Tei et al. developed optogenetic phospholipase D enzymes for precisely generating the pleiotropic lipid second-messenger phosphatidic acid on specific organelle membranes and applied these tools to elucidate that a PM pool of phosphatidic acid can attenuate Hippo signaling.
Abstract
Phosphatidic acid (PA) is both a central phospholipid biosynthetic intermediate and a multifunctional lipid second messenger produced at several discrete subcellular locations. Organelle-specific PA pools are believed to play distinct physiological roles, but tools with high spatiotemporal control are lacking for unraveling these pleiotropic functions. Here, we present an approach to precisely generate PA on demand on specific organelle membranes. We exploited a microbial phospholipase D (PLD), which produces PA by phosphatidylcholine hydrolysis, and the CRY2–CIBN light-mediated heterodimerization system to create an optogenetic PLD (optoPLD). Directed evolution of PLD using yeast membrane display and IMPACT, a chemoenzymatic method for visualizing cellular PLD activity, yielded a panel of optoPLDs whose range of catalytic activities enables mimicry of endogenous, physiological PLD signaling. Finally, we applied optoPLD to elucidate that plasma membrane, but not intracellular, pools of PA can attenuate the oncogenic Hippo signaling pathway. OptoPLD represents a powerful and precise approach for revealing spatiotemporally defined physiological functions of PA.
Introduction
Phosphatidic acid (PA) is a pleiotropic lipid second messenger with several physiological and pathological functions (Liu et al., 2013; Wang et al., 2006). PA can modify membrane charge and curvature and also engage and activate cytosolic effector proteins (Kooijman and Burger, 2009; Jang et al., 2012; Putta et al., 2016). These effects can lead to cellular changes, including in cytoskeletal organization, membrane trafficking, gene expression, growth, and migration. As such, dysregulation of PA homeostasis occurs in many diseases, including cancer, neurodegeneration, and infection (Gomez-Cambronero, 2014; Nelson and Frohman, 2015; Bruntz et al., 2014; Oliveira and Di Paolo, 2010).
PA is produced by three pathways: acylation of lysophosphatidic acid (LPA) by lysophosphatidic acid acyltransferases (LPAATs), phosphorylation of DAG by DAG kinases (DGKs), and hydrolysis of phosphatidylcholine (PC) by phospholipase Ds (PLDs; Bradley and Duncan, 2018; Selvy et al., 2011; Shulga et al., 2011). Pools of PA produced via these different routes are suggested to have distinct cellular functions. For example, PA produced by LPAAT in the ER functions as an intermediate in de novo phospholipid and triglyceride biosynthesis (Vance and Vance, 2004). PA produced by DGKs and PLDs on other organelle membranes can affect diverse and distinct processes, including actin polymerization, macropinocytosis, secretory vesicle formation, mTOR signaling, and, recently, the Hippo pathway (Selvy et al., 2011; Eichmann and Lass, 2015; Nelson and Frohman, 2015; Totaro and Piccolo, 2019; Fang et al., 2001; Foster, 2013; Yoon et al., 2015). In particular, Hippo signaling, which controls cell size and proliferation, was recently shown to be downregulated by PLD-derived PA (Han et al., 2018a).
Cells employ multiple routes to produce PA for several reasons. LPAATs, DGKs, and PLDs have different subcellular localizations, enabling production of local pools of PA on different organelle membranes (Bradley and Duncan, 2018; Eichmann and Lass, 2015; Selvy et al., 2011; Du et al., 2003, 2004; Shulga et al., 2011). Because of different substrate acyl tail compositions (LPA, DAG, and PC, respectively) and intrinsic acyl tail preferences, the enzymes produce different collections of PA species, some of which can differentially impact signaling pathways. Further, the availability of many isozymes from three different classes gives cells ample opportunities to exert control over PA production from diverse upstream stimuli. Given the central position that PA occupies in phospholipid metabolism (Vance and Vance, 2004; Vance, 2015), redundancy and many levels of regulation are a key feature of PA metabolism; however, these properties make it challenging to decipher specific biological functions of spatially segregated pools of PA.
PA levels can be manipulated using loss- and gain-of function approaches. Typical loss-of-function approaches involve LPAAT, DGK, or PLD inhibition, RNAi, or gene knockouts. The ability of PA-biosynthesizing enzymes to partially compensate for one another can, however, make it challenging to ascribe specific biological functions to subcellular pools of PA. For example, knockout or inhibition of PLD1 and PLD2, the two PLD isoforms responsible for PA production by hydrolysis of PC, results in modest to minimal changes, which can be cell-type and stimulus dependent, in the total PA levels (Su et al., 2009; Antonescu et al., 2010; Bohdanowicz et al., 2013; see Fig. S1, A and B). Loss-of-function studies can also be complicated by noncatalytic roles of some enzymes (Selvy et al., 2011).
Figure S1.
Characterization of PA levels, PLD levels, and PLD activity in PLD-deficient HEK 293T cells. (A and B) Quantification of PA levels in PLD-deficient HEK 293T cells. Lipid extracts from HEK 293T cells of the indicated genotype (DKO, PLD1/2 DKO; 1KO, PLD1 knockout; 2KO, PLD2 knockout) or WT cells treated with the PLD inhibitor (PLDi; 5-fluoro-2-indolyl des-chlorohalopemide, 750 nM for 1 h) were analyzed by electrospray ionization–time-of-flight MS, monitoring in negative mode, to determine levels of total PA (A) or individual PA species, indicated by number of carbons/number of double bonds in the combined acyl tails (B). Levels of total PA in each sample were normalized to phosphatidylinositol, which is a more abundant negatively charged lipid within the same sample. Horizontal bars indicate mean from three biological replicates, each of which was performed in technical triplicate. (C–E) Characterization of PLD levels and activity in PLD-knockout cell lines. (C) Western blot analysis of WT, PLD1 knockout (1KO), PLD2 knockout (2KO), and PLD1/2 DKO, probing for PLD1, PLD2, or tubulin as a loading control. (D) Quantification of PLD activity in the above cell lines using IMPACT. Cells were treated with 5 mM AzProp for 40 min, followed by lipid extraction and click chemistry tagging of azide-labeled transphosphatidylation products with BCN-BODIPY. The samples were analyzed by fluorescence-coupled HPLC. Horizontal bars indicate mean from three biological replicates. (E) Flow cytometry analysis of IMPACT-labeled WT or PLD DKO HEK 293T cells treated with or without PMA. Cells were treated with 1 mM AzProp with or without 750 nM PMA for 30 min and then tagged with BCN-BODIPY for 10 min. The cells were then trypsinized and analyzed by flow cytometry. Shown are violin plots of IMPACT-derived fluorescence, where white circles indicate mean and black boxes indicate 25 to 75% range. n = 6,000 cells analyzed per sample in the violin plot.
Complementary gain-of-function methods to manipulate PA levels present different challenges. One approach, adding PA-containing liposomes to the media, affords control of acyl chain composition, but not subcellular location. By contrast, overexpression of endogenous PA-producing enzymes gives some spatial control over PA localization and acyl tail composition. However, this approach offers poor temporal resolution, can produce ectopic pools of PA due to nonphysiological enzyme localization upon overexpression (Du et al., 2003), and can be complicated by noncatalytic functions of the enzymes. Finally, some PA-synthesizing enzymes have intrinsically low catalytic activity, requiring activation by additional endogenous factors (Powner and Wakelam, 2002; Du et al., 2000).
Critically, existing tools are not suitable for determining whether acute, local production of PA on specific organelle membranes is sufficient for driving individual physiological processes. This technological gap has hampered efforts to unravel the pleiotropy of PA, which would establish biological functions for spatiotemporally distinct pools of this lipid. Here, we develop a toolset for producing physiologically relevant pools of PA with spatial and temporal precision and apply it to define roles for organelle-specific PA in directing a critical signal transduction pathway. We first report the design, optimization, and validation of a suite of light-controlled, optogenetic PLDs (optoPLDs) capable of producing varying amounts of PA on specific organelle membranes. Using these optoPLDs, we then establish the sufficiency for a pool of PA at the plasma membrane (PM), but not on several intracellular organelles, in attenuating mammalian Hippo signaling. Because mammalian PLDs exhibit different localizations, durations, and strengths of activity when stimulated by several upstream factors, the tunable activity of the optoPLD system, in space, time, and amplitude, enables effective mimicry of the different activation states of endogenous, PLD-dependent PA signaling.
Results
Design of an optoPLD for spatiotemporal regulation of PA production
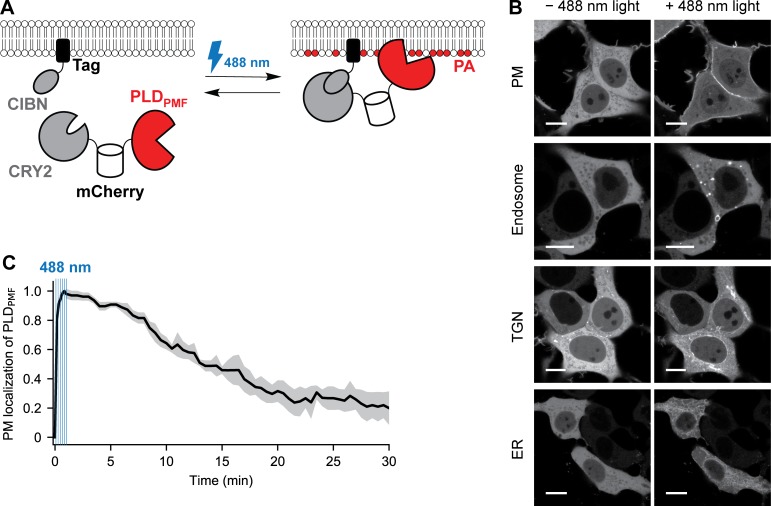
OptoPLD comprises two fusion proteins encoded on a single plasmid and connected by a P2A self-cleaving peptide: (1) CRY2 fused to an mCherry-tagged PLD and (2) CIBN fused to a variable organelle targeting tag (Fig. 1 A). Among several light-mediated heterodimerization systems, CRY2–CIBN was chosen because its dimerization is rapidly induced by nontoxic, 488-nm light, but its dissociation rate in the dark is relatively slow, making it ideal for sustained heterodimerization (and concomitant PA production) using intermittent and thus relatively minimal sample illumination (Kennedy et al., 2010; Idevall-Hagren et al., 2012; Benedetti et al., 2018; Niu et al., 2016).
Figure 1.
Design of an optoPLD for the spatiotemporal control of PA production. (A) Schematic depicting the design of optoPLD. A single plasmid encodes two chimeric proteins linked via a self-cleaving P2A peptide: (1) a fusion of CRY2, mCherry, and PLDPMF; and (2) a fusion of CIBN to an organelle targeting tag. Upon expression of optoPLD in cells, CRY2–CIBN heterodimerization induced by blue light (488 nm) causes recruitment of PLDPMF to the desired membrane. (B) Confocal images showing recruitment of optoPLD to different organelle membranes in HEK 293T cells. Shown is the CRY2-mCherry-PLDPMF localization before and after illumination with 488-nm light (five cycles of 2 s on, 2 s off). Scale bars: 10 µm. (C) Reversibility of optoPLD recruitment. HEK 293T cells were transfected with a version of optoPLD wherein PM-targeted CIBN is fused to iRFP, and colocalization of mCherry (PLDPMF) and iRFP (PM) fluorescence was analyzed by confocal microscopy, with the relative change in Pearson correlation coefficient (calculated as (r − rmin)/(rmax − rmin), where r is the Pearson correlation coefficient) plotted against time. CRY2–CIBN heterodimerization was induced by brief illumination with 488-nm light (six cycles of 2 s on, 8 s off in the first minute), as indicated by the blue vertical lines, and the dissociation of PLDPMF from the PM in the absence of blue light was observed over 30 min. Black line indicates mean (n = 7) and gray area indicates standard deviation.
For the PA-producing enzyme, we selected a PLD from Streptomyces sp. PMF (PLDPMF) for several reasons. We chose a PLD because its substrate, PC, is much more abundant than DGK and LPAAT substrates (DAG and LPA); consequently, activation of PA production would minimally impact other lipid levels. We selected a heterologous PLD to avoid effects from protein–protein interactions with mammalian proteins and because mammalian PLDs are subject to complex regulation, requiring activation by various factors. Among the many microbial PLDs, PLDPMF has only ∼20% sequence identity to human PLDs. This well-characterized enzyme exhibits high activity under physiological conditions (37°C, pH 7.4), and the availability of its crystal structure could enable protein-engineering efforts (Leiros et al., 2004).
To target CRY2-mCherry-PLDPMF to desired organelles, we fused CIBN to localization tags for the PM (CAAX motif), endosomes (2xFYVE domain of Hrs1), the TGN (sialyltransferase transmembrane domain), or the ER (Sac1 transmembrane domain). By encoding CRY2-mCherry-PLDPMF and CIBN tag on a bicistronic vector (Liu et al., 2017), we ensured their equimolar expression and eliminated the need to fluorescently tag CIBN (Fig. 1 A).
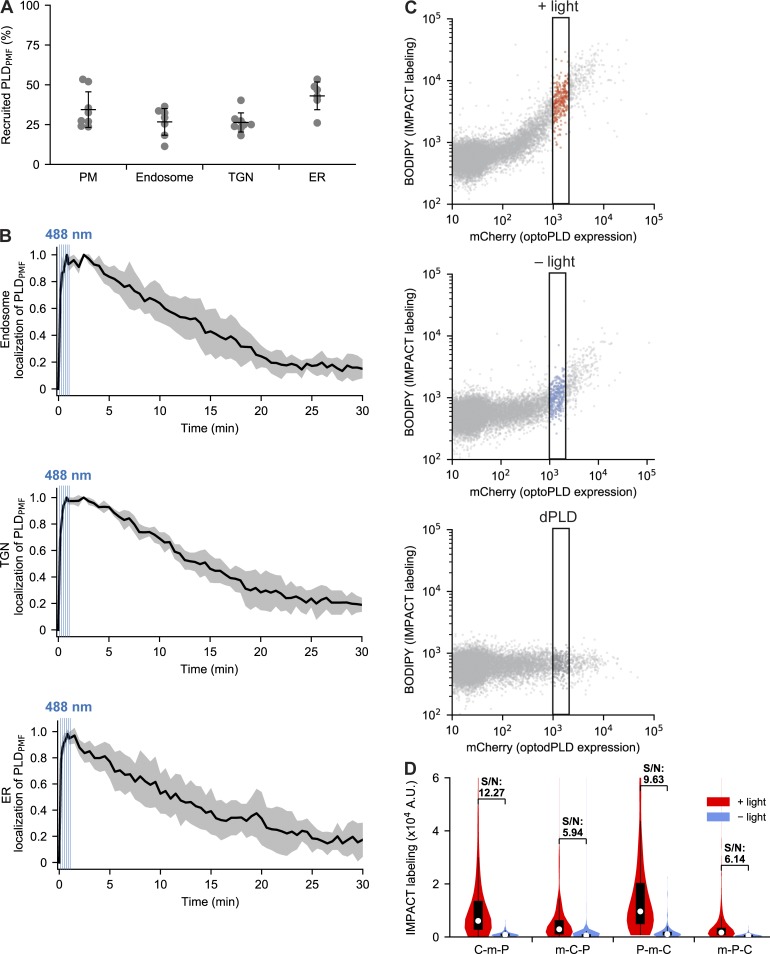
In the absence of blue light, CRY2-mCherry-PLDPMF was localized in the cytosol, but upon illumination with 488-nm light, it was recruited to the appropriate, CIBN-tagged membrane (Fig. 1 B). Colocalization analysis revealed that 25–45% of the total optoPLD relocalized from the cytosol to the desired membrane (Fig. S2 A). To quantify the turn-on/off kinetics of optoPLD photodimerization, we generated an infrared fluorescent protein (iRFP)–tagged CIBN-PM and examined colocalization with CRY2-mCherry-PLDPMF. The light-mediated PLDPMF recruitment to the PM was very rapid (t1/2 ∼5 s), and its dissociation in the dark was much slower (t1/2 ∼10–15 min; Fig. 1 C). Similar kinetics was observed for the association and dissociation of other organelle-targeted optoPLDs (Fig. S2 B). Thus, for sustained PLDPMF recruitment to target membranes, blue light pulses every 2–5 min was sufficient.
Figure S2.
Characterization and optimization of optoPLD. (A and B) Characterization of optoPLD recruitment properties and association/dissociation kinetics in HEK 293T cells. (A) Percentage of optoPLD that is recruited to different organelle membranes in HEK 293T cells after illumination with 488 nm light (six cycles of 2 s on, 8 s off). HEK 293T cells were cotransfected with the indicated optoPLD and iRFP-tagged organelle marker, which were generated by fusing iRFP to the same organelle-targeting tag used for optoPLD; i.e., PM: iRFP-CAAX; Endosome: iRFP-2xFYVE; TGN: iRFP-sialyltransferase(transmembrane domain); ER: iRFP-Sac1(transmembrane domain). After confocal microscopy imaging, the percentage of mCherry fluorescence appearing in iRFP-positive pixels was determined in ImageJ. Black horizontal bars indicate mean (n = 6–8) and vertical error bars indicate standard deviation. (B) Reversibility of optoPLD recruitment. Cells from A were analyzed by confocal microscopy, with the relative change in Pearson correlation coefficient (calculated as (r − rmin)/(rmax− rmin), where r is Pearson correlation coefficient) plotted against time. CRY2–CIBN heterodimerization was induced by brief illumination with 488-nm light (six cycles of 2 s on, 8 s off), as indicated by blue vertical bars, and the dissociation of PLDPMF from the organelle membrane was observed over the following 30 min. Black lines indicate mean (n = 5–7), and gray areas indicate standard deviation. (C and D) Optimization of optoPLD constructs using two-color flow cytometry analysis. (C) Two-color flow cytometry analysis of PLD DKO HEK 293T cells expressing PM-targeted optoPLD (the CRY2-mCherry-PLDPMF version). Cells were labeled via IMPACT by incubation with AzProp (2 mM) in the presence or absence of blue light (5-s illumination every 2 min for 40 min), followed by rinsing and click chemistry tagging with BCN-BODIPY (1 µM; 10 min followed by 10-min rinse at 37°C) and analysis by two-color flow cytometry, with PLD activity quantified in the IMPACT (green) channel and total optoPLD levels in the mCherry (red) channel. The indicated subpopulations with similar mCherry levels (1–2 × 103 fluorescence units, indicated by the black rectangle) were extracted for data analysis in all flow cytometry experiments to analyze PLD activity. (D) Comparison of IMPACT labeling intensities in PLD DKO HEK 293T cells transfected with PM-targeted optoPLD constructs containing either CRY2-mCherry-PLDPMF (C-m-P), mCherry-CRY2-PLDPMF (m-C-P), PLDPMF-mCherry-CRY2 (P-m-C), or mCherry-PLDPMF-CRY2 (m-P-C; and all expressing CIBN-CAAX separated by a P2A peptide), and labeled via IMPACT as described in C. Shown are violin plots of extracted cells with similar mCherry levels, where white circles indicate mean, black boxes indicate 25 to 75% range, and signal-to-noise (S/N) for ±488-nm light is indicated. Note that P-m-C exhibits the strongest labeling, but the signal-to-noise ratio was highest for C-m-P. n = 500–600 cells per experiment; shown is a representative biological experiment that was repeated three times.
A fluorescent PA biosensor reveals organelle-specific PA production by optoPLD
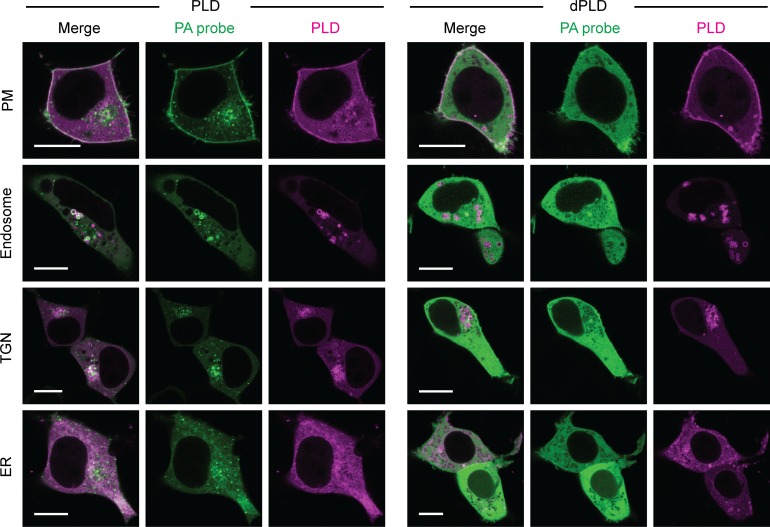
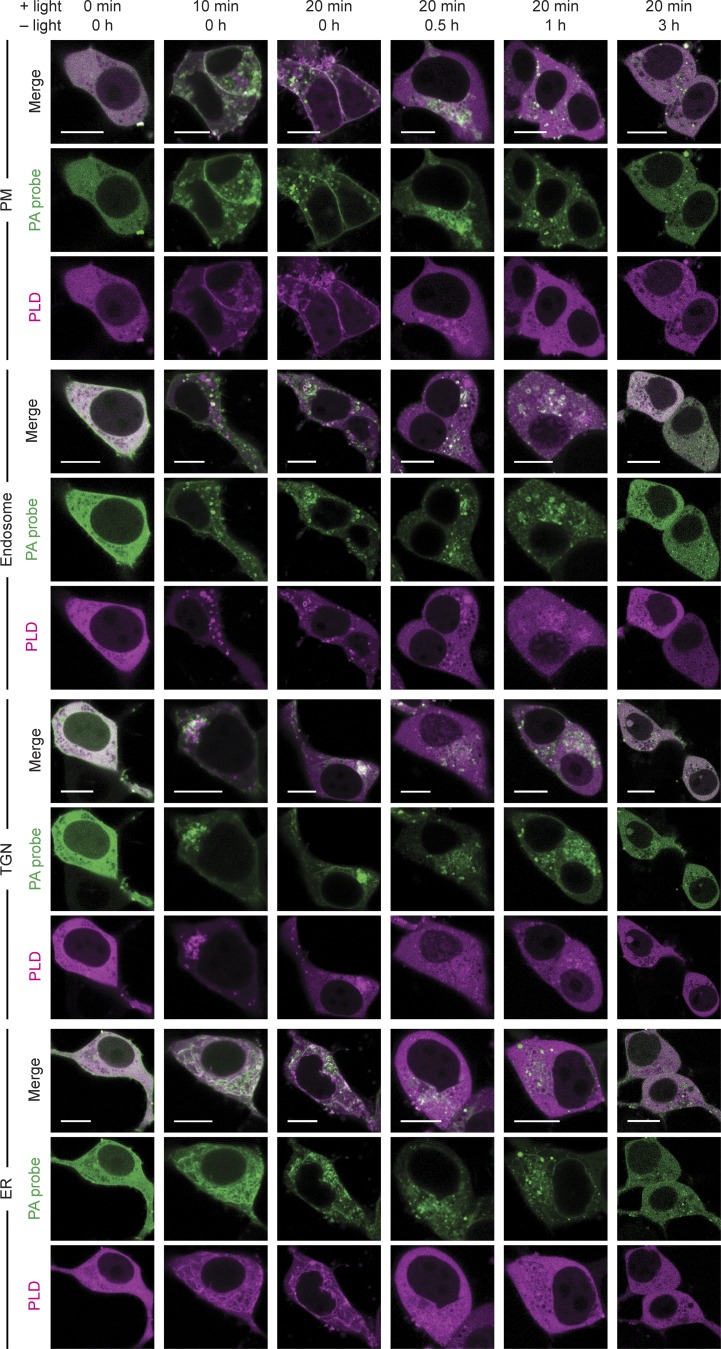
We next ascertained whether light-induced optoPLD recruitment to target membranes would result in PLD-catalyzed PA synthesis. Here, we used a genetically encoded biosensor derived from the Spo20 PA-binding domain, termed PASS (Lu et al., 2016; Zhang et al., 2014). We coexpressed GFP-PASS with each different organelle-targeted optoPLD in HEK 293T cells and monitored both PASS and optoPLD fluorescence by confocal microscopy after 30 min of intermittent blue light illumination (5-s pulses every 2 min). On each organelle membrane tested, we observed the colocalization of GFP-PASS with optoPLD (Fig. 2). Under identical conditions, expression of catalytic-dead optoPLD bearing the H170A mutation in PLDPMF (termed optodPLD) resulted in a mostly cytosolic localization of GFP-PASS, confirming that recruitment of PASS to the target membrane requires PLD activity (Fig. 2). We note that Spo20 has affinity for other anionic PM lipids, including phosphatidylinositol (4,5)-bisphosphate, which might explain a minor partial and optoPLD-independent localization of this probe at the PM (Horchani et al., 2014). A time-course experiment revealed that PASS accumulates on the target organelle membrane as little as 10 min after optoPLD activation and returns to the cytosol 1–3 h after optoPLD is inactivated by keeping the cells in the dark (Fig. S3). These results demonstrate the ability of optoPLD to produce spatiotemporally controlled pools of PA.
Figure 2.
A fluorescent PA biosensor reveals optoPLD-dependent PA production on different organelle membranes. HEK 293T cells cotransfected in serum-free media with a PA-binding probe (GFP-PASS) and either optoPLD (PLD, left) or the catalytically dead optodPLD (dPLD, right) targeted to the PM, endosomes, TGN, or ER were imaged via confocal microscopy after 30 min of intermittent 488-nm blue light illumination (5-s pulses every 2 min). Green, GFP-PASS; magenta, optoPLD fluorescence. Colocalization appears as white in the merged images. Scale bars: 10 µm.
Figure S3.
Time course of changes in PA sensor localization induced by optoPLD activation and inactivation. Visualization of the kinetics of PA production by optoPLD in HEK 293T cells using a PA-binding probe (iRFP-PASS). Cells cotransfected in serum-free media with iRFP-PASS and the indicated optoPLD were incubated with 488-nm light (+ light; 5-s illumination every 2 min) for 0–20 min, and then, after the 20-min time point, kept in the dark (− light) for 0–3 h. At each of the indicated time points, iRFP (PA-binding probe) and mCherry (optoPLD) fluorescence were analyzed by confocal microscopy. Scale bars: 10 µm.
OptoPLD generates physiologically relevant PA species
We determined that the PA generated by optoPLD comprised molecular species similar to PA generated by endogenous, mammalian PLDs. This issue is important because PA species with different acyl tail compositions can differentially affect signaling (Adachi et al., 2016; Yoon et al., 2015; Tanguy et al., 2018), and PLDPMF and mammalian PLDs have very different sequences, though they both share the characteristic HKD active site motifs (Selvy et al., 2011).
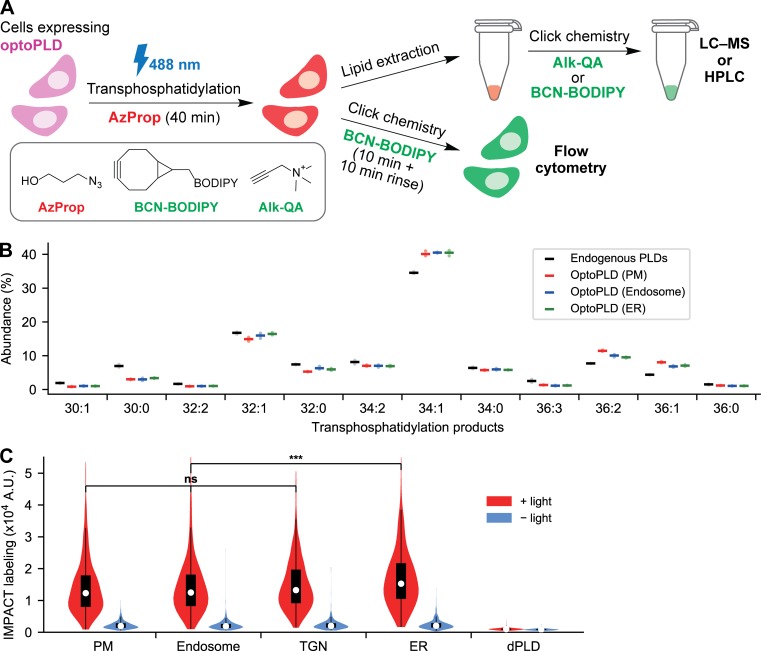
To characterize PA made by PLDs as opposed to total cellular PA, we took advantage of IMPACT (Imaging PLD Activity with Clickable alcohols via Transphosphatidylation), our recently reported chemoenzymatic method that generates tagged lipids that report on levels of PLD activity (Bumpus and Baskin, 2016, 2017; Bumpus et al., 2018). IMPACT relies on the ability of PLDs to catalyze transphosphatidylation reactions of PC with short primary alcohols to produce phosphatidyl alcohol lipid reporters of PLD activity (Brown et al., 2007; Morris et al., 1997).
In an IMPACT experiment, a functionalized primary alcohol such as 3-azido-1-propanol (AzProp) is used to produce an azido phosphatidyl alcohol (Fig. 3 A). A subsequent click chemistry tagging step with an alkyne-quaternary ammonium tag (Alk-QA) enables highly sensitive identification and quantification of the phosphatidyl alcohol species (varying by acyl tail composition) by using liquid chromatography–mass spectrometry (LC–MS) analysis. Importantly, the identities of the phosphatidyl alcohols match those of the natural PA species produced by the PLDs (Bumpus and Baskin, 2017; Brown et al., 2007). Alternatively, click chemistry tagging using a bicyclononyne-fluorophore conjugate (e.g., bicyclononane BODIPY fluorophore [BCN-BODIPY]) generates fluorescent phosphatidyl alcohols, which can be visualized by fluorescence microscopy in live cells or quantified by HPLC from lipid extracts (Fig. 3 B; Bumpus and Baskin, 2017).
Figure 3.
Visualization and quantification of optoPLD activity using IMPACT. (A) Schematic depicting IMPACT. Cells expressing PLDs are treated with AzProp to produce azido phosphatidyl alcohol lipids via transphosphatidylation. The lipids are then tagged with functional probes via a click chemistry reaction, which can be performed in lipid extracts with BCN-BODIPY or Alk-QA for, respectively, fluorescence-coupled HPLC or LC–MS analysis or in live cells for flow cytometry analysis. (B) Acyl chain compositions of individual phosphatidyl alcohol species produced by IMPACT of either endogenous, human PLDs in WT HEK 293T cells (black) or organelle-targeted optoPLDs in PLD1/2 DKO HEK 293T cells (red, PM; blue, endosomes; green, ER), analyzed by LC–MS. For analysis of endogenous PLD activity, WT HEK 293T cells were treated with AzProp (5 mM) for 40 min, followed by lipid extraction, click chemistry tagging with Alk-QA, and LC–MS analysis. For analysis of products of optoPLD activity, the same protocol was used except in PLD DKO HEK 293T cells expressing the indicated optoPLD and illuminated with 488-nm light during the AzProp labeling step (5-s illumination every 2 min for 40 min). Species with abundance <1% are not shown. Horizontal bars indicate mean from three biological replicates, each of which was performed in technical triplicate. (C) Flow cytometry analysis of IMPACT-labeled PLD DKO HEK 293T cells expressing optoPLDs targeted to different organelle membranes shows light-dependent PLD activity. Cells expressing the indicated optoPLD were treated with AzProp (2 mM) for 40 min either with or without blue light (5-s illumination every 2 min) and then tagged with BCN-BODIPY for 10 min. The cells were then trypsinized and analyzed by flow cytometry. Shown are violin plots of IMPACT-derived fluorescence from the population of cells expressing similar levels of optoPLD (based on mCherry fluorescence; see also Fig. S2 C; n = 550 cells analyzed per sample in the violin plot). Shown is a representative example biological experiment, which was repeated three times. ***, P < 0.001.
To eliminate background IMPACT signal from endogenous PLDs in our optoPLD validation experiments, we generated double knockout (DKO) HEK 293T cell lines of the two mammalian PLD isoforms, PLD1 and PLD2, using CRISPR/Cas9-mediated mutagenesis. Knockout was confirmed by Western blot and IMPACT using click chemistry tagging with BCN-BODIPY and HPLC and flow cytometry analyses (Fig. S1, C–E).
We expressed various optoPLDs in PLD DKO cells and performed IMPACT with click chemistry tagging using Alk-QA. LC–MS analysis revealed that optoPLDs targeted to the PM, endosomes, or ER produced transphosphatidylation products with acyl chain compositions similar to corresponding products of IMPACT labeling in WT HEK cells (i.e., coming from endogenous mammalian PLDs; Fig. 3 B). These data indicate that optoPLD has substrate preferences very similar to mammalian PLDs, which mirror the natural PA species produced by mammalian PLDs (Bumpus and Baskin, 2017). These LC–MS studies validate the potential of optoPLD to generate functionally relevant pools of PA in mammalian cells.
Quantification of optoPLD activity using IMPACT
To test whether optoPLD is equally active when targeted to different organelle membranes, we performed single-cell analysis of optoPLD activity using IMPACT. To eliminate undesired IMPACT signal from endogenous PLDs, we again expressed the optoPLDs in PLD DKO cells, which were treated with AzProp with or without blue light (5-s pulses every 2 min), followed by rinsing and BCN-BODIPY click chemistry tagging in live cells. To quantify the extent of optoPLD activity at each organelle, we performed two-color flow cytometry after IMPACT labeling, enabling us to normalize cellular IMPACT-derived fluorescence (BODIPY, green) to optoPLD expression level (mCherry, red; Fig. S2 C).
Gratifyingly, optoPLDs recruited to all of the tested organelle membranes exhibited high and roughly equivalent activity when stimulated with blue light (Fig. 3 C). The ER-targeted optoPLD showed a small but statistically significant increase, in line with the slightly higher abundance of the PLD substrate PC in this organelle membrane (van Meer and de Kroon, 2011). IMPACT labeling from optoPLD in the dark was still slightly higher than that from the catalytic-dead optodPLD, suggesting that cytosol-localized CRY2-mCherry PLDPMF had a small level of undesired background activity (Fig. 3 C).
To ensure that optoPLD had the highest possible signal to background, we optimized the interaction of PLDPMF with the membrane by switching the positions of CRY2, mCherry, and PLDPMF within the optoPLD constructs, creating mCherry-CRY2-PLDPMF, CRY2-mCherry-PLDPMF, PLDPMF-mCherry-CRY2, and mCherry-PLDPMF-CRY2. Each of these was expressed in PLD DKO cells with CIBN-PM, and two-color flow cytometry revealed that performed CRY2-mCherry-PLDPMF exhibited the highest signal to background (i.e., IMPACT signal under blue light illumination vs. in the dark; Fig. S2 D). We thus continued to use this version of optoPLD for all future studies.
Directed evolution of PLDPMF to develop optoPLDs with varying activities to mimic the range of endogenous PLD signaling
Mammalian PLD1 and PLD2 exhibit a wide range of physiologically relevant activities (i.e., between basal and fully activated states) as a result of their ability to be stimulated by several protein and lipid interactors (Du et al., 2000; Powner and Wakelam, 2002; Selvy et al., 2011; Petersen et al., 2016; Peng and Frohman, 2012; Bruntz et al., 2014). Because PLDs are regulated by changes in both subcellular localization and catalytic activity, we sought to develop a panel of optoPLDs with a range of activities to mimic the ability of cells to adjust endogenous PLD activity in different physiological contexts.
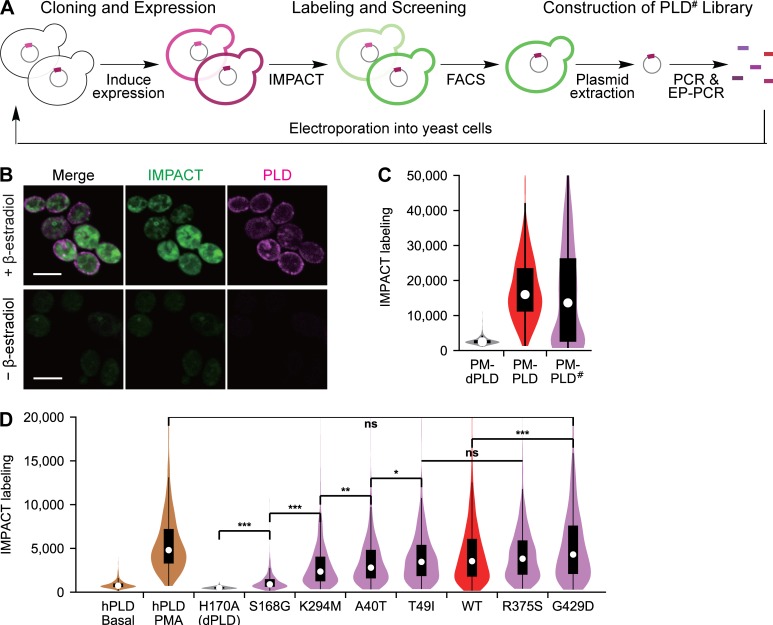
To accomplish this goal, we performed directed evolution of PLDPMF. We developed a modified version of yeast display (Liu, 2015), termed yeast membrane display, for high-throughput screening of PLDPMF mutants (termed PLD#). Because PLD is an enzyme that acts on membrane lipids, we reasoned that, by tethering PLD# members to an appropriate PC-rich organelle membrane within Saccharomyces cerevisiae, we could take advantage of IMPACT and FACS to enrich new PLD# enzymes with desired catalytic activities.
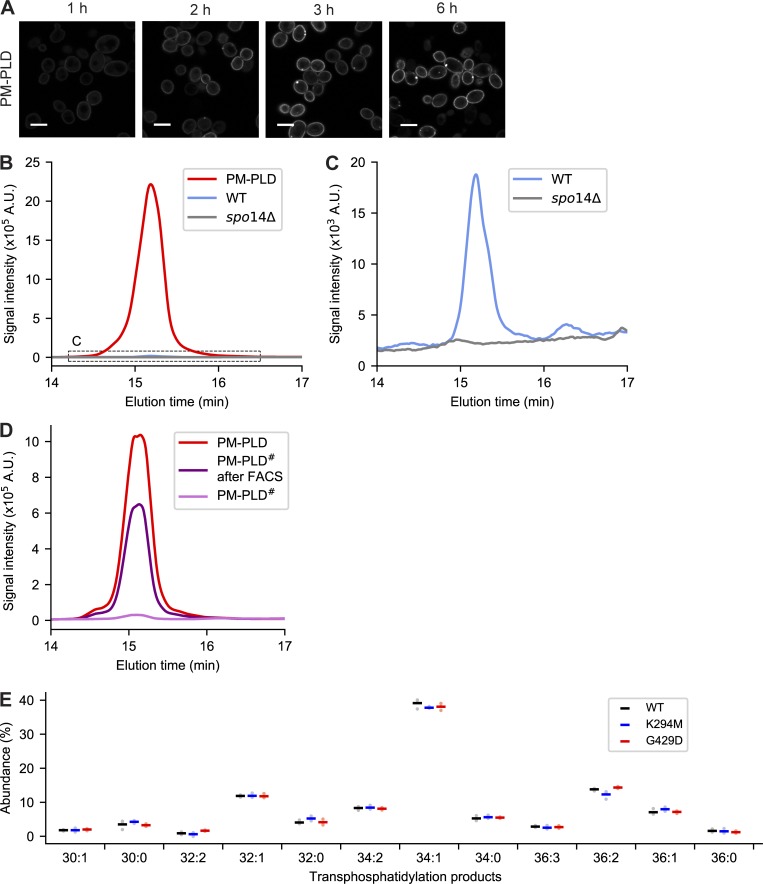
Our yeast membrane display platform involved three steps: (1) PLD# library construction, (2) cloning and expression in yeast, and (3) IMPACT labeling and FACS-based enrichment of yeast based on the expressed PLD# activity (Fig. 4 A). First, we built a construct to express PM-binding PLDPMF (PM-PLD), containing PLDPMF, mCherry and the tandem pleckstrin homology domain of phospholipase C (2xPLCδPH), which binds to phosphatidylinositol (4,5)-bisphosphate in the PM, in a β-estradiol-inducible expression system (Várnai and Balla, 1998; Stauffer et al., 1998; McIsaac et al., 2011). Using this system, maximal PM-PLD expression was observed within 3 h of β-estradiol treatment, with no observed toxicity (Fig. S4 A). To determine PM-PLD activity in yeast, we adapted IMPACT to work in this organism. Fortunately, S. cerevisiae treated with AzProp exhibited only very low endogenous transphosphatidylation activity compared with yeast expressing PM-PLD (Fig. S4, B and C). Thus, AzProp labeling of yeast, followed by paraformaldehyde fixation and tagging via click chemistry with BCN-BODIPY, enabled the selective fluorescent labeling of PM-PLD–expressing cells (Fig. 4 B).
Figure 4.
Directed evolution of a panel of optoPLDs with ranging activities. (A) Schematic depicting yeast membrane display-based directed evolution platform for discovery of PLD mutants (PLD#) with altered catalytic activities. PM-localized PLD# expression is induced by treatment of cells harboring a fusion of PLD# to 2xPLCδPH under an inducible promoter (PM-PLD#) with β-estradiol. Cells are fluorescently labeled using IMPACT via treatment with AzProp, fixation, and BCN-BODIPY tagging. Subsequent FACS selection is used to enrich cells expressing PM-PLD# with desired activities, followed by plasmid extraction and amplification of PLD# with further mutagenesis by error-prone PCR (EP-PCR) and cloning and transformation into yeast for subsequent rounds of selection. (B) IMPACT labeling of yeast expressing PM-PLD. Yeast cells harboring PM-PLD# were treated with or without β-estradiol for 3 h, treated with AzProp (10 mM) for 1 h, fixed with formaldehyde, tagged via click chemistry with BCN-BODIPY (1 µM) for 30 min, and imaged by confocal microscopy. Green shows IMPACT-derived fluorescence, and magenta shows PLD-derived fluorescence from its mCherry tag. Scale bars: 5 µm. (C) Flow cytometry analysis of IMPACT-labeled yeast expressing a catalytically dead PM-dPLD, PM-PLD, or a typical PM-PLD# library. n = 360 cells analyzed per sample in the violin plot. (D) PLD# mutants identified by directed evolution were cloned into PM-targeted optoPLD to generate optoPLD#s (violet) and expressed in PLD DKO HEK 293T cells. Their activities were quantified by IMPACT labeling followed by flow cytometry (performed as described in Fig. 3 C) and compared with WT optoPLD (red) and catalytically dead optodPLD (gray). As a comparison, the levels of endogenous, human PLD (hPLD) activity in WT HEK 293T cells are shown in the absence (basal) or presence (PMA) of strong stimulation with PMA (tan). See also Fig. S2 C for strategy to account for equal optoPLD expression levels. In this case, IMPACT was performed as in Fig. 3 C, except that the transphosphatidylation step involved treatment of cells with 2 mM AzProp in the absence of any 488-nm light illumination for 30 min in the presence of either DMSO control (basal) or 750 nM PMA stimulation. n = 820 cells analyzed per sample in the violin plot. Shown is a representative example biological experiment, which was repeated three times. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure S4.
Characterization of PM-targeted PLD in yeast. (A) Live-cell confocal microscopy imaging of S. cerevisiae harboring a plasmid encoding PM-targeted PLD (PLDPMF-mCherry-2xPLCδPH) under the control of the chimeric transcriptional activator Gal4dbd.ER.VP16 (GEV) and treated with β-estradiol (200 nM) for the indicated time to induce PLD expression. Shown is mCherry fluorescence. Scale bars: 5 µm. (B and C) Use of IMPACT, followed by fluorescence-coupled HPLC analysis, to quantify endogenous PLD activity in yeast due to Spo14 and exogenous PLD activity due to expression of PM-PLD. The indicated yeast populations (PM-PLD indicates WT yeast expressing PM-PLD) were labeled by IMPACT by incubation with AzProp (50 mM) for 1 h followed by lipid extraction and click chemistry tagging with BCN-BODIPY (1 µM) and HPLC analysis of the transphosphatidylation products. The curve in C indicates the zoomed-in region in B to show the very low but detectable extent of endogenous PLD activity due to the yeast PLD, Spo14. (D) Verification that yeast membrane display directed evolution system enriches PLD variants with desired activity. Shown are representative HPLC traces from yeast populations expressing either PM-PLD (WT PLD), the PM-PLD# library of PLD variants generated by error-prone PCR before IMPACT labeling and FACS enrichment, or the subset of FACS-enriched members of the PM-PLD# library. Note the substantial increase in PLD activity of the enriched mixed PM-PLD# population afforded by the IMPACT-based FACS enrichment strategy. Samples for HPLC analysis were generated as in B and C. (E) Acyl chain compositions of individual phosphatidyl alcohol species produced by IMPACT of different optoPLD#s in PLD DKO HEK 293T cells (black, WT; blue, K294M; and red, G429D), analyzed by LC–MS. The cells were treated with AzProp (5 mM) for 40 min with 488-nm light (5-s illumination every 2 min for 40 min), followed by lipid extraction, click chemistry tagging with Alk-QA, and LC–MS analysis. Species with abundance <1% are not shown. Horizontal bars indicate mean from three biological replicates, each of which was performed in technical triplicate.
To construct a diverse PLD# library, we performed random mutagenesis on PLDPMF using error-prone PCR and performed IMPACT-FACS–based enrichment of yeast expressing PM-PLD#. Yeast expressing the PM-PLD# library exhibited a broad range of IMPACT labeling, indicating that changes in PLD# activity caused by mutations are reflected in the IMPACT labeling efficiency (Fig. 4 C). After three enrichment rounds (Fig. S4 D) and subsequent DNA isolation and amplification, we determined the identities of the mutated residues in the enriched PLD# population by next-generation sequencing.
Among the many potential hits, we selected six highly reoccurring PLDPMF mutants for testing in mammalian cells: A40T, T49I, S168G, K294M, R375S, and G429D. We generated PM-targeted optoPLD# constructs with each of these six mutant PLDs, expressed them in PLD DKO HEK 293T cells, and evaluated their PLD activities using IMPACT followed by flow cytometry. Excitingly, we discovered that these six optoPLD#s exhibited a diversity of catalytic activities, ranging from as low as the basal activity of endogenous human PLDs in this cell line to as high as PLD activity elicited by very strong stimulation with PMA (Fig. 4 D). LC–MS analysis of the IMPACT-derived lipids produced by two representative optoPLD#s (G429D and K294M) established that these enzymes have very similar acyl tail substrate preferences as WT PLDPMF (Fig. S4 E).
These studies demonstrate that our optoPLD# collection spans the physiological range of endogenous PLD activity. Notably, the strongest optoPLD# (G429D) exhibited higher activity than WT PLDPMF, which is the first example to our knowledge of an evolved PLD variant with higher catalytic ability for PA generation than its WT counterpart. A key benefit of this collection of optoPLD#s is that PA formation can be tuned by simply swapping out the library member rather than by changing expression levels, which is harder to precisely control, particularly within a two-component, optogenetic heterodimerization system.
Visualization of optoPLD activity at the PM and ER using the highly active G429D variant and a real-time version of IMPACT
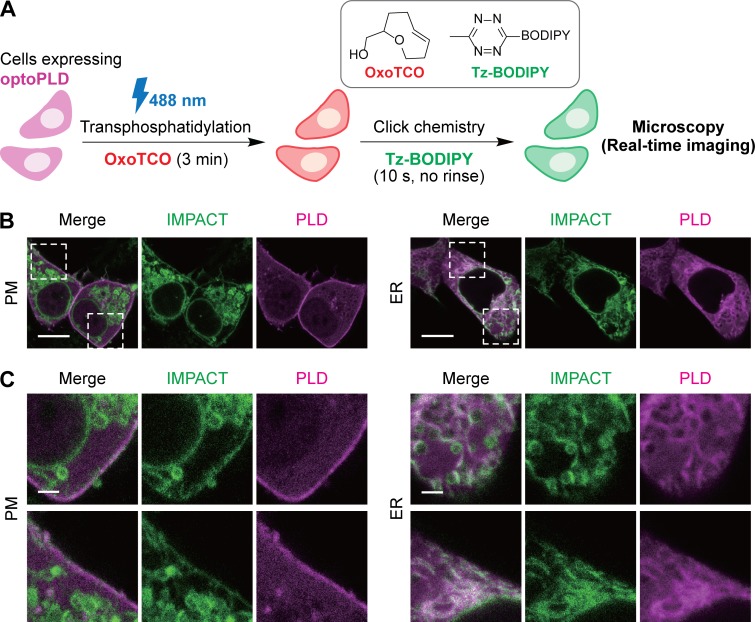
The availability of the G429D variant of PLDPMF allowed us to directly visualize the subcellular localizations of optoPLD activity using real-time (RT) IMPACT, a recently reported, highly time-resolved IMPACT variant (Liang et al., 2019). In RT-IMPACT, AzProp is replaced with an oxo-trans-cyclooctene alcohol (oxoTCO) for the transphosphatidylation step, and its corresponding click chemistry partner is a fluorogenic tetrazine-BODIPY (Tz-BODIPY) dye, enabling tagging via the tetrazine ligation click chemistry reaction (Liang et al., 2019; Lambert et al., 2017; Carlson et al., 2013; Devaraj et al., 2008; Blackman et al., 2008; Wu and Devaraj, 2016; Oliveira et al., 2017; Fig. 5 A). Due to the rapid kinetics and fluorescence turn-on of the tetrazine ligation, the fluorescent lipid products of IMPACT can be visualized within tens of seconds after the beginning of the click chemistry reaction. During the development of RT-IMPACT, we discovered that IMPACT-derived fluorescent lipids produced by PM-localized endogenous PLDs trafficked rapidly, over tens of seconds, from the PM to the ER; however, early time points of the click chemistry time-lapse movie could reveal accurate localizations of IMPACT lipid products before the bulk of this rapid diffusion and trafficking to other organelle membranes (Liang et al., 2019).
Figure 5.
Visualization of PM and ER localization of optoPLD activity using a real-time variant of IMPACT. (A) Schematic of a real-time variant of IMPACT (RT-IMPACT) for visualizing subcellular localizations of PLD activity. Cells expressing or harboring PLD enzymes are treated with oxoTCO to produce oxoTCO-containing phosphatidyl alcohol lipids via transphosphatidylation. These lipids are then tagged with a fluorogenic Tz-BODIPY conjugate via tetrazine ligation click chemistry, whose reaction progress can be visualized in real-time (due to fluorescence turn-on) by confocal microscopy. (B) Live-cell imaging of IMPACT-labeled PLD DKO cells expressing either PM- or ER-targeted optoPLDs shows optoPLD-dependent labeling on PM. Cells transfected with the indicated optoPLD were treated with oxoTCO (3 mM) for 3 min with blue light illumination (5 s every 1 min), followed by real-time imaging of the tetrazine ligation click chemistry reaction by addition of Tz-BODIPY (0.33 µM) and time-lapse monitoring by confocal microscopy. A single time point (10 s after the addition of Tz-BODIPY) is displayed. Green, IMPACT; magenta, PLD. (C) Zoomed-in images from B. Scale bars: 10 µm (B), 2 µm (C).
Having characterized the rapid, unidirectional PM-to-ER trafficking of the fluorescent lipid reporters of PLD activity and now having optoPLDs in hand, we were eager to use RT-IMPACT to establish that PM- and ER-targeted optoPLDs had activity on the expected organelle membrane. Because oxoTCO is a poorer PLD substrate compared with AzProp (Liang et al., 2019) and shorter transphosphatidylation durations are optimal in RT-IMPACT, to minimize trafficking of lipid reporters during this step, we used the highly active G429D optoPLD# to afford the highest possible IMPACT labeling.
Indeed, PLD DKO HEK 293T cells expressing PM-targeted optoPLD exhibited a substantial fraction of RT-IMPACT fluorescence at the PM, as well as some intracellular fluorescence likely at the ER (Liang et al., 2019), in an early time point of the time-lapse movie (Fig. 5 B). Interestingly, the internalization of these fluorescent lipids appears to be more rapid in HEK 293T cells than in HeLa or NIH 3T3 cells, which we used in the development of RT-IMPACT (Liang et al., 2019). Importantly, cells expressing ER-targeted optoPLD and labeled in an analogous manner exhibited strong ER signal and no fluorescence at the PM (Fig. 5, B and C). Thus, these studies, in conjunction with the PASS experiments (Fig. 2), support the conclusion that PM- and ER-targeted optoPLDs exhibit enzymatic activities at the expected and desired membrane locations.
Applying optoPLDs to understand roles of local PA production in modulating Hippo signaling
Having established optoPLDs as tools capable of generating organelle-specific pools of PA, we sought to demonstrate the ability of optoPLD-derived PA to affect a cellular signaling pathway. Among the many potential pathways influenced by PA signaling, we focused our efforts on the Hippo pathway, a mechanism by which cells can control growth and proliferation (Yu et al., 2015). Hippo signaling reduces growth and proliferation by activating the large tumor suppressor 1/2 (LATS1/2) kinases, which phosphorylate a key progrowth transcription factor, Yes-associated protein (YAP), and prevent its translocation to the nucleus.
A recent study demonstrated that PLD-derived PA attenuates Hippo signaling, leading to the decreased phosphorylation and correspondingly increased nuclear translocation of YAP (Han et al., 2018a). Different PA effectors were proposed as relevant PA effectors, including the LATS1/2 kinases themselves as well as an upstream factor, neurofibromin 2 (NF2)/merlin. The effects of PA on Hippo signaling were determined both by loss of function (inhibition and knockout of PLDs) and by gain of function (addition of PA-containing liposomes; Han et al., 2018a). Given the dynamic localizations of endogenous PLDs and the multiple proposed PA effectors in this pathway, we reasoned that Hippo signaling would represent an excellent test case for our optoPLDs.
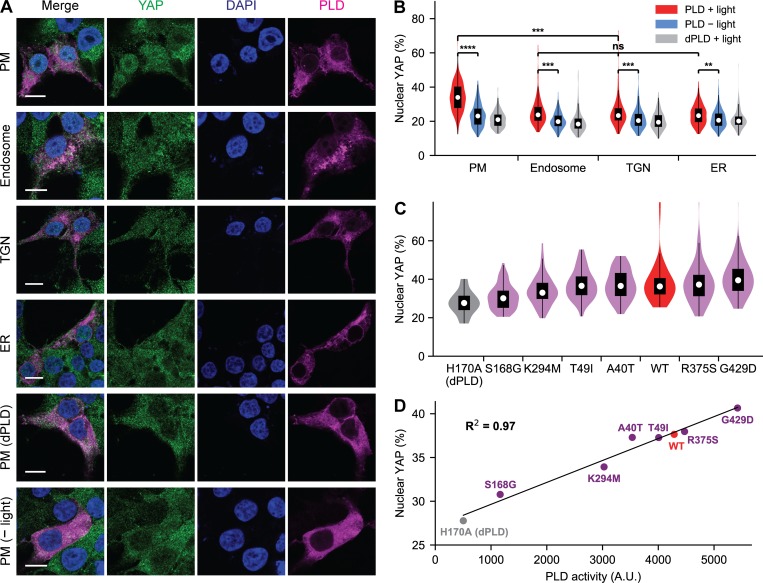
We first recapitulated a key finding of this study, demonstrating that addition of PA-containing liposomes to serum-starved HEK 293T cells caused a translocation of YAP from the cytosol to the nucleus, evaluated by immunofluorescence (IF; Fig. S5 A). We then used optoPLD to generate PA at the PM, the most likely location of action given the known site of action of NF2 in Hippo signaling (Yin et al., 2013). Excitingly, PA production by PM-targeted optoPLD induced a robust nuclear translocation of YAP, which we did not observe with either PM-optoPLD in the dark or using the catalytic-dead PM-optodPLD (Fig. 6 A). Intriguingly, when we replaced PM-optoPLD with versions targeted to other organelles (endosomes, TGN, or ER), we found that these optoPLDs were significantly less effective in downregulating the Hippo pathway (Fig. 6 B), despite their similar levels of PA-synthesizing activity (Fig. 3 C). These results both confirm the role of PA in allowing cells to attenuate Hippo signaling and, interestingly, suggest that local production of PA in the PM can control these signaling events.
Figure S5.
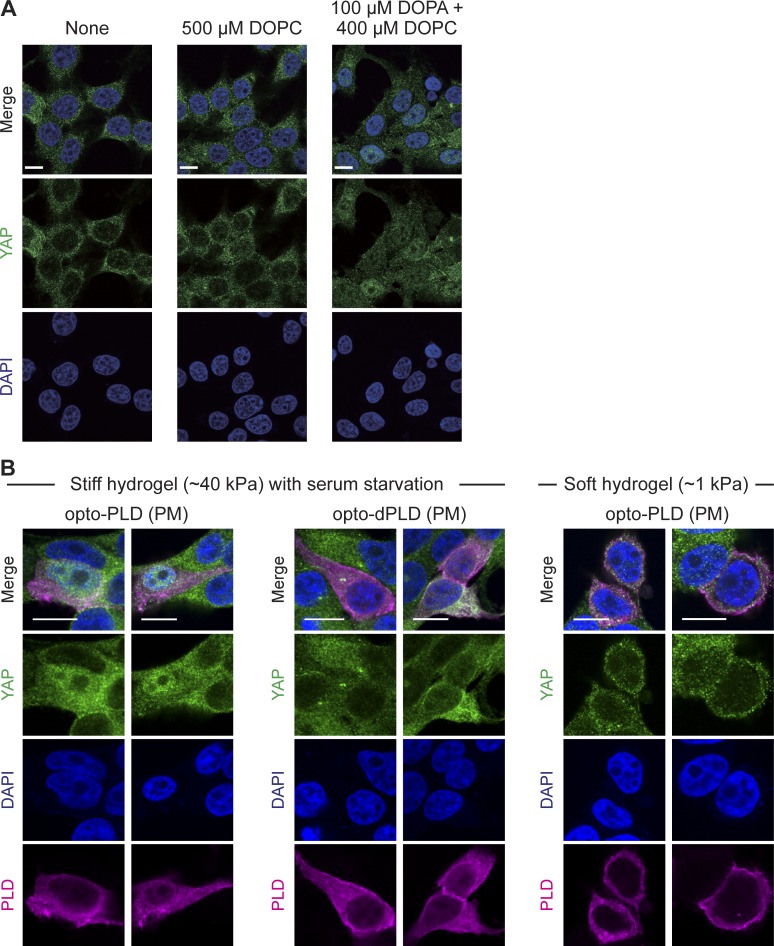
Effects of optoPLD on Hippo signaling depend on substrate stiffness. (A) Verification that application of exogenous PA to cells suppresses Hippo signaling. HEK 293T cells were serum starved for 20 h followed by addition of no reagent, control liposomes containing PC (DOPC), or liposomes containing 20% PA (dioleoylphosphatidic acid [DOPA]) and the balance as DOPC for 1 h. Cells were then fixed and immunostained for YAP, followed by confocal microscopy imaging. Note predominantly cytoplasmic localization of YAP under control conditions and nuclear localization of YAP when PA is added to cells, indicating a downregulation of Hippo signaling. (B) PM-targeting optoPLD downregulates the Hippo pathway in HEK 293T cells grown on stiff (∼40 kPa strength), but not soft (∼1 kPa), hydrogels. For experiments with high substrate stiffness (∼40 kPa), cells grown on stiff hydrogels were transfected with PM-targeting optoPLD or optodPLD, serum starved for 20 h to activate the Hippo pathway, illuminated with blue light for 30 min (5-s illumination every 2 min), fixed, and immunostained for YAP. Note that cells expressing optoPLD, but not optodPLD, exhibited nuclear localization of YAP, indicating a suppression of Hippo signaling by PM-targeted PA. For experiments with low substrate stiffness, cells grown on soft hydrogels (∼1 kPa) in full growth media were transfected with the PM-targeting optoPLD, illuminated with light for 30 min (5-s illumination every 2 min), fixed, and immunostained for YAP. Note that under these conditions, the low-stiffness substrate activates the Hippo pathway, which cannot be downregulated by optoPLD. Scale bars: 10 µm.
Figure 6.
Application of optoPLD to understand role of spatially defined PA pools in controlling Hippo signaling. (A) IF staining of YAP in serum-starved HEK 293T cells expressing optoPLD on the indicated organelle membrane. All samples except the bottom row were illuminated with blue light (5 s every 2 min) for 30 min. Green, YAP; magenta, optoPLD; blue, DAPI (nuclei). Scale bars: 10 µm. (B) PM-targeted optoPLD has the strongest effect on downregulating the Hippo pathway. Shown are violin plots to quantify, from IF images, the percentage of YAP IF signal in the nucleus (n = 200 cells from five biological replicates) in cells expressing optoPLD targeted to different organelles. (C) Members of the optoPLD# collection (violet) exhibit varying effects, when targeted to the PM, on causing nuclear translocation of YAP. Also included in the analysis are WT optoPLD (red) and catalytic-dead optodPLD (gray). Shown are violin plots to quantify, from IF images, percentage of YAP IF signal in the nucleus (n = 60 cells from three biological replicates). **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (D) Plot depicting the relationship between the PA-synthesizing enzymatic activity of a given PM-targeted optoPLD#, measured by IMPACT (x axis, data from Fig. 4 D) and its ability to cause nuclear translocation of YAP (y axis, data from C). The coefficient of determination (R2) is 0.97.
Finally, using the sensitive and quantitative IF-based assay for YAP localization, we examined whether our collection of optoPLD#s of varying activities could differentially influence Hippo signaling. Using PM-targeted versions of each of our six optoPLD#s, we found that those with lower PLD catalytic activity, determined by IMPACT, caused less YAP nuclear translocation and those with higher PLD activity caused more translocation, meaning that the highly active optoPLD# members were most effective in downregulating the Hippo pathway (Fig. 6 C). Remarkably, when we plotted the mean fraction of nuclear YAP against the mean intensity of IMPACT labeling (correlating to optoPLD catalytic activity), we observed a strong linear correlation (Fig. 6 D), indicating that by titrating more PA into the PM, we could precisely tune the strength of Hippo signaling. Altogether, these results demonstrate that the PA produced by optoPLD can downregulate the Hippo pathway in both a location- and dose-dependent manner.
Discussion
In this study, we set out to develop an addition to the toolkit for studying the cell biology of PA. There are excellent pharmacological and genetic knockout tools for decreasing the activity of PA-synthesizing enzymes from the PLD, DGK, and LPAAT families (Su et al., 2009; Scott et al., 2009; Lewis et al., 2009; O’Reilly et al., 2014; Purow, 2015; McCloud et al., 2018; Sakane et al., 2016; Hideshima et al., 2003). However, using these tools to controllably tune local pools of PA can be challenging due to dynamic and still controversial localizations of these enzymes, as well as evidence of crosstalk and compensation between PA-synthesizing pathways. To address these challenges, we sought to develop a complementary gain-of-function tool, optoPLD, to study PA signaling with spatiotemporal precision.
Several factors were critical to the design of optoPLD. First was the selection of PLDPMF as the desired exogenous PA-biosynthesizing enzyme, due to several factors, including its functionality in and orthogonality to eukaryotic cells. Second was the use of the CRY2–CIBN light-mediated dimerization system, which affords extremely fast turn-on kinetics and, equally desirable for sustained PA generation on the minute-to-hour timescale, relatively slow turn-off kinetics, necessitating only occasional and periodic illumination with low-toxicity, visible blue light (Kennedy et al., 2010; Tucker et al., 2014; Benedetti et al., 2018). We found dramatic differences in catalytic efficiencies of optoPLD constructs by varying the relative positions of CRY2, PLDPMF, and mCherry, highlighting the importance of the order of proteins in chimeric protein design in this context. We suggest that, of the four constructs we investigated, CRY2-mCherry-PLDPMF and PLDPMF-mCherry-CRY2 exhibited higher activity because positioning mCherry in between CRY2 (the membrane anchor, through its interaction with CIBN) and PLDPMF (the lipid-modifying enzyme) enhances the accessibility of PLDPMF to the membrane due to the structural features of mCherry, whose N- and C-termini extend from the same end of the β-barrel (Shu et al., 2006).
Another factor key to the development of optoPLD was IMPACT, a chemical biology tool that we had originally designed to study the activity and localization of endogenous mammalian PLDs (Bumpus and Baskin, 2016, 2017, 2018). Here, we confirmed that IMPACT could be applied to quantify the activity of an exogenous, bacterial PLD within mammalian cells. In addition, the application of IMPACT in a new organism (S. cerevisiae) was key to our efforts to use directed evolution to generate a panel of optoPLDs with varying catalytic activities. For these protein engineering studies, we developed a high-throughput selection strategy, yeast membrane display, wherein libraries of membrane-tethered PLDs expressed in yeast could be enriched according to PLD activity using IMPACT labeling followed by FACS. Through these directed evolution studies, we produced a panel of PLDPMF mutants (and, subsequently, optoPLD variants) with activities that were both lower and higher than WT PLDPMF. We envision that our yeast membrane display platform could be widely useful for the further evolution of PLDPMF mutants with varying and interesting catalytic properties.
As an application to validate the functionality of the PA produced by the optoPLDs, we revisited the recent discoveries concerning the roles of PLD and PA signaling in modulating the Hippo pathway, which controls cell growth and proliferation (Meng et al., 2018; Han et al., 2018a). In a detailed mechanistic study, Han et al. (2018a) proposed that PLD-derived PA downregulates Hippo signaling by binding to both NF2/merlin and LATS1/2, inhibiting the phosphorylation of YAP and ultimately leading to increases in YAP-dependent gene expression. Using similar experimental conditions, we recapitulated this finding by bulk addition of PA-containing liposomes to serum-starved HEK 293T cells. Excitingly, we found that optoPLD could, with organelle-level precision, cause a similar downregulation of Hippo signaling, but only when the optoPLD-generated PA was produced in the PM and not, at similar levels, on intracellular organelle locations (endosomes, TGN, or ER). This work implicates a pool of PA produced at the PM in allowing cells to downregulate, or bypass, the Hippo pathway.
Interestingly, concurrent work by Meng et al. examined the role of mechanotransduction on the Hippo pathway and concluded that PLD-derived PA had the opposite effect on Hippo signaling, namely that it promoted the pathway (Meng et al., 2018). Mechanistically, they proposed the Rab-like GTPase RAP2 as the relevant PA effector, but importantly, these studies were performed on cells grown in a low-stiffness environment (i.e., on collagen-coated hydrogels). Under those conditions, the Hippo pathway is controlled by the stiffness of the extracellular environment. In fact, the Hippo pathway is regulated by several upstream stimuli, including serum/growth factor availability, cell–cell contacts, and ECM stiffness, and the determination of the exact constellation of upstream factors that regulate Hippo signaling in different contexts is an interesting and active area of research (Meng et al., 2016).
Relevant to these studies, both serum starvation and low ECM stiffness activate the Hippo pathway and result in YAP exclusion from the nucleus. Although the Hippo activation caused by serum starvation was counteracted by optoPLD-derived PA, we found that the activation caused by low ECM stiffness overrode the effect of optoPLD (Fig. S5 B), consistent with the study by Meng et al. (2018). Thus our work using optoPLD lends support to the hypothesis emerging from these two studies that PA indeed has different effects on the Hippo pathway in different contexts and experimental conditions (Stampouloglou and Varelas, 2018; Han et al., 2018b). Future work using optoPLD in combination with other toolsets could reveal the causes and mechanisms underlying the differential roles of PA in modulating Hippo signaling.
In summary, optoPLD is a tool for precise, spatiotemporally controlled production of local pools of PA on individual organelle membranes that should complement existing toolsets for modulating PA using inhibition or knockout/knockdown of biosynthetic enzymes. OptoPLD interfaces with a variety of imaging-based tools for monitoring the dynamics of PA metabolism, including genetically encoded biosensors, FRET-based imaging of PLD–substrate binding events, and our recently developed chemoenzymatic method, IMPACT (Zhang et al., 2014; Kassas et al., 2012, 2017; Bumpus and Baskin, 2018; Kim et al., 2015; Lu et al., 2016; Hughes et al., 2002). We validated the physiological relevance of optoPLD-derived PA pools to confirm and elucidate differential roles of PA signaling in the modulation of Hippo signaling. We envision that our collection of optoPLDs, which in principle could be expanded to target additional organelles that are sites of PA-dependent signaling (e.g., lysosomes, mitochondria; Yoon et al., 2011, 2015; Yang and Frohman, 2012; Kameoka et al., 2018; Adachi et al., 2016), as well as potential implementation of PLDPMF in chemical-induced dimerization systems (Stanton et al., 2018; DeRose et al., 2013), could prove to be widely useful for understanding roles of spatially and temporally defined pools of PA in controlling the many signaling pathways modulated by this multifunctional lipid signaling agent.
Materials and methods
Reagents
The following reagents were obtained from commercial suppliers: Lipofectamine 2000, Invitrogen (catalog number 11668019); poly-L-lysine, Sigma-Aldrich (catalog number P2636), 3-aminopropyltriethoxysilane, TCI (catalog number A0439); tetrakis(acetonitrile)copper(I) tetrafluoroborate (CuBF4), Sigma-Aldrich (catalog number; glutaraldehyde solution, 50% in water, Beantown Chemical (catalog number 139070); dichlorodimethylsilane, VWR (catalog number 102572-962); acrylamide, 40% in aqueous solution, VWR (catalog number 97064-522); bis-acrylamide, 2% in aqueous solution, Alfa Aesar (catalog number J63265-K2); sulfo-SANPAH, VWR (catalog number 10189-584); tetramethylethylenediamine, VWR (catalog number 0761); ammonium persulfate, VWR (catalog number 0486); 5-gluoro-2-indolyl des-chlorohalopemide, Cayman Chemical (catalog number 13563); DGK inhibitor I (R59022), Enzo Life Sciences (catalog number ALX-430-027); PMA, ChemCruz (catalog number sc-3576); DTT, VWR (catalog number 97061-338); 8-oxo-dGTP, TriLink BioTechnologies (catalog numberN-2034); 2′-deoxy-P-nucleoside-5′-triphosphate, TriLink BioTechnologies (catalog number N-2037); β-estradiol: Sigma-Aldrich (catalog number E8875); 1,2-dioleoyl-sn-glycero-3-PC (DOPC), Cayman Chemical (catalog number 15098); 1,2-dioleoyl-sn-glycero-3-phosphate, Avanti Polar Lipids (catalog number 840875); mouse anti-PLD1, Santa Cruz Biotechnology (catalog number sc-28314, RRID:AB_677324); rabbit anti-PLD2, Cell Signaling Technology (catalog number 13904, RRID:AB_2798341); mouse anti-YAP, Santa Cruz Biotechnology (catalog number sc-101199, RRID:AB_1131430); goat anti-mouse HRP, Bio-Rad (catalog number 1706516, RRID:AB_11125547); donkey anti-Mouse Alexa Fluor 488, Invitrogen (catalog number A-21202, RRID:AB_141607); and goat anti-rabbit-HRP, Bio-Rad (catalog number 1706515, RRID:AB_11125142). The following compounds were prepared as described previously: BCN-BODIPY (Alamudi et al., 2016), AzProp (Yuan et al., 2012), oxoTCO ((R)-oxoTCO–C1; Lambert et al., 2017), and Tz-BODIPY (Carlson et al., 2013).
Plasmids and cloning
mCherry-CRY2-PLDPMF was made by swapping INPP5E in mCherry-CRY2-INPP5E (a gift from the De Camilli laboratory, Yale University, New Haven, CT) for PLDPMF (a coding sequence of PLD Streptomyces PMF, UniProt P84147_STRSM, primers: 5′-GCGATCGGGCGGATCTGCAGATTCAGCTACACCACA-3′ and 5′-GTGGTACCTTAAGCGTTGCAGATTCCTCTTG-3′) using PvuI/KpnI. The CRY2 sequence corresponds to UniProt Q96524, amino acids 1–498; the CIBN sequence corresponds to UniProt A0A178V495, amino acids 1–170. CRY2-mCherry-PLDPMF was cloned by first removing CRY2 from mCherry-CRY2-PLDPMF using double XhoI sites to clone mCherry-PLDPMF, followed by insertion of CRY2 using the megaprimer PCR method (primers: 5′-CGGTGGGAGGTCTATATAAGCAGAGCTGGTTTAGCATGAAGATGGACAAAAAGACTATAG-3′ and 5′-GGCGCTAGCGGATCTGACGGTTCCTGACCCTCCCGAGCCACCTGCTGCTCCGATCATG-3′). For CIBN-Sac1, CIBN (amplified from CIBN-GFP-CAAX, a gift from the De Camilli laboratory, primers: 5′-TTTAAACTTAAGCTTGGTACCATGAATGGAGCTATAGGAG-3′ and 5′-CGCCACCGGTCACATGAATATAATCCGTTTTC-3′) and Sac1 (UniProt Q9NTJ5; amino acids 521–587, amplified from CFP-FRB-Sac1, a gift from the De Camilli laboratory, primers: 5′-TATTCATGTGACCGGTGGCGCCTTCCTG-3′ and 5′-AGTGGATCCGAGCTCGGTACTCAGTCTATCTTTTCTTTCTGGACC-3′) were cloned into a KpnI-digested pCDNA5-FRT-TO vector (Thermo Fisher) using Gibson assembly. CIBN-CAAX (the CAAX amino acid sequence is LYKGKKKKKKSKTKCVIM), CIBN-TGN (the TGN sequence is UniProt Q64685, amino acids 2–37), and CIBN-2xFYVE (the FYVE sequence is UniProt O14964, amino acids 147–220) were cloned by swapping out Sac1 in CIBN-Sac1 for CAAX (amplified from CIBN-GFP-CAAX, primers: 5′-GTACCGGTGGATCAGGATCACTGTATAAGG-3′ and 5′-TGGGCCCCTACATAATTACACACTTTGTCTTTG-3′), TGN (constructed by overlap-extension PCR, primers: 5′-GTACCGGTATACATACTAATCTGAAAAAGAAGTTCAGCCTCTTCATCCTGGTCTTCCTGCTCTTCGCTGTGATC-3′ and 5′-TGGGCCCTTACAGTGTAAGGGCCTCGTAATCAGAGCCCTTCTTCCAAACGCAGATCACAGCGAAGAGCAGG-3′), or 2xFYVE (amplified from RFP-2xFYVE, a gift from the De Camilli laboratory, primers: 5′-GTACCGGTGGCGCCTTAGAATTCGAAAG-3′ and 5′-CATGATCTAGAGGGCCCTATTC-3′) using AgeI/ApaI. CIBN-iRFP-CAAX was cloned by replacing GFP in CIBN-GFP-CAAX with iRFP (Addgene; 31857) using the megaprimer PCR method (primers: 5′-GGAATTGGAGAAAACGGATTATATTCATGTGACCGGTGGATCAATGGCTGAAGGATCCG-3′ and 5′-ACACTTTGTCTTTGACTTCTTTTTCTTCTTTTTACCACCGGTCTCTTCCATCAC-3′). CRY2-mCherry-PLDPMF or mCherry-CRY2-PLDPMF were amplified by PCR with insertion of a P2A sequence (primers: 5′-TTTAAACTTAAGCTTGGTACGCCACCATGAAGATGGAC-3′ or 5′-TTTAAACTTAAGCTTGGTACGCCACCATGGTGAGCAAG-3′ and 5′-TATAGCTCCATTCATGGTACCGGGACCTGGGTTTTCCTCGACATCGCCAGCCTGCTTGAGCAGACTAAAGTTTGTAGCGCCGCTTCCAGCGTTGCAGATTCCTCTTG-3′), and the resultant product was cloned into KpnI-digested CIBN-X (X = Sac1, CAAX, TGN, or 2xFYVE) plasmids using Gibson assembly to clone CRY2-mCherry-PLDPMF-P2A-CIBN-X or mCherry-CRY2-PLDPMF-P2A-CIBN-X plasmids. For PLDPMF-mCherry-CRY2-P2A-CIBN-X plasmids, PLDPMF (primers: 5′-GCGTTTAAACTTAAGCTTGCTAAGCCACCATGGCAGATTC-3′ and 5′-AGATCCGCCTCCGGAAGATCCACCAGCGTTGCAGATTCCTCTTG-3′), mCherry-CRY2 (amplified from PLDPMF-mCherry-CRY2, primers: 5′-GGTGGATCTTCCGGAGGCGGATCTGTGAGCAAGGGCGAGGAG-3′ and 5′-TAGCGCCGCTTCCTGCTGCTCCGATCATGATC-3′), and P2A-CIBN-X (amplified from CRY2-mCherry-PLDPMF-P2A-CIBN-X, primers: 5′-CGGAGCAGCAGGAAGCGGCGCTACAAAC-3′ and 5′-CTAGAAGGCACAGTCGAGGCTGATCAGCGGGTTTAAACG-3′) were cloned into KpnI/ApaI-digested pCDNA5-FRT-TO vector using Gibson assembly. For mCherry-PLDPMF-CRY2-P2A-CIBN-X plasmids, PLDPMF (primers: 5′-CGCCGAGGGCCGCCACTC-3′ and 5′-TTCGAGATCTTCCGGAAGATCCACCAGC-3′) and CRY2-P2A (amplified from PLDPMF-mCherry-CRY2-P2A-CIBN-X, primers: 5′-ATCTTCCGGAAGATCTCGAAGCGCGGCC-3′ and 5′-TATAGCTCCATTCATGGTACCGGGAC-3′) were cloned into BsrGI/KpnI-digested mCherry-CRY2-PLDPMF-P2A-CIBN-X using Gibson assembly. A catalytically dead mutant of PLDPMF was made by mutating histidine 170 to alanine using site-directed mutagenesis (primers: 5′-AAAAACAGCTTTCTCCTGGAACGCTAGCAAAATCTTGGTTGTAGAC-3′ and 5′-GTCTACAACCAAGATTTTGCTAGCGTTCCAGGAGAAAGCTGTTTTT-3′). iRFP-PASS was cloned by swapping out GFP in GFP-PASS (a gift from the Du Laboratory, University of Texas Health Science Center at Houston, Houston, TX) for iRFP (amplified from iRFP-C1 [Addgene; 54786], primers: 5′-GTAGCTAGCGCCACCATGGCTGAAGG-3′ and 5′-GTCTGTACATCTCTTCCATCACGCCGATC-3′) using NheI and BsrGI. For Gal1-PLDPMF-mCherry-2xPLCδPH, Gal1 (amplified from pFA6a-TRP1-PGAL1-3HA [a gift from the Emr laboratory, Cornell University, Ithaca, NY], primers: 5′-GGATCCACTAGTTCTAGAGCGGCCGCTCCTTGACGTTAAAGTATAGAGGTATATTAACAATTTTTTG-3′ and 5′-CCATGACGTCCGGATTAGAAGCCGCCGAG-3′), PLDPMF-mCherry (amplified from PLDPMF-mCherry-CRY2, primers: 5′-TTCTAATCCGGACGTCATGGCAGATTCAGCTACAC-3′ and 5′-GGAGCCCGTGGGATCTGAGTCCGGACTTG-3′), 2xPLCδPH (amplified from pGPD-mCherry-2xPLCδPH [a gift from the Emr laboratory], primers: 5′-ACTCAGATCCCACGGGCTCCAGGATGAC-3′ and 5′-TATATGATCACTACTTCTGCCGCTGGTCCATG-3′), and Yps1term (amplified from S. cerevisiae genomic DNA, primers: 5′-GCAGAAGTAGTGATCATATAGTTCCATCTTTAC-3′ and 5′-TGGAGCTCCACCGCGGTGGCAGGCGTTCTGAAAAGAAG-3′) were cloned into NotI-digested pRS416 vector using Gibson assembly.
Mammalian cell culture and transfection
HEK 293T cells were grown in DMEM (Corning) supplemented with 10% FBS (Corning) and 1% penicillin/streptomycin (Corning) at 37°C in a 5% CO2 atmosphere. For serum starvation, HEK 293T cells were cultured in DMEM supplemented with 1% penicillin/streptomycin (serum-free medium). Cells were transfected 20–24 h after seeding using Lipofectamine 2000 (Thermo Fisher) by following the manufacturer’s protocol. Briefly, cells were incubated in regular growth media or serum-free media containing plasmids premixed with Lipofectamine 2000 (0.33 µg DNA and 0.75 µl Lipofectamine 2000 per well for a 24-well plate). Experiments were performed 20–22 h after transfection.
Generation of CRISPR/Cas9-based PLD1/2 knockout cell lines
A lentiviral plasmid for PLD1 knockout was made by cloning the sgRNA (5′-GTGAGCCCACAAATAGACGG-3′) into the LentiCRISPRv2-Puro vector (Addgene; 98290), and a lentiviral plasmid for PLD2 knockout was made by cloning the sgRNA (5′-GGCACCGAAAGATATACCAG-3′) into the LentiCRISPRv2-Blast vector (Addgene; 98293). 7.5 µg of each LentiCRISPR plasmid was cotransfected with packaging plasmids (5.6 µg of Gag and 1.8 µg of Env) into HEK 293TN cells seeded on a 10-cm dish using Lipofectamine 2000. The virus-containing medium was collected 48 h and 72 h after transfection, and the cell debris was removed by passing through a 0.45-µm filter. For lentiviral transduction, HEK 293T cells seeded on a 6-well plate were treated with transduction medium (freshly prepared by mixing 1.5 ml virus-containing medium, 0.5 ml DMEM media, and polybrene to a final concentration of 8 µg/ml) for 48 h. During the transduction, the transduction medium was replaced every 12 h. After transduction, the cells were placed in regular DMEM for 24 h and then reseeded on a 10-cm dish in DMEM containing 1 µg/ml puromycin (for PLD1 knockout), 5 µg/ml blasticidin (for PLD2 knockout), or 1 µg/ml puromycin + 5 µg/ml blasticidin (for PLD1/2 knockout, i.e., DKO). The cells were kept in the selective medium for 3–5 d until all cells on a nontransduced control plate had died. The selected cells were lysed in RIPA lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton-X, 0.5% sodium deoxycholate, 0.1% SDS, and 1 mM EDTA) for subsequent Western blot analysis, with detection by chemiluminescence using the Clarity Western ECL Substrate (Bio-Rad) and acquisition on a Bio-Rad ChemiDoc MP System.
Setup for optogenetics experiments
A homemade light box was built by attaching four strips of dimmable, 12 V blue-LED tape light (1000Bulbs.com; 2835–60-IP65-B1203) on the inside of a styrofoam box. For optogenetics experiments, the light box was placed in the CO2 incubator using an AC Outlet Power Bank (Omars; 24,000 mAh, 80 W) as a power supply. To switch the light on and off without disturbing the controlled atmosphere (i.e., temperature and CO2 levels) in the incubator, a Wi-Fi controlled outlet (ECOPlugs) was used to enable remote control via a smartphone. For optogenetic stimulation of cells, light illumination was applied for 5-s intervals every 2 min for the duration of the experiment unless otherwise noted.
Live-cell imaging and flow cytometry analysis of mammalian cells
1 d (20–24 h) before transfection, HEK 293T cells were seeded on 35-mm glass-bottom imaging dishes (MatTek Corporation) for live-cell imaging or 24-well plates (Corning) for flow cytometry, both of which were pretreated with 50 µg/ml poly-L-lysine for coating. For IMPACT labeling using AzProp and BCN-BODIPY (Bumpus and Baskin, 2017), cells were treated with 2 mM AzProp in regular growth media for 40 min at 37°C with or without light illumination as above and rinsed three times with PBS (pH 7.4). Then the cells were treated with 1 µM BCN-BODIPY for 10 min at 37°C, rinsed another three times with PBS, and then incubated in regular growth media for 10 min at 37°C to allow for remaining excess, unreacted BCN-BODIPY to be washed out. For imaging, media was replaced with imaging buffer (135 mM NaCl, 5 mM KCl, 20 mM Hepes, 1 mM CaCl2, 100 µM MgCl2, 1 mg/ml BSA, and 1 mg/ml glucose). For flow cytometry, cells were lifted with trypsin-EDTA (Corning; 0.25% trypsin and 0.1% EDTA). For RT-IMPACT labeling using oxoTCO and Tz-BODIPY (Liang et al., 2019), serum-starved cells were treated with 3 mM oxoTCO in serum-free media for 3 min at 37°C with light illumination as above and rinsed three times with PBS. After adding 100 µl of PBS onto the center of the imaging dish to keep the cells wet, cells were subjected to imaging with a time-lapse movie. Between the second and third frames of the movie, 100 µl of 0.66 µM Tz-BODIPY was added to initiate the click chemistry. Images were acquired at room temperature using Zeiss Zen Blue 2.3 on a Zeiss LSM 800 confocal laser scanning microscope equipped with Plan Apochromat objectives (40× 1.4 NA) and two gallium arsenide phosphide detectors. Solid-state lasers (405, 488, 561, and 640 nm) were used to excite DAPI, BODIPY, mCherry, and iRFP, respectively. Acquired images were processed using FIJI. For flow cytometry, cells were analyzed using a BD FACSAria Fusion Fluorescence Activated Cell Sorter. Acquired data were analyzed using FlowCytometryTools via Python.
HPLC analysis of PLD activity in mammalian cells
Fluorescence-based measurement of mammalian PLD activity was performed as described previously (Bumpus and Baskin, 2017). Cells were treated with 5 mM AzProp in regular growth media for 40 min at 37°C, rinsed once with PBS, and subjected to lipid extraction using a modified Bligh-Dyer extraction. Cells were scraped in 250 µl methanol, 125 µl acetic acid (20 mM in water), and 100 µl PBS using cell scrapers (Corning), and the suspension was transferred into 1.5-ml microcentrifuge tubes. After addition of 500 µl chloroform to the suspension, the mixture was vortexed vigorously for 5 min and centrifuged at 17,000 ×g for 1 min. The organic layer (bottom) was collected in a new tube and dried under a stream of N2 gas to form a lipid film. The film was dissolved in 40 µl chloroform/methanol/water (73:23:3) with 5 µM BCN-BODIPY and incubated overnight at 42°C. The solution was then diluted with 70 µl of a chloroform/methanol/water mixture (73:23:3), filtered through a syringe-driven filter unit (Millex; 0.45 µm), and subjected to analysis using a Shimadzu Prominence HPLC equipped with an in-line fluorescence detector. Separation was achieved using a Luna 3 µm Silica LC Column (Phenomenex; 150 × 4.6 mm) with a binary gradient elution system where solvent A was chloroform/methanol/ammonium hydroxide (95:7:0.5) and solvent B was chloroform/methanol/water/ammonium hydroxide (60:34:5:0.5).
LC–MS analysis of PLD activity in mammalian cells
Measurement of PLD transphosphatidylation activity or levels of cellular PA by LC–MS was performed as described previously (Bumpus and Baskin, 2017). Cells were treated with either 5 mM AzProp in regular growth media for 40 min at 37°C (for measuring transphosphatidylation activity) or with the indicated inhibitor (for measuring levels of PA), rinsed once with PBS, and subjected to Bligh-Dyer lipid extraction as described above. The lipid film was dissolved in 7 µl of chloroform, followed by addition of 30 µl of a click chemistry reaction master mix (prepared freshly by mixing 78 µl of 10 mM Alk-QA [Bumpus and Baskin, 2017] in degassed methanol, 2.95 µl of 25 mM CuBF4 in DMSO, and 312 µl of degassed ethanol). The tube was then flushed with argon gas and incubated overnight at 42°C. The incubated solution was diluted with 200 µl of a chloroform/methanol/water mixture (73:23:3), filtered, and subjected to high-resolution LC–MS analysis on an Agilent 6230 electrospray ionization–time-of-flight MS coupled to an Agilent 1260 HPLC equipped with a Luna 3 µm Silica LC Column (Phenomenex; 50 × 2 mm) using a binary gradient elution system where solvent A was chloroform/methanol/ammonium hydroxide (85:15:0.5) and solvent B was chloroform/methanol/water/ammonium hydroxide (60:34:5:0.5).
Yeast cell culture and transformation
S. cerevisiae (strain SEY6210) stably expressing Gal4dbd.ER.VP16 (GEV) was obtained from the Emr laboratory. Yeast were grown in yeast extract–peptone-dextrose (YPD) medium before transformation, and yeast nitrogen base (YNB; supplemented with necessitate amino acids) medium was used after transformation. Plasmids were transformed into yeast cells using electroporation by using slight modifications to a standard protocol (Richman et al., 2009). Briefly, an overnight yeast culture was diluted to approximately OD 0.1 in 100 ml of YPD and shaken for 5–6 h at 30°C. The cells were pelleted by centrifugation in a 50-ml conical tube (4,000 ×g for 3 min), resuspended in 10 ml LiOAc-TE buffer (0.1 M lithium acetate, 10 mM Tris-HCl, pH 8.0, and 1 mM EDTA), and gently shaken for 45 min at 30°C. The culture was added with 250 µl DTT (1 M in water), gently shaken for 15 min at 30°C, transferred into a 50-ml conical tube, diluted with 40 ml of water, and centrifuged at 4,000 ×g for 3 min. The cells were then rinsed by centrifugation and resuspension three times at 4°C with 25 ml of water, 3 ml of 1 M sorbitol, and finally 50 µl of 1 M sorbitol. To 40 µl of the obtained cell slurry, 50 ng of linearized vector and 50 ng of insert were added (total volume of DNA was <5 µl). The mixture was incubated on ice for 20 min, transferred into a prechilled 0.2-cm-gap electroporation cuvette, and pulsed at 1.5 kV, 25 µF, 200 Ω using a Bio-Rad Gene Pulser. Immediately after the pulse, the mixture was diluted with 1 ml of cold 1 M sorbitol and incubated, with shaking, in 10 ml of YPD + 1 M sorbitol for 3 h at 30°C to increase the survival rate of transformed yeast cells. Cells were then resuspended in 50 ml of YNB (supplemented with appropriate amino acids and lacking uracil) and shaken for 48 h at 30°C. The culture was then diluted by a factor of 1:100 in 50 ml YNB and shaken for another 24 h to reduce untransformed cells.
Imaging and FACS sorting of yeast
Transformed yeast cells were subcultured to OD ∼0.2 in 10 ml of YNB and shaken for 3–4 h at 30°C. When cell density was OD 0.8, 20 µl β-estradiol (100 µM in water, final concentration of 200 nM) was added to the culture to induce PLD expression. For IMPACT labeling, cells after the induction were harvested for OD 3 equivalent, resuspended in 1 ml YNB supplemented with 10 mM AzProp and 200 nM β-estradiol and shaken for 1 h at 30°C. The cells were then washed three times by centrifugation at 10,000 ×g for 1 min and resuspension in 0.1 M potassium phosphate buffer (prepared by mixing 0.1 M K2HPO4 and 0.1 M KH2PO4 to pH 6.5). Cells were fixed by resuspension of the pellet in 3.7% formaldehyde in 680 µl of potassium phosphate buffer and gentle shaking for 45 min at room temperature. The fixed cells were washed three times with 0.1 M potassium phosphate buffer, resuspended in 300 µl of potassium phosphate buffer supplemented with 1 µM BCN-BODIPY, and shaken for 30 min at 30°C for fluorescent tagging via click chemistry. The labeled cells were washed three times with 0.1 M potassium phosphate buffer and then imaged using a Zeiss LSM 800 confocal laser scanning microscope or sorted using a BD FACSAria Fusion Fluorescence Activated Cell Sorter. In FACS, 1–2 × 105 cells were collected for subsequent plasmid extraction.
Directed evolution of PLD variants from yeast by plasmid extraction and mutagenesis
Prior to plasmid extraction, yeast cells enriched by FACS were supplemented with a 1,000-fold excess of nontransformed yeast cells (e.g., 108 nontransformed cells for 105 of collected cells) to increase total DNA concentration in the solution and aid in efficient DNA recovery during the plasmid extraction. After centrifugation (10,000 ×g for 1 min), 200 µl of glass beads (Scientific Industries; SI-BG05) and 200 µl of miniprep buffer 1 (50 mM Tris-HCl, 10 mM EDTA, and 100 µg/ml RNaseA, pH 8.0) were added to the cell pellet, followed by vigorous vortexing for 10 min. The mixture was mixed with 200 µl of miniprep buffer 2 (0.2 M NaOH and 1% SDS), incubated for 10 min at room temperature, mixed with 280 µl of miniprep buffer 3 (4.2 M guanidine-HCl and 0.9 M potassium acetate, pH 4.8), and centrifuged for 10 min at 4°C. The supernatant was transferred into a miniprep column (BioBasic; EZ-10). After washing the column once with 750 µl of miniprep buffer 4 (10 mM Tris-HCl and 80% ethanol, pH 8.0), the DNA was eluted into 30 µl of water. The PLD fragment was amplified by a two-step, nested PCR sequence, using 1 µl of eluted DNA for a 20-µl reaction mixture for the first PCR reaction. The PLD fragment amplified by nested PCR was then mutagenized by error-prone PCR using dNTP analogues (8-oxo-dGTP and 2′-deoxy-P-nucleoside-5′-triphosphate) according to established procedures (Angelini et al., 2015).
HPLC analysis of PLD activity in yeast
Cells were treated with 50 mM AzProp in YNB for 1 h at 30°C and rinsed three times with 0.1 M potassium phosphate by centrifugation at 10,000 ×g for 1 min. To the cells were added 200 µl of methanol and 200 µl of glass beads, and the mixture was vortexed vigorously for 10 min. To the mixture was then added 100 µl of acetic acid (20 mM in water), 80 µl of 0.1 M potassium phosphate buffer, and 400 µl of chloroform, followed by further vortexing for 5 min. Afterward, the lipid extract was tagged with BCN-BODIPY by click chemistry and analyzed by HPLC as described previously for mammalian cells.
Preparation of 2D hydrogels
2D hydrogels of different stiffnesses were prepared following established procedures (Tse and Engler, 2010). Briefly, 125 µl of 0.1 M NaOH was evaporated on a 12-mm cover glass (Fisherbrand, Thermo Fisher) to coat the glass with a uniform NaOH film. The glass was then treated with 50 µl of 3-aminopropyltriethoxysilane for 5 min, rinsed extensively with water, incubated in 2 ml of 0.5% glutaraldehyde in PBS for 30 min, and dried under air after aspiration of the glutaraldehyde solution. In parallel, a microscope slide (Fisherbrand, Thermo Fisher) was treated with 200 µl dichlorodimethylsilane for 5 min, rinsed for 1 min with water, and wiped with a Kimwipe to dry. For preparing stiff hydrogels (∼40 kPa), 2 ml of 40% acrylamide and 2.4 ml of 2% bisacrylamide were mixed with 5.6 ml water. For preparing soft hydrogels (∼1 kPa), 1.25 ml of 40% acrylamide and 0.15 ml of 2% bisacrylamide were mixed with 8.6 ml of water. To the solution, 1/100th volume of 10% ammonium persulfate and 1/1,000th volume of tetramethylethylenediamine were added. A small amount (6.25 µl) of the gel solution was quickly applied onto the chloro-silanated microscope slide, and then the amino-silanated cover glass was placed on the top. The gel solution was allowed to polymerize for 15 min, and then the microscope slide was carefully removed to obtain a hydrogel that was adhered to the cover glass. The hydrogel was rinsed three times with water for 5 min each, treated with 125 µl of 0.4 mM sulfo-SANPAH in 50 mM Hepes under 365-nm UV illumination (Thermo Fisher; catalog number UVP95004207) for 15 min, rinsed three times with 50 mM Hepes, and incubated in 50 µg/ml collagen type I (Corning; catalog number 354236) in 50 mM Hepes at 37°C for 8–16 h. After rinsing two times with PBS, the collagen-coated hydrogel was submerged in PBS, placed in a tissue culture hood for 30 min under UV irradiation for sterilization, and used for cell culture.
IF imaging of YAP in mammalian cells
1 d (20–24 h) before transfection, HEK 293T cells were seeded on either 12-mm cover glass (Fisherbrand) pretreated with poly-L-lysine or homemade 2D hydrogels coated with collagen type I (described above). Approximately 20 h after transfection in serum-free media, optoPLD was activated by 488-nm light illumination (5 s of illumination at 2-min intervals) for 30 min at 37°C. The cells were then fixed in 4% formaldehyde for 10 min at room temperature, rinsed three times with PBS, permeabilized with 0.5% Triton X-100 in PBS for 5 min at room temperature, and blocked with 1% BSA and 0.1% Tween-20 in PBS (blocking buffer) for 30 min. The cells were then treated with a 1:100 dilution of anti-YAP antibody in blocking buffer for 1 h at room temperature, rinsed three times with 0.1% Tween-20 in PBS (PBS-T), treated with a 1:1,000 dilution of anti-mouse–Alexa Fluor 488 antibody conjugate in blocking buffer for 1 h at room temperature, and rinsed three times with PBS-T. After the final rinse, the cells were incubated in PBS-T for 5 min, mounted on microscope slides (Fisherbrand, Thermo Fisher) using ProLong Diamond Antifade Mountant with DAPI (Thermo Fisher), and incubated overnight at room temperature in the dark before imaging. Image acquisition by laser-scanning confocal microscopy was performed as described above by using solid-state lasers (405, 488, 561, and 640 nm) to excite DAPI, Alexa Fluor 488, mCherry, and Alexa Fluor 647, respectively. Acquired images were analyzed using FIJI and Python.
Statistical methods
For experiments involving quantification of comparisons between more than two independent groups, statistical significance was calculated using the Kruskal–Wallis test, followed by the Games–Howell post hoc test, operated using the StatsModels package in Python (for Kruskal–Wallis) and userfriendlyscience package in R (for Games–Howell). For confocal microscopy, colocalization was calculated in FIJI using Coloc 2 plugin to determine Pearson correlation coefficients. All experiments were performed in at least three biological replicates on different days. Exact numbers of replicate experiments and sample sizes are provided in each figure legend.
Online supplemental material
Fig. S1 includes MS data, HPLC traces, and Western blot data to characterize PA levels, PLD levels, and PLD activity in PLD-deficient HEK 293T cells. Fig. S2 includes confocal microscopy colocalization analysis to characterize optoPLD localization and flow cytometry plots of experiments to optimize optoPLD activity. Fig. S3 includes confocal micrographs of PA biosensor and optoPLD localization during a time course of optoPLD activation and inactivation. Fig. S4 includes confocal micrographs, HPLC traces, and MS data to characterize the PM-targeted PLD in yeast used for directed evolution. Fig. S5 includes confocal micrographs of IF staining to evaluate changes in YAP localization upon optoPLD activation in cells grown on hydrogels of different stiffnesses.
Supplementary Material
Acknowledgments
We thank the Cornell Biotechnology Resource Center Imaging Facility (National Science Foundation grant DBI-1428922). We thank Alex Batrouni, Jeff Jorgensen, Sho Suzuki, and Lu Zhu for helpful discussions; the Emr, Fromme, and Yu laboratories (Cornell University, Ithaca, NY) for equipment and reagents; and Jonathan Lee, Paris Ghazi, and Adam Wojno for assistance.
J.M. Baskin acknowledges support from a Beckman Young Investigator award, a Sloan Research Fellowship, and the National Science Foundation (CAREER CHE-1749919). R. Tei was supported by Funai Overseas and Cornell Fellowships.
The authors declare no competing financial interests.
Author contributions: R. Tei and J. Baskin conceived of the project, designed experiments, interpreted the results, and wrote the manuscript. R. Tei carried out all experiments and data analysis.
References
- Adachi Y., Itoh K., Yamada T., Cerveny K.L., Suzuki T.L., Macdonald P., Frohman M.A., Ramachandran R., Iijima M., and Sesaki H.. 2016. Coincident Phosphatidic Acid Interaction Restrains Drp1 in Mitochondrial Division. Mol. Cell. 63:1034–1043. 10.1016/j.molcel.2016.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamudi S.H., Satapathy R., Kim J., Su D., Ren H., Das R., Hu L., Alvarado-Martínez E., Lee J.Y., Hoppmann C., et al. 2016. Development of background-free tame fluorescent probes for intracellular live cell imaging. Nat. Commun. 7:11964 10.1038/ncomms11964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini A., Chen T.F., de Picciotto S., Yang N.J., Tzeng A., Santos M.S., Van Deventer J.A., Traxlmayr M.W., and Wittrup K.D.. 2015. Protein Engineering and Selection Using Yeast Surface Display. In Yeast Surface Display: Methods, Protocols, and Applications. B. Liu, editor. Springer, New York. 3–36. 10.1007/978-1-4939-2748-7_1 [DOI] [PubMed] [Google Scholar]
- Antonescu C.N., Danuser G., and Schmid S.L.. 2010. Phosphatidic acid plays a regulatory role in clathrin-mediated endocytosis. Mol. Biol. Cell. 21:2944–2952. 10.1091/mbc.e10-05-0421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti L., Barentine A.E.S., Messa M., Wheeler H., Bewersdorf J., and De Camilli P.. 2018. Light-activated protein interaction with high spatial subcellular confinement. Proc. Natl. Acad. Sci. USA. 115:E2238–E2245. 10.1073/pnas.1713845115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman M.L., Royzen M., and Fox J.M.. 2008. Tetrazine ligation: fast bioconjugation based on inverse-electron-demand Diels-Alder reactivity. J. Am. Chem. Soc. 130:13518–13519. 10.1021/ja8053805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohdanowicz M., Schlam D., Hermansson M., Rizzuti D., Fairn G.D., Ueyama T., Somerharju P., Du G., and Grinstein S.. 2013. Phosphatidic acid is required for the constitutive ruffling and macropinocytosis of phagocytes. Mol. Biol. Cell. 24:1700–1712: S1–S7. 10.1091/mbc.e12-11-0789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley R.M., and Duncan R.E.. 2018. The lysophosphatidic acid acyltransferases (acylglycerophosphate acyltransferases) family: one reaction, five enzymes, many roles. Curr. Opin. Lipidol. 29:110–115. 10.1097/MOL.0000000000000492 [DOI] [PubMed] [Google Scholar]
- Brown H.A., Henage L.G., Preininger A.M., Xiang Y., and Exton J.H.. 2007. Biochemical analysis of phospholipase D. Methods Enzymol. 434:49–87. 10.1016/S0076-6879(07)34004-4 [DOI] [PubMed] [Google Scholar]
- Bruntz R.C., Lindsley C.W., and Brown H.A.. 2014. Phospholipase D signaling pathways and phosphatidic acid as therapeutic targets in cancer. Pharmacol. Rev. 66:1033–1079. 10.1124/pr.114.009217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumpus T.W., and Baskin J.M.. 2016. A Chemoenzymatic Strategy for Imaging Cellular Phosphatidic Acid Synthesis. Angew. Chem. Int. Ed. Engl. 55:13155–13158. 10.1002/anie.201607443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumpus T.W., and Baskin J.M.. 2017. Clickable Substrate Mimics Enable Imaging of Phospholipase D Activity. ACS Cent. Sci. 3:1070–1077. 10.1021/acscentsci.7b00222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumpus T.W., and Baskin J.M.. 2018. Greasing the Wheels of Lipid Biology with Chemical Tools. Trends Biochem. Sci. 43:970–983. 10.1016/j.tibs.2018.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumpus T.W., Liang F.J., and Baskin J.M.. 2018. Ex Uno Plura: Differential Labeling of Phospholipid Biosynthetic Pathways with a Single Bioorthogonal Alcohol. Biochemistry. 57:226–230. 10.1021/acs.biochem.7b01021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson J.C.T., Meimetis L.G., Hilderbrand S.A., and Weissleder R.. 2013. BODIPY-tetrazine derivatives as superbright bioorthogonal turn-on probes. Angew. Chem. Int. Ed. Engl. 52:6917–6920. 10.1002/anie.201301100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRose R., Miyamoto T., and Inoue T.. 2013. Manipulating signaling at will: chemically-inducible dimerization (CID) techniques resolve problems in cell biology. Pflugers Arch. 465:409–417. 10.1007/s00424-012-1208-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj N.K., Weissleder R., and Hilderbrand S.A.. 2008. Tetrazine-based cycloadditions: application to pretargeted live cell imaging. Bioconjug. Chem. 19:2297–2299. 10.1021/bc8004446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du G., Altshuller Y.M., Kim Y., Han J.M., Ryu S.H., Morris A.J., and Frohman M.A.. 2000. Dual requirement for rho and protein kinase C in direct activation of phospholipase D1 through G protein-coupled receptor signaling. Mol. Biol. Cell. 11:4359–4368. 10.1091/mbc.11.12.4359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du G., Altshuller Y.M., Vitale N., Huang P., Chasserot-Golaz S., Morris A.J., Bader M.-F., and Frohman M.A.. 2003. Regulation of phospholipase D1 subcellular cycling through coordination of multiple membrane association motifs. J. Cell Biol. 162:305–315. 10.1083/jcb.200302033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du G., Huang P., Liang B.T., and Frohman M.A.. 2004. Phospholipase D2 localizes to the plasma membrane and regulates angiotensin II receptor endocytosis. Mol. Biol. Cell. 15:1024–1030. 10.1091/mbc.e03-09-0673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichmann T.O., and Lass A.. 2015. DAG tales: the multiple faces of diacylglycerol--stereochemistry, metabolism, and signaling. Cell. Mol. Life Sci. 72:3931–3952. 10.1007/s00018-015-1982-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Vilella-Bach M., Bachmann R., Flanigan A., and Chen J.. 2001. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 294:1942–1945. 10.1126/science.1066015 [DOI] [PubMed] [Google Scholar]
- Foster D.A. 2013. Phosphatidic acid and lipid-sensing by mTOR. Trends Endocrinol. Metab. 24:272–278. 10.1016/j.tem.2013.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cambronero J. 2014. Phosphatidic acid, phospholipase D and tumorigenesis. Adv. Biol. Regul. 54:197–206. 10.1016/j.jbior.2013.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H., Qi R., Zhou J.J., Ta A.P., Yang B., Nakaoka H.J., Seo G., Guan K.L., Luo R., and Wang W.. 2018a Regulation of the Hippo Pathway by Phosphatidic Acid-Mediated Lipid-Protein Interaction. Mol. Cell. 72:328–340.e8. 10.1016/j.molcel.2018.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H., Vargas R., Seo G., and Wang W.. 2018b Phosphatidic acid: a lipid regulator of the Hippo pathway. Mol. Cell. Oncol. 6:1558683 10.1080/23723556.2018.1558683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideshima T., Chauhan D., Hayashi T., Podar K., Akiyama M., Mitsiades C., Itsiades N.M., Gong B., Bonham L., de Vries P., et al. 2003. Antitumor Activity of Lysophosphatidic Acid Acyltransferase-β Inhibitors, a Novel Class of Agents, in Multiple Myeloma. Cancer Res. 63:8428–8436. [PubMed] [Google Scholar]
- Horchani H., de Saint-Jean M., Barelli H., and Antonny B.. 2014. Interaction of the Spo20 membrane-sensor motif with phosphatidic acid and other anionic lipids, and influence of the membrane environment. PLoS One. 9:e113484 10.1371/journal.pone.0113484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes W.E., Larijani B., and Parker P.J.. 2002. Detecting protein-phospholipid interactions. Epidermal growth factor-induced activation of phospholipase D1b in situ. J. Biol. Chem. 277:22974–22979. 10.1074/jbc.M201391200 [DOI] [PubMed] [Google Scholar]
- Idevall-Hagren O., Dickson E.J., Hille B., Toomre D.K., and De Camilli P.. 2012. Optogenetic control of phosphoinositide metabolism. Proc. Natl. Acad. Sci. USA. 109:E2316–E2323. 10.1073/pnas.1211305109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J.-H., Lee C.S., Hwang D., and Ryu S.H.. 2012. Understanding of the roles of phospholipase D and phosphatidic acid through their binding partners. Prog. Lipid Res. 51:71–81. 10.1016/j.plipres.2011.12.003 [DOI] [PubMed] [Google Scholar]
- Kameoka S., Adachi Y., Okamoto K., Iijima M., and Sesaki H.. 2018. Phosphatidic Acid and Cardiolipin Coordinate Mitochondrial Dynamics. Trends Cell Biol. 28:67–76. 10.1016/j.tcb.2017.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassas N., Tryoen-Tóth P., Corrotte M., Thahouly T., Bader M.-F., Grant N.J., and Vitale N.. 2012. Genetically encoded probes for phosphatidic acid. Methods Cell Biol. 108:445–459. 10.1016/B978-0-12-386487-1.00020-1 [DOI] [PubMed] [Google Scholar]
- Kassas N., Tanguy E., Thahouly T., Fouillen L., Heintz D., Chasserot-Golaz S., Bader M.-F., Grant N.J., and Vitale N.. 2017. Comparative Characterization of Phosphatidic Acid Sensors and Their Localization during Frustrated Phagocytosis. J. Biol. Chem. 292:4266–4279. 10.1074/jbc.M116.742346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M.J., Hughes R.M., Peteya L.A., Schwartz J.W., Ehlers M.D., and Tucker C.L.. 2010. Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods. 7:973–975. 10.1038/nmeth.1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Guzman-Hernandez M.L., Wisniewski E., and Balla T.. 2015. Phosphatidylinositol-Phosphatidic Acid Exchange by Nir2 at ER-PM Contact Sites Maintains Phosphoinositide Signaling Competence. Dev. Cell. 33:549–561. 10.1016/j.devcel.2015.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooijman E.E., and Burger K.N.J.. 2009. Biophysics and function of phosphatidic acid: a molecular perspective. Biochim. Biophys. Acta. 1791:881–888. 10.1016/j.bbalip.2009.04.001 [DOI] [PubMed] [Google Scholar]
- Lambert W.D., Scinto S.L., Dmitrenko O., Boyd S.J., Magboo R., Mehl R.A., Chin J.W., Fox J.M., and Wallace S.. 2017. Computationally guided discovery of a reactive, hydrophilic trans-5-oxocene dienophile for bioorthogonal labeling. Org. Biomol. Chem. 15:6640–6644. 10.1039/C7OB01707C [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiros I., McSweeney S., and Hough E.. 2004. The reaction mechanism of phospholipase D from Streptomyces sp. strain PMF. Snapshots along the reaction pathway reveal a pentacoordinate reaction intermediate and an unexpected final product. J. Mol. Biol. 339:805–820. 10.1016/j.jmb.2004.04.003 [DOI] [PubMed] [Google Scholar]
- Lewis J.A., Scott S.A., Lavieri R., Buck J.R., Selvy P.E., Stoops S.L., Armstrong M.D., Brown H.A., and Lindsley C.W.. 2009. Design and synthesis of isoform-selective phospholipase D (PLD) inhibitors. Part I: Impact of alternative halogenated privileged structures for PLD1 specificity. Bioorg. Med. Chem. Lett. 19:1916–1920. 10.1016/j.bmcl.2009.02.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D., Wu K., Tei R., Bumpus T.W., Ye J., and Baskin J.M.. 2019. A real-time, click chemistry imaging approach reveals stimulus-specific subcellular locations of phospholipase D activity. Proc. Natl. Acad. Sci. USA. 116:15453–15462. 10.1073/pnas.1903949116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B. 2015. Yeast surface display : methods, protocols, and applications. Humana Press, New York: 10.1007/978-1-4939-2748-7 [DOI] [Google Scholar]
- Liu Y., Su Y., and Wang X.. 2013. Phosphatidic Acid-Mediated Signaling. In Lipid-mediated Protein Signaling. Springer Netherlands, Dordrecht: 159–176. 10.1007/978-94-007-6331-9_9 [DOI] [Google Scholar]
- Liu Z., Chen O., Wall J.B.J., Zheng M., Zhou Y., Wang L., Ruth Vaseghi H., Qian L., and Liu J.. 2017. Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector. Sci. Rep. 7:2193 10.1038/s41598-017-02460-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Tay L.W.R., He J., and Du G.. 2016. Monitoring Phosphatidic Acid Signaling in Breast Cancer Cells Using Genetically Encoded Biosensors. Methods Mol. Biol. 1406:225–237. 10.1007/978-1-4939-3444-7_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloud R.L., Franks C.E., Campbell S.T., Purow B.W., Harris T.E., and Hsu K.L.. 2018. Deconstructing Lipid Kinase Inhibitors by Chemical Proteomics. Biochemistry. 57:231–236. 10.1021/acs.biochem.7b00962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIsaac R.S., Silverman S.J., McClean M.N., Gibney P.A., Macinskas J., Hickman M.J., Petti A.A., and Botstein D.. 2011. Fast-acting and nearly gratuitous induction of gene expression and protein depletion in Saccharomyces cerevisiae. Mol. Biol. Cell. 22:4447–4459. 10.1091/mbc.e11-05-0466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z., Moroishi T., and Guan K.L.. 2016. Mechanisms of Hippo pathway regulation. Genes Dev. 30:1–17. 10.1101/gad.274027.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z., Qiu Y., Lin K.C., Kumar A., Placone J.K., Fang C., Wang K.C., Lu S., Pan M., Hong A.W., et al. 2018. RAP2 mediates mechanoresponses of the Hippo pathway. Nature. 560:655–660. 10.1038/s41586-018-0444-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris A.J., Frohman M.A., and Engebrecht J.. 1997. Measurement of phospholipase D activity. Anal. Biochem. 252:1–9. 10.1006/abio.1997.2299 [DOI] [PubMed] [Google Scholar]
- Nelson R.K., and Frohman M.A.. 2015. Physiological and pathophysiological roles for phospholipase D. J. Lipid Res. 56:2229–2237. 10.1194/jlr.R059220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu J., Ben Johny M., Dick I.E., and Inoue T.. 2016. Following Optogenetic Dimerizers and Quantitative Prospects. Biophys. J. 111:1132–1140. 10.1016/j.bpj.2016.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly M.C., Oguin T.H. III, Scott S.A., Thomas P.G., Locuson C.W., Morrison R.D., Daniels J.S., Brown H.A., and Lindsley C.W.. 2014. Discovery of a highly selective PLD2 inhibitor (ML395): a new probe with improved physiochemical properties and broad-spectrum antiviral activity against influenza strains. ChemMedChem. 9:2633–2637. 10.1002/cmdc.201402333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira T.G., and Di Paolo G.. 2010. Phospholipase D in brain function and Alzheimer’s disease. Biochim. Biophys. Acta. 1801:799–805. 10.1016/j.bbalip.2010.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira B.L., Guo Z., and Bernardes G.J.L.. 2017. Inverse electron demand Diels-Alder reactions in chemical biology. Chem. Soc. Rev. 46:4895–4950. 10.1039/C7CS00184C [DOI] [PubMed] [Google Scholar]
- Peng X., and Frohman M.A.. 2012. Mammalian phospholipase D physiological and pathological roles. Acta Physiol. (Oxf.). 204:219–226. 10.1111/j.1748-1716.2011.02298.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen E.N., Chung H.W., Nayebosadri A., and Hansen S.B.. 2016. Kinetic disruption of lipid rafts is a mechanosensor for phospholipase D. Nat. Commun. 7:13873 10.1038/ncomms13873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powner D.J., and Wakelam M.J.O.. 2002. The regulation of phospholipase D by inositol phospholipids and small GTPases. FEBS Lett. 531:62–64. 10.1016/S0014-5793(02)03410-5 [DOI] [PubMed] [Google Scholar]
- Purow B. 2015. Molecular pathways: Targeting diacylglycerol kinase alpha in cancer. Clin. Cancer Res. 21:5008–5012. 10.1158/1078-0432.CCR-15-0413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putta P., Rankenberg J., Korver R.A., van Wijk R., Munnik T., Testerink C., and Kooijman E.E.. 2016. Phosphatidic acid binding proteins display differential binding as a function of membrane curvature stress and chemical properties. Biochim. Biophys. Acta. 1858:2709–2716. 10.1016/j.bbamem.2016.07.014 [DOI] [PubMed] [Google Scholar]
- Richman S.A., Kranz D.M., and Stone J.D.. 2009. Biosensor detection systems: engineering stable, high-affinity bioreceptors by yeast surface display. Methods Mol. Biol. 504:323–350. 10.1007/978-1-60327-569-9_19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakane F., Mizuno S., and Komenoi S.. 2016. Diacylglycerol Kinases as Emerging Potential Drug Targets for a Variety of Diseases: An Update. Front. Cell Dev. Biol. 4:82 10.3389/fcell.2016.00082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott S.A., Selvy P.E., Buck J.R., Cho H.P., Criswell T.L., Thomas A.L., Armstrong M.D., Arteaga C.L., Lindsley C.W., and Brown H.A.. 2009. Design of isoform-selective phospholipase D inhibitors that modulate cancer cell invasiveness. Nat. Chem. Biol. 5:108–117. 10.1038/nchembio.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvy P.E., Lavieri R.R., Lindsley C.W., and Brown H.A.. 2011. Phospholipase D: enzymology, functionality, and chemical modulation. Chem. Rev. 111:6064–6119. 10.1021/cr200296t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu X., Shaner N.C., Yarbrough C.A., Tsien R.Y., and Remington S.J.. 2006. Novel chromophores and buried charges control color in mFruits. Biochemistry. 45:9639–9647. 10.1021/bi060773l [DOI] [PubMed] [Google Scholar]
- Shulga Y.V., Topham M.K., and Epand R.M.. 2011. Regulation and functions of diacylglycerol kinases. Chem. Rev. 111:6186–6208. 10.1021/cr1004106 [DOI] [PubMed] [Google Scholar]
- Stampouloglou E., and Varelas X.. 2018. Phosphatidic Acid Signals via the Hippo Pathway. Mol. Cell. 72:205–206. 10.1016/j.molcel.2018.10.004 [DOI] [PubMed] [Google Scholar]
- Stanton B.Z., Chory E.J., and Crabtree G.R.. 2018. Chemically induced proximity in biology and medicine. Science. 359:eaao5902 10.1126/science.aao5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer T.P., Ahn S., and Meyer T.. 1998. Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr. Biol. 8:343–346. 10.1016/S0960-9822(98)70135-6 [DOI] [PubMed] [Google Scholar]
- Su W., Yeku O., Olepu S., Genna A., Park J.-S., Ren H., Du G., Gelb M.H., Morris A.J., and Frohman M.A.. 2009. 5-Fluoro-2-indolyl des-chlorohalopemide (FIPI), a phospholipase D pharmacological inhibitor that alters cell spreading and inhibits chemotaxis. Mol. Pharmacol. 75:437–446. 10.1124/mol.108.053298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanguy E., Kassas N., and Vitale N.. 2018. Protein–phospholipid interaction motifs: A focus on phosphatidic acid. Biomolecules. 8:E20 10.3390/biom8020020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totaro A., and Piccolo S.. 2019. Phosphatidic Acid Enters into the YAP/TAZ Arena. Trends Mol. Med. 25:5–7. 10.1016/j.molmed.2018.11.002 [DOI] [PubMed] [Google Scholar]
- Tse J.R., and Engler A.J.. 2010. Preparation of Hydrogel Substrates with Tunable Mechanical Properties. Curr. Protoc. Cell Biol. 47:10.16.1 10.1002/0471143030.cb1016s47 [DOI] [PubMed] [Google Scholar]
- Tucker C.L., Vrana J.D., and Kennedy M.J.. 2014. Tools for controlling protein interactions using light. Curr. Protoc. Cell Biol. 64:17–20. 10.1002/0471143030.cb1716s64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G., and de Kroon A.I.P.M.. 2011. Lipid map of the mammalian cell. J. Cell Sci. 124:5–8. 10.1242/jcs.071233 [DOI] [PubMed] [Google Scholar]
- Vance J.E. 2015. Phospholipid synthesis and transport in mammalian cells. Traffic. 16:1–18. 10.1111/tra.12230 [DOI] [PubMed] [Google Scholar]
- Vance J.E., and Vance D.E.. 2004. Phospholipid biosynthesis in mammalian cells. Biochem. Cell Biol. 82:113–128. 10.1139/o03-073 [DOI] [PubMed] [Google Scholar]
- Várnai P., and Balla T.. 1998. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J. Cell Biol. 143:501–510. 10.1083/jcb.143.2.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Devaiah S.P., Zhang W., and Welti R.. 2006. Signaling functions of phosphatidic acid. Prog. Lipid Res. 45:250–278. 10.1016/j.plipres.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Wu H., and Devaraj N.K.. 2016. Inverse Electron-Demand Diels-Alder Bioorthogonal Reactions. Top. Curr. Chem. (Cham). 374:3 10.1007/s41061-015-0005-z [DOI] [PubMed] [Google Scholar]
- Yang C.-Y., and Frohman M.A.. 2012. Mitochondria: signaling with phosphatidic acid. Int. J. Biochem. Cell Biol. 44:1346–1350. 10.1016/j.biocel.2012.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin F., Yu J., Zheng Y., Chen Q., Zhang N., and Pan D.. 2013. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell. 154:1342–1355. 10.1016/j.cell.2013.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon M.S., Du G., Backer J.M., Frohman M.A., and Chen J.. 2011. Class III PI-3-kinase activates phospholipase D in an amino acid-sensing mTORC1 pathway. J. Cell Biol. 195:435–447. 10.1083/jcb.201107033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon M.S., Rosenberger C.L., Wu C., Truong N., Sweedler J.V., and Chen J.. 2015. Rapid mitogenic regulation of the mTORC1 inhibitor, DEPTOR, by phosphatidic acid. Mol. Cell. 58:549–556. 10.1016/j.molcel.2015.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F.X., Zhao B., and Guan K.L.. 2015. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell. 163:811–828. 10.1016/j.cell.2015.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Fang X., Zhang L., Hong G., Lin Y., Zheng Q., Xu Y., Ruan Y., Weng W., Xia H., and Chen G.. 2012. Multi-responsive self-healing metallo-supramolecular gels based on “click” ligand. J. Mater. Chem. 22:11515–11518. 10.1039/c2jm31347b [DOI] [Google Scholar]
- Zhang F., Wang Z., Lu M., Yonekubo Y., Liang X., Zhang Y., Wu P., Zhou Y., Grinstein S., Hancock J.F., and Du G.. 2014. Temporal production of the signaling lipid phosphatidic acid by phospholipase D2 determines the output of extracellular signal-regulated kinase signaling in cancer cells. Mol. Cell. Biol. 34:84–95. 10.1128/MCB.00987-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.