Abstract
This work sought to assess optimal extraction conditions in the study of the metalloproteome of the dimorphic fungus Histoplasma capsulatum. One of the body’s responses to H. capsulatum infection is sequestration of zinc within host macrophage (MØ), as reported by Vignesh et al. (Immunity 39:697–710, 2013) and Vignesh et al. (PLOS Pathog 9:E1003815, 2013). Thus, metalloproteins containing zinc were of greatest interest as it plays a critical role in survival of the fungus. One challenge in metalloproteomics is the preservation of the native structure of proteins to retain non-covalently bound metals. Many of the conventional cell lysis, separation, and identification techniques in proteomics are carried out under conditions that could lead to protein denaturation. Various cell lysis techniques were investigated in an effort to both maintain the metalloproteins during lysis and subsequent analysis while, at the same time, serving to be strong enough to break the cell wall, allowing access to cytosolic metalloproteins. The addition of 1% Triton x-100, a non-ionic detergent, to the lysis buffer was also studied. Seven lysis methods were considered and these included: Glass Homogenizer (H), Bead Beater (BB), Sonication Probe (SP), Vortex with 1% Triton x-100 (V, T), Vortex with no Triton x-100 (V, NT), Sonication Bath, Vortex, and 1% Triton x-100 (SB, V, T) and Sonication Bath, Vortex, and no Triton x-100 (SB, V, NT). A Qubit® Assay was used to compare total protein concentration and inductively coupled plasma–mass spectrometry (ICP-MS) was utilized for total metal analysis of cell lysates. Size exclusion chromatography coupled to ICP-MS (SEC-HPLC-ICP-MS) was used for separation of the metalloproteins in the cell lysate and the concentration of Zn over a wide molecular weight range was examined. Additional factors such as potential contamination sources were also considered. A cell lysis method involving vortexing H. capsulatum yeast cells with 500 μm glass beads in a 1% Triton x-100 lysis buffer (V, T) was found to be most advantageous to extract intact zinc metalloproteins as demonstrated by the highest Zn to protein ratio, 1.030 ng Zn/μg protein, and Zn distribution among high, mid, and low molecular weights suggesting the least amount of protein denaturation.
Keywords: Cell lysis, Metalloproteomics, Histoplasma capsulatum, ICP-MS, Zinc
Introduction
The field of metallomics has emerged over the past few decades as the study of metals and their use for growth and survival within biological organisms [1–3]. Many times, metals are associated with a given protein and it is estimated that one third of all proteins contain a metal [4]. Within metallomics, the study of metal transport, storage, and use by proteins, known as metalloproteomics, has expanded protein studies considerably [2, 3].Investigations range from pure inquiry into the metalloproteome as a whole or targeting a specific metal, as the metal may be used by enzymes as cofactors to catalyze reactions, the metal may be necessary for protein function, or the metal may play key structural roles within a protein [5–8]. Other studies probe metal-protein binding as an assessment of environmental exposure [9–11] or disease pathogenesis [12, 13]. Still others seek to elucidate the mechanisms of metallodrugs and their fate within cells [14, 15].
Most microbial metalloproteomes are, as of yet, largely uncharacterized [16]. Histoplasma capsulatum, in particular, is a dimorphic fungus and causes a respiratory infection known as histoplasmosis which may develop into a progressive infection, especially for immunocompromised individuals [17]. One of the first lines of defense against H. capsulatum is macrophage (MΦ), yet H. capsulatum has the ability to avoid host immune defenses by replicating within the MΦ [18]. One of the body’s responses to this infection, using the H. capsulatum strain G217B, is the simultaneous sequestration of zinc and generation of reactive oxygen species (ROS) through NADPH within MΦ [18, 19]. This immune response indicates that the micronutrient zinc is essential to H. capsulatum.
In some metalloproteomic studies, the element of interest is bound within the primary structure of the protein, such as selenoproteins in which selenium replaces the sulfur in cysteine amino acids. However, in the vast majority of cases, the metal ion is held within the protein structure by non-covalent forces [20]. One of the greatest challenges in the experimental analysis of the metalloproteome of a given organism within the latter case is the development of an optimal cell lysis method [21]. The strength of the fungal cell wall varies between species and poses and added challenge in choosing a cell lysis method as compared to cells with weaker membrane structures such as mammalian cells [22]. When choosing a method for an organism that contains cell walls, the method must be strong to break the cell wall, allowing the release of the cytosolic proteins. At the same time, it cannot be so harsh that it denatures the protein as this leads to loss of the associated metal [20, 23]. As a result, established physical and chemical cell lysis techniques need to be carefully evaluated to determine if they can be used in such studies [21]. Klimek-Ochab showed fungal yeast may be better suited for extraction with glass beads, but few fungal species have been studied in depth [22].
Several cell lysis techniques have been developed over the years to disrupt cells, and the majority of them can be classified as chemical or mechanical [22]. Chemical cell lysis techniques often involve detergents, enzymes, and/or salts. Detergents are commonly used for cell lysis techniques to study proteins. Detergents contain hydrophobic tails and hydrophilic heads, allowing for the disruption of the hydrophobic-hydrophilic interface of biological membranes. Non-ionic detergents, such as Triton x-100, tend to be gentler and generally do not disrupt the native structure of proteins. Other non-ionic detergents may be suitable for metalloproteomic studies such as Triton x-114, Brij-35, Brij-58, Tween 20, Tween 80, octyl glucoside, and octyl thioglucoside. Salts are used in cell lysis buffers to maintain the pH and osmolarity of the cell lysate. Salts, such ethylenediaminetetraacetic acid (EDTA) disodium salt, are not suitable for metalloproteomic experiments due to metal-binding properties. Buffer agents such as tris(hydroxymethyl)aminomethane hydrochloride (Tris-HCl) and 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) are also often used in cell lysis solutions. The NP-40 lysis buffer, which can be used with the detergent Triton x-100, is fairly mild and is common for whole cell or soluble cytosolic proteins and was deemed suitable for this study. Several examples of mechanical cell lysis techniques include bead beater, homogenizer, French press, and freeze-thaw. Bead beaters use the mechanical shear of small glass beads, while homogenizers use the shear created by a ground glass tight fitting mortar and pestle. French press uses shear and decompression by forcing the sample through a small orifice under high pressure. Freeze-thaw involves rapid cycling of sample temperature to disrupt cells through ice crystal formation. Both the chemical and mechanical cell lysis methods described were originally designed for the study of proteins without consideration of metal-binding, and they may not be suitable for metalloproteomics experiments. Further, commercial cell lysis products contain proprietary formulas that may produce denaturing conditions and/or metal contamination. Therefore, it is essential to assess cell lysis methods and optimize protocols to preserve the metalloproteome.
In addition to preservation of the metalloproteome, the chosen cell lysis technique needs to be compatible with the analytical scheme. As noted by Hagege et al., losses of metal-protein complexes must be minimized by the separation technique [24]. Size exclusion chromatography (SEC) is routinely used to roughly separate metalloproteins by hydrodynamic radius [11, 15]. While not a particularly powerful separation technique, it can be carried out under non-denaturing conditions, although there may be components in the lysis buffer that interfere with the analysis such as certain detergents that absorb in the ultraviolet region when using UV detection. Further, SEC typically offers complete recovery of all species due to the absence of secondary separation principles such as absorption onto the stationary phase [25]. A common second dimension of separation is ion exchange chromatography (IEX). Lysis buffer components that are charged, such as the anionic detergent SDS, can interfere with this separation and should therefore be removed from all steps of sample preparation. For both separation procedures, inductively coupled plasma-mass spectrometry (ICP-MS) is commonly employed for detection due to its multi-element capability and low detection limits [11, 15].
In this work, several cell lysis techniques and two lysis buffers were investigated to evaluate the preservation of the zinc metalloproteome of H. capsulatum while maintaining compatibility with the analytical techniques employed. Investigation of the H. capsulatum zinc metalloproteome will expand knowledge of zinc pathways and the role of zinc in the survival of the fungus. Further characterization and quantification of the zinc metalloproteome could be used to assess changes to the fungus under zinc stress, mimicking the immune response. The cell lysis methods investigated were evaluated based the total protein concentration, the total concentration of metal, the molecular weight distribution of the metal, and additional factors pertinent to sample preparation.
Materials and methods
Reagents
All solutions were prepared using double de-ionized (DDI) water (18 MΩ cm−1, produced by the Nanopure treatment system purchased from Sybron Barnstead, Boston, MA, USA). Labware was acid washed with 10% nitric acid (Fisher Scientific, Fair Lawn, NJ, USA) to remove metals. Cells were washed with phosphate-buffered saline (PBS) without added calcium and magnesium (Mediatech, Inc. A Corning Subsidiary, Manassas, VA, USA). Lysis buffer components included sodium chloride (Fisher Scientific, Fair Lawn, NJ, USA), Triton x-100 (Sigma-Aldrich, St. Louis, MO, USA), tris(hydroxymethyl)aminomethane (Tris) (Acros Organics, Morris Plains, NJ, USA), and Mini EDTA-free Protease Inhibitor Cocktail Tablets (Roche Life Science, Indianapolis, IN, USA). The mobile phase for SEC contained 50 mM ammonium acetate (Fisher Scientific, Fair Lawn, NJ, USA) and 0.5% methanol (Fisher Scientific, Fair Lawn, NJ, USA) in DDI water, adjusted to pH 7.4 using ammonium hydroxide (Fisher Scientific, Fair Lawn, NJ, USA). The Gel Filtration Standard (Bio-Rad Laboratories, Inc., Hercules, CA, USA) used for SEC was a lyophilized mixture of molecular weight markers at 1.35, 17, 44, 158, and 670 kDa which was dissolved in DDI water. Total protein concentration was determined using a Qubit® Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA).
Instrumentation
Trace elemental analysis was conducted using an Agilent 8800 ICP-MS (Agilent Technologies, Santa Clara, CA, USA). The sample introduction system for the ICP-MS included a MicroMist nebulizer (Glass Expansion, Pocasset, MA), a Scott double-channel spray chamber (2 °C), and a shield torch. The ICP-MS was used in collision mode with helium gas at 3.0 mL/min to reduce isobaric interferences. Isotopes monitored included 55Mn, 56Fe, 59Co, 63Cu, and 66Zn. For total metal analysis, an Agilent ASX-500 Autosampler was employed and 45Sc was used as internal standards. For cell lysate separation, an Agilent 1100 HPLC (Agilent Technologies, Santa Clara, CA, USA) with a Zorbax GF-250 column (Agilent Technologies, Santa Clara, CA, USA) was used. The HPLC was equipped with a vacuum membrane degasser system, binary pump, cooled autosampler, a temperature-controlled column compartment (held at 25 °C), and a diode array detector where the UV absorbance was recorded at 280 nm. The integrated area of the Gel Filtration Standard was used to normalize the ICP-MS signal from day to day. The HPLC was coupled to the ICP-MS allowing for nearly simultaneous UV detection and metal detection. Total protein concentration was determined using a Qubit® 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA).
Preparation of H. capsulatum
H. capsulatum yeast (strain G217B) was prepared by inoculating 25 ml of Ham’s F-12 medium set at pH 7.5, as previously described [26], with 3 × 106 cells/ml from 5-day-old slants. After 30 h, samples were centrifuged for 5 min at 1500 g, the supernatant was removed, and the cells were rinsed with PBS, centrifuged again, and the supernatant was removed.
Cell lysis techniques
For each cell lysis technique, the yeast cell count was used and the ratio of yeast cells to volume of lysis buffer was held constant in order to normalize data and compare chromatographic areas.
Glass homogenizer
Glass homogenizers are primarily used for tissue homogenization, but have occasionally been used with yeast cells [27–29]. Due to the operator dependence and lack of reproducibility, it was expected that this method would not extract the greatest amount of protein or metal and was investigated as a comparison to other methods better suited for yeast cell lysis [27]. A 15-mL glass homogenizer (H) with tight fitting pestle was used to grind 3 × 108 yeast cells and 0.5 mL of lysis buffer (1% Triton x-100, 50 mM Tris, 100 mM NaCl, EDTA-free protease inhibitor) with 10 even rotations. The mixture was transferred to a 1.5-mL plastic vial and chilled on ice for 5 min. The homogenate was centrifuged for 5 min at 12,000 g and the supernatant transferred to an HPLC vial.
Bead beater
Bead beaters have been used extensively to disrupt microorganisms using small glass beads with several examples involving yeast [29–32]. A BioSpec 3110 BX Mini-BeadBeater (BB) (BioSpec, Bartlesville, OK) was used with 500 μm diameter glass beads and 2 mL screw cap vials with o-ring seals with straight walls and a sharp conical bottom. The vials were filled with 0.5 mL of lysis buffer (1% Triton x-100, 50 mM Tris, 100 mM NaCl, EDTA-free protease inhibitor), 3 × 108 yeast cells, and nitric acid-washed 500-μm glass beads to the brim of the vial. The Bead Beater was operated at 4800 rpm for 2 min, the vial was removed and put on ice for 2 min, and the cycle was repeated with 2 min on the BeadBeater, 2 min on ice, and finally 1 min on the BeadBeater and 1 min on ice. The homogenate was removed and centrifuged for 5 min at 12,000 g and the supernatant transferred to an HPLC vial.
Sonication probe
Cell lysis by sonication probe has been used in yeast using short pulse times to limit heat generation [33–37]. The Sonication Probe method was based on the work of Neppiras et al. [38] A Q125 Qsonica Sonicator (Qsonica, Newtown, CT) equipped with a 1/8″ diameter probe (SP) was operated at an amplitude of 20% of 65 W. A total of 3.0 × 108 yeast cells were combined with 0.5 mL of lysis buffer (1% Triton x-100, 50 mM Tris, 100 mM NaCl, EDTA-free protease inhibitor) in a 1.5-mL plastic vial that was suspended in an ice bath and subjected to a 1-s pulse followed by 10 s of rest and another 1-s pulse. The homogenate was centrifuged for 5 min at 12,000 g and the supernatant transferred to an HPLC vial.
Vortex with glass beads
There are several examples of the vortex and glass bead cell lysis technique for protein extraction in yeast [39–41]. The Vortex with Glass Beads method (V) involved 1 mL of 500 μm glass beads and 0.5 mL of lysis buffer combined with 3.0 × 108 yeast cells in a 1.5-mL plastic vial which was chilled at 4 °C for 15 min. Two lysis buffers were evaluated: one included detergent (1% Triton x-100, 50 mM Tris, 100 mM NaCl, EDTA-free protease inhibitor) (T) and the other contained no detergent (50 mM Tris, 100 mM NaCl, EDTA-free protease inhibitor) (NT). The vials were vortexed for 30 s, placed on ice for 30 s, and the cycle was repeated for a total of 10 min. The homogenate was removed and centrifuged for 5 min at 12,000 g and the supernatant transferred to an HPLC vial.
Sonication bath and vortex with glass beads
Sonication baths have been used with yeast to improve the reproducibility of protein extraction and was combined with the Vortex with Glass Bead method in this study to determine if it had a similar effect [39]. The Sonication Bath and Vortex with Glass Beads method (SB,V) involved 1 mL of 500 μm glass beads and 0.5 mL of lysis buffer combined with 3.0 × 108 yeast cells in a 1.5-mL vial which was chilled at 4 °C in a refrigerator for 15 min. Again, two lysis buffers were evaluated, one containing detergent (T) and the other with no detergent (NT). A Sonication Bath was chilled with ice while the samples chilled in the refrigerator and the ice was removed prior to sonicating the samples. The vials were placed in the Sonication Bath for 10 min and placed on ice for 2 min. The vials were then vortexed for 30 s, placed on ice for 30 s, and the cycle of vortexing and ice was repeated for a total of 10 min. The homogenate was removed, centrifuged for 5 min at 12,000 g, and the supernatant transferred to an HPLC vial.
Results and discussion
In this study, mechanical and chemical lysis techniques were investigated as well as various combinations of techniques. For the chemical lysis techniques, the NP-40 lysis buffer using the non-ionic detergent Triton x-100 was chosen to limit protein denaturation. In addition, to assess the impact of the detergent, a lysis buffer containing the same ingredients and no detergent was explored. Several mechanical techniques were also employed. Seven lysis methods were considered and these included: Glass Homogenizer (H), Bead Beater (BB), Sonication Probe (SP), Vortex with 1% Triton x-100 (V, T), Vortex with no Triton x-100 (V, NT), Sonication Bath, Vortex, and 1% Triton x-100 (SB, V, T) and Sonication Bath, Vortex, and no Triton x-100 (SB, V, NT).
Total protein concentration provides a limited amount of information to determine the most appropriate cell lysis method because it does not take into account if the proteins contain metals nor if they are denatured. Conventional cell lysis methods employ assays such as Bradford to determine total protein concentration. However, for our work, one of the disadvantages of such an assay is the incompatibility with detergents such as Triton x-100. Other assays are compatible with detergents, but do require significant dilution such as the Qubit® assay which was used in this work. Results for the concentration of protein by Qubit® assay are shown in Table 1. The methods that resulted in the greatest extracted protein concentration include Sonication Probe (SP) 215.73 μg/mL, Vortex, Triton x-100 method (V, T) 197.08 μg/mL, and Sonication Bath, Vortex and 1% Triton x-100 (SB, V, T) 165.71 μg/mL. An ANOVA was used to determine difference between groups (F(2,12) = 5.9984, p = 0.0156). A Tukey HSD post hoc test determined significant different between the Sonication Bath, Vortex, and 1% Triton x-100 method (SB, V, T) and the Sonication Probe method (SP) (p = 0.0129), but there was not a significant difference between the Sonication Probe method (SP) and the Vortex, 1% Triton x-100 method (V, T) (p = 0.4335) or Sonication Bath, Vortex, 1% Triton x-100 method (SB, V, T) and the Vortex, 1% Triton x-100 method (V, T) (p = 0.1214). The 280-nm signal collected using the UV detector of the HPLC might also be used for total protein comparison, as this wavelength is routinely used for protein concentration measurement, but it also presented a major challenge due to the Triton x-100 lysis buffer. Triton x-100 absorbs in the ultraviolet region of the spectrum leading to enhanced readings at this wavelength. Therefore, the UV signal was used to monitor the retention times of the protein standard mixture, but not used to compare protein concentration of lysate samples.
Table 1.
Comparison of total protein concentration in cell lysates by various cell lysis methods using Qubit® assay. All data is normalized to 3.0 × 108 H. capsulatum yeast and an HPLC injection volume of 100 μL. n = 5. Glass Homogenizer (H), Bead Beater (BB), Sonication Probe (SP), Vortex with 1% Triton x-100,(V, T), Vortex with no Triton x-100(V, NT), Sonication Bath, Vortex, and 1% Triton x-100 (SB, V, T) and Sonication Bath, Vortex, and no Triton x-100 (SB, V, NT)
| Cell lysis method | Total protein concentration, Qubit® Assay | ||
|---|---|---|---|
| Average protein concentration (μg/mL) | Standard deviation | % Relative standard deviation | |
| H | 116.16 | 21.39 | 18.42 |
| BB | 145.71 | 10.48 | 7.19 |
| SP | 215.73 | 33.46 | 15.51 |
| V, T | 197.08 | 16.39 | 8.32 |
| SB, V, T | 165.71 | 14.48 | 8.74 |
| V, NT | 145.31 | 22.53 | 15.51 |
| SB, V, NT | 137.68 | 14.46 | 10.50 |
Total metal analysis of the cell lysates was conducted by ICP-MS and is reported as ng Zn/100uL lysate in Table 2. The Vortex, Triton x-100 method (V, T) produced 18.51 ng Zn/100 μL lysate, the Sonication Bath, Vortex, Triton x-100 method (SB, V, T) produced 13.95 ng Zn/100 μL lysate, and the Sonication Probe method (SP) produced 10.92 ng Zn/100 μL lysate which demonstrate the highest zinc concentrations. An ANOVA was used to determine difference between groups (F(2,12) = 19.2197, p = 0.00002). A Tukey HSD post hoc test determined significant different between the Vortex, and 1% Triton x-100 method (V, T) and the Sonication Probe method (SP) (p = 0.0001) and the Sonication Bath, Vortex, and 1% Triton x-100 method(SB, V, T) (p = 0.0079), but there was not a significant difference between the Sonication Probe method (SP) and the Sonication Bath, Vortex, and 1% Triton x-100 method (SB, V, T) (p = 0.0720). However, this data is limited in value as it does not provide information regarding free and bound metals or molecular weight distribution.
Table 2.
Comparison of zinc in cell lysates by various cell lysis methods. All data is normalized to 3.0 × 108 H. capsulatum yeast and an HPLC injection volume of 100 μL. n = 5. Data in column 6 is the ratio of ng of Zn in a 100-μL volume was divided by the μg of protein in a 100-μL volume as determined by Qubit®. Glass Homogenizer (H), Bead Beater (BB), Sonication Probe (SP), Vortex with 1% Triton x-100, (V, T), Vortex with no Triton x-100 (V, NT), Sonication Bath, Vortex, and 1% Triton x-100 (SB, V, T),and Sonication Bath,Vortex, and no Triton x-100 (SB, V, NT)
| Cell lysis method | Total metal analysis, ICP-MS | Zn chromatogram, SEC-HPLC-ICP-MS | |||||
|---|---|---|---|---|---|---|---|
| Average ng Zn /100 μL cell lysate | Standard Deviation | % Relative Standard Deviation | Average ng Zn /100 μL cell lysate | Average ng Zn / μg protein | Standard Deviation | % Relative Standard Deviation | |
| H | 8.50 | 1.14 | 13.37 | 8.24 | 0.709 | 1.71 | 20.75 |
| BB | 9.54 | 1.18 | 12.39 | 7.66 | 0.526 | 1.14 | 14.88 |
| SP | 10.92 | 1.79 | 16.39 | 12.75 | 0.591 | 2.42 | 18.98 |
| V, T | 18.51 | 2.49 | 13.45 | 20.31 | 1.030 | 3.67 | 18.07 |
| SB, V, T | 13.95 | 1.41 | 10.11 | 15.88 | 0.958 | 2.55 | 16.06 |
| V, NT | 7.63 | 1.39 | 18.22 | 7.55 | 0.520 | 1.54 | 20.40 |
| SB, V, NT | 8.91 | 1.23 | 13.81 | 8.69 | 0.631 | 1.06 | 12.20 |
SEC-HPLC-ICP-MS was used for the chromatographic analysis of all lysis samples and the zinc signal was integrated using Origin Pro 9 (Origin Lab Corporation, Northampton, MA, USA) to compare the amount of bound zinc, indicating the strong possibility of zinc metalloproteins. Molecular weight markers from the protein standard mixture are indicated by arrows. A zinc calibration curve was analyzed by SEC-HPLC-ICP-MS to calculate the concentration of Zn in each zinc chromatogram, corresponding to 100 μL of cell lysate injected, reported as ng Zn/100 μL cell lysate. The average and standard deviation of replicates were calculated for all experiments and results can be found in Table 2. These data were then used to calculate the ratio of the zinc concentration by the protein concentration determined by the Qubit® assay. Summarily, the ng of Zn in a 100-μL volume was divided by the μg of protein in a 100-μL volume and these results are shown in Table 2, column 6. Based on this combination of data, there may be a greater probability of zinc proteins using the Vortex and Triton x-100 method (V, T) and the Sonication Bath, Vortex, and 1% Triton x-100 method (SB, V, T), but molecular weight distribution was deemed necessary to compare cell lysis methods.
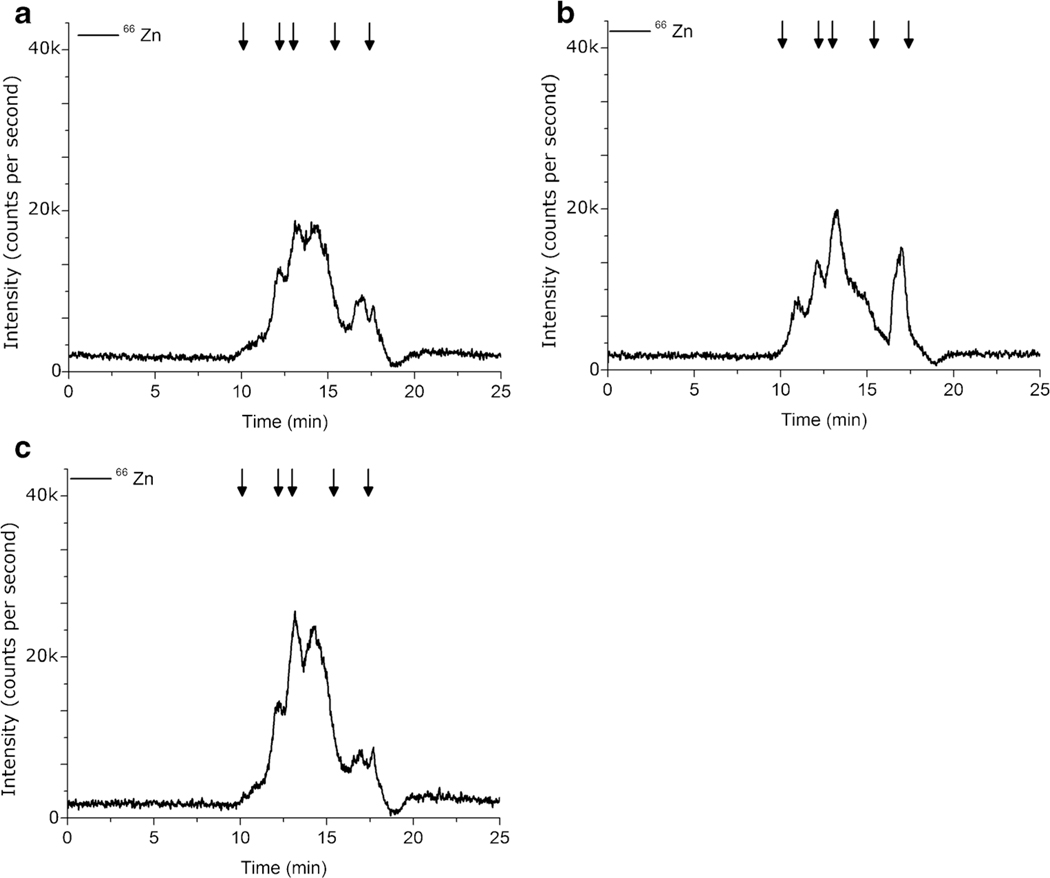
The first cell lysis technique investigated was a Glass Homogenizer (H) as it is common for tissue grinding, and in a way, mimics mortar and pestle action. However, with the small sample size of H. capsulatum yeast cells, the Glass Homogenizer was cumbersome and it was difficult to remove all of the cell lysate after grinding. Figure 1 shows the SEC-HPLC-ICP-MS signal intensity for 66Zn likely bound to proteins in the 1.35 to 670 kD molecular weight region extracted using the Glass Homogenizer (H) corresponding to 8.24 ng Zn/100 μL cell lysate as shown in Table 2.
Fig. 1.
Comparison of 66Zn SEC-HPLC-ICP-MS chromatograms using (a) Glass Homogenizer (H) (b) Bead Beater (BB) (c) Sonication Probe (SP). Lysis buffer contained 1% Triton x-100. Each sample contained 3.0 × 108 H. capsulatum yeast. n = 5. Arrows indicate molecular weight markers 670, 158, 44, 17, and 1.35 kDa
A Bead Beater (BB) was used with the lysis buffer containing 1% Triton x-100 to take advantage of the mechanical shear of small glass beads in conjunction with the lysing power of the detergent. This technique seemed promising as metal contamination could be controlled through acid washing the glass beads and the samples could be contained within individual vials for the duration of the lysis which limits sample transfer. This technique did result in 66Zn peaks at a range of molecular weights, as shown in Fig. 1b, but a surprisingly low amount of bound zinc, 7.66 ng Zn was extracted as shown in Table 2. While the concentrations of zinc by HPLC-SEC-ICP-MS in the cell lysates for the Glass Homogenizer (H) and the Bead Beater (BB) methods were similar as determined by a one-way ANOVA (F(1,8) = 0.3982, p = 0.5456), the chromatograms provide a comparison of molecular weight distribution. The Glass Homogenizer (H) zinc chromatogram, as shown in Fig. 1a, has very little high molecular weight bound zinc (8.85 to 11.13 min) as compared to the Bead Beater (BB) in Fig. 1b. This may indicate that the Bead Beater method (BB) preserves high molecular weight zinc proteins to a greater degree than the Glass Homogenizer method (H).
A Sonication Probe (SP) was used in combination with the 1% Triton x-100 lysis buffer. Initially, a longer pulse time was used (10 s pulse, 30 s rest, 10 s pulse) which resulted in a single large peak at 22 min, corresponding to approximately 1.35 kDa, as shown in Fig. 2. With a lack of mid and high low molecular weight proteins, it is highly unlikely that only low molecular weight proteins were extracted but, rather, the metals dissociated from the proteins in the extraction process. This was supported by examining the metal signal of 55Mn, 59Co, and 63Cu which have a similar pattern, as shown in Fig. 2. As a result, the pulse time was changed to 1 s followed by 10 s of rest and another 1-s pulse, which drastically reduced the amount of metalloprotein denaturation, as seen in Fig. 1. Short pulse times limited sample heating and the detergent did not lead to excessive foaming. A limited amount of high molecular weight bound zinc can be observed for the Sonication Probe (SP), which may indicate a certain amount of protein denaturation. The Sonication Probe method (SP) produced an integrated Zn concentration of 12.75 ng Zn/100 μL cell lysate.
Fig. 2.
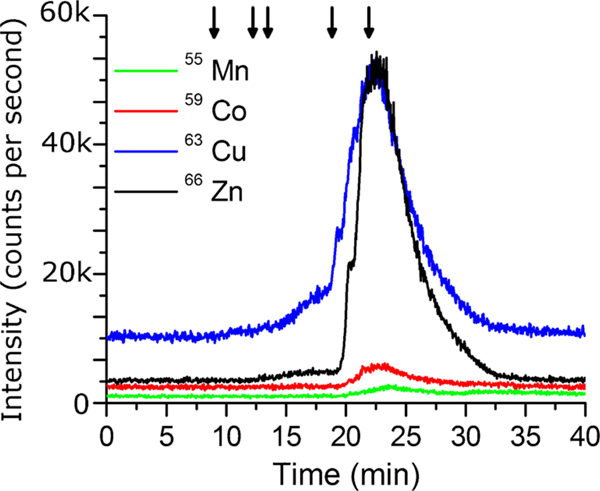
Comparison of Mn, Co, Cu, and Zn SEC-HPLC-ICP-MS signal using a Sonication Probe (SP) and lysis buffer with 1% Triton x-100.Data was collected using a TOSOH TSKgel G3000SW column (Tosoh Bioscience, King of Prussia, PA, USA) with a flow rate of 0.5 mL/min during method development. Samples were subjected to a 10 s pulse, 30 s rest, and a 10-s pulse. Arrows indicate molecular weight markers 670, 158, 44, 17, and 1.35 kDa
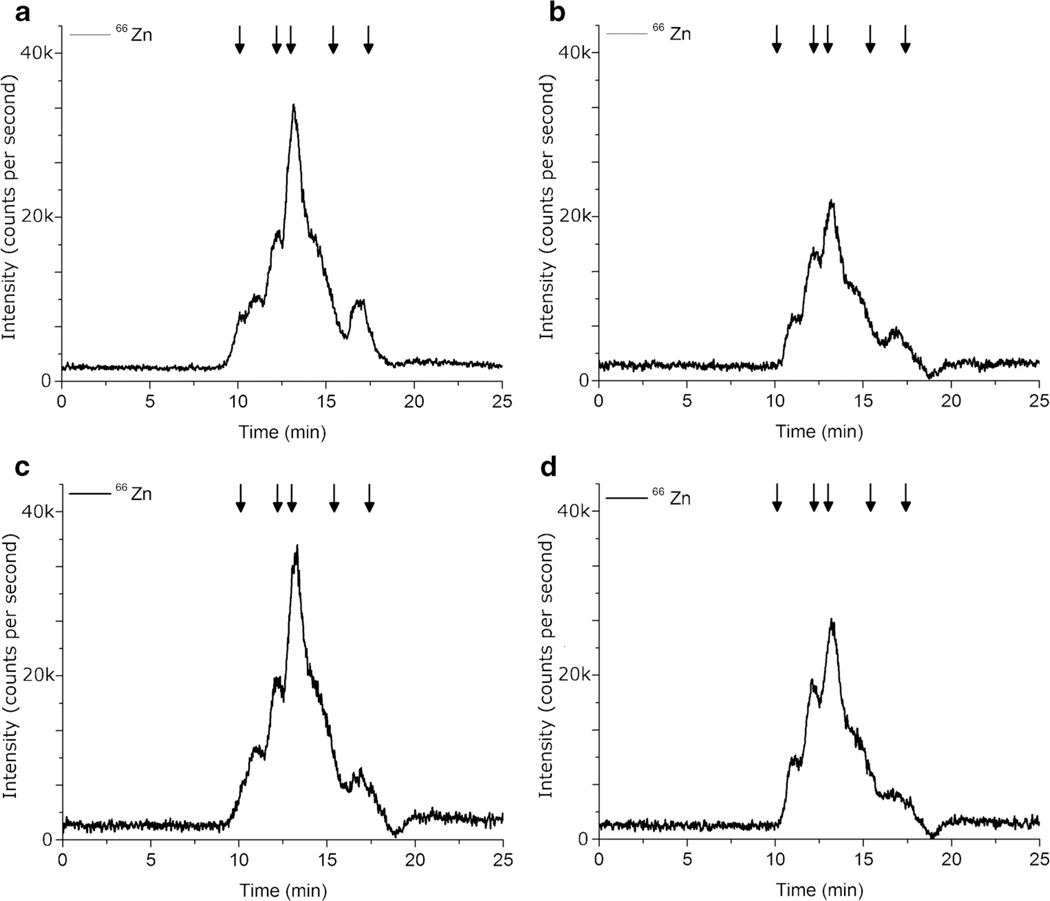
A hybrid Bead Beater method was developed to maximize simultaneous sample preparation and to take advantage of cell-bead interactions which may increase lysis efficiency of the cell wall in H. capsulatum. A vortex was used to mechanically agitate the sample with a lysis buffer containing 1% Triton x-100 (V,T) and glass beads with a 500-μm diameter were compared to 5 mm diameter glass beads (data not shown). A variety of containers were tested including 10 mL glass digestion vials, 15 mL plastic conical vials, 14 mL plastic round bottom capped test tubes, and 1.5 mL plastic conical vials (data not shown). The 500-μm diameter glass beads in 1.5 mL plastic conical vials, referred to as the Vortex with 1% Triton x-100 metho (V,T), produced the highest average integrated zinc concentration of 20.31 ng Zn/100 μL cell lysate and this was significantly higher than the Sonication Probe method (SP) as determined by one-way ANOVA (F(1,8) = 14.7872, p = 0.0049). The efficiency of this combination is likely due to the limited amount of available volume and small bead size, causing an increase in the collisions of the glass beads with the H. capsulatum yeast.
While the Sonication Probe (SP) technique may have been too harsh to keep the maximum amount of metalloproteins intact, a Sonication Bath is much gentler than a Sonication Probe. In an attempt to further improve the lysis efficiency, a Sonication Bath was employed prior to the vortex to take advantage of multiple mechanical techniques. Heating of the sample remained a concern so ice was added to the water in the Sonication Bath, while the samples remained under refrigeration and the ice was removed before the samples were sonicated. The resulting cold water bath provided an environment where heat generated through sonication was absorbed by the water bath, limiting the amount of thermal protein denaturation. Results from the Sonication Bath, Vortex, Triton x-100 method (SB, V, T) are show in Fig. 3 and Table 2 and demonstrate that despite employing a gentler sonication water bath to provide an additional mechanism, sonication by water bath does not provide an added advantage in the lysis of H. capsulatum. Although the total amount of Zn extracted determined by ICP-MS using the Vortex, Triton x-100 method (V, T) was significantly higher than the Sonication Bath, Vortex, Triton x-100 method, as determined by one-way ANOVA (F(1,8) = 12.6973, p = 0.0074), the results by HPLC-ICP-MS were not statistically different (F(1,8) = 4.9133, p = 0.0575).
Fig. 3.
Comparison of Zn SEC-HPLC-ICP-MS chromatograms using (a) Vortex, 1% Triton x-100 (V, T) (b) Sonication Bath, Vortex with 1% Triton x-100 (SB, V, T) (c) Vortex, and no detergent (V, NT) (d) Sonication Bath, Vortex, and no detergent (SB, V, NT). Each sample contained 3.0 × 108 H. capsulatum yeast. n = 5. Arrows indicate molecular weight markers 670, 158, 44, 17, and 1.35 kDa
To ensure that the use of Triton x-100 was more effective than no detergent, the two most promising techniques, Vortex and Sonication Bath then Vortex, were employed without the addition of detergent (V, NT and SB, V, NT) and results are shown in Fig. 3c, d. Concentrations of Zn by HPLC-SEC-ICP-MS are comparable, as determined by a one-way ANOVA (F(1,8) = 1.8385, p = 0.2122), at 7.55 ng Zn/100 μL cell lysate and 8.69 ng Zn/100 μL cell lysate, respectively, as shown in Table 2. While the zinc traces follow a similar pattern for each technique with and without detergent, using 1% Triton x-100 in the lysis buffer resulted in a significant increase in the amount of zinc extracted as compared to a simple salt buffer with no detergent using both techniques. The concentration of zinc by SEC-HPLC-ICP-MS for the Vortex, 1% Triton x-100 method (V, T) was significantly higher than the Vortex, no Triton method (V, NT) as determined by a one-way ANOVA (F(1,8) = 51.3928, p = 0.0001). Similarly, the concentration of zinc by SEC-HPLC-ICP-MS for the Sonication Bath, Vortex, 1% Triton x-100 method (SB, V, T) was significantly higher than the Sonication Bath, Vortex, no Triton method (SB, V, NT) as determined by a one-way ANOVA (F(1,8) = 33.8942, p = 0.0004).
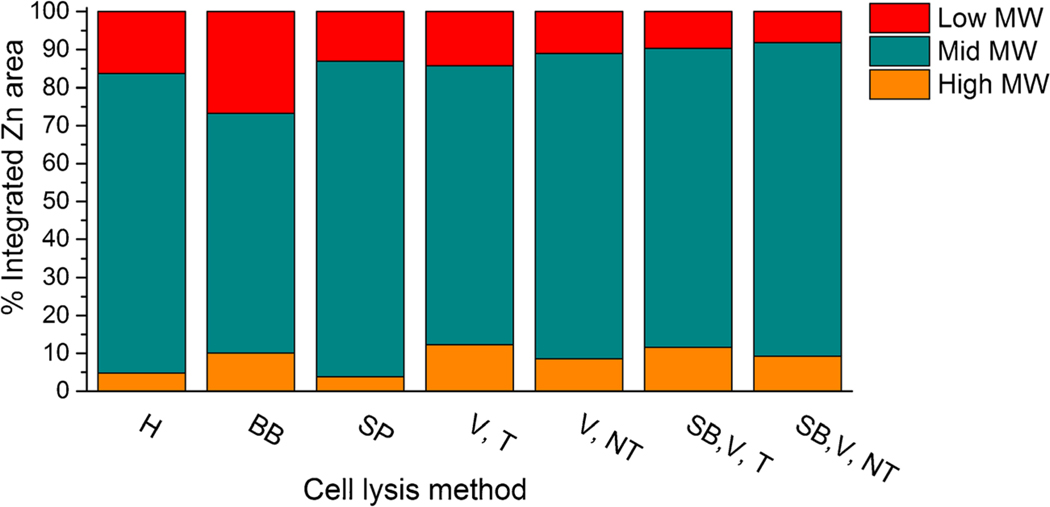
To further quantitatively probe the differences in molecular weight fractions produced by each of these techniques, specific regions in the zinc Size Exclusion Chromatograms were separately integrated. Three regions were examined, and results are reported as a percentage of the total zinc extracted which was set to 100% and graphically presented in Fig. 4. The Bead Beater method (BB) produced the largest percentage of low molecular weight zinc (15.8 to 18.8 min) which may indicate slight denaturation of zinc metalloproteins. This may be a result of sample heating when using the Bead Beater (BB) in this application.
Fig. 4.
Comparison of distribution of molecular weight (MW) bound zinc among various cell lysis methods. High MW: 8.85 to 11.13 min, Mid MW: 11.13 to 15.8 min, Low MW: 15.8 to 18.8 min. Glass Homogenizer (H), Bead Beater (BB), Sonication Probe (SP), Vortex with 1% Triton x-100, (V, T), Vortex with no Triton x-100 (V, NT), Sonication Bath, Vortex, and 1% Triton x-100 (SB, V, T), and Sonication Bath, Vortex, and no Triton x-100 (SB, V, NT)
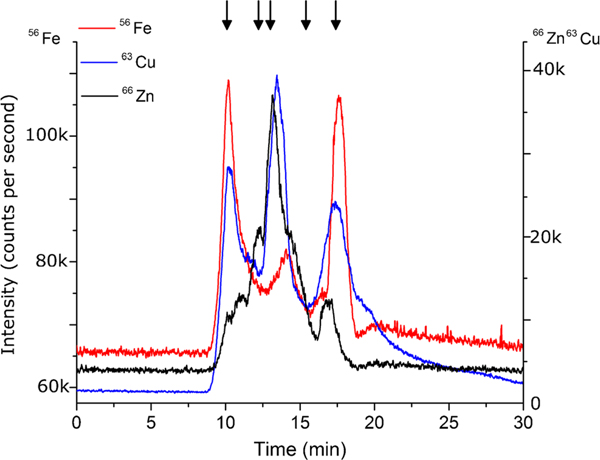
Comparing total protein concentration by Qubit® assay, zinc concentration by total metal analysis using ICP-MS, zinc concentration by SEC-HPLC-ICP-MS, molecular weight distribution, and other contributing factors such as potential contamination sources, vortexing H. capsulatum yeast cells with 500 μm glass beads in a 1% Triton x-100 lysis buffer (V, T) delivered the most potential zinc metalloproteins. This method produced the greatest concentration of zinc in the cell lysate by ICP-MS and HPLC-SEC-ICP-MS as well as one of the highest concentrations of protein by Qubit® Assay. Further, the ratio of zinc to total protein was the highest among the seven cell lysis methods studied, suggesting the greatest probability of zinc proteins. The molecular weight distribution of zinc for the Vortex, 1% Triton method (V, T) suggests limited protein denaturation and the sample preparation allows for less sample handling and contamination. Future studies will involve multi-dimensional separation and identification of metalloproteins in H. capsulatum and expansion to iron and copper proteins based on SEC-HPLC-ICP-MS results, as shown in Fig. 5. In addition, quantification of zinc metalloproteins in H. capsulatum grown under varying levels of zinc stress will provide insight into the response of the fungus during MΦ zinc sequestration.
Fig. 5.
Comparison of Fe, Cu, and Zn SEC-HPLC-ICP-MS chromatograms using the Vortex with 1% Triton x-100 method (V, T). Each sample contained 3.0 × 108 H. capsulatum yeast. n = 5. Arrows indicate molecular weight markers 670, 158, 44, 17, and 1.35 kDa
Conclusion
In order to address common challenges within metalloproteomics analysis, designing the cell lysis method is an important step that influences the entire analysis. For organisms with additional challenges such as the strong cell walls of fungi, cell lysis methods must be assessed to optimize the amount of metalloprotein extracted. Comparing the total protein concentration by Qubit® protein assay provides a simple comparison between cell lysis methods, but lacks information about protein-bound metals. Comparing the concentration of metal in cell lysates by total metal analysis using ICP-MS and the metal chromatogram by SEC-HPLC-ICP-MS allows for additional comparison between different cell lysis methods. Closer examination of the SEC metal chromatograms allows for comparison of molecular weight distribution which provides information regarding protein preservation. Additional factors such as sample heating and potential contamination sources also need to be considered when developing cell lysis methods. The combination of these factors can be used to assess cell lysis methods suitable for a particular sample. We found that a cooled non-ionic detergent based lysis buffer containing 1% Triton x-100 combined with 500 μm glass beads in a 1.5-mL conical plastic vial that is vortexed (V, T) provides the greatest amount of potential zinc proteins with a wide distribution of molecular weights in H. capsulatum. As the field of metalloproteomics continues to grow and more metalloproteomes are examined, optimizing cell lysis methods for different types of samples is as important as developing separation, identification, and quantification methods. We hope that this work provides one example of the process which can be adapted for future metalloproteomic studies.
Acknowledgments
The authors would like to thank the late Dr. Joseph A. Caruso for his guidance and support during the development of this project. The authors would also like to thank Agilent for instrument support. This work is supported by the National Institute of Allergy and Infectious Diseases RO1-AI-106269.
Footnotes
Compliance with ethical standards
Conflict of interest The authors declare that they have no conflict of interest.
References
- 1.Mounicou S, Szpunar J, Lobinski R. Metallomics: the concept and methodology. Chem Soc Rev. 2009;38:1119–38. [DOI] [PubMed] [Google Scholar]
- 2.Shi W, Chance MR. Metallomics and metalloproteomics. Cell Mol Life Sci. 2008;65:3040–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun H, Chai Z-F. Metallomics: an integrated science for metals in biology and medicine. Annu Rep Prog Chem Sect A. 2010;106:20–38. [Google Scholar]
- 4.Ibers JA, Holm RH. Modeling coordination sites in metallobiomolecules. Science. 1980;209:223–35. [DOI] [PubMed] [Google Scholar]
- 5.Bertini I, Rosato A. From genes to metalloproteins: a bioinformatic approach. Eur J Inorg Chem. 2007:2546–55. [Google Scholar]
- 6.Sevcenco A-M, Krijger GC, Pinkse MWH, Verhaert PDEM, Hagen WR, Hagedoorn P-L. Development of a generic approach to native metalloproteomics: application to the quantitative identification of soluble copper proteins in Escherichia coli. J Biol Inorg Chem. 2009;14:631–40. [DOI] [PubMed] [Google Scholar]
- 7.Sun X, Xiao C-L, Ge R, Yin X, Li H, Li N, et al. Putative copper-and zinc-binding motifs in Streptococcus pneumoniae identified by immobilized metal affinity chromatography and mass spectrometry. Proteomics. 2011;11:3288–98. [DOI] [PubMed] [Google Scholar]
- 8.Brown DA, Cook RA. Role of metal cofactors in enzyme regulation. Differences in the regulatory properties of the Escherichia coli nicotinamide adenine dinucleotide phosphate specific malic enzyme, depending on whether magnesium ion or manganese ion serves as divalent cation. Biochemistry. 1981;20(9):2503–12. doi: 10.1021/bi00512a022. [DOI] [PubMed] [Google Scholar]
- 9.Braga CP, Bittarello AC, Padilha CCF, Leite AL, Moraes PM, Buzalaf MAR, et al. Mercury fractionation in dourada (Brachyplatystoma rousseauxii) of the Madeira River in Brazil using metalloproteomic strategies. Talanta. 2015;132:239–44. [DOI] [PubMed] [Google Scholar]
- 10.Bucher G, Frelon S, Simon O, Lobinski R, Mounicou S. Development of non-denaturing off-gel isoelectric focusing for the separation of uranium–protein complexes in fish. Anal Bioanal Chem. 2014;406:3517–20. [DOI] [PubMed] [Google Scholar]
- 11.Lavradas RT, Rocha RCC, Saint TD, Pierre JMG, Hauser-Davis RA. Investigation of thermostable metalloproteins in Perna Perna musselsfrom differentially contaminated areas in Southeastern Brazil bybioanalytical techniques. J Trace Elem Med Biol. 2016;34:70–8. [DOI] [PubMed] [Google Scholar]
- 12.Gomes CPC, Freire MS, Pires BRB, Vasconcelos EAR, Rocha TL, MdFt G-d-S, et al. Comparative proteomical and metalloproteomical analyses of human plasma from patients with laryngeal cancer. Cancer Immunol Immunother. 2010;59:173–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heiss K, Junkes C, Guerreiro N, Swamy M, Camacho-Carvajall MM, Schamel WWA, et al. Subproteomic analysis of metal-interacting proteins in human B cells. Proteomics. 2005;5:3614–22. [DOI] [PubMed] [Google Scholar]
- 14.Ossipov K, Foteeva LS, Seregina IF, Perevalov SA, Timerbaev AR, Bolshov MA. Metallomics for drug development: serum protein binding and analysis of an anticancer tris(8-quinolinolato)gallium(III) drug using inductively coupled plasma mass spectrometry. Anal Chim Acta. 2013;785:22–6. [DOI] [PubMed] [Google Scholar]
- 15.Matczuk M, Kupiec M, Legat J, Pawlak K, Timerbaev AR, Jarosz M. A shotgun metalloproteomic approach enables identification of proteins involved in the speciation of a ruthenium anticancer drug in the cytosol of cancer cells. Analyst. 2015;140:3492–9. [DOI] [PubMed] [Google Scholar]
- 16.Cvetkovic A, Menon AL, Thorgersen MP, Scott JW, Poole FL II, Jenney FE Jr, et al. Microbial metalloproteomes are largely uncharacterized. Nature. 2010;266:779–82. [DOI] [PubMed] [Google Scholar]
- 17.Wheat LI, Freifeld AG, Kleiman MB, Baddley JW, McKinsey DS, Loyd JE, et al. Clinical practice guidelines for the management of patients with histoplasmosis: 2007 update by the infectious diseases society of america. Clin Infect Dis. 2007;45:807–25. [DOI] [PubMed] [Google Scholar]
- 18.Vignesh KS, Figueroa JAL, Porollo A, Caruso JA, Deepe GS. Zinc sequestration: arming phagocyte defense against fungal attack. PLoS Pathog. 2013;9:E1003815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vignesh K, Figueroa J, Porollo A, Caruso J, Deepe G. Granulocyte macrophage-colony stimulating factor induced Zn sequestration enhances macrophage superoxide and limits intracellular pathogen survival. Immunity. 2013;39:697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barnett JP, Scanlan DJ, Blindauer CA. Protein fractionation and detection for metalloproteomics: challenges and approaches. Anal Bioanal Chem. 2012;402:3311–22. [DOI] [PubMed] [Google Scholar]
- 21.Tran MQT, Nygren Y, Lundin C, Naredi P, Björn E. Evaluation of cell lysis methods for platinum metallomic studies of human malignant cells. Anal Biochem. 2010;396:76–82. [DOI] [PubMed] [Google Scholar]
- 22.Klimek-Ochab M, Brzezińska-Rodak M, Żymańczyk-Duda E, Lejczak B, Kafarski P. Comparative study of fungal cell disruption—scope and limitations of the methods. Folia Microbiol. 2011;56:469–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Magalhaes CS, Aurelio M, Arruda Z. Sample preparation for metalloprotein analysis: a case study using horse chestnuts. Talanta. 2007;71:1958–63. [DOI] [PubMed] [Google Scholar]
- 24.Hagège A, Huynh TNS, Hébrant M. Separative techniques for metalloproteomics require balance between separation and perturbation. Trends Anal Chem. 2015;64:64–74. [Google Scholar]
- 25.Michalke B, Nischwitz V. Review on metal speciation analysis in cerebrospinalfluid—current methods and results: a review. Anal Chim Acta. 2010;682:23–6. [DOI] [PubMed] [Google Scholar]
- 26.Allendoerfer R, Deepe GS Jr. Intrapulmonary response to Histoplasma capsulatum in gamma interferon knockout mice. Infect Immun. 1997;65:2564–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Lilley KS, Oliver SG. A protocol for the subcellular fractionation of Saccharomyces Cerevisiae using nitrogen cavitation and density gradient centrifugation. Yeast. 2014;31(4):127–35. doi: 10.1002/yea.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lundblad V, Struhl K. Yeast. Curr Protoc Mol Biol. Wiley, Inc.; 2001. [Google Scholar]
- 29.Burden DW. Guide to the homogenization of biological samples. Random Prim. 2008;7:1–14. [Google Scholar]
- 30.Gao Q, Madian AG, Liu X, Adamec J, Regnier FE. Coupling protein complex analysis to peptide based proteomics. J Chromatogr A. 2010;1217(49):7661–8. doi: 10.1016/j.chroma.2010.09.071. [DOI] [PubMed] [Google Scholar]
- 31.Sasidharan K, Amariei C, Tomita M, Murray DB. Rapid DNA, RNA and protein extraction protocols optimized for slow continuously growing yeast cultures: nucleotide and protein time-series extraction protocols. Yeast. 2012;29(8):311–22. doi: 10.1002/yea.2911. [DOI] [PubMed] [Google Scholar]
- 32.Forsburg SL, Rhind N. Basic methods for fission yeast. Yeast. 2006;23(3):173–83. doi: 10.1002/yea.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcerá A, Casas C, Herrero E. Expression of Candida albicans glutathione transferases is induced inside phagocytes and upon diverse environmental stresses: glutathione transferases in Candida albicans. FEMS Yeast Res. 2010;10(4):422–31. doi: 10.1111/j.1567-1364.2010.00613.x. [DOI] [PubMed] [Google Scholar]
- 34.Blechl AE, Thrasher KS, Vensel WH, Greene FC. Purification and characterization of wheat α-gliadin synthesized in the yeast, Saccharomyces cerevisiae. Gene. 1992;116(2):119–27. doi: 10.1016/0378-1119(92)90507-L. [DOI] [PubMed] [Google Scholar]
- 35.Capelo JL, Ximénez-Embún P, Madrid-Albarrán Y, Cámara C. Enzymatic probe sonication: enhancement of protease-catalyzed hydrolysis of selenium bound to proteins in yeast. Anal Chem. 2004;76(1):233–7. doi: 10.1021/ac034871d. [DOI] [PubMed] [Google Scholar]
- 36.Chisti Y, Moo-Young M. Disruption of microbial cells for intracellular products. Enzym Microb Technol. 1986;8:194–204. doi: 10.1016/0141-0229(86)90087-6. [DOI] [Google Scholar]
- 37.Feliu JX, Cubarsi R, Villaverde A. Optimized release of recombinant proteins by ultrasonication of E. coli cells. Biotechnol Bioeng. 1998;58(5):536–40. doi:. [DOI] [PubMed] [Google Scholar]
- 38.Neppiras EA, Hughes DE. Some experiments on the disintegration of yeast by high intensity ultrasound. Biotechnol Bioeng. 1964;6(3):247–70. doi: 10.1002/bit.260060302. [DOI] [Google Scholar]
- 39.Horvath A, Horvath A, Riezman H, Riezman H. Rapid protein extraction from Saccharomyces cerevisiae. Yeast. 1994;10(10): 1305–10. doi: 10.1002/yea.320101007. [DOI] [PubMed] [Google Scholar]
- 40.Papanayotou I, Sun B, Roth AF, Davis NG. Protein aggregation induced during glass bead lysis of yeast. Yeast. 2010;27(10):801–16. doi: 10.1002/yea.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu C, Apodaca J, Davis LE, Rao H. Proteasome inhibition in wild-type yeast Saccharomyces cerevisiae cells. BioTechniques. 2007;42(2):158–62. doi: 10.2144/000112389. [DOI] [PubMed] [Google Scholar]