Mast et al. review peroxisome biogenesis, interorganellar contacts, and the connections and shared molecular players acting in the formation of peroxisomes and lipid droplets at the ER.
Abstract
Peroxisomes play a central role in human health and have biochemical properties that promote their use in many biotechnology settings. With a primary role in lipid metabolism, peroxisomes share a niche with lipid droplets within the endomembrane-secretory system. Notably, factors in the ER required for the biogenesis of peroxisomes also impact the formation of lipid droplets. The dynamic interface between peroxisomes and lipid droplets, and also between these organelles and the ER and mitochondria, controls their metabolic flux and their dynamics. Here, we review our understanding of peroxisome biogenesis to propose and reframe models for understanding how peroxisomes are formed in cells. To more fully understand the roles of peroxisomes and to take advantage of their many properties that may prove useful in novel therapeutics or biotechnology applications, we recast mechanisms controlling peroxisome biogenesis in a framework that integrates inference from these models with experimental data.
Introduction
Peroxisomes are a nexus for harnessing the production and decomposition of hydrogen peroxide to metabolize fats and neutralize harmful molecules in cells. A typical human cell has 102–103 peroxisomes distributed throughout its cytoplasm, where they form contacts with other organelles, particularly the ER, lipid droplets, and mitochondria. Peroxisomes are typically spheroid, enclosed by a single lipid bilayer, and densely filled with enzymes for their varied metabolic roles (Lazarow and Fujiki, 1985; Smith and Aitchison, 2013). Substrates are delivered to the peroxisome by dedicated transport machineries, and evidence suggests that dynamic organelle contacts play an important role in the regulation of metabolite transfer and in the regulation of peroxisome biogenesis, growth, division, and turnover (Platta and Erdmann, 2007; Schrader et al., 2013; Shai et al., 2016).
Peroxisomes are essential for human vitality, health, and longevity. Mutations in the PEX genes required for peroxisome biogenesis result in frequently fatal genetic disorders known as the Zellweger spectrum of peroxisome biogenesis disorders (Lazarow and Moser, 1994; Wanders and Waterham, 2006; Braverman et al., 2016; Waterham et al., 2016). Defects in all facets of peroxisome biogenesis, from initial assembly to matrix protein import, and division, motility, and turnover of peroxisomes, impair efficient peroxisomal metabolism, especially of very long chain fatty acids (≥26 C; Wanders and Waterham, 2006; Wanders and Ferdinandusse, 2012; Waterham et al., 2016). The buildup of these fatty acids and other peroxisomal metabolites, including branched chain fatty acids, aberrantly esterified fatty acids, the ether lipid precursors of plasmalogens, and the steroid acid precursors of bile, is toxic (Wanders and Waterham, 2006). Defects in peroxisome biogenesis result in elevated amounts of cellular reactive oxygen species with wide-ranging pleiotropic effects (Fransen and Lismont, 2019; Lismont et al., 2019). Loss of peroxisomal integrity leads to additional cellular complications such as the mistargeting of peroxisomal proteins to other organelles, particularly mitochondria, impairing their functions (Goldfischer et al., 1973; Salpietro et al., 2015). Accordingly, the penetrance of any given peroxisome biogenesis disorder phenotype for any single mutant allele is diverse, and partial or complete peroxisome dysfunction has been linked to many additional human health concerns, including inherited neuropathologies, aging, cancer, heart disease, obesity, and diabetes (Espeel et al., 1995; Motley et al., 1996; Singh, 1997; Périchon et al., 1998; Song, 2002; Depreter et al., 2003; Klouwer et al., 2015; Braverman et al., 2016; Wangler et al., 2018).
In addition to their metabolic roles, peroxisomes serve as hubs for cellular signaling (Mast et al., 2015; Tripathi and Walker, 2016). Peroxisomal signaling and metabolism are likely integrated. For example, it has been suggested that peroxisome-localized tuberous sclerosis complex senses peroxisomal reactive oxygen species and in response, negatively regulates mammalian target of rapamycin complex 1 to promote autophagy and improve cell survival (Zhang et al., 2013). Similarly, peroxisome-localized mitochondrial antiviral signaling protein initiates a distinctive type-III interferon transcriptional response impacting innate immunity (Dixit et al., 2010; Odendall et al., 2014). We are just beginning to appreciate how the combination of peroxisomal signaling and metabolism contributes to a variety of processes in development and cellular immunity (Ferreira et al., 2016; Cook et al., 2019; Di Cara et al., 2019).
Several features of peroxisomes are attractive from a biotechnology perspective, especially with respect to their distinctive matrix protein import pathway (Cross et al., 2017). Unlike all other protein transporters in eukaryotic cells, peroxisomes import fully folded and even oligomeric protein complexes with bound cofactors into their lumen (Léon et al., 2006; Girzalsky et al., 2010). Efficient targeting and import into peroxisomes require only the presence of a simple tripeptide motif, e.g., Ser-Lys-Leu, located at the extreme C terminus of a protein (Gould et al., 1987, 1989). The ability to harness these import features would be of immense benefit to synthetic biology applications in which novel cocompartmentalization of disparate enzymes would support the creation of new, industrially useful metabolic pathways. An additional second peroxisomal targeting sequence can be attached to the N terminus of a protein to add an additional layer of regulation into the synthetic design of these metabolic networks (Swinkels et al., 1991). Such synthetic metabolic networks could be used in the production of drugs, flavor molecules, and other exotic compounds currently extracted using environmentally questionable practices (Gidijala et al., 2009; Stehlik et al., 2014; Hanko et al., 2018; Gao and Zhou, 2019).
In this review, we discuss exciting new insight into the initial steps required to form import-competent peroxisomes. Increasingly, it is clear that these steps are fundamental to all aspects of peroxisome biology, impacting metabolic function, matrix protein import, and peroxisome dynamics, including division, motility, inheritance, and turnover. Recent evidence suggests that the regulation of peroxisomal dynamics and function is also controlled by contact between peroxisomes and organelles, including the ER, mitochondria, and lipid droplets. In the case of lipid droplets, not only is contact with peroxisomes important but also features shared between peroxisomes and lipid droplets for membrane protein import and their sites of formation on the ER have been revealed.
The architecture of peroxisome biogenesis
Peroxisome proliferation and turnover influence their number and size, which can change dramatically and rapidly in response to stimuli or stressors. Following cytomegalovirus infection, human fibroblasts triple their number of peroxisomes within 96 h (Jean Beltran et al., 2018), while increases in peroxisome number and size are observed in rodent hepatocytes following treatment with xenobiotic peroxisome proliferators (i.e., substances foreign to the biological system; Hess et al., 1965). In the methylotrophic yeast Komagataella pastoris, formerly named Pichia pastoris, methanol dramatically increases the size of peroxisomes until they dominate the cytosolic volume (Gould et al., 1992); yet when this yeast metabolizes fatty acids, it is primarily the numbers of peroxisomes that are increased (Gould et al., 1992; Yan et al., 2008). To account for this plasticity, the cell must integrate and coordinate peroxisome growth and expansion of its membrane with processes controlling peroxisome abundance.
The number of any given organelle in a cell is controlled by biogenic and degradative processes. These include the following: de novo synthesis from component parts made elsewhere in the cell; fission of existing organelles to produce two or more smaller organelle progeny; homotypic fusion of two or more individual organelles, which reduces organelle copy number and increases organelle size; heterotypic fusion, whereby one organelle receives vesicles originating from another different organelle to facilitate the transfer of cargo but which also influences organelle size; maturation, where biochemical changes at the organelle convert it from one form to another; organelle segregation during cell division, a dilutive process that, when regulated, is known as organelle inheritance; and turnover, which is either by general and nonspecific, or by targeted and selective, autophagy (Nunnari and Walter, 1996; Warren and Wickner, 1996; Marshall, 2016). Many of the qualities that make each organelle distinct can be attributed to the unique combination of these processes underlying its steady-state distribution in the cell.
Peroxisomes do not synthesize their own biogenic material; proteins are posttranslationally incorporated into the peroxisome, and the synthesis of membrane lipids does not occur locally at the peroxisome. The lipid composition of the mammalian peroxisome is similar to that of the ER, being rich in phosphatidylcholine and phosphatidylethanolamine, and lacking in cardiolipin (Fujiki et al., 1982; Hardeman et al., 1990). In addition to vesicle fusion, peroxisomes acquire membrane through nonvesicular transport mechanisms. Contacts and nonvesicular membrane exchange between the ER and peroxisomes have been reported, although the process of nonvesicular membrane exchange between these two organelles appears to be highly inefficient under the experimental conditions explored (Raychaudhuri and Prinz, 2008). Such transfer mechanisms are likely critical for the efficient transfer of lipid metabolites to or from peroxisomes and for coordinating different aspects of peroxisome dynamics, as is discussed below.
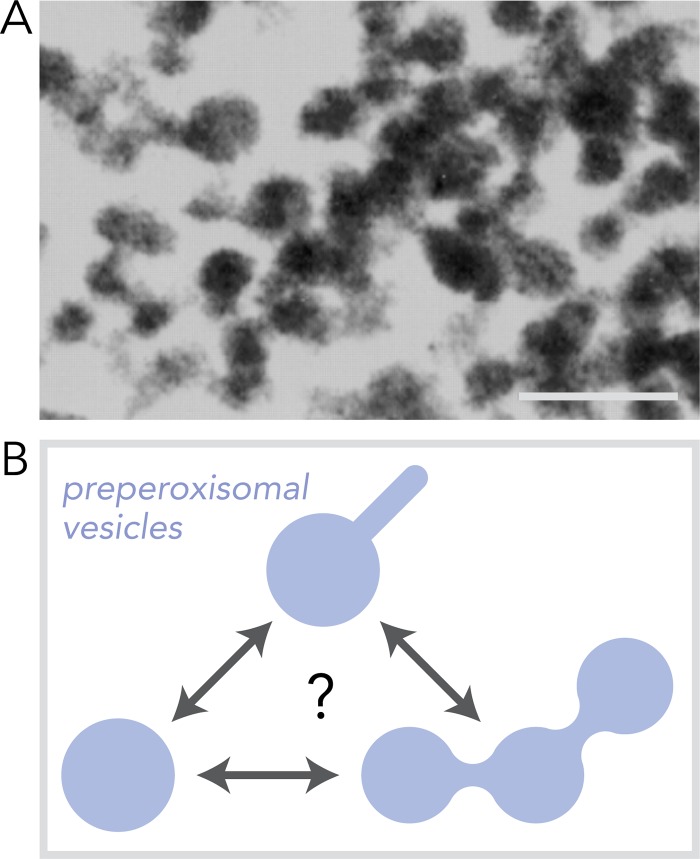
Peroxisomes can be made by the templated replication of existing peroxisomes (Lazarow and Fujiki, 1985; Motley and Hettema, 2007; Menendez-Benito et al., 2013). A single peroxisome becomes elongated, and its membrane and matrix constituents are apportioned between the two or more resulting “daughter” peroxisomes. To sustain replication, the peroxisome grows with the import of new matrix proteins and by the expansion of the peroxisomal membrane that is replenished by heterotypic fusion of preperoxisomal vesicles (PPVs; Hoepfner et al., 2005; Titorenko and Mullen, 2006). PPVs were first observed in now classic experiments that enriched for distinct populations of these vesicles by isopycnic density gradient subcellular fractionation (Titorenko et al., 2000). Pulse labeling of newly synthesized proteins with [35S]methionine enabled tracking of the formation of these PPVs and their fusion with and maturation into functional peroxisomes (Titorenko and Rachubinski, 1998; Titorenko et al., 2000). Isolated PPVs have diverse morphologies with asymmetric spherical profiles ∼80 nm in diameter connected to, and by, ∼20-nm-diameter tubules (Titorenko et al., 2000). These morphological features are suggestive of an asymmetric distribution of membrane protein and lipid constituents within the PPV that partition and segregate to these different membrane regions (Fig. 1).
Figure 1.
PPVs. (A) An electron micrograph of PPVs isolated from Y. lipolytica. Scale bar, 200 nm. (B) A schematic of preperoxisomal vesicles revealing the different morphologies observed within and between different PPVs. The different diameters of the membranes are proposed to reflect the asymmetric distribution of different classes of peroxisomal membrane proteins.
What are PPVs?
At least two populations of PPVs have been described and shown to bud from the ER during de novo peroxisome biogenesis. Enrichment of these two PPV populations by subcellular fractionation in the dimorphic yeast Yarrowia lipolytica confirmed the ER origin of both populations as they contained N-linked core-glycosylated peroxins Pex2 and Pex16 (Titorenko and Rachubinski, 1998; Titorenko et al., 2000). In Saccharomyces cerevisiae, Pex13 and Pex14 travel in vesicles separate from Pex10 and Pex12 from the ER to peroxisomes (van der Zand et al., 2012). In K. pastoris, these two populations of PPVs were observed by fluorescence microscopy and also produced by an in vitro budding assay (Agrawal et al., 2016). Protein segregation within the two PPV classes is thought to prevent premature assembly of peroxisome components, e.g., the matrix protein importomer and metabolite transporters, in the ER (van der Zand et al., 2012). Evidence suggests additional PPV subtypes also exist (Knoops et al., 2014; Wróblewska et al., 2017).
The mechanism of PPV formation has been partially dissected through use of an in vitro budding assay that recapitulates aspects of peroxisomal membrane protein (PMP) loading into nascent PPVs and budding of these PPVs from the ER (Lam et al., 2010; Agrawal et al., 2011, 2016; Mast et al., 2016, 2018). Analogous to the formation of COPII vesicles on the ER, cytosolic factors are recruited to the ER membrane surface, and energy consumption is required to sustain production of PPVs (Lam et al., 2010). However, unlike for COPII vesicle formation, PPV production uses ATP and not GTP hydrolysis, and PPVs are not coated.
While peroxisomes receive and fuse with PPVs, it is thought that mature peroxisomes do not fuse with each other. Assays that readily revealed mitochondrial fusion gave negative results for peroxisomes (Motley and Hettema, 2007; Motley et al., 2008). Still, pulse-chase experiments demonstrated that peroxisomes are a composite of old and new proteins, including membrane proteins (Motley and Hettema, 2007; Menendez-Benito et al., 2013). This finding reinforces the view that peroxisomes are formed by templated replication of the organelle. Because peroxisomes acquire protein content posttranslationally, this finding also suggests that PMPs must come from somewhere else, including such processes as vesicular transport or a dedicated posttranslational membrane protein import pathway. In the absence of peroxisomes, new peroxisomes can be formed by the same PPVs that support growth and division (Motley and Hettema, 2007; Motley et al., 2015; Mayerhofer et al., 2016). The fusogenic machinery for PPVs with themselves and with peroxisomes has not yet been identified. The asymmetric morphology of PPVs compared with the spherical morphology of peroxisomes may offer clues as to the sorts of proteins involved in PPV fusion.
The process of de novo peroxisome biogenesis from PPV precursors is comparatively inefficient, taking ∼4 h in yeast, and is typically not observed in cells containing functional peroxisomes (Hoepfner et al., 2005; Motley and Hettema, 2007; Mast et al., 2016, 2018). In human cells, de novo–formed peroxisomes have been observed by visualizing Pex16 tagged with photoactivatable GFP; new peroxisomes arising not only from fission but also from de novo biogenesis were detected in cells 24 h after transfection with the photoactivatable Pex16-GFP construct (Kim et al., 2006). De novo biogenesis has also been visualized in yeast cells deficient in peroxisome inheritance (Chang et al., 2009; Munck et al., 2009). Complementation of mutations that cause defects in peroxisome biogenesis can also reestablish the peroxisome compartment de novo in both mammalian and yeast cells (Brul et al., 1988; Hoepfner et al., 2005; Aranovich et al., 2014).
A one-way endosomal sorting complex required for transport (ESCRT) out of the ER
We recently discovered a novel role for ESCRT-III in PPV biogenesis (Mast et al., 2018). ESCRT-III, composed of Vps20, Snf7, Vps24, Did4, Ist1, Vps60, Did2, and Chm7 in yeast, is evolutionarily ancient and is a primary effector complex of membrane remodeling and vesicle/membrane scission in a host of contexts (Henne et al., 2011; Schöneberg et al., 2017; Vietri et al., 2020). Genetic screens had identified ESCRT-III mutants as being defective in peroxisome biogenesis (Smith et al., 2006; Saleem et al., 2010), but these findings were initially set aside because ESCRT-III mutants also exhibit growth defects on fermentable carbon sources (Smith et al., 2006). Furthermore, ESCRT-III deletion mutants also have import-competent peroxisomes, albeit at reduced numbers compared with the wild-type situation, and this fact was not initially reconciled with a role for ESCRT-III in peroxisome biogenesis (Saleem et al., 2010; Mast et al., 2018).
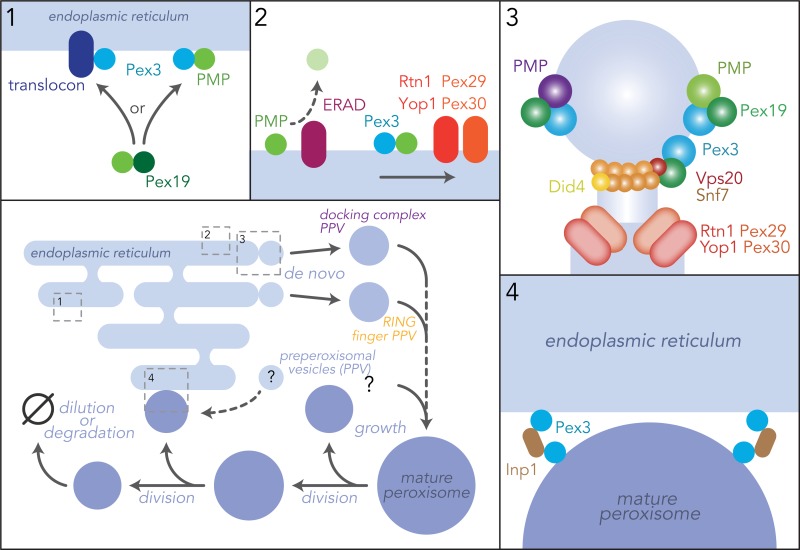
New quantitative phenotyping methodology for yeast cell fitness was necessary to separate the role of ESCRT-III in peroxisome biogenesis from its myriad other functions in cells (Mast et al., 2018). High-resolution, time-lapse microscopy of microcolony growth from populations of single yeast cells enabled a comparison across deletion strains of their standardized measures of cell growth parameters, and the fitness of any given mutant could be quantified as generally pleiotropic or condition specific (Herricks et al., 2017). In this analysis, ESCRT-III deletion strains showed severe growth phenotypes when exposed to a carbon source requiring peroxisome function over and above the general growth defects expected for such a widely engaged protein complex (Mast et al., 2018). De novo biogenesis was quantitatively assayed by controlled reintroduction of PEX19, an essential biogenesis factor. These studies, demonstrated a requirement for ESCRT-III components Vps20, Snf7, Did4, and Vps24 in the formation of functional peroxisomes from the ER (Mast et al., 2018). Genetic interactions between PEX19 and these ESCRT-III subunits positioned the function of ESCRT-III downstream of Pex19 in de novo peroxisome formation. Snf7 and Did4 dynamically localize to sites of de novo peroxisome biogenesis together with ER-localized Pex3, the master controller of de novo peroxisome biogenesis. Finally, in cell-free experiments, Vps20 and Snf7, with Pex19, were shown to be required for the release of Pex3-containing PPVs from the ER (Mast et al., 2018). Intriguingly, the energy requirement for PPV release was shown to be subsequent to the actual scission event, implicating its role in the recycling of scission components.
These findings suggest that ESCRT-III functions directly in the scission of PPVs by being corecruited with ER-localized peroxins acting in de novo peroxisome biogenesis. In this model, ESCRT-III would assemble at the PPV bud neck, polymerizing around its circumference to constrict the membrane for PPV scission (Fig. 2). Such a model is consistent with cryo-EM images of purified mammalian ESCRT-III components IST1 and CHMP1B, showing these proteins are capable of deforming and stabilizing membranes for normal-topology scission (McCullough et al., 2015). However, as this model is inconsistent with the canonical function of ESCRT-III in reverse-topology membrane scission (Schöneberg et al., 2017), other potential mechanisms of ESCRT-III function, acting directly or indirectly at the ER, may be at play in PPV budding, e.g., through an as-of-yet undiscovered novel topology for the canonical ESCRT-III proteins. Alternatively, the function of ESCRT-III could be indirect, e.g., by acting as an adaptor to recruit a different scission factor to the PPV exit site.
Figure 2.
A spatiotemporal model of peroxisome biogenesis and Pex3 function. Peroxisome proliferation occurs by two mechanisms: division of existing peroxisomes through fission and de novo formation from the ER. Peroxisomes are formed de novo from the ER through budding and then fusion of two classes of PPV (although there may be more). This mechanism separates RING finger and docking components of the import complex into different vesicles, which are not import competent until after fusion and assembly of a complete and functional import complex. Once mature, these peroxisomes can multiply by growth and incorporate proteins and membranes from the ER. These two arms of the biogenic cascade are mediated by Pex3, which has different functions depending on its location. (1) Pex3 facilitates PMP insertion into membranes, either with assistance of a membrane translocon, or, for a subset of membrane proteins, direct insertion into the membrane. (2) Pex3 chaperones PMPs, protecting them from quality control systems such as ER-associated protein degradation (ERAD), and assisting in their lateral sorting through the ER to sites of PPV formation. (3) Pex3 and Pex19 define sites of PPV budding and recruit the scission machinery, ESCRT-III, to release them from the ER. (4) Additional functions of Pex3, such as peroxisomal tethering to the ER, are mediated by its binding proteins, such as Inp1.
Several questions remain as to how ESCRT-III is recruited to the incipient PPV bud site. This process is likely regulated, as colocalization of Snf7 or Did4 with Pex3 was only increased under conditions favorable for the de novo biogenesis of peroxisomes (Mast et al., 2018). Recently, native Vps20, an ESCRT-III component, was shown to localize and enrich on ER membranes (Heinzle et al., 2019). Vps20 is myristoylated and required for the initiation of ESCRT-III polymerization (Henne et al., 2011; Schöneberg et al., 2017). Thus, Vps20 localization and/or myristoylation may play a role in the regulatory steps that initiate ESCRT-III recruitment to the budding PPV.
Deciphering the roles for peroxins in early peroxisome biogenesis
Peroxins are encoded by PEX genes and are responsible for the life cycle of the peroxisome (Distel et al., 1996). To date, 37 peroxins have been characterized. They are typically either peroxisomal membrane proteins or cytosolic chaperones, although a few are also residents of the ER (Farré et al., 2019). Any one given peroxin may have a myriad of molecular functions, presenting research conundrums (e.g., see Hettema et al., 2014; Costello and Schrader, 2018; Jansen and van der Klei, 2019). Of all the peroxins, Pex3 is most exemplary of this challenge.
Pex3 has been implicated in every step of the peroxisome biogenic cascade and is present in every organism where the presence of peroxisomes has been experimentally validated (Gabaldón et al., 2006; Schlüter et al., 2006; Mast et al., 2012). This situation was recently confirmed in the kinetoplastid parasite Trypanosoma brucei, where a highly divergent Pex3 was discovered and shown to be required for the biogenesis of peroxisomes, also known as glycosomes, in this organism (Banerjee et al., 2019; Kalel et al., 2019). Pex3 is critical for the biogenesis of the peroxisome, as cells lacking Pex3 lack any vestige of peroxisomes (Hettema et al., 2000; Shimozawa et al., 2000; Hoepfner et al., 2005). From processes of PMP synthesis and topogenesis in the ER (Schmidt et al., 2012) to the formation of PPVs (Mast et al., 2016, 2018), the stability of the peroxisome importomer (Baerends et al., 2000; Hettema et al., 2000; Wróblewska et al., 2017), peroxisome motility and inheritance (Chang et al., 2009; Munck et al., 2009), peroxisome–ER tethering (Knoblach et al., 2013), peroxisome–vacuole tethering (Wu et al., 2019), and peroxisome turnover (Motley et al., 2012), Pex3 seems to play a central role. So, what is Pex3 and what does it do?
Pex3 proteins are type-III transmembrane proteins (Fig. 3; Höhfeld et al., 1991). They possess a single transmembrane-spanning α helix near their N terminus that is anchored in the lipid bilayer by a stop-transfer sequence. The portion of Pex3 found on the lumenal side of the lipid bilayer contains a track of conserved basic amino acids that are important for its correct targeting to the ER (Baerends et al., 2000; Fakieh et al., 2013). Mutation of these basic residues results in mistargeting of Pex3 and its import into mitochondria (Fakieh et al., 2013). The portion of Pex3 found on the cytosolic side of the lipid bilayer, and which makes up most of the length of Pex3, is made up by a series of 10 α-helical segments that assemble into a helical bundle (Sato et al., 2010; Schmidt et al., 2010). Hydrophobic segments composed of amino acids from the first N-terminal α helix, located near the base of the cytosolic domain of Pex3, interact with the membrane and are important for Pex3 function (Schmidt et al., 2010; Chen et al., 2014).
Figure 3.
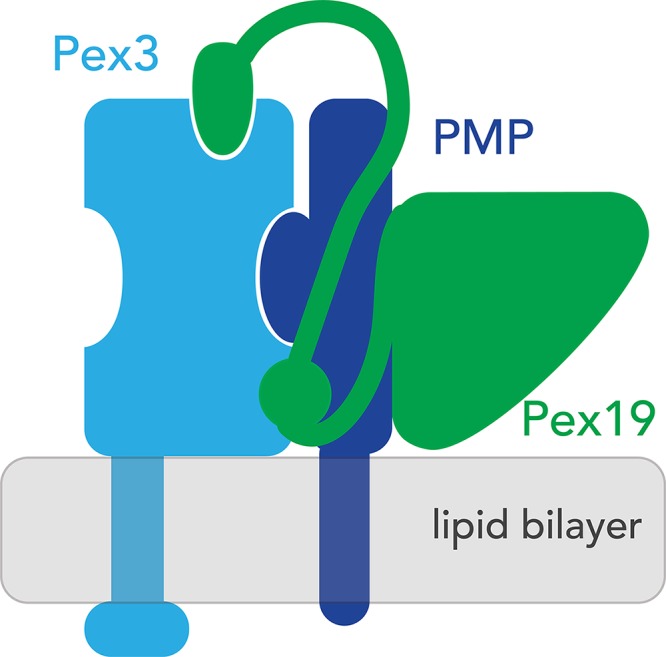
A model for Pex3 and Pex19 topology and chaperone function. Pex3 is a type-III transmembrane protein. It interacts with peroxisomal membrane proteins through binding sites located along the side of its cytosolic helical bundle. This interaction is initiated by Pex19, which first binds the PMP in the cytosol and then brings it into the proximity of Pex3.
The steady-state localization of Pex3 is at the peroxisome, and Pex3 serves as a marker of peroxisomes in cells (Hettema et al., 2000; Hoepfner et al., 2005). Under peroxisome-proliferating conditions, the levels of Pex3 slowly increase over time, but its transcription remains stable (Erdmann and Blobel, 1995; Smith et al., 2002; Knoblach and Rachubinski, 2010). Overexpression of Pex3 results in both general and organism-specific phenotypes. In S. cerevisiae, overexpressed Pex3 accumulates at the ER (Hoepfner et al., 2005; Breker et al., 2013), whereas in mammalian cells overexpressed Pex3 has been observed at the ER and mitochondria (Schmidt et al., 2012; Aranovich et al., 2014; Sugiura et al., 2017). In Y. lipolytica, overexpressed Pex3 proliferates and clusters peroxisomes and enhances class V myosin–directed transport of peroxisomes into growing daughter cells (Chang et al., 2009). In mammalian cells, the overexpression of Pex3 can also stimulate peroxisome turnover followed by the formation of new peroxisomes (Sugiura et al., 2017). Pex3 therefore marks peroxisomes and their sites of biogenesis. Without Pex3, peroxisomal membrane proteins are unstable and targeted for degradation; as a result, the peroxisomal compartment collapses and cannot be sustained (Ghaedi et al., 2000).
Pex3 is a critical receptor and partner for Pex19; together they chaperone most peroxisomal membrane proteins for insertion into the organelle (Fang et al., 2004; Matsuzaki and Fujiki, 2008). Pex19 surveils the cytosol for newly synthesized PMPs and binds to a hydrophobic membrane peroxisomal targeting sequence (mPTS) located near the transmembrane domain of most PMPs (Sacksteder et al., 2000; Jones et al., 2004). Sequence conservation of the mPTS is poor but has the general characteristic of a short hydrophobic α helix located next to a transmembrane segment (Rottensteiner et al., 2004). Lack of additional information to specify an mPTS likely stems from the fact that PMPs come in a variety of topologies from single-spanning to multispanning to C-terminally anchored membrane domains. Interestingly, Pex3 does not contain an mPTS and interacts with Pex19 through other mechanisms (Matsuzaki and Fujiki, 2008), as is discussed below.
Pex19 is composed of an unstructured N-terminal domain and a structured and globular C-terminal domain (Emmanouilidis et al., 2017). The N-terminal domain is highly flexible with unfolded disordered domains interspersed by five segments capable of forming amphipathic α helices (Schmidt et al., 2010; Chen et al., 2014). Almost all Pex19 proteins have a C-terminal farnesylation CAAX motif. When the motif is farnesylated, a hydrophobic pocket that forms part of the mPTS binding site incorporates the farnesyl moiety (Emmanouilidis et al., 2017). Farnesylation induces structural rearrangements within the C-terminal globular domain of Pex19, increasing its rigidity and enhancing its affinity for the mPTS of PMPs (Rucktäschel et al., 2009). These rearrangements have been proposed to function as an allosteric regulator of Pex19 binding of PMPs (Emmanouilidis et al., 2017). However, once Pex19 binds a PMP, it is not known how PMP release from Pex19 is mediated or coordinated with Pex19 function. Loss of the CAAX box impairs, but does not completely abolish, Pex19 function, as peroxisomes can still be formed in Pex19 CAAX mutants (Rucktäschel et al., 2009).
The interaction between Pex3 and Pex19 is dynamic. A high-affinity interaction between the first amphipathic segment of the N-terminal domain of Pex19 and the top of the helical bundle of Pex3 is essential for peroxisome biogenesis (Sato et al., 2008, 2010; Schmidt et al., 2010, 2012). A stretch of hydrophobic amino acids in the Pex3 interaction site of Pex19 induces and stabilizes the amphipathic segment of Pex19 to adopt a stable α-helical conformation (Schmidt et al., 2012). Pex19 can also associate with Pex3 through a second, lower affinity interaction that occurs between the fourth amphipathic helix of the N-terminal domain Pex19 and a groove located between two α helices near the base of the helical bundle of Pex3 (Chen et al., 2014). This two-step binding process reorients the C-terminal domain of Pex19 and its bound PMP cargo into close apposition with the hydrophobic base of Pex3 and the lipid bilayer to which Pex3 is anchored (Chen et al., 2014; Emmanouilidis et al., 2017). While this interaction is necessary for PMP topogenesis, it is hotly debated whether it is sufficient to orient the bound transmembrane domain of the PMP cargo protein for proper insertion into and translocation across the lipid bilayer. It remains difficult to understand how Pex3 and Pex19 function as a translocon in the classical sense, as these proteins lack any indication of the capability of forming a channel in membranes. Yet, it is conceivable that Pex3 and Pex19 could function as a membrane protein insertase for a subclass of membrane proteins either at the peroxisomal membrane (Liu et al., 2016), or at the ER, as discussed below. Data from systems-level experiments suggest that many PMPs are synthesized on ER-bound ribosomes (Jan et al., 2014; Kaewsapsak et al., 2017). Thus, while Pex3 and Pex19 are necessary for PMP integration into the membrane, whether they are sufficient for this process remains to be established. The sequential two-step amphipathic recruitment of Pex19 and the hydrophobic nature of the portion of Pex3 directly apposed to the lipid bilayer have been proposed to work together to overcome the energy barrier inherent in properly inserting a membrane protein (Chen et al., 2014).
Both Pex3 and Pex19 engage in promiscuous physical interactions with other PMPs. Pex3 engages with many PMPs through interactions mediated by grooves on the lateral sides of its helical bundle (Motley et al., 2012; Fakieh et al., 2013; Knoblach et al., 2013; Chen et al., 2014). Peripheral membrane proteins such as Inp1 and Atg36 bind to distinct surfaces on Pex3, and single point mutations in Inp1 and Atg36 that disrupt their binding to Pex3 ablate their functions in peroxisome inheritance and turnover, respectively (Munck et al., 2009; Motley et al., 2012; Knoblach et al., 2013). Likewise, the disordered N-terminal domain of Pex19 also interacts with other PMPs. In particular, the second amphipathic segment of Pex19 interacts with an FXXXF motif in Pex14 (Chen et al., 2014). Mutational analyses demonstrated that the separate functions of the N- and C-terminal domains of Pex19 can be, remarkably, reconstituted via bridging interactions between Pex3 and two other peroxins, Pex2 and Pex25 (Agrawal et al., 2017). This myriad of activities suggests a model in which Pex3 function is dependent on its location and local concentration (see text box).
A spatiotemporal model for Pex3-mediated peroxisome biogenesis
From the perspective of any given Pex3 molecule, we posit that its myriad functions can be delineated and accounted for by consideration of two parameters: concentration and location (Fig. 2). Thus, we propose that Pex3 first functions at its point of synthesis in the ER as a receptor for Pex19, assisting in the topogenesis of newly synthesized PMPs either directly or in association with other membrane protein insertion complexes including the Sec61 translocon, the Guided Entry of Tail–anchored proteins complex, or the ER membrane complex. Once a PMP has been inserted, Pex3 functions as a chaperone to traffic the PMP laterally within the ER to sites of PPV formation (Fakieh et al., 2013). The association of the PMP with Pex3 protects the PMP from quality control machinery such as ER-associated protein degradation at the ER and the ATAD/MSP1 complex at the peroxisome (Weir et al., 2017). This association is predicted to also prevent the assembly of a functional matrix protein translocation machinery at the ER through contact-dependent protein–protein inhibition. Finally, the PMP cargo, by its association with Pex3, is efficiently loaded into budding-competent PPVs. Fusion of distinct types of PPVs leads to rearrangement and assembly of the matrix protein importomer and the RING finger and retrotranslocation machinery associated with the translocon, liberating Pex3 to attract new binding partners for its additional roles at this new location. These include the role of Pex3 in peroxisome inheritance together with the peroxisome-anchoring protein Inp1 (Munck et al., 2009); its role in autophagy through interaction with Atg36 (Motley et al., 2012); and, e.g., its role in protecting some PMPs such as Pex15 from quality surveillance by the ATAD/MSP1 AAA-ATPase (Weir et al., 2017).
This model allows for several predictions, one being that both the global and local relative concentrations of Pex3 impact its function. Pex3 is not an abundant protein and, as described, is a chaperone for PMPs and a scaffold that nucleates peroxin and PMP complexes. Therefore, the numerous functions ascribed to Pex3 may result from its association with other peroxins. Altering Pex3 levels would alter the stoichiometry of the resulting complexes in a spatiotemporal manner. For example, overproduction of Pex3 would initially stimulate early steps in peroxisome biogenesis until the existing pool of PMPs in the ER is exhausted. Upon exhaustion of PMPs at the ER, excess Pex3 would become involved in other functions such as peroxisome motility via interaction with a class V myosin or degradation of peroxisomes via association with Atg36. Overproduction of Pex3 is known to lead to its mistargeting and the mistargeting of other peroxins to mitochondria, as the import and transport pathway for PMPs becomes overloaded (Aranovich et al., 2014). By comparison, an accumulation of Pex3 under conditions in which the levels of Pex3 and all of its binding partners increase stoichiometrically should lead to a more robust peroxisome biogenic response without accompanying pleiotropic effects.
A second prediction is that different pools of Pex3 in different membrane compartments associate with different binding proteins to affect the different spatiotemporal functions for Pex3. Different pools of Pex3 exist along a trafficking route that begins at the ER and ends at the peroxisome destined for degradation via autophagy (Aranovich et al., 2014; Mast et al., 2016; Mayerhofer et al., 2016). The function and stability of many PMPs depend on their physical association with Pex3 (Hettema et al., 2000; South et al., 2000; Weir et al., 2017). Thus, Pex3 at the ER fulfills a chaperone function by helping to laterally sort nascent PMPs from their site of synthesis or site of membrane insertion to sites of PPV egress (Fakieh et al., 2013). At the peroxisome, Pex3 protects PMPs; participates in additional processes, such as the recruitment of peroxisome motility proteins; and likely functions to assemble and organize higher order assemblies of peroxins involved in protein translocation, metabolite transfer, and import of a subclass of PMPs.
A third prediction is that mistargeting Pex3 to other membranes will result in an attempt by the cell to carry out the various processes of peroxisome biogenesis from that membrane. Peroxisome biogenesis “extra locum normalem” from the mitochondria, i.e., at a location where it does not normally occur, is initiated by forcing the direct targeting of Pex3 to the mitochondrial membrane (Rucktäschel et al., 2010). Pex3 is able to recruit Pex19, and even peripheral membrane proteins such as Inp1, when targeted to mitochondria (Rucktäschel et al., 2010; Knoblach et al., 2013). Pex3-laden vesicles have also been visualized budding from mammalian peroxisomes under conditions where Pex3 is overexpressed (Sugiura et al., 2017).
Many aspects of the pathways and functions of peroxins proposed in this model remain under-explored and in need of investigation. While we have specifically focused here on the roles of Pex3 to elaborate this spatiotemporal model of peroxisome biogenesis, additional peroxins are likely involved. For example, Pex16 and its homologues may also contribute to peroxisome biogenesis in a mechanism analogous and complementary to that of Pex3 in chaperoning, protecting, and reorganizing PMPs (Kim et al., 2006; Aranovich et al., 2014; Farré et al., 2017). Furthermore, roles for other peroxins in de novo biogenesis need to be further defined. For example, Pex11, and/or its orthologues, has been proposed to have a role in de novo peroxisome biogenesis in addition to its role in peroxisome division (Knoblach and Rachubinski, 2010; Huber et al., 2012; Chang et al., 2015).
This spatiotemporal model also highlights how care must be taken in interpreting data and peroxisome phenotypes arising from experiments that involve the manipulation of peroxin levels, in particular, even moderate overexpression of Pex3. This point is especially important when considering caveats for a purported role for mitochondria in peroxisome biogenesis. First, PMPs have only been observed on the outer mitochondrial membrane under conditions in which they are overexpressed or in which peroxisome biogenesis is defective due to mutation of one or more peroxins (Goldfischer et al., 1973; Aranovich et al., 2014; Sugiura et al., 2017). Second, the observed mitochondrial trafficking route of PMPs to peroxisomes is dispensable, as forcing overexpressed Pex3 into the ER by addition of an ER signal sequence to its N terminus still results in the complementation of the peroxisome biogenesis defects due to the absence of Pex3, and in the trafficking of Pex3 to peroxisomes (Toro et al., 2009; Aranovich et al., 2014; Sugiura et al., 2017). These observations demonstrate that the ER is necessary and sufficient for peroxisome biogenesis. However, as suggested by Sugiura et al. (2017), there may also be functional relevance for Pex3 or other peroxins at the mitochondrion in the mammalian system.
A nested coherent feedforward loop controls peroxisome proliferation
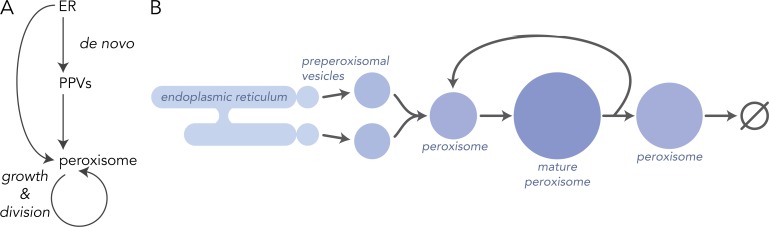
As previously proposed, the peroxisome biogenesis circuit resembles a coherent, nested, feed-forward network motif (Fig. 4; Mast and Aitchison, 2018). Network motifs define spatiotemporal relationships between cellular components that can be used for predicting the dynamics of their behavior. Here, the ER, PPVs, and peroxisomes are represented as the points, or nodes, of the network with the direction of flow for membrane and membrane proteins the connecting lines, or edges. The ER serves as source material for peroxisomal membrane and as a site for PMP insertion and maturation. PPVs that bud from the ER can form peroxisomes de novo or fuse with existing peroxisomes in the growth and division cycle of the organelle, thus the nested topology motif. As defined, an OR logic gate must control the switch between the de novo and the growth and division arms of the peroxisome biogenic pathway (Alon, 2006; Mast and Aitchison, 2018). However, the molecular players and mechanisms controlling this logic gate await discovery, although there are candidates (Saleem et al., 2008; Mast et al., 2016). In other settings, this simple network motif confers several distinctive features on the system. The first is the ability to rapidly control the proliferation response from “off/basal” to “on/induced” by increasing the production of peroxisome-destined membrane transport originating at the ER. Increased PPV production would increase peroxisome numbers by two mechanisms, either by enhancing the growth and division mechanism or by initiating the formation of peroxisomes de novo through fusion and maturation. Thus, the basal level of peroxisome biogenesis can be up-regulated rapidly in response to conditions requiring the organelle. The degree to which one pathway is used over the other would impact the heterogeneity of the protein content within the peroxisome population in cells. The second feature of the OR feed-forward network motif is robustness. In transcriptional networks, an OR input function serves as a filter by introducing a time delay to a noisy transcriptional induction signal supporting mRNA production in the absence of a signal for up to the equivalence of one cell division (Mangan and Alon, 2003). This rapid “on” and delayed “off” is consistent with experimental observations of peroxisome biogenesis (Saleem et al., 2008; Perry et al., 2009; Mast et al., 2016, 2018). To balance the combined de novo and division-based proliferative capacity of peroxisomes, peroxisome numbers are also controlled by dilutive and degradative processes (Fagarasanu et al., 2010; Motley et al., 2015).
Figure 4.
A nested feedforward network motif in peroxisome biogenesis. Network motifs describe the distinctive features of the relationship of cellular components that enable predictions of their behavior when stimulated or repressed. Here, the production of peroxisomes occurs not only by growth and division but also de novo biogenesis. Because the de novo biogenesis pathway feeds into the growth and division pathway, leading to the further proliferation of peroxisomes, this arrangement resembles a nested coherent feedforward network motif. See text for further details. (A) The nested feed-forward network motif regulating peroxisome biogenesis. (B) The biogenesis pathway of peroxisomes and their life cycle, spatially arranged to illustrate the topology of the nested feed-forward network motif.
Regulatory mechanisms of peroxisome formation and function: Control by interorganellar contacts
Membrane contacts between peroxisomes and other organelles have long been thought to function in the efficient interorganellar transport of metabolites. In a global analysis of the organelle interactome, spectral imaging of lysosomes, mitochondria, ER, peroxisomes, the Golgi, and lipid droplets revealed that peroxisomes associate most frequently with the ER, followed by mitochondria, lipid droplets, the Golgi, and lastly lysosomes (Valm et al., 2017).
Specific machineries responsible for facilitating the interactions between peroxisomes and other organelles involved in the β-oxidation of fatty acids have been discovered for peroxisome–lipid droplet contacts and peroxisome–mitochondrion contacts. Peroxisomal ABCD1 interacts with an isoform of the AAA-ATPase Spastin (M1-Spastin) to tether peroxisomes to lipid droplets and to recruit an ESCRT-III subcomplex composed of IST1 and CHMP1B (Chang et al., 2019). This contact site helps facilitate fatty acid transfer from the lipid droplet to the peroxisome and reduces the levels of peroxidated lipids in lipid droplets. In yeast, peroxisome–mitochondrion contact sites are mediated by either Fzo1 or Pex34 (Shai et al., 2018). Disruption of these contact sites by genetic perturbation reduced cellular fitness and mitochondrial ATP and CO2 production arising from fatty acid β-oxidation occurring in the peroxisome. Genome-wide screening using a split-Venus fluorescence protein tag strategy implicated additional peroxisomal and mitochondrial proteins in contact formation between these two organelles (Shai et al., 2018).
In addition to improving the metabolic flux between peroxisomes and other membrane-bound compartments, contact sites can also dynamically regulate peroxisome biogenesis. In mammalian cells, contacts between the peroxisome and ER are stabilized by peroxisome-localized ABCD5 and ER-localized VAP (Costello et al., 2017; Hua et al., 2017). One consequence of this interaction is contact-regulated transfer of phospholipids from the ER to the peroxisome that controls the expansion of the peroxisomal membrane and subsequent growth of peroxisomes (Costello et al., 2017). Thus, contact sites between the ER and peroxisomes can control the growth and division cycle of peroxisomes and the concentrations of peroxins on individual peroxisomes.
Membrane contacts can also directly impact peroxisome inheritance. Peroxisome anchoring in budding yeast ensures that the mother cell retains peroxisomes. Part of this anchoring mechanism is mediated by interaction between Pex3 proteins, in trans, bridged by the peripheral peroxisomal membrane protein Inp1 (Knoblach et al., 2013). Interestingly, this anchoring mechanism also impacts the levels of free Pex3 by sequestering both peroxisomal and ER Pex3 into anchoring complexes (Fig. 2). Thus, in addition to anchoring peroxisomes for retention by the mother cell, this contact site could serve to buffer local Pex3 levels at both the ER and peroxisomes. Pex3–Inp1–Pex3 complexes between the ER and peroxisomes in trans could therefore serve as a store of Pex3 to couple peroxisome biogenesis to the ebb and flow of total cellular levels of peroxisomes and Pex3 during the cell cycle.
Finally, the effect of ER architecture and architecture-forming proteins on peroxisome biogenesis is also mediated by interorganellar contact. Pex29 and Pex30 are membrane proteins that reside in distinct domains of the ER where they function to negatively regulate peroxisome biogenesis (Yan et al., 2008; Mast et al., 2016). Pex29 and Pex30 physically associate with each other and with the ER-shaping reticulon proteins Rtn1 and Yop1; members of the ER–plasma membrane tether/phosphatidylinositol 4-phosphate pathway, Scs2 and Tcb3; and Dpm1, the ER docking factor for the phosphatidylinositol 4-phosphate phosphatase Sac1 (David et al., 2013; Mast et al., 2016). The ER subdomains that these interacting proteins define form dynamic contact sites with peroxisomes that are stabilized when cells are grown in the presence of oleic acid, a carbon source requiring functional peroxisomes for its metabolism. Mutant cells lacking reticulon proteins or Pex29/30 have increased production of PPVs and exhibit greater numbers of peroxisomes per cell (Mast et al., 2016). Thus, the separate arms of the peroxisome biogenic program, de novo formation and growth and division, are likely coordinated and regulated by interorganellar contacts initiated by this subclass of ER-shaping membrane proteins.
Peroxisomes and lipid droplets: A shared beginning
Since the earliest electron micrographs revealed the bewildering complexity and meshwork of tightly juxtaposed membranes in eukaryotic cells, cell biologists have speculated about the shared spatial and functional roles of what Novikoff and Novikoff (1982) termed “a constellation” of organelles involved in lipid homeostasis.
Lipid droplets and peroxisomes function in tandem to balance the flux of fatty acids in the cell. Recently, the role of Pex3 and Pex19 in regulating the topogenesis of PMPs has been extended to include additional classes of membrane proteins, including the lipid droplet protein Ubx8 (Schrul and Kopito, 2016). Pex19 interacts with newly synthesized Ubx8 in the cytosol and recruits it to ER-localized Pex3 (Schrul and Kopito, 2016). Additionally, Pex19 and Pex3 have been implicated in the topogenesis of hairpin-containing membrane proteins, and this class of membrane proteins is frequently found on lipid droplets, which are enclosed by only a monolayer of phospholipid (hemi-membrane) surrounding a neutral lipid core (Yamamoto and Sakisaka, 2018).
A further intersection between lipid droplets and peroxisomes is a shared site for their biogenesis. Recent evidence from yeast suggests that the budding of lipid droplets and the budding of PPVs occur from subregions of the ER enriched for seipin components Fld1 and Ldb16 and the reticulon homology domain (RHD)–containing peroxin Pex30 (Joshi et al., 2016, 2018; Wang et al., 2018). Dysregulation of peroxisome and lipid droplet biogenesis is toxic to cells (Lockshon et al., 2007), and the subunits of seipin exhibit synthetic lethality with Pex30 (Joshi et al., 2018; Wang et al., 2018). Cells lacking Pex30 in combination with the absence of either Fld1 or Ldb16 have altered ER phospholipid metabolism and exhibit an overproliferation of ER membranes (Wang et al., 2018). Adjusting the phospholipid composition of the ER by removal of genes involved in these metabolic pathways reverses the defects in lipid droplet and peroxisome biogenesis. Thus, seipin and Pex30 work cooperatively to organize subregions of the ER permissive for the budding of nascent lipid droplets and peroxisomes (Joshi and Cohen, 2019). Similarly, the RHD-containing protein MCTP2 functions analogously to Pex30 in regulating lipid droplets in mammalian Cos7 cells and in the worm Caenorhabditis elegans (Joshi et al., 2018).
These findings suggest that the ER is partitioned by distinct classes of RHD-containing proteins to spatially segregate the ER for its different functions; one such RHD subfamily coordinates the biogenesis of peroxisomes and lipid droplets (Joshi and Cohen, 2019). These findings are appealing not only because lipid droplets and peroxisomes must coordinate metabolic functions but also because PPVs need to bud from sites devoid of ER matrix proteins, which are likely excluded by the high-curvature ER tubules mediated by the reticulons. In Y. lipolytica, the Pex30 homologue Pex23 is essential for peroxisome biogenesis and the metabolism of fatty acids (Brown et al., 2000). Loss of Pex23 results in the overproduction and accumulation of PPVs. Pex23, like Pex30, also localizes to the ER (Mast, 2013).
Pex23 proteins in metazoans and yeast share homology through their RHD (Mast et al., 2011; Joshi et al., 2016, 2018). In Drosophila, knockdown of the candidate Pex23 led to an increase in the number of peroxisomes per cell, but lipid droplets were not studied (Mast et al., 2011), whereas MCTP2 depletion in mammalian cells led to altered lipid droplet numbers and morphologies (Joshi et al., 2018). In addition to these roles, these RHD-containing proteins have been implicated in other cellular functions arising from ER contact with organelles such as endosomes and in coupling this contact to organelle motility and innate immune sensing (Joshi and Cohen, 2019). Understanding the dynamics of these RHD-containing proteins, individually and in concert with other proteins, in the context of peroxisome and lipid droplet formation is an important avenue for future research.
Conclusions and future directions
Understanding the complexity of the mechanisms of peroxisome biogenesis is not an unravelable Gordian knot (Costello and Schrader, 2018) but is actually an experimentally tractable and ultimately achievable goal. However, in pursuing this goal, we must remain mindful of the dynamics of the system with the influences of an ever-changing cellular environment on organelle dynamics. Furthermore, changes in both the stoichiometry of individual peroxins and their interacting partners will have oftentimes unexpected effects on the overall process of peroxisome biogenesis. This will necessitate robust quantitative methods that enable the study of peroxisome dynamics in response to defined and reproducible perturbations. These methods should ideally be combined with modeling approaches (Ratushny et al., 2012; Mast et al., 2014). Effective modeling of peroxisome biogenesis will incorporate large-scale global modeling approaches like those developed to model transcriptional regulation (Danziger et al., 2014). Beyond the immediate fundamental insights into peroxisome biogenesis, this integrated approach of modeling with systems cell biology will provide new opportunities for manipulating peroxisome biogenesis for therapeutic benefit and for harnessing the biotechnological potential of peroxisomes.
Acknowledgments
Research in J.D. Aitchison’s laboratory is supported by National Institutes of Health grants P41 GM109824 and R01 GM112108. Research in R.A. Rachubinski’s laboratory is supported by a foundation grant from the Canadian Institutes of Health Research.
The authors declare no competing financial interests.
Author contributions: The manuscript was written by F.D. Mast, R.A. Rachubinski, and J.D. Aitchison.
References
- Agrawal G., Joshi S., and Subramani S.. 2011. Cell-free sorting of peroxisomal membrane proteins from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 108:9113–9118. 10.1073/pnas.1018749108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal G., Fassas S.N., Xia Z.J., and Subramani S.. 2016. Distinct requirements for intra-ER sorting and budding of peroxisomal membrane proteins from the ER. J. Cell Biol. 212:335–348. 10.1083/jcb.201506141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal G., Shang H.H., Xia Z.J., and Subramani S.. 2017. Functional regions of the peroxin Pex19 necessary for peroxisome biogenesis. J. Biol. Chem. 292:11547–11560. 10.1074/jbc.M116.774067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon U. 2006. An Introduction to Systems Biology: Design Principles of Biological Circuits. Taylor & Francis Group CRC Press, Boca Raton, FL. [Google Scholar]
- Aranovich A., Hua R., Rutenberg A.D., and Kim P.K.. 2014. PEX16 contributes to peroxisome maintenance by constantly trafficking PEX3 via the ER. J. Cell Sci. 127:3675–3686. 10.1242/jcs.146282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerends R.J., Faber K.N., Kram A.M., Kiel J.A., van der Klei I.J., and Veenhuis M.. 2000. A stretch of positively charged amino acids at the N terminus of Hansenula polymorpha Pex3p is involved in incorporation of the protein into the peroxisomal membrane. J. Biol. Chem. 275:9986–9995. 10.1074/jbc.275.14.9986 [DOI] [PubMed] [Google Scholar]
- Banerjee H., Knoblach B., and Rachubinski R.A.. 2019. The early-acting glycosome biogenic protein Pex3 is essential for trypanosome viability. Life Sci Alliance. 2:e201900421 10.26508/lsa.201900421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braverman N.E., Raymond G.V., Rizzo W.B., Moser A.B., Wilkinson M.E., Stone E.M., Steinberg S.J., Wangler M.F., Rush E.T., Hacia J.G., and Bose M.. 2016. Peroxisome biogenesis disorders in the Zellweger spectrum: An overview of current diagnosis, clinical manifestations, and treatment guidelines. Mol. Genet. Metab. 117:313–321. 10.1016/j.ymgme.2015.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breker M., Gymrek M., and Schuldiner M.. 2013. A novel single-cell screening platform reveals proteome plasticity during yeast stress responses. J. Cell Biol. 200:839–850. 10.1083/jcb.201301120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T.W., Titorenko V.I., and Rachubinski R.A.. 2000. Mutants of the Yarrowia lipolytica PEX23 gene encoding an integral peroxisomal membrane peroxin mislocalize matrix proteins and accumulate vesicles containing peroxisomal matrix and membrane proteins. Mol. Biol. Cell. 11:141–152. 10.1091/mbc.11.1.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brul S., Wiemer E.A., Westerveld A., Strijland A., Wanders R.J., Schram A.W., Heymans H.S., Schutgens R.B., Van den Bosch H., and Tager J.M.. 1988. Kinetics of the assembly of peroxisomes after fusion of complementary cell lines from patients with the cerebro-hepato-renal (Zellweger) syndrome and related disorders. Biochem. Biophys. Res. Commun. 152:1083–1089. 10.1016/S0006-291X(88)80395-4 [DOI] [PubMed] [Google Scholar]
- Chang J., Mast F.D., Fagarasanu A., Rachubinski D.A., Eitzen G.A., Dacks J.B., and Rachubinski R.A.. 2009. Pex3 peroxisome biogenesis proteins function in peroxisome inheritance as class V myosin receptors. J. Cell Biol. 187:233–246. 10.1083/jcb.200902117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J., Klute M.J., Tower R.J., Mast F.D., Dacks J.B., and Rachubinski R.A.. 2015. An ancestral role in peroxisome assembly is retained by the divisional peroxin Pex11 in the yeast Yarrowia lipolytica. J. Cell Sci. 128:1327–1340. 10.1242/jcs.157743 [DOI] [PubMed] [Google Scholar]
- Chang C.L., Weigel A.V., Ioannou M.S., Pasolli H.A., Xu C.S., Peale D.R., Shtengel G., Freeman M., Hess H.F., Blackstone C., and Lippincott-Schwartz J.. 2019. Spastin tethers lipid droplets to peroxisomes and directs fatty acid trafficking through ESCRT-III. J. Cell Biol. 218:2583–2599. 10.1083/jcb.201902061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Pieuchot L., Loh R.A., Yang J., Kari T.M., Wong J.Y., and Jedd G.. 2014. Hydrophobic handoff for direct delivery of peroxisome tail-anchored proteins. Nat. Commun. 5:5790 10.1038/ncomms6790 [DOI] [PubMed] [Google Scholar]
- Cook K.C., Moreno J.A., Jean Beltran P.M., and Cristea I.M.. 2019. Peroxisome plasticity at the virus-host interface. Trends Microbiol. 27:906–914. 10.1016/j.tim.2019.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello J.L., and Schrader M.. 2018. Unloosing the Gordian knot of peroxisome formation. Curr. Opin. Cell Biol. 50:50–56. 10.1016/j.ceb.2018.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello J.L., Castro I.G., Hacker C., Schrader T.A., Metz J., Zeuschner D., Azadi A.S., Godinho L.F., Costina V., Findeisen P., et al. . 2017. ACBD5 and VAPB mediate membrane associations between peroxisomes and the ER. J. Cell Biol. 216:331–342. 10.1083/jcb.201607055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross L.L., Paudyal R., Kamisugi Y., Berry A., Cuming A.C., Baker A., and Warriner S.L.. 2017. Towards designer organelles by subverting the peroxisomal import pathway. Nat. Commun. 8:454 10.1038/s41467-017-00487-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danziger S.A., Ratushny A.V., Smith J.J., Saleem R.A., Wan Y., Arens C.E., Armstrong A.M., Sitko K., Chen W.M., Chiang J.H., et al. . 2014. Molecular mechanisms of system responses to novel stimuli are predictable from public data. Nucleic Acids Res. 42:1442–1460. 10.1093/nar/gkt938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David C., Koch J., Oeljeklaus S., Laernsack A., Melchior S., Wiese S., Schummer A., Erdmann R., Warscheid B., and Brocard C.. 2013. A combined approach of quantitative interaction proteomics and live-cell imaging reveals a regulatory role for endoplasmic reticulum (ER) reticulon homology proteins in peroxisome biogenesis. Mol. Cell. Proteomics. 12:2408–2425. 10.1074/mcp.M112.017830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depreter M., Espeel M., and Roels F.. 2003. Human peroxisomal disorders. Microsc. Res. Tech. 61:203–223. 10.1002/jemt.10330 [DOI] [PubMed] [Google Scholar]
- Di Cara F., Andreoletti P., Trompier D., Vejux A., Bülow M.H., Sellin J., Lizard G., Cherkaoui-Malki M., and Savary S.. 2019. Peroxisomes in immune response and inflammation. Int. J. Mol. Sci. 20:3877 10.3390/ijms20163877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distel B., Erdmann R., Gould S.J., Blobel G., Crane D.I., Cregg J.M., Dodt G., Fujiki Y., Goodman J.M., Just W.W., et al. . 1996. A unified nomenclature for peroxisome biogenesis factors. J. Cell Biol. 135:1–3. 10.1083/jcb.135.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit E., Boulant S., Zhang Y., Lee A.S., Odendall C., Shum B., Hacohen N., Chen Z.J., Whelan S.P., Fransen M., et al. . 2010. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 141:668–681. 10.1016/j.cell.2010.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmanouilidis L., Schütz U., Tripsianes K., Madl T., Radke J., Rucktäschel R., Wilmanns M., Schliebs W., Erdmann R., and Sattler M.. 2017. Allosteric modulation of peroxisomal membrane protein recognition by farnesylation of the peroxisomal import receptor PEX19. Nat. Commun. 8:14635 10.1038/ncomms14635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann R., and Blobel G.. 1995. Giant peroxisomes in oleic acid-induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J. Cell Biol. 128:509–523. 10.1083/jcb.128.4.509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeel M., Mandel H., Poggi F., Smeitink J.A., Wanders R.J., Kerckaert I., Schutgens R.B., Saudubray J.M., Poll-The B.T., and Roels F.. 1995. Peroxisome mosaicism in the livers of peroxisomal deficiency patients. Hepatology. 22:497–504. [PubMed] [Google Scholar]
- Fagarasanu A., Mast F.D., Knoblach B., and Rachubinski R.A.. 2010. Molecular mechanisms of organelle inheritance: lessons from peroxisomes in yeast. Nat. Rev. Mol. Cell Biol. 11:644–654. 10.1038/nrm2960 [DOI] [PubMed] [Google Scholar]
- Fakieh M.H., Drake P.J., Lacey J., Munck J.M., Motley A.M., and Hettema E.H.. 2013. Intra-ER sorting of the peroxisomal membrane protein Pex3 relies on its luminal domain. Biol. Open. 2:829–837. 10.1242/bio.20134788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Morrell J.C., Jones J.M., and Gould S.J.. 2004. PEX3 functions as a PEX19 docking factor in the import of class I peroxisomal membrane proteins. J. Cell Biol. 164:863–875. 10.1083/jcb.200311131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré J.C., Carolino K., Stasyk O.V., Stasyk O.G., Hodzic Z., Agrawal G., Till A., Proietto M., Cregg J., Sibirny A.A., and Subramani S.. 2017. A new yeast peroxin, Pex36, a functional homolog of mammalian PEX16, functions in the ER-to-peroxisome traffic of peroxisomal membrane proteins. J. Mol. Biol. 429:3743–3762. 10.1016/j.jmb.2017.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré J.C., Mahalingam S.S., Proietto M., and Subramani S.. 2019. Peroxisome biogenesis, membrane contact sites, and quality control. EMBO Rep. 20:e46864 10.15252/embr.201846864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira A.R., Magalhães A.C., Camões F., Gouveia A., Vieira M., Kagan J.C., and Ribeiro D.. 2016. Hepatitis C virus NS3-4A inhibits the peroxisomal MAVS-dependent antiviral signalling response. J. Cell. Mol. Med. 20:750–757. 10.1111/jcmm.12801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen M., and Lismont C.. 2019. Redox signaling from and to peroxisomes: Progress, challenges, and prospects. Antioxid. Redox Signal. 30:95–112. 10.1089/ars.2018.7515 [DOI] [PubMed] [Google Scholar]
- Fujiki Y., Fowler S., Shio H., Hubbard A.L., and Lazarow P.B.. 1982. Polypeptide and phospholipid composition of the membrane of rat liver peroxisomes: comparison with endoplasmic reticulum and mitochondrial membranes. J. Cell Biol. 93:103–110. 10.1083/jcb.93.1.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldón T., Snel B., van Zimmeren F., Hemrika W., Tabak H., and Huynen M.A.. 2006. Origin and evolution of the peroxisomal proteome. Biol. Direct. 1:8 10.1186/1745-6150-1-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., and Zhou Y.J.. 2019. Repurposing peroxisomes for microbial synthesis for biomolecules. Methods Enzymol. 617:83–111. 10.1016/bs.mie.2018.12.004 [DOI] [PubMed] [Google Scholar]
- Ghaedi K., Honsho M., Shimozawa N., Suzuki Y., Kondo N., and Fujiki Y.. 2000. PEX3 is the causal gene responsible for peroxisome membrane assembly-defective Zellweger syndrome of complementation group G. Am. J. Hum. Genet. 67:976–981. 10.1086/303086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidijala L., Kiel J.A., Douma R.D., Seifar R.M., van Gulik W.M., Bovenberg R.A., Veenhuis M., and van der Klei I.J.. 2009. An engineered yeast efficiently secreting penicillin. PLoS One. 4:e8317 10.1371/journal.pone.0008317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girzalsky W., Saffian D., and Erdmann R.. 2010. Peroxisomal protein translocation. Biochim. Biophys. Acta. 1803:724–731. 10.1016/j.bbamcr.2010.01.002 [DOI] [PubMed] [Google Scholar]
- Goldfischer S., Moore C.L., Johnson A.B., Spiro A.J., Valsamis M.P., Wisniewski H.K., Ritch R.H., Norton W.T., Rapin I., and Gartner L.M.. 1973. Peroxisomal and mitochondrial defects in the cerebro-hepato-renal syndrome. Science. 182:62–64. 10.1126/science.182.4107.62 [DOI] [PubMed] [Google Scholar]
- Gould S.G., Keller G.A., and Subramani S.. 1987. Identification of a peroxisomal targeting signal at the carboxy terminus of firefly luciferase. J. Cell Biol. 105:2923–2931. 10.1083/jcb.105.6.2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S.J., Keller G.A., Hosken N., Wilkinson J., and Subramani S.. 1989. A conserved tripeptide sorts proteins to peroxisomes. J. Cell Biol. 108:1657–1664. 10.1083/jcb.108.5.1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S.J., McCollum D., Spong A.P., Heyman J.A., and Subramani S.. 1992. Development of the yeast Pichia pastoris as a model organism for a genetic and molecular analysis of peroxisome assembly. Yeast. 8:613–628. 10.1002/yea.320080805 [DOI] [PubMed] [Google Scholar]
- Hanko E.K.R., Denby C.M., Sànchez I Nogué V., Lin W., Ramirez K.J., Singer C.A., Beckham G.T., Keasling J.D., and Keasling J.D.. 2018. Engineering β-oxidation in Yarrowia lipolytica for methyl ketone production. Metab. Eng. 48:52–62. 10.1016/j.ymben.2018.05.018 [DOI] [PubMed] [Google Scholar]
- Hardeman D., Versantvoort C., van den Brink J.M., and van den Bosch H.. 1990. Studies on peroxisomal membranes. Biochim. Biophys. Acta. 1027:149–154. 10.1016/0005-2736(90)90078-3 [DOI] [PubMed] [Google Scholar]
- Heinzle C., Mücke L., Brune T., and Kölling R.. 2019. Comprehensive analysis of yeast ESCRT-III composition in single ESCRT-III deletion mutants. Biochem. J. 476:2031–2046. 10.1042/BCJ20190141 [DOI] [PubMed] [Google Scholar]
- Henne W.M., Buchkovich N.J., and Emr S.D.. 2011. The ESCRT pathway. Dev. Cell. 21:77–91. 10.1016/j.devcel.2011.05.015 [DOI] [PubMed] [Google Scholar]
- Herricks T., Dilworth D.J., Mast F.D., Li S., Smith J.J., Ratushny A.V., and Aitchison J.D.. 2017. One-cell doubling evaluation by living arrays of yeast, ODELAY! G3 (Bethesda). 7:279–288. 10.1534/g3.116.037044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess R., Stäubli W., and Riess W.. 1965. Nature of the hepatomegalic effect produced by ethyl-chlorophenoxy-isobutyrate in the rat. Nature. 208:856–858. 10.1038/208856a0 [DOI] [PubMed] [Google Scholar]
- Hettema E.H., Girzalsky W., van Den Berg M., Erdmann R., and Distel B.. 2000. Saccharomyces cerevisiae pex3p and pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J. 19:223–233. 10.1093/emboj/19.2.223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema E.H., Erdmann R., van der Klei I., and Veenhuis M.. 2014. Evolving models for peroxisome biogenesis. Curr. Opin. Cell Biol. 29:25–30. 10.1016/j.ceb.2014.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner D., Schildknegt D., Braakman I., Philippsen P., and Tabak H.F.. 2005. Contribution of the endoplasmic reticulum to peroxisome formation. Cell. 122:85–95. 10.1016/j.cell.2005.04.025 [DOI] [PubMed] [Google Scholar]
- Höhfeld J., Veenhuis M., and Kunau W.H.. 1991. PAS3, a Saccharomyces cerevisiae gene encoding a peroxisomal integral membrane protein essential for peroxisome biogenesis. J. Cell Biol. 114:1167–1178. 10.1083/jcb.114.6.1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua R., Cheng D., Coyaud É., Freeman S., Di Pietro E., Wang Y., Vissa A., Yip C.M., Fairn G.D., Braverman N., et al. . 2017. VAPs and ACBD5 tether peroxisomes to the ER for peroxisome maintenance and lipid homeostasis. J. Cell Biol. 216:367–377. 10.1083/jcb.201608128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A., Koch J., Kragler F., Brocard C., and Hartig A.. 2012. A subtle interplay between three Pex11 proteins shapes de novo formation and fission of peroxisomes. Traffic. 13:157–167. 10.1111/j.1600-0854.2011.01290.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan C.H., Williams C.C., and Weissman J.S.. 2014. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science. 346:1257521 10.1126/science.1257521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R.L.M., and van der Klei I.J.. 2019. The peroxisome biogenesis factors Pex3 and Pex19: multitasking proteins with disputed functions. FEBS Lett. 593:457–474. 10.1002/1873-3468.13340 [DOI] [PubMed] [Google Scholar]
- Jean Beltran P.M., Cook K.C., Hashimoto Y., Galitzine C., Murray L.A., Vitek O., and Cristea I.M.. 2018. Infection-induced peroxisome biogenesis is a metabolic strategy for herpesvirus replication. Cell Host Microbe. 24:526–541.e7. 10.1016/j.chom.2018.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.M., Morrell J.C., and Gould S.J.. 2004. PEX19 is a predominantly cytosolic chaperone and import receptor for class 1 peroxisomal membrane proteins. J. Cell Biol. 164:57–67. 10.1083/jcb.200304111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A.S., and Cohen S.. 2019. Lipid droplet and peroxisome biogenesis: Do they go hand-in-hand? Front. Cell Dev. Biol. 7:92 10.3389/fcell.2019.00092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A.S., Huang X., Choudhary V., Levine T.P., Hu J., and Prinz W.A.. 2016. A family of membrane-shaping proteins at ER subdomains regulates pre-peroxisomal vesicle biogenesis. J. Cell Biol. 215:515–529. 10.1083/jcb.201602064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A.S., Nebenfuehr B., Choudhary V., Satpute-Krishnan P., Levine T.P., Golden A., and Prinz W.A.. 2018. Lipid droplet and peroxisome biogenesis occur at the same ER subdomains. Nat. Commun. 9:2940 10.1038/s41467-018-05277-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaewsapsak P., Shechner D.M., Mallard W., Rinn J.L., and Ting A.Y.. 2017. Live-cell mapping of organelle-associated RNAs via proximity biotinylation combined with protein-RNA crosslinking. eLife. 6:e29224 10.7554/eLife.29224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalel V.C., Li M., Gaussmann S., Delhommel F., Schäfer A.B., Tippler B., Jung M., Maier R., Oeljeklaus S., Schliebs W., et al. . 2019. Evolutionary divergent PEX3 is essential for glycosome biogenesis and survival of trypanosomatid parasites. Biochim Biophys Acta Mol Cell Res. 1866:118520 10.1016/j.bbamcr.2019.07.015 [DOI] [PubMed] [Google Scholar]
- Kim P.K., Mullen R.T., Schumann U., and Lippincott-Schwartz J.. 2006. The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J. Cell Biol. 173:521–532. 10.1083/jcb.200601036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klouwer F.C., Berendse K., Ferdinandusse S., Wanders R.J., Engelen M., and Poll-The B.T.. 2015. Zellweger spectrum disorders: clinical overview and management approach. Orphanet J. Rare Dis. 10:151 10.1186/s13023-015-0368-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblach B., and Rachubinski R.A.. 2010. Phosphorylation-dependent activation of peroxisome proliferator protein PEX11 controls peroxisome abundance. J. Biol. Chem. 285:6670–6680. 10.1074/jbc.M109.094805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblach B., Sun X., Coquelle N., Fagarasanu A., Poirier R.L., and Rachubinski R.A.. 2013. An ER-peroxisome tether exerts peroxisome population control in yeast. EMBO J. 32:2439–2453. 10.1038/emboj.2013.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Manivannan S., Cepinska M.N., Krikken A.M., Kram A.M., Veenhuis M., and van der Klei I.J.. 2014. Preperoxisomal vesicles can form in the absence of Pex3. J. Cell Biol. 204:659–668. 10.1083/jcb.201310148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S.K., Yoda N., and Schekman R.. 2010. A vesicle carrier that mediates peroxisome protein traffic from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 107:21523–21528. 10.1073/pnas.1013397107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarow P.B., and Fujiki Y.. 1985. Biogenesis of peroxisomes. Annu. Rev. Cell Biol. 1:489–530. 10.1146/annurev.cb.01.110185.002421 [DOI] [PubMed] [Google Scholar]
- Lazarow P.B., and Moser H.W.. 1994. Disorders of peroxisome biogenesis. In Metabolic and Molecular Bases of Inherited Diseases. Scriver C.R., Beaudet A.L., Sly W.S., and Valle D., editors. McGraw-Hill, New York. [Google Scholar]
- Léon S., Goodman J.M., and Subramani S.. 2006. Uniqueness of the mechanism of protein import into the peroxisome matrix: transport of folded, co-factor-bound and oligomeric proteins by shuttling receptors. Biochim. Biophys. Acta. 1763:1552–1564. 10.1016/j.bbamcr.2006.08.037 [DOI] [PubMed] [Google Scholar]
- Lismont C., Revenco I., and Fransen M.. 2019. Peroxisomal hydrogen peroxide metabolism and signaling in health and disease. Int. J. Mol. Sci. 20:E3673 10.3390/ijms20153673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Yagita Y., and Fujiki Y.. 2016. Assembly of peroxisomal membrane proteins via the direct Pex19p-Pex3p pathway. Traffic. 17:433–455. 10.1111/tra.12376 [DOI] [PubMed] [Google Scholar]
- Lockshon D., Surface L.E., Kerr E.O., Kaeberlein M., and Kennedy B.K.. 2007. The sensitivity of yeast mutants to oleic acid implicates the peroxisome and other processes in membrane function. Genetics. 175:77–91. 10.1534/genetics.106.064428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S., and Alon U.. 2003. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. USA. 100:11980–11985. 10.1073/pnas.2133841100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W.F. 2016. Cell Geometry: How Cells Count and Measure Size. Annu. Rev. Biophys. 45:49–64. 10.1146/annurev-biophys-062215-010905 [DOI] [PubMed] [Google Scholar]
- Mast F.D. 2013. A comprehensive assessment of peroxisome biology: ER-dependent peroxisome proliferation control, evolution of organelle inheritance in yeast and a Drosophila model system of Zellweger syndrome. PhD thesis. University of Alberta, Edmonton, Alberta, Canada. 311 pp. [Google Scholar]
- Mast F.D., and Aitchison J.D.. 2018. Characterization of peroxisomal regulation networks. Subcell. Biochem. 89:367–382. 10.1007/978-981-13-2233-4_16 [DOI] [PubMed] [Google Scholar]
- Mast F.D., Li J., Virk M.K., Hughes S.C., Simmonds A.J., and Rachubinski R.A.. 2011. A Drosophila model for the Zellweger spectrum of peroxisome biogenesis disorders. Dis. Model. Mech. 4:659–672. 10.1242/dmm.007419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mast F.D., Rachubinski R.A., and Dacks J.B.. 2012. Emergent complexity in Myosin V-based organelle inheritance. Mol. Biol. Evol. 29:975–984. 10.1093/molbev/msr264 [DOI] [PubMed] [Google Scholar]
- Mast F.D., Ratushny A.V., and Aitchison J.D.. 2014. Systems cell biology. J. Cell Biol. 206:695–706. 10.1083/jcb.201405027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mast F.D., Rachubinski R.A., and Aitchison J.D.. 2015. Signaling dynamics and peroxisomes. Curr. Opin. Cell Biol. 35:131–136. 10.1016/j.ceb.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mast F.D., Jamakhandi A., Saleem R.A., Dilworth D.J., Rogers R.S., Rachubinski R.A., and Aitchison J.D.. 2016. Peroxins Pex30 and Pex29 dynamically associate with reticulons to regulate peroxisome biogenesis from the endoplasmic reticulum. J. Biol. Chem. 291:15408–15427. 10.1074/jbc.M116.728154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mast F.D., Herricks T., Strehler K.M., Miller L.R., Saleem R.A., Rachubinski R.A., and Aitchison J.D.. 2018. ESCRT-III is required for scissioning new peroxisomes from the endoplasmic reticulum. J. Cell Biol. 217:2087–2102. 10.1083/jcb.201706044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki T., and Fujiki Y.. 2008. The peroxisomal membrane protein import receptor Pex3p is directly transported to peroxisomes by a novel Pex19p- and Pex16p-dependent pathway. J. Cell Biol. 183:1275–1286. 10.1083/jcb.200806062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayerhofer P.U., Bañó-Polo M., Mingarro I., and Johnson A.E.. 2016. Human peroxin PEX3 Is co-translationally integrated into the er and exits the ER in budding vesicles. Traffic. 17:117–130. 10.1111/tra.12350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J., Clippinger A.K., Talledge N., Skowyra M.L., Saunders M.G., Naismith T.V., Colf L.A., Afonine P., Arthur C., Sundquist W.I., et al. . 2015. Structure and membrane remodeling activity of ESCRT-III helical polymers. Science. 350:1548–1551. 10.1126/science.aad8305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez-Benito V., van Deventer S.J., Jimenez-Garcia V., Roy-Luzarraga M., van Leeuwen F., and Neefjes J.. 2013. Spatiotemporal analysis of organelle and macromolecular complex inheritance. Proc. Natl. Acad. Sci. USA. 110:175–180. 10.1073/pnas.1207424110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley A.M., and Hettema E.H.. 2007. Yeast peroxisomes multiply by growth and division. J. Cell Biol. 178:399–410. 10.1083/jcb.200702167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley A.M., Tabak H.F., Smeitink J.A., Poll-The B.T., Barth P.G., and Wanders R.J.. 1996. Non-rhizomelic and rhizomelic chondrodysplasia punctata within a single complementation group. Biochim. Biophys. Acta. 1315:153–158. 10.1016/0925-4439(95)00114-X [DOI] [PubMed] [Google Scholar]
- Motley A.M., Ward G.P., and Hettema E.H.. 2008. Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p. J. Cell Sci. 121:1633–1640. 10.1242/jcs.026344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley A.M., Nuttall J.M., and Hettema E.H.. 2012. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J. 31:2852–2868. 10.1038/emboj.2012.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley A.M., Galvin P.C., Ekal L., Nuttall J.M., and Hettema E.H.. 2015. Reevaluation of the role of Pex1 and dynamin-related proteins in peroxisome membrane biogenesis. J. Cell Biol. 211:1041–1056. 10.1083/jcb.201412066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munck J.M., Motley A.M., Nuttall J.M., and Hettema E.H.. 2009. A dual function for Pex3p in peroxisome formation and inheritance. J. Cell Biol. 187:463–471. 10.1083/jcb.200906161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikoff A.B., and Novikoff P.M.. 1982. Microperoxisomes and peroxisomes in relation to lipid metabolism. Ann. N. Y. Acad. Sci. 386(1 Peroxisomes a):138–152. [DOI] [PubMed]
- Nunnari J., and Walter P.. 1996. Regulation of organelle biogenesis. Cell. 84:389–394. 10.1016/S0092-8674(00)81283-0 [DOI] [PubMed] [Google Scholar]
- Odendall C., Dixit E., Stavru F., Bierne H., Franz K.M., Durbin A.F., Boulant S., Gehrke L., Cossart P., and Kagan J.C.. 2014. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat. Immunol. 15:717–726. 10.1038/ni.2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Périchon R., Bourre J.M., Kelly J.F., and Roth G.S.. 1998. The role of peroxisomes in aging. Cell. Mol. Life Sci. 54:641–652. 10.1007/s000180050192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R.J., Mast F.D., and Rachubinski R.A.. 2009. Endoplasmic reticulum-associated secretory proteins Sec20p, Sec39p, and Dsl1p are involved in peroxisome biogenesis. Eukaryot. Cell. 8:830–843. 10.1128/EC.00024-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta H.W., and Erdmann R.. 2007. Peroxisomal dynamics. Trends Cell Biol. 17:474–484. 10.1016/j.tcb.2007.06.009 [DOI] [PubMed] [Google Scholar]
- Ratushny A.V., Saleem R.A., Sitko K., Ramsey S.A., and Aitchison J.D.. 2012. Asymmetric positive feedback loops reliably control biological responses. Mol. Syst. Biol. 8:577 10.1038/msb.2012.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S., and Prinz W.A.. 2008. Nonvesicular phospholipid transfer between peroxisomes and the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 105:15785–15790. 10.1073/pnas.0808321105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottensteiner H., Kramer A., Lorenzen S., Stein K., Landgraf C., Volkmer-Engert R., and Erdmann R.. 2004. Peroxisomal membrane proteins contain common Pex19p-binding sites that are an integral part of their targeting signals. Mol. Biol. Cell. 15:3406–3417. 10.1091/mbc.e04-03-0188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rucktäschel R., Thoms S., Sidorovitch V., Halbach A., Pechlivanis M., Volkmer R., Alexandrov K., Kuhlmann J., Rottensteiner H., and Erdmann R.. 2009. Farnesylation of pex19p is required for its structural integrity and function in peroxisome biogenesis. J. Biol. Chem. 284:20885–20896. 10.1074/jbc.M109.016584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rucktäschel R., Halbach A., Girzalsky W., Rottensteiner H., and Erdmann R.. 2010. De novo synthesis of peroxisomes upon mitochondrial targeting of Pex3p. Eur. J. Cell Biol. 89:947–954. 10.1016/j.ejcb.2010.06.012 [DOI] [PubMed] [Google Scholar]
- Sacksteder K.A., Jones J.M., South S.T., Li X., Liu Y., and Gould S.J.. 2000. PEX19 binds multiple peroxisomal membrane proteins, is predominantly cytoplasmic, and is required for peroxisome membrane synthesis. J. Cell Biol. 148:931–944. 10.1083/jcb.148.5.931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem R.A., Knoblach B., Mast F.D., Smith J.J., Boyle J., Dobson C.M., Long-O’Donnell R., Rachubinski R.A., and Aitchison J.D.. 2008. Genome-wide analysis of signaling networks regulating fatty acid-induced gene expression and organelle biogenesis. J. Cell Biol. 181:281–292. 10.1083/jcb.200710009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem R.A., Long-O’Donnell R., Dilworth D.J., Armstrong A.M., Jamakhandi A.P., Wan Y., Knijnenburg T.A., Niemistö A., Boyle J., Rachubinski R.A., et al. . 2010. Genome-wide analysis of effectors of peroxisome biogenesis. PLoS One. 5:e11953 10.1371/journal.pone.0011953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salpietro V., Phadke R., Saggar A., Hargreaves I.P., Yates R., Fokoloros C., Mankad K., Hertecant J., Ruggieri M., McCormick D., and Kinali M.. 2015. Zellweger syndrome and secondary mitochondrial myopathy. Eur. J. Pediatr. 174:557–563. 10.1007/s00431-014-2431-2 [DOI] [PubMed] [Google Scholar]
- Sato Y., Shibata H., Nakano H., Matsuzono Y., Kashiwayama Y., Kobayashi Y., Fujiki Y., Imanaka T., and Kato H.. 2008. Characterization of the interaction between recombinant human peroxin Pex3p and Pex19p: identification of TRP-104 IN Pex3p as a critical residue for the interaction. J. Biol. Chem. 283:6136–6144. 10.1074/jbc.M706139200 [DOI] [PubMed] [Google Scholar]
- Sato Y., Shibata H., Nakatsu T., Nakano H., Kashiwayama Y., Imanaka T., and Kato H.. 2010. Structural basis for docking of peroxisomal membrane protein carrier Pex19p onto its receptor Pex3p. EMBO J. 29:4083–4093. 10.1038/emboj.2010.293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlüter A., Fourcade S., Ripp R., Mandel J.L., Poch O., and Pujol A.. 2006. The evolutionary origin of peroxisomes: an ER-peroxisome connection. Mol. Biol. Evol. 23:838–845. 10.1093/molbev/msj103 [DOI] [PubMed] [Google Scholar]
- Schmidt F., Dietrich D., Eylenstein R., Groemping Y., Stehle T., and Dodt G.. 2012. The role of conserved PEX3 regions in PEX19-binding and peroxisome biogenesis. Traffic. 13:1244–1260. 10.1111/j.1600-0854.2012.01380.x [DOI] [PubMed] [Google Scholar]
- Schmidt F., Treiber N., Zocher G., Bjelic S., Steinmetz M.O., Kalbacher H., Stehle T., and Dodt G.. 2010. Insights into peroxisome function from the structure of PEX3 in complex with a soluble fragment of PEX19. J. Biol. Chem. 285:25410–25417. 10.1074/jbc.M110.138503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöneberg J., Lee I.H., Iwasa J.H., and Hurley J.H.. 2017. Reverse-topology membrane scission by the ESCRT proteins. Nat. Rev. Mol. Cell Biol. 18:5–17. 10.1038/nrm.2016.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader M., Grille S., Fahimi H.D., and Islinger M.. 2013. Peroxisome interactions and cross-talk with other subcellular compartments in animal cells. Subcell. Biochem. 69:1–22. 10.1007/978-94-007-6889-5_1 [DOI] [PubMed] [Google Scholar]
- Schrul B., and Kopito R.R.. 2016. Peroxin-dependent targeting of a lipid-droplet-destined membrane protein to ER subdomains. Nat. Cell Biol. 18:740–751. 10.1038/ncb3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shai N., Yifrach E., van Roermund C.W.T., Cohen N., Bibi C., IJlst L., Cavellini L., Meurisse J., Schuster R., Zada L., et al. . 2018. Systematic mapping of contact sites reveals tethers and a function for the peroxisome-mitochondria contact. Nat. Commun. 9:1761 10.1038/s41467-018-03957-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shai N., Schuldiner M., and Zalckvar E.. 2016. No peroxisome is an island - Peroxisome contact sites. Biochim. Biophys. Acta. 1863:1061–1069. 10.1016/j.bbamcr.2015.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimozawa N., Suzuki Y., Zhang Z., Imamura A., Ghaedi K., Fujiki Y., and Kondo N.. 2000. Identification of PEX3 as the gene mutated in a Zellweger syndrome patient lacking peroxisomal remnant structures. Hum. Mol. Genet. 9:1995–1999. 10.1093/hmg/9.13.1995 [DOI] [PubMed] [Google Scholar]
- Singh I. 1997. Biochemistry of peroxisomes in health and disease. Mol. Cell. Biochem. 167:1–29. 10.1023/A:1006883229684 [DOI] [PubMed] [Google Scholar]
- Smith J.J., and Aitchison J.D.. 2013. Peroxisomes take shape. Nat. Rev. Mol. Cell Biol. 14:803–817. 10.1038/nrm3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.J., Marelli M., Christmas R.H., Vizeacoumar F.J., Dilworth D.J., Ideker T., Galitski T., Dimitrov K., Rachubinski R.A., and Aitchison J.D.. 2002. Transcriptome profiling to identify genes involved in peroxisome assembly and function. J. Cell Biol. 158:259–271. 10.1083/jcb.200204059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.J., Sydorskyy Y., Marelli M., Hwang D., Bolouri H., Rachubinski R.A., and Aitchison J.D.. 2006. Expression and functional profiling reveal distinct gene classes involved in fatty acid metabolism. Mol Syst Biol. 2:2006.0009. [DOI] [PMC free article] [PubMed]
- Song S. 2002. The role of increased liver triglyceride content: a culprit of diabetic hyperglycaemia? Diabetes Metab. Res. Rev. 18:5–12. 10.1002/dmrr.260 [DOI] [PubMed] [Google Scholar]
- South S.T., Sacksteder K.A., Li X., Liu Y., and Gould S.J.. 2000. Inhibitors of COPI and COPII do not block PEX3-mediated peroxisome synthesis. J. Cell Biol. 149:1345–1360. 10.1083/jcb.149.7.1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehlik T., Sandrock B., Ast J., and Freitag J.. 2014. Fungal peroxisomes as biosynthetic organelles. Curr. Opin. Microbiol. 22:8–14. 10.1016/j.mib.2014.09.011 [DOI] [PubMed] [Google Scholar]
- Sugiura A., Mattie S., Prudent J., and McBride H.M.. 2017. Newly born peroxisomes are a hybrid of mitochondrial and ER-derived pre-peroxisomes. Nature. 542:251–254. 10.1038/nature21375 [DOI] [PubMed] [Google Scholar]
- Swinkels B.W., Gould S.J., Bodnar A.G., Rachubinski R.A., and Subramani S.. 1991. A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 10:3255–3262. 10.1002/j.1460-2075.1991.tb04889.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko V.I., and Mullen R.T.. 2006. Peroxisome biogenesis: the peroxisomal endomembrane system and the role of the ER. J. Cell Biol. 174:11–17. 10.1083/jcb.200604036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko V.I., and Rachubinski R.A.. 1998. Mutants of the yeast Yarrowia lipolytica defective in protein exit from the endoplasmic reticulum are also defective in peroxisome biogenesis. Mol. Cell. Biol. 18:2789–2803. 10.1128/MCB.18.5.2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko V.I., Chan H., and Rachubinski R.A.. 2000. Fusion of small peroxisomal vesicles in vitro reconstructs an early step in the in vivo multistep peroxisome assembly pathway of Yarrowia lipolytica. J. Cell Biol. 148:29–44. 10.1083/jcb.148.1.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro A.A., Araya C.A., Córdova G.J., Arredondo C.A., Cárdenas H.G., Moreno R.E., Venegas A., Koenig C.S., Cancino J., Gonzalez A., and Santos M.J.. 2009. Pex3p-dependent peroxisomal biogenesis initiates in the endoplasmic reticulum of human fibroblasts. J. Cell. Biochem. 107:1083–1096. 10.1002/jcb.22210 [DOI] [PubMed] [Google Scholar]
- Tripathi D.N., and Walker C.L.. 2016. The peroxisome as a cell signaling organelle. Curr. Opin. Cell Biol. 39:109–112. 10.1016/j.ceb.2016.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valm A.M., Cohen S., Legant W.R., Melunis J., Hershberg U., Wait E., Cohen A.R., Davidson M.W., Betzig E., and Lippincott-Schwartz J.. 2017. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature. 546:162–167. 10.1038/nature22369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zand A., Gent J., Braakman I., and Tabak H.F.. 2012. Biochemically distinct vesicles from the endoplasmic reticulum fuse to form peroxisomes. Cell. 149:397–409. 10.1016/j.cell.2012.01.054 [DOI] [PubMed] [Google Scholar]
- Vietri M., Radulovic M., and Stenmark H.. 2020. The many functions of ESCRTs. Nat. Rev. Mol. Cell Biol. 21:25–42. 10.1038/s41580-019-0177-4 [DOI] [PubMed] [Google Scholar]
- Wanders R.J., and Ferdinandusse S.. 2012. Peroxisomes, peroxisomal diseases, and the hepatotoxicity induced by peroxisomal metabolites. Curr. Drug Metab. 13:1401–1411. 10.2174/138920012803762747 [DOI] [PubMed] [Google Scholar]
- Wanders R.J., and Waterham H.R.. 2006. Biochemistry of mammalian peroxisomes revisited. Annu. Rev. Biochem. 75:295–332. 10.1146/annurev.biochem.74.082803.133329 [DOI] [PubMed] [Google Scholar]
- Wang S., Idrissi F.Z., Hermansson M., Grippa A., Ejsing C.S., and Carvalho P.. 2018. Seipin and the membrane-shaping protein Pex30 cooperate in organelle budding from the endoplasmic reticulum. Nat. Commun. 9:2939 10.1038/s41467-018-05278-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangler M.F., Hubert L., Donti T.R., Ventura M.J., Miller M.J., Braverman N., Gawron K., Bose M., Moser A.B., Jones R.O., et al. . 2018. A metabolomic map of Zellweger spectrum disorders reveals novel disease biomarkers. Genet. Med. 20:1274–1283. 10.1038/gim.2017.262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G., and Wickner W.. 1996. Organelle inheritance. Cell. 84:395–400. 10.1016/S0092-8674(00)81284-2 [DOI] [PubMed] [Google Scholar]
- Waterham H.R., Ferdinandusse S., and Wanders R.J.. 2016. Human disorders of peroxisome metabolism and biogenesis. Biochim. Biophys. Acta. 1863:922–933. 10.1016/j.bbamcr.2015.11.015 [DOI] [PubMed] [Google Scholar]
- Weir N.R., Kamber R.A., Martenson J.S., and Denic V.. 2017. The AAA protein Msp1 mediates clearance of excess tail-anchored proteins from the peroxisomal membrane. eLife. 6:e28507 10.7554/eLife.28507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wróblewska J.P., Cruz-Zaragoza L.D., Yuan W., Schummer A., Chuartzman S.G., de Boer R., Oeljeklaus S., Schuldiner M., Zalckvar E., Warscheid B., et al. . 2017. Saccharomyces cerevisiae cells lacking Pex3 contain membrane vesicles that harbor a subset of peroxisomal membrane proteins. Biochim Biophys Acta Mol Cell Res. 1864:1656–1667. 10.1016/j.bbamcr.2017.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., de Boer R., Krikken A.M., Akşit A., Yuan W., and van der Klei I.J.. 2019. Peroxisome development in yeast is associated with the formation of Pex3-dependent peroxisome-vacuole contact sites. Biochim Biophys Acta Mol Cell Res. 1866:349–359. 10.1016/j.bbamcr.2018.08.021 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y., and Sakisaka T.. 2018. The peroxisome biogenesis factors posttranslationally target reticulon homology domain-containing proteins to the endoplasmic reticulum membrane. Sci. Rep. 8:2322 10.1038/s41598-018-20797-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M., Rachubinski D.A., Joshi S., Rachubinski R.A., and Subramani S.. 2008. Dysferlin domain-containing proteins, Pex30p and Pex31p, localized to two compartments, control the number and size of oleate-induced peroxisomes in Pichia pastoris. Mol. Biol. Cell. 19:885–898. 10.1091/mbc.e07-10-1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Kim J., Alexander A., Cai S., Tripathi D.N., Dere R., Tee A.R., Tait-Mulder J., Di Nardo A., Han J.M., et al. . 2013. A tuberous sclerosis complex signalling node at the peroxisome regulates mTORC1 and autophagy in response to ROS. Nat. Cell Biol. 15:1186–1196. 10.1038/ncb2822 [DOI] [PMC free article] [PubMed] [Google Scholar]