levels in NahG atg5 than in NahG plants (Fig. 3D). RPS14, RPL13, ARF1, COXII, and CAT were significantly more abundant in NahG atg5 plants, especially at 5 DAT. Even at 3 DAT, RPS14, ARF1, and CAT were present in NahG atg5 plants at higher levels than in the control. These data suggest that autophagy degrades various targets (proteins and organelles) as −Zn progresses.
Oxidative Stress Mediated by Light and Fe Is a Cause of −Zn-Induced Chlorosis in atg Mutant Plants
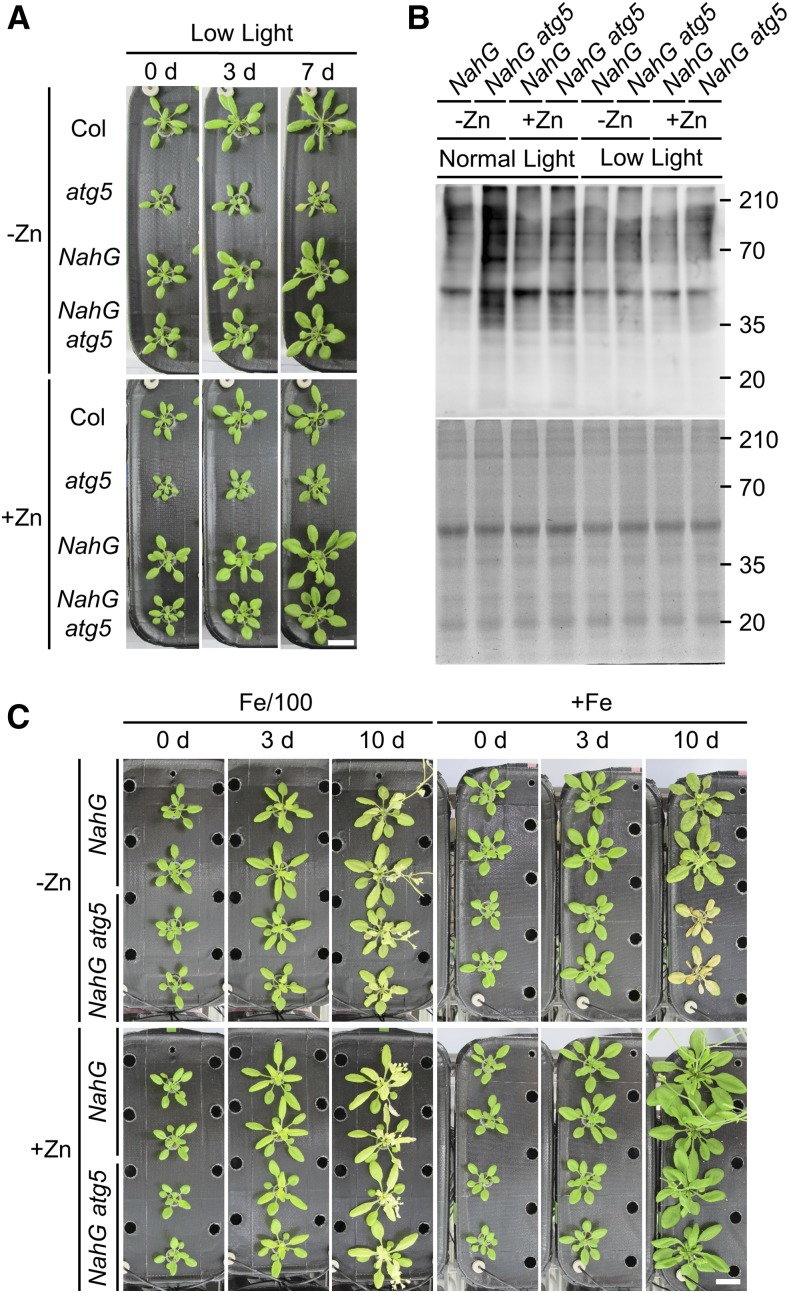
The severity of −Zn symptoms in plants depends on light intensity (Cakmak, 2000). As shown in Figure 2, atg5 and NahG atg5 plants developed –Zn-induced chlorosis at 3 and 5 DAT, respectively, under normal light intensity. To investigate the influence of light intensity on the onset of −Zn symptoms in atg mutant plants, we performed the same transplant experiment under low light intensity. For this experiment, plants were grown under normal light intensity for 21 d in +Zn, and then transferred to low light conditions simultaneously with transplantation to −Zn or +Zn media. −Zn -induced chlorosis in the atg mutant plants was observed later under low light intensity than under normal light (Fig. 4A): atg5 mutants developed −Zn-induced chlorosis at 7 DAT, and NahG atg5 plants did not exhibit chlorosis even at 7 DAT. These data indicate that severe −Zn symptoms in atg5 mutants were induced in a light-dependent manner, as in crops (Cakmak, 2000).
Figure 4.
−Zn symptoms are promoted by light and Fe ions. A, Phenotypes of Col, atg5, NahG, and NahG atg5 plants at 0, 3, and 7 DAT in hydroponic transplant experiments under low light conditions (17 μmol m−2 s−1). Plants grown for 21 d on +Zn under normal light intensity were transplanted to −Zn or +Zn media and grown under low light intensity. Scale bar = 2 cm. B, Detection of oxidized protein levels by western blot with anti-DNP antibody. Proteins were extracted from leaves at 3 DAT under normal or low light intensity. Upper representation shows the result of western blot. Lower representation shows a CBB-stained gel (loading control). C, Phenotypes of NahG and NahG atg5 plants in hydroponic transplant experiments under Fe-limited conditions. Plants grown for 21 d on +Zn media were transplanted to the following: −Zn, Fe-limited; −Zn, +Fe; +Zn, Fe-limited; or +Zn, +Fe. Photographs were taken at 0, 3, and 10 DAT. Scale bar = 2 cm.
Because the onset of –Zn-induced chlorosis was dependent on light intensity, we next investigated the mechanism underlying light-dependent chlorosis by comparing oxidized protein levels in leaves. To detect oxidized proteins, we performed anti–2,4-dinitrophenol (DNP) western blot. When extracted proteins are treated with 2,4-dinitrophenylhydrazine, DNP is added to their carbonylation sites, and DNP-modified proteins can be detected by western blot with anti-DNP antibody (Romero-Puertas et al., 2002). Because ATG-knockdown plants accumulate oxidized proteins at a certain level even under normal conditions (Xiong et al., 2007), we anticipated that it would be difficult to detect differences in oxidized protein levels between +Zn and −Zn conditions. Therefore, for these experiments, we used NahG and NahG atg5 plants because inhibition of SA signaling in atg mutants by NahG decreases oxidative stress and prolongs life (Yoshimoto et al., 2009). Under normal light intensity, −Zn induced the accumulation of higher levels of oxidized proteins in NahG atg5 plants compared to NahG plants, but this difference was not observed under +Zn (Fig. 4B). As expected, under low light intensity, the oxidized protein levels in NahG atg5 plants were not high even under −Zn; This was almost the same accumulation pattern observed in the control (+Zn and low light). These data indicate that the appearance of −Zn symptoms depends on oxidative stress levels.
Next, because Fe contents are elevated in aerial tissues under −Zn (Cakmak, 2000), we focused on the effect of Fe under −Zn. To investigate the relationships between −Zn phenotypes in atg mutant plants and Fe nutrition, we performed transplant experiments as described above, using media that were Zn-deficient and Fe-limited. In media containing 1% of the normal level of iron (Fe/100), −Zn-induced chlorosis in NahG atg5 plants was suppressed (Fig. 4C). The phenotype in NahG atg5 plants in −Zn Fe/100 was similar to that in NahG plants, and −Zn-induced chlorosis was not apparent. On Zn-deficient and Fe-sufficient media (−Zn, +Fe), NahG atg5 plants exhibited intense chlorosis throughout all leaves at 10 DAT. Additionally, NahG atg5 plants did not exhibit an obviously different phenotype compared to NahG plants under Fe starvation alone (+Zn Fe/100; Fig. 4C). These data suggest that Fe is an important determinant of −Zn-induced chlorosis in atg mutant plants, and that autophagy does not affect Fe starvation resistance in plants.
Reactive Oxygen Species Production and Homeostasis Are Not Controlled in atg Mutant Plants under −Zn
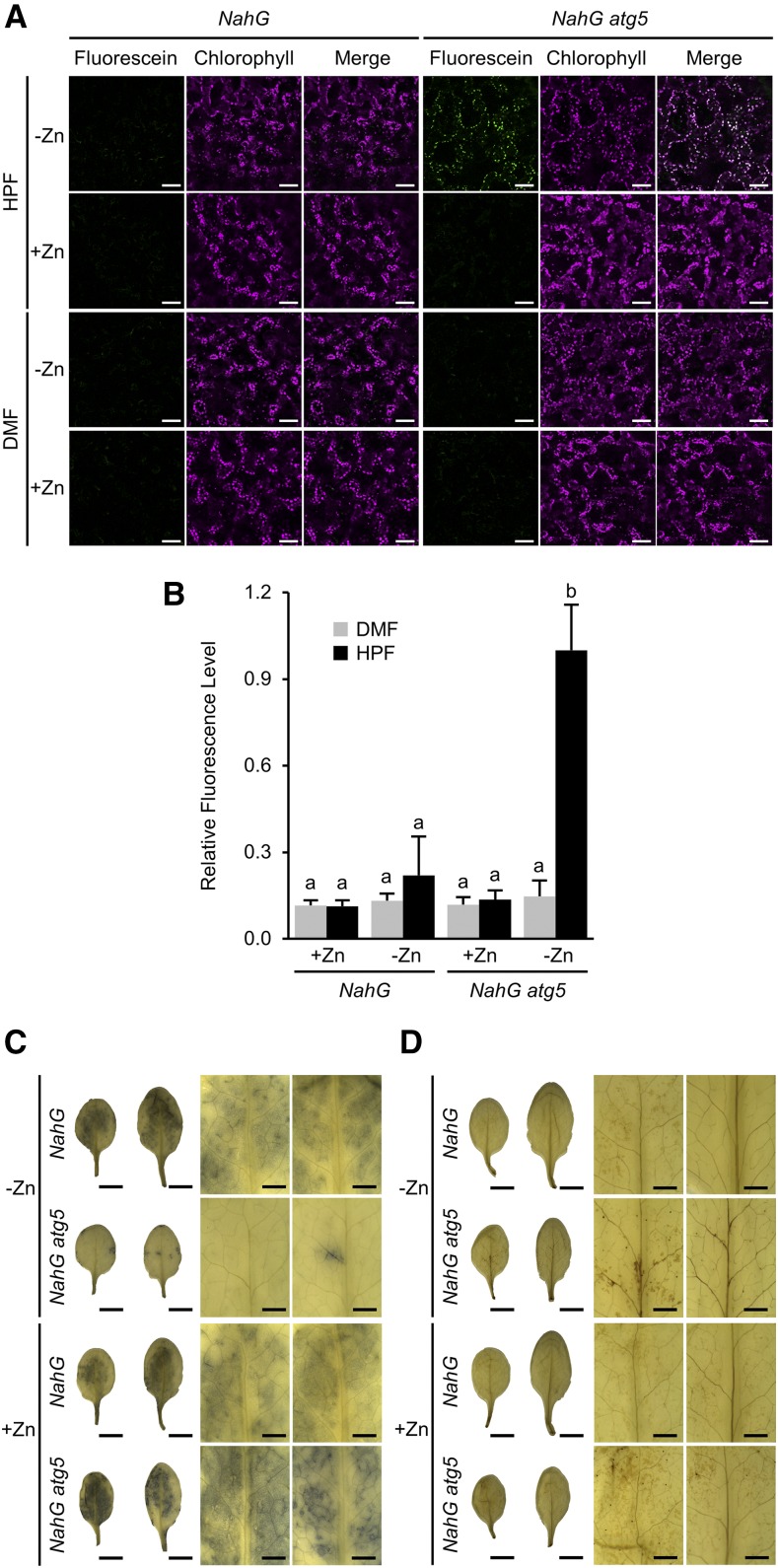
Because oxidative stress is enhanced under −Zn, we next sought to determine the levels of reactive oxygen species (ROS) in atg mutant plants under −Zn. First, we detected hydroxyl radicals (·OH) in leaves by fluorescence of 3′-(p-hydroxyphenyl) fluorescein (HPF). HPF produces fluorescein when it is oxidized by ROS, especially by ·OH, with quite high specificity. At 3 DAT in NahG atg5 plants under −Zn, we detected fluorescein fluorescence colocalized with autofluorescence of chlorophyll (Fig. 5A). In NahG plants or in NahG atg5 plants under +Zn, no such fluorescence was detected. Additionally, there was no fluorescence in leaves treated with n,n-dimethylformamide (DMF), which served as a control for the HPF solvent. Quantification of fluorescence intensity clearly showed that ·OH levels were significantly elevated in NahG atg5 plants under −Zn (Fig. 5B).
Figure 5.
Changes of ROS under −Zn in autophagy-defective plants. A, Detection of ·OH was performed using HPF. Fluorescence of fluorescein, generated from HPF by reaction with ·OH, is shown in green; autofluorescence of chlorophylls is shown in magenta. Merged images are shown in the right representations. Fluorescein fluorescence was increased in NahG atg5 plants at 3 DAT (from +Zn to −Zn), and was colocalized with chloroplasts. DMF, the solvent for HPF, was used as the negative control. Scale bars = 50 μm. B, Quantified data from (A). Relative intensity of green fluorescence was measured using the software ImageJ. Columns with the same letter are not significantly different (P < 0.01, Student’s t test). n = 4. Error bars show sd. C and D, Results of NBT (C) and DAB (D) staining of NahG and NahG atg5 leaves at 3 DAT under −/+Zn. Left show whole leaves digitally extracted from the images taken by stereo microscope and shown on a white background. Right shows higher magnifications of the left. For each magnification, the left column shows the lower leaves, and the right column shows the upper leaves. Scale bars = 5 mm (left, showing whole leaves) and 1 mm (right).
In atg5 mutants under −Zn, we observed marked elongation of stromules (stroma-filled tubules; Supplemental Fig. S5, A and B). Stromules elongate under various environmental stresses (Brunkard et al., 2015), suggesting that the chloroplasts in atg mutant plants under −Zn were experiencing stress. This result suggests that chloroplasts are responsible for stress development in the atg mutants under −Zn.
Next, we detected two other ROS species, superoxide (·O2−) and hydrogen peroxide (H2O2), using nitro blue tetrazolium and 3,3′-diaminobenzidine (DAB) staining, respectively (Doke, 1983; Thordal-Christensen et al., 1997). ·O2− accumulated at higher levels in NahG atg5 than in NahG plants under +Zn, whereas under −Zn, the level of ·O2− was dramatically lower in NahG atg5 plants (Fig. 5C). NahG atg5 plants contained slightly higher levels of H2O2 than NahG plants (Fig. 5D), consistent with our previous report (Yoshimoto et al., 2009). These data suggest that changes in ROS production—specifically, reduced ·O2− and elevated ·OH—are responsible for the more severe −Zn symptoms in atg mutant plants.
Autophagy Maintains Zn Pools under −Zn
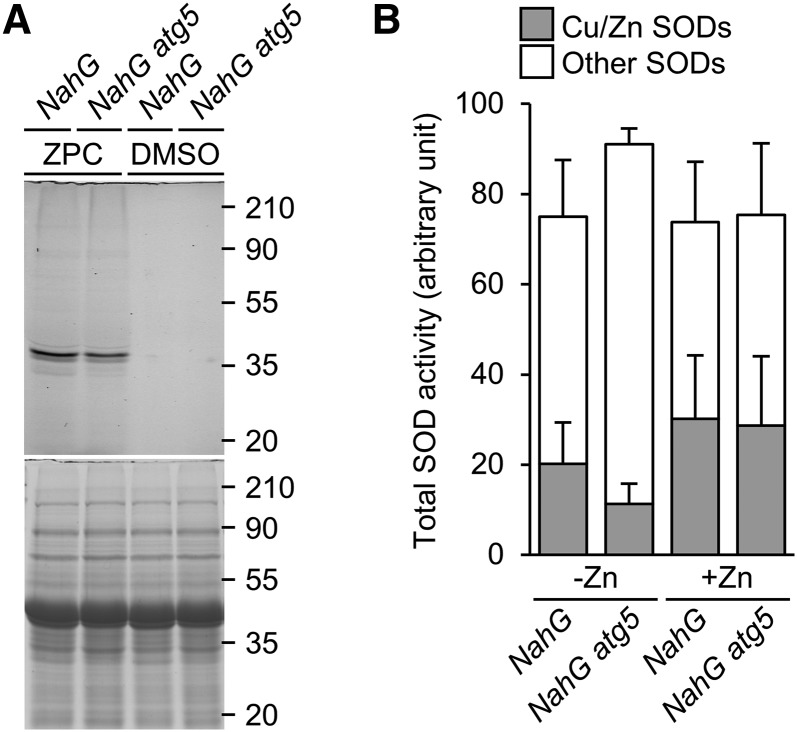
Finally, using the Zin-Pro Capture reagent (Funakoshi; Miki et al., 2016), we investigated how autophagy affects the level of free Zn ions in individual plants. When bound to mobile Zn ions, Zin-Pro Capture can react with proteins and bind fluorescein (Supplemental Fig. S6). We extracted proteins from plants infiltrated with Zin-Pro Capture, subjected them to SDS-PAGE, and observed the resultant gels on a fluorescence scanner. The levels of mobile Zn ions in NahG and NahG atg5 plants were estimated based on the levels of fluorescently labeled endogenous proteins. As shown in Figure 6A, the level of fluorescently labeled proteins was lower in NahG atg5 than in NahG plants, indicating that the level of mobile Zn ions inside plants was increased by autophagy under −Zn conditions.
Figure 6.
Decrease of mobile Zn levels in atg mutant plants causes decay of Zn-requiring enzyme activity. A, Proteins were extracted from Zin-Pro Capture (ZPC)-treated plants (NahG and NahG atg5, 3 DAT to −Zn), and subjected to SDS-PAGE; fluorescence was detected in-gel. The lower representation shows a CBB-stained gel, confirming equal protein loading. Approximate molecular mass (kilodaltons) is shown on the right. B, SOD activities of NahG and NahG atg5 plants at 5 DAT to −/+Zn. Gray parts indicate Cu/ZnSOD activities, and white parts indicate MnSOD and FeSOD activities. n = 4. Error bars show SD.
Next, we investigated the activity of SODs, as representative Zn-requiring metalloenzymes. Plants have Cu/ZnSODs that require both Zn and Cu, as well as FeSODs and MnSODs that require Fe and Mn, respectively, for their enzyme activities and/or maintenance of enzyme structures. The activities of Cu/ZnSODs and other SODs were measured separately using potassium cyanide (KCN), which is an inhibitor of Cu/ZnSOD. Cu/ZnSOD activities were calculated by subtraction of MnSOD and FeSOD activities, which were measured in reaction solutions with KCN, from total SOD activities. Under +Zn, the SOD activity in NahG atg5 plants was similar to that in NahG plants. By contrast, under −Zn, the activity of Cu/ZnSOD was dramatically lower in NahG atg5 than in NahG plants (Fig. 6B). Under −Zn, the abundance of Cu/ZnSOD proteins was rather higher in NahG atg5 than in NahG plants (Supplemental Fig. S7), indicating that a change in protein level was not the cause of the reduction in Cu/ZnSOD activity. These results indicate that an increase in Zn bioavailability, mediated by autophagy, helps maintain the activity of Zn-requiring enzymes under −Zn.
DISCUSSION
Zn Remobilization and Autophagy
Autophagy is involved in nitrogen remobilization in Arabidopsis (Guiboileau et al., 2012), rice (Oryza sativa; Wada et al., 2015), and maize (Zea mays; Li et al., 2015). In addition, the efficiency of Fe remobilization from leaves to seeds is reduced in Arabidopsis atg5 mutants (Pottier et al., 2019). Those facts led us to hypothesize that autophagy also contributes to Zn remobilization to seeds, as suggested in Pottier et al. (2019). That study reported that the Zn content in seed normalized against the Zn content in the whole plant is lower in atg5 mutants than in Col. By contrast, our data showed that seed weight and Zn content did not differ among atg2, atg5, and Col plants (Fig. 1, F and G). The discrepancy can be explained by the fact that the number of seeds per individual plant is smaller in atg mutants than in Col and that the Zn content of leaves and stems is higher in atg mutants than in Col (Pottier et al., 2019).
Relationships between SA and −Zn-Induced Chlorosis
Our biochemical and histochemical analyses revealed that oxidative stress is enhanced by the increase in ROS under −Zn (Figs. 4B and 5). SA induces ROS production, and ROS activates SALICYLIC ACID INDUCTION DEFICIENT2, a SA synthetase, through ENHANCED DISEASE SUSCEPTIBILITY1, thereby promoting SA biosynthesis during senescence and pathogen-induced PCD (Hofius et al., 2009; Yoshimoto et al., 2009). This SA-ROS amplification loop may function even under −Zn conditions. Indeed, as shown in Figure 2, overexpression of NahG is effective in suppressing –Zn-induced chlorosis. The symptoms of NahG atg5 plants were less severe than those of the atg5 single mutant: Chlorosis was observed starting at 3 DAT in atg5 mutants, but at 5 DAT in NahG atg5 plants (Fig. 2). This implies that the expression of NahG itself plays a role in suppressing chlorosis, consistent with the observation that NahG plants developed chlorosis later than Col plants. Zinc deficiency increases ROS levels, and SA further enhances this effect. Although autophagy functions as a suppressor of SA signaling, considering that there was a marked difference in –Zn-induced chlorosis between NahG and NahG atg5 plants, we concluded that SA is not the main cause of −Zn symptoms in atg mutant plants. When we measured SA contents in leaves and compared them among Col, atg5, NahG, and NahG atg5 plants under −/+Zn conditions, the SA contents of NahG atg5 plants under −Zn was much lower than those of Col under −Zn and even under +Zn (Supplemental Fig. S8), supporting our conclusion that excessive SA signaling is not a main cause of −Zn symptoms in the atg mutant.
Autophagic Defects Cause Damage to Chloroplasts under −Zn
In plants, −Zn symptoms manifest primarily as chlorosis, i.e. a reduction in the level of chlorophyll. This chlorosis was further enhanced in the atg mutants under −Zn, suggesting that chloroplasts are damaged and become less abundant in atg mutants (Fig. 1, A–C). Additionally, the levels of SMT1 and PsbA were decreased in NahG atg5 plants under −Zn (Fig. 3D). There are two possible reasons why these proteins might be less abundant in NahG atg5 plants. First, because Zn is important for transcriptional and translational activities, the expression of these proteins might decrease under −Zn. Second, −Zn might cause these proteins to be damaged so that they are broken down by non-autophagic pathways or cannot react with the antibodies we used. The second possibility is consistent with the data in Figure 1D, which show that chloroplasts were damaged and the levels of chloroplastic proteins were reduced in atg5 mutants under −Zn. The localization of ·OH in chloroplasts (Fig. 5A) is consistent with the idea that chloroplasts are the main onset place of −Zn symptoms.
As shown in Supplemental Figure S5, A and B, stromules significantly elongated under −Zn in atg5 plants. There are two possible causes for this phenotype. First, the chloroplasts could be experiencing stress. Consistent with this, stromule elongation is induced by various environmental stresses (Brunkard et al., 2015). Alternatively, partial degradation of chloroplasts may be promoted by −Zn. Rubisco-containing bodies (RCBs), small chloroplast-derived vesicles containing stromal proteins, have been proposed to be pinched off from stromules through sequestration by isolation membranes (Ishida et al., 2008); consequently, defects in autophagy promotes elongation of stromules. In other words, stromule elongations in atg5 mutants under −Zn mean that the RCB degradation pathway is activated under −Zn. To investigate whether such partial chloroplast degradation is induced under −Zn, we examined RCB accumulation inside vacuoles after treating plants expressing stroma-targeted GFP with ConA. Because ConA inhibits vacuolar acidification, RCB-derived vesicles (ABs) can be detected inside vacuolar lumens if autophagy is activated. Under dark, long-day, and Suc-added conditions, we detected no difference in accumulation of RCB-derived vesicles, irrespective of the presence or absence of Zn (Supplemental Fig. S9). These results suggest that chloroplast degradation by the RCB pathway is not enhanced under −Zn, in contrast to carbon starvation (Izumi et al., 2010). Based on these observations, we conclude that chloroplasts in the atg mutant plants are exposed to intense stress under −Zn.
Zinc deficiency-induced chlorosis in the atg mutant plants was suppressed under low light conditions (Fig. 4A). Additionally, oxidative stress in atg mutant plants was enhanced under −Zn in a light intensity-dependent manner (Fig. 4B). These results led us to hypothesize that −Zn symptoms in atg mutant plants were caused by light-dependent ROS production, which is largely a consequence of photosynthesis and photorespiration. To determine whether photorespiration is responsible for ROS production under −Zn, we grew plants under high CO2, which suppresses photorespiration (Reumann and Weber, 2006). We reasoned that if photorespiration is the source of ROS under −Zn, chlorosis should be suppressed under high CO2. However, when NahG and NahG atg5 plants were grown in +Zn media under ambient CO2 conditions (400 µL L−1) for 21 d, and then transferred to −Zn media under high CO2 conditions (3,000 µL L−1), −Zn symptoms were still observed in NahG atg5 plants (Supplemental Fig. S10). Indeed, the chlorosis in NahG atg5 plants was slightly accelerated relative to that observed in plants of the same genotype grown in ambient CO2 (Fig. 2A). These data suggest that the main cause of −Zn-induced chlorosis is photosynthesis-derived ROS. This is consistent with our observation that chloroplasts were significantly damaged (Fig. 1B, left, and C) and that ·OH accumulated exclusively in chloroplasts (Fig. 5A).
Bulk Autophagy Resupplies Zn in Plants under −Zn
Here, we propose a model of how autophagy helps plants tolerate −Zn (Fig. 7). When plants sense −Zn, they induce autophagy. This −Zn-induced autophagy seems to be bulk autophagy. Western blot analysis using antibodies against organelle marker proteins (Fig. 3D) revealed that multiple organelles and proteins are targets for autophagic degradation under −Zn. In yeast, bulk autophagy is induced under −Zn (Kawamata et al., 2017); therefore, it is reasonable that the −Zn- induced autophagy in plants is also bulk autophagy. In contrast to yeasts, however, plants are multicellular organisms, and may have different mechanisms for achieving selectivity in a tissue- or organ-specific manner. Therefore, future studies should investigate whether selective autophagic degradation occurs under −Zn.
Figure 7.
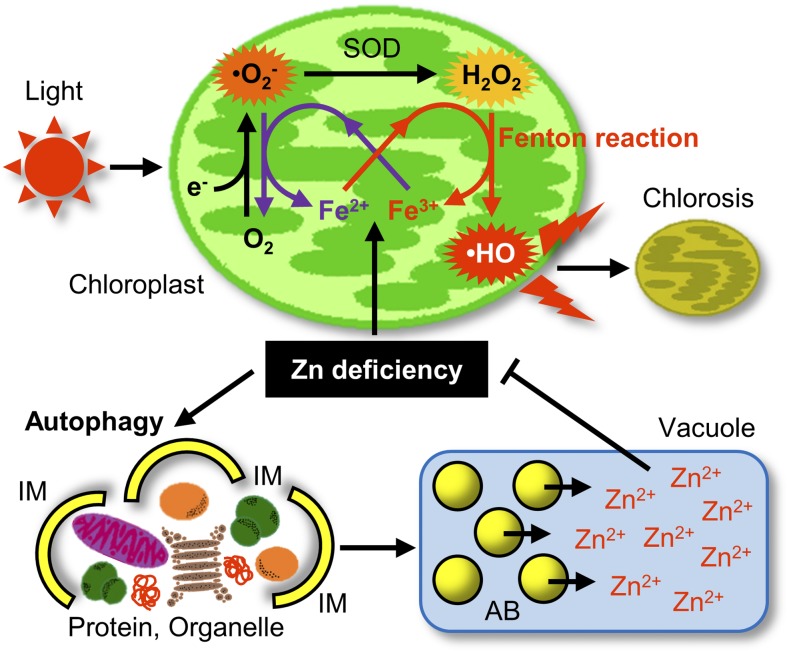
Model of autophagy-mediated −Zn tolerance mechanism in plants. During −Zn, autophagy is induced and supplies mobile Zn ions by degrading various proteins and organelles. This increases Zn bioavailability and alleviates −Zn. ⋅OH is generated in a light-dependent manner via the Fe-mediated Fenton-like reaction in chloroplasts. Production of ⋅OH is responsible for −Zn-induced chlorosis. IM, isolation membrane.
Zn-binding proteins or Zn-including organelles enclosed by the APs are transported to the vacuole to be degraded to release Zn ions. Because >5% of proteins bind Zn, the degradation of these intracellular components increases the level of free Zn ion (Fig. 6A), thereby alleviating −Zn. In other words, bulk autophagy promotes Zn bioavailability under −Zn.
Zinc supplied by autophagy is redistributed where it is needed. Cu/ZnSOD, a representative Zn-requiring enzyme, plays an important role in oxidative stress responses. Under −Zn, despite the high protein levels of Cu/ZnSOD in the atg mutant plants (Supplemental Fig. S7), the activity of Cu/ZnSOD was lower in atg mutant plants than in control plants (Fig. 6B), suggesting that the resupply of Zn to Zn enzymes by autophagy is important for resistance to −Zn.
−Zn Symptoms Are Due To a Fenton-like Reaction in atg Mutant Plants
Due to their reduced efficiency of Zn utilization, atg mutants exhibit more severe −Zn symptoms (Fig. 1, A–E). Cakmak (2000) predicted that Fe-mediated ·OH formation reactions are responsible for the symptoms of −Zn. We speculated that this reaction was facilitated by a defect in autophagy. The upper part of Figure 7 shows the possible reactions in the chloroplast that could produce ROS responsible for −Zn-induced chlorosis in an Fe- and light intensity-dependent manner. Under −Zn, Fe ions promote the Fenton-like reaction that generates ·OH from H2O2. In this reaction, Fe2+ is oxidized to Fe3+. As shown in Figure 5A, ·OH is produced in chloroplasts. On the other hand, Fe3+ is reduced to Fe2+ by oxidation of ·O2− to oxygen. The marked reduction in the level of ·O2− in the atg mutant plants under −Zn (Fig. 5C) supports this model. The suppression of chlorosis by Fe limitation under −Zn (Fig. 4C) suggests that the main cause of −Zn symptoms is ·OH production in chloroplasts via this loop involving Fe2+ and Fe3+. Furthermore, the suppression of chlorosis by reduced light intensity (Fig. 4A) suggests that this reaction occurs during photosynthesis. Therefore, we propose that −Zn-induced chlorosis is caused by chloroplast damage due to ROS production via this photosynthesis-dependent Fenton-like reaction.
CONCLUSION
In this study, we developed a hydroponic system that can precisely control nutrient conditions. Because Suc was not added in the media in this system, plants photosynthesize like they do in normal field conditions, allowing us to analyze only the effects of −Zn on phenotypes under more natural conditions. Additionally, the use of NahG atg5 plants allowed us to analyze true −Zn phenotypes excluding the effects of SA-dependent senescence and constitutive ROS accumulation in atg mutant plants. Thus, we clearly demonstrated that intracellular self-degradation by plant autophagy resupplies Zn ions from proteins and organelles to increase usable Zn under −Zn. Furthermore, here we also demonstrated the importance of the relationship between Zn and Fe. Namely, the main cause of −Zn-induced chlorosis is ⋅OH production mediated by Fe (Fenton-like reaction), which occurs in chloroplasts in a light-dependent manner. This was clearly demonstrated experimentally using autophagy-defective plants that show reduced Zn bioavailability. Our current findings would shed light on further mechanisms for plants to grow robustly under unfavorable soil environments.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 (Col) was used in this study. Details of atg2 (SALK_076727), atg5 (SAIL_129_B07), NahG, and NahG atg5 plants were described in our previous report (Yoshimoto et al., 2009). Plants were grown under long-day conditions (16-h light/8-h dark) at 22°C. Light intensity was 60 and 17 μmol m−2 s−1 in normal and low-light conditions, respectively. For high CO2 conditions, plants were grown under 3,000 µL L−1 CO2 (LPH-411SPC; NK System). Seeds were surface-sterilized, chilled at 4°C for 3 d in the dark, and then sown on solid media (for plate cultures) or rockwool (for hydroponic cultures). Plant growth media were based on MGRL media (Fujiwara et al., 1992). Media for −Zn were prepared without ZnSO4. Media for Fe limitation were prepared by reducing [FeSO4] to 0.086 μm and [Na2-EDTA] to 0.67 μm. Solid media plates were prepared by addition of 0.7% (w/v) agarose (A9539; Sigma-Aldrich) and 1.0% (w/v) Suc to MGRL liquid media. In hydroponic transplant experiments, the roots of plants grown for 21 d on +Zn media with aeration were well rinsed with −Zn media, and then transferred to −Zn media; images were acquired every day after transplantation. Two liters of liquid media were used per 12 individual plants. Hydroponic media were changed weekly. In the split-root assay, plants were vertically grown for 4 d under continuous light conditions on a +Zn plate gelled by 0.4% (w/v) Gelrite (075-05655; Fujifilm Wako Pure Chemical), then the main root was cut under the hypocotyl. Five d after cutting the root, split roots were transplanted to −Zn/+Zn-separated solid media gelled with agarose.
Measurements of Chlorophyll Contents
The fresh weight of shoots of plants grown for 14 d on agarose plates was measured. The shoots were immersed in DMF and placed at 4°C overnight in the dark. The absorbance of the resultant DMF solutions was measured at 664, 647, and 750 nm on a spectrophotometer (DS-11+; DeNovix) using glass cells (596492; Beckman Coulter), and the total amount of chlorophyll a and chlorophyll b was calculated by the equation of Porra (Porra et al., 1989). Finally, the amount of total chlorophyll per fresh weight (chlorophyll concentration) was calculated.
Measurements of Metal Contents
Contents of Zn in seeds were measured by inductively coupled plasma mass spectrometry (ICP-MS). Five-hundred seeds were counted and weighed after drying in a desiccator. Sample processing and metal measurements were performed as described in Horie et al. (2017).
Measurements of SA Contents
Freeze-dried samples (2–20 mg) were placed in 2-mL tubes and ground to powder with 3-mm zirconia beads using a tissue lyser (Qiagen). The samples were extracted with 1 mL of 80% (v/v) acetonitrile containing 1% (v/v) acetic acid and 5 ng of D6-SA (Isotec) for 2 h at 4°C in the dark. After centrifugation of the samples at 15,000g for 5 min, supernatants were transferred to fresh tubes and precipitates were extracted again with 1 mL of 80% (v/v) acetonitrile containing 1% (v/v) acetic acid for 2 h at 4°C in the dark. After centrifugation of the samples at 15,000g for 5 min, supernatants were combined with the first extracts and dried under a stream of N2 gas. The dried extracts were dissolved in 1 mL of water containing 1% (v/v) acetic acid and then applied to an Oasis WAX Column (1 cc; Waters), which had been pretreated with 1 mL of acetonitrile, 1 mL of KOH (0.1 N) and 1 mL of water containing 1% (v/v) acetic acid. The columns were washed with 1 mL of water containing 1% (v/v) acetic acid and then with 1 mL of 80% (v/v) acetonitrile containing 1% (v/v) acetic acid. Fractions containing SA were eluted with 2 mL of 80% (v/v) acetonitrile containing 5% (v/v) formic acid and dried under a stream of N2 gas. The dried samples were dissolved in 50 μL of water containing 1% (v/v) acetic acid and analyzed with liquid chromatography tandem-mass spectrometry as described in Kanno et al. (2016).
Immunoblot Analysis
Total protein lysates were prepared as described in Yoshimoto et al. (2004). Western blot using anti-PsbA, -ARF1, -COXII, -CAT, and -SMT1 antibodies was performed as described in Yoshimoto et al. (2014). Anti-RPS14 (AS12 2111), anti-RPL13 (AS13 2650), and anti-Cu/ZnSOD2 (CSD2; AS06 170) were acquired from Agrisera (Vännäs). Anti-GFP antibody (3H9-100) was acquired from Chromotek. The secondary antibodies were horseradish peroxidase–conjugated goat anti–rabbit IgG antibody (cat. no. 111-035-003; Jackson ImmunoResearch) and anti–rat IgG antibody (cat. no. 112-035-003; Jackson ImmunoResearch). Protein extraction, 2,4-dinitrophenylhydrazine treatment, and anti-DNP western blot for detection of oxidized protein were performed using the Oxidized Protein Western Blot Detection Kit (cat. no. ab178020; Abcam). Blots were visualized by chemiluminescence using ECL substrate (cat. no. 1705062, Clarity Max Western ECL Substrate; Bio-Rad) and recorded using a charge-coupled device imager (model no. LAS-4000; Fujifilm).
Fluorescence Microscopy
GFP fusion proteins, autofluorescence of chlorophyll, and fluorescein were visualized using a confocal laser scanning microscope (model no. FV1000; Olympus). Details of GFP-ATG8a and stroma-targeted GFP expressed plants were described in Yoshimoto et al. (2004) and Izumi et al. (2010). To observe RCB accumulation, 10 mm of MES-KOH at pH 5.7 containing 1 μm of ConA and 100 μm of E-64d were infiltrated into leaves of 3 DAT plants with a syringe and incubated for 20 h at 22°C. In Suc-added conditions, excised leaves were incubated in MGRL media containing 3% (w/v) Suc. For HPF treatments, a 5-mm stock solution of HPF (cat. no. H36004; Molecular Probes) was diluted in 10 mm of MES-KOH at pH 5.7 to prepare a 10-μm working solution. The solution was infiltrated into leaves with a syringe and incubated for 1.5 h in a growth chamber. The fluorescence intensities of HPF were quantified using the software ImageJ (National Institutes of Health; Schneider et al., 2012).
Histochemical Staining
For NBT staining, detached leaves from 3 DAT plants were submerged in 10 mm of KPO4 (K2HPO4 and KH2PO4) buffer (pH 7.8) containing 10 mm of NaN3, and then vacuum-infiltrated three times at 0.09 MPa for 20 s. Next, the buffer was removed, 0.1% (w/v) NBT (cat. no. 24720-01; Nacalai Tesque) in 10 mm of KPO4 buffer was added, and the leaves were incubated at room temperature for 30 min. For DAB staining, DAB (cat. no. 046-26861; Fujifilm Wako Pure Chemical) was added to water at 1 mg/mL and heated to 60°C, with addition of HCl to promote dissolution. After dissolution, NaOH was added to adjust the pH of the DAB solution, which was then cooled on ice. Detached leaves were vacuum-infiltrated with DAB solutions. Then, the solution was discarded, and the leaves were rinsed with tap water and incubated at room temperature for 7 h in the dark. After NBT and DAB staining, decolorization of chlorophyll was performed by boiling leaves with solutions containing 7% (v/v) lactate, 13% (v/v) glycerol, 7% (v/v) phenol, and 67% (v/v) ethanol for 2 min. Pictures were taken with a stereomicroscope.
Detection of Mobile Zn Ions in Plants by Zin-Pro Capture
A stock solution of Zin-Pro Capture (cat. no. FDV-0013A; Funakoshi) was prepared at 1 mm by dissolving in dimethyl sulfoxide. The working solution was 1 μm in 3 mm of MES-KOH at pH 7.0. The same amount of dimethyl sulfoxide was used as a control. The solution was vacuum-infiltrated into shoots of plants grown for 2 d under −Zn, which were then incubated at 22°C in the dark for 24 h. The samples were frozen in liquid nitrogen, homogenized in 50 mm of Tris-HCl at pH 7.5, and then centrifuged at 21,900 g for 5 min at 4°C to recover the supernatant. Protein concentration was measured using the BCA Protein Assay Kit (cat. no. 23227; Thermo Fisher Scientific), and equal quantities of proteins were subjected to SDS-PAGE. After electrophoresis, gels were scanned at 488 nm on a fluorescence imager (Fluorimager 595; Molecular Dynamics). Subsequently, CBB-R250 (Thermo Fisher Scientific) staining was performed, and a transmitted light image was acquired to confirm equal protein loading.
Measurements of SOD Activities
Leaves of 5 DAT plants were frozen in liquid nitrogen and homogenized in 50 mm of Tris-HCl (pH 7.5) containing 2% (w/v) polyvinylpolypyrrolidone, and then centrifuged at 14,000g for 10 min at 4°C. The supernatant was used for measurement of SOD activities using the SOD Assay Kit-WST (cat. no. S311; Dojindo). KCN was added to the samples at 10 mm to inhibit Cu/ZnSOD activities. After samples were aliquoted into a 96-well plate and reagents were added, they were incubated at 22°C for 20 min, and then absorbance was measured at 450 nm on a microplate reader (Epoch 2; BioTek). The inhibition rate was calculated from absorbance and used as an index of SOD activities (arbitrary unit). The inhibition rate is a measure of SOD activity because SOD inhibits the color reaction facilitated by ·O2−.
Statistical Analyses
Statistical analysis was performed with the software Microsoft Office Excel 2016. Student’s t test was used to compare samples as indicated in the figure legends.
Accession Numbers
Genetic information in this article can be obtained from The Arabidopsis Information Resource (https://www.arabidopsis.org/) under the following accession numbers: ARF1, AT1G23490; ATG2, AT3G19190; ATG5, AT3G51830; ATG8a, At4g21980; CAT, AT4G35090; COXII, ATMG00160; Cu/ZnSOD2, AT2G28190; PsbA, ATCG00020; RPL13, AT3G49010; RPS14, AT2G36160; SMT1, AT5G13710.
Supplemental Data
The following supplemental information is available.
Supplemental Figure S1. Temporal changes in the number of APs under −Zn.
Supplemental Figure S2. Accumulation of ABs under −Zn.
Supplemental Figure S3. Increase of autophagic degradation activity under −Zn.
Supplemental Figure S4. Autophagy is induced in the −Zn side but not in the +Zn side in the split-root assay.
Supplemental Figure S5. Marked elongation of stromules in atg5 mutants under −Zn.
Supplemental Figure S6. Visualization of mobile Zn ion contents in plants by Zin-Pro Capture.
Supplemental Figure S7. Cu/ZnSOD protein levels in NahG and NahG atg5 leaves.
Supplemental Figure S8. SA levels of Col, atg5, NahG, and NahG atg5 leaves under −/+Zn conditions.
Supplemental Figure S9. Zinc deficiency does not affect RCB accumulation.
Supplemental Figure S10. Zinc deficiency induces chlorosis even under high CO2 conditions.
Acknowledgments
We thank all members of the Yoshimoto Laboratory for providing a great deal of useful advice for experiments, as well as valuable discussion. We also thank the Suzukakedai Materials Analysis Division, Technical Department, Tokyo Institute of Technology, for ICP-MS analysis.
Footnotes
This work was partly supported by the Grant-in-Aid for Research Activity Start-up (grant no. 16H07255 to K.Y.); Grant-in-Aid for Scientific Research on Innovative Areas, Research in a Proposed Research Area (grant no. 19H05713 to K.Y.); the Institute of Science and Technology, Meiji University (Research Project Grant A to K.Y.); the Ministry of Education, Culture, Sports, Science and Technology Program for Strategic Research Foundation at Private Universities (grant no. S1411023); the Institut National de la Recherche Agronomique (INRA Package Program 2012–2015 to E.A.M., L.N. and K.Y.); and the National Research Agency, France (grant no. ANR-12-ADAPT-001-01-AUTOADAPT).
Articles can be viewed without a subscription.
References
- Assunção AG, Herrero E, Lin YF, Huettel B, Talukdar S, Smaczniak C, Immink RG, van Eldik M, Fiers M, Schat H, et al. (2010) Arabidopsis thaliana transcription factors bZIP19 and bZIP23 regulate the adaptation to zinc deficiency. Proc Natl Acad Sci USA 107: 10296–10301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham DC, MacIntosh GC (2017) Degradation of cytosolic ribosomes by autophagy-related pathways. Plant Sci 262: 169–174 [DOI] [PubMed] [Google Scholar]
- Brunkard JO, Runkel AM, Zambryski PC (2015) Chloroplasts extend stromules independently and in response to internal redox signals. Proc Natl Acad Sci USA 112: 10044–10049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakmak I. (2000) Tansley review No. 111: Possible roles of zinc in protecting plant cells from damage by reactive oxygen species. New Phytol 146: 185–205 [DOI] [PubMed] [Google Scholar]
- Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD (2002) The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 277: 33105–33114 [DOI] [PubMed] [Google Scholar]
- Doke N. (1983) Generation of superoxide anion by potato tuber protoplasts during the hypersensitive response to hyphal wall components of Phytophthora infestans and specific inhibition of the reaction by suppressors of hypersensitivity. Physiol Plant Pathol 23: 359–367 [Google Scholar]
- Eguchi M, Kimura K, Makino A, Ishida H (2017) Autophagy is induced under Zn limitation and contributes to Zn-limited stress tolerance in Arabidopsis (Arabidopsis thaliana). Soil Sci Plant Nutr 63: 342–350 [Google Scholar]
- Floyd BE, Morriss SC, MacIntosh GC, Bassham DC (2015) Evidence for autophagy-dependent pathways of rRNA turnover in Arabidopsis. Autophagy 11: 2199–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Hirai MY, Chino M, Komeda Y, Naito S (1992) Effects of sulfur nutrition on expression of the soybean seed storage protein genes in transgenic petunia. Plant Physiol 99: 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotz N, Fox T, Connolly E, Park W, Guerinot ML, Eide D (1998) Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency. Proc Natl Acad Sci USA 95: 7220–7224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiboileau A, Yoshimoto K, Soulay F, Bataillé MP, Avice JC, Masclaux-Daubresse C (2012) Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol 194: 732–740 [DOI] [PubMed] [Google Scholar]
- Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y (2002) Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 129: 1181–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius D, Schultz-Larsen T, Joensen J, Tsitsigiannis DI, Petersen NH, Mattsson O, Jørgensen LB, Jones JD, Mundy J, Petersen M (2009) Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137: 773–783 [DOI] [PubMed] [Google Scholar]
- Horie T, Kawamata T, Matsunami M, Ohsumi Y (2017) Recycling of iron via autophagy is critical for the transition from glycolytic to respiratory growth. J Biol Chem 292: 8533–8543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida H, Yoshimoto K, Izumi M, Reisen D, Yano Y, Makino A, Ohsumi Y, Hanson MR, Mae T (2008) Mobilization of Rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol 148: 142–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M, Wada S, Makino A, Ishida H (2010) The autophagic degradation of chloroplasts via rubisco-containing bodies is specifically linked to leaf carbon status but not nitrogen status in Arabidopsis. Plant Physiol 154: 1196–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T (2000) LC3, a mammalian homologue of yeast Apg8p, is processed and localized in autophagosome membranes after processing. EMBO J 19: 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Oikawa T, Chiba Y, Ishimaru Y, Shimizu T, Sano N, Koshiba T, Kamiya Y, Ueda M, Seo M (2016) AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat Commun 7: 13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Horie T, Matsunami M, Sasaki M, Ohsumi Y (2017) Zinc starvation induces autophagy in yeast. J Biol Chem 292: 8520–8530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y (1999) Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol 147: 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N (2011) Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 334: 941–948 [DOI] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, Peter M (2008) Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol 10: 602–610 [DOI] [PubMed] [Google Scholar]
- Li F, Chung T, Pennington JG, Federico ML, Kaeppler HF, Kaeppler SM, Otegui MS, Vierstra RD (2015) Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell 27: 1389–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liuzzi JP, Guo L, Yoo C, Stewart TS (2014) Zinc and autophagy. Biometals 27: 1087–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liuzzi JP, Yoo C (2013) Role of zinc in the regulation of autophagy during ethanol exposure in human hepatoma cells. Biol Trace Elem Res 156: 350–356 [DOI] [PubMed] [Google Scholar]
- Maret W. (2009) Molecular aspects of human cellular zinc homeostasis: Redox control of zinc potentials and zinc signals. Biometals 22: 149–157 [DOI] [PubMed] [Google Scholar]
- Merkulova EA, Guiboileau A, Naya L, Masclaux-Daubresse C, Yoshimoto K (2014) Assessment and optimization of autophagy monitoring methods in Arabidopsis roots indicate direct fusion of autophagosomes with vacuoles. Plant Cell Physiol 55: 715–726 [DOI] [PubMed] [Google Scholar]
- Miki T, Awa M, Nishikawa Y, Kiyonaka S, Wakabayashi M, Ishihama Y, Hamachi I (2016) A conditional proteomics approach to identify proteins involved in zinc homeostasis. Nat Methods 13: 931–937 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y (2011) The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27: 107–132 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta Bioenerg 975: 384–394 [Google Scholar]
- Pottier M, Dumont J, Masclaux-Daubresse C, Thomine S (2019) Autophagy is essential for optimal translocation of iron to seeds in Arabidopsis. J Exp Bot 70: 859–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Weber AP (2006) Plant peroxisomes respire in the light: some gaps of the photorespiratory C2 cycle have become filled—others remain. Biochim Biophys Acta 1763: 1496–1510 [DOI] [PubMed] [Google Scholar]
- Romero-Puertas MC, Palma JM, Gomez M, Del Rio LA, Sandalio LM (2002) Cadmium causes the oxidative modification of proteins in pea plants. Plant Cell Environ 25: 677–686 [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD (2005) Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol 138: 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen H, Zhang Z, Wei Y, Colloge DB (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11: 1187–1194 [Google Scholar]
- Vallee BL, Auld DS (1990) Zinc coordination, function, and structure of zinc enzymes and other proteins. Biochemistry 29: 5647–5659 [DOI] [PubMed] [Google Scholar]
- Wada S, Hayashida Y, Izumi M, Kurusu T, Hanamata S, Kanno K, Kojima S, Yamaya T, Kuchitsu K, Makino A, et al. (2015) Autophagy supports biomass production and nitrogen use efficiency at the vegetative stage in rice. Plant Physiol 168: 60–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Contento AL, Nguyen PQ, Bassham DC (2007) Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol 143: 291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K. (2012) Beginning to understand autophagy, an intracellular self-degradation system in plants. Plant Cell Physiol 53: 1355–1365 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y (2004) Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K (2009) Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Shibata M, Kondo M, Oikawa K, Sato M, Toyooka K, Shirasu K, Nishimura M, Ohsumi Y (2014) Organ-specific quality control of plant peroxisomes is mediated by autophagy. J Cell Sci 127: 1161–1168 [DOI] [PubMed] [Google Scholar]