Abstract
Coronaviruses are the well-known cause of severe respiratory, enteric and systemic infections in a wide range of hosts including man, mammals, fish, and avian. The scientific interest on coronaviruses increased after the emergence of Severe Acute Respiratory Syndrome coronavirus (SARS-CoV) outbreaks in 2002-2003 followed by Middle East Respiratory Syndrome CoV (MERS-CoV). This decade’s first CoV, named 2019-nCoV, emerged from Wuhan, China, and declared as ‘Public Health Emergency of International Concern’ on January 30th, 2020 by the World Health Organization (WHO). As on February 4, 2020, 425 deaths reported in China only and one death outside China (Philippines). In a short span of time, the virus spread has been noted in 24 countries. The zoonotic transmission (animal-to-human) is suspected as the route of disease origin. The genetic analyses predict bats as the most probable source of 2019-nCoV though further investigations needed to confirm the origin of the novel virus. The ongoing nCoV outbreak highlights the hidden wild animal reservoir of the deadly viruses and possible threat of spillover zoonoses as well. The successful virus isolation attempts have made doors open for developing better diagnostics and effective vaccines helping in combating the spread of the virus to newer areas.
Keywords: Coronavirus, 2019-nCoV, Severe Acute Respiratory Syndrome CoV, Middle East Respiratory Syndrome CoV, Public Health Emergency, genetic analyses, zoonoses, reservoir host, therapeutics, vaccines
1. Introduction
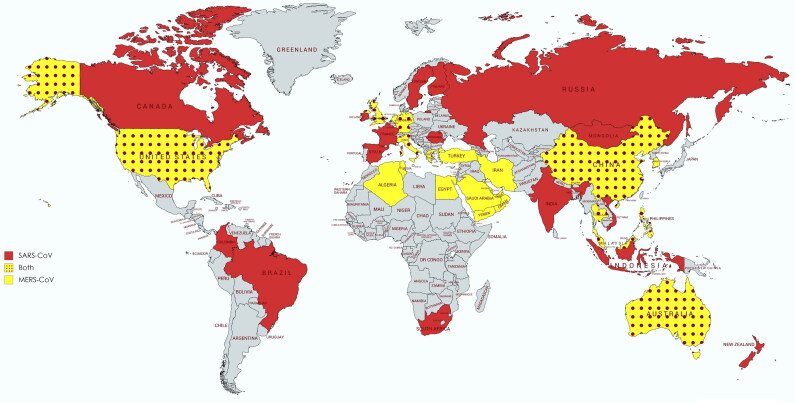
Coronaviruses (CoVs) are well-known causes of severe infections, respiratory, enteric and systemic, in humans and numerous animal hosts. The CoV infections have been reported in cattle, swine, horses, camels, rodents, cats, dogs, bats, palm civets, ferrets, mink, rabbits, snake, and several other wild animals and avian species (Fehr and Perlman 2015; Kahn and McIntosh 2005). The coronaviruses of relevant veterinary species are shown in Table 1 with organ affected and clinical signs. Though human CoVs were identified for the first time in the year 1960 from respiratory infections in adults as well as children, the major scientific interest in CoVs research grew only after the emergence of Severe Acute Respiratory Syndrome CoV (SARS-CoV) in the year 2002-2003 (Drosten et al. 2003; Ksiazek et al. 2003; Peiris et al. 2003). In this SARS-CoV epidemic, around 8000 confirmed human cases with 774 deaths (around 9.5% mortality rate) occurred that was a result of its global spread (Kahn and McIntosh 2005). Initially, the virus was detected in the caged Himalayan palm civets and these were thought to be the natural host of this virus (Guan et al. 2003). Following SARS-CoV incidence in 2003, a similar CoV named HKU3-1 to HKU3-3 were identified in the horseshoe bats (non-caged) in 2005 from Hong Kong (Lau et al. 2005). Since then, bats are considered to be the natural host and potential reservoir species that could be held responsible for any future CoVs epidemics and/or pandemics (Cui et al. 2019, Li et al. 2005). After the 2003 and 2005 SARS-CoV epidemics, an analogous virus emerged in the Middle East region of the world leading to severe respiratory illness and was named the Middle East Respiratory Syndrome CoV (MERS-CoV) (Zaki et al. 2012). The mortality was higher than previous SARS-CoV pandemic claiming around 919 lives out of the total 2521 human cases (around 35% mortality) (World Health Organization 2015). Notably, dromedary camels were connected with the transmission of MERS-CoV (Alagaili et al. 2014). Further, its origin was also traced from bats (Ithete et al. 2013). All these highly pathogenic human CoVs, SARS and MERS, show emergence over wider areas of the world posing high risk of human-to-human transmission and fatal consequences thereto (Figure 1).
Table 1.
Coronaviruses of relevant veterinary species.
| Host | Virus | Genus | Organ affected | Clinical sign |
|---|---|---|---|---|
| Bovine | Bovine coronavirus | Betacoronavirus | GI Tract Respiratory tract |
Calf diarrhea Winter dysentery (bloody diarrhea) in adult cattle Bovine respiratory disease complex (shipping fever) |
| Porcine | Transmissible gastroenteritis (TGE) virus | Alphacoronavirus | GI Tract | Profuse watery yellow diarrhea, Vomition, dehydration |
| Porcine respiratory coronavirus | Alphacoronavirus | Respiratory tract | Mild respiratory disease or subclinical | |
| Porcine epidemic diarrhea virus (PEDv) | Alphacoronavirus | GI Tract | Watery diarrhea, vomition, dehydration | |
| Porcine hemagglutinating encephalomyelitis virus | Betacoronavirus | GI Tract CNS |
Vomition, Diarrhea Wasting disease Encephalomyelitis |
|
| Porcine deltacoronavirus | Deltacoronavirus | GI Tract | Gastroenteritis | |
| Feline | Feline enteric coronavirus | Alphacoronavirus | GI Tract | Mild gastroenteritis and diarrhea |
| Feline infectious peritonitis virus | Alphacoronavirus | Respiratory tract Abdominal cavity CNS |
Pneumonia Peritonitis Neurological disorder |
|
| Canine | Canine coronavirus | Alphacoronavirus | GI Tract | Mild to severe gastroenteritis |
| Canine respiratory coronavirus | Betacoronavirus | Respiratory tract | Kennel cough Pneumonia |
|
| Equine | Equine coronavirus | Betacoronavirus | GI Tract | Gastroenteritis |
| Camel | Middle East respiratory syndrome (MERS) coronavirus | Betacoronavirus | Respiratory tract | Mild respiratory signs |
| Chicken | Avian infectious bronchitis virus | Gammacoronavirus | Trachea Kidney Reproductive tract |
Tracheobronchitis, Rales Nephritis Decreased egg production |
Figure 1.
World-map depicting countries with SARS-CoV and MERS-CoV outbreaks. Red and yellow colors represent the global distribution of only MERS-CoV and SARS-CoV, respectively. The yellow-red dotted pattern shows the incidence of both viruses in the countries.
This decade’s first CoV emergency was from Hubei province of China, and as on February 4, 2020, 425 deaths have been reported in China only (World Health Organization 2020b). Further, the spread of this novel coronavirus, named 2019-nCoV, has been noted in 24 countries till date. Considering the global threat of the 2019-nCoV, the World Health Organization (WHO) declared it as a ‘Public Health Emergency of International Concern’ on January 30th, 2020.
This rapid communication provides an overview of the recently emerging coronavirus (2019-nCoV) with regards to its current scenario, comparative analysis with respects to previously reported CoVs, evolutionary perspective based on genome analysis while covering the recent advances on vaccines and therapeutics in brief.
2. Coronaviruses
Coronaviruses (CoVs) constitute a large family of viruses found in nature. CoVs belongs to the family Coronaviridae and the order Nidovirales possessing a single-stranded, positive-sense RNA genome ranging from 26 to 32 kb in length (the largest genome of known RNA viruses) with G + C contents varying from 32 to 43%. Based on the genomic structure and phylogenetic analysis the subfamily Orthocoronavirinae consists of four genera namely Alphacoronavirus, Betacoronavirus, Gammacoronavirus and Deltacoronavirus. Among these Alphacoronavirus and Betacoronavirus infects only mammals and are responsible for respiratory infection in humans and enteritis in animals. Two major zoonotic pathogenic coronaviruses, SARS-CoV and MERS-CoV belong to the genus Betacoronavirus. The genus Betacoronavirus further distributed in five subgenus among which Sarbecovirus contains SARS-CoV and the novel coronavirus (2019-nCoV) (de Groot et al. 2012). The other subgenera under Betacoronavirus are Embecovirus, Hibecovirus, Merbecovirus, and Nobecovirus.
The nCoV contains at least six open reading frames (ORFs) and many other accessory genes like other CoVs. The 5′ terminal two-thirds of the genome contains two open reading frames (ORFs), ORF1 and ORF2 which encodes two polyproteins, pp1a and pp1ab, which is further cleaved into 11 and 16 proteins, respectively. These 16 mature proteins are responsible for several important functions in genome maintenance and virus replication. The structural proteins namely spike (S), an envelope protein (E), membrane protein (M) and nucleocapsid (N) are located at the one-third 3′ terminal of the genome. In addition to these genes, there are several accessory proteins which help in virus replication. The S gene is one of the most important genes for receptor binding and host specificity. Some coronaviruses have hemagglutinin-esterase (HE) protein in their virion.
3. The emergence of novel-Coronavirus (2019-nCoV)
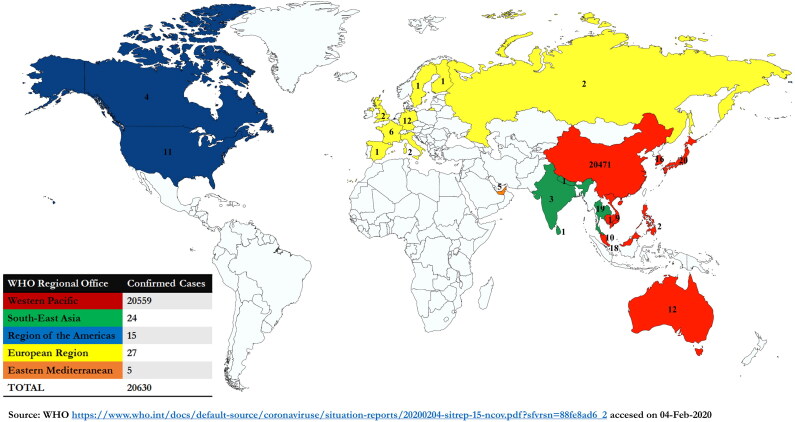
During the first week of December 2019, a few cases of pneumonia appeared in the city of Wuhan, Hubei province of China. The patients exhibited a history of visiting the local nearby Huanan seafood market which deals in the sale of different live animals, where zoonotic (animal-to-human) transmission suspected as the main route of disease origin (Hui et al. 2020). Firstly, the affected patients presented with pneumonia-like symptoms, followed by a severe acute respiratory infection. Some cases showed rapid development of acute respiratory distress syndrome (ARDS) followed by serious complications in the respiratory tract. On Jan 7th, 2020, it was confirmed by the Chinese Center for Disease Control and Prevention (CDC) that a new coronavirus has emerged and was named 2019-nCoV. As on February 4th 2020, China has confirmed 20471 cases with 425 deaths and 2788 severe cases of 2019-nCoV. In addition to China, 24 different countries from Europe, Northern America, Southeast Asia, Eastern Mediterranean, and Western Pacific Asia have reported the confirmed cases of this disease making the total tally of confirmed cases to 20630 worldwide (Figure 2). Although the mortality rate due to 2019-nCoV is comparatively lesser than the earlier outbreaks of SARS and MERS-CoVs, as well as this virus presents relatively mild manifestations, the total number of cases are increasing speedily and are crossing the old census. There is a high risk of human-to-human transmission which has also been reported in family clusters and medical workers. The infected patients with nCoV exhibit high fever and dyspnea with chest radiographs showing acute invasive lesions in both lungs.
Figure 2.
Countries, territories or regions with reported confirmed cases of 2019-nCoV, February 4th, 2020. Different colors indicate different geographical regions with the number of confirmed cases. In the table, region-wise total number of confirmed cases are depicted.
4. Phylogenetic analysis and sequence identity
Currently, knowledge about the origin of the virus, its particular receptor, clinical spectrum and host species preference is unknown. World over scientists are engaged in unraveling the genomic nature of this virus. As of now, around 15 complete 2019-nCoV genome sequences (10 China and 5 USA) have been reported. To note, a report based on the codon usage analysis speculated snakes to be its natural host, whereas several subsequent reports after that publication rejected this hypothesis. They found the nearest members to be Bat_SARS-like CoVs (Bat-SL-CoVs, MG772933 & MG772934) in phylogenetic analysis within the subgenus Sarbecovirus.
4.1. Analysis targeting the complete genome of 2019-nCoVs
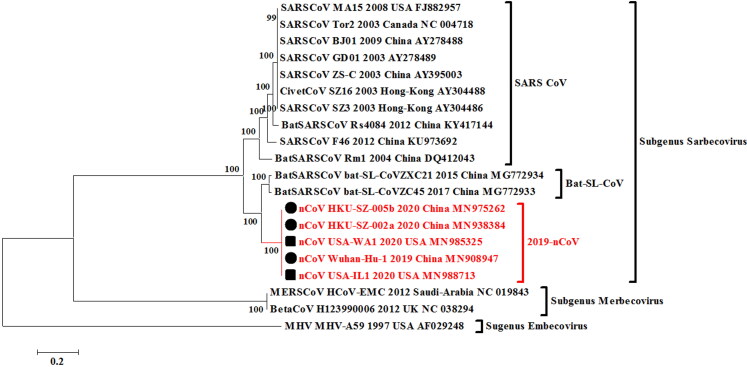
In this study, we also attempted to reveal the evolutionary perspective of the recently emerging 2019-nCoV based on the complete genome analysis. Phylogenetic analysis was done using the MEGA 7.0 version applying the Maximum likelihood method (ML) based General Time Reversible substitution model with the available whole genome sequences of 2019-nCoV available in the NCBI GenBank database till January 28th, 2020. Pairwise identity of the current 2019-nCoV outbreak sequences was calculated using the MegAlign software of DNASTAR. In the whole genome phylogenetic analysis, the 2019-nCoV strains from China and the USA clustered in a monophyletic clade (Figure 3). The nearest neighbors of the 2019-nCoV isolates from China and USA were two Bat_SARS-like coronaviruses (Bat-SL-CoVZC45, Accession no. MG772933 and Bat-SL-CoVZXC21, Accession no. MG772934). These two Bat_SARS-like CoVs shared a 100% bootstrap support with 2019-nCoV strains of the current outbreak. Using the MegAlign and MEGA 7.0 software based Clustal W alignments, the nucleotide sequence identity of 2019-nCoV strains revealed the highest similarity of greater than 88.2% with two Bat_SARS_like CoVs. These findings were in accordance with report of Zhu and colleagues where a nearby sequence identity of 86.9% with previously published Bat_SARS-like CoV was reported (Zhu et al. 2020). Contrarily, the genome of 2019-nCoV has also been reported to be 96% identical to the bat coronavirus based on Simplot analysis where it has been found closer to bat CoV isolate RaTG13 previously detected in Rhinolophus affinis (intermediate horseshoe bat) from Yunnan Province, indicating its origin from the bats (Zhou et al. 2020). Based on the available information it is rather early to predict the origin of this novel coronavirus without a comprehensive analysis of emerging nCoV strains from different parts of the world. To note, the sequence identity based on the complete genome sequences between current outbreaks 2019-nCoV isolates from China and the USA ranges 99.8 to 100% on the nucleotide level indicating their common origin of evolution.
Figure 3.
Phylogenetic analysis of 2019-nCoV isolates using complete genomes. The 2019-nCoV isolates analyzed with related CoVs from past human outbreaks and of animal origin. The solid-black circles are for nCoV isolates from China and solid-black squares are for the isolates from the USA.
4.2. Analysis targeting the spike (S)-gene of CoVs
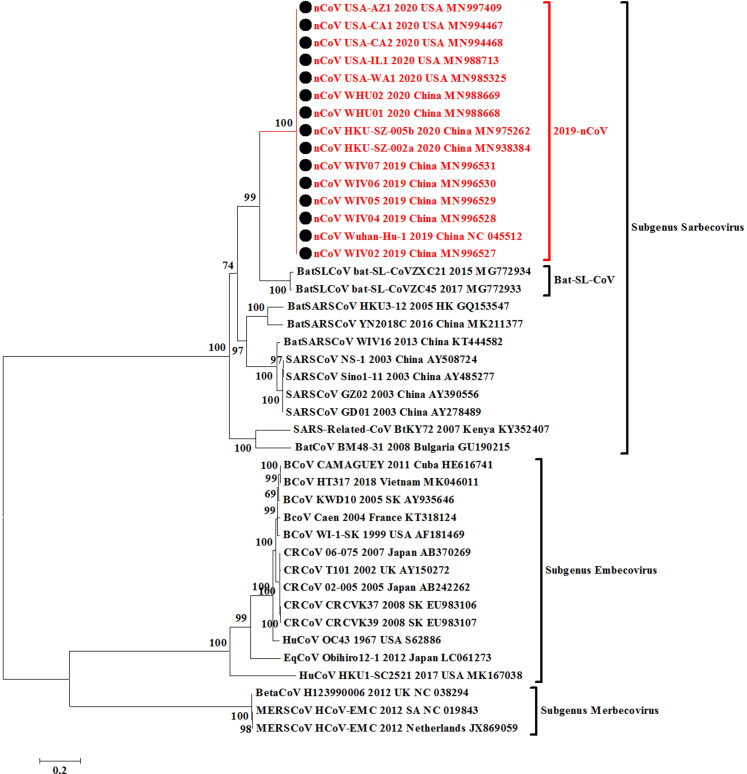
We furthermore extended the analysis targeting the Spike (S) glycoprotein gene of the CoVs from human SARS, animal-origin CoVs including MERSV (camel), bovine coronavirus, canine coronavirus, bat_coronaviruses and the current outbreak nCoVs from different regions. The sequences of nCoV available in the NCBI GenBank database till January 28th, 2020 were retrieved. Phylogenetic analysis was done using the MEGA 7.0 version applying the Maximum likelihood method (ML) based General Time Reversible substitution model with gamma distribution. Pairwise identity of the current 2019-nCoV outbreak sequences was calculated using the MegAlign software of DNASTAR. In the S-gene based phylogeny, 10 Chinese and 5 USA nCoV isolates (Figure 4) revealed that all the isolates are nearly identical across the S-gene based phylogeny constituting a monophyletic clade (Figure 4). The two Bat_SARS-like CoVs (Bat-SL-CoVZC45, MG772933 and bat-SL-CoVZXC21, MG772934) shared 100% bootstrap support with 2019-nCoV isolates of the current outbreaks. Based on the MegAlign and MEGA 7.0 software based Clustal W alignments, the sequence identity of 2019-nCoV strains revealed Bat SARS-like CoVs (Bat-SL-CoVZC45, MG772933 and bat-SL-CoVZXC21, MG772934) as the nearest neighbors with 77.6 to 78.2% sequence identity on nucleotide basis. In the phylogenetic tree, the current outbreak nCoV isolates were fairly distinct than the previously reported SARS-CoV or BatSARS-CoV strains but were clustering inside a common major clade which includes strains from subgenus Sarbecovirus. The per cent identity with other SARS-CoV or BatSARS-CoV strains was 70.8 to 74.7% (Supplementary data 1). The per cent similarity on nucleotide basis between nCoV isolates and canine respiratory coronaviruses (CRCoV) and bovine coronaviruses (BCoV) of subgenus Embecovirus ranged between 40.8 to 41.5%. Furthermore, the per cent identity of nCoV isolates was found lower (40.2%) with the mild respiratory human coronavirus isolate HCoV-OC43 of same subgenus Embecovirus containing animal-origin coronaviruses.
Figure 4.
Spike (S) glycoprotein gene-based phylogenetic analysis of 2019-nCoV isolates (10 Chinese and 5 USA isolates). The 2019-nCoV isolates analyzed with related CoVs from past human outbreaks and of animal-origin including MERSV (camel), bovine coronavirus, canine coronavirus, bat_coronaviruses. The solid-black circles are for nCoV isolates from China and the USA.
Additionally, for S-gene, per cent similarity range pattern was evaluated in amino acid based index where 2019-nCoV isolates from China and USA were 100% identical. The range varied between 81.2 to 81.8% and 77.0 to 78.1% for 2019-nCoV isolates with Bat_SARS-like CoVs and other SARS like CoVs, respectively. The per cent identity with human coronavirus isolate HCoV-OC43 was also found lower (28.0%) (Supplementary data 2).
Overall, the nucleotide and amino acid based per cent identities indicate toward highly diverged nature of novel coronaviruses. To note, in the phylogenetic tree the 2019-nCoV isolates classified in subgenus Sarbecovirus and the sequences from CRCoV, BCoV and HCoV-OC43 isolates clustered in subgenus Embecovirus.
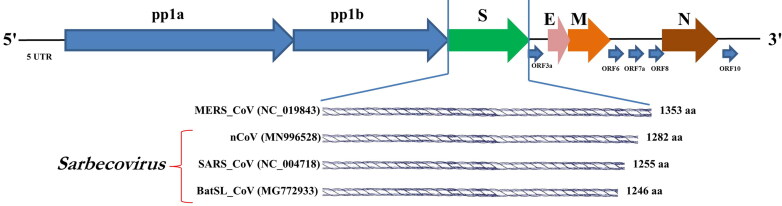
In the genomic analysis, the S gene of 2019-nCoVs was found to exhibit lower sequence identity with other Betacoronaviruses. Difference in spike protein encoded by 2019-nCoV compared with the Bat SARS-like CoVs, SARS-CoV, and MERS-CoV is depicted in Figure 5. The length of ‘S’ protein of nCoV is longer (1282 amino acids) than other two viruses (SARS-1255 amino acid and BatSL-1246 amino acids) under the same Sarbecovirus subgenus. The ‘S’ protein of nCoV has been detected with three short insertions at the N-terminal region, along with four changes in the receptor binding motif inside the receptor binding domain in comparison with SARS-CoV (Zhou et al. 2020). Several previous studies have shown the usage of different receptors for CoVs like human ACE2 for SARS-CoV and CD26 for MERS-CoV (Lu et al. 2020). Identical motifs and residues to that of SARS-CoVs have been identified in the nCoV, yet they show divergence based on phylogeny (Letko and Munster 2020). A recent finding proves that the binding capacity of nCoV ‘S’ protein with human ACE2 is as efficient as SARS-CoV, which further promotes the human-to-human transmission (Letko and Munster 2020).
Figure 5.
Genome organization of novel coronavirus. Comparative spike (S) protein sequence length of different Betacoronaviruses is depicted.
5. Advances and prospects in developing vaccines and therapeutics
Several attempts have been made to develop vaccines against human coronavirus infection in the past decades. But the degree of cross-protection provided by such vaccines is greatly limited due to the extensive diversity in antigenic variants even within the strains of a phylogenetic sub-cluster (Graham et al. 2013). As for MERS and SARS coronaviruses, there is no licensed specific antiviral treatment or vaccine available till now. However, few of the advances made in developing vaccines and therapeutics for SARS-CoV and MERS-CoV could be exploited for the countering 2019-nCoV. But since the efforts to design and develop any vaccine or antiviral agent to tackle the presently emerging coronavirus pathogen would take some time, therefore till then we need to rely extensively on enforcing highly effective prevention and control measures to minimize the risk of 2019-nCoV transmission and spread to the best feasible extent (Cheng et al. 2020). Majority of the vaccines that are being developed for coronaviruses targets the Spike glycoprotein or S protein (Graham et al. 2013). This is mainly because of the fact that S protein is the major inducer of neutralizing antibodies (Jiang et al. 2005). Several kinds of vaccines and antiviral drugs that are based on S protein have been previously evaluated. Among them, the S protein-based vaccines include full-length S protein vaccines, viral vector-based vaccine, DNA-based vaccine, recombinant S protein-based and recombinant RBD protein-based vaccines. Whereas S protein based antiviral therapies include RBD–ACE2 blockers, S cleavage inhibitors, fusion core blockers, neutralizing antibodies, protease inhibitors, S protein inhibitors, and small interfering RNAs (Du et al. 2009). Even though such therapeutic options have proven efficacy in the in vitro studies, however most of these haven’t undergone randomized animal or human trials and hence are of limited use in our present 2019-nCoV scenario. Remdesivir is a novel nucleotide analog prodrug that was intended to be used for the treatment of Ebola virus disease. It also has anti-coronavirus activity due to its inhibitory action on the SARS-CoV and MERS-CoV replication (Sheahan et al. 2017). At present, efforts are being made to identify and develop monoclonal antibodies that are specific and effective against 2019-nCoV. Combination therapy with 2019-nCoV specific monoclonal antibodies and remdesivir can be considered as the ideal therapeutic option for 2019-nCoV (Cohen 2020). Further evaluation is required before confirming the efficacy of such combination therapy. A variety of different therapeutic and vaccine designing approaches against coronaviruses are being explored and yet to be evaluated in terms of their potency, efficacy and safety, but hopefully the process of evaluation will be accelerated in the coming days (Cyranoski 2020; Lu 2020; Pillaiyar et al. 2020; Zaher et al. 2020).
6. Prevention and control measures
Seeing the possible role of animals in 2019-nCoV infection, WHO in its advice for public recommended to avoid the unprotected contact with both farm and wild animals (World Health Organization 2020a). The live-animal markets such as in China could provide chances to animal CoVs to get transmitted to humans and these markets may act as critical places for the origin of novel zoonotic pathogens and pose high public health risks during an outbreak.
The emergency pathogens could be counteracted by opting immediate and timely international collaborative efforts, cooperative efforts between human and animal health sectors. Other effective measures include One health approach, implementation of effective prevention and control strategies, rapid communication and networking, and exploring advances in science and technology for developing rapid and confirmatory diagnostics, enhancing disease surveillance and monitoring, implementation of strict biosecurity measures, and timely efforts toward designing appropriate and effective vaccines and therapeutics (Cheng et al. 2020; Cohen 2020; Cyranoski 2020; Lu 2020; Munjal et al. 2017; Singh et al. 2017).
In the present scenario of not having any direct acting anti-viral agent and vaccines, strict implementation of high vigilance for 2019-nCoV and appropriate prevention and control measures are of utmost importance to check the further spread and control of this virus (Cheng et al. 2020). Researchers and Authorities (WHO, CDC Atlanta and others) across the globe are working to combat the current ongoing 2019-nCoV outbreaks, identifying the possible origin of this novel virus, and to design and develop effective vaccines and therapeutics (Cohen 2020, Cyranoski 2020, Lu 2020, Mahase 2020). Studying the virus in details, its molecular biology and immunology, adaptive genetic changes, mutations and recombination events, elucidating clinical pathology and pathogenesis, identifying the route of origin, role of any mixing vessels (like birds, pigs, and mammals), jumping the species barrier, zoonotic potential, human-to-human transmission events, altogether would pave ways for designing effective prevention and control measures to counter 2019-nCoV
7. Conclusion and future prospects
The nCoV is the most recently emerging virus after the past episodes and panics haunted by Ebola, Zika and Nipah viruses, as well as earlier emergencies posed by Bird flu and Swine flu viruses. The emerging novel coronavirus (2019-nCoV) has become a global concern within a short span of time. Since its origin from Wuhan, Hubei province of China in the first week of December 2019, it has claimed 425 lives in China and infected 20630 in 24 countries including China. Additionally, new cases are emerging in different countries and three confirmed cases from India also emerged till compilation of this paper, February 4th, 2020. As of now deaths were reported only from China but now causality in Philippines reported for the first time outside China. The nCoV crisis has been declared as ‘Public Health Emergency of International Concern’ by WHO. The zoonotic route (animal-to-human) is suspected as the route of disease origin. Bats are considered as the natural reservoir hosts and play a crucial role in transmitting various viruses, including Ebola, Nipah, Coronavirus and others (Cui et al. 2019). A high diversity among zoonotic Alphacoronaviruses and SARS-CoV-related Betacoronaviruses has been found in the circulating bats of Western Europe (Gouilh et al. 2018). The genetic analyses predict bats as the most probable origin of 2019-nCoV. The diversity of CoVs in the bat population needs further investigation in details, as well as the surveillance and monitoring of bats becomes critical to prevent future outbreaks in animals and the public. The recent nCoV outbreak highlights the hidden wild animal reservoir of the deadly viruses and possible threat of spillover zoonoses. Successful virus isolation attempts have made doors open for developing better diagnostics and effective vaccines. A report emerged from China where the scientists claimed successful isolation of the 2019-nCoV virus in Vero and Huh7 cell lines from infected patients. Subsequently, in aiding the further research for designing rapid diagnostics and vaccine development for nCoV, scientists at The Peter Doherty Institute for Infection and Immunity at Melbourne, Australia, were also successful in growing the Wuhan coronavirus in cell culture. Further research is warranted on establishing animal models for the current 2019-nCoV unrevealing viral events of replication, transmission, and pathogenesis in humans. This could provide clues for discovering effective therapeutic regimens and vaccine testing purposes
Supplementary Material
Acknowledgements
All the authors acknowledge and thank their respective Institutes and Universities.
Author contributions
All the authors substantially contributed to the conception, design, analysis and interpretation of data, checking and approving the final version of the manuscript, and agree to be accountable for its contents.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Alagaili AN, Briese T, Mishra N, Kapoor V, Sameroff SC, Burbelo PD, de Wit E, Munster VJ, Hensley LE, Zalmout IS, et al. 2014. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. MBio. 5(2):e00884–00814. doi: 10.1128/mBio.00884-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng VC, Wong S-C, To KK, Ho P, Yuen K-Y.. 2020. Preparedness and proactive infection control measures against the emerging Wuhan coronavirus pneumonia in China. J Hosp Infect. doi: 10.1016/j.jhin.2020.01.010. [published online ahead of print, 2020 Jan 18] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. 2020. New coronavirus threat galvanizes scientists. Science, 367(6477): 492–493. doi: 10.1126/science.367.6477.492 [DOI] [PubMed] [Google Scholar]
- Cui J, Li F, Shi Z-L.. 2019. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyranoski D. 2020. This scientist hopes to test coronavirus drugs on animals in locked-down Wuhan. Nature. 577(7792):607. doi: 10.1038/d41586-020-00190-6 [DOI] [PubMed] [Google Scholar]
- de Groot RJ, Baker S, Baric R, Enjuanes L, Gorbalenya A, Holmes K, Perlman S, Poon L, Rottier P, Talbot P, Woo PC. 2012. Family coronaviridae. In: King AMQ, Adams MJ, Cartens EB, Lefkowitz EJ (eds.), Virus Taxonomy, the 9th report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, CA. doi: 10.1016/B978-0-12-384684-6.00068-9; p. 806–828. [DOI] [Google Scholar]
- Drosten C, Günther S, Preiser W, van der Werf S, Brodt H-R, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier R A.M, et al. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 348(20):1967–1976. doi: 10.1056/NEJMoa030747 [DOI] [PubMed] [Google Scholar]
- Du L, He Y, Zhou Y, Liu S, Zheng B-J, Jiang S.. 2009. The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat Rev Microbiol. 7(3):226–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr AR, Perlman S.. 2015. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouilh MA, Puechmaille SJ, Diancourt L, Vandenbogaert M, Serra-Cobo J, Roïg ML, Brown P, Moutou F, Caro V, Vabret A.. 2018. SARS-CoV related Betacoronavirus and diverse Alphacoronavirus members found in western old-world. Virology. 517:88–97. doi: 10.1016/j.virol.2018.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RL, Donaldson EF, Baric RS.. 2013. A decade after SARS: strategies for controlling emerging coronaviruses. Nat Rev Microbiol. 11(12):836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Zheng B, He Y, Liu X, Zhuang Z, Cheung C, Luo S, Li P, Zhang L, Guan Y.. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 302(5643):276–278. doi: 10.1126/science.1087139 [DOI] [PubMed] [Google Scholar]
- Hui D S, I Azhar E, Madani T A, Ntoumi F, Kock R, Dar O, Ippolito G, Mchugh T D, Memish Z A, Drosten C, et al. 2020. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health-The latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 91:264–266. doi: 10.1016/j.ijid.2020.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ithete NL, Stoffberg S, Corman VM, Cottontail VM, Richards LR, Schoeman MC, Drosten C, Drexler JF, Preiser W.. 2013. Close relative of human Middle East respiratory syndrome coronavirus in bat, South Africa. Emerg Infect Dis. 19(10):1697–1699. doi: 10.3201/eid1910.130946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, He Y, Liu S.. 2005. SARS vaccine development. Emerg Infect Dis. 11(7):1016–1020. doi: 10.3201/eid1107.050219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn JS, McIntosh K.. 2005. History and recent advances in coronavirus discovery. Pediatr Infect Dis J. 24(Supplement):S223–S227. doi: 10.1097/01.inf.0000188166.17324.60 [DOI] [PubMed] [Google Scholar]
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, et al. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 348(20):1953–1966. doi: 10.1056/NEJMoa030781 [DOI] [PubMed] [Google Scholar]
- Lau SK, Woo PC, Li KS, Huang Y, Tsoi H-W, Wong BH, Wong SS, Leung S-Y, Chan K-H, Yuen K-Y.. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci. 102(39):14040–14045. doi: 10.1073/pnas.0506735102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko MC, Munster V.. 2020. Functional assessment of cell entry and receptor usage for lineage B β-coronaviruses, including 2019-nCoV. bioRxiv. doi: 10.1101/2020.01.22.915660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H.. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science. 310(5748):676–679. [DOI] [PubMed] [Google Scholar]
- Lu H. 2020. Drug treatment options for the 2019-new coronavirus (2019-nCoV). Biosci Trends. doi: 10.5582/bst.2020.01020. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, et al. 2020. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet.395(10222): 391–393. doi: 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahase E. 2020. China coronavirus: what do we know so far? Bristish Med J. 368:m308. doi: 10.1136/bmj.m308 31980434 [DOI] [PubMed] [Google Scholar]
- Munjal A, Khandia R, Dhama K, Sachan S, Karthik K, Tiwari R, Malik Y S, Kumar D, Singh R K, Iqbal H M N, et al. 2017. Advances in developing therapies to combat Zika virus: current knowledge and future perspectives. Front Microbiol. 8:1469. doi: 10.3389/fmicb.2017.01469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris JSM, Lai ST, Poon LLM, Guan Y, Yam LYC, Lim W, Nicholls J, Yee WKS, Yan WW, Cheung MT, et al. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. The Lancet. 361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillaiyar T, Meenakshisundaram S, Manickam M.. 2020. Recent discovery and development of inhibitors targeting coronaviruses. Drug Discov Today. doi: 10.1016/j.drudis.2020.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan TP, Sims AC, Graham RL, Menachery VD, Gralinski LE, Case JB, Leist SR, Pyrc K, Feng JY, Trantcheva I.. 2017. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 9: 1–11. doi: 10.1126/scitranslmed.aal3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R K, Dhama K, Malik Y S, Ramakrishnan M A, Karthik K, Khandia R, Tiwari R, Munjal A, Saminathan M, Sachan S, et al. 2017. Ebola virus–epidemiology, diagnosis, and control: threat to humans, lessons learnt, and preparedness plans–an update on its 40 year’s journey. Vet Q. 37(1):98–135. doi: 10.1080/01652176.2017.1309474. [DOI] [PubMed] [Google Scholar]
- World Health Organization. 2015. Middle East respiratory syndrome coronavirus (MERS-CoV): summary of current situation, literature update and risk assessment. URL: https://apps.who.int/iris/handle/10665/179184 [Google Scholar]
- World Health Organization. 2020a. Novel Coronavirus (2019-nCoV) advice for the public. World Health Organization.URL. https://www.who.int/emergencies/diseases/novel-coronavirus-2019 [Google Scholar]
- World Health Organization. 2020b. Novel Coronavirus (2019-nCoV) situation reports. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/.
- Zaher NH, Mostafa MI, Altaher AY.. 2020. Design, synthesis and molecular docking of novel triazole derivatives as potential CoV helicase inhibitors. Acta Pharmaceutica. 70(2):145–159. doi: 10.2478/acph-2020-0024. [DOI] [PubMed] [Google Scholar]
- Zaki AM, Van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA.. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 367(19):1814–1820. [DOI] [PubMed] [Google Scholar]
- Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L.. 2020. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. bioRxiv. Preprint version, https://www.biorxiv.org/content/ 10.1101/2020.01.22.914952v1 [DOI] [Google Scholar]
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R.. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. doi: 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.