Abstract
For many crop pathogens including viruses, high genetic variation provides them with potential to adapt to and prevail in a changing environment. Understanding genetic variation in viruses and their significance is a key to elaborate virus epidemiology and evolution. While genetic variation of plant viruses has been documented to impact virus–host interactions, how it affects virus–insect vector interactions remains elusive. Here, we report the impact of mutations in the coat protein of squash leaf curl China virus (SLCCNV), a begomovirus, on the interaction between the virus and its whitefly vectors. We characterized mutations in the coat protein of SLCCNV and found that some residues exhibited higher mutation frequency than the others. We assayed the impact of mutation on infectivity using agroinoculation and found these mutations marginally affect virus infectivity. We further analyze their functions using virus acquisition and transmission trials and found some of mutations resulted in altered transmission of SLCCNV by different species of the whitefly Bemisia tabaci complex. We then identified the key amino acid residue(s) involved by constructing several mutant viruses and found that a single-residue mutation in the coat protein of SLCCNV was sufficient to significantly alter the whitefly transmission characteristics of SLCCNV. We examined the competition between different genotypes of SLCCNV in plant infection and whitefly transmission. We found that mutations in the coat protein did not alter the fitness of SLCCNV in plants, but they rendered the virus more competitive in transmission by certain species of whiteflies. Our findings indicate that mutations in the coat protein may play a key role in both the adaptation of begomoviruses to the changing vector populations and the evolution of begomoviruses.
Keywords: genetic variation, insect vector, virus transmission, coat protein, virus evolution
1. Introduction
In modern agricultural ecosystem, crop pathogens cause widespread disease epidemics and may evolve quickly in response to environmental changes (Mcdonald and Stukenbrock 2016). Among these pathogens, plant viruses represent one of the most important groups as they are able to adapt to changing environments rapidly (Harrison 2002; Jones 2009; Lefeuvre et al. 2019). For viruses, genetic variations such as mutation generate ensembles of nonidentical but genetically closely related individuals (Domingo, Sheldon, and Perales 2012). Selection pressure imposed on a virus by a given environment will change the virus population as it eliminates less fit individuals and by doing so favor the dominance of the fittest; in this way, viruses keep evolving and adapting to the changing environment, which provides them with substantial potential to outbreak and render many control strategies unsustainable (Duffy, Shackelton, and Holmes 2008; Domingo, Sheldon, and Perales 2012). Therefore, understanding genetic variation of viruses and its impact on their biological properties is a key to improved knowledge of virus epidemiology and evolution.
For most plant viruses, selection pressures are exerted mainly by infection and transmission (Lefeuvre et al. 2019). On the one hand, plant viruses need to overcome plant host resistance to achieve productive infections. On the other hand, since most of them rely on insect vectors to spread, for example, members of the genus Begomovirus (family Geminiviridae) are transmitted by whiteflies of the Bemisia tabaci complex, they need to adapt to insect vectors (Seal, vandenBosch, and Jeger 2006; Hogenhout et al. 2008). In terms of infection, genetic variation of viruses has been shown to be one major reason for the plants to lose their resistance to virus infection (Harrison 2002; Jones 2009). For example, a mutation in Cotton leaf curl Kokhran virus that truncated the transcriptional-activator protein from 134 amino acids to a 35-amino-acid peptide played a key role in the breakdown of resistance in certain cotton varieties (Briddon et al. 2014). For transmission, how genetic variation of viruses affects their adaptation to their insect vectors remains elusive.
In recent decades, begomoviruses, a group of small, nonenveloped viruses with single-stranded, circular DNA genome, have emerged as serious threats to the production of many crops (Navas-Castillo, Fiallo-Olivé, and Sánchez-Campos 2011). Begomoviruses are transmitted in a persistent, circulative manner by B. tabaci, which is a species complex consisting of many morphologically indistinguishable but genetically distinct and reproductively isolated cryptic species (Ghanim, Morin, and Czosnek 2001; Hogenhout et al. 2008; De Barro et al. 2011; Navas-Castillo, Fiallo-Olivé, and Sánchez-Campos 2011; Liu, Colvin, and De Barro 2012). Whiteflies of different species may transmit a given begomovirus with disparate efficiencies, and a given species of whitefly may vary substantially in its efficiency when transmitting different begomoviruses (Idris, Smith, and Brown 2001; Li et al. 2010; Wei et al. 2014; De Marchi et al. 2017; Pan et al. 2018a,b; Fiallo-Olivé et al. 2020). From the side of virus, change in transmission efficiency has been attributed to the variation in coat proteins. The region between two conserved peptides, namely GCEGPCKVQS and LYMACTHASN, in the coat proteins of begomoviruses, has been reported to determine their transmission characteristics by whitefly (Wei et al. 2014; Guo et al. 2018; Pan et al. 2018b). Although reports are available to show that mutation of a few residues may lead to loss or gain of transmissibility by a given whitefly species (Noris et al. 1998; Kheyr-Pour et al. 2000; Höhnle et al. 2001; Caciagli et al. 2009), little is known about the key amino acid residue(s) in the coat protein of begomoviruses that determine the interaction between begomoviruses and multiple whitefly species.
Genetic variation of begomoviruses can arise in their ssDNA genomes through mutation, recombination, and pseudo-recombination (Harrison and Robinson 1999; Seal, vandenBosch, and Jeger 2006). While recombination and pseudo-recombination have been shown to generate many new species or new strains of begomoviruses, they cannot occur without coinfection simultaneously with different viruses (Harrison and Robinson 1999; Seal, vandenBosch, and Jeger 2006). Mutations, which result from the activity of DNA polymerase, however, can occur in all begomoviruses. In many cases, mutation rates reported for begomoviruses are equivalent to that of RNA viruses, which are known to have probably the highest mutation rate in all organisms (Ge et al. 2007; Duffy, Shackelton, and Holmes 2008; Fondong and Chen 2011; Yang et al. 2017; Sánchez-Campos et al. 2018). Thus, theoretically, mutation may account for the generation of a large proportion of the genetic variation in begomoviruses. Indeed, through analysis of the genomic sequences of fifteen begomoviruses deposited in GenBank, Lima et al. (2017) found that the genetic variation of these begomoviruses was predominantly driven by mutation. Nevertheless, the biological significance of genetic variation of begomoviruses, especially these generated by mutation, remains largely uncharacterized.
Here, we explore the impact of mutation in the coat protein of begomoviruses on the interaction between viruses and their whitefly vectors. We first characterized mutations in the coat protein of squash leaf curl China virus (SLCCNV), a begomovirus that infects many species of plants in the family Cucurbitaceae, such as Cucurbita moschata and C. pepo, in many regions including Southern China, Pakistan, India, Vietnam, Thailand, and East Timor (Cai et al. 1994; Revill et al. 2003; Ito et al. 2008; Tahir, Haider, and Briddon 2010; Maina et al. 2017; Nagendran et al. 2017). Next, we examined the effects of mutations on SLCCNV infectivity and transmission by three whitefly species. We then identified the key amino acid residue(s) that presumably determine the specificity of SLCCNV transmission by whiteflies. Finally, we examined the competition among virus genotypes during infection and whitefly transmission. Our data provide novel knowledge and understanding with regard to the impact of mutations on the interaction between begomoviruses and their whitefly vectors.
2. Materials and Methods
2.1 Sequence analysis
The full-length amino acid sequences of forty-seven SLCCNV coat proteins, available in the GenBank, were used in the following analysis. Sequence alignment was then performed using DNAMAN 6.0 (Lynnon Corporation, USA). For analysis of mutation frequency (percentage mutation), all amino acid sequences were aligned with that of SLCCNV type isolate (GenBank accession code: AM260206) (Brown et al. 2015), and then mutation frequency for each residue was manually calculated. Since the region between two conserved peptides, namely GCEGPCKVQS (residue 66–75 in SLCCNV coat protein) and LYMACTHASN (residue 228–237), has been shown to determine the whitefly transmission characteristics of begomoviruses (Wei et al. 2014; Guo et al. 2018; Pan et al. 2018b), we analyzed the mutation frequency of amino acid residues between 76 and 227. For the percentage of virus genotypes, the combinations of four amino acid residues exhibiting the highest mutation frequencies were manually checked for each isolate.
2.2 Mutagenesis and construction of infectious clones
For the viruses used, all mutagenesis was performed based on SLCCNV isolate Guangxi2017 (GenBank accession codes: MG525551 [DNA-A] and MG525552 [DNA-B]), whose coat protein shares 100 per cent identity with the SLCCNV type isolate (GenBank accession code: AM260206 [DNA-A] and AM260208 [DNA-B]) (Brown et al. 2015). Briefly, SLCCNV DNA-A was amplified by polymerase chain reaction (PCR) using primers containing BamHI restriction site, namely SL-A-FLF and SL-A-FLR (Supplementary Table S1) and ligated into pGEM®-T Easy Vector (Promega, USA). To obtain the full-length DNA-A of mutant viruses, mutagenesis was performed using Fast Mutagenesis System (Transgen Biotech, China) with primers listed in Supplementary Table S1. The full-length genomes were then used to construct the corresponding infectious clones.
For the construction of infectious clone of SLCCNV DNA-A, 0.3 unit of the genome was released from T vector using EcoRI and BamHI digestion. The released fragment was purified and cloned into the binary vector pBINPLUS (van Engelen et al. 1995) to produce pBINPLUS-0.3A. Then, the complete genome of SLCCNV DNA-A was released from T vector by BamHI digestion and inserted into the unique BamHI site of pBINPLUS-0.3A to produce pBINPLUS-1.3A, which contains an 1.3-mer tandem repeat of SLCCNV DNA-A. For the construction of infectious clone of SLCCNV DNA-B, firstly DNA-B was amplified by PCR using primers containing HindIII restriction site, namely SL-B-FLF and SL-B-FLR (Supplementary Table S1) and ligated into pGEM®-T Easy Vector (Promega). After the verification by Sanger sequencing, 0.9 unit of the full-length genome was amplified with primers SL-B-0.9UF (a SalI restriction site was introduced) and SL-B-FLR. The amplified fragment was purified and digested with SalI and HindIII, and then introduced into the binary vector pBINPLUS to produce pBINPLUS-0.9B. The complete genome of SLCCNV DNA-B was released from T vector by HindIII digestion and then ligated into the unique HindIII restriction site of pBINPLUS-0.9B to produce pBINPLUS-1.9B, which contains an 1.9-mer tandem repeat of SLCCNV DNA-B. The infectious clones of SLCCNV DNA-A and DNA-B were obtained by introducing the plasmids (pBINPLUS-1.3A and pBINPLUS-1.9B) into agrobacteria (Agrobacterium tumefaciens) strain EHA105 by electrotransformation, respectively.
2.3 Plants and insects
Three species/cultivars of plants were used, namely cotton (Gossypium hirsutum L. cv. Zhemian 1793), zucchini (C. pepo cv. Faguodongkui), and squash (C. moschata cv. Mibennangua). All plants were grown in an insect-proof greenhouse under natural lighting and controlled temperature at 25 ± 3 °C. For zucchini and squash, agroinoculation and virus transmission were conducted when the plants reached one or two true leaf stage. In all experiments, zucchini plants were used unless specified otherwise.
Three species of whiteflies of the B. tabaci complex including MEAM1, Asia 1, and Asia II 1 were used. These species were chosen as they are abundant/common in the regions where SLCCNV occurs (De Barro et al. 2011; Hu et al. 2011; Prasanna et al. 2015; Götz and Winter 2016; Global Bemisia dataset version 15 May 2017, doi:10.4225/08/591a4018dfca8; Masood et al. 2017). Whiteflies were originally collected from the field between 2009 and 2012, and then one culture of each species was established and has been maintained thereafter on cotton plants in the laboratory. The mtCOI GenBank accession codes are KM821540 for MEAM1, KC540757 for Asia 1, and DQ309077 for Asia II 1. Every three generations, the purity of each whitefly population was monitored using mtCOI PCR-RFLP and sequencing as described by Qin et al. (2013). All experiments including insect rearing were conducted in insect-proof cages in a climate chamber at 26 ± 2 °C, 60–80 per cent relative humidity and 14/10-h light/dark cycles.
2.4 Agroinoculation and quantification of virus in plants
Agrobacteria containing infectious clones of DNA-A or DNA-B were first cultured separately until OD600 reached 1.0–1.4, and then they were centrifuged and resuspended in resuspension buffer (10 mM MgCl2, 10 mM MES, 200 μM acetosyringone). Resuspended agrobacteria containing equal amount of DNA-A and DNA-B were mixed, and then 1-ml syringes were used to introduce the agrobacteria into both true leaves and cotyledons of zucchini or squash plants. About 4 weeks post agroinoculation, visual inspection of typical symptom (Supplementary Fig. S1) was performed to verify the status of infection. For quantification of virus in plants, DNA was extracted using Plant Genomic DNA Kit (Tiangen, China) and real-time PCR was performed using SYBR Premix Ex Taq II (Takara, Japan) and CFX96™ Real-Time PCR Detection System (Bio-Rad, USA) with primers SL-A-RTF and SL-A-RTR for SLCCNV and Zu-Actin-F and Zu-Actin-R for zucchini actin (Supplementary Table S1).
2.5 Virus acquisition and transmission
For each of the three whitefly species, newly emerged whitefly adults (0–3 days) were collected and placed on virus-infected plants for virus acquisition. In the experiment using zucchini plants as the source of inoculum, we randomly collected ten female adults at designated acquisition access periods (AAPs), namely 0, 24, 48, 72, and 96 h, and then the adults were individually subjected to DNA extraction and PCR detection of SLCCNV. Total DNA was extracted with lysis buffer (50 mmol/l KCl, 10 mmol/l Tris, 0.45 per cent Tween20, 0.2 per cent gelatin, 0.45 per cent NP40, and 60 mg/ml protease K with pH at 8.4) as described in Pan et al. (2018a). Later, these samples were assayed for detectable SLCCNV DNA by PCR using SL-A-PCRF and SL-A-PCRR primers (Supplementary Table S1). Infected zucchini plants and squash plants were used as source of inoculum when zucchini plants and squash plants were used as test plants, respectively.
For virus transmission, cohorts of female adults of a given number, as designated in the design, were collected after 96 h virus acquisition and then caged on test plants to feed for a designated virus inoculation access periods. Unless specified otherwise, the number of whitefly adults used per test plant was 5, and the inoculation access period was 96 h. The leaf clip cages for enclosure of whiteflies on test plants are described in Ruan et al. (2007) . Imidacloprid (20 mg/l) was sprayed onto plants to kill the eggs after adult whiteflies were removed. When zucchini plants were used as test plants, for each combination of virus genotype and whitefly species, three replicates were conducted with each containing five to twelve plants. As for squash plants, nine to twelve plants were used for each combination of virus genotype and whitefly species. The test plants were cultured for another 4 weeks, and then virus infection status of the test plants was determined by PCR and symptom inspection (Supplementary Fig. S1).
2.6 Quantification of SLCCNV in whitefly whole body, honeydew, and organs
For virus quantification in whitefly whole body, adult whiteflies were collected in groups of twenty-five at each of the designated AAPs, and then DNA was extracted using lysis buffer as mentioned before. Samples of whitefly honeydew and organs were prepared as per Pan et al. (2018a,b). For honeydew, briefly, whiteflies were collected as groups of twenty each, and then released into leaf clip cages that were placed on the undersurface of zucchini leaves. The bottom of the leaf clip cages was covered with aluminum foil to collect honeydew produced by whiteflies during feeding. The honeydew was then collected with phosphate buffer saline and viral DNA was extracted using PureLink Viral RNA/DNA Mini Kit (Invitrogen, USA). Real-time PCR analysis was performed for these honeydew samples, and the copy number of virus in whitefly honeydew samples was calculated as normalized to a standard curve, which was made by real-time PCR reaction using serial dilution of plasmid extracted from the infectious clone of SLCCNV DNA-A. For organs, four midguts or primary salivary glands (PSGs) were collected as one sample and hemolymph of four whiteflies was collected as one sample. The number of replicates was 3–5 for whitefly whole body and 7–12 for organs. Real-time PCR was performed with primers SL-A-RTF and SL-A-RTR for SLCCNV and WF-Actin-F and WF-Actin-R for whitefly actin (Supplementary Table S1).
2.7 Immunofluorescence detection of SLCCNV in whitefly midguts and PSGs
Immunofluorescence detection of viruses was performed as described in Wei et al. (2014) with some modifications. At each of the designated AAPs, intact midguts and PSGs were dissected in phosphate buffer saline and incubated in 4 per cent paraformaldehyde (MultiSciences Biotech., China) at room temperature for 1 h. The samples were then washed and incubated in 0.2 per cent Triton X-100 for 30 min and further blocked with 1 per cent bull serum albumin dissolved in TBS-Tween 20 for 1 h at room temperature, whereafter anti-TYLCV CP monoclonal antibodies (provided by Professor Xueping Zhou, Institute of Biotechnology, Zhejiang University) were added with a dilution of 1:400, and the samples were incubated overnight at 4 °C. The next day, the samples were washed and incubated with 549-conjugated secondary antibodies (Earthox, China) with a dilution of 1:400. After 2-h incubation at room temperature, all samples were washed and placed on slides for inspection. The viral signal was examined under a Zeiss LSM710 confocal microscope (Zeiss, Germany).
2.8 Virus competition assay
To obtain plants infected by viruses of two genotypes, equal amounts of two resuspended infectious clones were mixed, and then agroinoculation was performed as mentioned above. Next, virus acquisition was conducted using plants infected by mixed infectious clones as the source of inoculum and virus transmission was then performed as described above. The percentage of each genotype in plants was determined as described by Tsetsarkin et al. (2007) with modification. First, genomic DNA was extracted from agroinoculated or whitefly-inoculated plants, and then DNA samples were subjected to PCR using SL-A-FLF containing a BamHI restriction site and SL-A-XbaI-R containing an XbaI restriction site. The PCR products were purified with QIAquick Gel Extraction Kit (QIAGEN, German) and subjected to BamHI and XbaI digestion. pBINPLUS plasmids were digested with the same enzymes and purified. Ligation was performed and the ligation product was transformed into Escherichia coli strain DH5α, and the recombinant plasmids from ten to seventeen clones of Escherichia coli were extracted and sequenced. The sequencing results were aligned using DNAMAN 6.0 (Lynnon Corporation), and the percentages of the two virus genotypes in each sample were manually calculated.
2.9 Statistical analysis
For the comparison of transmission efficiency, percentage data were arcsine square root transformed, and one-way analysis of variance (ANOVA) along with Fisher’s least significant difference tests was used. Kruskal–Wallis test was used to analyze the significance of differences in quantity of virus. All real-time data were calculated using 2−△Ct as normalized to plant or whitefly actin. To illustrate the differences between different whitefly species in a straightforward manner, the data of virus quantity in whitefly whole body, honeydew, and organs in each of the experiments were normalized to that of MEAM1 at each of the time points or whitefly organs. All data are presented as the mean ± standard errors of mean (mean ± SEM) and differences between treatments are considered significant when P < 0.05. All statistical analyses were conducted using SPSS 20.0 Statistics and EXCEL.
3. Results
3.1 Mutations in the coat protein of SLCCNV
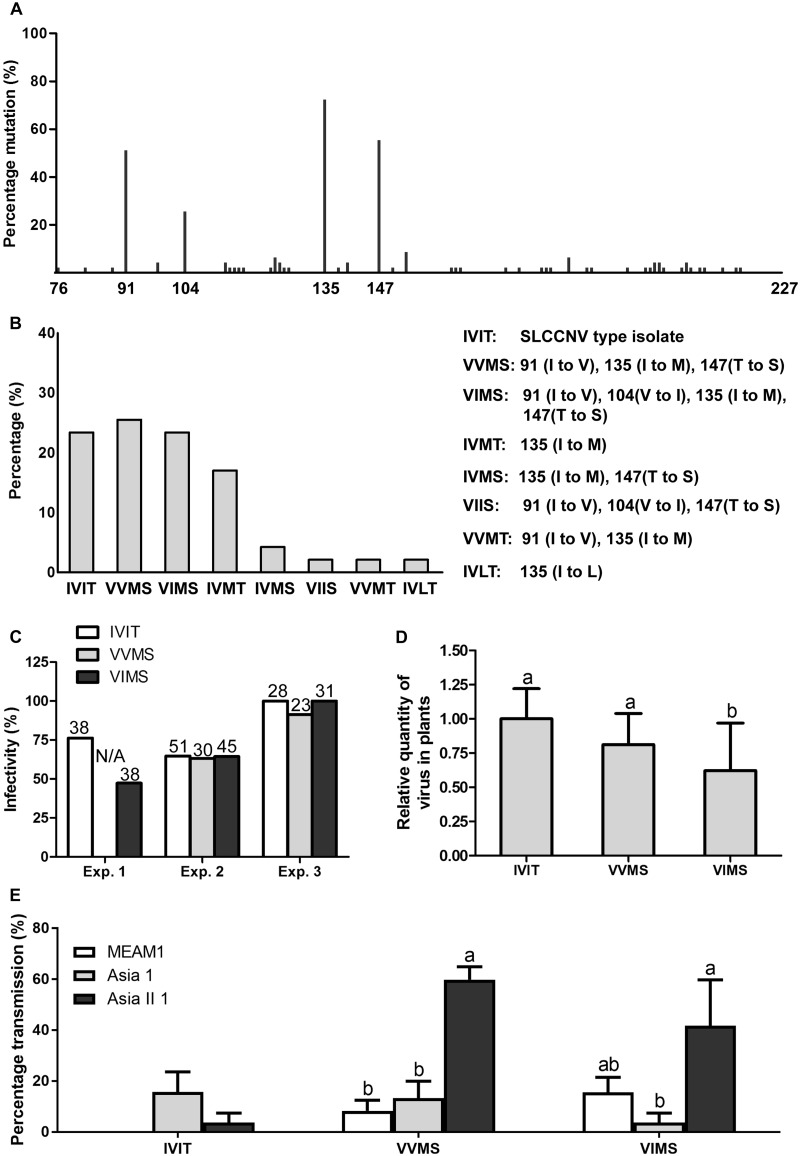
The mutation frequencies of each amino acid residue in SLCCNV coat protein were shown in Supplementary Fig. S2. As for the amino acid residues between 76 and 227 in the coat protein of SLCCNV, extensive divergence of mutation frequencies was found for different residues ranging from 0 to 72.3 per cent, and among them, four residues had the highest mutation frequencies, namely 91 (51.1 per cent), 104 (25.5 per cent), 135 (72.3 per cent), and 147 (55.3 per cent) (Fig. 1A). Next, we summarized all the combinations of these four residues in all SLCCNV isolates and found that three combinations had the highest abundance, namely IVIT (23.4 per cent), VVMS (25.5 per cent), and VIMS (23.4 per cent), accounting for over 70 per cent of all isolates analyzed (Fig. 1B). For these combinations, IVIT represented the combination of the four residues in the type isolate of SLCCNV, whereas VVMS represented a combination with a triple mutation at residues 91, 135, and 147, and VIMS represented a combination with a quadruple mutation at residues 91, 104, 135, and 147.
Figure 1.
Mutations in the coat protein of SLCCNV and their effects on infectivity and transmission of the virus by three whitefly species. Percentage of isolates divergent from the type isolate at each residue in all isolates (A), percentage of isolates exhibiting certain combinations in all isolates (B), percentage of symptomatic plants in all plants inoculated (C), data of relative quantities of virus in fifteen to seventeen symptomatic plants are normalized to that of IVIT and presented as mean ± SEM (D), and mean ± SEM of the percentage of virus-infected plants (symptomatic and PCR positive) in all plants tested (E). Figures above columns in (C) show the number of plants inoculated and N/A stands for not analyzed. Different letters above the columns in (D) and (E) indicate significant differences (Kruskal–Wallis test for (D), and one-way ANOVA for (E), P < 0.05).
3.2 Effects of mutations on virus infectivity and whitefly transmission
VVMS and VIMS induced similar symptoms on plants as IVIT (data not shown). The triple mutation (VVMS) did not influence the infectivity and quantity of virus in plants, whereas the quadruple mutation (VIMS) appeared to exert a deleterious effect on virus infection as the infectivity of VIMS was reduced in one of the three experiments and quantity of VIMS in plants was lower than that of IVIT (Fig. 1C and D). Similarly, when squash plants were used in the tests, the infection rate of plants by VIMS was 48 per cent, lower than that (71 per cent) by VVMS (Table 1).
Table 1.
Infectivity of SLCCNV genotypes and their transmission efficiency by three species of whiteflies of the B. tabaci complex when squash (C. moschata) plants were used as test plants.
| Virus genotype | Infectivitya | Transmission efficiencyb |
||
|---|---|---|---|---|
| MEAM1 | Asia 1 | Asia II 1 | ||
| IVIT | 11/20 | 0/9 | 6/11 | 4/11 |
| VVMS | 17/24 | 0/11 | 1/12 | 7/12 |
| VIMS | 13/27 | 1/10 | 1/9 | 5/10 |
aInfectivity is presented as the number of virus-infected (symptomatic) plants/total number of plants inoculated.
bTransmission efficiency is presented as virus-infected (symptomatic and PCR positive) plants/total number of plants tested.
Prior to transmission tests, virus acquisition was analyzed to identify the time point when whiteflies were maximally infected. When whiteflies were allowed to feed on infected plants for 96 h, all the three species became viruliferous (Supplementary Table S2). Consequently, 96 h was chosen as virus AAP for the transmission experiment. The transmission efficiencies of the three virus genotypes differed among the three whitefly species in the order of Asia 1 > Asia II 1 > MEAM1 for IVIT, Asia II 1 > Asia 1 > MEAM1 for VVMS, and Asia II 1 > MEAM1 > Asia 1 for VIMS (Fig. 1E). Although differences in transmission efficiency of IVIT between whitefly species were apparent, no statistical significance was detected. To examine whether these difference were of biological significance, two more transmission experiments were performed with different conditions and the data showed that IVIT was transmitted most efficiently by Asia 1, followed by Asia II 1 and MEAM1 (Supplementary Fig. S3). A similar pattern of transmission efficiency was found among the three species of whitefly when squash plants were used as test plants (Table 1).
3.3 Quantities and presence of SLCCNV genotype IVIT in whitefly
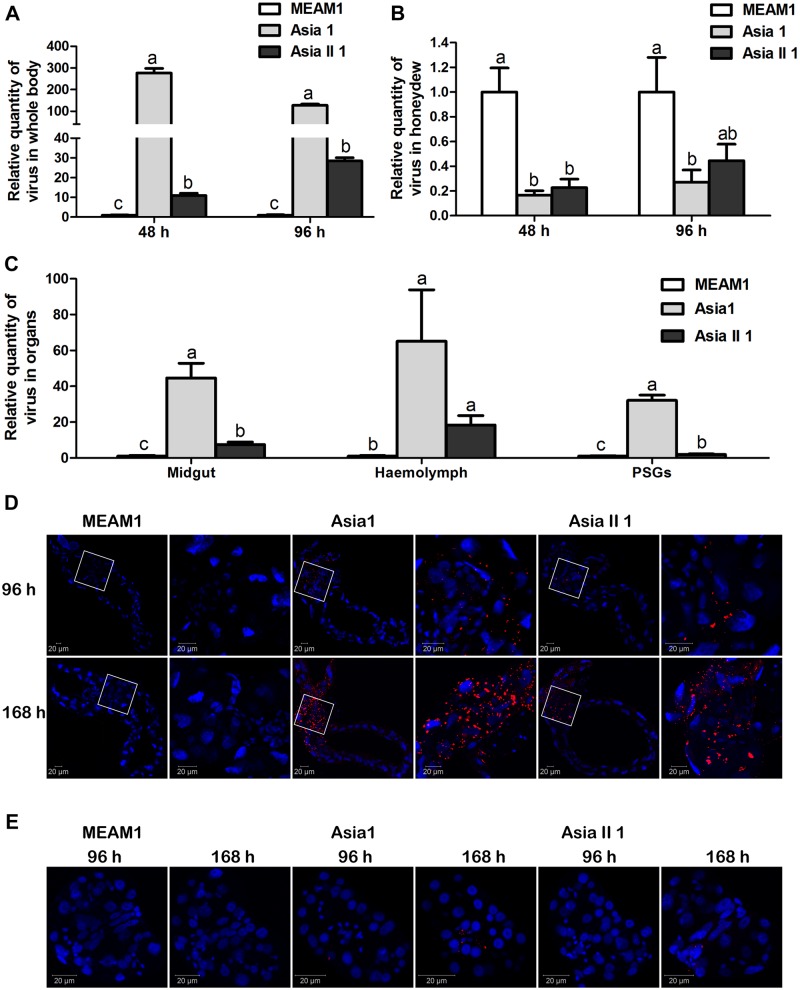
After 48- and 96-h feeding for virus acquisition, the relative quantity of virus in whitefly whole body differed significantly among the three whitefly species in the order of Asia 1 > Asia II 1 > MEAM1 (Fig. 2A), whereas the relative quantity of virus in honeydew among the whitefly species was found to be the opposite, although no significant difference was found between Asia 1 and Asia II 1 at either time point (Fig. 2B). It should be noted that the quantities of virus in the whole body of MEAM1, Asia 1, and Asia II 1 whiteflies increased with the increase of feeding time (data not shown), the pattern of which is not apparent in Fig. 2A as all data were normalized to that of MEAM1 at each of the time points. For organs, after 96-h virus acquisition, the relative quantity of virus in midguts, hemolymph, and PSGs likewise differed significantly among the three whitefly species in the order of Asia 1 > Asia II 1 > MEAM1 (Fig. 2C). Next, after MEAM1, Asia 1, and Asia II 1 whiteflies were given 96- and 168-h virus acquisition feeding, SLCCNV signals were detected in the midguts and PSGs using immunofluorescence. For midguts, no viral specific signal was detected in MEAM1 at the two designated time points, whereas for Asia 1 and Asia II 1, clear viral signals were found, and viral signals in midguts from Asia 1 were stronger than that of Asia II 1 at the two designated time points (Fig. 2D). In both Asia 1 and Asia II 1, viral signals increased with the increase of time for feeding, and the signals were enriched in the filter chamber. For PSGs, no viral specific signal was found in MEAM1 at the two designated time points. For Asia 1, clear viral signals were found after 96-h feeding and viral signals become stronger with the increase of time for feeding, whereas for Asia II 1, clear viral signals were found after 168-h feeding, and at this time point, signals in PSGs from Asia 1 were stronger than that of Asia II 1 (Fig. 2E). In both Asia 1 and Asia II 1, viral signals were specifically located in the central secretory regions along the ducts of PSG.
Figure 2.
Quantity of SLCCNV genotype IVIT in whole body, honeydew, and organs of MEAM1, Asia 1, and Asia II 1 whiteflies and immunofluorescence detection of virus in whitefly organs. Relative quantities of virus in whitefly whole body (A) or in honeydew (B) in each of the three species of whiteflies at each of the two time intervals of virus acquisition; relative quantities of virus in midguts, hemolymph, and PSGs (C) of the three species of whiteflies after 96-h feeding on virus-infected plants; viruses in midgut (D) and PSGs (E) of each of the three species of whiteflies after feeding on virus-infected plants for 96 and 168 h, as shown by immunofluorescence (nuclear: blue, virus: red). Data of relative quantities of virus in (A)–(C) are normalized to that of MEAM1 at each of the time points or whitefly organs and presented as mean ± SEM. Different letters above columns in each of the diagrams indicate significant differences (Kruskal–Wallis test, P < 0.05).
3.4 Quantities of SLCCNV genotypes VVMS and VIMS in whitefly
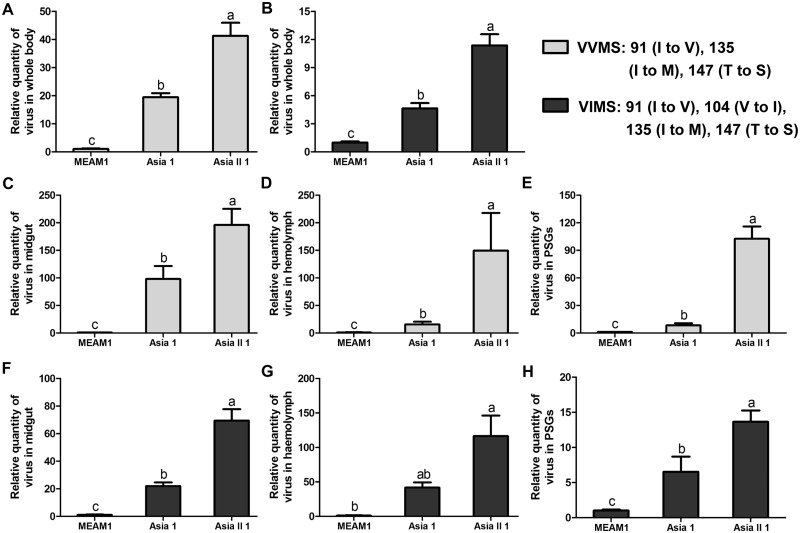
For both VVMS and VIMS, the relative quantities of virus in whitefly whole body differed significantly among the three whitefly species in the order of Asia II 1 > Asia 1 > MEAM1 (Fig. 3A for VVMS and Fig. 3B for VIMS). Also, the quantity of virus in midguts, hemolymph, and PSGs was analyzed, and the pattern was found to be similar to that of whole body (Fig. 3C–E: quantity of VVMS in midguts, hemolymph, and PSGs, respectively; Fig. 3F–H: quantity of VIMS in midguts, hemolymph, and PSGs, respectively).
Figure 3.
Quantities of SLCCNV genotypes VVMS and VIMS in the whole body and organs of MEAM1, Asia 1, and Asia II 1 whiteflies after feeding on virus-infected plants for 96 h. Quantities of VVMS in the whole body (A), midgut (C), hemolymph (D), and PSGs (E); Quantities of VIMS in the whole body (B), midgut (F), hemolymph (G), and PSGs (H). Data of relative quantities of virus in each of the diagrams are normalized to that of MEAM1 and presented as mean ± SEM. Different letters above columns in each of the diagrams indicate significant differences (Kruskal–Wallis test, P < 0.05).
3.5 Key amino acid residue(s) for altering SLCCNV transmission by whiteflies
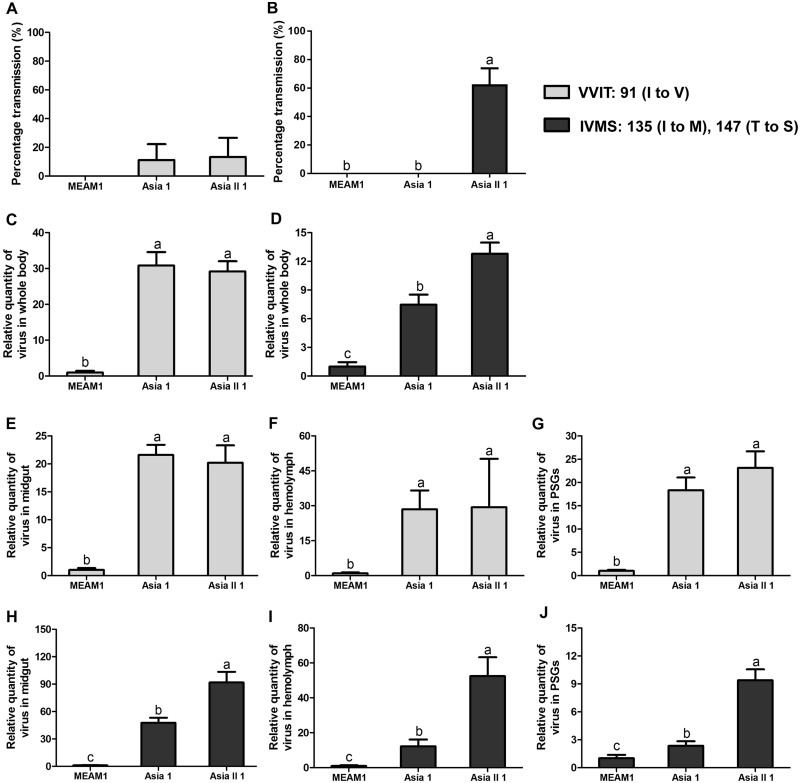
As mutation of three amino acids in the coat protein of SLCCNV changed the pattern of transmission efficiency of SLCCNV by three whitefly species and the capacity of viruses to transport across the midgut wall of different whitefly species (Table 1 and Figs 1–3), we then sought to identify the key amino acids residue(s) that was responsible for the altered whitefly transmission pattern. We first constructed two mutant viruses, namely VVIT and IVMS, of which the former has a mutation at residue 91 (I to V), and the latter has a double mutation at 135 (I to M) and 147 (T to S). While the transmission efficiency of VVIT by Asia 1 and Asia II 1 appeared similar (Fig. 4A), the transmission efficiency of IVMS by Asia II 1 was higher than that of Asia 1 (Fig. 4B). In addition, when whiteflies were allowed to acquire the viruses for 96 h, the quantity of virus in the whole body, midguts, hemolymph, and PSGs was analyzed, and the pattern was found to mirror that of transmission efficiency (Fig. 4C and E–G: quantity of VVIT in whole body, midguts, hemolymph, and PSGs respectively; Fig. 4D and H–J: quantity of IVMS in whole body, midguts, hemolymph, and PSGs, respectively).
Figure 4.
Transmission efficiency of SLCCNV genotype VVIT and IVMS by whiteflies and quantity of virus in whitefly whole body and organs. Transmission efficiency of VVIT (A) and quantities of VVIT in the whole body (C), midguts (E), hemolymph (F), and PSGs (G); transmission efficiency of IVMS (B) and quantities of IVMS in the whole body (D), midguts (H), hemolymph (I), and PSGs (J). Data of percentage transmission in (A) and (B) are presented as mean ± SEM, and data of relative quantities of virus in (C)–(J) are normalized to that of MEAM1 and presented as mean ± SEM. Different letters above the columns in each of the diagrams indicate significant differences (one-way ANOVA for (A) and (B), and Kruskal–Wallis test for (C)–(J), P < 0.05).
Since double mutation at residues 135 and 147 change the transmission efficiency of SLCCNV from Asia 1 > Asia II 1 to Asia 1 < Asia II 1, we next sought to examine whether mutation of a single amino acid residue was sufficient to induce the change of whitefly transmission. To this end, two mutant viruses, namely IVMT and IVIS, of which the former has a mutation at residue 135 (I to M) and the latter has a mutation at 147 (T to S), were constructed. IVMT seemed to be similar to IVIT, the type isolate with regard to whitefly transmission characteristics as the transmission efficiency of IVMS was in the order of Asia 1 > Asia II 1 > MEAM1 (Fig. 5A). IVIS seemed to be similar to VVMS with regard to whitefly transmission characteristics as the transmission efficiency of IVIS was in the order of Asia II 1 > Asia 1 > MEAM1 (Fig. 5B). The quantity of virus in the whole body, midguts, hemolymph, and PSGs was analyzed, and the pattern was analogous to that of transmission efficiency (Fig. 5C and E–G: quantity of IVMT in whole body, midguts, hemolymph, and PSGs respectively; Fig. 5D and H–J: quantity of IVIS in whole body, midguts, hemolymph, and PSGs, respectively).
Figure 5.
Transmission efficiency of SLCCNV genotype IVMT and IVIS by whiteflies and quantity of virus in whitefly whole body and organs. Transmission efficiency of IVMT (A) and quantities of IVMT in the whole body (C), midguts (E), hemolymph (F), and PSGs (G); transmission efficiency of IVIS (B) and quantities of IVIS in the whole body (D), midguts (H), hemolymph (I), and PSGs (J). Data of percentage transmission in (A) and (B) are presented as mean ± SEM, and data of relative quantities of virus in (C)–(J) are normalized to that of MEAM1 and presented as mean ± SEM. Different letters above the columns in each of the diagrams indicate significant differences (one-way ANOVA for (A) and (B), and Kruskal–Wallis test for (C)–(J), P < 0.05).
3.6 Competition between IVIT and VIMS during infection and transmission
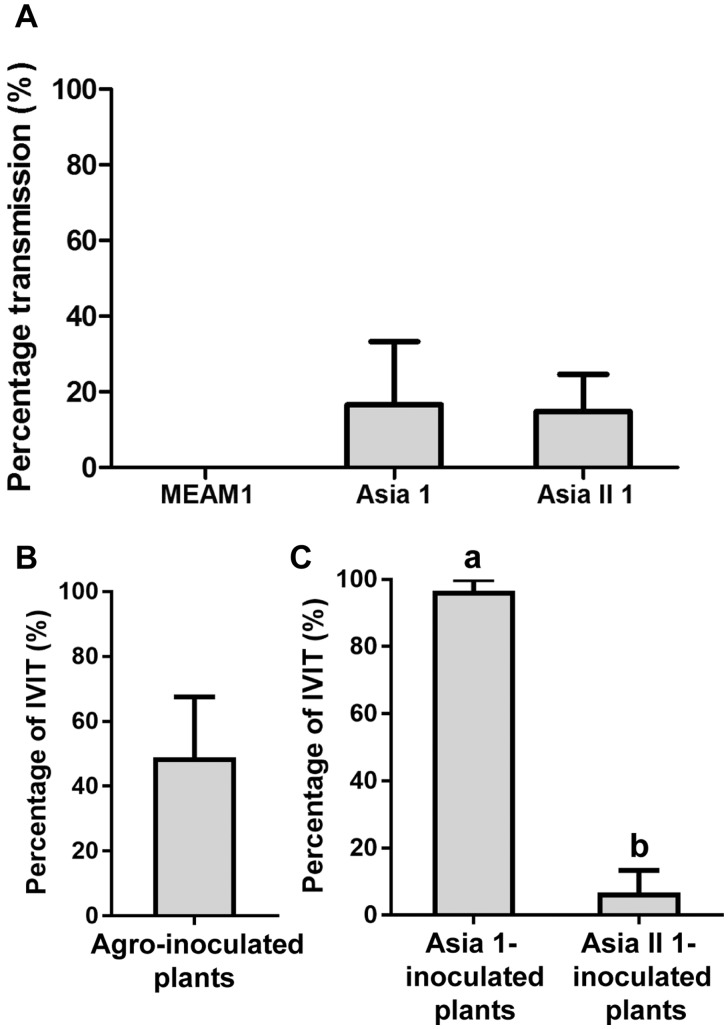
To explore the effects of mutations in the coat protein of SLCCNV on viral fitness, a viral competition assay was performed. First, mixed infection of SLCCNV genotypes IVIT and VIMS in zucchini plants was achieved by agroinoculation, and then the plants were presented to whiteflies to feed, which were later collected for virus transmission. While no transmission was found for MEAM1, similar transmission efficiency was found for Asia 1 and Asia II 1 (Fig. 6A). In agroinoculated plants, 48.5 per cent of the viruses were found to be IVIT, indicating a corresponding percentage of 51.5 per cent for VIMS (Fig. 6B). However, in plants inoculated by whiteflies, significant difference in virus percentage composition was found. For plants inoculated by Asia 1, 96.7 per cent of the viruses were found to be IVIT, while for plants inoculated by Asia II 1, only 6.7 per cent of the viruses were found to be IVIT (Fig. 6C).
Figure 6.
Transmission efficiency by whiteflies when zucchini plants infected by a mixed-virus infectious clone were used as source of inoculum and percentage of IVIT in plants. Efficiency of virus transmission by each of the three species of whiteflies MEAM1, Asia 1, and Asia II 1 (A); percentages of IVIT in the mixture of IVIT + VIMS in agroinoculated (B) and Asia 1-inoculated and Asia II 1-inoculated plants (C). Data are mean ± SEM and different letters above the columns in (A) and (C) indicate significant differences (one-way ANOVA, P < 0.05).
4. Discussion
In the present study, we first characterized the mutations in the residues between 76 and 227 in the coat protein of SLCCNV and found that several residues (91, 104, 135, and 147) exhibited much higher mutation frequencies than the others (Fig. 1A). Next, all combinations of amino acids in these residues, herein genotypes, were summarized (Fig. 1B) and three most abundant genotypes (IVIT, VVMS, and VIMS) were compared for their infectivity and whitefly transmission efficiency. While only a marginal difference was found for infectivity, these genotypes exhibited dramatic divergence in whitefly transmission; transmission efficiencies were in the order of Asia 1 > Asia II 1 > MEAM1 for IVIT, Asia II 1 > Asia 1 > MEAM1 for VVMS, and Asia II 1 > MEAM1 > Asia 1 for VIMS (Table 1 and Fig. 1C–E). The exploration of the key residue(s) responsible for the change of whitefly transmission revealed that a T to S substitution at residue 147 was sufficient to induce the change of whitefly transmission between Asia 1 and Asia II 1 (Figs 4 and 5). For all the combinations of virus genotypes and whitefly species that were used in the transmission experiments, quantification of virus in whitefly whole body and organs was performed, and there was a positive correlation between transmission efficiency and quantity of virus in whitefly whole body and organs except in one case (VIMS) (Figs 2–5). In our competition assay with two genotypes, namely IVIT and VIMS, their percentages in agroinoculated plants were similar, but in whitefly-inoculated plants the percentages were divergent with IVIT being dominant in Asia 1-inoculated plants and VIMS being dominant in Asia II 1-inoculated plants (Fig. 6).
Here, we found naturally occurring high frequencies of mutation in only a few amino acid residues in the coat protein of SLCCNV, a begomovirus. The mutation rates of begomoviruses have been reported to be in the order of 10−4 substitutions per site per year (subs/site/year), which were comparable to that reported in RNA viruses that are reported to have the highest mutation rates among all organisms (Ge et al. 2007; Fondong and Chen 2011; Yang et al. 2017; Sánchez-Campos et al. 2018). Moreover, the mutation of begomoviruses was supposed to be generated by the activity of DNA polymerase and is thus a random process (Duffy, Shackelton, and Holmes 2008). Therefore, the abundant presence of virus genotypes harboring these mutant residues suggests that they should confer considerable selective advantage under certain conditions. Here, our data showed that when viruses of different genotypes are transmitted by three whitefly species, their competence of transmission varies. Our competition assay showed that transmission by certain whitefly species favored the dominance of the most competent genotype. So together we suggest that these different genotypes of SLCCNV were generated by mutations and the persistence of these genotypes in the field might be a result of selection by different whitefly species.
This hypothesis suggests that there should be a clear association between whitefly species and virus genotypes in the field in most, if not all cases. Indeed, the limited data with regard to the association between whitefly species and genotype of SLCCNV seem to support this hypothesis. On the squash plants infected by SLCCNV isolate Guangxi2017 (GenBank accession codes for DNA-A: MG525551) (genotype: IVIT) in Guangxi, China, the whitefly population when sampled was dominated by Asia 1, the most competent whitefly vector of IVIT (of the fourteen whiteflies collected, ten were Asia 1, only three were Asia II 1, and one was Asia II 6) (data not shown). In Lahore, Punjab province, Pakistan, where Asia II 1 predominates (Masood et al. 2017), the only characterized virus isolate was found to be genotype VVMS (GenBank accession codes for DNA-A: AM286794), the most competent genotype for Asia II 1 (Tahir, Haider, and Briddon 2010). Of course, more detailed surveys of whitefly species and genotype of SLCCNV in the field should be carried out to further test our hypothesis. In addition, the evidence currently available does not rule out other unknown mechanisms involved in the persistence of these virus genotypes in the field.
Begomoviruses have long been observed to exhibit a geography-related other than host plant-related antigenic variation, based on which Harrison and Robinson (1999) suggested that this pattern of variation was caused by selection pressure exerted by the whitefly vector. This hypothesis was further supported by the phylogeographic concordance between whitefly mitochondrial cytochrome oxidase I (mtCOI) and coat proteins of begomoviruses (Brown and Idris 2005). At the same time, accumulating empirical evidence shows that while transmission efficiency varies for different combinations of virus and whitefly species/population, the begomovirus and whitefly species/population combinations that show the highest transmission efficiency tend to share a common geographical origin (Idris, Smith, and Brown 2001; Maruthi et al. 2002; Li et al. 2010; De Marchi et al. 2017; Pan et al. 2018a,b). For example, Maruthi et al. (2002) found that transmission efficiency was high when B. tabaci and cassava begomovirus isolates were collected from the same location, either Africa or India, were used in the transmission experiment, and much lower transmission efficiency was found when B. tabaci and virus isolates collected from different locations were used. Later, more case studies using specific viruses such as Tobacco curly shoot virus and Cotton leaf curl Multan virus further supported this claim (Idris, Smith, and Brown 2001; Li et al. 2010; De Marchi et al. 2017; Pan et al. 2018a,b). Here, our results showed that mutations in the coat protein of SLCCNV were able to change the whitefly transmission characteristics and by doing so allow SLCCNV to be readily vectored by different whitefly species. So combined, we suggest that mutations coupled with selection pressure exerted by whitefly may play important roles in shaping the evolutionary outcome of begomoviruses.
We found that the alteration of a single amino acid residue in the coat protein of SLCCNV was sufficient for changing its transmission characteristic by different whitefly species. Previously, mutations of several amino acid residues in coat protein were shown to lead to loss or gain of whitefly transmissibility in several begomoviruses (Noris et al. 1998; Kheyr-Pour et al. 2000; Höhnle et al. 2001; Caciagli et al. 2009). For example, a double mutation at residue 129 (Q129P) and 134 (Q134H) in coat protein of Tomato leaf curl Sardinia virus resulted in loss of whitefly transmissibility (Noris et al. 1998). And in Abutilon mosaic virus, mutation of two amino acids at residues 124 (Q124K) and 149 (H149Q) was sufficient to render Abutilon mosaic virus whitefly transmissible, and a further change at residue 174 (L174M) rendered the virus readily transmissible by whitefly (Höhnle et al. 2001). Since only a very limited number of studies are available, further exploration is warranted before drawing any conclusions with regard to the key residue(s) that determine the transmission of begomoviruses by whitefly. We found that the variation of transmission efficiencies of these virus genotypes by three whitefly species was associated with the differential efficiency of viruses to cross the midgut wall of different species of whiteflies. Further studies are warranted on the molecular mechanism underlying the differential capacity of viruses of different species/genotypes to cross the midgut of different whitefly species.
Taken together, we have found that naturally occurring mutations in the coat protein of a begomovirus significantly changed the interaction between the virus and different whitefly species, and we identified the key amino acid residue in the coat protein involved in differential begomovirus transmission by different whitefly species. Our results highlight the role of mutation in the evolution of begomoviruses, an important group of plant viruses, and open a new avenue for understanding whitefly-begomovirus interactions.
Data availability
Data in this study are available from the authors upon request.
Supplementary Material
Acknowledgements
We thank Myron Zalucki, the University of Queensland, Australia, for discussion and comments on an earlier version of this manuscript. Financial support for this study was provided by the China Postdoctoral Science Foundation (2018M642451).
Conflict of interest: None declared.
References
- Briddon R. W. et al. (2014) ‘Effects of Genetic Changes to the Begomovirus/Betasatellite Complex Causing Cotton Leaf Curl Disease in South Asia Post-Resistance Breaking’, Virus Research, 186: 114–9. [DOI] [PubMed] [Google Scholar]
- Brown J. K., Idris A. M. (2005) ‘Genetic Differentiation of Whitefly Bemisia tabaci Mitochondrial Cytochrome Oxidase I, and Phylogeographic Concordance with the Coat Protein of the Plant Virus Genus Begomovirus’, Annuals of the Entomological Society of America, 98: 827–37. [Google Scholar]
- Brown J. K. et al. (2015) ‘Revision of Begomovirus Taxonomy Based on Pairwise Sequence Comparisons’, Archives of Virology, 160: 1593–619. [DOI] [PubMed] [Google Scholar]
- Caciagli P. et al. (2009) ‘Virion Stability Is Important for the Circulative Transmission of Tomato Yellow Leaf Curl Sardinia Virus by Bemisia tabaci, but Virion Access to Salivary Glands Does Not Guarantee Transmissibility’, Journal of Virology, 83: 5784–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J. H. et al. (1994) ‘Studies on Chinese Squash Leaf Curl Virus: Biological and Serological Properties and Molecular Hybridization’ [in Chinese with English abstract], Virologica Sinica, 9: 222–5. [Google Scholar]
- De Barro P. J. et al. (2011) ‘Bemisia tabaci: A Statement of Species Status’, Annual Review of Entomology, 56: 1–19. [DOI] [PubMed] [Google Scholar]
- De Marchi B. R. et al. (2017) ‘Comparative Transmission of Five Viruses by Bemisia tabaci NW2 and MEAM1’, Tropical Plant Pathology, 42: 495. [Google Scholar]
- Domingo E., Sheldon J., Perales C. (2012) ‘Viral Quasispecies Evolution’, Microbiology and Molecular Biology Reviews, 76: 159–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S., Shackelton L. A., Holmes E. C. (2008) ‘Rates of Evolutionary Change in Viruses: Patterns and Determinants’, Nature Reviews Genetics, 9: 267–276. [DOI] [PubMed] [Google Scholar]
- Fiallo-Olivé E. et al. (2020) ‘Transmission of Begomoviruses and Other Whitefly-Borne Viruses: Dependence on the Vector Species’, Phytopathology, 110: 10–7. [DOI] [PubMed] [Google Scholar]
- Fondong V. N., Chen K. (2011) ‘Genetic Variability of East African Cassava Mosaic Cameroon Virus under Field and Controlled Environment Conditions’, Virology, 413: 275–82. [DOI] [PubMed] [Google Scholar]
- Ge L. M. et al. (2007) ‘Genetic Structure and Population Variability of Tomato Yellow Leaf Curl China Virus’, Journal of Virology , 81: 5902–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanim M., Morin S., Czosnek H. (2001) ‘Rate of Tomato Yellow Leaf Curl Virus Translocation in the Circulative Transmission Pathway of Its Vector, the Whitefly Bemisia tabaci’, Phytopathology, 91: 188–96. [DOI] [PubMed] [Google Scholar]
- Götz M., Winter S. (2016) ‘Diversity of Bemisia tabaci, in Thailand and Vietnam and Indications of Species Replacement’, Journal of Asia-Pacific Entomology, 19: 537–43. [Google Scholar]
- Guo T. et al. (2018) ‘The Level of Midgut Penetration of Two Begomoviruses Affects Their Acquisition and Transmission by Two Species of Bemisia tabaci’, Virology, 515: 66–73. [DOI] [PubMed] [Google Scholar]
- Harrison B. D. (2002) ‘Virus Variation in Relation to Resistance-Breaking in Plants’, Euphytica, 124: 181–92. [Google Scholar]
- Harrison B. D., Robinson D. (1999) ‘Natural Genomic and Antigenic Variation in Whitefly-Transmitted Geminiviruses (Begomoviruses)’, Annual Review of Phytopathology, 37: 369–98. [DOI] [PubMed] [Google Scholar]
- Hogenhout S. A. et al. (2008) ‘Insect Vector Interactions with Persistently Transmitted Viruses’, Annual Review of Phytopathology, 46: 327–59. [DOI] [PubMed] [Google Scholar]
- Höhnle M. et al. (2001) ‘Exchange of Three Amino Acids in the Coat Protein Results in Efficient Whitefly Transmission of a Nontransmissible Abutilon Mosaic Virus Isolate’, Virology, 290: 164–71. [DOI] [PubMed] [Google Scholar]
- Hu J. et al. (2011) ‘An Extensive Field Survey Combined with a Phylogenetic Analysis Reveals Rapid and Widespread Invasion of Two Alien Whiteflies in China’, PLoS One, 6: e16061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idris A. M., Smith S. E., Brown J. K. (2001) ‘Ingestion, Transmission, and Persistence of Chino Del Tomate Virus (CdTV), a New World Begomovirus, by Old and New World Biotypes of the Whitefly Vector Bemisia tabaci’, Annals of Applied Biology, 139: 145–54. [Google Scholar]
- Ito T. et al. (2008) ‘Yellow Leaf Curl Disease of Pumpkin in Thailand Is Associated with Squash Leaf Curl China Virus’, Plant Pathology, 57: 766. [Google Scholar]
- Jones R. (2009) ‘Plant Virus Emergence and Evolution: Origins, New Encounter Scenarios, Factors Driving Emergence, Effects of Changing World Conditions, and Prospects for Control’, Virus Research, 141: 113–30. [DOI] [PubMed] [Google Scholar]
- Kheyr-Pour A. et al. (2000) ‘Watermelon Chlorotic Stunt Virus from the Sudan and Iran: Sequence Comparisons and Identification of a Whitefly-Transmission Determinant’, Phytopathology, 90: 629–35. [DOI] [PubMed] [Google Scholar]
- Lefeuvre P. et al. (2019) ‘Evolution and Ecology of Plant Viruses’, Nature Reviews Microbiology, 17: 632–44. [DOI] [PubMed] [Google Scholar]
- Li M. et al. (2010) ‘Transmission of Tomato Yellow Leaf Curl Virus by Two Invasive Biotypes and a Chinese Indigenous Biotype of the Whitefly Bemisia tabaci’, International Journal of Pest Management, 56: 275–80. [Google Scholar]
- Lima A. T. M. et al. (2017) ‘The Diversification of Begomovirus Populations Is Predominantly Driven by Mutational Dynamics’, Virus Evolution, 3: vex005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. S., Colvin J., De Barro P. J. (2012) ‘Species Concepts as Applied to the Whitefly Bemisia tabaci Systematics: How Many Species Are There?’, Journal of Integrative Agriculture, 11: 176–86. [Google Scholar]
- Mcdonald B. A., Stukenbrock E. H. (2016) ‘Rapid Emergence of Pathogens in Agro-Ecosystems: Global Threats to Agricultural Sustainability and Food Security’, Philosophical Transactions of the Royal Society B: Biological Sciences, 371: 20160026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maina S. et al. (2017) ‘First Complete Squash Leaf Curl China Virus Genomic Segment DNA-A Sequence from East Timor’, Genome Announcements, 5: e0048317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruthi M. N. et al. (2002) ‘Coadaptation between Cassava Mosaic Geminiviruses and Their Local Vector Populations’, Virus Research, 86: 71–85. [DOI] [PubMed] [Google Scholar]
- Masood M. et al. (2017) ‘Diversity and Distribution of Cryptic Species of the Bemisia tabaci (Hemiptera: Aleyrodidae) Complex in Pakistan’, Journal of Economic Entomology, 110: 2295–300. [DOI] [PubMed] [Google Scholar]
- Nagendran K. et al. (2017) ‘The Occurrence and Distribution of Major Viruses Infecting Cucurbits in Tamil Nadu State, India’, Crop Protection, 99: 10–6. [Google Scholar]
- Navas-Castillo J., Fiallo-Olivé E., Sánchez-Campos S. (2011) ‘Emerging Virus Diseases Transmitted by Whiteflies’, Annual Review of Phytopathology, 49: 219–48. [DOI] [PubMed] [Google Scholar]
- Noris E. et al. (1998) ‘Amino Acids in the Capsid Protein of Tomato Yellow Leaf Curl Virus That Are Crucial for Systemic Infection, Particle Formation, and Insect Transmission’, Journal of Virology, 72: 10050–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L. L. et al. (2018a) ‘Differential Efficiency of a Begomovirus to Cross the Midgut of Different Species of Whiteflies Results in Variation of Virus Transmission by the Vectors’, Science China Life Sciences, 61: 1254–65. [DOI] [PubMed] [Google Scholar]
- Pan L. L. et al. (2018b) ‘Cotton Leaf Curl Disease: Which Whitefly Is the Vector?’, Phytopathology, 108: 1172–83. [DOI] [PubMed] [Google Scholar]
- Prasanna H. C. et al. (2015) ‘Cryptic Species Composition and Genetic Diversity within Bemisia tabaci Complex in Soybean in India Revealed by mtCOI DNA Sequence’, Journal of Integrative Agriculture, 14: 1786–95. [Google Scholar]
- Qin L. et al. (2013) ‘Identification of Nine Cryptic Species of Bemisia tabaci (Hemiptera: Aleyrodidae) from China by Using the mtCOI PCR-RFLP Technique’ [in Chinese with English abstract], Acta Entomologica Sinica, 56: 186–94. [Google Scholar]
- Revill P. A. et al. (2003) ‘The Complete Nucleotide Sequence of Two Distinct Geminiviruses Infecting Cucurbits in Vietnam’, Archives of Virology, 148: 1523–41. [DOI] [PubMed] [Google Scholar]
- Ruan Y.-M. et al. (2007) ‘Observing and Recording Copulation Events of Whiteflies on Plants Using a Video Camera’, Entomologia Experimentalis Et Applicata, 124: 229–33. [Google Scholar]
- Sánchez-Campos S. et al. (2018) ‘Differential Shape of Geminivirus Mutant Spectra across Cultivated and Wild Hosts with Invariant Viral Consensus Sequences’, Frontiers in Plant Science, 9: 932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seal S. E., vandenBosch F., Jeger M. J. (2006) ‘Factors Influencing Begomovirus Evolution and Their Increasing Global Significance: Implications for Sustainable Control’, Critical Reviews in Plant Sciences, 25: 23–46. [Google Scholar]
- Tahir M., Haider M. S., Briddon R. W. (2010) ‘First Report of Squash Leaf Curl China Virus in Pakistan’, Australasian Plant Disease Notes, 5: 21–4. [Google Scholar]
- Tsetsarkin K. A. et al. (2007) ‘A Single Mutation in Chikungunya Virus Affects Vector Specificity and Epidemic Potential’, PLoS Pathogens, 3: e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Engelen F. A. et al. (1995) ‘pBINPLUS: An Improved Plant Transformation Vector Based on pBIN19’, Transgenic Research, 4: 288–90. [DOI] [PubMed] [Google Scholar]
- Wei J. et al. (2014) ‘Specific Cells in the Primary Salivary Glands of the Whitefly Bemisia tabaci Control Retention and Transmission of Begomoviruses’, Journal of Virology, 88: 13460–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. L. et al. (2017) ‘Molecular Variation of Tomato Yellow Leaf Curl Virus in the Insect Vector Bemisia tabaci’, Scientific Reports, 7: 16427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data in this study are available from the authors upon request.