Coat disassembly, driven by the Hsc70 “uncoating ATPase” and mediated by auxilin, occurs within seconds after vesicle release. Using single-molecule imaging, He et al. find that auxilins are absent from assembling pits. Therefore, Hsc70 is not responsible for the clathrin exchange during pit formation.
Abstract
Clathrin-coated vesicles lose their clathrin lattice within seconds of pinching off, through the action of the Hsc70 “uncoating ATPase.” The J- and PTEN-like domain–containing proteins, auxilin 1 (Aux1) and auxilin 2 (GAK), recruit Hsc70. The PTEN-like domain has no phosphatase activity, but it can recognize phosphatidylinositol phosphate head groups. Aux1 and GAK appear on coated vesicles in successive transient bursts, immediately after dynamin-mediated membrane scission has released the vesicle from the plasma membrane. These bursts contain a very small number of auxilins, and even four to six molecules are sufficient to mediate uncoating. In contrast, we could not detect auxilins in abortive pits or at any time during coated pit assembly. We previously showed that clathrin-coated vesicles have a dynamic phosphoinositide landscape, and we have proposed that lipid head group recognition might determine the timing of Aux1 and GAK appearance. The differential recruitment of Aux1 and GAK correlates with temporal variations in phosphoinositide composition, consistent with a lipid-switch timing mechanism.
Graphical Abstract
Introduction
Endocytic clathrin coats assemble at the plasma membrane as coated pits and pinch off as coated vesicles. Delivery of recruited cargo then requires shedding of the clathrin lattice to liberate the enclosed vesicle (Kirchhausen et al., 2014). Coat disassembly, driven by the Hsc70 “uncoating ATPase” (Braell et al., 1984; Schlossman et al., 1984; Ungewickell, 1985), occurs a few seconds after vesicle release (Lee et al., 2006; Massol et al., 2006); the timing of Hsc70 recruitment depends in turn on arrival of a J-domain–containing protein, auxilin, immediately after the vesicle separates from the parent membrane (Lee et al., 2006; Massol et al., 2006). Human cells have two closely related auxilin isoforms (Eisenberg and Greene, 2007). Cyclin-G–dependent kinase (GAK; also called auxilin 2), expressed in all cells, has both a cyclin-G Ser/Thr–dependent kinase domain and a catalytically inactive, phosphatase and tensin-like (PTEN) N-terminal to its clathrin-binding and C-terminal J-domains (Guan et al., 2010). Auxilin 1 (Aux1), expressed principally in neurons, has PTEN-like, clathrin-binding, and J-domains, but lacks the N-terminal kinase.
To study uncoating in living cells, we expressed, from the endogenous locus, Aux1 or GAK bearing a genetically encoded fluorescent tag and followed recruitment to endocytic coated vesicles by total internal reflection fluorescence (TIRF) imaging with single-molecule sensitivity. The burst-like recruitment of Aux1 or GAK that led to uncoating, following scission of the membrane vesicle, was in all cases substoichiometric; uncoating with normal kinetics often occurred after just four to six molecules of either protein had accumulated. We also found that auxilins were absent from assembling pits, thus ruling out the possibility that earlier arrival could lead to Hsc70-driven clathrin exchange during coated pit formation or to uncoating of an incomplete lattice and hence to a futile assembly-disassembly cycle.
The phosphoinositide composition of an endocytic coated vesicle remains unchanged until the moment of separation from the plasma membrane but then undergoes a well-defined series of sequential modifications (He et al., 2017). Proposals for the mechanism by which the uncoating machinery distinguishes a pinched-off vesicle from maturing coated pit have invoked phosphoinositide recognition by PTEN-like domain and an enzymatic mechanism that alters vesicle lipid composition following budding from the parent membrane (Cremona et al., 1999; He et al., 2017). In the experiments reported here, recruitment of Aux1 and GAK followed these temporal variations in phosphoinositide composition, as dictated by the differential specificities of their PTEN-like domains. These observations suggest a coincidence-detection and lipid-switch timing mechanism that distinguishes a coated vesicle from a coated pit and that launches the uncoating process as soon as coated vesicle formation is complete.
Results
Dynamics of auxilin-mediated uncoating
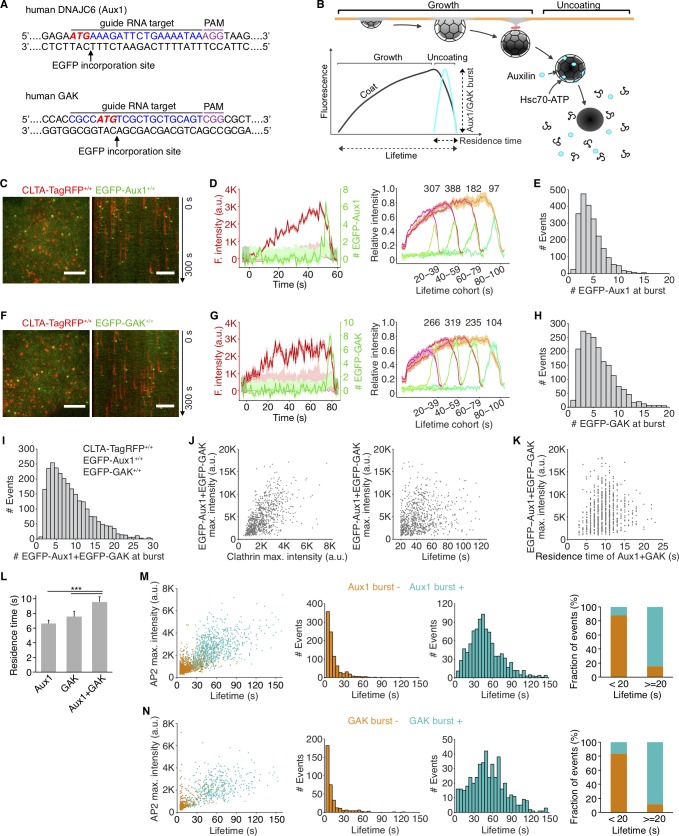
We established cell lines expressing fluorescently tagged Aux1 or GAK by homozygous replacement with a corresponding chimera bearing an N-terminal EGFP (EGFP-Aux1 or EGFP-GAK; Fig. 1 A and Fig. S1, A–C). The same cells also had either full replacement of clathrin light chain A (CLTA) with the fluorescent chimera CLTA-TagRFP or full replacement of adaptor protein 2 (AP2)-σ2 with AP2-σ2-TagRFP. SUM159 cells (Forozan et al., 1999), like HeLa and other nonneuronal lines (Borner et al., 2012; Hirst et al., 2008), express both Aux1 and GAK (Fig. S1, B and C). We verified that clathrin-mediated endocytic efficiency in the gene-edited cells resembled that of the parental cells (Fig. S1, D and E) and confirmed that the burst-like recruitment of EGFP-Aux1 and EGFP-GAK to coated vesicles was restricted to the time of clathrin uncoating (Fig. 1, B–H). Aux1 bursts and most GAK bursts occurred at the relatively immobile clathrin spots we have shown to be associated with endocytic events (Ehrlich et al., 2004). GAK, but not Aux1, also associates with more mobile, clathrin-coated structures emanating from the trans-Golgi network (TGN) and endosomes (Greener et al., 2000; Kametaka et al., 2007; Lee et al., 2005; Zhang et al., 2005); a few objects in the EGFP-GAK–expressing cells indeed appeared mobile in our TIRF microscopy time series. We confirmed this differential recruitment by full volume 3D live-cell lattice light-sheet microscopy (Fig. 2, A, B, and D).
Figure 1.
Recruitment of Aux1 and GAK to clathrin-coated vesicles in genome-edited cells. (A) CRISPR/Cas9 gene editing strategy used to incorporate EGFP at the N-terminus of Aux1 or GAK. The target sequence at the genomic locus of gene DNAJC6 (Aux1) recognized by the sgRNA, the protospacer adjacent motif (PAM), the start codon ATG (red), and the site of EGFP incorporation upon homologous recombination are highlighted. (B) Schematic representation of the bursts of Aux1/GAK during uncoating of clathrin-coated vesicles (modified from Massol et al., 2006). (C) Snapshot (left) and kymograph (right) from a representative TIRF microscopy time series showing transient burst of EGFP-Aux1 in coated vesicles containing CLTA-TagRFP in SUM159 cell double-edited for EGFP-Aux1+/+ and CLTA-TagRFP+/+. Time series with single molecule detection sensitivity for EGFP acquired for 300 s at 1-s intervals and 100-ms exposures. Kymograph shifted laterally by 5 pixels. Scale bars, 5 µm. (D) Left: Representative plot of a single endocytic event showing fluorescence (F.) intensity traces for CLTA-TagRFP and EGFP-Aux1 (arbitrary units for CLTA; number of molecules for Aux1) together with estimated uncertainties (1 SD, dark shade), corresponding local backgrounds (thin lines), and significance threshold above background (∼2 SD, light shade). Right: Averaged fluorescence intensity traces (mean ± standard error [SE]) for CLTA-TagRFP and EGFP-Aux1 from endocytic coated pits and vesicles automatically identified in eight cells and grouped in cohorts according to lifetimes. Numbers of traces analyzed are shown above each cohort. (E) Distribution of the maximum number of EGFP-Aux1 molecules recruited during the uncoating burst (2,198 traces from 17 cells). (F–H) Transient burst of EGFP-GAK in coated vesicles containing CLTA-TagRFP in SUM159 cell double-edited for EGFP-GAK+/+ and CLTA-TagRFP+/+. Scale bars, 5 µm. Cohorts in G are from eight cells and number of traces analyzed are shown above each cohort; distribution in H is from 1,935 traces from 16 cells. (I) Distribution of the maximum number of EGFP-Aux1 together with EGFP-GAK molecules recruited during uncoating of clathrin-coated vesicles (2,636 traces) from 31 cells triple-edited for EGFP-Aux1+/+, EGFP-GAK+/+, and CLTA-TagRFP+/+. (J) Scatter plots for the maximum (max.) fluorescence intensities of EGFP-Aux1 and EGFP-GAK (781 events) as a function of the maximum fluorescence intensity of CLTA-TagRFP (left; Pearson correlation coefficient r = 0.569) or lifetime of clathrin-TagRFP (right; Pearson correlation coefficient r = 0.212) from nine cells triple-edited for EGFP-Aux1+/+, EGFP-GAK+/+, and CLTA-TagRFP+/+. (K) Scatter plots of maximum fluorescence intensities of EGFP-Aux1 and EGFP-GAK (716 events) as a function of the duration of Aux1 and GAK bursts (Pearson correlation coefficient r = 0.132) from nine cells triple-edited for EGFP-Aux1+/+, EGFP-GAK+/+, and CLTA-TagRFP+/+. (L) Duration of Aux1 and GAK bursts during uncoating of coated vesicles in cells gene-edited for EGFP-Aux1+/+ (six cells, Aux1), EGFP-GAK+/+ (five cells, GAK), or EGFP-Aux1+/+ and EGFP-GAK+/+ (nine cells, Aux1 + GAK) together with CLTA-TagRFP+/+; ***, P < 0.001 by one-way ANOVA with Tukey’s comparison test. (M) Left: Scatter plot of maximum fluorescence intensities and lifetimes from AP2-TagRFP tracks with (996 traces) or without (919 traces) detectable EGFP-Aux1 burst, from nine cells double-edited for EGFP-Aux1+/+ and AP2-TagRFP+/+. Middle: Lifetime distributions from AP2-TagRFP tracks without or with detectable EGFP-Aux1 burst. Right: Fraction of AP2 tracks of lifetime shorter or longer than 20 s with or without detectable EGFP-Aux1 burst. (N) Left: Scatter plot of maximum fluorescence intensities and lifetimes of AP2-TagRFP tracks with (467 traces) or without (350 traces) detectable EGFP-GAK burst, from six cells double-edited for EGFP-GAK+/+ and AP2-TagRFP+/+. Middle: Lifetime distributions from AP2-TagRFP tracks without or with detectable EGFP-GAK burst. Right: Fraction of AP2 tracks of lifetime shorter or longer than 20 s with or without a detectable EGFP-GAK burst.
Figure S1.
Gene editing of SUM159 cells to express CLTA-TagRFP and EGFP-Aux1 or CLTA-TagRFP and EGFP-GAK. (A) CRISPR/Cas9 gene editing strategy used to incorporate EGFP at the N-terminus of Aux1 or GAK. The resulting DNA sequences, including the short linker between the C-terminus of EGFP and N-terminus of Aux1 or GAK, are shown. (B) Genomic PCR analysis showing biallelic integration first of EGFP into the DNAJC6 (Aux1) genomic locus to generate the clonal gene-edited cell line EGFP-Aux1+/+ (left) and then of TagRFP into the CLTA genomic locus of the same cells to generate the clonal double-edited cell line EGFP-Aux1+/+ CTLA-TagRFP+/+ (center). Right: Western blot analysis of cell lysates from the EGFP-Aux1+/+ cells probed with antibodies for Aux1/GAK and actin. Although the genomic PCR shows biallelic integration of EGFP sequence into the Aux1 genomic locus, the Western blot indicates expression of a small amount (∼15%) of untagged Aux1. The expression of EGFP-Aux1 in EGFP-Aux1+/+ cells was higher than endogenous Aux1 in the parental SUM159 cells; this up-regulation of EGFP-Aux1 expression is due to either single-cell cloning selection or the genome editing. (C) Genomic PCR analysis showing biallelic integration first of TagRFP into the CLTA genomic locus to generate the clonal gene-edited cell line CLTA-TagRFP+/+ (left) and then of EGFP into the GAK genomic locus of the same cells to generate the clonal double-edited cell line EGFP-GAK+/+ and CTLA-TagRFP+/+ (center). Right: Western blot analysis of cell lysates from the EGFP-GAK+/+ and CTLA-TagRFP+/+ cells probed with antibodies for GAK and actin. (D) Effect of expression of EGFP-Aux1 and CTLA-TagRFP on receptor-mediated uptake of transferrin. The histogram shows similar amounts of internalized Alexa Fluor 647–conjugated transferrin in parental and gene-edited EGFP-Aux1+/+ and CTLA-TagRFP+/+ cells probed by flow cytometry (n = 5 experiments, mean ± SD, P value by two-tailed t test). (E) Effect of expression of EGFP-GAK and CTLA-TagRFP on receptor-mediated uptake of transferrin (n = 5 experiments, mean ± SD, P value by two-tailed t test). (F) Scatter plots comparing maximum fluorescence intensities of EGFP-Aux1 and CLTA-TagRFP with each other (left; Pearson correlation coefficient r = 0.331) and maximum fluorescence intensity of EGFP-Aux1 with the lifetime of the endocytic coated structure in which it was found (right; Pearson correlation coefficient r = 0.115), from 938 traces in eight cells. Data from bottom surfaces of double-edited EGFP-Aux1+/+ and CLTA-TagRFP+/+ cells imaged at 1-s intervals for 300 s by TIRF microscopy. (G) Scatter plots comparing maximum (max.) fluorescence intensities of EGFP-GAK and CLTA-TagRFP with each other (left; Pearson correlation coefficient r = 0.373) and max. fluorescence intensity of EGFP-GAK with the lifetime of the endocytic coated structure in which it was found (right; Pearson correlation coefficient r = 0.153), from 900 traces in eight cells. Data from bottom surfaces of double-edited EGFP-GAK+/+ and CLTA-TagRFP+/+ cells imaged at 1-s intervals for 300 s by TIRF microscopy.
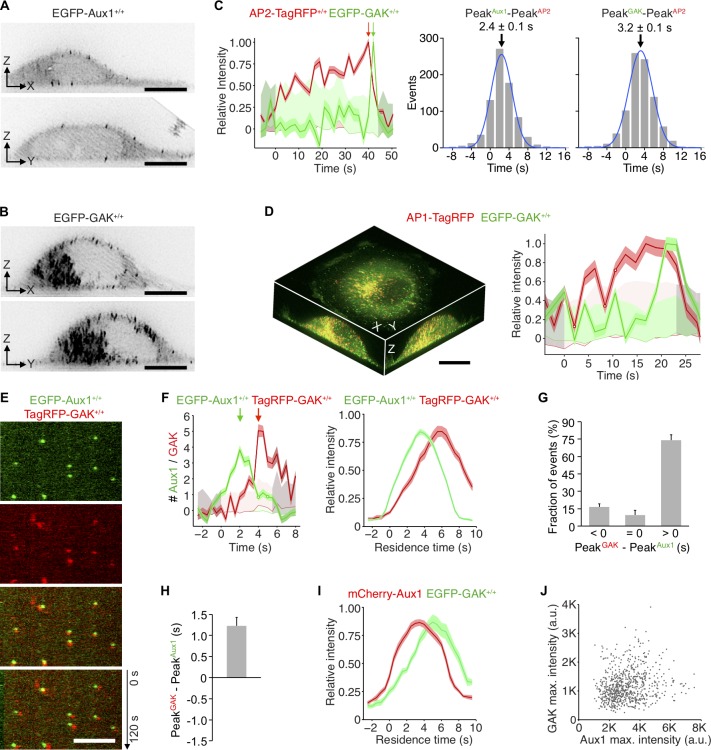
Figure 2.
Temporal and spatial distributions of Aux1 and GAK. (A and B) Maximum-intensity projections from a thin 2-µm optical section of a gene-edited cell expressing EGFP-Aux1 (A) or EGFP-GAK (B) recorded in 3D by lattice light-sheet microscopy. Scale bars, 10 µm. (C) Representative plot from 3D automated trackings of AP2-TagRFP and EGFP-Aux1 (872 traces from six cells) or AP2-TagRFP and EGFP-GAK (755 traces from six cells) in the double-edited cells imaged by lattice light-sheet microscopy. Distribution (fit with a single Gaussian) of the interval between maximum fluorescent signals for AP2 and Aux1 or AP2 and GAK (2.4 ± 0.1 and 3.2 ± 0.1 s, mean ± SE, respectively). (D) Distribution of GAK in cells gene-edited for EGFP-GAK+/+ and stably expressing AP1-TagRFP recorded in 3D by lattice light-sheet microscopy. Left: Maximum-intensity projections in X-Y, X-Z, and Y-Z of EGFP-GAK and AP1-TagRFP from a 3D rendered cell. Right: Representative plot of EGFP-GAK recruitment to a single AP1-positive carrier. Scale bar, 10 µm. (E) Snapshot from a representative TIRF microscopy time series showing the transient burst of EGFP-Aux1 and TagRFP-GAK+/+ on the plasma membrane of a cell double-edited for EGFP-Aux1+/+ and TagRFP-GAK+/+. Time series with single molecule detection sensitivity for EGFP and TagRFP acquired for 120 s at 1-s intervals using 100-ms exposures. Kymograph (bottom panel) shifted laterally by 5 pixels. Scale bar, 5 µm. (F) Representative plot (left) for a single endocytic event showing sequential recruitment of EGFP-Aux1 and TagRFP-GAK (recruitment peaks highlighted by arrows) imaged at 0.5-s intervals by TIRF microscopy. The traces (right) are averaged relative fluorescence intensity (mean ± SE) of EGFP-Aux1 and TagRFP-GAK for the cohort of EGFP-Aux1 bursts with residence times of 3–12 s (1,516 traces from eight cells). (G) The relative time differences (<0, 0, and >0 s) between peaks of EGFP-Aux1 and TagRFP-GAK recruitment (mean ± SD, n = 8 cells). (H) Distribution of interval between peaks of EGFP-Aux1 and TagRFP-GAK recruitment. The mean interval and SD from eight cells was 1.34 ± 0.26 s. (I) Averaged relative fluorescence intensity (mean ± SE) for bursts of duration of 3–12 s of transiently expressed mCherry-Aux1 and gene-edited EGFP-GAK (1,859 traces from eight cells). (J) Scatter plot of maximum (max.) fluorescence intensities of EGFP-Aux1 and TagRFP-GAK (750 traces from eight cells double-edited for EGFP-Aux1+/+ and TagRFP-GAK+/+, Pearson correlation coefficient r = 0.189).
In cells expressing AP2-σ2-TagRFP, nearly all (∼90%) AP2-containing structures with lifetimes <20 s incorporated relatively small amounts of AP2, failed to recruit EGFP-Aux1 or EGFP-GAK, and had a distinct quasi-exponential decay distribution of lifetimes, indicating a stochastic coat dissociation process (Fig. 1, M and N). They correspond to the early abortive coated pits previously described (Aguet et al., 2013; Ehrlich et al., 2004; Loerke et al., 2009). These characteristics provide a robust way to distinguish between dissociation of the lattices of abortive pits and disassembly of the lattices of coated vesicles and hence between an abortive pit (whatever its lifetime) and one that becomes a coated vesicle. They also show that the distinction between abortive and nonabortive events has a meaningful molecular signature.
As we and others have shown (Aguet et al., 2013; Ehrlich et al., 2004; Hong et al., 2015; Loerke et al., 2009), the interval between initiating an AP2-containing endocytic coated pit and pinching off as a coated vesicle ranges from 20 to 150 s. Most (∼90%) pits with lifetimes in this range incorporated greater amounts of AP2 than did the short-lived ones, had a multimodal lifetime distribution characteristic of a process with multiple steps, and showed at the time of uncoating a burst of EGFP-Aux1 or EGFP-GAK (Fig. 1, M and N). The multimodal lifetime distribution is a signature of productive coated pits (Aguet et al., 2013; Ehrlich et al., 2004; Loerke et al., 2009). The few longer-lived structures (∼10%) that failed to recruit auxilins had a quasi-exponential decay lifetime distribution (Fig. 1, M and N) and probably corresponded to the late abortives observed previously (Aguet et al., 2013; Ehrlich et al., 2004; Loerke et al., 2009). These characteristics also match the properties of abortive coated pits using dynamin as a surrogate marker (Aguet et al., 2013; Ehrlich et al., 2004; Loerke et al., 2009). We inferred from these observations that most endocytic clathrin-coated vesicles recruited both auxilins, and we confirmed this inference (see below) by observing concurrent recruitment of EGFP-Aux1 and TagRFP-GAK in double-edited SUM159 cells (Fig. 2 E).
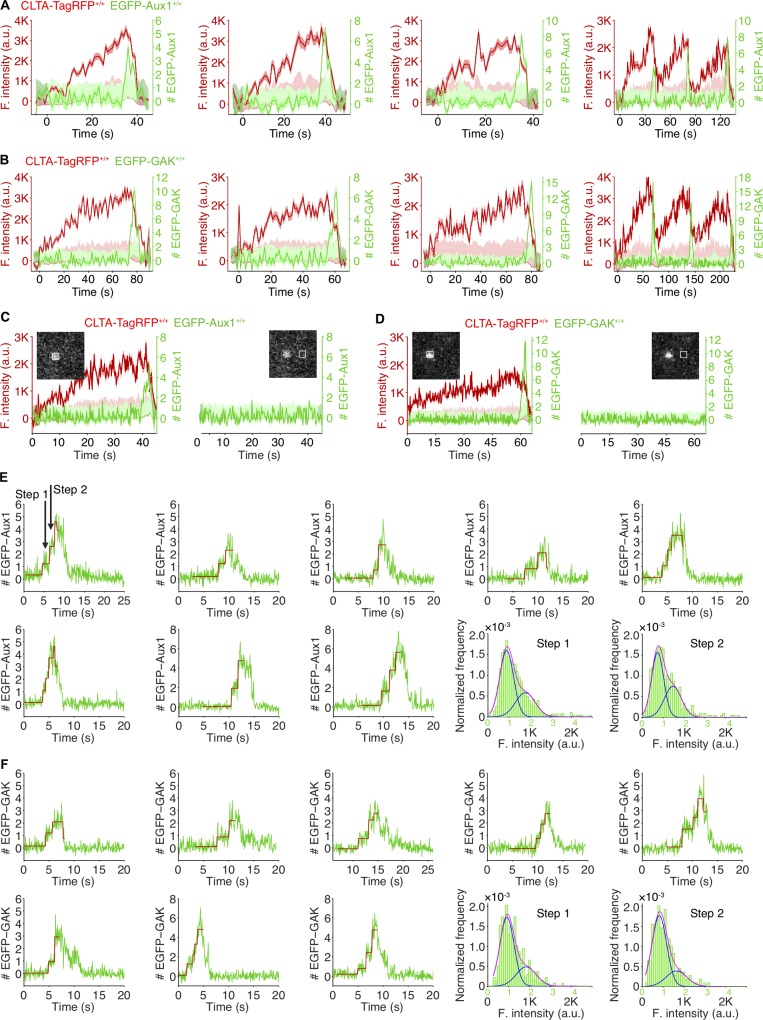
Auxilins are not recruited to assembling clathrin-coated pits
We did not detect EGFP-Aux1 or EGFP-GAK recruitment while coated pits were assembling, even with the single-molecule sensitivity of our TIRF microscopy (Fig. 1, C, D, F, and G; Fig. S2, A–D; Video 1; and Video 2); we saw burst recruitment only when assembly was complete. These results imply both that completion triggers recruitment and that an Aux1- or GAK-mediated process (and by inference activity of Hsc70, which has resisted attempts to append a fluorescent protein) cannot account for the observed partial exchange of clathrin between assembling endocytic coated pits and a cytosolic clathrin pool (Eisenberg and Greene, 2007; Wu et al., 2001). The lattice of an assembling coat is nonetheless competent to bind Aux1, since Aux1-based sensors for phosphatidylinositol-4-5-biphosphate (PtdIns(4,5)P2), the predominant lipid species in the plasma membrane, appear at coated pits in quantities that follow the clathrin content (He et al., 2017).
Figure S2.
Recruitment of Aux1 and GAK to clathrin-coated vesicles in genome-edited cells. (A) Representative plots of single endocytic events (first three panels) and hotspots (last panel) showing fluorescence intensity traces for CLTA-TagRFP and EGFP-Aux1 (arbitrary units for CLTA; number of molecules for Aux1) imaged at 1-s intervals by TIRF microscopy. (B) Representative plots of single endocytic events (first three panels) and hotspots (last panel) showing fluorescence intensity traces for CLTA-TagRFP and EGFP-GAK imaged at 1-s intervals by TIRF microscopy. (C) EGFP-Aux1 recruitment was not detected while coated pits were assembling. Representative plots of a single endocytic event showing fluorescence intensity traces for CLTA-TagRFP and EGFP-Aux1 (left) imaged at 250-ms intervals by TIRF microscopy. The EGFP-Aux1 signal was detected and measured as indicated in the insert image. Right: Fluorescence intensity fluctuations of the EGFP channel measured from the boxed area 12 pixels away from the detected EGFP-Aux1 burst signal. (D) EGFP-GAK recruitment was not detected while coated pits were assembling. (E) Stepwise recruitment of Aux1 to coated vesicles. Representative plots of EGPF-Aux1 burst-like recruitment (shown as number of molecules for Aux1) imaged at 62.5-ms intervals with TIRF microscopy; fit (red) obtained by applying a step-fitting function to estimate the average recruited molecules during the initiation phase of Aux1 burst-like recruitment. The last two panels show the histogram distributions (with Gaussian fitting) of EGFP-Aux1 molecules during the first step and second step of its recruitment. (F) Stepwise recruitment of GAK to coated vesicles.
Video 1.
Dynamics of EGFP-Aux1 recruitment to clathrin-coated vesicles. Bottom surface of a SUM159 cell gene-edited for EGFP-Aux1+/+ and CLTA-TagRFP+/+ was imaged by TIRF microscopy every 1 s for 200 s. To facilitate visualization, the EGFP channel was shifted laterally by 5 pixels in the right panel.
Video 2.
Dynamics of EGFP-GAK recruitment to clathrin-coated vesicles. Bottom surface of a SUM159 cell gene-edited for EGFP-GAK+/+ and CLTA-TagRFP+/+ was imaged by TIRF microscopy every 1 s for 200 s. To facilitate visualization, the EGFP channel was shifted laterally by 5 pixels in the right panel.
Recruitment specificity of auxilins
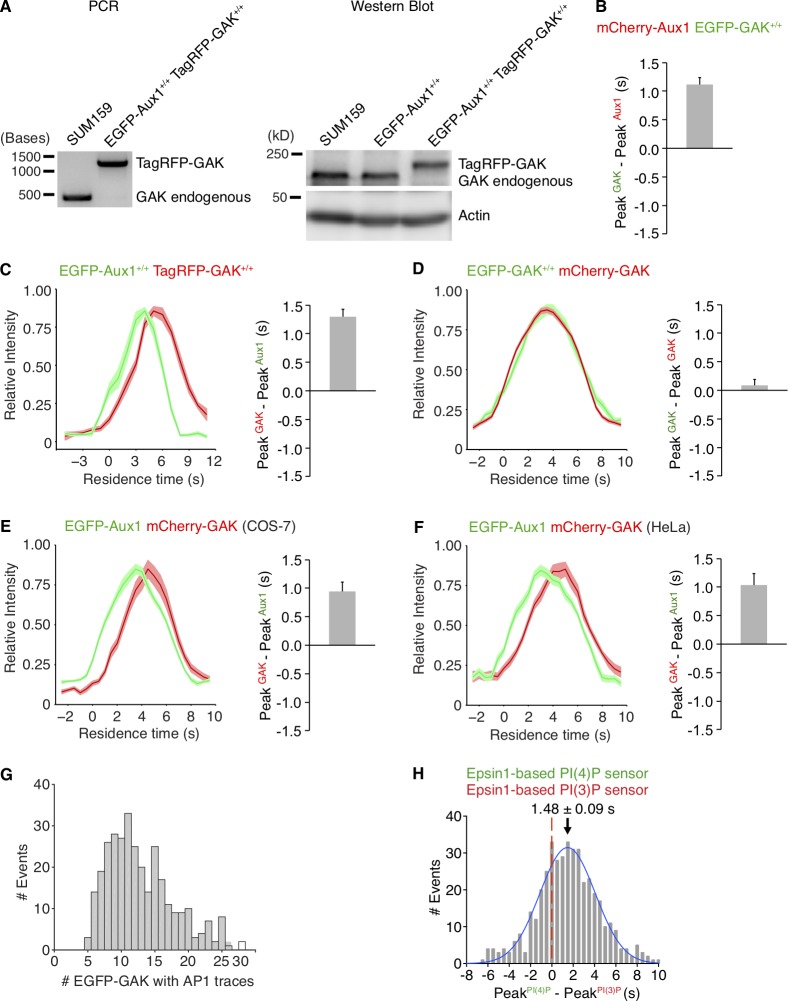
We probed recruitment specificity and burst dynamics of Aux1 and GAK by 3D tracking of EGFP-Aux1 or EGFP-GAK in gene-edited cells expressing AP2-σ2-TagRFP together with EGFP-Aux1 or EGFP-GAK (Fig. 2 C). Analysis of time series obtained by 3D live-cell lattice light-sheet microscopy showed that the time points for peak recruitment of Aux1 preceded those for GAK by ∼1 s (∼2.4- and ∼3.2-s peak recruitment after initiation of uncoating, respectively; Fig. 2 C). We found the same differential timing in gene-edited cells expressing both EGFP-Aux1 and TagRFP-GAK (Fig. 2, E–H; Fig. S3, A and C; and Video 3). The observed recruitment delays were not due to the fluorescent tags: Aux1 and GAK maintained their differential timing in cells gene edited to express EGFP-GAK and transiently expressing mCherry-Aux1 or mCherry-GAK (Fig. 2 I and Fig. S3, B and D). Moreover, we found the same relative recruitment dynamics of Aux1 and GAK in monkey COS-7 and human HeLa cells transiently expressing EGFP-Aux1 and mCherry-GAK (Fig. S3, E and F). We found no strong correlation between the maximum amplitudes of Aux1 and GAK in the same coated vesicle (Fig. 2 J).
Figure S3.
Sequential bursts of Aux1 and GAK during uncoating of clathrin-coated vesicles at the plasma membrane and recruitment of GAK to the intracellular clathrin-containing carriers. (A) The TagRFP sequence was inserted into the GAK genomic locus of the EGFP-Aux1+/+ cells to generate the double-edited cells EGFP-Aux1+/+ and TagRFP-GAK+/+, as confirmed by genomic PCR analysis (left) and Western blot analysis probed with antibodies for GAK and actin (right). (B) Gene-edited EGFP-GAK+/+ cells transiently expressing mCherry-Aux1 were imaged at 0.5-s intervals for 60 s by TIRF microscopy. The average time interval between the peaks of intensity for EGFP-GAK and mCherry-Aux1 is shown (mean ± SD, n = 8 cells). (C) Bottom surfaces of EGFP-Aux1+/+ and TagRFP-GAK+/+ cells were imaged at 1-s intervals for 120 s by TIRF microscopy. Left: Averaged fluorescence intensity traces (mean ± SE) of both EGFP-Aux1 (green) and TagRFP-GAK (red) for the EGFP-Aux1 3–12-s cohort (1,560 traces from 12 cells). Right: Average time interval between the peaks of intensity for EGFP-Aux1 and TagRFP-GAK (mean ± SD, n = 6 cells). (D) Gene-edited EGFP-GAK+/+ cells transiently expressing mCherry-GAK were imaged at 0.5-s intervals for 60 s by TIRF microscopy. Left: Averaged fluorescence intensity traces (mean ± SE) of EGFP-GAK (green) and mCherry-GAK (red) from the EGFP-GAK 3–12-s cohort (2,306 traces from 15 cells). Right: Average interval between the peak intensities of EGFP-GAK and mCherry-GAK (mean ± SD, n = 15 cells). (E) COS-7 cells transiently expressing EGFP-Aux1 and mCherry-GAK were imaged at 0.5-s intervals for 60 s by TIRF microscopy. Left: Averaged fluorescence intensity traces (mean ± SE) of EGFP-Aux1 (green) and mCherry-GAK (red) from the EGFP-Aux1 3–12-s cohort (656 traces from nine cells). Right: Average interval between the peak intensities of EGFP-Aux1 and mCherry-GAK (mean ± SD, n = 9 cells). (F) HeLa cells transiently expressing EGFP-Aux1 and mCherry-GAK were imaged at 0.5-s intervals for 60 s by TIRF microscopy. Left: Averaged fluorescence intensity traces (mean ± SE) of EGFP-Aux1 (green) and mCherry-GAK (red) from the EGFP-Aux1 3–12-s cohort (595 traces from 11 cells). Right: Average interval between the peak intensities of EGFP-Aux1 and mCherry-GAK (mean ± SD, n = 11 cells). (G) Gene-edited EGFP-GAK+/+ cells stably expressing AP1-TagRFP were imaged in 3D by lattice light-sheet microscopy. Distribution of the maximum number of EGFP-GAK molecules recruited to individual AP1-coated carriers (325 traces from 11 cells). (H) Bottom surfaces of cells transiently expressing Epsin1-based PtdIns(4)P sensor EGFP-P4M(DrrA)-Dlv2(508–736)-Epsin1(255–501) and PtdIns(3)P sensor mCherry-2xFYVE(Hrs)-Dlv2(508–736)-Epsin1(255–501) imaged by TIRF microscopy every 0.5 s for 100 s. Distribution (fit with a single Gaussian) for the interval between the peaks within single events showing that the Epsin1-based PtdIns(3)P sensor precedes the PtdIns(4)P sensor by 1.48 ± 0.09 s (mean ± SE, 436 traces from 23 cells).
Video 3.
Sequential recruitment of EGFP-Aux1 and TagRFP-GAK to clathrin-coated vesicles. Bottom surface of a SUM159 cell gene-edited for EGFP-Aux1+/+ and TagRFP-GAK+/+ was imaged by TIRF microscopy every 1 s for 120 s. To facilitate visualization, the TagRFP channel was shifted laterally by 5 pixels in the right panel.
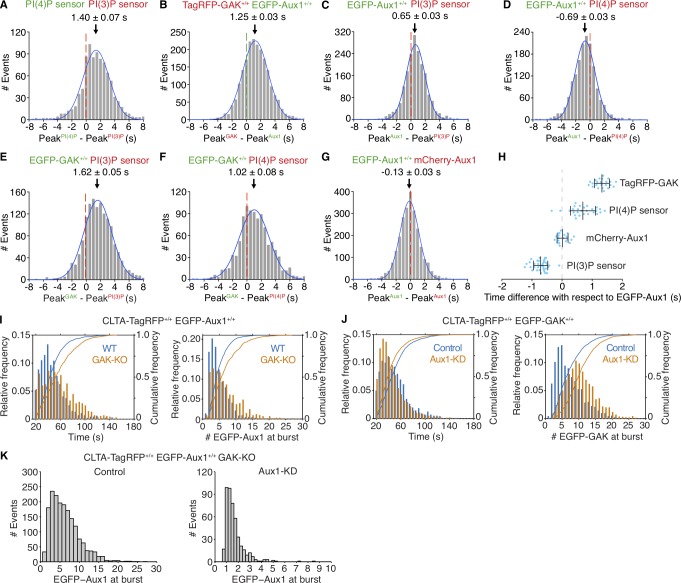
Why does Aux1 arrival precede that of GAK? Our study of phosphoinositide dynamics in endocytic compartments showed sequential bursts of Aux1-based phosphatidylinositol-3-phosphate (PtdIns(3)P) and phosphatidylinositol-4-phosphate (PtdIns(4)P) sensors, ∼1–2 s apart, during uncoating of endocytic clathrin-coated vesicles (He et al., 2017). The results in Fig. 3 (A–H) show a close correspondence between the arrival times at endocytic coated vesicles of a Aux1-based PtdIns(3)P sensor and Aux1 and between the (∼1–2 s later) arrival times of a Aux1-based PtdIns(4)P sensor and GAK. PtdIns(3)P and PtdIns(4)P sensors with an unrelated clathrin binder, Epsin1 replacing Aux1 (He et al., 2017), also showed sequential bursts ∼1–2 s apart (Fig. S3 H). These correlations suggest that phosphoinositide conversion determines the differential recruitment of Aux1 and GAK. This inference is also consistent with the results of in vitro lipid-protein overlay assays (Lee et al., 2006; Massol et al., 2006) and with the lipid dependence of Aux1- or GAK-mediated uncoating in the in vitro single-particle uncoating experiments described below.
Figure 3.
Recruitment of Aux1, GAK, and phosphoinositide sensors to clathrin-coated vesicles. (A–H) Bottom (adherent) surfaces of cells transiently expressing various combinations of Aux1, GAK, and Aux1-based PtdIns(3)P and PtdIns(4)P sensors imaged by TIRF microscopy every 0.5 s for 100 s. (A) Transient coexpression of Aux1-based PtdIns(4)P (EGFP-P4M(DrrA)-Aux1) and PtdIns(3)P (mCherry-2xFYVE(Hrs)-Aux1) sensors in parental SUM159 cells. Distribution (single Gaussian fit) for the interval between the peaks within single events: the PtdIns(3)P sensor precedes the PtdIns(4)P sensor by 1.40 ± 0.07 s (mean ± SE, 916 traces from 34 cells). (B) Distribution of interval between the peaks within single events of EGFP-Aux1 and TagRFP-GAK in cells double-edited for EGFP-Aux1+/+ and TagRFP-GAK+/+ (1.25 ± 0.03 s, mean ± SE, 2,033 traces from 23 cells). (C and D) Transient expression of PtdIns(3)P (mCherry-2xFYVE(Hrs)-Aux1; C) or PtdIns(4)P (mCherry-P4M(DrrA)-Aux1) sensor (D) in gene-edited EGFP-Aux1+/+ cells. Distributions of interval between burst peaks for Aux1 and phosphoinositide sensors in the same event. Aux1 and PtdIns(3)P sensor: 0.65 ± 0.03 s, mean ± SE, 1,863 traces in 35 cells; Aux1 and PtdIns(4)P sensor: −0.69 ± 0.03 s; 1,570 traces in 27 cells. (E and F) Transient expression of PtdIns(3)P (mCherry-2xFYVE(Hrs)-Aux1; E) or PtdIns(4)P (mCherry-P4M(DrrA)-Aux1) sensor (F) in gene-edited EGFP-GAK+/+ cells. Distributions of interval between burst peaks for GAK and phosphoinositide sensors in the same event. GAK and PtdIns(3)P sensor (1.62 ± 0.05 s; 1,435 traces in 36 cells); GAK and PtdIns(4)P sensor (1.02 ± 0.08 s; 1,020 traces in 34 cells). (G) Transient expression of mCherry-Aux1 in gene-edited EGFP-Aux1+/+ cells. Distribution of interval between burst peaks for mCherry-Aux1 and EGFP-Aux1 in the same event (−0.13 ± 0.03 s; 2,435 traces in 34 cells). (H) Interval between burst peaks of Aux1 in gene-edited EGFP-Aux1+/+ cells and of TagRFP-GAK+/+ gene edited in the same cells (n = 23 cells), or between the burst peaks of Aux1 in gene-edited EGFP-Aux1+/+ cells and transiently expressed PtdIns(4)P sensor (mCherry-P4M(DrrA)-Aux1, n = 27 cells), mCherry-Aux1 (n = 34 cells), or PtdIns(3)P sensor (mCherry-2xFYVE(Hrs)-Aux1, n = 35 cells). The values of each spot represent the average (mean ± SD) of the measurement obtained for a given single cell. The timing differences between the bursts for each group were statistically significant (P < 0.001 by one-way ANOVA with Tukey’s comparison test). (I) Effect of GAK knockout on recruitment of Aux1 to endocytic clathrin-coated vesicles in cells gene edited for EGFP-Aux1+/+ and CLTA-TagRFP+/+. Histogram and cumulative distributions showing increases in the number of EGFP-Aux1 molecules recruited during the burst (Cohen’s d = 0.45) and in the lifetime (Cohen’s d = 0.57) of clathrin-coated structures, determined in 1,272 traces from 14 WT cells and in 794 traces from 14 knockout (GAK-KO) cells. (J) Effect of Aux1 knockdown by shRNA on recruitment of GAK to endocytic clathrin-coated vesicles, in cells gene edited for EGFP-GAK+/+ and CLTA-TagRFP+/+. Histogram and cumulative distributions for the number of EGFP-Aux1 molecules recruited during the burst (Cohen’s d = 0.73) and the lifetimes of the clathrin-coated structures (Cohen’s d = 0.29), determined in 1,498 traces from 15 control cells and in 1,793 traces from 14 knockdown (Aux1-KD) cells. (K) Effect of GAK knockout and Aux1 knockdown by siRNA on recruitment of Aux1 to endocytic clathrin-coated vesicles, in cells gene edited for EGFP-Aux1+/+ and CLTA-TagRFP+/+ and knockout for GAK. Histogram distributions for the number of EGFP-Aux1 molecules recruited during the burst of clathrin-coated structures, determined in 1,794 traces from 20 control cells and in 465 traces from 47 knockdown (Aux1-KD) cells.
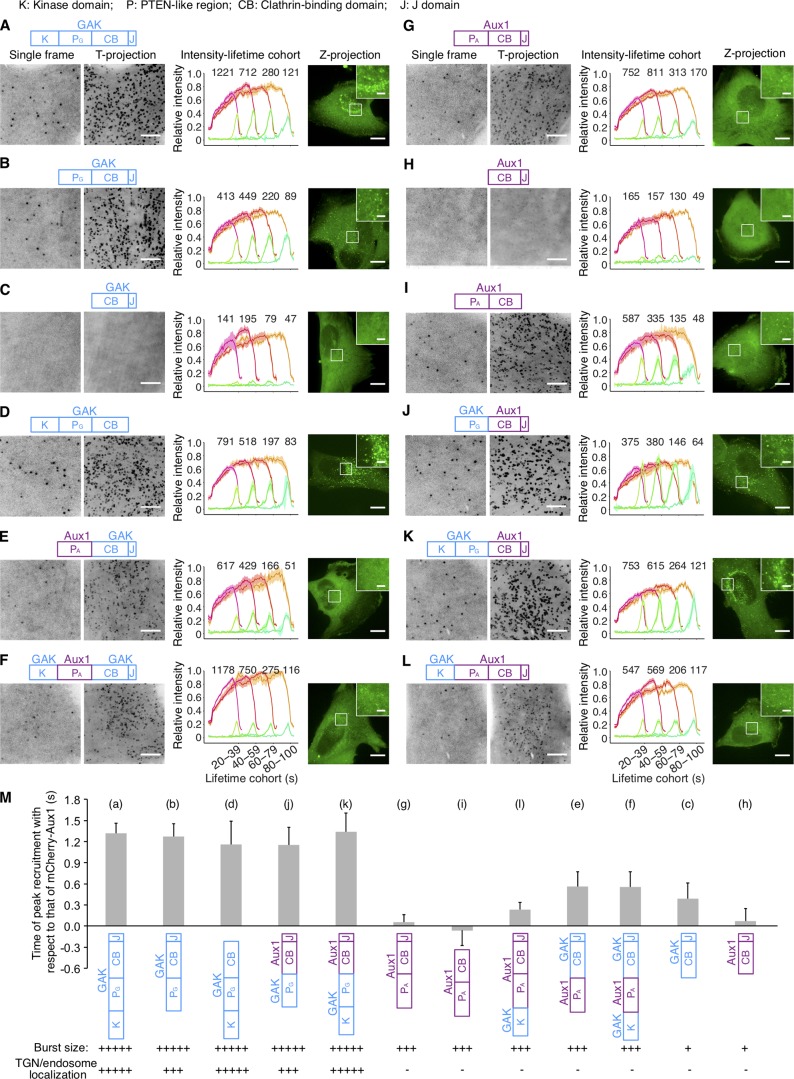
We swapped the PTEN-like domains of Aux1 and GAK and followed the intracellular location of the transiently expressed chimeric variants (Fig. 4). As confirmed above (Fig. 2, B and D), WT GAK appears in the perinuclear TGN and recycling endosomes, both of which are enriched in PtdIns(4)P (Kural et al., 2012; Wang et al., 2003), as well as in endocytic coated vesicles (Fig. 4 A). As shown previously (Guan et al., 2010; Lee et al., 2006; Massol et al., 2006), the PTEN-like domain was essential for efficiently targeting Aux1 or GAK to endocytic coated vesicles (Fig. 4, C and H). A GAK chimera with the PTEN-like domain of Aux1 instead of its own appeared exclusively in endocytic coated vesicles at the plasma membrane (Fig. 4, E and F), and the extent to which those vesicles recruited the GAK-Aux1 chimera was similar to the extent of recruitment of WT Aux1 (Fig. 4 G) but slightly less than that of WT GAK (Fig. 4 A); the arrival time of this chimera also corresponded to the arrival time for WT Aux1 (Fig. 4 M). The converse chimera, Aux1 with the PTEN-like domain of GAK, acquired the plasma membrane recruitment dynamics of WT GAK (Fig. 4, J and K). Although the kinase domain was not required for recruiting GAK to endocytic coated vesicles, its presence substantially enhanced perinuclear targeting (Fig. 4, A and B). The kinase also enhanced perinuclear targeting of chimeric Aux1 with the GAK PTEN-like domain (Fig. 4, J and K); adding it to WT Aux1 had no effect (Fig. 4 L).
Figure 4.
Influence of PTEN-like domains on Aux1 and GAK recruitment. (A–L) Bottom surfaces of gene-edited CLTA-TagRFP+/+ cells transiently expressing indicated EGFP-tagged constructs of Aux1 or GAK, imaged by TIRF microscopy every 1 s for 300 s. Each panel shows a schematic representation of the construct domain organization: K, kinase domain of GAK (amino acid residues 1–347); PA or PG, PTEN-like domain of GAK (360–766) or Aux1 (1–419); CB, clathrin-binding domain of GAK (767–1,222) or Aux1 (420–814); J, J domain of GAK (1,223–1,311) or Aux1 (815–910); a representative single frame and the corresponding maximum-intensity time projection of the time series as a function of time (T-projection). The plots show averaged fluorescence intensity traces (mean ± SE) of CLTA-TagRFP (red) and EGFP-fused constructs (green), from 6–13 cells per condition, grouped by cohorts according to lifetimes. The numbers of analyzed traces are shown above each cohort. The cells were also imaged in 3D by spinning-disk confocal microscopy; the images at the right of each panel show representative maximum-intensity z projections (Z-projection) from 34 sequential optical sections spaced at 0.3 µm (scale bars, 10 µm) and corresponding enlarged regions (scale bars, 2 µm). (M) Mean interval between the fluorescence maxima for the indicated EGFP-fused construct (diagram immediately below plot) and the mCherry-Aux1 burst (mean ± SD, n = 8–14 cells), in cells imaged at 0.5-s intervals for 60 s by TIRF microscopy. Below the construct schematics are qualitative estimates of the relative maximum amplitudes of fluorescence for the bursts at the plasma membrane and in regions of the TGN/endosome.
A few auxilins suffice to trigger uncoating
Previous in vitro ensemble studies have suggested that less than one auxilin per vertex is sufficient to elicit Hsc70-driven disassembly of synthetic coats (Böcking et al., 2011). To determine the requirements for Aux1 and GAK in vivo, we used the high sensitivity of calibrated oblique illumination TIRF microscopy (He et al., 2017) to follow recruitment of EGFP-Aux1 or EGFP-GAK to endocytic coated vesicles in gene-edited cells. The duration of the EGFP-Aux1 or EGFP-GAK bursts (6–8 s) was the same as for the corresponding ectopically expressed proteins (He et al., 2017; Lee et al., 2006; Massol et al., 2006; Fig. 1 L). Bursts had peak values of 3 ± 1 and 4 ± 2 molecules for Aux1 and GAK, respectively (Fig. 1, E and H). Detailed analysis of rapidly acquired EGFP-Aux1 or EGFP-GAK traces showed that the first recorded events were consecutive stepwise increases in fluorescence intensity (Fig. S2, E and F, selected examples). The analysis suggests that recruitment begins with arrival of a single auxilin followed by a second one (within the 62.5-ms time resolution of our measurements). The maximal occupancy of both auxilins during the burst ranged from 2 to 20, as determined by the peak fluorescence intensity of EGFP-Aux1 and EGFP-GAK inserted together in the same gene-edited cells (Fig. 1 I). The combined duration of their bursts was slightly longer (∼10 s) than each one alone (Fig. 1, K and L). We found no correlation between the number of recruited auxilins and the observed uncoating rate, nor did the peak level correlate with the size of the coat, as estimated from the peak clathrin light-chain fluorescence intensity (Fig. 1 J and Fig. S1, F and G). Most endocytic coated vesicles have between 36 and ∼100 vertices; uncoating proceeded even when only a relatively small proportion of the vertices have acquired an auxilin. We found a similarly substoichiometric GAK occupancy at perinuclear AP1-containing carriers; the amplitude of the GAK burst ranged from ∼5 to ∼25 (Fig. S3 G).
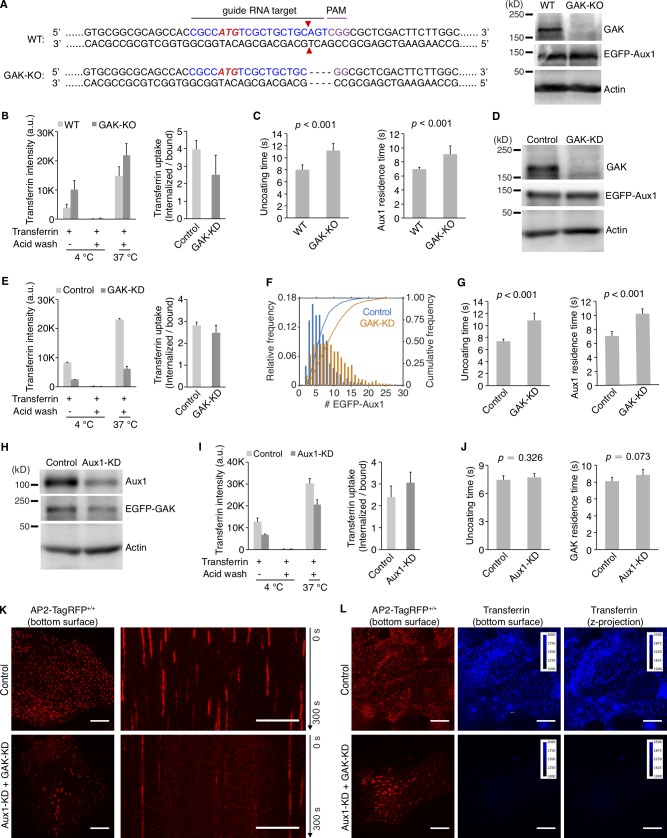
We examined the contribution of Aux1 or GAK alone to clathrin-mediated uptake of transferrin and to the kinetics of clathrin uncoating, by imaging cells lacking GAK (by CRISPR/Cas9-mediated knockout) or transiently depleted of Aux1 (by RNAi). Elimination of GAK (Fig. S4 A) had no significant effect on transferrin uptake (Fig. S4 B), while slightly increasing the lifetimes of clathrin-coated structures at the plasma membrane and the number of EGFP-Aux1 molecules recruited during the burst (Fig. 3 I). The expression level of Aux1 was unaffected (Fig. S4 A). The interval associated with the EGFP-Aux1 burst also increased slightly (Fig. S4 C). We obtained similar results with shRNA-mediated depletion of GAK (Fig. S4, D–G). Because we could not eliminate Aux1 by CRISPR/Cas9-mediated knockout, we used transient depletion with shRNA (Fig. S4 H). Aux1 depletion barely affected the transferrin uptake rate (Fig. S4 I) or the uncoating efficiency (Fig. 3 J and Fig. S4 J) but led to a small increase in the peak number of GAK molecules recruited during the uncoating burst (Fig. 3 J), mirroring the effect of GAK elimination.
Figure S4.
Effects on clathrin-mediated endocytosis of knockout or knockdown of GAK and knockdown of Aux1. (A) CRISPR/Cas9 gene editing strategy used to knock out GAK in cells gene edited for EGFP-Aux1+/+ and CTLA-TagRFP+/+. The double strand break (red triangles) induced by Cas9 resulted in elimination of four nucleotides (dotted lines). Loss of GAK expression was confirmed by Western blot with antibodies against GAK, Aux1/GAK, and actin (right). (B) Effect of GAK knockout on receptor-mediated uptake of transferrin (n = 3 experiments, mean ± SD). (C) Uncoating time and Aux1 residence time, in cells lacking GAK. Data from bottom surfaces of double-edited EGFP-Aux1+/+ and CLTA-TagRFP+/+ cells with GAK (n = 5 cells) or lacking GAK by knockout (n = 7 cells) imaged at 1-s intervals for 200 s by TIRF microscopy (mean ± SD, P values by two-tailed t test). (D) Western blot analysis of EGFP-Aux1+/+ and CLTA-TagRFP+/+ cells treated with lentivirus containing control shRNA (Control) or shRNA specific for GAK (GAK-KD), showing specific reduction of GAK expression 5 d after transduction. (E) Effect of GAK knockdown on receptor-mediated uptake of transferrin (n = 2 experiments, mean ± SD). (F) Influence of GAK depletion on Aux1 recruitment. Data from bottom surfaces of double gene-edited EGFP-Aux1+/+ and CLTA-TagRFP+/+ cells with GAK (1,058 traces, eight cells) or depleted of GAK by knockdown (1,380 traces, nine cells) imaged at 1-s intervals for 200 s by TIRF microscopy. The number of recruited EGFP-Aux1 molecules is significantly increased (Cohen’s d = 0.68). (G) Influence of GAK depletion on uncoating time (left) and Aux1 residence time (right). Data from bottom surfaces of double-edited EGFP-Aux1+/+ and CLTA-TagRFP+/+ cells with GAK (n = 5 cells) or depleted of GAK by knockdown (n = 5 cells) imaged at 1-s intervals for 200 s by TIRF microscopy (mean ± SD, P values by two-tailed t test). (H) Western blot analysis of parental SUM159 cells incubated with lentivirus containing control shRNA or shRNA specific for Aux1 (Aux1-KD) showing specific reduction of Aux1 expression 5 d after transduction. (I) Effect of Aux1 knockdown in gene-edited EGFP-GAK+/+ and CTLA-TagRFP+/+ cells on receptor-mediated uptake of transferrin (n = 2 experiments, mean ± SD). (J) Influence of Aux1 depletion on uncoating time (left) and GAK residence time (right). Data from bottom surfaces of double-edited EGFP-GAK+/+ and CLTA-TagRFP+/+ cells with Aux1 (n = 5 cells) or depleted of Aux1 by knockdown (n = 5 cells) imaged at 1-s intervals for 200 s by TIRF microscopy (mean ± SD, P values by two-tailed t test). (K) Bottom surfaces of AP2-TagRFP+/+ cells treated with lentivirus containing control shRNA or a mixture of shRNA targeting Aux1 and GAK (Aux1-KD + GAK-KD) imaged at 2-s intervals for 300 s by spinning-disk confocal microscopy. The representative images are from a single time point; the corresponding kymograph shows the entire time series. Scale bars, 10 µm. (L) AP2-TagRFP+/+ cells with or without double Aux1 + GAK knockdown incubated with 10 µg/ml Alexa Fluor 647–conjugated transferrin for 10 min at 37°C and then imaged in 3D using spinning-disk confocal microscopy (30 imaging planes spaced at 0.35 µm). Images from the bottom surface of control cells show diffraction-limited AP2-TagRFP spots associated with endocytic coated pits and coated vesicles; in cells depleted of Aux1 and GAK, the punctate distribution is replaced by characteristic larger patches. The images also show the extent of surface binding (bottom surface) and internalization (maximum z-projection of the 30 stacks) of transferrin in the control cells and its absence in the cells impaired in endocytosis due to the Aux1 and GAK depletion. Scale bars, 10 µm.
Combined depletion of GAK and Aux1 in gene-edited cells led to substantial loss of endocytic coated vesicles (Fig. S4 K) and inhibition of transferrin uptake (Fig. S4 L), as expected from knockdown experiments (Hirst et al., 2008). The remaining clathrin-coated structures recruited small bursts of one to two molecules of EGFP-Aux1 (Fig. 3 K), further suggesting that very few auxilins can recruit enough Hsc70 for uncoating.
Hsc70 dynamics
High-affinity engagement of substrate by Hsc70 requires both a bound J-domain and ATP hydrolysis (Böcking et al., 2011). Activated Hsc70 can therefore associate only at a vertex adjacent to its activating auxilin (Xing et al., 2010). We could not follow fluorescent-protein tagged Hsc70, however, because it is not recruited to clathrin coats (Lee et al., 2006; Video 4 and Video 5). The structure of Hsc70-bound coats suggests that a vertex can accommodate no more than one Hsc70 (Xing et al., 2010). Auxilin occupancy is therefore a first-level estimate of the number of Hsc70s required for functional uncoating in vivo. If the time for Hsc70 recruitment, clathrin binding, ATP hydrolysis, Hsc70 release, and auxilin exchange is shorter than the uncoating time, the peak steady-state level of auxilin could in principle underestimate the total number of auxilins that have participated. That is, dissociation of auxilin from the neighborhood of one vertex, having delivered its Hsc70, and vicinal rebinding (or binding of a different auxilin) at another vertex could result in an apparent steady-state occupancy of only one auxilin but delivery of Hsc70s to two distinct vertices. Some individual traces of the few events we could find in GAK knockout cells with essentially complete Aux1 knockdown suggested that two or three single Aux1 molecules might have arrived independently during the course of uncoating. From the various regimes we have examined, we estimate that maximal occupancy of no more than three to five auxilins (Fig. 1 I), and the inferred recruitment of <10 Hsc70s, is enough to dismantle coated vesicles in the size range (∼60 clathrin trimers) present in the cells we have used. Under normal conditions of Aux1 or GAK expression, additional Hsc70s might participate. A recent model derived from ensemble in vitro uncoating experiments posited that stoichiometric amounts of Aux1/GAK with respect to clathrin mediate a proposed sequential capture of up to three Hsc70 molecules to each triskelion (Rothnie et al., 2011). We have now ruled out this model by counting in vivo the number of molecules of Aux1/GAK recruited to a coated vesicle during uncoating (Fig. 1 I). Our in vivo data instead agree with earlier biochemical studies suggesting a “catalytic” role for Aux1 and GAK during uncoating (Ma et al., 2002).
Video 4.
EGFP-Hsc70 fails to be recruited by clathrin-coated vesicles. Bottom surface of a gene-edited AP2-TagRFP+/+ SUM159 cell transiently expressing EGFP-Hsc70 imaged by TIRF microscopy every 1 s for 112 s. The time series shows diffuse cytosolic fluorescence signal and absence of dots corresponding to clathrin-coated vesicles.
Video 5.
Hsc70-EGFP fails to be recruited by clathrin-coated vesicles. Bottom surface of a gene-edited AP2-TagRFP+/+ SUM159 cell transiently expressing Hsc70-EGFP imaged by TIRF microscopy every 1 s for 154 s. The time series shows diffuse cytosolic fluorescence signal and absence of spots corresponding to clathrin-coated vesicles.
Role of the PTEN-like domain
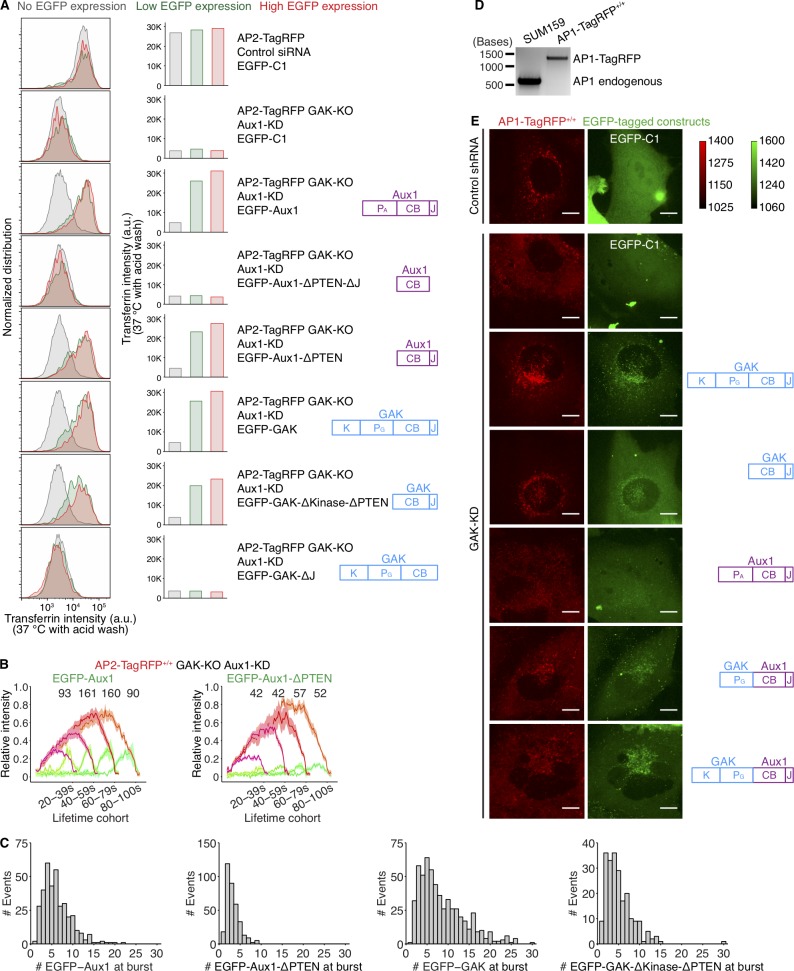
The experimental results in Fig. 4 show that the PTEN-like domains of Aux1 and GAK determine timing and amplitude of recruitment to coated vesicles. A truncated Aux1 that retains just the clathrin-binding and J domains (ΔPTEN Aux1) can nonetheless direct uncoating in vitro (Böcking et al., 2011), and ectopic expression of a GAK transgene encoding only the clathrin-binding and J-domains (ΔPTEN GAK) reverses the lethality of a conditional GAK knockout in liver or brain of mice and restores clathrin traffic in embryo fibroblasts derived from those mice (Park et al., 2015). We therefore compared the dynamics of recruitment and the compartment specificity of WT and ΔPTEN auxilins. Ectopic expression of various ΔPTEN variants of Aux1 or GAK, in cells devoid of GAK and depleted of Aux1, rescued transferrin uptake (Fig. S5 A). In these cells, ectopically expressed ΔPTEN EGFP-Aux1 or ΔPTEN EGFP-GAK also exhibited a burst of recruitment, with amplitude of 1–5 EGFPs, just after completion of coat assembly (Fig. S5, B and C). We chose for analysis cells with levels of ectopic expression comparable to those in gene-edited cells expressing the fluorescent auxilin under control of the endogenous promoter. Association with coated vesicles adequate to drive uncoating thus did not require PTEN-like domain recognition of phosphoinositides. The mean burst amplitude was comparable to the lowest amplitudes seen with full-length Aux1 or GAK, confirming that Hsc70 recruitment by relatively few auxilins can drive uncoating.
Figure S5.
Roles of the PTEN-like domain and clathrin-binding domain of auxilins in the endocytic and secretory pathways. (A) AP2-TagRFP+/+ cells with or without GAK (AP2-TagRFP GAK-KO) treated with control siRNA or siRNA targeting Aux1 for 3 d (two sequential transfections), then subjected to transient expression of the indicated EGFP-tagged constructs for additional 1 d followed by measurements of Alexa Fluor 647–conjugated transferrin uptake by flow cytometry. The plots (left) and equivalent histograms (right) show comparisons of the internalized transferrin (37°C with acid wash) in the absence or presence of low and high levels of ectopic expression of the indicated constructs. (B) The GAK-KO AP2-TagRFP+/+ cells were treated with siRNA targeting endogenous Aux1 and then transfected for transient expression of EGFP-tagged full-length Aux1 (left) or Aux1 lacking the PTEN-like domain (right). The cells were imaged at 1-s intervals for 300 s by TIRF microscopy. The averaged fluorescence intensity traces (mean ± SE) for AP2-TagRFP (red) and EGFP-tagged constructs (green) were identified in nine and seven cells, respectively, and then grouped in cohorts according to lifetimes. The numbers of analyzed traces are shown above each cohort. (C) The GAK-KO AP2-TagRFP+/+ cells were treated with siRNA targeting endogenous Aux1 and then transfected for transient expression of EGFP-tagged constructs as indicated. The cells with EGFP expression at a similar level as the endogenous auxilins were imaged at 1-s intervals for 300 s by TIRF microscopy. Distribution of the maximum number of EGFP-tagged molecules recruited during the uncoating burst (from left to right: 363 traces from five cells, 348 traces from six cells, 587 traces from five cells, 221 traces from five cells). (D) Genomic PCR analysis showing biallelic integration of TagRFP into the AP1S1 genomic locus to generate the clonal gene-edited cell line AP1-TagRFP+/+. (E) AP1-TagRFP+/+ cells were treated with lentivirus containing control shRNA or shRNA targeting GAK for 4 d, then subjected to transient expression of the indicated EGFP-tagged constructs for an additional 1 d, and volumetrically imaged by spinning-disk confocal microscopy (34 sequential optical sections spaced at 0.3 µm). Maximum-intensity z projections acquired using the same acquisition parameters as with gene-edited EGFP-GAK+/+ cells, making it possible to identify cells ectopically expressing at the same level as endogenous GAK. Scale bars, 10 µm.
The ΔPTEN EGFP-Aux1 in the experiments just described lacked the PTEN lipid sensor, yet it appeared only in fully assembled coated vesicles and not in coated pits. We showed (He et al., 2017) that probes with the Aux1 clathrin-binding domain and an unrelated sensor for PtdIns(4,5)P2 or PtdIns(4)P (phosphoinositides present in the plasma membrane) do appear in coated pits. Thus, during coated pit formation, recognition of the clathrin lattice appears to be necessary but not sufficient for auxilin recruitment; additional lipid-headgroup affinity is also required. After budding, however, clathrin-coated vesicles can recruit Aux1 or GAK lacking any PTEN-like domain. The average recruitment levels for these species in our experiments was substantially lower than for the corresponding WT proteins (Fig. S5 C), but it was nevertheless sufficient to elicit uncoating.
GAK lacking its PTEN-like domain can rescue the AP1 coated vesicle dispersal phenotype seen previously in HeLa cells depleted of full-length GAK (Kametaka et al., 2007; Lee et al., 2005; Fig. S5, D and E). Low-level ectopic expression of ΔPTEN GAK in cells depleted endogenous GAK led to recovery of the perinuclear distribution of AP1 seen in WT cells (Fig. 2 D and Fig. S5 E). A PTEN-less GAK thus appears to allow normal coated vesicle function in the secretory pathway.
Phosphoinositide binding preferences of Aux1 and GAK
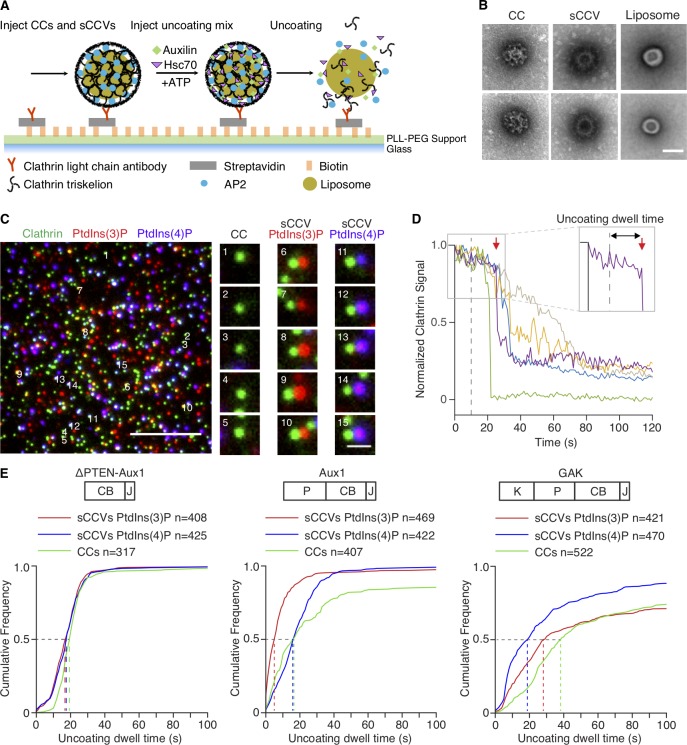
We modified our established in vitro uncoating assay (Böcking et al., 2011; see Materials and methods) to follow by single-particle TIRF microscopy the ATP-, Hsc70-, and auxilin-dependent release of fluorescent clathrin from synthetic clathrin/AP2-coated vesicles (sCCVs) and clathrin/AP2 coats immobilized on a glass coverslip (Fig. 5, A and B; Fig. S6; and Video 6). Because binding of the Aux1 or GAK PTEN-like domains with phosphoinositides is relatively weak, we turned to this assay when the conventional lipid-strip binding or vesicle flotation assays proved unreliable.
Figure 5.
Single-particle in vitro uncoating assay. (A) Schematic representation of the assay. The intensities of fluorescence from labeled clathrin and lipid dyes incorporated into liposomes surrounded by clathrin/AP2 coats were monitored by TIRF microscopy. Synthetic clathrin/AP2-coated vesicles (shown in the figure) and clathrin/AP2 coats were captured with a biotinylated monoclonal antibody specific for clathrin light chain on the surface of a poly(l-lysine)-poly(ethylene glycol)-Biotin-Streptavidin modified glass coverslip in a microfluidic chip. (B) Representative transmission EM images of negatively stained clathrin/AP2 coats (CC; left), synthetic clathrin/AP2 coated vesicles (sCCV; middle), or liposomes (right). Scale bar, 50 nm. (C) Representative TIRF image before initiation of uncoating. The snapshot combines images acquired in three different fluorescence channels (red and blue channels in enlarged images shifted right by 5 pixels) used to monitor the signal from coats and synthetic coated vesicles tagged with clathrin LCa-AF488 (green) and synthetic coated vesicles containing PtdIns(3)P or PtdIns(4)P and DiI (red) or DiD (blue), respectively. Scale bars, 5 µm (left) and 1 µm (right). (D) Representative uncoating profiles from single sCCVs. Plots show fluorescence intensity traces of the clathrin signal imaged at 1-s intervals starting 10 s before arrival of the uncoating mix (dashed vertical line at 0 s). Abrupt loss of signal in the green trace represents early release of the synthetic coated vesicle from the antibody on the glass surface. The enlarged boxed region (right corner) illustrates with a red arrow the onset of the uncoating reaction of the purple trace; the “uncoating dwell time” is the time it took to reach this point. (E) Cumulative distributions of uncoating dwell times of clathrin/AP2 and sCCVs containing PtdIns(4,5)P2 with either PtdIns(3)P or PtdIns(4)P obtained upon incubation with ΔPTEN-Aux1, full-length Aux1, or full-length GAK (P, PTEN-like domain; CB, clathrin-binding domain; J, J domain; K, kinase domain). The uncoating dwell times corresponding to 50% of the cumulative distributions (dashed lines) are from 3–10 independent experiments.
Figure S6.
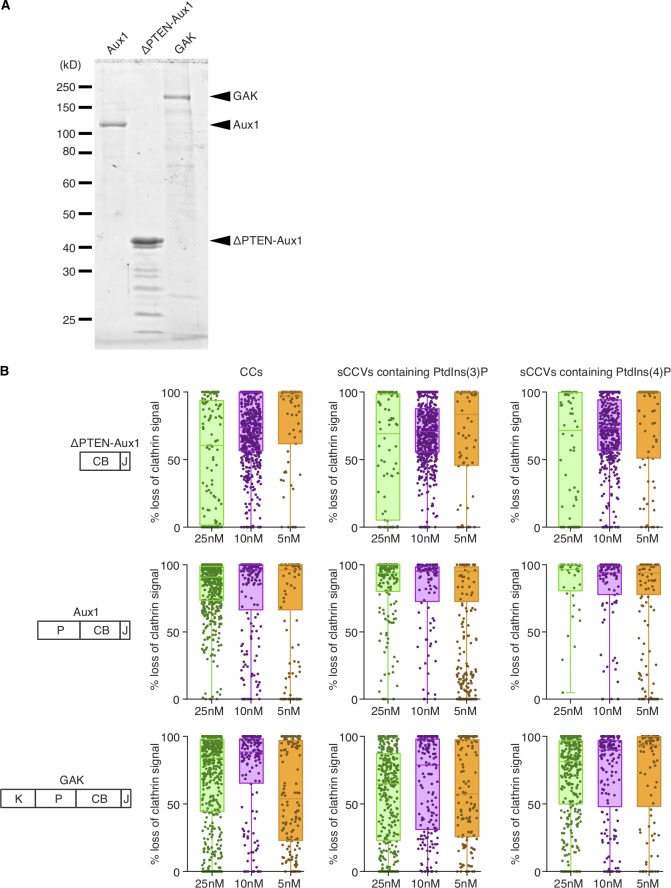
In vitro disassembly of clathrin/AP2 coats and sCCVs containing PtdIns(3)P or PtdIns(4)P. (A) SDS-PAGE (and Coomassie blue staining) of the recombinant full-length Aux1, ΔPTEN-Aux1, and full-length GAK. Molecular weight markers are shown. For GAK and ΔPTEN-Aux1, impurities (of high electrophoretic mobility relative to the target species) reduced the full-length target protein proportion to 60 and 50%, respectively (estimated by band densitometry). (B) Single-object uncoating efficiency determined from the loss of the clathrin LCa–Alexa Fluor 488 fluorescence signal as a function of ΔPTEN-Aux1, full-length Aux1, or full-length GAK concentration (5–25-nM range) added together with 1 µM Hsc70 and 5 mM ATP. Each sample included a mixture of clathrin/AP2 coats (CC) together with sCCVs containing PtdIns(4,5)P2 together with either PtdIns(3)P or PtdIns(4)P (distinguished by labeling with DiI or DiD lipid dyes). Data were acquired at 1-s intervals for 150 s using three-color TIRF microscopy; each dot in the box plots represents the final uncoating efficiency for a single object. Box plots include the median, and data are from three independent experiments.
Video 6.
Single-object in vitro disassembly of clathrin/AP2 coats and sCCVs. The still image imaged with TIRF at the beginning of the time series corresponds to clathrin/AP2 coats and sCCVs containing PtdIns(4,5)P2 together with either PtdIns(3)P or PtdIns(4)P distinguished by labeling with DiI (red) or DiD (blue) lipid dyes. Before the time series, the channels corresponding to clathrin LCa–Alexa Fluor 488 (green) were shifted 5 pixels with respect to the lipids. The time series follows the uncoating reaction and corresponds to the clathrin fluorescence signal as a function of 25 nM ΔPTEN-Aux1 added together with 1 µM Hsc70 and 5 mM ATP. Data were acquired at 1-s intervals for 150 s using three-color TIRF microscopy.
In previous in vitro single-molecule uncoating experiments, which were performed by saturating the coats with auxilin and then adding Hsc70, we found from kinetic modeling that uncoating occurred precipitously when the Hsc70 level had reached an occupancy of approximately one for every two vertices (Böcking et al., 2011). Since auxilin itself stabilizes coats (Ahle and Ungewickell, 1990), this level is likely to be substantially greater than the Hsc70 occupancy needed to uncoat with limiting auxilin present. To address this question directly, we modified our single-molecule uncoating reaction in two ways: first, by inducing uncoating with a mixture of 1 μM Hsc70 and 25 nM Aux1 or GAK (roughly physiological concentrations [Kulak et al., 2014]); and second, by including as substrates sCCVs containing PtdIns(3)P or PtdIns(4)P. This assay is a sensitive functional test of how the kinetics of uncoating depends on lipid composition. We found that Aux1 initiated uncoating of vesicles containing PtdIns(3)P more rapidly than uncoating of vesicles containing PtdIns(4)P; GAK had the opposite preference. The onset of uncoating mediated by Aux1, GAK, or ΔPTEN-Aux1 was the same when using as substrate clathrin/AP2 coats lacking any encapsulated liposome (Fig. 5). This functional assay was proven to be substantially more robust for the relatively low-affinity PTEN-like domains than liposome or lipid-strip binding assays.
We included trace amounts of one of two fluorescent lipid dyes in each type of liposome to identify it uniquely and to distinguish between sCCVs and membrane-free clathrin/AP2 coats; we labeled clathrin with a third fluorescent dye (Fig. 5 C). We determined the relative amounts of clathrin associated with each coat, before and during the uncoating reaction, as illustrated by the traces in Fig. 5 D, from which we obtained the “uncoating dwell time” (interval between the first exposure to the uncoating mixture and initiation of clathrin release; Fig. 5 D) and the “uncoating efficiency” (fraction of fluorescent clathrin released from a sCCV during the 150-s time series; Fig. S6 and Video 6).
Membrane-free clathrin/AP2 coats disassembled rapidly, as in our earlier work (Böcking et al., 2011, 2018). Uncoating of the sCCVs was generally slower, and we could often detect partial release of the clathrin coat in what appeared to be steps (Fig. 5 D). With a ΔPTEN-Aux1 fragment unable to recognize lipids but retaining its clathrin-, AP2-, and Hsc70-binding domains, dwell time and uncoating efficiency were equivalent for the clathrin/AP2 coats and for the PtdIns(3)P- and PtdIns(4)P-containing sCCVs (Fig. 5 E, left panel; and Fig. S6). Uncoating induced by full-length Aux1 initiated more rapidly for synthetic coated vesicles containing PtdIns(3)P (dwell time ∼5 s) than it did for synthetic coated vesicles containing PtdIns(4)P (dwell time ∼16 s; Fig. 5 E, middle panel), while uncoating induced by GAK initiated more rapidly for synthetic vesicles containing PtdIns(4)P (dwell time ∼20 s) than it did for those containing PtdIns(3)P (dwell time ∼29 s; Fig. 5 E, right panel). The observed dwell times, which we expect to depend on the kinetics of Aux1 or GAK recruitment, thus varied with lipid composition; the uncoating efficiency, which should depend only on the ultimate arrival of Hsc70, did not (Fig. S6). We conclude that the PTEN-like domains of Aux1 and GAK have phosphoinositide preferences for PtdIns(3)P and PtdIns(4)P, respectively.
Discussion
The experiments described here have yielded several unexpected findings. One concerns the absence of detectable Aux1 or GAK during the assembly phase of the coated pits. Past experiments done by ectopic expression of Aux1 or GAK often resulted in recruitment of Aux1 or GAK during the assembly phase; some have interpreted this recruitment as a way to explain the partial exchange of clathrin observed during coat assembly, concluding by inference that this is an Hsc70-mediated process. Here, we have combined single-molecule live-cell imaging sensitivity with physiological expression of fluorescently tagged Aux1 and GAK to show absence of Aux1 and GAK during coated pit assembly (Fig. S2, C–F). These observations resolve a long-standing discussion by demonstrating that Aux1 and/or GAK cannot explain the exchange of clathrin during pit formation (and by inference that Hsc70 likewise has no role).
A reasonable explanation for clathrin exchange is the dynamic equilibrium present at the edge of any growing 2D array. This mechanism is consistent with our finding that abortive-pit disassembly does not require the auxilin-dependent uncoating machinery. Past experiments (including some of our own) using ectopic expression of Aux1 or GAK have shown recruitment of Aux1 or GAK during the assembly phase (Chen et al., 2019; Massol et al., 2006): probably, in view of our present results, because of overexpression. A recent in vitro study of coated pit formation found that auxilin and Hsc70 were required for clathrin exchange during pit assembly (Chen et al., 2019). Our data show that this conclusion does not apply in vivo, since the single-molecule sensitivity of our measurements showed that auxilin was completely absent during growth of a coated pit. Moreover, the auxilin mutant used in the in vitro study, which fails to bind and recruit Hsc70, had previously been shown to cause coated vesicle accumulation in vivo (Morgan et al., 2001), as expected. Cargo loading is necessary in vivo for coated pits to become coated vesicles, and in its absence, coated pits abort (Cureton et al., 2009, 2012; Ehrlich et al., 2004). Thus, the in vitro observations are also at odds with the in vivo characteristics of the auxilin mutant used.
The second discovery concerned determination of the stoichiometry by which Aux1 and GAK are recruited to the clathrin/AP2 coat during uncoating. Understanding the extent of this recruitment is fundamental to understanding the mechanism of the uncoating process, and because Hsc70 is a “disassemblase” for many important cellular processes. We found that relatively few auxilins were sufficient for functional uncoating. Our finding, that in vivo uncoating requires relatively few copies of Aux1 or GAK, bears directly on models for the mechanism of the Hsc70-catalyzed uncoating process. Calibrated measurements showed that peak occupancy by three to four molecules of Aux1 or GAK yielded complete uncoating, and that only rarely was the Aux1 or GAK occupancy higher. Uncoating initiated with even fewer auxilins, and the maximum occupancy generally occurred after uncoating had begun, coinciding in average “cohort” traces with ∼50% loss of clathrin (Fig. 1, D and G). Moreover, when we eliminated GAK by gene editing and depleted Aux1 by siRNA, we found, in the cells with slightly incomplete knockdown, a maximal occupancy at any one time of just one to two Aux1 molecules per clathrin-coated vesicle, which nonetheless appeared to uncoat with normal kinetics.
During uncoating, loss of clathrin and loss of AP2 follow each other closely. AP2 adheres to the plasma membrane by virtue of its affinity of PtdIns(4,5)P2, which is hydrolyzed (to PtdIns(4)P) by synaptojanin (in coated pits) and OCRL (inositol polyphosphate 5-phosphatase; in coated vesicles; Chang-Ileto et al., 2011; He et al., 2017; Nández et al., 2014). But only after pinching off of the vesicle does cessation of rapid exchange allow the PtdIns(4,5)P2 concentration to fall and the PtdIns(4)P concentration to rise (He et al., 2017), reducing AP2 affinity for the vesicular membrane. Because AP2 also stabilizes clathrin association with the vesicle (and hence with other clathrins; Kirchhausen et al., 2014), PtdIns(4,5)P2 depletion after pinching may accelerate uncoating under conditions of limiting auxilin.
Auxilin binding requires contributions from three different clathrin trimers organized in the lattice of a coat (Fotin et al., 2004). Our results, together with our previous work (He et al., 2017), show that coincident recognition of this local structure and of the cognate lipid determines the timing of normal Aux1 and GAK recruitment. Nonetheless, deletion of the PTEN-like domain did not fully disable auxilin association. Interactions other than with PtdIns(3)P (for Aux1) and PtdIns(4)P (for GAK) must therefore contribute to the observed dependence on vesicle closure. One possibility could be a slightly different structure (e.g., a “tighter” one, due to closure) of the clathrin lattice associated with coated vesicles, resulting in enhanced accessibility of the Aux1/GAK binding regions in the clathrin terminal domain or in AP2 (Scheele et al., 2001). Our current data, however, offer no definitive evidence for the source of this redundancy.
Finally, concerning the dynamics of coated pit/coated vesicle formation, we have shown a straightforward way to distinguish abortive coated pits (i.e., those that fail to form coated vesicles) from coated pits that mature and become coated vesicles. The best currently available method relies on a detailed analysis of the distribution of coated pit lifetimes (Aguet et al., 2013). Because we have now shown that Aux1/GAK are recruited only to coated vesicles and not to assembling coated pits, we can simply segregate clathrin or AP2 traces by whether they end with an Aux1/GAK burst. This simple assignment, similar to the recruitment of dynamin (Aguet et al., 2013; Ehrlich et al., 2004), provides a robust way to distinguish between dissociation of the lattices of abortive pits and disassembly of the lattices of coated vesicles and hence between an abortive pit (whatever its lifetime) and one that proceeds to pinch off as a coated vesicle. We further note that the outcome of this analysis has shown in a simple way that the distinction between abortive and nonabortive events is a meaningful one.
Materials and methods
Antibodies
The antibody against Aux1/GAK was a kind gift from Sanja Sever (Massachusetts General Hospital, Harvard Medical School, Boston, MA; Newmyer et al., 2003). The antibody against GAK (M057-3) was purchased from MBL International.
Cell culture
The mostly diploid SUM159 human breast carcinoma cells (Forozan et al., 1999) kindly provided by J. Brugge (Harvard Medical School, Boston, MA) were routinely verified to be mycoplasma free using a PCR-based assay. SUM159 cells were grown at 37°C and 5% CO2 in DMEM/F-12/GlutaMAX (Gibco), supplemented with 5% FBS (Atlanta Biologicals), 100 U/ml penicillin and streptomycin (VWR International), 1 µg/ml hydrocortisone (Sigma-Aldrich), 5 µg/ml insulin (Sigma-Aldrich), and 10 mM Hepes (Mediatech), pH 7.4.
Plasmids, transfection, and ectopic expression
The DNA sequences encoding the full-length bovine Aux1 (910 residues, GenBank accession number NM_174836.2) or full-length human GAK (1,311 residues, GenBank accession number NM_005255.3) were amplified by PCR from full-length cDNA clones (Massol et al., 2006) and inserted into pEGFP-C1 or mCherry-C1 to generate the plasmids EGFP-Aux1, mCherry-Aux1, EGFP-GAK, and mCherry-GAK. The kinase domain (residues 1–347 of GAK), PTEN-like domain (residues 1–419 of Aux1 and residues 360–766 of GAK), clathrin-binding domain (residues 420–814 of Aux1 and residues 767–1,222 of GAK), and J domain (residues 815–910 of Aux1 and residues 1,223–1,311 of GAK) were amplified by PCR from the full-length cDNA clones (Massol et al., 2006) and inserted into pEGFP-C1 to generate the EGFP-fused Aux1 or GAK truncations. These DNA segments were also fused by overlap PCR and inserted into pEGFP-C1 to generate the EGFP-Aux1/GAK chimera. All constructs used the linker (5′-GGAGGATCCGGTGGATCTGGAGGTTCTGGTGGTTCTGGTGGTTCC-3′) placed between the DNA fragments and EGFP or mCherry. The amino acid sequences were SGLRSRA between EGFP and Hsc70 for EGFP-Hsc70 and ADPPVAT between Hsc70 and EGFP for Hsc70-EGFP.
Transfections were performed using TransfeX Transfection Reagent (ATCC) according to the manufacturer’s instructions. Cells with relatively low levels of protein expression were subjected to live cell imaging 16–20 h after transfection.
Genome editing of SUM159 cells to express EGFP-Aux1+/+, EGFP-GAK+/+, TagRFP-GAK+/+, or AP1-TagRFP+/+ using the CRISPR/Cas9 approach
SUM159 cells were gene edited to incorporate EGFP or TagRFP to the N-terminus of Aux1 or GAK, or the C-terminus of AP1-σ1 subunit, using the CRISPR/Cas9 approach (Ran et al., 2013). The target sequences overlapping the start codon ATG (bold) at the genomic locus recognized by the single guide RNA (sgRNA) are 5′-ATGAAAGATTCTGAAAATAA-3′ for DNAJC6 (encoding Aux1) and 5′-CGCCATGTCGCTGCTGCAGT-3′ for GAK. The target sequence overlapping the stop codon TAG (bold) at the genomic locus recognized by the sgRNA is 5′-GGTTTGGCATAGCCCCTGCT-3′ for AP1S1. The sgRNA containing the targeting sequence was delivered as PCR amplicons containing a PCR-amplified U6-driven sgRNA expression cassette (Ran et al., 2013).
The donor constructs EGFP-Aux1 and EGFP-GAK used as templates for homologous recombination to repair the Cas9-induced double-strand DNA breaks were generated by cloning into the pCR8/GW TOPO vector with two ∼800-nucleotide fragments of human genomic DNA upstream and downstream of the start codon of DNAJC6 or GAK and the open reading frame of EGFP by TA ligation cloning. The upstream and downstream genomic fragments were generated by PCR amplification reactions from the genomic DNA extracted from SUM159 cells using the QiaAmp DNA mini kit (Qiagen). The open reading frame encoding EGFP together with a flexible linker (5′-GGAGGTTCTGGTGGTTCTGGTGGTTCC-3′) was obtained by PCR from an EGFP expression vector.
The donor constructs TagRFP-GAK and AP1-TagRFP were generated by cloning into the pUC19 vector with two ∼800-nucleotide fragments of human genomic DNA upstream and downstream of the start codon of GAK or the stop codon of AP1S1 and the open reading frame of TagRFP using the Gibson Assembly Master Mix (New England BioLabs). The open reading frames encoding TagRFP together with a flexible linker (5′-GGAGGATCCGGTGGATCTGGAGGTTCT-3′) were obtained by PCR from a TagRFP expression vector.
Clonal cell lines expressing EGFP-Aux1+/+, EGFP-GAK+/+, TagRFP-GAK+/+, or AP1-TagRFP+/+ were generated as described (He et al., 2017). In brief, SUM159 were transfected with 800 ng each of the donor plasmid, the plasmid coding for the Streptococcus pyogenes Cas9, and the free PCR product using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instruction. Then the cells expressing EGFP or TagRFP chimeras were enriched by FACS using a FACSAria II instrument (BD Biosciences) and further subjected to single-cell sorting to select monoclonal cell populations. The cells with successful incorporation in the genomic locus of EGFP or TagRFP were screened by PCR using GoTaq Polymerase (Promega).
Knockout of GAK using the CRISPR/Cas9 approach
Knockout of GAK was performed using the CRISPR/Cas9 approach exactly as described before (He et al., 2017), except that the target sequence for GAK overlapping the start codon ATG (bold) is 5′-CGCCATGTCGCTGCTGCAGT-3′.
mRNA depletion of Aux1 and GAK by shRNA or siRNA knockdown
Lentivirus shRNA expressing 5′-CACTTATGTTACCTCCAGAAT-3′ or 5′-GAAGATCTGTTGTCCAATCAA-3′ was used to knock down the expression of Aux1 or GAK (Broad Institute The RNA Consortium Library) as described before (He et al., 2017); 5′-CCTAAGGTTAAGTCGCCCTCG-3′ was used as control. Alternatively, siRNAs were used to knockdown Aux1 or GAK using Lipofectamine RNAiMAX (Invitrogen). siGENOME SMARTpool (a mixture of four siRNAs) was used to knockdown GAK (M-005005-02-0005; Dharmacon); a single siGENOME siRNA was used to knockdown Aux1 (D-009885-02-0010; Dharmacon). A nontargeting siRNA (D-001210-03-05; Dharmacon) was used as a control. Knockdown of Aux1 or GAK by siRNA was achieved by two sequential transfections, the first one in cells after overnight plating and the second 2 d later, followed by analysis during the fourth day.
Transferrin uptake by flow cytometry
Transferrin uptake by a flow cytometry–based assay was done as described (Cocucci et al., 2014). Briefly, cells grown overnight in 12-well plates were washed with α-MEM and incubated for 10 min with 5 µg/ml Alexa Fluor 647–conjugated transferrin (transferrin-AF647; Life Technologies) at 4°C or 37°C. After incubation, cells cooled down on ice were rinsed with ice-chilled PBS. Surface-bound transferrin-AF647 was removed by two brief consecutive incubations with acid wash medium (150 mM NaCl, 1 mM MgCl2, 0.125 mM CaCl2, and 0.1 M glycine, pH 2.5). Cells were then suspended with 5 mM EDTA in PBS, rinsed by brief centrifugation with PBS, and suspended again in 250 µl PBS containing 1% BSA (all steps carried out at 4°C). The amount of internalized transferrin-AF647 was determined by flow cytometry using the 633-nm laser line of the FASCSCanto2 (BD Biosciences). Data are presented as total transferrin associated with the cells and as transferrin uptake (total at 37°C after acid wash/total at 4°C without acid wash).
Western blot analysis
Western blot analysis was performed as described previously (Cocucci et al., 2012). Briefly, protein samples were subjected to SDS-PAGE and transferred to nitrocellulose membranes (Whatman). The nitrocellulose membranes were then incubated for 1 h in Tris-buffered saline with Tween 20 (TBST) containing 3% BSA, followed by overnight incubation at 4°C with the anti-GAK antibody (1:500) or 4-h incubation at room temperature with the anti-Aux1/GAK antibody (1:500) diluted in TBST containing 3% BSA. After three consecutive 10-min washes with TBST, the membranes were incubated for 1 h at room temperature with the appropriate HRP-conjugated secondary antibody (Amersham Biosciences). After three additional 10-min washes with TBST, the membranes were imaged in the presence of LumiGLO Chemiluminescent Substrate (KPL) using a Las 3000 system (Fujifilm).
TIRF microscopy and spinning disk confocal microscopy: live-cell imaging and image analysis
The TIRF and spinning disk confocal microscopy experiments were as described (Cocucci et al., 2012). The single EGFP or TagRFP molecule calibration was performed as described (Cocucci et al., 2012, 2014). Recombinant EGFP made in Escherichia coli was used to determine the fluorescence intensity of a single EGFP molecule. The cytosol from gene-edited SUM159 cells expressing AP2-TagRFP+/+ was used to determine the fluorescence intensity of a single TagRFP molecule.
The detection and tracking of all fluorescent traces were performed using the cmeAnalysis software package (Aguet et al., 2013). For the automated detection, the minimum and maximum tracking search radii were 1 and 3 pixels, and the maximum gap length in a trajectory was 2 frames (He et al., 2017). Detection of an independent event was verified by establishing absence of significant signal during brief intervals preceding and following the first and last detected signals. The intensity-lifetime cohorts were generated as described (Aguet et al., 2013). Detection and tracking of clathrin-coated structures and the associated Aux1 or GAK were performed with clathrin or AP2 as the “master” and Aux1/GAK as the “slave” channel. The valid clathrin or AP2 traces with significant Aux1/GAK signal in the slave channel were selected automatically and verified manually. The amplitude of the 2-D Gaussian PSF fitting for the detected EGFP-Aux1 or EGFP-GAK was used to estimate the number of EGFP-Aux1 or EGFP-GAK molecules calibrated by the intensity of single EGFP. Detection and tracking of events in cells expressing only fluorescently tagged Aux1, GAK, or lipid sensors were performed with the following combinations of master and slave channels: PtdIns(4)P sensor/PtdIns(3)P sensor, PtdIns(3)P sensor/Aux1, PtdIns(4)P sensor/Aux1, PtdIns(3)P sensor/GAK, PtdIns(4)P sensor/GAK (Fig. 3, A, C, and D), EGFP-Aux1/TagRFP-GAK, and mCherry-Aux1/EGFP-Aux1 (Fig. 3, B and E). The validity of traces was verified manually. The frame associated with the maximum fluorescence intensities for these traces and the corresponding interval for each pair were determined automatically.
Lattice light-sheet microscopy: live-cell imaging and image analysis
To show the subcellular localization of Aux1 and GAK in the whole cell volume, the gene-edited EGFP-Aux1+/+ and CLTA-TagRFP+/+ cells, the gene-edited EGFP-GAK+/+ and CLTA-TagRFP+/+ cells, and the EGFP-GAK+/+ and AP1-TagRFP cells were imaged using lattice light-sheet microscopy with a dithered square lattice light-sheet (Aguet et al., 2016; Chen et al., 2014). The cells were plated on 5-mm coverslips (Bellco Glass) for ≥4 h before imaging and were imaged in FluoroBrite DMEM medium (Thermo Fisher Scientific) containing 5% FBS and 20 mM Hepes at 37°C. The cells were sequentially excited with a 488-nm laser (300 mW) and a 560-nm laser (10–50 mW) for ∼100 ms for each channel using a 0.35 inner and 0.4 outer numerical aperture excitation annulus. The 3D volumes of the whole cells were recorded by scanning the sample at 250-nm step sizes in the s-axis (corresponding to ∼131 nm along the z-axis), thereby capturing a volume of ∼50 × 50 × 75 µm (512 × 512 × 300 pixels).
To track the dynamic recruitment of Aux1 or GAK in 3D, the gene-edited EGFP-Aux1+/+ and AP2-TagRFP+/+ cells, and the gene-edited EGFP-GAK+/+ and AP2-TagRFP+/+ cells, and the EGFP-GAK+/+ and AP1-TagRFP cells were imaged using lattice light-sheet microscopy. The cells were excited with a 488-nm laser for ∼50 ms using a 0.505 inner and 0.6 outer numerical aperture excitation annulus. The 3D volumes of the imaged cells were recorded by scanning the sample every ∼2.1 s for 187 s at 500-nm step sizes in the s-axis (corresponding to ∼261 nm along the z-axis), thereby capturing a volume of ∼50 × 50 × 15 µm (512 × 512 × 40 pixels).
In vitro single-object uncoating
Protein production
The procedures to generate recombinant clathrin heavy chain produced in Sf9 cells and light chain produced in E. coli were as described (Böcking et al., 2011, 2018). Labeling of light chain with Alexa Fluor 488 and incorporation into recombinant clathrin triskelions were as described previously (Böcking et al., 2011). Recombinant Hsc70 and ΔPTEN-Aux1 were produced in E. coli and prepared as described previously (Rapoport et al., 2008).
The DNA sequences encoding full-length bovine Aux1 and full-length human GAK were flanked at the N-terminus (for Aux1) or C-terminus (for GAK) by 6x-His tags upon insertion into the pFastBac vector. Proteins were produced intracellularly in Sf9 cells following the Bac-to-Bac protocol (Thermo Fisher Scientific). Cells were lysed by sonication or using a ball bearing bore homogenizer. Lysates were ultracentrifuged, and the supernatant was applied to nickel-NTA resin. Proteins were eluted with imidazole. Aux1 was further purified by gel filtration chromatography and concentrated using Millipore centrifugal devices.
Preparation of YQRL peptidolipids
The CKVTRRPKASDYQRLNL peptidolipid was prepared by adapting a previously described procedure (Böcking et al., 2018; Kelly et al., 2014). Briefly, a mixture of 20 mg/ml of YQRL-containing peptide (prepared in 20 mM Hepes buffer, pH 7.4), DMSO, and maleimide-1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE; 1:1:2 vol/vol mixture respectively) was vortexed at 1,000 rpm for 2 h. The coupling reaction was quenched using 10 mM β-mercaptoethanol for 30 min. The YQRL peptidolipid was extracted by adding chloroform, methanol, and water (4:3:2.25 vol/vol mixture) followed by centrifugation at 1,000 rpm for 5 min. The organic phase containing the peptidolipid was dried under argon and stored in a sealed argon atmosphere-containing vial. The films were resuspended in chloroform and methanol mixture (2:1) at 2 mg/ml before use for liposome lipid film preparation.
Liposome preparation
All lipids (Avanti Polar Lipids) were mixed in 20:9:1 chloroform:methanol:water and dried to prepare composition specific lipid films. Before formation of the synthetic clathrin-coated vesicles, the lipid film aliquots were hydrated in coated vesicle formation buffer (20 mM MES hydrate, pH 6.5, 100 mM NaCl, 2 mM EDTA, and 0.4 mM DTT) to 300 µM final concentration, and liposomes were extruded with a 50-nm-diameter pore filter.
In vitro reconstitution of synthetic clathrin-coated vesicles
The following procedure, used to generate sCCVs (Böcking et al., 2018), was based on the coassembly of a clathrin and AP2 coat surrounding liposomes: a solution containing recombinant clathrin heavy chain and fluorescently labeled light chain (1:3 mol/mol ratio) and AP2 (3:1 wt/wt clathrin:AP2; 100 µg of clathrin heavy chain) was added to 15 µl of extruded liposomes (300 µmol lipid/300 µl) made of 86.9% 1,2-dioleoyl-sn-glycero-3-phosphocholine, 5% PtdIns(4,5)P2, 5% PtdIns(4)P or PtdIns(3)P, 3% YQRL DOPE peptidolipid, and 0.1% DiI or DiD lipid dye and dialyzed overnight at 4°C against coated vesicle formation buffer (80 mM MES, pH 6.5, 20 mM NaCl, 2 mM EDTA, and 0.4 mM DTT) using a Slide-A-Lyzer mini dialysis device (10K molecular weight cutoff, Thermo Fisher Scientific) followed by an additional 4 h of dialysis using fresh coated vesicle formation buffer. Large aggregates were removed by centrifugation using a benchtop Eppendorf centrifuge at 4°C for 10 min at maximum speed. The supernatant was then transferred to a fresh tube and centrifuged at high speed in a TLA-100.4 centrifuge (Beckman) at 70,000 rpm for 30 min. The sCCV-containing pellet was resuspended in coated vesicle formation buffer and centrifuged a second time at 70,000 rpm for 30 min. The pellet was resuspended using coated vesicle formation buffer (100 µl of buffer per 100 µg of clathrin heavy chain) and stored at 4°C for up to 1 wk.
Transmission EM
sCCVs were adsorbed for 60 s onto freshly glowed-discharged carbon-coated electron microscope grids, washed with a few drops of Milli-Q water, stained for 30 s with a few drops of 1.2% uranyl acetate, and blot dried. The samples were imaged on a Tencai G2 Spirit BioTWIN (FEI) at 23,000–49,000× magnification.
Microfluidic uncoating chamber preparation
Microfluidic chips (Böcking et al., 2011, 2018) with polydimethylsiloxane plasma bonded to glass coverslips suited for TIRF microscopy were used to efficiently deliver reagents to uncoat immobilized sCCVs. Glass coverslips (#1.5) were cleaned for a total of 20 min by sequential incubation in the following solvents: toluene, dichloromethane, ethanol, ethanol:hydochloric acid (1:1 vol/vol), and Milli-Q water. The clean coverslips were oxygen plasma treated and bonded to polydimethylsiloxane channels. These chips were immediately incubated with 1 mg/ml biotinylated poly(l-lysine)-poly(ethylene glycol) for 5 min, washed with Milli-Q water, and treated with streptavidin (20 µl of 1 mg/ml streptavidin dissolved in PBS added to 80 µl of 20 mM Tris, pH 7.5, 2 mM EDTA, and 50 mM NaCl) for 5 min. The chips were functionalized with CVC.6 biotinylated antibody specific for CLTA as previously described (Böcking et al., 2011, 2014).
In vitro uncoating of synthetic clathrin-coated vesicles
sCCVs were bound to the functionalized upper surface of the glass coverslip, and those that failed to attach washed away in flowing uncoating buffer (20 mM imidazole, pH 6.8, 100 mM KCl, 2 mM MgCl2, 5 mM protocatechuic acid, 50 nM protocatechuate-3,4-dioxygenase, 2 mM Trolox, and 8 mM 4-nitrobenzyl alcohol). Disassembly of the clathrin/AP2 coats was caused with uncoating buffer supplemented with 1 µM Hsc70, 5 mM ATP, 10 mM MgCl2, and the appropriate auxilins (typically 25 nM of ΔPTEN-Aux1, full-length Aux1, or full-length GAK) flowed through the chip at 20 µl/min.
The total internal fluorescence angle was set to the value that led to 80% of the maximal fluorescence signal observed for immobilized clathrin/AP2 coats and sCCVs before uncoating. The clathrin signal was monitored by exciting the Alexa Fluor 488 maleimide covalently linked to the clathrin light chain (Böcking et al., 2011). Time series starting 10 s before the uncoating mix and lasting 150 s were then recorded at intervals of 1 s. Liposomes containing PtdIns(4)P or PtdIns(3)P were independently labeled with DiI and DiD (Thermo Fisher Scientific) lipid dyes and detected by excitation at 561 and 640 nm, respectively. Signals from empty clathrin coats and PtdIns(3)P- and PtdIns(4)P-containing coated vesicles were classified using the 2D point source detector previously described (Aguet et al., 2013). The start of uncoating (the time point marking the onset of loss of the clathrin fluorescence signal) was manually curated for all traces included in the analysis.
Statistical tests
Because the large size of the sample sizes, the Cohen’s d effect size (Cohen, 1988) was used to report the practical significance of the difference in the magnitude between the recruited EGFP-Aux1 or EGFP-GAK molecules and the lifetime of coated pits before and after the knockout or knockdown of GAK and Aux1. To compare the means from the cells with different treatments, two-tailed t test or one-way ANOVA was used as indicated in figure legends.
Online supplemental material
Fig. S1 shows genome editing of SUM159 cells to express CLTA-TagRFP and EGFP-Aux1 or CLTA-TagRFP and EGFP-GAK. Fig. S2 shows recruitment of Aux1 and GAK to clathrin-coated vesicles in genome-edited cells. Fig. S3 shows sequential bursts of Aux1 and GAK during uncoating of clathrin-coated vesicles at the plasma membrane. Fig. S4 shows effects on clathrin-mediated endocytosis of knockout or knockdown of GAK and knockdown of Aux1. Fig. S5 shows roles of the PTEN-like domain and clathrin-binding domain of auxilins in the endocytic and secretory pathways. Fig. S6 shows in vitro disassembly of clathrin/AP2 coats and sCCVs containing PtdIns(3)P or PtdIns(4)P. Videos show the recruitment dynamics of EGFP-Aux1 (Video 1) or EGFP-GAK (Video 2) to clathrin-coated vesicles, the sequential recruitment of EGFP-Aux1 and TagRFP-GAK to clathrin-coated vesicles (Video 3), the inability to detect recruitment of EGFP-Hsc70 (Video 4) or Hsc70-EGFP (Video 5) to coated pits and coated vesicles, and single-object TIRF tracings of in vitro disassembly of clathrin/AP2 coats and sCCVs (Video 6).
Acknowledgments
We thank S. Sever for the Aux1/GAK antibody, Justin R. Houser for maintaining the TIRF and spinning disc microscopes, S.C. Harrison for discussions and editorial help, and members of our laboratory for help and encouragement.
T. Kirchhausen acknowledges support from the Janelia Visitor Program and E. Betzig, E. Marino, T. Liu, and W. R. Legant for help and advice in constructing and installing the lattice light-sheet microscope. E. Song was supported by grants from the Ministry of Science and Technology of the People’s Republic of China (2018YFA0507101 and 2016YFA0500203) and the National Natural Science Foundation of China (31770900 and 31270884). Construction of the lattice light-sheet microscope was supported by grants from Biogen and Ionis Pharmaceuticals to T. Kirchhausen. The research was supported by National Institutes of Health R01 GM075252 and National Institute of General Medical Sciences Maximizing Investigators’ Research Award GM130386.
The authors declare no competing financial interests.
Author contributions: K. He, E. Song, and T. Kirchhausen designed experiments; K. He, E. Song, and S. Dang generated the gene-edited cell lines and collected the imaging data using TIRF and spinning disk confocal microscopies; K. He, E. Song, and S. Dang analyzed the data collected using TIRF and spinning disk confocal microscopies; S. Upadhyayula and W. Skillern collected the imaging data using the lattice light-sheet microscope; S. Upadhyayula and K. He analyzed the imaging data from the lattice light-sheet microscope; K. He, E. Song, and S. Dang generated the constructs for ectopic expression of proteins; M. Ma, R. Gaudin, and E. Song generated the constructs for genome editing; S. Upadhyayula, K. Bu, B.R. Capraro, I. Rapoport, and I. Kusters participated in the preparation of reagents and acquisition of data associated with the in vitro single-object uncoating experiments. K. He and T. Kirchhausen contributed to the final manuscript in consultation with the authors.
References
- Aguet F., Antonescu C.N., Mettlen M., Schmid S.L., and Danuser G.. 2013. Advances in analysis of low signal-to-noise images link dynamin and AP2 to the functions of an endocytic checkpoint. Dev. Cell. 26:279–291. 10.1016/j.devcel.2013.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguet F., Upadhyayula S., Gaudin R., Chou Y.Y., Cocucci E., He K., Chen B.C., Mosaliganti K., Pasham M., Skillern W., et al. 2016. Membrane dynamics of dividing cells imaged by lattice light-sheet microscopy. Mol. Biol. Cell. 27:3418–3435. 10.1091/mbc.e16-03-0164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahle S., and Ungewickell E.. 1990. Auxilin, a newly identified clathrin-associated protein in coated vesicles from bovine brain. J. Cell Biol. 111:19–29. 10.1083/jcb.111.1.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böcking T., Aguet F., Harrison S.C., and Kirchhausen T.. 2011. Single-molecule analysis of a molecular disassemblase reveals the mechanism of Hsc70-driven clathrin uncoating. Nat. Struct. Mol. Biol. 18:295–301. 10.1038/nsmb.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böcking T., Aguet F., Rapoport I., Banzhaf M., Yu A., Zeeh J.C., and Kirchhausen T.. 2014. Key interactions for clathrin coat stability. Structure. 22:819–829. 10.1016/j.str.2014.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böcking T., Upadhyayula S., Rapoport I., Capraro B.R., and Kirchhausen T.. 2018. Reconstitution of Clathrin Coat Disassembly for Fluorescence Microscopy and Single-Molecule Analysis. Methods Mol. Biol. 1847:121–146. 10.1007/978-1-4939-8719-1_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner G.H., Antrobus R., Hirst J., Bhumbra G.S., Kozik P., Jackson L.P., Sahlender D.A., and Robinson M.S.. 2012. Multivariate proteomic profiling identifies novel accessory proteins of coated vesicles. J. Cell Biol. 197:141–160. 10.1083/jcb.201111049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braell W.A., Schlossman D.M., Schmid S.L., and Rothman J.E.. 1984. Dissociation of clathrin coats coupled to the hydrolysis of ATP: role of an uncoating ATPase. J. Cell Biol. 99:734–741. 10.1083/jcb.99.2.734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang-Ileto B., Frere S.G., Chan R.B., Voronov S.V., Roux A., and Di Paolo G.. 2011. Synaptojanin 1-mediated PI(4,5)P2 hydrolysis is modulated by membrane curvature and facilitates membrane fission. Dev. Cell. 20:206–218. 10.1016/j.devcel.2010.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B.C., Legant W.R., Wang K., Shao L., Milkie D.E., Davidson M.W., Janetopoulos C., Wu X.S., Hammer J.A. III, Liu Z., et al. 2014. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science. 346:1257998 10.1126/science.1257998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Yong J., Martínez-Sánchez A., Yang Y., Wu Y., De Camilli P., Fernández-Busnadiego R., and Wu M.. 2019. Dynamic instability of clathrin assembly provides proofreading control for endocytosis. J. Cell Biol. 218:3200–3211. 10.1083/jcb.201804136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocucci E., Aguet F., Boulant S., and Kirchhausen T.. 2012. The first five seconds in the life of a clathrin-coated pit. Cell. 150:495–507. 10.1016/j.cell.2012.05.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocucci E., Gaudin R., and Kirchhausen T.. 2014. Dynamin recruitment and membrane scission at the neck of a clathrin-coated pit. Mol. Biol. Cell. 25:3595–3609. 10.1091/mbc.e14-07-1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. 1988. Statistical power analysis for the behavioral sciences. Second edition L. Erlbaum Associates, Hillsdale, N.J. [Google Scholar]
- Cremona O., Di Paolo G., Wenk M.R., Lüthi A., Kim W.T., Takei K., Daniell L., Nemoto Y., Shears S.B., Flavell R.A., et al. 1999. Essential role of phosphoinositide metabolism in synaptic vesicle recycling. Cell. 99:179–188. 10.1016/S0092-8674(00)81649-9 [DOI] [PubMed] [Google Scholar]
- Cureton D.K., Massol R.H., Saffarian S., Kirchhausen T.L., and Whelan S.P.. 2009. Vesicular stomatitis virus enters cells through vesicles incompletely coated with clathrin that depend upon actin for internalization. PLoS Pathog. 5:e1000394 10.1371/journal.ppat.1000394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cureton D.K., Harbison C.E., Cocucci E., Parrish C.R., and Kirchhausen T.. 2012. Limited transferrin receptor clustering allows rapid diffusion of canine parvovirus into clathrin endocytic structures. J. Virol. 86:5330–5340. 10.1128/JVI.07194-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M., Boll W., Van Oijen A., Hariharan R., Chandran K., Nibert M.L., and Kirchhausen T.. 2004. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 118:591–605. 10.1016/j.cell.2004.08.017 [DOI] [PubMed] [Google Scholar]
- Eisenberg E., and Greene L.E.. 2007. Multiple roles of auxilin and hsc70 in clathrin-mediated endocytosis. Traffic. 8:640–646. 10.1111/j.1600-0854.2007.00568.x [DOI] [PubMed] [Google Scholar]
- Forozan F., Veldman R., Ammerman C.A., Parsa N.Z., Kallioniemi A., Kallioniemi O.P., and Ethier S.P.. 1999. Molecular cytogenetic analysis of 11 new breast cancer cell lines. Br. J. Cancer. 81:1328–1334. 10.1038/sj.bjc.6695007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotin A., Cheng Y., Grigorieff N., Walz T., Harrison S.C., and Kirchhausen T.. 2004. Structure of an auxilin-bound clathrin coat and its implications for the mechanism of uncoating. Nature. 432:649–653. 10.1038/nature03078 [DOI] [PubMed] [Google Scholar]
- Greener T., Zhao X., Nojima H., Eisenberg E., and Greene L.E.. 2000. Role of cyclin G-associated kinase in uncoating clathrin-coated vesicles from non-neuronal cells. J. Biol. Chem. 275:1365–1370. 10.1074/jbc.275.2.1365 [DOI] [PubMed] [Google Scholar]
- Guan R., Dai H., Harrison S.C., and Kirchhausen T.. 2010. Structure of the PTEN-like region of auxilin, a detector of clathrin-coated vesicle budding. Structure. 18:1191–1198. 10.1016/j.str.2010.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K., Marsland R. III, Upadhyayula S., Song E., Dang S., Capraro B.R., Wang W., Skillern W., Gaudin R., Ma M., and Kirchhausen T.. 2017. Dynamics of phosphoinositide conversion in clathrin-mediated endocytic traffic. Nature. 552:410–414. 10.1038/nature25146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J., Sahlender D.A., Li S., Lubben N.B., Borner G.H., and Robinson M.S.. 2008. Auxilin depletion causes self-assembly of clathrin into membraneless cages in vivo. Traffic. 9:1354–1371. 10.1111/j.1600-0854.2008.00764.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.H., Cortesio C.L., and Drubin D.G.. 2015. Machine-Learning-Based Analysis in Genome-Edited Cells Reveals the Efficiency of Clathrin-Mediated Endocytosis. Cell Rep. 12:2121–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kametaka S., Moriyama K., Burgos P.V., Eisenberg E., Greene L.E., Mattera R., and Bonifacino J.S.. 2007. Canonical interaction of cyclin G associated kinase with adaptor protein 1 regulates lysosomal enzyme sorting. Mol. Biol. Cell. 18:2991–3001. 10.1091/mbc.e06-12-1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly B.T., Graham S.C., Liska N., Dannhauser P.N., Honing S., Ungewickell E.J., and Owen D.J.. 2014. Clathrin adaptors. AP2 controls clathrin polymerization with a membrane-activated switch. Science. 345:459–463. 10.1126/science.1254836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T., Owen D., and Harrison S.C.. 2014. Molecular structure, function, and dynamics of clathrin-mediated membrane traffic. Cold Spring Harb. Perspect. Biol. 6:a016725 10.1101/cshperspect.a016725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak N.A., Pichler G., Paron I., Nagaraj N., and Mann M.. 2014. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods. 11:319–324. 10.1038/nmeth.2834 [DOI] [PubMed] [Google Scholar]
- Kural C., Tacheva-Grigorova S.K., Boulant S., Cocucci E., Baust T., Duarte D., and Kirchhausen T.. 2012. Dynamics of intracellular clathrin/AP1- and clathrin/AP3-containing carriers. Cell Reports. 2:1111–1119. 10.1016/j.celrep.2012.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.W., Zhao X., Zhang F., Eisenberg E., and Greene L.E.. 2005. Depletion of GAK/auxilin 2 inhibits receptor-mediated endocytosis and recruitment of both clathrin and clathrin adaptors. J. Cell Sci. 118:4311–4321. 10.1242/jcs.02548 [DOI] [PubMed] [Google Scholar]
- Lee D.W., Wu X., Eisenberg E., and Greene L.E.. 2006. Recruitment dynamics of GAK and auxilin to clathrin-coated pits during endocytosis. J. Cell Sci. 119:3502–3512. 10.1242/jcs.03092 [DOI] [PubMed] [Google Scholar]
- Loerke D., Mettlen M., Yarar D., Jaqaman K., Jaqaman H., Danuser G., and Schmid S.L.. 2009. Cargo and dynamin regulate clathrin-coated pit maturation. PLoS Biol. 7:e1000057 10.1371/journal.pbio.1000057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Greener T., Pacold M.E., Kaushal S., Greene L.E., and Eisenberg E.. 2002. Identification of domain required for catalytic activity of auxilin in supporting clathrin uncoating by Hsc70. J. Biol. Chem. 277:49267–49274. 10.1074/jbc.M203695200 [DOI] [PubMed] [Google Scholar]
- Massol R.H., Boll W., Griffin A.M., and Kirchhausen T.. 2006. A burst of auxilin recruitment determines the onset of clathrin-coated vesicle uncoating. Proc. Natl. Acad. Sci. USA. 103:10265–10270. 10.1073/pnas.0603369103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan J.R., Prasad K., Jin S., Augustine G.J., and Lafer E.M.. 2001. Uncoating of clathrin-coated vesicles in presynaptic terminals: roles for Hsc70 and auxilin. Neuron. 32:289–300. 10.1016/S0896-6273(01)00467-6 [DOI] [PubMed] [Google Scholar]
- Nández R., Balkin D.M., Messa M., Liang L., Paradise S., Czapla H., Hein M.Y., Duncan J.S., Mann M., and De Camilli P.. 2014. A role of OCRL in clathrin-coated pit dynamics and uncoating revealed by studies of Lowe syndrome cells. eLife. 3:e02975 10.7554/eLife.02975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmyer S.L., Christensen A., and Sever S.. 2003. Auxilin-dynamin interactions link the uncoating ATPase chaperone machinery with vesicle formation. Dev. Cell. 4:929–940. 10.1016/S1534-5807(03)00157-6 [DOI] [PubMed] [Google Scholar]
- Park B.C., Yim Y.I., Zhao X., Olszewski M.B., Eisenberg E., and Greene L.E.. 2015. The clathrin-binding and J-domains of GAK support the uncoating and chaperoning of clathrin by Hsc70 in the brain. J. Cell Sci. 128:3811–3821. 10.1242/jcs.171058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport I., Boll W., Yu A., Bocking T., and Kirchhausen T.. 2008. A motif in the clathrin heavy chain required for the Hsc70/auxilin uncoating reaction. Mol. Biol. Cell. 19:405–413. 10.1091/mbc.e07-09-0870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothnie A., Clarke A.R., Kuzmic P., Cameron A., and Smith C.J.. 2011. A sequential mechanism for clathrin cage disassembly by 70-kDa heat-shock cognate protein (Hsc70) and auxilin. Proc. Natl. Acad. Sci. USA. 108:6927–6932. 10.1073/pnas.1018845108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheele U., Kalthoff C., and Ungewickell E.. 2001. Multiple interactions of auxilin 1 with clathrin and the AP-2 adaptor complex. J. Biol. Chem. 276:36131–36138. 10.1074/jbc.M106511200 [DOI] [PubMed] [Google Scholar]
- Schlossman D.M., Schmid S.L., Braell W.A., and Rothman J.E.. 1984. An enzyme that removes clathrin coats: purification of an uncoating ATPase. J. Cell Biol. 99:723–733. 10.1083/jcb.99.2.723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungewickell E. 1985. The 70-kd mammalian heat shock proteins are structurally and functionally related to the uncoating protein that releases clathrin triskelia from coated vesicles. EMBO J. 4(13A):3385–3391. 10.1002/j.1460-2075.1985.tb04094.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.J., Wang J., Sun H.Q., Martinez M., Sun Y.X., Macia E., Kirchhausen T., Albanesi J.P., Roth M.G., and Yin H.L.. 2003. Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell. 114:299–310. 10.1016/S0092-8674(03)00603-2 [DOI] [PubMed] [Google Scholar]
- Wu X., Zhao X., Baylor L., Kaushal S., Eisenberg E., and Greene L.E.. 2001. Clathrin exchange during clathrin-mediated endocytosis. J. Cell Biol. 155:291–300. 10.1083/jcb.200104085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Böcking T., Wolf M., Grigorieff N., Kirchhausen T., and Harrison S.C.. 2010. Structure of clathrin coat with bound Hsc70 and auxilin: mechanism of Hsc70-facilitated disassembly. EMBO J. 29:655–665. 10.1038/emboj.2009.383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.X., Engqvist-Goldstein A.E., Carreno S., Owen D.J., Smythe E., and Drubin D.G.. 2005. Multiple roles for cyclin G-associated kinase in clathrin-mediated sorting events. Traffic. 6:1103–1113. 10.1111/j.1600-0854.2005.00346.x [DOI] [PubMed] [Google Scholar]