Nitrate enhances ABA accumulation through the transcriptional activation of TaBG1 by TaANR1, and this enhancement in turn affects nitrate uptake through the activation of TaNRT2s/NARs by TaWabi5.
Abstract
Nitrate is the preferred form of nitrogen for most plants, acting both as a nutrient and a signaling molecule. However, the components and regulatory factors governing nitrate uptake in bread wheat (Triticum aestivum), one of the world’s most important crop species, have remained unclear, largely due to the complexity of its hexaploid genome. Here, based on recently released whole-genome information for bread wheat, the high-affinity nitrate transporter2 (NRT2) and the nitrate-assimilation-related (NAR) gene family are characterized. We show that abscisic acid (ABA)- Glc ester deconjugation is stimulated in bread wheat roots by nitrate resupply following nitrate withdrawal, leading to enhanced root-tissue ABA accumulation, and that this enhancement, in turn, affects the expression of root-type NRT2/NAR genes. TaANR1 is shown to regulate nitrate-mediated ABA accumulation by directly activating TaBG1, while TaWabi5 is involved in ABA-mediated NO3− induction of NRT2/NAR genes. Building on previous evidence establishing ABA involvement in the developmental response to high-nitrate stress, our study suggests that ABA also contributes to the optimization of nitrate uptake by regulating the expression of NRT2/NAR genes under limited nitrate supply, offering a new target for improvement of nitrate absorption in crops.
Bread wheat (Triticum aestivum) is one of the most important staple crops globally, providing the majority of calories for some 30% of the world’s population. To meet this demand, approximately 100 kg/ha nitrogen (N) fertilizer is applied to wheat fields the world over, and this quantity is expected to increase three-fold by 2050 (West et al., 2014). The overuse of N fertilizers has led to low nitrogen-use efficiency in wheat fields and significant environmental costs (Chen et al., 2014; Coskun et al., 2017). Therefore, improvement of N use efficiency in wheat, via various avenues, such as enhancing the capacity for primary N acquisition from soil, is a critical task for plant scientists.
Nitrate (NO3−) from soil is the main source of N for most plants, including wheat (Andrews et al., 2013). Physiologically, two classes of NO3− transport systems, low-affinity transport systems and high-affinity transport systems, have been identified as the principal components governing NO3− uptake in plants (Kronzucker et al., 1995b; Noguero and Lacombe, 2016). Meanwhile, two gene families, encoding the nitrate transporter1/peptide transporter (NPFs, previously designated as NRT1s or PTRs) and the nitrate transporter2 (NRT2s), have been assigned to the physiological activities of low-affinity transport systems and high-affinity transport systems, respectively (Noguero and Lacombe, 2016; O’Brien et al., 2016). Among these, the NRT2 family has been shown to be critical to NO3− acquisition from soil under N-limitation conditions (Kiba and Krapp, 2016). The first NRT2 gene of land plants was identified in barley (Hordeum vulgare; Trueman et al., 1996), and, subsequently, as an outcome of whole-genome sequencing, seven NRT2 genes were discovered in the Arabidopsis (Arabidopsis thaliana) genome (Orsel et al., 2002), at least five in barley (Vidmar et al., 2000), and four in rice (Oryza sativa; Cai et al., 2008). Moreover, the NO3− transport capabilities of most NRT2 proteins in plants require the presence of a chaperone protein, NAR2, as part of a two-component high-affinity NO3− uptake system (Tong et al., 2005; Orsel et al., 2006). Previously, due to the lack of wheat genome information, only fragments of NRT2/NAR2 genes were isolated (Zhao et al., 2004; Yin et al., 2007; Taulemesse et al., 2015). However, the recent release of wheat whole-genome sequences (IWGSC, 2014, 2018) has opened a new era for genetic studies in bread wheat (Wang et al., 2015), giving an opportunity to deepen our understanding of the genetic basis that underpins wheat’s capacity for NO3− uptake (Wang et al., 2018a; Li et al., 2020).
Nitrate is not only one of the most significant macronutrients for plants, but also it acts as a signaling molecule (Kronzucker et al., 1995a; Zhang and Forde, 1998; Krouk et al., 2010). Therefore, cross talk between the NO3−-signaling pathway and other key signaling pathways, particularly phytohormone pathways, is an intriguing topic, and one of great importance (Krouk et al., 2011). For abscisic acid (ABA), one of the principal stress-related hormones, previous studies have indicated it is involved in root architecture (Signora et al., 2001) and seed germination (Matakiadis et al., 2009) under high concentrations (10 mm or higher) of NO3− provision. Ondzighi-Assoume et al. (2016) showed that the accumulation of ABA in Arabidopsis root tips was stimulated by additional NO3− treatment, resulting from the release of ABA from ABA-Glc ester (ABA-GE, an inactive form of conjugated ABA; Lee et al., 2006), instead of ABA biosynthesis. Even though ABA appears to be associated with high NO3− stress in Arabidopsis, the involvement of ABA under limited-NO3− conditions is less well established, and this relationship is especially important to clarify in crops. Historically, using a high-ABA hexaploid wheat breeding line, SQ1 (Quarrie, 1981), and the bread wheat model genotype Chinese Spring (CS) to generate a mapping population (Quarrie et al., 2005), multiple quantitative trait loci (QTL) associated with N-use efficiency were identified (Habash et al., 2007; Quraishi et al., 2011), of which some loci in the SQ1 background were activated by ABA and low NO3− (Quarrie et al., 2006, 2007), indicating ABA might contribute to key N-related traits in wheat. Moreover, it has been reported that certain NRT2/NAR2 genes in wheat can be up-regulated by ABA (Cai et al., 2007). Despite these discoveries, direct evidence, particularly with regard to the molecular regulatory components linking NO3− and ABA, has been lacking.
In this study, starting with a scan through the bread wheat genome information, an overview of the NRT2 and NAR gene families is presented, and the relationship between the NO3−- and ABA-signaling pathways is established. Moreover, regulatory factors governing NO3−-mediated ABA accumulation and ABA-mediated NO3− induction of NRT2/NAR genes in roots of bread wheat are identified. Our findings in wheat that the influence of ABA on NRT2/NAR gene expression leads to an alteration of NO3− influx offer a new potential target for improvement of NO3− uptake in crops.
RESULTS
Genome-wide Identification and Evolutionary Analysis of Wheat NRT2 and NAR Genes
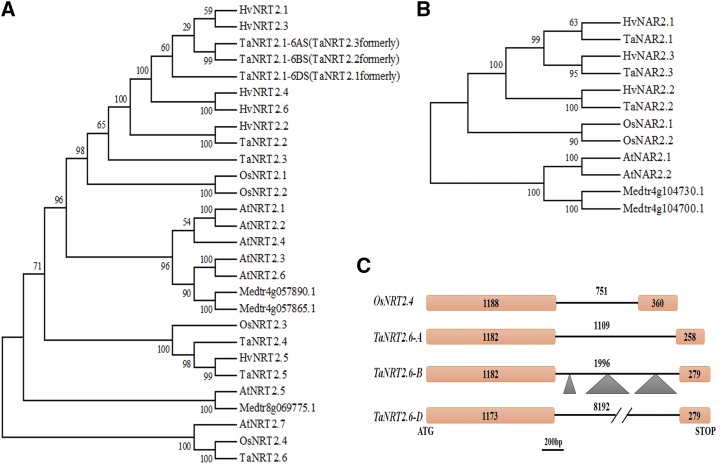
Using members of the NRT2 family in Arabidopsis, rice, and barley as templates (Supplemental Table S1), the genomic sequences, putative promoters, and coding sequences of six subgroups of NRT2 genes distributed on chromosomes 1S (short arm), 2S, 3L (long arm), 6S, 6S, and 7L were identified from the wheat genome databases (Supplemental Table S2). Theoretically, each subgroup of wheat NRT2 genes should contain three homoeoalleles in the A, B, and D subgenome, respectively, as bread wheat is a hexaploid species (Wang et al., 2015). However, no homeolog on chromosome 1AS to 1BS and 1DS was detectable in ref-sequence v1.1 of the wheat genome. A further search and cloning were carried out in the ancestral genomes of Triticum urartu (2n = 2x = 14, genome AA, the donor of A subgenome), Triticum monococcum (2n = 2x = 14, genome AmAm), and Triticum turgidum ssp. dicoccoides (2n = 4x = 28, genome AABB), and no counterpart was obtained, indicating that it was absent from the progenitor of the A subgenome of bread wheat. Moreover, TaNRT2.1, TaNRT2.2, and TaNRT2.3 (as designated formerly) were isolated in previous studies (Zhao et al., 2004; Yin et al., 2007), and, intriguingly, these emerged as homeologous genes located on 6BS, 6DS, and 6AS, respectively, through analysis of sequence alignment and subchromosomal location.
Since the coding sequences of homoeoalleles had high similarity, the D copy of each NRT2 subgroup was chosen, together with the NRT2 genes in Arabidopsis, Medicago truncatula (Pellizzaro et al., 2015), rice, and barley, to conduct the phylogenetic analyses based on deduced amino acid sequences (Fig. 1A). According to the phylogenetic relationship, wheat NRT2 genes were designated following the suggestion by Plett et al. (2010). TaNRT2.6 shared the highest identity with OsNRT2.4 (Fig. 1A), both of which uniquely contained one intron in the genomic sequences (Fig. 1C; Cai et al., 2008). However, compared with the 1109-bp-length intron of TaNRT2.6-7AL, the intron of TaNRT2.6-7BL comprised of 1996 bp was attributed to three additional insertions, while the length of TaNRT2.6-7DL intron was 8192 bp (Fig. 1C).
Figure 1.
Phylogenetic analysis and structure of wheat NRT2 and NAR genes. A, Phylogenetic analysis of wheat NRT2 proteins and NRT2 proteins from Arabidopsis, M. truncatula, O. sativa, and H. vulgare (gene IDs in Supplemental Tables S1 and S2). B, Phylogenetic analysis of wheat NAR proteins and NAR proteins from Arabidopsis, M. truncatula, O. sativa, and H. vulgare (gene IDs in Supplemental Tables S1 and S2). Since the sequences of homoeoalleles shared a high similarity, the D copy of each NRT2 and NAR subgroup was used (except for TaNRT2.1). Bootstrap support was indicated as numbers above the branches. The Arabidopsis sequences were used to root the tree. Os, O. sativa; Ta, wheat; Hv, H. vulgare; Medtr, M. truncatula; At, Arabidopsis. C, Genomic structures of TaNRT2.6s and OsNRT2.4. Orange box represents exon, black line represents intron, and triangle represents insertion. The number is the sequence length.
Similarly, three subgroups of NAR genes were identified genome-wide in bread wheat ‘CS’. Two subgroups were both located on chromosome 6L, while the third subgroup was on chromosomes 4AL, 5BL, and 5DL, due to translocation events between 4AL and 5AL during the evolution of wheat (Ma et al., 2013; IWGSC, 2014). All NAR genes contained one intron in the genomic sequences (Supplemental Table S2). The D copy of each NAR subgroup was aligned to generate the phylogenetic tree and designated accordingly (Fig. 1B).
Spatial and Nitrate-Responsive Expression Patterns of Wheat NRT2 and NAR Genes
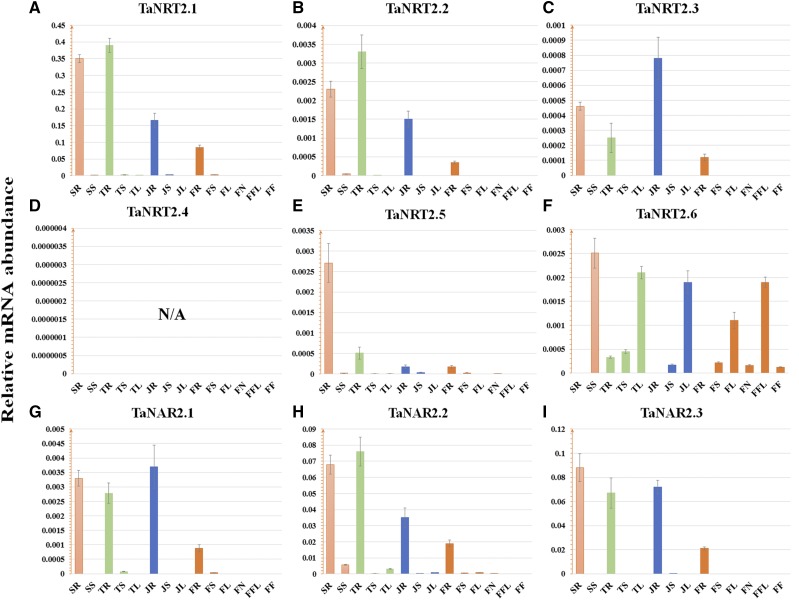
According to the classic Zadoks scale (Zadoks et al., 1974), transcript libraries of different tissues at different developmental stages were obtained. As shown in Figure 2, TaNRT2.1, TaNRT2.2, TaNRT2.3, TaNRT2.5, TaNAR2.1, and TaNAR2.3 were mainly expressed in roots, at four stages. The transcription level of TaNRT2.6 in leaves was substantially higher than that in roots and stems. Meanwhile, even though it exhibited higher expression in roots, TaNAR2.2 was also expressed moderately in leaves, indicating it was involved in the two-component uptake system both in roots and leaves. As for TaNRT2.4, overall expression was extremely low and barely detectable by reverse transcription quantitative PCR (RT-qPCR). TaNRT2.1 showed the highest expression level (at least 10-fold higher) compared to other TaNRT2 genes in roots (Fig. 2; Supplemental Fig. S1), indicating its predominant role among wheat NRT2 genes and explaining why this subgroup was first cloned without the availability of a genome sequence (Zhao et al., 2004; Yin et al., 2007).
Figure 2.
Transcription of wheat NRT2 and NAR genes in different organs at different developmental stages. A to F, Transcription of wheat NRT2 genes. G to I, Transcription of wheat NAR genes. According to the classic Zadoks scale (Zadoks et al., 1974), transcript libraries of different tissues were obtained at Z11 (seedling stage, column in orange), Z21 (tillering stage, column in green), Z32 (jointing stage, column in blue), and Z59 (flowering period, column in dark orange), respectively. SR and SS, Respectively, root and shoot at the seedling stage; TR, TS, and TL, respectively, root, shoot, and leaf at the tillering stage; JR, JS, and JL, respectively, root, stem, and leaf at the stem elongation stage; FR, FS, FL, FN, FFL, and FF, respectively, the root, stem, leaf, node, flag leaf, and inflorescence at the flowering period; N/A, no data available. TaEF1-α (M90077; Paolacci et al., 2009) was chosen as the endogenous control. Each bar represents the mean ± sd of at least three biological replicates.
Then, the expression patterns of these genes were examined when wheat seedlings were hydroponically challenged by 5-d nitrate deprivation, followed by 1-d nitrate resupply (2 mm was chosen as “full nitrate recovery” and 0.2 mm as “low nitrate provision”). Similar to the positive control TaNPF6.2 (Supplemental Fig. S2), previously shown to be nitrate-inducible in roots (Buchner and Hawkesford, 2014), expression of TaNRT2.1, TaNRT2.2, TaNRT2.3, TaNAR2.1, TaNAR2.2, and TaNAR2.3 was triggered in roots by nitrate resupply (Fig. 3A; Supplemental Fig. S2). Transcriptional inductions of these genes by 0.2 mm nitrate provision were less pronounced than those under full-nitrate treatment (Fig. 3A; Supplemental Fig. S2). By contrast, for TaNRT2.5 in roots, an up-regulation was observed under nitrate starvation, followed by a decline under nitrate resupply (Supplemental Fig. S2), indicating TaNRT2.5 is a nitrate-starvation-inducible gene. In leaves, TaNRT2.6 and TaNAR2.2 showed an obvious enhancement in transcription at 6 h and then a decline at 12 h under nitrate resupply (Supplemental Fig. S2).
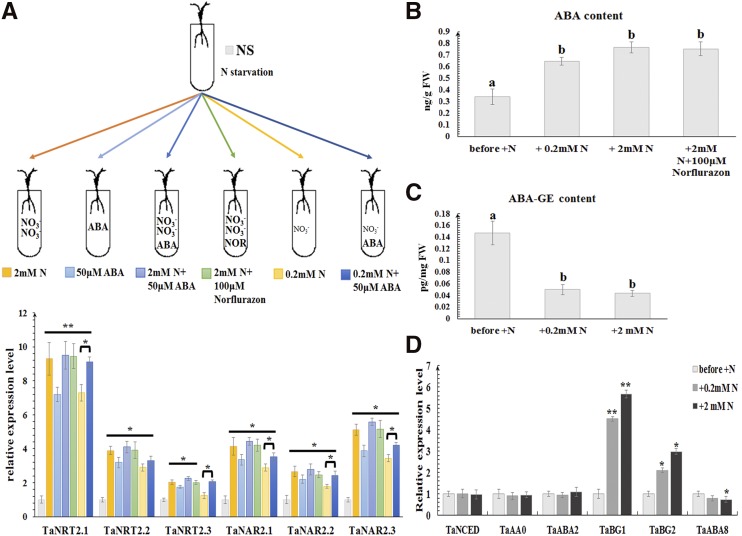
Figure 3.
The effect of ABA on root-type NRT2/NAR expression and of nitrate on ABA accumulation. A, Expression of wheat root-type NRT2/NAR genes under the treatment (at 1 h) of 2 mm nitrate, 50 μm ABA, 2 mm nitrate plus 50 μm ABA, 2 mm nitrate plus 100 μm norflurazon, 0.2 mm nitrate, and 0.2 mm nitrate plus 50 μm ABA, respectively, following nitrate withdrawal. Relative transcript abundance of every gene under nitrate starvation at the same time was calculated by giving the value 1. TaEF1-α (M90077; Paolacci et al., 2009) was chosen as the endogenous control. B, The exogenous ABA levels and (C) ABA-GE levels in roots of bread wheat under different nitrate treatments. D, The transcriptional levels of genes involved in ABA biosynthesis (TaNCED, TaAAO, and TaABA2) and catabolism (TaBG1, TaBG2, and TaABA8) in roots of bread wheat under different nitrate treatments. Each bar represents the mean ± sd of at least three biological replicates. Columns marked with asterisks (A and D) indicate significant differences (*P < 0.05; **P < 0.01) using Student’s t test. Different letters on top of the bars (B and C) indicate significance differences, based on the one-way Waller-Duncan test (P < 0.05). Letters shared in common indicate no significant difference.
The Involvement of ABA Accumulation via ABA-GE Deconjugation in the Nitrate Induction of NRT2 and NAR Genes in Wheat Roots
Since the root is the organ where nitrate is absorbed from soils, our study focused specifically on the root-type NRT2/NAR genes (TaNRT2.1, TaNRT2.2, TaNRT2.3, TaNAR2.1, TaNAR2.2, and TaNAR2.3). To investigate the transcriptional regulatory factors, cis elements were scanned against the promoters of these genes. Intriguingly, the ABA-responsive element (ABRE; ACGTG; Yamaguchi-Shinozaki and Shinozaki, 2005) appeared frequently (Supplemental Fig. S3; Supplemental Table S2), suggesting an ABA signal might be involved in the regulation of these genes. When exposed to 50 μm ABA instead of nitrate provision following nitrate withdrawal, all root-type NRT2/NAR genes were induced, although expression levels were not equivalent to those seen with nitrate induction (Fig. 3A), indicating that ABA can partially mimic the nitrate signal. Although the exposure to 50 μm ABA plus 2 mm nitrate caused a similar induction to that produced solely by 2 mm nitrate treatment, when treated with 50 μm ABA plus 0.2 mm nitrate, transcript abundances of the NRT2/NAR genes were significantly above those under 0.2 mm nitrate-only treatment (Fig. 3A), suggesting that ABA can amplify the nitrate signal under nitrate-limited conditions.
ABA content measurements showed endogenous ABA levels in wheat roots to be increased by nitrate provision, and the enhancement at 2 mm nitrate was stronger than at 0.2 mm (Fig. 3B). However, when 2 mm nitrate was provided together with 100 μm of the ABA synthesis inhibitor norflurazon, after nitrate withdrawal, similar ABA content (Fig. 3B) and expression of root-type NRT2/NAR genes (Fig. 3A) was found, indicating the accumulation of ABA stimulated by nitrate was not associated with de novo ABA synthesis. Then, the expression of a set of genes involved in ABA biosynthesis and metabolism (Lee et al., 2006; Seiler et al., 2011; Xu et al., 2012b; Ma et al., 2016) was examined. As shown in Figure 3D, only the transcription levels of TaBG1 and TaBG2, responsible for deconjugation of ABA-GE, were affected by nitrate. During nitrate starvation, the content of ABA-GE showed no significant change (Supplemental Fig. S4). However, ABA-GE content in wheat roots declined when nitrate was resupplied (Fig. 3C), suggesting that the deconjugation of ABA-GE is the main source of ABA accumulation stimulated by nitrate in wheat.
A previous study demonstrated that the addition of 10 mm nitrate to a control medium (0.5-strength Murashige and Skoog medium) already containing 20 mm nitrate, instead of the nitrate-free medium used in this study, can also strongly stimulate ABA accumulation via ABA-GE deconjugation in Arabidopsis root tips (Ondzighi-Assoume et al., 2016), encouraging us to examine whether our discovery in wheat also applies to Arabidopsis. Utilizing a specific root-nitrate-supply setup developed by our team (Supplemental Fig. S5A; Li et al., 2017), a similar result was found in Arabidopsis, in that AtBG1, but not AtABA1, AtABA2, AtCYP707A1, and AtCYP707A2, could be up-regulated in roots under nitrate resupply (Supplemental Fig. S5B). Furthermore, in the atbg1-1 and atbg1-2 mutants, the induction of AtNRT1.1, AtNRT2.1, AtNRT2.2, and AtNAR2.1 by nitrate resupply was suppressed, and the degree of expression change was significantly lower compared to that in the wild type Col-0 (Supplemental Fig. S5C). These results not only confirm that the deconjugation of ABA-GE triggered by nitrate signaling is the main source of ABA accumulation in roots both in wheat and Arabidopsis, but also indicate that ABA can, in turn, be involved in the activation of nitrate-uptake genes.
The Regulation of TaANR1 in Nitrate-Inducible ABA-GE Deconjugation and ABA Accumulation
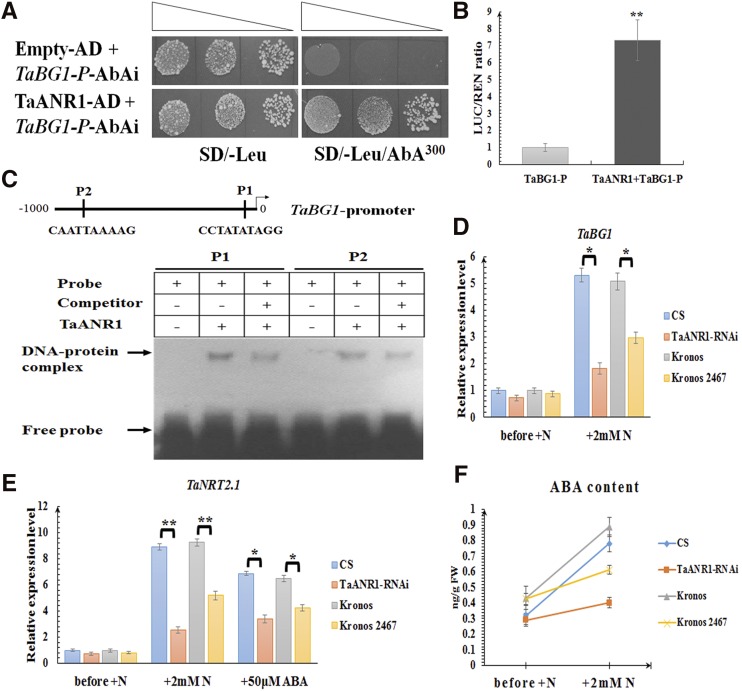
To identify the regulatory factors involved in nitrate-inducible ABA accumulation, a complementary DNA (cDNA) library of wheat roots that had been exposed to nitrate resupply was generated and subjected to yeast one-hybrid (Y1H) screening. Using the ∼500-bp upstream region of TaBG1 as a bait, 12 positive clones were successfully obtained (Supplemental Fig. S6A), among which the sequence of two clones were both identical to TaANR1 (Fig. 4A). Arabidopsis nitrate regulated1 (ANR1), encoding a MADS-box transcription factor, plays an important role in the response to external nitrate supply in Arabidopsis (Zhang and Forde, 1998). Although its exact role in wheat is not clear, a recent study revealed TaANR1 can interact with TaNUE1, a protein encoded by the candidate gene of a major QTL for nitrogen-use efficiency (Lei et al., 2018). An expression profile assembled from RNA-sequencing data (Ramírez-González et al., 2018) and confirmed by RT-qPCR concluded that TaANR1 was mainly expressed in roots through the wheat life cycle (Supplemental Fig. S6C). Moreover, it was strongly up-regulated by nitrate resupply in wheat roots, suggesting it might be involved in the nitrate signaling pathway (Supplemental Fig. S6B). Plant MADS-box proteins usually bind to CArG elements (C(A/T)8G or CC(A/T)6GG; de Folter and Angenent, 2006), and two CArG elements were identified in the promoter of TaBG1 (Fig. 4C). Using the electrophoretic mobility shift assay (EMSA), it was confirmed TaANR1 could bind to probes containing these two CArG elements in vitro (Fig. 4C). Moreover, the results from a dual luciferase assay, involving the addition of TaANR1 to enhance the luciferase signal driven by the promoter of TaBG1 (Fig. 4B), showed that TaANR1 could activate the promoter activity of TaBG1 in planta.
Figure 4.
The involvement of TaANR1 in nitrate-inducible ABA accumulation. A, Y1H assay showing that TaANR1 can bind to the promoter of TaBG1. SD/−Leu, SD medium without Leu; SD/−Leu/AbA300, SD medium without Leu supplemented with AbA at the concentration of 300 ng mL−1; P, promoter. Transformed yeast cells were dotted at 10−1 dilutions on the selective medium. B, Transient expression assay showing that TaANR1 can activate the expression of TaBG1. The gray column represents the relative LUC/REN ratio of the reporter in the presence of the empty effector (pBI221 empty vector) without TaANR1; and the black column represents the relative LUC/REN ratio of the reporter in the presence of the effector containing TaANR1. C, EMSA showing the relative binding affinity of TaANR1 to different CArG motifs harbored by the promoter of TaBG1. + and − indicate the presence and absence of the indicated probe or protein, respectively. P1, Probe 1; P2, probe 2. D, The expression levels of TaBG1 and (E) TaNRT2.1 in wild-type wheat lines, and the RNA interference (RNAi) and mutant (Kronos 2467) lines of TaANR1 under different conditions. The relative transcript abundance of every gene prior to adding nitrate was calculated and given the value 1. Columns marked with asterisks indicate significant differences (*P < 0.05; **P < 0.01) using Student’s t test. F, ABA levels in wild-type wheat lines, and the RNAi and mutant (Kronos 2467) lines of TaANR1 under different conditions. N, Nitrate. Each bar represents the mean ± sd of at least three biological replicates.
An analysis of the public wheat TILLING-mutant library (Krasileva et al., 2017) revealed accession Kronos 2467, a mutant harboring a splice-donor variant in TaANR1 that could lead to incorrect splicing resulting in a nonfunctional protein, was obtained (Supplemental Fig. S7B). Moreover, TaANR1-RNAi lines were generated in the background of cv CS (Supplemental Fig. S7A). Compared to the wild type, induction of TaBG1 (Fig. 4D) and TaNRT2.1 (Fig. 4E) by nitrate was greatly suppressed in Kronos 2467 and in the TaANR1-RNAi line, and the stimulation of ABA accumulation by nitrate was also attenuated (Fig. 4F). These outcomes demonstrated TaANR1 is a key component in the nitrate signaling transduction to stimulate ABA-GE deconjugation and ABA accumulation. Although treatment with 50 μm ABA could not fully restore the expression level of TaNRT2.1 to the level in the wild type, its expression was significantly up-regulated in Kronos 2467 and in the TaANR1-RNAi line (Fig. 4E), confirming that ABA acts downstream of the TaANR1-mediated nitrate signal pathway.
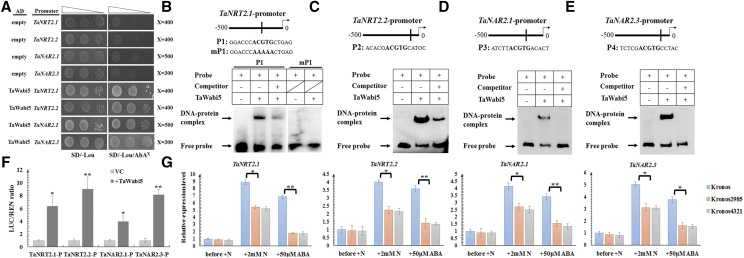
TaWabi5 Is Essential for ABA-Mediated Nitrate Induction of Root-Type NRT2/NAR Genes
A group of bZIP transcription factors, ABRE-binding factors (ABFs), have been shown to recognize and bind to ABREs and to function fundamentally in ABA-dependent gene expression (Yamaguchi-Shinozaki and Shinozaki, 2005). Since ABREs frequently appeared in the promoters of root-type NRT2/NAR genes in wheat, it prompted us to examine whether ABFs play a role in ABA-mediated nitrate induction of these genes. A set of ABF genes (including TaABF1-4, TaABI5, and TaWabi5) were identified from the wheat genome, and, in previous studies, two wheat ABF genes (TaABI5 and TaWabi5) were recognized as putative orthologs of the ABI5 gene (Kobayashi et al., 2008; Zhou et al., 2017). However, when subjected to nitrate resupply, only TaWabi5 showed an induction in expression (Supplemental Fig. S8A). TaWabi5 has been reported to be up-regulated by exogenous ABA treatment (Kobayashi et al., 2008), and, when conducting RT-qPCR on the samples also used in Figure 3A, the expression of TaWabi5 was found to be triggered by all treatments following nitrate withdrawal (Supplemental Fig. S8B). Moreover, based on spatial expression analysis, TaWabi5 showed a higher transcription level in roots (Supplemental Fig. S8C), while TaABI5 was mainly expressed in grains (Supplemental Fig. S8C), which agrees with a function in wheat seed dormancy revealed previously (Zhou et al., 2017). Therefore, TaWabi5 was chosen for further study.
The Y1H assay showed TaWabi5 could interact with the promoter of root-type NRT2/NAR genes (Fig. 5A). EMSA showed TaWabi5 could bind in vitro to probes containing ABRE in the promoter of these genes (Fig. 5, B–E); by contrast, when ABRE was mutated, TaWabi5 could not bind to the probe (Fig. 5B), concluding the binding site of TaWabi5 to the promoter of root-type NRT2/NAR genes was the same, as predicted. Furthermore, using the dual luciferase assay, it was verified that TaWabi5 could activate the transcriptions of root-type NRT2/NAR genes in planta (Fig. 5F). Then, two independent knockout lines (Kronos 2985 and 4321) of TaWabi5 were obtained and genotyped (Supplemental Fig. S7C). In these two mutants, the induction of root-type NRT2/NAR genes by nitrate was suppressed significantly, and, when treated with 50 μm ABA, the up-regulation of these genes seen in the wild type was even more strongly suppressed (Fig. 5G). These data show TaWabi5 is involved in ABA-mediated nitrate induction of root-type NRT2/NAR genes.
Figure 5.
The involvement of TaWabi5 in ABA-mediated nitrate induction of root-type NRT2/NAR genes. A, Y1H assay showing that TaWabi5 can bind to the promoter of root-type NRT2/NAR genes. SD/−Leu, SD medium without Leu; SD/−Leu/AbA300, SD medium without Leu supplemented with ABA at the concentration of 300 ng mL−1. Transformed yeast cells were dotted at 10−1 dilutions on the selective medium. B to E, EMSA showing the relative binding affinity of TaWabi5 to the ABRE motif harbored by the promoter of TaNRT2.1, TaNRT2.2, TaNAR2.1, and TaNAR2.3. + and − indicate the presence and absence of the indicated probe or protein, respectively. P1, Probe 1; mP1, the probe mutating ABRE motif. F, Transient expression assay showing that TaWabi5 can activate the expression of root-type NRT2/NAR genes. P, Promoter. G, The expression levels of TaNRT2.1, TaNRT2.2, TaNAR2.1, and TaNAR2.3 in wild-type and mutant wheat lines (Kronos 2985 and 4321) of TaWabi5 under different conditions. Relative transcript abundance before adding nitrate was calculated and given the value 1. N, Nitrate. Each bar represents the mean ± sd of at least three biological replicates. Columns marked with asterisks indicate significant differences (*P < 0.05; **P < 0.01) using Student’s t test.
Effect of ABA Accumulation on Nitrate Influx in Roots
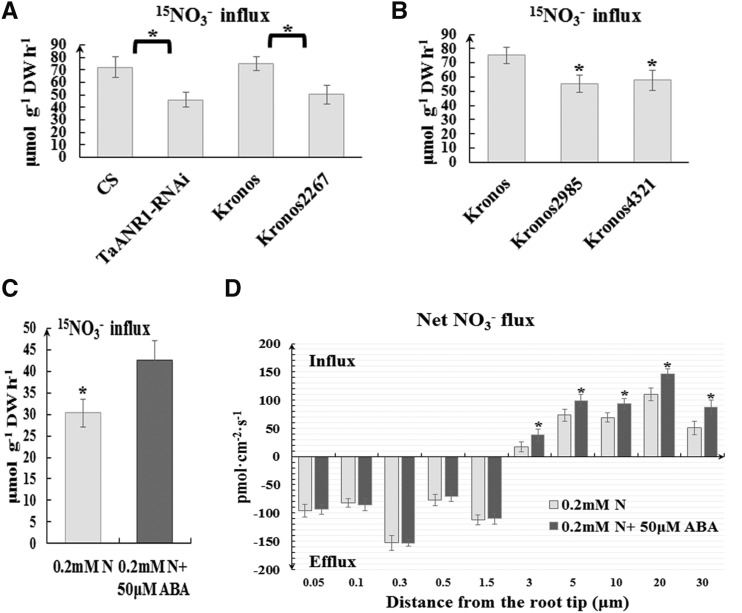
Transcription of the NRT2/NAR genes has been reported to correlate with NO3− uptake and with uptake efficiency in plants (Richard-Molard et al., 2008). Using an 15N-abundance assay, nitrate influx rates in Kronos 2467 and TaANR1-RNAi line were shown to be lower than in the wild type (Fig. 6A; Supplemental Fig. S9A). Similarly, in the knockout lines of TaWabi5, nitrate absorption was inferior (Fig. 6B; Supplemental Fig. S9B). These data indicate that the regulation of root-type NRT2/NAR genes by ABA accumulation can affect nitrate uptake in wheat.
Figure 6.
ABA can affect nitrate uptake in bread wheat. A, 15N-influx measurement in TaANR1-RNAi, Kronos 2467, and wild-type wheat lines. B, 15N-influx measurement in TaWabi5 mutant (Kronos 2985 and 4321) and wild-type wheat lines. After 5-d nitrate starvation, wheat seedlings were hydroponically treated with 2 mm 15NO3− for 1 h, and then whole seedlings were collected for 15N-influx measurements. C, 15N-influx measurement in wheat seedlings at 0.2 mm 15NO3− plus 50 μm ABA and at 0.2 mm 15NO3−. After 5-d nitrate starvation, wheat seedlings were hydroponically treated with 0.2 mm 15NO3− plus 50 μm ABA or 0.2 mm 15NO3− for 1 h, and then whole seedlings were collected for 15N-influx measurement. D, Temporal root net nitrate flux of wheat seedlings under 0.2 mm 15NO3− plus 50 μm ABA treatment and 0.2 mm 15NO3− treatment. After 1 h of treatment, seedlings were subjected to analysis of temporal root net nitrate flux using the high-resolution scanning ion-selective electrode technique. Each bar represents the mean ± sd of at least three biological replicates. Columns marked with asterisk indicate significant differences (*P < 0.05) using Student’s t test.
Treatment with 0.2 mm nitrate plus 50 μm ABA was capable of further enhancing the expression of root-type NRT2s and NARs relative to a simple 0.2 mm nitrate supply, encouraging us to investigate whether the exogenous application of ABA can enhance nitrate influx. A significant amount of nitrate was absorbed under 0.2 mm nitrate plus 50 μm ABA, as determined by the 15N-abundance assay (Fig. 6C; Supplemental Fig. S9C). Additionally, results from the noninvasive measurement of net NO3− fluxes via SIET (a high-resolution scanning ion-selective electrode technique) showed that the additional application of ABA mainly affected net nitrate influx, particularly so in the maturation zone of the root tip, but not nitrate efflux (Fig. 6D; Supplemental Fig. S9D).
DISCUSSION
Commonality and Specificity of Wheat NRT2 and NAR Genes Compared to Other Plant Species
In this study, six subgroups of NRT2 genes and three subgroups of NAR genes were identified by genome-wide scanning in bread wheat (Fig. 1, A and B). As expected of a hexaploid species, each subgroup consisted of three homoeoalleles distributed on the A, B, and D subchromosomes (Supplemental Table S2), except for TaNRT2.4, the 1AS counterpart of which was absent in bread wheat, as well as in the progenitor of its A genome, T. urartu. This is not surprising, given that about 10% of the genes in the bread wheat genome have been found to have lost their A subgenome homeologs (IWGSC, 2018). Intriguingly, the transcript level of TaNRT2.4 was the lowest among the NRT2 genes in bread wheat (Fig. 2; Supplemental Fig. S1). Moreover, the global transcript profile of wheat NRT2 and NAR genes in terms of tissue specificity (Fig. 2) and in response to treatments of nitrate withdrawal and subsequent nitrate resupply (Fig. 3A; Supplemental Fig. S2) showed that TaNRT2.1, TaNRT2.2, TaNRT2.3, TaNAR2.1, and TaNAR2.3 were principally responsive to nitrate in roots, TaNRT2.6 was leaf-specific, and TaNAR2.2 was significantly induced by nitrate in roots and moderately in shoots.
In the Arabidopsis genome, AtNRT2.1 (At1g08090) and AtNRT2.2 (At1g08100) are located in tandem on chromosome 1 (Zhuo et al., 1999). Moreover, OsNRT2.1 and OsNRT2.2 in rice (Cai et al., 2008), LjNRT2.1 and LjNRT2.2 in Lotus japonicus (Criscuolo et al., 2012), and MtNRT2.1 and MtNRT2.2 in M. truncatula (Pellizzaro et al., 2015) have been found to occur in the same tandem configuration, and this is also the case in maize (Zea mays) and Sorghum bicolor (Plett et al., 2010). Similarly, analysis of the genome data of bread wheat showed that TaNRT2.1 and TaNRT2.2 were closely distributed on the short arm of chromosome 6, although interval sequences were longer than those in the Arabidopsis and rice genomes, possibly due to the larger proportion of repetitive DNA in the wheat genome (IWGSC, 2014; Wang et al., 2015).
Phylogenetic analysis revealed TaNRT2.6 belonged to the same clade as AtNRT2.7 and OsNRT2.4 (Fig. 1A). AtNRT2.7 is uniquely expressed in leaves of Arabidopsis (Orsel et al., 2002), while OsNRT2.4 shows a divergent spatial expression pattern compared to other NRT2 genes in rice (Feng et al., 2011). In bread wheat, TaNRT2.6 was the only member of the NRT2 family with elevated expression in leaves, indicating genes in this clade play a distinct role against the background of other NRT2 genes. AtNRT2.7 and OsNRT2.4 are also expressed in reproductive organs (Chopin et al., 2007; Feng et al., 2011), and the transport functions of their translated proteins are independent of the presence of NAR proteins (Chopin et al., 2007; Wu et al., 2013). Whether TaNRT2.6 is similar in this regard remains unknown. Moreover, both TaNRT2.6 and OsNRT2.4 contained one intron in the genomic sequences (Fig. 1C), while no intron was present in any of the other NRT2 genes in bread wheat and rice (Cai et al., 2008). Intriguingly, in hexaploid wheat, the intron lengths of the three homoeoalleles of TaNRT2.6 were different: TaNRT2.6-7AL harbored an intron of 1109 bp, and TaNRT2.6-7BL one of 1996 bp, attributed to three additional insertions, while the length of TaNRT2.6-7DL intron was 8192 bp (Fig. 1C). The importance of intron variation among homoeoalleles in terms of both expression pattern and function has been reported in hexaploid wheat (Shitsukawa et al., 2007), and the case of TaNRT2.6s requires further investigation.
TaANR1 and TaWabi5 Are Essential Components Shaping the Two-Way Connection between the NO3− and ABA Signaling Pathways in Wheat Roots
A strong two-way connection has been found between nitrate and phytohormones. For example, the biosynthesis and transport of auxin and the biosynthesis of cytokinin are affected by NO3−, and the accumulation and distribution of auxin and cytokinin have both been shown to exert feedback on NO3− transport and related critical responses, such as the induction of NRT2 expression (Kiba et al., 2011; Krouk, 2016). This is not surprising, given nitrate’s dual nature as a nutrient and signaling molecule in plants (Kiba et al., 2010; Krouk et al., 2011; Krouk, 2016). The role of ABA has as well been examined, albeit predominantly in the context of the response to high nitrate (Signora et al., 2001; Matakiadis et al., 2009), whereas its interaction with nitrate in the context of nitrate withdrawal has been less well investigated. Differences in ABA content in Arabidopsis seedlings on nitrate-deficient medium compared to normal medium have been recorded, while the expression of AtNCED3, which encodes a key enzyme in ABA biosynthesis, was similar under the two conditions (Lu et al., 2015). Another study found nitrate could stimulate ABA accumulation in root tips of Arabidopsis, caused by ABA-GE deconjugation instead of ABA biosynthesis (Ondzighi-Assoume et al., 2016). Building on these discoveries in Arabidopsis, it was essential to further investigate the effect of nitrate on endogenous ABA content in crops and, moreover, to reveal more information on the consequence of this effect for nitrate uptake and on the nature of such a link.
Our study shows that nitrate provision following nitrate starvation can lead to a pronounced increase in the ABA content in wheat roots (Fig. 3B), mainly as a consequence of TaBG1-mediated release of ABA from the inactive ABA-GE form (Fig. 3, C and D). Furthermore, based on Y1H screening, TaANR1, an important component in the nitrate signaling pathway, was isolated and confirmed to be a direct regulatory factor of TaBG1 transcription under nitrate (Fig. 4). Recently, another MADS-box transcription factor in Arabidopsis, AtSVP, was found to bind to the promoter of AtBG1 and activate its expression in leaves under drought stress (Wang et al., 2018b), indicating that the regulation of BG1 by MADS-box transcription factors may be common in plants. Our results also point out that ABA accumulation via ABA-GE deconjugation could, in turn, contribute to the nitrate induction of root-type NRT2/NAR genes in wheat (Fig. 4E; Supplemental Fig. S5C). Considering the presence of the ABRE in the promoter regions of these genes (Supplemental Fig. S3), TaWabi5, an ABRE-binding factor that can be induced by ABA (Kobayashi et al., 2008) and nitrate (Supplemental Fig. S8), was identified and confirmed to activate the expression of root-type NRT2/NAR genes (Fig. 5). These lines of evidence not only describe a two-way connection between ABA and the NO3−-signaling pathway, but also shed light on the molecular mechanism underlying this cross talk (Fig. 7).
Figure 7.
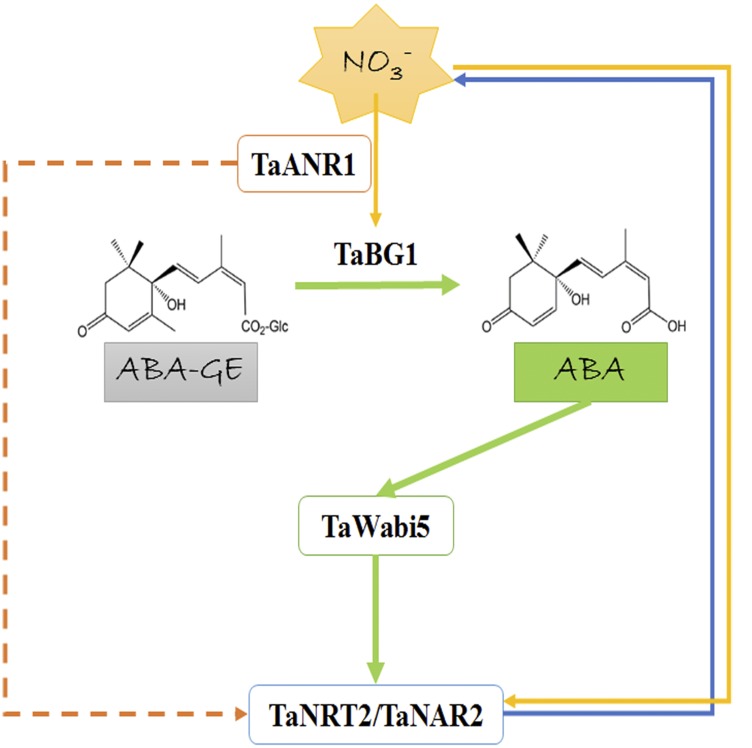
A proposed model for the two-way relationship between nitrate and ABA. When nitrate is resupplied after starvation, the transcription of TaANR1 is triggered. The products of TaANR1 are able to bind to the CArG motifs in the promoter of TaBG1 and activate its expression. TaBG1 can deconjugate ABA-GE, resulting in ABA accumulation. Then, the elevated ABA level causes an increase in TaWabi5 expression. TaWabi5 is capable of binding to the ABRE motifs in the promoter of root-type NRT2/NAR genes and activating their expression, thus leading to an acceleration in nitrate uptake.
Intriguingly, the nexus among ABA, ABA-GE, and nitrate under nitrate starvation appeared to be different from that under nitrate resupply. ABA content decreased during nitrate starvation, which was also the trend seen in nitrate content (Supplemental Fig. S4A). Similarly, a lower ABA content in Arabidopsis seedlings on nitrate-deficient medium compared to normal medium was also discovered (Lu et al., 2015). Considering the lower expression of root-specific NRT2 and NAR genes under nitrate starvation (Supplemental Fig. S2), these findings further support the mediator role of ABA in nitrate induction of these transporter genes. However, the content of ABA-GE showed no significant change during nitrate starvation (Supplemental Fig. S4B), indicating the decline in ABA content under this condition was not directly linked to ABA-GE content. Questions such as how the content of ABA is decreased during nitrate starvation warrant future investigation.
Apart from the transcriptional cross talk uncovered in this study, the two-way connection between the ABA and NO3−-signaling pathways has also been previously shown to occur at the posttranslational level (Krouk, 2017). For example, Léran et al. (2015) found AtABI2 played a positive role in nitrate uptake by mediating the phosphorylation of AtNRT1.1/AtNPF6.3. In that study, the authors suggested that the accumulation of ABA could potentially attenuate nitrate uptake by inhibition of AtABI2 activity, which would be especially beneficial for the survival of plants under stressful conditions, such as drought or salinity stress, since nitrate uptake and utilization are energy-intensive processes. In our study, ABA could enhance nitrate acquisition when plants were released from a status of nitrate starvation, which is also clearly beneficial for the survival. Taken together, these discoveries indicate ABA appears to fine-tune nitrate uptake through different mechanisms, depending on the growth conditions plants are subjected to.
Given the increasing number of NRT1/NPF (Buchner and Hawkesford, 2014) and NRT2 (in this study) genes recently identified in bread wheat and their functional verification (Li et al., 2020), more elements of the cross talk between ABA and nitrate acquisition and singling are likely to be uncovered in this important crop. In Arabidopsis, at least four nitrate transporters, including AtNPF4.6/NRT1.2/AIT1, were found to also function as ABA transporters (Kanno et al., 2012). The response of nitrate-mediated enhancement of seed germination was different between the atnpf4.6 mutant and the wild type. In M. truncatula, another nonorthologous NPF, MtNPF6.8/MtNRT1.3, also displayed this bifunctionality as a transporter of both nitrate and ABA (Pellizzaro et al., 2014). Intriguingly, it has been suggested that ABA is involved in nitrate inhibition of primary root growth in M. truncatula (Liang et al., 2007; Pellizzaro et al., 2014). Therefore, future investigations of such cotransporter activity of ABA and nitrate in bread wheat will broaden our understanding of the two-way connection between the ABA- and NO3−-signaling pathways.
ABA Accumulation Is Important in the Regulation of NO3− Uptake
Previous studies using a mapping population from a cross between cv CS and SQ1 (a high-ABA breeding line; Quarrie, 1981; Quarrie et al., 2005) identified multiple QTLs associated with N-use efficiency (Habash et al., 2007; Quraishi et al., 2011), among which some loci in the SQ1 background showed superior traits that could be activated by ABA and by limitations in NO3− (Quarrie et al., 2006, 2007). Our study shows that suppression of ABA accumulation (in the TaANR1 mutant and in RNAi lines) and suppression of the ABA-signaling pathway (in the TaWabi5 mutant lines) can inhibit nitrate uptake in wheat (Fig. 6, A and B). These lines of evidence suggest that the manipulation of ABA content or of ABA-signaling transduction might offer a new avenue toward the improvement of wheat nitrogen acquisition. Moreover, endogenous ABA levels appear to be an index for N-use efficiency, which might be helpful in breeding and selection of new, more N-use-efficient wheat lines. More investigations in wheat populations to verify the correlation between ABA accumulation and N-use efficiency more broadly are warranted.
Our study also shows that the application of exogenous ABA enhances NO3− uptake, particularly under NO3−-deficient conditions, in wheat. Compared to resupply with 0.2 mm following NO3− withdrawal, the provision of 0.2 mm NO3− plus 50 μm ABA could further increase the transcriptional levels of root-type NRT2/NAR genes (Fig. 3A) as well as NO3− influx (Fig. 6, C and D) in bread wheat. Importantly, multiple ABA-producing/utilizing rhizobacteria have been identified that could be applied to capitalize on these mechanisms and alleviate abiotic stress and promote crop growth under challenging field conditions (Dodd et al., 2010; Ilyas and Bano, 2010). As well, several ABA analogs that are cheaper and chemically more stable than ABA itself have been synthetized and have been field tested (Cao et al., 2017). Therefore, ABA-producing rhizobacteria and ABA analogs present promising alternatives to direct ABA applications together with nitrogen fertilizer to enhance nitrate-uptake capacity in wheat and other crops. It will be important to test and evaluate the practicality of such approaches in the field.
MATERIALS AND METHODS
Identification of NRT2/NAR/ABF Genes in the Wheat Genome Databases and Phylogenetic Analysis
The putative genomic and coding sequences of the wheat (Triticum aestivum) NRT2/NAR/ABF genes were scanned genome-wide and identified from the databases of the International Wheat Genome Sequencing Consortium (IWGSC), CerealsDB, NCBI, and Graingenes (IWGSC, 2014, 2018; Wang et al., 2015), based on a blast analysis of the NRT2/NAR/ABF genes in barley (Hordeum vulgare) and rice (Oryza sativa; Supplemental Table S1). The genomic structures of the NRT2/NAR genes were illustrated based on comparisons of genomic and coding sequences and by the Gene Structure Display Server (GSDS; http://gsds.cbi.pku.edu.cn/). The subchromosomal locations were determined using the IWGSC and CerealsDB databases (Supplemental Table S2). The putative ∼2000 bp upstream sequences (promoters) of the wheat NRT2/NAR genes were identified from the scaffold sequences of IWGSC and then submitted to PlanCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) for cis-elements analysis. To validate the sequences in silico, specific primers (Supplemental Table S3) were designed for PCR, and the amplified products were then resequenced.
The deduced NRT2s and NAR2s amino acid sequences were aligned by Clustal W (http://www.clustal.org) and then submitted to MEGA 6 to construct the phylogenetic trees based on the maximum-likelihood method (Hasegawa et al., 1991). The number of bootstrap replications was 1000.
Plant Materials and Growth Conditions
To correspond with the genome sequence in IWGSC, the T. aestivum ‘CS’ was chosen in this study. Additionally, the wheat mutants, Kronos 2467, 2985, and 4321, generated by Krasileva et al. (2017), were procured via the Chinese distribution site, Shandong Agricultural University. The RNAi line of TaANR1 was generated in the cv CS background; the sense and antisense fragments were designed according to Lei et al. (2018) and inserted into a ZmUbiquitin-promoter-containing vector, pTCK303 (Liu et al., 2014). Accession of Columbia (Col-0) was used as wild-type Arabidopsis (Arabidopsis thaliana), and all the mutant lines were in this background. The Arabidopsis AtBG1 (At1g52400) mutant lines (SALK_075731C and SALK_122533) were obtained from the ABRC stocks (Ondzighi-Assoume et al., 2016).
For tissue-specific RNA isolation, wheat seeds were germinated on wet filter paper at 20°C and transplanted into soil-filled pots for growth in controlled-environment rooms under a 12-h photoperiod (light/dark temperature 22/20°C), a relative humidity of 50%, and 300 μmol m−2 s−1 PAR (photosynthetically active radiation; Wang et al., 2014). According to Zadoks et al. (1974), different organs were collected at the seedling stage (Z11), the tillering stage (Z21), the jointing stage (Z32), and the flowering period (Z59), respectively.
For hydroponic experiments with wheat, after 5 d of germination on wet filter paper, seedlings were transferred and grown in modified half-strength Hoagland’s liquid medium containing 2 mm KNO3, 1 mm KH2PO4, 1 mm MgSO4, 1 mm CaCl2, 0.1 mm Fe-EDTA, 1 μm H3BO3, 1 μm ZnSO4, 0.5 μm CuSO4, 0.3 μm Na2MoO4, and 1 μm MnCl2. The medium was buffered at pH 6.0 using diluted 2 m NaOH and was replaced every 2 d. After 10 d, seedlings (apart from the +N control seedlings) were transferred into N-free medium (where KNO3 was replaced by KCl) for 5 d of a nitrogen starvation (NS) treatment. Samples were collected at NS-0d, NS-1d, NS-2d, and NS-5d. Consistent with our experimental objectives, subsequent treatments included (1) 2 mm KNO3 (or 15N-labeled KNO3) nitrogen recovery; (2) 0.2 mm KNO3 (or 15N-labeled KNO3) low-nitrogen provision; (3) 50 μm ABA (Cai et al., 2007); (4) 50 μm ABA plus 2 mm KNO3; (5) 50 μm ABA plus 0.2 mm KNO3 (or 15N-labeled KNO3); and (6) 100 μm ABA synthesis inhibitor norflurazon (Dong et al., 2013) plus 2 mm KNO3. Samples were collected at 0.5 h, 1 h, 6 h, and 12 h during treatment. The condition of the chamber was 14 h/10 h light/dark under a temperature regime of 22/20°C, a relative humidity of 50%, and 300 μmol m−2 s−1 PAR.
For Arabidopsis, seeds were germinated on half-strength Hoagland’s solid medium (nutrients presented above plus 1% [w/v] agar, pH 5.8) for 2 weeks. After a 4-d starvation on N-free medium, seedlings were transferred onto a specific root-nitrate-supplied system (Supplemental Fig. S5A; modified from Li et al. [2017]), in which the upper part (for leaves) was a N-free medium, while the bottom part (for roots) was a standard medium. The condition of the Arabidopsis chamber was 23°C and a photoperiod of 16 h light (100 μmol m−2 s−1)/8 h dark.
RNA Extraction and RT-qPCR Analysis
Total RNA was extracted from the samples collected using the TRIzol reagent (Takara). Total RNA quality and concentration were determined using an Agilent 2100 Bioanalyzer (Agilent Technologies). The first cDNA strand was synthesized using a PrimeScript RT reagent kit with gDNA Eraser (Takara). SYBR Premix Ex Taq II (Takara) was utilized for the RT-qPCR on the LightCycler 480 real-time PCR system (Roche Diagnostics), following the manufacturer’s instructions. For genes in bread wheat, degenerate primers covering A, B, and D copies were utilized. TaEF1-α (M90077; Paolacci et al., 2009) was chosen as the endogenous control. For Arabidopsis, AtActin2 (At3g18780) was the constitutive normalization control for RT-qPCR. All reactions were carried out four times for each of three independent biological samples. All the primers are as given in Supplemental Table S3.
Nitrate Content Analysis and 15N-Influx Measurement
Plants were treated with 0.1 mm CaSO4 for 1 min, were then transferred to complete nutrient medium containing NO3−/15NO3− at the indicated concentrations for 5 min, and finally to 0.1 mm CaSO4 for 1 min. NO3− content was measured as described by Cai et al. (2007), using a Skalar Four Flow Analyzer (San ++ System, Skalar).
For 15N-influx measurements, seedlings were collected, freeze-dried, and weighed. After grinding, the resultant powder was subjected to a Thermo Flash 2000 analyzer hyphenated to a Thermo Fisher Delta-V isotope-ratio mass spectrometer to determine total N and 15N abundance. Nitrate influx was calculated as 15N content per gram dry weight.
Net NO3− Flux Measurement at the Root Surface
A high-resolution scanning ion-selective electrode technique (BIO-003A system; Younger USA Science and Technology; Applicable Electronics; Science Wares) was used for net nitrate flux measurements in bread wheat roots, which was performed by Xuyue Science and Technology. The method was previously described in detail by Xu et al. (2012a) and Zhong et al. (2014).
Quantitative Analysis of Endogenous ABA and ABA-GE Content
For ABA content measurement, liquid chromatography-tandem mass spectrometry (LC-MS/MS) was performed by Zoonbio Biotechnology. The internal ABA standard was obtained from Sigma. High performance liquid chromatography analysis was performed using a ZORBAX SB-C18 (Agilent Technologies) column (2.1 mm × 150 mm; 3.5 mm).
For ABA-GE content measurement, 2H5-ABA-GE was obtained from the Plant Biology Institute of the National Research Council of Canada. Quantification of ABA-GE was performed using stable isotope dilution LC-MS/MS. Plant material (200 mg, fresh weight) was ground into fine powder in liquid nitrogen then extracted with ethyl acetate containing 2H5-ABA-GE (internal standard) at −20°C for 16 h. After centrifugation at 15,000g for 15 min, the supernatant was collected. After adding equivalent hexane, the sample was loaded onto a Sep-Pak Silica (Waters) cartridge, which was preconditioned with hexane:ethyl acetate (1/1, v/v). The cartridge was sequentially washed with hexane:ethyl acetate (1/1, v/v) and hexane:ethyl acetate (2/8, v/v) then eluted with ethyl acetate: methanol (8/2, v/v). The eluate was evaporated and redissolved in 20% (v/v) methanol for LC-MS/MS analysis. LC-MS/MS analysis was performed on a ultra performance liquid chromatography system (Waters) coupled to the 6500 Q-Trap system (AB SCIEX). LC separation used a BEH C18 column (1.7 mm, 2.1 × 100 mm; Waters) with a mobile phase of 0.05% (v/v) acetic acid (A) and 0.05% (v/v) acetic acid in acetonitrile (B). The gradient was set at an initial 2% B and then increased to 40% B within 8 min. ABA-GE undergoes in-source dissociation, and this was detected in MRM mode with transition 263/153.
Y1H Analysis
For Y1H screening, the Matchmaker Gold Yeast One-Hybrid Library Screening System (Clotech) was used. The promoter region (−1 ∼ −500) of TaBG1 was fused into the pHIS2 to generate the bait construct. The cDNA library of wheat roots constructed via SMART cDNA synthesis technology, was transferred into the bait yeast strains containing pHIS2-TaBG1, and was selected on synthetic dropout medium (SD) plates lacking Leu with or without 3-amino-1,2,4-triazole. The prey fragments from the positive colonies were identified by DNA sequencing.
For Y1H assay, the promoter region of TaBG1 or TaNRT2.1, TaNRT2.2, TaNAR2.1, and TaNAR2.3 was fused into the pABAi to generate the bait construct and then transformed into Y1HGold yeast strain (2nd Lab). The coding region of TaANR1 or TaWabi5 was fused into the pGADT7 vector as a prey construct. The prey construct and the empty construct were separately transformed into the bait strain. The transformed yeast cells were grown at 30°C for 4 d on SD plates lacking Leu with or without antibiotic (Wang et al., 2019).
EMSA
The recombinant protein of TaANR1-His was successfully expressed and purified from Escherichia coli Arctic-Express strains containing pCznI-TaANR (Supplemental Fig. S10A) and TaWabi5-His from Arctic-Express strains containing pET28a-TaWabi5 (Supplemental Fig. S10B). The promoter fragments of TaBG1, TaNRT2.1, TaNRT2.2, TaNAR2.1, and TaNAR2.3 shown in Figures 4C and 5, B–E, were biotin-labeled at their 3′ ends as EMSA probes. EMSA was performed following the protocol of chemiluminescent EMSA kit (LightShift, Thermo Scientific). Gel images were captured using the Gel Doc2000 (Bio-Rad).
Dual Luciferase Transient Expression Assay
The promoter region of TaBG1 or TaNRT2.1, TaNRT2.2, TaNAR2.1, and TaNAR2.3 was fused into the pGreenII 0800-Luc to generate the reporter construct. The coding region of TaANR1 or TaWabi5 was fused into the 35S::pBI221 vector as the effector construct. The empty vector was used as a control. The transient expression assay was performed in Arabidopsis mesophyll cell protoplasts, and the protoplasts were isolated following Liu et al. (2014). Each of the reporter constructs, together with the empty vector or either the 35S::TaANR1 effector or the 35S::TaWabi5 effector, was cotransformed into protoplasts and then incubated overnight. The relative LUC activity was determined by the LUC/REN ratio using a Dual-Luciferase Reporter Assay kit (Promega), according to the manufacturer's instructions.
Statistical Analyses
Data in this study were statistically analyzed by the Student’s t test or the one-way Waller-Duncan test.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers shown in Supplemental Tables S1 and S2.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Transcriptional comparison of wheat NRT2 and NAR genes in shoots (red) and roots (black) of young wheat seedlings (Zadoks scale = 11).
Supplemental Figure S2. Influence of nitrate starvation and nitrate resupply on nitrate content and gene expression of wheat NRT2 and NAR genes in roots and shoots of young wheat plants.
Supplemental Figure S3. The distributions of ABRE element in promoters of wheat NRT2 and NAR genes.
Supplemental Figure S4. The content of ABA, ABA-GE, and nitrate in wheat roots upon nitrate starvation.
Supplemental Figure S5. The involvement of ABA-GE deconjugation in nitrate induction of nitrate transporter genes in roots of Arabidopsis.
Supplemental Figure S6. Characterization of TaANR1.
Supplemental Figure S7. Information on RNAi and mutant lines used in this study.
Supplemental Figure S8. Characterization of TaABFs.
Supplemental Figure S9. The additional information showing the effect of ABA on nitrate uptake in bread wheat.
Supplemental Figure S10. The recombinant protein of TaANR1-His and TaWabi5-His expressed and purified from E. coli Arctic-Express.
Supplemental Table S1. Symbol numbers and gene ID of NRT2 and NAR genes in Arabidopsis, H. vulgare, and O. sativa.
Supplemental Table S2. Identification of NRT2 and NAR gene families in wheat genome.
Supplemental Table S3. Primers used in this study.
Footnotes
This work was supported by the Natural Science Fund of Jiangsu Province, China (no. BK20161092), the National Natural Science Foundation of China (nos. 31601306 and U1906202), the Innovation Program of the Chinese Academy of Sciences (no. ISSASIP1602), and the University of Melbourne. M.W. was also supported by Young Elite Scientists Sponsorship Program of the Chinese Ministry of Science and Technology (no. 2017QNRC001).
Articles can be viewed without a subscription.
References
- Andrews M, Raven J, Lea P (2013) Do plants need nitrate? The mechanisms by which nitrogen form affects plants. Ann Appl Biol 163: 174–199 [Google Scholar]
- Buchner P, Hawkesford MJ (2014) Complex phylogeny and gene expression patterns of members of the NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family (NPF) in wheat. J Exp Bot 65: 5697–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C, Wang JY, Zhu YG, Shen QR, Li B, Tong YP, Li ZS (2008) Gene structure and expression of the high-affinity nitrate transport system in rice roots. J Integr Plant Biol 50: 443–451 [DOI] [PubMed] [Google Scholar]
- Cai C, Zhao XQ, Zhu YG, Li B, Tong YP, Li ZS (2007) Regulation of the high-affinity nitrate transport system in wheat roots by exogenous abscisic acid and glutamine. J Integr Plant Biol 49: 1719–1725 [Google Scholar]
- Cao MJ, Zhang YL, Liu X, Huang H, Zhou XE, Wang WL, Zeng A, Zhao CZ, Si T, Du J, et al. (2017) Combining chemical and genetic approaches to increase drought resistance in plants. Nat Commun 8: 1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Cui Z, Fan M, Vitousek P, Zhao M, Ma W, Wang Z, Zhang W, Yan X, Yang J, et al. (2014) Producing more grain with lower environmental costs. Nature 514: 486–489 [DOI] [PubMed] [Google Scholar]
- Chopin F, Orsel M, Dorbe MF, Chardon F, Truong HN, Miller AJ, Krapp A, Daniel-Vedele F (2007) The Arabidopsis ATNRT2.7 nitrate transporter controls nitrate content in seeds. Plant Cell 19: 1590–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun D, Britto DT, Shi W, Kronzucker HJ (2017) Nitrogen transformations in modern agriculture and the role of biological nitrification inhibition. Nat Plants 3: 17074. [DOI] [PubMed] [Google Scholar]
- Criscuolo G, Valkov VT, Parlati A, Alves LM, Chiurazzi M (2012) Molecular characterization of the Lotus japonicus NRT1(PTR) and NRT2 families. Plant Cell Environ 35: 1567–1581 [DOI] [PubMed] [Google Scholar]
- de Folter S, Angenent GC (2006) trans meets cis in MADS science. Trends Plant Sci 11: 224–231 [DOI] [PubMed] [Google Scholar]
- Dodd I, Zinovkina N, Safronova V, Belimov A (2010) Rhizobacterial mediation of plant hormone status. Ann Appl Biol 157: 361–379 [Google Scholar]
- Dong W, Wang M, Xu F, Quan T, Peng K, Xiao L, Xia G (2013) Wheat oxophytodienoate reductase gene TaOPR1 confers salinity tolerance via enhancement of abscisic acid signaling and reactive oxygen species scavenging. Plant Physiol 161: 1217–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Yan M, Fan X, Li B, Shen Q, Miller AJ, Xu G (2011) Spatial expression and regulation of rice high-affinity nitrate transporters by nitrogen and carbon status. J Exp Bot 62: 2319–2332 [DOI] [PubMed] [Google Scholar]
- Habash DZ, Bernard S, Schondelmaier J, Weyen J, Quarrie SA (2007) The genetics of nitrogen use in hexaploid wheat: N utilisation, development and yield. Theor Appl Genet 114: 403–419 [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Saitou N (1991) On the maximum likelihood method in molecular phylogenetics. J Mol Evol 32: 443–445 [DOI] [PubMed] [Google Scholar]
- Ilyas N, Bano A (2010) Azospirillum strains isolated from roots and rhizosphere soil of wheat (Triticum aestivum L.) grown under different soil moisture conditions. Biol Fertil Soils 46: 393–406 [Google Scholar]
- International Wheat Genome Sequencing Consortium (IWGSC) (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345: 1251788. [DOI] [PubMed] [Google Scholar]
- Appels R, Eversole K, Feuillet C, Keller B, Rogers J, Stein N, Pozniak CJ, Choulet F, et al. ; Interational Wheat Genome Sequencing Consortium (IWGSC) (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361: eaar7191. [DOI] [PubMed] [Google Scholar]
- Kanno Y, Hanada A, Chiba Y, Ichikawa T, Nakazawa M, Matsui M, Koshiba T, Kamiya Y, Seo M (2012) Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc Natl Acad Sci USA 109: 9653–9658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T, Krapp A (2016) Plant nitrogen acquisition under low availability: Regulation of uptake and root architecture. Plant Cell Physiol 57: 707–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T, Kudo T, Kojima M, Sakakibara H (2011) Hormonal control of nitrogen acquisition: Roles of auxin, abscisic acid, and cytokinin. J Exp Bot 62: 1399–1409 [DOI] [PubMed] [Google Scholar]
- Kobayashi F, Maeta E, Terashima A, Takumi S (2008) Positive role of a wheat HvABI5 ortholog in abiotic stress response of seedlings. Physiol Plant 134: 74–86 [DOI] [PubMed] [Google Scholar]
- Krasileva KV, Vasquez-Gross HA, Howell T, Bailey P, Paraiso F, Clissold L, Simmonds J, Ramirez-Gonzalez RH, Wang X, Borrill P, et al. (2017) Uncovering hidden variation in polyploid wheat. Proc Natl Acad Sci USA 114: E913–E921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronzucker HJ, Glass ADM, Yaeesh Siddiqi M (1995a) Nitrate induction in spruce: An approach using compartmental analysis. Planta 196: 683–690 [Google Scholar]
- Kronzucker HJ, Siddiqi MY, Glass A (1995b) Kinetics of NO3− influx in spruce. Plant Physiol 109: 319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G. (2016) Hormones and nitrate: A two-way connection. Plant Mol Biol 91: 599–606 [DOI] [PubMed] [Google Scholar]
- Krouk G. (2017) Nitrate signalling: Calcium bridges the nitrate gap. Nat Plants 3: 17095. [DOI] [PubMed] [Google Scholar]
- Krouk G, Crawford NM, Coruzzi GM, Tsay Y-F (2010) Nitrate signaling: Adaptation to fluctuating environments. Curr Opin Plant Biol 13: 266–273 [DOI] [PubMed] [Google Scholar]
- Krouk G, Ruffel S, Gutiérrez RA, Gojon A, Crawford NM, Coruzzi GM, Lacombe B (2011) A framework integrating plant growth with hormones and nutrients. Trends Plant Sci 16: 178–182 [DOI] [PubMed] [Google Scholar]
- Léran S, Edel KH, Pervent M, Hashimoto K, Corratgé-Faillie C, Offenborn JN, Tillard P, Gojon A, Kudla J, Lacombe B (2015) Nitrate sensing and uptake in Arabidopsis are enhanced by ABI2, a phosphatase inactivated by the stress hormone abscisic acid. Sci Signal 8: ra43. [DOI] [PubMed] [Google Scholar]
- Lee KH, Piao HL, Kim H-Y, Choi SM, Jiang F, Hartung W, Hwang I, Kwak JM, Lee I-J, Hwang I (2006) Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126: 1109–1120 [DOI] [PubMed] [Google Scholar]
- Lei L, Li G, Zhang H, Powers C, Fang T, Chen Y, Wang S, Zhu X, Carver BF, Yan L (2018) Nitrogen use efficiency is regulated by interacting proteins relevant to development in wheat. Plant Biotechnol J 16: 1214–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Zhang L, Shi W (2017) Determination of the effects of local and systemic iron excess on lateral root initiation in Arabidopsis thaliana. Bio Protoc 7: e2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, He X, Chen Y, Jing Y, Shen C, Yang J, Teng W, Zhao X, Hu W, Hu M, et al. (2020) A wheat transcription factor positively sets seed vigour by regulating the grain nitrate signal. New Phytol 255: 1667–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Mitchell DM, Harris JM (2007) Abscisic acid rescues the root meristem defects of the Medicago truncatula latd mutant. Dev Biol 304: 297–307 [DOI] [PubMed] [Google Scholar]
- Liu S, Liu S, Wang M, Wei T, Meng C, Wang M, Xia G (2014) A wheat SIMILAR TO RCD-ONE gene enhances seedling growth and abiotic stress resistance by modulating redox homeostasis and maintaining genomic integrity. Plant Cell 26: 164–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Sasaki Y, Li X, Mori IC, Matsuura T, Hirayama T, Sato T, Yamaguchi J (2015) ABI1 regulates carbon/nitrogen-nutrient signal transduction independent of ABA biosynthesis and canonical ABA signalling pathways in Arabidopsis. J Exp Bot 66: 2763–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Ding H, Wang C, Qin H, Han Q, Hou J, Lu H, Xie Y, Guo T (2016) Alleviation of drought stress by hydrogen sulfide is partially related to the abscisic acid signaling pathway in wheat. PLoS One 11: e0163082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Stiller J, Berkman PJ, Wei Y, Rogers J, Feuillet C, Dolezel J, Mayer KF, Eversole K, Zheng YL, et al. (2013) Sequence-based analysis of translocations and inversions in bread wheat (Triticum aestivum L.). PLoS One 8: e79329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matakiadis T, Alboresi A, Jikumaru Y, Tatematsu K, Pichon O, Renou J-P, Kamiya Y, Nambara E, Truong H-N (2009) The Arabidopsis abscisic acid catabolic gene CYP707A2 plays a key role in nitrate control of seed dormancy. Plant Physiol 149: 949–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguero M, Lacombe B (2016) Transporters involved in root nitrate uptake and sensing by Arabidopsis. Front Plant Sci 7: 1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien JA, Vega A, Bouguyon E, Krouk G, Gojon A, Coruzzi G, Gutiérrez RA (2016) Nitrate transport, sensing, and responses in plants. Mol Plant 9: 837–856 [DOI] [PubMed] [Google Scholar]
- Ondzighi-Assoume CA, Chakraborty S, Harris JM (2016) Environmental nitrate stimulates abscisic acid accumulation in Arabidopsis root tips by releasing it from inactive stores. Plant Cell 28: 729–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsel M, Chopin F, Leleu O, Smith SJ, Krapp A, Daniel-Vedele F, Miller AJ (2006) Characterization of a two-component high-affinity nitrate uptake system in Arabidopsis. Physiology and protein-protein interaction. Plant Physiol 142: 1304–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsel M, Krapp A, Daniel-Vedele F (2002) Analysis of the NRT2 nitrate transporter family in Arabidopsis. Structure and gene expression. Plant Physiol 129: 886–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolacci AR, Tanzarella OA, Porceddu E, Ciaffi M (2009) Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol Biol 10: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzaro A, Clochard T, Cukier C, Bourdin C, Juchaux M, Montrichard F, Thany S, Raymond V, Planchet E, Limami AM, et al. (2014) The nitrate transporter MtNPF6.8 (MtNRT1.3) transports abscisic acid and mediates nitrate regulation of primary root growth in Medicago truncatula. Plant Physiol 166: 2152–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzaro A, Clochard T, Planchet E, Limami AM, Morère-Le Paven MC (2015) Identification and molecular characterization of Medicago truncatula NRT2 and NAR2 families. Physiol Plant 154: 256–269 [DOI] [PubMed] [Google Scholar]
- Plett D, Toubia J, Garnett T, Tester M, Kaiser BN, Baumann U (2010) Dichotomy in the NRT gene families of dicots and grass species. PLoS One 5: e15289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarrie S. (1981) Genetic variability and heritability of drought-induced abscisic acid accumulation in spring wheat. Plant Cell Environ 4: 147–151 [Google Scholar]
- Quarrie S, Kaminska A, Barnes J, Dodig D, Gennaro A (2007) A QTL for grain yield on 7AL of wheat is activated by ABA and low nutrient treatments during flag leaf ontogeny. Comp Biochem Physiol A Mol Integr Physiol 146: S253 [Google Scholar]
- Quarrie S, Pekic Quarrie S, Radosevic R, Rancic D, Kaminska A, Barnes JD, Leverington M, Ceoloni C, Dodig D (2006) Dissecting a wheat QTL for yield present in a range of environments: From the QTL to candidate genes. J Exp Bot 57: 2627–2637 [DOI] [PubMed] [Google Scholar]
- Quarrie SA, Steed A, Calestani C, Semikhodskii A, Lebreton C, Chinoy C, Steele N, Pljevljakusić D, Waterman E, Weyen J, et al. (2005) A high-density genetic map of hexaploid wheat (Triticum aestivum L.) from the cross Chinese Spring x SQ1 and its use to compare QTLs for grain yield across a range of environments. Theor Appl Genet 110: 865–880 [DOI] [PubMed] [Google Scholar]
- Quraishi UM, Abrouk M, Murat F, Pont C, Foucrier S, Desmaizieres G, Confolent C, Rivière N, Charmet G, Paux E, et al. (2011) Cross-genome map based dissection of a nitrogen use efficiency ortho-metaQTL in bread wheat unravels concerted cereal genome evolution. Plant J 65: 745–756 [DOI] [PubMed] [Google Scholar]
- Ramírez-González R, Borrill P, Lang D, Harrington S, Brinton J, Venturini L, Davey M, Jacobs J, Van Ex F, Pasha A, et al. (2018) The transcriptional landscape of polyploid wheat. Science 361: eaar6089. [DOI] [PubMed] [Google Scholar]
- Richard-Molard C, Krapp A, Brun F, Ney B, Daniel-Vedele F, Chaillou S (2008) Plant response to nitrate starvation is determined by N storage capacity matched by nitrate uptake capacity in two Arabidopsis genotypes. J Exp Bot 59: 779–791 [DOI] [PubMed] [Google Scholar]
- Seiler C, Harshavardhan VT, Rajesh K, Reddy PS, Strickert M, Rolletschek H, Scholz U, Wobus U, Sreenivasulu N (2011) ABA biosynthesis and degradation contributing to ABA homeostasis during barley seed development under control and terminal drought-stress conditions. J Exp Bot 62: 2615–2632 [DOI] [PubMed] [Google Scholar]
- Shitsukawa N, Tahira C, Kassai K, Hirabayashi C, Shimizu T, Takumi S, Mochida K, Kawaura K, Ogihara Y, Murai K (2007) Genetic and epigenetic alteration among three homoeologous genes of a class E MADS box gene in hexaploid wheat. Plant Cell 19: 1723–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signora L, De Smet I, Foyer CH, Zhang H (2001) ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J 28: 655–662 [DOI] [PubMed] [Google Scholar]
- Taulemesse F, Le Gouis J, Gouache D, Gibon Y, Allard V (2015) Post-flowering nitrate uptake in wheat is controlled by N status at flowering, with a putative major role of root nitrate transporter NRT2.1. PLoS One 10: e0120291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Zhou JJ, Li Z, Miller AJ (2005) A two-component high-affinity nitrate uptake system in barley. Plant J 41: 442–450 [DOI] [PubMed] [Google Scholar]
- Trueman LJ, Richardson A, Forde BG (1996) Molecular cloning of higher plant homologues of the high-affinity nitrate transporters of Chlamydomonas reinhardtii and Aspergillus nidulans. Gene 175: 223–231 [DOI] [PubMed] [Google Scholar]
- Vidmar JJ, Zhuo D, Siddiqi MY, Glass AD (2000) Isolation and characterization of HvNRT2.3 and HvNRT2.4, cDNAs encoding high-affinity nitrate transporters from roots of barley. Plant Physiol 122: 783–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Qin L, Xie C, Li W, Yuan J, Kong L, Yu W, Xia G, Liu S (2014) Induced and constitutive DNA methylation in a salinity-tolerant wheat introgression line. Plant Cell Physiol 55: 1354–1365 [DOI] [PubMed] [Google Scholar]
- Wang M, Wang S, Liang Z, Shi W, Gao C, Xia G (2018a) From genetic stock to genome editing: Gene exploitation in wheat. Trends Biotechnol 36: 160–172 [DOI] [PubMed] [Google Scholar]
- Wang M, Wang S, Xia G (2015) From genome to gene: A new epoch for wheat research? Trends Plant Sci 20: 380–387 [DOI] [PubMed] [Google Scholar]
- Wang M, Yuan J, Qin L, Shi W, Xia G, Liu S (2019) TaCYP81D5, one member in a wheat cytochrome P450 gene cluster, confers salinity tolerance via reactive oxygen species scavenging. Plant Biotechnol J 18: 791–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wang F, Hong Y, Yao J, Ren Z, Shi H, Zhu J-K (2018b) The flowering repressor SVP confers drought resistance in Arabidopsis by regulating abscisic acid catabolism. Mol Plant 11: 1184–1197 [Google Scholar]
- West PC, Gerber JS, Engstrom PM, Mueller ND, Brauman KA, Carlson KM, Cassidy ES, Johnston M, MacDonald GK, Ray DK, et al. (2014) Leverage points for improving global food security and the environment. Science 345: 325–328 [DOI] [PubMed] [Google Scholar]
- Wu P, Xu G, Lian X (2013) Nitrogen and phosphorus uptake and utilization In Zhang Q, and Wing RA, eds, Genetics and Genomics of Rice. Springer, New York, pp 217–226 [Google Scholar]
- Xu W, Shi W, Jia L, Liang J, Zhang J (2012a) TFT6 and TFT7, two different members of tomato 14-3-3 gene family, play distinct roles in plant adaption to low phosphorus stress. Plant Cell Environ 35: 1393–1406 [DOI] [PubMed] [Google Scholar]
- Xu Z-Y, Lee KH, Dong T, Jeong JC, Jin JB, Kanno Y, Kim DH, Kim SY, Seo M, Bressan RA, et al. (2012b) A vacuolar β-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis. Plant Cell 24: 2184–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (2005) Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci 10: 88–94 [DOI] [PubMed] [Google Scholar]
- Yin LP, Li P, Wen B, Taylor D, Berry JO (2007) Characterization and expression of a high-affinity nitrate system transporter gene (TaNRT2.1) from wheat roots, and its evolutionary relationship to other NTR2 genes. Plant Sci 172: 621–631 [Google Scholar]
- Zadoks JC, Chang TT, Konzak CF (1974) A decimal code for the growth stages of cereals. Weed Res 14: 415–421 [Google Scholar]
- Zhang H, Forde BG (1998) An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409 [DOI] [PubMed] [Google Scholar]
- Zhao XQ, Li YJ, Liu JZ, Li B, Liu QY, Tong YP, Li JY, Li ZS (2004) Isolation and expression analysis of a high-affinity nitrate transporter TaNRT2. 3 from roots of wheat. Acta Bot Sinica 46: 347–354 [Google Scholar]
- Zhong Y, Yan W, Chen J, Shangguan Z (2014) Net ammonium and nitrate fluxes in wheat roots under different environmental conditions as assessed by scanning ion-selective electrode technique. Sci Rep 4: 7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Yang J, Wang ZX, Wang JR (2017) Sequence analysis and expression profiles of TaABI5, a pre-harvest sprouting resistance gene in wheat. Genes Genomics 39: 161–171 [Google Scholar]
- Zhuo D, Okamoto M, Vidmar JJ, Glass AD (1999) Regulation of a putative high-affinity nitrate transporter (Nrt2;1At) in roots of Arabidopsis thaliana. Plant J 17: 563–568 [DOI] [PubMed] [Google Scholar]