Pemberton et al. characterize a molecular toolbox for the visualization and manipulation of phosphatidylinositol (PI) within intact cells. Results using these approaches define the steady-state distribution of PI across subcellular membrane compartments and provide new insights into the relationship between PI availability and polyphosphoinositide turnover.
Abstract
Phosphatidylinositol (PI) is an essential structural component of eukaryotic membranes that also serves as the common precursor for polyphosphoinositide (PPIn) lipids. Despite the recognized importance of PPIn species for signal transduction and membrane homeostasis, there is still a limited understanding of the relationship between PI availability and the turnover of subcellular PPIn pools. To address these shortcomings, we established a molecular toolbox for investigations of PI distribution within intact cells by exploiting the properties of a bacterial enzyme, PI-specific PLC (PI-PLC). Using these tools, we find a minor presence of PI in membranes of the ER, as well as a general enrichment within the cytosolic leaflets of the Golgi complex, peroxisomes, and outer mitochondrial membrane, but only detect very low steady-state levels of PI within the plasma membrane (PM) and endosomes. Kinetic studies also demonstrate the requirement for sustained PI supply from the ER for the maintenance of monophosphorylated PPIn species within the PM, Golgi complex, and endosomal compartments.
Introduction
The dynamic remodeling of cellular membranes relies on molecular mechanisms that exploit the unique physiochemical properties of individual lipid species (van Meer et al., 2008; Holthuis and Menon, 2014; Bigay and Antonny, 2012). Our current understanding of membrane composition has benefitted greatly from organelle separation techniques that are now being combined with comprehensive lipidomics (Shevchenko and Simons, 2010; Wenk, 2010), but these methods still lack information regarding the dynamics of lipid metabolism. Alternatively, advances in live-cell imaging using endogenous membrane-binding protein domains have allowed for the visualization of distinct lipid species with high spatial and temporal resolution (Várnai et al., 2017; Wills et al., 2018). However, despite this progress, imaging applications are currently limited to only a few classes of lipids and have been focused on low-abundance species that show changes as part of defined intracellular signaling responses. Visualization or manipulation of the structural lipids that form the bulk of eukaryotic membranes has received much less attention. In particular, among the core structural lipids, phosphatidylinositol (PI) is unique in that it also serves as the precursor for polyphosphoinositide (PPIn) lipids, which directly control important aspects of membrane trafficking and cellular metabolism (Balla, 2013). Consequently, defining the subcellular localization of PI is essential for understanding the central role of this lipid, and its phosphorylated derivatives, in cellular physiology.
In the present study, we used the well-characterized bacterial PI-specific PLCs (PI-PLC) to design molecular tools for the visualization and manipulation of PI content in the membranes of intact cells. Specifically, based on the structure and enzymology of different PI-PLC variants (Heinz et al., 1998; Roberts et al., 2018), we selected the Bacillus cereus PI-PLC (BcPI-PLC; Kuppe et al., 1989; Volwerk et al., 1989) to use as a platform for protein engineering efforts. Importantly, BcPI-PLC shows remarkable specificity for PI and does not hydrolyze phosphorylated PPIn species or other common structural phospholipids (Ikezawa and Taguchi, 1981). Variations in the fatty acyl chains are well tolerated by BcPI-PLC (Guther et al., 1994), which is not surprising given the conserved function of these secreted proteins as virulence factors that target the PI moiety of GPI-linked proteins present on the surface of host cells. Capitalizing on these features, our general strategy was twofold: first, we targeted residues within the conserved catalytic triad that would abolish enzymatic activity but maintain substrate coordination within the active site in order to map the steady-state subcellular distribution of PI. Second, we generated BcPI-PLC constructs with minimal interfacial binding, and therefore low basal catalytic activity from the cytosol, which could still rapidly hydrolyze PI when recruited in the proximity of membrane-embedded substrate. Combining the interfacially compromised BcPI-PLC variants with a chemically inducible protein heterodimerization system, the levels of DAG, which is the direct enzymatic product of PI-PLC–mediated PI hydrolysis, could be monitored as a proxy for the PI content within specific membrane compartments in live cells.
Using these two experimental strategies, we show that PI is present within membranes of the ER, but is also enriched in the cytosolic leaflets of the Golgi complex, peroxisomes, and outer mitochondrial membrane (OMM). Strikingly, we did not find significant amounts of PI within the plasma membrane (PM) or endosomal compartments in any of the mammalian cell types examined. Furthermore, depleting PI acutely within the ER reduced PPIn levels at the Golgi complex and had a larger impact on decreasing PPIn levels in the PM and endosomes than did hydrolysis of PI directly within these PPIn-containing membranes. These studies suggest that the sustained delivery of PI from the ER, rather than the absolute steady-state content of PI, is critical for the maintenance of monophosphorylated PPIn species within the PM, Golgi complex, and endosomal compartments. Our findings also support an important role for PI transfer, and possibly substrate channeling, for the spatial control of cellular PPIn metabolism.
Results
Visualizing the membrane distribution of PI using the BcPI-PLC scaffold
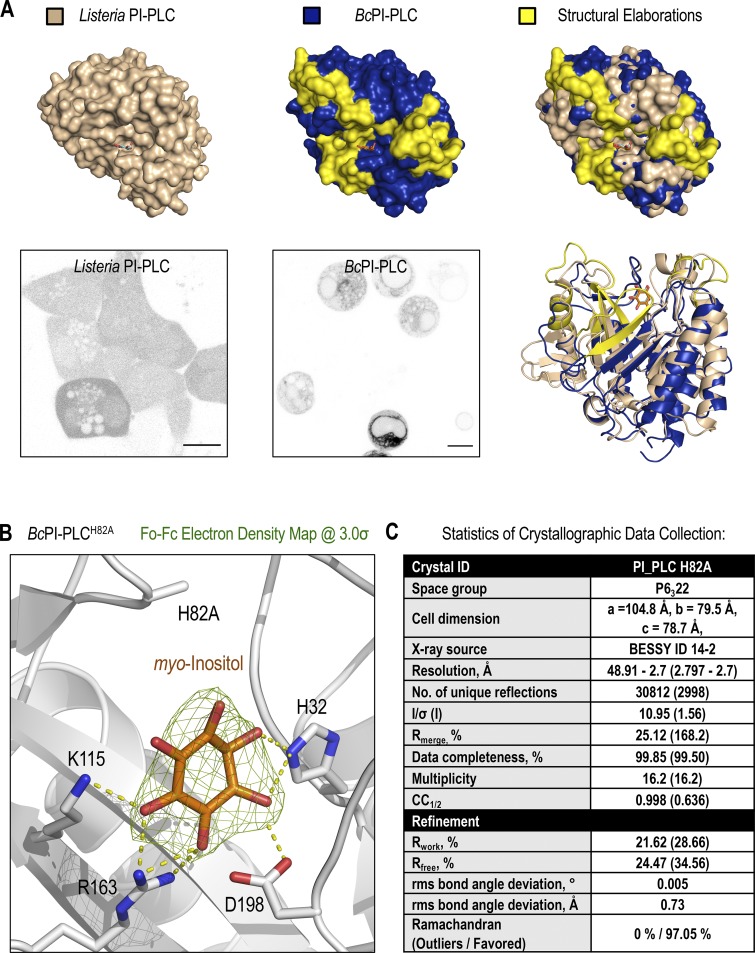
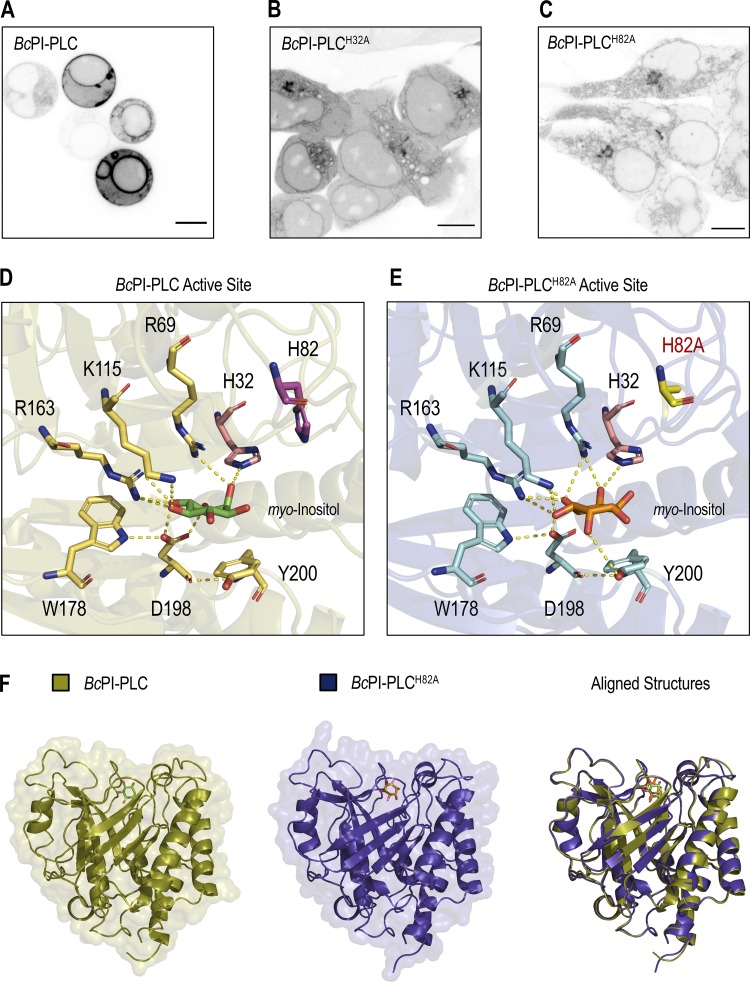
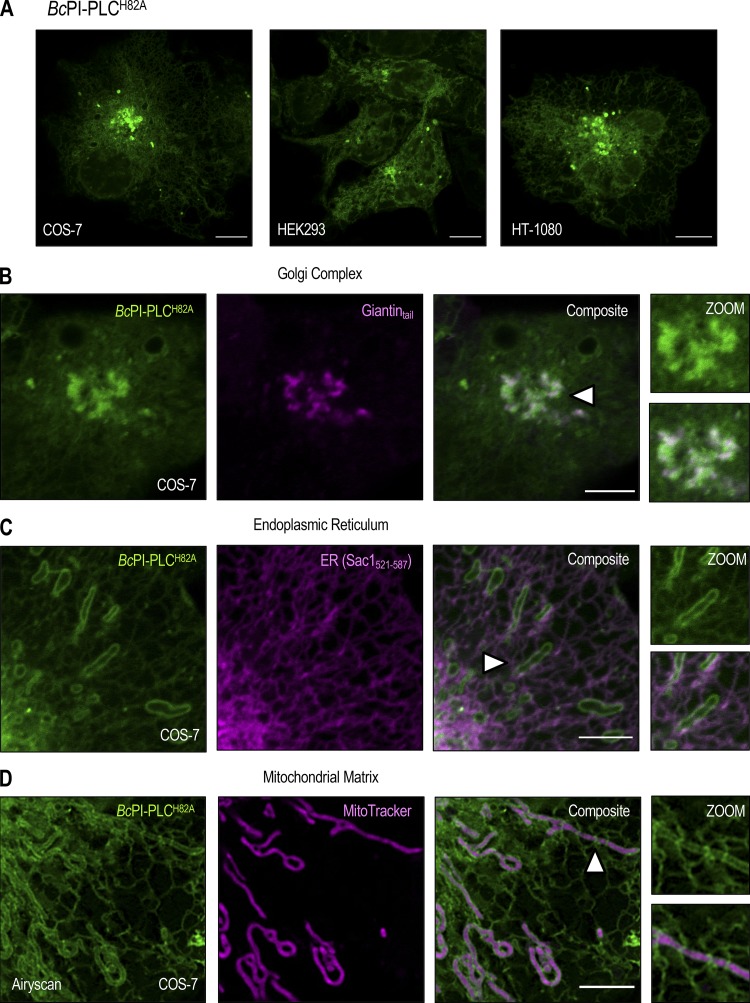
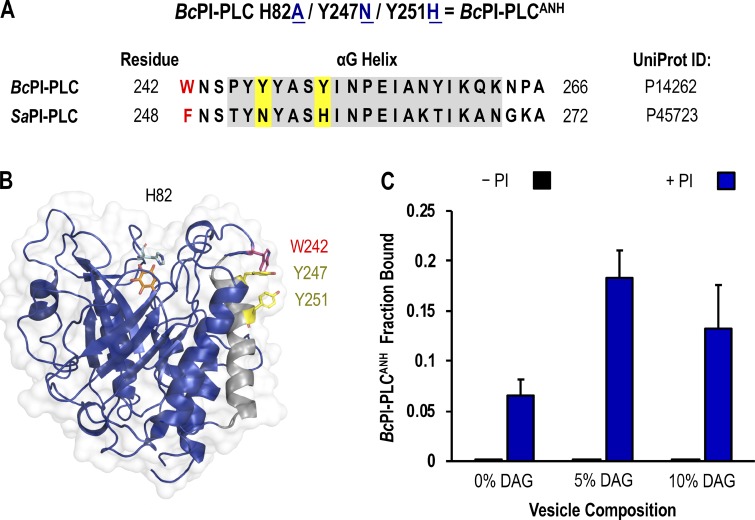
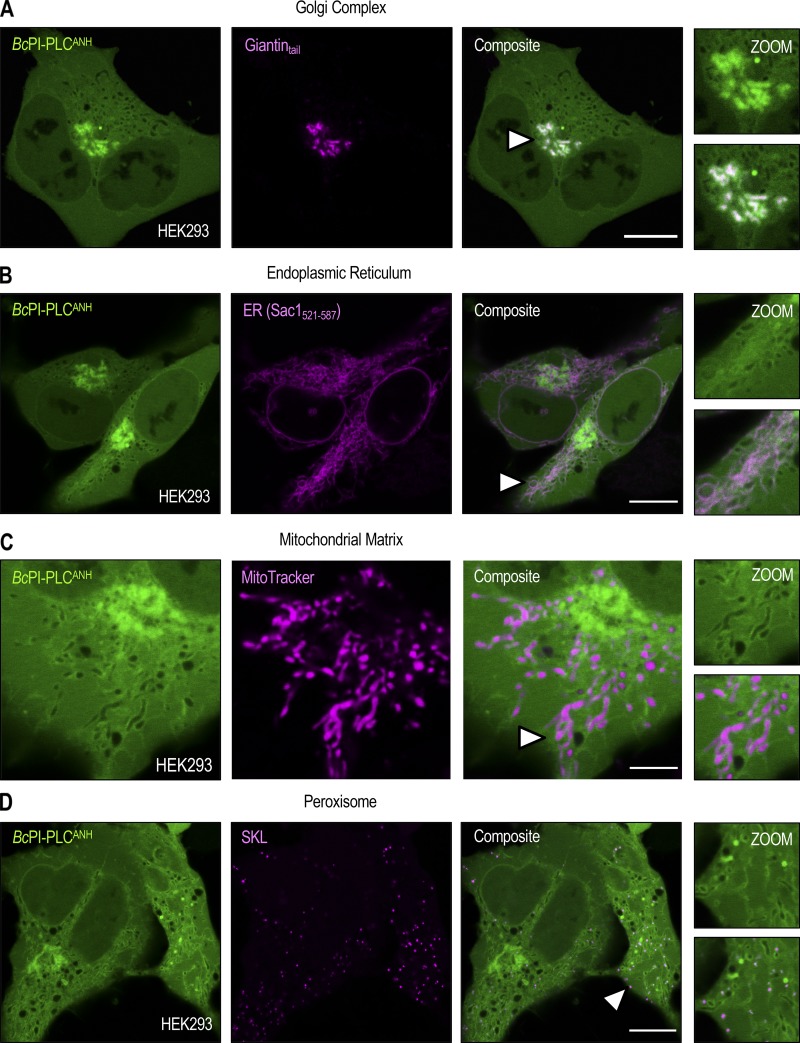
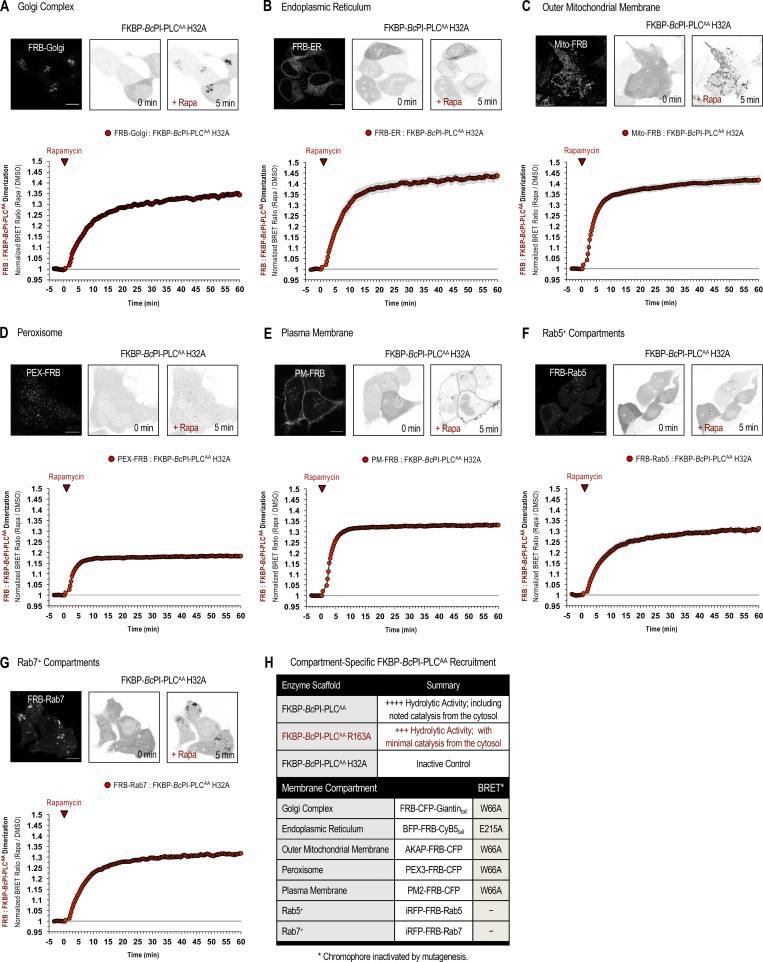
Unlike the low-abundance PPIn lipids, the intracellular distribution of PI has never been observed in live cells. Here, we devised a strategy to visualize the steady-state distribution of PI that exploits the substrate selectivity of bacterial PI-PLCs. Previous efforts using the PI-PLC from Listeria monocytogenes failed to show any membrane localization of the enzyme, presumably because of insufficient binding affinity (Fig. S1 A; Kim et al., 2011). Therefore, we turned to another PI-PLC enzyme from B. cereus that shows enhanced catalytic activity in vitro (Gandhi et al., 1993; Bruzik and Tsai, 1994) and, based on their structures (Heinz et al., 1995, 1996; Moser et al., 1997), possesses a more defined pocket for accommodating the inositol headgroup as well as protruding hydrophobic residues present at the membrane-binding interface (Fig. S1 A, highlighted in yellow). For expression in eukaryotic cells, the sequence of the BcPI-PLC (residues 32–329, with the N-terminal signal sequence removed) was codon optimized and prepared by custom synthesis for subcloning into mammalian expression vectors as a fusion to the C-terminus of EGFP. Expression of the active GFP–BcPI-PLC in mammalian cells was extremely cytotoxic, with only small necrotic cells remaining 16–20 h after transfection (Fig. 1 A). Previous studies of the BcPI-PLC enzymology identified H32 as the general base that is responsible for abstracting the hydrogen from the C2 hydroxyl on the inositol ring (Fig. 1 D; Gässler et al., 1997). Mutagenesis of H32 to alanine (H32A) eliminated the cytotoxicity associated with the fully active BcPI-PLC, which is consistent with earlier studies showing that this mutation effectively renders the enzyme catalytically inactive (Fig. S2 A; Gässler et al., 1997; Hondal et al., 1998). Importantly, GFP–BcPI-PLCH32A still showed a weak association with intracellular membranes that could reflect the sites of PI availability (Fig. 1 B). To enhance the relative affinity, we targeted a second histidine within the catalytic triad, H82, which, unlike H32, does not directly form hydrogen bonds with the inositol headgroup. Alanine substitution of this residue still completely abolishes the catalytic activity (Fig. S2 A; Gässler et al., 1997; Hondal et al., 1998), and x-ray crystallography confirmed that the BcPI-PLCH82A mutant showed no alterations to the overall architecture of the active site and retained the ability to coordinate the myo-inositol headgroup (Fig. 1, E and F; and Fig. S1 B). Expression of GFP–BcPI-PLCH82A showed a clear increase in membrane binding (Fig. 1 C), with specific associations of the probe to some, but not all, internal membrane compartments in multiple mammalian cell lines (Fig. 2 A). In addition to the reticular ER, the strongest signal was observed at the Golgi complex along with a clear enrichment within the OMM (Fig. 2, A–D). Due to the close apposition from the ER, the mitochondrial localization was confirmed using the high-resolution Airyscan detector (Huff, 2015; Scipioni et al., 2018) and MitoTracker fluorescent dye (Fig. 2 D; Chazotte, 2011). These high-contrast images also highlight an apparent concentration of the BcPI-PLCH82A probe at the ER–mitochondria interface, including at contacts where the ER appears to wrap around the mitochondrial membrane (Fig. 2 D, arrowhead). Notable, however, was the apparent absence of GFP–BcPI-PLCH82A from the PM (Figs. 2 A and 6 A) or endosomal structures (Figs. 2 A, 9 A, and 10 A). Still, we could observe GFP–BcPI-PLCH82A localization to small rounded structures, often associated with the mitochondria or ER, that colocalized with fluorescently tagged protein containing a peroxisomal targeting signal (mRFP-SKL; Fig. 5 A; Kim et al., 2006). Overall, the membrane distribution of GFP–BcPI-PLCH82A was consistent across representative mammalian cell lines, including the COS-7, HEK293, and HT-1080 lineages (Fig. 2 A).
Figure S1.
Structural features of the BcPI-PLC scaffold inform the rationale design of the BcPI-PLCH82A substrate trap. (A) Structural comparison of the L. monocytogenes (top row, left, beige; PDB accession no. 1AOD) and B. cereus (top row, center, blue and yellow; PDB accession no. 1PTG) PI-PLCs bound to myo-inositol. Notable differences on the membrane-oriented surface, including a general expansion of the PI-binding pocket in the BcPI-PLC, are highlighted in yellow and presented on a surface rendering of the binding pocket (top row, right). Ribbon representations of the aligned structures are shown on the bottom right alongside representative images of HEK293-AT1 cells expressing either the L. monocytogenes mRFP–PI-PLC (bottom row, left) or EGFP–BcPI-PLC (bottom row, center; scale bars, 10 µm). (B) Electron density map (Fo-Fc, 3.0σ) surrounding the myo-inositol headgroup within the BcPI-PLCH82A active site shows the intact coordination of the inositol ring. (C) Table listing statistics used for crystallographic data collection and refinement, which are also indexed in the associated PDB entry (accession no. 6S2A).
Figure 1.
Visualizing the subcellular distribution of PI using the BcPI-PLC scaffold. (A–C) Confocal images comparing the subcellular localization of the wild-type BcPI-PLC (A) with the H32A (B) and H82A (C) mutants in HEK293-AT1 cells (scale bars, 10 µm). (D and E) Enlarged views of the active sites from the myo-inositol–bound structures of BcPI-PLC (PDB accession no. 1PTG) and BcPI-PLCH82A (E; PDB accession no. 6S2A). The amino acid side chains coordinating the inositol headgroup are shown as stick representations, and the contacts made between these residues and the myo-inositol headgroup are shown as dashed yellow lines. (F) Structural comparison and threaded alignment (right) of the BcPI-PLC (gold, left) and BcPI-PLCH82A mutant (blue, center) bound to myo-inositol.
Figure S2.
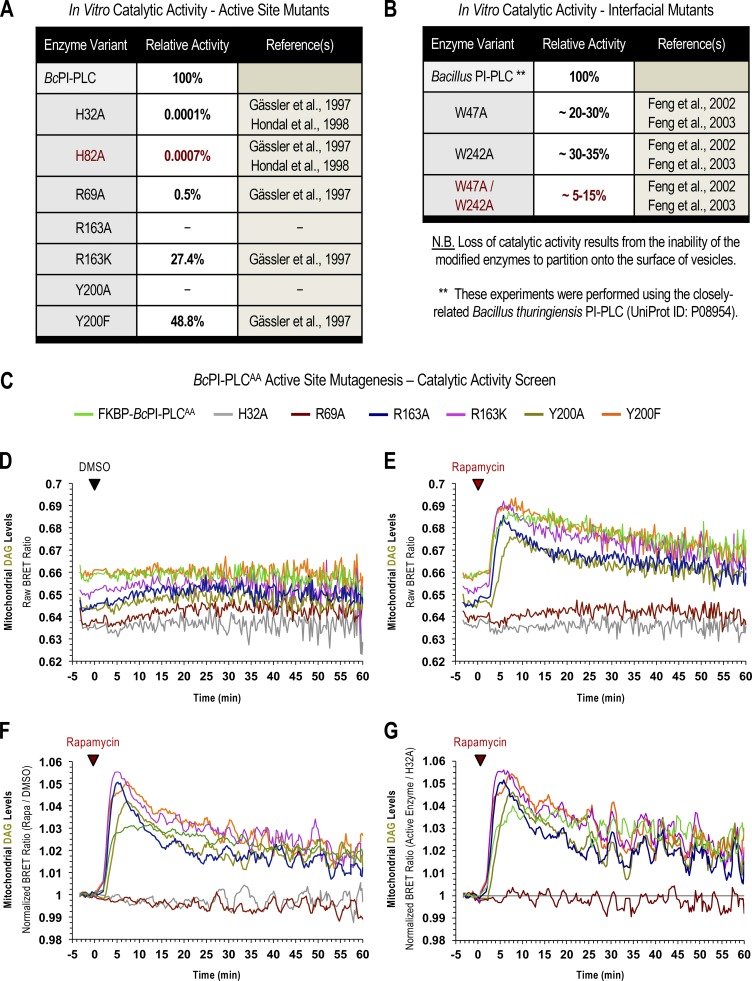
Tuning the catalytic activity of the FKBP–BcPI-PLCAA scaffold. (A and B) Published values of the relative in vitro catalytic activities of Bacillus PI-PLC mutants with residues altered within either the enzyme active site (A; adapted with permission from Gässler et al., 1997 and Hondal et al., 1998) or at the membrane-binding interface (B; adapted with permission from Feng et al., 2002, 2003). Please note that the values presented in B for the interfacial mutants were measured using the closely related Bacillus thuringiensis PI-PLC (UniProt ID: P08954). (C) List of the FKBP–BcPI-PLCs variants tested as part of the catalytic activity screen carried out using the mito-DAGBRET reporter. Briefly, mutagenesis of two interfacial tryptophan residues (W47A/W242A) render the resulting FKBP–BcPI-PLCAA enzyme entirely cytosolic, thereby significantly reducing the ability of the enzyme to hydrolyze membrane-embedded PI. To further tune the catalytic activity of the parent FKBP–BcPI-PLCAA scaffold, we screened active site mutants containing alanine or conservative substitutions of R69 (R69A, red traces), R163 (R163A, blue traces; R163K, magenta traces), or Y200 (Y200A, gold traces; Y200F, orange traces). These mutants were tested for their ability to generate DAG in the OMM using the compartment-selective mito-DAGBRET biosensor. As internal controls, we used the most active parent FKBP–BcPI-PLCAA scaffold (green traces) and the catalytically inactive H32A variant (gray traces). (D) The raw BRET ratios are shown for triplicate measurements treated with DMSO. Note the differing basal BRET ratios, which reflect the mitochondrial DAG content, as a readout of the background activity of the enzymes from the cytosol. (E) Rapamycin-induced recruitment of the BcPI-PLC variants to the mitochondria results in the rapid production of DAG, which reflects the relative activity of the associated enzyme active site. (F) Normalized BRET ratios are shown after dividing the rapamycin-treated wells by their time-matched DMSO controls. (G) Alternatively, the vehicle-normalized traces from each of the active enzymes can also be presented relative to the kinetics of the inactive FKBP–BcPI-PLCAA H32A control. After comparing the baseline values, as well as the rapid initial rises in DAG production after rapamycin treatment, we selected the FKBP–BcPI-PLCAA R163A, R163K, and Y200F mutants as the best of the available variants. These constructs possess a combination of low background activity before recruitment as well as robust catalysis after recruitment onto the membrane surface and were chosen for further exmaination in single-cell imaging studies.
Figure 2.
Steady-state localization of the BcPI-PLCH82A probe. (A) Comparison of the subcellular localization of the EGFP–BcPI-PLCH82A probe in COS-7 (left), HEK293-AT1 (center), and HT-1080 (right) cells (scale bars, 10 µm). (B–D) Confocal images of COS-7 cells coexpressing EGFP–BcPI-PLCH82A with the indicated organelle-specific markers or stably loaded with MitoTracker Red (D; Airyscan detector), which partitions into the mitochondrial matrix (scale bars, 5 µm). Enlarged views of the regions identified by the arrowheads are provided on the far right of each image series (inset, 7.5 µm).
Figure 6.
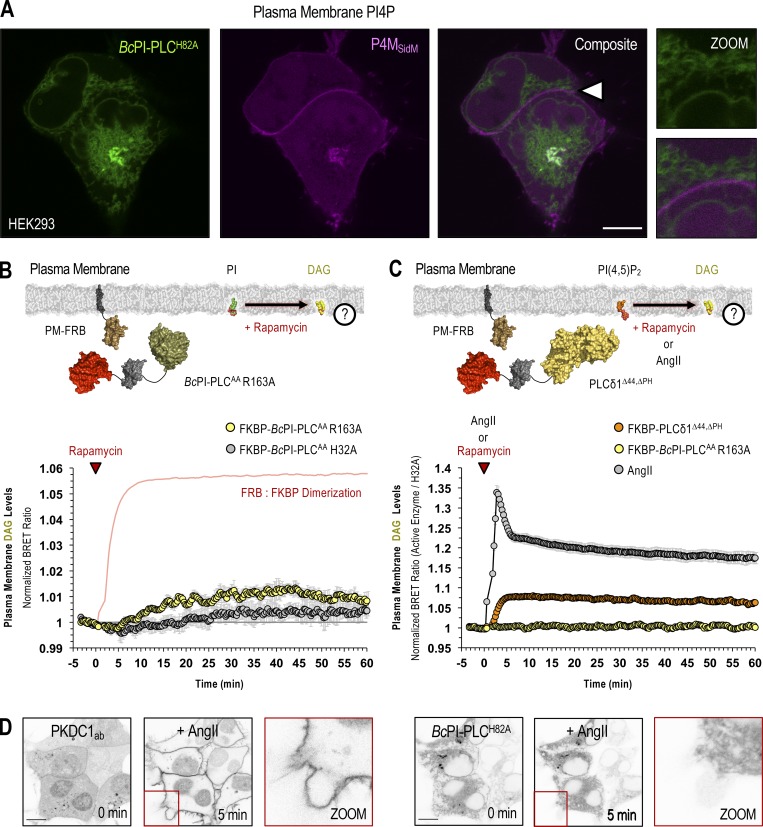
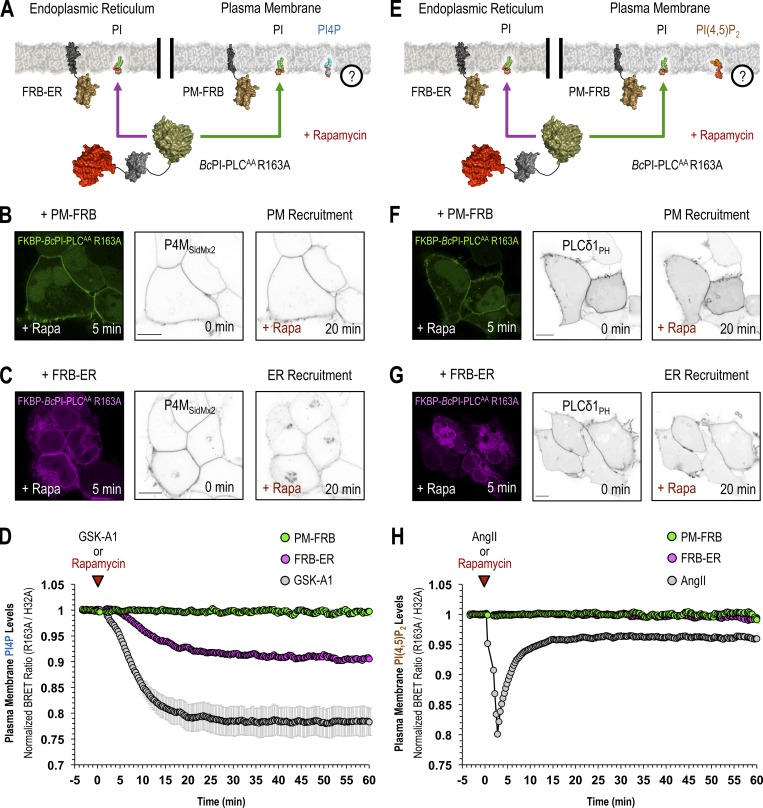
Steady-state levels of PI are low in the PM. (A) Confocal images of HEK293-AT1 cells coexpressing EGFP–BcPI-PLCH82A with the PI4P-binding probe, mCherry-P4MSidM (scale bar, 10 µm). An enlarged view of the region identified by the arrowhead is shown on the far right (inset, 10 µm). (B and C) For each BRET measurement, a schematic of the experimental design is provided above each quantified trace, with the question mark indicating the membrane lipid being measured. (B) Kinetics of DAG production within the cytosolic leaflet of the PM, measured by the PM-DAGBRET biosensor, after recruitment of FKBP–BcPI-PLCAA R163A to the PM. Please note that a time-matched but alternatively scaled trace shows the PM-FRB:FKBP–BcPI-PLCAA H32A dimerization kinetics (red line; see also Fig. S6 E). (C) Kinetics of DAG production within the PM, measured using the PM-DAGBRET biosensor, in response to stimulation with AngII (100 nM; gray trace) or following PM recruitment of an FKBP-tagged mammalian PLC (FKBP-PLCδ1Δ44,ΔPH; orange trace). For comparison, the normalized DAG response measured after the recruitment of FKBP–BcPI-PLCAA R163A, which is presented in B, is also included (yellow trace). BRET measurements are presented as mean values ± SEM from three independent experiments performed using triplicate wells. (D) Representative images from cells coexpressing the DAG-binding probe (mRFP-PKDC1ab; left panels) and EGFP–BcPI-PLCH82A (right panels; scale bars, 10 µm). Note the massive translocation of the DAG probe, but not the EGFP–BcPI-PLCH82A, from the cytosol to the PM after stimulation with AngII (100 nM). The enlarged image of the area marked by the red inset in the center panels is presented on the right of each image series (15-µm inset).
Figure 9.
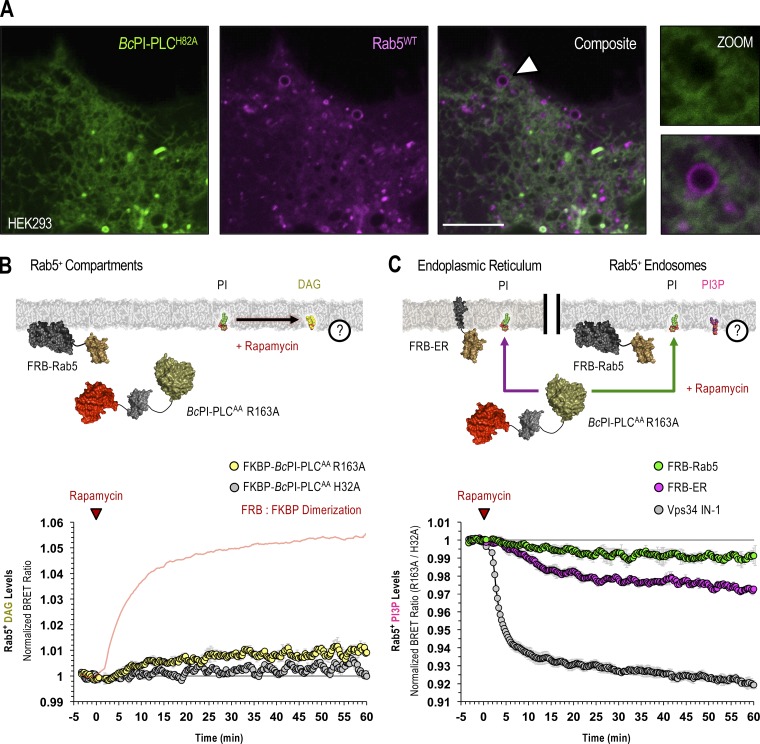
Maintenance of PI3P levels in Rab5-positive endosomes requires the delivery of PI from the ER. (A) Confocal images of HEK293-AT1 coexpressing the EGFP–BcPI-PLCH82A probe together with mCherry-Rab5WT (scale bars, 5 µm). An enlarged view of the region identified by the arrowhead is shown on the far right (inset, 2.5 µm). (B and C) For each BRET measurement, a schematic of the experimental design is provided above each quantified trace, with the question mark indicating the membrane lipid being measured. (B) Kinetics of DAG production in Rab5-positive compartments after recruitment of FKBP–BcPI-PLCAA R163A to Rab5-labeled membranes, as measured by the Rab5-DAGBRET biosensor. Please note that a time-matched but alternatively scaled trace shows the FRB-Rab5:FKBP–BcPI-PLCAA H32A dimerization kinetics (red line; see also Fig. S6 F). (C) Kinetics of PI3P levels within Rab5-positive compartments, measured using the biosensor (C; Rab5-PI3PBRET), in response to the recruitment of FKBP–BcPI-PLCAA R163A either directly to the surface of Rab5-labeled endosomes (green traces; FRB-Rab5) or to the ER (magenta traces; FRB-ER). Treatment with a selective class III PI 3-kinase inhibitor (VPS34-IN1, 300 nM; gray traces) is included as a positive control for the Rab5-specific PI3PBRET biosensor and to provide scale for any changes associated with the differential recruitment of FKBP–BcPI-PLCAA R163A to Rab5-positive endosomes or the ER. BRET measurements are presented as mean values ± SEM from three independent experiments performed using triplicate wells.
Figure 10.
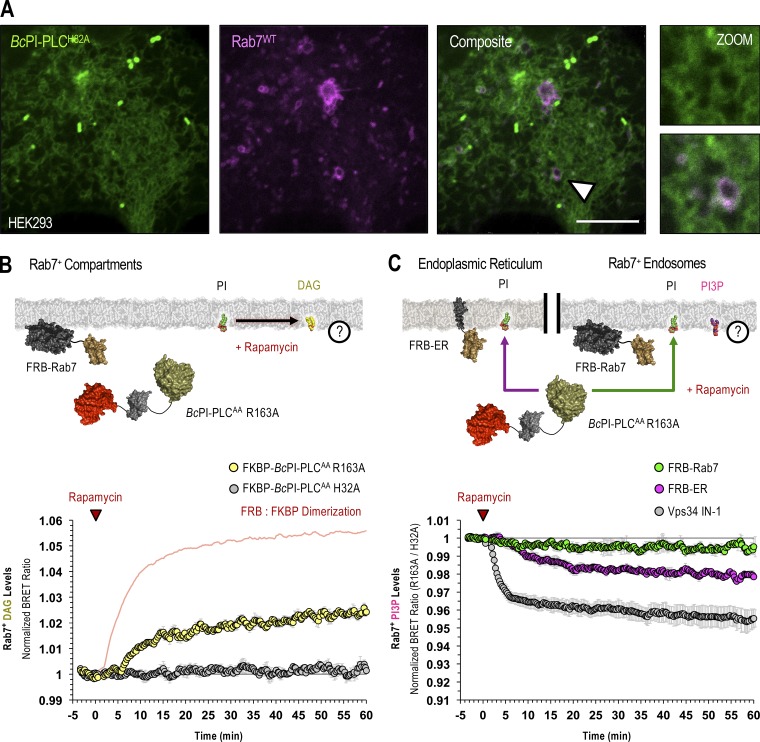
Maintenance of PI3P levels in Rab7-positive endosomes requires the delivery of PI from the ER. (A) Confocal images of HEK293-AT1 coexpressing the EGFP–BcPI-PLCH82A probe together with mCherry-Rab7WT (scale bars, 5 µm). An enlarged view of the region identified by the arrowhead is shown on the far right (inset, 2.5 µm). (B and C) For each BRET measurement, a schematic of the experimental design is provided above each quantified trace, with the question mark indicating the membrane lipid being measured. (B) Kinetics of DAG production in Rab7-positive compartments after recruitment of FKBP–BcPI-PLCAA R163A to Rab7-labeled membranes, as measured by the Rab7-DAGBRET biosensor. Please note that a time-matched but alternatively scaled trace shows the FRB-Rab7:FKBP–BcPI-PLCAA H32A dimerization kinetics (red line; see also Fig. S6 G). (C) Kinetics of PI3P levels within Rab7-positive compartments, measured using the Rab7-PI3PBRET biosensor, in response to the recruitment of FKBP–BcPI-PLCAA R163A either directly to the surface of Rab7-labeled endosomes (green traces; FRB-Rab7) or to the ER (magenta traces; FRB-ER). Treatment with a selective class III PI 3-kinase inhibitor (VPS34-IN1, 300 nM; gray traces) is included as a positive control for the Rab7-specific PI3PBRET biosensor and to provide scale for any changes associated with the differential recruitment of FKBP–BcPI-PLCAA R163A to Rab7-positive endosomes or the ER. BRET measurements are presented as mean values ± SEM from three independent experiments performed using triplicate wells.
Figure 5.
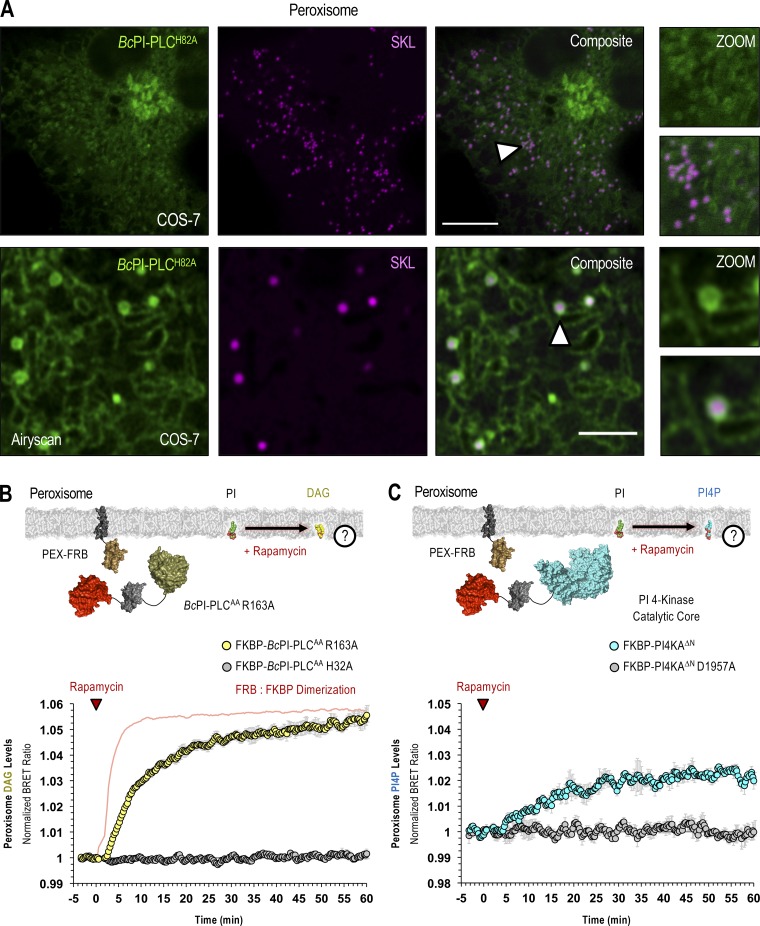
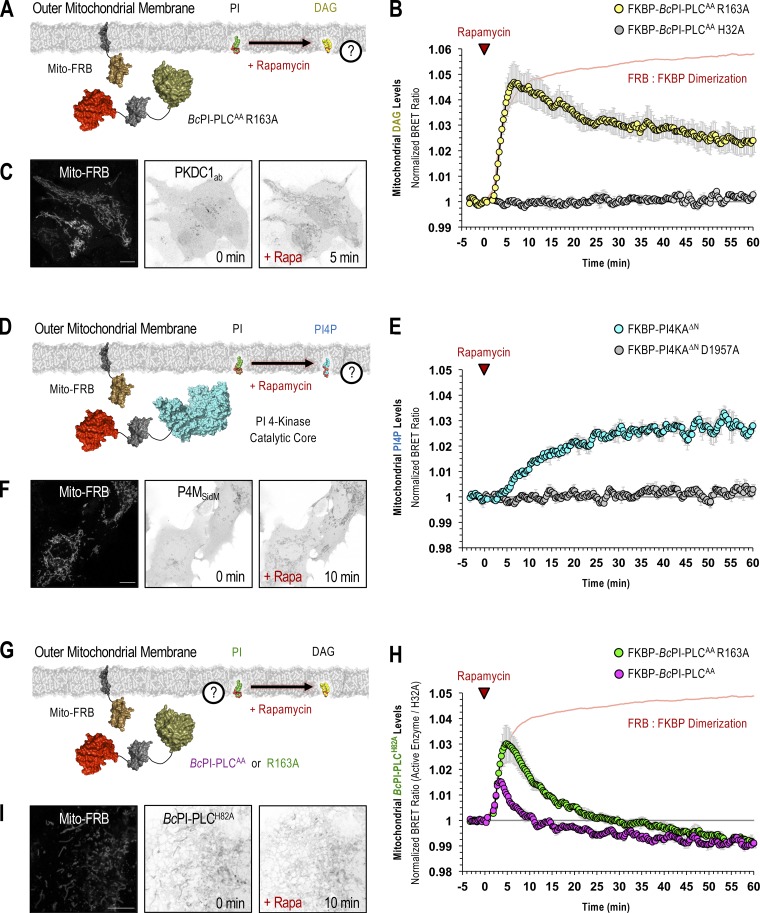
PI is enriched in the cytosolic leaflet of peroxisomes. (A) Representative images of COS-7 cells coexpressing EGFP–BcPI-PLCH82A and a luminally targeted peroxisomal marker (mRFP-SKL) obtained using either conventional confocal microscopy (top row; scale bar, 10 µm) or with the Airyscan detector (bottom row; scale bar, 2.5 µm). Enlarged views of the regions identified by the arrowheads are provided on the far right of each image series (inset, 10 and 2.5 µm, respectively). (B and C) For each BRET measurement, a schematic of the experimental design is provided above each quantified trace, with the question mark indicating the membrane lipid being measured. (B) Kinetics of DAG production in the cytosolic leaflet of peroxisomes after recruitment of FKBP–BcPI-PLCAA R163A, as measured by the PEX-DAGBRET biosensor. A time-matched but alternatively scaled trace shows the PEX-FRB:FKBP–BcPI-PLCAA H32A dimerization kinetics (red line; see also Fig. S6 D). (C) Kinetics of PI4P production within the cytosolic leaflet of peroxisomes following recruitment of FKBP-PI4KAΔN, as measured by the PEX-PI4PBRET biosensor. BRET measurements are presented as mean values ± SEM from three independent experiments performed using triplicate wells.
Next, we evaluated the in vitro binding of recombinant GFP–BcPI-PLC constructs to large unilamellar vesicles (LUVs) using fluorescence cross-correlation spectroscopy (FCCS; Sezgin and Schwille, 2011). These experiments revealed strong background binding of GFP–BcPI-PLCH82A to LUVs, which was not dependent on or enhanced by the presence of PI (data not shown). Attempts to decrease the phosphatidylcholine (PC) content of the prepared LUVs yielded unstable liposomes, which were severely aggregated, and prevented further analysis of the contribution of PC to the binding of GFP–BcPI-PLCH82A. Previous studies identified a cluster of tyrosine residues within the αG helix of the BcPI-PLC, including Y246, Y247, Y248, and Y251, that have been implicated in the binding of this enzyme to PC-rich vesicles through the formation of a cation-π box (Shi et al., 2009; Pu et al., 2010). Interestingly, unlike the BcPI-PLC, Staphylococcus aureus PI-PLC (SaPI-PLC) shows no binding to PC-rich vesicles (Cheng et al., 2012). A sequence alignment of these two bacterial PI-PLCs shows that while the tyrosine residues equivalent to Y246 and Y248 in the BcPI-PLC are conserved in the αG helix from SaPI-PLC, BcPI-PLC residues Y247 and Y251 have been replaced in the SaPI-PLC by an asparagine (N254) and histidine (H258), respectively (Fig. S3, A and B). Since the SaPI-PLC was shown to gain PC sensitivity by changing the residues in the N254 and H258 positions to tyrosines (SaPI-PLC N254Y/H258Y; Cheng et al., 2013), we reasoned that the PI-dependent component of the BcPI-PLCH82A membrane binding could be revealed by swapping Y247 and Y251 to the corresponding residues from the SaPI-PLC to diminish the PC sensitivity of the BcPI-PLC scaffold. For simplicity, the resulting BcPI-PLC H82A/Y247N/Y251H mutant will be referred to as BcPI-PLCANH. Indeed, GFP–BcPI-PLCANH did not show nonspecific associations with LUVs, and its bound fraction was increased by the introduction of 10% PI (Fig. S3 C). Importantly, DAG also enhanced binding of the GFP–BcPI-PLCANH to LUVs, but only if the prepared liposomes also contained PI (10%; Fig. S3 C). In light of these in vitro data, the cellular distribution of GFP–BcPI-PLCANH was also examined in HEK293-AT1 cells and showed a very similar distribution to that of the parent GFP–BcPI-PLCH82A construct, except with a larger fraction of the probe remaining within the cytosol (Fig. S4, A–D).
Figure S3.
In vitro binding studies using the BcPI-PLCANH mutant. (A) Sequence alignment of the αG helix region of BcPI-PLC (UniProt ID: P14262) and SaPI-PLC (UniProt ID: P45723) shows that two aromatic residues in the BcPI-PLC, Y247 and Y251, which are involved in forming the PC-coordinating cation-π box are replaced by asparagine (N254) and histidine (H258) residues, respectively, in the SaPI-PLC. As described in the Results section, we reasoned that changing Y247 and Y251 to the corresponding residues from SaPI-PLC could diminish the PC sensitivity of the BcPI-PLC scaffold to reveal the PI-dependent binding of the BcPI-PLCH82A mutant. For simplicity, the resulting BcPI-PLC H82A/Y247N/Y251H mutant is referred to as BcPI-PLCANH. (B) Ribbon representation of the BcPI-PLC (blue; PDB accession no. 1PTG) bound to myo-inositol is shown with the αG helix colored in gray and the side chains of residues Y247 and Y251 highlighted in yellow. Please note that these residues are situated on the outside of the molecule, away from the active site, and are positioned immediately below the W242 residue (red) that is also essential for interfacial binding. (C) The in vitro binding of recombinant GFP–BcPI-PLCANH to LUVs with varying DAG compositions (0%, 5%, and 10%) was measured using FCCS both in the absence (black bars) and presence (blue bars) of 10% liver PI. Binding measurements are presented as mean values ± SEM from three independent experiments.
Figure S4.
Steady-state localization of the BcPI-PLCANH probe. (A–D) Representative images of HEK293-AT1 cells coexpressing EGFP–BcPI-PLCANH with an integral Golgi-localized protein fragment (A; FRB-mCherry-Giantintail; scale bar, 10 µm), the C-terminal localization signal from the ER-resident protein Sac1 (B; mRFP-Sac1521-587; scale bar, 10 µm), MitoTracker Red (C; scale bar, 5 µm), or a fluorescently tagged consensus peroxisomal-targeting sequence (D; mRFP-SKL; scale bar, 10 µm). Enlarged views of the regions identified by the arrowheads are provided on the far right of each image series. Note that the size of the inset roughly matches the scale bar that is included for the corresponding organelle marker.
Acute manipulation of PI in defined membrane compartments
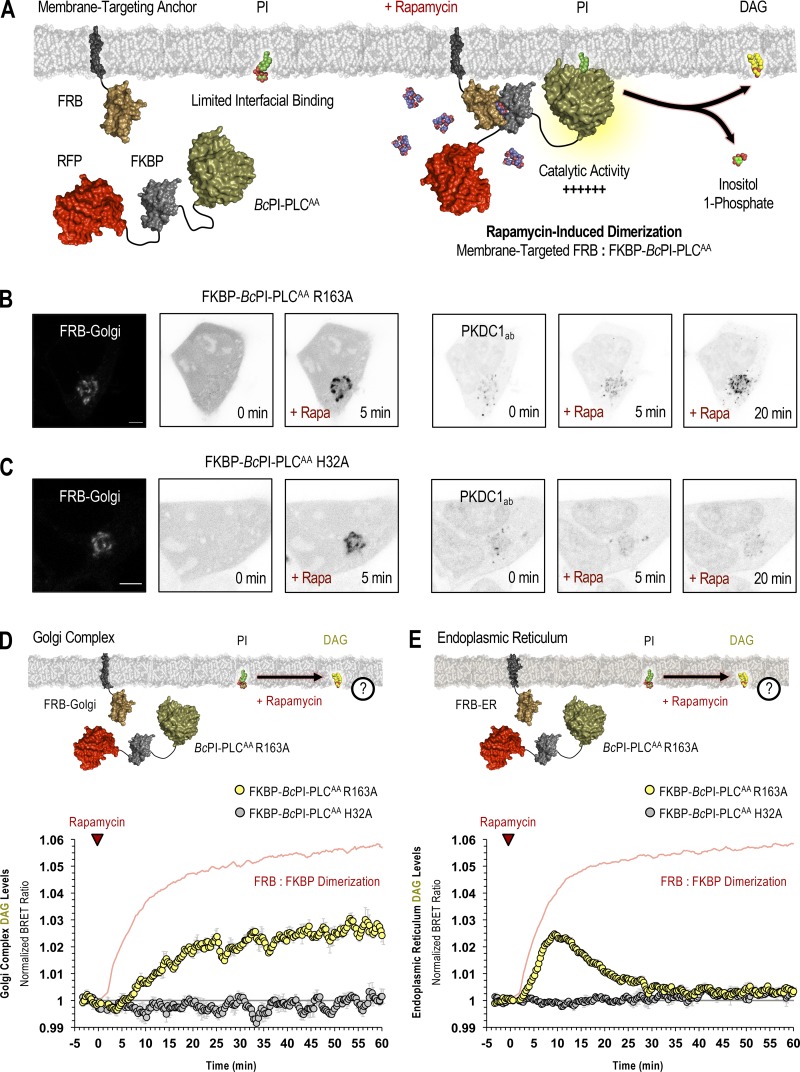
To validate these PI localization studies, we sought an alternative approach that involved the generation of an interfacially modified, but catalytically active, BcPI-PLC that could be acutely recruited from the cytosol onto specific membranes to hydrolyze the resident PI. Detection of the hydrolytic product, DAG, was used as a proxy for the relative abundance of PI within a particular membrane compartment, while enzyme recruitment involved a chemically inducible protein heterodimerization system that relies on the rapamycin-dependent association of FKBP12 with the FRB domain of mTOR (Choi et al., 1996; Liang et al., 1999). This system has been used in diverse cellular contexts to dynamically recruit FKBP-tagged proteins to specific organelles labeled with FRB-containing targeting proteins (Belshaw et al., 1996; Komatsu et al., 2010).
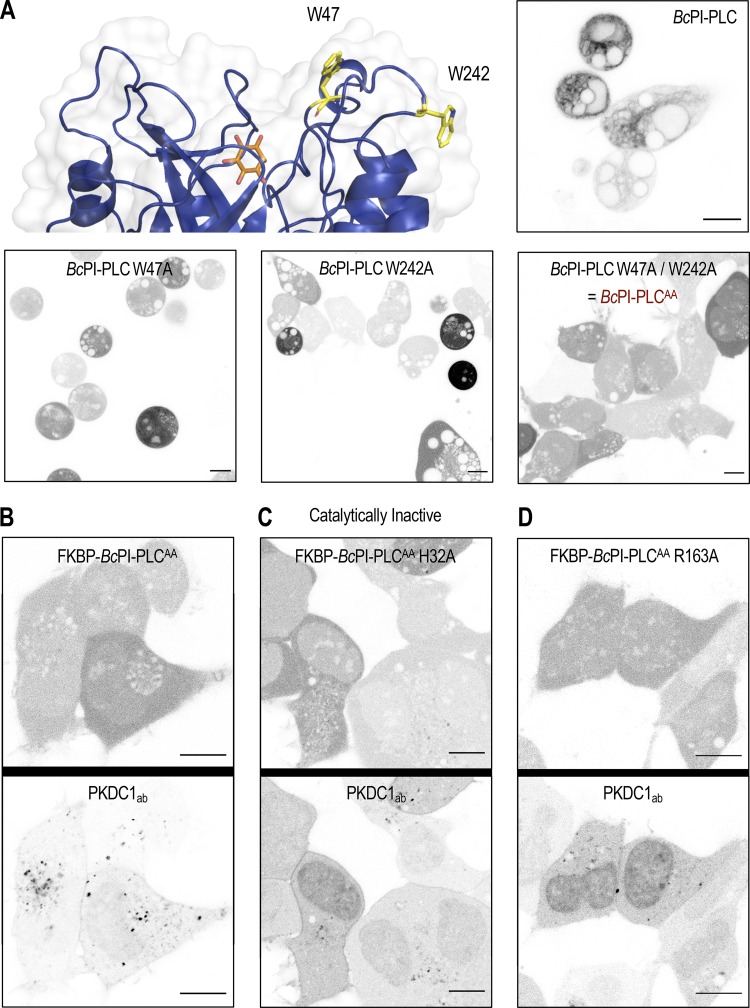
In vitro binding studies have previously identified two membrane-oriented tryptophan residues, namely W47 within helix B and W242 in the extended α7-βG loop (Fig. S5 A), as being critical for the membrane association and catalysis of BcPI-PLCs (Fig. S2 B; Feng et al., 2002, 2003; Guo et al., 2008). The catalytic activity of many phospholipase families displays marked interfacial activation, and the BcPI-PLCs specifically exhibit a substantial preference for substrates presented within an interface rather than as monomers in solution (Lewis et al., 1993; Zhou et al., 1997a, 1997b; Qian et al., 1998). Expression of GFP–BcPI-PLC with an alanine substitution of either W47 (W47A) or W242 (W242A) already failed to strongly interact with subcellular membranes, but these mutants still showed marked cytotoxicity (Fig. S5 A). However, mutagenesis of both W47 and W242 (W47A/W242A) prevented the membrane binding of BcPI-PLC and also greatly reduced the apparent cytotoxicity (Fig. S5 A). We could then tag the BcPI-PLC W47A/W242A (AA) mutant with the FKBP dimerization module to create an enzyme construct (FKBP–BcPI-PLCAA) that could be rapidly recruited to distinct organelle membranes with high spatial and temporal resolution (Fig. 3 A). The relative catalytic activity of the cytosolic FKBP–BcPI-PLCAA was assessed using an established biosensor to detect DAG (GFP-PKDC1ab; Kim et al., 2011), the direct product of PI hydrolysis. This experiment showed a reduced cytosolic fraction of the probe and the presence of small DAG-positive internal puncta in cells expressing FKBP–BcPI-PLCAA (Fig. S5 B). These alterations to the localization of the DAG sensor were not observed in cells expressing the catalytically inactive FKBP–BcPI-PLCAA H32A variant (Fig. S5 C). To further reduce the enzymatic activity and effectively enhance the dynamic range of the induced changes in DAG levels, we introduced mutations to the FKBP–BcPI-PLCAA scaffold at residues with accessory roles for catalysis: generating FKBP–BcPI-PLCAA R69A, R163A, R163K, Y200A, and Y200F variants. To compare the relative activities of these recruitable BcPI-PLC variants in live cells, we wanted to detect the localized production of DAG within a specific membrane compartment at the level of entire cell populations. For this, we worked to establish a semi-high throughput approach for the quantification of changes in subcellular membrane lipid compositions that complements the single-cell analyses that use confocal microscopy.
Figure S5.
Mutagenesis of the BcPI-PLC membrane-binding interface limits cytosolic activity. (A) An enlarged view of the membrane-oriented BcPI-PLC interface (top row, left) with the hydrophobic residues W47 and W242 highlighted (yellow sticks; PDB accession no. 1PTG). Images of HEK293-AT1 cells expressing the wild type or indicated mutants of EGFP–BcPI-PLC (scale bars, 10 μm). (B–D) FKBP-tagging of the cytosolic BcPI-PLCAA was done for acute recruitment to FRB-tagged membranes. Coexpression of the high-affinity DAG-binding probe (mRFP-PKDC1ab), which labels the direct hydrolytic product of BcPI-PLC activity, should reveal any residual activity of the modified enzymes from the cytosol. Representative images of the DAG-binding probe (bottom panels) coexpressed together with the parent mRFP-FKBP–BcPI-PLCAA scaffold or its mutated variants are shown (top panels; scale bars, 10 µm).
Figure 3.
Acute manipulation of PI content in membranes of the Golgi complex and ER. (A) Schematic depicting rapamycin-induced recruitment of the FKBP–BcPI-PLCAA scaffold onto FRB-labeled membrane compartments to locally hydrolyze PI and generate DAG. (B and C) Representative images showing recruitment of the catalytically active mRFP-FKBP–BcPI-PLCAA R163A (left side panels) or inactive mRFP-FKBP–BcPI-PLCAA H32A (C, left side panels) enzymes to the surface of the Golgi complex (scale bars, 10 µm) upon 5-min treatment with rapamycin (100 nM). Images on the right show the corresponding changes in the subcellular distribution of the DAG-binding probe (GFP-PKDC1ab) at 5 and 20 min following rapamycin-dependent enzyme recruitment. (D and E) For each BRET measurement, a schematic of the experimental design is provided above each quantified trace, with the question mark indicating the lipid being measured. Kinetics of DAG production at the Golgi complex (D) or ER (E) after recruitment of the FKBP–BcPI-PLCAA R163A to the corresponding membrane compartment, as measured using the Golgi-DAGBRET and ER DAGBRET biosensors, respectively. A time-matched but alternatively scaled trace shows the compartment-specific FRB:FKBP–BcPI-PLCAA H32A dimerization kinetics (red line; see also Fig. S6, A and B [FRB-Golgi and FRB-ER, respectively]). BRET measurements are presented as mean values ± SEM from three independent experiments performed using triplicate wells.
BRET-based biosensors allow for population-level analyses of membrane lipid dynamics within live cells
To allow for comparative measurements of localized BcPI-PLC activity in populations of live cells, we created a series of bioluminescence resonance energy transfer (BRET)-based biosensors to measure localized DAG levels by monitoring the resonance energy transfer between a high-affinity DAG-binding probe and organelle-specific membrane-targeted fluorophores. This methodology relies on a single-plasmid design that incorporates the self-cleaving viral 2A peptide from Thosea asigna (T2A; Donnelly et al., 2001; Szymczak et al., 2004; Liu et al., 2017), which allows for the uniform and roughly equimolar expression of both the organelle-targeted mVenus BRET acceptor and the luciferase-tagged lipid-binding probe that serves as the BRET donor (Várnai et al., 2017). Due to the prominent localization of the BcPI-PLCH82A probe to the OMM, as well as the low resting levels of DAG, we chose to validate the design of the BRET-based biosensors by monitoring acute DAG production within this compartment. The mitochondrial targeting sequence from A-kinase anchor protein 1 (AKAP; Csordás et al., 2010) was fused to mVenus, while super Renilla luciferase (sLuc) was conjugated to the PKDC1ab probe to produce the mito-DAGBRET biosensor. The resulting mito-DAGBRET reporter was then used in combination with a mitochondrial-targeted FRB construct and the cytosolic FKBP–BcPI-PLCAA enzyme, or its variants, to induce PI hydrolysis on the surface of mitochondria. To prevent any interference with measurements of the energy transfer process, the chromophores used in the membrane-targeting FRB constructs were mutated to eliminate their fluorescence. For the same reasons, the FKBP–BcPI-PLCAA construct was tagged with the BRET-compatible iRFP rather than with mRFP, which could potentially steal energy from the membrane-targeted mVenus. By changing either the membrane targeting motif or the lipid-binding reporter, we created a series of biosensors to monitor localized changes in the dynamics of various lipid species within specific membrane compartments of live cells.
Initial results using catalytic variants of the FKBP–BcPI-PLCAA recruitment system together with the mito-DAGBRET biosensor demonstrated the rapid production of DAG within the OMM following acute treatment with rapamycin (100 nM; Fig. S2, C–G). Relative to prerecruitment levels, or to DMSO-treated controls, the magnitude of the normalized DAG production was largest for the R163A, Y200F, R163K mutants. The baseline DAG levels measured using BRET were lowest for the FKBP–BcPI-PLCAA R163A variant, and this mutant also showed a relatively normal distribution of the DAG sensor compared with the changes observed in cells expressing the more active FKBP–BcPI-PLCAA (Fig. S5 D). Based on these results, we chose the FKBP–BcPI-PLCAA R163A for use in subsequent studies. In addition, for these and all remaining studies, recruitment of an inactive enzyme variant, FKBP–BcPI-PLCAA H32A, was used as a transfection control and for normalization of the BRET data (Fig. S2, F and G). Dimerization kinetics between the FRB-tagged membrane anchors and FKBP–BcPI-PLCAA H32A scaffold were measured separately using distinct BRET constructs that were designed for each organelle (Fig. S6, A–G). For simplicity, a complete list of the constructs used for organelle-specific recruitment of the FKBP–BcPI-PLCAA scaffold is included in Fig. S6 H, while the architecture and construction of these plasmids has been cataloged in the Materials and methods section and Table S1. Overall, in establishing this experimental strategy, the acute production of DAG by the recruited FKBP–BcPI-PLCAA, or the R163A variant, served as an indirect proxy of PI availability across the membrane compartments of intact cells.
Figure S6.
Kinetics of compartment-specific recruitment of the FKBP–BcPI-PLCAA scaffold. (A–G) Representative images of HEK293-AT1 cells are shown along with population-level BRET measurements defining the FRB:FKBP dimerization kinetics for the FKBP–BcPI-PLCAA H32A scaffold upon rapamycin-induced recruitment to the Golgi complex (A), ER (B), OMM (C), peroxisomes (D), PM (E), Rab5-positive compartments (F), or Rab7-positive compartments (G). For each image series, the localization of the FRB-tagged recruiter is shown (left panels) followed by images of the mRFP-FKBP–BcPI-PLCAA H32A protein before (center panels) and 5 min after (right panels) treatment with rapamycin (100 nM; scale bars, 10 µm). For each of these BRET measurements, the single-plasmid design of the biosensors was adapted such that sLuc was tagged with FKBP–BcPI-PLCAA H32A and the mVenus fluorescent protein was conjugated to FRB and the respective membrane-specific targeting sequence. For further details related to the design of these constructs, please refer to the Materials and methods section. BRET measurements are presented as mean values ± SEM from three independent experiments performed using triplicate wells. (H) Summary table listing the most relevant FKBP–BcPI-PLCAA variants and FRB-tagged organelle recruitment constructs for use in either imaging studies or BRET experiments.
PI is detected in the Golgi complex, ER, mitochondria, and peroxisomes
With the establishment of the recruitable FKBP–BcPI-PLCAA system, we applied this approach to various organelle membranes in order to validate the PI distributions mapped using the BcPI-PLCH82A and BcPI-PLCANH probes. Due to the prominent localization of the BcPI-PLC-based probes, we first tested the localization of the DAG sensor following recruitment of FKBP–BcPI-PLCAA R163A to the Golgi complex using the FRB-Golgi recruiter. Representative confocal images show efficient recruitment of the enzyme and a parallel increase in the DAG levels at the Golgi complex (Fig. 3 B). Recruitment of the inactive FKBP–BcPI-PLCAA H32A did not stimulate DAG production at the Golgi complex (Fig. 3 C). Enzyme recruitment experiments using the Golgi-DAGBRET sensor showed a relatively slow increase in the DAG levels at the Golgi complex (Fig. 3 D) when compared with the kinetics of the enzyme recruitment (red line; Fig. S6 A). This prolonged DAG response could reflect the high steady-state levels of DAG at the Golgi complex relative to the amount of DAG produced by the recruited FKBP–BcPI-PLCAA enzyme (Litvak et al., 2005). In contrast, recruitment of FKBP–BcPI-PLCAA R163A to the ER resulted in the rapid production of DAG within the cytosolic leaflet of the ER (ER-DAGBRET), which rapidly returned to basal levels within 30 min (Fig. 3 E). The transient nature of the DAG production observed in the ER likely reflects the rapid conversion of the DAG to other metabolites or possibly the flipping of this lipid into the luminal leaflet of the ER.
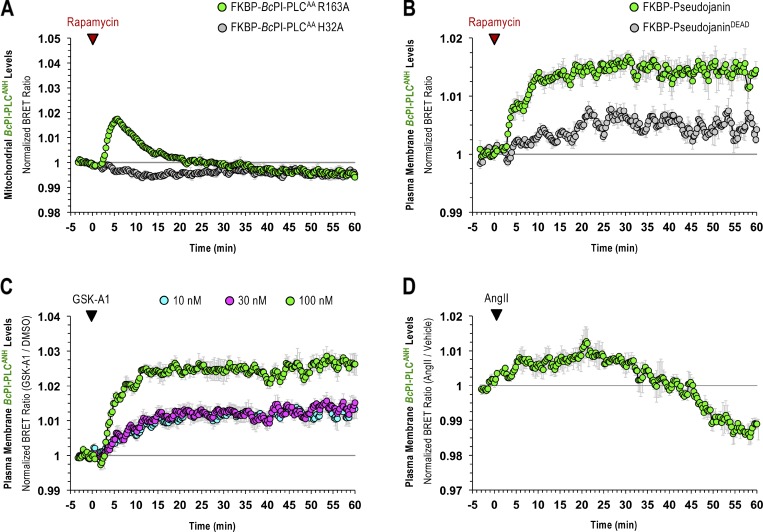
As already discussed, mitochondrial recruitment of FKBP–BcPI-PLCAA R163A yielded a rapid DAG response confirming the presence of PI within the OMM (Fig. 4, A–C). PI could also be converted to PI 4-phosphate (PI4P) in the OMM (Fig. 4, D–F) by recruiting the truncated catalytic core from PI 4-kinase IIIα (FKBP-PI4KAΔN; Hammond et al., 2014). Here, we also tested whether the localization of the putative PI-sensitive probes, BcPI-PLCH82A or BcPI-PLCANH, changed in response to recruitment of the FKBP–BcPI-PLCAA or its R163A mutant in BRET measurements. This analysis showed an extremely rapid rise in the BRET signal that was followed by a steady decrease below the initial baseline value. Notably, the magnitude of the increase measured in the mito-H82ABRET signal was smaller, and the subsequent clearance faster, when the more active FKBP–BcPI-PLCAA was used (Fig. 4, G–I). Similar kinetics were observed using BcPI-PLCANH as the reporter (mito-ANHBRET; Fig. S7 A). The increased initial localization of the BcPI-PLC-based probes to the OMM was attributed to the increased local DAG content, which enhanced the binding of the BcPI-PLCANH probe to PI-containing LUVs. Importantly, the catalytic activity of the BcPI-PLC has been shown to be enhanced by the accumulation of the enzymatic product, DAG, which facilitates interfacial binding (Ahyayauch et al., 2015). Despite this initial rise in membrane association, as the PI content of the mitochondrial membrane was reduced by the recruited FKBP–BcPI-PLCAA, the mito-H82ABRET and mito-ANHBRET BRET signals also dropped at a time when the DAG levels remained elevated (Fig. 4 B). Taken together, these data confirmed the presence of PI within the OMM. Additionally, in carrying out these experiments, we noted that acute hydrolysis of the mitochondrial pool of PI and the associated production of DAG, caused profound changes to the structure of the mitochondrial network. The ability to alter mitochondrial dynamics by initiating acute changes to the local membrane lipid composition represents an exciting application of these molecular tools that is being actively pursued.
Figure 4.
PI is enriched in the OMM. (A–H) For each BRET measurement, a schematic of the experimental design is provided to the left side of each quantified trace, with the question mark indicating the lipid being measured. (A and B) Kinetics of DAG production within the OMM following recruitment of FKBP–BcPI-PLCAA R163A to the mitochondria, as measured using the mito-DAGBRET biosensor. (C) Representative images of HEK293-AT1 cells showing the mitochondrial recruiter (mito-FRB, left) and the localization of the DAG-binding probe (EGFP-PKDC1ab) before and 5 min after rapamycin-induced recruitment of FKBP–BcPI-PLCAA R163A to the OMM (scale bar, 10 μm). (D and E) Kinetics of PI4P production within the OMM after recruitment of FKBP-PI4KAΔN to the mitochondria, as measured by the mito-PI4PBRET biosensor. (F) Representative images of cells showing the mitochondrial recruiter (mito-FRB, left) as well as the localization of the PI4P-binding probe (EGFP-P4MSidM) before and 10 min after rapamycin-induced recruitment of FKBP-PI4KAΔN to the OMM (scale bar, 10 μm). (G and H) Kinetics of BcPI-PLCH82A localization to the OMM after recruitment of either the parent FKBP–BcPI-PLCAA scaffold (magenta trace) or the modified FKBP–BcPI-PLCAA R163A (green trace) variant to the mitochondria, as measured using the mito-H82ABRET biosensor. (I) Representative images of cells showing the mitochondrial recruiter (mito-FRB, left) as well as the localization of the EGFP–BcPI-PLCH82A probe before and 10 min after rapamycin-induced recruitment of FKBP–BcPI-PLCAA R163A to the OMM. In B and H, time-matched but alternatively scaled traces show the mito-FRB:FKBP–BcPI-PLCAA H32A dimerization kinetics (red line; see also Fig. S6 C). BRET measurements are presented as mean values ± SEM from three independent experiments performed using triplicate wells (scale bar, 5 μm).
Figure S7.
BRET measurements using the BcPI-PLCANH probe. (A) Kinetics of BcPI-PLCANH localization to the OMM, as measured using the mito-ANHBRET biosensor, after mitochondrial recruitment of FKBP–BcPI-PLCAA R163A. (B) Kinetics of BcPI-PLCANH localization to the cytosolic leaflet of the PM, measured using the PM-ANHBRET biosensor, after PM recruitment of FKBP-Pseudojanin. (C) Kinetics of BcPI-PLCANH localization to the cytosolic leaflet of the PM, measured using the PM-ANHBRET biosensor, in response to treatments with 10 nM (blue trace), 30 nM (magenta trace), or 100 nM (green trace) of the PI4KA-selective inhibitor GSK-A1. (D) Kinetics of BcPI-PLCANH levels within the PM, measured using the PM-ANHBRET biosensor, after treatment with AngII (100 nM). BRET measurements are presented as mean values ± SEM from three independent experiments performed using triplicate wells.
Lastly, both the BcPI-PLCH82A (Fig. 5 A) and BcPI-PLCANH (Fig. S4 D) probes labeled the peroxisomes. This was also supported by experiments using FKBP-tagged enzymes as increases in the levels of DAG (Fig. 5 B) and PI4P (Fig. 5 C) in peroxisomal membranes were clearly observed after recruitment of FKBP–BcPI-PLCAA R163A or FKBP-PI4KAΔN, respectively, to the surface of peroxisomes using an FRB-tagged transmembrane segment from the peroxisome-targeted protein, PEX3 (PEX-FRB).
The steady-state level of PI is low within the PM
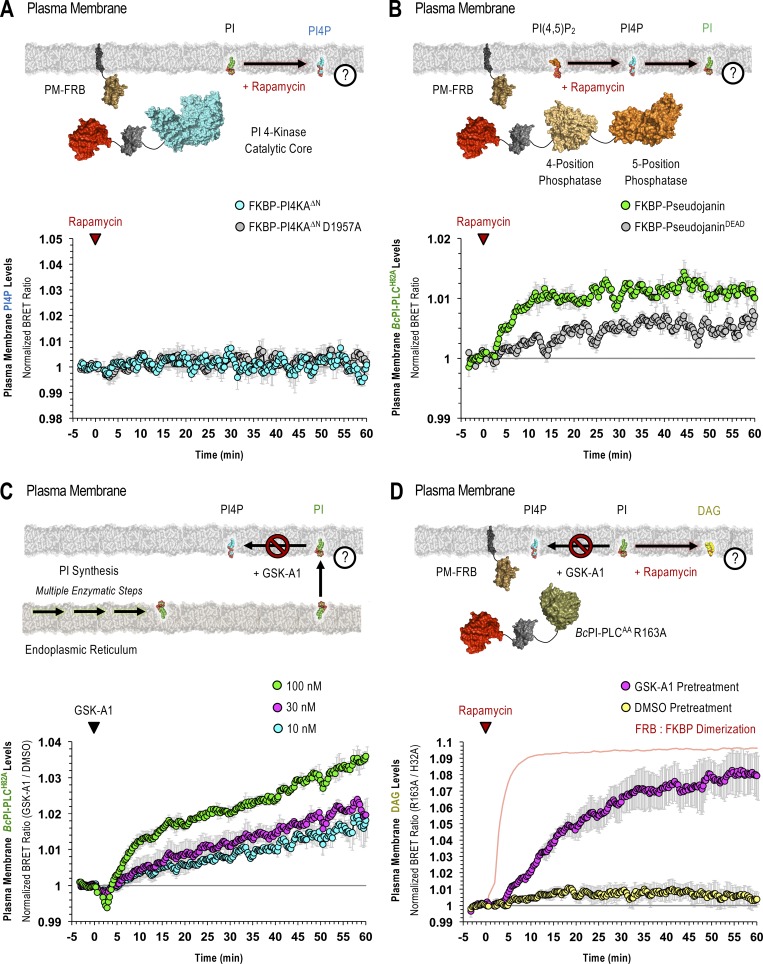
Perhaps the most striking observation using the GFP–BcPI-PLCH82A and BcPI-PLCANH probes was the lack of significant localization to the PM (Fig. 6 A and Fig. S4). Based on the well-documented importance and enrichment of PPIn lipids within the PM, this was unexpected as it has long been assumed, partly from studies of red blood cell membranes (King et al., 1987), that there is a significant reserve pool of PI present within the PM to support PPIn production. However, recent lipidomics analyses using enriched PM sheets already suggested that the PI content of the PM may be extremely low (Saheki et al., 2016). To investigate this question in detail using intact cells, we recruited the FKBP–BcPI-PLCAA R163A to the PM and measured local DAG production using the PM-DAGBRET biosensor. This analysis showed a minor increase of DAG within the PM that was only detectable using BRET-based measurements (Fig. 6 B). The ability of the PM-DAGBRET system to detect DAG increases in the PM was confirmed by stimulating PI(4,5)P2 hydrolysis either by activating Gαq-coupled AT1a receptors stably expressed in our HEK293-AT1 cell line using angiotensin II (AngII; 100 nM), or by recruiting a modified mammalian PLC enzyme (FKBP-PLCδ1Δ44,ΔPH) to the PM. Both of these manipulations evoked massive increases in the PM-DAGBRET signal that greatly surpassed the minor elevation observed upon recruitment of FKBP–BcPI-PLCAA R163A (Fig. 6 C). Notably, while stimulation with AngII (100 nM) generates a massive increase in the PM levels of DAG, this does not cause BcPI-PLCH82A association with the PM (Fig. 6 D). This suggests that, without significant amounts of PI present, acute DAG production or the introduction of membrane packing defects alone are not sufficient to localize BcPI-PLCH82A to membranes; this is also consistent with the in vitro binding studies performed using BcPI-PLCANH (Fig. S3). Furthermore, recruitment of FKBP-PI4KAΔN to the PM failed to increase the PM levels of PI4P (Fig. S8 A). These data were all consistent with low resting levels of PI within the inner leaflet of the PM. We then sought to monitor the PM content of PI in response to biochemical or pharmacological manipulations of PPIn-modifying enzymes. First, we used an established enzymatic system to rapidly dephosphorylate PPIn species within the PM and simultaneously monitored PI levels using the BcPI-PLCH82A or BcPI-PLCANH probes. Acute recruitment of the engineered 4- and 5-position PPIn phosphatase, FKBP-Pseudojanin (Hammond et al., 2012), caused an acute increase of the BcPI-PLCH82A (PM-H82ABRET; Fig. S8 B) or BcPI-PLCANH (PM-ANHBRET; Fig. S7 B) BRET signals within the PM relative to both the baseline and an inactive enzymatic control (FKBP-PseudojaninDEAD). Please note that the minor effect associated with the PM recruitment of FKBP-PseudojaninDEAD could be related to its residual 5-phosphatase activity.
Figure S8.
PI availability within the PM is controlled by delivery from the ER and local conversion to PPIn species. (A–D) For each BRET measurement, a schematic of the experimental design is provided to the left side of each quantified trace, with the question mark indicating the membrane lipid being measured. (A) Kinetics of PI4P production within the cytosolic leaflet of the PM after recruitment of FKBP-PI4KAΔN to the PM, as measured using the PM-PI4PBRET biosensor. (B) Kinetics of BcPI-PLCH82A localization to the cytosolic leaflet of the PM, measured using the PM-H82ABRET biosensor, after PM recruitment of the tandem PPIn phosphatase, Pseudojanin, or its inactive mutant. (C) Kinetics of BcPI-PLCH82A localization to the cytosolic leaflet of the PM in response to treatments with 10 nM (blue trace), 30 nM (magenta trace), or 100 nM (green trace) of the PI4KA-selective inhibitor GSK-A1, as measured using the PM-H82ABRET biosensor. (D) Kinetics of DAG production within the cytosolic leaflet of the PM, measured using the PM-DAGBRET biosensor, from cells pretreated with DMSO (yellow trace) or GSK-A1 (100 nM; magenta trace) for 30 min before recruitment of the FKBP–BcPI-PLCAA R163A enzyme to the PM. Please note that a time-matched but alternatively scaled trace shows the PM-FRB:FKBP–BcPI-PLCAA H32A dimerization kinetics (red line; see also Fig. S6 E). BRET measurements are presented as mean values ± SEM from three independent experiments performed using triplicate wells.
Next, we tested if the rapid conversion of PI to PI4P by PI4KA contributes to the low level of PI within the PM. We used the selective PI4KA inhibitor GSK-A1 (Bojjireddy et al., 2014), which gradually decreases PM levels of PI4P. BRET measurements showed that treatment with GSK-A1 resulted in a dose-dependent enrichment of BcPI-PLCH82A or BcPI-PLCANH within the PM (Figs. S7 C and S8 C). We then measured DAG levels in response to recruitment of FKBP–BcPI-PLCAA R163A after preincubation of cells with GSK-A1. In contrast to the minor increase in the DAG levels measured within the PM of DMSO-treated controls, the same enzyme recruitment evoked a rapid and sizeable increase in DAG content within the PM following a 30-min pretreatment of cells with GSK-A1 (100 nM; Fig. S8 D). These findings support the conclusion that BcPI-PLCH82A and BcPI-PLCANH are able to detect changes in membrane PI content, as well as suggest that the steady-state levels of PI are indeed low within the PM.
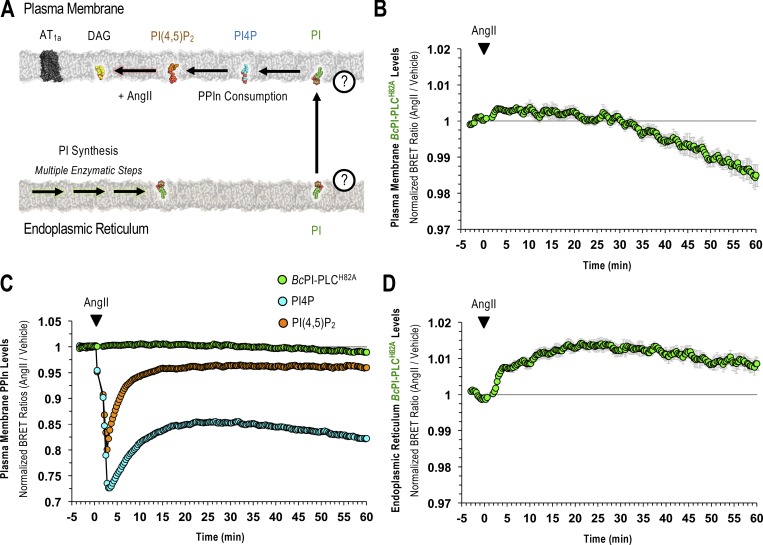
ER-derived PI feeds PI4P production within the PM and at the Golgi complex
The contribution of PI delivery from the ER for the maintenance of the PM pool of PPIn lipids is especially important if the resting level of PI within the PM is low. To determine the impact of acute PI depletion on PPIn levels, we monitored PI4P (PM-PI4PBRET) or PI(4,5)P2 (PM-PI(4,5)P2BRET) levels within the PM after recruitment of FKBP–BcPI-PLCAA R163A to either the PM or ER. Direct recruitment of FKBP–BcPI-PLCAA R163A to the PM failed to alter the resting levels of either PI4P or PI(4,5)P2. In contrast, recruitment of FKBP–BcPI-PLCAA R163A to the ER selectively reduced PI4P levels (Fig. 7, A–D) without altering PI(4,5)P2 content (Fig. 7, E–H). For comparison, treatments with GSK-A1 (100 nM), to inhibit PI4KA, or activation of endogenous PLC activity using AngII (100 nM) were used to achieve complete loss of PI4P or PI(4,5)P2 from the PM, respectively (Fig. 7, D and H). Representative confocal images further support the selective reduction of PI4P by ER recruitment of FKBP–BcPI-PLCAA R163A, as evidenced from the relocalization of the GFP-P4MSidMx2 probe from the PM toward the substantial PI4P pool at the Golgi complex (Fig. 7 C). Overall, these data suggest that PI4P production at the PM is highly dependent on PI supply from the ER and confirm that rapid depletion of PI4P within the PM does not necessarily yield comparable decreases in PI(4,5)P2, which is in agreement with earlier studies (Hammond et al., 2012; Nakatsu et al., 2012; Bojjireddy et al., 2014). Lastly, we examined whether the PM levels of PI, as reported by BcPI-PLCH82A and BcPI-PLCANH, show any changes after agonist stimulation. These studies using the PM-H82ABRET (Fig. S9) and PM-ANHBRET (Fig. S7 D) biosensors showed minor changes in the already very low PM signals detected by these probes. This contrasted the massive changes observed in both the PI4P and PI(4,5)P2 levels within the PM after AngII-evoked PLC activation (100 nM; Fig. S9 C). As shown with an expanded scale (Fig. S9 B), the localization of the BcPI-PLC–based probes initially showed a slight increase but soon declined below the initial levels measured within the PM, presumably because PPIn resynthesis begins to consume the resident PI. In contrast, we observed a sustained increase in ER-associated BcPI-PLCH82A localization after AngII stimulation (100 nM; Fig. S9 D), as monitored using the ER-H82ABRET probe, which is consistent with the well-documented increase in PI synthesis that follows treatment with AngII (Hunyady et al., 1982).
Figure 7.
PPIn production within the PM depends on PI delivery from the ER. (A–C and E–G) A schematic depicting the experimental design is provided above confocal images of representative cells showing the recruitment of FKBP–BcPI-PLCAA R163A to the PM (B and F, left panels, green) or to the ER (C and G, left panels, magenta) after a 5-min treatment with rapamycin (100 nM). The localization of PI4P (B and C; EGFP-P4MSidMx2) and PI(4,5)P2 (F and G; PLCδ1PH-EGFP) are provided before (center panels) and 20 min after (right panels) recruitment of FKBP–BcPI-PLCAA R163A to the respective membrane compartments (scale bars, 10 μm). (D and H) Kinetics of PI4P (D) or PI(4,5)P2 (H) levels within the cytosolic leaflet of the PM as measured using the PM-PI4PBRET or PM-PI(4,5)P2BRET biosensors, respectively, in response to recruitment of FKBP–BcPI-PLCAA R163A either directly to the PM (PM-FRB; green traces) or to the ER (FRB-ER; magenta traces). Treatment with GSK-A1 (100 nM; D, grey trace), which selectively inhibits PI4KA, or AngII (100 nM; H, grey trace), which stimulates PI(4,5)P2 hydrolysis, are included as positive controls for the PM-PI4PBRET and PM-PI(4,5)P2BRET biosensors, respectively, as well as to provide scale for any changes to PPIn levels that are associated with the membrane recruitment of FKBP–BcPI-PLCAA R163A. BRET measurements are presented as mean values ± SEM from three independent experiments performed using triplicate wells.
Figure S9.
PM and ER localization of the BcPI-PLCH82A probe in response to endogenous PLC activation. (A–C) A summary schematic of the experimental design is presented alongside the kinetics of BcPI-PLCH82A (B and C; PM-H82ABRET), PI4P (C; PM-PI4PBRET), and PI(4,5)P2 (C; PM-PI(4,5)P2BRET) levels within the PM after treatment with AngII (100 nM). Please note that the green BRET trace shown in B has been expanded in C to scale the magnitude of the changes observed. (D) In addition to the PM, the levels of BcPI-PLCH82A (ER-H82ABRET) within the ER are also shown after AngII stimulation (100 nM). BRET measurements are presented as mean values ± SEM from three independent experiments performed using triplicate wells.
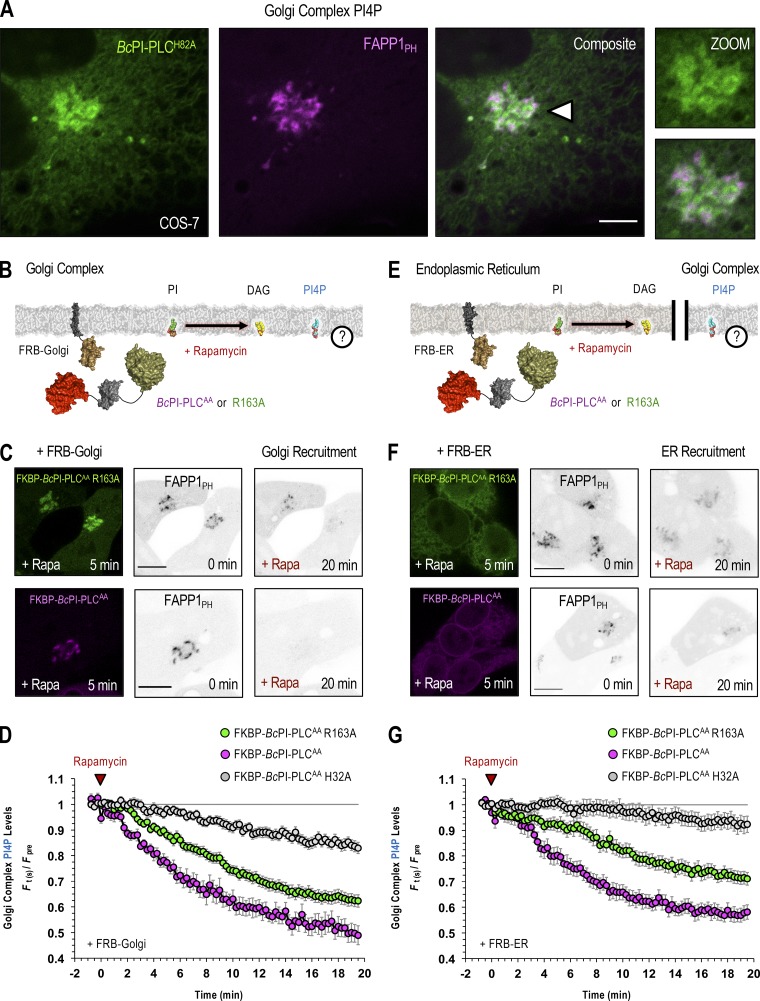
After examining the sensitivity of PPIn levels within the PM to targeted PI hydrolysis, we next investigated the turnover of the PI4P pool that is found at the Golgi complex. Changes in the PI4P levels were monitored after recruitment of FKBP–BcPI-PLCAA or the R163A variant to either to the surface of the Golgi complex or to the ER. Notably, for these studies, we followed the Golgi localization of the PH domain from PI4P-adaptor protein 1 (FAPP1PH-EGFP; Godi et al., 2004; Szentpetery et al., 2010) using confocal imaging (Fig. 8 A), which has been used reliably for PI4P measurements specifically at the Golgi complex. We chose this approach due to the fact that attempts to use unbiased PI4P sensors together with BRET were complicated by redistribution artifacts produced from the fraction of probe that monitors the PM pool of PI4P. Nevertheless, single-cell measurements using confocal microscopy showed that hydrolysis of PI directly at the Golgi complex or indirectly at the ER both caused a dissociation of the Golgi-localized FAPP1PH-GFP, and this was more pronounced when using the more active FKBP–BcPI-PLCAA enzyme (Fig. 8, B–G). These data suggest that PI4P levels at the Golgi complex are extremely sensitive to the local PI content, which is consistent with the relatively low affinity of the resident PI4KB toward PI (Downing et al., 1996; Zhao et al., 2000). Alternatively, it is also possible that only a fraction of the PI pool at the Golgi complex is dedicated to PI4P synthesis. It is interesting to note that the Golgi-specific localization of the FAPP1PH-GFP probe was also slightly reduced by the recruitment of the inactive FKBP–BcPI-PLCAA H32A directly to the surface of the Golgi, and to a lesser degree after recruitment to the ER (Fig. 8, D and G). This minor effect was attributed to the ability of the inactive enzyme to mask the available PI from Golgi-localized PI4K enzymes.
Figure 8.
PI4P levels at the Golgi complex are sensitive to the local availability of PI. (A) Confocal images of COS-7 cells coexpressing EGFP–BcPI-PLCH82A together with the PH domain of FAPP1 (FAPP1PH-EGFP; scale bar, 5 µm). An enlarged view of the region identified by the arrowhead is shown on the far right (inset, 10 µm). (B, C, E, and F) Schematics depicting the experimental design are provided above images of representative HEK293-AT1 cells showing the recruitment of FKBP–BcPI-PLCAA R163A (C and F; top row, left panels, green) or FKBP–BcPI-PLCAA (C and F; bottom row, left panels, magenta) to the Golgi complex (C; FRB-Golgi) or ER (F; FRB-ER) after a 5-min treatment with rapamycin (100 nM). Localization of the FAPP1PH-EGFP probe is shown before and 20 min after initiation of localized PI hydrolysis by recruitment of the BcPI-PLC mutants to the Golgi (C; FRB-Golgi) or ER (F; FRB-ER). Scale bars in C and F are 10 μm. (D and G) Pooled image analyses measuring changes in FAPP1PH-EGFP intensities at the perinuclear Golgi region after recruitment of FKBP–BcPI-PLCAA R163A (green traces; D, 46 cells; G, 42 cells), mRFP-FKBP–BcPI-PLCAA (magenta traces; D, 16 cells; G, 21 cells), or mRFP-FKBP–BcPI-PLCAA H32A (gray traces; D, 43 cells; G, 42 cells) to the respective compartments (D, Golgi; G, ER). Normalized intensities (F(t)/Fpre) of the FAPP1PH-EGFP signal at the perinuclear Golgi region, relative to the cytosolic fraction, are presented as mean values ± SEM from a minimum of four independent experiments. The pretreatment period used for normalization was defined as the average ratio of the Golgi/cytosolic signal intensity measured over the first four frames of each recording.
Endosomal PI3P levels depend on PI delivery from the ER
Apart from PI4P and PI(4,5)P2, monophosphorylated PI3P comprises the largest remaining PPIn pool at ∼20–30% of the total PI4P content in higher eukaryotes (Sarkes and Rameh, 2010). PI3P is produced primarily by class III phosphoinositide 3-kinase (Vps34) complexes (Volinia et al., 1995; Ohashi et al., 2019) and is enriched in early endosomes (Gillooly et al., 2000), as well as autophagosomes (Petiot et al., 2000). Importantly, we did not detect PI enrichment in the Rab5 or Rab7 endosomal compartments using the BcPI-PLCH82A probe (Figs. 9 A and 10 A). To define the relative contribution of local PI content and ER-derived PI for the maintenance of PI3P levels within Rab5- and Rab7-positive endosomal compartments, we again used recruitment of FKBP–BcPI-PLCAA R163A and measured local DAG production or changes in PI3P levels. A larger increase in DAG levels was measured in Rab7-positive membrane compartments compared with those labeled with Rab5 after recruitment of the FKBP–BcPI-PLCAA R163A enzyme (Figs. 9 B and 10 B). This suggested a somewhat larger PI content within Rab7-positive endosomes. Using biosensors incorporating the tandem FYVE domain from hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs; Gillooly et al., 2000), we then monitored changes in PI3P levels within both Rab5- and Rab7-positive endosomes after recruiting FKBP–BcPI-PLCAA R163A either directly to the surface of the respective endosomes or to the ER. Whereas only minor reductions In PI3P content were observed following the direct recruitment of FKBP–BcPI-PLCAA R163A to the respective endosome populations, recruitment of FKBP–BcPI-PLCAA R163A to the ER caused a larger reduction to PI3P content in both endosomal compartments (Figs. 9 C and 10 C). To compare the magnitude of these changes relative to the total PI3P content, we used a selective inhibitor of Vps34 (VPS34-IN1; Bago et al., 2014) as a positive control to eliminate PI3P from endosomal compartments (Figs. 9 C and 10 C).
Discussion
Within mammalian cell membranes, PI represents ∼10–15 mol% of total phospholipids (Vance, 2015), while the diverse PPIn species only comprise an estimated 2–5% of the available PI (Xu et al., 2003; Sarkes and Rameh, 2010). The relative abundance of PI and its role as a precursor for PPIn production has led to the assumption that PI is fairly abundant in membranes where the various PPIn lipids exist. Early studies designed to visualize the subcellular distribution of PI involved the synthesis of a fluorescent PI analogue and revealed accumulation of the tagged lipid within membranes corresponding to the ER, mitochondria, and perinuclear Golgi region (Uster and Pagano, 1986; Ting and Pagano, 1990). However, an examination of total lipid contents after the exogenous delivery of the labeled PI suggested that conversion of the fluorescently tagged PI to DAG was required for observing the labeling of intracellular membranes (Ting and Pagano, 1990). These complications, and the need to incorporate the highly polar fluorescent label into the fatty acid side chains, challenged the reliability of these early approaches for mapping the intracellular distribution of PI. Therefore, we looked for an alternative method for the visualization of membrane-embedded PI and eventually devised a protein-engineering platform that utilizes the bacterial enzyme BcPI-PLC to complete comprehensive investigations of PI distribution and availability in live cells.
Our efforts to map the cellular PI landscape relied on two complementary approaches. The first involved the rational design of two BcPI-PLC variants, BcPI-PLCH82A and the related BcPI-PLCANH, which both lack catalytic activity without interfering with the coordination of the lipid substrate within the active site. Second, we designed the interfacially compromised BcPI-PLCAA scaffold, which still has an intact active site but was cytosolic and, therefore, had minimal access to membrane-embedded PI. This modified BcPI-PLCAA enzyme could be recruited to specific membrane compartments to hydrolyze resident PI and locally generate DAG, which could be detected with an established probe and used as a proxy to assess the PI content of different membrane compartments. While both of these approaches have their own limitations, together, they still provide the most comprehensive description to date of the subcellular membrane distribution of PI in intact cells. It is important to note that these approaches only reflect the PI abundance within the cytoplasmic leaflet of organelle membranes. Overall, our results suggest that the resting levels of PI within the PM and endosomes are kept at low levels and, in addition to its presence within membranes of the ER, we also identify a relative enrichment of PI in membranes comprising the Golgi complex, mitochondria, and peroxisomes. This pattern of PI distribution is in good agreement with membrane fractionation studies that also define the ER and Golgi complex as containing the highest percentages of relative PI content among the various organelle compartments (∼9% of total phospholipids), exceeding those associated with the endolysosomal system, nucleus, and PM (∼4–7% of total phospholipids; Vance, 2015). That said, unlike the approaches used here, these values from fractionation studies refer to the sum of both the outer and inner leaflets of the respective membranes. Evidently, while bulk measurements show PI as a relatively minor component of mitochondrial membranes, estimated at ∼5–7% of the total lipid content, isolation of purified inner and outer membranes revealed an enrichment of PI in the OMM (∼9–13% of total phospholipids) relative to the inner membrane (∼2–5% of total phospholipids; Ardail et al., 1990; de Kroon et al., 1997; Daum and Vance, 1997). Based on the current study, within the cytosolic leaflet, the mitochondria appear to possess a PI content similar to that associated with bulk membranes of the ER, but likely lower than those of the Golgi complex.
Our findings that, when compared with the ER, PI concentrations are higher within the cytosolic leaflet of membranes corresponding to the Golgi complex, mitochondria, and peroxisomes raises the questions of how the enrichment of PI occurs and whether these PI pools serve any unrecognized functions. Although the Golgi has a high PI4P content that undergoes rapid turnover, this alone may not explain the high PI levels, since the PM and endosomes also have high PPIn levels and yet, limited amounts of PI were detected in these membrane compartments. Interestingly, a series of studies has defined an important regulatory role for DAG in the control of Golgi functions (Baron and Malhotra, 2002; Bossard et al., 2007; Fernández-Ulibarri et al., 2007; Asp et al., 2009), and it has been suggested that activation of PLC enzymes would generate DAG from PPIn species at the Golgi (Díaz Añel, 2007). Some of these studies also claim that the substrate used by the Golgi-resident PLC isoforms is PI4P (Zhang et al., 2013; Sicart et al., 2015), but direct hydrolysis of PI by some of the mammalian PLCs cannot be ruled out.
The physiological functions of the PI present within the mitochondria and peroxisomes are even more intriguing, especially given that enzymes directly using PI as a substrate have not been described to function within, or interact with, the OMM. It is notable, though, that multiple proteomic screens have identified PI4K isoforms within mitochondria-enriched datasets (Schon, 2007; Calvo et al., 2016; Hung et al., 2017). Downstream of PI, specific isoforms or alternatively spliced variants of the PPIn phosphatases synaptojanin 2 (Nemoto and De Camilli, 1999) and phosphatase and tensin homologue deleted on chromosome 10 (Bononi et al., 2013; Liang et al., 2014) have been shown to localize to the mitochondria. However, apart from scaffolding or other noncatalytic functions, the explicit roles for their lipid phosphatase activities remains largely uncharacterized. The localization of these enzymes is also interesting given that, using the available probes, no PPIn species has been detected on the outer surface of mitochondria and our efforts to find PPIn lipids associated with the OMM have been unsuccessful. That said, one study has explored a possible role for PPIn lipids within the OMM using a mitochondrial-targeted PLCδ1PH domain to mask any resident PI(4,5)P2 (Rosivatz and Woscholski, 2011). Based on the altered mitochondrial morphology that was associated with the overexpression of this construct, PI(4,5)P2 was proposed to serve as a local regulator of mitochondrial dynamics. The possibility remains that the mitochondrial-targeted PLCδ1PH domain may exert its effects indirectly, perhaps by altering interorganelle contacts or through localized buffering of inositol (1,4,5)-trisphosphate levels, which could change Ca2+ dynamics associated with ER–mitochondria contact sites. Similarly, very few studies have looked at the content or functions of PI and PPIn derivatives in peroxisomes. Fractionation studies showed that PI represents ∼5% of the total phospholipid content of the peroxisomes (Hardeman et al., 1990), and more recent measurements suggest that, among PPIn species, the levels of PI4P, PI(3,5)P2, and PI(4,5)P2 are enriched in peroxisomal membranes (Jeynov et al., 2006). The potential for a specific role for PPIn lipids in peroxisomes is supported by recent reports describing interaction between PI(4,5)P2 present on the peroxisome surface and the lysosomal protein synaptotagmin VII, which is important for establishing functionally relevant contact sites between lysosomes and peroxisomes (Chu et al., 2015; Hu et al., 2018). Undoubtedly, additional studies are needed to better define the role of PI and PPIn species within these diverse compartments.
Considering that the biosynthetic machinery required for PI production is almost exclusively localized to the ER, the low abundance of PI in membranes where PPIn species play critical regulatory roles, such as in the PM or endosomes, raises the question as to whether PI delivery to these compartments is part of the process by which PPIn generation is controlled. Our previous studies identified a mobile ER-derived compartment that contained the rate-limiting enzyme for PI production, PI synthase, which would be an ideal candidate to serve as a PI delivery platform (Kim et al., 2011). However, none of the BcPI-PLC–based probes used here were found to localize to the dynamic PI synthase–positive structures (data not shown). This finding may suggest that PI is rapidly transferred from this active compartment to acceptor membranes. In fact, it is interesting to note that, with the exception of the Golgi complex, PI appears to be enriched in membranes that lack significant local pools of PPIn lipids. Therefore, it is possible that the relatively low PI content associated with the PM and endosomal system reflects the fact that the delivered PI is quickly consumed for the production of phosphorylated PPIn derivatives. In this context, it is worth reiterating the fact that it was more effective to alter the levels of PI4P in the PM and PI3P in endosomes by consuming PI at the ER, rather than by trying to hydrolyze PI within the specific membranes where these PPIn lipids are made. These data are compatible with the idea of direct channeling of the PI substrate to the respective PPIn-producing kinases that are present in the membranes receiving PI and also highlight the importance of understanding how newly synthesized PI produced in the ER reaches other membranes. Defining the cargo selectivity and potential interplay between the diverse families of lipid transfer proteins that are thought to distribute PI between specific membranes represents a major goal for future studies. The inability of the FKBP–BcPI-PLCAA to intercept the delivered PI within the acceptor membrane is also not at odds with the proposed role for some lipid transfer proteins as molecular devices involed in substrate presentation to membrane-associated effectors, including examples involving PPIn kinases and both major eukaryotic superfamilies of putative PI-transfer proteins (Panaretou et al., 1997; Schaaf et al., 2008; Grabon et al., 2015). We hope that our findings using intact cells will inspire further investigations in pursuit of these important and exciting questions.
Lastly, while our studies suggest that PI is an important localization signal responsible for recruiting the BcPI-PLCH82A and BcPI-PLCANH probes to membrane compartments, PI binding may not be the sole determinant responsible for the membrane affinity of this probe. Briefly, in vitro studies have already shown that the dissociation constant (Kd) measured for the binding of Bacillus PI-PLCs to substrate-poor vesicles is significantly lower than the apparent catalytic rate, as inferred from the Michaelis constant (Km), associated with hydrolysis of membrane-embedded PI (Volwerk et al., 1994; Qian et al., 1998). These data highlight the fact that membrane recognition by the BcPI-PLC requires a combination of interfacial as well as active site binding events, in which the presence of non-substrate lipids, including PC and DAG, may also play important roles. In particular, although not sufficient for membrane localization, the sensitivity of the BcPI-PLC scaffold to membrane DAG content is apparent from our in vitro measurements and is likely related to the change in lipid order that occurs when DAG levels increase, which allows for hydrophobic elements present within the BcPI-PLC interface to target any packing defects that are introduced into the membrane (Lehto and Sharom, 2002; Ahyayauch et al., 2015). The relative concentration of PC has also been suggested to influence the structural dynamics or residency of the BcPI-PLCH82A probe at the membrane interface through specific interactions with residues within the αG helix (Zhang et al., 2004; Pu et al., 2009; Yang et al., 2015; Khan et al., 2016). Taken together, even though the enzyme variants presented here appear to faithfully map the subcellular PI landscape, it is important to emphasize that our conclusions regarding the membrane distribution of PI are based on a combination of approaches and not solely on the localization of the BcPI-PLCH82A or BcPI-PLCANH probes. Consequently, at present, we would still recommend using caution when interpreting the results of experiments that rely entirely on the membrane-binding behavior of the BcPI-PLC variants.
Summary
In this study, we define the steady-state subcellular distribution of PI and also introduce a powerful new strategy to dissect the complex trafficking events that distribute PI between membranes by selectively hydrolyzing distinct intracellular pools of PI. Additionally, it has not escaped our attention that the establishment of this organelle-targeted FKBP–BcPI-PLCAA recruitment system provides a unique opportunity to exploit the distinct biophysical properties of PI and DAG to examine the effects of selectively manipulating the local lipid composition on membrane dynamics and organelle structure. Overall, we hope these collective contributions help to place PI within the complex lipid landscape of eukaryotic membranes and provide new information relevant to understanding the basic molecular mechanisms that regulate the onset of PPIn-dependent pathologies, as well as other lipid-related metabolic disorders.
Materials and methods
Cell culture
COS-7 (CRL-1651; ATCC) or HEK293-AT1 cells, which stably express the rat AT1a AngII receptor (Hunyady et al., 2002), were cultured in DMEM-high glucose (GIBCO) containing 10% (volume/volume) FBS and supplemented with a 1% solution of penicillin/streptomycin (GIBCO). Alternatively, human HT-1080 fibrosarcoma cells (CCL-121; ATCC) were maintained using MEM (MilliporeSigma) supplemented with 2 mM L-glutamine and containing 10% (volume/volume) FBS as well as a 1% solution of penicillin/streptomycin. Each of these cell lines were maintained at 37°C and 5% CO2 in a humidified atmosphere. Cell lines are also regularly tested for Mycoplasma contamination using a commercially available detection kit (InvivoGen) and, after thawing, all cell cultures were treated with plasmocin (InvivoGen) at 500 µg/ml for the initial three passages (6–9 d) as well as supplemented with 5 µg/ml of the prophylactic for subsequent passages.
Reagents
All compounds were prepared in the indicated solvent and stored in small aliquots at −20°C. Rapamycin (MilliporeSigma; 100 µM stock) and VPS34-IN1 (Selleck Chemicals; 300 µM stock) were dissolved in DMSO. Production and validation of the PI4KA-selective inhibitor, GSK-A1, has been described previously (Bojjireddy et al., 2014) and stock solutions were prepared at 100 µM in DMSO. Coelenterazine h (Regis Technologies) was dissolved in 100% ethanol (volume/volume) at 5 mM. AngII (human octapeptide; Bachem) was first dissolved in ethanol at 1 mM before being prepared as 100-µM aliquots for storage by dilution with double-distilled water. MitoTracker Red (ThermoFisher Scientific) was prediluted 1:100 in DMSO from the concentrated stock for storage in small aliquots at −20°C. Diluted solutions of MitoTracker Red were added directly to the medium of transfected cells at a 1:1,000 dilution (1:100,000 final concentration) and allowed to equilibrate for 15–30 min at 37°C before imaging.
DNA constructs
In general, plasmids were constructed by standard restriction cloning using enzymes from New England Biolabs, while site-directed mutagenesis was done using the QuikChange II kit (Agilent). Complex reconfigurations of vector backbones and all point mutations were verified using standard Sanger sequencing (Macrogen). The design of the following plasmids have been described elsewhere: PLCδ1PH-EGFP (Várnai and Balla, 1998), FAPP1PH-EGFP (Balla et al., 2005), PM2-FRB-ECFP (Varnai et al., 2006), mRFP-FKBP-5-ptase-dom (Varnai et al., 2006), mRFP-ER(Sac1521-587) (Várnai et al., 2007), AKAP-FRB-ECFP (Csordás et al., 2010), mRFP-PI-PLC (L. monocytogenes; Kim et al., 2011), EGFP-PKDC1ab (Kim et al., 2011), EGFP-P4MSidM (Hammond et al., 2014), mCherry-P4MSidM (Hammond et al., 2014), iRFP-P4MSidM (Hammond et al., 2014), EGFP-P4MSidMx2 (Hammond et al., 2014), iRFP-FRB-Rab5 (Hammond et al., 2014), iRFP-FRB-Rab7 (Hammond et al., 2014), mCherry-FKBP-PI4KAΔN (Hammond et al., 2014), mCherry-FKBP-PI4KAΔN D1957A (Hammond et al., 2014), FRB-mCherry-Giantintail (Hammond et al., 2014), NES-mdsRed-Spo20DM (Kim et al., 2015), L10-mVenus-T2A-sLuc-D4H (Sohn et al., 2018), sLuc-P4MSidMx2-T2A-mVenus-Rab5 (Baba et al., 2019), sLuc-P4MSidMx2-T2A-mVenus-Rab7 (Baba et al., 2019), sLuc-FYVEEEA1-T2A-mVenus-Rab5 (Baba et al., 2019), and sLuc-FYVEEEA1-T2A-mVenus-Rab7 (Baba et al., 2019). Also, we would like to thank the laboratories of Bianxiao Cui (Stanford University, Stanford, CA; plasmid 102250; Addgene; Duan et al., 2015), Gerald Hammond (University of Pittsburgh, Pittsburgh, PA; mTagBFP2-FKBP-CyB5tail; Zewe et al., 2018), T. Kendall Harden (University of North Carolina at Chapel Hill, Chapel Hill, NC; pcDNA3.1-PLCδ1Δ44; Hicks et al., 2008), Takanari Inoue (Johns Hopkins University, Baltimore, MD; ECFP-FRB-Giantintail; Komatsu et al., 2010), Jennifer Lippincott-Schwartz (HHMI Janelia Research Campus, Ashburn, VA; mRFP-SKL; Kim et al., 2006), Robert Lodge (Institut de Recherches Cliniques de Montreal, Montreal, Canada; mCherry-Rab5WT and mCherry-Rab7WT; Hammond et al., 2014), Harald Stenmark (University of Oslo, Oslo, Norway; EGFP-FYVEHrsX2; Gillooly et al., 2000), and Peter Várnai (Semmelweis University Medical School, Budapest, Hungary; PLCδ1PH-mVenus, L10-mVenus-T2A-sLuc-P4MSidMx2, L10-mVenus-T2A-sLuc-PLCδ1PH, L10-FRB-T2A-mRFP-FKBP-Pseudojanin, and L10-FRB-T2A-mRFP-FKBP-PseudojaninDEAD; Tóth et al., 2016) for generously providing constructs. Alternatively, the cloning procedures used for generating the DNA constructs unique to this study are provided below, and the primers required for both PCR-mediated cloning or site-directed mutagenesis are listed in Table S1.
To facilitate the diverse cloning strategies needed for this study, BcPI-PLC (residues 32–329, with the N-terminal hydrophobic signal sequence removed; GenBank accession no. AAA22665.1) was codon optimized for mammalian expression and custom synthesized in the pUCminusMCS vector shuttle by Blue Heron Biotech. EGFP–BcPI-PLC was generated by inserting the BcPI-PLC sequence into the pEGFP-C1 vector (Clontech) using the flanking EcoRI and BamHI restriction sites included in the synthesized pUCminusMCS–BcPI-PLC construct. Site directed mutagenesis of the parent EGFP–BcPI-PLC backbone was done to introduce alanine substitutions at His32 (H32A), His82 (H82A), Trp47 (W47A), and Trp242A (W242A), as well as combined mutagenesis of residues Tyr247 (Y247N) and Tyr251 (Y251H) to generate BcPI-PLCANH and sequential mutagenesis of both interfacial tryptophan residues to generate EGFP–BcPI-PLCAA. To facilitate colocalization studies with the various EGFP–BcPI-PLC constructs, mRFP-PKDC1ab and AKAP-mRFP were made using the original cloning sites to digest the respective GFP-tagged variants and insert them into the empty pmRFP-C1 or pmRFP-N1 vectors (Clontech). Alternatively, the recruitable mRFP-FKBP–BcPI-PLCAA enzyme was created by replacing the type IV 5-phosphatase domain in the mRFP-FKBP-5-ptase-dom construct with the BcPI-PLCAA insert amplified from EGFP–BcPI-PLCAA using SacI and KpnI restriction sites. To alleviate any concerns related to potential interference with BRET measurements, we also made iRFP-FKBP–BcPI-PLCAA by replacing mRFP with the iRFP module from iRFP-P4MSidM using NheI and BspEI restriction sites. Site-directed mutagenesis of the iRFP-FKBP–BcPI-PLCAA backbone was used to generate the H32A inactive control as well as the R69A, R163A, R163K, Y200A, and Y200F active site mutants for screening. Similarly, for BRET measurements using recruitment of the truncated FKBP-PI4KAΔN (Δ1-1101; Harak et al., 2014), iRFP-FKBP-PI4KAΔN was generated from mCherry-FKBP-PI4KAΔN by replacing mCherry with the iRFP insert from iRFP-FKBP–BcPI-PLCAA using AgeI and HindIII restriction sites. Using the resulting iRFP-FKBP-PI4KAΔN construct, we also made a catalytically inactive mutant by replacing the PI4KAΔN with the PI4KAΔN D1957A insert from mCherry-FKBP-PI4KAΔN D1957A using HindIII and KpnI restriction sites. As a positive control for demonstrating DAG production within the PM, a truncated mammalian PLCδ1, lacking both a short segment of the X-Y domain linker (Δ44) and the pleckstrin homology (PH) domain (ΔPH), was tagged with FKBP by amplifying the PLCδ1Δ44,ΔPH insert from pcDNA3.1-PLCδ1Δ44 (Δ444–487; Hicks et al., 2008) and inserting it in place of the type IV 5-phosphatase domain in mRFP-FKBP-5-ptase-dom using PvuI and KpnI restriction sites. For use during BRET measurements, iRFP-FKBP-PLCδ1Δ44,ΔPH was then made from the mRFP-FKBP-PLCδ1Δ44,ΔPH by replacing mRFP with the iRFP module from iRFP-FKBP–BcPI-PLCAA using AgeI and NotI restriction sites. In addition to the FKBP-tagged enzymes, additional FRB-tagged constructs were also needed in order to target enzymes to the cytosolic leaflets of the ER and peroxisomes. The ER-targeted mTagBFP2-FRB-CyB5tail construct was created by amplifying the FRB insert from AKAP-FRB-ECFP and replacing the FKBP module from mTagBFP2-FKBP-CyB5tail using XhoI and EcoRI restriction sites. Peroxisomal targeting was done by creating the PEX-FRB-ECFP recruiter by amplifying the peroxisome-targeting transmembrane domain of PEX3 (residues 1–42; Duan et al., 2015) from PEX-mCherry-CRY2 and inserting this in place of the AKAP sequence in AKAP-FRB-ECFP using NheI and BamHI restriction sites. Additionally, mutagenesis of the ECFP (W66A; Heim et al., 1994) or mTagBFP2 (E215A; Subach et al., 2011) chromophores through alanine substitution was done to inactivate the fluorescence of the AKAP-FRB-ECFP, PEX-FRB-ECFP, ECFP-FRB-Giantintail, PM2-FRB-ECFP, and mTagBFP2-FRB-CyB5tail membrane recruitment constructs to prevent interference with BRET measurements.
The design of the organelle-specific BRET biosensors required several unique cloning strategies. To construct the mito-DAGBRET (AKAP-mVenus-T2A-sLuc-PKDC1ab), mito-PI4PBRET (AKAP-mVenus-T2A-sLuc-P4MSidMx2), and mito-H82ABRET (AKAP-mVenus-T2A-sLuc–BcPI-PLCH82A) biosensors, we first generated AKAP-mVenus by exchanging the PLCδ1PH domain from PLCδ1PH-mVenus with the mitochondrial-targeting sequence (N-terminal residues 34–63 of mAKAP1; Csordás et al., 2010) from AKAP-mRFP using AgeI and NotI restriction sites. Specifically for the mito-DAGBRET construct, we also needed to make an L10-mVenus-T2A-sLuc-PKDC1ab intermediate by replacing the P4MSidMx2 module of L10-mVenus-T2A-sLuc-P4MSidMx2 with the PKDC1ab domain from EGFP-PKDC1ab using BglII and BamHI restriction sites. We then used an NheI and BsrGI double digest to insert the AKAP-mVenus fragment in place of the L10-mVenus targeting sequence in both L10-mVenus-T2A-sLuc-PKDC1ab and L10-mVenus-T2A-sLuc-P4MSidMx2 to make the final AKAP-mVenus-T2A-sLuc-P4MSidMx2 and AKAP-mVenus-T2A-sLuc-PKDC1ab constructs. Alternatively, AKAP-mVenus-T2A-sLuc–BcPI-PLCH82A was generated by amplifying out BcPI-PLCH82A from EGFP–BcPI-PLCH82A and inserting it in place of the PKDC1ab module from AKAP-mVenus-T2A-sLuc-PKDC1ab using SpeI and BamHI restriction sites. The resulting AKAP-mVenus-T2A-sLuc-BcPI-PLCH82A construct was subsequently used to generate the mito-ANHBRET sensor by site-directed mutagenesis using a single Y247N/Y251H mutagenesis primer set. Production of the ER-DAGBRET (sLuc-PKDC1ab-T2A-mVenus-CyB5tail), Golgi-DAGBRET (sLuc-PKDC1ab-T2A-mVenus-Giantintail), and PEX-DAGBRET (PEX31-42-mVenus-T2A-sLuc-PKDC1ab) biosensors also required sequential steps. First, the CyB5tail (C-terminal residues 100–134; Zewe et al., 2018) or Giantintail (C-terminal residues 3140–3269; Komatsu et al., 2010) membrane-targeting fragments were amplified from mTagBFP2-FKBP-CyB5tail and ECFP-FRB-Giantintail, respectively, and inserted in place of Rab7 in sLuc-FYVEEEA1-T2A-mVenus-Rab7 using BspEI and KpnI restriction sites. The FYVEEEA1domain was then replaced with PKDC1ab in both the sLuc-FYVEEEA1-T2A-mVenus-CyB5tail and sLuc-FYVEEEA1-T2A-mVenus-Giantintail constructs by amplifying the PKDC1ab insert from AKAP-mVenus-T2A-sLuc-PKDC1ab and using PvuI and SalI restriction sites. Using the same cloning strategy, an ER-H82ABRET (sLuc–BcPI-PLCH82A-T2A-mVenus-CyB5tail) biosensor was made by amplifying BcPI-PLCH82A from an AKAP-mVenus-T2A-sLuc–BcPI-PLCH82A variant that lacks an internal PvuI site, which was removed by site-directed mutagenesis. Alternatively, we used a unique PvuI site present in the L10-mVenus-T2A-sLuc-Spo20(DM) construct to replace the PM-targeting sequence with the PEX31–42 fragment amplified from PEX-mCherry-CRY2 using NdeI and PvuI restriction sites. From the PEX31–42-mVenus-T2A-sLuc-Spo20(DM) intermediate, we replaced the phosphatidic acid–binding Spo2051–90 C54S/C82S (Spo20DM) cassette with the PKDC1ab domain from AKAP-mVenus-T2A-sLuc-PKDC1ab using BsrGI and BamHI restriction sites. Although not directly used in this study, it should be mentioned that the L10-mVenus-T2A-sLuc-Spo20(DM) construct was designed by amplifying the Spo20DM insert from NES-mdsRed-Spo20DM and replacing the P4MSidMx2 module in L10-mVenus-T2A-sLuc-P4MSidMx2 using BglII and EcoRI restriction sites. This construct was chosen for mutagenesis to introduce a unique PvuI site between the membrane-targeting and mVenus modules due to the relative abundance of cloning sites surrounding the Spo20DM cassette at the N-terminus of this construct. To measure potential changes in the PM localization of the PI-sensitive BcPI-PLCH82A probe, we generated the PM-H82ABRET (L10-mVenus-T2A-sLuc–BcPI-PLCH82A) biosensor using XhoI and EcoRI restriction sites to replace the cholesterol-binding D4H domain in the L10-mVenus-T2A-sLuc-D4H construct with the BcPI-PLCH82A insert amplified from EGFP–BcPI-PLCH82A. The resulting L10-mVenus-T2A-sLuc–BcPI-PLCH82A construct was subsequently used to generate the PM-ANHBRET sensor by site-directed mutagenesis using a single Y247N/Y251H mutagenesis primer set. Within the endosomal system, the Rab5-DAGBRET (sLuc-PKDC1ab-T2A-mVenus-Rab5) biosensor was made using PvuI and AgeI restriction sites to swap out the FYVEEEA1 domain in sLuc-FYVEEEA1-T2A-mVenus-Rab5 for the PKDC1ab domain from sLuc-PKDC1ab-T2A-mVenus-CyB5tail. Alternatively, for creating the Rab7-DAGBRET (sLuc-PKDC1ab-T2A-mVenus-Rab7) biosensor, the FYVEEEA1 domain from sLuc-FYVEEEA1-T2A-mVenus-Rab7 was exchanged for PKDC1ab from sLuc-PKDC1ab-T2A-mVenus-CyB5tail using PvuI and SalI restriction sites. The high-affinity Rab7-PI3PBRET (sLuc-FYVEHrsX2-T2A-mVenus-Rab7) biosensor was constructed by replacing the FYVEEEA1 module within sLuc-FYVEEEA1-T2A-mVenus-Rab7 with the FYVEHrsX2 insert amplified from EGFP-FYVEHrsX2 using PvuI and SalI restriction sites. Alternatively, to make the high-affinity Rab5-PI3PBRET (sLuc-FYVEHrsX2-T2A-mVenus-Rab5) biosensor, the FYVEHrsX2-T2A-mVenus insert from sLuc-FYVEHrsX2-T2A-mVenus-Rab7 was inserted in place of FYVEEEA1-T2A-mVenus in sLuc-FYVEEEA1-T2A-mVenus-Rab5 using PvuI and HindIII restriction sites.
Lastly, to monitor the FRB-dependent recruitment of the FKBP–BcPI-PLCAA scaffold at the level of entire cell populations, BRET constructs were designed to monitor the dimerization of FKBP–BcPI-PLCAA H32A with each of the FRB-Golgi, FRB-ER, mito-FRB, PEX-FRB, PM-FRB, FRB-Rab5, and FRB-Rab7 membrane recruiters. All of these constructs were built in several stages. First, the FKBP–BcPI-PLCAA H32A insert was amplified from iRFP-FKBP–BcPI-PLCAA H32A and moved in place of Spo20(DM) within L10-mVenus-T2A-sLuc-Spo20(DM) using BglII and KpnI restriction sites to generate L10-mVenus-T2A-sLuc-FKBP–BcPI-PLCAA H32A. Universal primers were then used to amplify the AKAP-FRB, PEX-FRB, and PM-FRB fragments from their respective CFP-tagged recruitment constructs before inserting them in place of L10 in the L10-mVenus-T2A-sLuc-FKBP–BcPI-PLCAA H32A backbone using NdeI and PvuI restriction sites to make AKAP-FRB-mVenus-T2A-sLuc-FKBP–BcPI-PLCAA H32A, PEX-FRB-mVenus-T2A-sLuc-FKBP–BcPI-PLCAA H32A, and PM-FRB-mVenus-T2A-sLuc-FKBP–BcPI-PLCAA H32A. For recruitment constructs tagged on the N-terminus, the sLuc-FKBP–BcPI-PLCAA H32A insert from L10-mVenus-T2A-sLuc-FKBP–BcPI-PLCAA H32A was amplified to replace the sLuc-FYVEEEA1 module in the sLuc-FYVEEEA1-T2A-mVenus-Rab7 construct using NheI and SalI restriction sites to generate sLuc-FKBP–BcPI-PLCAA H32A-T2A-mVenus-Rab7. Please note that the sLuc used for this cloning step has a silent mutation that removes an internal NheI site (Baba et al., 2019). From here, unique primer sets were used to amplify the FRB-Giantintail, FRB-CyB5tail, FRB-Rab5, and FRB-Rab7 inserts from the existing recruitment constructs and replace the Rab7 module within the sLuc-FKBP–BcPI-PLCAA H32A-T2A-mVenus-Rab7 backbone. Briefly, the exchange of Rab7 for FRB-Giantintail, FRB-CyB5tail, and FRB-Rab7 was done using BspEI and KpnI restriction sites, while insertion of FRB-Rab5 required a BspEI and SacII double-digest, to produce sLuc-FKBP–BcPI-PLCAA H32A-T2A-mVenus-FRB-Giantintail, sLuc-FKBP–BcPI-PLCAA H32A-T2A-mVenus-FRB-CyB5tail, sLuc-FKBP–BcPI-PLCAA H32A-T2A-mVenus-FRB-Rab5, and sLuc-FKBP–BcPI-PLCAA H32A-T2A-mVenus-Rab7. BRET measurements detailing the FRB:FKBP dimerization kinetics made using these constructs are provided as Fig. S6.
Live-cell confocal microscopy
For imaging studies, HEK293-AT1 cells (3 × 105 cells/dish) were plated with a final volume of 1.5 ml on 29-mm circular glass-bottom culture dishes (#1.5; Cellvis) precoated with 0.01% poly-L-lysine solution (MilliporeSigma), while both the COS-7 (105 cells/dish) and HT-1080 (1.5 × 105 cells/dish) cell lines were plated without any additional coating of the culture dishes. The cells were allowed to attach overnight before transfection with plasmid DNAs (0.1–0.2 µg/well) using Lipofectamine 2000 (2–5 µl/well; Invitrogen) within a small volume of Opti-MEM (200 µl; Invitrogen) according to the manufacturer’s instructions, but with the slight modification of removing the media containing the Lipofectamine-complexed DNA at 4–6 h after transfection and replacing it with complete culture medium. In general, cell densities were always kept between 50 and 80% confluence for the day of imaging. Also, please note that studies using the rapamycin-inducible protein heterodimerization system used a 1:2:1 ratio of plasmid DNA for transfection of the FKBP-tagged enzyme (0.1 µg), FRB-labeled recruiter (0.2 µg), and indicated protein or biosensor of interest (0.1 µg; total DNA: 0.4 µg/well). After 18–20 h of transfection, cells were incubated in 1 ml modified Krebs-Ringer solution (containing 120 mM NaCl, 4.7 mM KCl, 2 mM CaCl2, 0.7 mM MgSO4, 10 mM glucose, and 10 mM Hepes and adjusted to pH 7.4), and images were acquired at room temperature using either the Zeiss LSM 710 (63×/1.40 NA Plan-Apochromat objective) or Zeiss LSM 880 (63×/1.40 NA Plan-Apochromat differential interference contrast M27 oil objective) laser-scanning confocal microscopes (Carl Zeiss Microscopy). Image acquisition was performed using the ZEN software system (Carl Zeiss Microscopy), while image preparation and analysis was done using the open-source FIJI platform (Schindelin et al., 2012). Only linear adjustments to the displayed dynamic range were allowed and, unless explicitly labeled otherwise, all of the representative images shown are of HEK293-AT1 cells.
Measurements of membrane lipid composition using BRET-based biosensors within intact cells
The construction of the BRET-based lipid reporters has been detailed above. Briefly, the design of these biosensors allows for the quantitative measurement of membrane lipid composition within defined subcellular compartments at the population scale using intact cells. This methodology relies on a plasmid design that incorporates a self-cleaving viral 2A peptide to facilitate the production of two separate proteins in transfected cells at a fixed stoichiometry; specifically, a membrane-anchored BRET acceptor (mVenus) and a Luciferase-tagged peripheral lipid-binding probe that serves as the BRET donor. BRET measurements were made at 37°C using a Tristar2 LB 942 Multimode Microplate Reader (Berthold Technologies) with customized emission filters (540/40 nm and 475/20 nm). HEK293-AT1 cells (105 cells/well) were seeded in a 200 µl total volume to white-bottom 96-well plates precoated with 0.01% poly-L-lysine solution (MilliporeSigma) and cultured overnight. Cells were then transfected with 0.25 µg of the specified BRET biosensor using Lipofectamine 2000 (1 µl/well) within OPTI-MEM (40 µl) according to the manufacturer’s protocol, but once again with the slight modification of removing the media containing the Lipofectamine-complexed DNA and replacing it with complete culture medium at between 4 and 6 h after transfection. Where indicated, additional plasmids, including components of the rapamycin-inducible heterodimerization system, were transfected together with the BRET biosensor at an empirically determined ratio of 1:1:5 for the FRB (0.05 µg), FKBP (0.05 µg), and BRET (0.25 µg) constructs, respectively (0.35 µg/well total). Between 20 and 24 h after transfection, the cells were quickly washed before being incubated for 30 min in 50 µl of modified Krebs-Ringer buffer (containing 120 mM NaCl, 4.7 mM KCl, 2 mM CaCl2, 0.7 mM MgSO4, 10 mM glucose, and 10 mM Hepes and adjusted to pH 7.4) at 37°C in a CO2-independent incubator. After the preincubation period, the cell-permeable luciferase substrate, coelenterazine h (40 µl, final concentration 5 µM), was added, and the signal from the mVenus fluorescence and sLuc luminescence were recorded using 485- and 530-nm emission filters over a 4-min baseline BRET measurement (15 s/cycle). Following the baseline recordings, where indicated, the plates were quickly unloaded for the addition of various treatments, which were prepared in a 10-µl volume of the modified Krebs-Ringer solution and added manually. Detection time was always 500 ms for each wavelength, and measurements were initiated at the 0-min demarcation indicated on all graphs and continued for 60 min (15 s/cycle) after the addition of any treatments. All measurements were performed in triplicate wells and repeated in three independent experiments. From each well, the BRET ratio was calculated by dividing the 530- and 485-nm intensities, which were then normalized to the baseline measurement. To facilitate the pooling of data from individual wells and between replicate experiments, the raw BRET ratios were processed by using a simple moving average with a four-cycle interval across the BRET kinetic. The processed BRET ratios obtained from drug-treated wells were then normalized to internal vehicle controls. Additionally, where indicated, values obtained for the same BRET biosensor from experiments using multiple recruitable enzymes or different FRB-tagged recruiters were also normalized to control measurements made simultaneously using the corresponding catalytically inactive enzyme variants.
Protein expression and purification
The mutant BcPI-PLCH82A was cloned into a pHIS-GB1 expression vector with an N-terminal His6x-GB1 solubility tag and a tobacco etch virus (TEV) cleavage site (Baumlova et al., 2014) The protein was overexpressed in Escherichia coli BL21 Star cells and subsequently purified using affinity chromatography on a nickel-charged nitrilotriacetic acid resin column (Machery-Nagel). The solubility tag was cleaved with TEV protease and the protein further purified by size exclusion chromatography (Superdex75 GL300/10/300; GE Healthcare) at 4°C in a SEC buffer (20 mM Hepes and 3 mM β-mercaptoethanol, adjusted to pH 7.0). The purified protein was concentrated to 10 mg/ml and stored at −80°C.
Crystallization and crystallographic analysis
For crystallization, BcPI-PLCH82A (10 mg/ml) was supplemented with 100 mM myo-inositol. The crystals were obtained using the sitting-drop vapor-diffusion method at 18°C by mixing 300 nl protein–ligand complex with a reservoir solution (Morpheus screen; Molecular Dimensions). The diffraction dataset was collected at BESSY-II beamline (Mueller et al., 2012). The crystals belonged to the hexagonal P6322 space group and diffracted to 2.7 Å. Data were processed in XDSAPP (Krug et al., 2012), and the structure was solved by molecular replacement using Phaser (McCoy et al., 2007) and the previously solved structure of BcPI-PLC (PDB accession no. 1GYM; Heinz et al., 1996) as a search model. Refinement of the structure was performed using Phenix (Adams et al., 2010) or Coot (Debreczeni and Emsley, 2012) to Rwork= 21.62% and Rfree= 24.47%. Protein structures were retrieved using the PDB (Berman et al., 2003) and prepared using the PyMOL Molecular Graphics System Version 2.0 (Schrödinger, LLC). The statistics of crystallographic data collection and refinement are included as Fig. S1 C, while the solved BcPI-PLCH82A structure has been deposited in the PDB database under the accession no. 6S2A.
Preparation of LUVs
All lipids used for LUV preparation were purchased from Avanti Polar Lipids, with the lone exception of the headgroup-labeled lipid, Atto647N-DOPE, which was obtained from ATTO-TEC. LUVs were prepared by mixing lipid stocks in chloroform and then drying the comixture in a stream of nitrogen. The resulting lipid film was placed into a vacuum chamber for 1 h to remove any remaining solvent before being resuspended to a final lipid concentration of 1 mM in a buffer consisting of 10 mM Tris, pH 7.5, 150 mM KCl, 3 mM β-mercaptoethanol, and 1 mM EDTA. The lipid suspension was then extruded 21 times with an Avestin Europe extruder and 100-nm porous filter membranes. The general composition of the LUVs consisted of 40% POPC (1-palmitoyl-2-oleoyl-glycero-3-phosphocholine), 20% POPS (1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine), 40% liver phosphatidylethanolamine, and 0.01% Atto647N-DOPE. For binding studies using the BcPI-PLC mutants, 10% liver PI was added to lipid mixtures in place of 10% POPS, which was selected for substitution in order to balance the percentage of charged lipids present within the LUVs. Lastly, the effects of the DAG content of LUVs on BcPI-PLC binding were also determined by substituting either 5% or 10% of the total phosphatidylethanolamine with DAG in the LUV mixture.
FCCS
Images for FCCS measurements were acquired using a Zeiss LSM 780 (40×/1.20 NA C-Apochromat W Corr M27 Objective) laser-scanning confocal microscope (Carl Zeiss Microscopy) equipped with a fluorescence lifetime imaging upgrade kit from Picoquant consisting of two -single-photon avalanche diode detectors and a HydraHarp multichannel picosecond event timer. GFP excitation was performed using the 490-nm line of the InTune Laser System (Carl Zeiss Microscopy) pulsing at a 40-MHz repetition frequency, while Atto647N-DOPE was excited continuously at 633 nm. Fluorescence behind the pinhole was split on the external -single-photon avalanche diode, in front of which 525/41-nm and 679/41-nm emission band pass filters for the GFP and Atto-647N signals, respectively, were placed. For each measurement, 100 µl of the appropriate LUVs were mixed with either GFP–BcPI-PLCH82A or GFP–BcPI-PLCANH so that the final concentration of the protein was 50 nM and the total volume was 200 µl.
The data were processed by a custom-written script in MATLAB (MathWorks) according to the correlation algorithm described in Wahl et al. (2003). While correlation of the Atto-647N signal was used to check homogeneity of the liposomes, the correlation of the GFP signal was used to quantify the protein binding to the LUVs. The autocorrelation function, G(t), was fit by a mathematical model that assumes fast and slow diffusion of the bound/unbound protein, respectively:
where Ds and As are characteristic times and corresponding amplitudes of the two diffusion components, respectively, and S stands for the structural parameter. Autocorrelation functions for the GFP signal were globally fit so that τD for the free protein and the structure parameter were obtained from the measurement of the sole protein and fixed. τD for the slow component was set as a global parameter shared for all curves. As there was no significant change of the molecular brightness of GFP, which suggests multiple proteins per LUV, the amplitudes were directly used to calculate the bound fraction:
Online supplemental material
Fig. S1 compares the overall architecture of the L. monocytogenes PI-PLC and BcPI-PLC bound to myo-inositol, as well as shows the electron density map surrounding the inositol headgroup within the BcPI-PLCH82A active site. Fig. S2 provides tables cataloguing the in vitro catalytic activity of the BcPI-PLC mutants used in this study and also details screening efforts used to tune the catalysis of the FKBP–BcPI-PLCAA scaffold. Figs. S3, S4, and S7 show the in vitro binding, subcellular localization, and BRET responses measured using the BcPI-PLCANH mutant, respectively. Fig. S5 describes the generation of the FKBP–BcPI-PLCAA scaffold through the targeted mutagenesis of hydrophobic residues within the membrane-binding interface that function to limit the cytosolic activity of the modified enzyme. Fig. S6 presents microscopy and BRET measurements defining the FRB:FKBP dimerization kinetics associated with the localized recruitment of the FKBP–BcPI-PLCAA scaffold to defined membrane compartments. Fig. S8 provides additional enzyme recruitment and lipid-binding studies targeted at investigating the steady-state levels of PI in the PM as well as the flux of PI between the ER and PM. Fig. S9 shows the kinetics of BcPI-PLCH82A levels within the PM and ER in response to stimulation with AngII. Table S1 lists the primers and strategies used for PCR-mediated cloning or site-directed mutagenesis.
Supplementary Material
lists the primers and strategies used for PCR-mediated cloning or site-directed mutagenesis.
Acknowledgments
J.G. Pemberton would like to dedicate this article to his father, the late Dr. S. George Pemberton (University of Alberta, Canada), whose insightful scientific discussions, mentorship, and unwavering support were invaluable for the completion of this study. The lessons learned from him will never be forgotten.
The authors would like to thank Helmholtz-Zentrum Berlin (HZB) for the allocation of synchrotron radiation beamtime and the laboratories of Drs. Bianxiao Cui, Gerald Hammond, T. Kendall Harden, Takanari Inoue, Jennifer Lippincott-Schwartz, Harald Stenmark, and Peter Várnai for generously providing DNA constructs. We are also grateful to former lab members Drs. Takashi Baba (Akita University, Akita, Japan) and Gergö Gulyás (Semmelweis University Medical School) for their assistance with molecular cloning, and Dr. Tara Fischer (National Institute of Neurological Disorders and Stroke, Bethesda, MD) for her careful reading of the manuscript and lively discussions. Confocal imaging was performed at the Microscopy & Imaging Core of the Eunice Kennedy Shriver National Institute of Child Health and Human Development with the kind assistance of Dr. Vincent Schram and Lynne Holtzclaw.
This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development Intramural Research Program. Additional support to J.G. Pemberton was provided by an Eunice Kennedy Shriver National Institute of Child Health and Human Development visiting fellowship and a Natural Sciences and Engineering Research Council of Canada Banting postdoctoral fellowship. J. Humpolickova, A. Eisenreichova, and E. Boura were supported by the European Regional Development Fund (OP RDE, project Chemical biology for drugging undruggable targets “ChemBioDrug” CZ.02.1.01/0.0/0.0/16_019/0000729) and the Czech Academy of Sciences (RVO: 61388963).
The authors declare no competing financial interests.
Author contributions: J.G. Pemberton, Y.J. Kim, J. Humpolickova, D.J. Toth, E. Boura, and T. Balla conceived of the experiments and developed the methods. J.G. Pemberton, Y.J. Kim, J. Humpolickova, A. Eisenreichova, E. Boura, N. Sengupta, and T. Balla performed the experiments. J.G. Pemberton, J. Humpolickova, E. Boura, and T. Balla analyzed the data. T. Balla and E. Boura acquired financial support for this study. J.G. Pemberton wrote the first draft of the manuscript. J.G. Pemberton, Y.J. Kim, J. Humpolickova, E. Boura, and T. Balla reviewed and edited the manuscript.
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221. 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahyayauch H., Sot J., Collado M.I., Huarte N., Requejo-Isidro J., Alonso A., and Goñi F.M.. 2015. End-product diacylglycerol enhances the activity of PI-PLC through changes in membrane domain structure. Biophys. J. 108:1672–1682. 10.1016/j.bpj.2015.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardail D., Privat J.P., Egret-Charlier M., Levrat C., Lerme F., and Louisot P.. 1990. Mitochondrial contact sites. Lipid composition and dynamics. J. Biol. Chem. 265:18797–18802. [PubMed] [Google Scholar]
- Asp L., Kartberg F., Fernandez-Rodriguez J., Smedh M., Elsner M., Laporte F., Bárcena M., Jansen K.A., Valentijn J.A., Koster A.J., et al. 2009. Early stages of Golgi vesicle and tubule formation require diacylglycerol. Mol. Biol. Cell. 20:780–790. 10.1091/mbc.e08-03-0256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T., Toth D.J., Sengupta N., Kim Y.J., and Balla T.. 2019. Phosphatidylinositol 4,5-bisphosphate controls Rab7 and PLEKHM1 membrane cycling during autophagosome-lysosome fusion. EMBO J. 38:e100312 10.15252/embj.2019102837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bago R., Malik N., Munson M.J., Prescott A.R., Davies P., Sommer E., Shpiro N., Ward R., Cross D., Ganley I.G., and Alessi D.R.. 2014. Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase. Biochem. J. 463:413–427. 10.1042/BJ20140889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T. 2013. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93:1019–1137. 10.1152/physrev.00028.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla A., Tuymetova G., Tsiomenko A., Várnai P., and Balla T.. 2005. A plasma membrane pool of phosphatidylinositol 4-phosphate is generated by phosphatidylinositol 4-kinase type-III alpha: studies with the PH domains of the oxysterol binding protein and FAPP1. Mol. Biol. Cell. 16:1282–1295. 10.1091/mbc.e04-07-0578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron C.L., and Malhotra V.. 2002. Role of diacylglycerol in PKD recruitment to the TGN and protein transport to the plasma membrane. Science. 295:325–328. 10.1126/science.1066759 [DOI] [PubMed] [Google Scholar]
- Baumlova A., Chalupska D., Róźycki B., Jovic M., Wisniewski E., Klima M., Dubankova A., Kloer D.P., Nencka R., Balla T., and Boura E.. 2014. The crystal structure of the phosphatidylinositol 4-kinase IIα. EMBO Rep. 15:1085–1092. 10.15252/embr.201438841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshaw P.J., Ho S.N., Crabtree G.R., and Schreiber S.L.. 1996. Controlling protein association and subcellular localization with a synthetic ligand that induces heterodimerization of proteins. Proc. Natl. Acad. Sci. USA. 93:4604–4607. 10.1073/pnas.93.10.4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H., Henrick K., and Nakamura H.. 2003. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 10:980 10.1038/nsb1203-980 [DOI] [PubMed] [Google Scholar]
- Bigay J., and Antonny B.. 2012. Curvature, lipid packing, and electrostatics of membrane organelles: defining cellular territories in determining specificity. Dev. Cell. 23:886–895. 10.1016/j.devcel.2012.10.009 [DOI] [PubMed] [Google Scholar]
- Bojjireddy N., Botyanszki J., Hammond G., Creech D., Peterson R., Kemp D.C., Snead M., Brown R., Morrison A., Wilson S., et al. 2014. Pharmacological and genetic targeting of the PI4KA enzyme reveals its important role in maintaining plasma membrane phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate levels. J. Biol. Chem. 289:6120–6132. 10.1074/jbc.M113.531426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bononi A., Bonora M., Marchi S., Missiroli S., Poletti F., Giorgi C., Pandolfi P.P., and Pinton P.. 2013. Identification of PTEN at the ER and MAMs and its regulation of Ca(2+) signaling and apoptosis in a protein phosphatase-dependent manner. Cell Death Differ. 20:1631–1643. 10.1038/cdd.2013.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossard C., Bresson D., Polishchuk R.S., and Malhotra V.. 2007. Dimeric PKD regulates membrane fission to form transport carriers at the TGN. J. Cell Biol. 179:1123–1131. 10.1083/jcb.200703166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzik K.S., and Tsai M.D.. 1994. Toward the mechanism of phosphoinositide-specific phospholipases C. Bioorg. Med. Chem. 2:49–72. 10.1016/S0968-0896(00)82002-7 [DOI] [PubMed] [Google Scholar]
- Calvo S.E., Clauser K.R., and Mootha V.K.. 2016. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 44(D1):D1251–D1257. 10.1093/nar/gkv1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazotte B. 2011. Labeling mitochondria with MitoTracker dyes. Cold Spring Harb. Protoc. 2011:990–992. [DOI] [PubMed] [Google Scholar]
- Cheng J., Goldstein R., Stec B., Gershenson A., and Roberts M.F.. 2012. Competition between anion binding and dimerization modulates Staphylococcus aureus phosphatidylinositol-specific phospholipase C enzymatic activity. J. Biol. Chem. 287:40317–40327. 10.1074/jbc.M112.395277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Goldstein R., Gershenson A., Stec B., and Roberts M.F.. 2013. The cation-π box is a specific phosphatidylcholine membrane targeting motif. J. Biol. Chem. 288:14863–14873. 10.1074/jbc.M113.466532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Chen J., Schreiber S.L., and Clardy J.. 1996. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 273:239–242. 10.1126/science.273.5272.239 [DOI] [PubMed] [Google Scholar]
- Chu B.B., Liao Y.C., Qi W., Xie C., Du X., Wang J., Yang H., Miao H.H., Li B.L., and Song B.L.. 2015. Cholesterol transport through lysosome-peroxisome membrane contacts. Cell. 161:291–306. 10.1016/j.cell.2015.02.019 [DOI] [PubMed] [Google Scholar]
- Csordás G., Várnai P., Golenár T., Roy S., Purkins G., Schneider T.G., Balla T., and Hajnóczky G.. 2010. Imaging interorganelle contacts and local calcium dynamics at the ER-mitochondrial interface. Mol. Cell. 39:121–132. 10.1016/j.molcel.2010.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G., and Vance J.E.. 1997. Import of lipids into mitochondria. Prog. Lipid Res. 36:103–130. 10.1016/S0163-7827(97)00006-4 [DOI] [PubMed] [Google Scholar]
- de Kroon A.I., Dolis D., Mayer A., Lill R., and de Kruijff B.. 1997. Phospholipid composition of highly purified mitochondrial outer membranes of rat liver and Neurospora crassa. Is cardiolipin present in the mitochondrial outer membrane? Biochim. Biophys. Acta. 1325:108–116. 10.1016/S0005-2736(96)00240-4 [DOI] [PubMed] [Google Scholar]
- Debreczeni J.É., and Emsley P.. 2012. Handling ligands with Coot. Acta Crystallogr. D Biol. Crystallogr. 68:425–430. 10.1107/S0907444912000200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz Añel A.M. 2007. Phospholipase C beta3 is a key component in the Gbetagamma/PKCeta/PKD-mediated regulation of trans-Golgi network to plasma membrane transport. Biochem. J. 406:157–165. 10.1042/BJ20070359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly M.L., Hughes L.E., Luke G., Mendoza H., ten Dam E., Gani D., and Ryan M.D.. 2001. The ‘cleavage’ activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring ‘2A-like’ sequences. J. Gen. Virol. 82:1027–1041. 10.1099/0022-1317-82-5-1027 [DOI] [PubMed] [Google Scholar]
- Downing G.J., Kim S., Nakanishi S., Catt K.J., and Balla T.. 1996. Characterization of a soluble adrenal phosphatidylinositol 4-kinase reveals wortmannin sensitivity of type III phosphatidylinositol kinases. Biochemistry. 35:3587–3594. 10.1021/bi9517493 [DOI] [PubMed] [Google Scholar]
- Duan L., Che D., Zhang K., Ong Q., Guo S., and Cui B.. 2015. Optogenetic control of molecular motors and organelle distributions in cells. Chem. Biol. 22:671–682. 10.1016/j.chembiol.2015.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Wehbi H., and Roberts M.F.. 2002. Role of tryptophan residues in interfacial binding of phosphatidylinositol-specific phospholipase C. J. Biol. Chem. 277:19867–19875. 10.1074/jbc.M200938200 [DOI] [PubMed] [Google Scholar]
- Feng J., Bradley W.D., and Roberts M.F.. 2003. Optimizing the interfacial binding and activity of a bacterial phosphatidylinositol-specific phospholipase C. J. Biol. Chem. 278:24651–24657. 10.1074/jbc.M301207200 [DOI] [PubMed] [Google Scholar]
- Fernández-Ulibarri I., Vilella M., Lázaro-Diéguez F., Sarri E., Martínez S.E., Jiménez N., Claro E., Mérida I., Burger K.N., and Egea G.. 2007. Diacylglycerol is required for the formation of COPI vesicles in the Golgi-to-ER transport pathway. Mol. Biol. Cell. 18:3250–3263. 10.1091/mbc.e07-04-0334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi A.J., Perussia B., and Goldfine H.. 1993. Listeria monocytogenes phosphatidylinositol (PI)-specific phospholipase C has low activity on glycosyl-PI-anchored proteins. J. Bacteriol. 175:8014–8017. 10.1128/JB.175.24.8014-8017.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gässler C.S., Ryan M., Liu T., Griffith O.H., and Heinz D.W.. 1997. Probing the roles of active site residues in phosphatidylinositol-specific phospholipase C from Bacillus cereus by site-directed mutagenesis. Biochemistry. 36:12802–12813. 10.1021/bi971102d [DOI] [PubMed] [Google Scholar]
- Gillooly D.J., Morrow I.C., Lindsay M., Gould R., Bryant N.J., Gaullier J.M., Parton R.G., and Stenmark H.. 2000. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19:4577–4588. 10.1093/emboj/19.17.4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godi A., Di Campli A., Konstantakopoulos A., Di Tullio G., Alessi D.R., Kular G.S., Daniele T., Marra P., Lucocq J.M., and De Matteis M.A.. 2004. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat. Cell Biol. 6:393–404. 10.1038/ncb1119 [DOI] [PubMed] [Google Scholar]
- Grabon A., Khan D., and Bankaitis V.A.. 2015. Phosphatidylinositol transfer proteins and instructive regulation of lipid kinase biology. Biochim. Biophys. Acta. 1851:724–735. 10.1016/j.bbalip.2014.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Zhang X., Seaton B.A., and Roberts M.F.. 2008. Role of helix B residues in interfacial activation of a bacterial phosphatidylinositol-specific phospholipase C. Biochemistry. 47:4201–4210. 10.1021/bi702269u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guther M.L., de Almeida M.L., Rosenberry T.L., and Ferguson M.A.. 1994. The detection of phospholipase-resistant and -sensitive glycosyl-phosphatidylinositol membrane anchors by western blotting. Anal. Biochem. 219:249–255. 10.1006/abio.1994.1264 [DOI] [PubMed] [Google Scholar]
- Hammond G.R., Fischer M.J., Anderson K.E., Holdich J., Koteci A., Balla T., and Irvine R.F.. 2012. PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science. 337:727–730. 10.1126/science.1222483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond G.R., Machner M.P., and Balla T.. 2014. A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J. Cell Biol. 205:113–126. 10.1083/jcb.201312072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harak C., Radujkovic D., Taveneau C., Reiss S., Klein R., Bressanelli S., and Lohmann V.. 2014. Mapping of functional domains of the lipid kinase phosphatidylinositol 4-kinase type III alpha involved in enzymatic activity and hepatitis C virus replication. J. Virol. 88:9909–9926. 10.1128/JVI.01063-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardeman D., Zomer H.W., Schutgens R.B., Tager J.M., and van den Bosch H.. 1990. Effect of peroxisome proliferation on ether phospholipid biosynthesizing enzymes in rat liver. Int. J. Biochem. 22:1413–1418. 10.1016/0020-711X(90)90231-Q [DOI] [PubMed] [Google Scholar]
- Heim R., Prasher D.C., and Tsien R.Y.. 1994. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl. Acad. Sci. USA. 91:12501–12504. 10.1073/pnas.91.26.12501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz D.W., Ryan M., Bullock T.L., and Griffith O.H.. 1995. Crystal structure of the phosphatidylinositol-specific phospholipase C from Bacillus cereus in complex with myo-inositol. EMBO J. 14:3855–3863. 10.1002/j.1460-2075.1995.tb00057.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz D.W., Ryan M., Smith M.P., Weaver L.H., Keana J.F., and Griffith O.H.. 1996. Crystal structure of phosphatidylinositol-specific phospholipase C from Bacillus cereus in complex with glucosaminyl(alpha 1-->6)-D-myo-inositol, an essential fragment of GPI anchors. Biochemistry. 35:9496–9504. 10.1021/bi9606105 [DOI] [PubMed] [Google Scholar]
- Heinz D.W., Essen L.O., and Williams R.L.. 1998. Structural and mechanistic comparison of prokaryotic and eukaryotic phosphoinositide-specific phospholipases C. J. Mol. Biol. 275:635–650. 10.1006/jmbi.1997.1490 [DOI] [PubMed] [Google Scholar]
- Hicks S.N., Jezyk M.R., Gershburg S., Seifert J.P., Harden T.K., and Sondek J.. 2008. General and versatile autoinhibition of PLC isozymes. Mol. Cell. 31:383–394. 10.1016/j.molcel.2008.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis J.C., and Menon A.K.. 2014. Lipid landscapes and pipelines in membrane homeostasis. Nature. 510:48–57. 10.1038/nature13474 [DOI] [PubMed] [Google Scholar]
- Hondal R.J., Zhao Z., Kravchuk A.V., Liao H., Riddle S.R., Yue X., Bruzik K.S., and Tsai M.D.. 1998. Mechanism of phosphatidylinositol-specific phospholipase C: a unified view of the mechanism of catalysis. Biochemistry. 37:4568–4580. 10.1021/bi972646i [DOI] [PubMed] [Google Scholar]
- Hu A., Zhao X.T., Tu H., Xiao T., Fu T., Wang Y., Liu Y., Shi X.J., Luo J., and Song B.L.. 2018. PIP4K2A regulates intracellular cholesterol transport through modulating PI(4,5)P2 homeostasis. J. Lipid Res. 59:507–514. 10.1194/jlr.M082149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff J. 2015. The Airyscan detector from ZEISS: confocal imaging with improved signal-to-noise ratio and super-resolution. Nat. Methods. 12:i–ii. 10.1038/nmeth.f.388 [DOI] [Google Scholar]
- Hung V., Lam S.S., Udeshi N.D., Svinkina T., Guzman G., Mootha V.K., Carr S.A., and Ting A.Y.. 2017. Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. eLife. 6:e24463 10.7554/eLife.24463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunyady L., Balla T., Nagy K., and Spät A.. 1982. Control of phosphatidylinositol turnover in adrenal glomerulosa cells. Biochim. Biophys. Acta. 713:352–357. 10.1016/0005-2760(82)90253-3 [DOI] [PubMed] [Google Scholar]
- Hunyady L., Baukal A.J., Gaborik Z., Olivares-Reyes J.A., Bor M., Szaszak M., Lodge R., Catt K.J., and Balla T.. 2002. Differential PI 3-kinase dependence of early and late phases of recycling of the internalized AT1 angiotensin receptor. J. Cell Biol. 157:1211–1222. 10.1083/jcb.200111013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikezawa H., and Taguchi R.. 1981. Phosphatidylinositol-specific phospholipase C from Bacillus cereus and Bacillus thuringiensis. Methods Enzymol. 71:731–741. 10.1016/0076-6879(81)71086-3 [DOI] [Google Scholar]
- Jeynov B., Lay D., Schmidt F., Tahirovic S., and Just W.W.. 2006. Phosphoinositide synthesis and degradation in isolated rat liver peroxisomes. FEBS Lett. 580:5917–5924. 10.1016/j.febslet.2006.09.058 [DOI] [PubMed] [Google Scholar]
- Khan H.M., He T., Fuglebakk E., Grauffel C., Yang B., Roberts M.F., Gershenson A., and Reuter N.. 2016. A Role for Weak Electrostatic Interactions in Peripheral Membrane Protein Binding. Biophys. J. 110:1367–1378. 10.1016/j.bpj.2016.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P.K., Mullen R.T., Schumann U., and Lippincott-Schwartz J.. 2006. The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J. Cell Biol. 173:521–532. 10.1083/jcb.200601036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Guzman-Hernandez M.L., and Balla T.. 2011. A highly dynamic ER-derived phosphatidylinositol-synthesizing organelle supplies phosphoinositides to cellular membranes. Dev. Cell. 21:813–824. 10.1016/j.devcel.2011.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Guzman-Hernandez M.L., Wisniewski E., and Balla T.. 2015. Phosphatidylinositol-Phosphatidic Acid Exchange by Nir2 at ER-PM Contact Sites Maintains Phosphoinositide Signaling Competence. Dev. Cell. 33:549–561. 10.1016/j.devcel.2015.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C.E., Stephens L.R., Hawkins P.T., Guy G.R., and Michell R.H.. 1987. Multiple metabolic pools of phosphoinositides and phosphatidate in human erythrocytes incubated in a medium that permits rapid transmembrane exchange of phosphate. Biochem. J. 244:209–217. 10.1042/bj2440209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu T., Kukelyansky I., McCaffery J.M., Ueno T., Varela L.C., and Inoue T.. 2010. Organelle-specific, rapid induction of molecular activities and membrane tethering. Nat. Methods. 7:206–208. 10.1038/nmeth.1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug M., Weiss M.S., Heinemann U., and Meuller U.. 2012. XDSAPP: a graphical user interface for the convenient processing of diffraction data using XDS. J. Appl. Cryst. 45:568–572. 10.1107/S0021889812011715 [DOI] [Google Scholar]
- Kuppe A., Evans L.M., McMillen D.A., and Griffith O.H.. 1989. Phosphatidylinositol-specific phospholipase C of Bacillus cereus: cloning, sequencing, and relationship to other phospholipases. J. Bacteriol. 171:6077–6083. 10.1128/JB.171.11.6077-6083.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehto M.T., and Sharom F.J.. 2002. PI-specific phospholipase C cleavage of a reconstituted GPI-anchored protein: modulation by the lipid bilayer. Biochemistry. 41:1398–1408. 10.1021/bi011579w [DOI] [PubMed] [Google Scholar]
- Lewis K.A., Garigapati V.R., Zhou C., and Roberts M.F.. 1993. Substrate requirements of bacterial phosphatidylinositol-specific phospholipase C. Biochemistry. 32:8836–8841. 10.1021/bi00085a014 [DOI] [PubMed] [Google Scholar]
- Liang J., Choi J., and Clardy J.. 1999. Refined structure of the FKBP12-rapamycin-FRB ternary complex at 2.2 A resolution. Acta Crystallogr. D Biol. Crystallogr. 55:736–744. 10.1107/S0907444998014747 [DOI] [PubMed] [Google Scholar]
- Liang H., He S., Yang J., Jia X., Wang P., Chen X., Zhang Z., Zou X., McNutt M.A., Shen W.H., and Yin Y.. 2014. PTENα, a PTEN isoform translated through alternative initiation, regulates mitochondrial function and energy metabolism. Cell Metab. 19:836–848. 10.1016/j.cmet.2014.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak V., Dahan N., Ramachandran S., Sabanay H., and Lev S.. 2005. Maintenance of the diacylglycerol level in the Golgi apparatus by the Nir2 protein is critical for Golgi secretory function. Nat. Cell Biol. 7:225–234. 10.1038/ncb1221 [DOI] [PubMed] [Google Scholar]
- Liu Z., Chen O., Wall J.B.J., Zheng M., Zhou Y., Wang L., Ruth Vaseghi H., Qian L., and Liu J.. 2017. Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector. Sci. Rep. 7:2193 10.1038/s41598-017-02460-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., and Read R.J.. 2007. Phaser crystallographic software. J. Appl. Cryst. 40:658–674. 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser J., Gerstel B., Meyer J.E., Chakraborty T., Wehland J., and Heinz D.W.. 1997. Crystal structure of the phosphatidylinositol-specific phospholipase C from the human pathogen Listeria monocytogenes. J. Mol. Biol. 273:269–282. 10.1006/jmbi.1997.1290 [DOI] [PubMed] [Google Scholar]
- Mueller U., Darowski N., Fuchs M.R., Förster R., Hellmig M., Paithankar K.S., Pühringer S., Steffien M., Zocher G., and Weiss M.S.. 2012. Facilities for macromolecular crystallography at the Helmholtz-Zentrum Berlin. J. Synchrotron Radiat. 19:442–449. 10.1107/S0909049512006395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu F., Baskin J.M., Chung J., Tanner L.B., Shui G., Lee S.Y., Pirruccello M., Hao M., Ingolia N.T., Wenk M.R., and De Camilli P.. 2012. PtdIns4P synthesis by PI4KIIIα at the plasma membrane and its impact on plasma membrane identity. J. Cell Biol. 199:1003–1016. 10.1083/jcb.201206095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto Y., and De Camilli P.. 1999. Recruitment of an alternatively spliced form of synaptojanin 2 to mitochondria by the interaction with the PDZ domain of a mitochondrial outer membrane protein. EMBO J. 18:2991–3006. 10.1093/emboj/18.11.2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi Y., Tremel S., and Williams R.L.. 2019. VPS34 complexes from a structural perspective. J. Lipid Res. 60:229–241. 10.1194/jlr.R089490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaretou C., Domin J., Cockcroft S., and Waterfield M.D.. 1997. Characterization of p150, an adaptor protein for the human phosphatidylinositol (PtdIns) 3-kinase. Substrate presentation by phosphatidylinositol transfer protein to the p150.Ptdins 3-kinase complex. J. Biol. Chem. 272:2477–2485. 10.1074/jbc.272.4.2477 [DOI] [PubMed] [Google Scholar]
- Petiot A., Ogier-Denis E., Blommaart E.F., Meijer A.J., and Codogno P.. 2000. Distinct classes of phosphatidylinositol 3¢-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 275:992–998. 10.1074/jbc.275.2.992 [DOI] [PubMed] [Google Scholar]
- Pu M., Fang X., Redfield A.G., Gershenson A., and Roberts M.F.. 2009. Correlation of vesicle binding and phospholipid dynamics with phospholipase C activity: insights into phosphatidylcholine activation and surface dilution inhibition. J. Biol. Chem. 284:16099–16107. 10.1074/jbc.M809600200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu M., Orr A., Redfield A.G., and Roberts M.F.. 2010. Defining specific lipid binding sites for a peripheral membrane protein in situ using subtesla field-cycling NMR. J. Biol. Chem. 285:26916–26922. 10.1074/jbc.M110.123083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Zhou C., and Roberts M.F.. 1998. Phosphatidylcholine activation of bacterial phosphatidylinositol-specific phospholipase C toward PI vesicles. Biochemistry. 37:6513–6522. 10.1021/bi972650u [DOI] [PubMed] [Google Scholar]
- Roberts M.F., Khan H.M., Goldstein R., Reuter N., and Gershenson A.. 2018. Search and Subvert: Minimalist Bacterial Phosphatidylinositol-Specific Phospholipase C Enzymes. Chem. Rev. 118:8435–8473. 10.1021/acs.chemrev.8b00208 [DOI] [PubMed] [Google Scholar]
- Rosivatz E., and Woscholski R.. 2011. Removal or masking of phosphatidylinositol(4,5)bisphosphate from the outer mitochondrial membrane causes mitochondrial fragmentation. Cell. Signal. 23:478–486. 10.1016/j.cellsig.2010.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saheki Y., Bian X., Schauder C.M., Sawaki Y., Surma M.A., Klose C., Pincet F., Reinisch K.M., and De Camilli P.. 2016. Control of plasma membrane lipid homeostasis by the extended synaptotagmins. Nat. Cell Biol. 18:504–515. 10.1038/ncb3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkes D., and Rameh L.E.. 2010. A novel HPLC-based approach makes possible the spatial characterization of cellular PtdIns5P and other phosphoinositides. Biochem. J. 428:375–384. 10.1042/BJ20100129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf G., Ortlund E.A., Tyeryar K.R., Mousley C.J., Ile K.E., Garrett T.A., Ren J., Woolls M.J., Raetz C.R., Redinbo M.R., and Bankaitis V.A.. 2008. Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily. Mol. Cell. 29:191–206. 10.1016/j.molcel.2007.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon E.A. 2007. Appendix 5. Gene products present in mitochondria of yeast and animal cells. Methods Cell Biol. 80:835–876. 10.1016/S0091-679X(06)80044-0 [DOI] [PubMed] [Google Scholar]
- Scipioni L., Lanzanó L., Diaspro A., and Gratton E.. 2018. Comprehensive correlation analysis for super-resolution dynamic fingerprinting of cellular compartments using the Zeiss Airyscan detector. Nat. Commun. 9:5120 10.1038/s41467-018-07513-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezgin E., and Schwille P.. 2011. Fluorescence techniques to study lipid dynamics. Cold Spring Harb. Perspect. Biol. 3:a009803 10.1101/cshperspect.a009803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., and Simons K.. 2010. Lipidomics: coming to grips with lipid diversity. Nat. Rev. Mol. Cell Biol. 11:593–598. 10.1038/nrm2934 [DOI] [PubMed] [Google Scholar]
- Shi X., Shao C., Zhang X., Zambonelli C., Redfield A.G., Head J.F., Seaton B.A., and Roberts M.F.. 2009. Modulation of Bacillus thuringiensis phosphatidylinositol-specific phospholipase C activity by mutations in the putative dimerization interface. J. Biol. Chem. 284:15607–15618. 10.1074/jbc.M901601200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicart A., Katan M., Egea G., and Sarri E.. 2015. PLCγ1 participates in protein transport and diacylglycerol production triggered by cargo arrival at the Golgi. Traffic. 16:250–266. 10.1111/tra.12246 [DOI] [PubMed] [Google Scholar]
- Sohn M., Korzeniowski M., Zewe J.P., Wills R.C., Hammond G.R.V., Humpolickova J., Vrzal L., Chalupska D., Veverka V., Fairn G.D., et al. 2018. PI(4,5)P2 controls plasma membrane PI4P and PS levels via ORP5/8 recruitment to ER-PM contact sites. J. Cell Biol. 217:1797–1813. 10.1083/jcb.201710095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subach O.M., Cranfill P.J., Davidson M.W., and Verkhusha V.V.. 2011. An enhanced monomeric blue fluorescent protein with the high chemical stability of the chromophore. PLoS One. 6:e28674 10.1371/journal.pone.0028674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szentpetery Z., Várnai P., and Balla T.. 2010. Acute manipulation of Golgi phosphoinositides to assess their importance in cellular trafficking and signaling. Proc. Natl. Acad. Sci. USA. 107:8225–8230. 10.1073/pnas.1000157107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczak A.L., Workman C.J., Wang Y., Vignali K.M., Dilioglou S., Vanin E.F., and Vignali D.A.. 2004. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat. Biotechnol. 22:589–594. 10.1038/nbt957 [DOI] [PubMed] [Google Scholar]
- Ting A.E., and Pagano R.E.. 1990. Detection of a phosphatidylinositol-specific phospholipase C at the surface of Swiss 3T3 cells and its potential role in the regulation of cell growth. J. Biol. Chem. 265:5337–5340. [PubMed] [Google Scholar]
- Tóth J.T., Gulyás G., Tóth D.J., Balla A., Hammond G.R., Hunyady L., Balla T., and Várnai P.. 2016. BRET-monitoring of the dynamic changes of inositol lipid pools in living cells reveals a PKC-dependent PtdIns4P increase upon EGF and M3 receptor activation. Biochim. Biophys. Acta. 1861:177–187. 10.1016/j.bbalip.2015.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uster P.S., and Pagano R.E.. 1986. Synthesis and properties of fluorescent analogs of cytidine diphosphate-diacylglycerol and phosphatidylinositol. In Enzymes of Lipid Metabolism II. Freysz, L., Gatt, S., Massarelli, R., and Dreyfus, H., editors. Plenum Press, New York: 493–500. 10.1007/978-1-4684-5212-9_64 [DOI] [Google Scholar]
- van Meer G., Voelker D.R., and Feigenson G.W.. 2008. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9:112–124. 10.1038/nrm2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance J.E. 2015. Phospholipid synthesis and transport in mammalian cells. Traffic. 16:1–18. 10.1111/tra.12230 [DOI] [PubMed] [Google Scholar]
- Várnai P., and Balla T.. 1998. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J. Cell Biol. 143:501–510. 10.1083/jcb.143.2.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnai P., Thyagarajan B., Rohacs T., and Balla T.. 2006. Rapidly inducible changes in phosphatidylinositol 4,5-bisphosphate levels influence multiple regulatory functions of the lipid in intact living cells. J. Cell Biol. 175:377–382. 10.1083/jcb.200607116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Várnai P., Tóth B., Tóth D.J., Hunyady L., and Balla T.. 2007. Visualization and manipulation of plasma membrane-endoplasmic reticulum contact sites indicates the presence of additional molecular components within the STIM1-Orai1 Complex. J. Biol. Chem. 282:29678–29690. 10.1074/jbc.M704339200 [DOI] [PubMed] [Google Scholar]
- Várnai P., Gulyás G., Tóth D.J., Sohn M., Sengupta N., and Balla T.. 2017. Quantifying lipid changes in various membrane compartments using lipid binding protein domains. Cell Calcium. 64:72–82. 10.1016/j.ceca.2016.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S., Dhand R., Vanhaesebroeck B., MacDougall L.K., Stein R., Zvelebil M.J., Domin J., Panaretou C., and Waterfield M.D.. 1995. A human phosphatidylinositol 3-kinase complex related to the yeast Vps34p-Vps15p protein sorting system. EMBO J. 14:3339–3348. 10.1002/j.1460-2075.1995.tb07340.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volwerk J.J., Wetherwax P.B., Evans L.M., Kuppe A., and Griffith O.H.. 1989. Phosphatidylinositol-specific phospholipase C from Bacillus cereus: improved purification, amino acid composition, and amino-terminal sequence. J. Cell. Biochem. 39:315–325. 10.1002/jcb.240390311 [DOI] [PubMed] [Google Scholar]
- Volwerk J.J., Filthuth E., Griffith O.H., and Jain M.K.. 1994. Phosphatidylinositol-specific phospholipase C from Bacillus cereus at the lipid-water interface: interfacial binding, catalysis, and activation. Biochemistry. 33(12):3464–3474. 10.1021/bi00178a002 [DOI] [PubMed] [Google Scholar]
- Wahl M., Gregor I., Patting M., and Enderlein J.. 2003. Fast calculation of fluorescence correlation data with asynchronous time-correlated single-photon counting. Opt. Express. 11:3583–3591. 10.1364/OE.11.003583 [DOI] [PubMed] [Google Scholar]
- Wenk M.R. 2010. Lipidomics: new tools and applications. Cell. 143:888–895. 10.1016/j.cell.2010.11.033 [DOI] [PubMed] [Google Scholar]
- Wills R.C., Goulden B.D., and Hammond G.R.V.. 2018. Genetically encoded lipid biosensors. Mol. Biol. Cell. 29:1526–1532. 10.1091/mbc.E17-12-0738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Watras J., and Loew L.M.. 2003. Kinetic analysis of receptor-activated phosphoinositide turnover. J. Cell Biol. 161:779–791. 10.1083/jcb.200301070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Pu M., Khan H.M., Friedman L., Reuter N., Roberts M.F., and Gershenson A.. 2015. Quantifying transient interactions between Bacillus phosphatidylinositol-specific phospholipase-C and phosphatidylcholine-rich vesicles. J. Am. Chem. Soc. 137:14–17. 10.1021/ja508631n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zewe J.P., Wills R.C., Sangappa S., Goulden B.D., and Hammond G.R.. 2018. SAC1 degrades its lipid substrate PtdIns4P in the endoplasmic reticulum to maintain a steep chemical gradient with donor membranes. eLife. 7:e35588 10.7554/eLife.35588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wehbi H., and Roberts M.F.. 2004. Cross-linking phosphatidylinositol-specific phospholipase C traps two activating phosphatidylcholine molecules on the enzyme. J. Biol. Chem. 279:20490–20500. 10.1074/jbc.M401016200 [DOI] [PubMed] [Google Scholar]
- Zhang L., Malik S., Pang J., Wang H., Park K.M., Yule D.I., Blaxall B.C., and Smrcka A.V.. 2013. Phospholipase Cε hydrolyzes perinuclear phosphatidylinositol 4-phosphate to regulate cardiac hypertrophy. Cell. 153:216–227. 10.1016/j.cell.2013.02.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X.H., Bondeva T., and Balla T.. 2000. Characterization of recombinant phosphatidylinositol 4-kinase beta reveals auto- and heterophosphorylation of the enzyme. J. Biol. Chem. 275:14642–14648. 10.1074/jbc.275.19.14642 [DOI] [PubMed] [Google Scholar]
- Zhou C., Qian X., and Roberts M.F.. 1997a Allosteric activation of phosphatidylinositol-specific phospholipase C: specific phospholipid binding anchors the enzyme to the interface. Biochemistry. 36:10089–10097. 10.1021/bi970846o [DOI] [PubMed] [Google Scholar]
- Zhou C., Wu Y., and Roberts M.F.. 1997b Activation of phosphatidylinositol-specific phospholipase C toward inositol 1,2-(cyclic)-phosphate. Biochemistry. 36:347–355. 10.1021/bi960601w [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists the primers and strategies used for PCR-mediated cloning or site-directed mutagenesis.