Yang et al. demonstrated TORC1 inactivation not only up-regulates macroautophagy and vacuole recycling but also leads to the degradation of many vacuole membrane proteins. The degradation is initiated by ubiquitination and followed by ESCRT-dependent microautophagy. Furthermore, TORC1 directly regulates the activity of vacuole ubiquitination machinery.
Abstract
Cellular adaptation in response to nutrient limitation requires the induction of autophagy and lysosome biogenesis for the efficient recycling of macromolecules. Here, we discovered that starvation and TORC1 inactivation not only lead to the up-regulation of autophagy and vacuole proteins involved in recycling but also result in the down-regulation of many vacuole membrane proteins to supply amino acids as part of a vacuole remodeling process. Down-regulation of vacuole membrane proteins is initiated by ubiquitination, which is accomplished by the coordination of multiple E3 ubiquitin ligases, including Rsp5, the Dsc complex, and a newly characterized E3 ligase, Pib1. The Dsc complex is negatively regulated by TORC1 through the Rim15-Ume6 signaling cascade. After ubiquitination, vacuole membrane proteins are sorted into the lumen for degradation by ESCRT-dependent microautophagy. Thus, our study uncovered a complex relationship between TORC1 inactivation and vacuole biogenesis.
Introduction
The lysosome is a central catabolic station that breaks down and recycles cellular materials to maintain nutrient homeostasis (Lim and Zoncu, 2016; Settembre et al., 2013). Emerging evidence suggests that the lysosome is a dynamic organelle that constantly adjusts its membrane composition according to environmental cues. In yeast, changes in substrate concentration can lead to the degradation of their corresponding vacuole (yeast lysosome) membrane (VM) transporters. For example, depleting lysine from the cytoplasm triggers a selective degradation of the vacuolar lysine importer Ypq1 to stop lysine from being transported from the cytoplasm into the vacuole lumen (Li et al., 2015b; Sekito et al., 2014). Similarly, a low cytoplasmic Zn2+ level leads to the down-regulation of a vacuolar Zn2+ importer, Cot1 (Li et al., 2015a).
Our understanding of the underlying mechanisms for VM regulation is still incomplete. However, it has been shown that protein ubiquitination serves as a sorting signal to initialize the degradation process. In yeast, two independent vacuole E3 ligases are involved in the substrate-triggered degradation of transporters. Specifically, a cytosolic NEDD4 family E3 ligase, Rsp5, and its VM adapter, Ssh4, are responsible for the ubiquitination of Ypq1 (Li et al., 2015b), whereas Dsc, a multisubunit transmembrane ubiquitin ligase complex, is required for the ubiquitination of Cot1 (Li et al., 2015a; Yang et al., 2018). After ubiquitination, VM proteins are delivered into the lumen by the endosomal sorting complexes required for transport (ESCRT) pathway (Li et al., 2015a; Li et al., 2015b; Zhu et al., 2017). With this selective degradation mechanism, the VM composition is regulated accurately in response to specific transporting substrates. Besides this fine-level adjustment of individual transporters, how do other environmental stresses such as starvation regulate VM composition?
Beyond catabolism, the lysosome also plays a major role in cellular stress response. The evolutionarily conserved mechanistic target of rapamycin kinase complex 1 (MTORC1), a lysosome membrane–associated kinase complex, serves as the signaling hub to sense different stresses and regulate cellular metabolism accordingly (Laplante and Sabatini, 2009; Perera and Zoncu, 2016). In mammalian cells, under nutrient-rich conditions, MTORC1 is recruited to the lysosome membrane and activated to phosphorylate the ribosomal RPS6 kinases (RPS6KB1/S6K1, RPS6KB2/S6K2) and eukaryotic translation initiation factor 4E binding protein 1 to promote protein synthesis, leading to cell growth and proliferation (Holz et al., 2005). In addition, active MTORC1 inhibits the transcription of various stress response genes involved in autophagy, lysosome biogenesis, and ER stress by phosphorylating transcription factor EB (TFEB) and TFE3 (Rehli et al., 1999; Sardiello et al., 2009; Settembre et al., 2011; Settembre et al., 2012). Similarly, in yeast, active TORC1 directly phosphorylates Sch9, an analogue of the mammalian RPS6 kinases, to stimulate protein translation when nutrients are available (Jin et al., 2014; Urban et al., 2007). Meanwhile, TORC1 also inhibits autophagy through Atg13 phosphorylation, thereby inhibiting its ability to activate the Atg1 kinase (Fujioka et al., 2014; Kraft et al., 2012).
Conversely, when TORC1/MTORC1 is inactive under stress conditions, catabolic processes such as proteasome degradation, autophagy, and endocytosis of plasma membrane proteins are elevated. In addition, lysosome biogenesis is up-regulated to boost its degradative and recycling functions. In mammalian cells, when MTORC1 is inactive, dephosphorylated TFEB translocates into the nucleus to promote the transcription of numerous target genes, including those encoding lysosomal hydrolases, pumps, permeases, and lysosome positioning regulators (Sardiello et al., 2009; Settembre et al., 2011, Settembre et al.,2012; Martina et al., 2014; Willett et al., 2017; Puertollano et al., 2018). In yeast, many vacuolar proteases such as Prb1, Ape1, Cps1, and Pep4 are up-regulated to enhance the digestion function during starvation (Müller et al., 2015; Parzych and Klionsky, 2019). All these studies point to a direct correlation between MTORC1/TORC1 inactivation and the enhancement of lysosomal/vacuolar biogenesis. However, two recent studies suggested the complexity of their relationship in yeast. Sakai and colleagues observed that glucose depletion leads to the invagination and degradation of two VM proteins, Vph1 (a V0 subunit of the vacuolar ATPase) and Pho8 (vacuolar phosphatase), in an ESCRT-dependent manner (Oku et al., 2017). Similarly, De Virgilio and colleagues observed that rapamycin treatment leads to the ESCRT-dependent degradation of Pho8 (Hatakeyama et al., 2019). These observations indicated that remodeling the vacuole proteome after TORC1 inactivation might be more complex than previously anticipated and raised three interesting questions: First, is the down-regulation of VM proteins a general response to TORC1 inactivation? Second, what machinery is involved? And third, how does TORC1 regulate this machinery?
In this study, we report that TORC1 plays a critical role in regulating VM composition. In contrast to the simplified model that TORC1 inactivation leads to a global up-regulation of vacuole biogenesis, we discovered that TORC1 inactivation also triggers the concomitant down-regulation of numerous VM proteins. Further analysis revealed that multiple E3 ligase systems, including Rsp5, the Dsc complex, and a third E3 ligase, Pib1, function together on the VM to ubiquitinate proteins. Moreover, our results showed that TORC1 can regulate the activity of the vacuole ubiquitin machinery. Specifically, TORC1 regulates the production and assembly of the vacuolar Dsc complex. After ubiquitination, cargoes are sorted into the lumen for degradation in an ESCRT-dependent manner. This study thus extends our understanding of the complexity of how TORC1 regulates vacuole composition according to environmental cues.
Results
TORC1 inactivation triggers down-regulation of many VM proteins
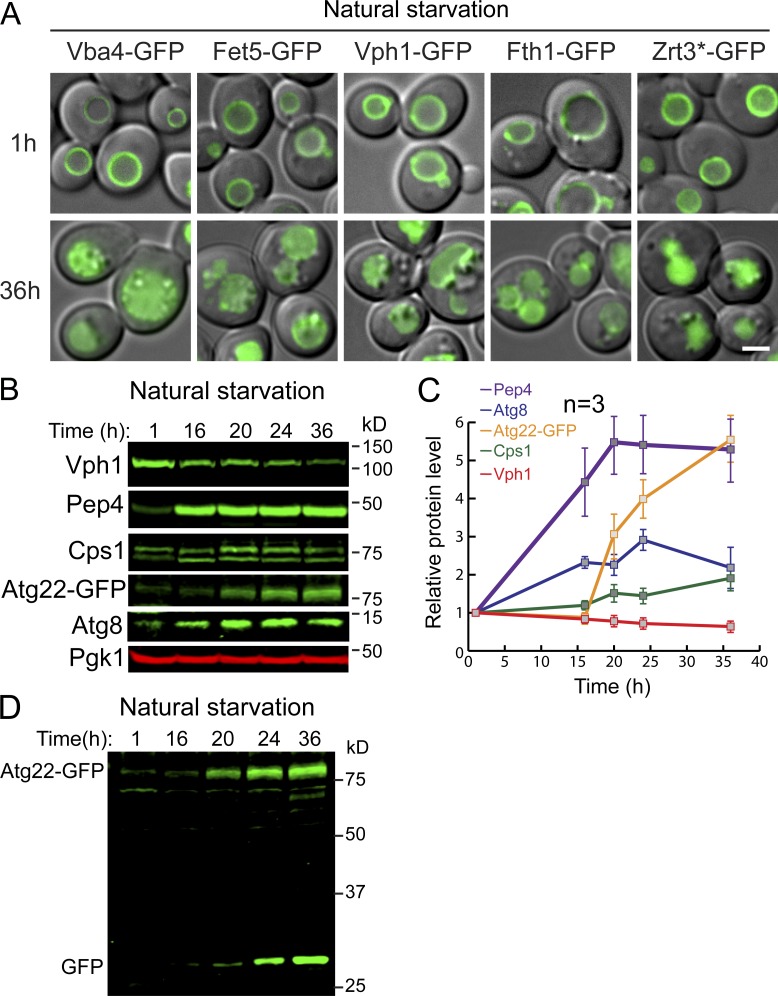
The TORC1 complex is responsible for regulating numerous stress responses according to environmental cues. However, how TORC1 regulates the vacuole protein composition is only partially understood. To answer this question, we set out to understand how VM composition responds to environmental stresses such as starvation and its relationship with TORC1 inactivation. As an initial test, we monitored the levels of five VM proteins, including Vba4 (a putative amino acid permease), Fet5 (a subunit of the putative iron transporter complex, which dimerizes with Fth1), Fth1, Vph1, and Zrt3* (a variant of the zinc exporter Zrt3; for details see Li et al., 2015a), after yeast cells enter the stationary phase. All five proteins were chromosomally tagged with GFP, and they properly localized to the VM (Fig. S1 A; Li et al., 2015a). As shown in Fig. 1, A and B, we took four time points (16, 20, 24, and 36 h) in the stationary phase and compared them to mid-log phase cells (1 h, with OD600 ∼0.7). All five proteins were significantly down-regulated after cells entered the stationary phase. During degradation, free GFP accumulated due to its resistance to vacuolar proteases (Fig. 1, A and B; and Fig. S1 A). Longer incubation in the stationary phase led to a higher level of protein degradation and more accumulation of free GFP (Fig. 1 A, 16–36 h). By contrast, very little degradation was observed in mid-log phase cells (Fig. 1 A, 1 h). Furthermore, acute nitrogen starvation caused a similar down-regulation of these proteins (data not shown). Together, our results suggested that long-term nutrient starvation can trigger the down-regulation of many VM proteins.
Figure S1.
TORC1 inactivation triggers a global down-regulation of VM proteins. (A) Subcellular localization of VM proteins in mid-log (1 h) or stationary (36 h) phase cells. (B) Western blots showing the changes of Vph1, Pep4, Cps1, Atg8, and Atg22-GFP in stationary phase cells. Samples were collected at the indicated time points, and 1 OD600 unit of cells was loaded in each lane. (C) Quantification of the protein levels in B. (D) Uncropped Atg22-GFP image from B. Scale bar, 2 µm. Related to Fig. 1.
Figure 1.
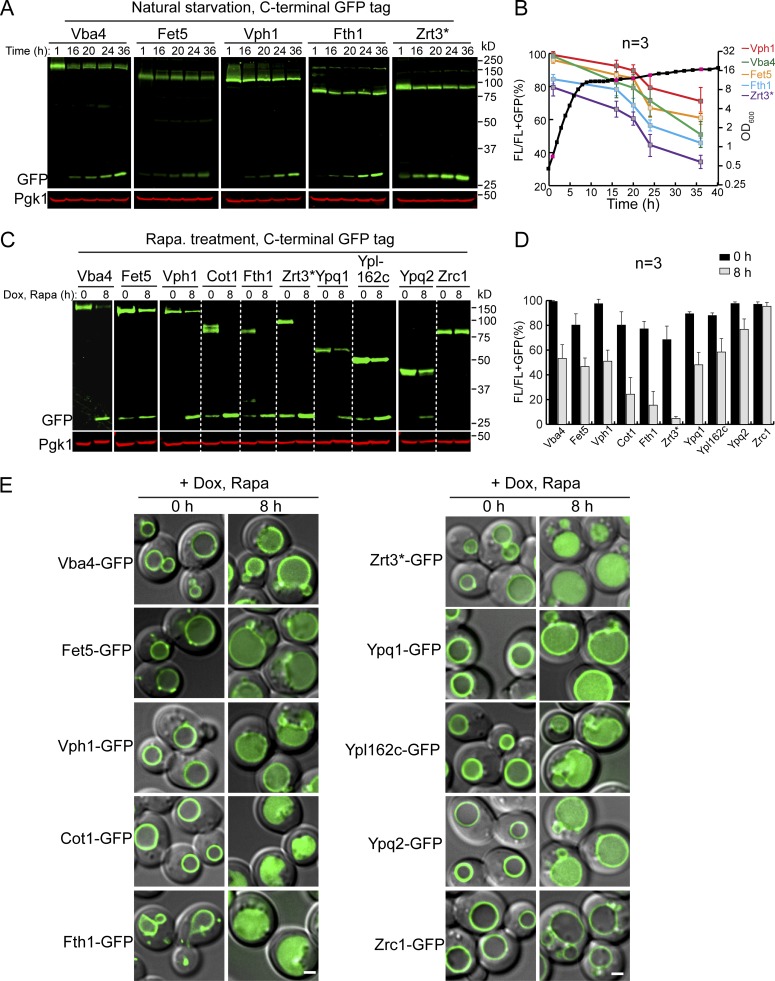
TORC1 inactivation triggers the down-regulation of many VM proteins. (A) Western blots showing the down-regulation of five VM proteins in stationary phase cells. (B) Quantification of the protein levels in A. The black curve represents the yeast growth curve in YPD medium at 28°C, and the red squares on the growth curve represent the time points chosen for Western blot analysis. The relative protein levels were calculated as FL/(FL + free GFP). The error bars represent SD (n = 3) for the entire article, unless stated otherwise. (C) Western blots showing the down-regulation of TET-OFF–controlled VM proteins after rapamycin treatment. (D) Quantification of the protein levels in C. (E) Merged images (differential interference contrast + GFP) to show subcellular localization of VM proteins before (0 h) and after (8 h) rapamycin treatment. Scale bar, 2 µm. Dox, doxycycline; FL, full-length protein fused with GFP; Rapa, rapamycin.
This result is a surprising finding, especially because it has been widely assumed that starvation promotes vacuole biogenesis and autophagy to boost the recycling of intracellular materials (Noda, 2017). Consistently, previous studies have shown that vacuolar hydrolases, components of the autophagic machinery, and the VM amino acid exporter Atg22 are indeed up-regulated under starvation conditions (Müller et al., 2015; Yang et al., 2006).
To verify those reported observations, we collected antibodies against endogenous vacuolar/autophagic proteins, including Vph1, Pep4, Cps1, and Atg8, and checked their response to natural starvation. Because of the lack of antibody, we chromosomally tagged Atg22 with GFP. As shown in Fig. S1, B and C, vacuolar proteases (Pep4 and Cps1) and autophagic machinery (Atg8 and Atg22) were indeed induced by starvation. In contrast, the protein level of untagged Vph1 decreased, which was consistent with the observation in Fig. 1 A where Vph1-GFP was partially degraded. Together, our data indicated that vacuoles undergo extensive remodeling in response to nutrient limitation. Proteins involved in the digestion and recycling functions are up-regulated, while many other membrane proteins are down-regulated. Interestingly, an accumulation of free GFP was also observed for Atg22-GFP (Fig. S1 D), indicating that even a protein directly involved in recycling can still be subjected to degradation after long-term starvation.
To directly test whether the down-regulation is due to TORC1 inactivation, we treated the mid-log phase cells with rapamycin. In this experiment, we expanded the substrate list to 10 GFP-tagged VM proteins by adding Cot1, Ypq1, Ypl162c (a putative transporter with unknown function), Ypq2 (a homologue of Ypq1), and Zrc1 (a zinc importer and a homologue of Cot1; Fig. 1, C–E). To focus on the preexisting pool of VM proteins, we expressed them under the control of a TET-OFF system (Garí et al., 1997). In this system, the gene transcription is controlled by the tetracycline operator (tetO) and can be supressed by the addition of either tetracycline or its derivatives. Yeast cells were preteated with doxycycline before rapamycin treatment. In this assay, 9 out of 10 VM proteins tested were down-regulated to different levels, and the accumulation of a lumenal GFP was observed (Fig. 1, C–E). The only exception was seen with Zrc1, which was unchanged after rapamycin treatment. This result suggested that maintaining the Zn2+ import capability is important for the recycling function of the vacuole, which contains many zinc-dependent metalloenzymes (Hecht et al., 2014; Hecht et al., 2013; Simm et al., 2007). Nevertheless, our results strongly suggested that TORC1 inactivation triggers the down-regulation of many VM proteins, in contrast to the long-held view that vacuole biogenesis will be globally up-regulated after TORC1 inactivation.
The degradation of VM proteins depends on lumenal proteases
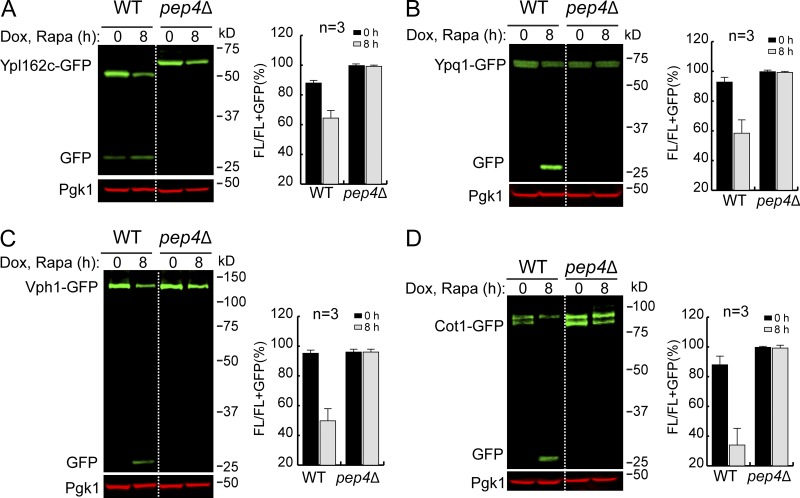
The accumulation of a lumenal GFP signal (Fig. 1 E) indicated that the degradation happens inside the vacuole. To confirm this, we chose four VM proteins (Ypl162c, Ypq1, Vph1, and Cot1) and performed the degradation assay in a pep4Δ strain. Pep4 is the master protease that processes and activates other vacuolar zymogens (Ammerer et al., 1986; Woolford et al., 1986). Deleting the PEP4 gene results in general defects in the vacuolar protease activity. As shown in Fig. 2, for all four tested substrates, full-length proteins were stabilized in the pep4Δ strain, and the accumulation of free GFP was entirely abolished. In addition, we observed a band shift for Ypl162c-GFP in the pep4Δ strain (Fig. 2 A), suggesting that after reaching the VM, Ypl162c undergoes a proteolytic cleavage for its maturation. In conclusion, these results indicated that the degradation depends on vacuolar protease activities.
Figure 2.
The degradation of VM proteins depends on lumenal proteases. (A–D) Western blots (left) and corresponding quantifications (right) showing the degradation of Ypl162c-GFP (A), Ypq1-GFP (B), Vph1-GFP (C), or Cot1-GFP (D) in WT and pep4Δ-strain cells. The error bars represent SD (n = 3). Dox, doxycycline; FL, full-length protein fused with GFP; Rapa, rapamycin.
The ESCRT machinery is required for invagination into the vacuole lumen
So far, three pathways have been suggested in yeast for delivering proteins to the vacuole lumen: macroautophagy, ESCRT-dependent sorting of ubiquitinated cargoes via the multivesicular body pathway or microautophagy, and the ESCRT-independent intralumenal fragment (ILF) pathway. Among them, the ILF pathway proposes that the single vesicle (named as an intralumenal fragment) created by vacuole–vacuole homotypic fusion is responsible for selectively sorting membrane proteins into the lumen for degradation (McNally et al., 2017). Although the sorting mechanism has not been addressed, the ILF pathway has been reported to be blocked by rapamycin treatment and stimulated by cycloheximide-triggered hyperactivation of TORC1 (McNally et al., 2017), hence making it unlikely to be responsible for internalizing VM proteins after TORC1 inactivation.
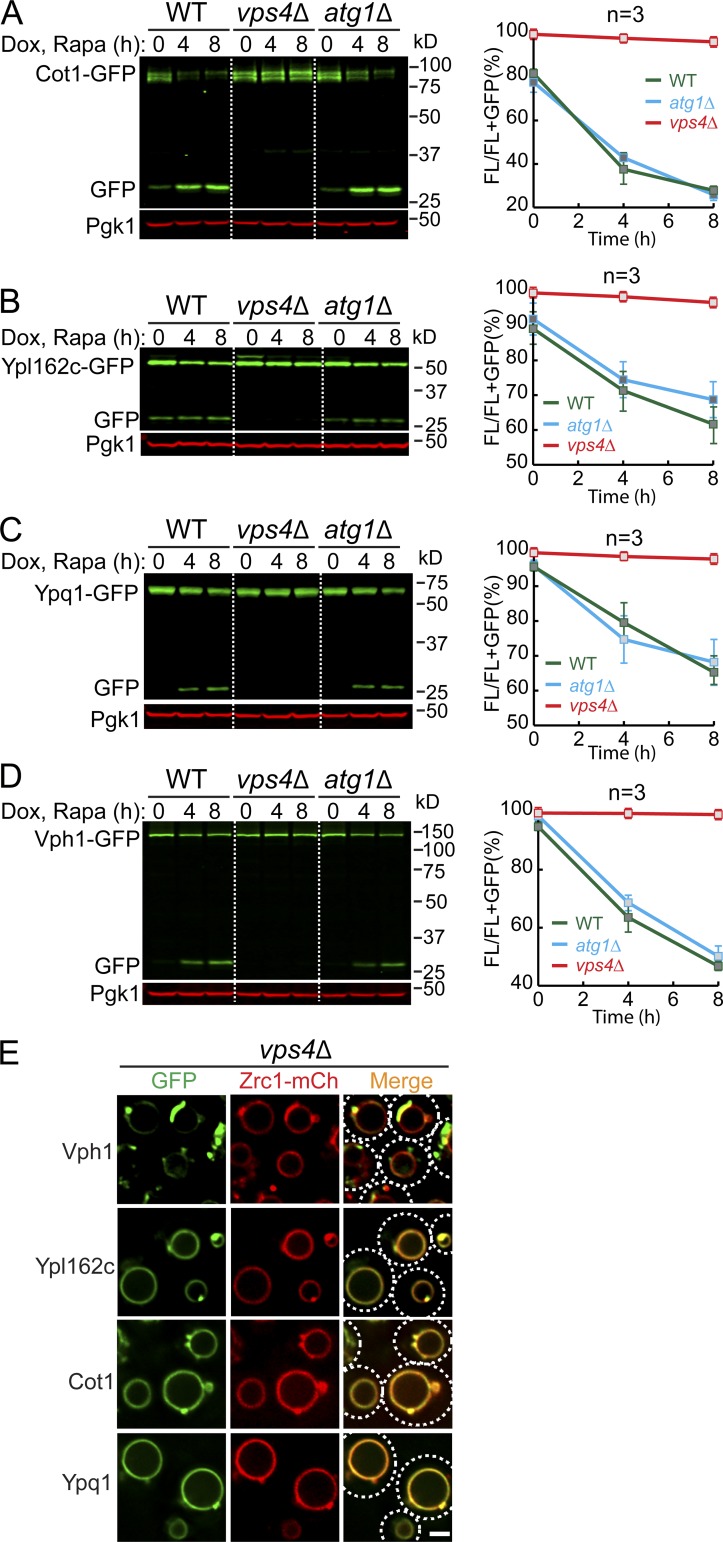
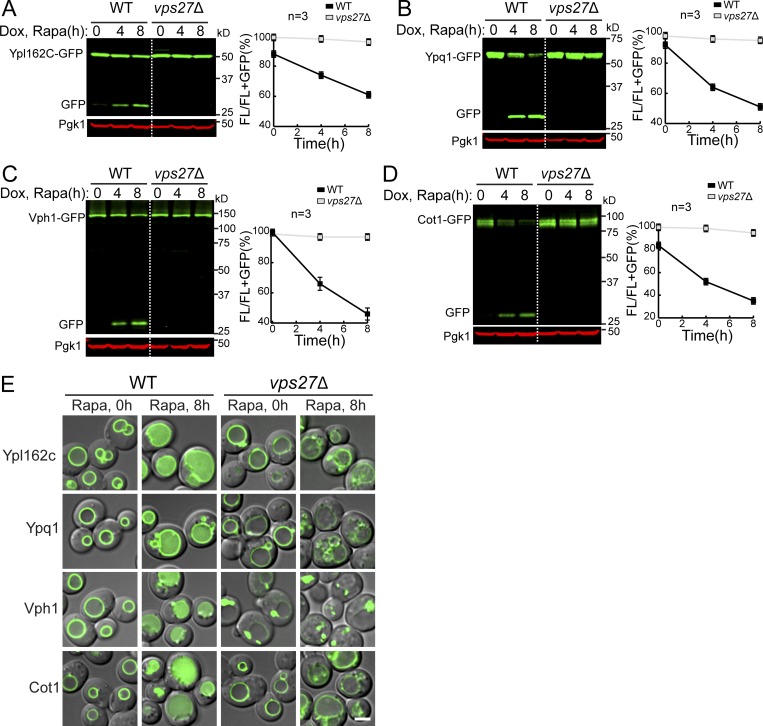
We then tested whether macroautophagy or ESCRT machinery was responsible for the degradation. Deleting ATG1, an essential gene for macroautophagy, had little effect on the degradation of all tested membrane substrates (Cot1, Vph1, Ypl162c, and Ypq1; Fig. S2, A–D). In contrast, all the substrates were stabilized when two independent ESCRT components, VPS4 (the AAA-ATPase that disassembles the ESCRT-III filaments) and VPS27 (a component of the ESCRT-0 subcomplex), were deleted (Figs. S2, A–D; and S3). These results showed that the ESCRT machinery, but not macroautophagy, is required for the degradation of VM proteins.
Figure S2.
ESCRT machinery, but not macroautophagy, is responsible for the degradation of VM proteins. (A–D) Western blots (left) and corresponding quantifications (right) showing the degradation of Cot1-GFP (A), Ypl162c-GFP (B), Ypq1-GFP (C), or Vph1-GFP (D) in WT, vps4Δ, and atg1Δ strain cells. The same volume of cells was loaded, with 0.5 OD600 unit of cells loaded at 0 h. (E) Subcellular localization of VM proteins in cells from the vps4Δ strain. Dashed circles highlight the yeast cell periphery. Scale bar, 2 µm. The error bars represent SD (n = 3). Dox, doxycycline; FL, full-length protein fused with GFP; mCh, mCherry; Rapa, rapamycin. Related to Fig. 3.
Figure S3.
ESCRT machinery is responsible for the degradation of VM proteins. (A–D) Western blots (left) and corresponding quantifications (right) showing the degradation of Ypl162c-GFP (A), Ypq1-GFP (B), Vph1-GFP (C), or Cot1-GFP (D) in WT and vps27Δ strain cells. The same volume of cells was loaded, with 0.5 OD600 unit of cells loaded at 0 h. (E) Subcellular localization of VM proteins in WT and vps27Δ strain cells before (0 h) or after (8 h) rapamycin treatment. Scale bar, 2 µm. Dox, doxycycline; FL, full-length protein fused with GFP; Rapa, rapamycin. Related to Fig. 3.
One caveat of using ESCRT deletion strains was the formation of class E compartments, which refers to the aberrant endosomal structures that are adjacent to vacuoles after the ESCRT machinery is inactivated. These aberrant endosomes can no longer efficiently fuse with the VM. As such, most proteins that travel through the multivesicular body pathway are trapped outside the vacuole, including Vph1-GFP (MacDonald et al., 2012; Yang et al., 2018; Zhu et al., 2017). As shown in Figs. S2 E and S3 E, in an ESCRT deletion strain, the majority of Vph1-GFP was trapped at the class E compartment. In contrast, membrane proteins that travel through the independent AP-3 pathway, including Ypl162c-GFP, Cot1-GFP, Ypq1-GFP, and Zrc1-mCherry, are mostly localized to the VM (Figs. S2 E and S3 E). This observation raises the possibility that the block of Vph1-GFP degradation is because it cannot reach the vacuole and thus cannot be recognized by the vacuole degradation machinery.
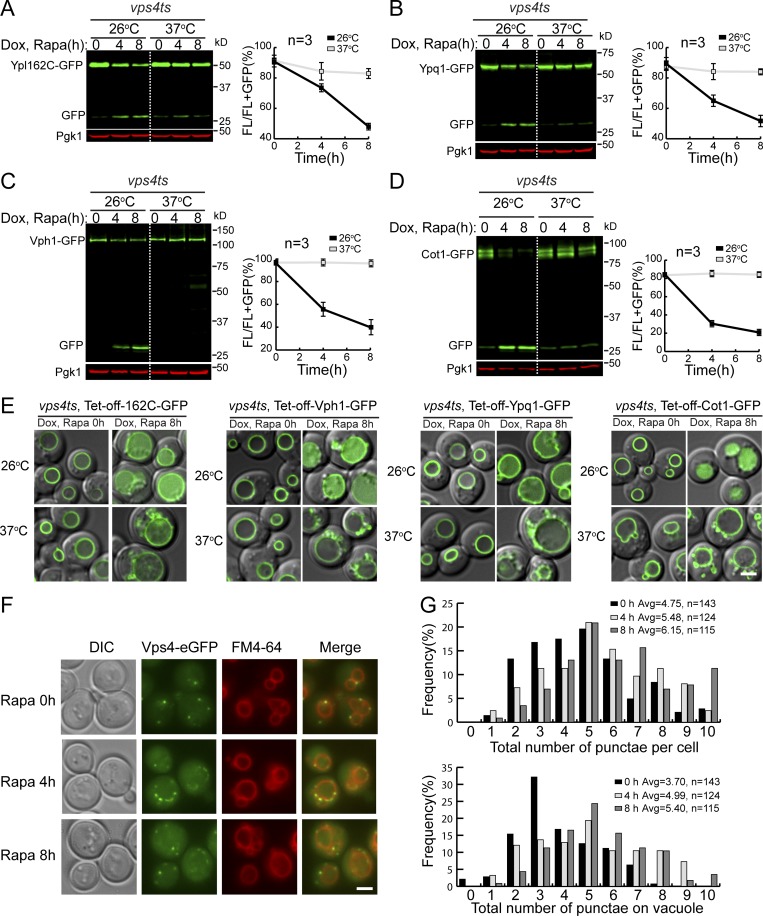
To exclude this possibility, we performed the degradation assay in a temperature-sensitive vps4ts strain (Babst et al., 1997). Yeast cells were first grown at the permissive temperature (26°C) to ensure all VM proteins, including substrates and the required degradation machinery, traffic normally to the vacuole. Next, the degradation assay was performed at both 26°C (permissive) and 37°C (nonpermissive) temperatures. At 26°C, the degradation of all tested substrates occurred normally (Fig. 3, A–E). However, at 37°C, their degradation was drastically reduced, and proteins were stabilized on the VM.
Figure 3.
The ESCRT machinery is required for the degradation of VM proteins. (A–D) Western blots (left) and corresponding quantifications (right) showing the degradation of Ypl162c-GFP (A), Ypq1-GFP (B), Vph1-GFP (C), or Cot1-GFP (D) in vps4ts cells at both 26 and 37°C. (E) Subcellular localization of VM substrates in vps4ts cells at both 26 and 37°C after rapamycin treatment. (F) Subcellular localization of Vps4-eGFP before (0 h) and after (4 h and 8 h) rapamycin treatment. (G) Quantification of the Vps4-eGFP puncta in F. Scale bar, 2 µm. The error bars represent SD (n = 3). DIC, differential interference contrast; Dox, doxycycline; FL, full-length protein fused with GFP; Rapa, rapamycin.
If the ESCRT machinery is responsible for increased degradation of VM proteins, one would expect to see more ESCRTs localize (or adjacent) to the VM after TORC1 inactivation. To test this, we tagged Vps4 with a functional 3HA-eGFP tag (Vps4-3HA-eGFP; Adell et al., 2017) and checked its localization. As shown in Fig. 3, F and G, after rapamycin treatment, more Vps4-GFP puncta were localized at or in the direct vicinity of the VM (4 h and 8 h vs. 0 h), supporting a model in which the ESCRT machinery was active at the membrane to sort cargoes.
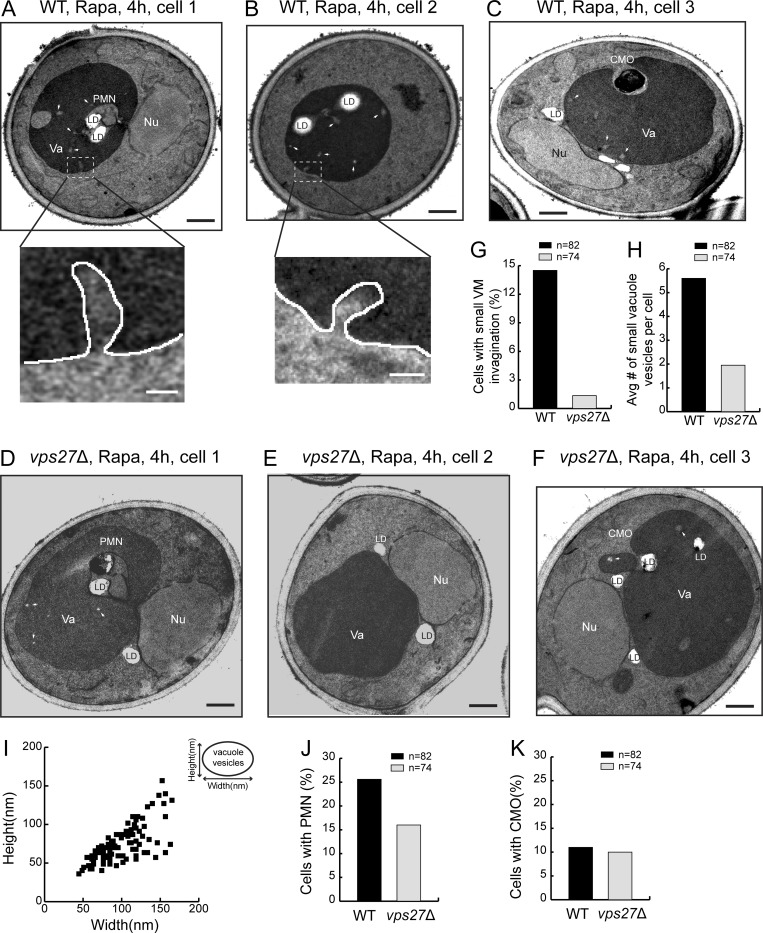
Last, we used transmission electron microscopy (TEM) to visualize the VM invagination after 4 h of rapamycin treatment. As shown in Fig. 4, A–C, three types of invagination were observed in WT cells. The most prominent group was the piecemeal microautophagy of the nucleus (PMN; ∼26%, n = 82; Fig. 4 J). Besides PMN that happens at the nucleus–vacuole junctions, we also observed cytoplasmic microautophagy of organelles (CMO) such as lipid droplets (∼10%, n = 82; Fig. 4 K). The third group was the small tubular invagination of VM (∼14.5%, n = 82; Fig. 4, A, B, and G, insets in A and B). In addition, we also observed small vesicles inside the vacuole (Fig. 4, A–F, arrows). After deleting VPS27, the small tubular microautophagy was nearly abolished (∼1.2%, n = 74; Fig. 4, D–G). Moreover, the number of small vesicles inside the vacuole was drastically reduced (Fig. 4 H). In contrast, the other two forms of microautophagy were either unaffected (for CMO) or reduced, but not abolished (for PMN). Combined with the Vps4 localization analysis, these data supported a model that the ESCRT localizes to the VM to directly invaginate cargo proteins as small vesicles (Fig. 4, A–C, and I).
Figure 4.
The ESCRT deletion abolished one form of microautophagy. (A–C) Representative TEM images showing three types of microautophagy, including PMN, CMO, and small VM invagination in WT cells after rapamycin (Rapa) treatment. Insets are zoomed-in images showing the small tubular invagination of VM. (D and E) Representative TEM images showing that after Rapa treatment, PMN and CMO still happened in vps27Δ cells, while the small VM invagination was nearly abolished. (G) Frequency of observing small VM invagination in WT and vps27Δ cells after Rapa treatment. (H) Number of small vacuole vesicles per cell in WT and vps27Δ cells after Rapa treatment. (I) Size distribution of small vacuole vesicles. (J) Frequency of observing PMN in WT and vps27Δ cells after Rapa treatment. (K) Frequency of observing CMO in WT and vps27Δ cells after Rapa treatment. LD, lipid droplet; Nu, nucleus; Va, vacuole. White arrowheads highlight small vacuole vesicles. Black scale bar, 0.5 µm; white scale bar, 50 nm.
Taken together, our results indicated that the ESCRT-mediated microautophagy, but not ILF or macroautophagy, is essential for the sorting of VM proteins into the lumen.
Multiple E3 ubiquitin ligases function at the vacuole
Ubiquitination is a prerequisite for cargo recognition by the ESCRT machinery, which implies that E3 ligases are important for the starvation-triggered VM degradation. As of now, two independent E3 ligase complexes, the Ssh4-Rsp5 complex and the Dsc complex, have been identified to ubiquitinate VM proteins (Li et al., 2015a; Li et al., 2015b; Yang et al., 2018). However, only three vacuole transporters (Ypq1, Cot1, and Zrt3*) have been shown to be their substrates. Our observation here expands their potential substrate repertoires.
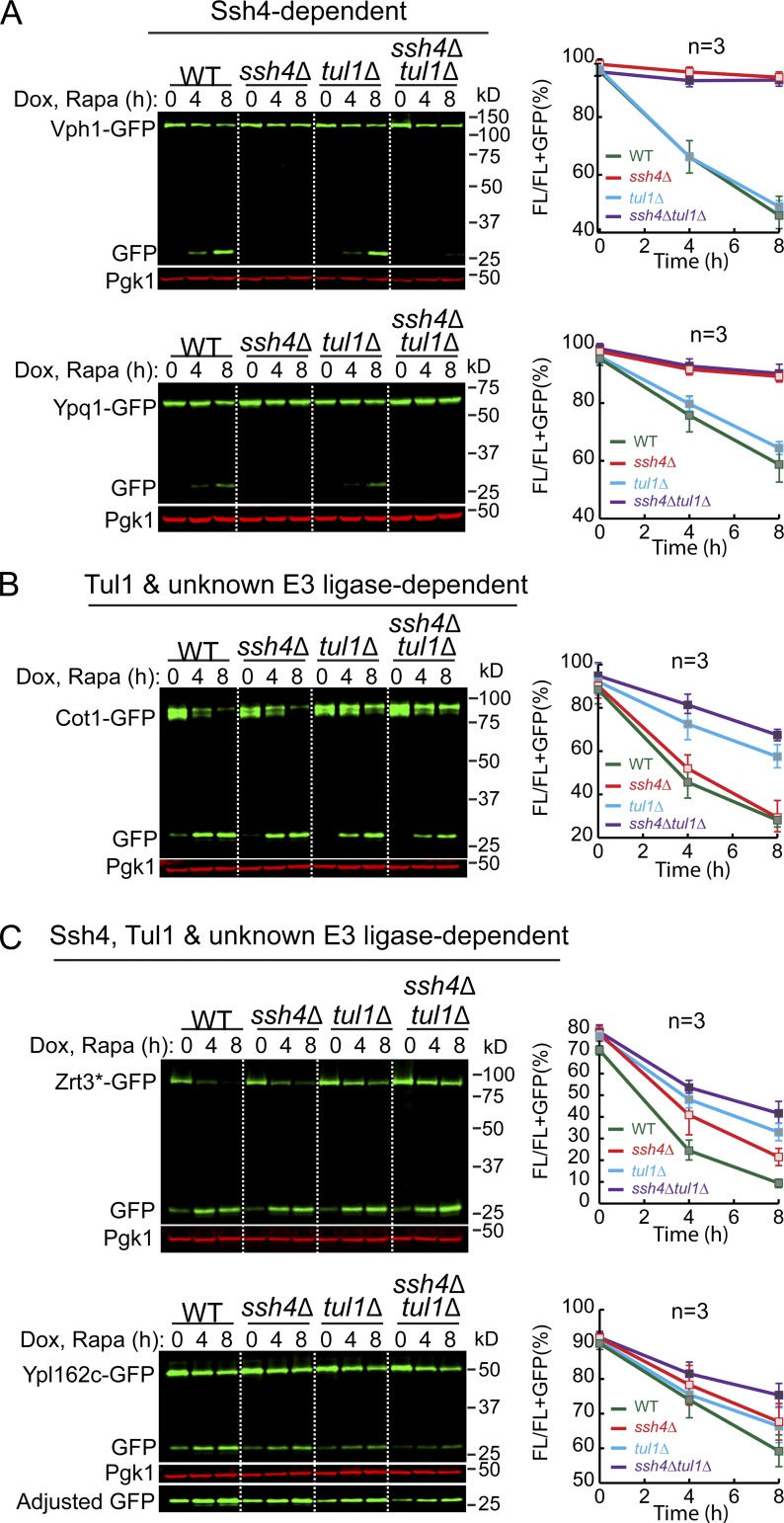
To evaluate the importance of these E3 ligases, we performed degradation assays in WT, ssh4Δ, tul1Δ (the E3 ligase in the Dsc complex), and ssh4Δ tul1Δ double mutant strains (Fig. 5 and Fig. S4). Interestingly, our tested substrates showed a diverse E3 ligase preference. Based on the E3 ligase dependence, we divided them into three groups: group A, Ssh4 dependent; group B, Tul1 and unknown E3 ligase dependent; and group C, Ssh4, Tul1, and unknown E3 ligase dependent.
Figure 5.
Multiple vacuole E3 ligases function downstream of the TORC1 kinase. (A–C) Western blots (left) and corresponding quantifications (right) showing the degradation of Vph1-GFP and Ypq1-GFP (A), Cot1-GFP (B), or Zrt3*-GFP and Ypl162c-GFP (C) in WT, ssh4Δ, tul1Δ, and ssh4Δ tul1Δ cells. The error bars represent SD (n = 3). Dox, doxycycline; FL, full-length protein fused with GFP; Rapa, rapamycin.
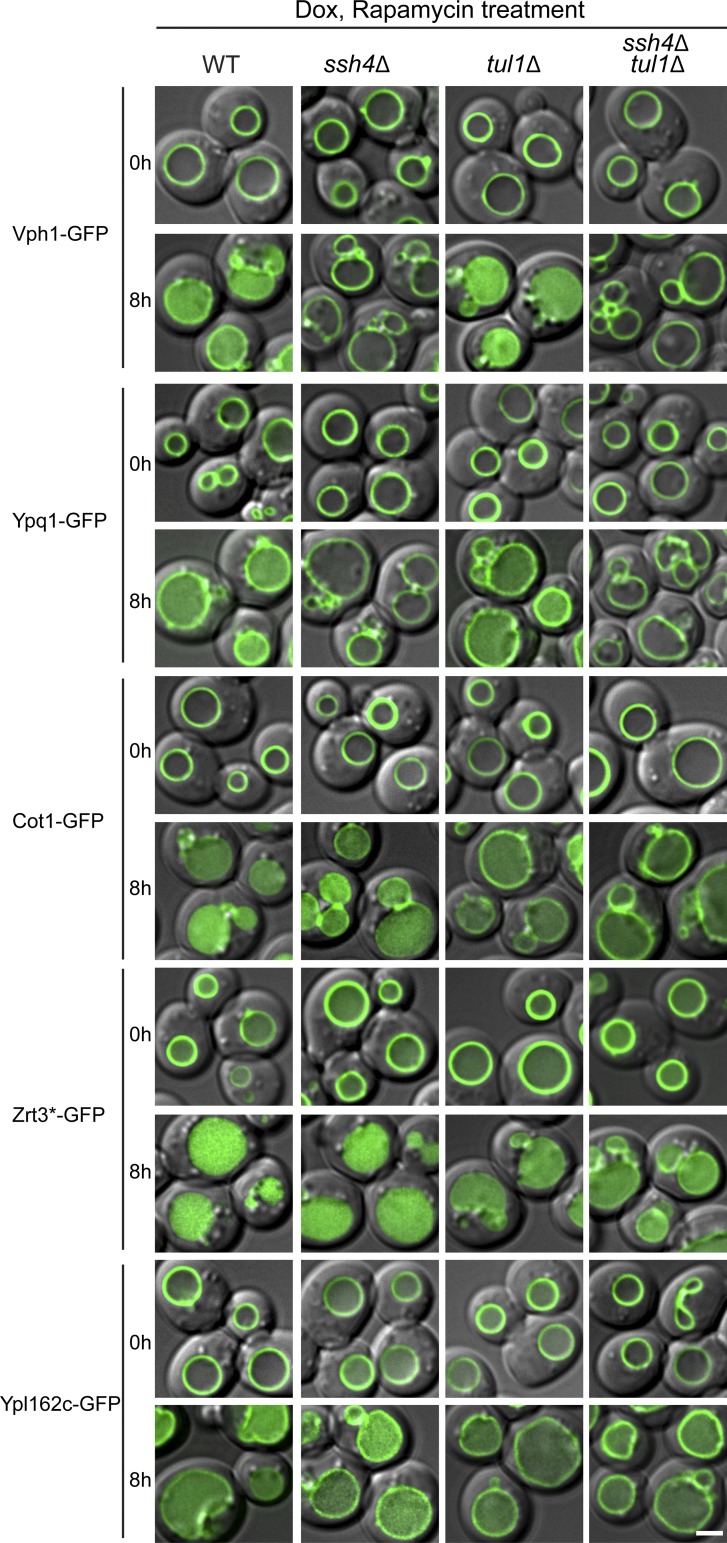
Figure S4.
Multiple vacuolar E3 ligases function downstream of the TORC1 kinase. Subcellular localization of VM proteins in WT, ssh4Δ, tul1Δ, and ssh4Δ tul1Δ strain cells before (0 h) or after (8 h) rapamycin treatment. Scale bar, 2 µm. Dox, doxycycline. Related to Fig. 5.
Group A contains two substrates: Vph1 and Ypq1. For both proteins, deleting SSH4 blocked degradation, whereas deleting TUL1 had no impact on the kinetics (Fig. 5 A and Fig. S4). In group B, the degradation of Cot1-GFP was significantly reduced in tul1Δ but nearly unaffected in the ssh4Δ strain (Fig. 5 B and Fig. S4), suggesting that Tul1, but not Ssh4, contributed to the degradation. However, a significant amount of Cot1-GFP was still degraded even in the ssh4Δ tul1Δ strain, indicating the existence of either a new ubiquitin ligase or a new Rsp5 adaptor. Of note, under zinc depletion conditions, the degradation of Cot1-GFP is mainly dependent on Tul1 and the Dsc complex (Li et al., 2015a), suggesting the action of different recognition mechanisms under these two conditions (i.e., rapamycin vs. Zn2+ depletion). Zrt3* and Ypl162c represented the Ssh4–, Tul1–, and unknown E3 ligase–dependent substrates (group C). As shown in Fig. 5 C, the degradation of full-length Zrt3*-GFP was partially reduced by either SSH4 or TUL1 deletion and was further decreased, but not completely abolished, in the ssh4Δ tul1Δ strain, again suggesting the existence of another E3 ligase/Rsp5 adaptor. Similarly, we found that the degradation of Ypl162c also involves an unknown E3 ligase/Rsp5 adaptor, with some contribution from Ssh4 and Tul1 (Fig. 5 C).
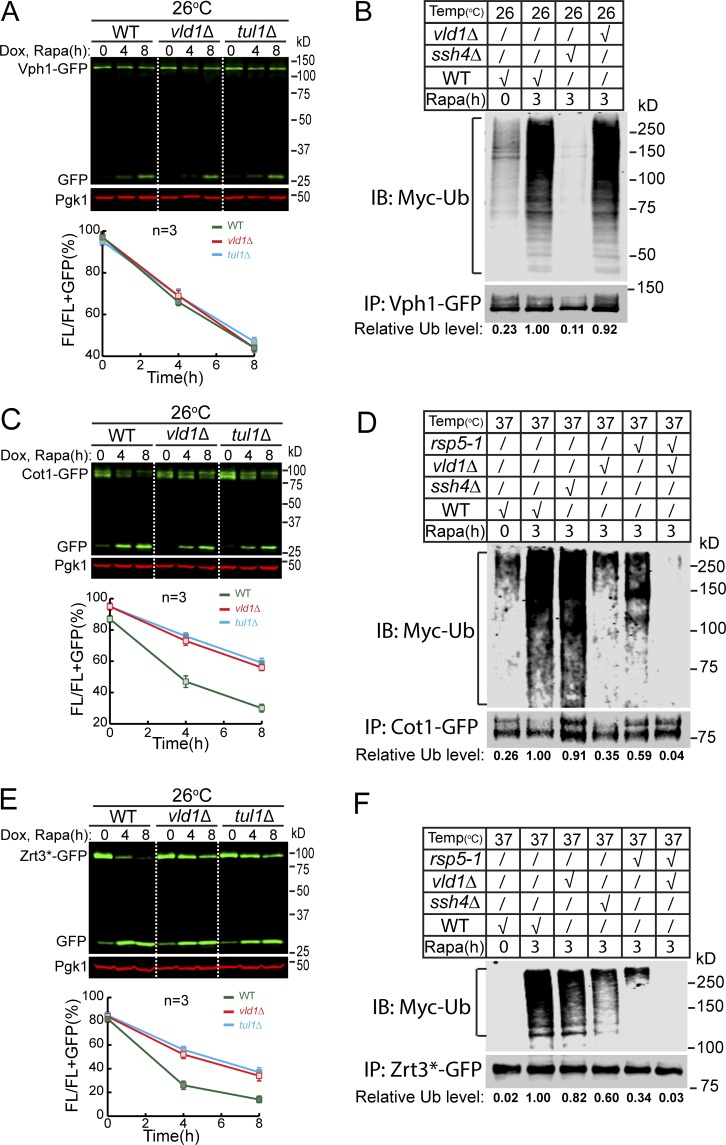
To directly demonstrate the role of vacuolar E3 ligases in cargo ubiquitination, we performed ubiquitin blots on Vph1 (group A), Cot1 (group B), and Zrt3* (group C). For Western blot detection, the ubiquitin was labeled with a MYC tag. To stabilize the ubiquitinated population, we deleted DOA4 that encodes a major deubiquitinase of the endomembrane system. Because doa4Δ tul1Δ is lethal (Li et al., 2015a; Tong et al., 2014), we generated a doa4Δ vld1Δ strain instead to study the role of the vacuolar Dsc complex. Vld1 is a bona fide Dsc component that guides the complex to the VM through the AP-3 pathway (Yang et al., 2018). Deleting VLD1 had a similar effect on VM degradation as TUL1 deletion (Fig. 6, A, C, and E).
Figure 6.
VM E3 ligases poly-ubiquitinate their membrane cargoes upon TORC1 inactivation. (A) Western blot (top) and quantification (bottom) showing the degradation of Vph1-GFP in WT, vld1Δ, and tul1Δ cells. (B) A representative Western blot (n = 3) showing the poly-ubiquitination of Vph1-GFP in WT, ssh4Δ, and vld1Δ cells. The relative ubiquitination level was normalized to the Vph1-GFP level. (C) Western blot (top) and quantification (bottom) showing the degradation of Cot1-GFP in WT, vld1Δ, and tul1Δ cells. (D) A representative Western blot (n = 2) showing the poly-ubiquitination of Cot1-GFP in WT, ssh4Δ, vld1Δ, rsp5-1, and rsp5-1 vld1Δ cells. (E) Western blot (top) and quantification (bottom) showing the degradation of Zrt3*-GFP in WT, vld1Δ, and tul1Δ cells. (F) A representative Western blot (n = 2) showing the poly-ubiquitination of Zrt3*-GFP in WT, ssh4Δ, vld1Δ, rsp5-1, and rsp5-1 vld1Δ cells. The error bars represent SD (n = 3). IB, immunoblot; Dox, doxycycline; FL, full-length protein fused with GFP; Rapa, rapamycin; Ub, ubiquitin.
As shown in Fig. 6, rapamycin treatment triggered the poly-ubiquitination of all three proteins. For Vph1, the deletion of SSH4 eliminated its ubiquitination, whereas deleting VLD1 had little effect (Fig. 6 B, compare the last two lanes). In contrast, VLD1 deletion led to a reduction (65%) of Cot1 ubiquitination, whereas SSH4 deletion had little effect (Fig. 6 D, lane 3 vs. lane 4). Interestingly, the temperature-sensitive rsp5-1 mutation at 37°C resulted in a strong reduction (41%), and further deletion of VLD1 abolished the Cot1 ubiquitination (Fig. 6 D, lane 5 vs. lane 6). The different effects between SSH4 and RSP5 mutants on Cot1 ubiquitination suggested the existence of a new Rsp5 adaptor, instead of a new E3 ligase. In the case of Zrt3*, deletion of either SSH4 or VLD1 caused a reduction (Fig. 6 F, 40 and 18%, respectively) of its ubiquitination. However, similar to Cot1, rsp5-1 mutant had a much stronger reduction (66%) than ssh4Δ (18%), and the double mutant of rsp5-1 vld1Δ completely abolished the ubiquitination, again suggesting the involvement of a new Rsp5 adaptor.
Our ubiquitin blots suggested the involvement of additional Rsp5 adaptors for Cot1 and Zrt3*. To confirm, we compared their degradation kinetics between the rsp5-1 tul1Δ strain and ssh4Δ tul1Δ strain at a nonpermissive temperature. As shown in Fig. 7, A and B, the rsp5-1 tul1Δ double mutant completely blocked the Cot1-GFP degradation, and no accumulation of free GFP was observed. By contrast, Cot1-GFP was still partially degraded in the ssh4Δ tul1Δ strain. Therefore, there must be a new Rsp5 adaptor to recognize Cot1.
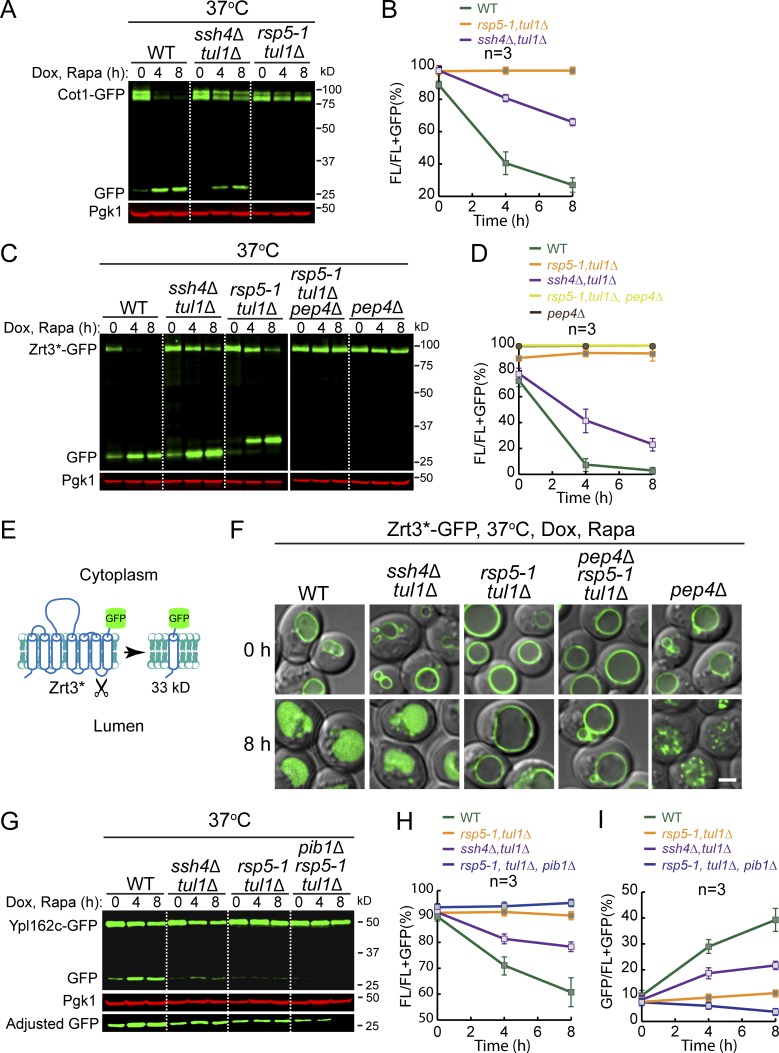
Figure 7.
Evidence for the existence of a new Rsp5 adaptor and identification of Pib1 as the third vacuole E3 ligase. (A) Western blot showing the degradation of Cot1-GFP in indicated strains. (B) Quantification of the protein levels in A. (C) Western blot showing the degradation of Zrt3*-GFP in indicated strains. (D) Quantification of the protein levels in C. Please note that the curves of pep4Δ rsp5-1 tul1Δ and pep4Δ samples are almost overlapping. rsp5-1 tul1Δ double mutant was quantified as (FL + 33 kD)/(FL + 33 kD + GFP). (E) A cartoon depicting the cleavage of Zrt3* between TM6 and TM7. (F) Subcellular localization of Zrt3*-GFP in the indicated strains before (0 h) and after (8 h) rapamycin treatment. (G) Western blot showing the degradation of Ypl162c-GFP in indicated strains. (H and I) Quantification of the full-length (H) or free GFP (I) levels in G. Scale bar, 2 µm. The error bars represent SD (n = 3). Dox, doxycycline; FL, full-length protein fused with GFP; Rapa, rapamycin.
The degradation of Zrt3*-GFP in the rsp5-1 tul1Δ double mutant is very intriguing. As shown in Fig. 7 C, the protein levels of the full-length protein were still decreasing in the double mutant at 37°C. However, no increase of free GFP was observed. Instead, an intermediate-sized band (∼33 kD) accumulated (Fig. 7 C, middle three lanes). These results suggested that the double mutant may have completely blocked the degradation of Zrt3*-GFP. However, Zrt3*-GFP may not be stable at high temperature and was cleaved by a lumenal protease. Based on the size of the cleavage product, the digestion might have happened at the lumenal loop between transmembrane helix 6 and 7 (Fig. 7 E). Consistent with this hypothesis, deletion of PEP4 in rsp5-1 tul1Δ, or deletion of PEP4 alone, abolished the accumulation of the 33-kD band, and no decrease of the full-length protein was observed (Fig. 7, C and D). Further supporting evidence was provided by imaging data. As shown in Fig. 7 F, no lumenal accumulation of the GFP signal was observed in the rsp5-1 tul1Δ cells, despite the fact that the 33-kD band accumulated based on the Western blot. Importantly, although the PEP4 single-deletion mutant and the triple mutant displayed a similar phenotype by Western blot, they exhibited different phenotypes by fluorescence microscopy. With the triple mutant, Zrt3*-GFP was completely stabilized on the VM. In contrast, in the pep4Δ mutant, Zrt3*-GFP was detected as small intravacuolar puncta, indicating that it was present on the membrane that had been invaginated, but that the resulting vesicles and their cargoes were not degraded. Together, we concluded that the rsp5-1 tul1Δ completely blocked the ubiquitination of Zrt3*-GFP. Furthermore, the difference between the rsp5-1 tul1Δ and ssh4Δ tul1Δ also suggested the existence of a new Rsp5 adaptor.
In summary, our analysis indicated that Rsp5 and the Dsc complex are the two major E3 ligases that function downstream of the TORC1 complex to regulate VM composition. In addition to Ssh4, there is strong evidence to suggest the existence of another Rsp5 adaptor, which will be characterized and reported elsewhere.
Identification of a third vacuole E3 ligase, Pib1
Next, we performed the E3 ligase deletion analysis for Ypl162c. Intriguingly, unlike Cot1 and Zrt3*, the degradation of Ypl162c was not completely blocked in the rsp5-1 tul1Δ double mutant at 37°C (Fig. 7, G–I), indicating the involvement of a third E3 ligase.
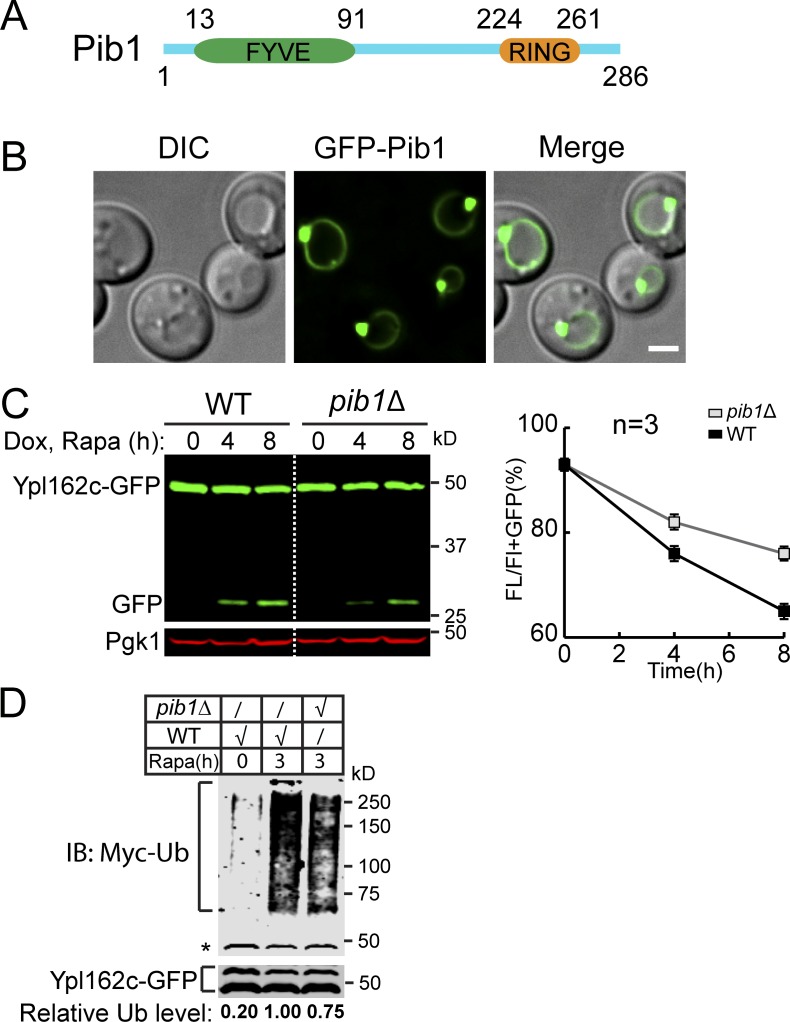
To identify the unknown E3 ligase, we generated a triple mutant strain by further deleting the PIB1 gene. We focused on Pib1 because this E3 ligase has been localized to the vacuole and endosome membrane, but its substrates were so far unknown (Burd and Emr, 1998; Shin et al., 2001). As shown in the last three lanes of Fig. 7 G, further deletion of PIB1 completely abolished the free GFP accumulation. Pib1 is a RING domain–containing E3 ligase with a FYVE domain close to its N terminus (Fig. S5 A). As reported, Pib1 is localized to the VM and endosomes (Fig. S5 B), presumably through its interaction with phosphatidylinositol 3-phosphate (Burd and Emr, 1998; Shin et al., 2001). Furthermore, both ubiquitination and degradation of Ypl162c-GFP were partially reduced in the single pib1Δ mutant (Fig. S5, C and D), indicating that Pib1 indeed participates in the ubiquitination of Ypl162c. Together, our data suggest that Pib1 plays a role in the regulation of VM proteins.
Figure S5.
Pib1 participates in the ubiquitination. (A) A schematic model to show the domain organization of Pib1. (B) Subcellular localization of GFP-Pib1. (C) Western blots (left) and quantification (right) comparing the degradation of Ypl162c-GFP in WT and pib1Δ strains. The same volume of cells was loaded, with 0.5 OD600 unit of cells loaded at 0 h. (D) A representative Western blot (n = 3) showing the poly-ubiquitination of Ypl162c-GFP in WT and pib1Δ cells at 28°C. The relative ubiquitin level was normalized to the Ypl162c-GFP level. The asterisk represents a background band. Scale bar, 2 µm. The error bars represent SD (n = 3). IB, immunoblot; DIC, differential interference contrast; Dox, doxycycline; FL, full-length protein fused with GFP; Rapa, rapamycin. Related to Fig. 7.
TORC1 regulates vacuolar E3 ligases
What are the underlying mechanisms for TORC1 to down-regulate VM proteins? We reasoned that there might be three different levels of regulation: (1) TORC1 may regulate the phosphorylation state of VM proteins, (2) TORC1 may regulate the activity of vacuolar ubiquitination machinery, and (3) TORC1 may regulate the assembly and disassembly of the ESCRT machinery. Recently, De Virgilio and colleagues reported that TORC1 negatively regulates ESCRT assembly through the phosphorylation of Vps27, a key component of ESCRT-0 (Hatakeyama et al., 2019). Upon starvation, Vps27 will be dephosphorylated to promote ESCRT assembly. This observation is consistent with the increase of VM protein degradation after TORC1 inactivation. However, whether TORC1 regulates the activity of vacuolar E3 ligases is unknown.
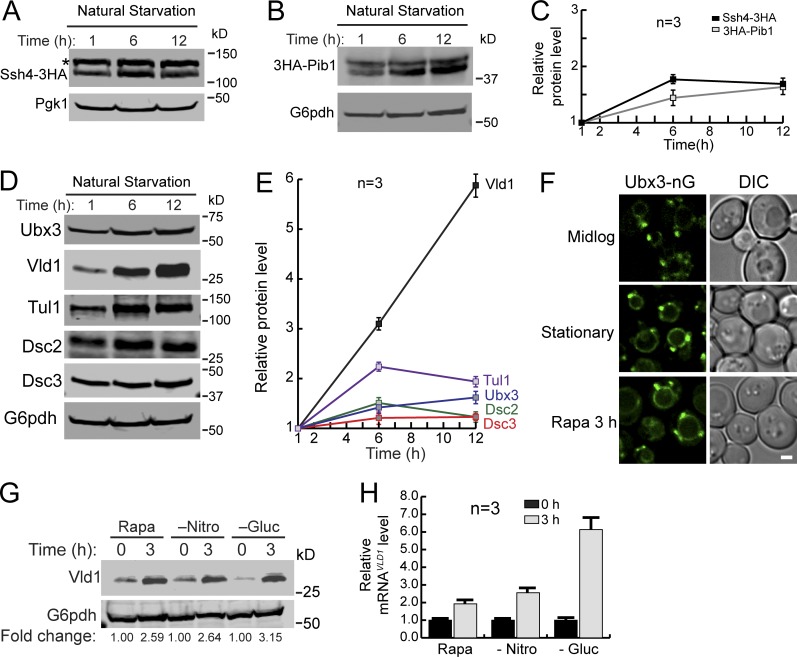
To address the relationship between TORC1 and vacuole E3 ligases, we measured the protein levels of the three identified E3 ligase systems after natural starvation. Consistent with the increasing demand for ubiquitinating VM proteins, the protein levels of Ssh4 and Pib1 were modestly increased after natural starvation (up to 1.8-fold; Fig. 8, A–C). A similar increase was also observed for most components of the Dsc complex, including Ubx3, Tul1, Dsc2, and Dsc3 (Fig. 8, D and E). Strikingly, the protein levels of Vld1 were dramatically elevated (sixfold at 12 h; Fig. 8, D and E).
Figure 8.
TORC1 regulates the activity of vacuole E3 ligases. (A and B) Western blot showing the protein level changes of Ssh4-mNeonGreen-3HA (A) or 3HA-Pib1 (B) in stationary phase cells. The asterisk represents a nonspecific band. (C) Quantification of the protein levels in A and B. (D) Western blots showing the protein level changes of different Dsc components in stationary phase cells. (E) Quantification of the protein levels in D. (F) Subcellular localizations of Ubx3-mNeonGreen (Ubx3-nG) in mid-log phase, stationary phase, and rapamycin-treated cells. (G) A representative Western blot (n = 3) showing the level of Vld1-3HA after rapamycin treatment, nitrogen starvation, or glucose starvation. (H) qRT-PCR showing the level of VLD1 mRNA after rapamycin treatment, nitrogen starvation, or glucose starvation. Scale bar, 2 µm. The error bars represent SD (n = 3). DIC, differential interference contrast; Gluc, glucose; Nitro, nitrogen; Rapa, rapamycin.
Because Vld1 serves as the vacuole trafficking adaptor of the Dsc complex, its up-regulation suggested that TORC1 can regulate the amount of the vacuolar Dsc complex by controlling Vld1 expression. Under nutrient-rich conditions, the Vld1 protein level was low with few vacuole Dsc complexes being assembled (Fig. 8, D–F). In contrast, TORC1 inactivation led to the overproduction of Vld1 and increased assembly of the vacuole Dsc complex, as evidenced by the vacuolar localization of Ubx3 (Fig. 8, D–F). To verify this model, we tested different conditions that can inactivate TORC1, including rapamycin treatment, nitrogen starvation, and glucose starvation. Strikingly, Vld1 protein up-regulation was observed under all conditions (Fig. 8 G). As the last test, we measured the VLD1 mRNA levels using quantitative real-time PCR (qRT-PCR). As shown in Fig. 8 H, the VLD1 mRNA levels were increased after TORC1 inactivation.
Taken together, we concluded that TORC1 inactivation leads to the up-regulation of vacuole E3 ligase systems. Focusing on the Dsc complex, we discovered that TORC1 regulates the amount of the vacuole-localized Dsc (vDsc) complex by controlling the expression of its trafficking adaptor, Vld1.
TORC1 regulates Vld1 expression through the Rim15-Ume6 signaling cascade
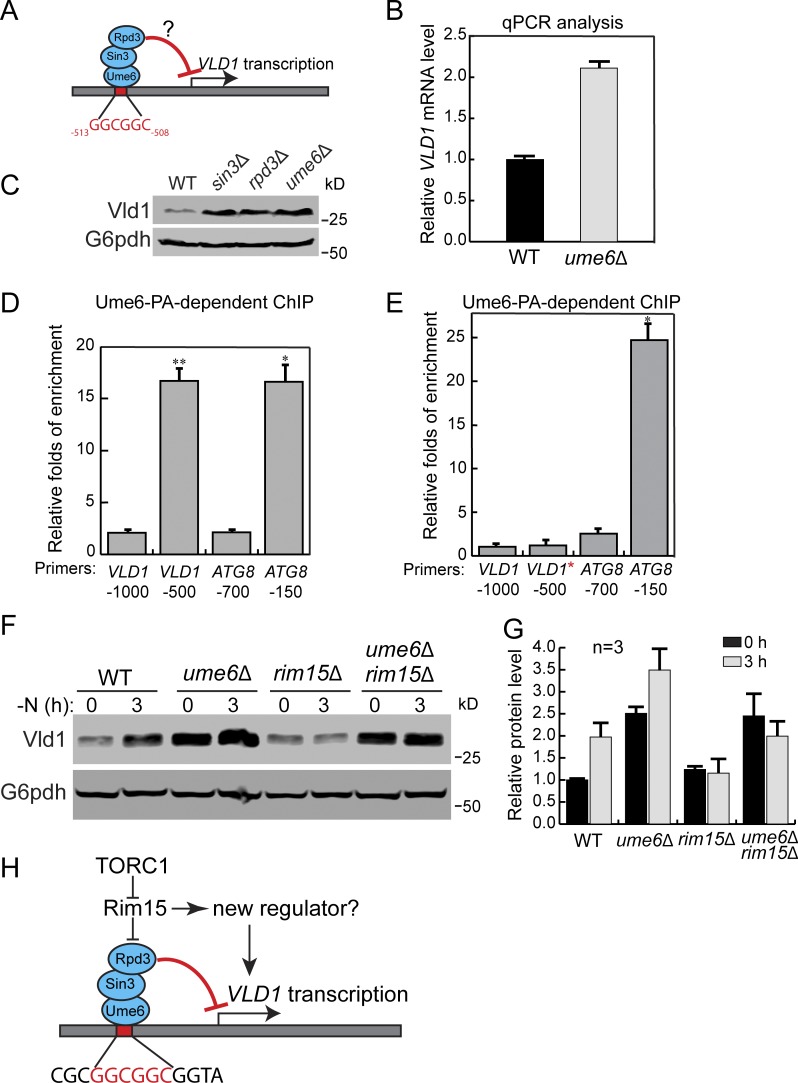
How does TORC1 control the expression of Vld1? The up-regulation of VLD1 mRNA after TORC1 inactivation (Fig. 8 H) suggested that it might occur via transcriptional regulation. Using bioinformatics analysis, we identified an upstream regulatory site 1 (URS1) sequence (GGCGGC) ∼500 bp upstream of the VLD1 start codon (Fig. 9 A), which is a putative binding site for Ume6. Ume6 is a transcription factor that forms a heterotrimeric complex with Sin3 and Rpd3 (Fig. 9 A; Williams et al., 2002). It has been reported that under nutrient-rich conditions, Ume6 can repress the expression of ATG8 by directly binding to the URS1 site of the ATG8 promoter (Backues et al., 2012; Bartholomew et al., 2012). Importantly, the activity of Ume6 is regulated by TORC1. Upon starvation, Ume6 is phosphorylated through a TORC1-Rim15 cascade to relieve its inhibition of gene transcription. To investigate the importance of Ume6 on VLD1 transcription, we first examined the VLD1 mRNA level. In mid-log phase cells, the VLD1 mRNA level was approximately twofold higher in the ume6Δ strain than in the WT strain (Fig. 9 B). Furthermore, up-regulation of Vld1 protein was detected after deleting either UME6, RPD3, or SIN3 (Fig. 9 C), indicating their roles in VLD1 repression. To test the direct binding of Ume6 to the VLD1 promoter, we applied the chromatin immunoprecipitation (ChIP) analysis using a protein A–tagged Ume6 (Ume6-PA) strain. In this assay, two regions from the ATG8 promoter served as controls: (1) a region with a confirmed URS1 binding motif (−150) as a positive control and (2) a region without a binding motif (−700) as a negative control (Fig. 9 D). On the VLD1 promoter, the enrichment of Ume6 was much higher in the URS1 region (−500) than in a region without the URS1 motif (−1,000) and was similar to the level of the positive control (Fig. 9 D). Furthermore, we mutated the URS1 motif (GGCGGC to AAAAAA) of the VLD1 promoter (VLD1*; Fig. 9 E) and performed another ChIP analysis. As expected, the URS1 mutation abolished the enrichment of Ume6 on the VLD1 promoter (Fig. 9 E). Taken together, these data suggest that the Ume6 ternary complex suppressed VLD1 transcription by directly binding to its URS1 motif.
Figure 9.
TORC1 regulates Vld1 through the Rim15-Ume6 signaling cascade. (A) A cartoon showing the interaction between the Ume6 complex with the putative binding motif in the VLD1 promoter region. (B) qRT-PCR showing the VLD1 mRNA level in WT and ume6Δ cells. (C) A representative Western blot (n = 3) showing the level of Vld1-3HA in WT, ume6Δ, sin3Δ, and rpd3Δ cells. (D) ChIP analysis showing the binding of Ume6 to the VLD1 promoter region. (E) ChIP analysis showing the disruption of Ume6 binding to the VLD1 promoter region after mutation. The enrichment values were normalized to the input DNA, and the error bars show the SEM of three independent experiments. The P value from two-sided t test is presented by asterisks: *P < 0.05, **P < 0.01. (F) Western blot showing the level of Vld1-3HA in WT, ume6Δ, rim15Δ, and ume6Δ rim15Δ cells. (G) Quantification of the protein levels in F. (H) A cartoon model showing that TORC1 regulates Vld1 expression through a Rim15-mediated signaling cascade. GGCGGC is the URS1 motif. PA, protein A; VLD1*, VLD1 promoter with URS1 motif mutated to AAAAAA. The error bars represent SD (n = 3).
As stated above, Ume6 activity is regulated by TORC1 through the Rim15 kinase (Bartholomew et al., 2012). Based on its phosphorylation state, Rim15 can shuttle between the nucleus and cytosol. Active TORC1 and its downstream effector Sch9 phosphorylate Rim15 and prevent it from entering the nucleus (Wanke et al., 2008; Wanke et al., 2005). In contrast, TORC1 inactivation leads to the dephosphorylation and activation of Rim15 (Pedruzzi et al., 2003). Active Rim15 then enters the nucleus to phosphorylate Ume6 and inhibits its repressor function (Bartholomew et al., 2012). Thus, we asked whether TORC1 is using this signaling cascade to regulate VLD1 transcription. To this end, we subjected cells to nitrogen starvation to inactivate TORC1 and checked Vld1 protein levels in the WT, ume6Δ, rim15Δ, and rim15Δ ume6Δ strains. In WT cells, an increase of Vld1 protein was detected after nitrogen starvation (Fig. 9, F and G). In rim15Δ cells, the basal level of Vld1 was unchanged; however, there was no up-regulation after nitrogen starvation, indicating that Rim15 works downstream of TORC1 as a positive regulator of VLD1 transcription. Further deletion of UME6 in the rim15Δ strain restored the Vld1 protein level after starvation, suggesting that Rim15 functions upstream of Ume6 (Fig. 9, F and G). Interestingly, although the Vld1 protein level was already increased in ume6Δ cells before nitrogen starvation, it could be further up-regulated after nitrogen starvation (Fig. 9, F and G). This result suggested that another regulator besides Ume6 may also function downstream of Rim15 to regulate VLD1 transcription (Fig. 9 H). Nevertheless, our results strongly support the hypothesis that TORC1 uses the Rim15-Ume6 signaling cascade to regulate VLD1 transcription.
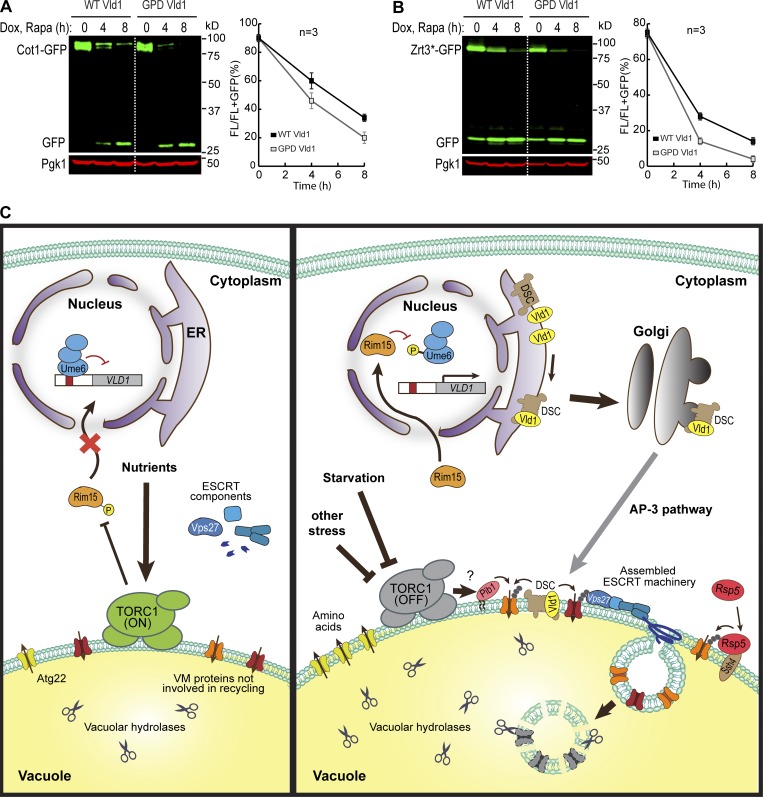
Last, we asked whether overexpression of Vld1 alone is sufficient to induce a constitutive degradation of VM proteins. Vld1 was overexpressed under the GPD promoter and two cargoes (Cot1 and Zrt3*) were tested. As shown in Fig. 10, A and B, before rapamycin treatment, protein levels of both tested substrates were similar between the WT and Vld1 overexpression strains. This indicates Vld1 overexpression alone was not sufficient to trigger a constitutive degradation when TORC1 is active. After rapamycin treatment, the degradation kinetics of both Cot1 and Zrt3* were slightly faster upon overexpression. These data support the hypothesis that the TORC1 regulation of VM proteins may be controlled at several different levels, and manipulating one condition is not sufficient to induce a dramatic change.
Figure 10.
A model summarizing how TORC1 regulates the VM composition via ubiquitin- and ESCRT-dependent microautophagy. (A and B) Western blot (left) and quantification (right) showing the degradation of Cot1-GFP (A) or Zrt3*-GFP (B) in WT and Vld1 overexpression strains. (C) The model. For details, please see the Discussion section. The error bars represent SD (n = 3). Dox, doxycycline; FL, full-length protein fused with GFP; Rapa, rapamycin.
Discussion
TORC1 regulation of the VM composition happens at different levels
In this study, we discovered that TORC1 inactivation leads to the down-regulation of many VM proteins through a ubiquitin- and ESCRT-dependent degradation pathway. This observation is inconsistent with the model that the entire process of vacuole biogenesis will be up-regulated upon TORC1 inactivation. We argue that although lumenal hydrolases and transporters involved in the recycling function are up-regulated, many other vacuolar proteins are down-regulated to supply additional amino acids for cell survival.
How does TORC1 regulate the VM composition? The latter must be controlled at several levels. First, TORC1 can affect the phosphorylation of VM proteins. In a large-scale proteomic study, Hall and colleagues reported that TORC1 activity can affect the phosphorylation state of vacuole transporters, including Fth1, Ccc1, Avt4, Bpt1, and Fun26 (Soulard et al., 2010). It is conceivable that phosphorylation may prime VM proteins for their degradation. Second, as uncovered by our study, under nutrient-rich conditions, active TORC1 inhibits the degradation of VM proteins by repressing the activity of ubiquitination machinery. Specifically, active TORC1 inhibits the transcription of VLD1 through the Rim15-Ume6 cascade. When TORC1 is inactive, the inhibition of VLD1 is relieved, resulting in the up-regulation of Vld1 and assembly of more vacuolar Dsc complexes. By controlling the assembly and trafficking of ubiquitin ligases, TORC1 can regulate the abundance of VM proteins in response to environmental cues (Fig. 10 C). Third, after ubiquitination, the ESCRT machinery is recruited to the VM to sort substrates into the lumen. Interestingly, De Virgilio and colleagues reported that active TORC1 directly phosphorylates Vps27 to inhibit its function on the VM (Hatakeyama et al., 2019). Upon starvation, the dephosphorylation of Vps27 can lead to more efficient assembly of the ESCRT complex on VM (Hatakeyama et al., 2019).
In summary, we propose that the VM composition is regulated by TORC1 in response to environmental cues. Instead of a simple model that TORC1 inactivation leads to the up-regulation of vacuole biogenesis, our study indicated that many membrane proteins are concomitantly degraded to recycle essential amino acids or possibly even “free up” space in the limiting membrane of the vacuole. The regulation may be achieved at three levels, including substrates, E3 ligases, and the ESCRT machinery. We are only at the beginning of understanding this complex relationship.
Different responses of vacuolar-type H+-ATPase (v-ATPase) to the (M)TORC1 inactivation between mammalian and yeast cells
It is intriguing to observe that in yeast, after TORC1 inactivation, Vph1 is down-regulated by ∼30–50%. Two other recent publications also made a similar observation (Hatakeyama and De Virgilio, 2019; Oku et al., 2017). This observation is surprising because, in mammalian cells, it is well established that the v-ATPase components are transcriptionally up-regulated by TFEB after MTORC1 inactivation (Sardiello et al., 2009). Considering that the v-ATPase is responsible for vacuole acidification, which is essential for the vacuole’s recycling function (Manolson et al., 1992), why is it down-regulated in yeast?
Three reasons might explain this difference. First, the yeast vacuole pH is maintained at ∼5–5.5 (Li and Kane, 2009), which is less acidic than the mammalian lysosome (pH 4.5–5; Mellman et al., 1986). This difference means the proton concentration inside the vacuole can be up to 10-fold lower than that in the lysosome. Consistent with the pH difference, GFP is quenched and quickly degraded in mammalian lysosomes, whereas in the yeast vacuole, GFP remains fluorescent and resistant to vacuolar proteases. As such, it may require less energy to maintain a proper vacuolar proton concentration. Second, Vph1 is an abundant protein (∼20,000 molecules/cell) in yeast (Belle et al., 2006). Accordingly, its partial degradation will not abolish the v-ATPase activity required for maintaining the proton gradient. Instead, this degradation may reduce ATP consumption by the v-ATPase besides supplying extra amino acids for cell survival. Third, consistent with the concept of preserving cellular ATP stores, it is well known that the yeast v-ATPase complex undergoes reversible dissociation between the V0 (proton translocation domain) and V1 (ATP hydrolysis domain) subcomplexes after glucose starvation and other stress conditions (Kane, 1995). In summary, the number of v-ATPase complexes might be more than what is required to maintain a functional vacuole pH when cells are shifted to starvation conditions. Instead, reducing ATP consumption and recycling enough amino acids might be much more pressing issues for yeast cell survival.
Materials and methods
Yeast strains, plasmids, media, and growth conditions
All yeast strains and plasmids used in this study are listed in Table S1. Both Difco Yeast Peptone Dextrose (YPD) broth and Difco Yeast Nitrogen Base (YNB) without amino acids and ammonium sulfate were purchased from Thermo Fisher Scientific. YNB without amino acids was purchased from Sigma-Aldrich. All yeast strains were grown at 26°C, unless indicated otherwise, in either YPD or YNB media before further analysis.
Growth curve analysis
Yeast cells were grown in YPD medium to mid-log phase (OD600 ∼0.5–0.7) at 28°C, which was arbitrarily defined as time point 1 h for the growth curve analysis. Next, the growth of the yeast cells was continued at 28°C for up to 40 h. The OD600 was measured every 1–2 h, and the same number of ODs of cells was collected at the indicated time points for further analysis.
Rapamycin-triggered degradation assay
For substrates that were tagged with GFP or 3×HA at genomic loci, yeast cells were grown in YPD medium to mid-log phase (OD600 ∼0.5–0.7), before being incubated with 500 ng/ml rapamycin. After an appropriate amount of time, typically 4–8 h, yeast cells were collected for further analysis. For substrates that were tested under the TET-OFF system, yeast cells were grown in YNB minus uracil medium to mid-log phase (OD600 ∼0.5–0.7). The cells were preincubated with 2 µg/ml doxycycline for an appropriate amount of time to allow complete ER exit (20 min for most of the substrates and 1 h for Vph1-GFP and Fth1-GFP). For the complete ER exit of Fet5-GFP, the plasmid was transformed into a 305-pGpd-Fth1 strain. Yeast cells were then incubated with 500 ng/ml rapamycin for an appropriate amount of time, typically 4–8 h, and collected for further analysis.
Nitrogen starvation assay
Yeast cells were grown in YPD medium to mid-log phase (OD600 ∼0.5–0.7) and then pelleted at 3,500 rpm for 5 min. After being washed with the nitrogen starvation medium (YNB without amino acids and ammonium sulfate, with 2% glucose) twice, cells were resuspended in the nitrogen starvation medium and incubated at 26°C for an appropriate amount of time (typically 3–4 h). Cells were then collected for further analysis.
Conventional TEM
Yeast cells were grown in YPD medium to mid-log phase (OD600 ∼0.5–0.7), before being incubated with 500 ng/ml rapamycin for 4 h. The samples were further processed in the University of Texas Southwestern Electron Microscopy Core Facility (Dallas, TX) using a published protocol (Hariri et al., 2019; Wright, 2000). Basically, cells were fixed with 2× prefix solution (4% glutaraldehyde in 0.2 M Pipes, 0.2 M sorbitol, 2 mM MgCl2, and 2 mM CaCl2) and then stained in uranyl acetate and embedded in Spurr resin. After being polymerized at 60°C overnight, the specimen blocks were sectioned at 70 nm with a diamond knife (Diatome) on an Ultracut UCT 6 ultramicrotome (Leica Microsystems). Sections were poststained with 2% uranyl acetate in water and lead citrate and were placed on copper grids (Thermo Fisher Scientific). TEM images were acquired on a Tecnai G2 spirit transmission electron microscope (FEI) equipped with a LaB6 source at 120 kV by using a Gatan Ultrascan charge-coupled device camera.
ChIP
ChIP was performed with some modifications from a previously published article (Aparicio et al., 2005). After the yeast cells were grown to OD600 ∼0.8 in YPD medium, formaldehyde was added for DNA–protein cross-linking. The DNA was then sheared by sonication, and the sheared chromatin was immunoprecipitated. Next, the protein–DNA complex was eluted, and the cross-linking was reversed. Finally, the purified DNA was examined by RT-qPCR analysis. The information for all primers is listed in Table S2.
RNA isolation and qRT-PCR
Total RNA samples were extracted from yeast cells using TRIzol (145105; Life Technologies) and PureLink RNA Mini Kit (1938678; Invitrogen). For qRT-PCR, ∼6 µg of RNA was applied for first-strand cDNA synthesis using PrimeScript RT Reagent Kit (AK6003; Takara) with oligo(dT) primers. PCR was then performed using the Power SYBR Green PCR Master Mix (1708558D; Thermo Fisher Scientific) with the primers targeting either UBC6 (internal control) or specific genes. For each sample, the relative transcript levels were determined by normalizing them to UBC6 levels. The information for all primers is listed in Table S2.
Microscopy and image processing
The microscopy and imaging processing were performed with a DeltaVision system (GE Healthcare Life Sciences) as described in Yang et al. (2018). The filter sets FITC (excitation, 475/28; emission, 525/48) and TRITC (excitation 542/27; emission 594/45) were used for GFP and mCherry, respectively. In brief, yeast cells were washed with MilliQ water and imaged immediately at room temperature. Image acquisition and deconvolution were performed with the softWoRx program. The images were further cropped and adjusted by using ImageJ (National Institutes of Health).
IP and detection of cargo ubiquitination
To stabilize ubiquitinated cargoes, the gene encoding the ubiquitin hydrolase Doa4 was deleted in either WT or E3 ligase mutant background. Transient overexpression of MYC-ubiquitin, which was under the control of a copper (CUP1)–inducible promoter, was induced by addition of 100 µM Cu2SO4 for 1 h (2 h for ssh4- or rsp5-1–related strains) before the cells were treated with rapamycin to trigger cargo ubiquitination. After 3 h of rapamycin treatment in the presence of 100 µM Cu2SO4, ∼50 OD600 units of cells were collected for the IP experiment.
The IP assay was adapted from Li et al., (2015a), with some modifications. Basically, yeast cells were resuspended in 500 µl IP buffer (50 mM Hepes-KOH, pH 6.8, 150 mM KOAc, 2 mM MgOAc, 1 mM CaCl2, and 15% glycerol) with 0.1% digitonin, supplemented with protease inhibitors and 50 mM N-ethylmaleimide. Whole-cell lysates were prepared by bead beating at 4°C for 10 min, followed by addition of 500 µl of 1.9% digitonin in IP buffer. Membranes were then solubilized by nutating lysates at 4°C for 50 min. After removing the pellet by spinning at 13,000 g for 10 min, the resulting lysate was incubated with 25 µl GFP-TRAP resin (Chromotek) at 4°C for 1 h. The resin was then washed four times with 0.1% digitonin in IP buffer, and bound proteins were eluted by incubating resin with sample buffer at 65°C for 5 min. The eluates were then analyzed by SDS-PAGE and probed with MYC or GFP antibody.
Sample preparation for Western blotting and antibodies
Briefly, yeast cells were treated with ice-cold 10% TCA and incubated on ice for at least 1 h. After washing with 0.1% TCA, the sample pellets were dissolved in 2× boiling buffer (50 mM Tris, pH 7.5, 1 mM EDTA, and 1% SDS), disrupted by glass beads using a vortex mixer for 5 min, and heated at 65°C for 5 min. After addition of 2× urea sample buffer (150 mM Tris, pH 6.8, 6 M urea, 6% SDS, 40% glycerol, 100 mM DTT, and 0.01% bromophenol blue), samples were mixed by vortex with glass beads for 5 min and incubated at 65°C for another 5 min. The supernatants were collected, subjected to SDS-PAGE, and transferred to nitrocellulose membranes for Western blotting analysis. Typically, 1 OD600 unit of cells was loaded in each lane. For TET-OFF system–related Western blotting, the same volume of cells was loaded, with 0.5 OD600 units of cells loaded at 0 h.
The following antibodies were used in this study: G6PDH (1:10,000, A9521; Sigma-Aldrich), Pgk1 (1:5,000, 22C5D8; Invitrogen), mouse anti-GFP (1:500, sc-9996; Santa Cruz Biotechnology), rabbit anti-GFP (1:3,000, TP401; Torrey Pines Biolabs), anti-HA (1:1,000, 16B12; BioLegend), mouse anti-MYC (1:500, 9E10; Santa Cruz Biotechnology), rabbit anti-MYC (1:2,000; Sigma-Aldrich), Vph1 (10D7; Invitrogen), Pep4 (1:10,000), Cps1 (1:5,000; Richter et al., 2007), and Atg8 (1:5,000). Antibodies against Dsc2, Dsc3, Ubx3, and Tul1 were generous gifts from Peter Espenshade (Johns Hopkins University, Baltimore, MD).
Western blotting quantification
The Western blot images were scanned using the Licor Odyssey CLx system and band intensities were quantified from the raw data files using the Image Studio Ver5.2 software. The degradation efficiency was calculated as full-length protein/(full-length protein + free GFP).
Online supplemental material
Fig. S1 shows TORC1 inactivation triggers a global down-regulation of VM proteins. Fig. S2 shows that ESCRT machinery, but not macroautophagy, is responsible for the degradation of VM proteins. Fig. S3 shows ESCRT machinery is responsible for the degradation of VM proteins. Fig. S4 shows multiple vacuolar E3 ligases function downstream of the TORC1 kinase. Fig. S5 shows that Pib1 participates in the ubiquitination. Table S1 lists all strains and plasmids used in this study. Table S2 lists all the primers used in this study.
Supplementary Material
lists yeast strains and plasmids used in this study.
lists primers used in this study.
Acknowledgments
We thank members of the M. Li laboratory for their technical support. We are also grateful to our colleagues in the Protein Folding and Disease Hub and the Molecular Cellular and Developmental Biology department, especially M. Duncan, H. Xu, and Y. Wang for the helpful discussion and critical reading of the manuscript. We thank K. Luby-Phelps and the University of Texas Southwestern Electron Microscopy Core Facility for expert technical assistance.
This research is supported by a startup fund and the MCubed 3.0 fund from the University of Michigan and National Institutes of Health grants GM133873 to M. Li and GM131919 to D.J. Klionsky, and by Fonds zur Förderung der Wissenschaftlichen Forschung grants Y444B12, P30263, P29583, and W1101-B18 to D. Teis.
The authors declare no competing financial interests.
Author contributions: Conceptualization, X. Yang, W. Zhang, and M. Li; Methodology, X. Yang, W. Zhang, X. Wen, and M. Li; Investigation, X. Yang, W. Zhang, X. Wen, P.J. Bulinski, D.A. Chomchai, S. Sprenger, F.M. Arines, Y.-Y. Liu, and M. Li; Original Draft, X. Yang, W. Zhang, and X. Wen; Editing, D.J. Klionsky, D. Teis, and M. Li; Funding Acquisition & Supervision, D.J. Klionsky, D. Teis, and M. Li.
References
- Adell M.A.Y., Migliano S.M., Upadhyayula S., Bykov Y.S., Sprenger S., Pakdel M., Vogel G.F., Jih G., Skillern W., Behrouzi R., et al. 2017. Recruitment dynamics of ESCRT-III and Vps4 to endosomes and implications for reverse membrane budding. eLife. 6:e31652 10.7554/eLife.31652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammerer G., Hunter C.P., Rothman J.H., Saari G.C., Valls L.A., and Stevens T.H.. 1986. PEP4 gene of Saccharomyces cerevisiae encodes proteinase A, a vacuolar enzyme required for processing of vacuolar precursors. Mol. Cell. Biol. 6:2490–2499. 10.1128/MCB.6.7.2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio O., Geisberg J.V., Sekinger E., Yang A., Moqtaderi Z., and Struhl K. (2005). Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr. Protoc. Mol. Biol. Chapter 21, Unit 21 23. [DOI] [PubMed] [Google Scholar]
- Babst M., Sato T.K., Banta L.M., and Emr S.D.. 1997. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 16:1820–1831. 10.1093/emboj/16.8.1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backues S.K., Lynch-Day M.A., and Klionsky D.J.. 2012. The Ume6-Sin3-Rpd3 complex regulates ATG8 transcription to control autophagosome size. Autophagy. 8:1835–1836. 10.4161/auto.21845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew C.R., Suzuki T., Du Z., Backues S.K., Jin M., Lynch-Day M.A., Umekawa M., Kamath A., Zhao M., Xie Z., et al. 2012. Ume6 transcription factor is part of a signaling cascade that regulates autophagy. Proc. Natl. Acad. Sci. USA. 109:11206–11210. 10.1073/pnas.1200313109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belle A., Tanay A., Bitincka L., Shamir R., and O’Shea E.K.. 2006. Quantification of protein half-lives in the budding yeast proteome. Proc. Natl. Acad. Sci. USA. 103:13004–13009. 10.1073/pnas.0605420103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C.G., and Emr S.D.. 1998. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol. Cell. 2:157–162. 10.1016/S1097-2765(00)80125-2 [DOI] [PubMed] [Google Scholar]
- Fujioka Y., Suzuki S.W., Yamamoto H., Kondo-Kakuta C., Kimura Y., Hirano H., Akada R., Inagaki F., Ohsumi Y., and Noda N.N.. 2014. Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat. Struct. Mol. Biol. 21:513–521. 10.1038/nsmb.2822 [DOI] [PubMed] [Google Scholar]
- Garí E., Piedrafita L., Aldea M., and Herrero E.. 1997. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 13:837–848. [DOI] [PubMed] [Google Scholar]
- Hariri H., Speer N., Bowerman J., Rogers S., Fu G., Reetz E., Datta S., Feathers J.R., Ugrankar R., Nicastro D., and Henne W.M.. 2019. Mdm1 maintains endoplasmic reticulum homeostasis by spatially regulating lipid droplet biogenesis. J. Cell Biol. 218:1319–1334. 10.1083/jcb.201808119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama R., and De Virgilio C.. 2019. A spatially and functionally distinct pool of TORC1 defines signaling endosomes in yeast. Autophagy. 15:915–916. 10.1080/15548627.2019.1580107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama R., Péli-Gulli M.P., Hu Z., Jaquenoud M., Garcia Osuna G.M., Sardu A., Dengjel J., and De Virgilio C.. 2019. Spatially distinct pools of TORC1 balance protein homeostasis. Mol. Cell. 73:325–338.e8. 10.1016/j.molcel.2018.10.040 [DOI] [PubMed] [Google Scholar]
- Hecht K.A., O’Donnell A.F., and Brodsky J.L.. 2014. The proteolytic landscape of the yeast vacuole. Cell. Logist. 4:e28023 10.4161/cl.28023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht K.A., Wytiaz V.A., Ast T., Schuldiner M., and Brodsky J.L.. 2013. Characterization of an M28 metalloprotease family member residing in the yeast vacuole. FEMS Yeast Res. 13:471–484. 10.1111/1567-1364.12050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz M.K., Ballif B.A., Gygi S.P., and Blenis J.. 2005. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 123:569–580. 10.1016/j.cell.2005.10.024 [DOI] [PubMed] [Google Scholar]
- Jin N., Mao K., Jin Y., Tevzadze G., Kauffman E.J., Park S., Bridges D., Loewith R., Saltiel A.R., Klionsky D.J., and Weisman L.S.. 2014. Roles for PI(3,5)P2 in nutrient sensing through TORC1. Mol. Biol. Cell. 25:1171–1185. 10.1091/mbc.e14-01-0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane P.M. 1995. Disassembly and reassembly of the yeast vacuolar H(+)-ATPase in vivo. J. Biol. Chem. 270:17025–17032. [PubMed] [Google Scholar]
- Kraft C., Kijanska M., Kalie E., Siergiejuk E., Lee S.S., Semplicio G., Stoffel I., Brezovich A., Verma M., Hansmann I., et al. 2012. Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg8 regulates autophagy. EMBO J. 31:3691–3703. 10.1038/emboj.2012.225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M., and Sabatini D.M.. 2009. mTOR signaling at a glance. J. Cell Sci. 122:3589–3594. 10.1242/jcs.051011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Koshi T., and Emr S.D.. 2015a Membrane-anchored ubiquitin ligase complex is required for the turnover of lysosomal membrane proteins. J. Cell Biol. 211:639–652. 10.1083/jcb.201505062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Rong Y., Chuang Y.S., Peng D., and Emr S.D.. 2015b Ubiquitin-dependent lysosomal membrane protein sorting and degradation. Mol. Cell. 57:467–478. 10.1016/j.molcel.2014.12.012 [DOI] [PubMed] [Google Scholar]
- Li S.C., and Kane P.M.. 2009. The yeast lysosome-like vacuole: endpoint and crossroads. Biochim. Biophys. Acta. 1793:650–663. 10.1016/j.bbamcr.2008.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C.Y., and Zoncu R.. 2016. The lysosome as a command-and-control center for cellular metabolism. J. Cell Biol. 214:653–664. 10.1083/jcb.201607005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald C., Stringer D.K., and Piper R.C.. 2012. Sna3 is an Rsp5 adaptor protein that relies on ubiquitination for its MVB sorting. Traffic. 13:586–598. 10.1111/j.1600-0854.2011.01326.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolson M.F., Proteau D., Preston R.A., Stenbit A., Roberts B.T., Hoyt M.A., Preuss D., Mulholland J., Botstein D., and Jones E.W.. 1992. The VPH1 gene encodes a 95-kDa integral membrane polypeptide required for in vivo assembly and activity of the yeast vacuolar H(+)-ATPase. J. Biol. Chem. 267:14294–14303. [PubMed] [Google Scholar]
- Martina J.A., Diab H.I., Lishu L., Jeong-A L., Patange S., Raben N., and Puertollano R.. 2014. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci. Signal. 7:ra9 10.1126/scisignal.2004754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally E.K., Karim M.A., and Brett C.L.. 2017. Selective lysosomal transporter degradation by organelle membrane fusion. Dev. Cell. 40:151–167. 10.1016/j.devcel.2016.11.024 [DOI] [PubMed] [Google Scholar]
- Mellman I., Fuchs R., and Helenius A.. 1986. Acidification of the endocytic and exocytic pathways. Annu. Rev. Biochem. 55:663–700. 10.1146/annurev.bi.55.070186.003311 [DOI] [PubMed] [Google Scholar]
- Müller M., Schmidt O., Angelova M., Faserl K., Weys S., Kremser L., Pfaffenwimmer T., Dalik T., Kraft C., Trajanoski Z., et al. 2015. The coordinated action of the MVB pathway and autophagy ensures cell survival during starvation. eLife. 4:e07736 10.7554/eLife.07736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T. 2017. Regulation of Autophagy through TORC1 and mTORC1. Biomolecules. 7:e52 10.3390/biom7030052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku M., Maeda Y., Kagohashi Y., Kondo T., Yamada M., Fujimoto T., and Sakai Y.. 2017. Evidence for ESCRT- and clathrin-dependent microautophagy. J. Cell Biol. 216:3263–3274. 10.1083/jcb.201611029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parzych K.R., and Klionsky D.J.. 2019. Vacuolar hydrolysis and efflux: current knowledge and unanswered questions. Autophagy. 15:212–227. 10.1080/15548627.2018.1545821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedruzzi I., Dubouloz F., Cameroni E., Wanke V., Roosen J., Winderickx J., and De Virgilio C.. 2003. TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol. Cell. 12:1607–1613. 10.1016/S1097-2765(03)00485-4 [DOI] [PubMed] [Google Scholar]
- Perera R.M., and Zoncu R.. 2016. The lysosome as a regulatory hub. Annu. Rev. Cell Dev. Biol. 32:223–253. 10.1146/annurev-cellbio-111315-125125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano R., Ferguson S.M., Brugarolas J., and Ballabio A.. 2018. The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J. 37:e98804 10.15252/embj.201798804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehli M., Den Elzen N., Cassady A.I., Ostrowski M.C., and Hume D.A.. 1999. Cloning and characterization of the murine genes for bHLH-ZIP transcription factors TFEC and TFEB reveal a common gene organization for all MiT subfamily members. Genomics. 56:111–120. 10.1006/geno.1998.5588 [DOI] [PubMed] [Google Scholar]
- Richter C., West M., and Odorizzi G.. 2007. Dual mechanisms specify Doa4-mediated deubiquitination at multivesicular bodies. EMBO J. 26:2454–2464. 10.1038/sj.emboj.7601692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M., Palmieri M., di Ronza A., Medina D.L., Valenza M., Gennarino V.A., Di Malta C., Donaudy F., Embrione V., Polishchuk R.S., et al. 2009. A gene network regulating lysosomal biogenesis and function. Science. 325:473–477. 10.1126/science.1174447 [DOI] [PubMed] [Google Scholar]
- Sekito T., Nakamura K., Manabe K., Tone J., Sato Y., Murao N., Kawano-Kawada M., and Kakinuma Y.. 2014. Loss of ATP-dependent lysine uptake in the vacuolar membrane vesicles of Saccharomyces cerevisiae ypq1∆ mutant. Biosci. Biotechnol. Biochem. 78:1199–1202. 10.1080/09168451.2014.918489 [DOI] [PubMed] [Google Scholar]
- Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P., et al. 2011. TFEB links autophagy to lysosomal biogenesis. Science. 332:1429–1433. 10.1126/science.1204592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Fraldi A., Medina D.L., and Ballabio A.. 2013. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 14:283–296. 10.1038/nrm3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Zoncu R., Medina D.L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M.C., et al. 2012. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31:1095–1108. 10.1038/emboj.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin M.E., Ogburn K.D., Varban O.A., Gilbert P.M., and Burd C.G.. 2001. FYVE domain targets Pib1p ubiquitin ligase to endosome and vacuolar membranes. J. Biol. Chem. 276:41388–41393. 10.1074/jbc.M105665200 [DOI] [PubMed] [Google Scholar]
- Simm C., Lahner B., Salt D., LeFurgey A., Ingram P., Yandell B., and Eide D.J.. 2007. Saccharomyces cerevisiae vacuole in zinc storage and intracellular zinc distribution. Eukaryot. Cell. 6:1166–1177. 10.1128/EC.00077-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulard A., Cremonesi A., Moes S., Schütz F., Jenö P., and Hall M.N.. 2010. The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol. Biol. Cell. 21:3475–3486. 10.1091/mbc.e10-03-0182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Z., Kim M.S., Pandey A., and Espenshade P.J.. 2014. Identification of candidate substrates for the Golgi Tul1 E3 ligase using quantitative diGly proteomics in yeast. Mol. Cell. Proteomics. 13:2871–2882. 10.1074/mcp.M114.040774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban J., Soulard A., Huber A., Lippman S., Mukhopadhyay D., Deloche O., Wanke V., Anrather D., Ammerer G., Riezman H., et al. 2007. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell. 26:663–674. 10.1016/j.molcel.2007.04.020 [DOI] [PubMed] [Google Scholar]
- Wanke V., Cameroni E., Uotila A., Piccolis M., Urban J., Loewith R., and De Virgilio C.. 2008. Caffeine extends yeast lifespan by targeting TORC1. Mol. Microbiol. 69:277–285. 10.1111/j.1365-2958.2008.06292.x [DOI] [PubMed] [Google Scholar]
- Wanke V., Pedruzzi I., Cameroni E., Dubouloz F., and De Virgilio C.. 2005. Regulation of G0 entry by the Pho80-Pho85 cyclin-CDK complex. EMBO J. 24:4271–4278. 10.1038/sj.emboj.7600889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett R., Martina J.A., Zewe J.P., Wills R., Hammond G.R.V., and Puertollano R.. 2017. TFEB regulates lysosomal positioning by modulating TMEM55B expression and JIP4 recruitment to lysosomes. Nat. Commun. 8:1580 10.1038/s41467-017-01871-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R.M., Primig M., Washburn B.K., Winzeler E.A., Bellis M., Sarrauste de Menthiere C., Davis R.W., and Esposito R.E.. 2002. The Ume6 regulon coordinates metabolic and meiotic gene expression in yeast. Proc. Natl. Acad. Sci. USA. 99:13431–13436. 10.1073/pnas.202495299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford C.A., Daniels L.B., Park F.J., Jones E.W., Van Arsdell J.N., and Innis M.A.. 1986. The PEP4 gene encodes an aspartyl protease implicated in the posttranslational regulation of Saccharomyces cerevisiae vacuolar hydrolases. Mol. Cell. Biol. 6:2500–2510. 10.1128/MCB.6.7.2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R. 2000. Transmission electron microscopy of yeast. Microsc. Res. Tech. 51:496–510. [DOI] [PubMed] [Google Scholar]
- Yang X., Arines F.M., Zhang W., and Li M.. 2018. Sorting of a multi-subunit ubiquitin ligase complex in the endolysosome system. eLife. 7:e33116 10.7554/eLife.33116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Huang J., Geng J., Nair U., and Klionsky D.J.. 2006. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol. Biol. Cell. 17:5094–5104. 10.1091/mbc.e06-06-0479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Jorgensen J.R., Li M., Chuang Y.S., and Emr S.D.. 2017. ESCRTs function directly on the lysosome membrane to downregulate ubiquitinated lysosomal membrane proteins. eLife. 6:e26403 10.7554/eLife.26403 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists yeast strains and plasmids used in this study.
lists primers used in this study.