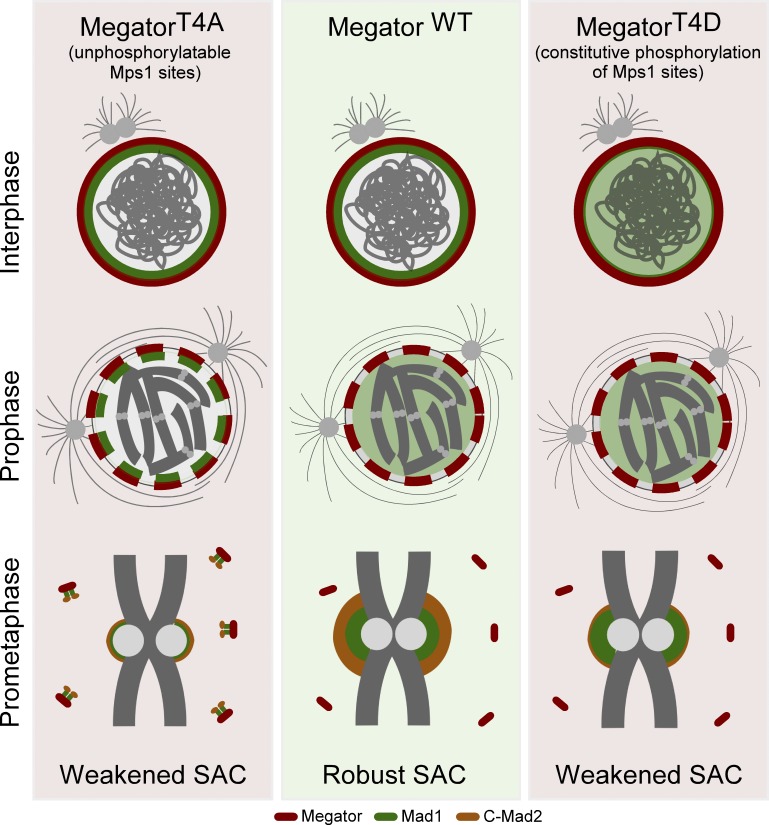
The spindle assembly checkpoint requires recruitment of Mad1-C-Mad2 to unattached kinetochores but also Mad1 binding to Megator/Tpr at nuclear pore complexes during interphase. Cunha-Silva et al. provide evidence that this spatiotemporal redistribution is due to Mps1-mediated dissociation of Mad1 from Megator/Tpr during prophase, which is critical for Mad1-C-Mad2 accumulation at unattached kinetochores and for the accuracy of chromosome segregation in vivo.
Abstract
The spindle assembly checkpoint (SAC) relies on the recruitment of Mad1-C-Mad2 to unattached kinetochores but also on its binding to Megator/Tpr at nuclear pore complexes (NPCs) during interphase. However, the molecular underpinnings controlling the spatiotemporal redistribution of Mad1-C-Mad2 as cells progress into mitosis remain elusive. Here, we show that activation of Mps1 during prophase triggers Mad1 release from NPCs and that this is required for kinetochore localization of Mad1-C-Mad2 and robust SAC signaling. We find that Mps1 phosphorylates Megator/Tpr to reduce its interaction with Mad1 in vitro and in Drosophila cells. Importantly, preventing Mad1 from binding to Megator/Tpr restores Mad1 accumulation at kinetochores, the fidelity of chromosome segregation, and genome stability in larval neuroblasts of mps1-null mutants. Our findings demonstrate that the subcellular localization of Mad1 is tightly coordinated with cell cycle progression by kinetochore-extrinsic activity of Mps1. This ensures that both NPCs in interphase and kinetochores in mitosis can generate anaphase inhibitors to efficiently preserve genomic stability.
Graphical Abstract
Introduction
The spindle assembly checkpoint (SAC) safeguards eukaryotic cells against chromosome missegregation by restraining the transition to anaphase in the presence of unattached kinetochores (KTs). Pivotal to this signaling pathway is the Mad1-C-Mad2 heterotetramer that catalyzes the structural conversion of open-Mad2 (O-Mad2) into closed-Mad2 (C-Mad2; De Antoni et al., 2005). This is the rate-limiting step in the assembly of the mitotic checkpoint complex (MCC), a diffusible tetrameric complex (C-Mad2-Cdc20-BubR1-Bub3) that inhibits anaphase-promoting complex/cyclosome–dependent sister chromatid separation and mitotic exit (De Antoni et al., 2005; Simonetta et al., 2009; Faesen et al., 2017). The strength of SAC response is dictated by the amount of Mad1-C-Mad2 present at KTs (Collin et al., 2013; Dick and Gerlich, 2013; Heinrich et al., 2014). However, robust SAC signaling also requires Mad1-C-Mad2 at nuclear pore complexes (NPCs) during interphase. Mad1 binds to the nuclear basket nucleoporin Megator/Tpr (Scott et al., 2005; Lee et al., 2008; De Souza et al., 2009; Lince-Faria et al., 2009; Schweizer et al., 2013; Rodriguez-Bravo et al., 2014), which regulates Mad1-C-Mad2 proteostasis to ensure that sufficient levels of this heterotetramer are available before mitosis (Schweizer et al., 2013). Moreover, it was proposed that Mad1-C-Mad2 at NPCs also converts O-Mad2 into C-Mad2, hence providing a scaffold for MCC assembly in interphase (Rodriguez-Bravo et al., 2014). This premitotic MCC is thought to operate as a timer to inhibit anaphase-promoting complex/cyclosome during early mitosis until KTs are able to instate efficient SAC activation (Sudakin et al., 2001; Meraldi et al., 2004; Malureanu et al., 2009; Maciejowski et al., 2010; Rodriguez-Bravo et al., 2014; Kim et al., 2018). Notwithstanding its importance for mitotic fidelity, how the subcellular redistribution of Mad1-C-Mad2 is coordinated with cell cycle progression remains elusive. Particularly, whether and how regulatory events at NPCs affect Mad1-C-Mad2 KT localization has not been established so far. We set out to address these questions in Drosophila melanogaster, where the multisequential phosphorylation cascade controlling Mad1 KT localization through the Mps1-Knl1-Bub1 pathway (London et al., 2012; Shepperd et al., 2012; Yamagishi et al., 2012; Primorac et al., 2013; London and Biggins, 2014; Vleugel et al., 2015; Mora-Santos et al., 2016; Faesen et al., 2017; Ji et al., 2017; Qian et al., 2017; Zhang et al., 2017; Rodriguez-Rodriguez et al., 2018) is inherently absent (Schittenhelm et al., 2009; Conde et al., 2013). This reduces KT-associated complexity, hence providing a simpler naturally occurring system to uncover the potential role of KT-extrinsic mechanisms in Mad1-C-Mad2 subcellular distribution throughout the cell cycle and their significance for SAC signaling and genomic integrity in vivo.
Results and discussion
Mps1 triggers Mad1 exclusion from NPCs during prophase
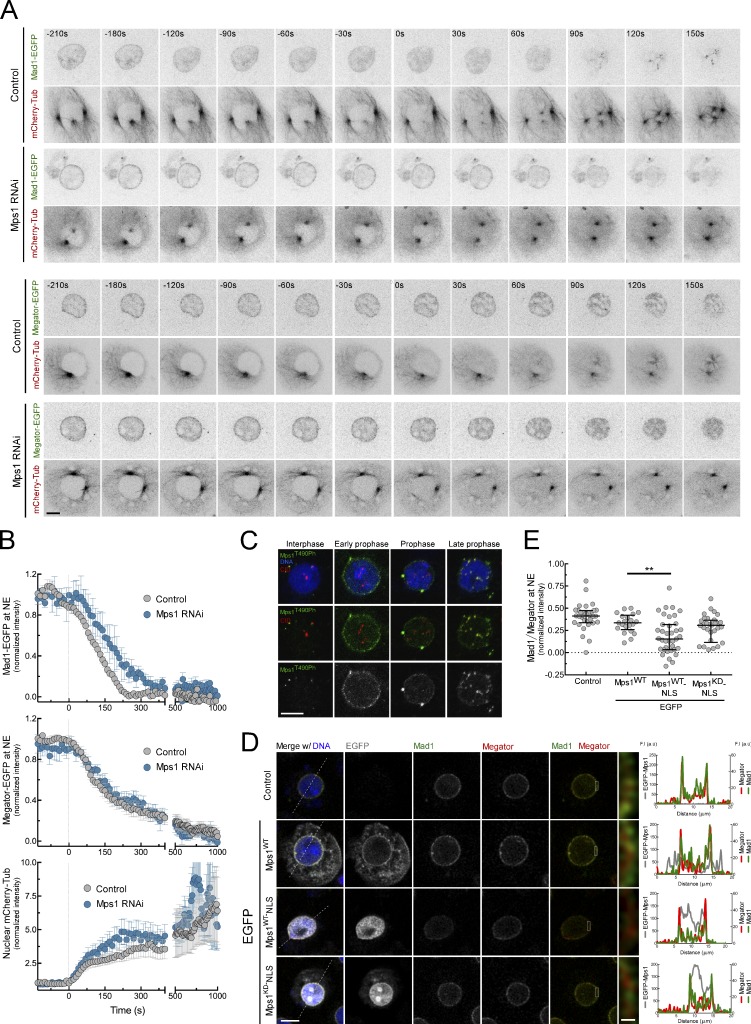
To investigate the subcellular redistribution of Mad1 during mitotic entry, we monitored with high-temporal resolution the localization of Mad1 and Megator in Drosophila S2 cells (Fig. 1, A and B). In control cells, Mad1–EGFP signal at the nuclear envelope (NE) begins to fade during early prophase, whereas Megator-EGFP persists until tubulin becomes detectable in the nucleus, an early event of nuclear envelope breakdown (NEB). However, Mps1 depletion significantly delays Mad1-EGFP dissociation from the NE, with no discernible impact on Megator-EGFP dynamics (Fig. 1, A and B), suggesting that Mad1 relocation from NPCs is triggered before NEB in an Mps1-dependent manner. Consistently, staining with a phospho-specific antibody that recognizes the activating autophosphorylation (T490Ph) of Mps1 T-loop (Jelluma et al., 2008; Moura et al., 2017) reveals that Mps1 is active at NPCs during prophase (Fig. 1 C). We then tested whether inducing premature activation of Mps1 in interphase displaces Mad1 from NPCs. As Mps1 is excluded from the nucleus until late G2/early prophase (Zhang et al., 2011; Jia et al., 2015), we promoted its nuclear import by fusing it with the SV40 large T-antigen nuclear localization signal (EGFP-Mps1WT-NLS). Overexpression of EGFP-Mps1WT-NLS elicits nuclear activation of Mps1 (Fig. S1 A) and decreases Mad1 levels at NPCs of interphase cells (Fig. 1, D and E), unlike catalytic dead EGFP-Mps1KD-NLS or EGFP-Mps1WT (Fig. 1, D and E), which, although active in the cytoplasm, fails to reach the nucleoplasmic side of NPCs (Fig. S1 A). Collectively, these results demonstrate that timely control of Mps1 nuclear import and activation triggers Mad1 dissociation from NPCs during early prophase before NEB.
Figure 1.
Msp1 promotes the dissociation of Mad1 from NPCs during prophase. (A and B) Mitotic progression (A) and quantification (B) of Mad1-EGFP (n ≥ 7 cells) or Megator-EGFP (n ≥ 6 cells) levels at NE and of mCherry-tubulin levels in the nucleus of control and Mps1-depleted Drosophila S2 cells. Time 0 corresponds to NEB. (C) Immunofluorescence images of Mps1T490Ph localization in interphase and prophase S2 cells. (D and E) Immunofluorescence images (D) and quantifications (E) of Mad1 levels at NE of interphase control S2 cells and interphase S2 cells expressing the indicated transgenes. Graphs in D represent the intensity profiles of EGFP-Mps1, Mad1, and Megator signal along the dotted lines (n ≥ 21 cells). In B, data are presented as mean ± SD; in E, data are presented as median with the interquartile range. **, P < 0.005 (Kruskal–Wallis, Dunn’s multiple comparison test). Scale bars, 5 µm (inset, 0.5 µm).
Figure S1.
Additional information related to Figs. 1, 2, and 3. (A) Immunofluorescence images of EGFP-Mps1, Mps1T490Ph, and Megator localization pattern in interphase control S2 cells and interphase S2 cells expressing EGFP-Mps1WT, EGFP-Mps1WT-NLS, or EGFP-Mps1KD-NLS. To prevent nuclear export, cultured cells were treated with 10 µM Leptomycin B for 3 h. Graphs represent the intensity profiles of GFP-Mps1, Mps1T490Ph, and Megator signal along the dotted lines. (B) Pull-downs of the indicated recombinant MBP-Megator fragments by bead-immobilized 6xHis-Mad11–493 or 6xHis-BubR1359–696 (negative control). Beads (B) and flow-through (FT) were blotted for the indicated proteins. (C) Pull-downs of recombinant purified MBP-Megator1,187–1,655/T2D, MBP-Megator1,187–1,655/T3D, MBP-Megator1,187–1,655/T4D and MBP-Megator1,187–1,655/T4A by bead-immobilized 6xHis-Mad11–493 or 6xHis-BubR11–358 (negative control). B and FT were blotted for the indicated proteins. (D) Quantification of MBP-Megator binding to 6xHis-Mad11–493 from pull-downs in C. The graph represents the ratio between the signal intensities of MBP-Megator and 6xHis-Mad11–493. The value obtained for MBP-Megator1,187–1,655/T4A was set to 1. (E and F) Representative immunofluorescence images (E) and corresponding quantifications (F) of Mad1 at the NE of interphase S2 cells depleted of endogenous Megator and expressing the indicated Megator-EGFP transgenes. Mad1 fluorescence intensities were determined relative to Nup107 signal (n ≥ 24 cells). In F, data are presented as mean ± SD. ***, P < 0.001 (Student’s t test). Scale bars, 5 µm (inset, 0.5 µm).
Mps1-mediated phosphorylation of Megator disrupts its interaction with Mad1
Since Megator localizes Mad1 to NPCs (Scott et al., 2005; Lee et al., 2008; De Souza et al., 2009; Lince-Faria et al., 2009; Schweizer et al., 2013; Rodriguez-Bravo et al., 2014), we next sought to examine whether Mps1 activity directly affects this interaction. We found that Megator coimmunoprecipitates with Mad1 in mitotically enriched S2 cell lysates only if Mps1 is depleted (Fig. 2 A). Moreover, knocking down Mps1 reduced Megator hyperphosphorylation (Fig. 2 B), which prompted us to test Megator as a direct substrate of Mps1. In vitro kinase assays and mass spectrometry (MS) analysis using three recombinant coiled-coil fragments of Megator, which we confirmed to directly interact with Mad1 N-terminus (Fig. S1 B; Lee et al., 2008), revealed that GST-Mps1 phosphorylates Megator on T1259, T1302, T1338, and T1390 (Fig. 2, C and D). Notably, 6xHis-Mad11–493 failed to bind as efficiently to MBP-Megator1,187–1,655/WT previously phosphorylated by GST-Mps1 or to phosphomimetic MBP-Megator1,187–1,655/T4D in which the four phosphorylation sites were converted to aspartates. Since Megator T1259 is adjacent to a conserved threonine in vertebrate orthologues and T1390 is well conserved among eukaryotes, we assessed whether phosphorylation of these two residues was sufficient to impair the interaction with Mad1. Contrasting with MBP-Megator1,187–1,655/T4D, the double phosphomimetic for T1259 and T1390 (MBP-Megator1,187–1,655/T2D) is efficiently pulled down by 6xHis-Mad11–493 (Fig. S1, C and D). However, introducing an additional phosphomimetic mutation on T1338 (MBP-Megator1,187–1,655/T3D) prevents binding to Mad1 N-terminus (Fig. S1, C and D). Thus, phosphorylation of Megator on T1259, T1338, and T1390 is sufficient to negatively regulate the association of its central coiled-coil domain with Mad1 N-terminus in vitro. Although T1338 is not conserved, it is replaced by an acidic residue in vertebrate Tpr, indicating that this phospho-regulation may be conserved in vertebrates.
Figure 2.
Msp1-mediated phosphorylation of Megator disrupts its interaction with Mad1. (A and B) Western blot analysis of Mad1 immunoprecipitates (IP; A) and Megator hyperphosphorylation (B) from lysates of asynchronous and mitotically enriched (+ colchicine) control and Mps1-depleted S2 cells. When indicated, lysates were treated with λ-phosphatase (λPP). (C) In vitro kinase assay with recombinant MBP-Megator fragments and GST-Mps1 in the presence of [γ-32P]ATP. (D) Schematic representation of Drosophila Megator (ELM resource) and Clustal Omega (EMBL-EBI) local sequence alignment for the indicated Megator/Tpr orthologues. Residues conservation: *, fully conserved;:, strongly similar properties;., weakly similar properties. Residues phosphorylated by Mps1 (P) were identified by MS analysis after in vitro kinase assay. (E and F) Pull-downs of recombinant MBP-Megator1,187–1,655 fragments by bead-immobilized 6xHis-Mad11–493 or 6xHis-BubR11–358 (negative control; E) and corresponding quantifications (F) from two independent experiments. B, beads; FT, flow-through. (G and H) Immunofluorescence images (G) and schematic representation (H) of EGFP-Megator1,187–1,655 clustering by LARIAT in mitotic S2 cells. EGFP-aPKC was used as negative control. (I and J) Quantification of Mad1 levels at EGFP-Megator1,187–1,655 clusters (n ≥ 114 clusters; I) and at KTs (n ≥ 64 KTs; J). In F, I, and J, data are presented as mean ± SD. **, P < 0.01; ****, P < 0.0001 (Kruskal–Wallis, Dunn’s multiple comparison test). Scale bars, 5 µm (inset, 1 µm).
To examine Mad1–Megator interaction in mitotic cells, we used LARIAT, a CRY2-based optogenetic tool that sequesters target proteins into clusters (Kennedy et al., 2010; Lee et al., 2014). This can uncover protein–protein interactions through coclustering (Osswald et al., 2019). We used CRY2 tagged with an anti-GFP nanobody to cluster WT, phosphodefective (T4A), and phosphomimetic (T4D) versions of EGFP-Megator1,187–1,655 (Fig. 2, G and H). Immunofluorescence analysis revealed limited association of Mad1 with EGFP-Megator1,187–1,655/WT clusters in the cytoplasm of colchicine-treated S2 cells (Fig. 2, G and I). However, Mad1 coclustering with EGFP-Megator1,187–1,655/WT increased significantly upon depletion of Mps1, and similar levels of Mad1 were observed at EGFP-Megator1,187–1,655/T4A clusters (Fig. 2, G and I). Importantly, Mad1 failed to associate with EGFP-Megator1,187–1,655/T4D clusters, even after Mps1 knockdown (Fig. 2, G and I). Collectively, these results demonstrate that Mps1-mediated phosphorylation of Megator on T1259, T1302, T1338, and T1390 prevents it from binding Mad1 during mitosis.
Recruitment of Mad1 to unattached KTs requires its dissociation from Megator
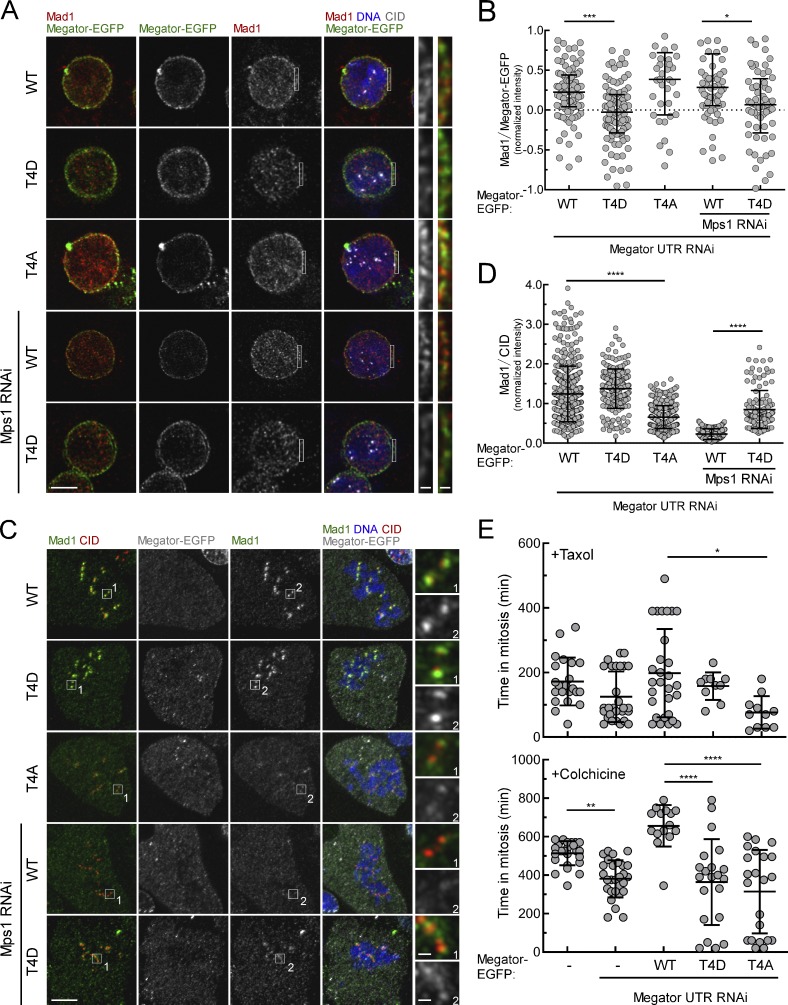
We then sought to examine the relevance of disengaging Mad1 from Megator in mitosis. In the LARIAT experiment, we observed that Mad1 levels at EGFP-Megator1,187–1,655 clusters and at unattached KTs are inversely correlated (Fig. 2, G–J), which suggested that retaining Mad1 with Megator during mitosis precludes its proper recruitment to KTs. To address this further, we generated stable S2 cell lines transfected with full-length Megator phosphomutants and depleted endogenous Megator. After 24-h induction, all transgenes were expressed at endogenous levels and localized at the NE of interphase cells (Fig. 3 A and Fig. 4, A and B). MegatorT4D-EGFP failed to recruit Mad1 to NPCs as efficiently as MegatorWT-EGFP (Fig. 3, A and B; and Fig. S1, E and F), further confirming that phosphorylation of Megator inhibits its interaction with Mad1. Conversely, MegatorT4A-EGFP was able to restore Mad1 association with NPCs (Fig. 3, A and B) but impaired Mad1 accumulation at unattached KTs. Expression of MegatorT4A-EGFP in colchicine-treated cells resulted in a twofold reduction of KT-associated Mad1 levels compared with MegatorWT-EGFP cells (Fig. 3, C and D). As expected, knocking down Mps1 abrogated Mad1 KT localization in MegatorWT-EGFP cells (Fig. 3, C and D). However, MegatorT4D-EGFP cells were still partially competent in recruiting Mad1 to unattached KTs upon depletion of Mps1 (Fig. 3, C and D). Collectively, these results support that full KT recruitment of Mad1 requires its dissociation from Megator driven by Mps1-mediated phosphorylation.
Figure 3.
Recruitment of Mad1 to unattached KTs and robust SAC signaling require phosphorylation of Megator by Mps1. (A and B) Immunofluorescence images (A) and quantifications (B) of Mad1 at NE of interphase S2 cells depleted of endogenous Megator and expressing Megator-EGFP transgenes (n ≥ 41 cells). (C and D) Immunofluorescence images (C) and corresponding quantification (D) of Mad1 at unattached KTs of S2 cells depleted of endogenous Megator and expressing Megator-EGFP transgenes (n ≥ 125 KTs). (E) Mitotic timings of control and Megator-depleted S2 cells expressing Megator-EGFP transgenes upon addition of taxol (100 nM; n ≥ 11 cells) or colchicine (30 µM; n ≥ 15 cells). In B, data are presented as median with interquartile range; in D and E, data are presented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (Kruskal–Wallis, Dunn’s multiple comparison test). Scale bars, 5 µm (inset, 0.5 µm).
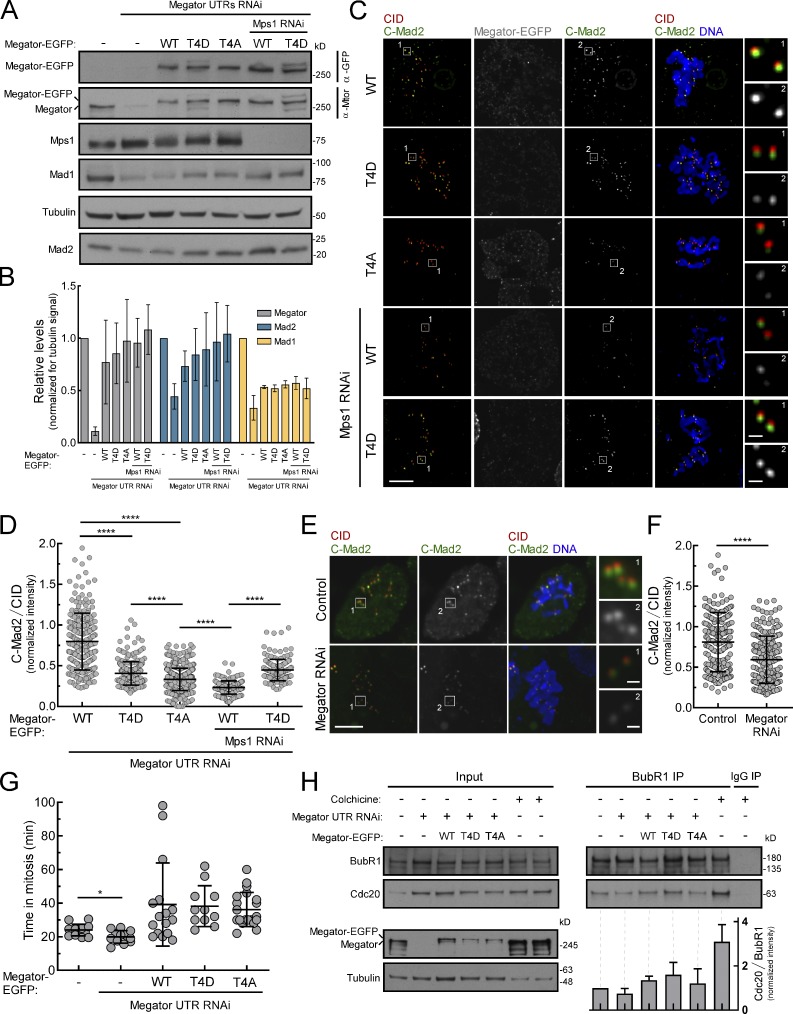
Figure 4.
Constitutive impairment of Mad1–Megator interaction reduces the levels of C-Mad2 at KTs. (A and B) Western blots (A) and quantifications (B) of Megator, Mad1, and Mad2 protein levels in lysates from control and Megator-depleted S2 cells expressing Megator-EGFP transgenes. The graph represents the signal intensities of Megator, Mad1, and Mad2 relative to tubulin from at least two independent experiments. The mean value for control parental cells was set to 1. (C and D) Immunofluorescence images (C) and quantifications (D) of C-Mad2 at unattached KTs of S2 cells depleted of endogenous Megator and expressing Megator-EGFP transgenes (n ≥ 148 KTs). (E and F) Immunofluorescence images (E) and quantifications (F) of C-Mad2 levels at unattached KTs of control and Megator-depleted S2 cells (n ≥ 224 KTs). (G) Mitotic timings of control and Megator-depleted S2 cells expressing Megator-EGFP transgenes under unperturbed (absence of spindle poisons) conditions (n ≥ 11 cells). (H) Immunoprecipitates (IP) of BubR1 from lysates of control and Megator-depleted S2 cells expressing Megator-EGFP transgenes. When indicated, cells were incubated with colchicine for 18 h. The graph represents the ratio between the signal intensities of Cdc20 and BubR1 present in BubR1 IPs from two independent experiments. The values for control parental cells were set to 1. In B, data are presented as mean ± SEM; in D, F, G, and H, data are presented as mean ± SD. *, P < 0.05; ****, P < 0.0001 (Kruskal–Wallis, Dunn’s multiple comparison test in D and Student’s t test in F and G). Scale bars, 5 µm (inset, 0.5 µm).
Dissociation of Mad1 from Megator in mitosis is required for functional SAC signaling
To examine the importance of Mps1-mediated phosphorylation of Megator for SAC signaling, we assessed by live-cell imaging the capacity of Megator phosphomutants to arrest in mitosis when incubated with spindle poisons (Fig. 3 E). In line with SAC proficiency, cells expressing MegatorWT-EGFP significantly delayed the transition to anaphase in the presence of colchicine (∼640 min) or taxol (∼180 min). In contrast, MegatorT4A-EGFP cells, although able to halt mitotic progression to some extent in response to unattached KTs (∼360 min in colchicine) or decreased microtubules dynamics (∼87 min in taxol), failed to maintain this arrest for the same amount of time as MegatorWT-EGFP cells (Fig. 3 E). This weakened SAC correlates with the observed reduction (∼50%) in Mad1 levels at unattached KTs (Fig. 3, C and D). Thus, we reason that phosphorylation of Megator by Mps1 is required to provide the KT with a sufficient amount of Mad1 that ensures a robust SAC response.
Constitutive phosphorylation of Megator reduces C-Mad2 levels at KTs and compromises SAC strength
Despite being fully competent in recruiting Mad1 to unattached KTs (Fig. 3, C and D), cells expressing MegatorT4D-EGFP also fail to arrest in mitosis in response to colchicine as efficiently as MegatorWT-EGFP cells (Fig. 3 E). Hence, constitutive phosphorylation of Megator is equally deleterious for SAC. Mad1 and Mad2 form a highly stable complex in vitro (Sironi et al., 2002; De Antoni et al., 2005; Vink et al., 2006) and interact with each other throughout the cell cycle (Chen et al., 1998; Chung and Chen, 2002; Fava et al., 2011; Schweizer et al., 2013). In human cells, Tpr has been proposed to protect Mad1-C-Mad2 from proteolytic degradation (Schweizer et al., 2013). We confirmed in Drosophila cells a significant reduction in the total levels of Mad1 and Mad2 following Megator depletion (Fig. 4, A and B; and Fig. S2 D). However, the levels of both proteins are partially rescued to a similar extent regardless of the Megator-EGFP transgene expressed (Fig. 4, A and B), which argues against altered proteostasis as the main cause underlying the weakened SAC function in MegatorT4D-EGFP cells.
Figure S2.
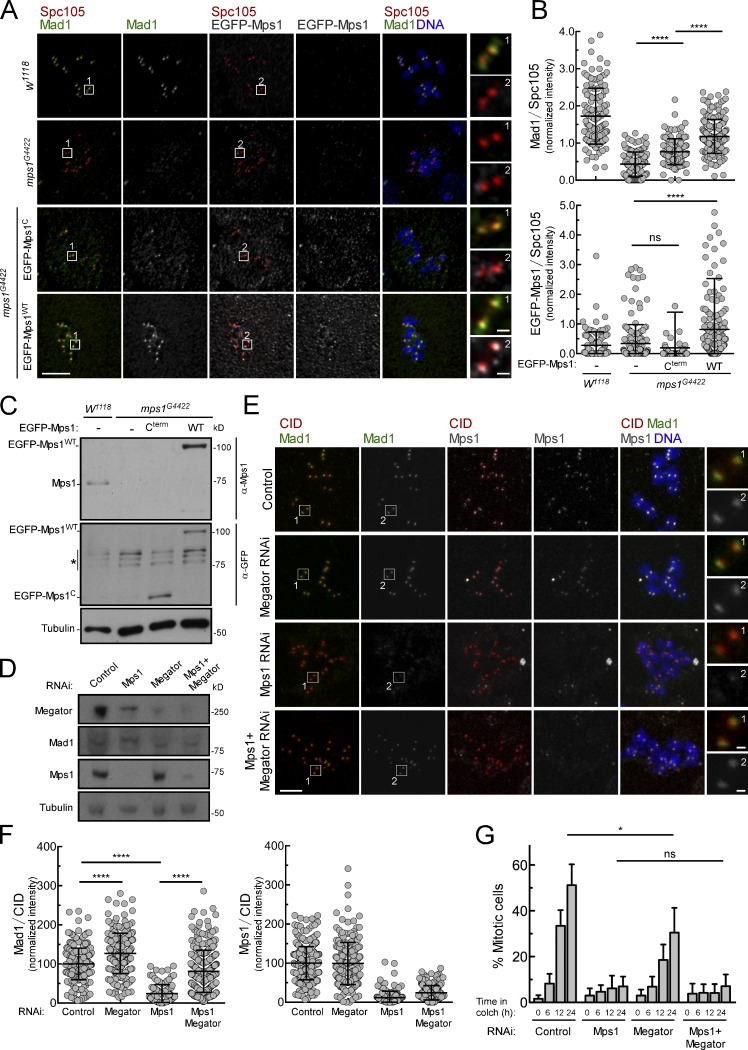
KT-extrinsic activity of Mps1 contributes to Mad1 KT recruitment. (A and B) Immunofluorescence images (A) and quantifications (B) of Mad1 and Mps1 levels at unattached KTs of neuroblasts from w1118 or homozygous mps1G4422 flies. When indicated, EGFP-Mps1-Cterm or EGFP-Mps1-WT transgenes were expressed under control of Mps1 native promoter in a homozygous mps1G4422 background. To generate unattached KTs, neuroblasts were incubated with colchicine (50 µM) for 90 min. Mad1 and Mps1 fluorescence intensities were determined relative to Spc105 signal (n ≥ 106 KTs). (C) Western blot analysis of endogenous Mps1, EGFP-Mps1-WT, and EGFP-Mps1-Cterm levels in total lysates of third instar larval brains from A. (D) Western blot analysis of Mps1, Megator, and Mad1 relative levels in control S2 cells and in cells depleted of the indicated proteins. Cells were incubated with MG123 (20 µM) for 1 h and with colchicine (30 µM) for 2 h. Asterisk denotes bands resulting from unspecific anti-GFP blotting. (E and F) Immunofluorescence images (E) and quantifications (F) of Mad1 and Mps1 levels at unattached KTs of control S2 cells and cells depleted of the indicated proteins. Cells were incubated with MG123 (20 µM) for 3 h and with colchicine (30 µM) for 2 h. Mad1 and Mps1 fluorescence intensities were determined relative to CID signal (n ≥ 109 KT for Mad1, n ≥ 139 KTs for Mps1). (G) Mitotic index quantification based on H3Ser10Ph staining of control S2 cells and cells depleted of the indicated proteins. Cells were incubated with colchicine (30 µM) for the indicated time periods. In B, F, and G, data are presented as mean ± SD. *, P < 0.05; ****, P < 0.0001 (Kruskal–Wallis, Dunn’s multiple comparison test). Scale bars, 5 µm (inset, 0.5 µm).
Using an antibody specific for C-Mad2 (Fava et al., 2011) we detected a significant decrease in C-Mad2 levels at unattached KTs of cells expressing MegatorT4D-EGFP compared with MegatorWT-EGFP cells (Fig. 4, C–F). We envisage that, in vivo, Megator/Tpr may function as a scaffold to assemble the Mad1-C-Mad2 complex before its targeting to KTs. Hence, while Mad1 dissociation from Megator is required for efficient KT recruitment of Mad1-C-Mad2, prematurely abrogating Mad1-Megator interaction during interphase is detrimental for SAC, as it may compromise the assembly of Mad1-C-Mad2 (Schweizer et al., 2013). Accordingly, a weakened SAC function is also observed in parental S2 cells and human cultured cells depleted of Megator (Figs. 3 E and Fig. S2 G) or Tpr (Schweizer et al., 2013; Rodriguez-Bravo et al., 2014), respectively. Furthermore, disrupting Mad1 localization from NPCs may also affect the formation of premitotic MCC (Rodriguez-Bravo et al., 2014), which is required for full SAC activity and to define the minimum length of time in mitosis in the absence of KT–microtubule attachment problems (Rodriguez-Bravo et al., 2014). Cells expressing MegatorT4D-EGFP, however, fail to exit faster from mitosis (Fig. 4 G), contrasting with Megator-depleted S2 cells (Fig. 4 G) or human cells in which Mad1–Tpr interaction is impaired (Rodriguez-Bravo et al., 2014). This prompted us to examine the assembly of interphase MCC in asynchronous cultured S2 cells (∼97% interphase cells). While depletion of Megator reduced the levels of BubR1-bound Cdc20 in interphase lysates, expression of MegatorT4D-EGFP restored premitotic MCC levels to a similar extent as MegatorWT-EGFP (Fig. 4 H). These data suggest that the residual pool of Mad1 still present at NPCs when MegatorT4D-EGFP is expressed (Fig. S1, E and F) may generate sufficient MCC during interphase to support normal mitotic timing in the absence of spindle poisons. These cells, however, fail to exhibit a robust KT-dependent SAC response (Fig. 3 E) due to limited association of C-Mad2 with KT-bound Mad1 (Fig. 4, C and D).
Mad1 release from NPCs rescues chromosome missegregation and aneuploidy in mps1-null neuroblasts
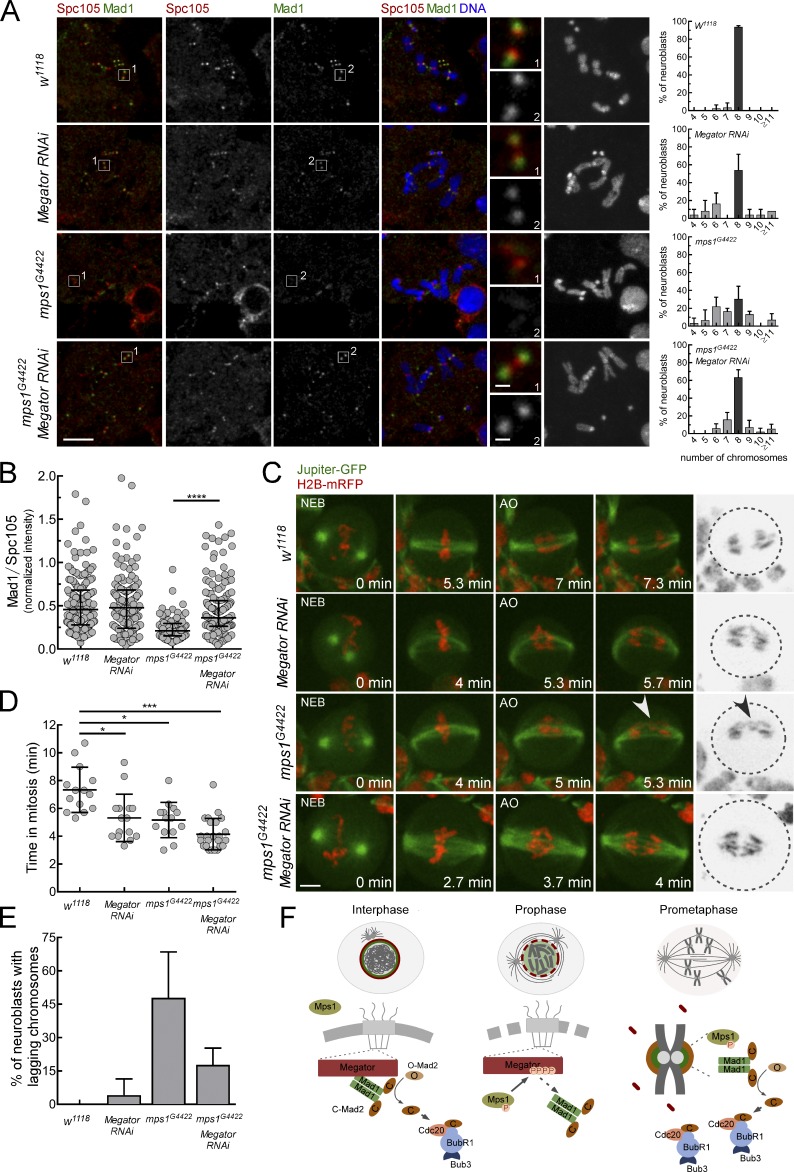
The results so far demonstrate that Mps1 abolishes Mad1 interaction with Megator to promote Mad1 KT recruitment. We next assessed whether this mechanism occurs in vivo and whether it is relevant for genomic stability. We used mps1-null mutant flies (mps1G4422; Bellen et al., 2011; Conde et al., 2013) to express a truncated version of Mps1 that lacks the N-terminus domain (gEGFP-Mps1-Cterm) required for KT targeting (Althoff et al., 2012) and examined its capacity to recruit Mad1 to unattached KTs. Notably, neuroblasts expressing the gEGFP-Mps1-Cterm transgene under control of mps1 promoter were still able to partially recruit Mad1 to unattached KTs (50% of the levels detected in control w1118 flies; Fig. S2, A–C), confirming in vivo a KT-extrinsic role for Mps1 in promoting Mad1 KT localization. Furthermore, RNAi-mediated repression of Megator restored Mad1 KT recruitment in a mps1G4422 genetic background in vivo (Fig. 5, A and B). Importantly, this concomitantly rescued the elevated rates of aneuploidy caused by loss of Mps1 (∼70% mitotic figures with aneuploid karyotypes in mps1G4422 versus ∼30% in mps1G4422 + Megator RNAi; Fig. 5 A). To test whether this recovery in genomic stability was due to improved fidelity of chromosome segregation, we monitored the mitotic progression of larval neuroblasts by live imaging (Fig. 5, C–E; and Videos 1, 2, 3, and 4). Interestingly, although UAS-MegatorRNAi and mps1G4422 neuroblasts have similarly decreased mitotic timings (∼5 min vs. ∼7.3 min in w1118 controls), the frequency of anaphases with lagging chromosomes is dramatically higher in mps1G4422 mutants (Fig. 5, C–E). Importantly, depletion of Megator significantly restores the accuracy of segregation in mps1G4422 neuroblasts, although failing to extend the time from NEB to anaphase onset (Fig. 5, C–E). A similar defect in SAC function was observed in cultured S2 cells codepleted of Mps1 and Megator: although proficient in Mad1 KT recruitment (Fig. S2, E and F), these cells failed to arrest in mitosis when challenged with colchicine (Fig. S2 G). This was expected due to compromised Mps1-mediated phosphorylation of Mad1, which is required to promote Cdc20-C-Mad2 interaction and consequently MCC assembly (Faesen et al., 2017; Ji et al., 2017). Hence, in Drosophila neuroblasts undergoing unperturbed mitosis, KT-associated Mad1 is able to efficiently safeguard anaphase fidelity and chromosomal euploidy independently of its SAC function. This is in line with previous studies reporting a role for Mad1 in preventing merotely independently of its interaction with Mad2 and SAC signaling (Emre et al., 2011; Akera et al., 2015). Collectively, these results strongly support that Mps1-mediated dissociation of Mad1 from Megator is a critical event for efficient KT localization of Mad1, the fidelity of chromosome segregation, and consequently, genome stability in vivo.
Figure 5.
Depletion of Megator restores Mad1 KT recruitment and mitotic fidelity in Drosophila mps1-null neuroblasts. (A and B) Immunofluorescence images with ploidy histograms (A) and quantifications (B) of Mad1 levels at unattached KTs of w1118, InscGal4>UAS-MegatorRNAi, homozygous mps1G4422, or homozygous mps1G4422;InscGal4>UAS-MegatorRNAi neuroblasts treated with colchicine (50 µM) for 90 min (n ≥ 91 KTs). (C–E) Mitotic progression (C), mitotic timing (D), and percentage of anaphases with lagging chromosomes (E) in neuroblasts with indicated genotypes coexpressing the microtubule-associated protein Jupiter-GFP and H2B-mRFP. The mitotic timing corresponds to the time from NEB to anaphase onset (AO; n ≥ 14 neuroblasts from at least two independent experiments). The arrowhead in C points to a lagging chromosome. (F) Model for the control of Mad1 subcellular redistribution during the G2/M transition. In interphase, inactive Mps1 (unphosphorylated T-loop) is in the cytoplasm and Mad1-C-Mad2 is docked to Megator at the nucleoplasmic side of NPCs, where it catalyzes the assembly of premitotic MCC. During prophase, active Mps1 (phosphorylated T-loop) becomes detectable in the nucleus and is now able to phosphorylate Megator, disrupting the interaction with Mad1. This ensures timely release of Mad1-C-Mad2 from NPCs, which enables Mad1-C-Mad2 to accumulate at prometaphase KTs and instate robust SAC signaling. In A–C and E, data are presented as mean ± SD. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001 (Kruskal–Wallis, Dunn’s multiple comparison test). Scale bars, 5 µm (inset, 0.5 µm).
Video 1.
Mitotic progression of w1118 neuroblasts. Related to Fig. 5, C–E. Mitotic progression of w1118 neuroblasts coexpressing Jupiter-GFP (green) and H2B-mRFP (red) was monitored by spinning disk confocal microscopy. H2B-mRFP signal is shown on the right. Frames were acquired every 20 s. NEB corresponds to time 0.0. Time is shown in minutes.
Video 2.
Mitotic progression of InscGal4>UAS-MegatorRNAineuroblasts. Related to Fig. 5, C–E. Mitotic progression of InscGal4>UAS-MegatorRNAi mutant neuroblasts coexpressing Jupiter-GFP (green) and H2B-mRFP (red) was monitored by spinning disk confocal microscopy. H2B-mRFP signal is shown on the right. Frames were acquired every 20 s. NEB corresponds to time 0.0. Time is shown in minutes.
Video 3.
Mitotic progression of mps1G4422 neuroblasts. Related to Fig. 5, C–E. Mitotic progression of homozygous mps1G4422 neuroblasts coexpressing Jupiter-GFP (green) and H2B-mRFP (red) was monitored by spinning disk confocal microscopy. H2B-mRFP signal alone is shown on the right. Frames were acquired every 20 s. NEB corresponds to time 0.0. Time is shown in minutes.
Video 4.
Mitotic progression of mps1G4422; InscGal4>UAS-MegatorRNAi neuroblasts. Related to Fig. 5, C–E. Mitotic progression of homozygous mps1G4422; InscGal4>UAS-MegatorRNAi neuroblasts coexpressing Jupiter-GFP (green) and H2B-mRFP (red) was monitored by spinning disk confocal microscopy. H2B-mRFP signal is show on the right. Frames were acquired every 20 s. NEB corresponds to time 0.0. Time is shown in minutes.
Depletion of Tpr partially restores Mad1 KT recruitment in human cells lacking Mps1 activity
Since two of the identified Mps1 sites on Megator (T1259 and T1390) are conserved in vertebrate Tpr, and a third one (T1338) occurs as glutamic acid, we tested whether an analogous mechanism operates in human cells. Reversine-mediated inactivation of Mps1 severely compromised Mad1 KT accumulation in HeLa and RPE cells treated with nocodazole (Fig. S3, A–D). However, siRNA-mediated depletion of Tpr was able to partially rescue the recruitment of Mad1 to unattached KTs of both cell lines in the presence of reversine (Fig. S3, A–D). These results, and the recent observation that Mad1 remains associated with Tpr during early mitosis in reversine-treated RPE cells (Jackman et al., 2019), strongly support that this KT-extrinsic mechanism orchestrated by Mps1 is evolutionarily conserved in vertebrates, where it likely coordinates with Cyclin B1–CDK1 (Jackman et al., 2019) the release of Mad1 from NPCs to enable its proper KT localization.
Figure S3.
Depletion of Tpr partially restores Mad1 KT recruitment in human cells lacking Mps1 activity. Immunofluorescence images (A and B) and quantifications (C and D) of Mad1 levels at unattached KTs of HeLa (A and C) or RPE1 cultured cells (B and D) in the indicated conditions. Cells were incubated with nocodazole (3.3 µM) and MG132 (1 µM) for 3 h (HeLa) or 6 h (RPE). When indicated, cells were treated with Tpr siRNA (50 nM) for 48 h and/or reversine (1 µM) for 1 h before nocodazole + MG132 incubation. Mad1 fluorescence intensities were determined relative to CenpC (n ≥ 4,081 KTs for HeLa cells, n ≥ 519 KTs for RPE cells). Data are presented as mean ± SD. ****, P < 0.0001 (Kruskal–Wallis, Dunn’s multiple comparison test). Scale bars, 5 µm (inset, 0.5 µm).
Our biochemical, cellular, and in vivo data concur to demonstrate that Mps1 activity at NPCs early in prophase sets the stage to enable appropriate recruitment of Mad1 to unattached/prometaphase KTs (Fig. 5 F). Phosphorylation of Megator by Mps1 abrogates Megator interaction with Mad1, which is essential for KT localization of Mad1-C-Mad2 at levels required to sustain robust SAC signaling and accurate chromosome segregation. During interphase, nuclear exclusion and reduced activity of Mps1 prevent the premature dissociation of Mad1 from NPCs. This is most likely important to facilitate Mad1-C-Mad2 interaction and MCC assembly before mitotic entry and KT maturation (Rodriguez-Bravo et al., 2014; Kim et al., 2018). Together, these observations establish that a key function of non-KT Mps1 is to coordinate Mad1 subcellular localization with cell cycle progression, so that both nuclear pores in interphase and KTs in mitosis generate anaphase inhibitors that preserve genomic stability.
Materials and methods
S2 cell cultures, RNAi-mediated depletion, and drug treatments
The Drosophila S2-DGRC cell line (#stock6) was acquired from the Drosophila Genomics Resource Center, Indiana University, and was not independently authenticated. The cell lines routinely tested negative for mycoplasma contamination. Cell cultures, RNAi synthesis, and RNAi treatment were performed as previously described (Conde et al., 2013). S2 cells were incubated with 30 µg of Mps1 RNAi or Megator UTRs RNAi for 120 h in 1.5 ml of Schneider’s insect medium (Sigma-Aldrich).
The PCR product used as template to produce Mps1 RNAi was amplified with the set of primers Mps1F (5′-TAATACGACTCACTATAGGGTCTTCCAAACACCTATGACCG-3′) and Mps1R (5′-TAATACGACTCACTATAGGGCGTTTAGATATCCCTGCACCA-3′). The PCR product used as template to produce Megator 5′ UTR RNAi was amplified with Mtor5′_F (5′-TAATACGACTCACTATAGGGATTTTTTCGCGTCCCTAAAG-3′) and Mtor5′_R (5′-TAATACGACTCACTATAGGGTTGATCCTTGTCAAGCAGC-3′). The PCR product used as template to produce Megator 3′ UTR RNAi was amplified with Mtor3′_F (5′-TAATACGACTCACTATAGGGGGCGAGGAGTTCGGCGGACC-3′) and Mtor3′_R (5′-TAATACGACTCACTATAGGGATCGACAAAATTACACATAT-3′). At selected time points, cells were collected and processed for immunofluorescence, time-lapse microscopy, immunoblotting, or immunoprecipitation. When required, cells were subjected to several drug treatments before being collected and processed for the desired analysis. To promote microtubule depolymerization, cells were incubated with 30 µM colchicine (Sigma-Aldrich). To decrease microtubule dynamics, cells were incubated with 100 nM taxol (Sigma-Aldrich). When required, 20 µM MG132 (Calbiochem) was added to inhibit the proteasome. For Fig. 2 (A, B, G, I, and J), cells were incubated with colchicine for 10 h, and MG132 was added 4 h before cell processing. For Fig. 3 (C and D) and Fig. 4 (C–F), cells were incubated with colchicine and MG132 for 30 min before fixation. For Fig. 3 E, cells were added immediately before live-imaging acquisition. For Fig. 4 (A, B, and H), when indicated, cells were treated with colchicine for 18 h before lysis. For experiments in Fig. 1 (D and E) and Fig. S1 A, cells were incubated with 10 µM Leptomycin B (Sigma-Aldrich) for 3 h to block Crm1-mediated nuclear export.
Constructs and S2 cell transfection
Recombinant plasmids pHWG[blast]-MegatorWT, pHWG[blast]-MegatorT4A, pHWG[blast]-MegatorT4D, pHGW[blast]-Megator1,187–1,655/WT, pHGW[blast]-Megator1,187–1,655/T4A, pHGW[blast]-Megator1,187–1,655/T4D, and pHGW[blast]-Mps1WT were generated using the Gateway Cloning System (Invitrogen). Megator, Megator1,187–1,655, or Mps1 cDNAs were amplified by PCR and inserted into modified versions of pENTR-entry vector through FastCloning (Li et al., 2011). To generate pENTR-Megator1,187–1,655/T4A and pENTR-Megator1,187–1,655/T4D, codons corresponding to T1259, T1302, T1338, and T1390 of pENTR-Megator1,187–1,655 were converted to codons for either alanine (A) or aspartate (D), respectively, by several cycles of site-directed mutagenesis with primers harboring the desired mutations. To generate pENTR-MegatorT4A and pENTR-MegatorT4D, the fragment corresponding to aa 1,187–1,655 on pENTR-MegatorWT was replaced by Megator1,187–1,655/T4A or Megator1,187–1,655/T4D PCR products, respectively, through FastCloning (Li et al., 2011). PCRs were performed using Phusion polymerase (New England Biolabs). PCR products were digested with DpnI restriction enzyme (New England Biolabs), used to transform competent bacteria and selected for positives. Subsequently, pENTR-Megator1,187–1,655 constructs and pENTR-Mps1 were recombined with pHGW[blast] (blasticidinR; N-terminal EGFP tag) and pENTR-Megator constructs with pHWG (blasticidinR; C-terminal EGFP tag) using Gateway LR Clonase II (Invitrogen), according to the manufacturer’s instructions. pHGW-Mps1WT-NLS was produced by PCR amplification of pHGW-Mps1WT with primers harboring SV40 large T-antigen NLS sequence. pHGW-Mps1KD-NLS was produced by site-directed mutagenesis of pHGW-Mps1WT-NLS with primers harboring the mutation to convert D478 to A478. PCRs were performed with Phusion polymerase, followed by digestion with DpnI restriction enzyme (New England Biolabs). The constructs H2B-mCherry, H2B-GFP, mCherry–α-tubulin, Mad1-EGFP, pHW-CIB-MP-HRW-CRY2-VHH, and pHGW-aPKC have been previously described (Conde et al., 2013; Moura et al., 2017; Osswald et al., 2019). Plasmids were transfected into S2 cells using Effectene Transfection Reagent (Qiagen), according to the manufacturer’s instructions. Transiently expressing cells were harvested 4–5 d after transfections. Stable cell lines were obtained by selection in medium with 25 µg/ml blasticidin. To induce expression of pHW-CIB-MP-HRW-CRY2-VHH, pHGW[blast], or pHWG[blast] constructs, cells were incubated for 30 min at 37°C 24 h before processing. Cells transfected with pHW-CIB-MP-HRW-CRY2-VHH were maintained in the dark until processing.
Live-cell imaging
Live analysis of mitosis was performed in S2 cell lines and neuroblasts expressing the indicated constructs. S2 cells were plated on glass-bottom dishes (MatTek) coated with Concanavalin A (0.25 mg/ml; Sigma-Aldrich). Third-instar larvae brains were dissected in PBS and mounted in PBS between coverslips of different sizes. The preparation was squashed and sealed with Halocarbon oil 700 (Sigma-Aldrich). 4D datasets were collected at 25°C with a spinning disc confocal system (Revolution; Andor) equipped with an electron multiplying charge-coupled device camera (iXonEM+; Andor) and a CSU-22 unit (Yokogawa) based on an inverted microscope (IX81; Olympus). Two laser lines (488 and 561 nm) were used for near-simultaneous excitation of EGFP and mCherry or RFP. The system was driven by iQ software (Andor). Time-lapse imaging of z stacks with 0.8-µm steps for S2 cells and 0.5-µm steps for neuroblasts were collected, and image sequence analysis, video assembly, and fluorescence intensity quantification were performed using ImageJ (National Institutes of Health). Quantification of Mad1-EGFP and Megator-EGFP levels at the NE and mCherry-tubulin at the nucleus was performed on single Z stacks from images acquired with fixed exposure settings. Mad1-EGFP and Megator-EGFP intensities at the NE were determined for each time point (t), using the following formula:
where Bi is the integrated density of a region of interest (ROI) harboring the nucleus (including outer nuclear membrane); Ba is the area of the ROI harboring the nucleus; Si is the integrated density of an ROI encompassing the nucleoplasm; Sa is the area of the ROI harboring the nucleoplasm; bm is the mean intensity of an ROI outside the cell (background); Cit is the integrated density of an ROI harboring the cell at time t; and Cit0 is the integrated density of an ROI harboring the cell on the first frame. mCherry-tubulin intensities in the nucleus were determined for each time point (t), using the following formula:
where Nm is the mean intensity of an ROI inside the nucleus; bm is the mean intensity of the background; Cit is the integrated density of an ROI harboring the cell at time t; and Cit0 is the integrated density of an ROI harboring the cell on the first frame. The changes in fluorescence intensity over time were plotted as normalized signal relative to the mean signal measured before NEB.
Immunofluorescence analysis
For immunofluorescence analysis of S2 cells, 105 cells were centrifuged onto slides for 5 min at 1,500 rpm (Cytospin 2, Shandon). For LARIAT experiments, cells were irradiated with blue light for 30 min before centrifugation to trigger the formation of protein clusters through blue light–mediated heterodimerization between the cryptochrome CRY2 fused to an anti-GFP nanobody and a multimeric version of the cryptochrome-interacting basic helix-loop-helix 1. Afterward, cells were fixed in 4% PFA in PBS for 12 min and further extracted for 8 min with 0.1% Triton X-100 in PBS. Alternatively, cells were simultaneously fixed and extracted in 3.7% formaldehyde (Sigma-Aldrich) and 0.5% Triton X-100 in PBS for 10 min followed by three washing steps of 5 min with PBST (PBS with 0.05% Tween20). For immunofluorescence analysis of Drosophila neuroblasts, third-instar larval brains were dissected in PBS and incubated with 50 µM colchicine for 90 min. Afterward, the brains were fixed in 1.8% formaldehyde (Sigma-Aldrich) and 45% glacial acetic acid for 5 min, squashed between slide and coverslip, and immersed in liquid nitrogen. Subsequently, coverslips were removed, and the slides were incubated in cold ethanol for 10 min and washed in PBS with 0.1% Triton X-100. Immunostaining was performed as previously described (Moura et al., 2017). Images were collected in a Zeiss Axio Imager microscope (Carl Zeiss) or a Leica TCS II scanning confocal microscope (Leica Microsystems). For immunofluorescence quantification, the mean pixel intensity was obtained from raw images acquired with fixed exposure acquisition settings. Fluorescence intensities at the NE were obtained from single Z stack projections. The NE was defined based on Megator or Megator-EGFP staining, by subtracting an ROI containing the nucleoplasm from an ROI harboring the entire nucleus (outer membrane) after subtraction of background intensities estimated from regions outside the cell. Mad1 fluorescence intensities were determined relative to Nup107, Megator, or Megator-EGFP. Fluorescence intensities of LARIAT-mediated clustered proteins and KT proteins were obtained from maximum projected images. For Mad1 and EGFP-Megator1,187–1,655, the fluorescence intensities were quantified for individual clusters selected manually by mRFP-Cry2 staining. After subtraction of background intensities estimated from regions inside the cell with no clusters, the intensity of Mad1 was determined relative to Megator-EGFP signal. For KT proteins, the fluorescence intensity was quantified for individual KTs selected manually by Mad1, CID, or Spc105 staining. The size of the ROI was predefined so that each single KT could fit into it. After subtraction of background intensities, estimated from regions outside the cell, the intensity of the proteins was determined relative to cytoplasmic Mad1, CID, or Spc105.
S2 cell lysates, immunoprecipitation, and Western blotting
For Drosophila brain lysates, at least 10 third-instar larvae brains were dissected in PBS, transferred to Laemmli buffer (4% SDS, 10% β-mercaptoethanol, 0.125 M Tris-HCl, 20% glycerol, and 0.004% bromophenol blue), and boiled at 95°C for 5 min. S2 cell lysates for immunoprecipitation and Western blot analysis were obtained from nontransfected S2 cells or S2 cells expressing Megator-EGFP transgenes treated with colchicine and MG132 when indicated. For Western blot of total S2 cell lysates, 106 cells were harvested through centrifugation at 5,000 rpm for 10 min. The resulting pellet was resuspended in Laemmli sample buffer and boiled at 95°C for 5 min.
For immunoprecipitation assays, cells were harvested through centrifugation at 5,000 rpm for 10 min at 4°C and washed with 2 ml PBS supplemented with 1× protease inhibitor cocktail (Roche). The cell pellet was resuspended in lysis buffer (150 mM KCl, 75 mM Hepes, pH 7.5, 1.5 mM EGTA, 1.5 mM MgCl2, 15% glycerol, 0.1% NP-40, 1× protease inhibitor cocktail [Roche], and 1× phosphatase inhibitor cocktail 3 [Sigma-Aldrich]) before freezing disruption in liquid nitrogen. Cell lysates were then clarified through centrifugation at 10,000 rpm for 10 min at 4°C and concentration determined by measuring absorbance at 280 nm in the Nanodrop 1000 (Thermo Fisher Scientific). Lysates containing 800 or 1,000 µg of protein in a total volume of 400 µl lysis buffer were precleared by incubation with 15 or 20 µl Protein A magnetic beads (New England Biolabs), respectively, for 1 h at 4°C under agitation. Precleared extracts were incubated with rabbit anti-Mad1 (1:100) or anti-BubR1 (1:100) overnight, at 4°C under agitation. Afterward, the mixture was incubated with 40 or 50 µl of Protein A magnetic beads for 90 min, at 4°C with agitation. Magnetic beads were collected and washed four times with 500 µl of lysis buffer. Magnetic beads were resuspended in Laemmli sample buffer and boiled at 95°C for 5 min. To confirm protein hyperphosphorylation status, 50 µg of mitotic cell lysates were treated with 400 U of λ-phosphatase (New England Biolabs) at 30°C for 1 h in a total volume of 50 µl PMP phosphatase buffer (50 mM Hepes, pH 7.5, 100 mM NaCl, 2 mM DTT, 0.01% Brij 35, and 1 mM MnCl2; New England Biolabs).
Samples were resolved by SDS-PAGE and transferred to a nitrocellulose membrane, using the iBlot Dry Blotting System (Invitrogen) according to the manufacturer’s instructions. Transferred proteins were confirmed by Ponceau staining (0.25% Ponceau S in 40% methanol and 15% acetic acid). The membrane was blocked for 1 h at room temperature with 5% powder milk prepared in PBST and subsequently incubated with primary antibodies diluted in blocking solution overnight at 4°C under agitation. Membranes were washed three times for 10 min with PBST and incubated with secondary antibodies (diluted in blocking solution) for 1 h at room temperature with agitation. Secondary antibodies conjugated to HRP (Santa Cruz Biotechnology) or VeriBlot for IP Detection Reagent (ab131366; Abcam) were used according to the manufacturer’s instructions. Blots were developed with ECL Chemiluminescent Detection System (Amersham) according to the manufacturer’s protocol and detected on x-ray film (Fuji Medical). When required, proteins were resolved in 4–20% Mini-Protean TGX Precast Gel (Bio-Rad) and transferred to nitrocellulose membrane overnight in 48 mM Tris, 39 mM glycine, 0.037% SDS, and 20% methanol, pH 8.3, at 20 V and 4°C.
Production and purification of recombinant proteins
To generate 6xHis-Mad11–493, 6xHis-BubR11–358, and 6xHis-BubR1359–696 constructs for expression in bacteria, PCR products with the corresponding coding sequence were cloned into NdeI/XhoI or SalI/XhoI, sites of pET30a (+) vector (Novagen). TOP10 competent cells were transformed and selected for positives. The recombinant construct was used to transform BL21-star competent cells, and protein expression was induced with 0.05 mM IPTG at 15°C overnight. Cells were harvested and lysed in bacterial lysis buffer (50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole, pH 8.0) supplemented with 1 mM PMSF (Sigma-Aldrich) and 0.4 mg/ml Lysozyme (Sigma-Aldrich), sonicated, and clarified by centrifugation at 4°C. Recombinant 6xHis-Mad11–493 and 6xHis-BubR11–358 were purified with Novex Dynabeads (Invitrogen) in bacterial lysis buffer.
To generate recombinant MBP-Megator fragments (aa 1–402; 403–800), PCR products harboring the coding sequences for fragments 1–402 and 403–800 of Megator were cloned between the EcoRI/PstI and KpnI/SbfI restriction sites of pMal-c2 (New England Biolabs) vector, respectively. A Megator fragment with the coding sequence for aa 1,187–1,655 was isolated through cDNA restriction with EcoRI and inserted at the EcoRI restriction site of pMal-c2. Megator1,187–1,655/T4A and Megator1,187–1,655/T4D were inserted into pMal-c2 vector through FastCloning (Li et al., 2011) using Phusion Polymerase (New England Biolabs). To generate pMal-c-Megator1,187–1,655/T2D and pMal-c-Megator1,187–1,655/T3D, the codons corresponding to T1259 and T1390 or to T1259, T1338, and T1390 of pMal-c-Megator1,187–1,655/WT were converted to codons for aspartate by several cycles of site-directed mutagenesis with primers harboring the desired mutations, using Phusion Polymerase (New England Biolabs). These constructs were used to transform TOP10 competent bacteria, and cells were selected for the incorporation of plasmids. Selected recombinant constructs were used to transform BL21-star competent cells, and protein expression was induced with 0.05 mM IPTG at 15°C overnight. Pellets of these cultures were lysed in column buffer (200 mM NaCl, 20 mM Tris-HCl, 1 mM EDTA, and 1 mM DTT, pH 7.4) supplemented with 1% Triton X-100 (Sigma-Aldrich), 1 mM PMSF (Sigma-Aldrich), and 0.4 mg/ml of lysozyme (Sigma-Aldrich), sonicated, and clarified by centrifugation at 4°C. Recombinant MBP-Megator fragments were purified with amylose magnetic beads (New England Biolabs) and eluted in Column Buffer supplemented with 10 mM Maltose. The purified recombinant proteins (eluted or bound to magnetic beads) were resolved by SDS-PAGE, and their relative amounts were determined after Coomassie blue staining. Similar amounts of protein were used in the subsequent assays.
In vitro kinase assays, MS analysis, and pull-down assays
For in vitro kinase assays, recombinant fragments of MBP-Megator were incubated with 0.05 µg HsMps1/TTK (SignalChem) in a total volume of 30 µl kinase reaction buffer (5 mM morpholino propane sulfonic acid, pH 7.2, 2.5 mM β-glycerol-phosphate, 5 mM MgCl2, 1 mM EGTA, 0.4 mM EDTA, 0.25 mM DTT, and 100 µM ATP supplemented with 1× phosphatase inhibitor cocktail 3 [Roche]). Reactions were performed at 30°C for 30 min and analyzed by autoradiography, subjected to MS analysis, or used in pull-down assays. For detection of 32P incorporation, the kinase reaction buffer was supplemented with 10 µCi [γ-32P]ATP (3,000 Ci/mmol and 10 mCi/ml), and the reaction was stopped by addition of Laemmli sample buffer, boiled for 5 min at 95°C, and resolved by SDS-PAGE. After drying at 80°C under vacuum, the gel was exposed to x-ray films (Fuji Medical). For identification of phosphorylated residues, the reaction was stopped by addition of 6 M urea and analyzed by liquid chromatography (LC) coupled with MS. Samples were digested with LysC/Trypsin and/or GluC and prepared for LC-MS/MS analysis as previously described (Rappsilber et al., 2007). Peptides (100 ng) were separated on an EASY-nLC 1000 HPLC system (Thermo Fisher Scientific) for 1 h from 5–60% acetonitrile with 0.1% formic acid and directly sprayed via a nano-electrospray source in a quadrupole Orbitrap mass spectrometer (Q Exactive; Thermo Fisher Scientific; Michalski et al., 2011). The Q Exactive was operated in data-dependent mode, acquiring one survey scan and subsequently 10 MS/MS scans (Olsen et al., 2007). Resulting raw files were processed with MaxQuant software (v1.5.2.18) using a reduced database containing only the proteins of interest and giving phosphorylation on serine, threonine, and tyrosine as variable modification (Cox and Mann, 2008). A false discovery rate cutoff of 1% was applied at the peptide and protein levels and the phosphorylation site decoy fraction.
For pull-down assays, 6xHis-Mad11–493 or 6xHis-BubR11–358 bound to Novex Dynabeads (Invitrogen) was incubated with the MPB-Megator1,187–1,655 constructs in a final volume of 50 µl column buffer (250 mM NaCl, 20 mM Tris-HCl, pH 7.4, 1 mM EDTA, 1 mM DTT, 0.05% Tween20 [Sigma-Aldrich], 1× protease inhibitor cocktail (Roche), and 1× phosphatase inhibitor cocktail 3 [Sigma-Aldrich]) for 90 min at room temperature with agitation. The magnetic beads (with bound protein) were collected and washed three times with 500 µl column buffer, resuspended in Laemmli sample buffer and boiled at 95°C for 5 min. After removal of the magnetic beads, samples were resolved by SDS-PAGE and probed for proteins of interest through Western blotting. Quantification of MBP-Megator binding to 6xHis-Mad11–493 was represented as the ratio between the chemiluminescence signal intensities of MBP-Megator and 6xHis-Mad11–493. The values obtained for MBP-Megator1,187–1,655/WT were set to 1.
siRNA-mediated depletion, drug treatments, and immunofluorescence analysis in human cultured cells
HeLa and RPE1 cells were cultured in DMEM supplemented with 10% FBS (Invitrogen) in a 37°C incubator with 5% CO2. Depletion of Tpr was performed using a siRNA directed against the following target sequence: 5′-GCACAACAGGAUAAGGUUA-3′ (Sigma-Aldrich). siRNA transfection was performed using Lipofectamine RNAiMAX (Invitrogen) with 50 nM Tpr siRNA in final concentration for 48 h. Microtubule depolymerization in HeLa and RPE1 cells was induced by nocodazole (Sigma-Aldrich) at 3.3 µM for 3 or 6 h, respectively. To inhibit the proteasome, induce a metaphase arrest, and prevent mitotic exit due to a compromised SAC, cells were treated with 5 µM MG132 (EMD Millipore). For Mps1 inhibition, cells were treated with 1 µM reversine (Sigma-Aldrich) for 1 h before the addition of the other drugs (nocodazole and MG132).
For immunofluorescence assays, cells were fixed with either 4% PFA for 10 min or ice-cold ethanol for 3 min. In the case of the PFA fixation, cells were subsequently extracted in 0.3% Triton X-100 in 1× PBS for 10 min. After fixation and extraction, cells were blocked for 1 h with 10% FBS in PBS. Next, cells were incubated with primary antibodies at 4°C overnight and with the respective secondary antibodies for 1 h at room temperature. DNA was counterstained with DAPI (1 µg/ml; Sigma-Aldrich) before coverslips were mounted in 90% glycerol, 10% Tris, pH 8.5, and 0.5% N-propyl gallate on glass slides. Images were acquired on an AxioImager Z1 (63× plan-Apochromatic oil differential interference contrast objective lens, 1.4 NA; all from Carl Zeiss) equipped with a charge-coupled device camera (ORCA-R2; Hamamatsu Photonics) using Zen software (Carl Zeiss). 41 0.2-µm separated z-planes covering the entire mitotic cell were collected. Images were blind deconvolved using Autoquant X (Media Cybernetics), analyzed in ImageJ, and represented with a maximum-intensity projection of a deconvolved Z stack. For quantification measurements, all images compared were acquired using identical acquisition settings. Image analysis was performed using Imaris (Bitplane, Scientific Solutions). Briefly, individual KTs were automatically detected and represented as spots using centromere protein C (CenpC) staining. KT protein accumulation was measured by quantifying pixel gray levels for each channel within the selected spots. Background fluorescence was measured using ImageJ outside the KT region and subtracted. Fluorescent intensity measurements were normalized to the CenpC signals. The results are reported after normalization to relative fluorescence levels in control samples.
Antibodies
The following primary antibodies were used for immunofluorescence studies: rat anti-CID (Rat4) used at 1:250, rabbit anti-phosphorylated Thr676-Mps1 (T676; a gift from Geert Kops, Hubrecht Institute, Utrecht, Netherlands; Jelluma et al., 2008) used at 1:2,000, chicken anti-GFP (ab13970; Abcam) used at 1:2,000 for S2 cells and 1:1,000 for neuroblasts, mouse anti-Megator (gift from Jørgen Johansen and Kristen Johansen, Iowa State University, Ames, IA; Qi et al., 2004; RRID:AB_2721935) used at 1:20, rabbit anti-Mad1 (Rb1; Conde et al., 2013) used at 1:2,500 for S2 cells and 1:1,000 for neuroblasts, mouse anti-C-Mad2 (Sigma-Aldrich) used at 1:50 for S2 cells, rat anti-Spc105 used at 1:250, guinea pig anti-Mps1 (Gp15; a gift from Scott Hawley, Stowers Institute for Medical Research, Kansas City, MO; RRID:AB_2567774) used at 1:250, rabbit anti-phosphorylated ser10-Histone H3 (p-H3; Millipore; RRID:AB_565299) used at 1:5,000, rat anti-Mad1 (Rat1) used at 1:1,000, rabbit anti-Nup107 (gift from Valérie Doye, Institut Jacques Monod CNRS-Université Paris Diderot, Paris, France; Katsani et al., 2008) used at 1:250, mouse anti-Mad1 (Merck Millipore) used at 1:500 in human cells, and human anti-CenpC (MBL) used at 1:500. The following primary antibodies were used for Western blotting studies: mouse anti–α-tubulin DM1A (Sigma-Aldrich; RRID:AB_477593) used at 1:10,000, rabbit anti-Cyclin B (gift from C. Lehner, Institute for Molecular Life Sciences-University of Zurich, Zurich, Switzerland) used at 1:10,000, guinea pig anti-Mps1 (Gp15; a gift from Scott Hawley; RRID:AB_2567774) used at 1:5,000, mouse anti-Megator (gift from Jørgen Johansen and Kristen Johansen; Qi et al., 2004; RRID:AB_2721935) used at 1:100, rabbit anti-Mad1 (Rb1; Conde et al., 2013) used at 1:2,000, rabbit anti-Mad2 (Rb1223) used at 1:100, rabbit anti-BubR1 (Rb666; Conde et al., 2013) used at 1:3,500, mouse anti-Cdc20 (mAb 20.B.9; a gift from S. Artavanis-Tsakonas, Harvard Medical School, Boston, MA; Dawson et al., 1995) used at 1:10, mouse anti-MBP (New England Biolabs; RRID:AB_ 1559738) used at 1:5,000, and mouse anti-His-Tag (05-949; Millipore) used at 1:2,500.
Fly stocks
All fly stocks were obtained from Bloomington Stock Center unless stated otherwise. The mps1 mutant allele mps1G4422 (Bellen et al., 2011) has been described and characterized before (Conde et al., 2013; Resende et al., 2018). w1118 was used as WT control. Fly stocks harboring gEGFP-MPS1WT and gEGFP-MPS1325–630 under control of Mps1 cis-regulatory region were kindly provided by Christian Lehner (Althoff et al., 2012).
Statistical analysis
All statistical analysis was performed with Prism V7.0f (GraphPad Software).
Online supplemental material
Fig. S1 shows that Mps1 promotes Mad1 dissociation from Megator. Fig. S2 shows experiments with neuroblasts and cultured S2 cells demonstrating that Mps1-mediated dissociation of Mad1 from Megator is required for KT recruitment of Mad1. Fig. S3 shows experiments supporting that Mps1-mediated dissociation of Mad1 from Tpr contributes for Mad1 KT localization in human cells. The videos show the mitotic progression of w1118 (Video 1), InscGal4>UAS-MegatorRNAi (Video 2), homozygous mps1G4422 (Video 3), or homozygous mps1G4422;InscGal4>UAS-MegatorRNAi (Video 4) neuroblasts.
Supplementary Material
Acknowledgments
We thank Geert Kops, Thomas Maresca, Christian Lehner, Eurico Morais-de-Sá, Helder Maiato, Jørgen Johansen, and Kristen Johansen for providing antibodies, constructs, and fly stocks.
This work was funded by national funds through Fundação para a Ciência e a Tecnologia under the project IF/01755/2014/CP1241/CT0004. This work has also received funding from the project Norte-01-0145-FEDER-000029—Advancing Cancer Research: From basic knowledge to application, supported by Norte Portugal Regional Operational Program (NORTE 2020) under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund. C. Conde is supported by a Fundação para a Ciência e a Tecnologia investigator position and funding (IF/01755/2014).
The authors declare no competing financial interests.
Author contributions: conceptualization, M. Osswald and C. Conde; formal analysis, S. Cunha-Silva, M. Osswald, C.E. Sunkel, and C. Conde; investigation, S. Cunha-Silva, M. Osswald, J. Goemann, J. Barbosa, L.M. Santos, P. Resende, T. Bange, C. Ferrás, and C. Conde; writing, C. Conde; supervision, C.E. Sunkel and C. Conde; funding acquisition, C.E. Sunkel and C. Conde.
References
- Akera T., Goto Y., Sato M., Yamamoto M., and Watanabe Y.. 2015. Mad1 promotes chromosome congression by anchoring a kinesin motor to the kinetochore. Nat. Cell Biol. 17:1124–1133. 10.1038/ncb3219 [DOI] [PubMed] [Google Scholar]
- Althoff F., Karess R.E., and Lehner C.F.. 2012. Spindle checkpoint-independent inhibition of mitotic chromosome segregation by Drosophila Mps1. Mol. Biol. Cell. 23:2275–2291. 10.1091/mbc.e12-02-0117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H.J., Levis R.W., He Y., Carlson J.W., Evans-Holm M., Bae E., Kim J., Metaxakis A., Savakis C., Schulze K.L., et al. 2011. The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics. 188:731–743. 10.1534/genetics.111.126995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.H., Shevchenko A., Mann M., and Murray A.W.. 1998. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J. Cell Biol. 143:283–295. 10.1083/jcb.143.2.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung E., and Chen R.-H.. 2002. Spindle checkpoint requires Mad1-bound and Mad1-free Mad2. Mol. Biol. Cell. 13:1501–1511. 10.1091/mbc.02-01-0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin P., Nashchekina O., Walker R., and Pines J.. 2013. The spindle assembly checkpoint works like a rheostat rather than a toggle switch. Nat. Cell Biol. 15:1378–1385. 10.1038/ncb2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde C., Osswald M., Barbosa J., Moutinho-Santos T., Pinheiro D., Guimarães S., Matos I., Maiato H., and Sunkel C.E.. 2013. Drosophila Polo regulates the spindle assembly checkpoint through Mps1-dependent BubR1 phosphorylation. EMBO J. 32:1761–1777. 10.1038/emboj.2013.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., and Mann M.. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Dawson I.A., Roth S., and Artavanis-Tsakonas S.. 1995. The Drosophila cell cycle gene fizzy is required for normal degradation of cyclins A and B during mitosis and has homology to the CDC20 gene of Saccharomyces cerevisiae. J. Cell Biol. 129:725–737. 10.1083/jcb.129.3.725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Antoni A., Pearson C.G., Cimini D., Canman J.C., Sala V., Nezi L., Mapelli M., Sironi L., Faretta M., Salmon E.D., and Musacchio A.. 2005. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr. Biol. 15:214–225. 10.1016/j.cub.2005.01.038 [DOI] [PubMed] [Google Scholar]
- De Souza C.P., Hashmi S.B., Nayak T., Oakley B., and Osmani S.A.. 2009. Mlp1 acts as a mitotic scaffold to spatially regulate spindle assembly checkpoint proteins in Aspergillus nidulans. Mol. Biol. Cell. 20:2146–2159. 10.1091/mbc.e08-08-0878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick A.E., and Gerlich D.W.. 2013. Kinetic framework of spindle assembly checkpoint signalling. Nat. Cell Biol. 15:1370–1377. 10.1038/ncb2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emre D., Terracol R., Poncet A., Rahmani Z., and Karess R.E.. 2011. A mitotic role for Mad1 beyond the spindle checkpoint. J. Cell Sci. 124:1664–1671. 10.1242/jcs.081216 [DOI] [PubMed] [Google Scholar]
- Faesen A.C., Thanasoula M., Maffini S., Breit C., Müller F., van Gerwen S., Bange T., and Musacchio A.. 2017. Basis of catalytic assembly of the mitotic checkpoint complex. Nature. 542:498–502. 10.1038/nature21384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fava L.L., Kaulich M., Nigg E.A., and Santamaria A.. 2011. Probing the in vivo function of Mad1:C-Mad2 in the spindle assembly checkpoint. EMBO J. 30:3322–3336. 10.1038/emboj.2011.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich S., Sewart K., Windecker H., Langegger M., Schmidt N., Hustedt N., and Hauf S.. 2014. Mad1 contribution to spindle assembly checkpoint signalling goes beyond presenting Mad2 at kinetochores. EMBO Rep. 15:291–298. 10.1002/embr.201338114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman M., Marcozzi C., Pardo M., Yu L., Tyson A.L., Choudhary J.S., and Pines J.. 2019. Cyclin B1-Cdk1 binding to Mad1 links nuclear pore disassembly to chromosomal stability. bioRxiv. Preprint 701474 10.1101/701474 [DOI] [Google Scholar]
- Jelluma N., Brenkman A.B., McLeod I., Yates J.R. III, Cleveland D.W., Medema R.H., and Kops G.J.P.L.. 2008. Chromosomal instability by inefficient Mps1 auto-activation due to a weakened mitotic checkpoint and lagging chromosomes. PLoS One. 3:e2415 10.1371/journal.pone.0002415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z., Gao H., Jia L., Li B., and Yu H.. 2017. A sequential multi-target Mps1 phosphorylation cascade promotes spindle checkpoint signaling. eLife. 6:e22513 10.7554/eLife.22513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Zhang X., Wang W., Bai Y., Ling Y., Cao C., Ma R.Z., Zhong H., Wang X., and Xu Q.. 2015. A putative N-terminal nuclear export sequence is sufficient for Mps1 nuclear exclusion during interphase. BMC Cell Biol. 16:6 10.1186/s12860-015-0048-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsani K.R., Karess R.E., Dostatni N., and Doye V.. 2008. In vivo dynamics of Drosophila nuclear envelope components. Mol. Biol. Cell. 19:3652–3666. 10.1091/mbc.e07-11-1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M.J., Hughes R.M., Peteya L.A., Schwartz J.W., Ehlers M.D., and Tucker C.L.. 2010. Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods. 7:973–975. 10.1038/nmeth.1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Han J.S., Ly P., Ye Q., McMahon M.A., Myung K., Corbett K.D., and Cleveland D.W.. 2018. TRIP13 and APC15 drive mitotic exit by turnover of interphase- and unattached kinetochore-produced MCC. Nat. Commun. 9:4354 10.1038/s41467-018-06774-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Park H., Kyung T., Kim N.Y., Kim S., Kim J., and Heo W.D.. 2014. Reversible protein inactivation by optogenetic trapping in cells. Nat. Methods. 11:633–636. 10.1038/nmeth.2940 [DOI] [PubMed] [Google Scholar]
- Lee S.H., Sterling H., Burlingame A., and McCormick F.. 2008. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 22:2926–2931. 10.1101/gad.1677208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Wen A., Shen B., Lu J., Huang Y., and Chang Y.. 2011. FastCloning: a highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC Biotechnol. 11:92 10.1186/1472-6750-11-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lince-Faria M., Maffini S., Orr B., Ding Y., Cláudia Florindo C.E., Sunkel A., Tavares J., Johansen K.M., Johansen, and Maiato H.. 2009. Spatiotemporal control of mitosis by the conserved spindle matrix protein Megator. J. Cell Biol. 184:647–657. 10.1083/jcb.200811012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N., and Biggins S.. 2014. Mad1 kinetochore recruitment by Mps1-mediated phosphorylation of Bub1 signals the spindle checkpoint. Genes Dev. 28:140–152. 10.1101/gad.233700.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N., Ceto S., Ranish J.A., and Biggins S.. 2012. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr. Biol. 22:900–906. 10.1016/j.cub.2012.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski J., George K.A., Terret M.E., Zhang C., Shokat K.M., and Jallepalli P.V.. 2010. Mps1 directs the assembly of Cdc20 inhibitory complexes during interphase and mitosis to control M phase timing and spindle checkpoint signaling. J. Cell Biol. 190:89–100. 10.1083/jcb.201001050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malureanu L.A., Jeganathan K.B., Hamada M., Wasilewski L., Davenport J., and van Deursen J.M.. 2009. BubR1 N terminus acts as a soluble inhibitor of cyclin B degradation by APC/C(Cdc20) in interphase. Dev. Cell. 16:118–131. 10.1016/j.devcel.2008.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., Draviam V.M., and Sorger P.K.. 2004. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell. 7:45–60. 10.1016/j.devcel.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Michalski A., Damoc E., Hauschild J.-P., Lange O., Wieghaus A., Makarov A., Nagaraj N., Cox J., Mann M., and Horning S.. 2011. Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol. Cell. Proteomics. 10:M111.011015 10.1074/mcp.M111.011015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Santos M.D., Hervas-Aguilar A., Sewart K., Lancaster T.C., Meadows J.C., and Millar J.B.A.. 2016. Bub3-Bub1 Binding to Spc7/KNL1 Toggles the Spindle Checkpoint Switch by Licensing the Interaction of Bub1 with Mad1-Mad2. Curr. Biol. 26:2642–2650. 10.1016/j.cub.2016.07.040 [DOI] [PubMed] [Google Scholar]
- Moura M., Osswald M., Leça N., Barbosa J., Pereira A.J., Maiato H., Sunkel C.E., and Conde C.. 2017. Protein Phosphatase 1 inactivates Mps1 to ensure efficient Spindle Assembly Checkpoint silencing. eLife. 6:e25366 10.7554/eLife.25366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J.V., Macek B., Lange O., Makarov A., Horning S., and Mann M.. 2007. Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods. 4:709–712. 10.1038/nmeth1060 [DOI] [PubMed] [Google Scholar]
- Osswald M., Santos A.F., and Morais-de-Sá E.. 2019. Light-Induced Protein Clustering for Optogenetic Interference and Protein Interaction Analysis in Drosophila S2 Cells. Biomolecules. 9:61 10.3390/biom9020061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primorac I., Weir J.R., Chiroli E., Gross F., Hoffmann I., van Gerwen S., Ciliberto A., and Musacchio A.. 2013. Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. eLife. 2:e01030 10.7554/eLife.01030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H., Rath U., Wang D., Xu Y.-Z., Ding Y., Zhang W., Blacketer M.J., Paddy M.R., Girton J., Johansen J., and Johansen K.M.. 2004. Megator, an essential coiled-coil protein that localizes to the putative spindle matrix during mitosis in Drosophila. Mol. Biol. Cell. 15:4854–4865. 10.1091/mbc.e04-07-0579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J., García-Gimeno M.A., Beullens M., Manzione M.G., Van der Hoeven G., Igual J.C., Heredia M., Sanz P., Gelens L., and Bollen M.. 2017. An attachment-independent biochemical timer of the spindle assembly checkpoint. Mol. Cell. 68:715–730.e5. 10.1016/j.molcel.2017.10.011 [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Mann M., and Ishihama Y.. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2:1896–1906. 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- Resende L.P., Monteiro A., Brás R., Lopes T., and Sunkel C.E.. 2018. Aneuploidy in intestinal stem cells promotes gut dysplasia in Drosophila. J. Cell Biol. 217:3930–3946. 10.1083/jcb.201804205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Bravo V., Maciejowski J., Corona J., Buch H.K., Collin P., Kanemaki M.T., Shah J.V., and Jallepalli P.V.. 2014. Nuclear pores protect genome integrity by assembling a premitotic and Mad1-dependent anaphase inhibitor. Cell. 156:1017–1031. 10.1016/j.cell.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Rodriguez J.A., Lewis C., McKinley K.L., Sikirzhytski V., Corona J., Maciejowski J., Khodjakov A., Cheeseman I.M., and Jallepalli P.V.. 2018. Distinct roles of RZZ and Bub1-KNL1 in mitotic checkpoint signaling and kinetochore expansion. Curr. Biol. 28:3422–3429.e5. 10.1016/j.cub.2018.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittenhelm R.B., Chaleckis R., and Lehner C.F.. 2009. Intrakinetochore localization and essential functional domains of Drosophila Spc105. EMBO J. 28:2374–2386. 10.1038/emboj.2009.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer N., Ferrás C., Kern D.M., Logarinho E., Cheeseman I.M., and Maiato H.. 2013. Spindle assembly checkpoint robustness requires Tpr-mediated regulation of Mad1/Mad2 proteostasis. J. Cell Biol. 203:883–893. 10.1083/jcb.201309076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R.J., Lusk C.P., Dilworth D.J., Aitchison J.D., and Wozniak R.W.. 2005. Interactions between Mad1p and the nuclear transport machinery in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 16:4362–4374. 10.1091/mbc.e05-01-0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepperd L.A., Meadows J.C., Sochaj A.M., Lancaster T.C., Zou J., Buttrick G.J., Rappsilber J., Hardwick K.G., and Millar J.B.A.. 2012. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr. Biol. 22:891–899. 10.1016/j.cub.2012.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetta M., Manzoni R., Mosca R., Mapelli M., Massimiliano L., Vink M., Novak B., Musacchio A., and Ciliberto A.. 2009. The influence of catalysis on mad2 activation dynamics. PLoS Biol. 7:e1000010 10.1371/journal.pbio.1000010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi L., Mapelli M., Knapp S., DeAntoni A., Jeang K.-T., and Musacchio A.. 2002. The Mad1-Mad2 complex: implications of a “safety belt” binding mechanism for the spindle checkpoint. EMBO J. 21:2496–2506. 10.1093/emboj/21.10.2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V., Chan G.K.T., and Yen T.J.. 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154:925–936. 10.1083/jcb.200102093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink M., Simonetta M., Transidico P., Ferrari K., Mapelli M., De Antoni A., Massimiliano L., Ciliberto A., Faretta M., Salmon E.D., and Musacchio A.. 2006. In vitro FRAP identifies the minimal requirements for Mad2 kinetochore dynamics. Curr. Biol. 16:755–766. 10.1016/j.cub.2006.03.057 [DOI] [PubMed] [Google Scholar]
- Vleugel M., Omerzu M., Groenewold V., Hadders M.A., Lens S.M.A., and Kops G.J.P.L.. 2015. Sequential multisite phospho-regulation of KNL1-BUB3 interfaces at mitotic kinetochores. Mol. Cell. 57:824–835. 10.1016/j.molcel.2014.12.036 [DOI] [PubMed] [Google Scholar]
- Yamagishi Y., Yang C.-H., Tanno Y., and Watanabe Y.. 2012. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat. Cell Biol. 14:746–752. 10.1038/ncb2515 [DOI] [PubMed] [Google Scholar]
- Zhang G., Kruse T., López-Méndez B., Sylvestersen K.B., Garvanska D.H., Schopper S., Nielsen M.L., and Nilsson J.. 2017. Bub1 positions Mad1 close to KNL1 MELT repeats to promote checkpoint signalling. Nat. Commun. 8:15822 10.1038/ncomms15822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yin Q., Ling Y., Zhang Y., Ma R., Ma Q., Cao C., Zhong H., Liu X., and Xu Q.. 2011. Two LXXLL motifs in the N terminus of Mps1 are required for Mps1 nuclear import during G(2)/M transition and sustained spindle checkpoint responses. Cell Cycle. 10:2742–2750. 10.4161/cc.10.16.15927 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.