Sánchez-Martín and Komatsu preview work from Itakura and colleagues that describes a mechanism that mediates degradation of misfolded extracellular proteins.
Abstract
The mechanisms of quality control for extracellular proteins are still poorly understood. In this issue, Itakura et al. (2020. J. Cell. Biol. https://doi.org/10.1083/jcb.201911126) show that upon binding to misfolded proteins, the extracellular chaperone clusterin is internalized via the heparan sulfate receptor to undergo lysosomal degradation.
Once proteins are folded into their functional structure, they need to retain their shape against destabilizing stressors such as changes in the pH, temperature, or exposure to environmental electrophiles such as methylmercury and cadmium. The regulation of intracellular protein quality is achieved by three sequential steps: helping the folding of nascent proteins; refolding or disaggregating misfolded proteins; and degrading proteins that are no longer able to be refolded. The first two steps rely on chaperones, while the latter one is performed by the main degradative pathways: the ubiquitin–proteasome and autophagy–lysosomal systems. While the mechanisms of intracellular protein quality control are well known, the quality control machinery for extracellular proteins, which are subjected to more stringent environmental conditions, is largely hidden in a veil of mystery. In the current issue, Itakura et al. identified a mechanism by which aberrant extracellular proteins are transported into cells in a receptor-mediated manner to degrade them in the lysosome (1).
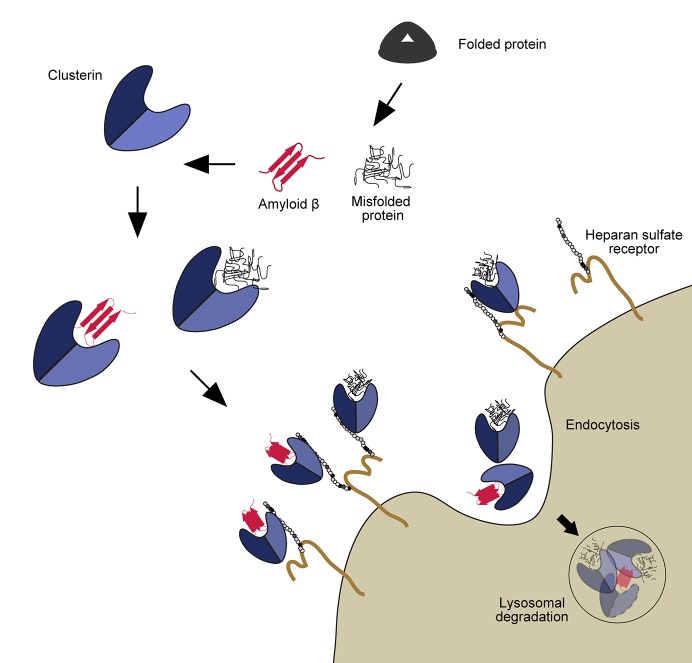
Clusterin is the main extracellular chaperone and is induced in stress conditions to prevent the aggregation of misfolded proteins by keeping them in large soluble complexes (2). To monitor the dynamics of clusterin, the authors used a traditional assay to measure lysosomal degradation in which the GFP signal is quenched by acidification but the RFP signal remains, so the ratio between them can be quantified (3). Using clusterin-RFP-GFP (clusterin-RG) and a model substrate, heat-shocked firefly luciferase (Luc), they found that clusterin is internalized in the cell after its binding to misfolded Luc, and then they are delivered to the lysosome. This degradation system was observed in a variety of cancer cell lines including U2OS, HeLa, and A549, suggesting that it is a general mechanism. The next question is how substrate-binding clusterin is distinguished from free clusterin and endocytosed into the cell. Previous studies pointed to low density lipoprotein receptor–related protein 2 as a possible receptor for clusterin (4, 5). Meanwhile, the genome-wide CRISPR screening employed by Itakura et al. specified a series of genes encoding enzymes related to heparan sulfate (HS) synthesis as essential genes for lysosomal degradation of clusterin-RG. They hypothesized that heparan sulfate proteoglycans (HSPGs) act as receptors for clusterin. HSPG is a group of molecules in which HS is covalently bound to a core protein and is present on the cell membrane surface and extracellular matrix of almost all animal cells. As expected, the authors showed that HS binds to clusterin through electrostatic interactions mediated by negatively charged HS and positively charged residues of clusterin, which are well conserved among species. Which kind of extracellular substrates are targeted by this pathway? Intravascular hemolysis, which is robustly induced by heat stress and autoantibodies, causes the release of proteins from red blood cells to extracellular spaces. Haptoglobin, a specific chaperone for hemoglobin, entraps hemoglobin released from red blood cells, and the complex is subsequently internalized into the macrophages. However, the fate of other released proteins remained unclear. The authors showed that clusterin-RG has an ability to bind to hemolysis-derived proteins and to deliver them into lysosomes in an HS-dependent fashion. They also find that Alzheimer’s disease–associated peptide Amyloid β, whose deposition is known to colocalize with HSPGs, promoted the delivery of clusterin-RG into lysosomes. These data raise the possibility that chaperone- and receptor-mediated extracellular protein degradation (CRED) is a universal pathway that plays a protective role against pathogenic aggregation of misfolded proteins (Fig. 1).
Figure 1.
Model of the CRED pathway. Stressors induce the misfolding of extracellular proteins. These aberrant proteins are recognized by the chaperon clusterin. Cargo-bound clusterin can interact then with heparan sulfate proteoglycans in the surface of the cells to undergo endocytosis and finally be degraded in the lysosome.
This exciting discovery by Itakura and colleagues shows the first features of a novel mechanism of extracellular protein quality control. However, some challenging questions remain to be addressed. First, how can clusterin recognize such a variety of diverse substrates? Although the exposure of hydrophobic regions might be enough for clusterin binding, it is also possible that different cofactors can drive clusterin toward specific substrates. Structural studies could possibly shed light on this question, as well as further characterizing the binding mechanism of clusterin to both substrates and the HS. In human pathological conditions, CRED could become a double-edged sword, analogous to intracellular protein degradation systems (6, 7). For example, the induction of CRED could become a new therapeutic approach in diseases in which the protein clearance systems are unable to keep degenerated proteins under control (e.g., Huntington’s disease, Alzheimer’s disease, and other proteinopathies). However, it might also prove to be deleterious in patients suffering from metastatic tumors, where clusterin is overexpressed and promotes tumor cell survival through a mechanism not yet fully understood (8). Thus, further in vivo studies are needed to clarify the physiological role of CRED and determine whether it is beneficial or deleterious in different pathogenic situations.
Acknowledgments
M. Komatsu is supported by a Japan Society for the Promotion of Science (an A3 foresight program) Grant-in-Aid for Scientific Research on Innovative Areas (19H05706) and Grant-in-Aid for Scientific Research (B) (18H02611) and the Takeda Science Foundation.
The authors declare no competing financial interests.
References
- 1.Itakura E., et al. J. Cell Biol. 2020 doi: 10.1083/jcb.201911126. [DOI] [Google Scholar]
- 2.Wyatt A.R., et al. J. Biol. Chem. 2009 doi: 10.1074/jbc.M109.033688. [DOI] [Google Scholar]
- 3.Kimura S., et al. Autophagy. 2007 doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 4.Bell R.D., et al. J. Cereb. Blood Flow Metab. 2007 doi: 10.1038/sj.jcbfm.9600419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammad S.M., et al. J. Biol. Chem. 1997 doi: 10.1074/jbc.272.30.18644. [DOI] [PubMed] [Google Scholar]
- 6.Leidal A.M., et al. Nat. Cell Biol. 2018 doi: 10.1038/s41556-018-0235-8. [DOI] [PubMed] [Google Scholar]
- 7.Poillet-Perez L., and White E. Genes Dev. 2019 doi: 10.1101/gad.325514.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng M., et al. Cancer Manag. Res. 2019 doi: 10.2147/CMAR.S196273. [DOI] [Google Scholar]