Itakura et al. identify a protein quality control system for aberrant extracellular proteins, including misfolded proteins and amyloid β. This extracellular proteostasis pathway involves the cell-surface heparan sulfate receptor, which mediates internalization of aberrant extracellular proteins in complex with an extracellular chaperone.
Abstract
The accumulation of aberrant proteins leads to various neurodegenerative disorders. Mammalian cells contain several intracellular protein degradation systems, including autophagy and proteasomal systems, that selectively remove aberrant intracellular proteins. Although mammals contain not only intracellular but also extracellular proteins, the mechanism underlying the quality control of aberrant extracellular proteins is poorly understood. Here, using a novel quantitative fluorescence assay and genome-wide CRISPR screening, we identified the receptor-mediated degradation pathway by which misfolded extracellular proteins are selectively captured by the extracellular chaperone Clusterin and undergo endocytosis via the cell surface heparan sulfate (HS) receptor. Biochemical analyses revealed that positively charged residues on Clusterin electrostatically interact with negatively charged HS. Furthermore, the Clusterin–HS pathway facilitates the degradation of amyloid β peptide and diverse leaked cytosolic proteins in extracellular space. Our results identify a novel protein quality control system for preserving extracellular proteostasis and highlight its role in preventing diseases associated with aberrant extracellular proteins.
Introduction
Protein deposition diseases are associated with the accumulation of aberrant proteins. The protein deposits consist of misfolded or aggregate-prone proteins. Various stresses, including heat shock or pathological conditions, generate misfolded proteins that induce toxicity. Although cells have developed elaborate protein quality control systems against various substrates (Wolff et al., 2014), the failure of these protein quality control systems perturbs protein homeostasis (proteostasis) and contributes to protein deposition diseases, such as neurodegenerative diseases, including Alzheimer’s disease, Huntington’s disease, Parkinson’s disease, amyotrophic lateral sclerosis, and transmissible spongiform encephalopathies (Kaushik and Cuervo, 2015). Thus, proteostasis regulators are attractive targets for pharmacological intervention (Lai and Crews, 2017; Powers et al., 2009).
ATP-dependent molecular chaperones interact with misfolded intracellular proteins, and the energy from ATP binding and hydrolysis is used to either refold or disaggregate the misfolded proteins (Klaips et al., 2018). Misfolded proteins that cannot be productively folded are targeted to one of the cell’s many protein degradation pathways that mainly culminate in either the ubiquitin-proteasome system or autophagy (Ciechanover and Kwon, 2017; Dikic and Elazar, 2018; Itakura et al., 2012; Kwon and Ciechanover, 2017; Levine and Kroemer, 2019). These intracellular protein degradation pathways selectively recognize misfolded proteins through various molecular mechanisms and transport these proteins to degradative compartments. Misfolded proteins in organelles, such as the ER, are also recognized via different mechanisms for refolding or degradation (Walter and Ron, 2011). Damaged organelles, such as mitochondria, are also distinguished from intact organelles and degraded by autophagy (Gatica et al., 2018; Sica et al., 2015). Thus, the misfolded proteins in cells are almost exclusively targeted via the protein quality control systems to maintain proteostasis (Wolff et al., 2014). Proteins in multicellular organisms function not only intracellularly but also extracellularly. Secreted proteins collectively constitute ∼11% of the human proteome (Uhlén et al., 2015). These proteins play essential roles in physiological and pathological processes. As with intracellular proteins, extracellular proteins are damaged by heat stress, oxidative stress, and pathological conditions. Furthermore, extracellular fluids are subjected to shear stress, and acidosis and alkalosis disturb extracellular pH (Wyatt et al., 2013). Thus, extracellular proteins are exposed to more stringent conditions than intracellular proteins. In addition, Alzheimer’s disease, the most prevalent cause of dementia, affecting 47.5 million people worldwide (Hung and Fu, 2017), is mainly characterized by amyloid β (Aβ) deposits in the extracellular space. There is currently no cure for Alzheimer’s disease. However, the mechanisms underlying the protein degradation pathway for aberrant extracellular proteins are poorly understood.
Previous studies proposed that extracellular chaperons stabilize stressed proteins. The major extracellular chaperone in body fluids of vertebrates is Clusterin (Wyatt et al., 2013), which binds to stressed extracellular proteins (Poon et al., 2000; Wojtas et al., 2017). Due to the lack of ATPase activity among extracellular chaperones, including Clusterin, and the low concentration of ATP in the extracellular space in vertebrates (Poon et al., 2000), proteins in the extracellular space cannot be refolded. It has been suggested that irreversible binding of Clusterin to stressed proteins stabilizes them to prevent their aggregation (Humphreys et al., 1999; Wyatt et al., 2013). Meanwhile, the half-life of secreted proteins in vivo is short (Price et al., 2010). Inspired by the mechanisms of intracellular degradation, we hypothesized that misfolded extracellular proteins may engage chaperone-like proteins that facilitate their degradation through an unidentified cell surface receptor.
Here, we demonstrate the chaperone- and receptor-mediated extracellular protein degradation (CRED) pathway for aberrant extracellular proteins. Clusterin interacted with various misfolded proteins or Aβ and selectively internalized these proteins into the cell for lysosomal degradation. Genome-wide screening and biochemical analyses revealed that the cell surface heparan sulfate (HS) receptor leads to the degradation of the Clusterin complex through electrostatic interactions. We show that the CRED pathway is a general extracellular protein quality control system for various misfolded proteins in diverse tissues. Our discovery of a receptor-mediated extracellular protein degradation pathway provides a novel concept in cell biology.
Results
Clusterin–misfolded protein complexes are selectively degraded in lysosomes
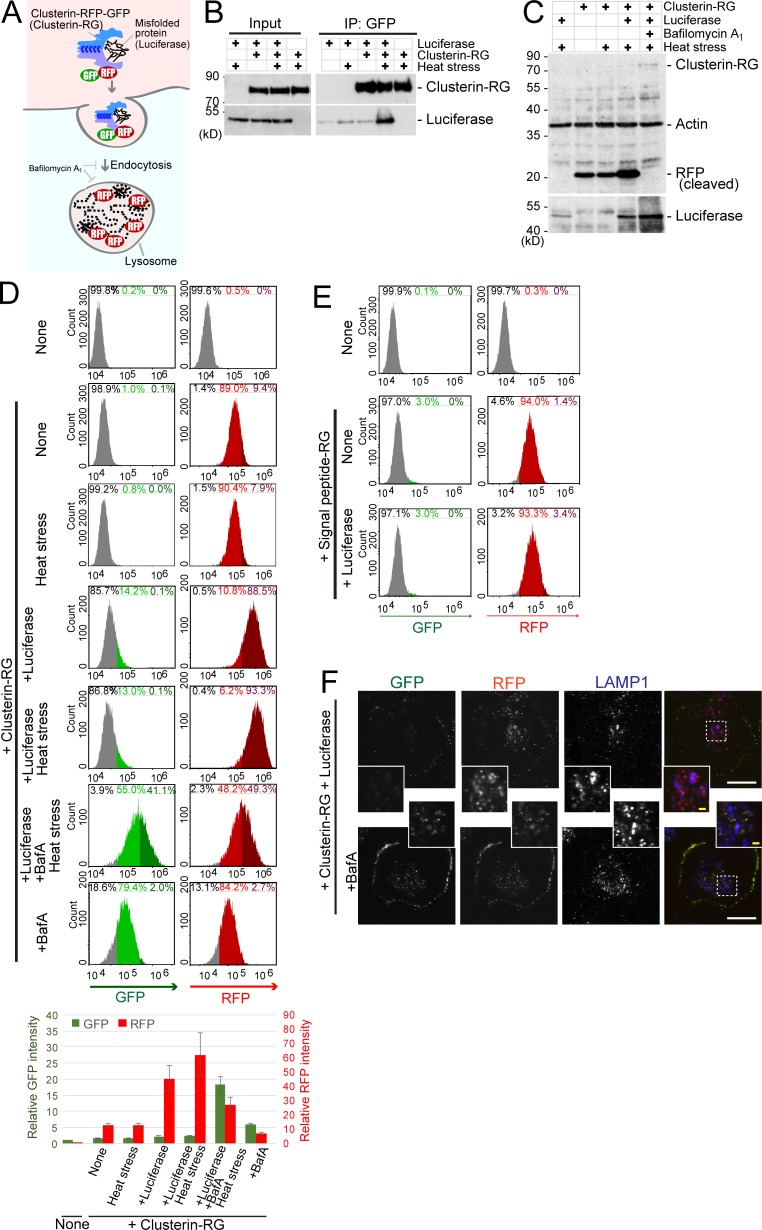
To determine if the extracellular Clusterin–substrate complex undergoes degradation in lysosomes, we developed an internalization assay for Clusterin tandemly fused to RFP and GFP (Clusterin-RFP-GFP, or CluRG for short). If CluRG reaches the lysosome, Clusterin and GFP are degraded but RFP, which is protease and pH resistant (Katayama et al., 2008; Kimura et al., 2007), remains intact and fluorescent (Fig. 1 A). Basically, this internalization assay enables simple quantification of lysosomal degradation of Clusterin. Firefly luciferase (Luc) is used as a model substrate that is thermodynamically unstable at 42°C (Glover and Lindquist, 1998; Rodrigo-Brenni et al., 2014).
Figure 1.
A novel method to monitor cumulative lysosomal degradation of an extracellular protein. (A) Schematic of the Clusterin-RFP-GFP (Clusterin-RG/CluRG) internalization assay. After internalization, lysosomal enzymes degrade Clusterin and GFP, whereas RFP, which is resistant to proteases and acidic pH, accumulates in lysosomes. The blue C-shaped structures in the Clusterin structure represent disulfide bonds between the α and β subunits. (B) Clusterin selectively binds to a misfolded protein. Purified CluRG was mixed with or without recombinant Luc in serum-free medium and preincubated at 4°C or 42°C for 20 min (heat stress). Samples were subjected to immunoprecipitation (IP) with anti-GFP Sepharose at 4°C. (C) Increased lysosomal degradation of Clusterin by misfolded proteins. Purified CluRG was mixed with or without recombinant Luc in serum-free medium and preincubated at 4°C or 42°C for 20 min (heat stress). HEK293 cells were cultured in the medium with or without Bafa for 14 h at 37°C and analyzed by immunoblotting. (D) Misfolded protein–dependent Clusterin internalization. Cells were treated as in C and analyzed by flow cytometry. The bar graph shows the relative fluorescence intensities (GFP, green left axis; RFP, red right axis) in a cell normalized to those in nontreated cells (n = 3). The data are presented as the mean ± SEM. Note that a 14-h incubation at 37°C moderately induces the misfolding of Luc without preheat stress. (E) Misfolded protein-independent RFP-GFP internalization. Purified SP-RG was mixed with or without recombinant Luc in serum-free medium and preincubated at 42°C for 20 min. HEK293 cells were cultured in the medium for 14 h at 37°C and analyzed by flow cytometry. (F) Lysosomal accumulation of Clusterin. Purified CluRG was mixed with recombinant Luc in serum-free medium and preincubated at 42°C for 20 min. HEK293 cells were cultured in the medium with or without BafA for 14 h at 37°C, immunolabeled for LAMP1 (a lysosomal marker), and imaged by confocal microscopy. Main scale bar, 10 µm. Inset scale bar, 2 µm.
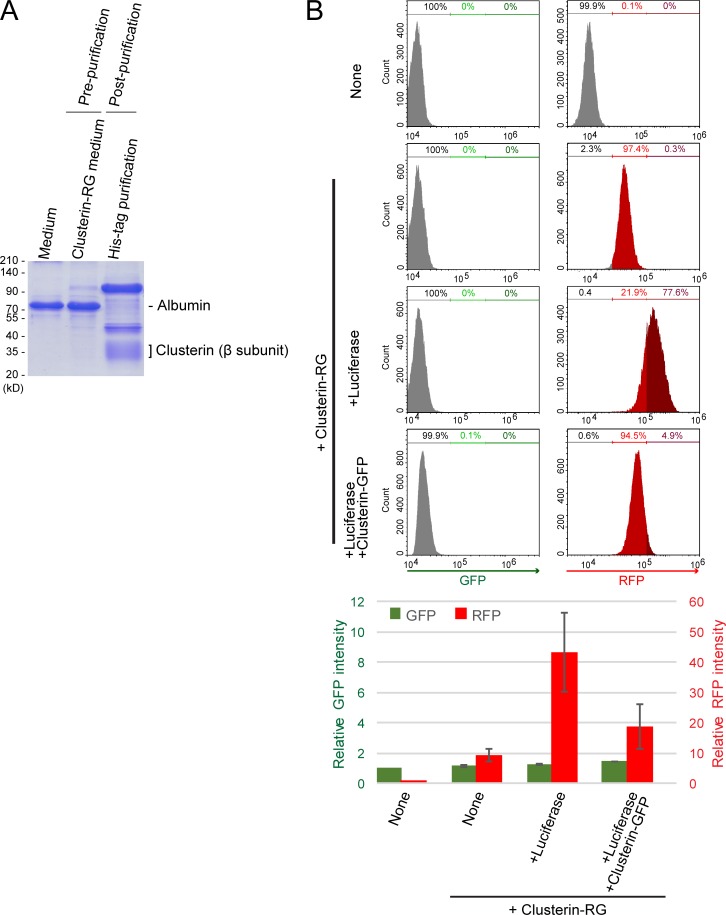
We purified secreted CluRG from conditioned medium of CluRG-expressing human cells (Fig. S1 A) and demonstrated in pulldown assays that it interacts selectively with Luc induced to misfold at 42°C (heat shock; Fig. 1 B). CluRG was mixed into serum-free medium at a concentration of 5 µg/ml (∼10 times lower than the physiological Clusterin plasma concentration, 35–105 µg/ml; Murphy et al., 1988), preincubated with Luc at 42°C for 20 min, and then added to human embryonic kidney (HEK) 293 cells for 14 h at 37°C. Both cleaved RFP and Luc were observed by immunoblotting in cell lysates, indicating uptake of both CluRG and Luc (Fig. 1 C). In the presence of the vacuolar proton ATPase inhibitor bafilomycin A1 (BafA), intact CluRG was observed instead of cleaved RFP, and Luc levels were increased. Less RFP was recovered with the cells in the absence of Luc, and less Luc was recovered in the absence of CluRG. These results suggest that the CluRG–Luc complex is preferentially internalized for lysosomal degradation.
Figure S1.
Competitive inhibition of Clusterin internalization. (A) Recombinant CluRG (His-tagged) was purified from mammalian cells (see Materials and methods). Conditioned medium was collected from HEK293 cells expressing CluRG (prepurification). CluRG in the conditioned medium was affinity purified via the His tag (postpurification) and analyzed by Coomassie brilliant blue staining. (B) Purified CluRG was mixed with or without recombinant Luc in serum-free medium and preincubated at 42°C for 20 min. HEK293 cells were cultured in the medium with or without a 10-fold excess of Clusterin-GFP with Luc for 14 h at 37°C and analyzed by flow cytometry. The bar graph shows the relative fluorescence intensities (GFP, green left axis; RFP, red right axis) in a cell normalized to those in nontreated cells (n = 3). The data are presented as the mean ± SEM.
Quantification of CluRG internalization assay by flow cytometry also showed that the intracellular RFP signal is increased in the presence of misfolded Luc (Fig. 1 D). As expected, GFP was stabilized by BafA treatment (Fig. 1 D); the decrease in RFP under these conditions is likely a consequence of BafA partially inhibiting endocytosis. The cells engulf RFP-GFP fusion protein without Clusterin (Fig. 1 E), suggesting that proteins are nonspecifically internalized via fluid phase endocytosis. The internalization of an RFP-GFP protein was unaffected by misfolded Luc, illustrating a crucial role for Clusterin (Fig. 1 E). Consistent with this idea, the uptake of CluRG could be competitively inhibited by excess Clusterin-GFP (Fig. S1 B), suggesting that Clusterin-dependent uptake was a saturable process.
Microscopy of the cells after CluRG-Luc uptake showed that RFP was colocalized with lysosomes, whereas GFP was presumably degraded or quenched by the acidic pH (Fig. 1 F). Treatment with BafA stabilized GFP, which was seen together with RFP in lysosomes. In addition, a substantial amount of CluRG was observed on the cell surface (Fig. 1 F, bottom), implying that endocytosis inhibition by BafA trapped CluRG in complex with putative cell surface receptors. These results strongly suggest that there is a receptor-mediated degradation pathway for extracellular Clusterin-misfolded protein complex.
The selective extracellular protein degradation is a general protein quality control system
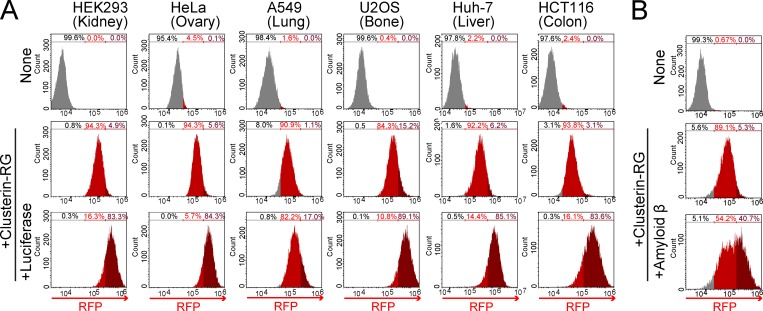
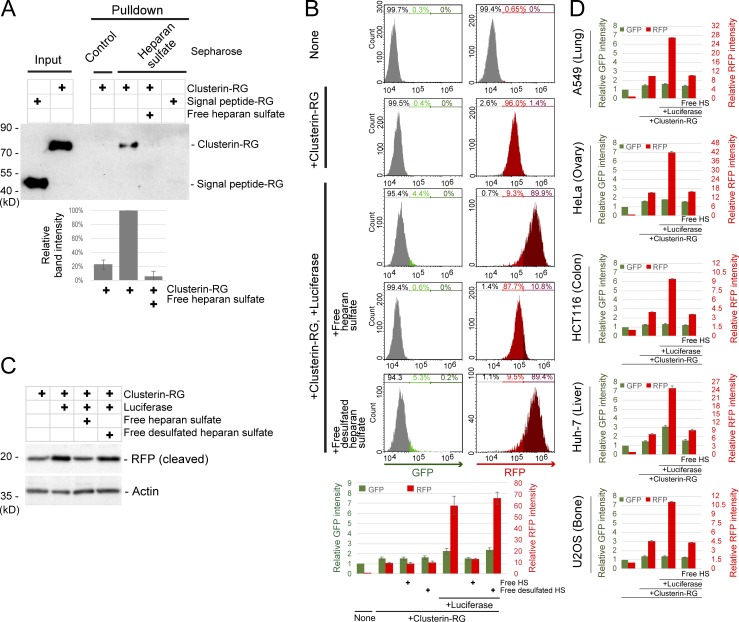
To investigate whether the Clusterin-mediated degradation is a general system among mammalian tissues, CluRG internalization assays were performed using human cell lines derived from various tissues including the kidney, ovary, lung, bone, liver, and colon. All of these cell lines showed increased internalization of CluRG in the presence of misfolded Luc (Fig. 2 A). We next examined the Alzheimer’s disease–associated peptide Aβ, which is known to interact with Clusterin (Matsubara et al., 1996; Narayan et al., 2011; Zlokovic et al., 1996). Preincubation of Aβ also increased Clusterin internalization in HEK293 cells (Fig. 2 B), suggesting that Clusterin-mediated degradation is a ubiquitous system.
Figure 2.
Selective extracellular protein degradation is ubiquitous in diverse tissues and for Aβ. (A) Clusterin-mediated degradation in various cell lines. Purified CluRG was mixed with or without recombinant Luc in serum-free medium and preincubated at 42°C for 20 min. The cells were cultured in medium for 14 h at 37°C and analyzed by flow cytometry. (B) Aβ-dependent Clusterin internalization. Purified CluRG was mixed with or without recombinant Aβ in serum-free medium and preincubated at 37°C. HEK293 cells were cultured in medium with or without free HS for 14 h at 37°C and analyzed by flow cytometry.
HS biosynthesis enzymes are essential for the degradation of the Clusterin complex
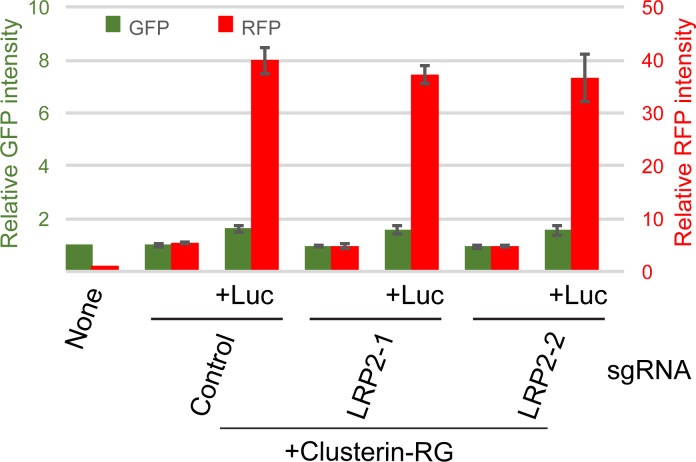
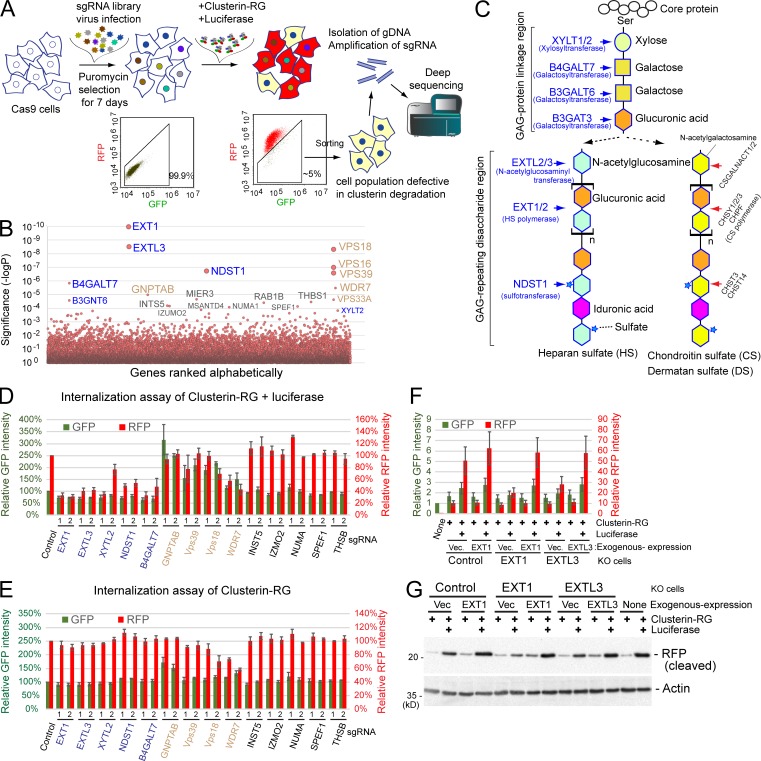
Although previous studies suggested that low density lipoprotein receptor-related protein 2 (LRP2) and Plexin A4 bind to Clusterin (Bell et al., 2007; Hammad et al., 1997; Kang et al., 2016; Zlokovic et al., 1996), the tissue distribution of LRP2 and Plaxin A4 is restricted (Lundgren et al., 1997; Pontén et al., 2009). Indeed, CRISPR-mediated LRP2 gene knockout (KO) HEK293 cells did not show any significant reduction in the uptake of the CluRG–Luc complex (Fig. S2). To identify the putative clearance receptors, we performed a genome-wide CRISPR screen (Shalem et al., 2014) for genes whose disruption impeded CluRG-Luc uptake (Fig. 3 A). Sequencing of cells with a low RFP/GFP ratio revealed an enrichment of guide RNAs targeting ∼20 different genes (Fig. 3 B). Strikingly and unexpectedly, many of the genes are implicated in HS biosynthesis (Sarrazin et al., 2011; Fig. 3 C). Although B4GALT7 and XYLT2 catalyze the initial steps for HS, chondroitin sulfate (CS) and dermatan sulfate (DS), CS- and DS-specific genes were not identified in the screen (Fig. 3 B).
Figure S2.
LRP2 is not involved in the internalization of the Clusterin–misfolded protein complex. Purified CluRG was mixed with or without recombinant Luc in serum-free medium and incubated at 42°C for 20 min. HEK293 cells expressing Cas9 with control or LRP2 sgRNA were cultured in the mixtures for 14 h at 37°C and analyzed by flow cytometry. The bar graph shows the relative fluorescence intensities (GFP, green left axis; RFP, red right axis) in a cell normalized to those in nontreated cells (n = 3). The data are presented as the mean ± SEM.
Figure 3.
HS biosynthesis enzymes are essential for the degradation of the Clusterin-substrate complex. (A) Schematic representation of the screening. The GeCKO v2 sgRNA library was delivered into Cas9-expressing HEK293 cells by lentiviral infection. After 1 wk of culture, KO cells were treated with heat-stressed CluRG–Luc complex for 14 h, and those in the bottom 5% of the RFP/GFP ratio (cell population defective in Clusterin degradation) were sorted using a cell sorter. After this enrichment process was repeated twice, PCR amplification of the sgRNA coding sequence integrated into the chromosomes was conducted for next-generation sequencing. (B) CRISPR screening of Clusterin degradation-deficient cells. A modified robust rank aggregation algorithm was used to rank sgRNAs based on P values. HS biosynthesis enzyme genes (blue) and endolysosomal genes (brown) are indicated (B–E). (C) Schematic representation of the synthesis of the GAG backbones of HS or CS/DS chains. (D and E) HS synthesis enzymes are essential for misfolded protein–dependent Clusterin internalization. Purified CluRG was mixed with recombinant Luc in serum-free medium and preincubated at 42°C for 20 min. HEK293 cells expressing Cas9 with the indicated sgRNAs (two per candidate) were cultured in the medium for 14 h at 37°C and analyzed by flow cytometry (D). Cells were treated as in D except without Luc and analyzed by flow cytometry (E). The bar graph shows the relative fluorescence intensities (GFP, green left axis; RFP, red right axis) in a cell normalized to those in control cells (n = 3). The data are presented as the mean ± SEM. (F and G) Loss of Clusterin internalization in EXT1 or EXTL3 KO cells. Purified CluRG was mixed with or without recombinant Luc in serum-free medium and preincubated at 42°C for 20 min. Control, EXT1 KO, or EXTL3 KO HEK293 cells (generated by CRISPR) introduced with EXT1, EXTL3 or empty vector were cultured in the medium for 14 h at 37°C and analyzed by flow cytometry. The bar graph shows the relative fluorescence intensities (GFP, green left axis; RFP, red right axis) in a cell normalized to those in nontreated cells (n = 3). The data are presented as the mean ± SEM (F). Cells were treated as in F and analyzed by immunoblotting (G).
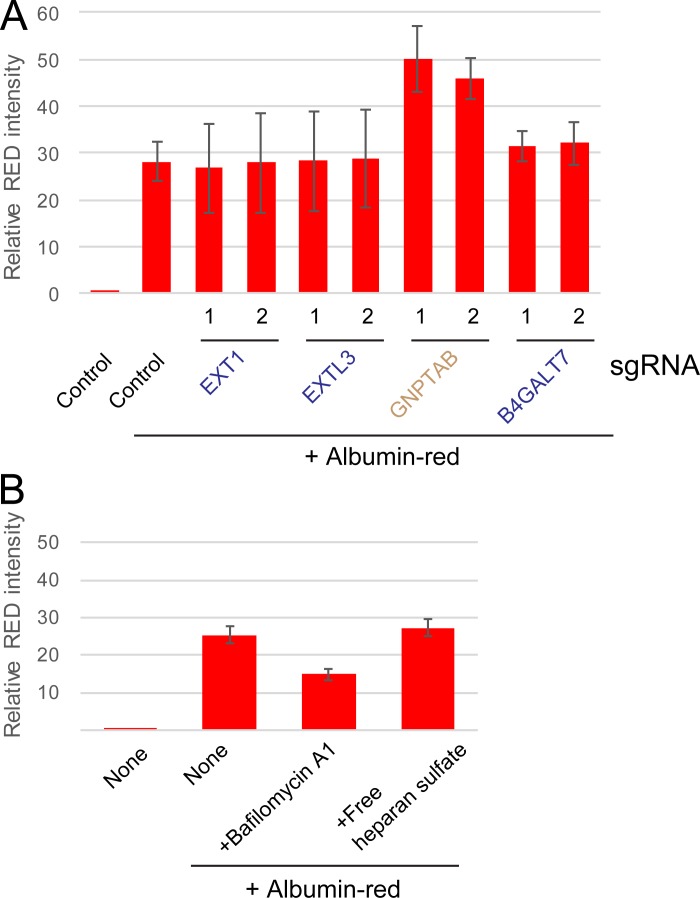
To validate the role of HS biosynthesis, we generated KO cell lines using single guide RNA (sgRNA) against individual HS pathway genes (EXT1, EXTL3, NDST1, B4GALT7, and XYLT2) and examined them by the CluRG uptake assay. Each KO cell line showed markedly reduced CluRG-Luc uptake (Fig. 3 D) with no effect on the uptake of either CluRG alone (Fig. 3 E) or albumin-red (Fig. S3 A). Exogenous expression of the knocked-out gene in each of these cell lines restored CluRG-Luc uptake (Fig. 3, F and G), verifying that the effects are on-target and reversible. Thus, HS pathway disruption selectively impairs endocytosis of Clusterin when it contains a substrate, but not endocytosis in general. In contrast, KO cells corresponding to other hits from the screen (Vps18, Vps39, WDR7, and GNPTAB) exhibited an increased GFP signal in the CluRG uptake assay, indicating their role in lysosomal degradation, but not internalization (Fig. 3, D and E). This effect is not specific to CluRG, as indicated by albumin-red stabilization in cells lacking GNPTAB, which functions in the transport of lysosomal enzymes (Fig. S3 A).
Figure S3.
HS synthesis enzymes are not required for endocytosis. (A) HEK293 cells expressing Cas9 with the indicated sgRNAs (two per candidate) were cultured with or without albumin-red for 14 h and analyzed by flow cytometry. The bar graph shows the relative fluorescence intensities in a cell normalized to those in nontreated cells (n = 3). The data are presented as the mean ± SEM. (B) HS is not required for albumin endocytosis. HEK293 cells were cultured with or without albumin-red in the presence of BafA or free HS for 14 h and analyzed by flow cytometry. The bar graph shows the relative fluorescence intensities in a cell normalized to those in nontreated cells (n = 3). The data are presented as the mean ± SEM.
HS chain electrostatically interacts with Clusterin
As nearly all vertebrate cells express a small set of cell-surface HS proteoglycans that are receptors for several growth factors and viruses (Liu and Thorp, 2002; Pillay et al., 2016; Xu and Esko, 2014; Yayon et al., 1991), we postulated that HS acts as a Clusterin receptor. To determine whether Clusterin directly binds to HS, we used a pulldown assay with HS-coated Sepharose. CluRG but not RG was precipitated by HS-coated Sepharose, and this interaction was competed by free HS (Fig. 4 A). Free HS also markedly interfered with the internalization of CluRG-Luc into the cells (Fig. 4 B) and delivery to lysosomes (Fig. 4 C) but not with the internalization of albumin or CluRG alone (Fig. S3 B and Fig. 4 B). Desulfated HS did not inhibit CluRG-Luc internalization (Fig. 4, B and C). As mentioned above, HS is a ubiquitous cell surface proteoglycan. Consistent with this, competitive inhibition by free HS was observed in other cell lines derived from different tissues (Fig. 4 D). Together, these results suggest that HS interacts with Clusterin by electrostatic interaction through the negatively charged sulfate residues on the HS chain.
Figure 4.
Clusterin directly interacts with HS. (A) In vitro binding of Clusterin and HS. Purified CluRG or SP-RG in the presence or absence of free HS was subjected to a pulldown assay with HS-conjugated or control Sepharose at 4°C. The bar graph shows the relative band intensity normalized to that of CluRG by HS-conjugated Sepharose pulldown (n = 3). The data are presented as the mean ± SEM. (B and C) Competitive inhibition of Clusterin internalization by free HS. Purified CluRG was mixed with or without recombinant Luc in serum-free medium and preincubated at 42°C for 20 min. HEK293 cells were cultured in the medium with or without free HS or free desulfated HS for 14 h at 37°C and analyzed by flow cytometry. The bar graph shows the relative fluorescence intensities (GFP, green left axis; RFP, red right axis) in a cell normalized to those in nontreated cells (n = 3). The data are presented as the mean ± SEM (B). HEK293 cells were treated as in B and analyzed by immunoblotting (C). (D) The Clusterin–HS pathway in various cell lines. Purified Clusterin WT-RG was mixed with or without recombinant Luc in serum-free medium and preincubated at 42°C for 20 min. The cells were cultured in medium with or without free HS for 14 h at 37°C and analyzed by flow cytometry. The bar graph shows the relative fluorescence intensities (GFP, green left axis; RFP, red right axis) in the cells normalized to that in nontreated cells (n = 3). The data are presented as mean ± SEM.
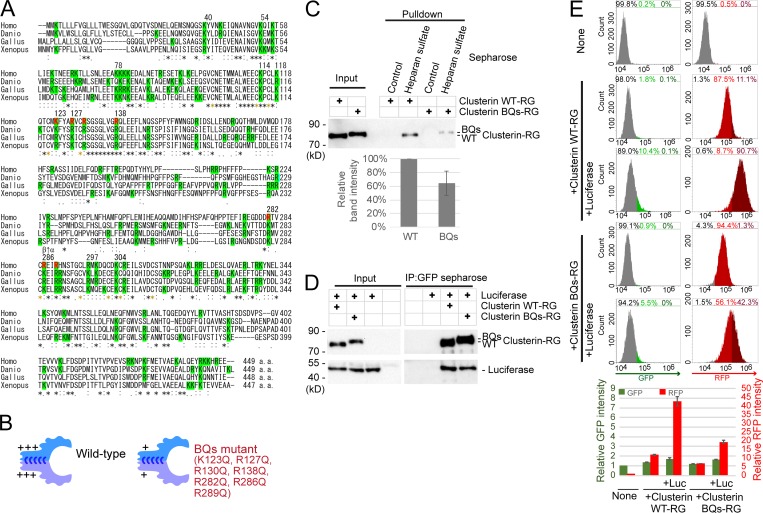
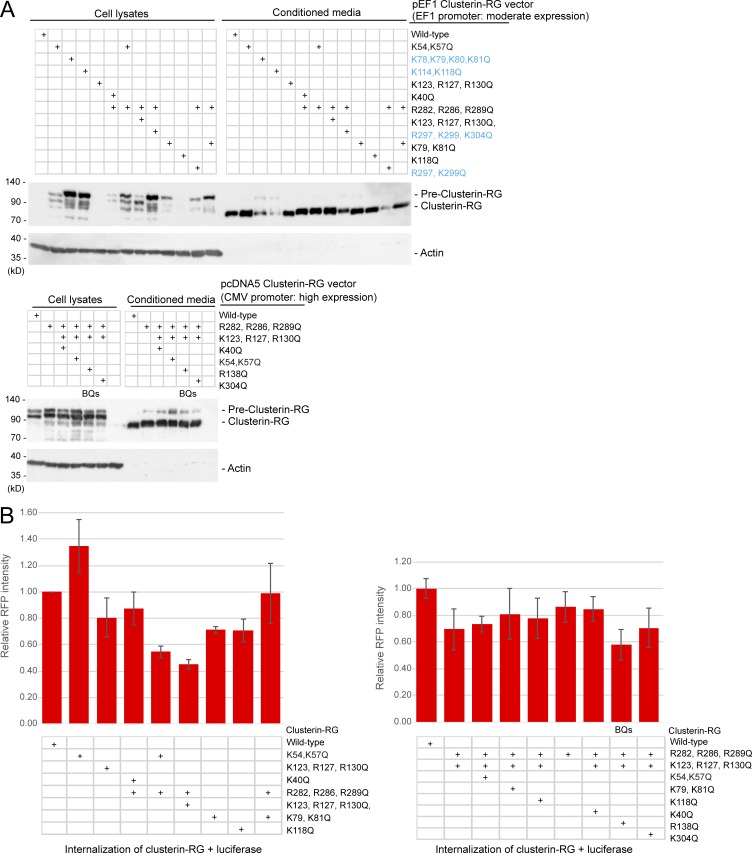
The HS binding sites in HS-binding proteins typically contain four to seven basic amino acids (Xu and Esko, 2014). 16 lysine and arginine residues in vertebrate Clusterin are completely conserved (Fig. 5 A). Mutagenesis identified K and R residues whose mutation to Q are compatible with Clusterin secretion (Fig. S4 A). Based on this result, we prepared recombinant CluRG containing seven basic residue mutations (K123Q, R127Q, R130Q, R138Q, R282Q, R286Q, and R289Q) and analyzed the functionality of this variant (called BQs; Fig. 5 B). Secretion of CluRG-BQs from cells is not impaired (Fig. S4 A), suggesting that the mutant is structurally intact. CluRG-BQs displayed ∼50% reduced binding to HS (Fig. 5 C) but no impairment in Luc binding (Fig. 5 D). Consistent with a key role for HS binding in CluRG-Luc internalization, CluRG-BQs-Luc showed significantly decreased internalization and lysosomal degradation (Fig. 5 E and Fig. S4 B). Thus, CluRG-Luc delivery to lysosomes can be impaired by genetic disruption of HS biosynthesis, competition by free HS, or a Clusterin mutant that reduces HS interaction. These findings strongly indicate that HS on cell-surface proteoglycans is the receptor for uptake of Clusterin with misfolded protein.
Figure 5.
Electrostatic interactions between HS and Clusterin. (A) Multiple sequence alignment of different vertebrate Clusterin sequences. Identical residues in all sequences are indicated by asterisks (*), conserved substitutions are indicated by colons (:), and conserved cysteine residues for disulfide bonds are indicated by yellow asterisks. Basic amino acid resides are highlighted in green. Residues for BQs mutant are indicated by red. Homo, Homo sapiens; Danio, Danio rerio; Gallus, Gallus gallus; Xenopus, Xenopus laevis. The arrow between β and α indicates the cleavage site producing the β and α subunits. (B) Diagrams of WT Clusterin and the BQs mutant. The BQs mutant exhibited decreased positive charges. (C) Weak binding of Clusterin BQs to HS. Purified Clusterin WT-RG or Clusterin-BQs-RG was subjected to pulldown assays with HS-conjugated or control Sepharose at 4°C. The bar graph shows the relative band intensity normalized to that of Clusterin WT-RG by HS-conjugated Sepharose pulldown (n = 3). The data are presented as the mean ± SEM. (D) Normal binding of Clusterin BQs to misfolded proteins. Purified Clusterin WT-RG or Clusterin BQs-RG was mixed with recombinant Luc and incubated at 42°C for 20 min. Samples were subjected to immunoprecipitation with anti-GFP Sepharose at 4°C. (E) Inefficient internalization of Clusterin BQs. Purified Clusterin WT-RG or Clusterin BQs-RG was mixed with or without recombinant Luc in serum-free medium and preincubated at 42°C for 20 min. HEK293 cells were cultured in the medium for 14 h at 37°C and analyzed by flow cytometry. The bar graph shows the relative fluorescence intensities (GFP, green left axis; RFP, red right axis) in a cell normalized to those in nontreated cells (n = 4). The data are presented as the mean ± SEM.
Figure S4.
The conserved basic amino acids in Clusterin for secretion and internalization. (A) Some basic residues in Clusterin are required for secretion. HEK293 cells transiently transfected with the indicated CluRG vectors were cultured for 3 d, and conditioned medium and cell lysates were analyzed by immunoblotting. Clusterin mutants colored in blue showed reduced cleavage efficiency and secretion. Note that the expression of CluRG by the cytomegalovirus (CMV) promoter increased pre-Clusterin expression in the cell due to overexpression. (B) The indicated Clusterin-RG mutants were mixed with recombinant Luc in serum-free medium and incubated at 42°C for 20 min. HEK293 cells were cultured in the mixtures for 14 h at 37°C and analyzed by flow cytometry. The bar graph shows the relative RFP intensity in a cell normalized to that of Clusterin WT-RG (n = 3). The data are presented as the mean ± SEM. The combination of the R282Q, R286Q, R289Q, K123Q, R127Q, R130Q, and R138Q mutations (BQs) most significantly reduced Clusterin internalization.
The Clusterin–HS pathway is a versatile degradation system for a wide variety of substrates
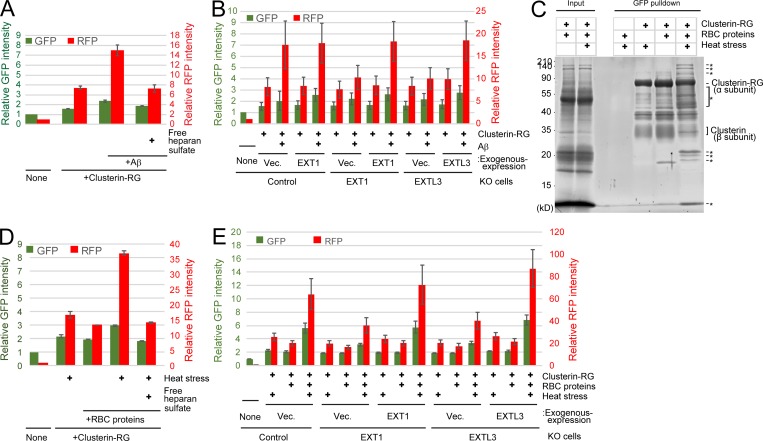
To analyze Clusterin–HS-mediated degradation of potentially physiological substrates, we focused on the Alzheimer’s disease–associated peptide Aβ. CluRG was preincubated with Aβ in serum-free medium and then incubated with cells with or without free HS. CluRG uptake was increased by the presence of Aβ but inhibited by free HS (Fig. 6 A). Furthermore, EXT1 or EXTL3 KO cells exhibited markedly decreased internalization of the CluRG–Aβ complex (Fig. 6 B). These data are consistent with the observation that HS proteoglycans colocalize with amyloid deposits (Snow et al., 1988) and that the injection of Clusterin into the rat brain prevents Aβ-induced neuronal degeneration in vivo (Cascella et al., 2013).
Figure 6.
The Clusterin–HS pathway is a universal degradation system for a wide variety of substrates. (A) Clusterin–HS pathway–dependent Aβ degradation. Purified CluRG was mixed with or without recombinant Aβ in serum-free medium and preincubated at 37°C. HEK293 cells were cultured in medium with or without free HS for 14 h at 37°C and analyzed by flow cytometry. (B) Medium was prepared as in A. Control, EXT1 KO, or EXTL3 KO HEK293 cells (generated by CRISPR) introduced with EXT1, EXTL3, or empty vector (Vec.) were cultured in medium for 14 h at 37°C and analyzed by flow cytometry. The bar graph shows the relative fluorescence intensities (GFP, green left axis; RFP, red right axis) in the cells normalized to that in nontreated cells (n = 3). The data are presented as mean ± SEM. (C) Coimmunoprecipitation of CluRG with various RBC proteins. Purified CluRG was mixed with or without RBC proteins and preincubated at 4°C or 50°C for 60 min (heat stress). Samples were subjected to a pulldown assay using an anti-GFP sepharose antibody at 4°C, separated by SDS-PAGE, and detected with SYPRO-Ruby stain. The asterisks indicate heat stress–dependent Clusterin-binding proteins. (D and E) HS is essential for internalization of Clusterin–RBC protein complexes. Purified CluRG was mixed with or without RBC proteins in serum-free medium and preincubated at 50°C for 60 min (heat stress). Control (D), EXT1 KO, or EXTL3 KO (E) HEK293 cells introduced with EXT1, EXTL3, or empty vector were cultured in medium with or without free HS for 14 h at 37°C and analyzed by flow cytometry. The bar graph shows the relative fluorescence intensity (GFP, green left axis; RFP, red right axis) in the cells normalized to that in nontreated cells (n = 3). The data are presented as mean ± SEM.
We further searched ubiquitous substrates for the Clusterin–HS pathway. Intravascular hemolysis is the destruction of RBCs in the circulation with the release of intracellular proteins including hemoglobin into the plasma. Approximately 10–20% of normal RBC destruction is intravascular (Quigeley et al., 2014), and increased hemolysis due to thermal injury, infectious agents, chemicals, venoms, or autoantibodies leads to pathophysiologies including hemolytic anemia. Haptoglobin, a specific chaperone for hemoglobin, captures the released hemoglobin from damaged RBCs, and the hemoglobin–haptoglobin complex is internalized into the macrophage (Kristiansen et al., 2001). Although adaptive responses to released hemoglobin are well characterized, the other intracellular proteins released from damaged RBCs are not understood (Kato, 2009). If the Clusterin–HS pathway is a general degradation pathway for various misfolded proteins, Clusterin is expected to bind to stressed RBC proteins. To test this hypothesis, CluRG was mixed with RBC proteins and then subjected to heat stress. A variety of RBC proteins were pulled down with CluRG under heat stress (Fig. 6 C), suggesting that Clusterin interacts with various misfolded cytoplasmic proteins. A CluRG internalization assay was performed using the CluRG–RBC protein mixture. The RFP signal in the cells increased in the presence of heat-stressed RBC proteins (Fig. 6 D). Importantly, free HS completely suppressed the increase by competitive inhibition (Fig. 6 D). Furthermore, EXT1 or EXTL3 is required for the internalization of CluRG with stressed RBC proteins (Fig. 6 E). Taken together, these results demonstrate that the Clusterin–HS pathway is a general protein quality control system for misfolded extracellular proteins.
Discussion
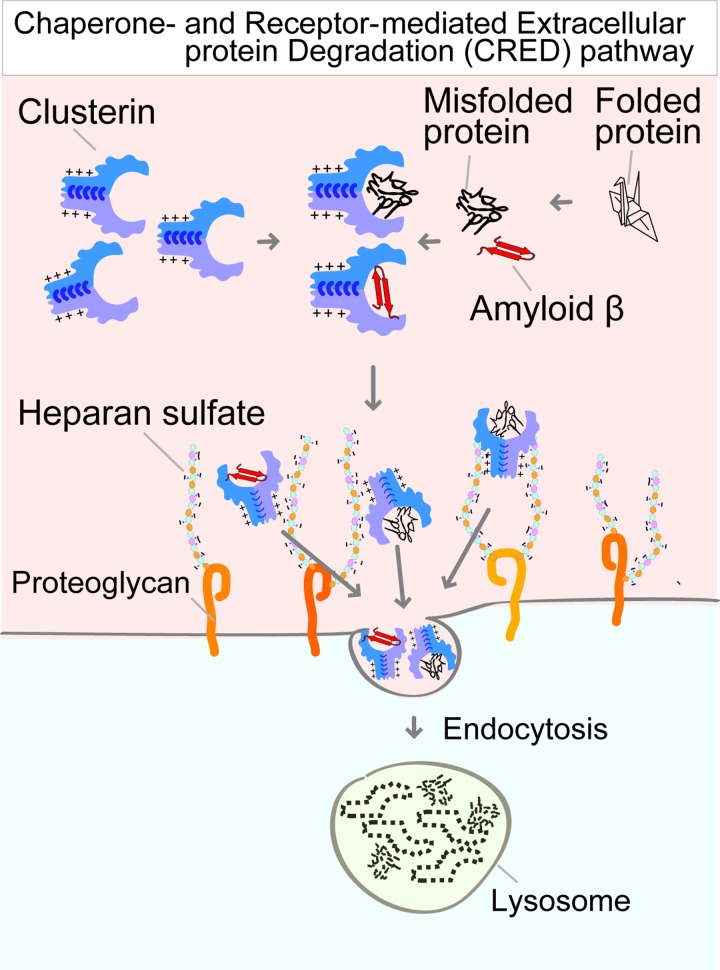
The results of this study have led to the identification of the receptor of an uncharacterized pathway for CRED (Fig. 7). More than 10 yr ago, Wilson and colleagues hypothesized that extracellular chaperones protect stressed proteins and might lead degradation via an unknown cell-surface receptor (Wyatt et al., 2013; Yerbury et al., 2005). The receptor candidate was LRP2, which has been shown to interact with Clusterin (Hammad et al., 1997; Kounnas et al., 1995). However, several studies have shown that LRP2–Clusterin is involved in leptin signaling and the efflux of Aβ across the blood–brain barrier (Byun et al., 2014; Nelson et al., 2017; Wojtas et al., 2017), suggesting that LRP2–Clusterin plays other physiological roles but does not function in the degradation of Clusterin. Indeed, CRISPR-mediated LRP2 KO cells did not display any defect in the internalization of the Clusterin-misfolded protein complexes (Fig. S2). We combined a novel quantitative fluorescence assay and genome-wide screening and identified the essential role of HS synthesis enzymes in the CRED pathway. The Clusterin-misfolded protein complex directly interacted with HS. The interaction is dependent on electrostatic interactions between positively charged residues on Clusterin and negatively charged sulfate on HS. Free HS competitively inhibits the internalization of the Clusterin complex. Our mechanistic analyses of Clusterin and HS delineate a working framework for the principal step in the CRED pathway and suggest that Clusterin with HS maintains extracellular protein homeostasis via the degradation of aberrant extracellular proteins.
Figure 7.
Model of the CRED pathway. Stresses induce the generation of extracellular aberrant proteins, which selectively interact with Clusterin. Positively charged residues on Clusterin electrostatically bind to the negatively charged HS chain on proteoglycan. The Clusterin–protein complex on the HS chain is then delivered into the lysosome via endocytosis and is digested into amino acids.
Previous studies indicated that Clusterin inhibits aggregations of stressed proteins, including glutathione S-transferase, catalase, BSA, citrate synthase, fibrinogen, and ovotransferrin, after heating at 45 to ∼60°C in the presence or absence of DTT (Humphreys et al., 1999; Poon et al., 2000; Wyatt et al., 2009), whereas the binding region in the substrates has not been elucidated. In this study, we used Luc as a heat-sensitive substrate (Gupta et al., 2011; Schröder et al., 1993). Luc is known to have hydrophobic domains that are exposed by heat stress at 42°C and targeted by intracellular chaperones (Rodrigo-Brenni et al., 2014). We demonstrated that Clusterin directly bound to misfolded Luc (Fig. 1 B). Aβ also contains a short hydrophobic region. In addition, Clusterin interacted with heat-stressed RBC proteins (Fig. 6 C), indicating that Clusterin might recognize a substrate with an exposed hydrophobic domain.
We elucidated the direct binding between Clusterin and HS by electrostatic interactions (Fig. 5), which dominate the interaction of HS-binding proteins with HS (Xu and Esko, 2014). The binding residues in Clusterin are conserved among vertebrates (Fig. 5 A), indicating that the degradation of extracellular proteins via the CRED pathway is evolutionarily conserved. Comparative analyses of several HS-binding proteins defined two HS-binding motifs, XBBXBX and XBBBXXBX (B represents a basic residue, and X represents any other residue; Cardin and Weintraub, 1989). Basic residues in Clusterin are critical for HS binding (Fig. 5), whereas the HS-binding residues in Clusterin did not correlate with the binding motifs. Indeed, HS-binding sites in many proteins do not include binding motifs (Billings and Pacifici, 2015). In addition, hydrogen bonding and Van der Waals interactions also contribute to HS binding in some cases (Xu and Esko, 2014). As topology defines most HS-binding sites, the structural analysis of Clusterin will clearly identify the set of binding resides in Clusterin. In addition, apolipoprotein E is a chaperone for Aβ and also known to interact with HS (Ji et al., 1993), implying that HS might be a common receptor for extracellular chaperones.
Extracellular proteases, which may also function in protein degradation, mainly cleave substrates for activation or inactivation rather than complete degradation (Overall and Blobel, 2007; Saido and Leissring, 2012). In contrast, Clusterin delivers substrates into lysosomes, where they are degraded and recycled as amino acids. Although the degradation of not only substrates but also Clusterin in the CRED pathway is apparently an inefficient system, Clusterin, which is a chaperone lacking ATPase activity, cannot refold and release a substrate by conformational change. Furthermore, endosomal recycling (for example, transferrin) is dependent on specific binding motifs that harbor histidine residues as a pH-inducible switch responsible for iron release in an acidic environment (Eckenroth et al., 2011). In contrast, substrate release from Clusterin presumably requires complex processes, since binding patterns between Clusterin and substrates are different for each misfolded protein and may mainly involve hydrophobic interactions. Therefore, the codegradation of substrate with Clusterin, resulting in the recycling of amino acids, is sufficiently efficient.
The codegradation of Clusterin–substrate complexes is analogous to the mechanism used by another extracellular chaperone, haptoglobin, which specifically binds to plasma hemoglobin for lysosomal degradation via CD163 scavenger receptor–mediated internalization in macrophages (Kristiansen et al., 2001). In contrast to the substrate-specific haptoglobin system, the CRED pathway is conserved in all human tissues tested (Fig. 5 E) and targets a wide variety of misfolded proteins derived from RBCs (Fig. 6). This is consistent with the fact that Clusterin KO mice develop autoimmune myocarditis via myocardial myosin injection (McLaughlin et al., 2000) and immune-complex deposition in glomeruli (Rosenberg et al., 2002). In addition, intracellular proteins released from ruptured RBCs are subjected to heat stress, oxidative stress, shear stress, and acidosis during circulation, indicating that the CRED pathway is important for degrading damaged or unwanted endogenous proteins in body fluids.
Clusterin interacts with oligomeric Aβ to form stable complexes (Narayan et al., 2011), and several mutations in the Clusterin gene have been identified as a genetic risk association of Alzheimer’s disease (Bettens et al., 2012), while the analysis of Clusterin KO mice in a mouse model of Alzheimer’s disease is complex. The loss of Clusterin in mice was shown to reduce amyloid plaque formation and neuritic toxicity (DeMattos et al., 2002), and further studies revealed that Clusterin KO mice accumulate Aβ plaques in the cerebrovasculature instead of the cortex and hippocampus (Wojtas et al., 2017), suggesting that Clusterin may physiologically play a role in the transport of Aβ across the blood–brain barrier rather than in protein degradation in the brain (Boland et al., 2018; Nelson et al., 2017). Our results demonstrated that the CRED pathway internalizes the Clusterin–Aβ complex (Fig. 6). However, the internalization efficiency might not be high compared with that of the Clusterin–Luc complex in our assay (Figs. 1 and 6). The development of a modified Clusterin mutant with an enhanced ability to degrade Aβ (e.g., an increased binding affinity to Aβ and HS) would be beneficial for the treatment of Alzheimer’s disease.
The CRED pathway exhibited selectivity for Clusterin complexes over free Clusterin (Fig. 1), whereas the binding affinity between Clusterin and HS, at least in vitro, was not affected by the presence or absence of misfolded Luc (not depicted). It is possible that an unidentified coreceptor regulates receptor-mediated endocytosis of the Clusterin complexes, analogous to other HS-binding growth factors and viruses that rely on coreceptors (Liu and Thorp, 2002; Pillay et al., 2016; Yayon et al., 1991). Defining the mechanism underlying the selective internalization of Clusterin complexes is therefore an important future goal.
Thus, given the widespread expression of HS-containing proteoglycans and Clusterin, as well as extensive substrates, the CRED pathway is a general extracellular protein quality control system responsible for clearance of body fluids. Our results provide insights into the basic molecular mechanism underlying extracellular proteostasis and provide new avenues for the possible treatment or prevention of diseases associated with aberrant extracellular proteins such as neurodegenerative diseases and autoimmune disease.
Materials and methods
Cell culture
HEK293FT, Flp-in T-Rex HEK293, HeLa, A549, U2OS, Huh-7, and HCT116 cells were cultured in DMEM supplemented with 10% FBS and 50 µg/ml penicillin/streptomycin in a humidified 5% CO2 incubator. Flp-in T-Rex HEK293 cells were maintained in the presence of 100 µg/ml zeocin and 15 µg/ml blasticidin. To generate stably doxycycline (dox)-inducible Clusterin-RG–, signal peptide (SP)-RG–, or Cas9-expressing cells, pcDNA5 FRT TO Clusterin-RG, pcDNA5 FRT TO SP-RG, or pcDNA5 FRT TO FLAG-Cas9, respectively, was cotransfected with pOG44, encoding the FLP recombinase, into Flp-in T-Rex HEK293 cells, and positive integrants were selected by resistance to 100 µg/ml hygromycin. Stable cells were maintained in the presence of 15 µg/ml blasticidin and 100 µg/ml hygromycin. Dox at 100 ng/ml was used for induction of the integrated gene at the FRT site.
Plasmids
To generate the C-terminal RFP-GFP-His-tagged Clusterin α subunit (Clusterin-RG/CluRG), full-length human Clusterin cDNA, which encodes a precursor polypeptide that is internally cleaved into the β and α subunits in the ER, was amplified by PCR from total cDNA of HEK293 cells. RFP/mCherry, GFP/sfGFP, and SP were amplified by PCR from plasmids encoding mCherry, sfGFP-His, and bovine preprolactin, respectively. The PCR products were cloned into the pcDNA5 FRT TO vector to generate pcDNA5 FRT TO Clusterin-RG-His, pcDNA5 FRT TO Clusterin-GFP-His, or pcDNA5 FRT TO SP-RG. Clusterin BQ mutants were derived by multisite-directed mutagenesis using Gibson assembly. The pcDNA5 FRT TO FLAG-Cas9 vector was previously described (Itakura et al., 2016). To generate pcDNA5-FRT-TO neo EXT1 or EXTL3, EXT1 and EXTL3 were amplified by PCR from total cDNA of HEK293 cells. The PCR products were cloned into the pcDNA5-FRT-TO neo vector (Addgene; 41000).
Antibodies
Rabbit polyclonal anti-LAMP1 antibodies were a gift from Y. Tanaka (Kyushu University, Fukuoka, Japan). Rabbit polyclonal anti-Luc (cat. no. PM016) and mouse monoclonal anti-RFP (cat. no. M204-3) antibodies were purchased from MBL. Mouse monoclonal anti-GFP (clone mFX75, cat. no. 012–22541) and mouse monoclonal anti-β-actin (clone 2F3, cat. no. 013–24553) antibodies were purchased from Wako. GFP-nanobody Sepharose was prepared by conjugating GFP-nanobody protein purified from pOPINE Nanobody (Addgene; 49172) to N-hydroxysuccinimide (NHS)–activated Sepharose 4 Fast Flow (GE).
Production of secreted Clusterin-RG and SP-RG
Purification of secreted Clusterin-RG and SP-RG was performed as previously described (Itakura et al., 2017), with modification. 80%-confluent Flip-in T-Rex HEK293 cells stably expressing inducible Clusterin-RG or SP-RG in 100-mm dishes were washed with PBS and cultured in 10 ml of advanced DMEM/F12 (Thermo Fisher Scientific) without FBS in the presence of 100 ng/ml dox for 4 d without changing the culture medium. Harvested conditioned medium was centrifuged at 2,000 g for 5 min, and the supernatants were passed through a Ni-NTA agarose column (Wako). The column was washed four times with PBS containing 10 mM imidazole and eluted with PBS containing 200 mM imidazole. Imidazole in the eluted fraction was removed by ultrafiltration using a Nanosep 3K (PALL). The 100 ml of conditioned medium yielded 0.5 mg of purified protein.
Clusterin internalization assay
Purified Clusterin-RG or SP-RG was mixed in serum-free advanced DMEM/F12 at a concentration of 0.06 µM (Clusterin medium). For heat stress, Clusterin medium with or without 2.2 µM recombinant Luc (Promega) was incubated at 42°C for 20 min. For heat stress of RBC proteins, Clusterin medium with or without 10 µg/ml RBC proteins was incubated at 50°C for 60 min. Subconfluent cell cultures in 12-well plates were washed in serum-free DMEM to remove bovine Clusterin derived from FBS and were then cultured in 500 µl of Clusterin medium with or without 0.1 µM BafA (LC Laboratories), 80 µg/ml free HS (Iduron), or 80 µg/ml free desulfated HS (Iduron) for 14 h. Cells were trypsinized with EDTA and recovered by detachment from the dish for analysis by flow cytometry or immunoblotting.
Aβ internalization assay
Binding of Clusterin with Aβ was performed as previously described, with modifications (Hammad et al., 1997). Purified Clusterin-RG (0.1 µM) in serum-free advanced DMEM/F12 with or without 20 µg/ml Aβ (Aβ-Protein Human, 1-40, Peptide Institute, cat. no. 4307-v) was incubated at 37°C for 2 d. The mixtures were incubated with cells as described above.
Clusterin-RG and Luc binding assay
Purified 15 nM Clusterin-RG was mixed in DMEM with or without 2.6 nM recombinant Luc and incubated at 4°C or 42°C for 20 min. The mixtures were incubated with 10 µl of GFP-nanobody sepharoses for 4 h at 4°C. The sepharoses were then washed four times with cold PBS and transferred to fresh tubes before elution with SDS sample buffer.
Albumin internalization assay
To generate fluorescently labeled albumin (albumin-red), 10 mg of BSA (fatty acid-free; Wako) was mixed with 0.58 mg of ATTO 565-NHS-Ester (ATTO-TEC) and incubated for 1 h. The reaction was terminated by separation of the albumin from unreacted dye by Sephadex G-25 chromatography in PBS. For the endocytosis assay, cells washed with PBS were incubated with 2 µg/ml albumin-red in advanced DMEM/F12 for 14 h and then analyzed by flow cytometry.
Flow cytometry
Trypsinized cells were passed through a 70-µm cell strainer and resuspended in 5% newborn calf serum and 1 µg/ml DAPI in PBS for flow cytometric analysis using a CytoFLEX S flow cytometer equipped with NUV 375-nm (DAPI), 488-nm (GFP), and 561-nm (mCherry) lasers (Beckman Coulter). Dead cells were detected by DAPI staining. In each sample, 10,000 cells were acquired.
Immunoblotting
Cells were washed with cold PBS and lysed in lysis buffer (1% Triton X-100, 50 mM Tris/HCl, pH 7.5, 1 mM EDTA, and 150 mM NaCl) supplemented with protease inhibitor cocktail (EDTA-free; Nacalai Tesque) and 1 mM PMSF for 15 min at 4°C. Lysates were clarified by centrifugation at 20,630 g for 5 min, and 6× SDS sample buffer was added. Samples were boiled at 95°C for 5 min before SDS-PAGE. 20 µg of protein per lane was separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Millipore). Immunoblot analysis was performed with the indicated antibodies, and the immunoreactive proteins were visualized using an ImmunoStar Zeta (Wako).
Lentiviral infection
Stable cell lines were generated using a lentiviral expression system. HEK293FT cells were transiently cotransfected with lentiviral vector, psPAX2, and pCMV VSV-G using PEI MAX reagent (Polysciences). 4 h after transfection, the medium was replaced with fresh culture medium. After culture for 72 h, the growth medium containing lentivirus was collected, followed by centrifugation at 2,000 rpm for 5 min to pellet cell debris. For CRISPR screening, the virus-containing medium was frozen in aliquots at −80°C. Cells were incubated with the collected virus-containing medium supplemented with 10 mg/ml polybrene for 48 h. Uninfected cells were removed using 1 µg/ml puromycin (InvivoGen) or 5 µg/ml blasticidin S (Wako).
Genome-wide CRISPR screen
The Genome-scale CRISPR KO (GeCKO) v2 human library was purchased from Addgene and amplified according to the protocol provided by the Zhang laboratory (Shalem et al., 2014; https://www.addgene.org/pooled-library/zhang-human-gecko-v2/). A genome-wide screen was performed as described previously (Joung et al., 2017). Lentivirus was first tested to achieve an MOI of 0.3 in Flp-in T-Rex HEK293 cells. A total of 4.0 × 107 HEK293 cells stably expressing Cas9 (in ten 10-cm dishes) were transduced with GeCKO v2 library B. Cells were treated with puromycin (Sigma-Aldrich) 24 h after transduction and maintained with 100 ng/ml dox (for Cas9 induction) for 7 d. Subsequently, 4.0 × 107 cells were collected to obtain input genomic DNA for analysis. The remaining cells were split for the Clusterin internalization assay. The cells transduced with the GeCKO library were cultured in Clusterin-Luc medium for 14 h and then trypsinized and resuspended in DMEM containing 10% FBS without phenol red. Cell sorting was performed using a Cell Sorter SH800 (Sony). We sorted the cells in the bottom 5% of the RFP/GFP ratio (Fig. 3 A). In the screening, we prepared 10 individual 10-cm dishes with 8.0 × 106 cells per dish, for a total of 8.0 × 107 cells. The sorted cells were cultured for 10 d and then subjected to a second sort in an identical manner. Following the second sort, the cells were expanded to 4.0 × 107 cells and harvested, and genomic DNA was extracted using a Blood Genomic DNA Extraction Mini Kit (Favorgen).
Library preparation for next-generation sequencing was performed as described previously (Joung et al., 2017). Briefly, sgRNA inserts were PCR amplified from 130 µg of genomic DNA using KAPA HiFi DNA Polymerase (Kapa Biosystems). The resulting PCR products (from unsorted cells and sorted cells) were purified and sequenced on a NEXTseq 500 instrument (Illumina) with a single-end 75-bp run (DNAFORM). Data analysis was performed using Model-based Analysis of Genome-wide CRISPR/Cas9 KO (MAGeCK; Li et al., 2014).
Generation of KO cells using CRISPR
Each sgRNA oligo was designed as described previously (Shalem et al., 2014; Fig. S5) and cloned into lentiGuide-puro (Addgene; 52963). HEK293 cells stably expressing FLAG-Cas9 were infected with lentivirus harboring sgRNA at an MOI of 0.5. Puromycin was added to cells after 24 h of transduction, and cells were maintained with 100 ng/ml dox for 7 d. After culture for >7 d, cells were used as a KO cell line.
Figure S5.
sgRNA sequences used in this study.
Establishment of EXT1 or EXTL3 KO-rescue cells
EXT1 or EXTL3 KO cells were generated by lentiGuide-puro gEXT1-1 or gEXTL3-1, respectively, as described above. The FLAG-Cas9 sequence at the FRT site of the KO cells was retargeted with either the empty pcDNA5-FRT-TO neo vector or pcENA5-FRT-TO neo containing EXT1 or EXTL3 (no tag). After G418 selection, the cell line transfected with the empty vector was used as the KO cell line, while the cell lines harboring EXT1 or EXTL3 were used as the rescue cell lines. For experiments, these cell lines were used under dox treatment. Note that exogenous C-terminal HA-tagged EXT1-HA and EXTL3-HA did not restore the reduction in Clusterin incorporation into EXT1 or EXTL3 KO cells, respectively (not depicted).
Preparation of HS-conjugated Sepharose
1 mg of HS (Iduron; GAG-HS01) was conjugated to a 0.5-ml bed volume of NHS-activated Sepharose 4 Fast Flow. After washing with blocking buffer (50 mM Tris-HCl, pH 7.5, and 1 M NaCl) and glycine buffer (0.1 M glycine, pH 2.3), the HS-conjugated Sepharose was stored in storage buffer (20 mM Tris-HCl, pH 7.5, and 0.15 M NaCl) at 4°C. Control Sepharose was prepared in the same way except for the absence of HS.
HS binding assay
Purified 0.06 µM Clusterin-RG was mixed in PBS with or without 2.2 µM recombinant Luc and incubated at 42°C for 20 min (heat stress). The 0.5-ml Clusterin mixtures were incubated with 15 µl of Sepharose (either control or HS-conjugated Sepharose) for 4 h at 4°C. The Sepharose samples were then washed four times with cold PBS and transferred to fresh tubes, after which they were eluted in SDS sample buffer.
Immunofluorescence
Cells were plated on coverslips and fixed in 3.7% formaldehyde in PBS for 15 min. For immunostaining, fixed cells were permeabilized with 50 µg/ml digitonin in PBS for 5 min, blocked with 10% newborn calf serum in PBS for 30 min, and incubated with primary antibodies for 1 h. After washing, cells were incubated with Alexa Fluor 647–conjugated goat anti-rabbit IgG secondary antibodies (Thermo Fisher Scientific) for 1 h. The stained cells were observed under a confocal laser microscope (FV1000 IX81; Olympus) using a 100× oil-immersion objective lens with NA of 1.40. The images were acquired using FV10-ASW 2.1 imaging software.
Preparation of RBC proteins
Blood samples were collected from C57BL/6 mice in EDTA tubes and centrifuged at 1,000 g for 2 min to separate plasma and RBCs. RBCs were washed with PBS, resuspended in an equal volume of homogenization buffer (20 mM Hepes, pH 7.4, 1 mM EDTA, 1 mM PMSF, and protease inhibitor cocktail), and disrupted using a narrow-gauge syringe. The homogenized cells were centrifuged at 541,000 g (Hitachi S110AT) for 30 min to remove cell debris. The concentration of RBC total proteins was determined by the Bradford method and was typically 50–100 mg/ml.
Immunoprecipitation of CluRG with cytosol
CluRG (1 µg/ml final concentration) was mixed with 30 µg/ml RBC proteins (final concentration) in 20 mM Hepes, pH 7.4, buffer. To induce heat stress, the mixture was incubated at 50°C for 60 min and then centrifuged at 20,000 g for 10 min to remove debris. GFP-nanobody sepharoses (15 µl) were added to the mixture and incubated for 1 h at 4°C. The sepharoses were washed five times with PBS and transferred to fresh tubes before elution with SDS sample buffer.
Online supplemental material
Fig. S1 shows the competitive inhibition of Clusterin internalization. Fig. S2 shows that LRP2 is not involved in the internalization of the Clusterin–misfolded protein complex. Fig. S3 shows that HS synthesis enzymes are not required for endocytosis. Fig. S4 shows the conserved basic amino acids in Clusterin for secretion and internalization. Fig. S5 shows sgRNA sequences used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Yoshitaka Tanaka for anti-LAMP1 antibodies, Atsushi Iwama (University of Tokyo, Tokyo, Japan), and Yaeko Nakajima-Takagi (University of Tokyo) for cell sorting support; and Ramanujan S. Hegde (Medical Research Council Laboratory of Molecular Biology, Cambridge, UK) for the writing of this manuscript.
This work was supported by KAKENHI (grants 19K22413, 16H06167, and 16H01194 to E. Itakura), the Japan Foundation for Applied Enzymology (to E. Itakura), the Naito Foundation (to E. Itakura), the Heiwa Nakajima Foundation (to E. Itakura), the Takeda Science Foundation (to E. Itakura), and the Uehara Memorial Foundation (to E. Itakura).
The authors declare no competing financial interests.
Author contributions: E. Itakura and M. Chiba performed the experiments. E. Itakura designed the overall study, interpreted the data, and wrote the manuscript. T. Murata contributed to cell sorting and the writing. A. Matsuura contributed to the writing and data interpretation. All authors discussed the results and approved the manuscript.
References
- Bell R.D., Sagare A.P., Friedman A.E., Bedi G.S., Holtzman D.M., Deane R., and Zlokovic B.V.. 2007. Transport pathways for clearance of human Alzheimer’s amyloid beta-peptide and apolipoproteins E and J in the mouse central nervous system. J. Cereb. Blood Flow Metab. 27:909–918. 10.1038/sj.jcbfm.9600419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettens K., Brouwers N., Engelborghs S., Lambert J.C., Rogaeva E., Vandenberghe R., Le Bastard N., Pasquier F., Vermeulen S., Van Dongen J., et al. 2012. Both common variations and rare non-synonymous substitutions and small insertion/deletions in CLU are associated with increased Alzheimer risk. Mol. Neurodegener. 7:3 10.1186/1750-1326-7-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billings P.C., and Pacifici M.. 2015. Interactions of signaling proteins, growth factors and other proteins with heparan sulfate: mechanisms and mysteries. Connect. Tissue Res. 56:272–280. 10.3109/03008207.2015.1045066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland B., Yu W.H., Corti O., Mollereau B., Henriques A., Bezard E., Pastores G.M., Rubinsztein D.C., Nixon R.A., Duchen M.R., et al. 2018. Promoting the clearance of neurotoxic proteins in neurodegenerative disorders of ageing. Nat. Rev. Drug Discov. 17:660–688. 10.1038/nrd.2018.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun K., Gil S.Y., Namkoong C., Youn B.S., Huang H., Shin M.S., Kang G.M., Kim H.K., Lee B., Kim Y.B., and Kim M.S.. 2014. Clusterin/ApoJ enhances central leptin signaling through Lrp2-mediated endocytosis. EMBO Rep. 15:801–808. 10.15252/embr.201338317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardin A.D., and Weintraub H.J.. 1989. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis. 9:21–32. 10.1161/01.ATV.9.1.21 [DOI] [PubMed] [Google Scholar]
- Cascella R., Conti S., Tatini F., Evangelisti E., Scartabelli T., Casamenti F., Wilson M.R., Chiti F., and Cecchi C.. 2013. Extracellular chaperones prevent Aβ42-induced toxicity in rat brains. Biochim. Biophys. Acta. 1832:1217–1226. 10.1016/j.bbadis.2013.04.012 [DOI] [PubMed] [Google Scholar]
- Ciechanover A., and Kwon Y.T.. 2017. Protein Quality Control by Molecular Chaperones in Neurodegeneration. Front. Neurosci. 11:185 10.3389/fnins.2017.00185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMattos R.B., O’dell M.A., Parsadanian M., Taylor J.W., Harmony J.A., Bales K.R., Paul S.M., Aronow B.J., and Holtzman D.M.. 2002. Clusterin promotes amyloid plaque formation and is critical for neuritic toxicity in a mouse model of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 99:10843–10848. 10.1073/pnas.162228299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I., and Elazar Z.. 2018. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 19:349–364. 10.1038/s41580-018-0003-4 [DOI] [PubMed] [Google Scholar]
- Eckenroth B.E., Steere A.N., Chasteen N.D., Everse S.J., and Mason A.B.. 2011. How the binding of human transferrin primes the transferrin receptor potentiating iron release at endosomal pH. Proc. Natl. Acad. Sci. USA. 108:13089–13094. 10.1073/pnas.1105786108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatica D., Lahiri V., and Klionsky D.J.. 2018. Cargo recognition and degradation by selective autophagy. Nat. Cell Biol. 20:233–242. 10.1038/s41556-018-0037-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover J.R., and Lindquist S.. 1998. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 94:73–82. 10.1016/S0092-8674(00)81223-4 [DOI] [PubMed] [Google Scholar]
- Gupta R., Kasturi P., Bracher A., Loew C., Zheng M., Villella A., Garza D., Hartl F.U., and Raychaudhuri S.. 2011. Firefly luciferase mutants as sensors of proteome stress. Nat. Methods. 8:879–884. 10.1038/nmeth.1697 [DOI] [PubMed] [Google Scholar]
- Hammad S.M., Ranganathan S., Loukinova E., Twal W.O., and Argraves W.S.. 1997. Interaction of apolipoprotein J-amyloid beta-peptide complex with low density lipoprotein receptor-related protein-2/megalin. A mechanism to prevent pathological accumulation of amyloid beta-peptide. J. Biol. Chem. 272:18644–18649. 10.1074/jbc.272.30.18644 [DOI] [PubMed] [Google Scholar]
- Humphreys D.T., Carver J.A., Easterbrook-Smith S.B., and Wilson M.R.. 1999. Clusterin has chaperone-like activity similar to that of small heat shock proteins. J. Biol. Chem. 274:6875–6881. 10.1074/jbc.274.11.6875 [DOI] [PubMed] [Google Scholar]
- Hung S.Y., and Fu W.M.. 2017. Drug candidates in clinical trials for Alzheimer’s disease. J. Biomed. Sci. 24:47 10.1186/s12929-017-0355-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Chen C., and de Bono M.. 2017. Purification of FLAG-tagged Secreted Proteins from Mammalian Cells. Bio Protoc. 7:e2430 10.21769/BioProtoc.2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Kishi-Itakura C., and Mizushima N.. 2012. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 151:1256–1269. 10.1016/j.cell.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Itakura E., Zavodszky E., Shao S., Wohlever M.L., Keenan R.J., and Hegde R.S.. 2016. Ubiquilins Chaperone and Triage Mitochondrial Membrane Proteins for Degradation. Mol. Cell. 63:21–33. 10.1016/j.molcel.2016.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z.S., Brecht W.J., Miranda R.D., Hussain M.M., Innerarity T.L., and Mahley R.W.. 1993. Role of heparan sulfate proteoglycans in the binding and uptake of apolipoprotein E-enriched remnant lipoproteins by cultured cells. J. Biol. Chem. 268:10160–10167. [PubMed] [Google Scholar]
- Joung J., Konermann S., Gootenberg J.S., Abudayyeh O.O., Platt R.J., Brigham M.D., Sanjana N.E., and Zhang F.. 2017. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc. 12:828–863. 10.1038/nprot.2017.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.S., Kurti A., Wojtas A., Baker K.E., Liu C.C., Kanekiyo T., Deming Y., Cruchaga C., Estus S., Bu G., and Fryer J.D.. 2016. Identification of plexin A4 as a novel clusterin receptor links two Alzheimer’s disease risk genes. Hum. Mol. Genet. 25:3467–3475. 10.1093/hmg/ddw188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama H., Yamamoto A., Mizushima N., Yoshimori T., and Miyawaki A.. 2008. GFP-like proteins stably accumulate in lysosomes. Cell Struct. Funct. 33:1–12. 10.1247/csf.07011 [DOI] [PubMed] [Google Scholar]
- Kato G.J. 2009. Haptoglobin halts hemoglobin’s havoc. J. Clin. Invest. 119:2140–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S., and Cuervo A.M.. 2015. Proteostasis and aging. Nat. Med. 21:1406–1415. 10.1038/nm.4001 [DOI] [PubMed] [Google Scholar]
- Kimura S., Noda T., and Yoshimori T.. 2007. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 3:452–460. 10.4161/auto.4451 [DOI] [PubMed] [Google Scholar]
- Klaips C.L., Jayaraj G.G., and Hartl F.U.. 2018. Pathways of cellular proteostasis in aging and disease. J. Cell Biol. 217:51–63. 10.1083/jcb.201709072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kounnas M.Z., Loukinova E.B., Stefansson S., Harmony J.A., Brewer B.H., Strickland D.K., and Argraves W.S.. 1995. Identification of glycoprotein 330 as an endocytic receptor for apolipoprotein J/clusterin. J. Biol. Chem. 270:13070–13075. 10.1074/jbc.270.22.13070 [DOI] [PubMed] [Google Scholar]
- Kristiansen M., Graversen J.H., Jacobsen C., Sonne O., Hoffman H.J., Law S.K., and Moestrup S.K.. 2001. Identification of the haemoglobin scavenger receptor. Nature. 409:198–201. 10.1038/35051594 [DOI] [PubMed] [Google Scholar]
- Kwon Y.T., and Ciechanover A.. 2017. The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends Biochem. Sci. 42:873–886. 10.1016/j.tibs.2017.09.002 [DOI] [PubMed] [Google Scholar]
- Lai A.C., and Crews C.M.. 2017. Induced protein degradation: an emerging drug discovery paradigm. Nat. Rev. Drug Discov. 16:101–114. 10.1038/nrd.2016.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., and Kroemer G.. 2019. Biological Functions of Autophagy Genes: A Disease Perspective. Cell. 176:11–42. 10.1016/j.cell.2018.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Xu H., Xiao T., Cong L., Love M.I., Zhang F., Irizarry R.A., Liu J.S., Brown M., and Liu X.S.. 2014. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15:554 10.1186/s13059-014-0554-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., and Thorp S.C.. 2002. Cell surface heparan sulfate and its roles in assisting viral infections. Med. Res. Rev. 22:1–25. 10.1002/med.1026 [DOI] [PubMed] [Google Scholar]
- Lundgren S., Carling T., Hjälm G., Juhlin C., Rastad J., Pihlgren U., Rask L., Akerström G., and Hellman P.. 1997. Tissue distribution of human gp330/megalin, a putative Ca(2+)-sensing protein. J. Histochem. Cytochem. 45:383–392. 10.1177/002215549704500306 [DOI] [PubMed] [Google Scholar]
- Matsubara E., Soto C., Governale S., Frangione B., and Ghiso J.. 1996. Apolipoprotein J and Alzheimer’s amyloid beta solubility. Biochem. J. 316:671–679. 10.1042/bj3160671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin L., Zhu G., Mistry M., Ley-Ebert C., Stuart W.D., Florio C.J., Groen P.A., Witt S.A., Kimball T.R., Witte D.P., et al. 2000. Apolipoprotein J/clusterin limits the severity of murine autoimmune myocarditis. J. Clin. Invest. 106:1105–1113. 10.1172/JCI9037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy B.F., Kirszbaum L., Walker I.D., and d’Apice A.J.. 1988. SP-40,40, a newly identified normal human serum protein found in the SC5b-9 complex of complement and in the immune deposits in glomerulonephritis. J. Clin. Invest. 81:1858–1864. 10.1172/JCI113531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan P., Orte A., Clarke R.W., Bolognesi B., Hook S., Ganzinger K.A., Meehan S., Wilson M.R., Dobson C.M., and Klenerman D.. 2011. The extracellular chaperone clusterin sequesters oligomeric forms of the amyloid-β(1-40) peptide. Nat. Struct. Mol. Biol. 19:79–83. 10.1038/nsmb.2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson A.R., Sagare A.P., and Zlokovic B.V.. 2017. Role of clusterin in the brain vascular clearance of amyloid-β. Proc. Natl. Acad. Sci. USA. 114:8681–8682. 10.1073/pnas.1711357114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overall C.M., and Blobel C.P.. 2007. In search of partners: linking extracellular proteases to substrates. Nat. Rev. Mol. Cell Biol. 8:245–257. 10.1038/nrm2120 [DOI] [PubMed] [Google Scholar]
- Pillay S., Meyer N.L., Puschnik A.S., Davulcu O., Diep J., Ishikawa Y., Jae L.T., Wosen J.E., Nagamine C.M., Chapman M.S., and Carette J.E.. 2016. An essential receptor for adeno-associated virus infection. Nature. 530:108–112. 10.1038/nature16465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontén F., Gry M., Fagerberg L., Lundberg E., Asplund A., Berglund L., Oksvold P., Björling E., Hober S., Kampf C., et al. 2009. A global view of protein expression in human cells, tissues, and organs. Mol. Syst. Biol. 5:337 10.1038/msb.2009.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon S., Easterbrook-Smith S.B., Rybchyn M.S., Carver J.A., and Wilson M.R.. 2000. Clusterin is an ATP-independent chaperone with very broad substrate specificity that stabilizes stressed proteins in a folding-competent state. Biochemistry. 39:15953–15960. 10.1021/bi002189x [DOI] [PubMed] [Google Scholar]
- Powers E.T., Morimoto R.I., Dillin A., Kelly J.W., and Balch W.E.. 2009. Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 78:959–991. 10.1146/annurev.biochem.052308.114844 [DOI] [PubMed] [Google Scholar]
- Price J.C., Guan S., Burlingame A., Prusiner S.B., and Ghaemmaghami S.. 2010. Analysis of proteome dynamics in the mouse brain. Proc. Natl. Acad. Sci. USA. 107:14508–14513. 10.1073/pnas.1006551107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigeley J.G., Means R.T., and Glader B.. 2014. The birth, life, and death of red blood cells: erythropoiesis, the mature red blood cell, and cell destruction. In Wintrobe’s Clinical Hematology. 14th edition Wloters Kluwer Health, Alphen aan den Rijn, Netherlands. [Google Scholar]
- Rodrigo-Brenni M.C., Gutierrez E., and Hegde R.S.. 2014. Cytosolic quality control of mislocalized proteins requires RNF126 recruitment to Bag6. Mol. Cell. 55:227–237. 10.1016/j.molcel.2014.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M.E., Girton R., Finkel D., Chmielewski D., Barrie A. III, Witte D.P., Zhu G., Bissler J.J., Harmony J.A., and Aronow B.J.. 2002. Apolipoprotein J/clusterin prevents a progressive glomerulopathy of aging. Mol. Cell. Biol. 22:1893–1902. 10.1128/MCB.22.6.1893-1902.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saido T., and Leissring M.A.. 2012. Proteolytic degradation of amyloid β-protein. Cold Spring Harb. Perspect. Med. 2:a006379 10.1101/cshperspect.a006379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrazin S., Lamanna W.C., and Esko J.D.. 2011. Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 3:a004952 10.1101/cshperspect.a004952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder H., Langer T., Hartl F.U., and Bukau B.. 1993. DnaK, DnaJ and GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage. EMBO J. 12:4137–4144. 10.1002/j.1460-2075.1993.tb06097.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G., and Zhang F.. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 343:84–87. 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sica V., Galluzzi L., Bravo-San Pedro J.M., Izzo V., Maiuri M.C., and Kroemer G.. 2015. Organelle-Specific Initiation of Autophagy. Mol. Cell. 59:522–539. 10.1016/j.molcel.2015.07.021 [DOI] [PubMed] [Google Scholar]
- Snow A.D., Mar H., Nochlin D., Kimata K., Kato M., Suzuki S., Hassell J., and Wight T.N.. 1988. The presence of heparan sulfate proteoglycans in the neuritic plaques and congophilic angiopathy in Alzheimer’s disease. Am. J. Pathol. 133:456–463. [PMC free article] [PubMed] [Google Scholar]
- Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. 2015. Proteomics. Tissue-based map of the human proteome. Science. 347:1260419 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- Walter P., and Ron D.. 2011. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 334:1081–1086. 10.1126/science.1209038 [DOI] [PubMed] [Google Scholar]
- Wojtas A.M., Kang S.S., Olley B.M., Gatherer M., Shinohara M., Lozano P.A., Liu C.C., Kurti A., Baker K.E., Dickson D.W., et al. 2017. Loss of clusterin shifts amyloid deposition to the cerebrovasculature via disruption of perivascular drainage pathways. Proc. Natl. Acad. Sci. USA. 114:E6962–E6971. 10.1073/pnas.1701137114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff S., Weissman J.S., and Dillin A.. 2014. Differential scales of protein quality control. Cell. 157:52–64. 10.1016/j.cell.2014.03.007 [DOI] [PubMed] [Google Scholar]
- Wyatt A.R., Yerbury J.J., and Wilson M.R.. 2009. Structural characterization of clusterin-chaperone client protein complexes. J. Biol. Chem. 284:21920–21927. 10.1074/jbc.M109.033688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt A.R., Yerbury J.J., Ecroyd H., and Wilson M.R.. 2013. Extracellular chaperones and proteostasis. Annu. Rev. Biochem. 82:295–322. 10.1146/annurev-biochem-072711-163904 [DOI] [PubMed] [Google Scholar]
- Xu D., and Esko J.D.. 2014. Demystifying heparan sulfate-protein interactions. Annu. Rev. Biochem. 83:129–157. 10.1146/annurev-biochem-060713-035314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yayon A., Klagsbrun M., Esko J.D., Leder P., and Ornitz D.M.. 1991. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell. 64:841–848. 10.1016/0092-8674(91)90512-W [DOI] [PubMed] [Google Scholar]
- Yerbury J.J., Stewart E.M., Wyatt A.R., and Wilson M.R.. 2005. Quality control of protein folding in extracellular space. EMBO Rep. 6:1131–1136. 10.1038/sj.embor.7400586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlokovic B.V., Martel C.L., Matsubara E., McComb J.G., Zheng G., McCluskey R.T., Frangione B., and Ghiso J.. 1996. Glycoprotein 330/megalin: probable role in receptor-mediated transport of apolipoprotein J alone and in a complex with Alzheimer disease amyloid beta at the blood-brain and blood-cerebrospinal fluid barriers. Proc. Natl. Acad. Sci. USA. 93:4229–4234. 10.1073/pnas.93.9.4229 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.