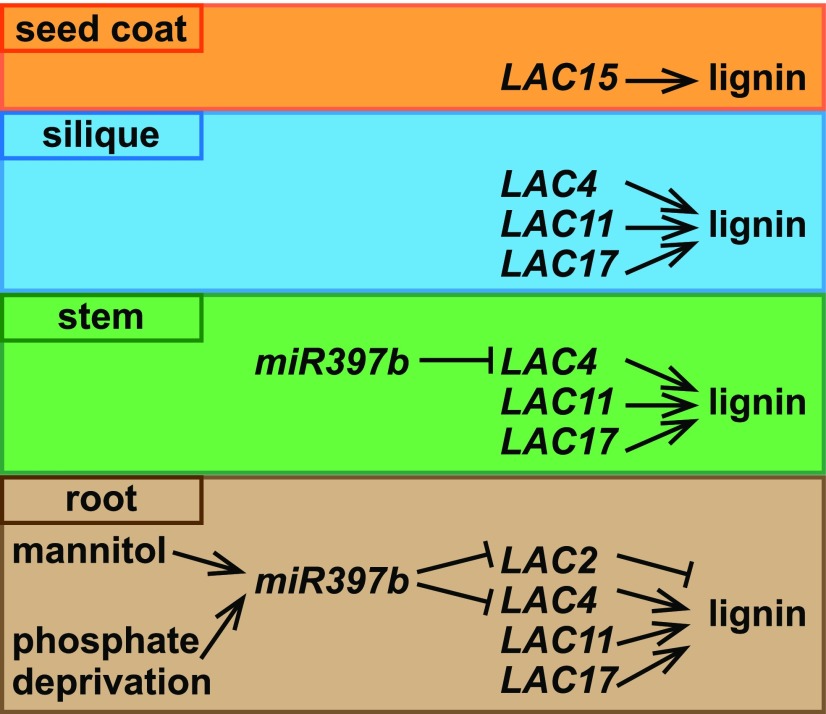
Lignin is a complex aromatic biopolymer in the secondary cell wall of vascular plants. It is crucial for providing physical strength and pathogen resistance to the plant. Lignified xylem vessels are also necessary for efficient water transport. Lignin is formed by oxidative polymerization of monomers including p-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol. These monolignols are synthesized in the cytosol and transported into the cell wall, where they are polymerized and incorporated into p-hydroxyphenyl, guaiacyl, and syringyl units of lignin, respectively (Wang et al., 2015). Monolignol polymerization is catalyzed by oxidative enzymes such as hydrogen peroxide-dependent peroxidases and oxygen-dependent laccases (Tobimatsu and Schuetz, 2019). Laccases are a form of multicopper oxidoreductases, which are present in both eukaryotes and prokaryotes. Arabidopsis (Arabidopsis thaliana) has 17 laccases, some of which have been demonstrated to act in lignin polymerization in various tissue types (Fig. 1; Wang et al., 2015).
Figure 1.
LACCASEs regulate lignin polymerization in various organs of Arabidopsis. Figure by Yunqing Yu.
In this issue of Plant Physiology, Khandal et al. (2020) report that LACCASE2 (LAC2) is a negative regulator of lignin deposition in root vascular tissues under abiotic stress. In wild-type seedlings, the expression of LAC2 is drastically decreased by mannitol treatment, accompanied by increased lignin content in roots and arrested root growth. Under normal conditions, the lac2 mutants have shorter primary roots than the wild type and enhanced lignin deposition in root xylem, consistent with the negative role of LAC2 in lignin deposition (Khandal et al., 2020). Intriguingly, other characterized LAC genes are all positive regulators of lignin polymerization. For example, the lac4 lac17 double mutants contain 20% to 40% less lignin in the Arabidopsis stem compared to the wild type (Berthet et al., 2011). Additional disruption of LAC11 along with LAC4 and LAC17 results in severely reduced lignification in multiple organs including stem, root, and siliques, and a tiny plant stature (Berthet et al., 2011; Zhao et al., 2013). Furthermore, LAC15 is expressed mainly in the seed coat, and the lac15 mutant has decreased extractable lignin and increased soluble proanthocyanidins, resulting in altered seed color (Fig. 1; Pourcel et al., 2005; Liang et al., 2006). In this study, Khandal et al. (2020) showed that expression of these other LAC genes is not significantly changed in the lac2 background compared to the wild type, refuting the idea that the lac2 phenotype is due to overexpression of other LAC genes.
LAC genes are posttranscriptionally regulated by microRNAs (Wang et al., 2015). An earlier study examined 2-week–old seedlings and found that miR397b negatively regulates the transcript levels of several LAC genes, including LAC2, LAC4, and LAC17. miR397b-overexpression lines exhibited reduced lignification in the vascular and interfascicular tissues of stems compared to the wild type (Wang et al., 2014). By contrast, Khandal et al. (2020) focused on root tissues and showed that LAC2 and LAC4, but not LAC17, are regulated by miR397b, with LAC2 being the dominant target. Furthermore, the miR397b overexpression lines exhibit more lignin deposition in root xylem compared to the wild type. Consistent with these results, overexpression lines either blocking miR397b function by expressing a short tandem target mimic (STTM) or mutating the miR397b target site of LAC2 (rLAC2) both result in increased LAC2 transcript abundance, reduced lignification in root xylem, and longer primary roots (Khandal et al., 2020). These results indicate that miR397b may have affinity for different LAC targets depending on tissue types and/or developmental stages.
Abiotic stress is often associated with inhibition of root growth and promotion of root xylem lignification. However, whether laccases are involved in this process had not been investigated previously. Khandal et al. (2020) compared root growth and lignification in response to dehydration and phosphate deprivation between the genotypes with lower LAC2 expression (lac2 and the miR397b overexpression line, miR397bOX) and those with higher LAC2 expression (LAC2OX and STTM), and revealed a correlation between root developmental response and LAC2 expression. Specifically, LAC2 expression is inhibited by both mannitol and phosphate deprivation treatments, whereas miR397b expression is enhanced by both treatments. Likewise, lac2 and miR397bOX lines are hypersensitive to both treatments, whereas LAC2OX and STTM lines are hyposensitive. Nonetheless, the hyposensitive lines still accumulate considerable lignin under stress conditions compared to normal growth conditions, suggesting that miR397b-regulated LAC2 is partially responsible for lignin deposition induced by dehydration and phosphate deprivation (Khandal et al., 2020).
Despite growing genetic evidence of laccases being under both developmental and environmental control in lignin formation, their exact role in lignin polymerization is still poorly understood. For example, although fungal laccases have been reported to catalyze lignin degradation (Barros et al., 2015), no such evidence has been demonstrated in plants, and thus it is not clear how Arabidopsis LAC2 inhibits lignin deposition. Laccases may be monomeric, dimeric, and tetrameric glycoproteins, and have broad in vitro substrates (Berthet et al., 2012). Disruption of LAC2 may thus affect the catalytic activity of the laccase complex, or may enhance lignification indirectly by catalyzing other metabolic reactions that promote monolignol biosynthesis. Metabolic profiling of intermediates in the lignin biosynthetic pathway may further elucidate the role of laccases in lignin formation.
References
- Barros J, Serk H, Granlund I, Pesquet E (2015) The cell biology of lignification in higher plants. Ann Bot 115: 1053–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthet S, Demont-Caulet N, Pollet B, Bidzinski P, Cézard L, Le Bris P, Borrega N, Hervé J, Blondet E, Balzergue S, Lapierre C, Jouanin L (2011) Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23: 1124–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthet S, Thevenin J, Baratiny D, Demont-Caulet N, Debeaujon I, Bidzinski P, Leple J-C, Huis R, Hawkins S, Gomez LD, et al. (2012) Role of plant laccases in lignin polymerization In Jouanin L, and Lapierre C, eds, Advances in Botanical Research, Vol Vol 61 Elsevier Academic Press, New York, pp 145–172 [Google Scholar]
- Khandal H, Singh AP, Chattopadhyay D (2020) MicroRNA397b-LACCASE2 module regulates root lignification under water- and phosphate deficiency. Plant Physiol 182: 1387–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M, Davis E, Gardner D, Cai X, Wu Y (2006) Involvement of AtLAC15 in lignin synthesis in seeds and in root elongation of Arabidopsis. Planta 224: 1185–1196 [DOI] [PubMed] [Google Scholar]
- Pourcel L, Routaboul J-M, Kerhoas L, Caboche M, Lepiniec L, Debeaujon I (2005) TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 17: 2966–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobimatsu Y, Schuetz M (2019) Lignin polymerization: How do plants manage the chemistry so well? Curr Opin Biotechnol 56: 75–81 [DOI] [PubMed] [Google Scholar]
- Wang CY, Zhang S, Yu Y, Luo YC, Liu Q, Ju C, Zhang YC, Qu LH, Lucas WJ, Wang X, Chen YQ (2014) MiR397b regulates both lignin content and seed number in Arabidopsis via modulating a laccase involved in lignin biosynthesis. Plant Biotechnol J 12: 1132–1142 [DOI] [PubMed] [Google Scholar]
- Wang J, Feng J, Jia W, Chang S, Li S, Li Y (2015) Lignin engineering through laccase modification: A promising field for energy plant improvement. Biotechnol Biofuels 8: 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Nakashima J, Chen F, Yin Y, Fu C, Yun J, Shao H, Wang X, Wang Z-Y, Dixon RA (2013) Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25: 3976–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]