ZmEHD1 regulates auxin homeostasis by mediating clathrin-mediated endocytosis and is crucial for maize normal development.
Abstract
The roles of C-terminal Eps15 homology domain (EHD) proteins in clathrin-mediated endocytosis in plants are poorly understood. Here, we isolated a maize (Zea mays) mutant, designated ehd1, which showed defects in kernel development and vegetative growth. Positional cloning and transgenic analysis revealed that ehd1 encodes an EHD protein. Internalization of the endocytic tracer FM4-64 was substantially reduced in the ehd1 mutant and ZmEHD1 knockout mutants. We further demonstrated that ZmEHD1 and the ZmAP2 σ subunit physically interact at the plasma membrane. Auxin distribution and ZmPIN1a-YFP localization were altered in the ehd1 mutant. Kernel indole-3-acetic acid levels were substantially lower in the ehd1 mutant than in wild-type maize. Exogenous application of 1-naphthaleneacetic acid, but not GA3 or 2-naphthaleneacetic acid, rescued the seed germination and seedling emergency phenotypic defects of ehd1 mutants. Taken together, these results indicate that ZmEHD1 regulates auxin homeostasis by mediating clathrin-mediated endocytosis through its interaction with the ZmAP2 σ subunit, which is crucial for kernel development and vegetative growth of maize.
Endocytosis, which can be clathrin dependent or clathrin independent, is the internalization of plasma membrane (pm) proteins or uptake of extracellular materials for transport to the endosome (Murphy et al., 2005). Clathrin-mediated endocytosis (CME) is the major gateway into plant cells (Fan et al., 2015). Although detailed information is available on the roles of endocytosis in animals, researchers have only recently documented that CME is critical for various developmental processes in plants, including cell polarity determination (Van Damme et al., 2011; Wang et al., 2013), male reproductive organ development (Blackbourn and Jackson, 1996; Kim et al., 2013), gametophyte development (Backues et al., 2010), and embryogenesis (Kitakura et al., 2011; Fan et al., 2013). CME is also important in plant responses to abiotic and biotic stresses through internalization of PM-resident transporters or receptors, such as the boron transporter (BOR1; Takano et al., 2010), the iron-regulated transporter (IRT1; Barberon et al., 2011), the auxin efflux transporter PIN-FORMED (PINs; Dhonukshe et al., 2007), the brassinosteroid receptor BRASSINOSTEROID INSENSITIVE1 (BRI1; Di Rubbo et al., 2013), the bacterial flagellin receptor FLAGELIN SENSING2 (FLS2; Robatzek et al., 2006), and the ethylene-inducing xylanase receptor (LeEIX2; Bar and Avni, 2009).
CME is a complex process that can be divided into at least four steps: the budding of a vesicle, the packaging of cargo into the vesicle, the release of the mature vesicle from the PM, and the fusing of the vesicle with the endosome (Sigismund et al., 2012). Because clathrin cannot directly bind to the PM or to cargoes, the formation of clathrin-coated endocytic vesicles initiates the association of adaptor protein complex 2 (AP2) with the PM, and thus, AP2 plays a central role in the initiation of CME. The AP2 complex forms a heterotetrameric complex containing two large subunits (β1–5 and either γ, α, δ, ε, or ζ), a medium subunit (μ1–5), and a small subunit (σ1–5; McMahon and Boucrot, 2011). Our understanding of AP2 in plants has been enhanced by the following recent findings: knockdown of the two Arabidopsis (Arabidopsis thaliana) AP2A genes or overexpression of a dominant-negative version of the medium AP2 subunit, AP2M, was shown to substantially impair BRI1 endocytosis and to enhance brassinosteroid signaling (Di Rubbo et al., 2013); α and μ adaptins were shown to be crucial for pollen production and viability, as well as elongation of staminal filaments and pollen tubes by modulating the amount and polarity of PINs (Kim et al., 2013); and σ adaptin was found to play important roles in the assembly of a functional AP2 complex, with ap2 σ loss-of-function Arabidopsis exhibiting defects in multiple aspects of plant growth and development (Fan et al., 2013).
In addition to the classical AP2 complex, other accessory proteins, including the C-terminal Eps15 homology domain (EHD) proteins, also connect the cargo and membrane lipid to form the CME vesicle (Deo et al., 2018). The EHD family contains one member in Drosophila melanogaster and Caenorhabditis elegans and four orthologs in mouse (Mus musculus) and human (Homo sapiens). RME1 was shown to be a key player controlling the recycling of internalized receptors from the endocytic recycling compartment to the PM in C. elegans (Lin et al., 2001). The ortholog in Drosophila, Past1, was shown to be involved in endocytosis (Olswang-Kutz et al., 2009). By homologous cloning, two EHD genes were isolated from Arabidopsis (Bar et al., 2008). Despite high sequence similarity, the two AtEHD proteins regulated endocytosis in distinct patterns. Over-expression of AtEHD2 had an inhibitory effect on endocytosis, while downregulation of AtEHD1 caused retardation of the entry of endocytosed material into plant cells (Bar et al., 2008). However, the mechanisms by which EHD family members are linked to CME in plants have not been characterized.
Maize (Zea mays) ranks first in total production among major staple cereals and is also an important raw material for biofuel and many other industrial products (McLaren, 2005). To better understand maize endosperm filling and maturation, we characterized a maize mutant, ehd1, that is impaired in kernel development and vegetative growth. Positional cloning of ehd1 and transgenic analysis identified the causative locus as a gene that encodes an EHD-containing protein and that is herein designated as ZmEHD1. We show that ZmEHD1 is involved in CME through interaction with the ZmAP2 σ subunit. We further demonstrate that ZmEHD1 affects the expression of genes involved in auxin-related processes and that exogenous 1-naphthaleneacetic acid (1-NAA) application rescues the fertility of the ehd1 mutant. Our results indicate that by regulating auxin homeostasis, ZmEHD1-mediated CME is crucial for kernel development and vegetative growth of maize.
RESULTS
The ehd1 Mutant Is Impaired in Kernel Development and Vegetative Growth
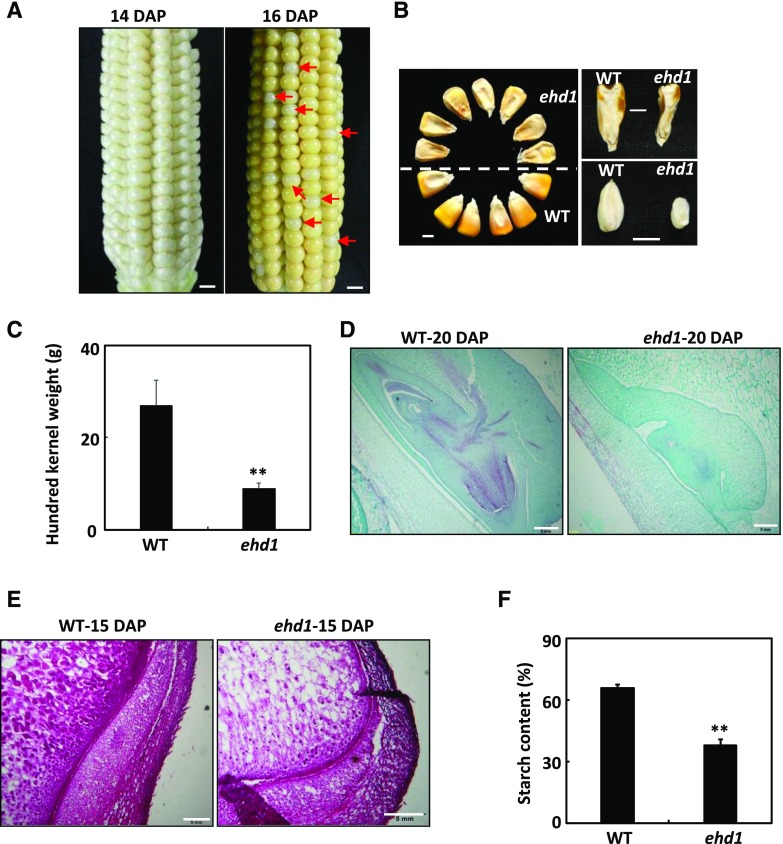
The ehd1 mutant was originally isolated as a shrunken kernel mutant in the screening of natural mutagenesis lines defective in the filling of maize grains. At 14 d after pollination (DAP), the kernels of the ehd1 homozygous mutant (ehd1/ehd1) and the wild type (EHD1/EHD1 and EHD1/ehd1) resembled each other (Fig. 1A). At 16 DAP, the wild-type kernels were canary yellow, while the ehd1 mutants were ivory white (Fig. 1A). At maturity, both the endosperm and embryo of the ehd1 mutant were shrunken (Fig. 1B), which led to a significant reduction in the 100-kernel weight (Fig. 1C). To analyze the kernel development of the ehd1 mutant and the wild type, we examined immature embryos at 20 DAP by light microscopy. In contrast to the wild-type embryo, the embryo of the ehd1 mutant was at the early stage of leaf primordium formation (Fig. 1D). Endosperm development and texture were also observed by light microscopy. Compared to wild-type endosperm cells, those of the ehd1 mutant had fewer starch granules (Fig. 1E). Biochemical analysis indicated that the total starch content in ehd1 seeds was ∼58% of that in wild-type seeds (Fig. 1F).
Figure 1.
Phenotypes of the maize ehd1 mutant kernels. A, F2 ears of ehd1 × Xun9058 at 14 or 16 DAP. Red arrows indicate the ehd1 kernels. Scale bars = 1 cm. B, Mature kernels of the wild type (WT) and ehd1 mutant randomly selected from F2 ears of ehd1 × Xun9058. Scale bars = 0.5 cm. C, 100-grain weight of the wild type and the ehd1 mutant. Values are means ± se (n = 4). **P < 0.01 indicates a significant difference between the wild type and the ehd1 mutant as determined by Student’s t test. D, Comparison of wild-type and ehd1 embryos at 20 DAP. The sections were stained with Safranin and Fast Green. Scale bars = 0.5 mm. E, Microstructure of wild-type and ehd1 endosperms at 15 DAP. The sections were stained with fuchsin. Scale bars = 0.5 mm. F, The starch contents in mature kernels of wild-type and ehd1 mutant maize. **P < 0.01 indicates a significant difference between the wild type and the ehd1 mutant as determined by Student’s t test.
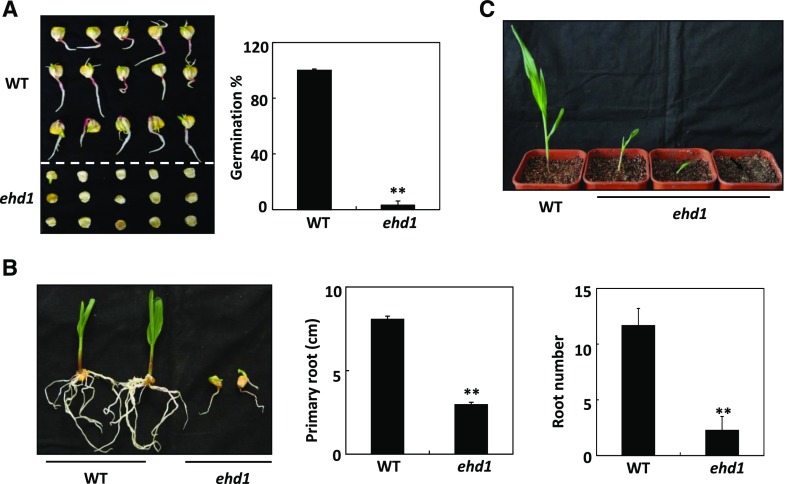
A paper-culture system was used to measure the germination rates of the wild type and the ehd1 mutant. The germination rate of the ehd1 mutant was only ∼3%, which was far lower than that of the wild type (Fig. 2A). Primary root length and root number were reduced by ∼61% and ∼73%, respectively, in the ehd1 mutant relative to the wild type (Fig. 2B). Most of the ehd1 seedlings died before the first two leaves had completely expanded (Fig. 2C). These results indicated that ZmEHD1 is essential for maize kernel development and vegetative growth.
Figure 2.
ZmEHD1 is required for normal growth and development of maize. A, Germination of the wild type (WT) and the ehd1 mutant. Values are means ± se of ∼100 seeds from three independent experiments. B, Root number and root elongation of the wild type and the ehd1 mutant. Values are means ± se (n = 5). **P < 0.01 indicates a significant difference between the wild type and the ehd1 mutant. C, Phenotypes of wild-type and ehd1 seedlings (n = 30). Representative photograph is shown.
The mutant was crossed to the inbred line Xun9058. The hybrid F1 displayed normal kernels like those of Xun9058. Due to the low germination rate of the ehd1 mutant, the normal kernels from F2 ears were planted. Among the 244 F2 individuals, 79 had the normal kernel phenotype (EHD1/EHD1), and 165 had the segregation phenotype (EHD1/ehd1). The segregation ratio followed the 1:2 theoretical ratio predicted by a χ2 test at a 0.05 level (Supplemental Table S1). For each F2 individual with kernel phenotype segregation, the pattern of shrunken and normal kernel phenotypes fit a 1:3 segregation ratio (Supplemental Table S2). An F3 population of 376 individuals derived from normal kernels of F2 selfed ears was also planted to validate the kernel phenotype segregation. Of this population, 130 individuals had the normal kernel phenotype (EHD1/EHD1) and 246 had the segregation phenotype (EHD1/ehd1; Supplemental Table S1). These results indicate that this shrunken kernel mutant is controlled by a single recessive gene.
Positional Cloning of ZmEHD1
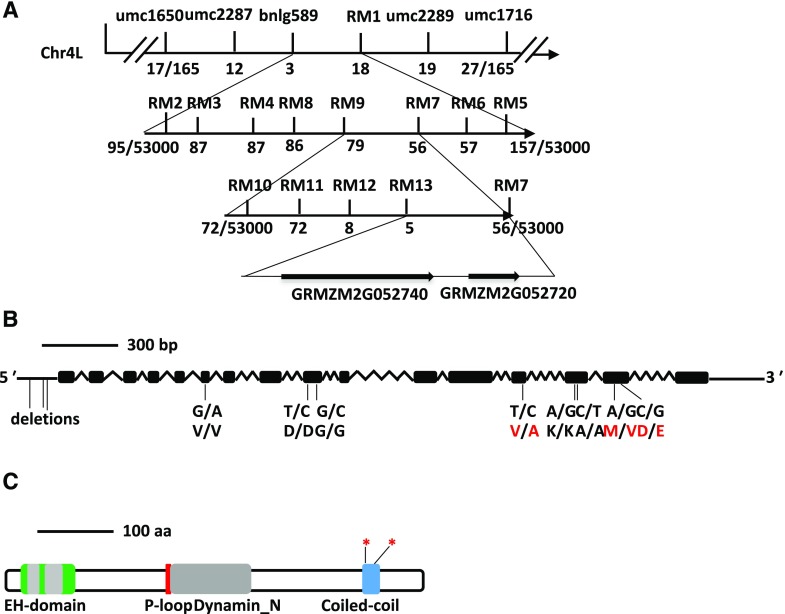
Preliminary genetic mapping of ZmEHD1 was carried out by bulked segregant analysis using the F2 population, which contained 165 individuals that differed in seed size in the ears. The ZmEHD1 gene was first mapped to a 2,339-kb region between the simple sequence repeat (SSR) markers umc1650 (17 recombinants) and umc1716 (27 recombinants) on the long arm of chromosome 4 (Fig. 3A). Because the germination rate of the ehd1 mutant was rather low, the expanded F3 mapping population containing ∼53,000 normal kernels (EHD1/EHD1 and EHD1/ehd1) was obtained to increase mapping resolution as described by Zuo et al. (2015). Thirteen polymorphic SSR markers were developed (Supplemental Table S3), and ZmEHD1 was eventually mapped to an ∼6-kb region between the SSR markers RM7 (56 recombinants) and RM13 (5 recombinants; Fig. 3A). This region contains two predicted open reading frames (ORFs; Fig. 3A). Sequence analysis revealed three deletions in the 5′ untranslated region and eight single-nucleotide polymorphisms in the ORF of GRMZM2G052740 (Fig. 3B). Among the latter group, three led to amino acid replacement between the ehd1 mutant and the wild type (Fig. 3B). In addition to GRMZM2G052740, GRMZM2G052720 was also located in this region. Its candidacy as a ZmEHD1 gene was excluded because there was no sequence difference between the wild type and the ehd1 mutant. Thus, we inferred that GRMZM2G052740 was the candidate gene for the ZmEHD1 locus.
Figure 3.
Map-based cloning of ZmEHD1. A, Schematic representation of the positional cloning of the ZmEHD1 gene on chromosome 4. The SSR markers, umc1716 and umc1650, were used for rough mapping. Recombinants are indicated below each SSR marker. B, Gene structure of ZmEHD1. Black boxes indicate exons, and lines between black boxes represent introns. The positions of mutations are marked. The mutations that led to the amino acid change between the ehd1 mutant and the wild type are indicated with red letters. C, Schematic representation of the predicted structure of ZmEHD1. The regions encoding the potential protein domains are shown. The positions of mutations on the coiled-coil domain are marked by asterisks.
The genomic DNA sequence of GRMZM2G052740 spanned ∼4.4-kb and generated a transcript that included 16 exons (Fig. 3B). The corresponding 2,209-bp complementary DNA (cDNA) sequence encoded a polypeptide of 547 amino acids with a molecular mass of ∼61 kD (Supplemental Fig. S1). BLASTP analysis in GenBank indicated that GRMZM2G052740 is closed related to the Arabidopsis EHD-containing proteins, AtEHD1 and AtEHD2 (Supplemental Fig. S1). The ZmEHD1 protein shares the typical structure of the EHD family; it has an EH domain with two calcium-binding EF-hands (15–39 amino acids and 49–84 amino acids), a P-loop (GQYSTGKT), a dynamin-type guanine nucleotide-binding domain, and a coiled-coil domain (Fig. 3C).
Validating That ZmEHD1 Is the Causative Locus by ZmEHD1 Loss-of-Function Mutants
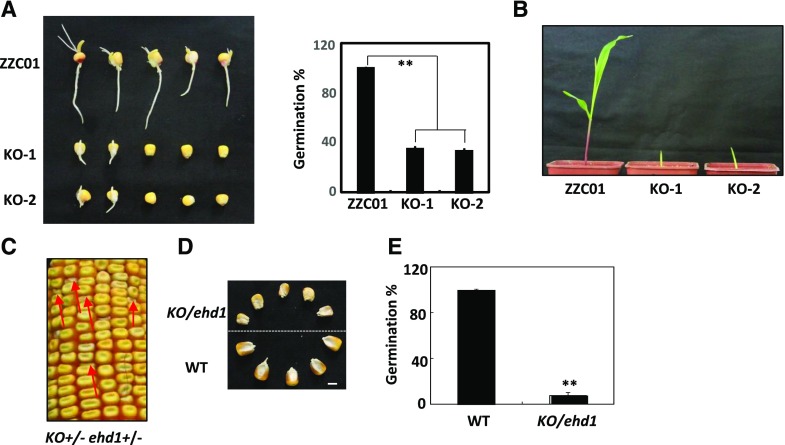
To determine whether disruption of the EHD1 locus is responsible for the ehd1 phenotypes, we used the CRISPR/Cas9 system to generate ZmEHD1 loss-of-function mutants (knockouts [KOs]). Cas9-free transgenic plants were identified for the phenotypic analysis. The given gene sequence in ZmEHD1 loss-of-function mutants revealed deletion of 775-bp or 785-bp fragments at the coding sequence, which resulted in frameshifts (Supplemental Fig. S2). ZmEHD1 loss of function also led to decreased germination rates and retarded vegetative development (Fig. 4, A and B).
Figure 4.
Transgenic validation of ZmEHD1. A, Germination of inbred line ZZC01 (wild type [WT]) and ZmEHD1 KO mutants. Values are means ± se of ∼100 seeds from three independent experiments. A lsd test was used to assess differences between the wild type and the ZmEHD1 KO mutant. **P < 0.01 (Student’s t test) indicates a significant difference between the wild type and KO mutants. B, Phenotypes of the wild type and KO mutants (n = 20). A representative photograph is shown. C, Heterozygous KO(+/−) × ehd1(+/−) were used in an allelism test. The red arrows indicate the ehd1 kernels. D, Mature kernels of the wild type and KO/ehd1 mutant randomly selected from ears of KO(+/−) × ehd1(+/−). Scale bars = 0.5 cm. E, Germination of the wild type and KO/ehd1 mutant. Values are means ± se of ∼140 seeds from three independent experiments. An lsd test was used to assess differences between the wild type and the KO/ehd1 mutant. **P < 0.01 (Student’s t test) indicates a significant difference between the wild type and the KO/ehd1 mutant.
An allelism test was performed by crossing KO-1 F1 (KO+/−) and ehd1 heterozygotes (ehd1+/−). The shrunken kernel phenotype (KO/ehd1) and the wild-type phenotype (EHD1/EHD1; EHD1/KO; EHD1/ehd1) in the hybrid F1 ears (5 ears, 1,452 individuals) displayed a 1:3 segregation ratio (Fig. 4, C and D; Supplemental Table S4), indicating that KO cannot complement ehd1. Meanwhile, the germination rate was much lower in the KO/ehd1 mutant than in the wild type (Fig. 4E). These results indicate that ZmEHD1 is the locus affected by ehd1.
The ehd1 and KO Mutants Are Defective in Endocytosis
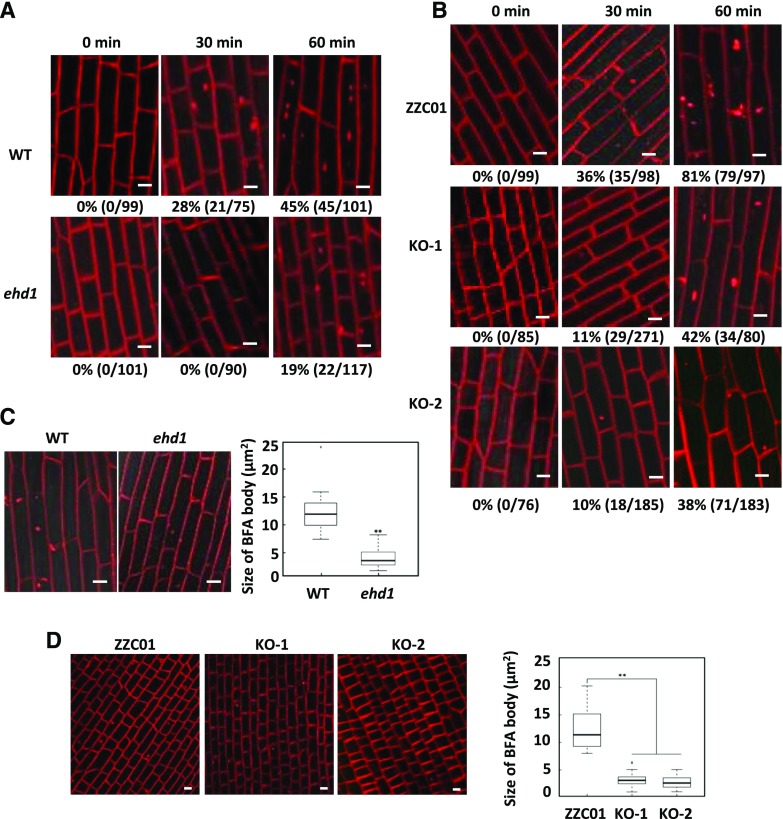
EHD has usually been identified among proteins involved in endocytosis, vesicle transport, and signal transduction (Lin et al., 2001). To test whether the ZmEHD1 protein is involved in CME in maize, we examined the uptake of N-(3-triethylammonium-propyl)-4-(4-diethylaminophenylhexatrienyl) pyridinium dibromide (FM4-64), a commonly used endocytosis tracer, in the ehd1 mutant and the wild type. Because maize seedlings are relatively large, we could not place the whole plant on the microscope platform as done with 3-d-old Arabidopsis plants in a previous study (Fan et al., 2013). Instead, we detached similar parts of roots from the ehd1 mutant and the wild-type seedlings that had been treated with 5 μm FM4-64 for 10 min to monitor FM4-64 endocytosis. At 30 min after labeling, FM4-64-labeled fluorescent puncta were detected in 28% of the wild-type root cells (21 of 75 cells), while no FM4-64-labeled fluorescent puncta were detected in ehd1 root cells (0 of 90 cells; Fig. 5A). Although FM4-64-labeled fluorescent puncta could be detected in root cells of both the wild type and the ehd1 mutant at 60 min after labeling, they were detected in only 19% of ehd1 root cells (22 of 117 cells) but in 45% of wild-type root cells (45 of 101 cells; Fig. 5A). The effects of ZmEHD1 on endocytosis were further confirmed by the delayed internalization of FM4-64 in ZmEHD1 KO mutants (Fig. 5B).
Figure 5.
Endocytosis of FM4-64 is reduced in ehd1 and in ZmEHD1 KO mutants. A and B, Three-dimensional reconstructions of z-stacks (60 μm with 2.4-μm steps) were obtained in ehd1 (A) and the ZmEHD1 KO mutants (B). Boxplots of FM-64-labeled BFA bodies in the ehd1 mutant (C) and ZmEHD1 KO mutants (D). Representative photographs at indicated durations are shown. Scale bars = 10 µm. Numbers below the photographs indicate the rate of FM4-64-labeled fluorescent puncta (% and number of FM4-64-labeled fluorescent puncta/total cell number). **P < 0.01 indicates a significant difference from the wild type according to Student’s t test.
To further investigate whether ZmEHD1 was involved in the vesicle traffic pathway, we inhibited endocytic recycling of FM-64 using the fungal toxin brefeldin A (BFA). The accumulation of FM-64 in BFA bodies was clearly observed in the wild type when treated with BFA (Fig. 5, C and D). In contrast, the aggregation of FM-64 in BFA bodies remarkably decreased in the ehd1 mutant and ZmEHD1 KO mutants (Fig. 5, C and D). Compared to the wild type, the sizes of FM-64-labeled BFA bodies in the ehd1 mutant and ZmEHD1 KO mutants decreased by ∼67% and ∼76%, respectively (Fig. 5, C and D). These results suggested that ZmEHD1 is important for endocytic transport.
Subcellular Localizations of ZmEHD1 and the ZmAP2 σ Subunit
In its drastically reduced fertility and endocytosis, the ehd1 mutant resembles Arabidopsis ap2 σ mutants, in which CME and multiple stages of plant development are impaired (Fan et al., 2013). We hypothesized that ZmEHD1 is involved in CME by virtue of its interaction with the ZmAP2 σ subunit. To test this hypothesis, we cloned the cDNA sequence of the maize AP2 σ subunit (GRMZM2G052713) and introduced it into the Arabidopsis ap2 σ loss-of-function mutant. Arabidopsis ap2 σ homozygous for the T-DNA insertion was identified by PCR (Supplemental Fig. S3A). Reverse-transcription quantitative PCR (RT-qPCR) showed that the ZmAP2 σ subunit was expressed in the Arabidopsis ap2 σ mutant (Supplemental Fig. S3B). The transformation of the Arabidopsis ap2 σ mutant with the ZmAP2 σ subunit rescued the developmental defects, such as reduced leaf size and fertility (Supplemental Fig. S3C). These results suggested that ZmAP2 σ subunit has similar functions as AtAP2 σ subunit in CME.
Before testing the possible relationship between ZmEHD1 and the ZmAP2 σ subunit, we first examined their subcellular localizations. GFPs fused C-terminally to ZmEHD1 and the ZmAP2 σ subunit was transiently expressed in maize mesophyll protoplasts under the control of the CaMV 35S promoter. ZmEHD1-GFP and ZmAP2 σ subunit-GFP signals were detected only in the PM (Supplemental Fig. S4). We also cloned the ZmEHD1 CDS from the ehd1 mutant and examined the subcellular localizations of ZmEHD1mut and ZmAP2 σ subunit protein in ehd1 mesophyll protoplasts. ZmEHD1mut-GFP and ZmAP2 σ subunit-GFP signals were diffused throughout the cells, including PM cells (Supplemental Fig. S4). Thus, we deduced that the subcellular localizations of ZmEHD1 and the ZmAP2 σ subunit at least partly overlap at the PM and that the mutation in ZmEHD1 affected the subcellular localizations of ZmEHD1 and the ZmAP2 σ subunit.
ZmEHD1 Interacts with the ZmAP2 σ Subunit in Yeast and Plants
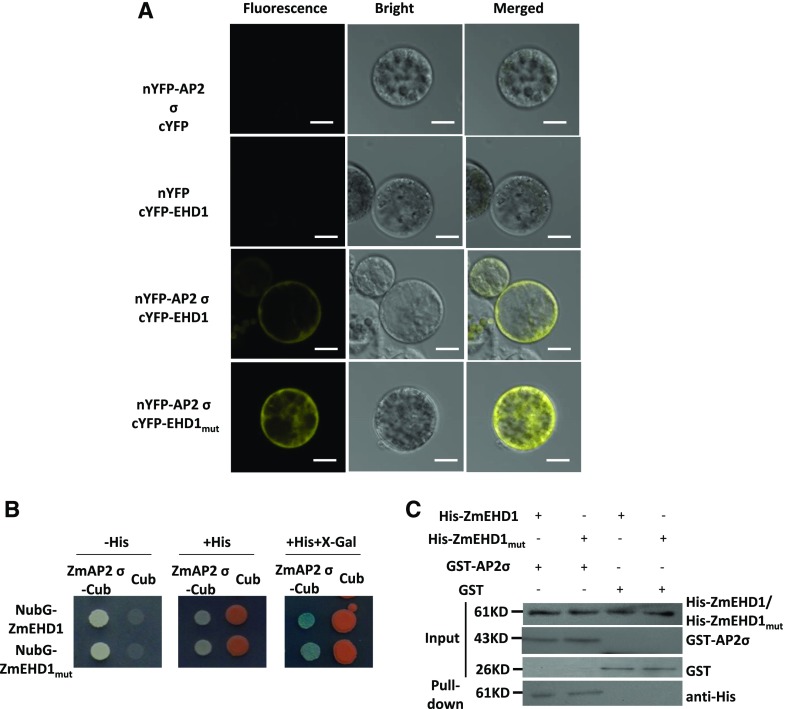
The potential interaction between ZmEHD1 and the ZmAP2 σ subunit was first evaluated by a bimolecular fluorescence complementation (BiFC) assay in maize mesophyll protoplasts. The ZmEHD1, ZmEHD1mut, and ZmAP2 σ subunit constructs were introduced into maize mesophyll protoplasts. Reconstituted yellow fluorescent protein (YFP) signals were observed in the PM when both N-terminal YFP (nYFP)-ZmAP2 σ subunit and C-terminal YFP (cYFP)-ZmEHD1 constructs were coexpressed (Fig. 6A). Reconstituted YFP signals were also observed when the nYFP-ZmAP2 σ subunit and cYFP-ZmEHD1mut constructs were coexpressed (Fig. 6A). Consistent with the subcellular localization of ZmEHD1mut, the reconstituted YFP signals were diffused throughout the cells when the nYFP-ZmAP2 σ subunit and cYFP-ZmEHD1mut constructs were coexpressed (Fig. 6A). We also used a split-ubiquitin membrane-based yeast two-hybrid system to verify the interaction between the ZmAP2 σ subunit and ZmEHD1, as well as ZmEHD1mut. Both ZmEDH1 and ZmEHD1mut interacted directly with ZmAP2 σ subunit in yeast (Saccharomyces cerevisiae). The interaction resulted in survival in a medium lacking His and β-galactosidase activity (Fig. 6B). Pull-down experiments using purified His-ZmEHD1/His-ZmEHD1mut to isolate the GST-ZmAP2 σ subunit also confirmed the physical interaction between ZmEHD1/ZmEHD1mut and the ZmAP2 σ subunit. His-ZmEHD1/His-ZmEHD1mut was clearly detected in the pull-down fraction (Fig. 6C). These results strongly suggested that ZmEHD1 directly interacts with the ZmAP2 σ subunit at the PM in maize.
Figure 6.
ZmEHD1 directly interacts with the ZmAP2 σ subunit in plants and yeast. A, Interaction between ZmEHD1 and the ZmAP2 σ subunit in maize mesophyll protoplasts observed by BiFC assays. Representative photographs are shown. Scale bars = 10 μm. B, Interaction between ZmEHD1 and the ZmAP2 σ subunit as indicated by split-ubiquitin yeast two-hybrid assays. The ZmAP2 σ subunit was used as the fused bait protein (AP2 σ-Cub), and ZmEHD1 was used as the fused prey protein (NubG-ZmEHD1). The presence or absence of His or X-Gal is indicated. C, Pull-down assays for the interaction between ZmEHD1 and the ZmAP2 σ subunit. The ZmEHD1/ZmEHD1mut in the pulled-down fraction was detected by immunoblot using anti-His antibody.
Transcriptome Profiling of the ehd1 Mutant
To gain insight into the molecular events involved in the ZmEHD1-mediated signaling pathway, we compared the whole-transcriptome profiles of endosperms of the ehd1 mutant and the wild type at 15 DAP using RNA sequencing (RNA-seq). Each sample was represented by two biological replicates, and the libraries were sequenced by Illumina high-throughput sequencing technology. These four RNA libraries yielded >0.5 billion raw reads, and ∼97% of the raw reads remained after adaptor polluted reads, Ns reads, and low-quantity reads were trimmed. Of the remaining reads, >0.12 billion could be perfectly mapped to maize B73 RefGen_V3.27 (ftp://ftp.ensemblgenomes.org/pub/plants/release-27/fasta/zea_mays). Sequences that could not be mapped to the maize genome were discarded, and only those perfectly mapped were analyzed further. The abundance of each gene was expressed by reads per kilobase per million mapped reads (Wer et al., 2012). We calculated the Pearson’s correlation coefficients (R value) of the two biological replicates for each treatment to investigate the variability between the replicates. The R value of both comparisons exceeded 0.99 (Supplemental Fig. S5), indicating a high correlation between biological replicates.
Based on the criteria that the log2 fold-change ratio was ≥1 and that the adjusted P value was ≤0.05, 4,760 genes, including ZmEHD1, were identified as differentially expressed genes (DEGs) by DEGseq software (Wang et al., 2010). Among the DEGs, 2,208 were up-regulated and 2,552 genes were downregulated in the ehd1 mutant relative to the wild type (Supplemental Tables S5 and S6). The results of RNA-seq were confirmed by RT-qPCR. In agreement with our RNA-seq data, the expression levels of randomly selected GRMZM2G088273, GRMZM2G346897, GRMZM2G067929, and GRMZM2G418119 were lower in the ehd1 mutant than in the wild type (Supplemental Fig. S6A). As expected, GRMZM2G156877 and GRMZM2G420988 were expressed at higher levels in the ehd1 mutant than in the wild type (Supplemental Fig. S6A), demonstrating the reliability of our RNA-seq data. We then performed Gene Ontology (GO; http://bioinfo.cau.edu.cn/agriGO/) analysis to determine the molecular events related to the DEGs. GO analysis indicated that the 4,760 DEGs were highly enriched in biological processes involved in response to stress (GO:0006950, P = 1.2e−12), integral to the membrane (GO:0016021, P = 1e−5), in intracellular membrane-bounded organelles (GO:0043231, P = 0.0002), in vacuoles (GO:0005773, P = 0.0007), in starch metabolic processes (GO:0005982, P = 1.6e−5), and in carbohydrate metabolic processes (GO:0005975, P = 4.16e−5).
Interestingly, some of the DEGs have known or presumed functions associated with the auxin-mediated signaling pathway (GO:0009734, P = 1.2e−25) and response to auxin stimulus (GO:0009733, P = 2.1e−25), including AUXIN RESPONSE FACTOR (ARF), AUX/IAA transcription factor, small auxin up-regulated RNA (SAUR), indole-3-acetaldehyde oxidase, auxin transporters, and efflux carrier (Table 1). We randomly chose four of the DEGs listed in Table 1 (GRMZM2G082943, GRMZM2G365188, GRMZM5G809195, and GRMZM2G019799) and validated their difference in expression levels in the ehd1 mutant versus the wild type by RT-qPCR (Supplemental Fig. S6B). These results indicated that ZmEHD1 at least partially affected auxin homeostasis in maize.
Table 1. Differentially expressed genes involved in auxin-related processes in the ehd1 mutant.
| Gene Identification | Annotationa | Log2 Fold Change (ehd1 versus Wild Type) | Adjusted P Value |
|---|---|---|---|
| GRMZM2G126260 | PIN10a | −5.53 | 0.0007 |
| GRMZM2G149481 | Auxin transporter-like protein3 | 3.40 | 0.0023 |
| GRMZM2G127949 | Auxin transporter-like protein1 | 1.94 | 2.10e−16 |
| GRMZM2G019799 | Indole-3-acetaldehyde oxidase-like | 1.39 | 0.0006 |
| GRMZM2G082943 | ZAR9 | −2.46 | 2.04e−12 |
| GRMZM2G065244 | Auxin-induced in root cultures protein 12-like | 1.01 | 0.0158 |
| GRMZM2G079957 | AUX17 | −1.17 | 0.0061 |
| GRMZM2G130953 | AUX22 | 1.80 | 0.0004 |
| GRMZM2G081158 | Auxin response factor 25-like | 1.36 | 2.61e−6 |
| GRMZM2G475263 | Auxin response factor1 | 1.15 | 7.27e−11 |
| GRMZM2G028980 | Auxin response factor6 isoform X1 | 1.53 | 1.62e−12 |
| GRMZM5G808366 | Auxin response factor5 | 1.99 | 0.0038 |
| GRMZM2G137413 | Auxin response factor14 | 2.06 | 5.04e−41 |
| GRMZM2G064371 | Auxin binding protein-like protein | 1.38 | 0.0001 |
| GRMZM2G365188 | SAUR23 | −3.37 | 0.0004 |
| GRMZM2G414727 | SAUR56 | −2.95 | 1.12e−26 |
| GRMZM2G059138 | SAUR33 | −2.01 | 7.63e−8 |
| GRMZM2G121309 | IAA7 | 1.16 | 5.26e−8 |
| GRMZM2G030465 | IAA9 | 3.19 | 0.0146 |
| GRMZM2G077356 | IAA13 | 2.90 | 9.83e−18 |
| GRMZM5G809195 | IAA14 | −2.12 | 0.0046 |
| GRMZM2G115357 | IAA24 | −1.25 | 8.09e−7 |
| GRMZM2G048131 | IAA26 | inf | 0.0002 |
Annotation is based on maize IDs
Auxin Distribution and ZmPIN1a-YFPLlocalization in the ehd1 Mutant
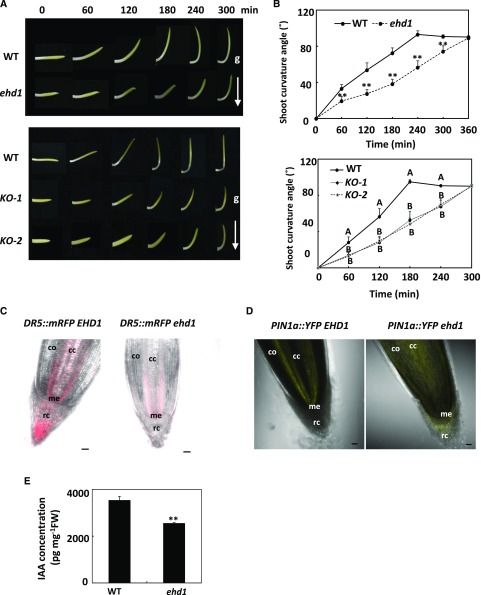
To test the hypothesis that ZmEHD1 might affect auxin homeostasis in maize, we first explored the gravitropic response by examining mesocotyl-coleoptiles. We placed the upright mesocotyl-coleoptiles in the horizontal direction. It took <4 h for the wild type to recover vertical growth, in contrast to ∼5 h for the ehd1 and ZmEHD1 KO mutants under the same conditions (Fig. 7, A and B). To further demonstrate that ZmEHD1 regulates auxin homeostasis in maize, we first crossed the auxin-responsive ZmDR5::RFP reporter maize with the ehd1 mutant to examine auxin distribution in the root tips of the ehd1 mutant. The RFP signals were substantially reduced in the root caps in the ehd1 mutant compared to wild-type maize (Fig. 7C; Supplemental Fig. S7). We also crossed the ZmPIN1a::YFP line with the ehd1 mutant. The YFP signals of PIN1a were mainly detected in the central cylinder of wild-type maize (Fig. 7D). In contrast, the YFP signals in the roots of ehd1 mutant showed more diffuse localization than in wild-type maize, and mainly accumulated in the root cortex (Fig. 7D). The free indole-3-acetic acid (IAA) content in kernels of the ehd1 mutant and the wild type at 15 DAP were also determined. The free IAA content in the kernels of the ehd1 mutant was 2,558 petagram mg−1 fresh weight, which was ∼ 30% lower than that in the WT (Fig. 7E). These results further suggested that auxin homeostasis was altered in the ehd1 mutant.
Figure 7.
Auxin distribution and ZmPIN1a-YFP localization were altered in the ehd1 mutant relative to the wild type (WT). A, The response of horizontally placed mesocotyl-coleoptiles to gravity is delayed in the ehd1 mutant. B, Characterization of the gravitropic response rates of mesocotyl-coleoptiles in the ehd1 mutant. Error bars represent ses (n = 15). **P < 0.01 (Student’s t test) indicates a significant difference between the wild type and the ehd1 mutant. A lsd test was used to assess differences between the wild type and the ZmEHD1 KO mutant. C, The fluorescence of ZmDR5::mRFP in the wild type and the ehd1 mutant. D, Fluorescence of ZmPIN1a::YFP in the wild type and the ehd1 mutant. Scale bars = 50 μm. cc, Central cylinder; me, root meristem; rc, root cap; co, cortex. E, Free IAA content in wild-type maize and ehd1 mutant kernels at 15 DAP. Error bars represent ses (n = 4). **P < 0.01 indicates a significant difference from the wild type according to Student’s t test.
1-NAA Application Rescues the Fertility of the ehd1 Mutant
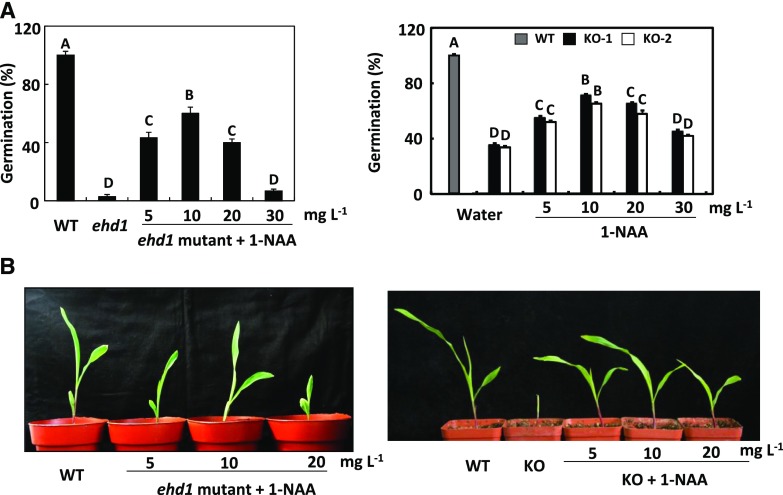
To further verify that the rather low fertility of the ehd1 mutant and the ZmEHD1 KO mutants was caused by auxin homeostasis, we attempted to rescue the phenotypic defects of the ehd1 mutant and the ZmEHD1 KO mutants by exogenous application of the active auxin compound 1-NAA. Seeds of the mutants were rinsed in water or in one of four increasing concentrations of 1-NAA for 12 h and then germinated in the paper-culture system. Little or no phenotypic rescue of the ehd1 mutant was observed after treatment with 30 mg L−1 1-NAA or water alone (Fig. 8, A and B). However, the germination rates of the ehd1 mutant and the ZmEHD1 KO mutants were significantly increased under treatment with low concentrations of 1-NAA (Fig. 8A). Also, when treated with low concentrations of 1-NAA, all mutants survived after their first two leaves had completely expanded (Fig. 8B). We also incubated the seeds of the ehd1 mutant and ZmEHD1 KO mutants with GA3 and the inactive auxin compound 2-NAA. In contrast to 1-NAA, exogenous GA3 and 2-NAA had no effect on the germination of the ehd1 mutant and the ZmEHD1 KO mutants (Supplemental Figs. S8 and S9). Overall, these results demonstrated that the phenotypic defects of the ehd1 mutant could be rescued by exogenous application of 1-NAA.
Figure 8.
Rescue of the ehd1 phenotype by exogenous 1-NAA. A, Germination of the ehd1 mutant and ZmEHD1 KO mutants after treatment with water alone or with different concentrations of 1-NAA. Values are means ± ses of ∼100 seeds from three independent experiments. A lsd test was used to assess differences between treatments. Means with the same letter are not significantly different at P < 0.01. B, Phenotypes of ehd1 and KO seedlings treated with different concentrations of 1-NAA.
DISCUSSION
In this study, we characterized the ehd1 mutant, which is impaired in kernel development and vegetative growth, and used positional cloning and transgenic validation to verify that the ehd1 gene encodes an EHD protein. The results of an FM4-64 uptake experiment, a split-ubiquitin membrane-based yeast two-hybrid system, BiFC, and a pull-down assay indicated that ZmEHD1 is involved in CME through its interaction with the ZmAP2 σ subunit. Additionally, the transcriptome profiling of the ehd1 mutant, the auxin distribution and ZmPIN1a-YFP localization in the ehd1 mutant, and the rescue of the mutant phenotypes following exogenous application of 1-NAA revealed that ZmEHD1-mediated endocytosis mainly affects auxin homeostasis in maize.
In Arabidopsis, the AP2 σ subunit is primarily recruited from the cytoplasm to the PM to initiate clathrin-coated endocytic vesicle formation (Fan et al., 2013, 2015). The developmental defects of ap2 σ loss-of-function Arabidopsis, such as reduced leaf size and fertility, could be rescued by ZmAP2 σ cDNA, suggesting that the ZmAP2 σ subunit has functions similar to those of the AtAP2 σ subunit in CME. According to the publicly available maize microarray database (Waters et al., 2011), ZmEHD1 is expressed in various tissues and its mRNA abundance is high in immature cobs (V18), embryos, and endosperms at 12 and 14 DAP. The AP2 σ subunit was also ubiquitously expressed in various tissues throughout maize development (Sekhon et al., 2011), which is consistent with the expression pattern of ZmEHD1. Based on the direct interaction between the ZmAP2 σ subunit and ZmEHD1 at the PM, as demonstrated in this study by BiFC, pull-down assay, and a split-ubiquitin membrane-based yeast two-hybrid system, we suspected that ZmEHD1 contributes to CME by interacting with the ZmAP2 σ subunit.
The naturally occurring ehd1 mutant had amino acid substitutions in three positions. Of these, the V/A substitution was in a linker region. The other two mutations were in the coiled-coil domain of ZmEHD1. The coiled-coil domain was responsible for protein-protein interactions (Sharma et al., 2008; Bar et al., 2009). Here, we showed that the subcellular location of ZmEHD1mut was obviously different from that of ZmEHD1. Though ZmEHD1mut could directly interact with the ZmAP2 σ subunit, the PM was not the main location for interaction between ZmEHD1mut and the ZmAP2 σ subunit. The impaired clathrin-coated endocytic vesicle formation and/or recycling of the ZmAP2 σ subunit from the endosome to the PM should be major contributors to phenotypes observed in the ehd1 mutant.
The patterning processes of auxin and its intracellular accumulation are determined by auxin transporters, including AUXIN RESISTANT1/LIKE AUX influx proteins (Kerr and Bennett, 2007), the ATP-BINDING cassette subfamily B transporters (Geisler and Murphy, 2006), and PIN efflux carriers (Tanaka et al., 2006). PIN-mediated auxin transport is important for plant growth and development (Benková et al., 2003; Friml et al., 2003; Ding et al., 2011). In Arabidopsis, CME constitutes the predominant pathway for internalization and recycling of PIN proteins (Dhonukshe et al., 2007). The YFP signals of ZmPIN1a in the roots of the ehd1 mutant showed different localization from those in wild-type maize, suggesting that the EHD1 mutation affected the distribution of ZmPIN1a. There are eight PIN genes in Arabidopsis. In contrast, 12 PIN genes and two PIN-like genes were identified in maize (Forestan et al., 2012). ZmPIN10a transcript is localized in both the basal endosperm transfer layer and the embryo-surrounding region endosperm cellular domains at 5 DAP, and is also detectable in the chalazal and pedicel kernel tissues at 12 DAP (Forestan et al., 2012). ZmPIN10a transcript was significantly downregulated in the endosperms of the ehd1 mutant compared to wild-type maize, indicating the importance of ZmPIN10a in maize kernel development.
Arabidopsis ARF2 controls seed size by repressing cell proliferation (Schruff et al., 2006); in rice (Oryza sativa), activation of BIG GRAIN1 (BG1) substantially improves grain size by regulating auxin transport (Liu et al., 2015); TGW6, an IAA-Glc hydrolase, negatively regulates endosperm development of rice (Ishimaru et al., 2013). Analyses of a seed-specific viable maize mutant, defective endosperm 18 (de18), demonstrated that the ZmYUC1 gene (which is critical for IAA biosynthesis) is essential for normal endosperm development in maize (Bernardi et al., 2012). Thus, auxin biosynthesis, transport, and signaling might coordinate to regulate vegetative and reproductive development in maize (Gallavotti et al., 2008; Li and Li, 2016). Using ZmDR5::RFP reporter maize, we demonstrated that less auxin accumulates in ehd1 mutant compared to wild-type maize. The current results also showed that ARFs, indole-3-acetaldehyde oxidase, auxin transporters, and efflux carrier are differentially expressed in the ehd1 mutant compared to the wild type. Importantly, exogenous application of 1-NAA rescues the phenotypes of the ehd1 mutant. These results suggest that the growth defects of the ehd1 mutant result from the loss of auxin homeostasis.
MATERIALS AND METHODS
Plant Materials
The maize (Zea mays) ZmPIN1a::YFP and ZmDR5::mRFP lines were kindly provided by Fang Yang (Huazhong Agricultural University). The maize mutant ehd1 was isolated by screening for natural mutants defective in grain filling. To construct a mapping population, the mutant was crossed with inbred line Xun9058 in the winter of 2011 in Sanya, Hainan Province. Xun9058 is the male parent of the elite hybrid Xundan20 and has been widely used in the breeding of hybrid maize in China. F2 individuals were obtained by selfing the F1 plants in the summer of 2012 in Zhengzhou, Henan Province. The site (113°42′E, 34°48′N) is located in central China and has an average annual temperature of 14.3°C and an average annual rainfall of 640.9 mm. A F2 population with 165 individuals was used to map the preliminary location of the ZmEHD1 gene. Normal kernels from F3 segregation ears were used for fine mapping of the candidate gene. The F3 population contains ∼53,000 individuals. A small piece of each F3 kernel was chipped and genotyped before planting. At harvest stage, the ear phenotype was investigated to verify the kernel segregation of each individual. Two markers, bnlg589 and RM1, were closely linked to the locus to determine recombined individuals.
From the F2 generation, the normal kernels from segregation ears were analyzed and selfed. This process was continued for five generations to construct nearly isogenic lines, which were used as the wild type.
Molecular Markers
Bulked segregant analysis was used to detect the genetic linkage of the ZmEHD1 gene (Michelmore et al., 1991), and 1,082 SSR markers from the maize genome database (www.maizegdb.org) were tested for polymorphism in the two parents and the two bulks. To develop markers for fine mapping, the sequence of the B73 reference genome between markers bnlg589 and umc2289 on chromosome 4 was downloaded from MaizeGDB (http://www.maizegdb.org/). SSR-Hunter software was used to find SSRs. After BLAST was performed (http://blast.ncbi.nlm.nih.gov/Blast.cgi), SSRs with a single copy were developed as markers with PRIMER 5.0. The SSRs were tested by PAGE for polymorphism in the ehd1 mutant, Xun9058, pooled samples of normal kernels (EHD1/EHD1 and EHD1/ehd1), or shrunken kernels (ehd1/ehd1). SSRs with polymorphism were used for subsequent fine mapping. The sequences of the primers used for mapping are listed in Supplemental Table S3.
Transgenic Functional Validation
The CRISPR/Cas9-mediated ZmEHD1 editing was performed as described by Xing et al. (2014), with some modifications. In brief, two guide RNAs that directly target sequences located at nucleotides 16 and 791 of ZmEHD1 were produced using primers listed in Supplemental Table S3. Thereafter, the fragments were cloned into the pBUE411 vector using the BsaI restriction site by T4 ligase reaction. The plasmid contained the Streptomyces hygroscopicus phosphinothricin acetyltransferase gene (bar) under the control of a CaMV 35S promoter and was electroporated into Agrobacterium tumefaciens EHA105. Immature embryos of the maize inbred line ZZC01 were transformed by cocultivation with EHA105 at the Life Science and Technology Center of China National Seed Group. Transformants were selected with gradually increasing concentrations of Bialaphos. T2 homozygous lines were sequenced to ensure that ZmEHD1 was knocked out. Cas9-free plants were identified for phenotypic analysis.
For Arabidopsis transformation, cDNAs of the ZmAP2 σ subunit without the 3′ untranslated region were amplified by PCR. The corresponding products were introduced into the pENTR/D-TOPO vector (Invitrogen) and cloned into pMDC32 by LR reactions (Invitrogen). Transgenic plants were selected with the use of 35 μg mL−1 hygromycin. The sequences of the primer pairs used in the experiments are listed in Supplemental Table S3.
Subcellular Localization of ZmEHD1 and the ZmAP2 σ Subunit
The full-length ZmEHD1, ZmEHD1mut, and ZmAP2 σ subunit coding region was amplified and cloned into the pRTL2 vector, which was fused to a GFP protein in the C terminus using the EcoRI restriction site by In-Fusion reaction. The expression plasmids were transiently expressed in wild-type and ehd1 mesophyll protoplasts as described previously (Cai et al., 2017). The images were collected with a Zeiss LSM700 confocal microscope.
Split-Ubiquitin Yeast Two-Hybrid Assay
The split-ubiquitin two-hybrid system was used to detect the interactions between membrane proteins. The assay was carried out according to the instructions provided with the DUALmembrane Kit (Dualsystems Biotech). The full-length coding regions of the ZmAP2 σ subunit, ZmEHD1, and ZmEHD1mut were amplified and cloned into the vectors pST3-STE and pPR3-N NubG by In-Fusion reaction using the SfiI restriction site. Vectors were cotransformed into yeast strain NMY51. The interactions were assessed by the growth of yeast colonies on synthetic minimal medium containing 7.5 mm 3-aminnotriazole without Leu, Trp, His, and Ade, and also by chloroform overlay β-galactosidase plate assay (Duttweiler, 1996).
BiFC assays
The coding sequences of the ZmAP2 σ subunit, ZmEHD1, and ZmEHD1mut were amplified using specific primers listed in Supplemental Table S3, and were cloned into the pSPYNE-35S or pSPYCE-35S binary vectors using the BamHI restriction site. The various combinations of ZmAP2 σ subunit, ZmEHD1, and ZmEHD1mut expression vectors were transiently expressed in maize mesophyll protoplasts. YFP images were obtained with a Zeiss LSM700 confocal microscope.
Pull-Down Analysis
The full-length coding sequences of ZmEHD1, ZmEHD1mut, and ZmAP2 σ subunit were individually subcloned into the pET-28a(+) and pGEX-4T-1 vectors by In-Fusion reaction using the BamHI restriction site. The resulting constructs were verified by sequencing. His-ZmEHD1, His-ZmEHD1mut, GST, and GST-ZmAP2 σ subunit were expressed in Escherichia coli BL21.
For the in vivo pull-down analysis, 10 μg of purified His-ZmEHD1, His-ZmEHD1mut was incubated with GST-ZmAP2 σ subunit or GST bound to glutathione-Sepharose 4B (GE Healthcare) for 4 h at 4°C on a rotary shaker. Precipitated beads were washed six times with washing buffer. Washed beads were boiled with 50 μL of 1× SDS sample buffer for 5 min and subjected to SDS-PAGE and immunoblot analysis.
FM4-64 Internalization Assay
FM4-64 internalization assays were carried out as described by Fan et al. (2013), with some modifications, to evaluate the rate of endocytosis. Wild-type and ehd1 seedlings were incubated in half-strength Hoagland’s nutrient solution containing 5 μm FM4-64 for 10 min at 22°C. The roots were then cut and transferred to glass slides. The FM4-64 internalization was monitored at the indicated durations at 22°C with a Zeiss LSM700 confocal microscope.
RNA-Seq Analysis
At 15 DAP, total RNA was extracted from the endosperms of the ehd1 mutant and the wild type with TRIZOL reagent (Invitrogen), and 3 μg of total RNA was used as input material for construction of the RNA libraries. The RNA-seq libraries were constructed with NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs). In brief, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. The enriched mRNA was then fragmented. The random hexamers were used for first-strand cDNA synthesis. After second-strand cDNA synthesis, terminal repair, and poly(A) tail and sequencing oligonucleotide adaptors ligation, the fragments were purified and subsequently amplified by PCR. The insert size was assessed with the Agilent Bioanalyzer 2100 system (Agilent Technologies). Finally, the libraries of inserting cDNAs 200 bp in size were generated and sequenced with the IlluminaHiseq 2500 platform (ANROAD).
The raw reads were produced after exclusion of low-quality reads and 5′ and 3′ adaptor contaminants. The unique RNAs were aligned to the maize RefGen_V3.27 (ftp://ftp.ensemblgenomes.org/pub/plants/release-27/fasta/zea_mays). Only perfectly matching sequences were considered for further analysis. The count information was used to determine normalized gene expression levels as reads per kilobase per million mapped reads (Wagner et al., 2012). Multiple testing with the Benjamini-Hochberg procedure for false discovery rate (FDR) was taken into account by using an adjusted P-value. Genes were considered to be differentially expressed if the Log2 fold-change ratio was ≥1 and if the adjusted P-value was <0.05 according to the DEGseq method (Wang et al., 2010).
Free IAA Analysis
At 15 DAP, kernels of the wild type and the ehd1 mutant were collected and frozen in liquid nitrogen. A 200-mg (fresh weight) sample of wild-type and ehd1 kernels was finely ground in liquid nitrogen and then extracted with 1 mL of methanol containing antioxidant and 2H2-IAA (internal standard; CDN Isotopes) at 4°C for 24 h. After centrifugation, the extract was purified with an Oasis Max solid-phase extract cartridge (150 mg/6 cc; Waters). IAA was quantified using ultra-performance liquid chromatography (UPLC) tandem mass spectrometry consisting of a UPLC system (ACQUITY UPLC; Waters) and a triple quadruple tandem mass spectrometer (Quattro Premier XE; Waters), as described by Wang et al. (2015). Four independent biological replicates and two technical repeats were performed for the wild type and the ehd1 mutant.
Starch Content Measurement
To measure starch content, 2.5 g of wild-type and 2.5 g of ehd1 kernels were finely ground. Total starch was quantified using the Ewers polarimetric method as described by Mitchell (1990). Three independent biological replicates were performed for the wild type and the ehd1 mutant.
Phytohormone Treatments
For phytohormone treatments, wild-type and ehd1 mutant seeds were immersed in water and the indicated concentrations of 1-NAA and 2-NAA for 12 h or GA3 for 24 h (Fig. 8; Supplemental Figs. S8 and S9). The seeds were then kept at 22°C in the dark for 120 h for germination. A seed was scored as germinated if its radicle protruded through the seed coat.
Gravitropism Analysis
The sterilized seeds were sown in sand and grown in the dark at 28°C for 36 h. Gravitropism analysis was performed as described by Dong et al. (2013).
Statistical Analyses
Differences in phenotypic values were examined by Student’s t tests or one-way ANOVA.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers GRMZM2G052740 (ZmEHD1), At3g20290 (AtEHD1), At4g05520 (AtEHD2), GRMZM2G052713 (ZmAP2 σ subunit), GRMZM2G098643 (ZmPIN1a), and At1g47380 (AtAP2 σ subunit).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignment with AtEHD1, AtEHD2, ZmEHD1, and ZmEHDmut.
Supplemental Figure S2. Target deletion of ZmEHD1 in ZmEHD1 loss-of-function mutants observed by alignment and RT-qPCR.
Supplemental Figure S3. Rescue of Arabidopsis ap2 σ mutant by ZmAP2 σ cDNA.
Supplemental Figure S4. The different subcellular localization of ZmEHD1mut protein and the ZmAP2 σ subunit in wild-type maize and the ehd1 mutant.
Supplemental Figure S5. Pearson’s correlation coefficients (R values) of biological replicates in wild-type maize and the ehd1 mutant.
Supplemental Figure S6. Validation of RNA-seq by RT-qPCR.
Supplemental Figure S7. Boxplot of the ZmDR5::mRFP fluorescence replicates in the wild type and the ehd1 mutant.
Supplemental Figure S8. Germination of the ehd1 mutant and the ZmEHD1 KO mutant treated with water alone or with different concentrations of 2-NAA.
Supplemental Figure S9. Germination of the ehd1 mutant and the ZmEHD1 KO mutant treated with water alone or with different concentrations of GA3.
Supplemental Table S1. Ear performance of F2 and F3 individuals evaluated in the fields.
Supplemental Table S2. Kernel performance of the F2 population evaluated in the fields.
Supplemental Table S3. Oligos used as primers in the experiment.
Supplemental Table S4. Allelism test between KO+/− and ehd1+/−.
Supplemental Table S5. List of genes with lower expression in ehd1 mutants.
Supplemental Table S6. List of genes with higher expression in ehd1 mutants.
Acknowledgments
We thank Dr. Jinxing Lin for the seeds of Arabidopsis ap2 σ loss-of-function mutant and Dr. Youfa Cheng for useful comments.
Footnotes
This work was supported by the National Key Research and Development Program of China (grant 2016YFD0101002 to W.-X.L.) and the National Basic Research Program of China (grant 31370303 to J.T.).
References
- Backues SK, Korasick DA, Heese A, Bednarek SY (2010) The Arabidopsis dynamin-related protein2 family is essential for gametophyte development. Plant Cell 22: 3218–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar M, Aharon M, Benjamin S, Rotblat B, Horowitz M, Avni A (2008) AtEHDs, novel Arabidopsis EH-domain-containing proteins involved in endocytosis. Plant J 55: 1025–1038 [DOI] [PubMed] [Google Scholar]
- Bar M, Avni A (2009) EHD2 inhibits ligand-induced endocytosis and signaling of the leucine-rich repeat receptor-like protein LeEix2. Plant J 59: 600–611 [DOI] [PubMed] [Google Scholar]
- Bar M, Sharfman M, Schuster S, Avni A (2009) The coiled-coil domain of EHD2 mediates inhibition of LeEix2 endocytosis and signaling. PLoS One 4: e7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberon M, Zelazny E, Robert S, Conéjéro G, Curie C, Friml J, Vert G (2011) Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc Natl Acad Sci USA 108: E450–E458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Bernardi J, Lanubile A, Li QB, Kumar D, Kladnik A, Cook SD, Ross JJ, Marocco A, Chourey PS (2012) Impaired auxin biosynthesis in the defective endosperm18 mutant is due to mutational loss of expression in the ZmYuc1 gene encoding endosperm-specific YUCCA1 protein in maize. Plant Physiol 160: 1318–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackbourn HD, Jackson AP (1996) Plant clathrin heavy chain: Sequence analysis and restricted localisation in growing pollen tubes. J Cell Sci 109: 777–786 [DOI] [PubMed] [Google Scholar]
- Cai M, Li S, Sun F, Sun Q, Zhao H, Ren X, Zhao Y, Tan BC, Zhang Z, Qiu F (2017) Emp10 encodes a mitochondrial PPR protein that affects the cis-splicing of nad2 intron 1 and seed development in maize. Plant J 91: 132–144 [DOI] [PubMed] [Google Scholar]
- Deo R, Kushwah MS, Kamerkar SC, Kadam NY, Dar S, Babu K, Srivastava A, Pucadyil TJ (2018) ATP-dependent membrane remodeling links EHD1 functions to endocytic recycling. Nat Commun 9: 5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P, Aniento F, Hwang I, Robinson DG, Mravec J, Stierhof YD, Friml J (2007) Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr Biol 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Di Rubbo S, Irani NG, Kim SY, Xu ZY, Gadeyne A, Dejonghe W, Vanhoutte I, Persiau G, Eeckhout D, Simon S, et al. (2013) The clathrin adaptor complex AP-2 mediates endocytosis of brassinosteroid insensitive1 in Arabidopsis. Plant Cell 25: 2986–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Galván-Ampudia CS, Demarsy E, Łangowski Ł, Kleine-Vehn J, Fan Y, Morita MT, Tasaka M, Fankhauser C, Offringa R, et al. (2011) Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat Cell Biol 13: 447–452 [DOI] [PubMed] [Google Scholar]
- Dong Z, Jiang C, Chen X, Zhang T, Ding L, Song W, Luo H, Lai J, Chen H, Liu R, et al. (2013) Maize LAZY1 mediates shoot gravitropism and inflorescence development through regulating auxin transport, auxin signaling, and light response. Plant Physiol 163: 1306–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duttweiler HM. (1996) A highly sensitive and non-lethal β-galactosidase plate assay for yeast. Trends Genet 12: 340–341 [DOI] [PubMed] [Google Scholar]
- Fan L, Hao H, Xue Y, Zhang L, Song K, Ding Z, Botella MA, Wang H, Lin J (2013) Dynamic analysis of Arabidopsis AP2 σ subunit reveals a key role in clathrin-mediated endocytosis and plant development. Development 140: 3826–3837 [DOI] [PubMed] [Google Scholar]
- Fan L, Li R, Pan J, Ding Z, Lin J (2015) Endocytosis and its regulation in plants. Trends Plant Sci 20: 388–397 [DOI] [PubMed] [Google Scholar]
- Forestan C, Farinati S, Varotto S (2012) The maize PIN gene family of auxin transporters. Front Plant Sci 3: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Gallavotti A, Barazesh S, Malcomber S, Hall D, Jackson D, Schmidt RJ, McSteen P (2008) sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc Natl Acad Sci USA 105: 15196–15201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Murphy AS (2006) The ABC of auxin transport: The role of p-glycoproteins in plant development. FEBS Lett 580: 1094–1102 [DOI] [PubMed] [Google Scholar]
- Ishimaru K, Hirotsu N, Madoka Y, Murakami N, Hara N, Onodera H, Kashiwagi T, Ujiie K, Shimizu B, Onishi A, et al. (2013) Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat Genet 45: 707–711 [DOI] [PubMed] [Google Scholar]
- Kerr ID, Bennett MJ (2007) New insight into the biochemical mechanisms regulating auxin transport in plants. Biochem J 401: 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Xu ZY, Song K, Kim DH, Kang H, Reichardt I, Sohn EJ, Friml J, Juergens G, Hwang I (2013) Adaptor protein complex 2-mediated endocytosis is crucial for male reproductive organ development in Arabidopsis. Plant Cell 25: 2970–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitakura S, Vanneste S, Robert S, Löfke C, Teichmann T, Tanaka H, Friml J (2011) Clathrin mediates endocytosis and polar distribution of PIN auxin transporters in Arabidopsis. Plant Cell 23: 1920–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Li Y (2016) Signaling pathways of seed size control in plants. Curr Opin Plant Biol 33: 23–32 [DOI] [PubMed] [Google Scholar]
- Lin SX, Grant B, Hirsh D, Maxfield FR (2001) Rme-1 regulates the distribution and function of the endocytic recycling compartment in mammalian cells. Nat Cell Biol 3: 567–572 [DOI] [PubMed] [Google Scholar]
- Liu L, Tong H, Xiao Y, Che R, Xu F, Hu B, Liang C, Chu J, Li J, Chu C (2015) Activation of Big Grain1 significantly improves grain size by regulating auxin transport in rice. Proc Natl Acad Sci USA 112: 11102–11107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren JS. (2005) Crop biotechnology provides an opportunity to develop a sustainable future. Trends Biotechnol 23: 339–342 [DOI] [PubMed] [Google Scholar]
- McMahon HT, Boucrot E (2011) Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol 12: 517–533 [DOI] [PubMed] [Google Scholar]
- Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88: 9828–9832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell GA. (1990) Methods of starch analysis. Starch 42: 131–134 [Google Scholar]
- Murphy AS, Bandyopadhyay A, Holstein SE, Peer WA (2005) Endocytotic cycling of PM proteins. Annu Rev Plant Biol 56: 221–251 [DOI] [PubMed] [Google Scholar]
- Olswang-Kutz Y, Gertel Y, Benjamin S, Sela O, Pekar O, Arama E, Steller H, Horowitz M, Segal D (2009) Drosophila Past1 is involved in endocytosis and is required for germline development and survival of the adult fly. J Cell Sci 122: 471–480 [DOI] [PubMed] [Google Scholar]
- Robatzek S, Chinchilla D, Boller T (2006) Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev 20: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, Scott RJ (2006) The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133: 251–261 [DOI] [PubMed] [Google Scholar]
- Sekhon RS, Lin H, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM (2011) Genome-wide atlas of transcription during maize development. Plant J 66: 553–563 [DOI] [PubMed] [Google Scholar]
- Sharma M, Naslavsky N, Caplan S (2008) A role for EHD4 in the regulation of early endosomal transport. Traffic 9: 995–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigismund S, Confalonieri S, Ciliberto A, Polo S, Scita G, Di Fiore PP (2012) Endocytosis and signaling: Cell logistics shape the eukaryotic cell plan. Physiol Rev 92: 273–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Dhonukshe P, Brewer PB, Friml J (2006) Spatiotemporal asymmetric auxin distribution: A means to coordinate plant development. Cell Mol Life Sci 63: 2738–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J, Tanaka M, Toyoda A, Miwa K, Kasai K, Fuji K, Onouchi H, Naito S, Fujiwara T (2010) Polar localization and degradation of Arabidopsis boron transporters through distinct trafficking pathways. Proc Natl Acad Sci USA 107: 5220–5225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme D, Gadeyne A, Vanstraelen M, Inzé D, Van Montagu MC, De Jaeger G, Russinova E, Geelen D (2011) Adaptin-like protein TPLATE and clathrin recruitment during plant somatic cytokinesis occurs via two distinct pathways. Proc Natl Acad Sci USA 108: 615–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP, Kin K, Lynch VJ (2012) Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci 131: 281–285 [DOI] [PubMed] [Google Scholar]
- Wang B, Chu J, Yu T, Xu Q, Sun X, Yuan J, Xiong G, Wang G, Wang Y, Li J (2015) Tryptophan-independent auxin biosynthesis contributes to early embryogenesis in Arabidopsis. Proc Natl Acad Sci USA 112: 4821–4826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Yan X, Chen Q, Jiang N, Fu W, Ma B, Liu J, Li C, Bednarek SY, Pan J (2013) Clathrin light chains regulate clathrin-mediated trafficking, auxin signaling, and development in Arabidopsis. Plant Cell 25: 499–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Feng Z, Wang X, Wang X, Zhang X (2010) DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26: 136–138 [DOI] [PubMed] [Google Scholar]
- Waters AJ, Makarevitch I, Eichten SR, Swanson-Wagner RA, Yeh CT, Xu W, Schnable PS, Vaughn MW, Gehring M, Springer NM (2011) Parent-of-origin effects on gene expression and DNA methylation in the maize endosperm. Plant Cell 23: 4221–4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo W, Chao Q, Zhang N, Ye J, Tan G, Li B, Xing Y, Zhang B, Liu H, Fengler KA, et al. (2015) A maize wall-associated kinase confers quantitative resistance to head smut. Nat Genet 47: 151–157 [DOI] [PubMed] [Google Scholar]