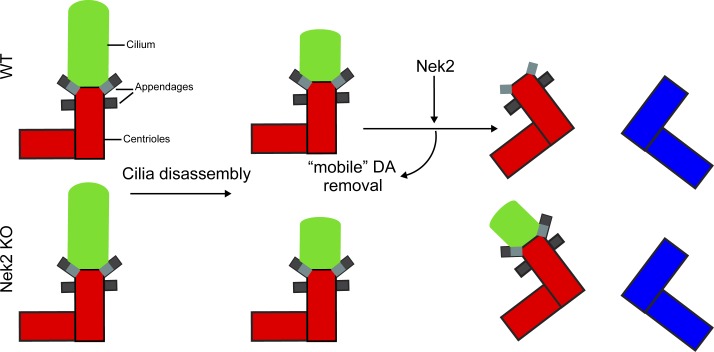
Viol et al. show that the conserved Never-in-mitosis-A–related kinase-2 (Nek2) removes distal appendage components from the mother centriole prior to mitosis in every cell cycle. These findings suggest that Nek2 prevents cilia maintenance during mitosis via distal appendage regulation.
Abstract
Distal appendages (DAs) of the mother centriole are essential for the initial steps of ciliogenesis in G1/G0 phase of the cell cycle. DAs are released from centrosomes in mitosis by an undefined mechanism. Here, we show that specific DAs lose their centrosomal localization at the G2/M transition in a manner that relies upon Nek2 kinase activity to ensure low DA levels at mitotic centrosomes. Overexpression of active Nek2A, but not kinase-dead Nek2A, prematurely displaced DAs from the interphase centrosomes of immortalized retina pigment epithelial (RPE1) cells. This dramatic impact was also observed in mammary epithelial cells with constitutively high levels of Nek2. Conversely, Nek2 knockout led to incomplete dissociation of DAs and cilia in mitosis. As a consequence, we observed the presence of a cilia remnant that promoted the asymmetric inheritance of ciliary signaling components and supported cilium reassembly after cell division. Together, our data establish Nek2 as an important kinase that regulates DAs before mitosis.
Graphical Abstract
Introduction
The centrosome is the main microtubule-organizing center of most animal cells. Each centrosome comprises a pair of orthogonally arranged centrioles surrounded by a well-ordered matrix of pericentriolar material. Several proteins including microtubule nucleators, cell cycle regulators, and signaling molecules represent essential elements of pericentriolar material. Centrosomes perform important functions related to the control of chromosome segregation, cell cycle progression, and cell motility and polarization. In addition, centrioles are the seeding points for the formation of cilia. The primary, nonmotile cilium is a microtubule-based projection that serves as an “antenna” for detecting and responding to external signals (Nigg and Raff, 2009). Cilia can be assembled on almost all cell types in the human body (Olsen, 2005). Dysfunctions in cilia formation lead to impaired cell signaling and/or embryonic development and a wide range of diseases known as ciliopathies (Reiter and Leroux, 2017), whereas loss of cilia, or centrosomes, is associated with multiple types of cancer (Hassounah et al., 2013; Emoto et al., 2014; Nobutani et al., 2014).
Primary cilia formation occurs when cells enter the G0/G1 phase of the cell cycle (Seeley and Nachury, 2010). As the primary cilium is an important signaling hub, the control of cilia assembly and disassembly is critical. In most differentiated somatic cells, the primary cilium is fully disassembled before mitosis before being reassembled following division in G1 of the next cell cycle (Pugacheva et al., 2007; Ford et al., 2018). By contrast, certain neuronal stem cells only partially disassemble the cilium in mitosis. In those cells, a ciliary remnant is retained at one of the two centrosomes during mitosis, to be inherited by one of the two daughter cells after cell division (Paridaen et al., 2013). The molecular mechanisms accounting for synchronizing cilium assembly/disassembly with the cell cycle are only partially understood (Wang and Dynlacht, 2018).
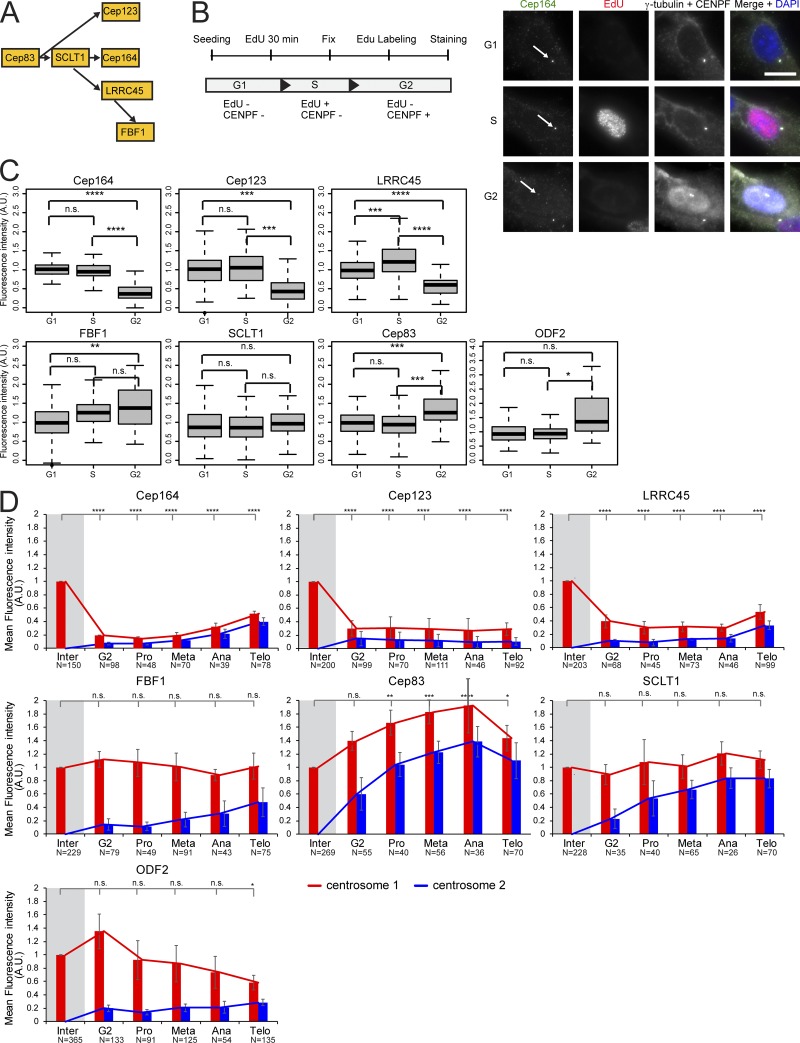
Cilia formation requires the older mother centriole (M-centriole) that is generated one cell cycle earlier than the younger daughter centriole (D-centriole). Only the M-centriole associates with a subset of proteins that form macromolecular structures at the subdistal and distal ends of this centriole. These components appear in electron micrographs as spikelike protrusions that originate from the centriole wall that are referred to as subdistal appendages (SDAs) and distal appendages (DAs). Several components of SDAs and DAs have been identified. SDA formation requires the protein ODF2 that recruits additional SDA components, including Cep170, Cep128, CCDC120, CCDC68, centriolin, and ninein (Mazo et al., 2016). SDAs are involved in microtubule anchoring and centriole/cilia positioning (Vorobjev and Nadezhdina, 1987; Hung et al., 2016; Mazo et al., 2016). DAs comprise the proteins C2CD3 and Cep83 that are essential for the recruitment of additional DAs, such as Cep123/Cep89, SCLT1, LRRC45, Cep164, and FBF1 (Fig. 1 A; Tanos et al., 2013; Ye et al., 2014; Kurtulmus et al., 2018; Wang et al., 2018). DAs are essential at initial stages of cilia formation in order to establish the interaction between the basal body (a modified M-centriole) and the ciliary membrane compartment (Schmidt et al., 2012; Tanos et al., 2013).
Figure 1.
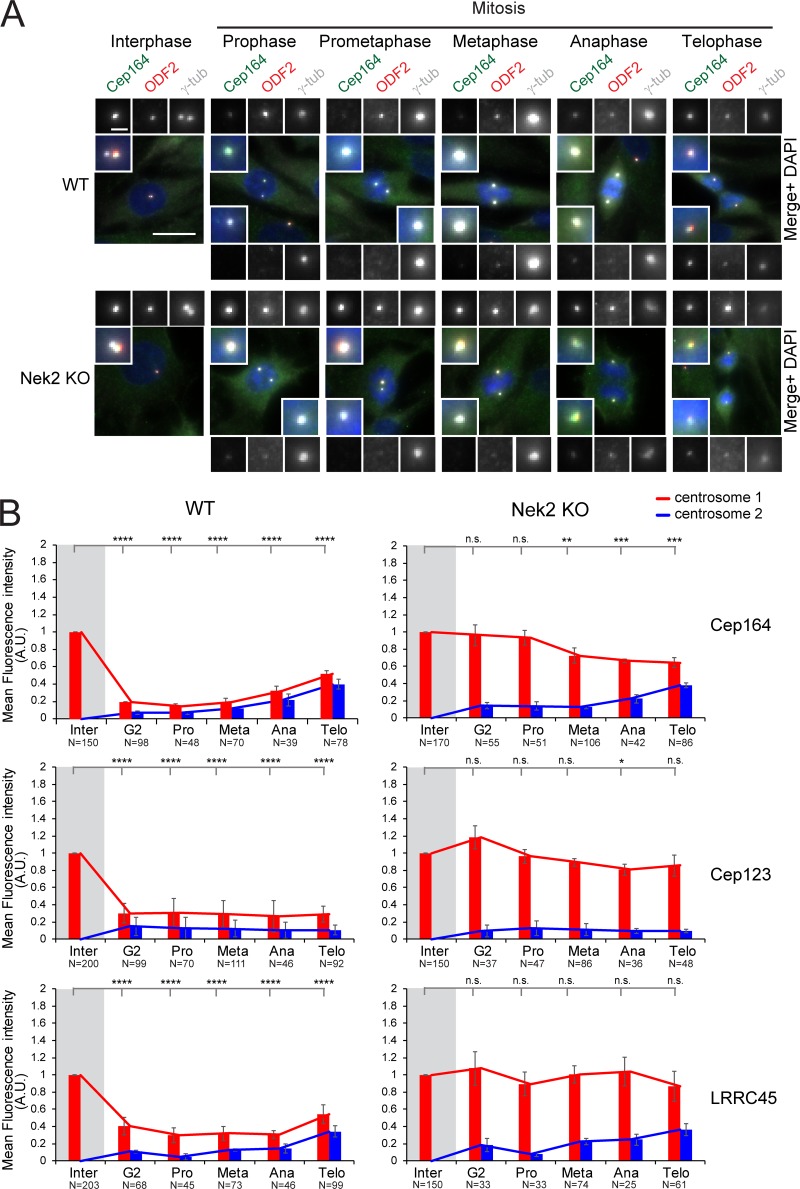
Localization of DAs at the centrosome is cell cycle dependent. (A) Hierarchy of DAs (Tanos et al., 2013; Kurtulmus et al., 2018). (B) Experimental setup to classify cells in G1, S, and G2 phases based on EdU and CENPF staining as depicted. Cells were treated with EdU and stained for Cep164. The arrows mark centrosomal Cep164. γ-tubulin labels centrosomes, and DAPI stained the DNA. Scale bar, 20 µm. (C) Quantification of B. Boxplots show the fluorescence intensity of DAs and ODF2 at centrosomes in A.U. The G1 population was used for normalization. Four independent experiments were done for LRRC45 and three independent experiments for all other proteins. Boxes show interquartile range, lines inside the box represent the median, and whiskers show minimum and maximum values excluding outliers. n = 149 (G1), 107 (S), and 32 (G2) cells per sample. (D) Quantification of cell cycle–dependent behavior of DAs and ODF2 in RPE1 cells. The levels of the indicated protein were measured at each centrosome (centrosomes 1 and 2) during interphase (inter), G2, prometaphase (pro), metaphase (meta), anaphase (ana), and telophase (telo) and were normalized to the average of interphase (marked by gray shading). Bar graphs represent mean ± SD. Numbers below the bars represent total number of cells analyzed in three or four independent experiments. A.U., arbitrary units; n.s., not significant. Significance probability values are: n.s., P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
The establishment of appendage proteins requires 1.5 generations after formation of the new centriole as the D-centriole is converted into an M-centriole that can function as a basal body during ciliation (Nigg and Stearns, 2011). Once assembled, at least some appendage proteins, such as ODF2, remain at the M-centriole throughout the cell cycle (Lange and Gull, 1995), which suggests that cilia disassembly or lack of cilia in mitosis is not regulated at the level of the presence or absence of centriolar appendage proteins. The general validity of this concept was, however, challenged by a number of reports showing that the level of some appendage components drastically decreases during mitosis, in a cell type–dependent manner (Schmidt et al., 2012; Sillibourne et al., 2013; Kong et al., 2014). The underlying molecular mechanism and functional significance of this mitotic regulation remain to be established.
In this study, we investigate the role of the mitotic kinase Nek2 in DA regulation. The first established function of Nek2 was the promotion of centrosome separation at the G2/M transition, when the levels and activity of this kinase peak (Fry et al., 1998b, 2012; Hames et al., 2001; Hames and Fry, 2002; Whitfield et al., 2002; Faragher and Fry, 2003). Nek2 was also shown to regulate cilia disassembly via phosphorylation of the microtubule-depolymerizing kinesin Kif24, to prevent the outgrowth of cilia in proliferating cells (Kim et al., 2015). Nek2 localizes to the proximal ends of both M- and D-centrioles (Fry et al., 1998a). In addition, Nek2 associates with the distal end of the M-centriole and the basal body. Its accumulation at the distal part of the M-centriole is cell cycle dependent and appears to peak in late G2 (Spalluto et al., 2012; Kim et al., 2015). Furthermore, Nek2 phosphorylates the M-centriole–specific component ninein-like protein (Nlp), a protein implicated in microtubule anchoring that is released from the centrosome upon mitotic entry (Casenghi et al., 2003; Rapley et al., 2005). These observations suggest that Nek2 could regulate additional appendage components at the M-centriole at the G2/M transition.
Here, we show that Nek2 is required for the release of a subset of DAs including Cep123, Cep164, and LRRC45 from the M-centriole at the onset of mitosis. In the absence of Nek2, ciliary disassembly is incomplete, leading to the persistence of a ciliary remnant at the older centrosome of mitotic cells. This facilitates cilia elongation shortly after division. Our data thus define Nek2 as an important kinase that controls DAs and timing of cilia formation.
Results
A subset of DAs is displaced from centrosomes in mitosis
To investigate the behavior of DAs on the two centrosomes during mitosis, we quantified the levels of DAs at various cell cycle phases (Fig. 1 and Fig. S1, A and B). Because DAs were previously shown to behave differently depending on the cell type (Schmidt et al., 2012; Sillibourne et al., 2013; Kong et al., 2014), we performed our analysis using human immortalized retina pigment epithelial cells (RPE1) and primary human CD34+ hematopoietic stem and progenitor cells (HSPCs) isolated from healthy donors (Figs. 1 and S1 C). This allowed us to analyze the behavior of DAs throughout the cell cycle in differentiated versus immature/stem cells. We used HSPCs because they are one of the few primary multipotent cells that can readily be identified and isolated from the bone marrow of healthy human subjects.
Figure S1.
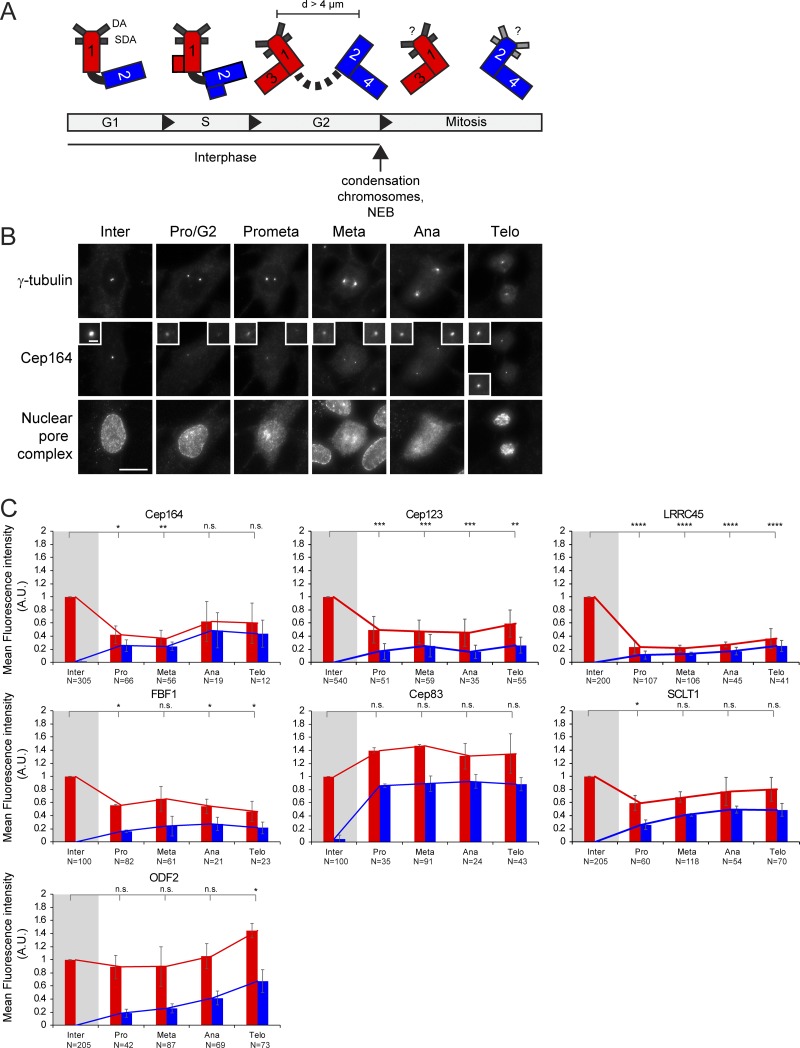
Cell cycle behavior of appendage proteins in primary HSPCs. (A) Centrioles and centrosomes are asymmetric. At G1 phase, the sole centrosome is composed of an M-centriole (1) and a D-centriole (2). At the beginning in S phase and extending in the G2 phase, two new D-centrioles (3 and 4) are generated. The D-centriole (2) acquires appendage proteins later in the cell cycle, whereas the newly formed D-centrioles (3 and 4) are devoid of appendages. Consequently, the centrosome containing the grandmother centriole (1) is the older centrosome in mitosis. The bar indicates the inter-centrosomal distance (d) in G2 phase. (B) Representative immunofluorescence images showing the localization of the DA protein Cep164 at the centrosome in different cell cycle phases in RPE1 cells. γ-tubulin was used as a centrosome marker. The insets represent magnifications of the centrosome signals. Nuclear pore complex antibody Mab414 was used for nuclear envelope staining. Cep164 release from the centrosome starts before nuclear envelope breakdown (NEB; G2/Pro). Scale bars, 20 µm (panel) and 2 µm (inset). (C) Quantification of cell cycle–dependent behavior of DA proteins and ODF2 in primary HSPCs. Appendage proteins were labeled with specific antibodies. γ-tubulin serves as a centrosome marker. The levels of the indicated appendage protein were measured at each centrosome (centrosome 1 and centrosome 2) during interphase (inter), G2, prometaphase (pro), metaphase (meta), anaphase (ana), and telophase (telo) and normalized to the average of interphase (marked by gray shading). Graphs depict fluorescence average intensity in a.u. Graphs show average ± SD of at least two independent experiments. Graphs represent mean ± SD of at least two independent experiments. Numbers below the bars represent total number of cells analyzed for each condition. A.U., arbitrary units; n.s., not significant. Significance probability values are: n.s., P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
In agreement with previous work, the levels of Cep164 and Cep123 at the M-centriole were drastically reduced before mitosis in RPE1 cells (Fig. S1 B; Schmidt et al., 2012; Sillibourne et al., 2013; Kong et al., 2014). Specifically, removal of Cep164 and Cep123 from the M-centriole occurred at the G2 phase, marked by centrosome separation and nuclear accumulation of the centromere protein F (CENPF; Fig. 1, B and C; Liao et al., 1995). Reduced levels were maintained from prophase to telophase (Fig. 1 D). Similar results were obtained for HSPCs (Fig. S1 C). LRRC45 signals emulated those of Cep164 in declining in intensity at one centrosome before mitosis in RPE1 cells and HSPCs (Fig. 1, C and D; and Fig. S1 C). Notably, FBF1 showed a different behavior in RPE1 cells compared with HSPCs. During mitosis, FBF1 levels decreased in HSPCs but maintained interphase levels at one centrosome in RPE1 cells (Figs. 1 D and S1 C) to define a clear cell type–specific regulation for FBF1 recruitment at DAs. Finally, the levels of Cep83 and SCLT1 did not decrease at the M-centriole during mitosis in RPE1 cells (Fig. 1, C and D) or HSPCs (Fig. S1 C) and resembled the behavior of the SDA ODF2 (Figs. 1 D and S1 C).
In conclusion, our data show that the mitotic behavior of appendage proteins reflects the hierarchical network of their assembly (Fig. 1 A). The assembly of appendage proteins, which were released from the centrosome in mitosis (Cep164, FBF1, Cep123, and LRRC45), has been shown to depend on the presence of components that stay on the M-centrosome in mitosis (Cep83 and SCLT1; Tanos et al., 2013; Kurtulmus et al., 2018). Together, our data illustrate the differential centrosome binding of some cohorts of DAs during the cell cycle and suggest a common mode of regulation of appendage dynamics with some cell type–specific components, such as FBF1.
The kinase Nek2 colocalizes with appendages
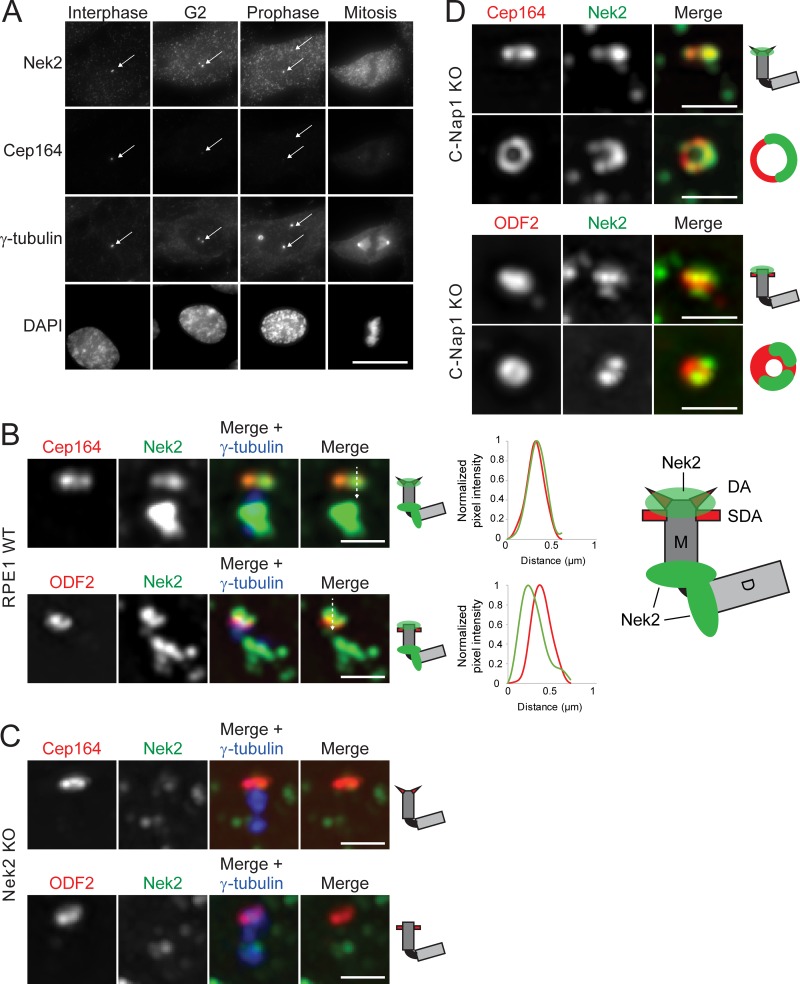
To understand the molecular mechanisms involved in the regulation of DAs during mitosis, we next asked whether the G2/M Nek2 kinase is involved. Concomitant with appendage release, Nek2 accumulates in cells and at centrosomes before mitosis (Fig. 2 A; Fry et al., 2012; Spalluto et al., 2012; Kim et al., 2015).
Figure 2.
Co-localization of the kinase Nek2 with appendages. (A) Representative images show localization of Nek2 and Cep164 in RPE1 cells. γ-tubulin labels centrosomes (indicated by arrows), and DAPI stained the DNA. Scale bar, 20 µm. (B and C) Representative 3D-SIM images of Cep164 and ODF2 costained with Nek2 in RPE1 WT (B) and RPE1 Nek2 KO (C) cells. The association of Nek2 with centrioles is represented in the cartoons. Line graphs show the plot profile of Cep164 or ODF2 (red) and Nek2 (green), plotted according to the line shown in the merge picture. Scale bar, 1 µm. (D) Representative 3D-SIM images of Cep164 and ODF2 costained with Nek2 in RPE1 C-Nap1 KO cells. Scale bar, 1 µm. D, D-centriole; M, M-centriole.
To define the localization overlap of Nek2 with DAs, we performed 3D structural illumination microscopy (3D-SIM). We found that in addition to its proximal pool, Nek2 fully colocalized with Cep164 in 49% of RPE1 cells (Fig. 2 B). A partial colocalization with the SDA ODF2 was observed in 60% of the cells (Fig. 2 B). The Nek2 signal was specific as it was absent in Nek2 knockout (KO) cells (Figs. 2 C and S2 A). As the intensity of the Nek2 signal was much stronger on the proximal versus distal pool (Fig. 2 B), we monitored Nek2 localization in RPE1 C-Nap1 KO cells, which lack the proximal Nek2 pool (Fig. 2 D; Spalluto et al., 2012; Hardy et al., 2014; Panic et al., 2015). The colocalization of Nek2 with Cep164 or ODF2 in interphase cells persisted (Fig. 2 D), indicating that Nek2 binds to the distal ends of the M-centriole independently of the proximal pool. When the plane of observation was aligned with the centriole cross-sectional axis, Nek2 appeared as dots or incomplete rings (Fig. 2 D), to suggest that Nek2 association with appendages might be transient. Together, the localization pattern of Nek2 supports the view of Nek2 being a potential kinase that regulates DAs.
Figure S2.
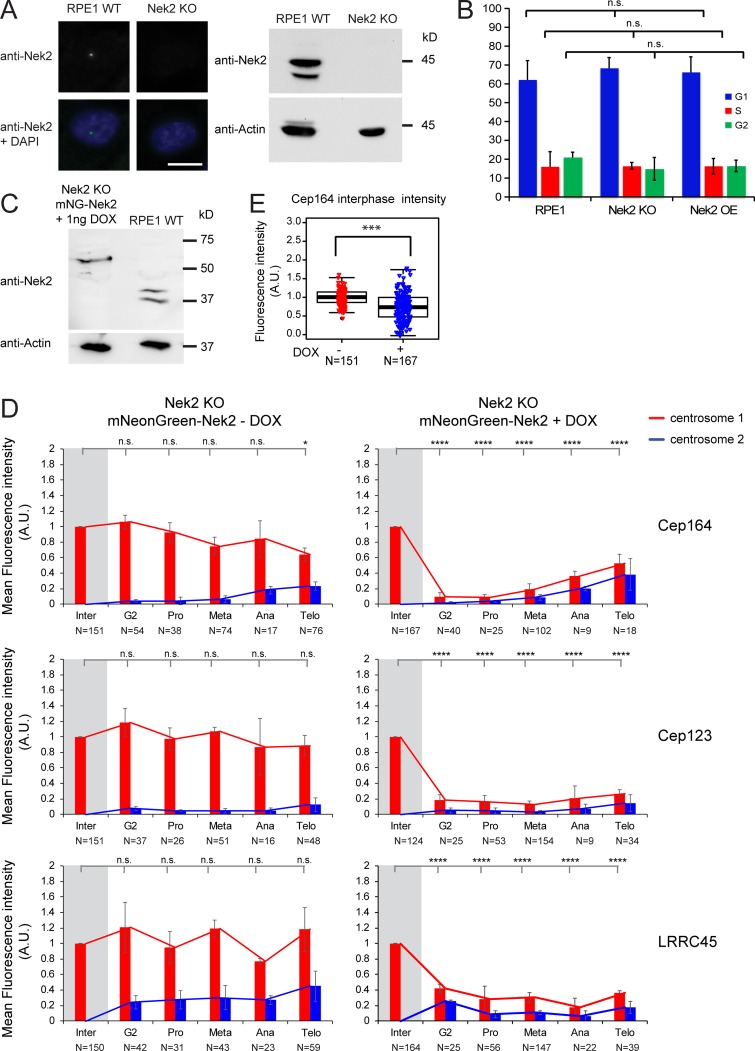
Phenotypical rescue of Nek2 KO cells. (A) Analysis of Nek2 in Nek2 KO cells. Immunofluorescence and Western blot analysis of RPE1 WT and RPE1 Nek2 KO cells stained with anti-Nek2 antibodies. DNA was stained with DAPI, and γ-tubulin serves as a centrosome marker. Actin was used as a loading control. Scale bar, 20 µm. (B) Quantification of the FACS profile of RPE1 WT, Nek2 KO, and DOX-treated Nek2A-mNeonGreen–overexpressing cells stained with propidium iodide for DNA content analysis. Results represent mean ± SD from four independent experiments. (C) Western blot analysis of RPE1 WT and RPE1 Nek2 KO cells carrying mNeonGreen-Nek2A under control of the DOX-inducible promoter treated with 1 ng/ml DOX stained with anti-Nek2 antibodies. The lower Nek2 band (∼45 kD) is the size of endogenous Nek2, and the upper band corresponds to mNeonGreen-Nek2A. Actin was used as a loading control. (D) Quantification of cell cycle–dependent behavior of Cep164, Cep123, and LRRC45 in RPE1 Nek2 KO cells carrying mNeonGreen-Nek2A under control of the DOX-inducible promoter. Cells were incubated with solvent only (− DOX) or 1 ng/ml DOX (+ DOX) for 24 h and analyzed by immunofluorescence. Appendage proteins were labeled with specific antibodies. γ-tubulin serves as a centrosome marker. The levels of the indicated appendage protein were measured at each centrosome (centrosome 1 and centrosome 2) during interphase (inter), G2, prometaphase (pro), metaphase (meta), anaphase (ana), and telophase (telo) and normalized to the average of interphase (marked by gray shading). Graphs depict fluorescence average intensity in a.u. Bar graphs represent mean ± SD. N represents the total number of cells analyzed for each condition in three independent experiments. (E) Box/dot plots show quantifications of the fluorescence intensity of Cep164 in RPE1 Nek2 KO cells carrying mNeonGreen-Nek2A under control of the DOX-inducible promoter using solvent only (− DOX) and 1 ng DOX for 24 h (+ DOX). Dots represent individual cells, boxes show interquartile range, lines inside the box represent the median, and whiskers show minimum and maximum values excluding outliers. N represents the total number of cells analyzed for each condition in three independent experiments. A.U., arbitrary units; n.s., not significant. Significance probability values are: n.s., P > 0.05; *, P ≤ 0.05; ** P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Nek2 removes DAs from M-centrioles before mitosis
Next, we asked whether Nek2 is involved in the release of Cep164, Cep123, and LRRC45 before mitosis. For this, we first monitored the fluorescence levels of these proteins throughout the cell cycle in RPE1 Nek2 KO cells. The KO of Nek2 did not interfere with the cell cycle profile of the cells (Fig. S2 B). In contrast to WT cells, high levels of Cep164, Cep123, and LRRC45 remained at the M-centriole in all Nek2 KO cells during mitosis (Fig. 3, A and B). The mitotic attachment of Cep164, Cep123, and LRRC45 on the M-centrosome in Nek2 KO cells was reverted upon expression of low levels of Nek2A in the Nek2 KO background (Fig. S2, C and D), which only moderately decreased the interphase levels of appendages in comparison with control cells (20% reduction; Fig. S2 E). This indicates that the regulation arose from a specific property of Nek2. Our data establish a key role for Nek2 in the release of DAs before mitosis.
Figure 3.
DAs are not removed from the M-centriole in mitotic Nek2 KO cells. (A) Representative pictures show Cep164 and ODF2 costaining in RPE1 WT and Nek2 KO cells throughout the cell cycle. γ-tubulin was used as a centrosome marker. DNA was stained with DAPI. The insets in each panel show the enlargement of the centrosome area, and the enlargements on the top or bottom of each panel show each single staining. Scale bars, 20 µm (panel) and 2 µm (inset). (B) Quantification of cell cycle–dependent behavior of indicated DAs in RPE1 WT and Nek2 KO cells (done as described in Fig. 1 D). The quantifications for RPE1 WT cells are identical to those in Fig. 1 D, as experiments were done together. Bar graphs represent mean ± SD. Numbers below the bars represent total number of cells analyzed in three independent experiments. A.U., arbitrary units; n.s., not significant. Significance probability values are: n.s., P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
DAs are released from interphase centrosomes by higher levels of Nek2
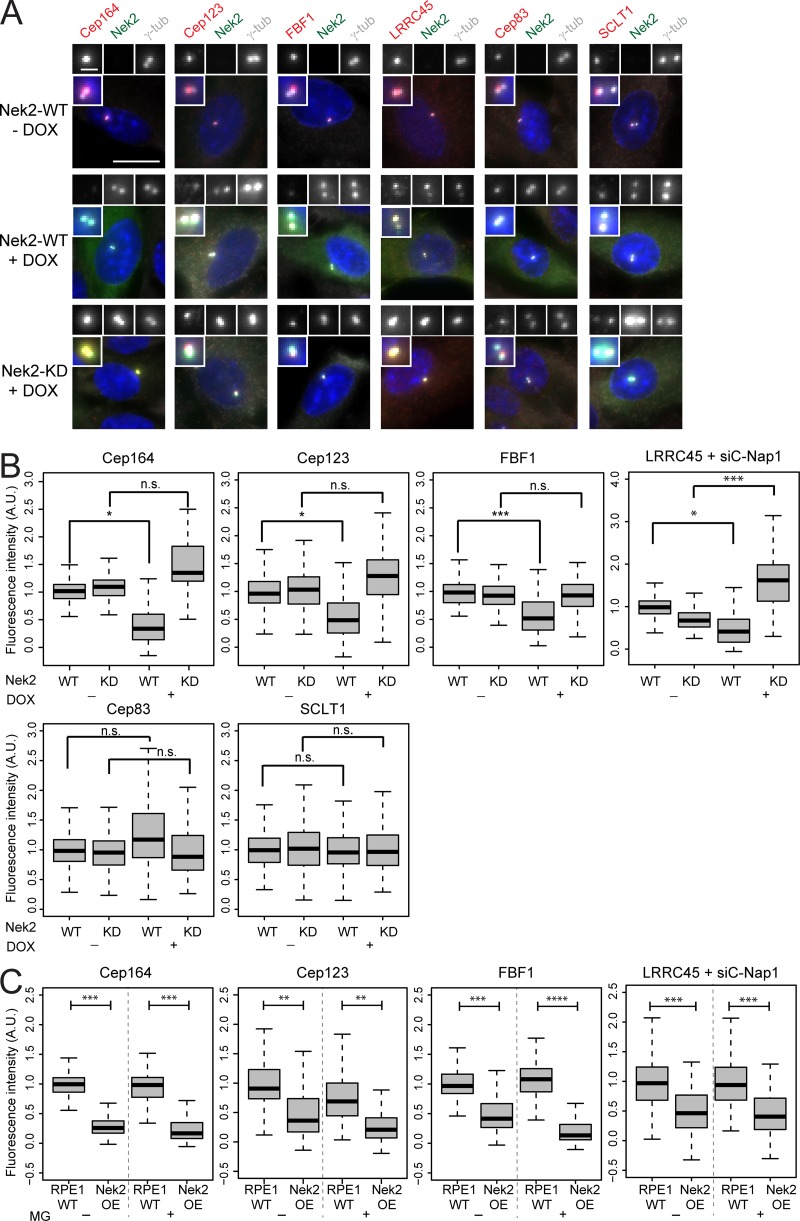
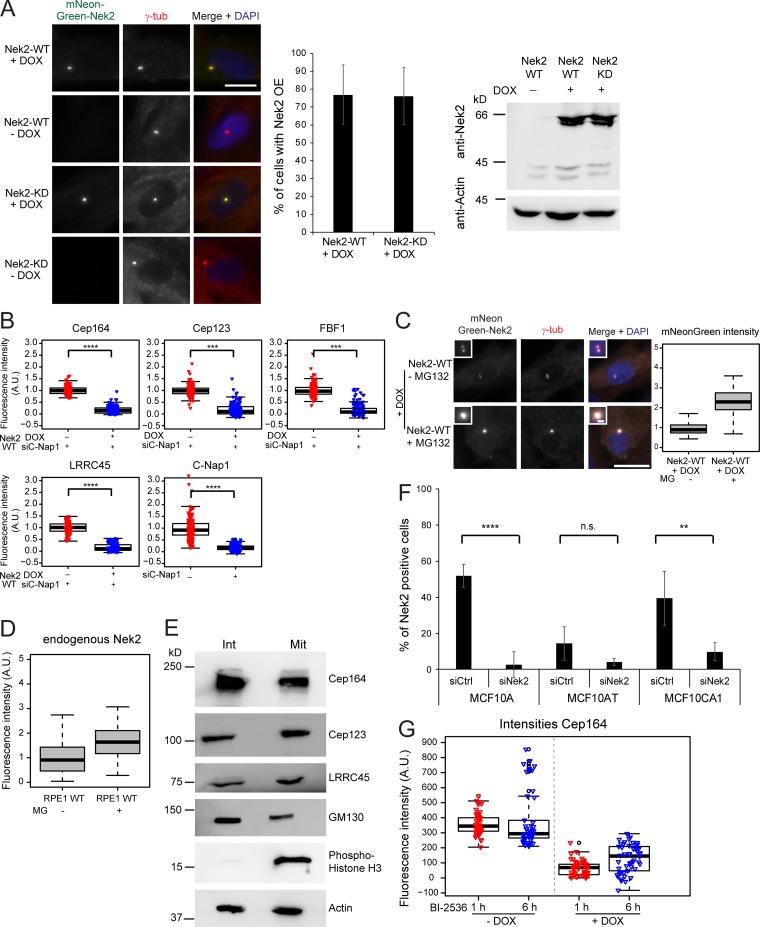
As Nek2 levels peak at G2/M transition (Hames and Fry, 2002), we asked whether overexpression of active Nek2A prematurely displaces DAs from the M-centriole. To address this question, we made use of RPE1 cells stably expressing doxycycline (DOX)–inducible Nek2A or kinase-dead Nek2A (Nek2A-KD) with a K37R substitution (Fry et al., 1995) fused to mNeonGreen (Fig. 4, A and B; and Fig. S3 A). Expression of mNeonGreen-Nek2A and mNeonGreen-Nek2A-KD was observed in ∼75% of cells only in the presence of DOX (Figs. 4 A and S3 A). The overexpression of active, but not inactive, Nek2A caused a significant reduction in Cep164, Cep123, FBF1, and LRRC45 signals at the M-centriole in interphase (Fig. 4 B) in the majority of the Nek2A-overexpressing interphase cells (65–70%). For LRRC45, we analyzed the protein levels in RPE1 C-Nap1–depleted cells, thereby allowing us to exclusively detect the distal pool of LRRC45 (He et al., 2013; Kurtulmus et al., 2018). The analysis of DAs upon C-Nap1 knockdown also confirmed that Nek2A overexpression induced the release of Cep164, Cep123, and FBF1 from centrosomes in the absence of C-Nap1 (Fig. S3 B). This excludes the involvement of Nek2’s proximal pool in DA regulation. Notably, the overexpression of Nek2A-KD led to a significant increase of LRRC45, but no other DAs at the interphase centriole (Fig. 4 B), to reveal a dominant impact of Nek2A upon LRRC45. The removal of DAs by Nek2 did not arise from a pleiotropic impact on all DAs because the levels of Cep83 and SCLT1 remained unchanged upon Nek2A overproduction (Fig. 4, A and B). Also, this effect of Nek2 upon DAs was not due to a shift in cell cycle distribution, as no significant difference was observed in the FACS profile of Nek2-overexpressing and WT cells (Fig. S2 B).
Figure 4.
DAs are released from interphase centrosomes after ectopic Nek2 overexpression. (A) Immunofluorescence of the analyzed appendages in RPE1 cells carrying mNeonGreen-Nek2A (WT) and mNeonGreen-Nek2A-K37R (KD mutant) under control of the DOX-inducible promoter. γ-tubulin labels centrosomes, and DAPI stained the DNA. Insets inside each panel show a magnification of the centrosomal area and enlargements for the indicated staining are shown on top of each panel. Scale bars, 20 µm (panel) and 2 µm (inset). (B) Boxplots show the quantification of DAs at the centrosome in control (− DOX) and Nek2A-overexpressing (+ DOX) samples of A. The control Nek2 WT−DOX sample was used for normalization of three independent experiments (n = 150 cells per condition). For LRRC45 analysis, C-Nap1 was depleted using CEP250 siRNA before DOX treatment and siC-Nap1 (WT−DOX) samples were used for normalization. (C) RPE1 WT and mNeonGreen-Nek2A–expressing cells were treated with the proteasome inhibitor MG132 (+ MG) or solvent only (− MG) during the last 4 h of DOX treatment and analyzed as described in B. n = 150 cells per condition in three independent experiments. Boxes show interquartile range, lines inside the box represent the median, and whiskers show minimum and maximum values excluding outliers. A.U., arbitrary units; n.s., not significant. Significance probability values are: n.s., P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Figure S3.
Analysis of Nek2A overexpression and DAs. (A) Analysis of Nek2A overexpression. Representative images of RPE1 cells carrying either mNeonGreen-Nek2A WT or mNeonGreen-Nek2A-KD under control of the DOX-inducible promoter. Cells were treated with solvent only (− DOX) or DOX (+ DOX) and analyzed by immunofluorescence using γ-tubulin (γ-tub; as a centrosome marker) and direct mNeonGreen fluorescence to detect Nek2. DAPI stained the DNA. Scale bar, 20 µm. Graphs show the quantification of Nek2A-WT– and Nek2A–KD–overexpressing cells. Bar graphs represent mean ± SD. Only cells treated with DOX were analyzed. n = 335 for mNeonGreen-Nek2A (Nek2-WT) and n = 424 for mNeonGreen-Nek2A-KD RPE1 cells in three independent experiments. Western blot analysis of RPE1 cells carrying either mNeonGreen-Nek2A WT or mNeonGreen-Nek2A-KD under control of the DOX-inducible promoter treated with solvent (− DOX) or DOX (+ DOX) and stained with anti-Nek2 antibodies. The upper band corresponds to mNeonGreen-Nek2A, and the lower and weaker Nek2 band is the size of endogenous Nek2. Actin was used as a loading control. (B) Box/dot plots show quantifications of the fluorescence intensity of the indicated proteins upon C-Nap1 siRNA treatment and DOX induction of mNeonGreen-Nek2A (Nek2-WT). Dots represent individual cells, boxes show interquartile range, lines inside the box represent the median, and whiskers show minimum and maximum values excluding outliers. Data from three independent experiments are shown. n = 150 cells per condition. (C) Proteasome inhibition increases the levels of Nek2 at centrosomes. RPE1 cells carrying mNeonGreen-Nek2A were treated with DOX as described in A in the absence or presence of the proteasome inhibitor MG132 (MG). Cells were subjected to immunofluorescence analysis using anti–γ-tubulin (γ-tub) antibodies (centrosome marker). mNeonGreen-Nek2A was visualized by direct fluorescence. DAPI stained the DNA. The boxplot shows the fluorescence intensity in a.u. of mNeonGreen-Nek2A. Boxes show interquartile range, lines inside the box represent the median, and whiskers show minimum and maximum values excluding outliers. n = 50 cells per condition. As published (Spalluto et al., 2012), Nek2 accumulates at the centrosome after proteasome inhibition. Scale bars, 20 µm (panel) and 2 µm (inset). (D) RPE1 cells were treated with solvent (− MG132 [MG]) or MG132 and analyzed by immunofluorescence using anti-Nek2 and anti–γ-tubulin (γ-tub) antibodies. The boxplot shows the fluorescence intensity in a.u. of Nek2 at centrosomes. Boxes show interquartile range, lines inside the box represent the median, and whiskers show minimum and maximum values excluding outliers. n = 50 cells per condition. (E) Western blot analysis of RPE1 cells in interphase (Int) and mitosis (Mit). Mitotic cells were enriched by mitotic shake off. Cells were lysed and analyzed with the indicated antibodies. The appearance of slower migrating forms of GM130 as well as Histone H3 serve as a marker for mitotic cells. Actin was used as a loading control. (F) Quantification of Nek2 knockdown in MCF10A, MCF10AT, and MCF10CA1 cells. The indicated cell lines were treated with control (siCtrl) or Nek2 siRNA and Nek2 centrosomal levels were analyzed by immunofluorescence using anti-Nek2 and γ-tubulin antibodies. The graph shows the percentage of Nek2-positive interphase cells for each condition. Bar graphs represent mean ± SD. n > 450 cells were analyzed per cell type and treatment in a total of four independent experiments (three in the case of MCF10CA1). (G) RPE1 expressing mNeonGreen-Nek2A under control of the DOX-inducible promoter were treated with solvent only (− DOX) or DOX (+ DOX) in the presence of absence of the Plk1 inhibitor BI-2536 for 1 or 6 h. Cells were analyzed by immunofluorescence using anti-Cep164 and γ-tubulin antibodies. γ-tubulin serves as a centrosome marker. The DNA was stained with DAPI. Boxplots show the quantification of Cep164 signal at the centrosome. Dots represent individual cells, boxes show interquartile range, lines inside the box represent the median, and whiskers show minimum and maximum values excluding outliers. Data from one experiment are shown. n = 150 cells per experimental condition. A.U., arbitrary units; n.s., not significant. Significance probability values are: n.s., P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
We next considered the possibility that proteolysis of the phosphorylated proteins by the proteasome could account for the Nek2-driven dissociation of DA proteins from centrioles. However, the intensity of DA signals was still significantly decreased upon Nek2A overexpression when the proteasome was inhibited with MG132 (Fig. 4 C). As described previously (Hames et al., 2005), centrosomal intensity of endogenous and overexpressed Nek2A rose upon MG132 treatment (Fig. S3, C and D). This shows that, despite the lack of impact upon the DA components, MG132 treatment had been effective. These data indicate that protein degradation is unlikely to account for DA removal following Nek2A overproduction. Furthermore, DA protein levels did not decrease in mitotic extracts of RPE1 cells, arguing against a global protein degradation of DAs in mitosis (Fig. S3 E). It is therefore most likely that it is the physical dissociation of the proteins from the DA that accounts for their removal.
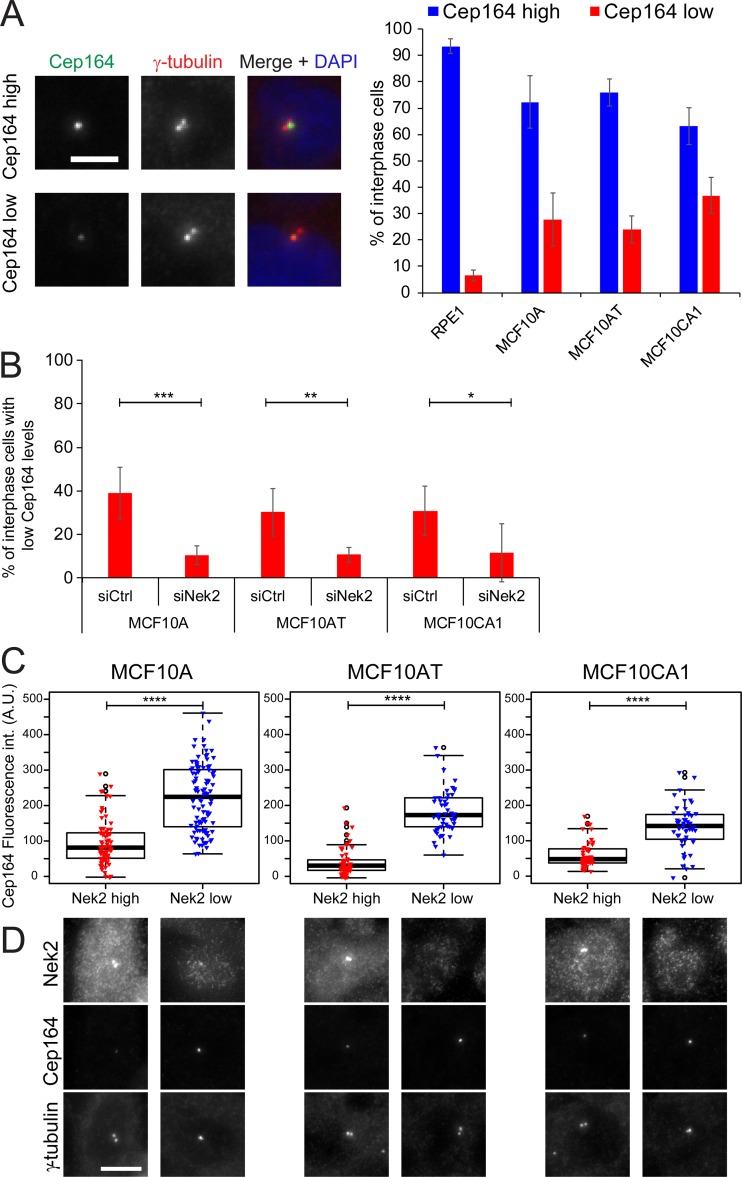
We next asked whether the elevation of Nek2 expression in cell lines other than RPE1 affected DAs. To this end, we focused our attention on mammary epithelial cell lines because of the recorded up-regulation of Nek2 in those cells (Hayward et al., 2004; Zhou et al., 2013; Kim et al., 2015). We analyzed Cep164 centrosomal protein levels in MCF10A (a near-diploid and nontransformed human mammary epithelial cell line; Soule et al., 1990), MCF10AT, and MCF10CA1 lines. MCF10AT and MCF10CA1 lines were derived from MCF10A cells that had been transformed with oncogenic Ras (V12G) and clonally selected after passage in mice. They represent a model for premalignant mammary cells (MCF10AT) and invasive, metastatic carcinoma (MCF10CA1; Dawson et al., 1996; Santner et al., 2001).
Compared with the intensity of signals in RPE1 cells (Fig. 5 A), there was a marked increase in the proportion of cells exhibiting lower levels of Cep164 at interphase centrosomes in MCF10A, MCF10AT, and MCF10CA1 cells. The decrease in Cep164 levels in MCF10-derived cell lines was a direct impact arising from Nek2 function because depletion of Nek2 using specific siRNA reverted the phenotype (Figs. 5 B and S3 F). This impact of Nek2 upon appendages became more apparent when Nek2-positive and -negative cells within the same line were scored separately (Fig. 5 C), as Nek2-positive cells, in which Nek2 accumulated at centrosomes (Fig. 5 D), had significantly diminished levels of Cep164. Thus, high Nek2 levels repress DA association with the M-centriole.
Figure 5.
Interphase Cep164 levels are affected in Nek2-overexpressing breast cell lines. (A) Immunofluorescence analysis of Cep164 centrosomal levels in RPE1, MCF10A, MCF10AT, and MCFCA1 interphase cells. DNA was stained with DAPI, and γ-tubulin labels centrosomes. Percentage of interphase cells with high and low Cep164 levels was quantified. Representative images are shown to the left. Bar graphs represent mean ± SD. n > 900 cells per cell type in five independent experiments. Scale bar, 5 µm. (B) Percentage of cells with low Cep164 centrosomal levels in interphase in control and Nek2 siRNA-treated cells. Cells were analyzed as in A. Bar graphs represent mean ± SD. n > 300 cells per cell type and condition in four independent experiments for MCF10A and MCF10AT and three experiments for MCF10CA1. (C) Correlative analysis of Cep164 and centrosomal Nek2 levels. The box/dot plots show Cep164 fluorescent intensity (int.) measured at the centrosome in interphase cells with high or low Nek2 centrosomal signals (as shown in D). The graphs show one representative experiment. Dots represent individual cells, boxes show interquartile range, lines inside the box represent the median, and whiskers show minimum and maximum values excluding outliers. n = 80 cells per sample. Similar results were obtained in five independent experiments. (D) Pictures show representative images for Nek2-positive and low/negative cells. Specific antibodies against Nek2, Cep164, and γ-tubulin were used. Scale bar, 10 µm. A.U., arbitrary units; n.s., not significant. Significance probability values are: n.s., P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
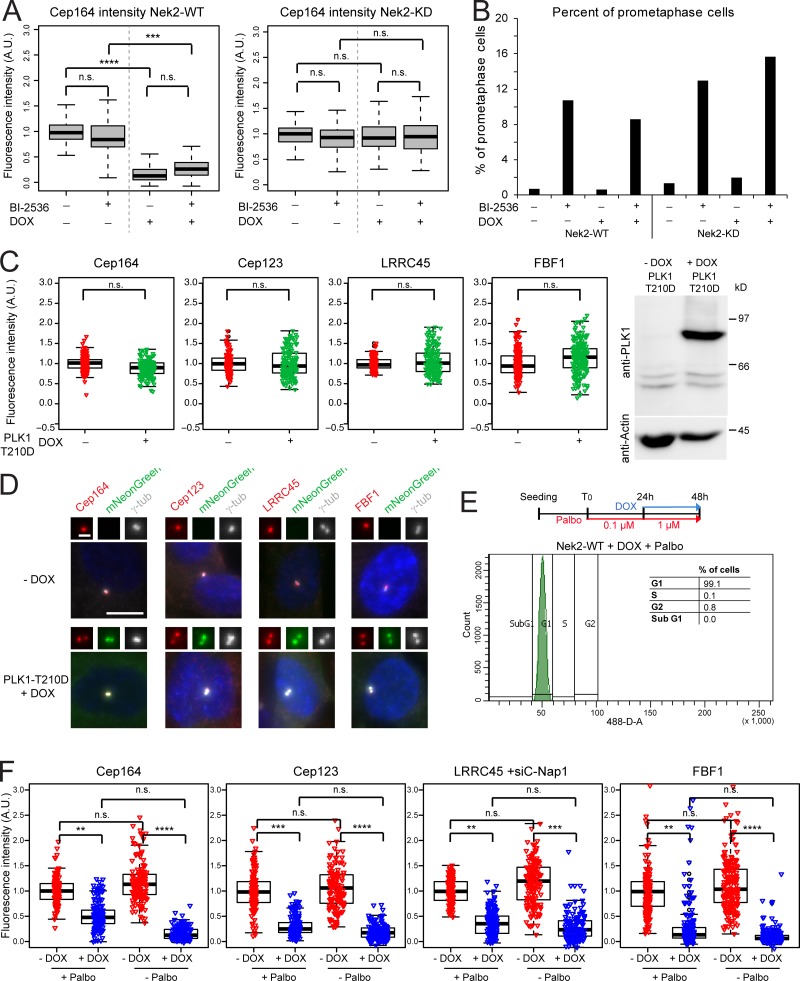
Previously, Nek2 and Plk1 were proposed to work synergistically to remove the centrosomal component Nlp from centrosomes before mitosis (Rapley et al., 2005). By contrast, we find no reliance upon Plk1 function for the impact of Nek2 upon DA loss from centrioles as the presence or absence of the Plk1-kinase inhibitor BI-2536 did not alter the outcome of Nek2 induction (Figs. 6 A and S3 G), yet it led to an accumulation of cells in prometaphase, as reported previously (Fig. 6 B; Sumara et al., 2004). Consistently, overexpression of hyperactive Plk1 had no impact upon the level of Cep164, Cep123, LRRC45, or FBF1 at interphase centrosomes (Fig. 6, C and D), indicating that Plk1 does not phenocopy Nek2 in respect to DA removal. Furthermore, Nek2A overexpression in G1-arrested cells was sufficient to displace DAs similarly to the effect observed in nonarrested interphase cells (Fig. 6, E and F). These data suggest that Nek2 exerts its function over DAs in the absence of most other mitotic kinases (Fig. 6, E and F).
Figure 6.
Nek2-dependent release of DAs is independent of Plk1. (A) RPE1 expressing mNeonGreen-Nek2A (Nek2-WT) or mNeonGreen-Nek2A-KD mutant (Nek2A-KD) under control of the DOX-inducible promoter were treated with solvent only (− DOX) or DOX (+ DOX) in the presence or absence of the Plk1 inhibitor BI-2536. Cells were analyzed using anti-Cep164 antibodies. Boxplots show the quantification of Cep164 signal at the centrosome after normalization to the − DOX − BI-2536 control. Boxes show interquartile range, lines inside the box represent the median, and whiskers show minimum and maximum values excluding outliers. Data from three independent experiments are shown. n = 250 cells per condition. (B) Percentage of prometaphase cells for experiment shown in A was quantified as a control for Plk1 inhibition (Sumara et al., 2004). n = 70 cells per condition. (C) Box/dot plots show the normalized intensities of the indicated proteins in RPE1 cells carrying the hyperactive mutant PLK1-T210D under control of the DOX-inducible promoter. Cells were treated with solvent (− DOX) or DOX (+ DOX). Dots represent individual cells, boxes show interquartile range, lines inside the box represent the median, and whiskers show minimum and maximum values excluding outliers. The graphs show the data from three independent experiments. n = 50 cells were analyzed per condition and experiment. Western blot shows the levels of PLK1-mNeonGreen ± DOX. Actin was used as a loading control. (D) Images show the signal of the indicated DAs (red) in RPE1 cells without (− DOX) or with (+ DOX) overexpression of mNeonGreen-PLK1-T210D. The insets represent magnification of the centrosome area. Scale bar, 10 µm. γ-tub, γ-tubulin. (E) Schematic diagram of the experimental procedure and quantification of the average of cells in G1 phase in three independent experiments analyzed by FACS (n = 37,487 cells). Graph shows a representative FACS plot for propidium iodide detection area (488-D-A). (F) RPE1 mNeonGreen-Nek2A–expressing cells were treated as described in E, and the indicated protein intensities at the centrosome were quantified. Dots represent individual cells, boxes show interquartile range, lines inside the box represent the median, and whiskers show minimum and maximum values excluding outliers. n = 150 cells per condition and protein in three independent experiments. γ-tubulin and DAPI serve as markers for centrosomes and nuclei, respectively. A.U., arbitrary units; n.s., not significant. Significance probability values are: n.s., P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Regulation of DAs by Nek2 does not require ODF2
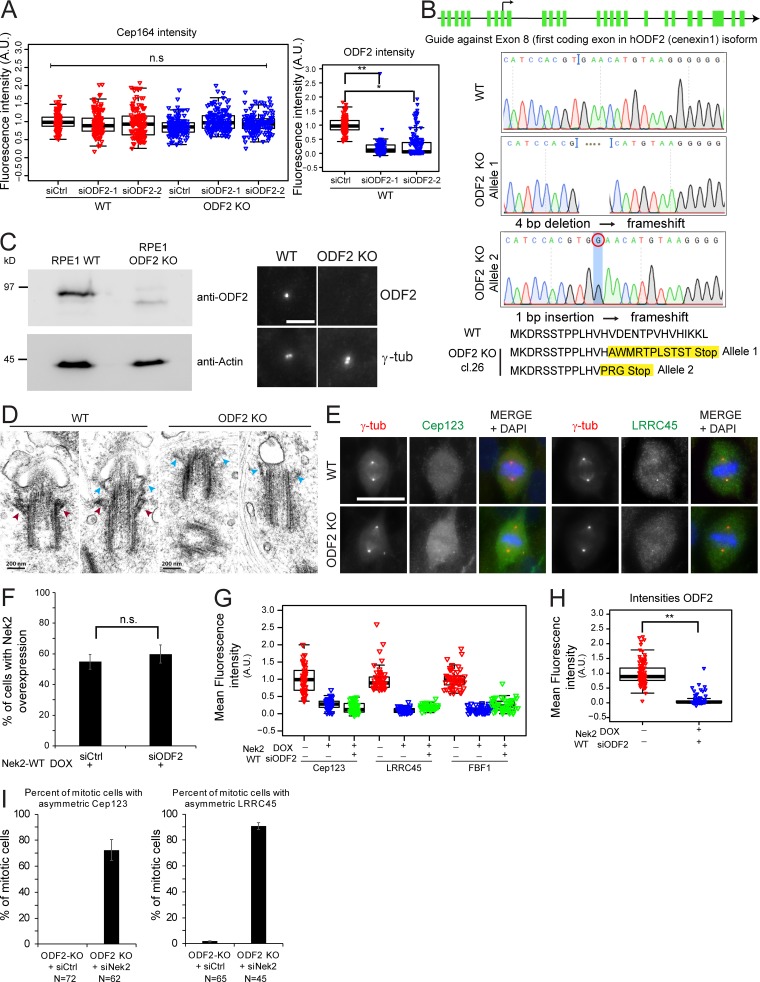
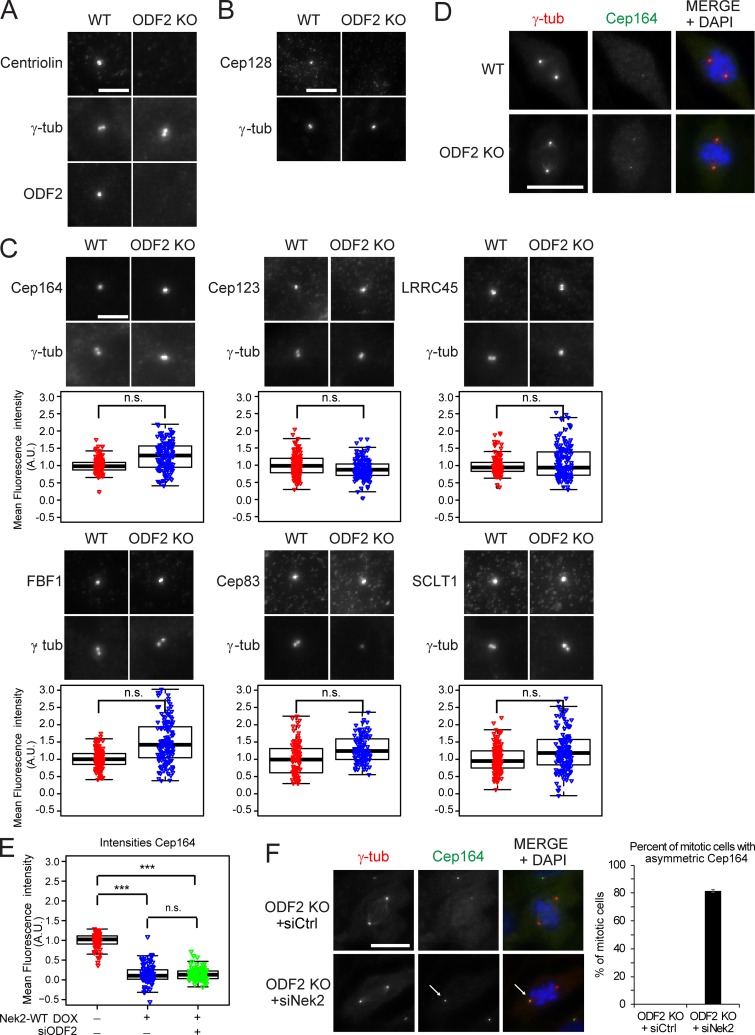
ODF2 was proposed to be required for DA establishment in mouse embryonic stem cells (Ishikawa et al., 2005); however, depletion of ODF2 using siRNAs did not influence Cep164 centrosome association in RPE1 cells (Fig. S4 A; Kuhns et al., 2013). To clarify this discrepancy, we used CRISPR/Cas9 to produce an RPE1 ODF2 KO cell line (Fig. S4, B and C). Electron microscopy confirmed that the absence of ODF2 disrupted SDA formation (Fig. S4 D), while immunofluorescence showed that this loss of SDAs was accompanied by a drastic reduction in centriolin and Cep128 levels at centrosomes (Fig. 7, A and B; Ishikawa et al., 2005; Tateishi et al., 2013; Mazo et al., 2016). Furthermore, RT-PCR of RPE1 Nek2 KO cells indicated that all sequenced transcripts contained the mutated exon (data not shown), excluding the possibility of exon skipping (Uddin et al., 2015; Rodriguez-Rodriguez et al., 2018; Meraldi, 2019; Zhang et al., 2019). Interestingly, the levels of DAs at interphase centrosomes remained unchanged in ODF2 KO cells (Fig. 7 C), even after treatment of these cells with ODF2 siRNAs (Fig. S4 A), further excluding the possibility that partial ODF2 mutants would compensate for the lack of ODF2. Furthermore, Cep164, Cep123, and LRRC45 centrosomal levels decreased during mitosis in ODF2 KO cells to the same degree as in WT cells (Figs. 7 D and S4 E). The most logical conclusion that ODF2 is not required for DA regulation is supported by the decline in the levels of Cep164, Cep123, LRRC45, and FBF1 upon Nek2A overexpression in ODF2-depleted cells (Fig. 7 E; and Fig. S4, F–H). Likewise, Cep164 persisted at the M-centriole during mitosis upon Nek2 knockdown in ODF2 KO cells (Figs. 7 F and S4 I). Thus, we conclude that ODF2 is dispensable for DA formation and regulation by Nek2 in RPE1 cells.
Figure S4.
DAs regulation in ODF2 KO cells. (A) Box/dot plots show the quantification of Cep164 and ODF2 intensity at centrosomes in RPE1 WT and ODF2 KO cells using two different ODF2 siRNAs as indicated. Dots represent individual cells, boxes show interquartile range, lines inside the box represent the median, and whiskers show minimum and maximum values excluding outliers. n = 150 cells were analyzed per condition for both stainings. (B) Schematic representation of the ODF2 KO strategy using CRISPR/Cas9 targeting exon 8 the first coding exon for the hODF2 (cenexin1) isoform. The line represents the region of chromosome 9 that carry the ODF2 gene. The green bars represent the exons of ODF2. Sequencing results and the resulting frameshift mutations, which led to premature stop of translation, are shown. Cl.26, clone 26. (C) Western blot and immunofluorescence analysis of RPE1 WT and ODF2 KO cells using anti-ODF2 antibodies. Actin was used as a loading control, and γ-tubulin (γ-tub) was used as a centrosome marker. Scale bar, 5 µm. (D) Electron micrographs show longitudinal serial sections of RPE1 WT and RPE1 ODF2 KO cells. Cells were serum starved for 48 h before fixation for transmission electron microscopy analysis to induce ciliogenesis and facilitate the recognition of the M-centriole via the associated ciliary membrane. Red arrowheads indicate SDAs, and blue arrowheads show DAs. Early stages of ciliogenesis are shown. Approximately 10 centrioles were analyzed per cell line. Scale bar, 200 nm. (E) Representative images of fixed mitotic RPE1 WT and RPE1 ODF2 KO cells stained using the indicated antibodies for DAs. γ-tubulin (γ-tub; red) and DAPI (blue) serve as markers for centrosomes and nuclei, respectively. Scale bar, 20 µm. (F) Levels of Nek2A overexpression upon ODF2 depletion. Quantification of RPE1 mNeonGreen-Nek2A–positive cells (overexpressed by DOX addition as described in Fig. 7 E) upon control (siCtrl) or ODF2 siRNA treatment. Bar graphs represent mean ± SD. n = 229 (siCtrl) and n = 265 (siODF2) in two independent experiments. (G) Effect of Nek2A overexpression in the presence or absence of ODF2. RPE1 mNeonGreen-Nek2A cells were treated with solvent (− DOX) or DOX (+ DOX) to induced Nek2A overexpression as well as control (− siODF2) or ODF2 siRNA (+ siODF2) to deplete ODF2. Box/dot plots show quantification of the indicated appendage intensities at centrosomes. One representative experiment is shown. Dots represent individual cells, boxes show interquartile range, lines inside the box represent the median, and whiskers show minimum and maximum values excluding outliers. n = 50 cells per condition. Cep164 levels are shown in Fig. 7 E. (H) Box/dot plots show the quantification of ODF2 intensity at centrosomes for G and Fig. 7 E. n = 98 cells per condition. (I) RPE1 ODF2 KO cells were stained with Cep123 or LRRC45 antibodies upon control (Ctrl) and Nek2 siRNA treatment. γ-tubulin and DAPI serve as markers for centrosomes and nuclei, respectively. The graphs show the percentage of mitotic cells in which Cep123 or LRRC45 associates preferentially at one centrosome. Bar graphs represent mean ± SD. Please see Fig. 6 E for quantifications of Cep164 from the same experiment. A.U., arbitrary units; n.s., not significant. Significance probability values are: n.s., P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Figure 7.
Nek2-induced centrosomal appendage release is independent of ODF2. (A and B) Images of RPE1 WT and ODF2 KO cells stained with centriolin, ODF2 (A) and Cep128 (B) antibodies. Scale bar, 5 µm. (C) Images show RPE1 WT and ODF2 KO interphase cells stained for the indicated DAs. Box/dot plots show the normalized quantification of DA signal at the centrosome in RPE1 WT and ODF2 KO cells. Dots represent individual cells, boxes show interquartile range, lines inside the box represent the median, and whiskers show minimum and maximum values excluding outliers. Combined data from three independent experiments are shown (n = 150 cells per condition). Scale bar, 5 µm. (D) Representative images of mitotic RPE1 WT and ODF2 KO cells stained for Cep164. Scale bar, 20 µm. (E) Effect of Nek2A overexpression upon Cep164 in the absence of ODF2. Box/dot plots show quantification of Cep164 intensity in RPE1 mNeonGreen-Nek2A cells treated with control or ODF2 siRNA in the presence (+ DOX) or absence (− DOX) of DOX. Combined data from three independent experiments are shown (n = 150 cells per condition). (F) Images of mitotic RPE1 ODF2 KO cells treated with control (Ctrl) or Nek2 siRNA and stained with Cep164 antibodies. The graph shows the percentage of mitotic cells in which Cep164 associates at one centrosome (arrow in ODF2 KO siNek2 sample, asymmetric Cep164). Bar graphs represent mean ± SD. Scale bar, 20 µm. n = 128 (siCtrl) and n = 108 (siNek2) in two independent experiments. γ-tubulin (γ-tub) and DAPI serve as markers for centrosomes and nuclei, respectively. A.U., arbitrary units; n.s., not significant. Significance probability values are: n.s., P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Impairment of ciliogenesis upon Nek2 overexpression correlates with reduction of DAs
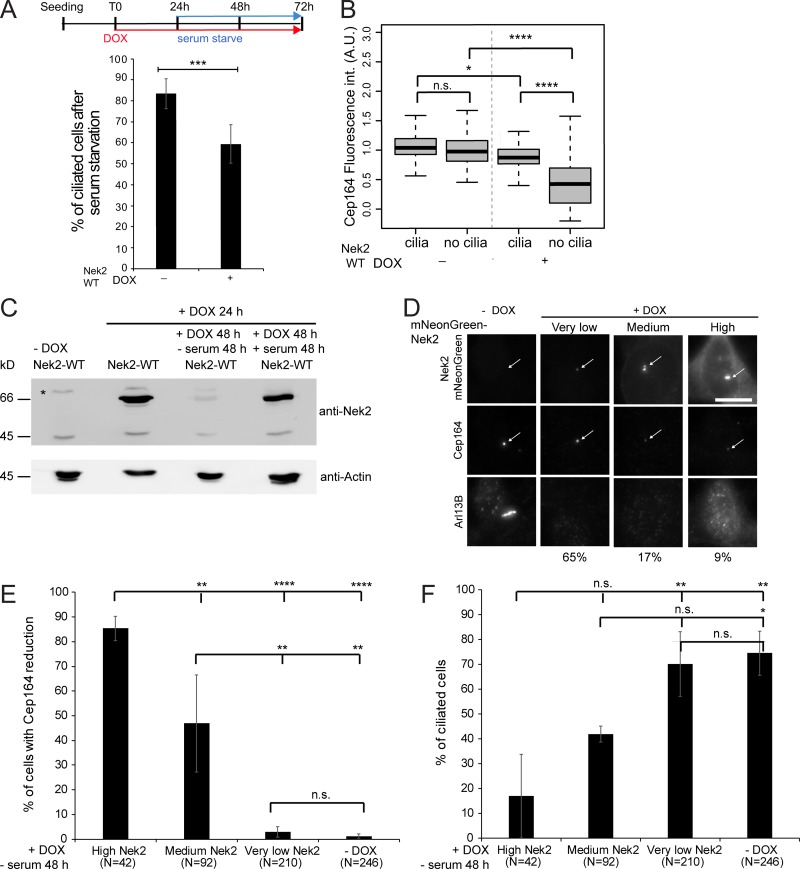
DAs are required for the initial steps of cilia formation (Graser et al., 2007; Schmidt et al., 2012; Tanos et al., 2013). We postulated that high levels of Nek2 might decrease DAs to levels that are unable to support ciliogenesis. To investigate the correlation between Nek2 overexpression, appendage loss, and ciliation, we overexpressed Nek2A in serum-containing medium and induced ciliogenesis after serum withdrawal. In agreement with previous reports (Spalluto et al., 2012; Kim et al., 2015; DeVaul et al., 2017), we observed a significant reduction in the frequency of ciliation when serum starvation was preceded by overexpression of Nek2A (Fig. 8 A). In line with Kim et al. (2015), ciliation was not decreased when Nek2A overexpression was induced after cells had been serum starved (unpublished data), reinforcing the conclusion that Nek2 is unlikely to act on assembled cilia (Kim et al., 2015; DeVaul et al., 2017). Upon Nek2A induction, Cep164 levels at DAs in nonciliated cells were significantly reduced below the levels seen in either ciliated or nonciliated cells in which Nek2A had not been induced (Fig. 8 B).
Figure 8.
DA release is implicated in reducing ciliation upon Nek2 overexpression. (A) Schematic diagram of the experimental procedure in Fig. 8, A–F (each block represents 24 h). Cells were fixed for immunofluorescence and stained for Arl13B (cilia marker), Cep164, and γ-tubulin (basal body markers). The graph shows the percentage of ciliated cells in six independent experiments. Bar graphs represent mean ± SD. n > 250 cells per condition. (B) Quantification of A for Cep164 centrosomal levels in ciliated and nonciliated cells in the presence or absence of DOX. Boxplot shows the data of six independent experiments. Boxes show interquartile range, lines inside the box represent the median, and whiskers show minimum and maximum values excluding outliers. n = 195 cells per condition. int., intensity. (C) Representative Western blot shows mNeonGreen-Nek2A levels detected with anti-Nek2 antibodies upon mNeonGreen-Nek2A (Nek2-WT) overexpression by DOX and serum starvation as indicated. Actin serves as a loading control. Asterisk indicates an unspecific band. The lower Nek2 band (∼45 kD) is the size of endogenous Nek2, and the upper band corresponds to mNeonGreen-Nek2A. (D) Correlative analysis of Nek2 and Cep164 centrosomal levels. Cells were treated as in A and analyzed by immunofluorescence using Cep164 and Arl13B antibodies. mNeonGreen-Nek2A was visualized by direct fluorescence. Cells were categorized as carrying low, medium, and high centrosomal Nek2A levels upon DOX induction. The arrows mark centrosomal mNeonGreen-Nek2A and Cep164. Scale bar, 20 µm. Percentages below the pictures indicate the proportion of cells found for each classification. (E and F) Quantification of D showing the percentage of cells with reduced centrosomal Cep164 levels (E) and ciliated cells (F) in each indicated category in four independent experiments. Bar graphs represent mean ± SD. The number of inspected cells (N) in E and F is indicated below the graphs. A.U., arbitrary units; n.s., not significant. Significance probability values are: n.s., P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
To understand why loss of DAs and cilia was only seen in such a small percentage of Nek2A-overproducing cells, we analyzed Nek2 protein levels at both the level of total cell lysates and at the single cell level. The overproduced Nek2A levels were drastically reduced in the absence of serum (Fig. 8 C), as previously noted for endogenous Nek2 (Kim et al., 2015). In agreement with low Nek2 levels upon serum starvation, we could not detect overexpressed Nek2A in the majority of cells (Fig. 8 D). Only ∼30% of the cells had Nek2 at the centrosomes at medium-to-high levels (Fig. 8 D). The reduction of Cep164 at DAs and loss of cilia were only pronounced in cells with higher Nek2 centrosomal levels (Fig. 8, E and F), explaining the low penetrance of this phenotype upon serum starvation.
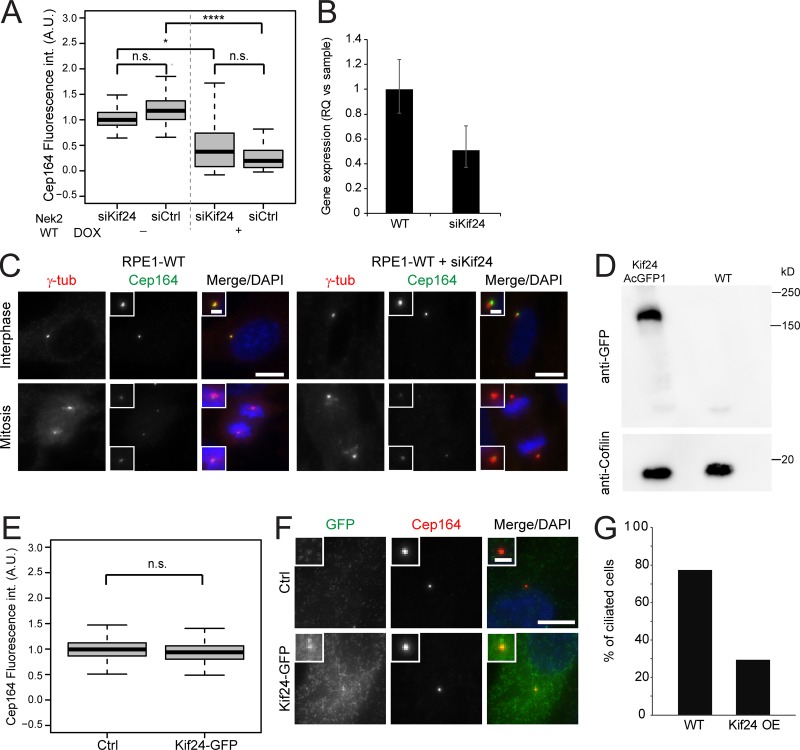
Nek2 has been shown to regulate cilium disassembly via phosphorylation of Kif24 to prevent the outgrowth of cilia in proliferating cells (Kim et al., 2015). We next asked whether Nek2 could regulate DAs via Kif24. To address this hypothesis, we asked whether centriolar removal of Cep164 upon Nek2A overexpression required Kif24. We found that Cep164 declined to similar levels following Nek2 overexpression in interphase cells, irrespective of whether cells had been treated with control or Kif24 siRNA (Fig. 9, A and B). Furthermore, the mitotic decline in Cep164 centrosomal levels in RPE1 cells was independent of Kif24 depletion (Fig. 9 C). Finally, Kif24 overexpression had no impact upon Cep164 levels (Fig. 9, D and F), yet reduced ciliogenesis (Fig. 9 G), as reported previously (Kobayashi et al., 2011; Kim et al., 2015). We therefore concluded that Kif24 is dispensable for appendage regulation by Nek2.
Figure 9.
Effect of Kif24 upon appendages. (A) RPE1 mNeonGreen-Nek2A (Nek2 WT) cells were treated with control (siCtrl) or Kif24 siRNA before addition of solvent (− DOX) or doxycycline (+ DOX). Box/dot plots show quantification of the indicated appendage intensities at centrosomes. Boxplots show fluorescence intensity (int.) measurements for Cep164 in interphase cells. Boxes show interquartile range, lines inside the box represent the median, and whiskers show minimum and maximum values excluding outliers. Data of three independent experiments are shown. n = 175 cells per condition. (B) qPCR was used to quantify Kif24 mRNA levels in RPE1 cells treated with control or Kif24 siRNAs. Results show the RQ value for the respective treatment, and error bars show the respective maximum and minimum RQ value for each sample. One representative experiment of two is shown. (C) Representative images show Cep164 staining in interphase and mitotic RPE1 cells treated with control or Kif24 siRNA. The insets show magnification of the centrosome area. Scale bars, 10 µm (panel) and 2 µm (inset). γ-tub, γ-tubulin. (D) Western blot shows AcGFP1-Kif24 levels using anti-GFP antibodies. Cofilin was used as a loading control. (E) Analysis of Cep164 upon AcGFP1-Kif24 expression under the strong cytomegalovirus promoter. Fluorescence intensity (int.) measurement of Cep164 in control (Ctrl) and AcGFP1-Kif24–transfected RPE1 cells. Boxes show interquartile range, lines inside the box represent the median, and whiskers show minimum and maximum values excluding outliers. Data from three independent experiments are shown. n = 150 per condition. (F) Representative images show AcGFP1-Kif24 and Cep164 centrosomal staining in control (Ctrl) and AcGFP1-Kif24–transfected cells. The insets show enlargements of the centrosomal area. Scale bar, 10 µm (panel) and 2 µm (inset). (G) Ciliation was quantified in control (Ctrl) and AcGFP1-Kif24–transfected RPE1 cells. Cells were transfected with AcGFP1-Kif24 for 48 h and serum starved for 24 h and stained with anti-GFP and anti-Cep164 antibodies. n = 62 (Ctrl) and n = 29 (AcGFP1-Kif24). n.s., not significant. A.U., arbitrary units. Significance probability values are: n.s., P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
A ciliary remnant in mitotic Nek2 KO cells leads to the asymmetric inheritance of ciliary signaling components and asynchronous cilium reassembly
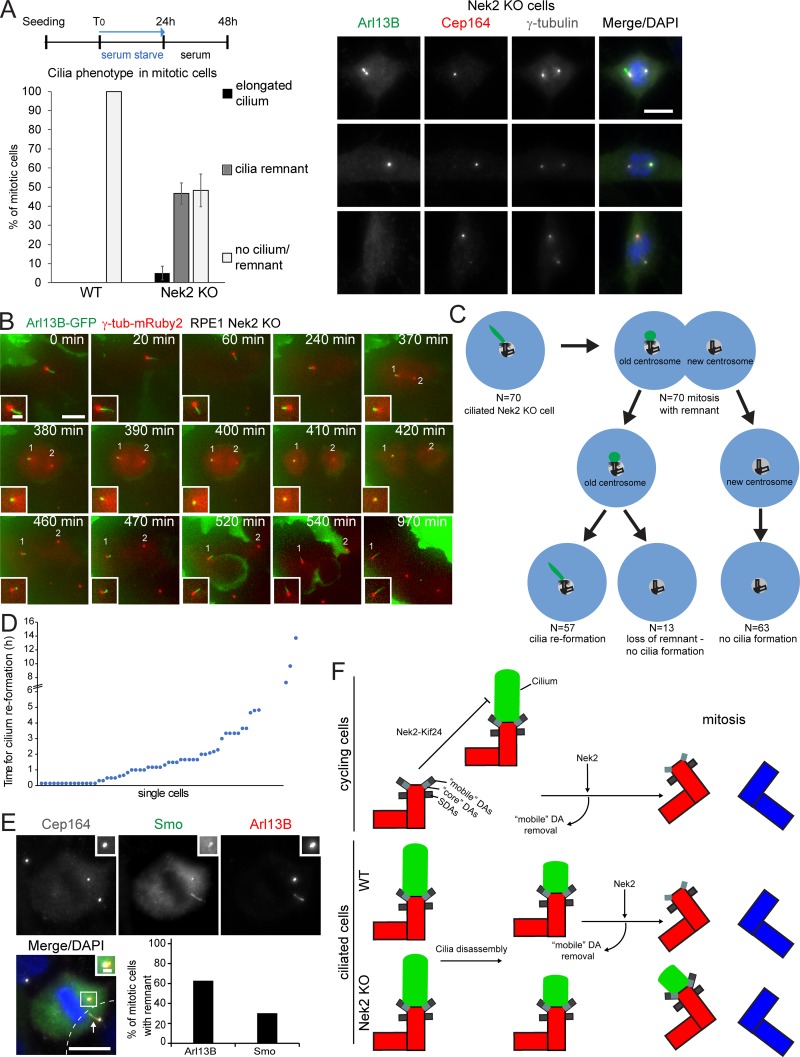
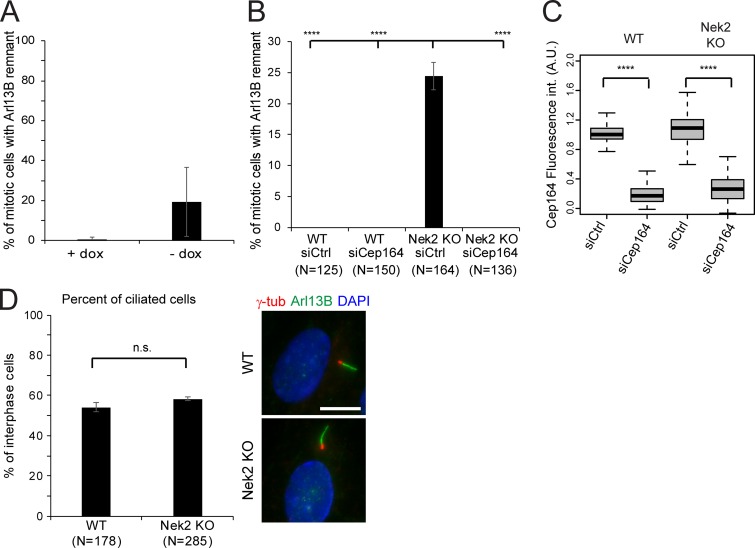
As the primary cilium disassembles before mitosis and assembles again in G1 following division (Wang and Dynlacht, 2018), we asked whether the persistence of DAs in mitosis would influence this process. To this end, we analyzed the behavior of mitotic Nek2 KO cells that retain DAs at their M-centriole. While no cilium or cilia remnant was observed during mitosis in WT cells, ∼20% of mitotic Nek2 KO cells showed the cilia membrane marker Arl13B as a dot-like staining at the M-centriole decorated by Cep164 in cycling cells cultured in serum-containing media (data not shown). The number of mitotic cells with a ciliary remnant increased further after serum-starved ciliated cells were restimulated with serum to induce cilia loss. In this case, no cilia or cilia remnant was observed in mitotic WT cells, yet nearly half of the mitotic cells retained a cilia membrane dot (marked by Arl13B) on the M-centrosome (marked by Cep164) in Nek2 KO cells (Fig. 10 A). This indicates that an Arl13B-associated ciliary membrane (referred as ciliary remnant) remains associated with the M-centriole during mitosis, as previously reported for cells with reduced levels of Nek2 (Spalluto et al., 2012). The phenotype was specific to Nek2 loss, as it could be rescued by the inducible expression of Nek2A in the Nek2 KO background (Fig. S5 A). Furthermore, no mitotic remnant was detected after depletion of Cep164 (Fig. S5, B and C), confirming that Arl13B docked at the M-centriole in a Cep164-dependent manner. This phenotype was not a consequence of differences in ciliogenesis because RPE1 WT and Nek2 KO interphase cells displayed a similar capacity for ciliation (Fig. S5 D).
Figure 10.
The ciliary remnant in mitotic Nek2 KO cells leads to asynchronous cilium reassembly. (A) Schematic diagram of the experimental procedure and quantification of RPE1 WT and Nek2 KO mitotic cells analyzed using Cep164, Arl13B, and γ-tubulin antibodies. Bar graphs represent mean ± SD. n = 161 (WT) and 232 (Nek2 KO) mitotic cells in three independent experiments. Representative cells are shown to the right. Scale bar, 10 µm. (B) Representative still images of live-cell imaging performed with RPE1 Nek2 KO cells stably expressing Arl13B-GFP and γ-tubulin-mRuby2 (Video 1). Cells were treated as detailed in Fig. 8 A. The insets show magnifications of the basal body area. The numbers inside the panels indicate the position of the two centrosomes. Scale bars, 10 µm (panel) and 2 µm (inset). (C and D) Quantification of Arl13B-GFP remnant inheritance of B. Only ciliated cells that entered mitosis during the time of inspection were analyzed as depicted in C. N indicates the number of inspected cells in three independent experiments. The time of cilia reformation after cell division is plotted for individual cells in D. (E) Nek2 KO cells were treated as shown in Fig. 10 A and serum restimulated in the presence or absence of SAG. Representative image shows Nek2 KO cells stained with Arl13B and Smo as cilia marker and γ-tubulin as basal body marker. DNA was stained with DAPI. The cilium from a neighboring cell (cell boundaries indicated by dashed lines) is marked by an arrow. The graph shows the percentage of mitotic cells with Arl13B- and Smo-positive remnants in n = 43 cells. Scale bars, 10 µm (panel) and 2 µm (inset). (F) Schematic model for the function of Nek2 in cilia and appendage regulation. In cycling cells, Nek2 inhibits cilia assembly in a Kif24-dependent manner and removes DAs from the M-centriole before mitosis independently of Kif24. In ciliated RPE1 cells, cilia disassemble before mitosis. In WT cells, the displacement of “mobile” DAs prevents ciliary vesicle attachment in mitosis. In the absence of Nek2, maintenance of DAs results in a mitotic cilia remnant with the consequence of premature cilia reformation in the next cell cycle. A.U., arbitrary units; n.s., not significant. Significance probability values are: n.s., P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Figure S5.
The ciliary remnant in mitotic Nek2 KO cells can be rescued upon siRNA of Cep164 and expression of Nek2 in the Nek2 KO background. (A) Quantification of mitotic cells with Arl13B remnants at centrosomes in RPE1 Nek2 KO cells expressing DOX-inducible mNeonGreen-Nek2A. Cells were analyzed in the presence or absence of DOX. Bar graphs represent mean ± SD. n = 306 and 292 mitotic cells were analyzed without and with DOX treatment, respectively in three independent experiments. (B) Quantification of mitotic cells with Arl13B remnants at centrosomes in RPE1 WT and Nek2 KO cells treated with control (Ctrl) or Cep164 siRNA as indicated. The graph shows the average and SD of three independent experiments. The graph represents mean ± SD of three independent experiments. Numbers below the bars represent the total number of cells analyzed for each condition. (C) Boxplots represent the quantification of Cep164 appendage signal at the centrosome in RPE1 WT and Nek2 KO cells treated with control (Ctrl) or Cep164 siRNA as indicated. Data from three independent experiments are shown. Boxes show interquartile range, lines inside the box represent the median, and whiskers show minimum and maximum values excluding outliers. n = 150 cells were analyzed for each condition. (D) Ciliation was examined in RPE1 WT and Nek2 KO cells after 48 h of serum starvation. Results show the mean ± SD of three independent experiments. Results show the average and SD of three independent experiments. Numbers below the bars represent the total number of cells analyzed for each condition. Representative images are shown on the right. Arl13B was used as a cilia and γ-tubublin (γ-tub) as a basal body marker. DNA was stained with DAPI. Scale bar, 10 µm. A.U., arbitrary units; n.s., not significant. Significance probability values are: n.s., P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
To understand how cilia behave in Nek2 KO cells, we performed live-cell imaging. The videos revealed that ciliated Nek2 KO cells shortened the cilium to a remnant during mitosis, from which a new cilium was formed after division (Fig. 10 B and Video 1). We determined that the daughter cell that retained the cilia remnant reformed a cilium in 2 ± 2.6 h after cell division (Fig. 10, C and D). We did not observe cilia reformation in the daughter cell that inherited the younger centrosome (not decorated by Cep164) within a time period up to 19.3 h. These data suggest that the presence of DAs during mitosis in Nek2 KO cells might contribute to cilia remnant maintenance during mitosis and faster cilia reformation after cell division. This mechanism reflects asymmetric cell division since it gives rise to two different daughter cells.
Video 1.
Inheritance of a ciliary remnant in mitotic Nek2 KO cells and subsequent cilia reformation after mitosis. Live-cell imaging related to Fig. 10 B. Time-lapse epifluorescence microscopy was performed with RPE1 Nek2 KO cells stably expressing Arl13B-GFP (green) and γ-tubulin-mRuby2 (red). Cells were serum starved for 24 h and subsequently serum restimulated for 24 h before inspection. Cells were inspected for 24 h with a time interval of 10 min. Speed = 3 frames per second.
Finally, we asked whether the mitotic cilia remnant in RPE1 Nek2 KO cells was able to recruit signaling components. The membrane protein Smoothened (Smo), one of the hedgehog (Hh) pathway components, localizes to cilia when stimulated by Hh ligand or by pathway agonists such as N-methyl-N′-(3-pyridinylbenzyl)-N′-(3-chlorobenzo[b]thiophene-2-carbonyl)1,4-diamino-cyclo-hexane (SAG; Chen et al., 2002). In addition, overexpressed Smo enters the cilium in nonstimulated RPE1 cells and, similar to Arl13B, serves as a cilia membrane marker (Rohatgi et al., 2009). Like Arl13B, the mitotic cilia remnant was visible during live-cell imaging of Nek2 KO cells using overexpressed Smo-EGFP (Video 2). Immunofluorescence using an antibody that recognizes endogenous Smo indicated that Smo decorated the mitotic remnant upon SAG activation (Fig. 10 E). We thus conclude that the mitotic cilia remnant can recapitulate the cilia property of retaining signaling molecules.
Video 2.
The ciliary remnant in mitotic Nek2 KO cells is positive for Smo. Live-cell imaging related to Fig. 10 E. Nek2 KO cells expressing Tet3G-Smo-EGFP (green) were treated with DOX for 24 h to induce Smo expression, serum starved for 24 h, and subsequently incubated in medium containing serum in the presence or absence of 0.4 µM SAG. After 24 h, cells were analyzed by time-lapse epifluorescence microscopy and inspected for 24 h with a time interval of 10 min. Speed = 3 frames per second. 20 nM sirTubulin (red) and 5 µM verapamil were added 3 h before imaging.
Discussion
DA behavior during mitosis
The importance of DAs for cilia biogenesis is well documented (Graser et al., 2007; Schmidt et al., 2012; Tanos et al., 2013), yet their behavior and function during mitosis remain poorly understood. The systematic analysis of DAs in RPE1 cells and HSPCs identified common modes of mitotic behavior for most of the DAs analyzed, including Cep83, SCLT1, FBF1, LRRC45, Cep123, and Cep164. Of these DA components, Cep83 and SCLT1 remained at the older centrosome throughout the cell cycle, to emulate the behavior of the SDA marker ODF2 (Fig. 1; Kong et al., 2014). In contrast, LRRC45, Cep123, and Cep164 were displaced from the M-centriole before mitosis. Interestingly, this behavior of DAs reflects the mode of their assembly (Tanos et al., 2013; Kurtulmus et al., 2018): a stable core composed of Cep83 and SCLT1 recruits the components decreasing in mitosis (Cep164, Cep123, and LRRC45). Similar results were recently reported in a study applying super-resolution microscopy (Bowler et al., 2019).
We propose that DAs can be subdivided into two categories: “core components” with scaffolding functions and “mobile components,” the latter of which are released from the older centrosome in every cell cycle. This suggests a dynamic cell cycle–dependent regulation of DA composition. Such a regulation would allow cells to differentially modulate the recruitment of DAs to mitotic centrosomes depending on the cell type or cellular environment. Cep164 is a good example for a DA shown to behave differently between two cell types (Graser et al., 2007; Schmidt et al., 2012; Kong et al., 2014). Another example is FBF1, which remains at the older centrosome in RPE1 cells (Fig. 1; Tanos et al., 2013), but not in HSPCs (Fig. S1 C). In this context, FBF1 is removed from mitotic M-centrioles as reported for HeLa cells (Fig. S1 C; Kong et al., 2014). The physiological significance of this fascinating cell type–specific regulation remains to be determined. It is puzzling that FBF1 stays at the M-centriole in mitosis in RPE1 cells; yet, the FBF1-targeting protein LRRC45 (Kurtulmus et al., 2018) is partly removed from this structure. This suggests that either residual levels of LRRC45 are sufficient to keep FBF1 at mitotic centrioles or FBF1 might be recruited to mitotic centrioles in an LRRC45-independent manner. Indeed, FBF1 was recently reported to localize at the proximal ends of the centriole in RPE1 and HeLa cells (Inoko et al., 2018). Super-resolution microscopy also indicated that FBF1 localization differs from that of other DAs as FBF1 forms bladelike structures that intercalate with DAs (Yang et al., 2018). Therefore, the hierarchical appendage formation model might not apply to FBF1, depending on the cell type or cell cycle stage.
DA regulation by Nek2
Several lines of evidence support a key role for Nek2 kinase in DA regulation. First, Nek2 increases in levels and activity concomitantly with the decrease of appendage levels in the G2/M transition (Fry et al., 1998b, 2012; Hames et al., 2001; Hames and Fry, 2002; Whitfield et al., 2002). Second, besides its localization at the proximal ends of both M- and D-centrioles (Fry et al., 1998a), Nek2 has also been mapped to colocalize with the distal ends of the centriole by super-resolution microscopy (Fig. 2). Furthermore, we found that overexpression of active Nek2A, but not Nek2A-KD, prematurely displaced Cep164, Cep123, and LRRC45 from M-centrioles in interphase. This displacement was independent of C-Nap1, indicating that Nek2 controls DAs independent of Nek2 regulation of the centrosome linker. Finally, in the absence of Nek2, DAs (Cep164, Cep123, and LRRC45) remained at the M-centriole throughout mitosis.
The mechanism by which Nek2 removes DAs remains to be unraveled. As proteasome inhibition did not prevent the release of DAs upon Nek2A overexpression, we reason that displacement rather than degradation underlies the regulation of DAs by Nek2. This assumption is further supported by the observation that DAs are not degraded in mitosis (Fig. S3 E; Schmidt et al., 2012).
Nek2 might, however, not be the only kinase regulating DAs. In the case of Nlp, Rapley et al. (2005) showed that Nek2 was joined by hyperactive Plk1 in displacing Nlp from the centrosome when overexpressed. In our analysis, Plk1 inhibition had no impact on DA removal by overproduced Nek2A. In addition, overexpression of hyperactive Plk1 did not change DA levels in interphase, suggesting that this process is primarily dependent on Nek2. However, a role of Plk1 in mitosis cannot be excluded. Indeed, a recent study reports that inhibition of Plk1 increased the levels of mitotic DAs to some extent (Bowler et al., 2019). These results point toward a regulation of DAs by multiple mitotic kinases.
Nek2 is a proto-oncogene that is overexpressed in several forms of cancer, including breast cancer (Hayward et al., 2004; Zhou et al., 2013; Cappello et al., 2014). By analyzing mammary epithelial MCF10-derived cell lines with increased levels of Nek2 but decreased primary cilia (Yuan et al., 2010; Kim et al., 2015), we detected a higher percentage of interphase cells with reduced Cep164 levels compared with RPE1 cells. Cep164 levels could be partly restored upon Nek2 depletion, implying that high Nek2 levels were indeed the cause of reduced centrosomal Cep164. It is thus tempting to speculate that DA mis-regulation might be one underlying mechanism that contributes to cilia loss in tumors overexpressing Nek2.
Influence of SDAs in DA regulation
ODF2 has been previously shown to be required for DA assembly in mouse embryonic F9 cells (Ishikawa et al., 2005; Tateishi et al., 2013); however, the analysis of other cell types did not confirm this observation after ODF2 depletion (Kuhns et al., 2013; Tanos et al., 2013), suggesting that residual ODF2 levels could be sufficient for DA assembly. Using RPE1 ODF2 KO cells, we now show that DA assembly and regulation by Nek2 are independent of SDAs in RPE1 cells. In ODF2 KO cells, the levels of DAs at the M-centriole during interphase were not decreased in comparison with those of WT cells. This implies that the dependency of ODF2 for DA formation is most likely cell type dependent.
Consequences of impaired DA regulation
Why should DAs be released from the M-centriole during mitosis? One possibility is that DAs perform functions outside the centrosome. For instance, Cep164 was proposed to be involved in DNA damage control, genomic stability, and cell cycle progression (Sivasubramaniam et al., 2008; Pan and Lee, 2009; Chaki et al., 2012; Slaats et al., 2014). However, centrosome-independent functions of Cep164 have been disputed in a recent publication using RPE1 Cep164 KO cells (Daly et al., 2016). Another reason could be related to their role in cilia biogenesis. Our data show that in RPE1 WT cells, the cilium is fully disassembled before mitosis, as no residual ciliary membrane marker could be observed at mitotic centrosomes. This scenario was different in Nek2 KO cells. In Nek2 KO cells, the percentage of mitotic cells in which the older centrosome was decorated with the cilia marker Arl13b increased considerably. This is in agreement with a previous report showing that siRNA-mediated depletion of Nek2 compromised the cells’ ability to resorb cilia in G2 and prophase (Spalluto et al., 2012). Kim et al. (2015) showed that Nek2 prevents outgrowth of cilia in proliferating cells via phosphorylation of the kinesin motor protein Kif24, thereby likely blocking cilia formation at the G2/M transition. However, in our study the impact of Nek2 upon DAs was not dependent on the activity of Kif24. These data suggest that the Nek2-dependent appendage release at the G2/M transition might constitute a fail-safe mechanism that prevents the persistence of ciliation throughout mitosis and that most likely operates in parallel to the Nek2–Kif24 pathway. We propose that Nek2 has a dual function in controlling ciliogenesis: it regulates Kif24 to inhibit cilia assembly (Kim et al., 2015), while also promoting the mitotic displacement of DAs to ensure the detachment of ciliary vesicles during mitosis (Fig. 10 F).
Our live-cell imaging analysis established that cilia growth from older centrosomes associated with a ciliary remnant seems to initiate faster than a cilium is nucleated from the “naked” older centrosomes that lack a remnant (Anderson and Stearns, 2009). We therefore suppose that, by regulating the extension of cilia disassembly, cells could modulate the speed of cilia regrowth in daughter cells just after cell division. The retention of DAs in mitosis could be part of such a mechanism. Interestingly, neural stem cells were shown to keep a ciliary remnant at the older centrosome in mitosis. The inheritance of the ciliary remnant seems to give cells a temporal advantage in reforming the cilium and becoming responsive to Hh ligands (Paridaen et al., 2013). The centrosomal association of the ciliary membrane in dividing neural stem cells decreased at the later stages of neurogenesis when cells differentiate (Paridaen et al., 2013). The molecular mechanisms underlying this change in behavior are unknown. It would be interesting to determine whether Nek2 controls the behavior of daughter cells by modulating the levels of DAs during mitosis in differentiated versus stem cells.
Materials and methods
Plasmid constructions
Plasmids were L13-Arl13B-GFP (no. 40879; Addgene; Larkins et al., 2011); pRetroX-TRE3G (Takara Bio) with murine Smo-EGFP, monomeric mNeonGreen, and pQCXIZ-TUBG1-mRuby2 (a gift of Elmar Schiebel, University of Heidelberg, Heidelberg, Germany); and AcGFP1-Kif24 (Miyamoto et al., 2015). pRetroX-TRE3G-mNeonGreen-PLK1-T210D was generated by subcloning PLK1-T210D from pCMV-3Tag-1C-PLK1-T210D (Schmidt et al., 2012) into pRetroX-TRE3G-mNeonGreen. For expression of gRNAs and Cas9, the two oligos containing the forward and reverse guide sequences targeting exon 8 of ODF2 isoform 9 (also known as cenexin1, NM_002540.4) were 5′-CACCGTCCCCCCTTACATGTTCACG-3′ and 5′-AAACCGTGAACATGTAAGGGGGGAC-3′. Briefly, oligos were annealed by incubating 1 µl of each oligo (100 µM) with 0.5 µl of T4 Polynucleotide kinase (no. M0201S; New England Biolabs) and 1 µl of 10× ligation buffer in a total of 10 µl for 30 min at 37°C. The reaction was further incubated at 95°C for 5 min with subsequent cooling to 25°C at a cooling rate of 5°C/min. Annealed oligos (1 µl from 1:200 diluted annealing reaction) were ligated into the PX458 plasmid (no. 48138; Addgene) and digested with BbsI using the standard New England Biolabs T4 ligase protocol.
Cell lines
Cell lines used in this study were hTERT-immortalized RPE1 (ATCC CRL-4000), HEK293T cells (ATCC CRL-3216), and GP2-293 (Takara Bio). MCF10A, MCF10CA1 (CA1d), and MCF10AT1 (ATk.cl2; Santner et al., 2001) were a courtesy of Christian Conrad and Katharina Jechow (German Cancer Research Centre and BioQuant Center, Heidelberg University, Heidelberg, Germany).
RPE1 cells stably expressing Tet3G were a gift of Elmar Schiebel. RPE1 Nek2 KO cells (clones 18 and 27), RPE1 Nek2 KO clone 27 with Tet3G, RPE1 Nek2 KO clone 27 with Tet3G pRetrox-Tre3G-mNeonGreen-Nek2A, RPE1 Tet3G pRetrox-Tre3G-mNeonGreen-Nek2A, and RPE1 Tet3G pRetrox-Tre3G-mNeonGreen-Nek2A-KD cell lines are described in Hata et al. (2019). RPE1 C-Nap1 (CEP250) KO cell lines were described previously (Panic et al., 2015).
Stable cell lines expressing L13-Arl13B-GFP were generated using lentiviral transduction. For this, HEK293T cells were transiently transfected with L13-Arl13B-GFP, vesicular stomatitis virus glycoprotein envelope expressing plasmid pMD2.G (no. 12259; Addgene), and lentiviral packaging plasmid psPAX2 (no. 12260; Addgene). Cells were harvested 48 h after transfection. The supernatant was filtered, supplemented with Polybrene at a final concentration of 4 µg/ml, and used to transduce RPE1 and RPE1 Nek2 KO cells. After 2–4 d, GFP-positive cells were selected by FACS (FACSAria II SORP cell sorter; BD Biosciences) and collected as a pool.
Retroviral transduction was used to generate stable RPE1 or RPE1 Nek2 KO L13-Arl13B-GFP cell lines carrying pQCXIZ-TUBG1-mRuby2 (γ-tubulin-mRuby2) and RPE1 or RPE1 Nek2 KO Tet3G pRetroX-TRE3G cell lines carrying Smo-EGFP as well as mNeonGreen-PLK1-T210D. For this, the plasmid carrying the gene of interest was cotransfected with pMDG2 into the HEK293-based retroviral packaging cell line GP2-293. Media were changed after 24 and 48 h. The virus-containing media were harvested and filtered using a 0.45-µm filter (Millipore). Four parts of filtered medium were supplemented with two parts fresh media, one part FBS, and 4 µg/ml Polybrene (Sigma-Aldrich). RPE1 and RPE1 Nek2 KO cells were preseeded in a six-well plate and were infected by adding the virus-containing solution three times every 6 h. Cells were split 24 h after the first transduction. GFP- and mRuby-positive cells were selected by FACS and collected as a pool.
Construction and characterization of ODF2 KO cells
RPE1 ODF2 KO clones were generated by transfecting 106 cells with 15 µg of the gRNA-containing plasmid using the electroporation NEPA21 Transfection System (Nepa Gene) according to the manufacturer’s instructions. 48 h after transfection, GFP-positive cells were single sorted in 96-well plates containing 50% normal and 50% conditioned medium. After expansion, genomic DNA was extracted using the GeneJET Genomic DNA Purification Kit (Thermo Fisher Scientific) and sequenced.
ODF2 KO cells were further characterized by RT-PCR. Briefly, total RNAs were purified from the RPE1 WT and ODF2 KO cell line using the NucleoSpin RNA Plus Purification Kit (Macherey-Nagel) and reverse transcribed to first-strand cDNAs using ReverseAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) and the oligo(dT)18 primer according to the manufacturer’s recommendations. The resulting products were then used as templates for subsequent PCR with a forward primer (5′-AAGGAAAAAAGCGGCCGCAATGAAGGACCGCTCTTCAAC-3′) and a reverse primer (5′-CTTCGCCTCCGTGATCTTCA-3′) that were designed to target the full-length cenexin1 (isoform 9). The obtained PCR fragments were cloned into pJET 2.1 (CloneJET Kit; Thermo Fisher Scientific) for sequence analyses with pJET1.2 forward and reverse primers. Reads were trimmed and aligned in SnapGene (GSL Biotech LCC). Among 23 independent clones sequenced per genotype, isoform 9/NM_002540.5 (also known as human cenexin1), isoform 13/NM_001351578.1 (human cenexin1 variant 1), isoform 8/NM_001242354.1, and isoform 18/NM_001351587.2 were found.
Isolation of human HSPCs
All experiments involving the use of human HSPCs were approved by the Ethics Committee of the Medical Faculty, University of Heidelberg (Heidelberg, Germany) and performed after obtaining informed consent from all voluntary donors in accordance with relevant guidelines and regulations. Human HSPCs were derived from umbilical cord blood or from healthy allogeneic stem cell donors. The latter had received a mobilization regimen with granulocyte–colony-stimulating factor (10 µg/kg body wt/d subcutaneously for 5 d). A sample of 60 ml of peripheral blood was taken for this study before leukapheresis. HSPCs were isolated as described previously (Wein et al., 2010; Ludwig et al., 2014). Briefly, mononuclear cells were isolated by density gradient centrifugation using the Ficoll-Hypaque technique (Merck KGaA). CD34+ cells from the mononuclear cell fraction were enriched by labeling with magnetic microbeads and sorted twice using an affinity column with the AutoMACS ystem (Miltenyi Biotec GmbH).
Cell culture
All cell lines were grown at 37°C under 5% CO2. RPE1 and derived cell lines were grown in DMEM/F12 (Sigma-Aldrich) supplemented with 10% FBS (Biochrom), 2 mM l-glutamine (Thermo Fisher Scientific), and 0.348% sodium bicarbonate (Sigma-Aldrich). HEK293T and GP2-293 cells were cultured in DMEM with high glucose supplemented with 10% FBS. MCF10A, MCF10A, and MCF10AT1 cell lines were maintained in DMEM/F12 supplemented with 0.1 µg/ml cholera toxin, 10 µg/ml insulin, 0.5 µg/ml hydrocortisone, 0.02 µg/ml EGF, and 5% horse serum. Primary CD34+ cells (HSPCs) were provided by the Department of Internal Medicine, University Hospital Heidelberg (Heidelberg, Germany). Primary HSPCs were cultured in Stemline II Hematopoietic Cell Expansion Medium (Sigma-Aldrich), supplemented with l-glutamine (2 mmol/liter; Thermo Fisher Scientific), thrombopoietin (100 ng/ml; R&D Systems), stem cell factor (100 ng/ml; R&D Systems), granulocyte–colony-stimulating factor (100 µg/ml; R&D Systems), and Flt-3 Ligand (500 ng/ml; R&D Systems). HSPCs were expanded for 4 d after isolation on retronectin-coated dishes (T100B Recombinant Human Fibronectin Fragment; Takara Bio). The coating was performed by incubating the dish for 2 h at 37°C with retronectin at a final concentration of 20 µg/ml in PBS, followed by washing in PBS.
Cell culture treatments and transfection
RPE1 cells were transiently transfected with plasmid DNA by either electroporation using the NEPA21 Transfection System (Nepa Gene) or FuGENE 6 (Promega) according to the manufacturer’s protocol and fixed 24–48 h after transfection. Transient transfection in HEK293T and GP2-293 cells was performed using polyethyleneimine (PEI 25000; Polysciences) as described previously (Longo et al., 2013).
RPE1 cells were seeded to subconfluence (25,000 cells/well in a 24-well plate) and incubated in serum-free medium for 48–72 h to induce cilia formation. To induce cilia disassembly, cells were serum starved for 24 h, followed by 24 h of serum restimulation. Expression of Tet-on-inducible constructs was induced by the addition of DOX (Sigma-Aldrich) at a concentration of 10 ng/ml for 24 and 72 h as indicated in the respective figure legends. In the rescue cell line Nek2 KO with Tet3G pRetrox-Tre3G-mNeonGreen-Nek2A, Nek2A expression was induced by the addition of DOX at a concentration of 1 ng/ml for 24 h. SAG (no. 4366; Tocris) was used at a final concentration of 0.4 µM for 24 h to activate Hh signaling. For visualization of tubulin during live-cell imaging, 20 nM sirTubulin (Cytoskeleton Kit 251SC006; tebu-bio) and 5 µM verapamil (Cytoskeleton Kit 251SC006; tebu-bio) were added 3 h before imaging. Cells were treated with MG132 (SMQ-SIH-537; Biozol) for 4 h at a final concentration of 20 µM to inhibit the proteasome. Cells were treated with BI-2536 (Boehringer Ingelheim) at a concentration of 200 nM for 1 or 6 h to inhibit Plk1. For G1 arrest, cells were treated with the Cdk4/6 inhibitor palbociclib (no. S1116; Selleckchem) at a concentration of 0.1 µM for 24 h and for an additional 24 h at a concentration of 1 µM as indicated in the respective figure legends. All drugs were dissolved in DMSO, and solvent only was used as a negative control. Upon Nek2 overexpression using 10 ng/µl DOX, only cells with medium or high Nek2 levels (detected by immunofluorescence) were considered for appendage quantification, if not stated otherwise.
RNAi
Transfections of siRNA were performed using Lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. 30,000–40,000 cells were seeded per well in a 24-well plate and analyzed 72 h after the initial transfection. The following siRNAs (50 nM final concentration) were used: siLuciferase (siControl), 5′-UCGAAGUAUUCCGCGUACG-3′ (Dharmacon ON-TARGET plus/Ambion Silencer Select; Knodler et al., 2010); siNek2, 5′-GAUGCAAUUUGGUCAUUAAUU-3′ and 5′-GAAAGGCAAUACUUAGAUGUU-3′ (Dharmacon ON-TARGET plus; Kim et al., 2015); siCep164, 5′-GGUGACAUUUACUAUUUCA-3′ (Ambion Silencer Select; Graser et al., 2007); siHsODF2-1, 5′-AGACUAAUGGAGCAACAAG-3′ (Ambion Silencer Select; Soung et al., 2009); siODF2-2, 5′-GGAUCUUUAUGUCGCUGAATT-3′ (Ambion Silencer Select); siKif24, 5′-GGAACACCCTGGAGAATAGTT-3′ and 5′-GAGTTGAGCTCTCCTTTGGTT (Dharmacon ON-TARGET plus; Kobayashi et al., 2011); and siC-Nap1 (human CEP250), 5′-GAGCAGAGCUACAGCGAAU-3′ and 5′-AAGCUGACGUGGUGAAUAA-3′ (Dharmacon ON-TARGET plus; Panic et al., 2015).
Real-time quantitative PCR (qPCR)
Real-time qPCR was used to analyze the knockdown efficiency of Kif24 depletion. Briefly, RPE1 cells treated with control and Kif24 siRNA were detached by trypsin treatment and centrifuged to form a cell pellet. RNA was isolated using the RNeasy Mini Kit (no. 74104; Qiagen) according to manufacturer’s instructions. Total RNA was quantified on an ND-1000 spectrophotometer (Nanodrop). Real-time qPCR was performed with 100 ng of RNA using One-Step SYBR PrimeScript Real-Time PCR Kit II (Perfect Real Time no. RR086A; Takara Bio) with ROX reference dye on a 7300 Real-Time PCR System (Applied Biosystems) according to the manufacturer’s protocols. Forward and reverse primer pairs for Kif24 were 5′-GCATGAGAAGAAAGAAGCAGTTGACC-3′ and 5′-GGTGCACGCCTCACCAAAGA-3′ and for GAPDH were 5′-TGCACCACCAACTGCTTAGC-3′ and 5′-GGCATGGACTGTGGTCATGAG-3′. Primer pairs were reconstituted in nuclease-free water. Results were exported to Excel (Microsoft) for analysis. All the corresponding real-time qPCR data were analyzed using the ΔΔCT method and normalized against GAPDH (housekeeping gene; Milstein et al., 2013). The RQ value (fold change) was calculated by using the formula 2(−ΔΔCT).
Antibodies
The following primary antibodies were used: mouse anti-Nek2 (1:250, D-8, sc-55601; Santa Cruz Biotechnology), mouse anti-Nek2 (immunofluorescence 1:100, Western blot 1:500, no. 610593; BD Biosciences), rabbit anti-ARL13B (1:500, no. 17711-1-AP; Proteintech), mouse anti–γ-tubulin clone GTU-88 (1:500, no. T6557; Sigma-Aldrich), rabbit anti–γ-tubulin (1:500, no. T5192; Sigma-Aldrich), rabbit anti-Cep83 (CCDC41, 1:400, no. HPA038161; Sigma-Aldrich), rabbit anti-SCLT1 (1:250, no. HPA036561; Sigma-Aldrich), rabbit anti-FBF1 (1:500, no. HPA023677; Sigma-Aldrich), mouse anti-actin (1:1,000, MAB1501; EMD Millipore), rabbit anti-Cofilin (1:3,000, ab42824; Abcam), mouse anti-GFP (1:800, no. 11814460001; Roche), anti–nuclear pore complex (mAB414, ab24609; Abcam), goat anti-C-Nap1 (1:1,000; Panic et al., 2015), guinea pig anti-Cep164N (immunofluorescence 1:500) and rabbit anti-Cep164M (Western blot 1:400; Schmidt et al., 2012), guinea pig anti-ODF2 (1:500; Kuhns et al., 2013), guinea pig anti-LRRC45 (immunofluorescence 1:400, Western blot 1:500; Kurtulmus et al., 2018), guinea pig anti–Cep123-N (immunofluorescence 1:500, Western blot 1:1,000; Kurtulmus et al., 2018), rabbit anti-centriolin (1:100; a gift of Elmar Schiebel), rabbit anti-Cep128 (1:500, ab118797; Abcam), mouse anti–Plk1 F-8 (1:500, sc-17783; Santa Cruz Biotechnology), rabbit anti–Phospho-Histone3(Ser10) (1:1,000, no. 06-570; Millipore), mouse anti–Smo E-5 (1:100, sc-166685; Santa Cruz Biotechnology), and anti-CENPF (1:100, clone 11; BD Biosciences). Secondary antibodies for immunofluorescence analysis were coupled to Alexa Fluor 488, 546, 594, or 647 dyes (Thermo Fisher Scientific). Antibodies for Western blot analysis were coupled to HRP (Dianova).
Microscopy-based analysis
Cells were grown on coverslips (no. 1.5; Thermo Fisher Scientific) and fixed in methanol at −20°C for 5 min or prefixed with 3% PFA for 3 min before methanol fixation. Cells were blocked with blocking solution containing 3% IgG-free BSA (Jackson ImmunoResearch) and 0.1% Triton X-100 (Sigma-Aldrich) in PBS for 30 min and incubated with primary antibodies dissolved in blocking solution for 1 h at room temperature. The cells were incubated with conjugated secondary antibodies diluted in blocking solution containing 0.1% DAPI (Sigma-Aldrich) for 1 h at room temperature. Coverslips were mounted with Mowiol (EMD Millipore).
For the staining of HSPCs, cells were fixed in suspension as described above. The washing steps were performed by centrifugation (at 650 g for 5 min at room temperature). After fixation, cells were resuspended in blocking solution and incubated for 1 h at room temperature. Approximately 20–30 µl of cell suspension was dropped per well on Teflon-coated slides (HTC supercured; Thermo Fisher Scientific) and allowed to dry in a laminar flow hood. Dried cells were washed with 30 µl of water and subsequently stained as described above for adherent cells.
Cell cycle stages were determined using 5-ethynyl-2′-deoxyuridine (EdU) and CENPF stainings. The EdU assay was performed using the Click-iT EdU Alexa Fluor 555 imaging kit (Life Technologies). Briefly, cells were treated with 10 µM EdU for 30 min before fixation with methanol at −20°C for 5 min. The Click-IT reaction was performed according to the manufacturer’s recommendations, and samples were subsequently subjected to indirect immunofluorescence analysis using antibodies against CENPF. CENPF and EdU double-negative cells were classified as G1 phase cells. EdU-positive and CENPF-negative cells were classified as S phase cells. CENPF-positive and EdU-negative cells were classified as G2–early prophase cells. Cells with condensed chromosomes and intact nuclei (assessed with DAPI staining) were classified as prophase cells. Mitotic cells were identified by immunofluorescence microscopy based on cell rounding, chromosome morphology (condensed chromosomes), nuclear envelope breakdown, or intercentrosomal distances.
For live-cell imaging, cells were seeded in CELLview culture dishes (Greiner Bio-One). Experiments were performed over a 24-h period at 37°C with 5% CO2. Seventeen Z-stacks of 1.25-µm spacing were taken every 10 min for 24 h. Images were acquired using an Axio Observer Z1 fluorescence motorized microscope (Zeiss) equipped with 63× NA 1.4 Plan-Apochromat oil immersion objective and AxioCam MRm charge-coupled device camera using Zeiss Efficient Navigation software. For 3D-SIM, cells were processed for immunofluorescence microscopy as described above, except that cells were mounted in Prolong Gold antifade reagent (Molecular Probes). Samples were analyzed with a 3D-SIM Ti inverted microscope (Nikon) equipped with three lasers (488, 561, and 647 nm), an Apo total internal reflection fluorescence 100 × 1.49 NA oil immersion objective (Nikon), and an iXon3 DU-897E single-photon detection electron multiplying charge-coupled device camera (Andor). After image capture, raw images were reconstructed in the NIS-Elements program of the microscope to obtain 3D-SIM images.
Image processing and analysis
Images were processed in Fiji (Schindelin et al., 2012) and Illustrator CS5 (Adobe). For figures, the maximum projection of representative images was generated using Fiji. Image brightness and contrast were adjusted equally in Fiji. Quantification of fluorescence intensity was performed using maximum projection of images. Centrosomes were segmented based on γ-tubulin staining. For signal intensity measurements, the mean fluorescence intensity of an area of 25 square pixels around the centrosome was calculated. The mean fluorescence intensity of an area adjacent to the centrosome was used for background correction. In all quantifications of mitotic cells, centrosome 1 was considered the centrosome carrying higher levels of appendages (M-centrosome, as defined by higher levels of ODF2). Intercentriolar distances were manually measured using Fiji. Intensity measurements from replicate experiments were normalized to the average of each repetition and combined. Statistical tests were performed in Excel (Microsoft), R (Synergy Software), and Prism 8 (GraphPad). The data were plotted as boxplots using R and as bar graphs using Excel. Co-localization of signals in 3D-SIM images was analyzed using the Plot Profile tool in Fiji, and graphs were normalized and plotted in Excel.
FACS analysis
Cell cycle profile of control, Nek2 KO, and Nek2A-overexpressing RPE1 cells were determined by analyzing total DNA content stained with propidium iodide. Briefly, cells were fixed with ice-cold ethanol (70%) and incubated with staining solution (50 µg/ml propidium iodide, 0.08% Triton X-100, 0.2 mg/ml RNase A, and 1 mM EDTA in PBS) for 30 min. Cells were subjected to analysis on a BD FACSCanto II flow cytometer and BD FACSDiva 8 software (BD Biosciences).
Electron microscopy
Cells were grown on 12-mm coverslips, rinsed twice with PBS, and fixed at room temperature for 30 min in 2.5% glutaraldehyde in 50 mM cacodylate buffer containing 2% sucrose. Cells were subsequently washed with 50 mM cacodylate buffer and postfixed with 2% osmium tetroxide in 50 mM cacodylate buffer for 45 min on ice and in darkness. Cells were rinsed with water and stained overnight at 4°C with 0.5% uranyl acetate in water. Cells were rinsed with water and dehydrated with ethanol in a stepwise manner using 40, 60, 70, 80, 90, 95, and 100% water-free ethanol. Capsules filled with Spurr resin were immediately placed on top of each coverslip and polymerized for 24 to 48 h at 60°C. The embedded cells were sectioned using a Reichert Ultracut S microtome (Leica Instruments) to a thickness of 70 nm. Poststaining was performed with 3% uranyl acetate in water and lead citrate. Sections were imaged in a JE-1400 transmission electron microscope (JEOL Ltd.) at 80 kV, equipped with a 4k × 4k digital camera (F416; TVIPS). Micrographs were adjusted in brightness and contrast using ImageJ (Schneider et al., 2012).
Cell lysis and Western blot
Cells were scrapped out of the dishes, transferred into 1.5-ml tubes, and washed with PBS by centrifugation. Pellets were lysed in SDS sample buffer (63 mM Tris-HCl, pH 6.8, 10% [vol/vol] glycerol, 2% [wt/vol] SDS, 0.01% [wt/vol] bromphenol blue, and 100 mM DTT) containing 0.1% Benzonase solution (101654; Merck) at 37°C for 30 min. For the mitotic shake-off analysis (Fig. S3 E), cells were treated with 50 nM CENP-E inhibitor in DMSO or DMSO only (control) for 16 h. Mitotic cells were collected by shaking off the dishes (Jackman and O’Connor, 1998). Cell pellets were lysed in RIPA buffer (25 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, and 0.1% Benzonase). Lysates were cleared by centrifugation (20,000 g for 5 min at room temperature), heated at 95°C for 5 min, and subjected to SDS-PAGE. For analysis of mNeonGreen-PLK1-T210D (Fig. 6 C), cells were lysed in 8 M urea containing 0.1% Benzonase for 1 h at room temperature, and SDS sample buffer was added before loading. After transfer to a nitrocellulose or polyvinylidene fluoride membrane, immunoreactive proteins were detected with the respective antibodies and visualized with enhanced chemiluminescence (Pierce no. 32106; Thermo Fisher Scientific).
Statistical analysis
Statistical analysis was applied for the mean values of each repetition. Two-tailed Student’s t tests were applied in case two conditions were compared (Figs. 5 C; 6 C; 7 C; 9 E; S2 E; S3 B; S4, F and H; and S5 D). When more than two conditions were compared, one-way ANOVA tests were applied followed by post hoc tests. Tukey’s post hoc test was applied when every condition was compared with every other condition (Figs. 1 C; 6, A and F; 7 E; 8, B, E, and F; 9 A; S2 B; S4 A; and S5 B); Dunnett’s post hoc test was applied if several conditions were compared with one control condition (Figs. 1 D; 3 B; S1 C; and S2 D). Holm–Šídák post hoc test was applied to compare selected experimental pairs of conditions (Figs. 4, B and C; 5 B; S3 F; and S5 C). Significance probability values are as follows: NS, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. The sampling sizes for quantifications are indicated in the respective figures or figure legends. Boxplots show the lower and upper quartiles; the whiskers show the minimum and maximal values excluding outliers. Outliers were calculated as values greater or lower than 1.5 times the interquartile range. Box/dot plots show outliers. The line inside the box indicates the median. Bar graphs show the mean ± SD of the independent experiments.
Online supplemental material
Fig. S1 illustrates a scheme for centrosome maturation, representative images for the cell cycle–dependent behavior of the DA Cep164, and the cell cycle behavior of appendage proteins in primary HSPCs. Fig. S2 shows the characterization of the Nek2 KO cell line and the phenotypical rescue of Nek2 KO cells. Fig. S3 demonstrates the analysis of Nek2A overexpression and DAs. Fig. S4 shows the characterization of ODF2 KO cells and the effect of ODF2 KO on DA regulation. Fig. S5 demonstrates that the ciliary remnant in mitotic Nek2 KO cells can be rescued upon siRNA of Cep164 and expression of Nek2 in the Nek2 KO background. Video 1 shows representative mitotic progression of Nek2 KO cells stably expressing Arl13B-GFP and γ-tubulin-mRuby2. Video 2 shows representative mitotic progression of Nek2 KO cells overexpressing Smo-EGFP.
Supplementary Material
Acknowledgments
We thank Marko Panic and Elmar Schiebel (University of Heidelberg, Heidelberg, Germany), Christian Conrad and Katharina Jechow (German Cancer Research Center, Heidelberg, Germany), Patrick Horn and Caroline Pabst (University of Heidelberg, Heidelberg, Germany) for sharing cell lines and reagents; Iain Hagan (Cancer Research UK Manchester Institute, Manchester, UK) for sharing protocols; Angela Lenze (University of Heidelberg, Heidelberg, Germany) for purification of CD34+ HSPCs; Astrid Hofmann (member of the Pereira lab) for excellent technical assistance; Volker Eckstein and Monika Langlotz (University of Heidelberg, Heidelberg, Germany) for FACS and help with cell cycle analysis; Damir Krunic and Felix Bestvater (German Cancer Research Center, Heidelberg, Germany) and Ulrike Engel (University of Heidelberg, Heidelberg, Germany) for providing access to microscopes, help with imaging, and image processing; and Elmar Schiebel, Iain Hagan, and members of the G. Pereira laboratory for critical comments on the paper.
This project was funded by the collaborative research grant of the Deutsche Forschungsgemeinschaft (SFB873 Project A14) to G. Pereira. Core funding for microscopy and FACS was provided by the Deutsche Forschungsgemeinschaft SFB873 projects Z03 and Z02, respectively. L. Viol is a member of the Heidelberg Biosciences International Graduate School and funded by the Deutsche Forschungsgemeinschaft SFB873. G. Pereira holds a Heisenberg Professorship from the Deutsche Forschungsgemeinschaft (PE1883/3).
The authors declare no competing financial interests.
Author contributions: L. Viol performed and analyzed all the experiments and assembled figures, except Fig. S4 D. A. Neuner performed and analyzed the electron microscopy experiment reported in Fig. S4 D. S. Hata and A. Pastor-Pediro constructed cell lines overexpressing Nek2 and Nek2 KO cell line. F. Murke, P. Wuchter, A.D. Ho, and B. Giebel provided tools and cell lines. S. Hata contributed to the conceptualization of the paper. G. Pereira conceptualized and supervised the project and wrote the paper, with contributions from all authors.
References
- Anderson C.T., and Stearns T.. 2009. Centriole age underlies asynchronous primary cilium growth in mammalian cells. Curr. Biol. 19:1498–1502. 10.1016/j.cub.2009.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler M., Kong D., Sun S., Nanjundappa R., Evans L., Farmer V., Holland A., Mahjoub M.R., Sui H., and Loncarek J.. 2019. High-resolution characterization of centriole distal appendage morphology and dynamics by correlative STORM and electron microscopy. Nat. Commun. 10:993 10.1038/s41467-018-08216-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappello P., Blaser H., Gorrini C., Lin D.C.C., Elia A.J., Wakeham A., Haider S., Boutros P.C., Mason J.M., Miller N.A., et al. . 2014. Role of Nek2 on centrosome duplication and aneuploidy in breast cancer cells. Oncogene. 33:2375–2384. 10.1038/onc.2013.183 [DOI] [PubMed] [Google Scholar]
- Casenghi M., Meraldi P., Weinhart U., Duncan P.I., Körner R., and Nigg E.A.. 2003. Polo-like kinase 1 regulates Nlp, a centrosome protein involved in microtubule nucleation. Dev. Cell. 5:113–125. 10.1016/S1534-5807(03)00193-X [DOI] [PubMed] [Google Scholar]
- Chaki M., Airik R., Ghosh A.K., Giles R.H., Chen R., Slaats G.G., Wang H., Hurd T.W., Zhou W., Cluckey A., et al. . 2012. Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell. 150:533–548. 10.1016/j.cell.2012.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.K., Taipale J., Young K.E., Maiti T., and Beachy P.A.. 2002. Small molecule modulation of Smoothened activity. Proc. Natl. Acad. Sci. USA. 99:14071–14076. 10.1073/pnas.182542899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly O.M., Gaboriau D., Karakaya K., King S., Dantas T.J., Lalor P., Dockery P., Krämer A., and Morrison C.G.. 2016. CEP164-null cells generated by genome editing show a ciliation defect with intact DNA repair capacity. J. Cell Sci. 129:1769–1774. 10.1242/jcs.186221 [DOI] [PubMed] [Google Scholar]
- Dawson P.J., Wolman S.R., Tait L., Heppner G.H., and Miller F.R.. 1996. MCF10AT: a model for the evolution of cancer from proliferative breast disease. Am. J. Pathol. 148:313–319. [PMC free article] [PubMed] [Google Scholar]
- DeVaul N., Koloustroubis K., Wang R., and Sperry A.O.. 2017. A novel interaction between kinase activities in regulation of cilia formation. BMC Cell Biol. 18:33 10.1186/s12860-017-0149-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto K., Masugi Y., Yamazaki K., Effendi K., Tsujikawa H., Tanabe M., Kitagawa Y., and Sakamoto M.. 2014. Presence of primary cilia in cancer cells correlates with prognosis of pancreatic ductal adenocarcinoma. Hum. Pathol. 45:817–825. 10.1016/j.humpath.2013.11.017 [DOI] [PubMed] [Google Scholar]
- Faragher A.J., and Fry A.M.. 2003. Nek2A kinase stimulates centrosome disjunction and is required for formation of bipolar mitotic spindles. Mol. Biol. Cell. 14:2876–2889. 10.1091/mbc.e03-02-0108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford M.J., Yeyati P.L., Mali G.R., Keighren M.A., Waddell S.H., Mjoseng H.K., Douglas A.T., Hall E.A., Sakaue-Sawano A., Miyawaki A., et al. . 2018. A cell/cilia cycle biosensor for single-cell kinetics reveals persistence of cilia after G1/S transition is a general property in cells and mice. Dev. Cell. 47:509–523. 10.1016/j.devcel.2018.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A.M., Mayor T., Meraldi P., Stierhof Y.D., Tanaka K., and Nigg E.A.. 1998a C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J. Cell Biol. 141:1563–1574. 10.1083/jcb.141.7.1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A.M., Meraldi P., and Nigg E.A.. 1998b A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J. 17:470–481. 10.1093/emboj/17.2.470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A.M., O’Regan L., Sabir S.R., and Bayliss R.. 2012. Cell cycle regulation by the NEK family of protein kinases. J. Cell Sci. 125:4423–4433. 10.1242/jcs.111195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A.M., Schultz S.J., Bartek J., and Nigg E.A.. 1995. Substrate specificity and cell cycle regulation of the Nek2 protein kinase, a potential human homolog of the mitotic regulator NIMA of Aspergillus nidulans. J. Biol. Chem. 270:12899–12905. 10.1074/jbc.270.21.12899 [DOI] [PubMed] [Google Scholar]
- Graser S., Stierhof Y.-D., Lavoie S.B., Gassner O.S., Lamla S., Le Clech M., and Nigg E.A.. 2007. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 179:321–330. 10.1083/jcb.200707181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hames R.S., and Fry A.M.. 2002. Alternative splice variants of the human centrosome kinase Nek2 exhibit distinct patterns of expression in mitosis. Biochem. J. 361:77–85. 10.1042/bj3610077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hames R.S., Crookes R.E., Straatman K.R., Merdes A., Hayes M.J., Faragher A.J., and Fry A.M.. 2005. Dynamic recruitment of Nek2 kinase to the centrosome involves microtubules, PCM-1, and localized proteasomal degradation. Mol. Biol. Cell. 16:1711–1724. 10.1091/mbc.e04-08-0688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hames R.S., Wattam S.L., Yamano H., Bacchieri R., and Fry A.M.. 2001. APC/C-mediated destruction of the centrosomal kinase Nek2A occurs in early mitosis and depends upon a cyclin A-type D-box. EMBO J. 20:7117–7127. 10.1093/emboj/20.24.7117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy T., Lee M., Hames R.S., Prosser S.L., Cheary D.-M., Samant M.D., Schultz F., Baxter J.E., Rhee K., and Fry A.M.. 2014. Multisite phosphorylation of C-Nap1 releases it from Cep135 to trigger centrosome disjunction. J. Cell Sci. 127:2493–2506. 10.1242/jcs.142331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassounah N.B., Nagle R., Saboda K., Roe D.J., Dalkin B.L., and McDermott K.M.. 2013. Primary cilia are lost in preinvasive and invasive prostate cancer. PLoS One. 8:e68521 10.1371/journal.pone.0068521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata S., Pastor Peidro A., Panic M., Liu P., Atorino E., Funaya C., Jäkle U., Pereira G., and Schiebel E.. 2019. The balance between KIFC3 and EG5 tetrameric kinesins controls the onset of mitotic spindle assembly. Nat. Cell Biol. 21:1138–1151. 10.1038/s41556-019-0382-6 [DOI] [PubMed] [Google Scholar]
- Hayward D.G., Clarke R.B., Faragher A.J., Pillai M.R., Hagan I.M., and Fry A.M.. 2004. The centrosomal kinase Nek2 displays elevated levels of protein expression in human breast cancer. Cancer Res. 64:7370–7376. 10.1158/0008-5472.CAN-04-0960 [DOI] [PubMed] [Google Scholar]
- He R., Huang N., Bao Y., Zhou H., Teng J., and Chen J.. 2013. LRRC45 is a centrosome linker component required for centrosome cohesion. Cell Rep. 4:1100–1107. 10.1016/j.celrep.2013.08.005 [DOI] [PubMed] [Google Scholar]
- Hung H.-F., Hehnly H., and Doxsey S.. 2016. The mother centriole appendage protein Cenexin modulates lumen formation through spindle orientation. Curr. Biol. 26:793–801. 10.1016/j.cub.2016.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoko A., Yano T., Miyamoto T., Matsuura S., Kiyono T., Goshima N., Inagaki M., and Hayashi Y.. 2018. Albatross/FBF1 contributes to both centriole duplication and centrosome separation. Genes Cells. 23:1023–1042. 10.1111/gtc.12648 [DOI] [PubMed] [Google Scholar]
- Ishikawa H., Kubo A., Tsukita S., and Tsukita S.. 2005. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat. Cell Biol. 7:517–524. 10.1038/ncb1251 [DOI] [PubMed] [Google Scholar]
- Jackman J., and O’Connor P.M.. 1998. Methods for synchronizing cells at specific stages of the cell cycle. Curr. Protoc. Cell Biol. 8.3.1–8.3.20. https://doi.org/. 10.1002/0471143030.cb0803s00 [DOI] [PubMed]
- Kim S., Lee K., Choi J.H., Ringstad N., and Dynlacht B.D.. 2015. Nek2 activation of Kif24 ensures cilium disassembly during the cell cycle. Nat. Commun. 6:8087 10.1038/ncomms9087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler A., Feng S., Zhang J., Zhang X., Das A., Peranen J., and Guo W.. 2010. Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc. Natl. Acad. Sci. USA. 107:6346–6351. 10.1073/pnas.1002401107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Tsang W.Y., Li J., Lane W., and Dynlacht B.D.. 2011. Centriolar kinesin Kif24 interacts with CP110 to remodel microtubules and regulate ciliogenesis. Cell. 145:914–925. 10.1016/j.cell.2011.04.028 [DOI] [PubMed] [Google Scholar]
- Kong D., Farmer V., Shukla A., James J., Gruskin R., Kiriyama S., and Loncarek J.. 2014. Centriole maturation requires regulated Plk1 activity during two consecutive cell cycles. J. Cell Biol. 206:855–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhns S., Schmidt K.N., Reymann J., Gilbert D.F., Neuner A., Hub B., Carvalho R., Wiedemann P., Zentgraf H., Erfle H., et al. . 2013. The microtubule affinity regulating kinase MARK4 promotes axoneme extension during early ciliogenesis. J. Cell Biol. 200:505–522. 10.1083/jcb.201206013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtulmus B., Yuan C., Schuy J., Neuner A., Hata S., Kalamakis G., Martin-Villalba A., and Pereira G.. 2018. LRRC45 contributes to early steps of axoneme extension. J. Cell Sci. 131:jcs223594 10.1242/jcs.223594 [DOI] [PubMed] [Google Scholar]
- Lange B.M., and Gull K.. 1995. A molecular marker for centriole maturation in the mammalian cell cycle. J. Cell Biol. 130:919–927. 10.1083/jcb.130.4.919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkins C.E., Aviles G.D.G., East M.P., Kahn R.A., and Caspary T.. 2011. Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. Mol. Biol. Cell. 22:4694–4703. 10.1091/mbc.e10-12-0994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H., Winkfein R.J., Mack G., Rattner J.B., and Yen T.J.. 1995. CENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosis. J. Cell Biol. 130:507–518. 10.1083/jcb.130.3.507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo P.A., Kavran J.M., Kim M.-S., and Leahy D.J.. 2013. Transient mammalian cell transfection with polyethylenimine (PEI). In Laboratory Methods in Enzymology: DNA . Lorsch E., editor. Academic Press, New York: 227–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A., Saffrich R., Eckstein V., Bruckner T., Wagner W., Ho A.D., and Wuchter P.. 2014. Functional potentials of human hematopoietic progenitor cells are maintained by mesenchymal stromal cells and not impaired by plerixafor. Cytotherapy. 16:111–121. 10.1016/j.jcyt.2013.07.007 [DOI] [PubMed] [Google Scholar]
- Mazo G., Soplop N., Wang W.-J., Uryu K., and Tsou M.-F.B.. 2016. Spatial control of primary ciliogenesis by subdistal appendages alters sensation-associated properties of cilia. Dev. Cell. 39:424–437. 10.1016/j.devcel.2016.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P. 2019. Bub1-the zombie protein that CRISPR cannot kill. EMBO J. 38:e101912 10.15252/embj.2019101912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstein S., Nguyen M., Meyers R., and de Fougerolles A.. 2013. Measuring RNAi knockdown using qPCR. Meth. Enzymol. 533:57–77. 10.1016/B978-0-12-420067-8.00006-4 [DOI] [PubMed] [Google Scholar]
- Miyamoto T., Hosoba K., Ochiai H., Royba E., Izumi H., Sakuma T., Yamamoto T., Dynlacht B.D., and Matsuura S.. 2015. The microtubule-depolymerizing activity of a mitotic kinesin protein KIF2A drives primary cilia disassembly coupled with cell proliferation. Cell Rep. 10:664–673. 10.1016/j.celrep.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A., and Raff J.W.. 2009. Centrioles, centrosomes, and cilia in health and disease. Cell. 139:663–678. 10.1016/j.cell.2009.10.036 [DOI] [PubMed] [Google Scholar]
- Nigg E.A., and Stearns T.. 2011. The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat. Cell Biol. 13:1154–1160. 10.1038/ncb2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobutani K., Shimono Y., Yoshida M., Mizutani K., Minami A., Kono S., Mukohara T., Yamasaki T., Itoh T., Takao S., et al. . 2014. Absence of primary cilia in cell cycle-arrested human breast cancer cells. Genes Cells. 19:141–152. 10.1111/gtc.12122 [DOI] [PubMed] [Google Scholar]
- Olsen B. 2005. Nearly all cells in vertebrates and many cells in invertebrates contain primary cilia. Matrix Biol. 24:449–450. 10.1016/j.matbio.2005.09.003 [DOI] [PubMed] [Google Scholar]
- Pan Y.-R., and Lee E.Y.-H.P.. 2009. UV-dependent interaction between Cep164 and XPA mediates localization of Cep164 at sites of DNA damage and UV sensitivity. Cell Cycle. 8:655–664. 10.4161/cc.8.4.7844 [DOI] [PubMed] [Google Scholar]
- Panic M., Hata S., Neuner A., and Schiebel E.. 2015. The centrosomal linker and microtubules provide dual levels of spatial coordination of centrosomes. PLoS Genet. 11:e1005243 10.1371/journal.pgen.1005243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paridaen J.T.M.L., Wilsch-Bräuninger M., and Huttner W.B.. 2013. Asymmetric inheritance of centrosome-associated primary cilium membrane directs ciliogenesis after cell division. Cell. 155:333–344. 10.1016/j.cell.2013.08.060 [DOI] [PubMed] [Google Scholar]
- Pugacheva E.N., Jablonski S.A., Hartman T.R., Henske E.P., and Golemis E.A.. 2007. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 129:1351–1363. 10.1016/j.cell.2007.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapley J., Baxter J.E., Blot J., Wattam S.L., Casenghi M., Meraldi P., Nigg E.A., and Fry A.M.. 2005. Coordinate regulation of the mother centriole component nlp by nek2 and plk1 protein kinases. Mol. Cell. Biol. 25:1309–1324. 10.1128/MCB.25.4.1309-1324.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter J.F., and Leroux M.R.. 2017. Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 18:533–547. 10.1038/nrm.2017.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Rodriguez J.-A., Lewis C., McKinley K.L., Sikirzhytski V., Corona J., Maciejowski J., Khodjakov A., Cheeseman I.M., and Jallepalli P.V.. 2018. Distinct roles of RZZ and Bub1-KNL1 in mitotic checkpoint signaling and kinetochore expansion. Curr. Biol. 28:3422–3429. 10.1016/j.cub.2018.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R., Milenkovic L., Corcoran R.B., and Scott M.P.. 2009. Hedgehog signal transduction by Smoothened: pharmacologic evidence for a 2-step activation process. Proc. Natl. Acad. Sci. USA. 106:3196–3201. 10.1073/pnas.0813373106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner S.J., Dawson P.J., Tait L., Soule H.D., Eliason J., Mohamed A.N., Wolman S.R., Heppner G.H., and Miller F.R.. 2001. Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Res. Treat. 65:101–110. 10.1023/A:1006461422273 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. . 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K.N., Kuhns S., Neuner A., Hub B., Zentgraf H., and Pereira G.. 2012. Cep164 mediates vesicular docking to the mother centriole during early steps of ciliogenesis. J. Cell Biol. 199:1083–1101. 10.1083/jcb.201202126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley E.S., and Nachury M.V.. 2010. The perennial organelle: assembly and disassembly of the primary cilium. J. Cell Sci. 123:511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillibourne J.E., Hurbain I., Grand-Perret T., Goud B., Tran P., and Bornens M.. 2013. Primary ciliogenesis requires the distal appendage component Cep123. Biol. Open. 2:535–545. 10.1242/bio.20134457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasubramaniam S., Sun X., Pan Y.-R., Wang S., and Lee E.Y.-H.P.. 2008. Cep164 is a mediator protein required for the maintenance of genomic stability through modulation of MDC1, RPA, and CHK1. Genes Dev. 22:587–600. 10.1101/gad.1627708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaats G.G., Ghosh A.K., Falke L.L., Le Corre S., Shaltiel I.A., van de Hoek G., Klasson T.D., Stokman M.F., Logister I., Verhaar M.C., et al. . 2014. Nephronophthisis-associated CEP164 regulates cell cycle progression, apoptosis and epithelial-to-mesenchymal transition. PLoS Genet. 10:e1004594–e1004594. 10.1371/journal.pgen.1004594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soule H.D., Maloney T.M., Wolman S.R., Peterson W.D., Brenz R., McGrath C.M., Russo J., Pauley R.J., Jones R.F., and Brooks S.C.. 1990. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 50:6075–6086. [PubMed] [Google Scholar]
- Soung N.-K., Park J.-E., Yu L.-R., Lee K.H., Lee J.-M., Bang J.K., Veenstra T.D., Rhee K., and Lee K.S.. 2009. Plk1-dependent and -independent roles of an ODF2 splice variant, hCenexin1, at the centrosome of somatic cells. Dev. Cell. 16:539–550. 10.1016/j.devcel.2009.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalluto C., Wilson D.I., and Hearn T.. 2012. Nek2 localises to the distal portion of the mother centriole/basal body and is required for timely cilium disassembly at the G2/M transition. Eur. J. Cell Biol. 91:675–686. 10.1016/j.ejcb.2012.03.009 [DOI] [PubMed] [Google Scholar]
- Sumara I., Gime J.F., Gerlich D., Hirota T., Kraft C., De Torre C., Ellenberg J., and Peters J.. 2004. Roles of Polo-like Kinase 1 in the assembly of functional mitotic spindles. Curr. Biol. 14:1712–1722. [DOI] [PubMed] [Google Scholar]
- Tanos B.E., Yang H.J., Soni R., Wang W.J., Macaluso F.P., Asara J.M., and Tsou M.F.B.. 2013. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 27:163–168. 10.1101/gad.207043.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi K., Yamazaki Y., Nishida T., Watanabe S., Kunimoto K., Ishikawa H., and Tsukita S.. 2013. Two appendages homologous between basal bodies and centrioles are formed using distinct Odf2 domains. J. Cell Biol. 203:417–425. 10.1083/jcb.201303071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin B., Chen N.-P., Panic M., and Schiebel E.. 2015. Genome editing through large insertion leads to the skipping of targeted exon. BMC Genomics. 16:1082 10.1186/s12864-015-2284-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev I.A., and Nadezhdina E.S.. 1987. The centrosome and its role in the organization of microtubules. Int. Rev. Cytol. 106:227–293. 10.1016/S0074-7696(08)61714-3 [DOI] [PubMed] [Google Scholar]
- Wang L., and Dynlacht B.D.. 2018. The regulation of cilium assembly and disassembly in development and disease. Development. 145:dev151407 10.1242/dev.151407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Failler M., Fu W., and Dynlacht B.D.. 2018. A distal centriolar protein network controls organelle maturation and asymmetry. Nat. Commun. 9:3938 10.1038/s41467-018-06286-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wein F., Pietsch L., Saffrich R., Wuchter P., Walenda T., Bork S., Horn P., Diehlmann A., Eckstein V., Ho A.D., and Wagner W.. 2010. N-Cadherin is expressed on human hematopoietic progenitor cells and mediates interaction with human mesenchymal stromal cells. Stem Cell Res. (Amst.). 4:129–139. 10.1016/j.scr.2009.12.004 [DOI] [PubMed] [Google Scholar]
- Whitfield M.L., Sherlock G., Saldanha A.J., Murray J.I., Ball C.A., Alexander K.E., Matese J.C., Perou C.M., Hurt M.M., Brown P.O., and Botstein D.. 2002. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell. 13:1977–2000. 10.1091/mbc.02-02-0030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T.T., Chong W.M., Wang W.-J., Mazo G., Tanos B., Chen Z., Tran T.M.N., Chen Y.-D., Weng R.R., Huang C.-E., et al. . 2018. Super-resolution architecture of mammalian centriole distal appendages reveals distinct blade and matrix functional components. Nat. Commun. 9:2023 10.1038/s41467-018-04469-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Zeng H., Ning G., Reiter J.F., and Liu A.. 2014. C2cd3 is critical for centriolar distal appendage assembly and ciliary vesicle docking in mammals. Proc. Natl. Acad. Sci. USA. 111:2164–2169. 10.1073/pnas.1318737111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan K., Frolova N., Xie Y., Wang D., Cook L., Kwon Y.-J., Steg A.D., Serra R., and Frost A.R.. 2010. Primary cilia are decreased in breast cancer: analysis of a collection of human breast cancer cell lines and tissues. J. Histochem. Cytochem. 58:857–870. 10.1369/jhc.2010.955856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Kruse T., Guasch Boldú C., Garvanska D.H., Coscia F., Mann M., Barisic M., and Nilsson J.. 2019. Efficient mitotic checkpoint signaling depends on integrated activities of Bub1 and the RZZ complex. EMBO J. 38:e100977 10.15252/embj.2018100977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Yang Y., Xia J., Wang H., Salama M.E., Xiong W., Xu H., Shetty S., Chen T., Zeng Z., et al. . 2013. NEK2 induces drug resistance mainly through activation of efflux drug pumps and is associated with poor prognosis in myeloma and other cancers. Cancer Cell. 23:48–62. 10.1016/j.ccr.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.