A full-length transcriptome-based proteogenomic data set reveals the complexity of rice gene arrangement and the transcriptome’s coding ability.
Abstract
Rice (Oryza sativa) molecular breeding has gained considerable attention in recent years, but inaccurate genome annotation hampers its progress and functional studies of the rice genome. In this study, we applied single-molecule long-read RNA sequencing (lrRNA_seq)-based proteogenomics to reveal the complexity of the rice transcriptome and its coding abilities. Surprisingly, approximately 60% of loci identified by lrRNA_seq are associated with natural antisense transcripts (NATs). The high-density genomic arrangement of NAT genes suggests their potential roles in the multifaceted control of gene expression. In addition, a large number of fusion and intergenic transcripts have been observed. Furthermore, 906,456 transcript isoforms were identified, and 72.9% of the genes can generate splicing isoforms. A total of 706,075 posttranscriptional events were subsequently categorized into 10 subtypes, demonstrating the interdependence of posttranscriptional mechanisms that contribute to transcriptome diversity. Parallel short-read RNA sequencing indicated that lrRNA_seq has a superior capacity for the identification of longer transcripts. In addition, over 190,000 unique peptides belonging to 9,706 proteoforms/protein groups were identified, expanding the diversity of the rice proteome. Our findings indicate that the genome organization, transcriptome diversity, and coding potential of the rice transcriptome are far more complex than previously anticipated.
Rice (Oryza sativa) is a model monocot and one of the most important crop species globally. Functional studies using rice cultivars have been largely facilitated by the release of its genome sequences and subsequent transcriptomic profiling (Ouyang et al., 2007). The representative japonica (geng) rice genome was released in the early 21st century, and initial genome annotation was based on multiple approaches, including ab initio prediction, paralog comparison, and transcript libraries (e.g. cDNA and ESTs; Ouyang et al., 2007). In recent years, this annotation has been continuously updated using next-generation sequencing (short-read RNA sequencing [srRNA_seq])-based transcriptome data sets in popular databases such as Phytozome (Ouyang et al., 2007; Wang et al., 2018b).
When srRNA_seq became widespread during the past decade, pervasive transcription, a mechanism originally defined to generate unknown noncoding RNAs, was proposed for nearly all sequenced species (Mills et al., 2016). The complexity of the RNA landscape revealed by high-throughput sequencing techniques came as a major surprise. In particular, natural antisense transcripts (NATs), which were initially regarded as transcriptional noise, are among the most interesting elements (Mills et al., 2016). NATs are defined as a pair of transcription units located in different strands of DNA with overlapping loci coordinates (Pelechano and Steinmetz, 2013). This type of genomic organization was initially identified in viruses in 1969 (Bøvre and Szybalski, 1969) and was subsequently observed to be a common feature in prokaryotic bacteria and eukaryotic organisms (Wek and Hatfield, 1986; Wong et al., 1987). In recent years, comprehensive transcriptome studies have revealed an ever-increasing percentage of loci involved in this genomic organization, suggesting that NATs are highly prevalent in eukaryotes. According to a current research summary, approximately 50% to 70% of mammalian loci and 20% to 70% of plant loci have antisense transcripts in the opposite strand (Katayama et al., 2005). Although NATs have recently drawn increasing attention, their functional significance is only just beginning to be understood (Xu et al., 2017). In addition, the genomic arrangement of NATs reveals potential functional correlations between these gene pairs (Pelechano and Steinmetz, 2013). For example, NATs have been demonstrated to play crucial roles at both transcriptional and posttranscriptional levels under a variety of abiotic and biotic stresses (Werner, 2005), with described functions including roles in activating or silencing other members of NAT pairs (Prescott and Proudfoot, 2002; Modarresi et al., 2012), mRNA processing and splicing (Morrissy et al., 2011), the maintenance of RNA stability (Su et al., 2012), the direction of chromatin remodeling (Swiezewski et al., 2009), induction of the formation of small interfering RNA (Borsani et al., 2005), and translational control (Faghihi and Wahlestedt, 2009). Given the considerable number of NATs identified in animals and plants, it is perhaps unsurprising that the biological functions of most NATs remain to be elucidated by mechanistic studies.
In addition to NATs, specialized transcripts such as fusion genes have emerged from transcriptome studies and opened a new research horizon. By definition, fusion transcripts are chimeric mRNAs created by the fusion of parts of different genes. Fusion events commonly result from genomic translocation, chromosomal deletion, and inversion, or by trans-splicing mechanisms (Weirather et al., 2015). Trans-splicing, which is often observed in lower eukaryotes, had been considered rare in higher eukaryotic organisms (McManus et al., 2010). To date, the cellular functions of these transcripts have been well characterized in mammalian tumorigenesis (Edwards, 2010; Edwards and Howarth, 2012); however, cases in other higher eukaryotes, including plants, are rarely reported.
In comparison with NATs and fusion genes, posttranscriptional regulation methods such as alternative transcription start (ATS), alternative splicing (AS), and alternative polyadenylation (APA), as well as their resulting mRNA isoforms, have been well established in recent years (Abdelghany et al., 2016; Wang et al., 2016). It has been documented that 50% of genes have ATS, over 95% of genes exhibit AS, and 75% of genes have APA in humans (Pan et al., 2008; Reddy et al., 2013). Furthermore, approximately 15% of human diseases are caused by mutations that affect splicing machinery (Eckardt, 2013). Hence, these three mechanisms are proposed to interdependently expand the transcriptome coding ability and proteome diversity based on the limited information stored in eukaryotic genomes (Abdelghany et al., 2016). At the transcriptional level, the potential roles of ATS and APA in delicately controlling translation efficiency and mRNA stability are well documented (Reyes and Huber, 2018). Eukaryotic genes typically consist of multiple exons and introns. In vertebrates, an average of 7.8 to nine introns per gene have been observed (Mourier and Jeffares, 2003), suggesting that AS could greatly increase the repertoire of translated proteins involved in every aspect of developmental and environmental responses (Kalsotra and Cooper, 2011; Laloum et al., 2018). However, the question of whether a transcribed mRNA isoform can be translated is still under open debate (Tress et al., 2017). That said, a considerable number of isoforms have been found to be associated with ribosomes or proteins, as evidenced by proteomic studies, suggesting their coding potential under normal conditions or stress treatment (Zhu et al., 2017). Although a number of functional studies have characterized the mRNA isoforms in animals and plants in order to reveal their potential roles in signal transduction and cellular activities (Rühl et al., 2012; Duan et al., 2016; Hwang et al., 2018), the functional significance of the vast majority of isoforms remains poorly understood. In addition to transcriptional and posttranscriptional control, eukaryotes can further increase their coding potential to generate proteins or short peptides by using alternative open reading frames or small open reading frames located in the second or third frame of the same transcript, respectively. These translational mechanisms are defined as alternative translation initiation (ATI; Sonenberg and Hinnebusch, 2009). Additionally, the usage of non-AUG or noncanonical start codons has been demonstrated by parallel analysis of ribosome sequencing and proteomic profiling, further enhancing eukaryotic and prokaryotic genome coding potential, respectively (Ingolia et al., 2011; Menschaert et al., 2013; Bouthier de la Tour et al., 2015; Lomsadze et al., 2018).
Proteogenomics is an analytical approach to integrate genomic, transcriptomic, and proteomic data for comprehensive analysis. The first proteogenomic work was carried out in Arabidopsis (Arabidopsis thaliana) for its genome annotation (Castellana et al., 2008). Subsequently, this approach has been applied to the model legume Medicago truncatula and grapevine (Vitis vinifera; Volkening et al., 2012; Chapman and Bellgard, 2017). Proteogenomics has been carried out not only in plants but also in animals and microorganisms (Jaffe et al., 2004; Kumar et al., 2016; Locard-Paulet et al., 2016). In addition to aiding the curation of genome annotation, proteogenomics can be used to detect processed signal peptides, to identify specialized transcripts and their protein products, to discover protein maturation events, and to reveal leaderless mRNA and its mechanism during translation initiation (de Groot et al., 2014; Kucharova and Wiker, 2014).
The aforementioned genomic features and specialized transcripts are efficiently detected by srRNA_seq with sufficient sequencing depth. However, the main limitation of this technology is the dependence on bioinformatic assembly of transcripts from short sequencing reads (75–150 bp) by available computational tools (Conesa et al., 2016). For instance, although srRNA_seq can accurately detect AS events or splicing sites, it is challenging to determine the combinatory usage of splicing junctions or assemble full-length transcript isoforms and fusion transcripts using this method (Wang et al., 2016, 2018a). Furthermore, the lengths of transcripts assembled by srRNA_seq can be further limited by the computational algorithm, which subsequently leads to inaccurate annotation of gene models and their genomic coordinates. This seriously hampers the identification of NATs. With the development of technology for single-molecule long-read RNA sequencing (lrRNA_seq) from Pacific Biosciences, researchers are now able to obtain full-length transcripts as a single read without further assembly (Deveson et al., 2018). Recent transcriptome studies have demonstrated the utility of this technology in providing superior information on transcript isoforms in yeast, humans, and plants (Sharon et al., 2013; Abdel-Ghany et al., 2016; Wang et al., 2016, 2018a; Kuang et al., 2017). These studies have suggested that even in the highly characterized human transcriptome, the identification of genes and splice isoforms is far from complete (Sharon et al., 2013; Wang et al., 2016). In addition, most studies have been inspired by the diversity and complexity of various types of transcripts, such as splicing isoforms and fusion transcripts, or by posttranscriptional regulations such as ATS and APA, and little attention has been paid to the study of genomic arrangements, such as NATs. Furthermore, although studies have questioned the coding potential of these transcripts, no direct experiments have been carried out.
Recent studies have applied srRNA_seq-based proteogenomics on rice and lrRNA_seq for rice transcriptome analysis (Ren et al., 2019; Zhang et al., 2019). In this study, we performed a comprehensive analysis of lrRNA_seq-based transcriptome and proteomic data sets simultaneously to provide direct proteomic evidence for rice. In order to systematically characterize transcript isoforms, we chose six tissue types at different developmental stages from japonica (geng) rice ‘Nipponbare,’ including seeds, seedlings, roots, leaves, stems, and flowers, for library construction and lrRNA_seq. Meanwhile, parallel srRNA_seq using an Illumina HiSeq 4000 platform was carried out for comparison. We demonstrate that 58.5% of the genes form NAT pairs and 72.9% of the genes have transcript isoforms. This suggests that lrRNA_seq has a superior ability to reveal complex genomic arrangements and transcriptome dynamics. Furthermore, the coding potential and characteristics of the rice transcriptome and proteome were assessed using both data sets alongside parallel qualitative proteomic experiments and data entries in public databases. Our findings indicate that it is common for rice transcripts to not only use all three frames to encode proteins but to also use multiple transcripts to encode a single protein. In summary, our data demonstrate that the lrRNA_seq-assisted proteogenomic approach can be applied to eukaryotic organisms in order to identify genomic arrangement, transcriptome diversity, and coding ability, which complements current transcriptomic approaches and contributes to a better understanding of the systems-level control of a wide range of biological processes.
RESULTS
Analytical Pipeline of lrRNA_Seq-Based Proteogenomics
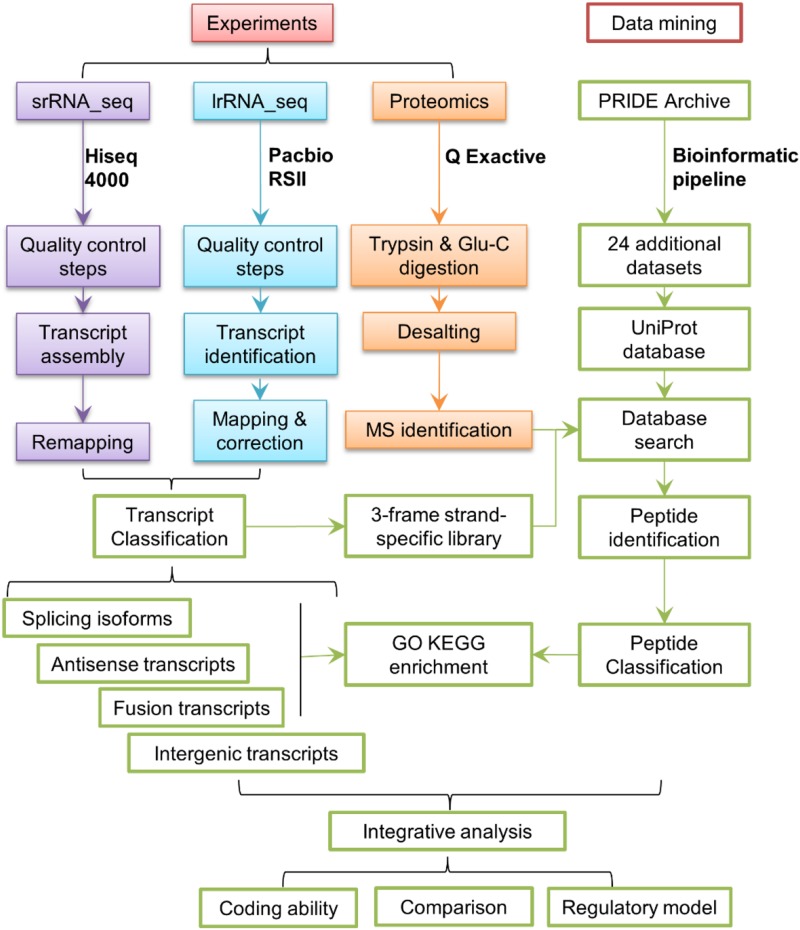
A schematic view of the analytical pipeline used in this study is shown in Figure 1, which was modified based on a previous study in Arabidopsis (Zhu et al., 2017). Since transcripts of srRNA_seq and lrRNA_seq were assembled by different bioinformatic pipelines (Supplemental Tables S1 and S2), we remapped the assembled srRNA_seq transcripts together with lrRNA_seq transcripts using GMAP (Abdel-Ghany et al., 2016) for normalization. The resulting gff files were used for subsequent specialized transcript identification and comparison between these two data sets. Pipeline refinements upon the identification of AS events, fusion and intergenic transcripts, and NATs were conducted as detailed in “Materials and Methods.” Proteomic profiling was conducted similarly to previous protocols with minor modification by using a second digestion enzyme, Glu-C, as an independent method to improve protein coverage. In addition, 24 protein data sets deposited in the Proteomics Identifications (PRIDE) archive were added for the subsequent peptide search. Due to the usage of a strand-specific library, a three-frame library was constructed instead of the six-frame library used in previous studies, which consequently halved the computing power required for the database search. Integrative analysis, such as coding ability assessment and comparison between srRNA_seq and lrRNA_seq, was carried out using methods custom made for this study.
Figure 1.
Schematic view of the experimental and analytical pipelines used in this study. srRNA_seq and lrRNA_seq were performed by using HiSeq 4000 and Pacific Biosciences RSII platforms. Proteomic analysis was performed by using the Q-Exactive platform. Data mining was carried out by using data sets deposited online. Major steps of the analytical pipeline are shown.
General Features and Transcript Identification
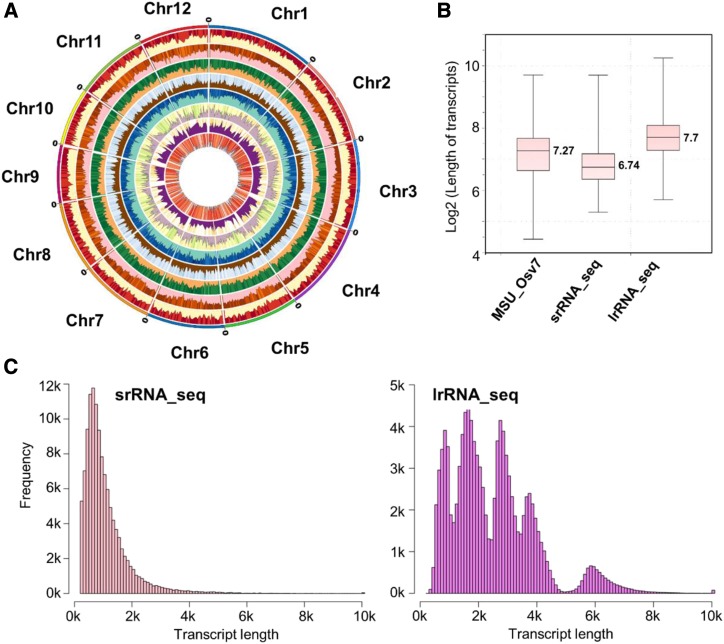
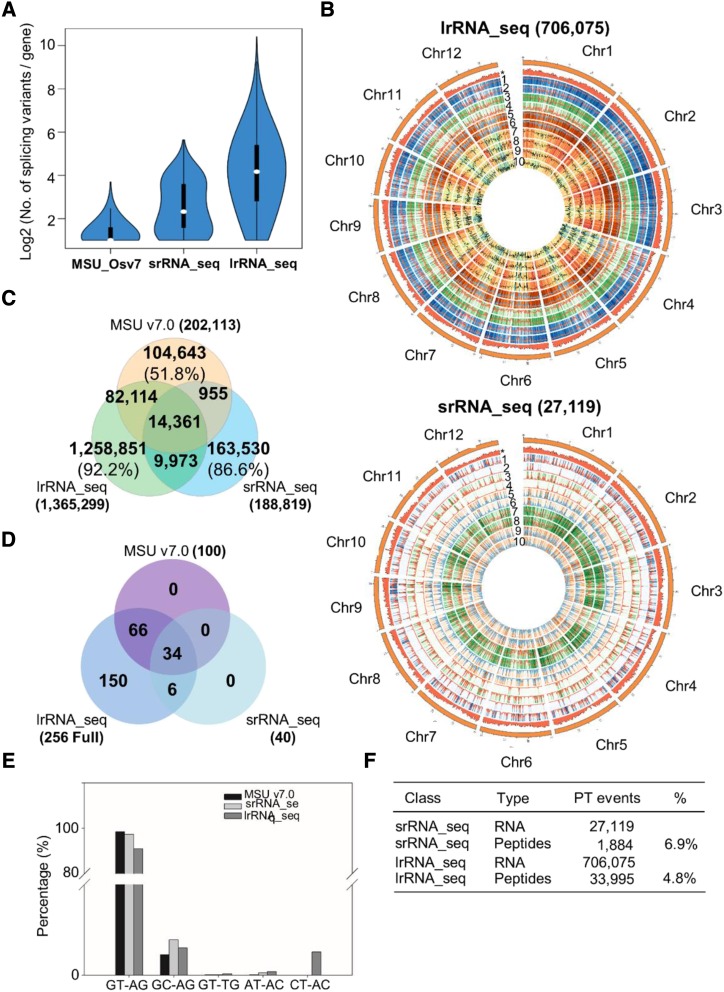
To ensure the coverage and identification of low-abundance transcripts, both srRNA_seq and lrRNA_seq were conducted with sufficient sequencing depth (Supplemental Tables S1 and S2). In general, lrRNA_seq is superior to srRNA_seq in transcript identification and characterization. A total of 120,958 and 1,100,036 unique transcripts were identified by srRNA_seq and lrRNA_seq, respectively (Table 1). Subsequently, 120,905 transcripts from srRNA_seq and 906,456 transcripts from lrRNA_seq were mapped to 15,451 and 34,674 loci, respectively (Table 1). In comparison with the Phytozome rice annotation database (MSU_Osv7), srRNA_seq detected 65,723 unannotated transcripts from 5,686 unannotated loci, whereas lrRNA_seq identified 102,614 transcripts from 11,023 unannotated loci. For transcript isoform identification, 6,384 loci with 16,617 splice isoforms were recorded in the current rice annotation. srRNA_seq assembled 104,942 isoforms from 13,745 loci, with 6,540 of these loci being present in the current rice annotation. lrRNA_seq identified 867,136 isoforms from 32,780 loci, with over eight times more transcripts and a 2.4-fold increase in loci characterization (Table 1). Additionally, 52,840 transcripts from 7,205 unannotated loci and 65,942 transcripts from 7,505 unannotated loci were identified by srRNA_seq and lrRNA_seq, respectively (Table 1). With regard to specialized transcripts, lrRNA_seq identified 11 times, 6.5 times, and 3.6 times more NATs, fusion transcripts, and intergenic transcripts than srRNA_seq, respectively (Table 1). The genome-wide coverage and frequency of the aforementioned transcripts are shown in a Circos diagram (Fig. 2A). In addition to the advantage of detecting a much greater number of transcripts, lrRNA_seq was additionally better at finding longer transcripts due to its longer read length. For example, the median value of the transcript length from srRNA_seq was 845 bp, whereas this value reached 2,206 bp for lrRNA_seq-identified transcripts (Fig. 2B). This further increased the median length of transcripts in the current rice annotation from 1,435 to 2,206 bp. Similar results can be obtained by comparing the length distribution of the total transcripts generated by both RNA sequencing techniques (Fig. 2C), suggesting that a greater number of longer transcripts (>5 kb) were characterized using lrRNA_seq.
Table 1. Comparison of the existing database with srRNA_seq and lrRNA_seq.
| Type | MSU_Osv7 | srRNA_Seq | lrRNA_Seq | Fold |
|---|---|---|---|---|
| Traditional gene models | ||||
| No. of loci | 42,189 | 15,451 | 34,674 | 2.24 |
| No. of mapped transcripts | 52,424 | 120,950 | 906,456 | 7.49 |
| Novel loci | 0 | 5,686 | 11,023 | 1.94 |
| Novel transcripts | 0 | 65,723 | 102,614 | 1.56 |
| Unmapped transcripts | 0 | 8 | 193,580 | – |
| No. of transcripts (total) | 52,424 | 120,958 | 1,100,036 | 9.09 |
| Loci with splicing variants | 6,384 | 13,745 | 32,780 | 2.38 |
| Total splicing isoforms | 6,384 | 104,942 | 867,136 | 8.26 |
| MSU loci with splicing variants | 6,384 | 6,540 | 20,142 | 3.08 |
| MSU splicing variants | 16,617 | 52,102 | 801,194 | 15.38 |
| Novel loci with splicing variants | – | 7,205 | 7,505 | – |
| Novel splicing variants | – | 52,840 | 65,942 | – |
| Specialized transcripts | ||||
| NATs | 21,759 | 78,833 | 899,359 | 11.41 |
| Fusion transcripts | 0 | 1,192 | 7,804 | 6.55 |
| Intergenic transcripts | 0 | 28,422 | 31,095 | 1.09 |
Figure 2.
Comparison of transcript properties between srRNA_seq and lrRNA_seq. A, Circos diagram of specialized transcripts identified by srRNA_seq and lrRNA_seq. 1, Total transcripts identified by srRNA_seq; 2, total transcript identified by lrRNA_seq; 3, intergenic transcripts identified by srRNA_seq; 4, intergenic transcripts identified by lrRNA_seq; 5, NATs identified by srRNA_seq; 6, NATs identified by lrRNA_seq; 7, fusion transcripts identified by srRNA_seq; 8, fusion transcripts identified by lrRNA_seq. B, Box plot of transcript lengths summarized in the three data sets using MSU_Osv7 annotation, srRNA_seq, and lrRNA_seq. C and D, Histogram plots showing the frequency of transcript lengths between srRNA_seq (C) and lrRNA_seq (D).
Comparative Analysis of Fusion and Intergenic Transcripts
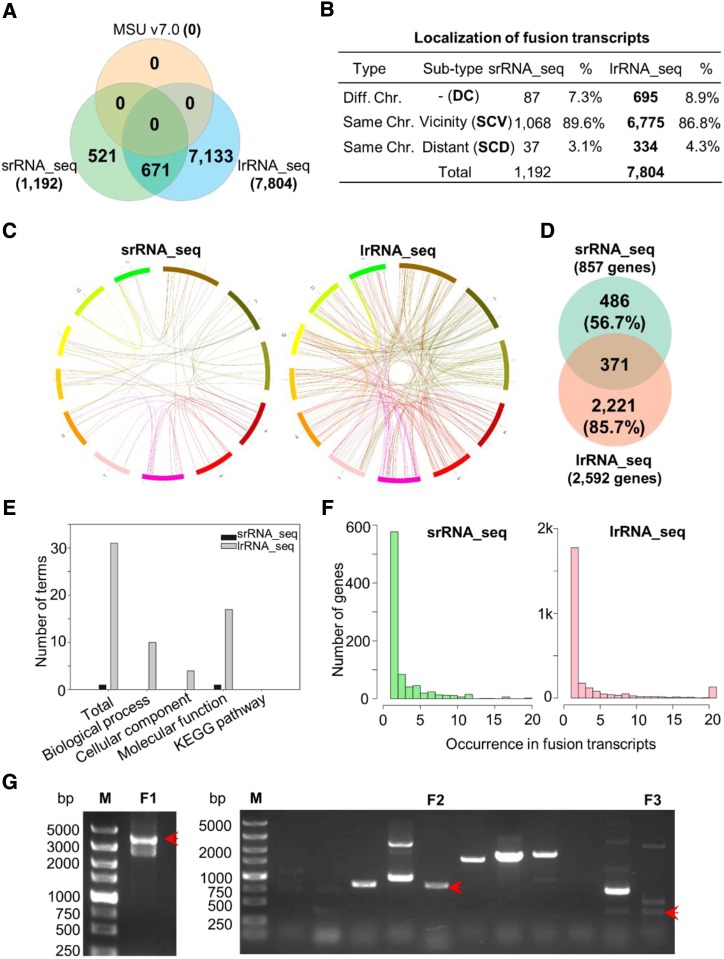
Single-molecule transcriptome analysis in humans and plants has demonstrated that transcript fusion events appear to be more common than previously thought (Weirather et al., 2015; Wang et al., 2016). Given that these chimeric transcripts are able to further expand the transcriptional diversity in eukaryotic genomes, we additionally analyzed fusion transcripts in our rice samples. The identification of fusion events by srRNA_seq is questionable in its reliability due to the number of assembly steps required. Although lrRNA_seq identified seven times more fusions than did srRNA_seq, a considerable amount of srRNA_seq-identified fusions were validated by lrRNA_seq (Fig. 3A). Subtype statistics revealed that most of the identified chimeric transcripts (∼90%) were intrachromosomal fusions, resulting from the joining of two adjacent genes (Fig. 3B; Supplemental Fig. S1A). Only a small proportion of transcripts (∼4%) and genes (∼10%–15%) were detected to be interchromosomally fused by both sequencing approaches (Fig. 3B; Supplemental Fig. S1A), which is similar to the results obtained previously in cancer cells (Okonechnikov et al., 2016). Moreover, no preference of chromosome usage could be observed within the identified fusion transcripts (Fig. 3C). In total, 857 and 2,592 fusion-related genes were identified, respectively, by srRNA_seq and lrRNA_seq, with approximately 56.7% and 85.7% uniquely identified by each sequencing approach (Fig. 3D). Among these transcripts, the majority consisted of two genes, and approximately 1.5% and 2.8% consisted of three genes in the srRNA_seq and lrRNA_seq data sets, respectively (Supplemental Fig. S1B). Furthermore, the internal organization of the fusion transcripts determined using the sense or antisense strand varied between these two data sets (Supplemental Fig. S1C). With a higher number of identified transcripts, more Gene Ontology (GO) terms were enriched in the lrRNA_seq data set (Fig. 3E; Supplemental Table S3). In addition, some genes were found at a high frequency as important building blocks for the construction of a variety of fusion transcripts (Fig. 3F) and may hence play pivotal biological functions. Three fusion transcripts identified by lrRNA_seq were validated by reverse transcription quantitative PCR (RT-qPCR) and subsequent DNA sequence analysis (Fig. 3G), confirming our confidence of this approach in fusion transcript identification.
Figure 3.
Comparative analysis of fusion transcripts. A, Venn diagram showing the overlapping and unique fusion transcripts identified by srRNA_seq and lrRNA_seq. B, Summary of fusion transcript subtypes. C, Circos representations of fusion transcripts consisting of two genes. D, Venn diagram presenting the overlapping and unique genes involved in fusion transcript formation. E, GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of fusion genes. F, Loci frequency present in fusion transcripts. G, RT-qPCR validation of fusion transcripts. Red arrows indicate the validated fusion transcripts. F1 to F3, Three fusion transcripts; M, marker.
Intergenic transcripts are transcripts mapped to intergenic regions that are frequently regarded to be noncoding transcripts (Chang et al., 2014). Interestingly, here the number of transcripts identified by the two methods was highly similar: 28,422 and 31,095 intergenic transcripts were identified by srRNA_seq and lrRNA_seq, respectively. Their potential coding abilities were assessed by classic long noncoding RNA (lncRNA) analysis. In general, 5,364 and 5,637 transcripts were considered to be lncRNA according to previous descriptions (Supplemental Fig. S2; Chang et al., 2014). However, determination of whether they can be translated or not requires further protein evidence.
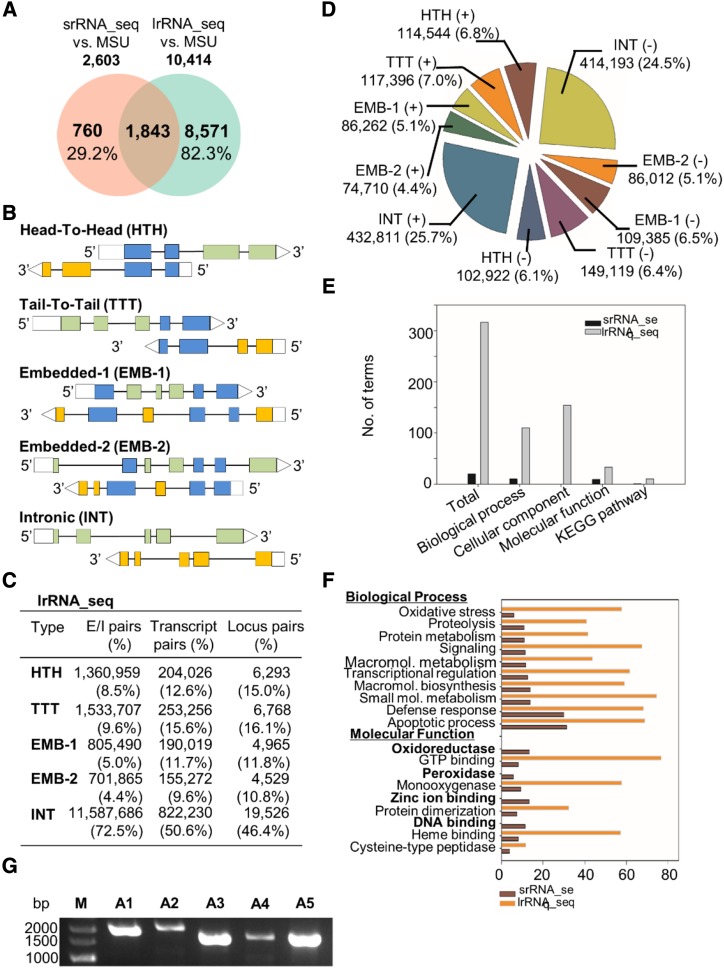
NATs Reveal the Complex Linear Arrangement of the Rice Genome
A previous report stated that by using tilling arrays, approximately 23.8% of annotated rice genes could be identified as NATs (Li et al., 2006). Here, using innovative lrRNA_seq with its wide coverage of transcripts, we were able to classify 58.5% of the annotated genes as NATs (Table 1). A total of 2,603 and 10,414 NAT genes identified by srRNA_seq and lrRNA_seq, respectively, overlapped with the current rice annotation (Fig. 4A). Furthermore, we summarized the previous categorization of NATs into five subtypes based on their relative orientations and regions of overlap (Fig. 4B; Yuan et al., 2015): head-to-head, tail-to-tail, embedded-1, embedded-2, and intronic. These five subtypes were further assessed at three levels: exon/intron pairs, transcript pairs, and locus pairs (Fig. 4C; Supplemental Fig. S3, A and B). Among these, intronic was the most abundant type in all three data sets, whereas the other four subtypes were present in comparable percentages (Fig. 4C; Supplemental Fig. S3, A and B). In addition, a different statistical approach was used to characterize the NAT subtypes as sense or antisense strands. No preference of strand usage for NATs was observed in the lrRNA_seq data set (Fig. 4D). Although srRNA_seq-identified NATs were uniquely enriched in several GO terms such as oxidoreductase, zinc ion binding, and DNA binding, lrRNA_seq-identified NATs were much more enriched in GO and KEGG terms (Fig. 4, E and F; Supplemental Fig. S3C; Supplemental Tables S4 and S5) due to the higher number of identified transcripts and NAT genes resulting from the use of this innovative technology. Five of these transcripts were validated by an independent RT-qPCR analysis (Fig. 4G), proving the validity of our approach in the identification of NATs.
Figure 4.
Comparison of NATs identified by srRNA_seq and lrRNA_seq. A, Venn diagram showing the overlapped and unique transcripts present in the current annotation in comparison with the srRNA_seq and lrRNA_seq data sets. B, Classification of five subtypes of NATs. C, Summary of NATs identified by lrRNA_seq at the levels of exon/intron pairs, transcript pairs, and locus pairs. D, Summary of NAT subtypes in two strands of genomic DNA. E and F, GO and KEGG enrichment analyses of NATs. G, RT-qPCR validation of antisense transcripts. A1 to A5, Antisense transcripts; M, marker.
Diversity of Posttranscriptional Events and Splicing Site Usage
An increasing number of reports indicate that posttranscriptional events, such as ATS, AS, and APA, are coordinately responsible for the majority of transcript diversity (Reyes and Huber, 2018). As described previously, lrRNA_seq presented the most diverse and abundant transcript isoforms in comparison with srRNA_seq and the current rice genome annotation (Fig. 5A). A total of 27,119 and 706,075 posttranscriptional events were identified in the srRNA_seq and lrRNA_seq data sets, respectively (Supplemental Fig. S4). In comparison with srRNA_seq, the lrRNA_seq results had a higher number of posttranscriptional events both on a per transcript and per locus basis (Supplemental Fig. S4A). Previously, we proposed that two types of AS events, named alternative first exon (AFE) and alternative last exon (ALE), are the two most abundant AS events in rice and Arabidopsis (Zhu et al., 2017). Some of these AS types were coordinated by non-AS events, such as ATS in AFE or APA in ALE. Thus, we further defined these two events by removing events purely caused by ATS and APA at diverse genomic positions (Supplemental Table S6; i.e. AFE was a type of posttranscriptional event with coordinative effects between ATS and AS, whereas ALE was a combined posttranscriptional event with APA and AS). Hence, in addition to traditional AS types, 10 posttranscriptional events were defined in this study to facilitate further analysis. Circos representation suggested that lrRNA_seq was powerful for identifying these genome-wide posttranscriptional events with a higher frequency and density than that afforded by the srRNA_seq (Supplemental Fig. S5B). However, the compositions of these events varied between the two sequencing techniques. Four AS types, intron retention (IR), multiple intron retention (MIR), exon skipping (SKIP), and multiple exon skipping (MSKIP), were increased in percentage in the lrRNA_seq results (Supplemental Fig. S4, B and C), suggesting that the longer read length of lrRNA_seq may greatly facilitate the identification of these four AS types. By contrast, four posttranscriptional events, ATS, APA, AFE, and ALE, were largely reduced in percentage within the lrRNA_seq data sets (Supplemental Fig. S4, B and C), suggesting that they were overrepresented in the srRNA_seq due to the inability to detect all AS types.
Figure 5.
Identification of ATS, AS, and APA. A, Violin plot of splicing variants identified in MSU_Osv7 annotation, srRNA_seq, and lrRNA_seq. B, Circos representations of posttranscriptional events identified in srRNA_seq and lrRNA_seq. *, Density of transcripts recorded in MSU_Os7 annotation; 1, IR; 2, MIR; 3, SKIP; 4, MSKIP; 5, alternative exon 5′ (AE5′); 6, alternative exon 3′ (AE3′); 7, ATS; 8, APA; 9, AFE; 10, ALE. C to E, Exon comparisons (C), paired splicing site comparisons (D), and statistical analysis of paired splicing sites (E) among MSU_Os7 annotation, srRNA_seq, and lrRNA_seq. F, Summary of identified posttranscriptional events and peptides in srRNA_seq and lrRNA_seq.
In addition to alternative spliced isoform analysis, we further compared all exons annotated in the three data sets (Supplemental Fig. S5C). The current rice annotation, srRNA_seq, and lrRNA_seq annotated 202,113, 188,819, and 1,365,299 exons, respectively. Approximately 86.6% and 92.2% of exons were uniquely present in data sets of srRNA_seq and lrRNA_seq, respectively (Supplemental Fig. S5C), highlighting the complexity of the posttranscriptional control of mRNA. Traditionally, the choices of splice sites are recognized to strongly contribute to exon variability (Zhu et al., 2017). Thus, we performed single splice site analysis to reveal the genome-wide splice site conservation. Similar to previous results (Chen et al., 2019b), the conventional 5′-splice site (5′-ss; GT) was present at approximately 60% in both srRNA_seq and lrRNA_seq. However, the percentage of conventional 3′-splice site (3′-ss; AG) was largely reduced in the lrRNA_seq data sets, along with an increase in all types of nonconventional 3′-ss sequences (Supplemental Fig. S4D), implying that these nonconventional 3′-ss are more likely to be detected in lrRNA_seq with its longer read length. Thus, both 5′-ss and 3′-ss were less conserved (Supplemental Fig. S4E) than previously anticipated, suggesting a higher variability in the splice choices than previously envisaged in eukaryotic genomes. Therefore, we employed a paired splice site assay to locate 5′-ss/3′-ss positions and sequences simultaneously at a single intron. Findings from this analysis suggested that the Phytozome annotation exhibited 100 and srRNA_seq had 40 types of 5′-ss and 3′-ss sequence combinations (Fig. 5D). Surprisingly, all 256 combinations of splice site sequences were observed in the lrRNA_seq data set. Another interesting finding was that, besides conventional U2 (GT-AG) and U12 (AT-AC) complexes, a third splicing combination (GC-AG) accounted for a considerable percentage in all the splice sites identified in this assay (Fig. 5E). However, the underlying mechanisms and responsible protein complex of this combination remain to be elucidated. Furthermore, proteomic identification using the AS event library suggested that approximately 6.9% (1,884) and 4.8% (33,995) of posttranscriptional events identified from srRNA_seq and lrRNA_seq could be translated to peptides (Fig. 5F). This number is slightly lower in comparison with previous examples reported in Arabidopsis and rice (Zhu et al., 2017; Chen et al., 2019b).
Proteogenomic Analysis Suggests Multiple Mechanisms for Enhancing Genome Coding Ability
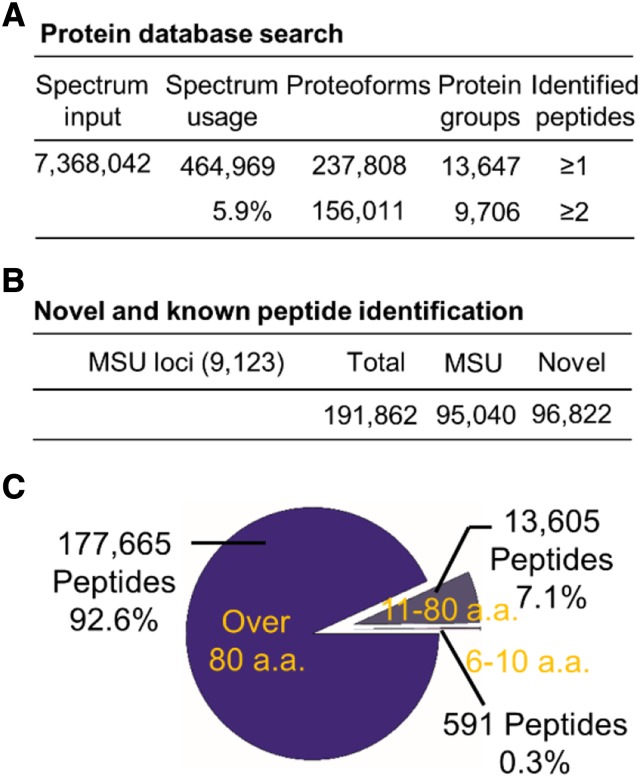
The pervasive transcription of eukaryotic genomes has been documented for years, but whether these transcripts can be translated is still a matter of debate (Jensen et al., 2013; Wade and Grainger, 2014). To address this question, we conducted large-scale profiling of the rice proteome to assess the potential coding ability of the rice genome. Together with 24 previously published data sets (Supplemental Table S7), 7,368,042 spectra were included in the initial input file (Fig. 6A). Approximately 5.9% (464,969 spectra) were positively matched to peptide sequences from a customized three-frame translated database generated by combining both srRNA_seq and lrRNA_seq transcripts (Fig. 6A). In total, 9,706 proteoforms/protein groups (false discovery rate < 0.01; Meier et al., 2018) were identified with at least two peptide sequences (Fig. 6A). In general, 191,862 peptides were found to be translated from annotated loci and unannotated loci with at least two unique peptide sequences for each loci (Fig. 6B; Nesvizhskii, 2014). Among these, 92.6% of the peptides were found to be regular proteins larger than 80 amino acids (Fig. 6C), ∼6.6% of peptides belonged to small proteins between 11 and 80 amino acids, and ∼0.3% of peptides were from small peptide-encoding loci (six to 10 amino acids; Fig. 6C).
Figure 6.
Assessment of coding potential by proteogenomics. A, Basic parameters used in proteomic database search. B, Summary of known and unannotated peptides. C, Distribution of identified proteoforms/protein groups and peptides. a.a., Amino acids.
DISCUSSION
In the past decade, srRNA_seq has become an essential technique for characterizing eukaryotic transcriptomes. Given the complexity of eukaryotic transcriptomes, using srRNA_seq is akin to putting pieces of a jigsaw puzzle together to see the whole picture. Thus, the development of computational algorithms for reliable full-length transcript reconstruction represents a major challenge (Steijger et al., 2013; Tilgner et al., 2013). By contrast, lrRNA_seq has a number of advantages that may allow it to supersede srRNA_seq. For example, the production of nearly full-length reads greatly reduces the computing power required for transcript assembly. Simultaneously, lrRNA_seq is powerful for revealing the higher complexity of eukaryotic genomes and has become the gold standard for genome reannotation due to its wide coverage of full-length transcriptomes (Sharon et al., 2013; Wang et al., 2016). In addition, as lrRNA_seq is a long-read-directed technology, it will facilitate the discovery of long transcripts and low-abundance sequences (Wang et al., 2016). However, both srRNA_seq and lrRNA_seq are able to uniquely identify a batch of transcripts (Figs. 3A, 4A, and 5C). For this reason, we maximized the sampling diversity by using rice samples at different developmental stages to ensure transcript coverage. We also used srRNA_seq as a complementary data set in parallel with the lrRNA_seq-based proteogenomic analysis. In this way, we analyzed the rice transcriptome with sufficient depth and transcript length (Supplemental Tables S1 and S2). This data set has the potential to become a useful resource for studying transcriptional and posttranscriptional regulation and genome annotation or to provide database updates. This is exemplified by the fact that it allowed the discovery of a large number of unannotated genes, along with their AS isoforms and coding proteins, suggesting their authenticity as protein-coding loci. Furthermore, the expansion of the transcript population may facilitate biological interpretation during developmental processes and stress responses (Figs. 3E and 4E) by leading to the discovery of unannotated structural or regulatory components of such processes.
The Universality of NATs Implies High Complexity and Divergence in Transcriptional and Posttranscriptional Regulation
Using srRNA_seq approaches, studies have demonstrated that NATs are universal components of eukaryotic genomes (Balbin et al., 2015), participating in diverse biological processes and stress responses (Xu et al., 2017). Previously, approximately 20% of the genes in rice were thought to be NATs (Li et al., 2006). In this study, we found that nearly 60% of genes can be classified as NAT pairs, suggesting the superior coverage of lrRNA_seq in NAT identification (Table 1). Furthermore, since some NATs could be involved in multiple NAT pairs and a large number of transcript isoforms were identified by lrRNA_seq, the ratio of NATs to NAT pairs is much larger than 2:1, suggesting that in excess of 30% of the rice genome is represented by NATs. Hence, our findings represent the most comprehensive study of antisense transcripts in rice according to current transcriptome analyses. Given the large percentage of genes that have at least one antisense sequence, several regulatory mechanisms have been proposed. For example, studies in both animals and plants have suggested that NATs are connected to chromatin modifications (Modarresi et al., 2012). In particular, deposition of the transcriptional repressive marker H3K27me3 is a prerequisite to activate the expression of COOLAIR, an antisense gene of the flowering locus FLC (Swiezewski et al., 2009). Additionally, small interfering RNA generation sites have been found to be clustered in overlapping genomic regions of NATs (Borsani et al., 2005; Zhang et al., 2013), suggesting a role for NATs in regulating small RNA biogenesis.
In some studies, NATs are classified into three categories according to their coding ability (Wang et al., 2014). Most NATs are considered to be noncoding loci as reported by genome-wide studies in animals and plants (Katayama et al., 2005; Wang et al., 2014). However, low coding potentials demonstrated by previous research, largely based on prediction and examples of protein-encoding antisense genes, have also been documented (Suenaga et al., 2014). Our previous proteogenomic work in Arabidopsis identified 960 potential NATs with coding ability, and a majority of these genes were not annotated (Zhu et al., 2017). There is no comprehensive proteomic assessment of the bona fide coding ability of rice NATs. Here, we identified 200,830 proteins potentially encoded from 899,359 NATs using lrRNA_seq-assisted proteogenomics, accounting for approximately 84.5% of identified proteoforms. This result suggests that these NATs do indeed have considerable coding ability in rice.
As described earlier in this article, pervasively transcribed NATs are able to regulate gene expression via both transcriptional and posttranscriptional mechanisms (Pelechano and Steinmetz, 2013). Therefore, the niche of a particular NAT pair needs to be taken into account as a whole unit in functional studies. This is particularly the case in the use of T-DNA or CRISPR mutants in plant functional genomics, where T-DNA insertion or CRISPR editing will likely affect multiple NAT loci in close vicinity to the target gene. This scenario will be further complicated when these NATs contain transcript isoforms. Furthermore, some antisense transcripts may have trans-functions in genes or gene products different from those of their sense partner (Camblong et al., 2009), leading to a more complicated scenario. Thus, a comprehensive pipeline for systematic characterization of NAT function should be developed for both animals and plants. Bioinformatic tools are needed for functional annotation and conservation evaluation of NATs among eukaryotic organisms (Pelechano and Steinmetz, 2013). Importantly, the modification of specific gene expression by its antisense transcripts could be developed into a potential technique as our understanding of NAT regulation improves (Modarresi et al., 2012). In summary, the regulatory mechanisms of NATs will likely become routine research topics in future functional studies across eukaryotic organisms. Progress in this field will help yield deeper understanding of gene regulation, interactions among close or overlapping loci, and the evolution of the genomic arrangement and decoding process.
The Diversity of Transcript Isoforms Expands the Complexity of the Regulatory Hierarchy from Transcription to Posttranscription
The posttranscriptional mechanisms responsible for generating transcript isoforms have been extensively investigated (Zhu et al., 2017; Reyes and Huber, 2018). Recent advancement in this field indicates that together with AS, ATS and APA coordinately contribute to the diversity of transcript isoforms, especially in humans (de Klerk and ’t Hoen, 2015). Thus, comprehensive analysis including these posttranscriptional events has been carried out in this study. Here, we have classified these posttranscriptional events into 10 subtypes (Fig. 5B; Supplemental Fig. S4, B and C). Among these subtypes, six (IR, MIR, SKIP, MSKIP, AE5′, and AE3′) were pure AS events. Two events, namely ATS and APA, were pure posttranscriptional regulations. The remaining two events, AFE and ALE, were a combination of ATS/AS and APA/AS, respectively (Reyes and Huber, 2018). These findings are different from examples in animal studies, where ATS and APA contribute to isoform diversity more than AS (de Klerk and ’t Hoen, 2015; Anvar et al., 2018). ATS- and APA-related events only accounted for 13% of the total posttranscriptional events in the rice lrRNA_seq data set (Supplemental Fig. S4C). By contrast, intron-retention events, IR and MIR, accounted for 56.5% of the total posttranscriptional events, further demonstrating the important function of lrRNA_seq in modeling rice transcript diversity. SKIP/MSKIP and AE5′/AE3′ accounted for 16.3% and 14.1% of the total posttranscriptional events, respectively (Supplemental Fig. S4C). However, the underlying mechanism of these event types in regulating transcript diversity remains unclear.
Given that AS has a major contribution (greater than 85%) to the transcript diversity of the rice transcriptome, the mechanism for splice site selection was further analyzed. Conventionally, two types of spliceosome responsible for splice site identification have been reported. One is defined as a U2 complex with canonical sequences of GT (5′-ss) and AG (3′-ss), and the other is named as a U12 complex with canonical sequences of AT (5′-ss) and AC (3′-ss; Lorkovic et al., 2005; Will and Lührmann, 2011). Previous srRNA_seq-based transcriptome studies have indicated that U2 complex sequences accounted for approximately 99% of the total identified splice sites, showing a high degree of conservation (Will and Lührmann, 2011). However, by using lrRNA_seq, we suggested that 91% of the total splice sites with GT-AG pair sequences (Fig. 5E) were possibly processed by conventional U2 splicing machinery, whereas the single GT and AG percentage dropped to 60% in AS transcripts (Supplemental Fig. S4D), indicating that alternatively spliced transcripts may prefer to use noncanonical splice sites. Furthermore, two new pair sequences, GC-AG and CT-AC, were found to account for 1.5% and 1.3% of the total splice sites, respectively. This value is much higher than that of the minor U12 splicing complex (∼0.2%) in the lrRNA_seq data set (Fig. 5E), suggesting the presence of an uncharacterized splicing complex or recognition mechanisms. Proteins that can directly bind RNA sequences to regulate the splice site recognition process are defined as splicing factors (Kalyna et al., 2006). Previous biochemical and structural analyses have demonstrated that U1 and U2/U6 complexes may be responsible for the selection of splice site sequences (Golovkin and Reddy, 1996; Shi, 2017). In comparison with Arabidopsis (Zhu et al., 2017), rice splice sites showed less conservation at both 5′ and 3′ positions (Supplemental Fig. S4D). Subsequent evaluation of splicing-related proteins suggested that rice splice components exhibit more splice isoforms than do those of Arabidopsis (Supplemental Fig. S5), implying that rice may have a higher complexity of splicing machinery and corresponding splicing mechanisms. However, the exact mechanism of this molecular process remains to be further investigated in various plant developmental stages and under conditions of stress.
lrRNA_Seq-Based Proteogenomics Expand Current Knowledge of Protein Translation and Transcript Classification
Transcript isoforms have been profiled by either by srRNA_seq or lrRNA_seq in a number of eukaryotic organisms. However, whether these isoforms can be truly functional at the protein level is still under debate. Although case studies have demonstrated the specific functions of transcript isoforms in animals and plants (Wang et al., 2015; Hwang et al., 2018), several reports have proposed that the majority of these isoforms will not be translated and will be degraded by RNA surveillance (Bitton et al., 2015). Thus, the roles of these transcript isoforms have been suggested to be similar to those of noncoding transcripts (Kuang et al., 2017). In addition, another hypothesis has been proposed suggesting that these isoforms may function as a reservoir of divergent transcripts for the evolution of new genes or neofunctionalization of existing genes (Wu et al., 2011). To assess the coding ability of these isoforms, we applied a proteogenomic analytical pipeline based on the combined data sets from both srRNA_seq and lrRNA_seq. In total, we identified 191,862 peptides of 9,706 proteoforms/protein groups from three-frame translations of 906,456 transcripts (Table 1; Fig. 6A). Previous results have indicated that thousands of unannotated proteins can be identified using self-constructed protein databases translated from srRNA_seq-assembled transcripts (Zhu et al., 2017; Chen et al., 2019b). Similarly, an additional 96,822 unannotated peptides were translated from unannotated coding loci (Fig. 6B). These unannotated proteins will not be detected using the conventional Uniprot protein database, indicating the superior power of proteogenomics in unannotated protein identification. In addition, previously defined lncRNA may have the ability to encode proteins or peptides (Supplemental Fig. S2), and a large number of splicing isoforms may not be translated. Furthermore, transcripts could be translated using alternative frames or under various developmental/stress conditions. Due to the limited throughput and coverage of current mass spectrometry (MS)-based proteomics, we estimate that a large number of proteins or peptides remain to be discovered. Given the complexity of genome coding ability, we propose that caution should be taken in defining noncoding transcripts. Additional criteria may be needed to accurately classify coding and noncoding transcripts in the future.
Proteogenomics Facilitates Decoding of Eukaryotic Genome
Proteogenomics has long been used for omics-based comprehensive analysis in eukaryotic organisms (Castellana et al., 2014; Zhang et al., 2014). Single profiling techniques, such as transcriptome or proteome, will be reinforced when they are integrated into one proteogenomic pipeline. For instance, pure transcriptomic data do not provide direct evidence to assess the coding ability of the corresponding transcript isoforms. In contrast, single proteomic identification is usually composed of incomplete information for genome annotation. Hence, integrative analysis using both transcriptome and proteome data are more likely to identify dynamic variant proteins encoded by transcript isoforms and unannotated proteins encoded by ATI under specific conditions, providing additional insight into eukaryotic genome coding abilities in response to external stimuli (Zhang et al., 2016). However, the analytical pipeline requires further improvement by using emerging innovative biotechnologies. For example, the high error rate of lrRNA_seq restrains further construction of three-frame protein databases. Thus, enhancement of sequencing accuracy is the basis for improving whole-genome reannotation. Furthermore, using one combined library (e.g. 0.5–10 k) instead of five separate libraries will increase the coverage of transcripts, especially for sizes between the current library selection boundaries (Fig. 2C). Moreover, the lower quantification accuracy of lrRNA_seq and current MS-based proteomics results from their relatively low throughput and coverage. Therefore, solving the problems of complex isoform quantification at both transcriptional and protein levels will deepen our insights into the eukaryotic decoding process. Last, but not least, in previous studies, approximately 40% to 50% of raw spectra could be used by searching against either the Uniprot or frame database (Zhu et al., 2017; Chen et al., 2019b). Similarly, in this work, we used the same stringent criteria as two previous works for database search. The low spectra usage in this study may be due to the incompatibility of raw data generated by different tandem mass spectrometry (MS/MS) platforms and the search engine ProteinPilot v5.0 developed by AB Sciex. Subsequently, we have applied two additional databases, the Uniprot protein database (downloaded on November 9, 2018; 97,832 entries) and the AS events library (21,015,710 entries), for protein/peptide identification under the same searching criteria. Approximately 10.8% and 5.1% of the total spectra were matched to the Uniprot and AS events database, respectively. It appears that the spectra usage of the frame database (5.9%; 310,391,750 entries) and the newly prepared AS events database (5.1%) was slightly lower than the traditional Uniprot database, suggesting the validity of our database search criteria for protein identification. However, how to increase the percentage of spectra usage in such studies remains to be elucidated.
CONCLUSION
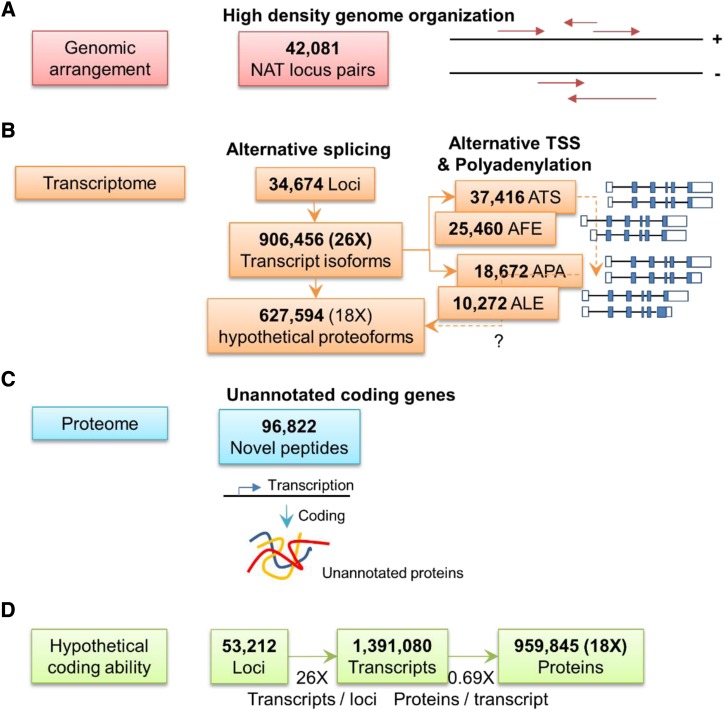
It has been estimated that the human transcriptome contains over 80,000 transcripts with the potential to be translated into 250,000 proteins (de Klerk and ’t Hoen, 2015; Reyes and Huber, 2018). In this study, lrRNA_seq-based proteogenomics further expanded our knowledge of the complexity of the rice genome and its coding potential (Fig. 7). First, the high-density arrangement of NATs in the rice genome elicits extensive undiscovered transcriptional or posttranscriptional regulation mechanisms. Second, the interdependent coordination among the three posttranscriptional mechanisms, ATS, AS, and APA, increases the rice transcriptome by 26 times in the form of transcript isoforms. Third, taking into consideration the hypothetical proteins translated by aforementioned transcript isoforms, we estimated that there is an approximately 18-fold increase in the number of translated proteins compared with the 53,212 annotated loci in the rice genome (Fig. 7D). This estimation largely agrees with previous results using srRNA_seq, but newly discovered mechanisms suggest an incredible level of complexity in how genetic information is stored and decoded in rice genomes. The unannotated loci identified in this comprehensive study also provide public information for rice genome reannotation. Moreover, the integrative analytical pipeline developed herein will likely serve as a valuable tool for both srRNA_seq- and lrRNA_seq-based proteogenomics in eukaryotic organisms.
Figure 7.
Modeling and estimation of genome coding ability and functional regulation as revealed by lrRNA_seq. A, Schematic showing the high-density genomic arrangement of 42,081 NATs. B, Transcriptome diversity and potential coding ability. A total of 906,456 transcripts were identified from 34,674 loci by lrRNA_seq with the potential to encode 627,594 different proteoforms. In addition, thousands of ATS, APA, AFE, and ALE events were identified by lrRNA_seq. They may be responsible for transcript stability and translational efficiency. C, The newly identified peptides (96,822) by proteogenomics contribute to protein diversity of the eukaryotic genome. D, An estimation of the rice genome coding ability, showing a 26-fold increase in transcript isoforms with respect to 53,212 identified loci. The estimated proteins decreased by 0.8-fold due to ATI and translational redundancy. In total, a 21-fold increase from loci to protein products is estimated.
MATERIALS AND METHODS
Plant Materials and Total RNA Extraction
Field-grown rice (Oryza sativa ‘Nipponbare/Geng’) tissues including dry seeds, 14-d-old seedlings, mature plant flag leaves, stems, roots, and flowers were harvested and frozen in liquid nitrogen for subsequent RNA sequencing and proteomic experiments. The RNeasy Mini Kit (Qiagen) bench protocol was used for plant total RNA extraction.
srRNA_Seq, Data Filtering, and Read Mapping
Generally, approximately 1 µg of plant total RNA was used for library construction using a TruSeq RNA Sample Prep Kit v2 (Illumina) following the manufacturer’s bench protocol. A strand-specific library (∼250 bp) was generated according to a previous description (Chen et al., 2019b). Subsequently, an Agilent 2100 Bioanalyzer and RT-qPCR were used to check the library quality and quantity, respectively. The purified library was subjected to paired-end sequencing (2 × 101 bp) using an Illumina HiSeq 4000 platform (BGI). For subsequent bioinformatic analysis, raw reads from all samples were assessed by quality-control steps to obtain clean reads (Supplemental Table S1). The rice reference genome annotation file (Osativa_323_v7.0.gene_exons.gff3) was downloaded from Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html). The mapping and assembly pipeline used was similar to that previously described for srRNA_seq (Zhu et al., 2017). The assembled transcripts were used for subsequent specialized transcript characterization.
Single-Molecule lrRNA_Seq and Data Analysis
The library construction steps and sequencing strategies were described previously (Zhu et al., 2017) and performed with minor modifications (Supplemental Table S2). In general, five libraries (i.e. 0.5–1 k, 1–2 k, 2–3 k, 3–6 k, and 5–10 k) were generated and sequenced using 16 SMRT cells for each tissue type on a Pacific Biosciences RSII platform (BGI). The resulting raw data were processed by the ToFu pipeline as described on the company Web site [https://github.com/PacificBiosciences/cDNA_primer/wiki/tofu-Tutorial-(optional).-Removing-redundant-transcripts]. Both high- and low-quality full-length transcripts were subjected to base correction by two rounds of BLAST against the Phytozome reference genome and cDNA sequences for subsequent bioinformatic analysis.
Transcript Remapping and Identification of AS
The soft-masked rice genome sequences were downloaded from Phytozome v12.1.6 (https://phytozome.jgi.doe.gov/pz/portal.html; last accessed on May 3, 2018) and indexed using gmap_build (version 2018-03-25). Remapping of the previously genome-guided assembled transcripts (total 120,958) from Illumina stranded paired-end reads (srRNA_seq data set) and Pacific Biosciences full-length transcripts (total 1,100,036) from the lrRNA_seq data set to the rice genome was performed using GMAP (Abdel-Ghany et al., 2016) with the following parameters: –no-chimaeras–cross-species–min-identity 0.98–allow-close-indels 2 -n 1 -z sense_force, where only the transcripts aligned with a minimum identity of 0.98 and correct strand information were included for subsequent analyses.
Further filtering was performed by comparison with the extant rice gene models, retaining transcripts that contained at least one correct junction or covered an intact exon. Then, AS events were analyzed using ASprofile (https://ccb.jhu.edu/software/ASprofile/) according to a previous description (Zhu et al., 2017). A Circos diagram was drafted using the AS frequency mapped on the rice genome with a 300-kb sliding window. Additionally, the splice site statistics and conservation analysis were summarized and constructed using the online software WebLogo v3 (http://weblogo.threeplusone.com/; Crooks et al., 2004). Splicing variants were identified by using full-length transcripts after two rounds of correction against the cv Nipponbare reference genome and cDNAs as described previously (Reyes and Huber, 2018). Redundant transcripts were then removed based on BLAST results filtered by parameters as 98% identity and more than three mismatches. In addition, unannotated transcripts of srRNA_seq and lrRNA_seq data sets were identified by performing comparisons using the same criteria against the Phytozome annotation of rice transcripts. After the removal of redundancy, the remaining transcripts was characterized as unannotated transcripts (Supplemental Table S8).
Characterization of Natural Antisense, Fusion, and Intergenic Transcripts
NATs were identified according to previous methods with minor modifications (Wang et al., 2014; Xu et al., 2017). In general, transcripts located in different strands of genomic DNA with overlapping coordinates were used for NAT characterization.
Fusion transcripts were analyzed using previously described procedures with minor modifications (Weirather et al., 2015; Wang et al., 2016). In short, transcripts mapped to two or more places on the rice genome were selected for further analysis.
Intergenic transcripts were identified by choosing transcripts mapped to intergenic regions (class u transcripts by GMAP).
GO and Pathway Enrichment
Generally, GO functional enrichment was conducted using the AgriGOv2 annotation database (http://systemsbiology.cau.edu.cn/agriGOv2/download.php). KEGG pathway enrichment analysis was carried out according to the Kobas database (http://kobas.cbi.pku.edu.cn). Significant GO and KEGG terms were identified using the following parameters: gene number (>5) and adjusted P value (<0.05).
Plant Protein Extraction, Processing, and MS/MS Analysis
Plant total proteins were extracted according to a previous description (Chen et al., 2014, 2019a; Zhu et al., 2018) for proteomic identification. In general, approximately 10-g rice tissue samples were ground in liquid nitrogen for total protein extraction. Trypsin or Glu-C digestion was performed on two parallel batches of the samples. The resulting peptides were separated and detected using a Q-Exactive tandem mass spectrometer equipped with an Orbitrap analyzer (Thermo Fisher Scientific). In brief, mixed peptides were subsequently fractionated by using a C18-Gemini column (4.6 mm × 250 mm, 5 μm particle size) on the Shimadzu LC-20AB system. An elution gradient of ∼60 min was used for peptide separation with 5% (v/v) acetonitrile (pH 9.8) as mobile phase A and 95% (v/v) acetonitrile (pH 9.8) as mobile phase B. The gradient elution profile was composed of 5% mobile phase B for 10 min, 5% to 35% mobile phase B for 40 min, 35% to 95% mobile phase B for 1 min, maintained at 100% mobile phase B for 3 min, and ending with 5% mobile phase B for 10 min. The flow rate was adjusted to 1 mL min−1, and UV absorbance (214 nm) was monitored. A total of 20 fractions were collected and then freeze-dried via speed-vacuum method. Liquid chromatography-MS/MS detection was carried out on a Q-Exactive mass spectrometer (Thermo Fisher Scientific) equipped with a nanoESI source. Generally, fractionated peptides were first loaded onto a trap column and then eluted into a self-packed C18 analytical column (3 μm particle size, 75 μm × 150 mm). A constant flow rate was set at 300 nL min−1, and mobile phase B (0.1% [v/v] formic acid and 98% [v/v] acetonitrile) was used to establish a 65-min gradient, which consisted of 5% B during 0 to 8 min, 8% to 35% B during 8 to 43 min, 35% to 60% B during 43 to 48 min, 60% to 80% B during 48 to 50 min, 80% B during 50 to 55 min, and a final step of 5% B during 55 to 65 min. MS scans were carried out using the data-dependent acquisition mode with the following parameters: the ion source voltage was set to 1.6 kV; each scan cycle consisted of one full-scan mass spectrum (with mass-to-charge ratio [m/z] ranging from 350 to 1,600 m/z and charge states from 2 to 7) followed by 20 MS/MS events (with m/z starting from 100 m/z); the resolutions of MS and MS/MS were set to 70,000 and 17,500, respectively; the threshold count was set to 10,000 to activate MS/MS accumulation, and former target ion exclusion was set for 15 s; higher-energy collisional dissociation collision energy was set to 27; automatic gain controls of MS and MS/MS were set to 3E6 and 1E6, respectively.
In addition to the 24 data sets from the PRIDE database (Supplemental Table S7), 7,368,042 high-quality raw spectra were used for subsequent proteogenomic analysis (Fig. 6A). All raw spectral data were processed using the same quality parameters.
Database Construction and MS Data Set Searching
A self-constructed virtual peptide library (155,195,875 entries) was generated based on previously developed protocols with minor modifications. Briefly, three-frame translations of strand-specific transcripts from both srRNA_seq and lrRNA_seq were performed. Redundant peptide sequences were removed, and the sequences were combined. Peptide entries longer than six amino acids were filtered for inclusion in the final virtual library. In total, peptides generated from 1,221,140 transcripts containing six to 10 amino acids (52,664,121 entries), 11 to 80 amino acids (96,670,677 entries), and more than 80 amino acids (5,861,077 entries) were used for subsequent protein identification. The AS events library (21,015,710 entries) was generated as described previously (Zhu et al., 2017). In general, the strand-specific cDNA sequences of identified posttranscriptional events and their junctions underwent three-frame translation to generate target entries in this library. The database search was carried out according to a previous description (Chen et al., 2014). Briefly, ProteinPilot software v5.0 (AB Sciex) was used for peptide and protein identification with global false discovery rate < 0.01. Proteoforms/protein groups with at least two unique peptides at the 95% confidence level were summarized as conservative/minimum number of proteoforms for further proteogenomic analysis (Supplemental Table S9).
RT-qPCR Validation of Select Transcripts
Approximately 5 μg of total RNA from rice was extracted and reverse transcribed into cDNA following the bench protocol of the Superscript First-Strand Synthesis System (Invitrogen). RT-qPCR was carried out following a previous experimental description (Zhu et al., 2017). Transcript-specific primers used in RT-qPCR are summarized in Supplemental Table S10.
Accession Numbers
The data from srRNA_seq and lrRNA_seq have been uploaded to the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under Bioproject PRJNA482217. We have submitted our proteomic raw data into the PRIDE database with accession number PXD013462.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Characterization and comparison of fusion transcripts between srRNA_seq and lrRNA_seq.
Supplemental Figure S2. Identification of intergenic transcripts and lncRNA.
Supplemental Figure S3. Statistics and functional analysis of NATs.
Supplemental Figure S4. Comparison of posttranscriptional events and single splice site analysis between srRNA_seq and lrRNA_seq.
Supplemental Figure S5. Comparison of rice and Arabidopsis splicing factor transcript isoforms.
Supplemental Table S1. Basic sequencing information for srRNA_seq.
Supplemental Table S2. Basic sequencing information for lrRNA_seq.
Supplemental Table S3. GO enrichment of fusion transcripts identified by lrRNA_seq.
Supplemental Table S4. GO enrichment of NATs identified by lrRNA_seq.
Supplemental Table S5. Functional annotation of NATs identified by lrRNA_seq.
Supplemental Table S6. Identification and annotation of ATS and APA.
Supplemental Table S7. List of protein data sets used for protein database search.
Supplemental Table S8. Annotation file of unannotated transcripts identified from the lrRNA_seq data set.
Supplemental Table S9. List of identified proteoforms/protein groups and their supporting information.
Supplemental Table S10. Primers used in this study.
Footnotes
This work was supported by the Natural Science Foundation of Guangdong Province (grant no. 2018A030313030), the Funds of Shandong “Double Top” Program, the National Natural Science Foundation of China (grant nos. NSFC81401561 and 91535109), the Shenzhen Virtual University Park Support Scheme to the CUHK Shenzhen Research Institute (grant no. YFJGJS1.0), the Natural Science Foundation of Hunan Province (grant no. 2019JJ50263), the Hong Kong Research Grant Council (grant nos. AoE/M-05/12, AoE/M-403/16, GRF14160516, 14177617, and 12100318), and the European Union H2020 project PlantaSYST (SGA-CSA no. 739582 under FPA no. 664620).
References
- Abdel-Ghany SE, Hamilton M, Jacobi JL, Ngam P, Devitt N, Schilkey F, Ben-Hur A, Reddy AS (2016) A survey of the sorghum transcriptome using single-molecule long reads. Nat Commun 7: 11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anvar SY, Allard G, Tseng E, Sheynkman GM, de Klerk E, Vermaat M, Yin RH, Johansson HE, Ariyurek Y, den Dunnen JT, et al. (2018) Full-length mRNA sequencing uncovers a widespread coupling between transcription initiation and mRNA processing. Genome Biol 19: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbin OA, Malik R, Dhanasekaran SM, Prensner JR, Cao X, Wu YM, Robinson D, Wang R, Chen G, Beer DG, et al. (2015) The landscape of antisense gene expression in human cancers. Genome Res 25: 1068–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitton DA, Atkinson SR, Rallis C, Smith GC, Ellis DA, Chen YY, Malecki M, Codlin S, Lemay JF, Cotobal C, et al. (2015) Widespread exon skipping triggers degradation by nuclear RNA surveillance in fission yeast. Genome Res 25: 884–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK (2005) Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouthier de la Tour C, Blanchard L, Dulermo R, Ludanyi M, Devigne A, Armengaud J, Sommer S, De GA (2015) The abundant and essential HU proteins in Deinococcus deserti and Deinococcus radiodurans are translated from leaderless mRNA. Microbiology 161: 2410–2422 [DOI] [PubMed] [Google Scholar]
- Bøvre K, Szybalski W (1969) Patterns of convergent and overlapping transcription within the b2 region of coliphage λ. Virology 38: 614–626 [DOI] [PubMed] [Google Scholar]
- Camblong J, Beyrouthy N, Guffanti E, Schlaepfer G, Steinmetz LM, Stutz F (2009) Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes Dev 23: 1534–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellana NE, Payne SH, Shen Z, Stanke M, Bafna V, Briggs SP (2008) Discovery and revision of Arabidopsis genes by proteogenomics. Proc Natl Acad Sci USA 105: 21034–21038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellana NE, Shen Z, He Y, Walley JW, Cassidy CJ, Briggs SP, Bafna V (2014) An automated proteogenomic method uses mass spectrometry to reveal novel genes in Zea mays. Mol Cell Proteomics 13: 157–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CY, Lin WD, Tu SL (2014) Genome-wide analysis of heat-sensitive alternative splicing in Physcomitrella patens. Plant Physiol 165: 826–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman B, Bellgard M (2017) Plant proteogenomics: Improvements to the grapevine genome annotation. Proteomics 17: 1700197. [DOI] [PubMed] [Google Scholar]
- Chen MX, Sun C, Zhang KL, Song YC, Tian Y, Chen X, Liu YG, Ye NH, Zhang J, Qu S, et al. (2019a) SWATH-MS-facilitated proteomic profiling of fruit skin between Fuji apple and a red skin bud sport mutant. BMC Plant Biol 19: 445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MX, Zhu FY, Wang FZ, Ye NH, Gao B, Chen X, Zhao SS, Fan T, Cao YY, Liu TY, et al. (2019b) Alternative splicing and translation play important roles in hypoxic germination in rice. J Exp Bot 70: 817–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Chan WL, Zhu FY, Lo C (2014) Phosphoproteomic analysis of the non-seed vascular plant model Selaginella moellendorffii. Proteome Sci 12: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Szcześniak MW, Gaffney DJ, Elo LL, Zhang X, et al. (2016) A survey of best practices for RNA-seq data analysis. Genome Biol 17: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: A sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot A, Roche D, Fernandez B, Ludanyi M, Cruveiller S, Pignol D, Vallenet D, Armengaud J, Blanchard L (2014) RNA sequencing and proteogenomics reveal the importance of leaderless mRNAs in the radiation-tolerant bacterium Deinococcus deserti. Genome Biol Evol 6: 932–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Klerk E, ’t Hoen PA (2015) Alternative mRNA transcription, processing, and translation: Insights from RNA sequencing. Trends Genet 31: 128–139 [DOI] [PubMed] [Google Scholar]
- Deveson IW, Brunck ME, Blackburn J, Tseng E, Hon T, Clark TA, Clark MB, Crawford J, Dinger ME, Nielsen LK, et al. (2018) Universal alternative splicing of noncoding exons. Cell Syst 6: 245–255.e5 [DOI] [PubMed] [Google Scholar]
- Duan L, Xiao W, Xia F, Liu H, Xiao J, Li X, Wang S (2016) Two different transcripts of a LAMMER kinase gene play opposite roles in disease resistance. Plant Physiol 172: 1959–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckardt NA. (2013) The Plant Cell reviews alternative splicing. Plant Cell 25: 3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards PA. (2010) Fusion genes and chromosome translocations in the common epithelial cancers. J Pathol 220: 244–254 [DOI] [PubMed] [Google Scholar]
- Edwards PA, Howarth KD (2012) Are breast cancers driven by fusion genes? Breast Cancer Res 14: 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Wahlestedt C (2009) Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol 10: 637–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovkin M, Reddy AS (1996) Structure and expression of a plant U1 snRNP 70K gene: Alternative splicing of U1 snRNP 70K pre-mRNAs produces two different transcripts. Plant Cell 8: 1421–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Cao D, Na Y, Kim DY, Zhang T, Yao J, Oh H, Hu J, Zheng H, Yao Y, et al. (2018) Far Upstream Element-Binding Protein 1 regulates LSD1 alternative splicing to promote terminal differentiation of neural progenitors. Stem Cell Rep 10: 1208–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe JD, Berg HC, Church GM (2004) Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics 4: 59–77 [DOI] [PubMed] [Google Scholar]
- Jensen TH, Jacquier A, Libri D (2013) Dealing with pervasive transcription. Mol Cell 52: 473–484 [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Cooper TA (2011) Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet 12: 715–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyna M, Lopato S, Voronin V, Barta A (2006) Evolutionary conservation and regulation of particular alternative splicing events in plant SR proteins. Nucleic Acids Res 34: 4395–4405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. (2005) Antisense transcription in the mammalian transcriptome. Science 309: 1564–1566 [DOI] [PubMed] [Google Scholar]
- Kuang Z, Boeke JD, Canzar S (2017) The dynamic landscape of fission yeast meiosis alternative-splice isoforms. Genome Res 27: 145–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharova V, Wiker HG (2014) Proteogenomics in microbiology: Taking the right turn at the junction of genomics and proteomics. Proteomics 14: 2660–2675 [DOI] [PubMed] [Google Scholar]
- Kumar D, Mondal AK, Kutum R, Dash D (2016) Proteogenomics of rare taxonomic phyla: A prospective treasure trove of protein coding genes. Proteomics 16: 226–240 [DOI] [PubMed] [Google Scholar]
- Laloum T, Martín G, Duque P (2018) Alternative splicing control of abiotic stress responses. Trends Plant Sci 23: 140–150 [DOI] [PubMed] [Google Scholar]
- Li L, Wang X, Stolc V, Li X, Zhang D, Su N, Tongprasit W, Li S, Cheng Z, Wang J, et al. (2006) Genome-wide transcription analyses in rice using tiling microarrays. Nat Genet 38: 124–129 [DOI] [PubMed] [Google Scholar]
- Locard-Paulet M, Pible O, Gonzalez de Peredo A, Alpha-Bazin B, Almunia C, Burlet-Schiltz O, Armengaud J (2016) Clinical implications of recent advances in proteogenomics. Expert Rev Proteomics 13: 185–199 [DOI] [PubMed] [Google Scholar]
- Lomsadze A, Gemayel K, Tang S, Borodovsky M (2018) Modeling leaderless transcription and atypical genes results in more accurate gene prediction in prokaryotes. Genome Res 28: 1079–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic ZJ, Lehner R, Forstner C, Barta A (2005) Evolutionary conservation of minor U12-type spliceosome between plants and humans. RNA 11: 1095–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus CJ, Duff MO, Eipper-Mains J, Graveley BR (2010) Global analysis of trans-splicing in Drosophila. Proc Natl Acad Sci USA 107: 12975–12979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier F, Geyer PE, Virreira Winter S, Cox J, Mann M (2018) BoxCar acquisition method enables single-shot proteomics at a depth of 10,000 proteins in 100 minutes. Nat Methods 15: 440–448 [DOI] [PubMed] [Google Scholar]
- Menschaert G, Van Criekinge W, Notelaers T, Koch A, Crappé J, Gevaert K, Van Damme P (2013) Deep proteome coverage based on ribosome profiling aids mass spectrometry-based protein and peptide discovery and provides evidence of alternative translation products and near-cognate translation initiation events. Mol Cell Proteomics 12: 1780–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills JD, Chen BJ, Ueberham U, Arendt T, Janitz M (2016) The antisense transcriptome and the human brain. J Mol Neurosci 58: 1–15 [DOI] [PubMed] [Google Scholar]
- Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, van der Brug MP, Wahlestedt C (2012) Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol 30: 453–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissy AS, Griffith M, Marra MA (2011) Extensive relationship between antisense transcription and alternative splicing in the human genome. Genome Res 21: 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourier T, Jeffares DC (2003) Eukaryotic intron loss. Science 300: 1393. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI. (2014) Proteogenomics: Concepts, applications and computational strategies. Nat Methods 11: 1114–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonechnikov K, Imai-Matsushima A, Paul L, Seitz A, Meyer TF, Garcia-Alcalde F (2016) InFusion: Advancing discovery of fusion genes and chimeric transcripts from deep RNA-sequencing data. PLoS ONE 11: e0167417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, et al. (2007) The TIGR Rice Genome Annotation Resource: Improvements and new features. Nucleic Acids Res 35: D883–D887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Pelechano V, Steinmetz LM (2013) Gene regulation by antisense transcription. Nat Rev Genet 14: 880–893 [DOI] [PubMed] [Google Scholar]
- Prescott EM, Proudfoot NJ (2002) Transcriptional collision between convergent genes in budding yeast. Proc Natl Acad Sci USA 99: 8796–8801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS, Marquez Y, Kalyna M, Barta A (2013) Complexity of the alternative splicing landscape in plants. Plant Cell 25: 3657–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z, Qi D, Pugh N, Li K, Wen B, Zhou R, Xu S, Liu S, Jones AR (2019) Improvements to the rice genome annotation through large-scale analysis of RNA-seq and proteomics data sets. Mol Cell Proteomics 18: 86–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A, Huber W (2018) Alternative start and termination sites of transcription drive most transcript isoform differences across human tissues. Nucleic Acids Res 46: 582–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rühl C, Stauffer E, Kahles A, Wagner G, Drechsel G, Rätsch G, Wachter A (2012) Polypyrimidine tract binding protein homologs from Arabidopsis are key regulators of alternative splicing with implications in fundamental developmental processes. Plant Cell 24: 4360–4375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon D, Tilgner H, Grubert F, Snyder M (2013) A single-molecule long-read survey of the human transcriptome. Nat Biotechnol 31: 1009–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. (2017) Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat Rev Mol Cell Biol 18: 655–670 [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG (2009) Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steijger T, Abril JF, Engström PG, Kokocinski F, Hubbard TJ, Guigó R, Harrow J, Bertone P (2013) Assessment of transcript reconstruction methods for RNA-seq. Nat Methods 10: 1177–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su WY, Li JT, Cui Y, Hong J, Du W, Wang YC, Lin YW, Xiong H, Wang JL, Kong X, et al. (2012) Bidirectional regulation between WDR83 and its natural antisense transcript DHPS in gastric cancer. Cell Res 22: 1374–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suenaga Y, Islam SM, Alagu J, Kaneko Y, Kato M, Tanaka Y, Kawana H, Hossain S, Matsumoto D, Yamamoto M, et al. (2014) NCYM, a cis-antisense gene of MYCN, encodes a de novo evolved protein that inhibits GSK3β resulting in the stabilization of MYCN in human neuroblastomas. PLoS Genet 10: e1003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C (2009) Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462: 799–802 [DOI] [PubMed] [Google Scholar]
- Tilgner H, Raha D, Habegger L, Mohiuddin M, Gerstein M, Snyder M (2013) Accurate identification and analysis of human mRNA isoforms using deep long read sequencing. G3 (Bethesda) 3: 387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tress ML, Abascal F, Valencia A (2017) Alternative splicing may not be the key to proteome complexity. Trends Biochem Sci 42: 98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkening JD, Bailey DJ, Rose CM, Grimsrud PA, Howes-Podoll M, Venkateshwaran M, Westphall MS, Ané JM, Coon JJ, Sussman MR (2012) A proteogenomic survey of the Medicago truncatula genome. Mol Cell Proteomics 11: 933–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade JT, Grainger DC (2014) Pervasive transcription: Illuminating the dark matter of bacterial transcriptomes. Nat Rev Microbiol 12: 647–653 [DOI] [PubMed] [Google Scholar]
- Wang B, Tseng E, Regulski M, Clark TA, Hon T, Jiao Y, Lu Z, Olson A, Stein JC, Ware D (2016) Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat Commun 7: 11708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Chung PJ, Liu J, Jang IC, Kean MJ, Xu J, Chua NH (2014) Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res 24: 444–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wang P, Liang F, Ye Z, Li J, Shen C, Pei L, Wang F, Hu J, Tu L, et al. (2018a) A global survey of alternative splicing in allopolyploid cotton: Landscape, complexity and regulation. New Phytol 217: 163–178 [DOI] [PubMed] [Google Scholar]
- Wang W, Mauleon R, Hu Z, Chebotarov D, Tai S, Wu Z, Li M, Zheng T, Fuentes RR, Zhang F, et al. (2018b) Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557: 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Ji H, Yuan B, Wang S, Su C, Yao B, Zhao H, Li X (2015) ABA signalling is fine-tuned by antagonistic HAB1 variants. Nat Commun 6: 8138. [DOI] [PubMed] [Google Scholar]
- Weirather JL, Afshar PT, Clark TA, Tseng E, Powers LS, Underwood JG, Zabner J, Korlach J, Wong WH, Au KF (2015) Characterization of fusion genes and the significantly expressed fusion isoforms in breast cancer by hybrid sequencing. Nucleic Acids Res 43: e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek RC, Hatfield GW (1986) Nucleotide sequence and in vivo expression of the ilvY and ilvC genes in Escherichia coli K12: Transcription from divergent overlapping promoters. J Biol Chem 261: 2441–2450 [PubMed] [Google Scholar]
- Werner A. (2005) Natural antisense transcripts. RNA Biol 2: 53–62 [DOI] [PubMed] [Google Scholar]
- Will CL, Lührmann R (2011) Spliceosome structure and function. Cold Spring Harb Perspect Biol 3: a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong F, Yuh ZT, Schaefer EL, Roop BC, Ally AH (1987) Overlapping transcription units in the transient receptor potential locus of Drosophila melanogaster. Somat Cell Mol Genet 13: 661–669 [DOI] [PubMed] [Google Scholar]
- Wu DD, Irwin DM, Zhang YP (2011) De novo origin of human protein-coding genes. PLoS Genet 7: e1002379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wang Q, Freeling M, Zhang X, Xu Y, Mao Y, Tang X, Wu F, Lan H, Cao M, et al. (2017) Natural antisense transcripts are significantly involved in regulation of drought stress in maize. Nucleic Acids Res 45: 5126–5141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C, Wang J, Harrison AP, Meng X, Chen D, Chen M (2015) Genome-wide view of natural antisense transcripts in Arabidopsis thaliana. DNA Res 22: 233–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi Z, Chambers MC, Zimmerman LJ, Shaddox KF, Kim S, et al. (2014) Proteogenomic characterization of human colon and rectal cancer. Nature 513: 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Sun M, Wang J, Lei M, Li C, Zhao D, Huang J, Li W, Li S, Li J, et al. (2019) PacBio full-length cDNA sequencing integrated with RNA-seq reads drastically improves the discovery of splicing transcripts in rice. Plant J 97: 296–305 [DOI] [PubMed] [Google Scholar]
- Zhang H, Liu T, Zhang Z, Payne SH, Zhang B, McDermott JE, Zhou JY, Petyuk VA, Chen L, Ray D, et al. (2016) Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell 166: 755–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lii Y, Wu Z, Polishko A, Zhang H, Chinnusamy V, Lonardi S, Zhu JK, Liu R, Jin H (2013) Mechanisms of small RNA generation from cis-NATs in response to environmental and developmental cues. Mol Plant 6: 704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu FY, Chen MX, Chan WL, Yang F, Tian Y, Song T, Xie LJ, Zhou Y, Xiao S, Zhang J, et al. (2018) SWATH-MS quantitative proteomic investigation of nitrogen starvation in Arabidopsis reveals new aspects of plant nitrogen stress responses. J Proteomics 187: 161–170 [DOI] [PubMed] [Google Scholar]
- Zhu FY, Chen MX, Ye NH, Shi L, Ma KL, Yang JF, Cao YY, Zhang Y, Yoshida T, Fernie AR, et al. (2017) Proteogenomic analysis reveals alternative splicing and translation as part of the abscisic acid response in Arabidopsis seedlings. Plant J 91: 518–533 [DOI] [PubMed] [Google Scholar]