Abstract
Fibrosis of the kidney is the final common pathway leading to end stage renal failure. By analyzing kidneys of patients and animal models with fibrosis we observed a significant mitochondrial defect, including the loss of the mitochondrial transcription factor A (TFAM) in kidney tubule cells. Here, we generated mice with tubule-specific deletion of TFAM (Ksp-Cre/Tfam flox/flox). While these mice developed severe mitochondrial loss and energetic deficit (ATP level decline) by 6 weeks of age, kidney fibrosis, immune cell infiltration and progressive azotemia causing death was only observed around 12 weeks of age. Mechanistic studies demonstrated that in the TFAM KO mice aberrant packaging of the mitochondrial DNA (mtDNA) resulted in escape of the mtDNA into the cytosol of the renal cells, activation of the cytosolic cGAS-STING (Stimulator of interferon genes) DNA sensing pathway, and thus cytokine expression and immune cell recruitment. Genetic deletion or pharmacological inhibition of STING ameliorated kidney fibrosis in mouse models of chronic kidney disease, demonstrating that in addition to its essential role in metabolism TFAM sequesters mtDNA to prevent the activation of innate immune pathways and fibrosis.
Keywords: Chronic kidney disease, renal fibrosis, TFAM (mitochondrial transcription factor A), mitochondrial DNA, innate immunity, cGAS-STING pathway
Introduction
One in 10 people in the world suffer from chronic kidney disease (CKD)(Collins et al., 2010; Levin et al., 2017). CKD is the 10th leading cause of death worldwide (Levin et al., 2017), and diabetic kidney disease (DKD) and hypertensive kidney disease (HKD) account for more than 75% of the incidence of this condition (Foley, 2010; Kovesdy et al., 2013; Zelnick et al., 2017). Over the last two decades, no new drugs have been approved to specifically prevent CKD or to improve kidney function (Breyer and Susztak, 2016; Tieu et al., 2016). The lack of progress, likely stems from our poor understanding of the mechanism of kidney dysfunction (Breyer and Susztak, 2016; Huang and Susztak, 2016; Reidy et al., 2014).
While studies on the genetics of chronic and diabetic kidney disease have not yielded common genes and mechanisms for CKD, unbiased microarray-based gene expression analysis have indicated dysregulation of a few common pathways. This includes the unexpected activation of immune cells, genes and pathways in DKD and HKD (Beckerman et al., 2017; Qiu et al., 2018; Woroniecka et al., 2011). As HKD and DKD do not respond to conventional immunosuppressive drugs such as glucocorticoids, these diseases were traditionally grouped under non-immune mediated CKD. Recent human and mouse studies, however, have demonstrated the functional role of inflammation in DKD (Fischer et al., 2010; Lemos et al., 2018; Rodwell et al., 2004; Rudnicki et al., 2007; Wu et al., 2018). For example, blockade of monocyte chemoattractant protein 1 (CCL2) has reduced albuminuria in phase II clinical trials (https://clinicaltrials.gov/) in patients with DKD. Tumor necrosis factor receptor 1 (TNFRSF1A) remains one of the best biomarkers for progressive functional decline in patients with diabetes (Niewczas et al., 2019). The role of several other cytokines such as interleukin-6 (IL6), chemokine (C-C motif) ligand 2 (CCL2), chemokine (C-C motif) ligand 5 (CCL5), and interleukin-17 (IL17) have been demonstrated in animal model systems (Pichler et al., 2017). While increasing evidence suggest the role of inflammation in DKD, the upstream regulators of immune system activation in DKD remains unclear. Another key question has been the relationship between immune activation and metabolic defects. A large body of literature indicates the role of low-grade inflammation and various cytokines as key drivers of metabolic alterations in a wide range of cell types (Bonnet and Scheen, 2018; Wada and Makino, 2016). It is however unclear whether the inflammatory process is the key driver of the metabolic dysfunction observed in CKD.
The function of the kidney is to maintain electrolyte, water and acid-base balance while eliminating toxic waste from the body. Fundamentally, the glomerulus is an ~60 kDa size-selective filter. This broad permeability requires long tubule segments to reclaim >99% of the filtered small molecule, which is a highly energetic process. Most kidney tubule segments are mitochondrial-enriched and depend almost exclusively on fatty acid oxidation (FAO) and ensuing mitochondrial oxidative phosphorylation (OXPHOS) as their sole energy source to meet their functional needs (Han et al., 2015; Kang et al., 2015; Mandel, 1985; Tran et al., 2016). Mitochondrial biogenesis requires the coordinated expression of the nuclear- and mitochondrial-encoded subunits of the mitochondrial respiratory complexes (Tanaka et al., 2019). TFAM is a key regulator of mitochondrial gene expression (Scarpulla, 2008). It is a downstream target of key metabolic regulators such as peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC1α) and peroxisome proliferator-activated receptor alpha (PPARα). TFAM is the most abundant protein associated with mitochondrial DNA (mtDNA) and is crucial for maintaining mtDNA structure, transcription, and replication (Campbell et al., 2012). Global Tfam knockout is embryonically lethal (Larsson et al., 1998). However, tissue-specific ablation of Tfam has been used to model mitochondrial dysfunction observed in different human diseases (West et al., 2015; Li et al., 2000; Peralta et al., 2012; Vernochet et al., 2012; Wang et al., 1999).
A correlation between kidney function, fatty acid oxidation and tubule cell energetics have been demonstrated repeatedly, suggesting that changes in tubule epithelial metabolism directly contribute to CKD and fibrosis development (Han et al., 2015; Huang et al., 2018; Kang et al., 2015; Tran et al., 2016). In mice, functional and structural parameters can be improved by tubule-specific transgenic overexpression of key transcriptional regulators of metabolism, such as PPARα, or PGC1α or by pharmacological activators of PPARα, such as fenofibrate (Han et al., 2017; Kang et al., 2015). Consistently, genetic deletion of Estrogen Related Receptor gamma (ESRRγ), a key transcription factor that regulates metabolism, in kidney tubules causes severe structural defect without leading to end stage kidney failure (Zhao et al., 2018). On the other hand, surprisingly, genetic deletion of PPARα, PPARγ, farnesoid X receptor (FXR), liver X Receptor (LXR), ESRRα or PGC1α does not lead to observable renal phenotype at baseline (Guan, 2004; Kamijo et al., 2007; Park et al., 2006; Ruan et al., 2008; Su et al., 2017; Tran et al., 2011; Tsushida et al., 2018; Zhang et al., 2005; Zhang et al., 2014). It is difficult to reconcile the lack of phenotype development in these animal models with the critical role of metabolism in CKD and fibrosis development. The activation of compensatory pathways or an underappreciated metabolic reserve of kidney tubules could underlie the preserved phenotypes, but these observations clearly indicate the need for further studies to elucidate the relationship between kidney tubule cell metabolism and CKD.
In this study, we aimed to understand pathways that control tubule metabolism, and their relationship to changes in cytokine expression and immune cell activation. To address these issues, we generated mice with genetic deletion of Tfam specifically in kidney tubule cells to mimic the mitochondrial and metabolic defect observed in kidney fibrosis. We show that these animals develop a metabolic defect around 6 weeks of age, characterized by reduced mitochondrial number and reduced concentration of ATP. Despite overt mitochondrial dysfunction, the animals survive until ~12 weeks of age. At this point, cytokine activation and immune cell infiltration can be detected. We show that in the absence of TFAM, mtDNA escapes into the cytoplasm and activates the innate immune pathway STING, which strongly contributes to cell death and kidney failure. Genetic deletion or pharmacological inhibitors of STING ameliorated both TFAM loss-induced and FA-induced kidney fibrosis. Furthermore small molecular inhibitors also showed effectiveness. Our results indicate a key role of activation of the innate viral response pathway, rather than compromised bioenergetics per se, in mediating the detrimental consequences of metabolic dysfunction in the kidney.
Results
Kidneys of patients with CKD and mouse kidney fibrosis models are characterized by reduced expression of TFAM, mitochondrial genes and activation of immune pathways
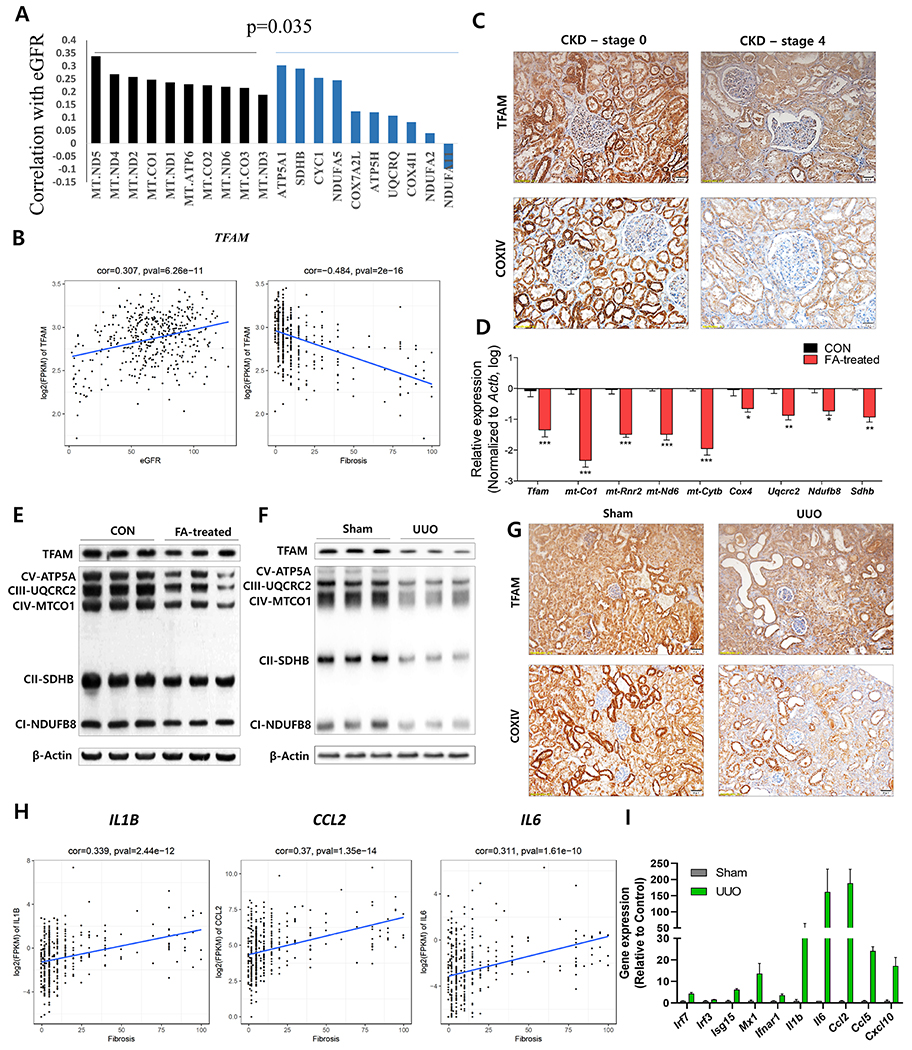
Here, we analyzed 433 microdissected human kidney tubule samples collected from subjects with normal kidney function in the presence and absence of hypertension (HTN), diabetes (DM), and subjects with HKD and DKD. Clinical demographics of the subjects and histological analysis of kidney samples can be found in Supplemental Table 1. Gene expression was quantified by RNA-sequencing. We found that the expression of all mitochondrial-encoded genes significantly correlated with estimated glomerular filtration rate (eGFR) (Figure 1A). On the other hand, not all nuclearly-encoded mitochondrial genes correlated with kidney function (Figure 1A). Overall, a significant difference was detected between the correlation of mitochondrially and nuclearly-encoded genes and kidney function (i.e., eGFR) (Figure 1A, Supplemental Figure 1A). The expression of TFAM, which plays key roles in the maintenance of mtDNA and control mitochondrially-encoded gene expression, positively correlated with eGFR and negatively correlated with the extent of fibrosis (Figure 1B, 1C). Protein level of Cytochrome C Oxidase Subunit IV (COXIV), that labels mitochondria, indicated a strong signal in kidney tubules and confirmed lower mitochondrial content in CKD (Figure 1C).
Figure 1. Mouse and human kidney disease is characterized a lower expression of TFAM and metabolic genes and an higher level of inflammatory genes.
(A) Correlation between kidney function (eGFR) and transcript levels of mitochondrial (Black) and nuclear-encoded mitochondrial genes (Blue). (B) Relative transcript levels of TFAM (FPKM) and kidney function (eGFR) kidney fibrosis as analyzed in 433 microdissected human kidney samples. (C) TFAM and COXIV expressions in human chronic kidney diseases (stage 0 and stage 4) were visualized by immunohistochemical (IHC) staining. Scale bar = 20 μm. (D) Relative mRNA levels of Tfam and mitochondrial OXPHOS genes (mt-Co1, mt-Rnr2, mt-Nd6, mt-Cytb, Cox4, Uqcrc2, Ndufb8, and Sdhb) in FA-induced mice kidney fibrosis model (red) as compared to control (Black). * P < 0.05, ** P < 0.01, *** P < 0.001 . (E) Protein levels of TFAM and mitochondrial OXPHOS proteins (CV-ATP5A, CIII-UQCRC2, CIV-MTCO1, CII-SDHB, and CI-NDUFB8) in FA-induced mice kidney fibrosis model and (F) UUO-induced mice kidney fibrosis models. β-actin was used as a loading control. (G) TFAM and COXIV expressions in UUO-induced mice kidney fibrosis model were visualized by IHC staining. Scale bar = 20 μm. (H) Transcript levels (RNA-sequencing) of IL6, CCL2, and IL1B, correlated with the degree of kidney fibrosis in 433 microdissected human kidney samples. (I) Proinflammatory gene expression (RNA-seq) in UUO-induced kidney fibrosis mice.
Next, we also analyzed mouse models of renal fibrosis and CKD, including folic acid (FA)- and unilateral ureter obstruction (UUO)-induced pathology. Transcript levels of Tfam and other mitochondrial targets were significantly lower in both models (Figure 1D, Supplemental Figure 1C). Immunoblotting analysis showed lower TFAM and OXPHOS-related proteins in both fibrosis models as compared to controls (Figure 1E, 1F). Immunohistochemistry further confirmed the lower protein levels of TFAM and COXIV in renal tubule cells (Figure 1G, Supplemental Figure 1B). Concurrently, the expression of inflammatory cytokines such as IL1B, IL6 and CCL2 were found to be more in human CKD samples and animal models of kidney fibrosis and their levels strongly and significantly correlated with kidney function and fibrosis (Figure 1H, 1I, Supplemental Figure 1D). Collectively, we concluded that TFAM and other mitochondrial proteins were significantly lower in CKD patients and mouse models of kidney fibrosis, while expression of inflammatory cytokines were elevated in diseased kidneys.
Tubule-specific TFAM deletion in mice causes renal failure
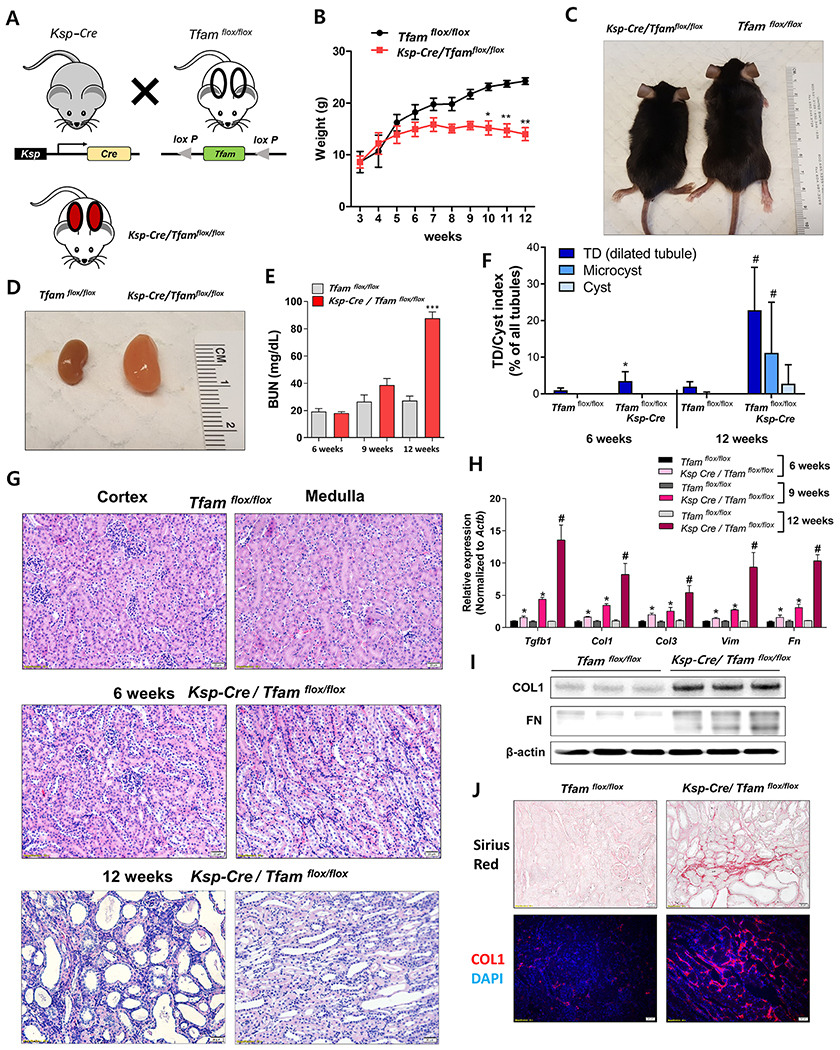
To understand whether TFAM loss, observed in patients with CKD, actually contributes to kidney disease development, we deleted the mitochondrial regulator TFAM in kidney tubules (Figure 2A) by crossbreeding Ksp-Cre and Tfamflox/flox mice. Ksp-Cre/Tfamflox/flox mice showed growth retardation (Figure 2C) and presented with significantly lower weight by 10 weeks of age compared to control (Wt/Tfamflox/flox) littermates (Figure 2B, 2C). Ksp-Cre/Tfamflox/flox mice developed severe azotemia with changes in serum blood urea nitrogen (BUN), sodium, potassium, and total CO2 levels (Figure 2E, Supplemental Figure 2B) at 12 week of age and did not survive beyond 16 weeks of age (Supplemental Figure 2C). At 12 week of age, the kidney weight was higher (Figure 2D). Histological analysis indicated significant epithelial atrophy, dilated tubules and interstitial fibrosis (Figure 2F, 2G, Supplemental Figure 2A). To exclude the possibility that this was a cystic kidney disease model, we measured tubule diameters by an objective point counting method. Overall, we detected no true cysts (Figure 2F), although 20% of the tubules showed dilation, establishing that the Ksp-Cre/Tfamflox/flox is a model of kidney fibrosis with significant tubule epithelial atrophy.
Figure 2. Tubule-specific TFAM deletion in mice causes renal failure.
(A) Experimental scheme for generating the Ksp-Cre/Tfamflox/flox mice. (B) Weight changes of Ksp-Cre/Tfamflox/flox mice and WT/Tfamflox/flox mice. * P < 0.05, ** P < 0.01. (C) Representative images of mice and (D) kidneys of Ksp-Cre/Tfamflox/flox mice and WT/Tfamflox/flox mice taken at 12 weeks of age. (E) Blood urea nitrogen (BUN) levels of Ksp-Cre/Tfamflox/flox and WT/Tfamflox/flox mice ***P < 0.001. (F) Quantification of tubule dilation (TD), microcysts, and cysts in Ksp-Cre/Tfamflox/flox and WT/Tfamflox/flox mice kidney. *P < 0.05 vs. 6 weeks WT mice. #P < 0.05 vs. 12 weeks WT mice. (G) Representative haematoxylin and eosin (H&E) staining of 6 and 12 weeks old Ksp-Cre/Tfamflox/flox and WT/Tfamflox/flox kidneys. Scale bar = 20 μm. (H) Relative mRNA levels of fibrosis-associated genes (Tgfb1, Col1, Col3, Vim, and Fn1) in 6 weeks, 9 weeks and 12 weeks of Ksp-Cre/Tfamflox/flox mice and WT/Tfamflox/flox mice kidneys. * P < 0.05 vs. 6 weeks WT/Tfamflox/flox. * P < 0.01 vs. 9 weeks WT/Tfamflox/flox. # P < 0.05 vs. 12 weeks WT/Tfamflox/flox. (I) Protein levels of collagen1 and fibronectin in 12 weeks of Ksp-Cre/Tfamflox/flox mice and WT/Tfamflox/flox mice kidney. β-actin was used as a loading control. (J) Representative images of Sirius-red staining of 12 weeks of Ksp-Cre/Tfamflox/flox mice and WT/Tfamflox/flox mice kidneys. Representative immunofluorescence images of Collagen I (COL1) staining (Red) and DAPI (Blue) in 12 weeks of Ksp-Cre/Tfamflox/flox and WT/Tfamflox/flox mice kidneys.
To further substantiate the presence of kidney fibrosis, we performed Sirius-red staining showing elevated in profibrotic collagen deposition in Tfam-deficient mice (Figure 2J). Expression of genes associated with kidney fibrosis such as Tgfb1, Col1, Col3, Vim, and Fn showed minor changes at 6 weeks of age, but their levels were only marginally changed at 9 weeks, but significantly elevated in kidneys of 12-week-old Ksp-Cre/Tfamflox/flox mice (Figure 2H). The elevation in COL1 and FN were further confirmed by western blotting and immunostaining (Figure 2I, 2J). Taken together, Tfam-deficient mice showed tubule atrophy, progressive profibrotic collagen deposition, with lower kidney function leading to kidney failure and death at 12-16 week of age, indicating that TFAM loss per se is sufficient to drive kidney fibrosis and CKD.
Ksp-Cre/Tfamflox/flox kidneys are characterized by severe metabolic defects
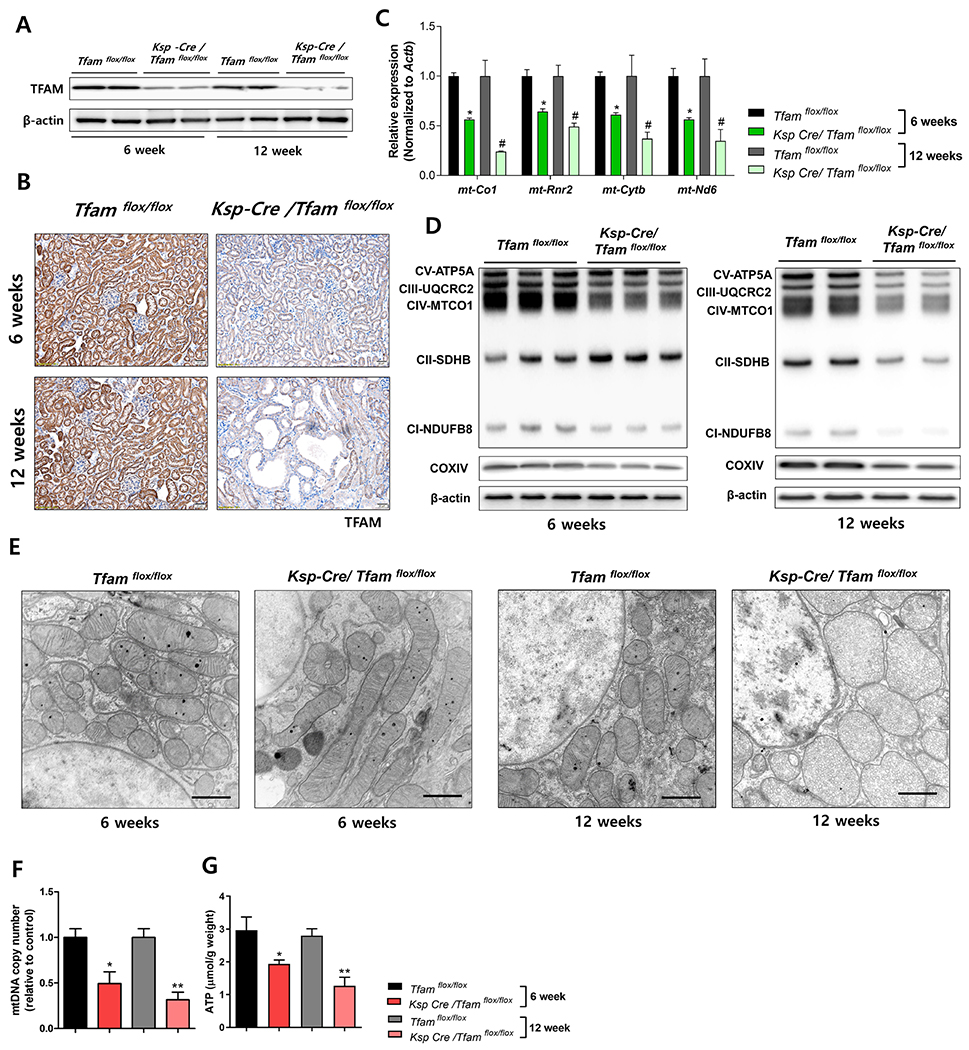
Next, we analyzed metabolic changes in Ksp-Cre/Tfamflox/flox kidneys. We found that protein levels of TFAM were already less at 6 weeks of age as detected by western blotting and IHC (Figure 3A, 3B), confirming the successful generation of the tubule conditional knock-out mice. Also, transcript levels of mitochondrial genes, mt-Co1, mt-Rnr2, mt-Cytb, and mt-Nd6 were lower at 6-week-old mice (Figure 3C). Immunoblotting analysis confirmed the lower levels of OXPHOS proteins in kidneys of 6-week-old mice (Figure 3D, Supplemental Figure 2D). Interestingly, protein levels of some nuclear-encoded mitochondrial genes, such as Atp5a and Sdhd, were found to be more at only 6 weeks of age. (Figure 3D, Supplemental Figure 2D). Electron microscopical analysis of mitochondrial structure showed progressively severe structure defects, including enlarged mitochondria at 6 weeks of age and severely disorganized mitochondria at 12 weeks of age (Figure 3E).
Figure 3. Ksp-Cre/Tfamflox/flox kidneys are characterized by severe metabolic defects.
(A) TFAM protein expression in whole kidney lysates of 6 and 12 weeks of Ksp-Cre/Tfamflox/flox and WT/Tfamflox/flox mice. β-actin was used as a loading control. (B) Representative IHC images of TFAM. Scale bar = 20 μm. (C) Relative mRNA levels of mitochondrially-encoded genes (mt-Co1, mt-Rnr2, mt-Nd6, and mt-Cytb) in 6 and 12 weeks of Ksp-Cre/Tfamflox/flox mice and WT/Tfamflox/flox mice kidneys. * P < 0.05 vs. 6 weeks WT/Tfamflox/flox. # P < 0.05 vs. 12 weeks WT/Tfamflox/flox. (D) Protein levels of mitochondrial OXPHOS proteins (CV-ATP5A, CIII-UQCRC2, CIV-MTCO1, CII-SDHB, CI-NDUFB8, and COXIV) at 6 weeks and 12 weeks of Ksp-Cre/Tfamflox/flox mice and WT/Tfamflox/flox mice kidney. β-actin was used as a loading control. (E) Representative transmission electron microscopy (TEM) images of Ksp-Cre/Tfamflox/flox mice and WT/Tfamflox/flox mice kidney tubules (6 and 12 weeks). Scale bar = 500 nm. (F) mtDNA copy number was quantified by qPCR. * P < 0.05 vs. 6 weeks WT/Tfamflox/flox. ** P < 0.01 vs. 12 weeks WT/Tfamflox/flox. (G) ATP concentration per total protein amount was measured. * P < 0.05 vs. 6 weeks WT/Tfamflox/flox. ** P < 0.01 vs. 12 weeks WT/Tfamflox/flox.
Immunohistological analysis of mitochondrial proteins such as COXIV confirmed the lower mitochondrial number in Ksp-Cre/Tfamflox/flox kidneys (Supplemental Figure 2E). We found that mtDNA copy number was significantly less in 6-week-old mice in absence of TFAM (Figure 3F). ATP content was also significantly lower at 6-week-old Tfam knock-out kidneys and that showed no further significant decline by 12 weeks of age (Figure 3G). In summary, tubule-specific Tfam deletion causes depletion of mtDNA, loss of mitochondrial gene expression, mitochondrial respiratory chain, aberrant ultrastructure and low cellular ATP levels by 6 weeks of age, with no clear worsening by 12 weeks of age, when fibrosis and azotemia developed.
To recapitulate the in vivo changes, we generated an in vitro cell-based model by culturing primary renal TECs from kidneys of wild-type (WT) and WT/Tfamflox/flox mice. TECs were infected with Cre-recombinase (Ad-Cre-GFP) adenovirus to genetically delete Tfam (Supplemental Figure 3A). Cre infection significantly lower Tfam and other mitochondrial gene expression levels (Supplemental Figure 3B), and corresponding protein levels (Supplemental Figure 3C). Loss of TFAM in cultured TECs resulted in disorganized mitochondrial structure as shown by MitoTracker staining and electron microscopical analysis (Supplemental Figure 3D, 3E). Cellular ATP content was lower in Tfam-deficient TECs (Supplemental Figure 3F). Thus, TFAM loss in primary culture of TECs lead to severe mitochondrial structure changes and metabolic alterations resulting in lower ATP content, recapitulating our in vivo findings.
Imbalance between proliferation and apoptosis of tubules leads to epithelial dysfunction
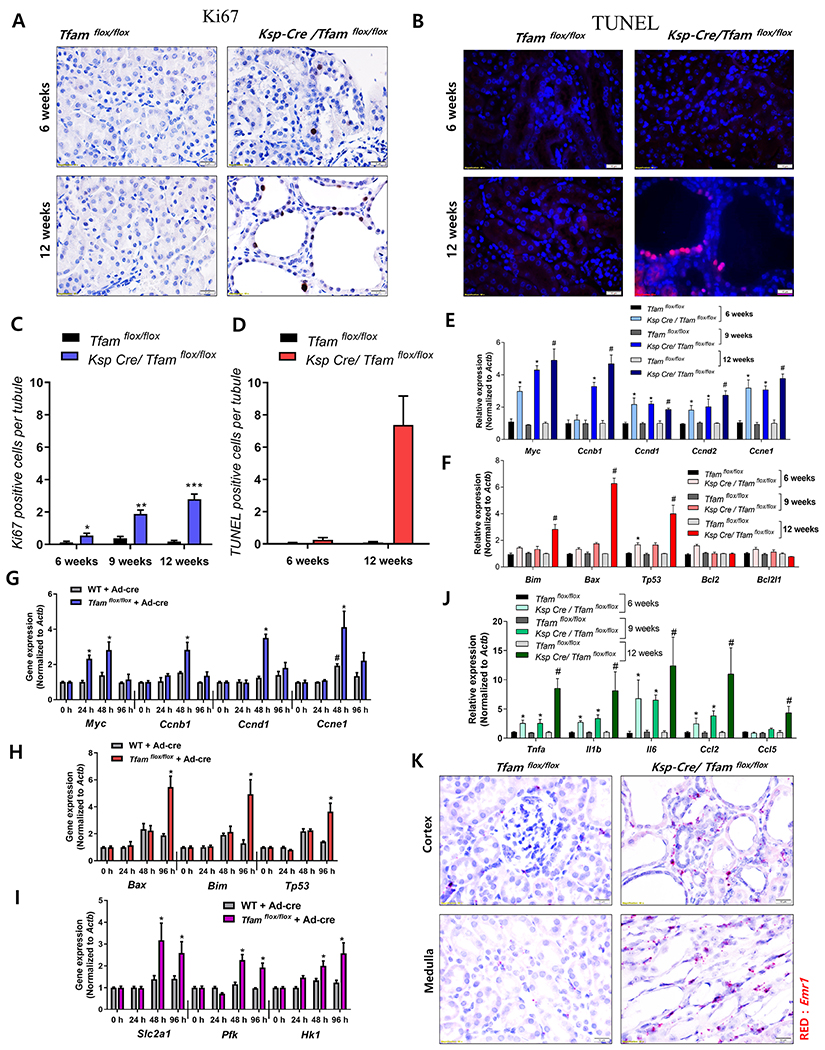
To understand the mechanism of Tfam-deficiency-induced kidney disease, we examined cell proliferation and apoptosis in renal tubule cells. We observed a higher level of tubule cell proliferation as assessed by more Ki67-positive cells in Tfam-deficient mice as early as 6 weeks of age (Figure 4A, 4C, Supplemental Figure 4A). Expression of Myc and cyclins were consistent with tubule cell proliferation (Figure 4E). Interestingly, although mtDNA copy number and ATP content was already reduced at 6 weeks of age, there was no detectable change in tubule apoptosis as detected by TUNEL assay at this age (Figure 4B, 4D). At 12 week of age, TUNEL positive cell number significantly elevated, specifically affecting the dilated, abnormal appearing segments (Figure 4B, 4D). Expression of pro-apoptotic genes such as Bax, Bim and Tp53 were higher correlating with the elevation in apoptosis rate (Figure 4F).
Figure 4. Early wave of proliferation is followed by higher level cell death in Ksp-Cre/Tfamflox/flox mice.
(A) Representative Ki67 immunostaining of kidney section of control and Ksp-Cre/Tfamflox/flox mice. Scale bar = 10 μm. (B) Representative images showing TUNEL staining of control and Ksp-Cre/Tfamflox/flox mice kidneys. Scale bar = 10 μm. (C) Quantification of Ki67-positive cells. * P < 0.05 vs. 6 WT/Tfamflox/flox. ** P < 0.01 vs. 9 weeks WT/Tfamflox/flox. *** P < 0.001 vs. 12 weeks WT/Tfamflox/flox. (D) Quantification of TUNEL positive cells. *** P < 0.001 vs. 12 weeks WT/Tfamflox/flox. (E) Relative mRNA levels of genes associated with cell proliferation (Myc, Ccnb1, Ccnd1, Ccnd2, and Ccne1) in control and Ksp-Cre/Tfamflox/flox mice. * P < 0.05 vs. 6 weeks WT/Tfamflox/flox. *P <0.01 vs. 9 weeks WT/Tfamflox/flox. # P < 0.05 vs. 12 weeks WT/Tfamflox/flox. (F) Relative mRNA levels of apoptosis-related genes (Bim, Bax, Tp53, Bcl2, and Bcl2l1) in control and TFAM deficient mice. * P < 0.05 vs. 6 weeks WT/Tfamflox/flox. *P <0.01 vs. 9 weeks WT/Tfamflox/flox. # P < 0.05 vs. 12 weeks WT/Tfamflox/flox. (G) Relative mRNA levels of proliferation associated genes (Myc, Ccnb1, Ccnd1, and Ccne1) in control and TFAM deficient primary cultured tubule cells. # P < 0.05 vs. wild type cells at 0 h. * P < 0.05 vs. Cre adenovirus treated cells at 0 h. (H) Relative mRNA levels of apoptosis associated genes (Bax, Bim, and Tp53) in primary cultured control and TFAM knock-out cells. * P < 0.05 vs. Cre adenovirus treated cells at 0 h. (I) Relative mRNA levels of glycolysis associated genes (Pfk, Slc2a1, and Hk1) in primary cultured control and TFAM knock-out cells. * P < 0.05 vs. Cre adenovirus treated cells at 0 h. (J) Relative mRNA levels of pro-inflammatory cytokines (Tnfa, Il1b, Il6, Ccl2, and Ccl5) in 6 weeks, 9 weeks and 12 weeks of Ksp-Cre/Tfamflox/flox mice and WT/Tfamflox/flox mice kidneys. * P < 0.05 vs. 6 weeks WT/Tfamflox/flox. *P <0.05 vs. 9 weeks WT/Tfamflox/flox. # P < 0.05 vs. 12 weeks WT/Tfamflox/flox. (K) Representative images of in situ hybridization with Emr1 (red) probe in Tfam-deficient kidney cortex and medulla. Scale bar = 10 μm.
Next, we studied control and Tfam-null primary culture TECs. Consistent with the in vivo studies, the initial wave of proliferation was followed by a higher rate of cell death (Figure 4G, 4H, Supplemental Figure 4B). The wave of proliferation correlated with an elevated expression of glycolytic genes as observed in other systems (Figure 4I). In summary, Tfam-deficient tubule cells higher reliance on glycolysis and an initial wave of proliferation was followed by an in apoptosis rate. In Tfam-knockout mice, the wave of apoptosis was observed much later (12 weeks) than the metabolic defect (6 and 9 weeks). Apoptosis correlated with kidney fibrosis and kidney disease development, but not with the metabolic derangement.
RNA-sequencing reveals higher inflammation in TFAM-deficient kidney
Our studies indicated a severe metabolic defect and low ATP level in Tfam-deficient mice already at 6 week of age; however, higher apoptosis rate, fibrosis and animal demise was only prominent at 12 weeks of age. We therefore performed RNA-sequencing analysis to comprehensively define cellular pathways affected by Tfam-deficiency. We analyzed whole kidney samples of Ksp-Cre/Tfamflox/flox mice and Wt/Tfamflox/flox mice (n = 3 per group at 12 weeks of age. We defined differentially expressed genes with a log fold change above 1.2, with FDR-adjust P < 0.05, yielding differences in 2,372 protein-coding genes between mutant and wild type mice (Supplemental Table 2). Gene ontology enrichment tools revealed differential regulation of key pathways (Supplemental Figure 4C, 4D), such as expression of genes encoding metabolic function were less in Tfam knock-out mice (Supplemental Figure 4C, 4D). Our unbiased results indicated that transcript levels of mitochondrial-encoded genes were lower (Supplemental Figure 4E); however, many mitochondrial genes encoded by the nuclear genome were found to have higher expression (Supplemental Figure 4F). Interestingly, immune- and inflammation-related pathways were among the top pathways enriched in Tfam-deficient kidneys (Supplemental Figure 4D, Supplemental Figure 5B). Gene set enrichment analysis (GSEA) confirmed the strong enrichment for inflammatory genes among the differentially expressed pathways (Supplemental Figure 4G). Transcript levels of pro-inflammatory cytokines (Tnfa, Il1b, Il6, Ccl2, and Ccl5) (Figure 4J) as well as inflammatory cell markers (Lyz2, Emr1, and Cd68) were higher in Tfam-deficient kidney (Supplemental Figure 5A). These changes were similar to that observed in patients with DKD and HKD (Figure 1). Gene expression analysis showed much higher expression in NF-κB and its target gene expression and lesser change in IRF3 and its target gene expression (Supplemental Figure 5B–5E). Furthermore, changes were not limited to cytokines, as we observed more infiltrating immune cells in kidney samples. Emr1 (F40/80) is an important marker expressed by macrophages. Using in situ hybridization, we found more Emr1 positive cells in the interstitial region (Figure 4K). Emr1 and Col1a2 positive cells localized to the same region, indicating the importance of inflammation in fibrosis development (Supplemental Figure 5F). Time-course analysis indicated significant changes in inflammatory cytokine expression between 6 and 12-weeks old animals and immune gene expression strongly correlated with the functional and structural kidney damage (Figure 4J, 2G, 2H). In summary, RNA-sequencing highlighted significant changes in inflammation related pathways in 12-week old Tfam-deficient kidneys coinciding with the timing with the functional and structural kidney damage.
Tfam deficiency leads to direct activation of cGAS-STING, NF-κB and cytokine release
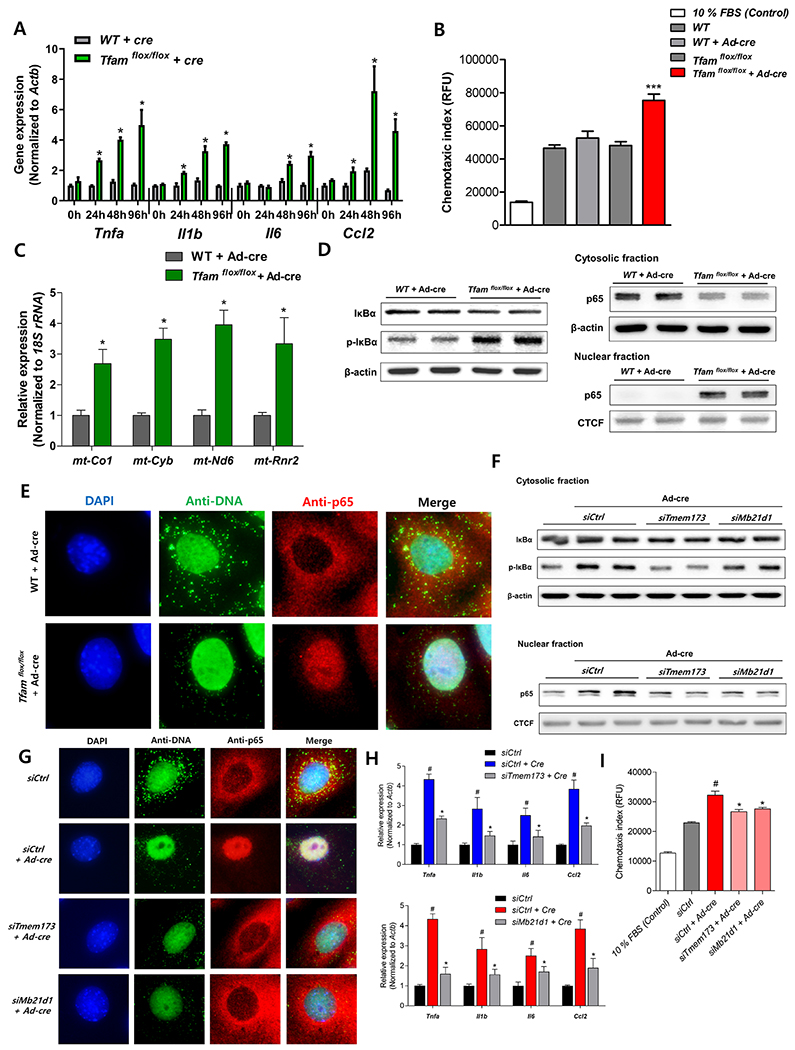
Next, we examined whether the absence of TFAM in tubule cells can directly lead to cytokine activation by turning to our primary TEC system. We found that Tfam-deficiency in TECs leads to an early but brief interferon response followed by significant higher level in proinflammatory cytokines Il6, Tnfa, Il1b and Ccl2 levels, indicating that cytokine activation is an autonomous effect induced by TFAM loss (Figure 5A, Supplemental Figure 6A). To examine, whether cytokines/chemokines are responsible for recruiting macrophages, we measured macrophage chemotaxis index using TECs culture medium. The culture medium from Tfam-deficient tubule cells (Tfamflox/flox infected with Cre-recombinase) resulted in a significantly higher level in the macrophage chemotaxis index compared to that from control cells (Figure 5B).
Figure 5. TFAM deficiency causes cytosolic leak of mtDNA and cGAS-STING, NF-κB activation.
(A) Relative mRNA levels of pro-inflammatory cytokine genes (Tnfa, Il1b, Il6, and Ccl2) in Tfam-deficient primary cultured TECs. * P < 0.05 vs. control group. (B) Macrophage chemotaxis index. Raw 267.4 macrophage cells were incubated with medium of control and Tfam-deficient primary tubule cells. *** P < 0.001 vs. Cre-treated group. (C) Cytosolic translocation of mtDNA was quantified by qPCR. Cytosol fractions of control and Tfam-deficient primary cultured TECs were analyzed * P < 0.05 vs. control group. (D) Protein levels of IκBα, phosphorylated IκBα, and p65 were determined in cytosolic fraction, protein level of p65 was determined in nuclear fraction in primary cultured cell in different condition. β-actin and CTCF were used as the loading control for cytosolic and nuclear fractions, respectively. (E) Microscopy images of control and Tfam knock-down primary cultured TECs stained with anti-DNA and anti-p65 antibodies and DAPI. Scale bar = 10 μm. (F) Cytosolic protein levels of IκBα and phosphorylated IκBα and nuclear p65 was determined in primary Tfam-null TECs at baseline or in the combination of siRNA mediated Mb21d1(cGAS) and Tmem173 (STING) knock-down. β-actin and CTCF were used as loading control for cytosolic and nuclear fractions, respectively. (G) Representative immunofluorescence images of primary cultured TECs stained with anti-DNA and anti-p65 antibodies and DAPI. All cells were incubated with AdCre to delete Tfam and control siRNA or Mb21d1(cGAS)/Tmem173 (STING) knock-down. Scale bar = 10 μm. (H) Relative mRNA levels of proinflammatory cytokine genes (Tnfa, Il1b, Il6, and Ccl2). Cells were incubated with AdCre to delete Tfam and control siRNA or Mb21d1(cGAS)/Tmem173 (STING) knock-down. # P < 0.05 vs. control group. * P < 0.05 vs. Cre-treated group. (I) Macrophage chemotaxis index from media obtained from TECs. TECs were incubated with AdCre to knock-out Tfam and control siRNA or Mb21d1(cGAS)/Tmem173 (STING) knock-down. # P < 0.05 vs. control group. * P < 0.05 vs. Cre-treated group.
To understand the mechanism of Tfam-induced cytokine release, we hypothesized that TFAM deficiency leads to abnormal mtDNA packing and cytosolic translocation of mtDNA. Cytosolic mtDNA is recognized by the cGAS-STING pathway that could activate IRF and NF-κB. In wild type TECs, intact mtDNA structures were observed when stained with anti-DNA antibody (Supplemental Figure 6B). However, ablation of TFAM lead to disorganized mtDNA architecture (Supplemental Figure 6B, Figure 5E). To examine whether TFAM deficiency can result in mtDNA release into the cytoplasm, we isolated cytosolic fractions lacking cross contamination from membranes of control and Tfam knock-out cells and quantified mtDNA amount using quantitative PCR (Supplemental Figure 6G). We found significant enrichment for mtDNA , such as mt-Co1, mt-Cyb, mt-Nd6, and mt-Rnr2 in the cytosolic compartments of Tfam knock-out cells when compared to control cells (Figure 5C).
Higher level of NF-κB was observed in Tfam knock-out mice, therefore we examined whether Tfam-deletion and cytosolic translocation of mtDNA leads to NF-κB activation. TFAM depletion higher level the phosphorylation of IκBα and lead to higher amount in nuclear translocation of the p65 subunit of NF-κB as analyzed by immunoblotting (Figure 5D). Cytosolic p65 levels were lower at the same time (Figure 5D, 5E). In accordance, with in vitro experiments, Ksp-Cre/Tfamflox/flox kidneys also showed more nuclear p65 levels in TECs (Supplemental Figure 6C), as well as higher amount of IκBα phosphorylation (Supplemental Figure 6D). In some cellular system, cGAS-STING activation leads to IRF3 activation as well, however in cultured TECs we failed to observe significant IRF3 activation (Supplemental Figure 5B–5E).
To demonstrate the role of the cytosolic DNA sensor cGAS-STING in TFAM-induced cytokine expression, we performed Mb21d1(cGAS) and Tmem173 (STING) silencing experiments in Tfam-deficient cells. Using an siRNA approach, we efficiently silenced Mb21d1(cGAS) and Tmem173 (STING) in primary cultured TECs (Supplemental Figure 6E, 6F). Knockdown of cGAS or STING abrogated the phosphorylation of IκBα and restricted the nuclear translocation of p65 (Figure 5F, 5G), however, mitochondrial alterations still persisted (Figure 5G). cGAS and STING knock-down in primary TECs reduced the expression of proinflammatory genes indicating the role of STING in the process (Figure 5H). Furthermore, to demonstrate the functional role of cGAS-STING signaling, we found that the culture medium from STING-silenced TECs showed significantly lower chemotaxis index (Figure 5I). Collectively, these data suggest that TFAM deficiency directly induces proinflammatory cytokine expression in TECs, which is mediated by STING-dependent NF-κB activation and likely induced by cytosolic translocation mtDNA.
STING inhibition blocks TFAM deficiency-induced kidney failure and fibrosis
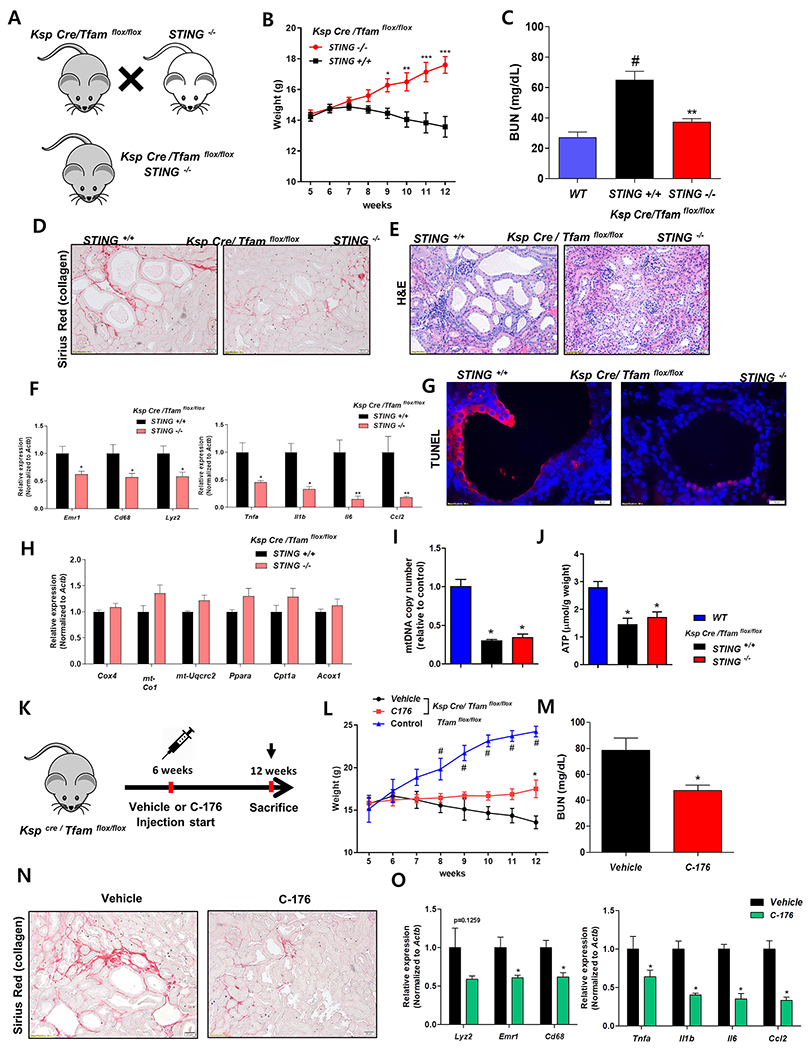
We hypothesized that the elevated STING dependent inflammatory phenotype plays a key role in TFAM deficiency-induced renal failure and fibrosis development. To investigate the functional role of STING, we crossed Ksp-Cre/ Tfam flox/flox with STING knockout mice (Figure 6A). Compared to the TFAM deficient mice, Ksp-Cre/ Tfam flox/flox STING knockout mice showed improved growth and development (Figure 6B). The degree of azotemia was lower in STING knock-out mice compared to Ksp-Cre/Tfamflox/flox mice (Figure 6C). Structural damage such as tubule dilation was significantly attenuated, and fibrosis was markedly reduced in STING knockout mice compared with Ksp-Cre/ Tfam flox/flox mice (Figure 6D, 6E, Supplemental Figure 7A–7C). Consistent with the morphologic alterations, inflammation was lower in the STING knockout mice. Cytokine levels and inflammatory cell markers were significantly lower in absence of STING (Figure 6F). The level of mitochondrial and metabolic gene expression, mitochondrial copy number, and tissue ATP level were not improved in absence of STING (Figure 6H–6J) indicating that the effect of STING on kidney function and structure improvement was not mediated by metabolic improvement. TFAM deficiency-induced apoptotic cell death was attenuated by STING depletion. TUNEL staining showed significantly lower level cell death in STING knockout mice (Figure 6G, Supplemental Figure 7D).
Figure 6. Genetic deletion of pharmacological inhibition of STING attenuates renal disease of TFAM-deficient kidneys.
(A) Experimental scheme for the generation Ksp-Cre/Tfamflox/floxSTING−/− mice. (B) Weight changes. *P <0.05. ** P < 0.01. *** P< 0.001 vs. STING +/+ group. (C) Blood urea nitrogen (BUN) levels at 12 weeks of age in control Ksp-Cre/Tfamflox/flox and Ksp-Cre/Tfamflox/floxSTING−/− mice. ** P < 0.01 vs. STING +/+ group. (D) Representative images of Sirius-red staining in 12 weeks of Ksp-Cre/Tfamflox/flox and Ksp-Cre/Tfamflox/floxSTING−/− mice. (E) Representative images of hematoxylin and eosin staining of 12 weeks of Ksp-Cre/Tfamflox/flox and Ksp-Cre/Tfamflox/floxSTING−/− mice. (F) Relative mRNA levels of inflammatory cell marker genes (Lyz2, Emr1, and Cd68) and pro-inflammatory cytokines (Tnfa, Il1b, Il6, and Ccl2) in Ksp-Cre/Tfamflox/flox and Ksp-Cre/Tfamflox/floxSTING−/− mice. *P <0.05. ** P < 0.01 vs. STING +/+ group. (G) Representative TUNEL staining of Ksp-Cre/Tfamflox/flox and Ksp-Cre/Tfamflox/floxSTING−/− mice. Scale bar = 10 μm. (H) Relative mRNA levels of mitochondrial genes (Cox4, mt-Cox1, and mt-Uqcrc2) and lipid metabolism associated genes (Ppara, Cpt1a, and Acox1) in Ksp-Cre/Tfamflox/flox and Ksp-Cre/Tfamflox/floxSTING−/− mice. (I) mtDNA copy number and (J) ATP concentration per protein was measured in Ksp-Cre/Tfamflox/flox and Ksp-Cre/Tfamflox/floxSTING−/− mice. * P < 0.05 vs. wild type kidneys. (K) Study design of STING inhibitor (C-176) experiment. (L) Weight changes of control and Ksp-Cre/Tfamflox/flox mice treated with C-176 or vehicle. * P < 0.05 vs. vehicle treated mice, # P < 0.05 vs. vehicle treated mice. (M) Blood urea nitrogen levels were measured in Ksp-Cre/Tfamflox/flox mice treated with C-176 or vehicle. * P < 0.05 vs. vehicle treated mice. (N) Representative images of Sirius-red staining in 12 weeks old Ksp-Cre/Tfamflox/flox mice treated with C-176 or vehicle. Scale bar = 20μm. (O) Relative mRNA levels of inflammatory marker genes (Lyz2, Emr1, and Cd68) and pro-inflammatory cytokine genes (Tnfa, Il1b, Il6, and Ccl2) in Ksp-Cre/Tfamflox/flox mice treated with C-176 or vehicle. * P < 0.05 vs. vehicle treated mice.
Furthermore, to understand the therapeutic potential of STING inhibition, we examined the effect of a recently reported STING-specific inhibitor: C-176 (Haag et al., 2018). Tfam-deficient mice were injected with C-176 starting at 6 weeks of age (Figure 6K). STING inhibition attenuated the growth retardation, functional and structural changes of kidney fibrosis induced by Tfam-deficiency (Figure 6L–6M, Supplemental Figure 7E–7G). STING inhibition showed lower inflammation and apoptotic cell death similar to STING knockout (Figure 6O, Supplemental Figure 7H, 7I).
Collectively, genetic deletion or pharmacological inhibition of STING significantly ameliorated Tfam deficiency-induced kidney fibrosis, suggesting the importance of TFAM in regulating inflammatory signaling by sequestering mtDNA.
Limiting STING activity ameliorates fibrosis in mouse CKD models
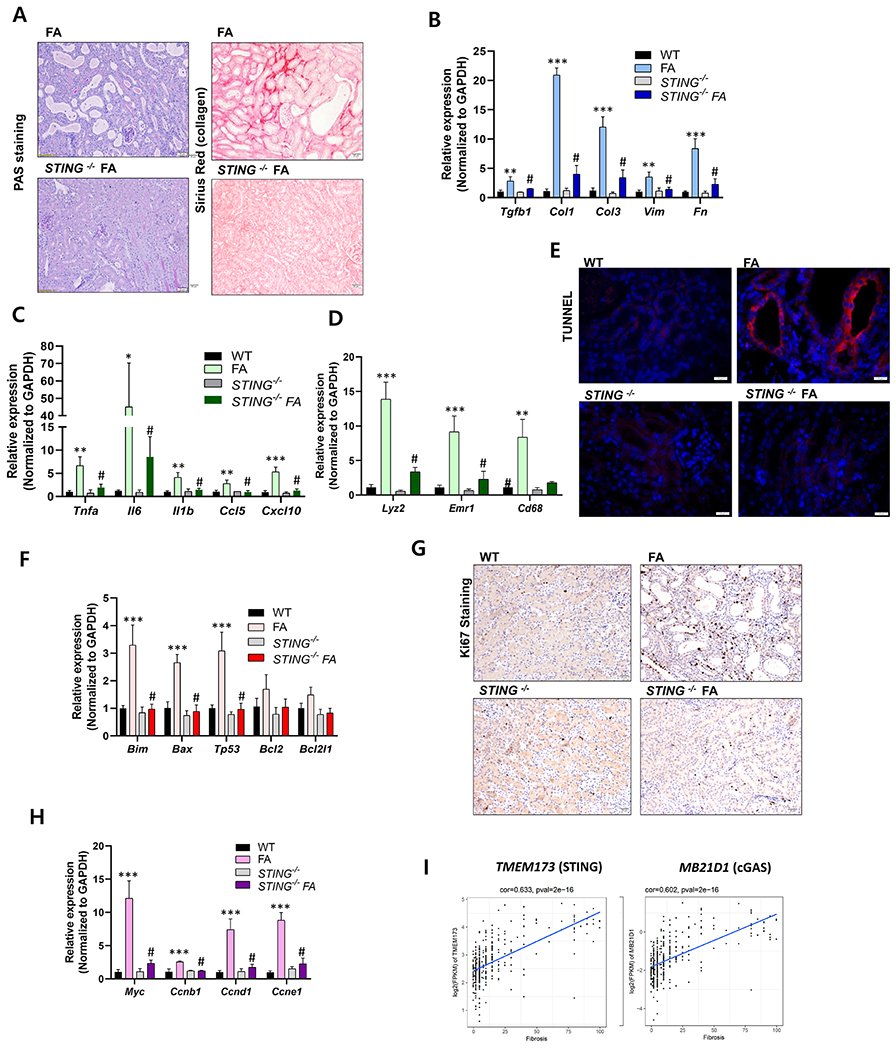
To understand the therapeutic potential of STING inhibition in kidney fibrosis, we examined the renal phenotype of the STING knock-out mice following folic acid (FA)-induced kidney injury. FA-injected STING−/− mice showed lesser tubular atrophy, tubule dilation and fibrosis when compared to wild type (Figure 7A). Expression of pro-fibrotic, Tgfβ, Col1, Col3, Vim and Fn (Figure 7B), and inflammatory markers such as Tnfa, Il1b, Ccl5, and Cxcl10 (Figure 7C), and immune cell markers Lyz2, Emr1, Cd68 (Figure 7D) were markedly lower in FA-injected STING knock-out mice when compared to wild type. We observed less cell death (Figure 7E, 7F) and cell proliferation in FA-injected STING−/− mice (Figure 7G, 7H).
Figure 7. STING plays and important role in kidney fibrosis.
(A) Periodic acid-Schiff (PAS) staining and Picrosirius-red staining of FA-injected WT and STING−/− mice. (B) Relative mRNA levels of fibrosis-associated genes (Tgfb1, Col1, Col3, Vim, and Fn1), ** P < 0.01, *** P< 0.001 vs. WT. # P < 0.01 vs. FA mice. (n = 3) (C) pro-inflammatory cytokines (Tnfa, Il1b, Il6, CCL5 and Cxcl10), (D) inflammatory cell marker genes (Lyz2, Emr1, and Cd68) in Wt and STING−/− mice injected with vehicle or FA. ** P < 0.01, *** P< 0.001 vs. WT. # P < 0.01 vs. FA mice. (E) Representative images showing TUNEL staining of WT and STING−/− mice injected with vehicle or FA. Scale bar = 10μm (F) Relative mRNA levels of apoptosis-related genes (Bim, Bax, Tp53, Bcl2, and Bcl2l1) in kidneys of WT and STING−/− mice injected with vehicle/FA. *** P < 0.001 vs. WT. # P < 0.01 vs. FA mice. (G) Ki67 positive staining (Scale bar = 20 μm) and (H) relative mRNA levels of cell proliferative markers (Myc, Ccnb1, Ccnd1, and Ccne1) of WT and STING−/− mice injected with vehicle/FA kidneys. *** P < 0.001 vs. WT. # P < 0.01 vs. FA mice. (I) Transcript level (RNA-sequencing) of STING (TMEM173) and cGAS (MB21D1), correlates with the degree of kidney fibrosis in 433 microdissected human kidney tubule samples.
We also studied the effect of C-176 in the FA-induced kidney fibrosis model (Supplemental Figure 8A). STING inhibition notably lowered FA-induced renal injury (Supplemental Figure 8B-8D, 8G). We observed lower expression of pro-inflammatory and immune cell markers, and in cell proliferation (Supplemental Figure 8E, 8F). Therefore, we concluded that the genetic deletion of STING and to lesser degree the STING inhibitor ameliorated kidney fibrosis in the FA-induced model of kidney disease.
To understand the clinical relevance of cGAS-STING activation in patients with CKD, we again examined gene expression levels in 433 human kidney tubule samples. We found a strong correlation between cGAS and STING expression and kidney function and kidney fibrosis, indicating the broad significance of this pathway activation (Figure 7I).
Discussion
Here, we report that mitochondrial/metabolic dysfunction of TECs, a key hallmark of CKD, is a significant contributor to immune activation and results in kidney disease development via cGAS-STING activation. Metabolic dysfunction and higher cytokine expression and immune cell infiltration have long been observed in kidneys of patients with DKD and HKD (Beckerman et al., 2017; Fischer et al., 2010; Kang et al., 2015; Lemos et al., 2018; Rodwell et al., 2004; Rudnicki et al., 2007; Woroniecka et al., 2011). Blocking cytokine activation, by targeting MCP1 or JAK has shown some benefit in patients with kidney disease; (Pichler et al., 2017); however, the upstream regulatory network of cytokine activation has not been established. There is a large body of literature on the role of inflammatory cytokines regulating metabolism; however, the mechanism of immune activation remains poorly understood.
Using a large collection of microdissected human kidney tubule samples, we show that TFAM levels were lower in patients with CKD and its levels correlate with the degree of fibrosis. We therefore hypothesized that TFAM could be an important regulator of mitochondrial/metabolic function. We modeled the metabolic dysfunction observed in CKD by deleting Tfam in renal TECs and observed TFAM loss resulted in severe renal disease characterized by tubule atrophy accumulation of collagen, and infiltration of immune cells. Despite the key role of TFAM in regulating mitochondrial structure and function, the diseased phenotype development observed in the Ksp-Cre/ Tfam flox/flox mice was slower than we expected. While the genetic deletion occurred during embryonic development we observed no clear functional and structural changes until 10-12 weeks of age. These observations are in accordance with other studies performed by using Tfam knock-out animals (Li et al., 2000; Vernochet et al., 2012; Wang et al., 1999). Another surprising finding is that animals tolerated the severe metabolic defect we observed at 6 weeks of age including mitochondrial loss, OXPHOS and FAO defect and low ATP content. Traditionally, it has been proposed that such changes (for example observed in acute kidney injury) cause significant functional defect in the kidney (Ferenbach and Bonventre, 2015). Our results indicate that renal tubule cells have a much greater metabolic plasticity than expected. These results clearly need further exploration by titrating and correlating metabolic output and kidney function.
To our surprise, mitochondrial dysfunction caused by Tfam-deficiency not only resulted in metabolic dysregulation, but also directly lead to cytokines and chemokines release and immune cell activation. Mechanistically, cytosolic translocation of mtDNA lead to NF-κB activation via the cGAS-STING pathway, linking metabolic insufficiency to enhanced inflammation. Genetic or pharmacological inhibition of STING significantly ameliorated inflammation and improved kidney function and reduced structural damage (fibrosis) in the kidney. Indeed, the functional decline we observed in the kidney, correlated with the immune cell activation and not with the metabolic defect. The functional and structural kidney damage was observable at 12 weeks of age while the metabolic defect was obvious at 6 weeks and did not appear to be more severe at 12 weeks. These results indicate that the role of TFAM is not restricted to mitochondrial function but also branched out to maintain homeostasis between inflammation and kidney fibrosis.
It has been previously reported that abnormal mtDNA packing caused by TFAM deficiency promotes cGAS-STING dependent IRF3 activation leading to higher level of antiviral innate immune response in mouse embryonic fibroblasts (West et al., 2015). In Tfam-deficient kidney TECs, we also found that Tfam-deficiency induced aberrant mtDNA packing and activation of cGAS-STING signaling pathway. However, the expression IRF3 targeted antiviral genes were only slightly more. Because cytosolic mtDNA can also induce cGAS-STING dependent NF-κB activation, we further focused on NF-κB induced classical inflammatory cytokines changes (Fang et al., 2017). Ablation of STING significantly lower NF-κB activation and cytokine-expression induced by Tfam deficiency, implicating the importance of NF-κB dependent inflammatory signaling in the development of kidney disease. As mitochondria are intracellular organelles that originate from bacterial symbiont, TFAM plays a key role in maintaining this immune tolerance.
Recent studies indicated that activation of cGAS-STING pathway plays a very important role in immune cell activation in cancer cells. The higher STING activity has successfully reduced tumor burden in mice and it has been successfully tested in early stages of human studies (Chen et al., 2016; Ng et al., 2018). Our study indicates a deleterious role of hyperactive STING signaling leading to immune cell activation and higher cytokine activity in kidney disease and fibrosis. Therefore, these results raise concerns about the usage of the recent developing potential therapeutic STING activators as drugs for patients who are at risk of kidney disease.
There has been a large body of literature in the renal and metabolism field indicating the key role of immune cells, including macrophages, and inflammatory cytokines in altering metabolism (Gaber et al., 2017; Gleeson and Sheedy, 2016; Osborn and Olefsky, 2012; Ralston et al., 2017). Indeed, an higher amount of inflammation has been considered a key component of metabolic alteration. Our studies, here highlight an important key inverse relationship between STING and TFAM, where STING is an important downstream regulator of the metabolic defect and its ablation can profoundly influence kidney function whereas energy content or metabolic alterations remain unchanged.
In conclusion, we examined the role of TFAM and the relationship between metabolic and immune signals in kidney disease and fibrosis. We observed that tubule-specific TFAM loss causes renal failure not only by inducing a severe metabolic and energy defect but also by the translocation of mtDNA to the cytosol leading to cGAS-STING-dependent NF-κB activation. Here, we concluded that STING activation plays an important downstream role in mediating kidney disease development and limiting STING activity can ameliorates kidney disease development.
Limitations of study
While we observed a severe metabolic and mitochondrial defect in patients with CKD, at present it is difficult to ascertain the degree of mtDNA leak in patients with CKD. Furthermore, our data highlighted the activation of cGAS-STING pathway in kidneys of Tfam-deficient mice; however, it is still unclear whether STING activation is limited to tubule epithelial cells or can also be observed in other cell types. It will be interesting to study the role of other upstream regulators of STING that might have significant effect on its activity during kidney progression. We believe further studies will be required to understand the metabolic and immune communications in kidney disease pathogenesis.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by Dr. Katalin Susztak (ksusztak@pennmedicine.upenn.edu).
EXPERIMENTAL MODEL DETAILS
Animals
Mice with Tfam-floxed alleles were purchased from Jackson Lab (Stock#026123). Tfamflox/flox mice were crossed with transgenic mice expressing Cre-recombinase under the cadherin 16 promoter (Ksp-Cre) (Jackson Lab, stock#012237). Age-matched (6 and 12 weeks) Ksp-Cre/Tfamflox/flox and WT/Tfamflox/flox male mice were used to investigate the role of TFAM-deficiency on kidney disease. To examine the role of STING pathway, STING−/− mice were purchased from Jackson Lab (Stock#025805), and further crossed with Ksp-Cre/Tfamflox/wt mice to generate Ksp-Cre/Tfamflox/flox with STING−/−. The efficacy study of STING inhibitor in Ksp-Cre/Tfamflox/flox mice was conducted as follow: mice (4 weeks of age) were injected with 7.5 μl (750nmol) of C-176 per mouse (Vitas-M laboratory, STK016322) or DMSO dissolved in 85 μl corn oil once per day until 12 weeks. For mice fibrosis model, eight-week-old (C57BL/6) male mice were injected with a single dose of folic acid (FA) (250 mg/kg dissolved in 0.3 M NaHCO3) or vehicle intraperitoneally. Blood nitrogen urea (BUN) and sodium, potassium, and total carbon dioxide levels were measured using an i-STAT1 CHEM8+ cartridge analyzer immediately at the time of blood sampling according to manufacturer’s protocol.
For all studies, littermates were used, and mice were maintained under controlled environmental conditions under a 12-h light/dark cycle and allowed ad libitum access to water and a standard laboratory diet. Water and cages were autoclaved. Cages were changed once weekly, and the health status of the mice was monitored by University Laboratory Animal Resources (ULAR) veterinary care. For all the mice experiments, mice were randomly assigned to experimental groups, unless stated otherwise. All experiments in animals were reviewed and approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania, and were performed in accordance with the institutional guidelines.
METHOD DETAILS
Cell culture
Tubular epithelial cells (TECs) were isolated from WT, WT/Tfamflox/flox male mice kidneys. Briefly, kidneys were dissected, placed in 0.1 ml ice-cold Dulbecco’s phosphate buffered saline (DPBS) (Cellgro) and minced into pieces (~ 1 mm3). Fragments were transferred to 50 ml tube containing 10 ml of RPMI medium with 100 μl of Collagenase IV (1 mg/ml, Sigma Aldrich) and digested for 30 minutes at 37 °C. Afterwards, 100 μl of fetal bovine serum (FBS) were added to stop the Collagenase IV activity. Cells were further sieved through 100 μm nylon mesh, followed by 40 μm nylon mesh. Cells were centrifuged for 10 minutes at 3000 rpm. The pellet was resuspended in 1 ml of sterile RBS lysis buffer and incubated 5 minutes on ice. DPBS was added followed by centrifugation for 10 minutes at 3000 rpm. The pellet was than resuspended in 10 % FBS contained RPMI media before plating. Cells were cultured in RPMI containing 10% FBS and penicillin/streptomycin in a humidified atmosphere containing 5% CO2 at 37°C. Cells were further transfected with Ad5CMV-eGFP (Ad-GFP) and Ad5CMVCre-eGFP (Ad-Cre-GFP) (University of Iowa Gene Transfer Vector Core, Iowa City, IA) according to experimental design. Infection efficiency was estimated under fluorescence microscope by the presence of GFP-positive cells (data not shown). STING (Tmem173) siRNA were purchased from Integrated DNA Technologies (IDT) to examine the role of STING. siRNA and negative control siRNA transfections were performed using Lipofectamine 3000. For transfection, cells were seeded in 6-well plates, grown for overnight until 60–70% confluent, and then transfected with 50 nM (final concentration) STING siRNA. Transfection efficiency was determined under fluorescence microscope by the presence of Cy3 transfection control. Cells were harvested and scraped off 48 hours post infection under different condition.
Protein extraction
All solutions, tubes, and centrifuges were maintained at 4 °C. RIPA buffer (Cell signaling, #9806). Protease cocktail (Complete mini, Roche) was used to extract total protein lysates from tissues or cells according to the manufacturer’s instructions. To separate cytosolic and nuclear proteins, the Nuclei Pure Prep kit (Sigma, NUC-201) was used. Briefly, cells or tissues were homogenized using lysis buffer with 1 % Triton X-100 and protease inhibitor cocktail, incubated on ice for 15 min, and centrifuged at 13,000 rpm for 15 min at 4 °C. Supernatants were used as cytosolic fractions and pellets were considered as nuclei. Successful nuclei isolation was determined by staining with Trypan blue solution (Sigma, T8154). Pellets were resuspended in RIPA buffer with protease inhibitors, incubated on ice for 30 minutes with vortex every 5 minutes, and centrifuged at 13,000 rpm for 15 min at 4 °C. The resulting supernatants were used as nuclear fractions. Protein concentrations were measured based on a BCA assay (Pierce, #23225).
Western blotting
Western blot assays were performed to detect protein expression in kidneys or cells. Briefly, whole cell, nuclei or cytosol lysates (10 ~ 50 μg of protein) were heated (95 °C) for 5 minutes in laemmli sample buffer (Biorad). Proteins were then separated by polyacrylamide gel electrophoresis in acrylamide gels (8~15%) and transferred using a Bio-Rad western system to polyvinylidene difluoride (PVDF; Biorad) membranes, which were immediately placed in 5% non-fat milk in Tris-buffered saline (TBS, 50 mM Tris, pH 7.6, 150 mM NaCl)-Tween (0.1% Tween20) buffer for blocking (1 h at 25 °C). Membranes were then washed 3 times in TBS-Tween buffer for 10 minutes, followed by incubation with specific primary antibodies (dilution 1:500 to 1:2000) at 4 °C overnight. Membranes were then washed 3 times for 10 min in TBS-Tween buffer, and incubated with a horseradish peroxidase-conjugated anti-mouse antibody (1:10,000), or anti-rabbit antibody (1:10,000) at 25°C for 1 h. Resulting immunoblots were visualized using ECL Western Blotting Substrate (Pierce) and a LI-COR chemiluminescence imager (LI-COR), according to the manufacturers’ instructions.
RNA extraction and qPCR
To isolate total RNA, kidney tissue samples or cells were homogenized in TRIZOL. Aliquots of 0.2 ml chloroform per 1 ml homogenate were added and samples were shaken vigorously and incubate on ice for 15 min. The aqueous phases were transferred to fresh tubes, to which an equal volume of isopropanol was added. Samples were then incubated at 4°C for 15 min and centrifuged at 12,000 rpm at 4°C for 15 min. Supernatants were removed and RNA pellets were washed once with 75% ethanol by vortexing and then centrifuged at 8,000 rpm at 4°C for 8 min. Pellets were dried for 10–15 min and dissolved in Nuclease free water. RNA (2 μg) was reverse-transcribed using a cDNA synthesis kit (Applied Biosystems). The cDNA was stored at −20°C until use. The qPCR was performed using SYBR green dye (Applied Biosystems) under the ViiA 7 System (Life Technologies). The data were normalized and analyzed using the ΔΔCt method. The primers used are listed in Supplemental Table 3.
Human sample analysis
Kidney samples were obtained from routine surgical nephrectomies. Samples were de-identified and clinical information was collected via an honest broker. Pathology examination was performed by local nephropathologist. The study was approved by the institutional review board (IRB) of the University of Pennsylvania. The kidney tissue was immediately placed and stored in RNAlater (Ambion) according to manufacturer’s instruction. The tissue was manually microdissected under a microscope in RNAlater for glomerular and tubular compartments. Dissected glomerular and tubular compartments were homogenized, and RNA was prepared using RNAeasy mini columns (Qiagen, Valencia, CA) according to the manufacturer’s instructions. RNA quality was assessed with the Agilent Bioanalyzer 2100 and RIN scores above 7 were used. Libraries were prepared using the Illumina TruSeq RNA Preparation Kit. Samples were sequenced using Illumina HiSeq for single-end 100 bp. Trimmed reads were aligned to the human genome (hg19/GRCh37) with STAR-2.4.1d. The read-count of each sample was obtained using HTSeq-0.6.1 (htseq-count). Normalized fragment per kilobase million values were used to perform association analysis with fibrosis and sclerosis using linear regression.
RNA-sequencing of mouse kidney samples
Total RNA was isolated from 3 mutation mice and 3 wild type mice. RNA quality was assessed with the Agilent TapeStation and quantified with both NanoDrop and Qubit. Only samples with RIN scores above 6 and a minimum total RNA of 100ng were used for library preparation. Strand-specific, poly A-selected RNA-seq libraries were generated using the Illumina TruSeq stranded mRNA protocol. Libraries were pooled and sequenced on an Illumina HiSeq in the 2x150bp configuration.
We first assessed sequence quality using FastQC and then trimmed the adaptor and lower-quality bases using Trim-galore, RNA-seq reads were aligned to the mouse genome (mm10) using STAR (v2.4.1d) based on GENCODE vM18 annotation. Gene-level expression was estimated by uniquely mapped reads as transcripts per million (TPM) by HTSeq 0.11.0 and our Perl scripts. Differential gene experience analysis was implemented by DESeq2. We next adopted DAVID (Database for Annotation, Visualization and Integrated Discovery) and GSEA (gene set enrichment analysis) for functional annotation of the differentially expressed genes.
Histological analysis
Kidneys were fixed in 10% neutral formalin and paraffin-embedded sections were stained with hematoxylin and eosin (H&E) or Periodic acid Schiff (PAS) to analyze the histology of samples. Sirius-red staining was performed to determine the degree of fibrosis. Immunohistochemical analyses were performed to measure the expression of proteins in the kidney. Briefly, paraffin-embedded sections were incubated in indicated primary antibodies, and visualized using diaminobenzidine (DAB) substrates. Sections were further counterstained with hematoxylin.
In situ hybridization
In situ hybridization was performed using formalin-fixed paraffin-embedded tissue samples and the RNAscope® 2.5 HD Duplex Detection Kit (bio-techne, 322436). We followed the manufacturer’s original protocol. The following probes were used for the RNAscope assay: Mm-Emr1 cat# 317969-C2, Mm-Col1a1 cat# 319379.
Immunofluorescence
Primary cultured TECs were seeded in a covered glass-bottom-dish. Cells were fixed in 10 % ice-cold neutral formalin for 10 minutes and washed with PBS. Fixed cells were then exposed to 0.5 % Triton-X 100 in PBS for 10 minutes for permeabilization. Cells were blocked in 3 % BSA/0.1% Triton X-100 in PBS at room temperature for 30 minutes, and incubated with anti-DNA antibody, anti-p65 antibody diluted in blocking buffer (1 % BSA in PBS) at 4 ℃ overnight. After washing with PBS, cells were incubated with secondary antibody and counterstained with DAPI. Images were obtained using a microscope.
Transmission Electron Microscopy (TEM)
Mitochondrial structure was further examined by standard transmission electron microscopy. Fresh kidney or primary cultured TECs were fixed with a mixture of 2% paraformaldehyde and 2.5% glutaraldehyde overnight, washed, dehydrated, and embedded in resin according to standard procedures. Embedded samples were analyzed by a JEOL 1010 electron microscope (Tokyo, Japan).
Measurement of cytosolic mitochondrial DNA
Mitochondrial DNA in the cytosolic fraction was detected by cell compartment fractionation followed by qPCR (Bronner et al., 2015; Bronner and O’Riordan, 2016). Briefly, cells were lysed by mild detergent (0.1% NP-40) and incubated on ice for 15 minutes. Lysates were centrifuged at 13,000 rpm for 15 min at 4 °C. DNeasy Blood and Tissue KIT were used to purify cytosolic mitochondrial DNA from supernatant cytosolic fraction according to manufacturer’s instructions. The qPCR was performed using SYBR green dye (Applied Biosystems) in the ViiA 7 System (Life Technologies). The data were normalized and analyzed using the ΔΔCt method. The primers used and programs run are provided in Supplemental Table 3.
Chemotaxis assay
To measure macrophage migration, Cytoselect 96-well Cell migration assay KIT (Cell Biolabs, CBA-105) was utilized according to manufacturer’s instruction. Briefly, Raw 267.4 macrophages (ATCC) were incubated with medium harvested from primary cultured TECs. After 24 hours’ incubation at 37 °C, cells passed through polycarbonate membranes were dissociated from membrane by the addition of cell detachment buffer. Migratory cells are lysed and quantified using fluorescent dye. The chemotaxis index was calculated by fluorescent intensity read at 480 nm/520 nm.
ATP measurement
The ATP concentration in cells and tissue samples were measured using an ATP Fluorometric Assay Kit (BioVision) according to manufacturer’s protocol.
Quantification of dilated tubules
We used PAS-stained kidney section images and overlaid a grid measuring 13.625 μm. Then, we calculated the number of dots inside the lumen. The degree of tubule dilation was determined using the previously published criteria: (1) normal, one dot; (2) dilated tubule, two dots; (3) microcyst, three to ten dots; and (4) cyst, > 10 dots.
TUNEL assay
TUNEL staining was performed to detect cell death on paraffin-fixed kidney sections and cells following the manufacturer’s instructions (AB66110, Abcam).
QUANTIFICATION AND STATISTICAL ANALYSIS
Student’s t-test was used to analyze differences between two groups, and One-way or Two-way ANOVA was used to analyze intergroup differences with a Bonferroni multiple comparison post-test. P-values less than 0.05 were considered statistically significant. The analysis was performed using GraphPad Prism 5 (GraphPad software). Densitometry results of western blotting were quantified using ImageJ software. All data are presented as mean ± SEM and other details such as the number of replicates and the level of significance is mentioned in figure legends and supplementary tables.
DATA AND SOFTWARE AVAILABILITY
The accession number for the RNA-seq data reported in this paper is GSE115098 and GSE134950.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-TFAM antibody | Sigma | Cat#SAB1401383 |
| Anti-COX IV antibody | Abcam | Cat#Ab16056 |
| Total OXPHOS Rodent WB Antibody Cocktail | Abcam | Cat#ab110413 |
| Anti-Collagen I antibody | Abcam | Cat# ab34710 |
| Anti-Fibronectin antibody | Abcam | Cat#Ab2413 |
| IκBα (L35A5) Mouse mAb | Cell Signaling | Cat#4814 |
| Phospho-IκBα (Ser32) (14D4) Rabbit mAb | Cell Signaling | Cat#2859 |
| Anti-NF-kB p65 antibody | Abcam | Cat# ab16502 |
| Anti-CTCF Antibody | Millipore Sigma | Cat# 07-729 |
| Monoclonal Anti-β-Actin antibody | Sigma | Cat#A5441 |
| Antibodies for Immunohistochemistry (IHC) | ||
| TFAM Antibody | Proteintech | Cat#22586-1-AP |
| Anti-COX IV antibody | Abcam | Cat#Ab16056 |
| Anti-NF-kB p65 antibody | Abcam | Cat# ab16502 |
| Ki-67 (D3B5) Rabbit mAb | Cell Signaling | Cat# #12202 |
| Antibodies for Immunofluorescence (IF) | ||
| Anti-NF-kB p65 antibody | Abcam | Cat# ab16502 |
| Anti-DNA Antibody | Millipore Sigma | Cat# CBL186 |
| Secondary Antibodies | ||
| Anti-rabbit IgG, HRP-linked Antibody | Cell Signaling | Cat#7074 |
| Anti-mouse IgG, HRP-linked Antibody | Cell Signaling | Cat#7076 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody | Invitrogen | Cat#A-31572 |
| Chicken anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody | Invitrogen | Cat#A-21200 |
| Bacterial and Virus Strains | ||
| Ad5CMVCre | University of Iowa Gene Transfer Vector Core | N/A |
| Ad5CMV-eGFP | University of Iowa Gene Transfer Vector Core | N/A |
| Biological Samples | ||
| Human kidney samples | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| C-176 | Vitas-M laboratory | Cat#STK016322 |
| Collagenase IV | Life Technologies | Cat#17104019 |
| Lipofectamine 3000 | ThermoFisher | Cat#11668027 |
| Trypan blue solution | Sigma | Cat#T8154 |
| Protease inhibitor cocktail | Roche | Cat#11836153001 |
| RIPA buffer | Cell signaling | Cat#9806 |
| Folic acid | Fisher Scientific | Cat#AC216630500 |
| Critical Commercial Assays | ||
| Nuclei pure prep isolation kit | Sigma | Cat#NUC-201 |
| BCA Protein Assay Kit | Thermo Scientific | Cat#23225 |
| cDNA Reverse Transcription Kit | Applied Biosystems | Cat#4368813 |
| Rneasy Mini Kit | Qiagen | Cat#74106 |
| RNAscope® 2.5 HD Duplex Detection Kit | Acdbio | Cat#322430 |
| DNeasy Blood & Tissue Kits | Qiagen | Cat#69506 |
| Cytoselect 96-well Cell migration assay KIT | Cell Biolabs | Cat#CBA-105 |
| ATP Colorimetric/Fluorometric Assay Kit | BioVision | Cat#K354 |
| Deposited Data | ||
| Human Kidneys: RNA-seq | GEO | GSE115098 |
| Tfam−/− mice Kidneys: RNA-seq | GEO | GSE134950 |
| Experimental Models: Cell Lines | ||
| Raw264.7 cell line | ATCC | Cat#TIB-71™ |
| Experimental Models: Organisms/Strains | ||
| B6.Cg-Tg(Cdh16-cre)91Igr/J | Jackson Laboratory | Cat#012237 |
| B6.Cg-Tfam<tm1.1Ncdl>/J | Jackson Laboratory | Cat#026123 |
| B6(Cg)-Tmem173<tm1.2Camb>/J | Jackson Laboratory | Cat#025805 |
| Oligonucleotides | ||
| Primers for qPCR, see Table S3 | This paper | N/A |
| siRNA targeting sequence: Tmem173 (Duplex 1): GAAUCGGGUUUAUUCCAACAGCGTC | IDT | N/A |
| siRNA targeting sequence: Tmem173 (Duplex 2): UUCUUAGCCCAAAUAAGGUUGUCGCAG | IDT | N/A |
| Software and Algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij |
| Prism 5 | Graphpad Software | https://www.graphpad.com/scientific-software/prism |
| STAR v2.4.1d | Cold Spring Harbor Laboratory | http://code.google.com/p/rna-star/ |
| David v6.8 | Laboratory of Human Retrovirology and Immunoinformatics (LHRI) | https://david.ncifcrf.gov/ |
| GSEA Desktop v3.0 | Broad Institute, Inc.,Massachusetts Institute of Technology | http://software.broadinstitute.org/gsea/index.jsp |
Acknowledgement
Work in the Susztak lab is supported by NIDDK R01DK076077, R01 DK087635 and DP3 DK108220. J.P. is supported by American Diabetes Association Training grant #1-17-PDF-036. L.P. is supported by the Office of the Assistant Secretary of Defense for Health Affairs through the Peer Reviewed Medical Research Program under Award W81XWH-16-1-0400, NIH DK111495, and pilot awards from the Diabetes Research Center at the University of Pennsylvania NIH DK19525.
Declaration of interests
The Susztak lab is supported by Boehringer Ingelheim, Lilly, Regeneron, Bayer, GSK, Merck, ONO Pharma, Gilead and Celgene for work that is not related to the current manuscript.
References
- Beckerman P, Qiu C, Park J, Ledo N, Ko YA, Park AD, Han SY, Choi P, Palmer M, and Susztak K (2017). Human Kidney Tubule-Specific Gene Expression Based Dissection of Chronic Kidney Disease Traits. EBioMedicine 24, 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet F, and Scheen AJ (2018). Effects of SGLT2 inhibitors on systemic and tissue low-grade inflammation: The potential contribution to diabetes complications and cardiovascular disease. Diabetes Metab 44, 457–464. [DOI] [PubMed] [Google Scholar]
- Breyer MD, and Susztak K (2016). The next generation of therapeutics for chronic kidney disease. Nat Rev Drug Discov 15, 568–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner DN, Abuaita BH, Chen X, Fitzgerald KA, Nuñez G, He Y, Yin XM, O’Riordan MX (2015). Endoplasmic Reticulum Stress Activates the Inflammasome via NLRP3- and Caspase-2-Driven Mitochondrial Damage 43, 451–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner DN and O’Riordan MX (2016). Measurement of Mitochondrial DNA Release in Response to ER Stress. Bio-protocol 6, e1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CT, Kolesar JE, and Kaufman BA (2012). Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number. Biochim Biophys Acta 1819, 921–929. [DOI] [PubMed] [Google Scholar]
- Chen Q, Sun L, and Chen ZJ (2016). Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol 17, 1142–1149. [DOI] [PubMed] [Google Scholar]
- Collins AJ, Gilbertson DT, Snyder JJ, Chen SC, and Foley RN (2010). Chronic kidney disease awareness, screening and prevention: rationale for the design of a public education program. Nephrology (Carlton) 15 Suppl 2, 37–42. [DOI] [PubMed] [Google Scholar]
- Fang R, Wang C, Jiang Q, Lv M, Gao P, Yu X, Mu P, Zhang R, Bi S, Feng JM, et al. (2017). NEMO-IKKbeta Are Essential for IRF3 and NF-kappaB Activation in the cGAS-STING Pathway. J Immunol 199, 3222–3233. [DOI] [PubMed] [Google Scholar]
- Ferenbach DA, and Bonventre JV (2015). Mechanisms of maladaptive repair after AKI leading to accelerated kidney ageing and CKD. Nat Rev Nephrol 11, 264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer H, Lutay N, Ragnarsdottir B, Yadav M, Jonsson K, Urbano A, Al Hadad A, Ramisch S, Storm P, Dobrindt U, et al. (2010). Pathogen specific, IRF3-dependent signaling and innate resistance to human kidney infection. PLoS Pathog 6, e1001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley RN (2010). Temporal trends in the burden of chronic kidney disease in the United States. Curr Opin Nephrol Hypertens 19, 273–277. [DOI] [PubMed] [Google Scholar]
- Gaber T, Strehl C, and Buttgereit F (2017). Metabolic regulation of inflammation. Nat Rev Rheumatol 13, 267–279. [DOI] [PubMed] [Google Scholar]
- Gleeson LE, and Sheedy FJ (2016). Metabolic reprogramming & inflammation: Fuelling the host response to pathogens. Semin Immunol 28, 450–468. [DOI] [PubMed] [Google Scholar]
- Guan Y (2004). Peroxisome proliferator-activated receptor family and its relationship to renal complications of the metabolic syndrome. J Am Soc Nephrol 15, 2801–2815. [DOI] [PubMed] [Google Scholar]
- Haag SM, Gulen MF, Reymond L, Gibelin A, Abrami L, Decout A, Heymann M, van der Goot FG, Turcatti G, Behrendt R, Ablasser A (2018). Targeting STING with covalent small-molecule inhibitors. Nature 559, 269–273. [DOI] [PubMed] [Google Scholar]
- Han SH, Malaga-Dieguez L, Chinga F, Kang HM, Tao J, Reidy K, and Susztak K (2015). Deletion of Lkb1 in Renal Tubular Epithelial Cells Leads to CKD by Altering Metabolism. J Am Soc Nephrol 27, 439–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SH, Wu MY, Nam BY, Park JT, Yoo TH, Kang SW, Park J, Chinga F, Li SY, and Susztak K (2017). PGC-1alpha Protects from Notch-Induced Kidney Fibrosis Development. J Am Soc Nephrol 28, 3312–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Park J, Qiu C, Chung KW, Li SY, Sirin Y, Han SH, Taylor V, Zimber-Strobl U, and Susztak K (2018). Jagged1/Notch2 controls kidney fibrosis via Tfam-mediated metabolic reprogramming. PLoS Biol 16, e2005233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, and Susztak K (2016). Epithelial Plasticity versus EMT in Kidney Fibrosis. Trends Mol Med 22, 4–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamijo Y, Hora K, Kono K, Takahashi K, Higuchi M, Ehara T, Kiyosawa K, Shigematsu H, Gonzalez FJ, and Aoyama T (2007). PPARalpha protects proximal tubular cells from acute fatty acid toxicity. J Am Soc Nephrol 18, 3089–3100. [DOI] [PubMed] [Google Scholar]
- Kang HM, Ahn SH, Choi P, Ko YA, Han SH, Chinga F, Park AS, Tao J, Sharma K, Pullman J, et al. (2015). Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat Med 21, 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovesdy CP, Bleyer AJ, Molnar MZ, Ma JZ, Sim JJ, Cushman WC, Quarles LD, and Kalantar-Zadeh K (2013). Blood pressure and mortality in U.S. veterans with chronic kidney disease: a cohort study. Ann Intern Med 159, 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, and Clayton DA (1998). Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet 18, 231–236. [DOI] [PubMed] [Google Scholar]
- Lemos DR, McMurdo M, Karaca G, Wilflingseder J, Leaf IA, Gupta N, Miyoshi T, Susa K, Johnson BG, Soliman K, et al. (2018). Interleukin-1beta Activates a MYC-Dependent Metabolic Switch in Kidney Stromal Cells Necessary for Progressive Tubulointerstitial Fibrosis. J Am Soc Nephrol 29, 1690–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin A, Tonelli M, Bonventre J, Coresh J, Donner JA, Fogo AB, Fox CS, Gansevoort RT, Heerspink HJL, Jardine M, et al. (2017). Global kidney health 2017 and beyond: a roadmap for closing gaps in care, research, and policy. Lancet 390, 1888–1917. [DOI] [PubMed] [Google Scholar]
- Li H, Wang J, Wilhelmsson H, Hansson A, Thoren P, Duffy J, Rustin P, and Larsson NG (2000). Genetic modification of survival in tissue-specific knockout mice with mitochondrial cardiomyopathy. Proc Natl Acad Sci U S A 97, 3467–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel LJ (1985). Metabolic substrates, cellular energy production, and the regulation of proximal tubular transport. Annu Rev Physiol 47, 85–101. [DOI] [PubMed] [Google Scholar]
- Ng KW, Marshall EA, Bell JC, and Lam WL (2018). cGAS-STING and Cancer: Dichotomous Roles in Tumor Immunity and Development. Trends Immunol 39, 44–54. [DOI] [PubMed] [Google Scholar]
- Niewczas MA, Pavkov ME, Skupien J, Smiles A, Md Dom ZI, Wilson JM, Park J, Nair V, Schlafly A, Saulnier PJ, et al. (2019). A signature of circulating inflammatory proteins and development of end-stage renal disease in diabetes. Nat Med 25,805–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn O, and Olefsky JM (2012). The cellular and signaling networks linking the immune system and metabolism in disease. Nat Med 18, 363–374. [DOI] [PubMed] [Google Scholar]
- Park CW, Kim HW, Ko SH, Chung HW, Lim SW, Yang CW, Chang YS, Sugawara A, Guan Y, and Breyer MD (2006). Accelerated diabetic nephropathy in mice lacking the peroxisome proliferator-activated receptor alpha. Diabetes 55, 885–893. [DOI] [PubMed] [Google Scholar]
- Peralta S, Wang X, and Moraes CT (2012). Mitochondrial transcription: lessons from mouse models. Biochim Biophys Acta 1819, 961–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler R, Afkarian M, Dieter BP, and Tuttle KR (2017). Immunity and inflammation in diabetic kidney disease: translating mechanisms to biomarkers and treatment targets. Am J Physiol Renal Physiol 312, F716–F731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu C, Huang S, Park J, Park Y, Ko YA, Seasock MJ, Bryer JS, Xu XX, Song WC, Palmer M, et al. (2018). Renal compartment-specific genetic variation analyses identify new pathways in chronic kidney disease. Nat Med 24, 1721–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston JC, Lyons CL, Kennedy EB, Kirwan AM, and Roche HM (2017). Fatty Acids and NLRP3 Inflammasome-Mediated Inflammation in Metabolic Tissues. Annu Rev Nutr 37, 77–102. [DOI] [PubMed] [Google Scholar]
- Reidy K, Kang HM, Hostetter T, and Susztak K (2014). Molecular mechanisms of diabetic kidney disease. J Clin Invest 124, 2333–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodwell GE, Sonu R, Zahn JM, Lund J, Wilhelmy J, Wang L, Xiao W, Mindrinos M, Crane E, Segal E, et al. (2004). A transcriptional profile of aging in the human kidney. PLoS Biol 2, e427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan X, Zheng F, and Guan Y (2008). PPARs and the kidney in metabolic syndrome. Am J Physiol Renal Physiol 294, F1032–1047. [DOI] [PubMed] [Google Scholar]
- Rudnicki M, Eder S, Perco P, Enrich J, Scheiber K, Koppelstatter C, Schratzberger G, Mayer B, Oberbauer R, Meyer TW, et al. (2007). Gene expression profiles of human proximal tubular epithelial cells in proteinuric nephropathies. Kidney Int 71, 325–335. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC (2008). Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev 88, 611–638. [DOI] [PubMed] [Google Scholar]
- Su W, Huang SZ, Gao M, Kong XM, Gustafsson JA, Xu SJ, Wang B, Zheng F, Chen LH, Wang NP, et al. (2017). Liver X receptor beta increases aquaporin 2 protein level via a posttranscriptional mechanism in renal collecting ducts. Am J Physiol Renal Physiol 312, F619–F628. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Nishimura A, Nishiyama K, Goto T, Numaga-Tomita T, and Nishida M (2019). Mitochondrial dynamics in exercise physiology. Pflugers Arch. [DOI] [PubMed] [Google Scholar]
- Tieu A, House AA, and Urquhart BL (2016). Drug Disposition Issues in CKD: Implications for Drug Discovery and Regulatory Approval. Adv Chronic Kidney Dis 23, 63–66. [DOI] [PubMed] [Google Scholar]
- Tran M, Tam D, Bardia A, Bhasin M, Rowe GC, Kher A, Zsengeller ZK, Akhavan-Sharif MR, Khankin EV, Saintgeniez M, et al. (2011). PGC-1alpha promotes recovery after acute kidney injury during systemic inflammation in mice. J Clin Invest 121, 4003–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran MT, Zsengeller ZK, Berg AH, Khankin EV, Bhasin MK, Kim W, Clish CB, Stillman IE, Karumanchi SA, Rhee EP, et al. (2016). PGC1alpha drives NAD biosynthesis linking oxidative metabolism to renal protection. Nature 531, 528–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsushida K, Tanabe K, Masuda K, Tanimura S, Miyake H, Arata Y, Sugiyama H, and Wada J (2018). Estrogen-related receptor alpha is essential for maintaining mitochondrial integrity in cisplatin-induced acute kidney injury. Biochem Biophys Res Commun 498, 918–924. [DOI] [PubMed] [Google Scholar]
- West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, MacDuff DA, et al. (2015). Mitochondrial DNA Stress Primes the Antiviral Innate Immune Response. Nature 520(7548): 553–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernochet C, Mourier A, Bezy O, Macotela Y, Boucher J, Rardin MJ, An D, Lee KY, Ilkayeva OR, Zingaretti CM, et al. (2012). Adipose-specific deletion of TFAM increases mitochondrial oxidation and protects mice against obesity and insulin resistance. Cell Metab 16, 765–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada J, and Makino H (2016). Innate immunity in diabetes and diabetic nephropathy. Nat Rev Nephrol 12, 13–26. [DOI] [PubMed] [Google Scholar]
- Wang J, Wilhelmsson H, Graff C, Li H, Oldfors A, Rustin P, Bruning JC, Kahn CR, Clayton DA, Barsh GS, et al. (1999). Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nat Genet 21, 133–137. [DOI] [PubMed] [Google Scholar]
- Woroniecka KI, Park AS, Mohtat D, Thomas DB, Pullman JM, and Susztak K (2011). Transcriptome analysis of human diabetic kidney disease. Diabetes 60, 2354–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Malone AF, Donnelly EL, Kirita Y, Uchimura K, Ramakrishnan SM, Gaut JP, and Humphreys BD (2018). Single-Cell Transcriptomics of a Human Kidney Allograft Biopsy Specimen Defines a Diverse Inflammatory Response. J Am Soc Nephrol 29, 2069–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelnick LR, Weiss NS, Kestenbaum BR, Robinson-Cohen C, Heagerty PJ, Tuttle K, Hall YN, Hirsch IB, and de Boer IH (2017). Diabetes and CKD in the United States Population, 2009–2014. Clin J Am Soc Nephrol 12, 1984–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang A, Kohan DE, Nelson RD, Gonzalez FJ, and Yang T (2005). Collecting duct-specific deletion of peroxisome proliferator-activated receptor gamma blocks thiazolidinedione-induced fluid retention. Proc Natl Acad Sci U S A 102, 9406–9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Huang S, Gao M, Liu J, Jia X, Han Q, Zheng S, Miao Y, Li S, Weng H, et al. (2014). Farnesoid X receptor (FXR) gene deficiency impairs urine concentration in mice. Proc Natl Acad Sci U S A 111, 2277–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Lupino K, Wilkins BJ, Qiu C, Liu J, Omura Y, Allred AL, McDonald C, Susztak K, Barish GD, et al. (2018). Genomic integration of ERRgamma-HNF1beta regulates renal bioenergetics and prevents chronic kidney disease. Proc Natl Acad Sci U S A 115, E4910–E4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNA-seq data reported in this paper is GSE115098 and GSE134950.