Abstract
Mitochondrial DNA (mtDNA) is a circular genome of 16 kb that is present in multiple copies in mitochondria. mtDNA codes for genes that contribute to mitochondrial structure and function. A long-standing question has asked whether mtDNA is epigenetically regulated similarly to the nuclear genome. Recently published data suggest that unlike the nuclear genome where CpG methylation is the norm, mtDNA is methylated predominantly at non-CpG cytosines. This raises important methodological considerations for future investigations. In particular, existing bisulphite PCR techniques may be unsuitable due to primers being biased towards amplification from unmethylated mtDNA. Here, we describe how this may have led to previous studies underestimating the level of mtDNA methylation and reiterate methodological strategies for its accurate assessment.
Keywords: non-CpG methylation, mtDNA methylation, bisulphite sequencing, mitochondrial DNA, PCR bias
Non-CpG Methylation Biases Bisulphite PCR towards Unmethylated Alleles with Standard Primer Design
The existence of mitochondrial DNA (mtDNA) methylation has been controversial for decades (reviewed in [1–3]). However, in the last 10 years there has been a growing consensus for its existence, and further, its functional relevance, including in environmental responses [4–12]. Three recent publications [13–15] are strong contributions to this shift in opinion as they provide technical advancements, as well as information on regional CpG and non-CpG mtDNA methylation, regulation by methyltransferases and associations with disease. The strength of evidence in these studies stems from innovative new techniques and inclusion of controls which are crucial for investigating this unique, small, circular genome of prokaryotic origin that exists within eukaryotic cells. The aim of this perspective is not to add to the number of recent reviews on mtDNA methylation but to highlight a technical problem which stems from discoveries in the three recent publications. This methodological issue has to our knowledge not previously been highlighted in relation to mtDNA methylation.
The most widely used technique for the study of mtDNA methylation is targeted/locus-specific/amplicon bisulphite sequencing [4–7, 9, 10, 16–19]. This technique allows identification of individual nucleotide methylation state within a chosen locus as an unmethylated cytosine is represented by a thymine following PCR, whereas a methylated cytosine is protected from bisulphite conversion and remains a cytosine. Targeted bisulphite sequencing was used in 11 of the 16 publications that investigated mtDNA methylation listed in Pubmed in the period between the 1 August 2018 and 28 October 2019. Several studies [8, 13, 15, 19] observed lower levels of mtDNA methylation with this technique than were detected in the same samples with whole-genome bisulphite sequencing (WGBS). WGBS utilizes bisulphite treatment followed by shotgun next-generation sequencing. Indeed, the observations of low DNA methylation levels with targeted bisulphite sequencing have even been used as evidence to support the idea that mtDNA methylation is at extremely low levels, or non-existent [7, 8, 16–18]. To explain this difference between targeted bisulphite sequencing and WGBS, Patil et al. [13] suggested that it could be due to PCR-based sequencing assays (pyrosequencing, methylation-specific PCR) usually only assaying methylation at CpG sites. Therefore, the high levels of methylation at non-CpG (CpA, CpT, CpC) sites is not detected [10, 13].
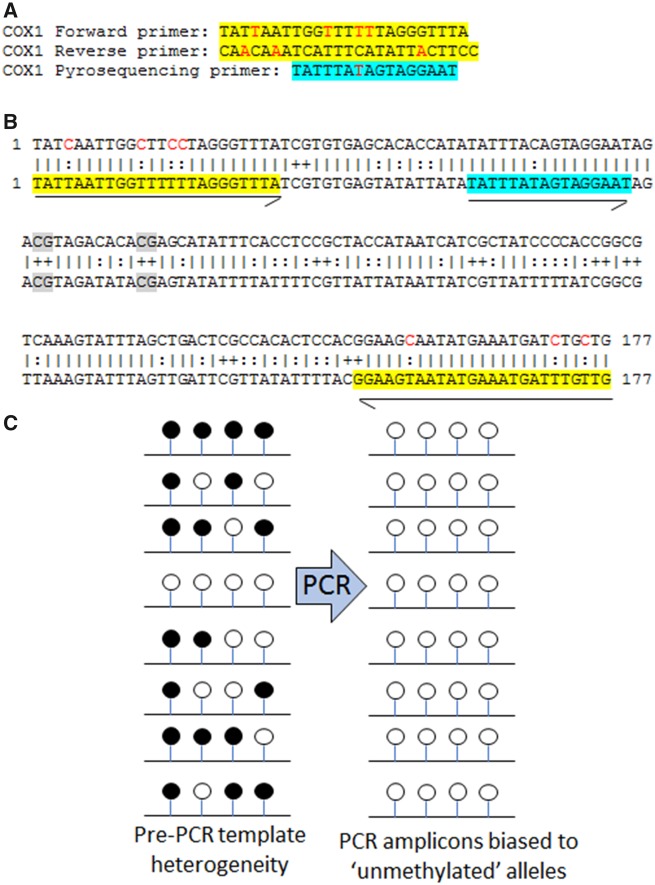
Another reason that may make targeted bisulphite sequencing a less accurate method for measuring methylation in mtDNA is the inherent bias of these assays when a DNA template has high levels of non-CpG methylation (Fig. 1). As DNA methylation in the mammalian nuclear genome is predominantly at CpG dinucleotides, current bisulphite PCR primer design methodologies [20] ensure that the primers contain thymines that correspond to converted non-CpG cytosines (CpA, CpC, CpT). These thymines are intended to promote amplification from fully converted templates. However, the assumption that all non-CpG cytosines are converted makes most targeted bisulphite sequencing primers unsuitable in regions that do in fact contain significant levels of non-CpG methylation [21]. In these regions, standard bisulphite sequencing primers will selectively amplify from the templates with the least methylation (Fig. 1C). For example, in the recent study by Patil et al., the forward and reverse primers used to assay the MT-COX1 gene region in mtDNA contain four and three non-CpG cytosines, respectively (Fig. 1A and B), which would explain why they observed lower methylation in the bisulphite sequencing assay than with WGBS.
Figure 1:
PCR from bisulphite-converted DNA in regions with non-CpG methylation is biased towards amplification of unmethylated alleles if the primer contains cytosines. (A) Typical primers used for bisulphite sequencing with bases that hybridize to converted non-CpG cytosines (red text). (B) Alignment of unconverted (top) and converted (bottom) sequences with non-CpG cytosines indicated with colons and CpGs with plus signs. Locations of PCR and pyrosequencing primers highlighted in yellow and blue, respectively, and indicated with arrows. Converted non-CpG cytosines in primers in red text. (C) Schematic showing selective amplification in PCR from unmethylated alleles in regions with a high frequency of non-CpG methylation. Methylated/unconverted and unmethylated/converted cytosines indicated with black and white lollipops, respectively
Methodological Approaches to Compensate for the Presence of Non-CpG Methylation
Unlike most mammalian nuclear genomes, non-CpG methylation is abundant in plant genomes. Accordingly, plant researchers have proposed methods [22] to allow bisulphite sequencing in regions with non-CpG methylation that could be applied to mammalian mtDNA. These methodological changes include using primers that contain no cytosines or are degenerate in that they contain C/T at non-CpG cytosines (or G/A nucleotides on the complementary strand) so that 100% complementary primers are available for any potential combination of bases across the primer region. However, degenerate primers will have different proportions of cytosines and therefore differences in binding affinities and in optimal annealing temperatures. This means that higher PCR annealing temperatures are likely to ultimately report higher levels of methylation than lower annealing temperatures. Therefore, each primer pair must be tested with control templates [19, 22] of known percentages of C and T (or G and A) to identify the annealing temperature required to faithfully replicate the methylation level of the template.
A further risk of degenerate primers is that the introduced variation in primer sequence increases the possibility of non-specific amplification. If the mtDNA purification procedures do not adequately exclude the nuclear genome, degenerate primers increase the likelihood of amplifying mtDNA-like sequences from nuclear DNA [23]. One solution to this problem may be the use of sequencing technologies such as PacBio and Oxford Nanopore as the longer reads can be unambiguously identified as being from mtDNA [24].
General Checklist for Best Practice Methodologies for Studying mtDNA Methylation
The unique challenges for analysing the epigenetic state of mtDNA have, throughout the development of the field, inspired innovative work to surmount those challenges. In Table 1, we list recommendations of the current best practices for DNA methylation analysis in mtDNA. For guidance purposes, we have only indicated a few recent studies that best demonstrate or describe these issues.
Table 1:
best practices for assaying mtDNA methylation
| All methodologies | |
| Confirm results with multiple techniques e.g. WGBS, MeDIP, mass spectrometry, PacBio, Nanopore | [13, 14, 19] |
| Enrich mitochondria or mtDNA prior to assaying to avoid unintentional detection of the nuclear genome | [13, 14, 19] |
| Present evidence of success of enrichment (e.g. qPCR of mtDNA vs nuclear DNA) | [13, 14] |
| Bisulphite methodologies | |
| Spike sample with control DNA for evaluation of conversion efficiency | [14, 19] |
| Linearize mtDNA prior to bisulphite conversion to avoid secondary structure effects | [13, 14, 16, 19, 25] |
| In WGBS separately analyse and present data from light and heavy strands | [13, 14] |
| Measure CpG and non-CpG methylation | [10, 13, 14] |
| Differentiate between 5mC and 5hmC | [14, 19, 26] |
| Use PCR primers or controls to account for non-CpG methylation | No examples of primers at present. Suitable positive and negative controls in [13, 19, 22] |
In summary, given the rapidly increasing interest in the field of mtDNA methylation, more research is urgently required to investigate the underlying mechanisms and the functional role of methylation in development, environmental responses and disease. Targeted bisulphite sequencing has proven to be a quick and reliable method for assaying methylation levels in the nuclear genome. However, the discovery of widespread non-CpG methylation in mtDNA, means that adaptations to currently used methodologies are required to make targeted bisulphite sequencing a viable tool for researching this unique genome. Finally, as non-CpG methylation is present at significant levels in the nuclear genome in the brain and pluripotent cells [21], the approaches described here may also be adopted more broadly for the analysis of nuclear DNA.
Conflict of interest statement. None declared.
References
- 1. Maresca A, Zaffagnini M, Caporali L, Carelli V, Zanna C.. DNA methyltransferase 1 mutations and mitochondrial pathology: is mtDNA methylated? Front Genet 2015;6:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Byun HM, Baccarelli AA.. Environmental exposure and mitochondrial epigenetics: study design and analytical challenges. Hum Genet 2014;133:247–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van der Wijst MGP, Rots MG.. Mitochondrial epigenetics: an overlooked layer of regulation? Trends Genet 2015;31:353–6. [DOI] [PubMed] [Google Scholar]
- 4. Gao J, Wen S, Zhou H, Feng S.. De-methylation of displacement loop of mitochondrial DNA is associated with increased mitochondrial copy number and nicotinamide adenine dinucleotide subunit 2 expression in colorectal cancer. Mol Med Rep 2015;12:7033–8. [DOI] [PubMed] [Google Scholar]
- 5. Tong H, Zhang L, Gao J, Wen S, Zhou H, Feng S.. Methylation of mitochondrial DNA displacement loop region regulates mitochondrial copy number in colorectal cancer. Mol Med Rep 2017;16:5347–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van der Wijst MGP, van Tilburg AY, Ruiters MHJ, Rots MG.. Experimental mitochondria-targeted DNA methylation identifies GpC methylation, not CpG methylation, as potential regulator of mitochondrial gene expression. Sci Rep 2017;7:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu B, Du Q, Chen L, Fu G, Li S, Fu L, Zhang X, Ma C, Bin C.. CpG methylation patterns of human mitochondrial DNA. Sci Rep 2016;6:23421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Morris MJ, Hesson LB, Poulos RC, Ward RL, Wong JWH, Youngson NA.. Reduced nuclear DNA methylation and mitochondrial transcript changes in adenomas do not associate with mtDNA methylation. Biomark Res 2018;6:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pirola CJ, Gianotti TF, Burgueño AL, Rey-Funes M, Loidl CF, Mallardi P, Martino JS, Castaño GO, Sookoian S.. Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut 2013;62:1356–63. [DOI] [PubMed] [Google Scholar]
- 10. Bellizzi D, D'Aquila P, Scafone T, Giordano M, Riso V, Riccio A, Passarino G.. The control region of mitochondrial DNA shows an unusual CpG and non-CpG methylation pattern. DNA Res 2013;20:537–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lambertini L, Byun HM.. Mitochondrial epigenetics and environmental exposure. Curr Environ Health Rep 2016;3:214–24. [DOI] [PubMed] [Google Scholar]
- 12. Armstrong DA, Green BB, Blair BA, Guerin DJ, Litzky JF, Chavan NR, Pearson KJ, Marsit CJ.. Maternal smoking during pregnancy is associated with mitochondrial DNA methylation. Environ Epigenet 2016;2:dvw020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Patil V, Cuenin C, Chung F, Aguilera JRR, Fernandez-Jimenez N, Romero-Garmendia I, Bilbao JR, Cahais V, Rothwell J, Herceg Z.. Human mitochondrial DNA is extensively methylated in a non-CpG context. Nucleic Acids Res 2019;47:10072–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dou X, Boyd-Kirkup JD, McDermott J, Zhang X, Li F, Rong B, Zhang R, Miao B, Chen P, Cheng H, Xue J, Bennett D, Wong J, Lan F, Han J-DJ.. The strand-biased mitochondrial DNA methylome and its regulation by DNMT3A. Genome Res 2019;29:1622–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sirard MA. Distribution and dynamics of mitochondrial DNA methylation in oocytes, embryos and granulosa cells. Sci Rep 2019;9:11937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Owa C, Poulin M, Yan L, Shioda T.. Technical adequacy of bisulfite sequencing and pyrosequencing for detection of mitochondrial DNA methylation: sources and avoidance of false-positive detection. PLoS One 2018;13:e0192722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mechta M, Ingerslev LR, Fabre O, Picard M, Barrès R.. Evidence suggesting absence of mitochondrial DNA methylation. Front Genet 2017;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hong EE, Okitsu CY, Smith AD, Hsieh C-L.. Regionally specific and genome-wide analyses conclusively demonstrate the absence of CpG methylation in human mitochondrial DNA. Mol Cell Biol 2013;33:2683–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sun X, Vaghjiani V, Jayasekara WSN, Cain JE, St. John JC.. The degree of mitochondrial DNA methylation in tumor models of glioblastoma and osteosarcoma. Clin Epigenet 2018;10:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li LC, Dahiya R.. MethPrimer: designing primers for methylation PCRs. Bioinformatics 2002;18:1427–31. [DOI] [PubMed] [Google Scholar]
- 21. Patil V, Ward RL, Hesson LB.. The evidence for functional non-CpG methylation in mammalian cells. Epigenetics 2014;9:823–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. How-Kit A, Daunay A, Mazaleyrat N, Busato F, Daviaud C, Teyssier E, Deleuze J-F, Gallusci P, Tost J.. Accurate CpG and non-CpG cytosine methylation analysis by high-throughput locus-specific pyrosequencing in plants. Plant Mol Biol 2015;88:471–85. [DOI] [PubMed] [Google Scholar]
- 23. Mishmar D, Ruiz-Pesini E, Brandon M, Wallace DC.. Mitochondrial DNA-like sequences in the nucleus (NUMTs): insights into our African origins and the mechanism of foreign DNA integration. Hum Mutat 2004;23:125–33. [DOI] [PubMed] [Google Scholar]
- 24. Jenjaroenpun P, Wongsurawat T, Pereira R, Patumcharoenpol P, Ussery DW, Nielsen J, Nookaew I.. Complete genomic and transcriptional landscape analysis using third-generation sequencing: a case study of Saccharomyces cerevisiae CEN.PK113-7D. Nucleic Acids Res 2018;46:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mechta M, Ingerslev LR, Barres R.. Methodology for accurate detection of mitochondrial DNA methylation. J Vis Exp 2018;135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM.. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc Natl Acad Sci USA 2011;108:3630–5. [DOI] [PMC free article] [PubMed] [Google Scholar]