Conspectus
Bioluminescence is widely used for real-time imaging in living organisms. This technology features a light-emitting reaction between enzymes (luciferases) and small molecule substrates (luciferins). Photons produced from luciferase-luciferin reactions can penetrate through heterogeneous tissue, enabling readouts of physiological processes. Dozens of bioluminescent probes are now available and many are routinely used to monitor cell proliferation, migration, and gene expression patterns in vivo.
Despite the ubiquity of bioluminescence, traditional applications have been largely limited to imaging one biological feature at a time. Only a handful of luciferase-luciferin pairs can be easily used in tandem, and most are poorly resolved in living animals. Efforts to develop spectrally distinct reporters have been successful, but multispectral imaging in large organisms remains a formidable challenge due to interference from surrounding tissue. Consequently, a lack of well-resolved probes has precluded multi-component tracking. An expanded collection of bioluminescent probes would provide insight into processes where multiple cell types drive physiological tasks, including immune function and organ development.
We aimed to expand the bioluminescent toolkit by developing substrate-resolved imaging agents. The goal was to generate multiple orthogonal (i.e., non-cross-reactive) luciferases that are responsive to unique scaffolds and could be used concurrently in living animals. We adopted a parallel engineering approach to genetically modify luciferases to accept chemically modified luciferins. When the mutants and analogs are combined, light is produced only when complementary enzyme-substrate partners interact. Thus, the pairs can be distinguished based on substrate selectivity, regardless of the color of light emitted. Sequential administration of the luciferins enables the unique luciferases to be illuminated (and thus resolved) within complex environments, including whole organisms.
This Account describes our efforts to develop orthogonal bioluminescent probes, crafting custom luciferases (or “biological flashlights”) that can selectively process luciferin analogs (or “batteries”) to produce light. In the first section, we describe synthetic methods that were key to accessing diverse luciferin architectures. The second section focuses on identifying complementary luciferase enzymes via a combination of mutagenesis and screening. To expedite the search for orthogonal enzymes and substrates, we developed a computational algorithm to sift through large data sets. The third section features examples of the parallel engineering approach. We identified orthogonal enzyme-substrate pairs comprising two different classes of luciferins. The probes were vetted both in cells and whole organisms. This expanded collection of imaging agents is applicable to studies of immune function and other multi-component processes. The final section of the Account highlights ongoing work toward building better bioluminescent tools. As ever-brighter and more selective probes are developed, the frontiers of what we can “see” in vivo will continue to expand.
Graphical Abstract
Introduction
Bioluminescence is a powerful imaging modality for monitoring molecular and cellular features in real time.1–3 This technology relies on a chemical transformation: the luciferase-catalyzed oxidation of small molecule substrates (luciferins). Bioluminescent reactions release light that can penetrate skin and some tissues, enabling sensitive imaging in vivo.2 Virtually no background signal exists, as mammals do not emit large numbers of detectable photons. Several luciferases have been co-opted as biological “flashlights”—powered by luciferin “batteries”—in whole organisms. The most popular pair originates from the firefly.4–6 Firefly luciferase (Fluc) catalyzes the oxidation of D-luciferin (D-luc), releasing primarily yellow-green light (Figure 1A). The reaction also generates a sufficient number of red (i.e., tissue-penetrant) photons, enabling physiological processes to be noninvasively tracked.1 Indeed, Fluc and D-luc have been used for decades to monitor gene expression, cell proliferation, and other activities in live rodents.2,7–8
Figure 1.
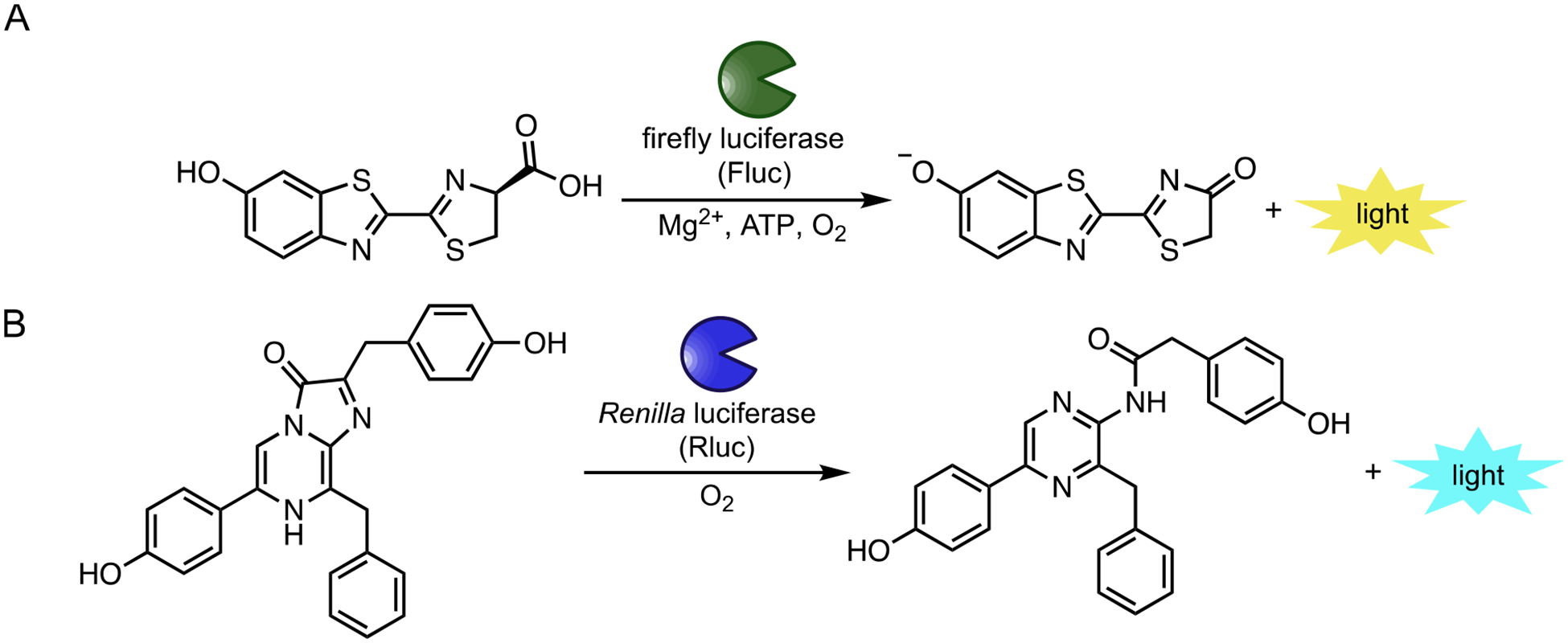
Popular bioluminescent enzymes and substrates. (A) Insect luciferases (including Fluc) catalyze the oxidation of D-luciferin (D-luc), resulting in photon production. (B) Marine luciferases (including Rluc) produce blue light via oxidation of coelenterazine substrates
While both versatile and ubiquitous, bioluminescence has been largely restricted to monitoring one feature at a time in whole organisms.1 Only a handful of luciferase-luciferin pairs have been optimized for imaging in vivo, and most cannot be easily distinguished in thick tissue.6 For example, dozens of unique luciferases have been characterized from the insect family alone.9 All use D-luciferin in the light-emitting reaction and exhibit broad emission spectra, though, making them difficult to resolve in heterogeneous environments.10 More spectrally discrete probes have been generated via enzyme engineering and modifications to the D-luciferin core.6,10 However, distinguishing these probes by wavelength alone remains challenging in small animals. Only a tiny fraction of the photons (>650 nm) can effectively escape tissue for detection, and the perceived color is dependent on the depth of the source.
Instead of color resolution, multi-component bioluminescence can be readily achieved via substrate resolution, using luciferases that respond to unique luciferins. For example, luciferases that process D-luciferin (e.g., Fluc) are easily distinguished from those (e.g., Renilla luciferase or Gaussia luciferase) that use coelenterazine, the bioluminescent substrate found in marine species (Figure 1B). In fact, Fluc and Renilla luciferase (Rluc) have been used in tandem for decades and remain a popular pair for two-component imaging.11–12 Rluc emits primarily blue light, further enabling it to be spectrally distinguished from Fluc in vitro.13 Blue light is poorly tissue penetrant, though, so Rluc and related marine luciferases have historically been less employed in vivo. Red-shifted variants of these enzymes are available for cellular imaging,14–17 and some of these reporters can enable more sensitive imaging in rodents and other organisms.18–20 The requisite small molecules still have liabilities, though, owing to poor pharmacokinetic properties.21–22 While efforts to develop more soluble and bioavailable analogs have been fruitful,20,23 multi-component bioluminescence imaging in vivo with these and other probes is far from routine.
We were inspired to expand the set of biological flashlights and batteries for multicellular imaging, building on the platform of orthogonal substrate usage. Hundreds of unique luciferase-luciferin pairs exist among diverse phyla.9,24–26 In theory, these pairs could be co-opted for multiplexed imaging, using distinct luciferins to selectively illuminate the cognate luciferases. Many naturally occurring bioluminescent probes are ill suited for use in vivo, though, owing to poor substrate accessibilities, non-optimal photon outputs, or other factors. Instead of re-purposing these native biomolecules, we focused on crafting artificial enzymes and substrates. The most tractable and tissue-penetrant bioluminescent pair—Fluc and D-luciferin—provided an initial blueprint. We aimed to reengineer Fluc to accept chemically unique luciferins (Figure 2). Robust signal would only be observed when complementary (i.e., “orthogonal”) enzyme-substrate partners interact. This approach would more rapidly expand the collection of bioluminescent probes and enable multi-component imaging in small animals.
Figure 2.
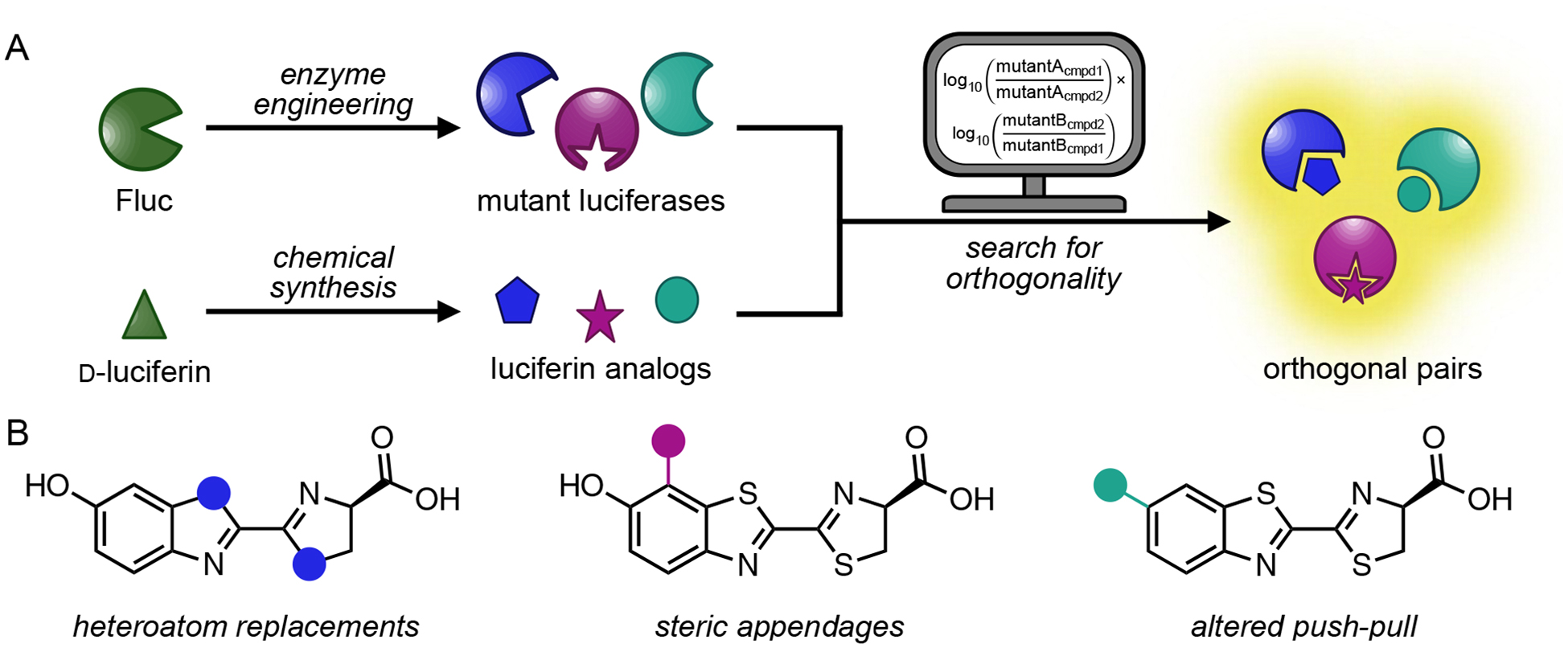
Bioluminescent probes for multi-component imaging. (A) Parallel engineering of luciferase enzymes and luciferin analogs to generate orthogonal pairs. A computational algorithm assists in the search for compatible imaging probes. (B) Sample classes of luciferin analogs targeted for orthogonal probe development. Blue, magenta, and teal dots represent unique modifications to the luciferin core. Part (A) was adapted with permission from ref. 67. Copyright 2017 American Chemical Society.
This Account highlights our efforts to produce orthogonal luciferase-luciferin pairs. We used a parallel engineering strategy, modifying Fluc and D-luc in tandem. Key to the success of the approach was accessing an array of structurally diverse luciferin analogs. From this pool, luciferases that were uniquely responsive to individual luciferins could be identified via iterative screening and computational analysis. The first part of the Account outlines our approach to rapidly access luciferin analogs. The second part describes our enzyme engineering efforts, including methods to produce and screen mutant libraries. We also discuss computing methods to efficiently identify substrate-selective luciferases. We showcase how concurrent enzyme and substrate engineering was used to generate different classes of orthogonal bioluminescent probes. Finally, we discuss ongoing challenges in building designer flashlights and batteries. The continued refinement of existing probes—and the development of new ones—will enable multi-component studies. Such expanded imaging capabilities are likely to spur new discoveries in a variety of disciplines.
Designing and synthesizing luciferin “batteries”
Generating orthogonal bioluminescent probes requires facile access to unique luciferin scaffolds. High levels of structural diversity were envisioned to expedite the production of orthogonal pairs. The more differentiated the substrates, the easier the search for exquisitely selective enzymes. Gram-scale quantities of analog were also desired, as enzyme screens and directed evolution methods typically require large quantities of substrate. At the outset of our work, traditional methods to produce D-luciferin were lengthy, low yielding, and refractory to late-stage modification.27 Most syntheses relied on the formation of a key cyanobenzothiazole intermediate, followed by condensation with D-cysteine (Figure 3A). This latter step enabled efficient installation of the requisite stereocenter, but accessing the benzothiazole itself required multiple steps and harsh conditions.28–29 Many of the steps were also incompatible with functional groups that we intended to append to the luciferin core.
Figure 3.
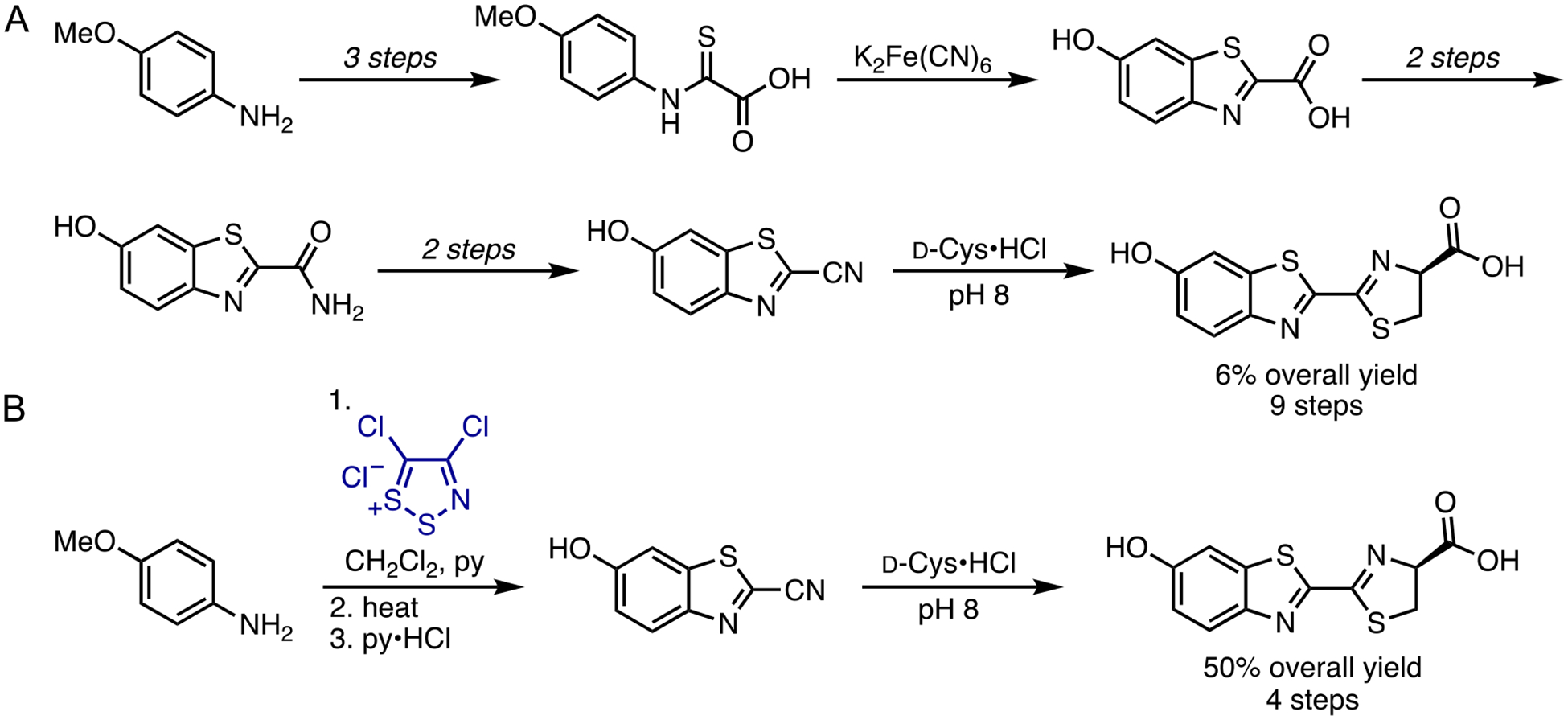
Methods to produce D-luc. (A) Traditional syntheses to form the key benzothiazole core. (B) A streamlined method to produce D-luc using Appel’s salt (blue). Both routes comprise a condensation reaction with D-cysteine as the final step.
To circumvent the limitations of traditional luciferin synthesis, we devised an alternative method to access the cyanobenzothiazole intermediate. The route features a dithiazolium chloride reagent, known as Appel’s salt (Figure 3B).30 This material has been used to produce a wide variety of substituted heterocycles, including quinazolines and benzothiazoles, from aromatic amines.31–34 We reasoned that Appel’s salt could similarly provide rapid access to cyanobenzothiazoles, key precursors en route to luciferin scaffolds. Indeed, many substituted anilines could be readily condensed with Appel’s salt and subsequently fragmented with DBU, phosphines, or other nucleophiles, to provide cyanothioformamides in high yield.35–36 These intermediates were readily cyclized via C-H activation to provide cyanobenzothiazoles. The fragmentation and cyclization steps could also be performed in a single pot,37 further simplifying the process. Condensation of the cyanobenzothiazole intermediate en route to D-luc provided the desired substrate in 40–60% overall yield. In addition to improving the yield, the streamlined route was scalable. We have routinely synthesized the cyanobenzothiazole intermediate in >10 g quantities.
The Appel’s salt methodology not only enabled facile access to D-luc, but also several analogs for orthogonal probe development. We and others have used the chemistry to produce a variety of heterocyclic probes (Figure 4A).35,37–39 Some examples include benzimidazole and benzoxazole luciferins. These scaffolds were easily accessed from the corresponding ortho-substituted anilines via Appel’s salt condensation and cyclization.35,38 Alkyl-substituted luciferins were also readily synthesized from commercially available starting materials (Figure 4B).35 Moreover, a large number of analogs could be prepared via late-state functionalization of the cyanobenzothiazole intermediates.40–41 Being able to rapidly access a diverse array of luciferin architectures was critical to the development of substrate-selective bioluminescent tools.
Figure 4.
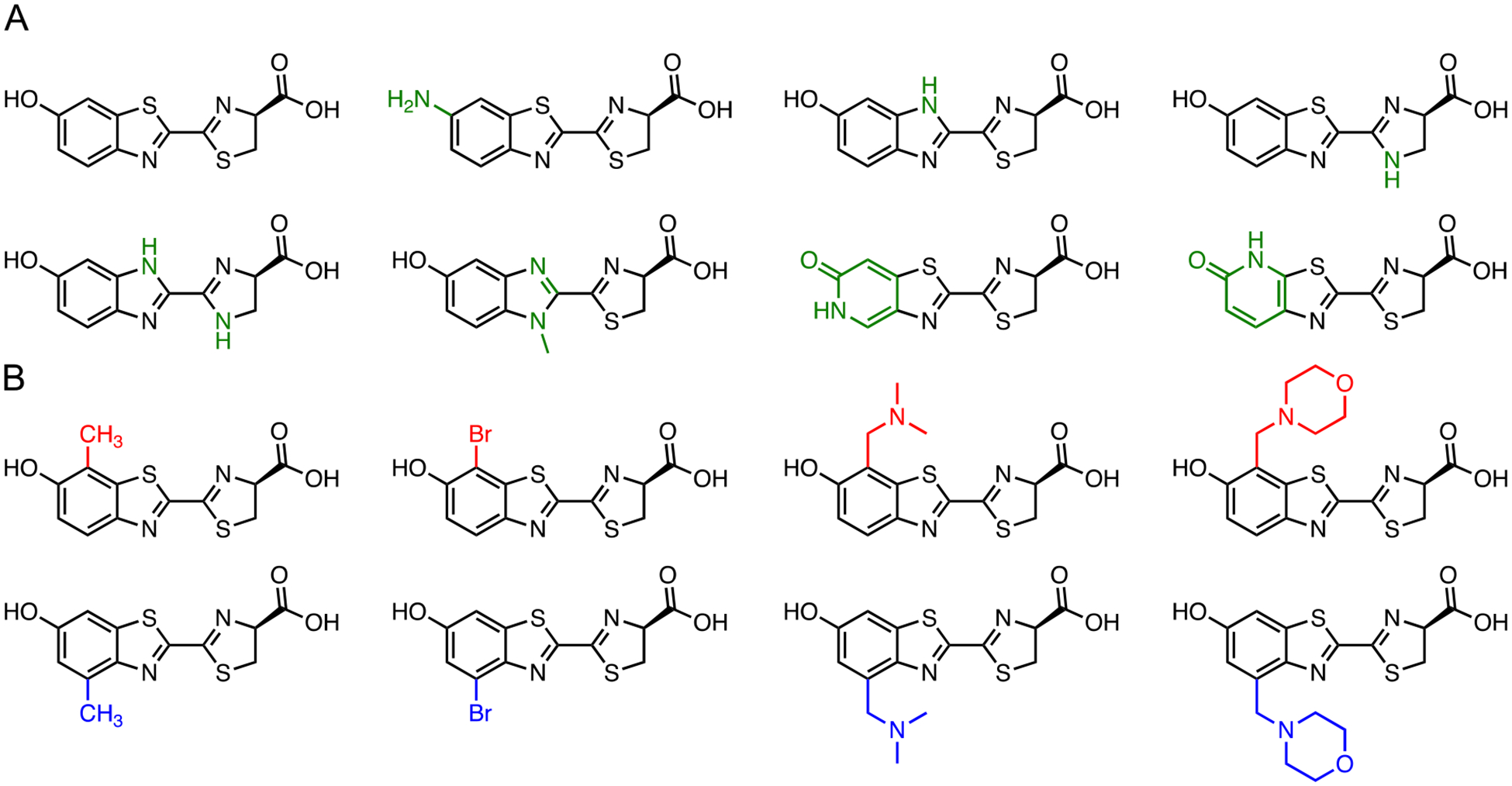
Diverse luciferin architectures synthesized with Appel’s salt. Representative scaffolds with (A) unique heteroaromatic cores (green) and (B) steric modifications are pictured. Unique C7′ modifications are shown in red and C4′ modifications are shown in blue
To determine which luciferin analogs would be most useful for orthogonal imaging, we benchmarked their activities using standard light emission assays. All of the substrates exhibited reduced photon outputs with Fluc compared to the native substrate, D-luc.35,40 Low levels of light emission were encouraging, as they suggested that the analogs were occluded from the active site or poorly turned over. However, it was also possible that the luciferins were simply weak light emitters and thus not good candidates for imaging in vivo. To discriminate among these possibilities, we used a traditional chemiluminescence assay.40–41 The assay mimics the Fluc reaction (but in the absence of enzyme), providing a readout on the inherent light-emitting ability of a luciferin. While most of the molecules were only weakly bioluminescent, chemiluminescence measurements revealed that they were capable of robust emission. The analogs were thus good candidates for orthogonal probe development. We just needed to identify mutant luciferases capable of tapping into their light-emitting potential.
Identifying luciferase “flashlights”
We envisioned that substrate-selective luciferases could be found by iteratively screening large pools of mutant enzymes. Similar methods have been used to engineer luciferases with improved thermostability, altered wavelengths of emission, and other properties. Altered substrate specificity has been achieved for some marine luciferases, using screens of mutants with designer coelenterazine analogs.18,23,42–44 Changing the substrate bias of Fluc is perhaps less straightforward, as the enzyme carries out a multi-step reaction and few crystal structures exist to guide engineering efforts.45–47 Docking analyses and other biochemical studies suggested that certain amino acids were prime targets for mutagenesis, although none were “gatekeeper” residues per se.48–49 Fluc can also tolerate a variety of modified substrates in the light-emitting reaction, suggesting that exquisite selectivity might be difficult to achieve.
While identifying multiple orthogonal enzymes and substrates would require a larger number of screens, the process seemed straightforward: generate large collections of mutant luciferases, screen for functional enzymes via light emission, and check for luciferin specificity among the “hits”.40 Further refinements in light output or substrate selectivity could then be achieved via additional rounds of mutagenesis and screening (i.e., directed evolution). Indeed, we have employed this general workflow to generate dozens of orthogonal bioluminescent probes. The paragraphs that follow provide additional details on the screens, and the next two sections showcase its application toward identifying substrate-selective luciferases with different classes of luciferins.
Luciferase screening required access to unique, yet functional, mutant enzymes. Toward this end, we employed site-saturation mutagenesis and other techniques to introduce diversity within the enzyme framework. Residues were initially targeted based on sequence homology within the insect luciferase family, previous biochemical assays, and proximity to bound luciferins (Figure 5).50–52 Mutations at these sites could potentially modulate analog binding or turnover. Libraries were produced using standard cloning methods and codon compression techniques.53–55 Library sizes were purposefully kept small (19–6,860) to enable complete coverage in screening.
Figure 5.
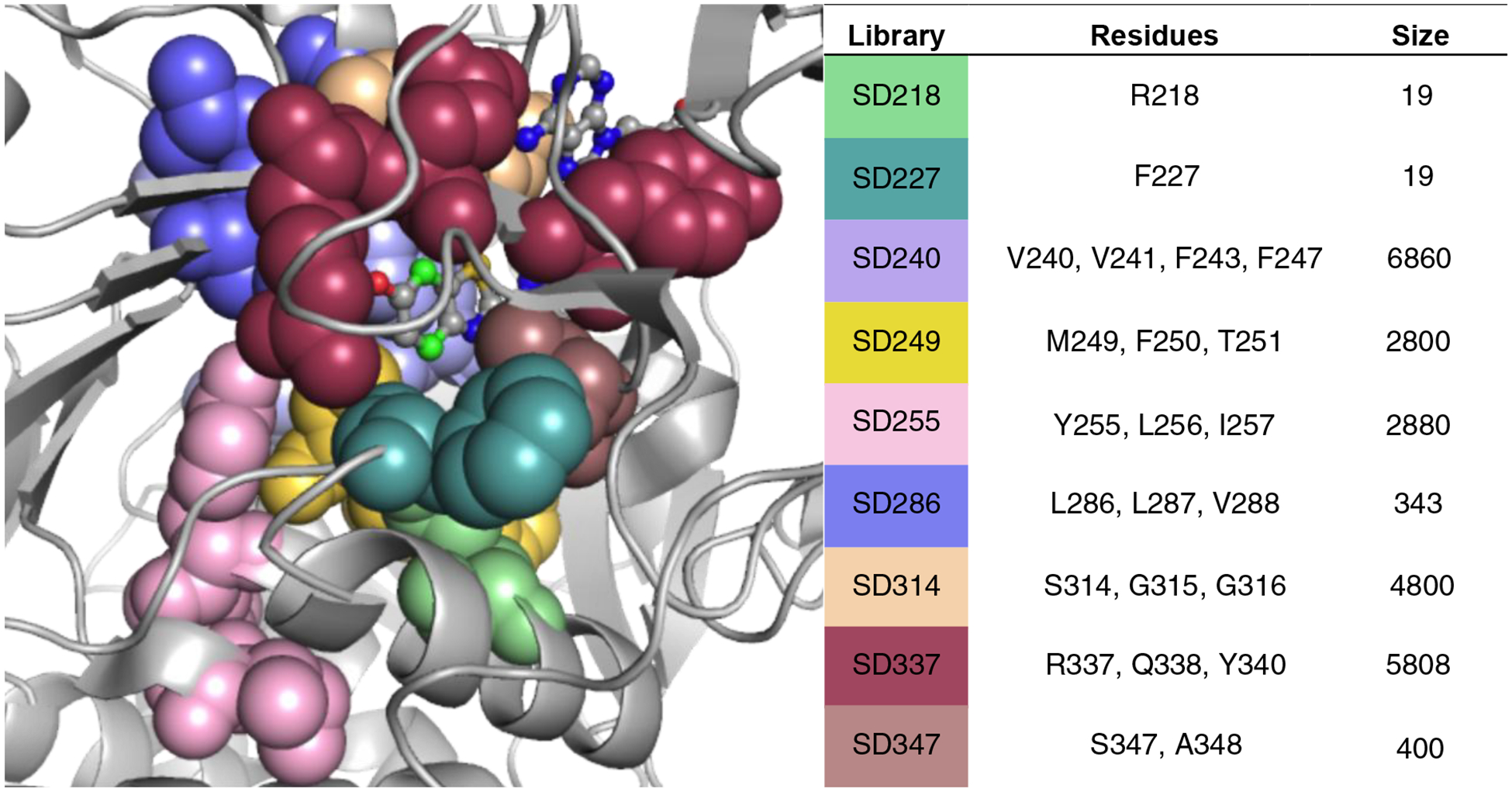
Luciferases screened to identify orthogonal probes. Mutant libraries were prepared using site-saturation mutagenesis. Library sizes varied based on the degree of saturation enforced at each residue. Such determinations were made based on sequence conservation among the insect luciferase family and residue location. Targeted residues are colored, and most lie in close proximity to the bound luciferin (PDB: 4G36).
The mutant libraries were subjected to a two-tier screen to identify orthogonal pairs. Mutants were initially examined on-plate following transformation of library DNA into E. coli. In some cases, the bacteria were grown on luciferin-embedded agar. In other cases, the analog was directly sprayed onto the cells. Both approaches consumed large amounts of luciferin, so having access to multi-gram quantities of analog was critical. Light-emitting colonies were ultimately detected using a cooled CCD camera. Glowing cells were collected and expanded in 96-well plates. The mutant “hits” were then treated with various luciferins in a secondary assay to identify substrate-selective luciferases.40 This two-tiered approach enabled efficient identification of orthogonal probes as the on-plate analyses rapidly eliminated the vast majority of non-functional enzymes. The initial screen also weeded out luciferins that were cell impermeable, toxic, or otherwise poorly suited for biological imaging. The key parameter—substrate orthogonality—could then be assayed in a more controlled secondary screen, normalizing for differences in expression levels and compound transport. Head-to-head comparisons of analog utilization by mutant enzymes could also be performed.
Finding flashlights for electronically modified batteries
Our initial efforts to craft orthogonal bioluminescent tools focused on D-luciferin analogs comprising different heteroatoms. We were attracted to these compounds as they were easily accessible and likely to be competent light emitters. Indeed, Branchini and others previously demonstrated that quinoline and other heterocyclic variants could function as bioluminescent substrates with Fluc.56–57 Inspired by this work, we explored whether simple heteroatom replacements to the D-luciferin core could elicit orthogonality. Benzimidazole and imidazoline scaffolds were first targeted. These molecules comprised nitrogen atoms in place of sulfurs. We hypothesized that such swaps would sufficiently perturb Fluc processing, but that proper binding and light emission could be restored with a mutant. When the nitrogenous luciferins were incubated with Fluc, reduced photon outputs were observed. The analogs also emitted distinct colors (Figure 6A), similar to other heterocyclic luciferins.56–57
Figure 6.
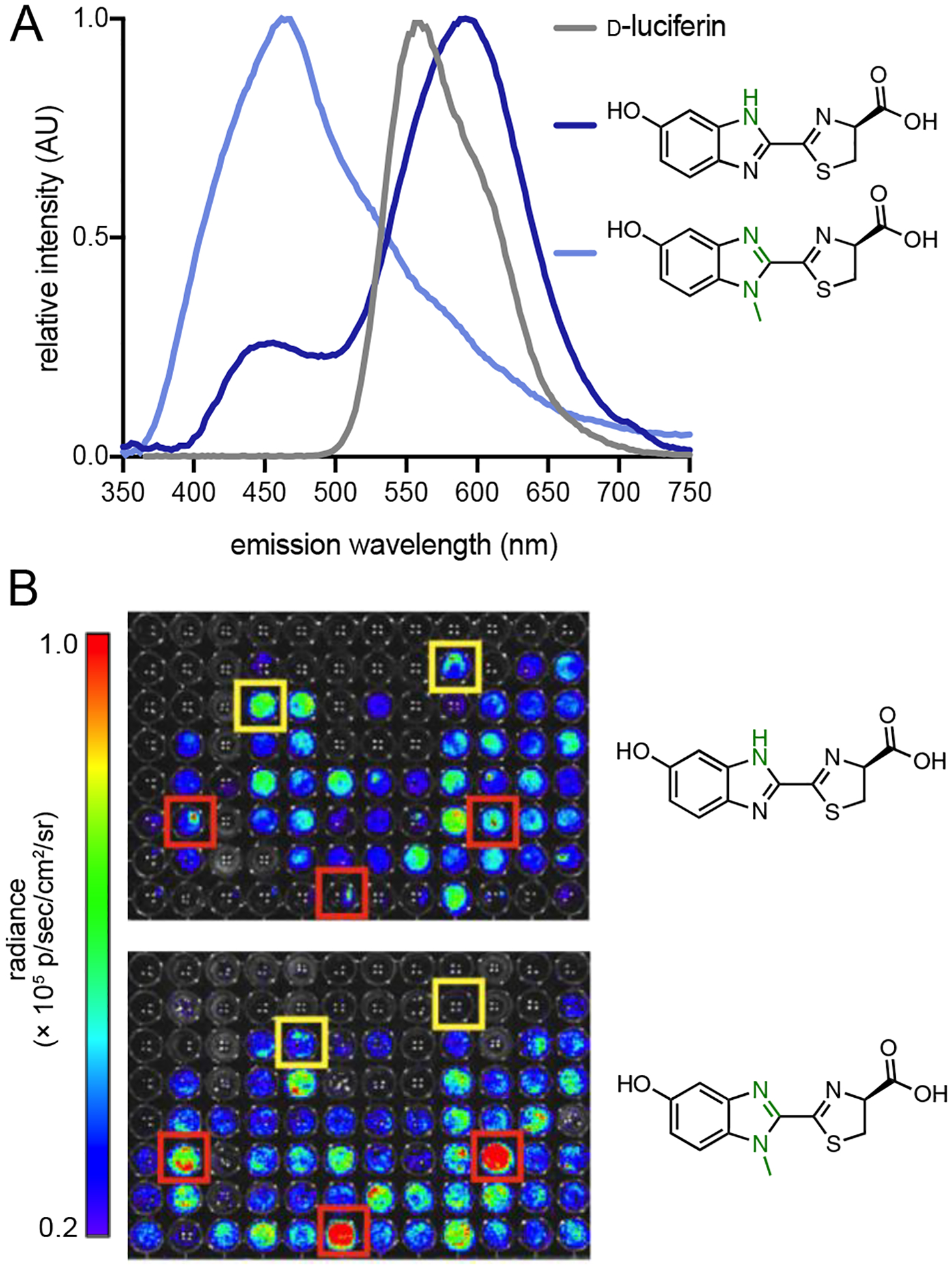
Benzimidazole luciferins exhibit unique patterns of light emission with Fluc and mutant luciferases. (A) Bioluminescent spectra of the analogs (blue) and D-luciferin (gray). (B) Unique patterns of light emission were observed. Mutants were arrayed across 96-well plates and treated with either the benzimidazole (top) or N-methylated (bottom) variant. Sample bioluminescence images are shown. Yellow and red boxes indicate enzymes that preferentially process the benzimidazole and N-methylated analogs, respectively. Part (A) was adapted with permission from ref. 35. Copyright 2012 American Chemical Society.
To determine if complementary luciferases could be identified for these scaffolds, we screened the analogs against a small library of mutant enzymes (where mutations were confined to the luciferin binding site of Fluc). Unique patterns of light emission were observed (Figure 6B), suggesting that some luciferases could differentially process the substrates.58 The experiment also indicated that even subtle structural modifications would be sufficient to elicit orthogonality. Further efforts to characterize and optimize the orthogonal enzymes were complicated, though, owing to the suboptimal permeability and weak light output of the benzimidazole analogs.
In a quest to find not only orthogonal, but also robust light emitters, we turned our attention to pyridone scaffolds. These motifs are commonly found in natural products and drug-like molecules, suggesting that they are sufficiently biocompatible and cell permeable.59–60 We also reasoned that pyridone analogs could function as canonical luciferins. D-Luc and related molecules undergo proton transfer reactions in the excited state.61–62 In the case of the pyridone, deprotonation would provide a fully aromatic luminophore (Figure 7A). We synthesized two pyridone analogs (5′-PyrLuc and 7′-PyrLuc) using the Appel’s salt methodology.63 Both isomers were found to be competent light emitters with Fluc. Importantly, the analogs were significantly more robust than the benzimidazole probes. The pyridones were also sufficiently bright and cell permeable to image Fluc-expressing cells. While suitable for biological imaging, the pyridone luciferins were surprisingly poor binders of Fluc (>100-fold higher apparent KM values than D-luc). We aimed to identify luciferases that could better process the analogs, and library screens revealed two mutants that exhibited 10–80 fold improved photon outputs (Figure 7B).63 The KM values measured for both enzymes, though, were similar to that of Fluc, suggesting that the mutated residues improved substrate turnover rather than binding.
Figure 7.
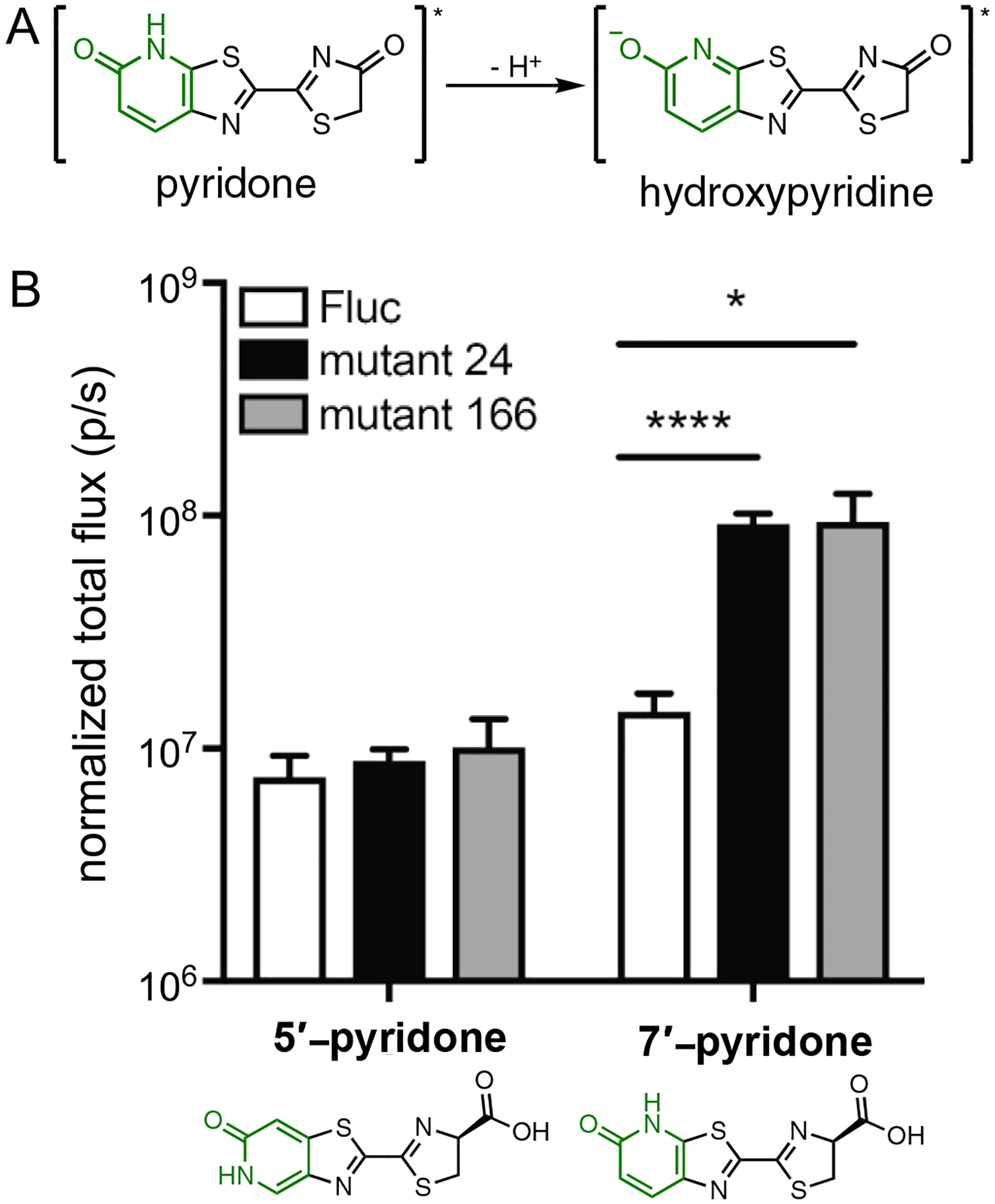
Pyridone luciferins exhibit unique patterns of light emission with Fluc and mutant luciferases. (A) Excited state deprotonation of a pyridone analog to provide a functional light emitter. (B) HEK293 cells expressing mutant luciferases or Fluc were incubated with pyridone analogs (1 mM) and photon outputs were measured. Luciferase expression was normalized to a GFP reporter. Asterisks denote values from one-tailed unpaired t-tests (* p<0.05, **** p<0.0001). Adapted with permission from ref. 63. Copyright 2018 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
We further attempted to identify orthogonal luciferases capable of selectively processing the pyridone analogs. No such enzymes were found in our initial screens. However, mutants were identified that could readily discriminate the 7′-pyridone analog from more structurally divergent luciferins. When the pyridone scaffold was screened alongside 10 other luciferins, two mutants were identified that could distinguish 7′-PyrLuc from a brominated luciferin, 4′-BrLuc.64 Subsequent mutagenesis studies revealed that residues N229, S239, G246, and F250 were responsible for substrate discrimination. Mutants comprising G246A, in particular, were found to be selective for 4′-BrLuc. Other mutations that conferred selectivity for the brominated analog (N229T and F250A/C/G) were also identified. Interestingly, mutations at N229 and F250 alone did not result in substantially altered substrate preference. The combination exhibited a significant change in luciferin use. This example highlights the power of functional screens, as the key mutations at N229 and F250 would have been unlikely predictions from purely structure-guided approaches.
Finding flashlights for sterically modified batteries
Perhaps a more fruitful and conceptually straightforward approach to developing orthogonal pairs involves placing “bumps” on substrates, and identifying mutant enzymes (with “holes”) to accommodate the modifications. While the bump-hole analogy does not perfectly apply to luciferases and luciferins (see above), steric modifications to D-luc were envisioned to perturb Fluc processing. Mutants that tolerate the extra bulk could restore light production. When contemplating which sites to modify on D-luciferin, we shied away from C6′. Fluc is known to tolerate many appendages at this site,65–66 so adding diversity here was less likely to result in selective enzymes. We instead focused on C4′ and C7′. These positions lie in close proximity to enzyme residues (based on docking studies, Figure 8B). Thus, steric modifications at these sites could potentially engender a clash with the native enzyme, but be tolerated by a more promiscuous mutant.
Figure 8.
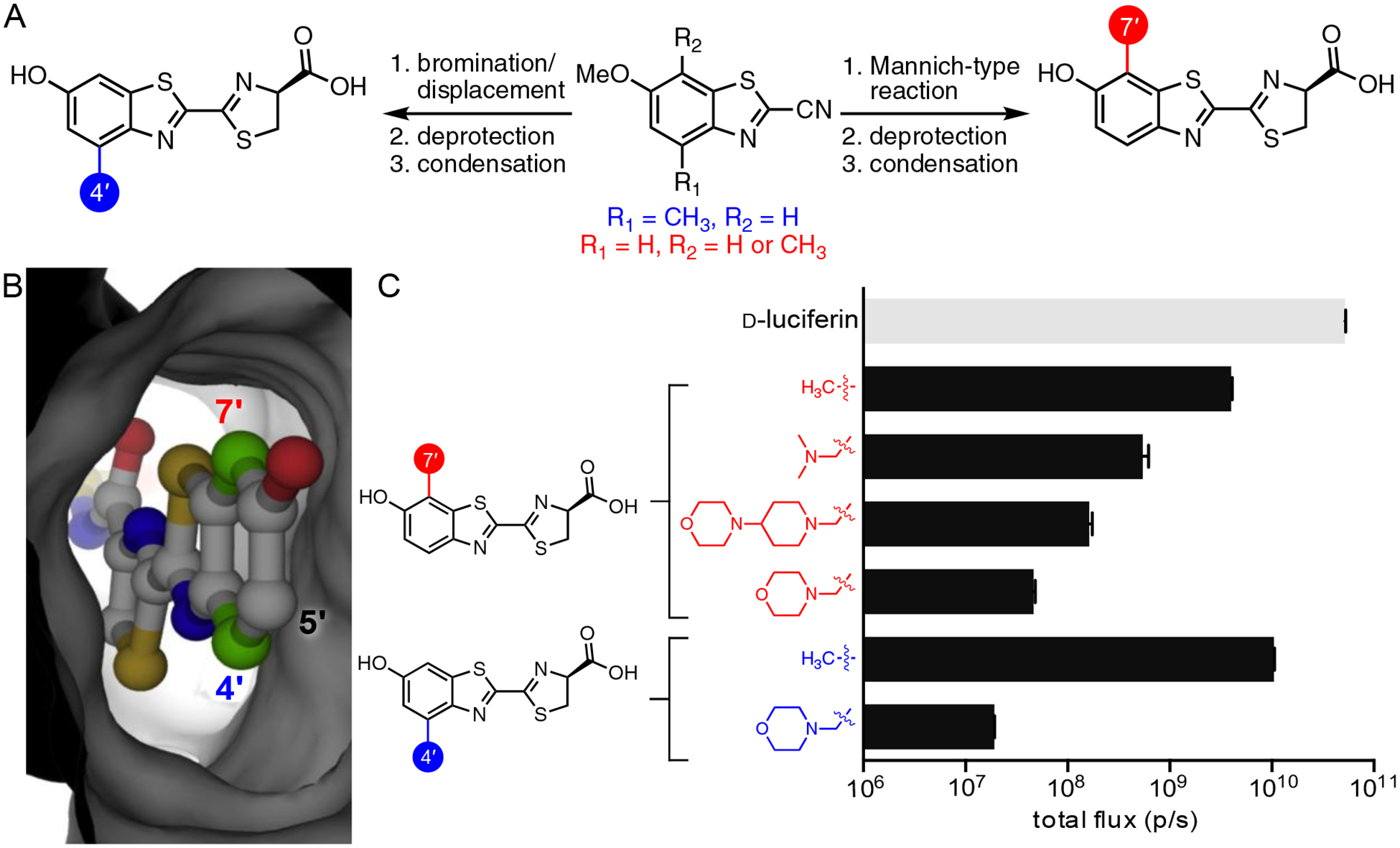
Sterically modified luciferins as orthogonal probes. (A) Divergent synthesis of luciferin analogs. Common benzothiazole intermediates were used to access an array of C4′- and C7′- modified luciferins. (B) Modeling studies suggested that C4′ and C7′ are prime targets for modification. These positions lie in close proximity to the Fluc backbone (PDB: 4G36). (C) Luciferin analogs exhibit varying levels of light output with Fluc. Analogs (100 μM) were incubated with Fluc (1 μg) and photon outputs were measured. Emission intensities are plotted as total photon flux values on a log scale. Adapted with permission from ref. 40. Copyright 2017 American Chemical Society.
Similar to earlier examples, identifying orthogonal pairs with sterically modified analogs required large quantities of candidate substrates. Both C4′- and C7′-modified luciferins were readily accessed from common cyanobenzothiazole intermediates (prepared via Appel’s salt, Figure 8A). C4′-modified substrates were ultimately accessed via a benzylic bromination/SN2 displacement sequence. C7′-modified substrates were prepared using Mannich chemistry. Both routes enabled halogens and a variety of other modifications to be efficiently installed.40 Nearly all of the C4′ and C7′ analogs emitted light with Fluc, although signals from bulkier compounds were quite dim (Figure 8C). Even these weak-emitting compounds were good candidates for orthogonal probe development, though, based on chemiluminescence assays.40
To search for enzymes that could process the steric analogs, we again screened mutant libraries using a two-tiered approach. Functional mutants were first identified on agar plates, and then secondary assays were used to analyze the substrate preference of the “hits”. After an initial screen of ~140 mutants with 6 compounds (Figure 5), we could already identify enzyme pairs with unique substrate preference.40 In most cases, mutants that could process C4′-modified analogs were dim with C7′-modified compounds (and vice versa). The differential between “matched” and “unmatched” partners was typically >10-fold. This result was encouraging, as further rounds of mutagenesis could only improve the selectivity.
The most resolved pair identified from the initial screen comprised 4′-MorphoLuc and 7′-DMAMeLuc. These substrates were preferentially processed by mutants 81 and 37, respectively (Figure 9A). Selective light emission was verified in vitro, using purified enzymes. Orthogonal light emission was also observed in cellulo. Cell populations expressing either mutant 81 or 37 were treated with the individual luciferins.40 Light emission was observed only from cells expressing the complementary luciferase. The utility of the engineered pair was further assessed in vivo. Two different cell populations, each expressing either mutant 81 or 37, were implanted in opposing flanks of a mouse (Figure 9B). Upon injection of 4′-MorphoLuc, cells expressing mutant 81 were visible. Once the signal dissipated, 7′-DMAMeLuc was administered, and cells expressing mutant 37 were illuminated.67
Figure 9.
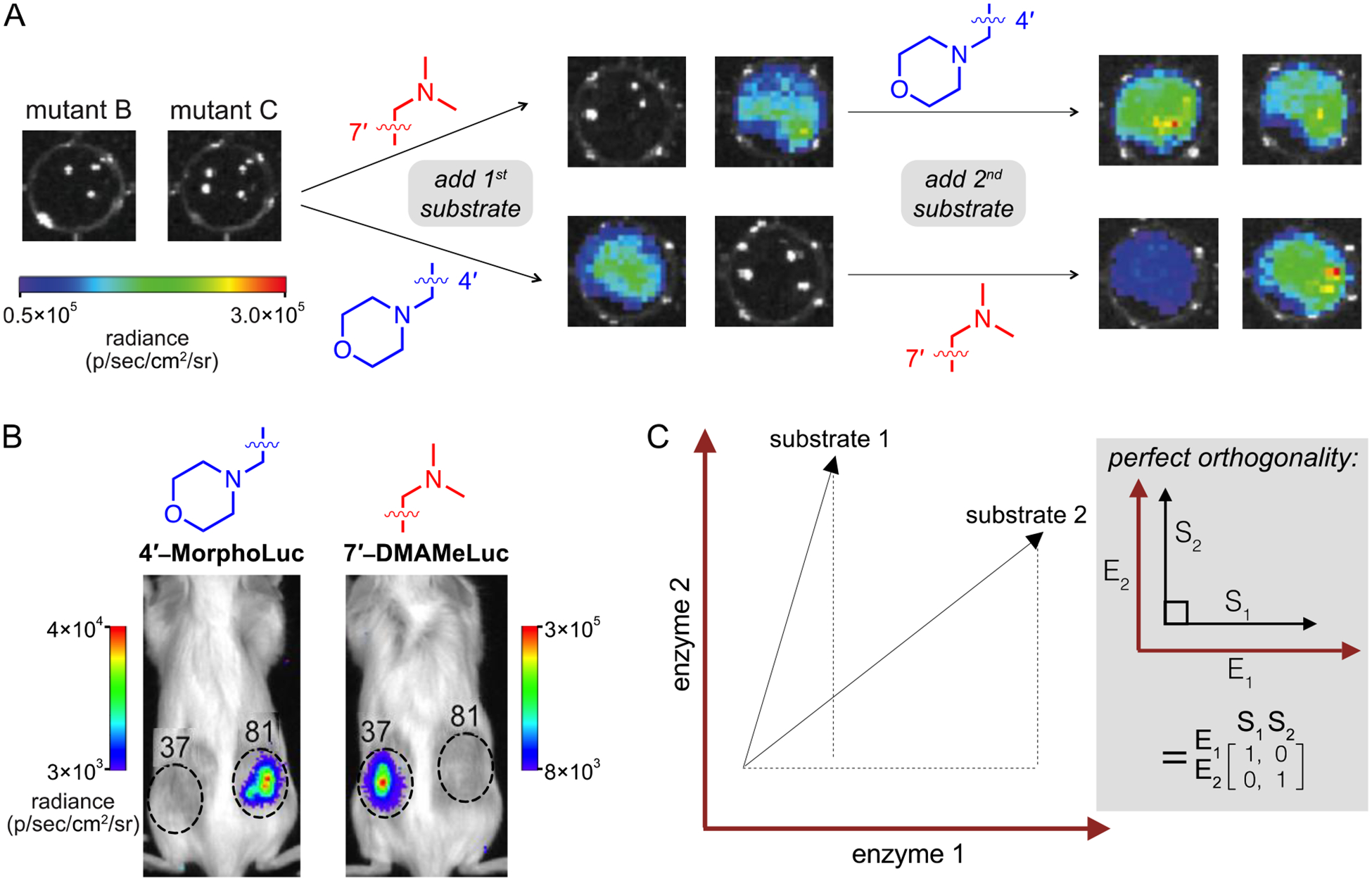
Multi-component imaging with orthogonal bioluminescent tools. (A) Orthogonal imaging in vitro. DB7 cells expressing mutant luciferases were placed in 96-well plates (1.5 × 105 cells/well) and imaged upon serial addition of luciferin analogs (750 μM). Representative bioluminescence images are shown. (B) Orthogonal imaging in vivo. FVB/NJ mice were injected with DB7 cells stably expressing luciferase mutants (37 and 81, which prefer 7′-DMAMeLuc and 4′-MorphoLuc, respectively). Inoculation sites are indicated with dashed circles. Luciferins were administered intraperitoneal and light emission was measured. (C) Matrix analysis of light emission data. The photon output from each enzyme (E)-substrate (S) pair can be represented as a vector. Perfect orthogonality is defined as the identity matrix. Matrices can be readily expanded to search for orthogonal triplets and other higher order sets. Part (A) was adapted with permission from ref. 40. Copyright 2017 American Chemical Society. Parts (B) and (C) were adapted with permission from ref. 67. Copyright 2017 American Chemical Society.
While the initial orthogonal pairs could be differentiated in a variety of environments, they exhibited weaker photon outputs compared to native bioluminescent systems. We have since identified “brighter” orthogonal imaging tools from library screens. One pair comprises 4′-BrLuc and D-luc, two compounds that are selectively processed by mutants 51 and 93, respectively. Both substrates are robust light emitters, cell permeable, and stable in vivo. Indeed, this combination of analogs enabled more facile two-component imaging in rodents. Cells expressing either mutant 51 or 93 were implanted in mice.67 Sequential administration of the complementary luciferins resulted in selective illumination of each “matched” pair. The light emission intensities were markedly higher than other orthogonal probes, and within the same order of magnitude as Fluc/D-luc. Further improvements in light output can likely be achieved via directed evolution.
While our screening studies have revealed a number of orthogonal pairs, the origins of selectivity remain a mystery in most cases. Structural analyses are underway, but insect luciferases are notoriously difficult to crystallize. Sequencing analyses revealed that certain mutations were hotspots for specific substrates. For example, F250M mutations typically favored 4′-MorphoLuc.40,64 This result could suggest that structural changes create more room for steric appendages. Our data set also revealed that the “best” orthogonal pairs comprise structurally divergent compounds (i.e., C4′-modified luciferins with C7′-modified luciferins).67 These substrates likely interact differently with the enzyme, making it easier to achieve orthogonality. Selectivity is thus achieved by destroying light emission with “unmatched” partners, versus making the complementary pair brighter. This trend suggests that the fastest way to expand the collection of orthogonal probes is to screen among more and more structurally diverse luciferins and luciferases.
Expediting the search for viable orthogonal flashlights
The orthogonal probes described thus far were handpicked from a large compilation of screening data. Such manual searches could potentially miss key enzyme-substrate pairs, and they become unwieldy as the data set expands. For example, screening a relatively small collection of mutants (~150) and analogs (~10) generated >800,000 possible combinations; a data set of this size was impractical to analyze manually.67 To expedite the search, we turned to a data-mining approach. We first defined substrate specificity as a numerical quantity (e.g., “orthogonality”) using a simple mathematical relationship (Figure 2A).67 This equation formed the basis of a computer script to cross-compare light emission values within the data set and rank pairs based on “orthogonality scores”. Higher numbers correlated with more substrate-specific pairs. This fast and simple algorithm rank-ordered 1,000 combinations in fewer than 30 minutes. Over 300 pairs exhibiting >25-fold substrate selectivity were identified, and 175 were validated in lysate, confirming the success of the approach.67 Importantly, the algorithm provides a method to rapidly hone in on promising orthogonal probes and continuously mine an ever-expanding data set. As new candidate pairs are generated from future screens, they can be assayed against previous hits.
The search for additional orthogonal probes is also benefitting from access to more functional luciferase variants. Active-site targeted libraries comprise a high number of non-functional variants, as many mutations have a deleterious impact on enzyme catalysis.68 Eliminating such mutants from candidate libraries enables more rapid identification of desirable enzymes. The Leconte laboratory recently applied a statistical coupling analysis (SCA) method to produce luciferase pools enriched with functional mutants.64 SCA identifies amino acids that are functionally important and likely possess synergistic interactions, which are desirable for library design.69 Residues for mutagenesis were identified from sequence alignments of luciferase homologues. In total, 14 residues located in and around the Fluc active site were targeted. Intriguingly, many of these residues had been previously identified as “hot spots” for orthogonal enzyme development.40,67 The SCA-focused libraries were screened with a panel of luciferin analogs, and luciferases were identified that were not only substrate specific, but that also exhibited other desirable features, including red-shifted emission and thermostability.64
Expanding the number of viable flashlights
Multi-component imaging requires more than two orthogonal enzymes and substrates. When we attempted to mine our screening data for a triplet set of luciferases and luciferins, we exceeded the computing capacity of standard processors. Searching for an orthogonal triplet alone (via orthogonality score) required more than one billion comparisons. To simplify the search, we turned to a matrix algebra approach.67 Enzyme-substrate combinations were represented as vectors in n dimensions (Figure 9C). Perfect orthogonality was then defined as the identity matrix. Triplet combinations that were closest to perfection (as assessed by root-mean-square distance) ranked highest. Using this approach, the algorithm predicted 6,171 potential triplet sets that were mutually orthogonal. The top triplet set comprised two C4′-modified luciferins, both with unique steric perturbations, and a C7′-modified analog (Figure 10). When the luciferins were incubated with their “matched” luciferases, selective light emission was observed. The matrix-mining algorithm is further applicable to searching for quadruplet, quintet, and other higher order sets, and work along these lines is ongoing.
Figure 10.
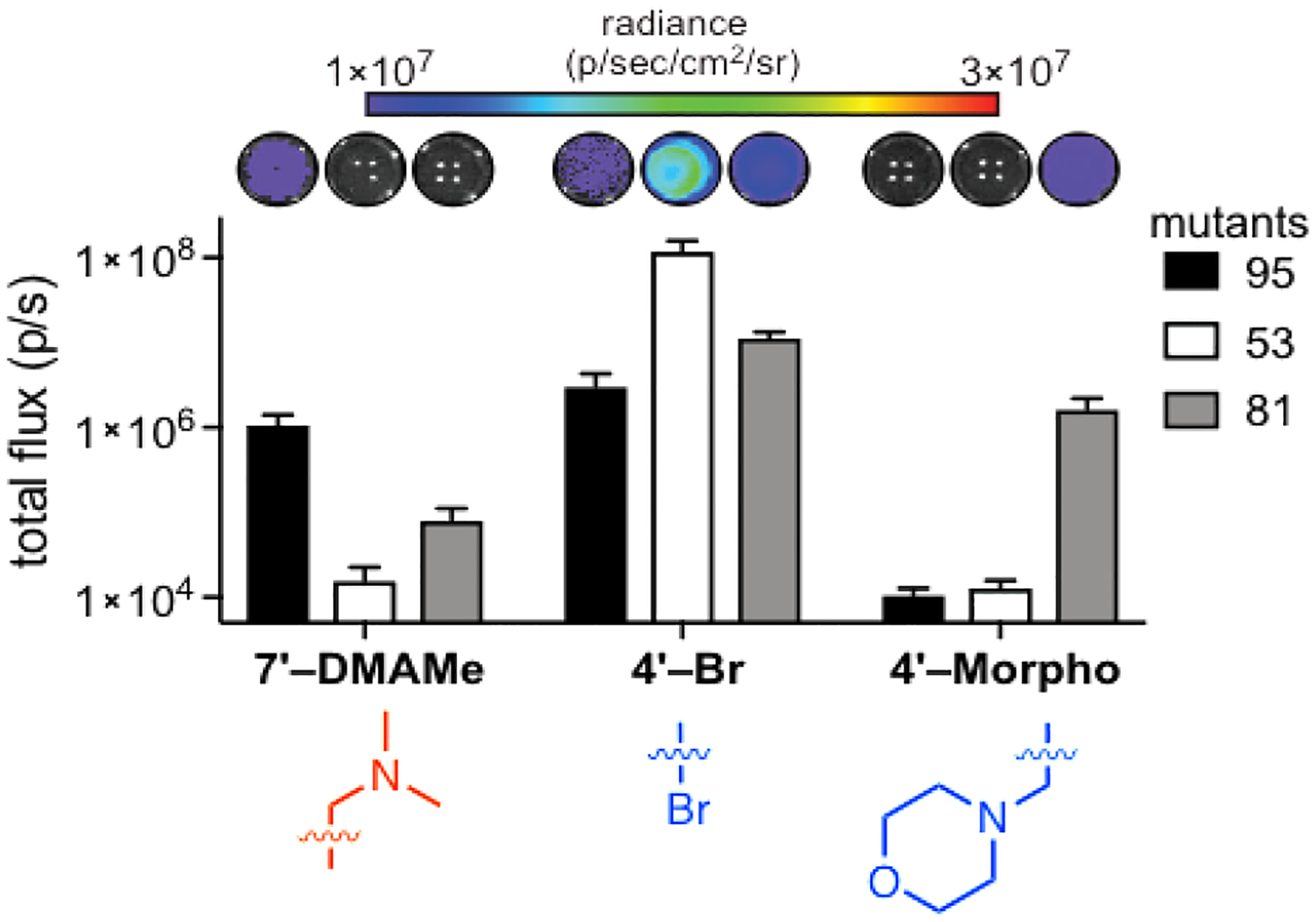
Orthogonal triplet set identified from computational analyses. Sterically modified luciferins (250 μM) were incubated with mutant luciferases 95, 53, and 81 (in bacterial lysate). Photon outputs were measured and the error bars represent the standard error of the mean for n = 3 experiments. Adapted with permission from ref. 67. Copyright 2017 American Chemical Society.
Towards building better and bri flashlights
Although we were able to identify luciferase mutants that confer substrate bias, the gains often came at the expense of other enzymatic parameters, including thermostability and turnover.40,64 An ongoing challenge is to improve the fitness of the engineered enzymes. Directed evolution approaches will be useful in this regard,70 and many of the orthogonal pairs are currently being optimized for enhanced brightness and stability. Machine learning approaches and deep mutational scanning analyses will also provide insight into how mutations affect the fitness landscape of luciferases and thus guide probe development.
Designing bioluminescent tools that are “brighter” and emit more red-shifted light will be advantageous for in vivo imaging. The Fluc-catalyzed oxidation of D-luc and most other analogs emits only a fraction for tissue-penetrant light. More deep tissue targets could be visualized if the orthogonal probes produced substantially more red photons.10 Several groups have addressed this issue by expanding the π-conjugation of D-luc.71–74 Many of these analogs are poorly processed by Fluc, though, such that the gains in red color are offset by low photon counts. In some cases, the intensities can be recovered using mutant enzymes.72,75 Such optimized bioluminescent probes have recently enabled noninvasive imaging of brain tissue in marmosets, highlighting the power of parallel engineering.75 Ongoing efforts to enhance substrate turnover rates will further enhance the brightness and sensitivity of the designer probes.
Additional gains in orthogonal probe development will come from screening more drug-like luciferins. Relatively high concentrations of D-luc and related analogs are typically required for bioluminescence imaging. More bioavailable luciferins would thus be advantageous. Brominated luciferins are attractive probes for this purpose, as they are “bright” and quite cell permeant.41 Swapping the hydroxyl group on D-luc for an amino or alkylamino group is also attractive. Miller and coworkers have shown that cyclic aminoluciferin analogs are robust emitters with favorable biodistribution properties.76–77 Small modifications to the aminoluciferin cores were also readily discriminated by mutant luciferase enzymes, enabling further orthogonal probe development.78–79 These and other structurally diverse luciferases20 will accelerate the search for larger collections of substrate-resolved luciferins.
Multiplexed bioluminescence will also benefit from improved imaging protocols. Our initial approach involves sequential administration of substrates, with a long delay between each delivery. Images are typically acquired with open filters (to capture all photons), such that the second compound cannot be administered until signal from the first substrate clears. In small animal models, the time delay is typically >8 hours. Methods to speed the delivery are necessary, and work along these lines is ongoing in our laboratory and others. We anticipate that modern machine learning algorithms can be used to deconvolute patterns of bioluminescent signal in a streamlined fashion. Such approaches could shorten imaging times from days to minutes, and eliminate the need for perfect orthogonality.
Summary and outlook
Bioluminescence is a cornerstone technology for noninvasive imaging in vivo. This technique has been widely employed to track cell proliferation, gene expression, and enzymatic activities, among other features. The list of imaging targets will continue to grow as new and improved bioluminescent probes are developed. The past decade alone has witnessed a surge in engineered luciferases and luciferins for numerous applications.
Despite recent advances, bioluminescence has been difficult to employ for multi-component imaging. A lack of suitable probes has largely relegated the technique to visualizing one cell or feature at a time. To expand the number of probes, we have adopted a parallel engineering approach. This strategy involves crafting Fluc mutants that can selectively process structurally diverse luciferins (i.e., orthogonal pairs). As a starting point, we focused on electronically and sterically modified luciferins. We developed scalable and divergent methods to access a variety of luciferin analogs. Most were poor bioluminescent light emitters, but inherently capable of robust emission. Enzymes to selectively process the analogs were identified by screening mutant luciferase libraries. The search for such biological “flashlights” and “batteries” was enhanced using computational algorithms, and the pairs characterized to date are functional in a wide range of environments.
Identifying expanded sets of orthogonal bioluminescent tools will benefit from advances on several fronts. Robust and scalable syntheses are necessary to access an even greater number of diverse luciferin architectures. More functional libraries and screening strategies are also required to expedite the search for bright, unique probes. A larger collection of optimized luciferase-luciferin pairs will enable multiple cell types and biological features to be visualized in tandem. Such studies promise to illuminate more complex facets of biology. Moreover, lessons learned from bioluminescent probe development should be broadly applicable to other families of enzymes, where access to unique substrates and orthogonal functions is desired.
ACKNOWLEDGMENTS
S.J.W. is a National Science Foundation Graduate Research Fellow. Our work on bioluminescent probes is supported by an award from the National Institutes of Health (R01GM107630 to J.A.P.) We thank Zi Yao and other members of the Prescher laboratory for helpful discussions and manuscript edits.
BIOGRAPHICAL INFORMATION
Sierra J. Williams obtained her Bachelor’s of Science in Chemistry from Temple University in 2016. She is currently a Ph.D. candidate in the Prescher laboratory at UC Irvine. Her thesis research involves engineering novel bioluminescent probes for multi-component imaging.
Jennifer A. Prescher is a Professor of Chemistry, Molecular Biology and Biochemistry, and Pharmaceutical Sciences at UC Irvine. She earned her Bachelor’s of Science in Chemistry at the University of Wisconsin—La Crosse. She obtained her Ph.D. in Chemistry at UC Berkeley in 2006. At Berkeley, Prescher worked with Prof. Carolyn Bertozzi to designing and employ bioorthogonal chemistries to monitor changes in cell surface glycosylation. Prescher conducted postdoctoral research in molecular imaging with Prof. Christopher Contag at Stanford University. In 2010, she joined the faculty at UC Irvine. Her current research focuses on developing chemical tools and noninvasive imaging strategies to interrogate biological processes.
REFERENCES
- 1.Paley MA; Prescher JA Bioluminescence: a versatile technique for imaging cellular and molecular features. MedChemComm 2014, 5, 255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prescher JA; Contag CH Guided by the light: Visualizing biomolecular processes in living animals with bioluminescence. Curr. Opin. Chem. Biol 2010, 14, 80–89. [DOI] [PubMed] [Google Scholar]
- 3.Yeh H-W; Ai H-W Development and applications of bioluminescent and chemiluminescent reporters and biosensors. Annu. Rev. Anal. Chem 2019, 12, 129–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yao Z; Zhang BS; Prescher JA Advances in bioluminescence imaging: New probes from old recipes. Curr. Opin. Chem. Biol 2018, 45, 148–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thorne N; Inglese J; Auld DS Illuminating insights into firefly luciferase and other bioluminescent reporters used in chemical biology. Chem. Biol 2010, 17, 646–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rathbun CM; Prescher JA Bioluminescent probes for imaging biology beyond the culture dish. Biochemistry 2017, 56, 5178–5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Contag CH; Spilman SD; Contag PR; Oshiro M; Eames B; Dennery P; Stevenson DK; Benaron DA Visualizing gene expression in living mammals using a bioluminescent reporter. Photochem. Photobiol 1997, 66, 523–531. [DOI] [PubMed] [Google Scholar]
- 8.Close DM; Xu T; Sayler GS; Ripp S In vivo bioluminescent imaging (BLI): Noninvasive visualization and interrogation of biological processes in living animals. Sensors 2011, 11, 180–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaskova ZM; Tsarkova AS; Yampolsky IV 1001 Lights: Luciferins, luciferases, their mechanisms of action and applications in chemical analysis, biology and medicine. Chem. Soc. Rev 2016, 45, 6048–6077. [DOI] [PubMed] [Google Scholar]
- 10.Zhao H; Doyle TC; Coquoz O; Kalish F; Rice BW; Contag CH Emission spectra of bioluminescent reporters and interaction with mammalian tissue determine the sensitivity of detection in vivo. J. Biomed. Opt 2005, 10, 1–9. [DOI] [PubMed] [Google Scholar]
- 11.Chan CT; Reeves RE; Geller R; Yaghoubi SS; Hoehne A; Solow-Cordero DE; Chiosis G; Massoud TF; Paulmurugan R; Gambhir SS Discovery and validation of small-molecule heat-shock protein 90 inhibitors through multimodality molecular imaging in living subjects. Proc. Natl. Acad. Sci. U. S. A 2012, 109, E2476–E2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan F; Wood KV Bioluminescent assays for high-throughput screening. Assay Drug Dev. Tech 2007, 5, 127–136. [DOI] [PubMed] [Google Scholar]
- 13.Gammon ST; Leevy WM; Gross S; Gokel GW; Piwnica-Worms D Spectral unmixing of multicolored bioluminescence emitted from heterogeneous biological sources. Anal. Chem 2006, 78, 1520–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hiblot J; Yu Q; Sabbadini MDB; Reymond L; Xue L; Schena A; Sallin O; Hill N; Griss R; Johnsson K Luciferases with tunable emission wavelengths. Angew. Chem. Int. Ed 2017, 56, 14556–14560. [DOI] [PubMed] [Google Scholar]
- 15.Loening AM; Wu AM; Gambhir SS Red-shifted Renilla reniformis luciferase variants for imaging in living subjects. Nat. Methods 2007, 4, 641–643. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki K; Kimura T; Shinoda H; Bai G; Daniels MJ; Arai Y; Nakano M; Nagai T, Five colour variants of bright luminescent protein for real-time multicolour bioimaging. Nature Communications 2016, 7, 13718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takai A; Nakano M; Saito K; Haruno R; Watanabe TM; Ohyanagi T; Jin T; Okada Y; Nagai T Expanded palette of Nano-lanterns for real-time multicolor luminescence imaging. Proc. Natl. Acad. Sci. U. S. A 2015, 112, 4352–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chu J; Oh Y; Sens A; Ataie N; Dana H; Macklin JJ; Laviv T; Welf ES; Dean KM; Zhang F; Kim BB; Tang CT; Hu M; Baird MA; Davidson MW; Kay MA; Fiolka R; Yasuda R; Kim DS; Ng HL; Lin MZ A bright cyan-excitable orange fluorescent protein facilitates dual-emission microscopy and enhances bioluminescence imaging in vivo. Nat. Biotechnol 2016, 34, 760–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeh H-W; Karmach O; Ji A; Carter D; Martins-Green MM; Ai H-W Red-shifted luciferase–luciferin pairs for enhanced bioluminescence imaging. Nat. Methods 2017, 14, 971–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeh H-W; Xiong Y; Wu T; Chen M; Ji A; Li X; Ai H-W ATP-independent bioluminescent reporter variants to improve in vivo imaging. ACS Chem. Biol 2019, 14, 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao H; Doyle TC; Wong RJ; Cao Y; Stevenson DK; Piwnica-Worms D; Contag CH Characterization of coelenterazine analogs for measurements of Renilla luciferase activity in live cells and living animals. Mol. Imaging 2004, 3, 43–54. [DOI] [PubMed] [Google Scholar]
- 22.Yeh H-W; Wu T; Chen M; Ai H-W Identification of factors complicating bioluminescence imaging. Biochemistry 2019, 58, 1689–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall MP; Unch J; Binkowski BF; Valley MP; Butler BL; Wood MG; Otto P; Zimmerman K; Vidugiris G; Machleidt T; Robers MB; Benink HA; Eggers CT; Slater MR; Meisenheimer PL; Klaubert DH; Fan F; Encell LP; Wood KV Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol 2012, 7, 1848–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martini S; Haddock SHD Quantification of bioluminescence from the surface to the deep sea demonstrates its predominance as an ecological trait. Sci. Rep 2017, 7, 45750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsarkova AS; Kaskova ZM; Yampolsky IV A tale of two luciferins: Fungal and earthworm new bioluminescent systems. Acc. Chem. Res 2016, 49, 2372–2380. [DOI] [PubMed] [Google Scholar]
- 26.Kotlobay AA; Dubinnyi MA; Purtov KV; Guglya EB; Rodionova NS; Petushkov VN; Bolt YV; Kublitski VS; Kaskova ZM; Ziganshin RH; Nelyubina YV; Dorovatovskii PV; Eliseev IE; Branchini BR; Bourenkov G; Ivanov IA; Oba Y; Yampolsky IV; Tsarkova AS Bioluminescence chemistry of fireworm Odontosyllis. Proc. Natl. Acad. Sci. U. S. A 2019, 201902095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White EH; McCapra F; Field GF The structure and synthesis of firefly luciferin. J. Am. Chem. Soc 1963, 85, 337–343. [Google Scholar]
- 28.Ciuffreda P; Casati S; Meroni G; Santaniello E A new synthesis of dehydroluciferin [2-(6′-hydroxy-2′-benzothiazolyl)-thiazole-4-carboxylic acid] from 1,4-benzoquinone. Tetrahedron 2013, 69, 5893–5897. [Google Scholar]
- 29.Würfel H; Weiss D; Beckert R; Güther A A new application of the “mild thiolation” concept for an efficient three-step synthesis of 2-cyanobenzothiazoles: A new approach to firefly-luciferin precursors. J. Sulfur Chem 2012, 33, 9–16. [Google Scholar]
- 30.Appel R; Janssen H; Siray M; Knoch F Synthesis and reactions of 4,5-dichloro-1,2,3-dithiazolium-chloride. Chem. Ber 1985, 118, 1632–1643. [Google Scholar]
- 31.Cuadro AM; Alvarez-Buila J 4,5-Dichloro-1,2,3-dithiazolium chloride (Appel’s Salt): Reactions with N-nucleophiles. Tetrahedron 1994, 50, 10037–10046. [Google Scholar]
- 32.Chabane H; Lamazzi C; Thiéry V; Guillaumet G; Besson T Synthesis of novel 2-cyanothiazolocarbazoles analogues of ellipticine. Tetrahedron Lett 2002, 43, 2483–2486. [Google Scholar]
- 33.Pereira MDF; Picot L; Guillon J; Léger J-M; Jarry C; Thiéry V; Besson T Efficient synthesis of novel pentacyclic 6,7-dihydro-5a,7a,13,14-tetraaza-pentaphene-5,8-diones. Tetrahedron Lett 2005, 46, 3445–3447. [Google Scholar]
- 34.Akhavan-Tafti HD S. R; Wang G; Eickholt RA; Gupta RK; Kaanumalle LS U. S. Patent 2011/0014599 A1, January 20, 2011.
- 35.McCutcheon DC; Paley MA; Steinhardt RC; Prescher JA Expedient synthesis of electronically modified luciferins for bioluminescence imaging. J. Am. Chem. Soc 2012, 134, 7604–7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michaelidou SS; Koutentis PA The conversion of 2-(4-chloro-5H-1,2,3-dithiazolylideneamino)benzonitriles into 3-aminoindole-2-carbonitriles using triphenylphosphine. Tetrahedron 2009, 65, 8428–8433. [Google Scholar]
- 37.McCutcheon DC; Porterfield WB; Prescher JA Rapid and scalable assembly of firefly luciferase substrates. Org. Biomol. Chem 2015, 13, 2117–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woodroofe CC; Meisenheimer PL; Klaubert DH; Kovic Y; Rosenberg JC; Behney CE; Southworth TL; Branchini BR Novel heterocyclic analogues of firefly luciferin. Biochemistry 2012, 51, 9807–9813. [DOI] [PubMed] [Google Scholar]
- 39.Steinhardt RC; O’Neill JM; Rathbun CM; McCutcheon DC; Paley MA; Prescher , J. A. Design and synthesis of an alkynyl luciferin analogue for bioluminescence imaging. Chem. Eur. J 2016, 22, 3671–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones KA; Porterfield WB; Rathbun CM; McCutcheon DC; Paley MA; Prescher JA Orthogonal luciferase–luciferin pairs for bioluminescence imaging. J. Am. Chem. Soc 2017, 139, 2351–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steinhardt RC; Rathbun CM; Krull BT; Yu JM; Yang Y; Nguyen BD; Kwon J; McCutcheon DC; Jones KA; Furche F; Prescher JA Brominated luciferins are versatile bioluminescent probes. ChemBioChem 2017, 18, 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shakhmin A; Hall MP; Machleidt T; Walker JR; Wood KV; Kirkland TA Coelenterazine analogues emit red-shifted bioluminescence with NanoLuc. Org. Biomol. Chem 2017, 15, 8559–8567. [DOI] [PubMed] [Google Scholar]
- 43.Jiang T; Du L; Li M Lighting up bioluminescence with coelenterazine: Strategies and applications. Photochem. Photobiol. Sci 2016, 15, 466–480. [DOI] [PubMed] [Google Scholar]
- 44.Kim SB; Torimura M; Tao H Creation of artificial luciferases for bioassays. Bioconjugate Chem 2013, 24, 2067–2075. [DOI] [PubMed] [Google Scholar]
- 45.Branchini BR; Southworth TL; Murtiashaw MH; Wilkinson SR; Khattak NF; Rosenberg JC; Zimmer M Mutagenesis evidence that the partial reactions of firefly bioluminescence are catalyzed by different conformations of the luciferase C-terminal domain. Biochemistry 2005, 44, 1385–1393. [DOI] [PubMed] [Google Scholar]
- 46.Sundlov JA; Fontaine DM; Southworth TL; Branchini BR; Gulick AM Crystal structure of firefly luciferase in a second catalytic conformation supports a domain alternation mechanism. Biochemistry 2012, 51, 6493–6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakatsu T; Ichiyama S; Hiratake J; Saldanha A; Kobashi N; Sakata K; Kato H Structural basis for the spectral difference in luciferase bioluminescence. Nature 2006, 440, 372–376. [DOI] [PubMed] [Google Scholar]
- 48.Branchini BR; Magyar RA; Murtiashaw MH; Anderson SM; Zimmer M Site-directed mutagenesis of histidine 245 in firefly luciferase: A proposed model of the active site. Biochemistry 1998, 37, 15311–15319. [DOI] [PubMed] [Google Scholar]
- 49.Branchini BR; Magyar RA; Murtiashaw MH; Portier NC The role of active site residue arginine 218 in firefly luciferase bioluminescence. Biochemistry 2001, 40, 2410–2418. [DOI] [PubMed] [Google Scholar]
- 50.Viviani VR The origin, diversity, and structure function relationships of insect luciferases. Cell. Mol. Life Sci 2002, 59, 1833–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Viviani VR; Amaral DT; Neves DR; Simões A; Arnoldi FGC The luciferin binding site residues C/T311 (S314) influence the bioluminescence color of beetle luciferases through main-chain interaction with oxyluciferin phenolate. Biochemistry 2013, 52, 19–27. [DOI] [PubMed] [Google Scholar]
- 52.Viviani VR; Neves DR; Amaral DT; Prado RA; Matsuhashi T; Hirano T Bioluminescence of beetle luciferases with 6′-amino-D-luciferin analogues reveals excited keto-oxyluciferin as the emitter and phenolate/luciferin binding site interactions modulate bioluminescence colors. Biochemistry 2014, 53, 5208–5220. [DOI] [PubMed] [Google Scholar]
- 53.Pines G; Pines A; Garst AD; Zeitoun RI; Lynch SA; Gill RT Codon compression algorithms for saturation mutagenesis. ACS Synth. Biol 2015, 4, 604–614. [DOI] [PubMed] [Google Scholar]
- 54.Quan J; Tian J Circular polymerase extension cloning for high-throughput cloning of complex and combinatorial DNA libraries. Nat. Protoc 2011, 6, 242–251. [DOI] [PubMed] [Google Scholar]
- 55.Ness JE; Kim S; Gottman A; Pak R; Krebber A; Borchert TV; Govindarajan S; Mundorff EC; Minshull J Synthetic shuffling expands functional protein diversity by allowing amino acids to recombine independently. Nat. Biotechnol 2002, 20, 1251–1255. [DOI] [PubMed] [Google Scholar]
- 56.Branchini BR; Hayward MM; Bamford S; M. Brennan PM; Lajiness EJ Naphthyl- and quinolylluciferin: Green and red light emitting firefly luciferin analogues. Photochem. Photobiol 1989, 49, 689–695. [DOI] [PubMed] [Google Scholar]
- 57.Takakura H; Sasakura K; Ueno T; Urano Y; Terai T; Hanaoka K; Tsuboi T; Nagano T Development of luciferin analogues bearing an amino group and their application as BRET donors. Chem. Asian J 2010, 5, 2053–2061. [DOI] [PubMed] [Google Scholar]
- 58.Paley MA Expanding the bioluminescent toolkit for in vivo imaging. Ph.D. Dissertation, University of California, Irvine, Irvine, California, 2014. [Google Scholar]
- 59.e Candia M; Fossa P; Cellamare S; Mosti L; Carotti A; Altomare C Insights into structure–activity relationships from lipophilicity profiles of pyridin-2(1H)-one analogs of the cardiotonic agent milrinone. Eur. J. Pharm. Sci 2005, 26, 78–86. [DOI] [PubMed] [Google Scholar]
- 60.Dragovich PS; Prins TJ; Zhou R; Brown EL; Maldonado FC; Fuhrman SA; Zalman LS; Tuntland T; Lee CA; Patick AK; Matthews DA; Hendrickson TF; Kosa MB; Liu B; Batugo MR; Gleeson JPR; Sakata SK; Chen L; Guzman MC; Meador JW; Ferre RA; Worland ST Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 6. Structure−activity studies of orally bioavailable, 2-pyridone-containing peptidomimetics. J. Med. Chem 2002, 45, 1607–1623. [DOI] [PubMed] [Google Scholar]
- 61.Bhattacharya B; Samanta A Excited-state proton-transfer dynamics of 7-hydroxyquinoline in room temperature ionic liquids. J. Phys. Chem. B 2008, 112, 10101–10106. [DOI] [PubMed] [Google Scholar]
- 62.Kuchlyan J; Banik D; Roy A; Kundu N; Sarkar N Excited-state proton transfer dynamics of firefly’s chromophore D-luciferin in DMSO–water binary mixture. J. Phys. Chem. B 2014, 118, 13946–13953. [DOI] [PubMed] [Google Scholar]
- 63.Zhang BS; Jones KA; McCutcheon DC; Prescher JA Pyridone luciferins and mutant luciferases for bioluminescence imaging. ChemBioChem 2018, 19, 470–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu MD; Warner EA; Morrissey CE; Fick CW; Wu TS; Ornelas MY; Ochoa GV; Zhang BS; Rathbun CM; Porterfield WB; Prescher JA; Leconte AM Statistical coupling analysis-guided library design for the discovery of mutant luciferases. Biochemistry 2018, 57, 663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li J; Chen L; Du L; Li M Cage the firefly luciferin! – A strategy for developing bioluminescent probes. Chem. Soc. Rev 2013, 42, 662–676. [DOI] [PubMed] [Google Scholar]
- 66.Woodroofe CC; Shultz JW; Wood MG; Osterman J; Cali JJ; Daily WJ; Meisenheimer PL; Klaubert DH N-Alkylated 6′-aminoluciferins are bioluminescent substrates for Ultra-Glo and QuantiLum luciferase: New potential scaffolds for bioluminescent assays. Biochemistry 2008, 47, 10383–10393. [DOI] [PubMed] [Google Scholar]
- 67.Rathbun CM; Porterfield WB; Jones KA; Sagoe MJ; Reyes MR; Hua CT; Prescher JA Parallel screening for rapid identification of orthogonal bioluminescent tools. ACS Cent. Sci 2017, 3, 1254–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bloom JD; Arnold FH In the light of directed evolution: Pathways of adaptive protein evolution. Proc. Natl. Acad. Sci. U. S. A 2009, 106, 9995–10000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Socolich M; Lockless SW; Russ WP; Lee H; Gardner KH; Ranganathan R Evolutionary information for specifying a protein fold. Nature 2005, 437, 512–518. [DOI] [PubMed] [Google Scholar]
- 70.Brustad EM; Arnold FH Optimizing non-natural protein function with directed evolution. Curr. Opin. Chem. Biol 2011, 15, 201–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kuchimaru T; Iwano S; Kiyama M; Mitsumata S; Kadonosono T; Niwa H; Maki S; Kizaka-Kondoh S A luciferin analogue generating near-infrared bioluminescence achieves highly sensitive deep-tissue imaging. Nat. Commun 2016, 7, 11856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hall MP; Woodroofe CC; Wood MG; Que I; van’t Root M; Ridwan Y; Shi C; Kirkland TA; Encell LP; Wood KV; Löwik C; Mezzanotte L Click beetle luciferase mutant and near infrared naphthyl-luciferins for improved bioluminescence imaging. Nat. Commun 2018, 9, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Anderson JC; Grounds H; Jathoul AP; Murray JAH; Pacman SJ; Tisi L Convergent synthesis and optical properties of near-infrared emitting bioluminescent infra-luciferins. RSC Adv 2017, 7, 3975–3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Iwano S; Obata R; Miura C; Kiyama M; Hama K; Nakamura M; Amano Y; Kojima S; Hirano T; Maki S; Niwa H Development of simple firefly luciferin analogs emitting blue, green, red, and near-infrared biological window light. Tetrahedron 2013, 69, 3847–3856.. [Google Scholar]
- 75.Iwano S; Sugiyama M; Hama H; Watakabe A; Hasegawa N; Kuchimaru T; Tanaka KZ; Takahashi M; Ishida Y; Hata J; Shimozono S; Namiki K; Fukano T; Kiyama M; Okano H; Kizaka-Kondoh S; McHugh TJ; Yamamori T; Hioki H; Maki S; Miyawaki A Single-cell bioluminescence imaging of deep tissue in freely moving animals. Science 2018, 359, 935–939. [DOI] [PubMed] [Google Scholar]
- 76.Reddy GR; Thompson WC; Miller SC Robust light emission from cyclic alkylaminoluciferin substrates for firefly luciferase. J. Am. Chem. Soc 2010, 132, 13586–13587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sharma DK; Adams ST Jr.; Liebmann KL; Miller SC Rapid access to a broad range of 6′-substituted firefly luciferin analogues reveals surprising emitters and inhibitors. Org. Lett 2017, 19, 5836–5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mofford DM; Reddy GR; Miller SC Aminoluciferins extend firefly luciferase bioluminescence into the near-infrared and can be preferred substrates over D-luciferin. J. Am. Chem. Soc 2014, 136, 13277–13282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Adams ST Jr.; Mofford DM; Reddy GSKK; Miller SC Firefly luciferase mutants allow substrate-selective bioluminescence imaging in the mouse brain. Angew. Chem. Intl. Ed 2016, 55, 4943–4946. [DOI] [PMC free article] [PubMed] [Google Scholar]


