Several previously unknown low-abundant lipid droplet proteins are identified in Arabidopsis thaliana seeds and seedlings by quantitative proteomics combined with two cell biological approaches.
Abstract
The developmental program of seed formation, germination, and early seedling growth requires not only tight regulation of cell division and metabolism, but also concerted control of the structure and function of organelles, which relies on specific changes in their protein composition. Of particular interest is the switch from heterotrophic to photoautotrophic seedling growth, for which cytoplasmic lipid droplets (LDs) play a critical role as depots for energy-rich storage lipids. Here, we present the results of a bottom-up proteomics study analyzing the total protein fractions and LD-enriched fractions in eight different developmental phases during silique (seed) development, seed germination, and seedling establishment in Arabidopsis (Arabidopsis thaliana). The quantitative analysis of the LD proteome using LD-enrichment factors led to the identification of six previously unidentified and comparably low-abundance LD proteins, each of which was confirmed by intracellular localization studies with fluorescent protein fusions. In addition to these advances in LD protein discovery and the potential insights provided to as yet unexplored aspects in plant LD functions, our data set allowed for a comparative analysis of the LD protein composition throughout the various developmental phases examined. Among the most notable of the alterations in the LD proteome were those during seedling establishment, indicating a switch in the physiological function(s) of LDs after greening of the cotyledons. This work highlights LDs as dynamic organelles with functions beyond lipid storage.
While the sporophyte of angiosperms is photoautotrophic during most of its life cycle, it is largely heterotrophic during its initial formation, including embryo development and early seedling establishment. Toward that end, the growing embryo is protected during seed formation by the mother plant, which provides it with nutrients for embryo growth and the accumulation of storage compounds needed later for seed germination and seedling establishment. In Arabidopsis (Arabidopsis thaliana) seeds, a combination of storage proteins and lipids accumulates in the embryo, with a minor proportion also being deposited in the endosperm (Penfield et al., 2005). This leads to seeds that contain up to 40% of their dry weight in lipids (Baud et al., 2002), most of which are in the form of neutral lipids, such as triacylglycerols (TAGs) and sterol esters, and that are compartmentalized and stored in cytoplasmic lipid droplets (LDs). LDs in seeds typically range in size from 0.5 to 2 μm and comprise up to ∼60% of the volume of a mature embryonic cell, making them, along with storage vacuoles, the most abundant organelles (Mansfield and Briarty, 1992; Tzen et al., 1993; Kretzschmar et al., 2018). The proper mobilization of these storage lipids within LDs during germination and seedling establishment is essential for providing the growing plant with carbon and energy. However, while the major function of LDs in seeds is related to energy storage, which is relatively well understood in terms of the metabolic pathways involved, the roles of LDs after seedling establishment, and later in development in vegetative tissues (i.e. leaves, roots, and stems), are far less understood.
A variety of proteins are known to be associated with the surface of LDs in plant cells and to carry out specific structural and/or enzymatic functions (Chapman et al., 2019). Among these so-called LD proteins, three major families, oleosins, steroleosins, and caleosins, have been relatively well characterized in terms of their association with LDs and participation in LD biology. Oleosins, for instance, were the first family of plant LD proteins discovered (Qu et al., 1986; Vance and Huang, 1987) and are predominantly expressed in the seeds and pollen, where they are the most abundant proteins on LDs (Huang, 2017). The functions of oleosins include roles in the formation of nascent LDs at the endoplasmic reticulum (ER), as well as preventing the fusion and coalescence of mature LDs (Siloto et al., 2006; Shimada et al., 2008). Steroleosins (also referred to as HYDROXYSTEROID DEHYDROGENASES [HSDs]) are homologs of metazoan sterol dehydrogenases (Lin et al., 2002), which mediate the homeostasis of steroid-derived hormones (Chapman et al., 2012). While the substrates and products of steroleosins remain to be determined, they are thought to play a role in brassinosteroid metabolism (Li et al., 2007; Baud et al., 2009). Caleosins are calcium-binding and heme-containing peroxygenases that, like oleosins, accumulate primarily in seeds (Naested et al., 2000; Hanano et al., 2006), although their function is not known. In leaves, however, caleosins appear to work in close coordination with another LD protein, α-dioxygenase (α-DOX), to produce 2-hydroxy-octadecatrienoic acid that can act as a phytoalexin, indicating that LDs serve as a production site for antimicrobial compounds within the plant cell (Shimada et al., 2014). Other LD proteins involved in the generation of oxylipins are lipoxygenases in cucumber (Cucumis sativus; Rudolph et al., 2011) and a lipase from tomato (Solanum lycopersicum; Garbowicz et al., 2018). Homologs of the latter in castor bean (Ricinus communis) and Arabidopsis localize to LDs and catalyze the hydrolysis of lipids, including TAG (Eastmond, 2004; Müller and Ischebeck, 2018).
With ongoing advances in proteomic techniques, it is becoming increasingly easier to detect proteins that are of relatively low abundance. This has facilitated the discovery of LD proteins in recent years, and in doing so has presented an increasingly complex picture of the functional roles of LDs in plants (Pyc et al., 2017b). For instance, the identification of the LD-ASSOCIATED PROTEIN (LDAP) family has provided insights into the function of LDs in vegetative tissues. Originally identified in the LD proteome of the mesocarp of avocado (Persea americana; Horn et al., 2013), LDAPs were subsequently shown to be ubiquitously expressed in Arabidopsis and to influence LD abundance in leaves (Gidda et al., 2016), as well as conferring resistance to drought stress (Kim et al., 2016). Additionally, an interacting protein of the LDAPs, referred to as LDIP (LDAP-INTERACTING PROTEIN) has since been identified (Pyc et al., 2017a). While the physiological role of LDIP remains to be determined, its ubiquitous expression, LD localization, and unique LD phenotype in the leaves of mutant plants (i.e. enlarged LDs) suggest that it is important in LD biogenesis and/or turnover. Another recently discovered plant LD protein is a member of the UBX domain-containing (PUX) protein family, PUX10, which was shown to localize to LDs in the tobacco (Nicotiana tabacum) pollen tube and Arabidopsis seeds, and appears to play a role in the polyubiquitination pathway for degradation of other LD proteins (Deruyffelaere et al., 2018; Kretzschmar et al., 2018). Evidence for this latter conclusion include a delay in the turnover of LD proteins, including the oleosins, during seed germination in the pux10 mutant, which is consistent with the oleosins being a substrate for the ubiquitin-proteasomal pathway (Hsiao and Tzen, 2011; Deruyffelaere et al., 2015).
Despite the above-mentioned examples, only a few other plant LD proteins have been identified, which is conspicuous considering the relatively large number of LD proteins that have been found in other evolutionarily diverse organisms, such as yeast and animals, and the correspondingly wide range of cellular functions ascribed to these proteins (Du et al., 2013; Bersuker et al., 2018; Zhang and Liu, 2019). Moreover, given the general conservation that appears to exist in LD functioning and biogenesis/turnover among different organisms (Chapman et al., 2019), including plant versus nonplant species, there are likely many as yet unidentified plant LD proteins that carry out comparable, as well as plant-specific, functions.
Proteomic approaches have been used with a variety of plant species as a means to inventory proteins involved in specific processes during growth and development, in certain organs and tissues, and/or in response to an environmental cue (Eldakak et al., 2013). Also, seeds of several plant species have been the focus of previous proteomics-based studies (Hajduch et al., 2005; Li et al., 2016; Wang et al., 2016). In studies on Arabidopsis seeds, for example, often two or more conditions are compared to each other, such as the influence of different hormones (Chibani et al., 2006; Yin et al., 2015; Li et al., 2016) or stress (Lee et al., 2015; Fercha et al., 2016; Xu et al., 2017). Similarly, proteomics approaches have been used to study Arabidopsis seed dormancy, ripening, and aging (Gallardo et al., 2001; Chibani et al., 2006; Kubala et al., 2015; Nguyen et al., 2015; Yin et al., 2015), seed development (Hajduch et al., 2010; Lorenz et al., 2018), germination (Quan et al., 2013; Durand et al., 2019), and seedling establishment. However, none of these latter studies provided a comparative analysis of proteomes from these different developmental stages, nor a proteome of any specific organelle(s), such as LDs.
Here, we present the results of a study using a state-of-the-art proteomics platform to survey tissues from two stages during Arabidopsis silique development and six stages during seed germination and early seedling establishment. Additionally, we analyzed the proteome of an LD-enriched fraction for each developmental stage and discovered six proteins that localize to LDs, as confirmed by fluorescence microscopy. Our proteomics survey also allowed us to follow the dynamics of LD protein composition through silique development, germination, and seedling establishment, which will undoubtedly serve to help inform further research aimed at understanding the molecular mechanisms underlying LD (protein) biology in plants and will provide a better understanding of these developmental stages in Arabidopsis in general.
RESULTS
A Proteomic Analysis of Arabidopsis Tissues from Silique Development to Seedling Establishment
In order to gain more insight into the proteins mediating the processes underlying silique development, seed germination, and seedling establishment in Arabidopsis, we investigated the proteome at each of these stages using label-free tandem mass spectrometry (MS/MS). More specifically, proteins were isolated from two phases of silique growth, corresponding roughly to seed maturation (referred to as Phase I siliques, 7–14 d after fertilization [DAF]) and desiccation (i.e. Phase II siliques, 14–21 DAF), as previously defined (Fig. 1A; Baud et al., 2002). Additionally, proteomes during seed germination and seedling establishment were monitored using rehydrated mature seeds, stratified seeds, and seedlings at 24, 36, 48, and 60 h after stratification (Fig. 1A). In the latter experiments, seeds had completed germination, as defined by radicle emergence after 24 h in long-daylight conditions at 22°C. By 48 h, seedling cotyledons had emerged and opened, and by 60 h, they were dark green.
Figure 1.
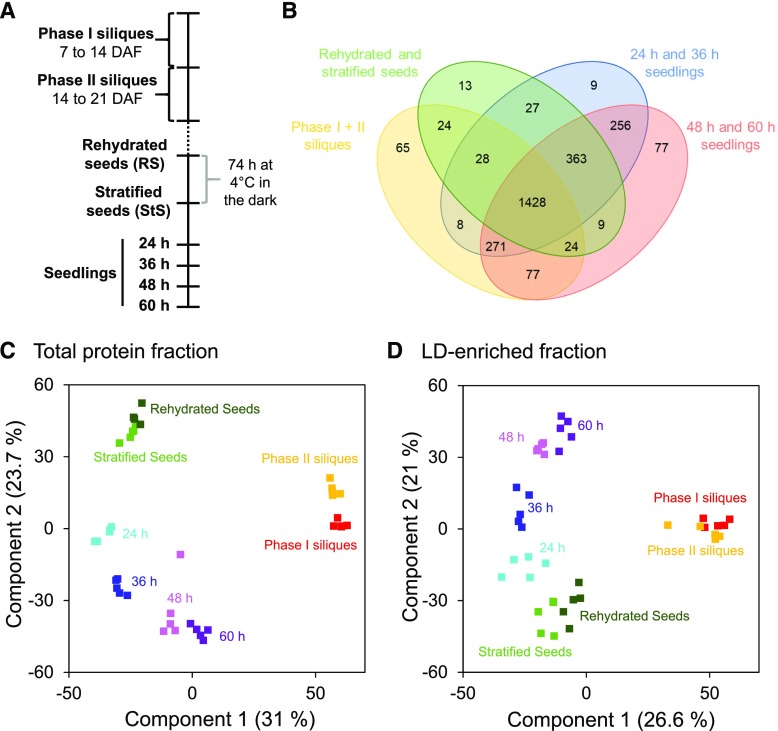
Graphical representation of proteomic data derived from Arabidopsis siliques, seeds, and seedlings. A, Tissues were collected from two developmental stages of silique development (phase I, 7–14 DAF; phase II, 14–21 DAF) and six stages of seed germination and seedling establishment (rehydrated seeds, stratified seeds, and seedlings from 24 to 60 h of growth). B, Venn diagram of the distribution of all detected proteins from each of the different developmental stages examined. Proteins identified with at least two peptides using the iBAQ algorithm were grouped into four groups, as depicted. Common proteins were identified via InteractiVenn (Heberle et al., 2015). C and D, PCA was performed to compare the distribution of the five biological replicates at each developmental stage for both the total protein and LD-enriched fractions. Numbers in brackets give the percentage of the total variance represented by Components 1 and 2, respectively.
For each developmental stage examined, total cellular extracts and LD-enriched fractions were generated in five biological replicates. As described in more detail in the “Materials and Methods” section, peptides derived from both fractions were analyzed by liquid chromatography (LC)-MS/MS, and the MS raw data files were processed using MaxQuant software to identify proteins (Supplemental Datasets S1–S4). Protein intensities were determined by both the intensity-based absolute quantification (iBAQ) and the label-free quantification (LFQ) algorithms (Cox and Mann, 2008; Cox et al., 2014). iBAQ and LFQ values were then calculated as per mille of all intensities in each sample to obtain relative iBAQ (riBAQ) and relative LFQ (rLFQ) values. When calculating enrichment factors for protein identification (i.e. the relative protein abundance in the LD-enriched fraction divided by the relative abundance in the total fraction), riBAQ values were used. For quantitative comparison of the total proteome of different stages (i.e. protein dynamics), rLFQ values were used. In total, we detected 2,696 protein groups identified by at least two peptides, based on the iBAQ data processing algorithm (Table 1). The number of protein groups was lowest in older (Phase II) siliques and rehydrated seeds and highest during the late stages of seedling establishment.
Table 1. Summary of proteins identified in the total protein and LD-enriched protein fractions derived from Arabidopsis siliques, seeds, and seedlings, and the percentage of LD proteins within all samples.
iBAQ-processed proteomic data of both the total protein fraction and the LD-enriched fraction was filtered for at least two peptides per protein group. Then, the relative abundance of LD-associated proteins in the LD-enriched fraction was calculated based on their iBAQ scores. LD proteins were chosen taking into account protein families known and identified in this work.
| Developmental Stage | Total Protein Fraction | LD-Enriched Fraction | % of LD Proteins in LD-Enriched Fraction |
|---|---|---|---|
| Phase I siliques | 1,723 | 1,266 | 16.8 ± 4.4 |
| Phase II siliques | 1,417 | 1,337 | 17.5 ± 1.1 |
| Rehydrated seeds | 1,425 | 1,353 | 31.8 ± 1.5 |
| Stratified seeds | 1,511 | 1,024 | 31.8 ± 8 |
| 24 h seedlings | 2,004 | 1,158 | 24.3 ± 7.9 |
| 36 h seedlings | 2,197 | 1,368 | 25.9 ± 5.1 |
| 48 h seedlings | 2,218 | 1,478 | 28.5 ± 4.5 |
| 60 h seedlings | 2,198 | 1,723 | 34.1 ± 12.2 |
As shown in Figure 1B, more than half of the total protein groups detected (i.e. 1,428 of 2,696) were shared between all of the developmental stages examined, while a wide range of other proteins were unique for certain stages. In principal component analysis (PCA) plots, the total protein and LD-enriched protein groups from the five biological replicates of each stage clustered closely together (Fig. 1, C and D; Supplemental Datasets S5–S10), indicating good reproducibility of protein sampling and processing. The PCA plots also reflected the unique proteome of siliques compared to the other tissues, based on their separation in component 1 for both the total protein and LD-enriched protein fractions (Fig. 1, C and D). This unique proteome is likely based, in part, on the inclusion of silique wall material in these samples. Further, the distribution along component 2 indicated in both total and LD-enriched protein fractions that the younger siliques (Phase I) are more similar to older (60 h) green seedlings, while the older siliques (Phase II) are more similar to rehydrated or stratified seeds. The fact that siliques are more similar to seedlings than seeds in component 2 may be because of the high content of silique wall cells in addition to developing seed cells. Likewise, along component 2, the seeds and seedlings are distributed in a stage-dependent manner, with rehydrated seeds and 60 h seedlings being the most distinct (Fig. 1, C and D).
Unique Proteins Exist in the Seed and Early Seedling Total Protein Proteomes
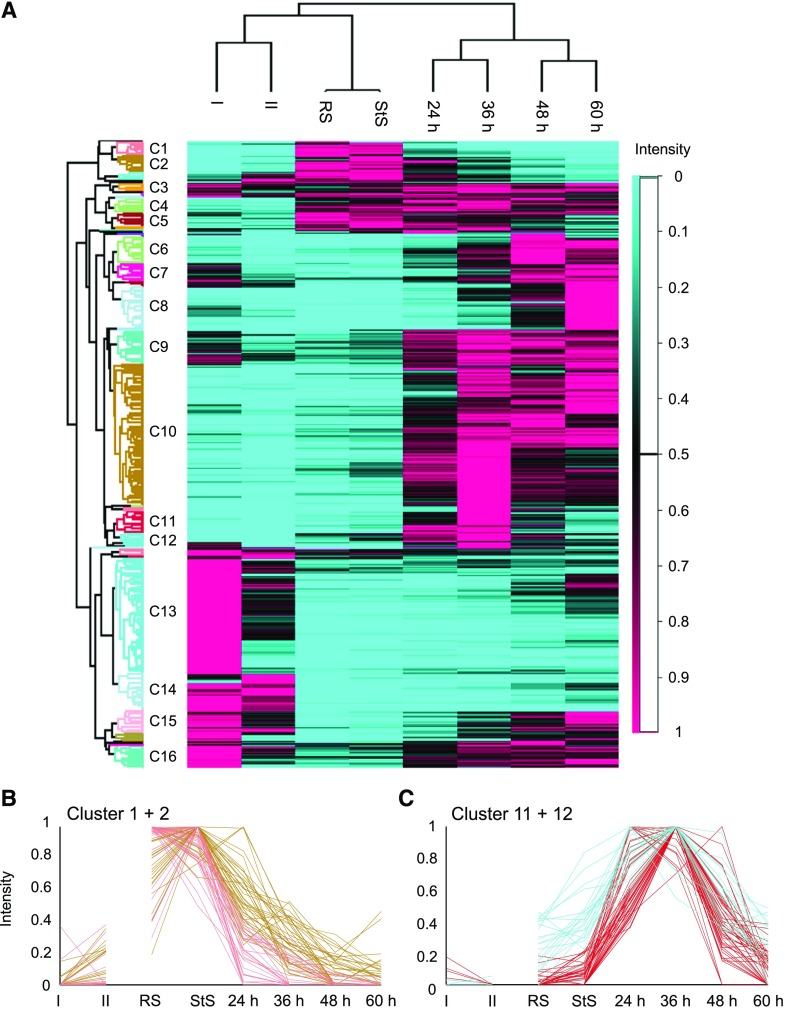
One objective for generating an extensive proteomics dataset for Arabidopsis during seed development, germination, and post-germinative growth was to identify developmental-stage-specific proteins potentially involved in distinct cellular processes during these time points in the plant’s life cycle, such as during seed desiccation and the switch in the seedling from hetero- to photoautotrophic growth. Toward that end, we performed hierarchical clustering after data normalization to analyze our total proteome datasets for proteins that were increased in abundance during these developmental phases (Supplemental Datasets S11 and S12). In total, 40 clusters were defined, and clusters containing >20 proteins are presented as a heat map in Figure 2A. Notably, four of the clusters contain proteins that are highest in abundance during either the two seed stages (i.e. Fig. 2, A and B, RS and StS, Clusters 1 and 2) or the two earliest stages of seedling establishment (i.e. 24 and 36 h after stratification; Clusters 11 and 12; Fig. 2, A and C). In Clusters 1 and 2, for instance, we found 71 proteins (Supplemental Dataset S12), including several late-embryogenesis-abundant proteins, two cell wall-modifying enzymes, and several proteins of unknown function. While seed storage proteins like CRUCIFERIN2 and CRUCIFERIN3 also have their highest intensities during the seed stages, their degradation is slower than that of the proteins present in Clusters 1 and 2. Clusters 11 and 12 contained 70 proteins (Supplemental Dataset S12), including several involved in either β-oxidation, such as LONG-CHAIN ACYL-COA SYNTHETASE7 (LACS7), ACYL-COA OXIDASE1 (ACX1), and enoyl-CoA hydratase ABNORMAL INFLORESCENCE MERISTEM1 (AIM1), or lipid metabolism, such as CYCLOARTENOL SYNTHASE (CAS) and OIL BODY LIPASE1 (OBL1). On the other hand, glyoxylate cycle enzymes, such as the peroxisomal NAD-MALATE DEHYDROGENASE1 or MALATE SYNTHASE, were grouped in Cluster 10, which, like Clusters 11 and 12, was also highest in abundance at 36 h after stratification, but remained so during later stages of seedling establishment.
Figure 2.
Identification of protein abundance clusters in total protein fractions derived from Arabidopsis siliques, seeds, and seedlings. A, Hierarchical clustering analysis of the normalized protein abundance over each of the developmental stages examined. In total, 40 row clusters were defined. Clusters harboring >20 proteins are labeled C1–C16. B and C, The expression profiles of all proteins in Clusters 1 and 2 (B; pink and brown lines; seed-specific proteins) and Clusters 11 and 12 (C; red and blue lines; early seedling establishment proteins) are shown. As silique samples also contain proteins derived from the silique wall, they should not necessarily be considered seed precursors, as indicated by the interrupted line between the II and RS samples. I, Phase I siliques; II, phase II siliques; RS, rehydrated seeds; StS, stratified seeds; 24 to 60 h, seedlings 24 to 60 h after stratification.
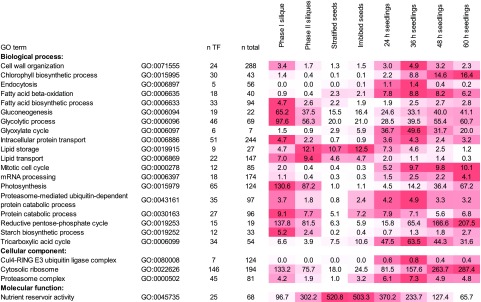
We also applied a gene ontology (GO) term analysis of our proteomics datasets (Fig. 3; Supplemental Datasets S13–S15) by assigning all of the associated GO terms to every protein identified in the total protein fractions for each developmental stage. Thereafter, rLFQ intensities of all proteins assigned to each GO term were summed. As presented in Figure 3, the GO term analysis revealed some important general trends. For instance, proteins involved in the “fatty acid biosynthesis process” were more abundant during silique (seed) development, and proteins associated with “nutrient reservoir activity” were of highest abundance during the seed stages. Further, proteins involved in GO-term-defined processes related to seedling establishment were upregulated during this phase (i.e. 24- to 60-h seedlings). These include proteins involved in “Fatty acid β-oxidation” and the “Glyoxylate cycle,” as well as the “Proteasome-mediated ubiquitin-dependent protein catabolic process,” which, as mentioned in the the first section. reflects, at least in part, the proteasomal degradation of LD proteins but also of other proteins. There was also a higher abundance of proteins with GO terms associated with photoautotrophic growth (e.g. “Chlorophyll biosynthetic process” and “Reductive pentose-phosphate cycle”) at the later stages of seedling establishment (i.e. 48- and 60-h seedlings).
Figure 3.
Changes in protein intensity of functional groups, based on GO terms, in the total protein fractions derived from Arabidopsis siliques, seeds, and seedlings. Proteins were assigned to GO terms and the relative abundance (rLFQ) of all proteins within a GO term is shown for each stage examined. “n total” corresponds to the total number of genes assigned to one GO term, “n TF” to the number of proteins detected in the total cellular fraction assigned to this GO term. Darker red represents higher total intensities compared to the other growth stages. Shown are selected GO terms.
We next analyzed the subcellular localization of the proteins. For this, we used the proteomics-confirmed annotation dataset at the Plant Proteome Database (Sun et al., 2009). This dataset contains 78 different subcellular annotations that we summed up to 10 different localizations: plastid, ER, vacuole, peroxisome, Golgi apparatus, mitochondrion, nucleus, cytoplasm, plasma membrane, and plastoglobule. The LD annotation was performed by us based on previous studies and taking into account LD proteins identified in this study (see later). Overall, the largest changes in abundance were observed for the proteins annotated as plastidial (Table 2; Supplemental Datasets S16–S18), with their abundance changing ∼10-fold between siliques and seeds, and between seeds and 60-h seedlings. On the other hand, the abundance of LD proteins nearly triples from younger to older siliques, is highest in rehydrated and stratified seeds, and decreases progressively in seedlings (Table 2; Supplemental Fig. S1).
Table 2. Annotation of subcellular localization of proteins in the total protein fractions derived from Arabidopsis siliques, seeds, and seedlings.
All proteins were annotated with 78 subcellular localizations obtained from the Plant Proteome Database and combined into 11 groups. The riBAQ intensities (in per mille) of the proteins were added up for 11 different subcellular compartments.
| Localization | Phase I Siliques | Phase II Siliques | Rehydrated Seeds | Stratified Seeds | 24 h Seedlings | 36 h Seedlings | 48 h Seedlings | 60 h Seedlings |
|---|---|---|---|---|---|---|---|---|
| Plastid | 571.3 | 392.3 | 44.1 | 40.0 | 52.4 | 154.0 | 334.3 | 477.5 |
| ER | 8.0 | 5.1 | 2.2 | 2.6 | 8.8 | 13.2 | 13.1 | 15.9 |
| Vacuole | 14.8 | 16.1 | 4.8 | 5.5 | 7.1 | 9.9 | 12.1 | 23.6 |
| Peroxisome | 15.1 | 9.8 | 9.6 | 10.0 | 22.7 | 26.6 | 26.4 | 21.2 |
| Golgi apparatus | 0.3 | 0.2 | 0.5 | 0.6 | 1.1 | 1.4 | 0.7 | 0.5 |
| Mitochondrion | 22.8 | 13.5 | 9.0 | 10.7 | 32.0 | 39.5 | 33.9 | 34.5 |
| Nucleus | 29.1 | 18.7 | 17.3 | 19.3 | 29.2 | 34.3 | 35.8 | 26.8 |
| Cytoplasm | 67.8 | 28.3 | 40.3 | 48.3 | 148.1 | 195.7 | 205.9 | 183.5 |
| Plastoglobule | 12.3 | 14.1 | 0.8 | 0.8 | 0.9 | 2.0 | 5.1 | 6.4 |
| Plasma membrane | 15.4 | 9.1 | 2.5 | 3.0 | 6.3 | 8.9 | 9.3 | 9.2 |
| LD | 9.1 | 25.3 | 57.4 | 69.8 | 37.9 | 18.8 | 8.3 | 4.8 |
Calculation of Enrichment Factors Enables the Identification of Low-Abundant LD Proteins in Arabidopsis
The second main objective of this work was to identify previously unidentified proteins associated with plant LDs, since further characterization of these proteins would undoubtedly aid in our understanding of LD functions and/or biogenesis, maintenance, and turnover. Our LD enrichment protocol (see “Materials and Methods” for details) avoided the use of harsh chemicals or extensive washing of the LDs in an effort to preserve weaker protein associations to the LDs and, thus, allowing us to potentially identify new, low-abundant LD proteins, albeit at the cost of perhaps including non-LD protein contaminants.
As shown in Table 1, >1,000 proteins were identified in each of the LD-enriched fractions from the different developmental stages examined. Known LD proteins (and those proteins described in this study) constituted between ∼17% (in the siliques) and >30% (in seeds and 60-h seedlings) of the total protein content in the LD-enriched fractions (Table 1). To distinguish putative LD proteins from contaminating proteins, we calculated the LD-enrichment factor for each protein, i.e. the ratio of the protein’s relative intensity in the LD-enriched fraction to its relative intensity in the total protein fraction (Supplemental Dataset S19). Further, in order to ensure that no other subcellular compartment(s) copurified with LDs, we also calculated the enrichment factors of proteins from different subcellular compartments (Supplemental Fig. S2). Overall, the LD-enrichment factors indicated that while occasionally some organelles copurified with LDs (i.e. enrichment factor >1), most had enrichment factors <1, and the enrichment factors were consistently the highest for LDs (ranging from 4.7 to 122.7; Supplemental Fig. S2). As LDs are ER derived (Chapman et al., 2019), our data could help to identify ER proteins that are localized at ER-LD junction sites. On the other hand, it is important to rule out ER proteins that are simply copurifying with LDs. We believe this to be unlikely, as the overall enrichment factors of previously annotated ER proteins only reached a highest value of 2.1 in 60-h seedlings, while LD proteins had an overall enrichment factor of 122.7 in this stage (Supplemental Fig. S2). Also, none of the annotated ER proteins showed large enrichment factors that would make them strong candidates to be LD proteins (Supplemental Fig. S3).
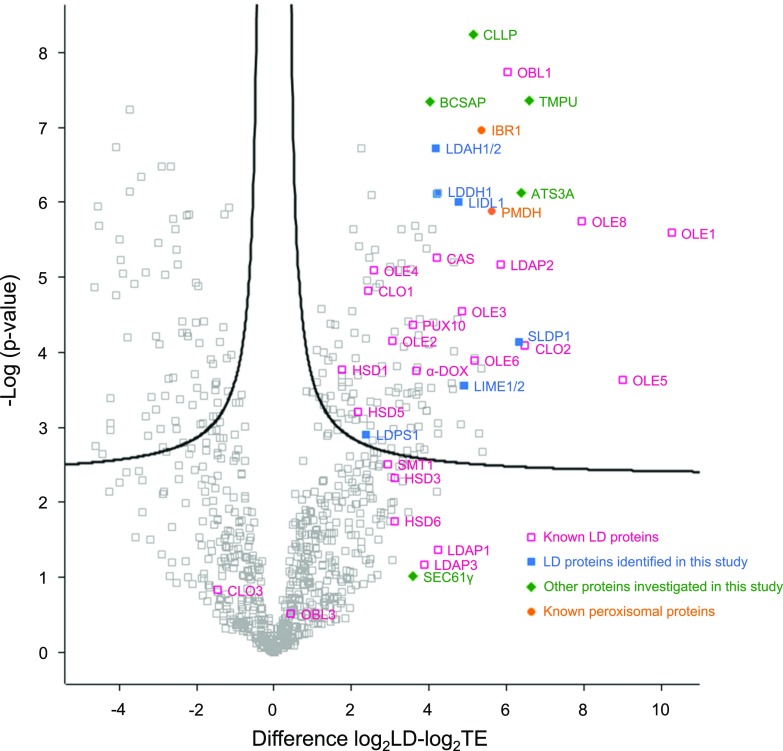
To narrow down candidate LD proteins from the extensive proteome of the LD-enriched fraction(s), we calculated enrichment factors for each protein and tested whether this enrichment was statistically significant. In doing so, we obtained a P-value based on on a two-sided t test (Supplemental Dataset 19). For each protein, we chose the stage where its abundance was relatively highest, only considering proteins that were identified in at least four of the five replicates in this stage, and had a riBAQ of at least 0.1 ‰. These data were used to generate the volcano plot shown in Figure 4. In total, 291 proteins significantly enriched in the LD-enriched fraction were found (refer to proteins on the right side of the plot in Fig. 4). Among these were most previously known LD proteins, including oleosin, steroleosin, and caleosin family members, as well as the LDAPs. Also prevalent in the LD-enriched fraction were several promising candidate LD proteins, which were chosen based on their strong enrichment and high P-value. On the other hand, overall abundance was less of a factor for selection, and several of the candidate LD proteins had a riBAQ of <0.4 ‰ in the LD-enriched fraction (Table 3) and were not among the top 100 most abundant proteins therein (based on data included in Supplemental Dataset S4).
Figure 4.
Enrichment analysis of proteins in the LD-enriched fractions derived from Arabidopsis siliques, seeds, and seedlings. A volcano plot was constructed to visualize proteins that are significantly LD enriched (upper right). The developmental stage (siliques, seeds, and seedlings) with the highest abundance (riBAQ) was chosen for each protein and the log2-transformed values and P-values were calculated at this stage. Only proteins detected in four of the five replicates and with an riBAQ >0.1 are included in this figure. As depicted in the legend, known LD and peroxisomal proteins are indicated in blue and orange, respectively, and the proteins chosen for further study that did or did not localize to LDs in pollen tubes and tobacco leaves are shown in blue and green, respectively. Black lines indicate a false discovery rate of 0.001. Abbreviations: ATS3A, EMBRYO-SPECIFIC PROTEIN; BCSAP, BRISC COMPLEX SUBUNIT ABRO1-LIKE PROTEIN; CCLP, CURCULIN-LIKE LECTIN FAMILY PROTEIN; SMT, STEROL METHYLTRANSFERASE; TMPU, TRANSMEMBRANE PROTEIN OF UNKNOWN FUNCTION.
Table 3. Arabidopsis proteins chosen as LD protein candidates.
In part, proteins were named in this study. ATS3A, EMBRYO-SPECIFIC PROTEIN; BCSAP, BRISC COMPLEX SUBUNIT ABRO1-LIKE PROTEIN; CCLP, CURCULIN-LIKE LECTIN FAMILY PROTEIN; TMPU, TRANSMEMBRANE PROTEIN OF UNKNOWN FUNCTION.
| Protein Name | AGI Codea | Homolog | Descriptionb | Stage with Highest Abundance in LD Fraction | Abundance at Highest Stage (riBAQ ‰) | Enrichment Factorc | P-Value (−Log)c | Subcellular Localizationd |
|---|---|---|---|---|---|---|---|---|
| ATS3A | AT2G41475.1 | AT5G62200 | Embryo-specific protein 3 | 48 h seedlings | 1.17 ± 0.54 | 84.4 | 6.12 | Non-LD foci |
| BCSAP | AT3G08780.1 | – | BRISC complex subunit Abro1-like protein | 60 h seedlings | 0.17 ± 0.05 | 16.3 | 7.34 | Cytoplasm |
| CCLP | AT1G78830.1 | – | Curculin-like lectin family protein | 60 h seedlings | 0.36 ± 0.1 | 35.0 | 8.24 | Non-LD foci |
| LDAH1 | AT1G10740.1* | AT1G23330* | α/β-hydrolase | 60 h seedlings | 0.19 ± 0.07 | 19.1 | 6.72 | LD |
| LDDH1 | AT1G75180.1 | AT1G19400 | Dehydrogenase | 60 h seedlings | 0.26 ± 0.04 | 18.4 | 6.11 | LD |
| LDPS | AT3G19920.1 | AT5G64230 | Unknown | Stratified seeds | 0.31 ± 0.19 | 7.4 | 2.36 | LD |
| LIDL1 | AT1G18460.1 | AT1G19360 | Lipase family protein | 60 h seedlings | 0.31 ± 0.15 | 26.9 | 6.01 | LD |
| LIME1 | AT4G33110.1 | AT4G33120 | Coclaurine methyltransferase | 48 h seedlings | 1.12 ± 0.62 | 30.3 | 3.55 | LD |
| OBAP1A | AT1G05510.1 | AT5G45690 | Oil body-associated protein | Rehydrated seeds | 1.95 ± 1.19 | 0.22 | 2.65 | Cytoplasm |
| SEC61γ | AT5G50460.1* | AT4G24920* | Subunit of SEC61 translocon | Stratified seeds | 0.96 ± 1.2 | 11.9 | 1.02 | ER/LD |
| SLDP1 | AT1G65090.3 | AT5G36100 | Unknown | 60 h seedlings | 2.1 ± 0.45 | 17.5 | 6.31 | LD |
| TMPU | AT1G27290.1 | – | Transmembrane protein of unknown function | 60 h seedlings | 1.12 ± 0.47 | 97.0 | 7.36 | ER subdomains |
Gene locus identifier according to the Arabidopsis Genome Initiative. The given splice variant corresponds to the one tested for subcellular localization. Arabidopsis Genome Initiative codes marked with an asterisk were not identified unambiguously.
Description based on TAIR10 annotation.
Enrichment factors and P-values were calculated from the given developmental stages at which the protein had the highest riBAQ intensity value in the LD-enriched fraction. Missing values were imputed with a value of 0.01.
Subcellular localizations found in this study
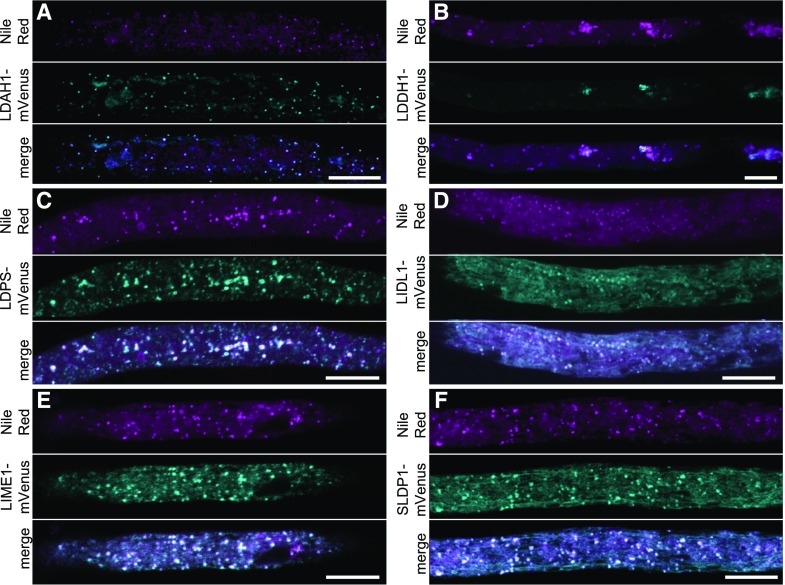
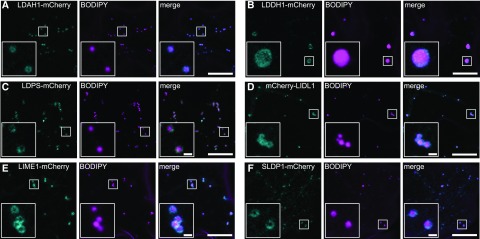
The subcellular localization of the candidate LD proteins was subsequently assessed using two independent plant cell systems that are both well established for the study of protein trafficking and localization, including to LDs: N. tabacum pollen tubes transformed by particle bombardment (Fig. 5; Supplemental Fig. S4; Müller et al., 2017) and Nicotiana benthamiana leaves transformed by Agrobacterium tumefaciens infiltration (Fig. 6; Cai et al., 2015; Gidda et al., 2016). In both cases, full-length open reading frames (ORFs) encoding candidate LD proteins were cloned as mVenus or mCherry fluorescent protein fusions and transiently expressed, and subcellular localization then was assessed by confocal laser-scanning microscopy. LD localization of fusion proteins was determined by staining of LDs with the neutral lipid stains Nile red (Greenspan et al., 1985) or BODIPY493/503 (Listenberger and Brown, 2007).
Figure 5.
Subcellular localization of selected candidate LD proteins in N. tabacum pollen tubes. A to F, Candidate proteins fused to mVenus at their C termini were transiently expressed in N. tabacum pollen tubes (cyan channel). LDs were stained with Nile red (magenta channel). In the merge channel, colocalization appears white. Note, in B, that expression of LDDH1-mVenus led to clustering of LDs. Bars = 10 μm.
Figure 6.
Subcellular localization of selected LD protein candidates in N. benthamiana leaves. A to F, Candidate proteins fused to mCherry at their N or C termini were transiently expressed in N. benthamiana leaves (cyan channel). LDs were stained with BODIPY 493/503 (magenta channel). In the corresponding merge channel, note that a torus fluorescence pattern attributable to the expressed fusion protein encircles the BODIPY-stained LDs. Boxes denote portions of the cells shown at higher magnification in the insets. Bars = 10 μm (1 μm in insets).
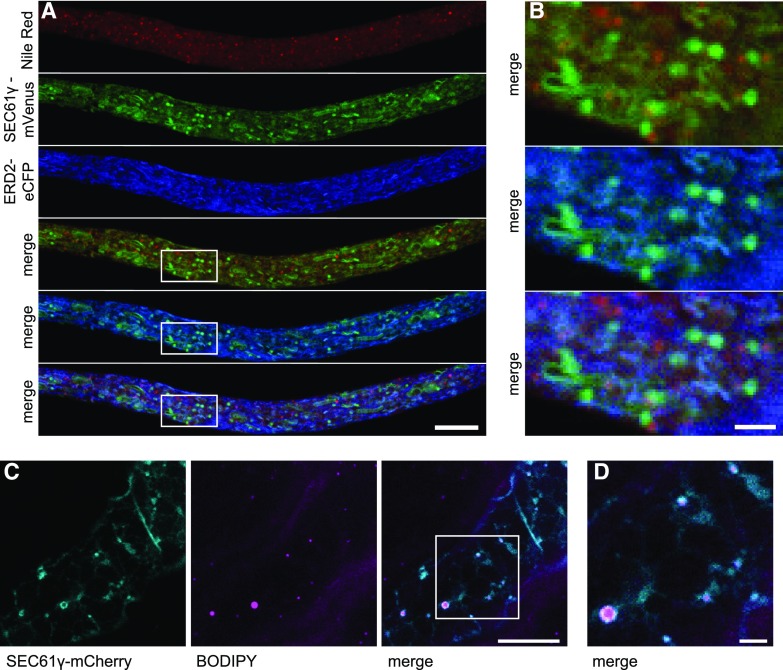
In total, the LD localization of six proteins was confirmed using both plant cell systems (Figs. 5 and 6). We termed these proteins LD-ASSOCIATED LIPASE1 (LIDL1), LD METHYLTRANSFERASE1 (LIME1), LD PROTEIN OF SEEDS (LDPS), SEED LD PROTEIN1 (SLDP1), LD-ASSOCIATED HYDROLASE1 (LDAH1), and LD DEHYDROGENASE1 (LDDH1), taking into account their functional annotations at The Arabidopsis Information Resource (TAIR) and their expression patterns based on the Arabidopsis eFP Browser tool at the Bio-Analytic Resource for Plant Biology (https://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi). Other proteins, although initially selected as promising candidates due to their highly significant, strong enrichment in LD-enriched fractions (Fig. 4), did not localize to LDs, based on experiments in pollen tubes (Supplemental Fig. S4), although at least one of these proteins, SEC61γ, which is a subunit of the Sec translocon at the ER membrane (Spiess et al., 2019), appeared to localize to regions of the ER that in some instances were in close proximity to or encircled LDs (Fig. 7). These results with SEC61γ are intriguing because they are reminiscent of the localization of SEIPIN in plant cells, an ER membrane protein involved in the formation of nascent LDs (Cai et al., 2015), suggesting that SEC61γ might also function at ER-LD junctions.
Figure 7.
Subcellular localization of SEC61γ in tobacco pollen tubes and N. benthamiana leaves. SEC61γ fused at its C terminus to mVenus or mCherry was transiently expressed in N. tabacum pollen tubes (A and B) or in N. benthamiana leaves (C). LDs were stained with Nile red (A and B) or BODIPY 493/503 (C and D). SEC61γ was cotransformed with the ER marker ERD2-CFP (A and B) and partially colocalizes with the ER. SEC61γ also accumulates at potential ER-LD contact sites (B–D). The box in C indicates the area of the cells shown at higher magnification in D. Bars = 10 μm (A and C) or 2 μm (B and D).
Another candidate protein assessed in terms of its subcellular localization was the OIL BODY-ASSOCIATED PROTEIN1A (OBAP1A). OBAPs were described previously to be localized to LDs, based on studies with a maize (Zea mays) protein isoform (López-Ribera et al., 2014). However, all three Arabidopsis OBAPs identified in this study (i.e. OBAP1A, OBAP2, and OBAP3) were only slightly enriched in the LD-enriched fraction during the silique stage and were strongly depleted in LDs during all other phases examined, including those stages where their abundance in total protein fractions was highest (i.e. rehydrated and stratified seeds; Supplemental Fig. S4A; refer also to Fig. 4). These results are consistent with our previous reported Arabidopsis seedling LD proteomes (Pyc et al., 2017a; Kretzschmar et al., 2018), in which OBAPs were not enriched. Consistent with these observations, when transiently expressed as either C- or N-terminal-tagged mVenus fusion proteins in tobacco pollen tubes, OBAP1A displayed exclusively a diffuse fluorescence, indicative of its localization to the cytoplasm, and there were no obvious associations with any distinct subcellular compartment(s) (Supplemental Fig. S4B). Similarly, OBAP1A-mCherry transiently expressed in N. benthamiana leaf cells localized throughout the cell (i.e. cytoplasm) and did not appear to localize to BODIPY-stained LDs (Supplemental Fig. S4C).
The LD Proteome Changes during Seedling Establishment
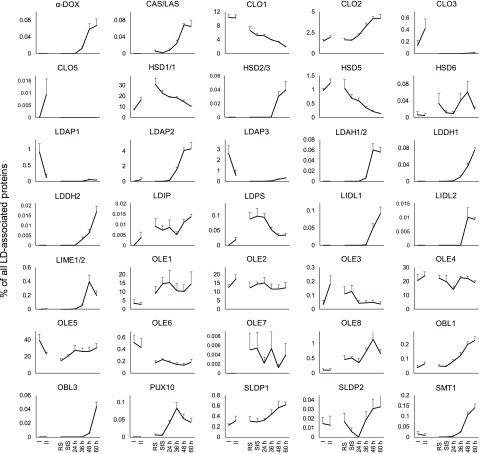
In addition to the discovery of LD proteins (Figs. 5 and 6), we analyzed the dynamics of LD proteins in general, in terms of their relative abundance throughout the various stages of seed development, germination, and post-germinative growth. As presented in Table 1 and Supplemental Figure S1, the abundance of both previously known and newly identified LD proteins in the total proteome increased during seed development (i.e. in siliques), was relatively high in seeds (rehydrated and stratified seeds), and then progressively decreased in seedlings after germination, until 60 h after stratification when it was highest. Next, in order to further assess changes in the composition of the LD proteome during development, the abundance of all LD proteins was added in each stage, and the individual fraction for each protein was calculated (Supplemental Dataset S20).
Collectively, the most abundant oleosin protein family members (i.e. OLE1, OLE2, OLE4, and OLE5) contributed a relatively constant proportion of proteins in the LD proteome throughout seedling establishment (Fig. 8). By contrast, the prevalence of the most abundant caleosin, CLO1, and steroleosin, HSD1, continuously decreased during the same time period, while CLO2 and HSD2/3 increased. In general, >85% of the LD proteome during all of the developmental stages examined consisted of six proteins: OLE1, OLE2, OLE4, OLE5, CLO1, and HSD1. Less abundant LD proteins, on the other hand, displayed a wide range of dynamics (Fig. 8). For instance, while some of these proteins, such as LDIP, SLDP1, LDPS, and OBL1, were detected in the LD proteomes at most or all time points, their relative abundance varied considerably. The proportion of OBL1, for example, increases steadily over the course of seedling establishment, while that of LDPS decreases (Fig. 8). Several other proteins, e.g. LIDL1 and LIDL2, LIME1/2, LDAH1/2, and LDDH1/2, contribute only to the seedling LD proteomes, but not those of seeds or siliques. None of the LDAP protein isoforms, LDAP1–LDAH3, were detected in the seed LD proteomes, but were most abundant in siliques (LDAP1 and LDAP3) and seedlings (LDAP2; Fig. 8). Interestingly, α-DIOXYGENASE1, which was previously described to be specifically enriched at LDs during senescence (Brocard et al., 2017) and pathogen attack (Shimada et al., 2014), was present in the seedling LD proteome at 36 h and onward, suggesting that it might also have functions during early plant growth.
Figure 8.
Dynamic composition of the LD proteome derived from Arabidopsis siliques, seeds, and seedlings. The riBAQ intensities of LD-associated proteins in the LD-enriched fraction were calculated as a percentage of the riBAQ of all known LD-associated proteins. This way, the contribution of each protein to the complete LD proteome and the dynamic changes in the abundance of the LD proteins can be observed. Protein isoform numbers separated by a slash indicate that these proteins could not be distinguished based on the proteomic data. Two highly similar genes of the steroleosin family are both annotated as HSD1. I, Phase I siliques; II, phase II siliques; RS, rehydrated seeds; StS, stratified seeds; 24 to 60 h, seedlings 24 to 60 h after stratification. As silique samples also contain proteins derived from silique wall, they should not necessarily be considered to be seed precursors, as indicated by the interrupted line between the II and RS samples. n = 5 per stage. Error bars represent the sd.
Phosphorylation and Ubiquitination of LD Proteins
It is well established that posttranslational modifications can influence the activity, localization, and/or fate (e.g. turnover) of a protein (Arsova et al., 2018). Two such modifications, phosphorylation and ubiquitination, have previously been identified on oleosins, caleosins, and steroleosins, and their ubiquitination has been implicated in their degradation (Hsiao and Tzen, 2011; Deruyffelaere et al., 2015; Kretzschmar et al., 2018). To assess whether other LD proteins are targets of similar posttranslational modifications, possibly as a means of (protein) regulation in the cell, we analyzed our proteomic dataset for peptides with appended phosphate and/or ubiquitin moieties. Overall, modifications were detected on 182 proteins, including the LD proteins CLO1, HSD1, LDAP2, OLE2, and OLE4 (Table 4; Supplemental Datasets S21–S30).
Table 4. Posttranslational modifications detected on LD proteins and ubiquitin.
Sites marked with an asterisk were not identified unambiguously. For a complete list of modified proteins and modification sites, see Supplemental Datasets 21–30. RS, rehydrated seeds; StS, stratified seeds.
| Protein | Ubiquitination | Phosphorylation | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RS | StS | 24-h Seedlings | 36-h Seedlings | 48-h Seedlings | 60-h Seedlings | RS | StS | 24-h Seedlings | 36-h Seedlings | 48-h Seedlings | 60-h Seedlings | |
| OLE2 | – | – | – | – | K146 | – | S18 | – | – | – | – | – |
| OLE4 | K157* | K157* | K43 | K157* | K157* | K157* | Y153 | Y153 | S40 | – | – | – |
| K159* | K159* | K157* | K159* | K159* | K159* | Y164 | – | Y42 | – | – | – | |
| K168 | – | K159* | K168 | K168 | K168 | – | – | – | – | – | – | |
| – | – | K168 | – | – | – | – | – | – | – | – | – | |
| CLO1 | – | K4 | – | – | – | – | – | – | – | – | – | – |
| HSD1 | – | – | – | K289 | – | – | S340 | S348 | – | T283 | – | – |
| – | – | – | K295 | – | – | T343 | – | – | – | – | – | |
| – | – | – | – | – | – | S348 | – | – | – | – | – | |
| LDAP2 | – | – | – | K108 | – | – | – | – | – | T118 | – | S128 |
| – | – | – | K125 | – | – | – | – | – | S122 | – | – | |
| UBQ13 | – | – | K48 | K6 | K48 | – | – | – | S246 | T7 | S246 | – |
| – | – | K63 | K11 | K63 | – | – | – | S247 | T9 | S247 | – | |
| – | – | – | K48 | – | – | – | – | – | S246 | T249 | – | |
| – | – | – | K63 | – | – | – | – | – | S247 | – | – | |
DISCUSSION
Growth Stage-Specific Proteomes Provide Extensive and Useful Data
We present here a quantitative proteomic dataset, generated with an Orbitrap MS, for Arabidopsis silique development, seed germination, and seedling establishment. Previous works investigating Arabidopsis seed germination (Gallardo et al., 2001; Galland et al., 2014) identified 67 and 475 proteins, respectively. Our data are consistent with those studies, but offer coverage of a relatively larger number of proteins across various developmental stages, including seedling establishment. In another recent study on the seed proteome of Arabidopsis seeds, 3,243 proteins were identified (Li et al., 2019). While the number of proteins in our study is lower, we also used more stringent criteria for protein selection (such as a minimum of two peptides per protein instead of one peptide) and focused on changes in protein abundance throughout several developmental phases. More specifically, in order to detect relative differences in abundance of individual proteins between different samples, the corresponding MS raw datasets were analyzed with the MaxQuant software (https://www.maxquant.org/). MaxQuant integrates and compares the area of peptide peaks within MS1 spectra and considers the monoisotopic peak, as well as isotopic peaks. This integration is considered more accurate than peptide-count-based quantification (Cox et al., 2014; Wang et al., 2019a). Isotope labeling‐based methods have advantages in relative and absolute quantification of proteins in comparison to label-free quantification, as used in this study, as peptides from different samples can be mixed and analyzed in a single run. However, the number of isotope tags that can be used to differentiate between peptides from different samples is usually ≤10 (Wang et al., 2019a), limiting the number of comparable conditions (in this study, for example, 16 conditions were analyzed).
MaxQuant implies two quantification algorithms. In the simpler iBAQ algorithm, the sum of the MS1 intensities of peptides from one protein is divided by the number of theoretically possible peptides of reasonable length, thereby giving protein intensities normalized to the theoretical peptide number. The intensities of a given protein can be also compared between samples (Krey et al., 2014). By contrast, the LFQ algorithm uses elaborated normalization strategies between samples on the level of peptide intensities within and across MS1 chromatograms (Cox et al., 2014). It leads to smaller variations of relative quantifications between biological replicates in general, as well as in our samples (Supplemental Datasets S2 and S4).
While the coverage of our proteomics dataset is relatively lower than that of datasets derived using transcriptomic-based techniques (Narsai et al., 2011), transcript abundance does not necessarily correlate with protein abundance, especially during developmental processes when transcripts can exist before they are actually translated. For example, one of the proteins we identified, AT4G27450, which is annotated (by TAIR) to be of unknown function, is present at the transcript level during seed maturation (based on eFP Browser; Nakabayashi et al., 2005; Schmid et al., 2005), but is not detectable at the protein level. Instead, AT4G27450 protein abundance increases sharply in seedlings 24 h after stratification and decreases thereafter (Fig. 2, Cluster 11; Supplemental Datasets S11 and S12). Since AT4G27450 transcripts are not present in vegetative tissues, except during anoxia stress (based on the eFP Browser), it is also a good candidate to play a distinct role(s) during seed germination, which could be further addressed by studying knockout lines. Other proteins from Clusters 11 and 12 that increased transiently during seedling establishment could also play important roles during seed germination. Examples of proteins from Clusters 1 and 2 (Fig. 2; Supplemental Datasets S11 and S12) that are almost exclusively present in imbibed seeds and then decrease in abundance include putative oxidoreductase (At1g54870) and β-glucosidase (AT3G21370) enzymes (Supplemental Datasets S11 and S12), which are highly abundant (18.5‰ and 7.3‰, respectively (Supplemental Datasets S11 and S12), indicative of important roles during seed desiccation or germination that could be further studied.
Proteomics Combined with a Cell-Biological Approach Was Used to Identify Low-Abundance LD Proteins
The number of studies to date investigating the protein composition of LDs in plants, including senescent Arabidopsis leaves (Brocard et al., 2017) and a variety of algal species (Moellering and Benning, 2010; Siegler et al., 2017; Lupette et al., 2019; Wang et al., 2019b), has increased concomitantly with the use of MS-based proteomic techniques in general. However, in those studies, as well as in this one, not all proteins detected in LD-enriched fractions are necessarily bona fide LD proteins, as other proteins may copurify with LDs or are just not sufficiently depleted, which is an intrinsic shortcoming of MS-based analysis of LDs. To address this important potential caveat, we employed several measures of “quality control” in order to minimize false-positive LD proteins. First, in addition to sampling the LD-enriched fraction, we took a sample of the total cellular protein extract prior to LD enrichment. With both the LD-enriched proteome and the total cellular proteome we were able to calculate LD-enrichment factors. We then used the iBAQ algorithm for candidate discovery, since the LFQ algorithm, as discussed above, leads to a larger number of missing values, causing low-abundance proteins to be missing from the total cellular extracts and disallowing calculation of an enrichment factor.
Using enrichment factors, we could identify contaminants in the LD-enriched fractions that were abundant therein, but did not have a high enrichment factor. On the other hand, we were also able to identify proteins of relatively low abundance (e.g. LDAH1 is only the 283rd most abundant protein in the LD fraction in 60-h seedlings) as good candidates for a cell-biological verification. These latter studies were carried out using two different model plant cell systems (Figs. 5 and 6) that have been successfully used elsewhere to assess the subcellular localization of fluorescent protein-tagged LD proteins, tobacco pollen tubes and N. benthamiana leaves (Cai et al., 2017; Müller et al., 2017; Pyc et al., 2017a; Siegler et al., 2017; Kretzschmar et al., 2018).
Overall, we confirmed six candidate LD proteins (Fig. 5 and 6), and we also presented the subcellular localization of several proteins that did not localize to LDs (Supplemental Fig. S4) despite being excellent candidates based on their prevalence in the LD-enriched fractions (Fig. 4). This indicates that candidate protein selection based on enrichment factors can still yield false positives, probably caused by noise and/or artificial protein coenrichment. Importantly, apparent non-LD localization could also occur because these candidate proteins need specific binding partners for LD association that are either present in insufficient amounts or absent in pollen tubes and leaves. Furthermore, the interaction of proteins with LDs might be more transient, yielding an enrichment in isolated LD fractions, but not readily apparent (via microscopy) in plant cell-based expression studies. We also provided evidence that proteins previously annotated as being localized to LDs, based on their homology to proteins in other species, might not in fact be associated with LDs in Arabidopsis. A pertinent example is OBAP1A, a transiently expressed (fusion) protein that was not enriched in the LD fractions of seeds and seedlings and did not localize to LDs in pollen tubes or leaves (Supplemental Fig. S5). These observations are in contrast to the reported LD localization of maize OBAP1 in N. benthamiana leaves, which shares ∼64% amino acid sequence identity with Arabidopsis OBAP1A (López-Ribera et al., 2014), suggesting that the two proteins have diverged in terms of their ability to target to LDs.
The LD Proteome Varies Across Developmental Stages and Tissues
The considerable changes in the protein composition of LDs over the time frame of seed formation and maturation, germination, and seedling establishment (Fig. 8) highlights the organelle’s dynamic nature in terms of its functioning throughout these various stages of growth and development. The main role of LDs during seed development, for instance, is to serve as cellular depots for newly synthesized storage lipids (e.g. TAGs), which, during germination, are subsequently mobilized via β-oxidation and used as an energy source for early seedling growth. Thereafter, during greening of the cotyledons in seedlings and the onset of photoautotrophic growth (i.e. in 48- and 60-h seedlings), LDs are no longer primarily required for the compartmentalization of stored lipids for heterotrophic growth and thus likely play roles in other cellular processes. Consistent with the latter notion, the LD proteome in the 48- and 60-h seedlings underwent among some of the most rapid and pronounced changes (Fig. 8), and many of the 35 LD proteins identified are only found in these two stages. Indeed, this diversification possibly represents a transition from seed-type LDs to vegetative-type LDs, wherein functions other than energy storage are carried out. One such previously described function for LDs in vegetative tissues is pathogen defense, which is mediated, in part, by CLO3 and α-DOX (Shimada et al., 2014; Brocard et al., 2017). Further, RNA sequencing data indicate that the newly identified LD proteins LIDLs, LDAHs, LDDHs, and LIME1 are all ubiquitously expressed in vegetative tissues, while LIME2 is expressed primarily in roots (Supplemental Dataset S31; data based on trava.org; Klepikova et al., 2016). Further research on these and other LD proteins will undoubtedly add to the growing list of functions related to vegetative-type LDs.
The changes in the LD proteome observed across the different stages of growth and development that we examined (Fig. 8) may require not only the synthesis of proteins, but also their degradation. For instance, during germination and seedling establishment, the overall amount of LD proteins decreases (Supplemental Fig. S1) and the bulk of the proteins is degraded within the first 36 h after stratification. During these time points, the LD protein PUX10, which is known to be involved in the degradation of other LD proteins via the ubiquitin-proteasome degradation pathway, shows the highest relative abundance on LDs (Fig. 8; Deruyffelaere et al., 2018; Kretzschmar et al., 2018). Our data also show that several LD proteins harbor ubiquitination sites (Table 3). Furthermore, on ubiquitin itself, we detected ubiquitinations on lysines 48 and 63 (K48 and K63), which is indicative of polyubiquitin chains associated with protein degradation (Komander and Rape, 2012) only during the earlier stages of seedling establishment (i.e. 24- to 48-h seedlings; Table 3).
The Identification of LD Proteins Inspires Future Investigation into the Roles of Plant LDs
We successfully identified members of six protein families that have not been described previously as localized to LDs in plant cells or been studied with respect to their physiological function. Five of these proteins, namely LIDL1, LIME1, LDDH1, LDAH1, and SLDP1, each have one homolog (45% to 90% identity at the amino acid sequence level) that was either found separately in our proteomics dataset enriched in the LD fraction or was indistinguishable from its counterpart based on the detected peptide sequences (Supplemental Dataset S4). The function of the six identified proteins can in part be hypothesized by considering their homology to other related and characterized proteins, including those with known or putative enzymatic function(s).
The LIME1/LIME2 proteins, for example, share homology with a protoberberine methyltransferase (Liscombe and Facchini, 2007) and a coclaurine N-methyltransferase from poppy (Papaver somniferum), which catalyzes the synthesis of methylate coclaurine, an intermediate in the morphine biosynthetic pathway (Onoyovwe et al., 2013). Morphine is a hydrophobic compound found in the latex of the opium poppy, and latex particles are a class of LDs that contain polyisoprenoids instead of TAGs and sterol esters (Nawamawat et al., 2011). Thus, the LIME1/LIME2 proteins in Arabidopsis might catalyze a step in the synthesis of secondary metabolites, which are stored within LDs due to their high hydrophobicity. Similarly, the LD protein LDPS may also participate in plant secondary metabolism. LDPS is annotated as a BTB/POZ-domain-containing protein, the domain of which is thought to mediate protein-protein interactions (Collins et al., 2001). The homolog of LDPS, AT5G64230.1, which shares 33% identity at the protein level, is annotated as a 1,8-cineole synthase but has not been studied in terms of whether it actually functions in the monoterpene biosynthetic pathway. Nonetheless, this sequence similarity suggests that LDPS is also involved in monoterpene metabolism in plant cells, perhaps serving in the proper compartmentalization of hydrophobic monoterpenes in LDs.
The yeast (Saccharomyces cerevisiae) homolog of Arabidopsis LIDL1 and LIDL2, which are putative lipases, has been shown to have sterol esterase activity, and loss-of-function yeast mutants accumulate sterol esters (Athenstaedt et al., 1999; Jandrositz et al., 2005), suggesting that LIDL1 and LIDL2 may be LD-specific sterol esterases. This latter hypothesis is notable because sterol esters are also present in the LD core, where they are found in minor amounts in Arabidopsis seed LDs (Bouvier-Navé et al., 2010) but can make up a much higher proportion of the stored neutral lipid in other tissues in plants or in other organisms (Onal et al., 2017; Rotsch et al., 2017). Whether LIDL1 and LIDL2 are responsible for the metabolism of stored sterol esters in Arabidopsis is an open question. It also remains to be determined whether LDAH1 and LDDH1 participate in plant metabolism, as indicated by their annotation as an α/β-hydrolase and a putative erythronate-4-phosphate dehydrogenase, respectively. Interestingly, LDDH1 was found previously to be associated with plastids (Teresinski et al., 2019) and we detected it only after the greening of the (60 h) seedling cotyledons. This could indicate that LDDH1 is associated with plastid-LD contact sites. LDDH1 may also act in concert with LDAH1, as these two genes are coexpressed based on ATTED-II version 9.2 (Obayashi et al., 2018) and GENEVESTIGATOR 7.2.6 (Hruz et al., 2008).
SLDPs are uncharacterized proteins, and with the exception of a hydrophobic domain near their N termini that might serve as a membrane anchor, they possess no conserved domains/motifs or significant sequence similarity to other characterized proteins in plants or in nonplant species, such as yeast and mammals. SLDP2, like SLDP1, was found enriched in the LD fraction, but was much less abundant and therefore did not meet the criteria to be considered a strong candidate LD protein (Fig. 4).
While some of the candidate proteins investigated did not localize to LDs (Supplemental Fig. S4), despite being excellent candidates based on their prevalence in the LD-enriched fractions (Fig. 4), it is still possible that these proteins are important for LD biology. One such protein is SEC61γ, which was enriched in the LD proteome and localized to regions of the ER that were often in close contact with LDs (Fig. 7). The SEC61 complex is important for both the incorporation of nascent proteins into the ER (Spiess et al., 2019) and also their retrotranslocation (Scott and Schekman, 2008). Therefore, it is plausible that this complex is also involved in the targeting and/or degradation of LD proteins, as was previously discussed in a study where oleosin was ectopically expressed in yeast (Beaudoin et al., 2000).
In conclusion, the discovery of LD proteins in this study opens up the possibility for uncovering new aspects of plant LD function, biogenesis, and turnover. The existence of several proteins conserved across kingdoms implies that at least some aspects of LD biology are conserved, yet on the other hand, there are also plant-specific LD proteins, suggesting that plant LDs serve additional, unique roles not found in other organisms (Chapman et al., 2019). Undoubtedly, insights into such functions will be gained by identifying and confirming additional LD proteins in plants, and the combined MS-based proteomics and cell-biological approach used here will continue to serve toward that end. Furthermore, our understanding of LDs will increase as these proteins are investigated further.
MATERIALS AND METHODS
Plant Materials
Tobacco (Nicotiana tabacum) and Arabidopsis (Arabidopsis thaliana, ecotype Columbia) plants were grown as described previously (Kretzschmar et al., 2018). For the two silique development phases (I and II), complete siliques were harvested, and 2 g of the older siliques, or 3 g of the younger siliques, were pooled for one biological replicate. For rehydrated seeds, stratified seeds, and seedlings 24 and 36 h post-stratification, 100 mg of dry seed material per biological replicate was used. For 48- and 60-h seedlings, dry seed starting material was increased to 160 mg per biological replicate. Rehydrated seeds were incubated in water for 30 min without surface sterilization. For stratified seeds and post-stratification seedling time points, surface-sterilized seeds (sterilized with 6% [w/v] sodium hypochlorite and 0.1% [v/v] Triton X-100) were spread on half-strength Murashige and Skoog media (Murashige and Skoog, 1962) and incubated in the dark at 4°C for 74 h. Then, stratified seed samples were harvested, and all other plants were transferred into a 22°C 16-h light/8-h dark cycle growth chamber with 150 µmol photons m−2 s−1 daytime light strength; post-stratification seed/seedling time points (24–60 h) indicate the time spent in this condition.
Isolation of Total and LD-Enriched Protein Fractions
After harvesting, each sample was mixed with appropriate amounts (2 mL for rehydrated seeds, 3 mL for stratified seeds – 48 h samples, 3.5 mL for 60 h, 15 mL for younger siliques, 20 mL for older siliques) of grinding buffer [10 mm sodium phosphate buffer, pH 7.4, 200 μm phenylmethylsulfonyl fluoride, 0.5 mm dithiobis (succinimidyl propionate)] and ground with sand to homogeneity with a precooled (on ice) mortar and pestle. The homogenates were centrifuged for 1 min at 100g. For the total protein sample, 100 µL of the homogenate was mixed with 900 μL 96% (v/v) ethanol to precipitate proteins. For enrichment of LDs, the homogenate was subjected to three consecutive 20,000g centrifugations for 15 min at 4°C. After each centrifugation step, the resulting fat pad was taken off the aqueous phase and transferred into a fresh aliquot of grinding buffer, where it was resuspended. After the third centrifugation step, the fat pad was resuspended in 1 mL 96% (v/v) ethanol.
Proteomic Sample Preparation and LC-MS/MS Analysis of Peptides
Proteins were isolated and their concentrations were determined. They were then subjected to in-gel tryptic-digestion as described previously (Kretzschmar et al., 2018), but with 20 μg of protein digested per replicate. Peptides were then subjected to LC-MS/MS analysis modified from what was described previously (Schmitt et al., 2017). First, 2 μL peptide samples were separated by nano-flow LC on a RSLCnano Ultimate 3000 system (Thermo Fisher Scientific). The peptides were loaded with 0.07% (v/v) trifluoroacetic acid on an Acclaim PepMap 100 precolumn (100 μm × 2 cm, C18, 3 μm, 100 Å; Thermo Fisher Scientific) with a flow rate of 20 μL/min for 3 min. Then, peptides were separated by reverse-phase chromatography on an Acclaim PepMan RSLC column (75 μm × 50 cm, C18, 3 μm, 100 Å; Thermo Fisher Chemical) with a flow rate of 300 nL/min. The peptides were eluted with the following gradient: 96% solvent A (0.1% [v/v] formic acid) and 4% solvent B (80% [v/v] acetonitrile and 0.1% [v/v] formic acid) to 10% solvent B over 2 min, to 30% solvent B over 58 min, to 45% solvent B over 22 min, and to 90% solvent B over 12 min. All solvents and acids were of Optima LC-MS quality and purchased from Thermo Fisher Scientific. Eluted peptides were ionized online by nano-electrospray ionization with a Nanospray Flex Ion Source (Thermo Fisher Scientific) at 1.5 kV (liquid junction) and analyzed with a Q Exactive HF mass spectrometer (Thermo Fisher Scientific). Full scans were recorded in a mass range of 300–1,650 m/z at a resolution of 30,000 followed by data-dependent top 10 higher-energy collisional dissociation fragmentation (dynamic exclusion enabled). LC-MS method programming and data acquisition were performed with XCalibur 4.0 software (Thermo Fisher Scientific).
MS Data Processing
MS and MS/MS raw data were processed for feature detection, peptide identification, and protein group assembly with the MaxLFQ algorithm in MaxQuant software version 1.6.2.10 (Cox and Mann, 2008; Cox et al., 2014). Default settings were used and are specified in the metadata file (Supplemental Table S1). Additionally, label-free quantification was enabled in group-specific parameter settings. In global parameter settings, “match between runs” and iBAQ were enabled. The TAIR10 protein database was used as reference. Only proteins identified by at least two peptides were considered. When comparing and relating LD-enriched fractions to total protein fractions, riBAQ values were used, as this algorithm picks up smaller values more often. Enrichment factors were calculated by dividing the average riBAQ value of the LD-enriched fraction by the riBAQ value of the total cellular extract. For quantitative comparison of the total proteome of different time points, rLFQ values are shown, as these display smaller variations between replicates in our hands.
To identify posttranslational modifications, the data were searched for phosphorylation of Ser, Thr, and Tyr, and for ubiquitination of Lys. Settings are specified in the metadata file (Supplemental Table S2). Data analysis was performed in Perseus 1.6.2.2 (Tyanova et al., 2016) and in Excel 2016 (Microsoft).
The Venn diagram was created with InteractiVenn (Heberle et al., 2015). For PCA, proteins were only taken into account if they were found in all replicates of at least one of the stages. The rLFQ values were log2-transformed. After imputation of missing values by normal distribution (width, 0.9; down shift, 1.8 for total cellular extracts and 2.1 for LD-enriched fractions), PCA was performed with Perseus using default settings. Projections were exported and the data were graphed in Excel 2016.
For cluster analysis, LFQ-processed data were filtered for at least four valid values in at least one of the eight time points. For each protein, the maximum value was set to 1, and the remaining time points were calculated as fractions of this value. The resulting data matrix was used for hierarchical clustering in Perseus 1.6.2.2, with Euclidean distances and preprocessing with k-means.
GO terms were assigned based on the annotation of the TAIR homepage as of December 13, 2018. Annotations of protein localization were obtained from the Plant Proteome Database (Sun et al., 2009) as of February 26, 2019. LD localization was reassigned based on results of this study and Kretzschmar et al. (2018). The volcano plot was created with Perseus. Proteins were included if the average riBAQ intensity was >0.1 in at least one stage and if the protein was detected in at least four of the five biological replicates of at least one stage. Missing values were imputed by 0.01 before log2 transformation and calculation of P-values and false discovery rate using the following settings: Test, t test; side, both; number of randomizations, 250; s0 (artificial within groups variance), 0.1; and false discovery rate, 0.001.
Molecular Cloning and Plasmid Construction
Complementary DNA was prepared from RNA extracted from 10 mg mature (dry) seeds, 50 mg 7-d-old seedlings, or 50 mg inflorescences using a Spectrum Plant Total RNA Kit (Sigma-Aldrich). All constructs were amplified using Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific) according to the manufacturer’s protocol. Molecular cloning into the Gateway vectors pUC-LAT52-mVenusC-GW and pUC-LAT52-mVenusN-GW (Müller et al., 2017), as well as the vector pMDC32-ChC, was performed as depicted in Müller et al. (2017). The use of CFP-SKL for marking peroxisomes (Müller et al., 2017) and the use of ERD2-CFP (ARABIDOPSIS ENDOPLASMIC RETICULUM RETENTION DEFECTIVE2) for marking the ER (Kretzschmar et al., 2018) has been described previously.
The pMDC32-ChC plant expression binary vector, encoding the monomeric Cherry fluorescent protein ORF adjacent to a 5′ recombination site that allows for mCherry to be translationally fused to the C terminus protein of interest, was constructed in the following manner. First the mCherry ORF was amplified using the mCherry-FP-PacI and mCherry-RP-SacI primers (Supplemental Table S3) and pRTL2-Cherry (Gidda et al., 2011) as template DNA. The resulting PCR products were then digested with PacI and SacI and inserted into similarly digested pMDC32 (Curtis and Grossniklaus, 2003), yielding pMDC32-ChC1. Thereafter, the Cmr/ccdB region of pMDC32-ChC1 was amplified using the primers ccdB-FP-KpnI and ccdB-RP-PacI (Supplemental Table S3), which resulted in the removal of a stop codon upstream of the Cherry ORF and reinsertion into KpnI-PacI-digested pMDC32-ChC1, yielding pMDC32-ChC.
Custom oligonucleotide primers were synthesized by Sigma-Aldrich; a complete list of all primers is given in Supplemental Table S3. All plasmids constructed in this study, including their promoter and cloning regions and any fusion protein ORFs, were verified by automated sequencing performed at Microsynth AG or the University of Guelph Advanced Analysis Centre Genomics Facility.
Particle Bombardment and Microscopy of N. tabacum Pollen Tubes
Pollen grains were transformed by particle bombardment, in vitro cultivated on microscope slides, stained with Nile red (Sigma-Aldrich), and analyzed by confocal laser-scanning microscopy as described by Müller et al. (2017) and Kretzschmar et al. (2018), or with a Zeiss LSM780 (Carl Zeiss) using similar settings. For each construct, at least 10 pollen tubes were imaged.
Transient Transformation and Microscopy of Nicotiana benthamiana Leaves
N. benthamiana plants were grown in soil with a 16-h light/8-h dark day/night cycle at 22°C. Leaves of ∼4-week-old plants were infiltrated with Agrobacterium tumefaciens (strain LBA4404) harboring the selected expression vector as described in Pyc et al. (2017b). All infiltrations were performed with pORE04-35S:p19 containing the tomato bushy stunt virus gene P19 in order to enhance transgene expression (Petrie et al., 2010).
A. tumefaciens-infiltrated leaves were processed for confocal laser scanning microscopy imaging, including staining of LDs with BODIPY 493/503 (Invitrogen), as previously described (Gidda et al., 2016). Micrographs of leaves were acquired using a Leica SP5 confocal laser scanning microscope (Leica Microsystems). Excitations and emission signals for fluorescent proteins and BODIPY were collected sequentially as single optical sections in double-labeling experiments like those described in Gidda et al. (2016); single-labeling experiments showed no detectable crossover at the settings used for data collection. All fluorescence images of cells shown are representative of at least two separate experiments, including at least three separate transformations of leaf cells.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AT3G01420 (αDOX); AT2G41475 (ATS3A); AT3G08780 (BCSAP); AT2G07050 (CAS); AT1G78830 (CCLP); AT4G26740 (CLO1); AT5G55240 (CLO2); AT2G33380 (CLO3); AT1G70680 (CLO5); AT1G29330 (ERD2); At5G50600/AT5G50700 (HSD1/1); AT3G47350 (HSD2); AT3G47360 (HSD3); AT4G10020 (HSD5); AT5G50770 (HSD6); AT3G45130 (LAS); AT1G10740 (LDAH1); AT1G23330 (LDAH2); AT1G67360 (LDAP1); AT2G47780 (LDAP2); AT3G05500 (LDAP3); AT1G75180 (LDDH1); AT1G19400 (LDDH2); AT5G16550 (LDIP); AT1G18460 (LIDL1); AT1G73920 (LIDL2); AT3G19920 (LDPS); AT4G33110 (LIME1); AT4G33120 (LIME2); AT3G20820 (LRR); AT1G05510 (OBAP1A); AT3G14360 (OBL1); AT1G45201 (OBL3);. AT4G25140 (OLE1); AT5G40420 (OLE2); AT5G51210 (OLE3); AT3G27660 (OLE4); AT3G01570 (OLE5); AT1G48990 (OLE6); AT2G25890 (OLE7); AT3G18570 (OLE8); AT4G10790 (PUX10); AT5G50460 (SEC61γ); AT1G65090 (SLDP1); AT5G36100 (SLDP2); AT5G13710 (SMT1); and AT1G27290 (TMPU).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Abundance of LD proteins within the total protein fraction from Arabidopsis siliques, seeds, and seedlings.
Supplemental Figure S2. Enrichment of organelle-specific proteins in the LD-enriched fractions from Arabidopsis siliques, seeds, and seedlings.
Supplemental Figure S3. Enrichment analysis of proteins in the LD-enriched fractions derived from Arabidopsis siliques, seeds, and seedlings with highlighted LD and ER proteins.
Supplemental Figure S4. Subcellular localization of other selected, candidate proteins in N. tabacum pollen tubes.
Supplemental Figure S5. Abundance in total and LD-enriched protein fractions and subcellular localization of the Arabidopsis OBAP1A protein.
Supplemental Table S1. Metadata file for LC-MS/MS data processing with MaxQuant.
Supplemental Table S2. Metadata file for LC-MS/MS data processing with MaxQuant checking for posttranslational modifications.
Supplemental Table S3. Primers used for Gateway molecular cloning and sequencing.
Supplemental Dataset S1. Proteins found in siliques and seedlings—Raw LFQ values.
Supplemental Dataset S2. Proteins found in siliques and seedlings—Normalized and sorted LFQ values.
Supplemental Dataset S3. Proteins found in siliques and seedlings—Raw iBAQ values.
Supplemental Dataset S4. Proteins found in siliques and seedlings—Normalized and sorted iBAQ values.
Supplemental Dataset S5. Proteins found in siliques and seedlings—Imputed log2-transformed LFQ values of total protein fraction.
Supplemental Dataset S6. Loadings of PCA plot created with Supplemental Dataset 5 (total fraction).
Supplemental Dataset S7. Projections of PCA plot created with Supplemental Dataset 5 (total fraction).
Supplemental Dataset S8. Proteins found in siliques and seedlings—Imputed log2-transformed LFQ values of lipid droplet-enriched fractions.
Supplemental Dataset S9. Loadings of PCA plot created with Supplemental Dataset 8 (lipid droplet-enriched fraction).
Supplemental Dataset S10. Projections of PCA plot created with Supplemental Dataset 8 (lipid droplet-enriched fraction).
Supplemental Dataset S11. Proteins found in the total protein fraction of siliques and seedlings—Normalized and sorted LFQs with at least four valid values in at least one condition.
Supplemental Dataset S12. Data set used to create the heat map presented in Figure 2.
Supplemental Dataset S13. Sums of rLFQ values of proteins associated with the same GO ID.
Supplemental Dataset S14. Phase-dependent averages of sums of rLFQ values of proteins associated with the same GO ID.
Supplemental Dataset S15. Selected GO ID sums.
Supplemental Dataset S16. Subcellular localization of proteins.
Supplemental Dataset S17. Sums of riBAQ values of proteins with the same subcellular localization.
Supplemental Dataset S18. List of curated subcellular localization acquired from the Plant Proteome Data Base.
Supplemental Dataset S19. Results matrix from the enrichment analysis.
Supplemental Dataset S20. Contribution of LD proteins to the total LD proteome.
Supplemental Dataset S21. Modified proteins identified in Phase I silique samples.
Supplemental Dataset S22. Modified proteins identified in Phase II silique samples.
Supplemental Dataset S23. Modified proteins identified in rehydrated seed samples.
Supplemental Dataset S24. Modified proteins identified in stratified seed samples.
Supplemental Dataset S25. Modified proteins identified in 24-h seedling samples.
Supplemental Dataset S26. Modified proteins identified in 36-h seedling samples.
Supplemental Dataset S27. Modified proteins identified in 48-h seedling samples.
Supplemental Dataset S28. Modified proteins identified in 60-h seedling samples.
Supplemental Dataset S29. All modified proteins identified across samples, including their modified sites.
Supplemental Dataset S30. All modified LD proteins identified across samples, including their modified sites.
Supplemental Dataset S31. Gene expression levels of LD proteins identified in this study.
Acknowledgments
We thank Ivo Feussner for all his support and many helpful discussions, Dr. Christiane Gatz and Dr. Alexander Stein for their valuable advice, and Dr. Kent Chapman and Dr. John Dyer for their insightful discussions about this work. We are grateful also to Dr. Jörg Großhans and Dr. Steven Johnsen for granting access to their confocal microscopes, Dr. Florian Wegwitz and Johannes Sattmann for their assistance, Dr. Leonie Steinhorst and Dr. Jörg Kudla for generating and providing plasmids, and Dr. Ivo Feussner, Dr. Jennifer Popko, and Dr. Frederike Ruhe for providing the plasmid pCambia33.1Gs mCherry-LIDL1. Technical assistance was provided by Siqi Sun and Antony Grüness.
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (DFG; IS 273/2-2 to T.I., INST1525/16-1 FUGG to Jörg Großhans, and DFG-GZ: INST 186/1230-1 FUGG to Stefanie Pöggeler), the Studienstiftung des Deutschen Volkes (to F.K.K. and P.S.), the U.S. Department of Energy, Office of Science, BES-Physical Biosciences Program (DE-SC0016536, in part to R.T.M.) to support N. benthamiana experiments, and the Natural Sciences and Engineering Research Council of Canada (RGPIN-2018-04629 to R.T.M.).
Articles can be viewed without a subscription.
References
- Arsova B, Watt M, Usadel B (2018) Monitoring of plant protein post-translational modifications using targeted proteomics. Front Plant Sci 9: 1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athenstaedt K, Zweytick D, Jandrositz A, Kohlwein SD, Daum G (1999) Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J Bacteriol 181: 6441–6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Boutin JP, Miquel M, Lepiniec L, Rochat C (2002) An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 40: 151–160 [Google Scholar]
- Baud S, Dichow NR, Kelemen Z, d’Andréa S, To A, Berger N, Canonge M, Kronenberger J, Viterbo D, Dubreucq B, et al. (2009) Regulation of HSD1 in seeds of Arabidopsis thaliana. Plant Cell Physiol 50: 1463–1478 [DOI] [PubMed] [Google Scholar]
- Beaudoin F, Wilkinson BM, Stirling CJ, Napier JA (2000) In vivo targeting of a sunflower oil body protein in yeast secretory (sec) mutants. Plant J 23: 159–170 [DOI] [PubMed] [Google Scholar]
- Bersuker K, Peterson CWH, To M, Sahl SJ, Savikhin V, Grossman EA, Nomura DK, Olzmann JA (2018) A proximity labeling strategy provides insights into the composition and dynamics of lipid droplet proteomes. Dev Cell 44: 97–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier-Navé P, Berna A, Noiriel A, Compagnon V, Carlsson AS, Banas A, Stymne S, Schaller H (2010) Involvement of the phospholipid sterol acyltransferase1 in plant sterol homeostasis and leaf senescence. Plant Physiol 152: 107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard L, Immel F, Coulon D, Esnay N, Tuphile K, Pascal S, Claverol S, Fouillen L, Bessoule J-J, Bréhélin C (2017) Proteomic analysis of lipid droplets from Arabidopsis aging leaves brings new insight into their biogenesis and functions. Front Plant Sci 8: 894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Goodman JM, Pyc M, Mullen RT, Dyer JM, Chapman KD (2015) Arabidopsis SEIPIN proteins modulate triacylglycerol accumulation and influence lipid droplet proliferation. Plant Cell 27: 2616–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, McClinchie E, Price A, Nguyen TN, Gidda SK, Watt SC, Yurchenko O, Park S, Sturtevant D, Mullen RT, et al. (2017) Mouse fat storage-inducing transmembrane protein 2 (FIT2) promotes lipid droplet accumulation in plants. Plant Biotechnol J 15: 824–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman KD, Aziz M, Dyer JM, Mullen RT (2019) Mechanisms of lipid droplet biogenesis. Biochem J 476: 1929–1942 [DOI] [PubMed] [Google Scholar]
- Chapman KD, Dyer JM, Mullen RT (2012) Biogenesis and functions of lipid droplets in plants. J Lipid Res 53: 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibani K, Ali-Rachedi S, Job C, Job D, Jullien M, Grappin P (2006) Proteomic analysis of seed dormancy in Arabidopsis. Plant Physiol 142: 1493–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins T, Stone JR, Williams AJ (2001) All in the family: The BTB/POZ, KRAB, and SCAN domains. Mol Cell Biol 21: 3609–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13: 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deruyffelaere C, Bouchez I, Morin H, Guillot A, Miquel M, Froissard M, Chardot T, D’Andrea S (2015) Ubiquitin-mediated proteasomal degradation of oleosins is involved in oil body mobilization during post-germinative seedling growth in Arabidopsis. Plant Cell Physiol 56: 1374–1387 [DOI] [PubMed] [Google Scholar]
- Deruyffelaere C, Purkrtova Z, Bouchez I, Collet B, Cacas J-L, Chardot T, Gallois J-L, D’Andrea S (2018) PUX10 is a CDC48A adaptor protein that regulates the extraction of ubiquitinated oleosins from seed lipid droplets in Arabidopsis. Plant Cell 30: 2116–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Barisch C, Paschke P, Herrfurth C, Bertinetti O, Pawolleck N, Otto H, Rühling H, Feussner I, Herberg FW, Maniak M (2013) Dictyostelium lipid droplets host novel proteins. Eukaryot Cell 12: 1517–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand TC, Cueff G, Godin B, Valot B, Clément G, Gaude T, Rajjou L (2019) Combined proteomic and metabolomic profiling of the Arabidopsis thaliana vps29 mutant reveals pleiotropic functions of the retromer in seed development. Int J Mol Sci 20: 1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond PJ. (2004) Cloning and characterization of the acid lipase from castor beans. J Biol Chem 279: 45540–45545 [DOI] [PubMed] [Google Scholar]
- Eldakak M, Milad SIM, Nawar AI, Rohila JS (2013) Proteomics: A biotechnology tool for crop improvement. Front Plant Sci 4: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fercha A, Capriotti AL, Caruso G, Cavaliere C, Stampachiacchiere S, Zenezini Chiozzi R, Laganà A (2016) Shotgun proteomic analysis of soybean embryonic axes during germination under salt stress. Proteomics 16: 1537–1546 [DOI] [PubMed] [Google Scholar]
- Galland M, Huguet R, Arc E, Cueff G, Job D, Rajjou L (2014) Dynamic proteomics emphasizes the importance of selective mRNA translation and protein turnover during Arabidopsis seed germination. Mol Cell Proteomics 13: 252–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SPC, Puype M, Demol H, Vandekerckhove J, Job D (2001) Proteomic analysis of arabidopsis seed germination and priming. Plant Physiol 126: 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbowicz K, Liu Z, Alseekh S, Tieman D, Taylor M, Kuhalskaya A, Ofner I, Zamir D, Klee HJ, Fernie AR, et al. (2018) Quantitative trait loci analysis identifies a prominent gene involved in the production of fatty acid-derived flavor volatiles in tomato. Mol Plant 11: 1147–1165 [DOI] [PubMed] [Google Scholar]
- Gidda SK, Park S, Pyc M, Yurchenko O, Cai Y, Wu P, Andrews DW, Chapman KD, Dyer JM, Mullen RT (2016) Lipid droplet-associated proteins (LDAPs) are required for the dynamic regulation of neutral lipid compartmentation in plant cells. Plant Physiol 170: 2052–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidda SK, Shockey JM, Falcone M, Kim PK, Rothstein SJ, Andrews DW, Dyer JM, Mullen RT (2011) Hydrophobic-domain-dependent protein-protein interactions mediate the localization of GPAT enzymes to ER subdomains. Traffic 12: 452–472 [DOI] [PubMed] [Google Scholar]
- Greenspan P, Mayer EP, Fowler SD (1985) Nile red: A selective fluorescent stain for intracellular lipid droplets. J Cell Biol 100: 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajduch M, Ganapathy A, Stein JW, Thelen JJ (2005) A systematic proteomic study of seed filling in soybean. Establishment of high-resolution two-dimensional reference maps, expression profiles, and an interactive proteome database. Plant Physiology 137: 1397–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajduch M, Hearne LB, Miernyk JA, Casteel JE, Joshi T, Agrawal GK, Song Z, Zhou M, Xu D, Thelen JJ (2010) Systems analysis of seed filling in Arabidopsis: Using general linear modeling to assess concordance of transcript and protein expression. Plant Physiol 152: 2078–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanano A, Burcklen M, Flenet M, Ivancich A, Louwagie M, Garin J, Blée E (2006) Plant seed peroxygenase is an original heme-oxygenase with an EF-hand calcium binding motif. J Biol Chem 281: 33140–33151 [DOI] [PubMed] [Google Scholar]
- Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R (2015) InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics 16: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn PJ, James CN, Gidda SK, Kilaru A, Dyer JM, Mullen RT, Ohlrogge JB, Chapman KD (2013) Identification of a new class of lipid droplet-associated proteins in plants. Plant Physiol 162: 1926–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv Bioinforma 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao ESL, Tzen JTC (2011) Ubiquitination of oleosin-H and caleosin in sesame oil bodies after seed germination. Plant Physiol Biochem 49: 77–81 [DOI] [PubMed] [Google Scholar]
- Huang AHC. (2017) Plant lipid droplets and their associated oleosin and other proteins: Potential for rapid advances. Plant Physiol 176: 1894–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jandrositz A, Petschnigg J, Zimmermann R, Natter K, Scholze H, Hermetter A, Kohlwein SD, Leber R (2005) The lipid droplet enzyme Tgl1p hydrolyzes both steryl esters and triglycerides in the yeast, Saccharomyces cerevisiae. Biochim Biophys Acta 1735: 50–58 [DOI] [PubMed] [Google Scholar]
- Kim EY, Park KY, Seo YS, Kim WT (2016) Arabidopsis small rubber particle protein homolog SRPs play dual roles as positive factors for tissue growth and development and in drought stress responses. Plant Physiol 170: 2494–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepikova AV, Kasianov AS, Gerasimov ES, Logacheva MD, Penin AA (2016) A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J 88: 1058–1070 [DOI] [PubMed] [Google Scholar]
- Komander D, Rape M (2012) The ubiquitin code. Annu Rev Biochem 81: 203–229 [DOI] [PubMed] [Google Scholar]
- Kretzschmar FK, Mengel LA, Müller AO, Schmitt K, Blersch KF, Valerius O, Braus GH, Ischebeck T (2018) PUX10 is a lipid droplet-localized scaffold protein that interacts with CELL DIVISION CYCLE48 and is involved in the degradation of lipid droplet proteins. Plant Cell 30: 2137–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krey JF, Wilmarth PA, Shin JB, Klimek J, Sherman NE, Jeffery ED, Choi D, David LL, Barr-Gillespie PG (2014) Accurate label-free protein quantitation with high- and low-resolution mass spectrometers. J Proteome Res 13: 1034–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubala S, Garnczarska M, Wojtyla Ł, Clippe A, Kosmala A, Żmieńko A, Lutts S, Quinet M (2015) Deciphering priming-induced improvement of rapeseed (Brassica napus L.) germination through an integrated transcriptomic and proteomic approach. Plant Sci 231: 94–113 [DOI] [PubMed] [Google Scholar]
- Lee J, Lee W, Kwon SW (2015) A quantitative shotgun proteomics analysis of germinated rice embryos and coleoptiles under low-temperature conditions. Proteome Sci 13: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Asami T, Wu X, Tsang EWT, Cutler AJ (2007) A putative hydroxysteroid dehydrogenase involved in regulating plant growth and development. Plant Physiol 145: 87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P-C, Ma J-J, Zhou X-M, Li G-H, Zhao C-Z, Xia H, Fan S-J, Wang X-J (2019) Arabidopsis MDN1 is involved in the establishment of a normal seed proteome and seed germination. Front Plant Sci 10: 1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q-F, Xiong M, Xu P, Huang L-C, Zhang C-Q, Liu Q-Q (2016) Dissection of brassinosteroid-regulated proteins in rice embryos during germination by quantitative proteomics. Sci Rep 6: 34583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L-J, Tai SSK, Peng C-C, Tzen JTC (2002) Steroleosin, a sterol-binding dehydrogenase in seed oil bodies. Plant Physiol 128: 1200–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscombe DK, Facchini PJ (2007) Molecular cloning and characterization of tetrahydroprotoberberine cis-N-methyltransferase, an enzyme involved in alkaloid biosynthesis in opium poppy. J Biol Chem 282: 14741–14751 [DOI] [PubMed] [Google Scholar]
- Listenberger LL, Brown DA (2007) Fluorescent detection of lipid droplets and associated proteins. Curr Protoc Cell Biol 35: 24.2.1-24.2.11 [DOI] [PubMed] [Google Scholar]
- López-Ribera I, La Paz JL, Repiso C, García N, Miquel M, Hernández ML, Martínez-Rivas JM, Vicient CM (2014) The evolutionary conserved oil body associated protein OBAP1 participates in the regulation of oil body size. Plant Physiol 164: 1237–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz C, Brandt S, Borisjuk L, Rolletschek H, Heinzel N, Tohge T, Fernie AR, Braun H-P, Hildebrandt TM (2018) The role of persulfide metabolism during Arabidopsis seed development under light and dark conditions. Front Plant Sci 9: 1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupette J, Jaussaud A, Seddiki K, Morabito C, Brugière S, Schaller H, Kuntz M, Putaux JL, Jouneau PH, Rébeillé F, et al. (2019) The architecture of lipid droplets in the diatom Phaeodactylum tricornutum. Algal Res 38: 101415 [Google Scholar]
- Mansfield SG, Briarty LG (1992) Cotyledon cell development in Arabidopsis thaliana during reserve deposition. Can J Bot 70: 151–164 [Google Scholar]
- Moellering ER, Benning C (2010) RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot Cell 9: 97–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller AO, Blersch KF, Gippert AL, Ischebeck T (2017) Tobacco pollen tubes—A fast and easy tool for studying lipid droplet association of plant proteins. Plant J 89: 1055–1064 [DOI] [PubMed] [Google Scholar]
- Müller AO, Ischebeck T (2018) Characterization of the enzymatic activity and physiological function of the lipid droplet-associated triacylglycerol lipase AtOBL1. New Phytol 217: 1062–1076 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Naested H, Frandsen GI, Jauh GY, Hernandez-Pinzon I, Nielsen HB, Murphy DJ, Rogers JC, Mundy J (2000) Caleosins: Ca2+-binding proteins associated with lipid bodies. Plant Mol Biol 44: 463–476 [DOI] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: Epigenetic and genetic regulation of transcription in seed. Plant J 41: 697–709 [DOI] [PubMed] [Google Scholar]
- Narsai R, Law SR, Carrie C, Xu L, Whelan J (2011) In-depth temporal transcriptome profiling reveals a crucial developmental switch with roles for RNA processing and organelle metabolism that are essential for germination in Arabidopsis. Plant Physiol 157: 1342–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawamawat K, Sakdapipanich JT, Ho CC, Ma Y, Song J, Vancso JG (2011) Surface nanostructure of Hevea brasiliensis natural rubber latex particles. Colloids Surf A Physicochem Eng Asp 390: 157–166 [Google Scholar]
- Nguyen T-P, Cueff G, Hegedus DD, Rajjou L, Bentsink L (2015) A role for seed storage proteins in Arabidopsis seed longevity. J Exp Bot 66: 6399–6413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Aoki Y, Tadaka S, Kagaya Y, Kinoshita K (2018) ATTED-II in 2018: A plant coexpression database based on investigation of the statistical property of the mutual rank index. Plant Cell Physiol 59: e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onal G, Kutlu O, Gozuacik D, Dokmeci Emre S (2017) Lipid droplets in health and disease. Lipids Health Dis 16: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onoyovwe A, Hagel JM, Chen X, Khan MF, Schriemer DC, Facchini PJ (2013) Morphine biosynthesis in opium poppy involves two cell types: Sieve elements and laticifers. Plant Cell 25: 4110–4122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Graham S, Graham IA (2005) Storage reserve mobilization in germinating oilseeds: Arabidopsis as a model system. Biochem Soc Trans 33: 380–383 [DOI] [PubMed] [Google Scholar]
- Petrie JR, Shrestha P, Liu Q, Mansour MP, Wood CC, Zhou X-R, Nichols PD, Green AG, Singh SP (2010) Rapid expression of transgenes driven by seed-specific constructs in leaf tissue: DHA production. Plant Methods 6: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyc M, Cai Y, Gidda SK, Yurchenko O, Park S, Kretzschmar FK, Ischebeck T, Valerius O, Braus GH, Chapman KD, et al. (2017a) Arabidopsis lipid droplet-associated protein (LDAP)-interacting protein (LDIP) influences lipid droplet size and neutral lipid homeostasis in both leaves and seeds. Plant J 92: 1182–1201 [DOI] [PubMed] [Google Scholar]
- Pyc M, Cai Y, Greer MS, Yurchenko O, Chapman KD, Dyer JM, Mullen RT (2017b) Turning over a new leaf in lipid droplet biology. Trends Plant Sci 22: 596–609 [DOI] [PubMed] [Google Scholar]
- Qu R, Wang SM, Lin YH, Vance VB, Huang AH (1986) Characteristics and biosynthesis of membrane proteins of lipid bodies in the scutella of maize (Zea mays L.). Biochem J 235: 57–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan S, Yang P, Cassin-Ross G, Kaur N, Switzenberg R, Aung K, Li J, Hu J (2013) Proteome analysis of peroxisomes from etiolated Arabidopsis seedlings identifies a peroxisomal protease involved in β-oxidation and development. Plant Physiol 163: 1518–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotsch AH, Kopka J, Feussner I, Ischebeck T (2017) Central metabolite and sterol profiling divides tobacco male gametophyte development and pollen tube growth into eight metabolic phases. Plant J 92: 129–146 [DOI] [PubMed] [Google Scholar]
- Rudolph M, Schlereth A, Körner M, Feussner K, Berndt E, Melzer M, Hornung E, Feussner I (2011) The lipoxygenase-dependent oxygenation of lipid body membranes is promoted by a patatin-type phospholipase in cucumber cotyledons. J Exp Bot 62: 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schmitt K, Smolinski N, Neumann P, Schmaul S, Hofer-Pretz V, Braus GH, Valerius O (2017) Asc1p/RACK1 connects ribosomes to eukaryotic phosphosignaling. Mol Cell Biol 37: e00279-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DC, Schekman R (2008) Role of Sec61p in the ER-associated degradation of short-lived transmembrane proteins. J Cell Biol 181: 1095–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada TL, Shimada T, Takahashi H, Fukao Y, Hara-Nishimura I (2008) A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J 55: 798–809 [DOI] [PubMed] [Google Scholar]
- Shimada TL, Takano Y, Shimada T, Fujiwara M, Fukao Y, Mori M, Okazaki Y, Saito K, Sasaki R, Aoki K, et al. (2014) Leaf oil body functions as a subcellular factory for the production of a phytoalexin in Arabidopsis. Plant Physiol 164: 105–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegler H, Valerius O, Ischebeck T, Popko J, Tourasse NJ, Vallon O, Khozin-Goldberg I, Braus GH, Feussner I (2017) Analysis of the lipid body proteome of the oleaginous alga Lobosphaera incisa. BMC Plant Biol 17: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siloto RMP, Findlay K, Lopez-Villalobos A, Yeung EC, Nykiforuk CL, Moloney MM (2006) The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess M, Junne T, Janoschke M (2019) Membrane protein integration and topogenesis at the ER. Protein J 38: 306–316 [DOI] [PubMed] [Google Scholar]
- Sun Q, Zybailov B, Majeran W, Friso G, Olinares PDB, van Wijk KJ (2009) PPDB, the plant proteomics database at Cornell. Nucleic Acids Res 37: D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teresinski HJ, Gidda SK, Nguyen TND, Howard NJM, Porter BK, Grimberg N, Smith MD, Andrews DW, Dyer JM, Mullen RT (2019) An RK/ST C-terminal motif is required for targeting of OEP7.2 and a subset of other Arabidopsis tail-anchored proteins to the plastid outer envelope membrane. Plant Cell Physiol 60: 516–537 [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13: 731–740 [DOI] [PubMed] [Google Scholar]
- Tzen J, Cao Y, Laurent P, Ratnayake C, Huang A (1993) Lipids, proteins, and structure of seed oil bodies from diverse species. Plant Physiol 101: 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance VB, Huang AH (1987) The major protein from lipid bodies of maize. Characterization and structure based on cDNA cloning. J Biol Chem 262: 11275–11279 [PubMed] [Google Scholar]
- Wang X, Shen S, Rasam SS, Qu J (2019a) MS1 ion current-based quantitative proteomics: A promising solution for reliable analysis of large biological cohorts. Mass Spectrom Rev 38: 461–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wei H, Mao X, Liu J (2019b) Proteomics analysis of lipid droplets from the oleaginous alga Chromochloris zofingiensis reveals novel proteins for lipid metabolism. Genomics Proteomics Bioinformatics 17: 260–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ma X, Zhang X, He X, Li H, Cui D, Yin D (2016) ITRAQ-based proteomic analysis of the metabolic mechanisms behind lipid accumulation and degradation during peanut seed development and postgermination. J Proteome Res 15: 4277–4289 [DOI] [PubMed] [Google Scholar]
- Xu E, Chen M, He H, Zhan C, Cheng Y, Zhang H, Wang Z (2017) Proteomic analysis reveals proteins involved in seed imbibition under salt stress in rice. Front Plant Sci 7: 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, He D, Gupta R, Yang P (2015) Physiological and proteomic analyses on artificially aged Brassica napus seed. Front Plant Sci 6: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Liu P (2019) The new face of the lipid droplet: Lipid droplet proteins. Proteomics 19: e1700223. [DOI] [PubMed] [Google Scholar]