Abstract
Deletion of dystrobrevin binding protein 1 has been linked to Hermansky-Pudlak syndrome type 7 (HPS-7), a rare disease characterized by oculocutaneous albinism and retinal dysfunction. We studied dysbindin-1 null mutant mice (Dys−/−) to shed light on retinal neurodevelopment defects in HPS-7. We analyzed the expression of a focused set of miRNAs in retina of wild type (WT), Dys+/− and Dys−/− mice. We also investigated the retinal function of these mice through electroretinography (ERG). We found that miR-101-3p, miR-137, miR-186-5p, miR-326, miR-382-5p and miR-876-5p were up-regulated in Dys−/−mice retina. Dys−/− mice showed significant increased b-wave in ERG, compared to WT mice. Bioinformatic analysis highlighted that dysregulated miRNAs target synaptic plasticity and dopaminergic signaling pathways, affecting retinal functions of Dys−/− mice. Overall, the data indicate potential mechanisms in retinal neurodevelopment of Dys−/− mice, which may have translational significance in HSP-7 patients, both in terms of diagnostic/prognostic biomarkers and novel pharmacological targets.
Subject terms: Predictive markers, Hereditary eye disease
Introduction
Dystrobrevin binding protein 1 gene (DTNBP1) encodes dysbindin-1, a ubiquitous protein that regulates membrane localization of synaptic proteins, through the regulation of synaptic vesicles and receptors recycling1,2. Dysbindin-1 is widely expressed in the brain, both in neurons and glial cells1,3–5. Dysbindin-1 is also expressed in the eye6 and mutations leading to DTNBP1 deletion have been associated with the subtype 7 of Hermansky-Pudlak syndrome (HPS-7)7,8.
Hermansky-Pudlak syndromes (HPS) are heterogeneous genetic disorders characterized by pulmonary fibrosis, abnormalities in platelet aggregation and oculocutaneous albinism7,8. Pulmonary fibrosis is the most serious complication of HPS, which cannot be effectively treated with steroids or pirfenidone, when insufficient residual lung function occurs9.
The zebrafish fade out (fad) locus mutant has been reported as lower vertebrate model of HSP with retinal morphology and function characterization10. Recently, DTNBP1 knock-out mice (Dys−/−) showed ocular albinism related to a drop out of melanosomes in retinal pigmented epithelium and choroid, compared to wild type mice (WT)11. Additionally, retinal melanosomes in Dys−/− mice were found to have irregular shape and small dimensions7.
Retinal function has not yet been evaluated in Dys−/− mice. Visual dysfunction, i.e. decreased ERG response was found in patients with HPS syndromes12 and ocular albinism13. Interestingly, visual dysfunctions have also been found in schizophrenic patients and individuals bearing dysbindin mutations, associated with increased risk of schizophrenia development6,14. Furthermore, several mutations at DTNBP1 have been associated with human intelligence15–18, and cognitive responses to antipsychotics in animal models and patients with schizophrenia19,20. Dysregulation of dysbindin-1 expression and function, related to gene mutation, have detrimental effects on neurodevelopment1.
On the premises that the eye is the embryonic extension of the brain21 and ocular albinism has been associated to neurodevelopmental disorders22,23, we focused our research on the effects of dysbindin-1 deletion in the retina, by using Dys−/− mice as a novel animal model of HPS-7. Because the role of dysbindin in neurodevelopment has been widely investigated, we also explored the effect of dysbindin mutation on miRNAs expression; this kind of approach has been previously used on several animal paradigms such as fragile X mental retardation protein knockout mice (FMRP−/−)24, apolipoprotein E-knockout mice (APOE−/−)25, PSEN1/PSEN2 double knockout26, PTEN knockout mice27.
Expression analysis of 13 miRNAs, a selected set obtained from preliminary in-silico analysis, was carried out in retina and serum of Dys+/−, Dys−/− and WT mice. On the basis of ERG abnormalities previously found in children and young adults with ocular albinism13 and HPS syndromes12, ERG analysis on Dys−/− mice was also carried out. We identified 6 out of 13 miRNAs (miR-101-3p, miR-137, miR-186-5p, miR-326, miR-382-5p and miR-876-5p) up-regulated in Dys−/− mice retina. Accordingly, abnormalities in ERG b-wave amplitude were detected in Dys−/− mice. This study was aimed at identification of novel miRNAs as prognostic/diagnostic tools for HPS-7, as well as, new potential pharmacological targets for HPS-7 complications, such as pulmonary fibrosis.
Results
Prediction of miRNA dysregulated in Dys−/− mice
High-throughput expression analysis of miRNAs is expensive and may require large number of samples28; for this reason, we first carried out an integrated bioinformatic approach to select miRNAs potentially dysregulated in Dys−/− mice (Tables 1 and 2). With this bioinformatic approach we mined 40 miRNAs. We also predicted the pathways29 that can be dysregulated by these retrieved miRNAs. Subsequently, miRNAs were rescored based on their regulatory potential on pathways linked to albinism or neurodevelopment (see methods section). After rescoring, the expression of the top scored 13 miRNAs (miR-377-3p, miR-876-5p, miR-224-5p, miR-326, miR-330-5p, miR-155-5p, miR-590-3p, miR-101-3p, miR-137, miR-186-5p, miR-382-5p, miR-146a-5p, and miR-27a-3p) was evaluated in retina and serum of Dys−/−, Dys+/− and WT mice.
Table 1.
Prediction of miRNA-binding sites modification upon dysbindin gene mutations.
| miRNA | SNP (DTNBP1) | mirSVR | Effect |
|---|---|---|---|
| miR-1193 | rs742106 | Create | |
| miR-1246 | rs742106 | −0.500 | Enhance |
| miR-1293 | rs742106 | −1.014 | Break |
| miR-3167 | rs742106 | −0.833 | Enhance |
| miR-377-3p | rs1047631 | −1.072 | Decrease |
| miR-4275 | rs1047631 | Create | |
| miR-432-3p | rs742106 | Create | |
| miR-4483 | rs742106 | Break | |
| miR-4495 | rs1047631 | Break | |
| miR-4511 | rs2056943 | Create | |
| miR-4694-3p | rs742106 | Enhance | |
| miR-4760-3p | rs2056943 | Break | |
| miR-4782-5p | rs742106 | Create | |
| miR-5706 | rs742106 | Create | |
| miR-876-5p | rs742106 | −0.830 | Decrease |
Table 2.
Prediction of miRNAs targeting genes associated with albinism.
| Genes | Top scored miRNAs |
|---|---|
| Melanine Biosynthesis | |
| TYR | miR-326, miR-330-5p, miR-328, miR-506, miR-124, miR-378 |
| TYRP1 | miR-155, miR-590-3p, miR-128, miR-154, miR-365 |
| OCA2 | miR-495, miR-101, miR-590-3p, miR-212 |
| SLC45A2 | miR-154 |
| Melanocyte development | |
| DTNBP1 | miR-224 |
| PAX3 | miR-1, miR-206, miR-613 |
| MITF | miR-137, miR-186, miR-152 |
| SRY | miR-209, miR-219-5p, miR-487, miR-155 |
| SOX10 | miR-590-3p, miR-221, miR-222 |
| EDNRB | miR-590-5p (3p), miR-382, miR-146a |
| EDN3 | miR-496, miR-186, miR-27a |
Expression pattern of miRNAs in serum and retina
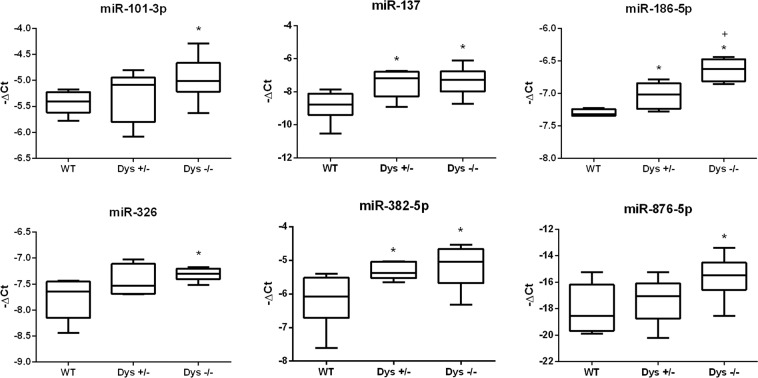
We evaluated the expression of miR-377-3p, miR-876-5p, miR-224-5p, miR-326, miR-330-5p, miR-155-5p, miR-590-3p, miR-101-3p, miR-137, miR-186-5p, miR-382-5p, miR-146a-5p, and miR-27a-3p in serum and retina of WT, Dys+/− and Dys−/− mice. Expression analysis of these miRNAs in serum did not show any significative difference between experimental groups (data not shown). On the contrary, we found 6 out of 13 miRNA up-regulated in retina of Dys−/− compared to WT mice: miR-101-3p, miR-137, miR-186-5p, miR-326, miR-382-5p and miR-876-5p (Fig. 1). Moreover, dysbindin deletion in mutant mice (Dys+/− and Dys−/−) led to gene dose effect on miRNAs dysregulation, which attained statistical significance for miR-186-5p. Moreover, a loss of one copy of the dysbindin gene significantly led to increased expression of miR-137 and miR-382-5p in Dys+/−, compared to WT mice.
Figure 1.
miRNAs dysregulated in the retina of WT, Dys+/− and Dys−/− mice. −ΔCt distribution box-plot. *p < 0.05 Dys+/− or Dys−/− vs. WT mice; +p < 0.05 Dys−/− vs. Dys+/− mice (N = 6 mice per group, each run in triplicate).
Post-hoc bioinformatic analysis
In order to understand the biological impact of dysregulated miRNAs in the Dys−/− retina, we analyzed potential miRNA-pathway interactions by accessing to the DIANA miRPath v. 3 tool, using as input miR-101-3p, miR-137, miR-186-5p, miR-326, miR-382-5p, and miR-876-5p and iteratively applying the Tarbase, microT-CDS and Targetscan prediction algorithms30. On the basis of the experimentally validated miRNA:mRNA interactions, the application of the Tarbase algorithm to the DIANA miRPath v. 3 tool gave a list of pathways regulated only by miR-101-3p, miR-186-5p, and miR-382-5p (Table 1S, supplemental information). These results are consistent with the main function of dysbindin, in fact, miR-101-3p, miR-186-5p and miR-382-5p were predicted to control “endocytosis” and “protein processing in the endoplasmatic reticulum”. Furthermore, miR-101-3p and miR-186-5p can control the expression of genes belonging to the “melanogenesis” pathway (p-value 0.033, Fig. 1S, supplemental information). Interestingly, Tarbase prediction (Table 1S, supplemental information) showed that target genes of miR-101-3p and miR-186-5p control the expression of genes belonging to the “long term potentiation” (LTP) and “long term depression” (LTD) pathways. This result is consistent with evidences about the role of dysbindin in synaptic plasticity31. In order to predict which pathways can be targeted by all 6 microRNAs up-regulated in the Dys−/− retina, microT-CDS and Targetscan algorithms were further applied; data are shown in Tables 3 and 4, respectively.
Table 3.
Pathways dysregulated in the Dys−/− retina – microT-CDS prediction.
| KEGG pathway | p-value | #genes | #miRNAs |
|---|---|---|---|
| Prion diseases | 1.44E-17 | 3 | 3 |
| MAPK signaling pathway | 2.68E-07 | 58 | 6 |
| Axon guidance | 0.000285 | 32 | 6 |
| Endocytosis | 0.000792 | 38 | 6 |
| Long-term potentiation | 0.001331 | 20 | 5 |
| Rap1 signaling pathway | 0.001331 | 41 | 6 |
| Thyroid hormone signaling pathway | 0.001403 | 23 | 6 |
| Oxytocin signaling pathway | 0.001742 | 32 | 6 |
| mRNA surveillance pathway | 0.002901 | 22 | 6 |
| Calcium signaling pathway | 0.003031 | 35 | 6 |
| mTOR signaling pathway | 0.00307 | 18 | 6 |
| Fatty acid elongation | 0.015193 | 4 | 3 |
| Glutamatergic synapse | 0.015193 | 21 | 5 |
| HTLV-I infection | 0.015193 | 47 | 6 |
| Adherens junction | 0.017904 | 15 | 6 |
| cGMP-PKG signaling pathway | 0.017904 | 33 | 6 |
| FoxO signaling pathway | 0.017904 | 30 | 6 |
| Wnt signaling pathway | 0.020194 | 28 | 6 |
| cAMP signaling pathway | 0.022403 | 37 | 6 |
| T cell receptor signaling pathway | 0.022579 | 22 | 6 |
| Ubiquitin mediated proteolysis | 0.024099 | 29 | 6 |
| VEGF signaling pathway | 0.024486 | 15 | 5 |
| Insulin signaling pathway | 0.024486 | 28 | 6 |
| Bacterial invasion of epithelial cells | 0.024486 | 17 | 6 |
| Transcriptional misregulation in cancer | 0.026035 | 31 | 6 |
| Neurotrophin signaling pathway | 0.026125 | 24 | 6 |
| Vasopressin-regulated water reabsorption | 0.026702 | 8 | 6 |
| Amyotrophic lateral sclerosis (ALS) | 0.026758 | 13 | 6 |
| Protein processing in endoplasmic reticulum | 0.034352 | 29 | 6 |
| Amphetamine addiction | 0.040134 | 15 | 4 |
| AMPK signaling pathway | 0.041784 | 24 | 6 |
| TGF-beta signaling pathway | 0.041784 | 20 | 5 |
| Circadian entrainment | 0.045922 | 18 | 6 |
Table 4.
Pathways dysregulated in the Dys−/− retina – Targetscan prediction.
| KEGG pathway | p-value | #genes | miRNAs |
|---|---|---|---|
| Adherens junction | 3.03E-05 | 10 | miR-101-3p/miR-326 |
| Morphine addiction | 0.010632 | 7 | miR-101-3p/miR-326 |
| Notch signaling pathway | 0.014221 | 9 | miR-326 |
| Endocytosis | 0.014221 | 20 | miR-101-3p/miR-326 |
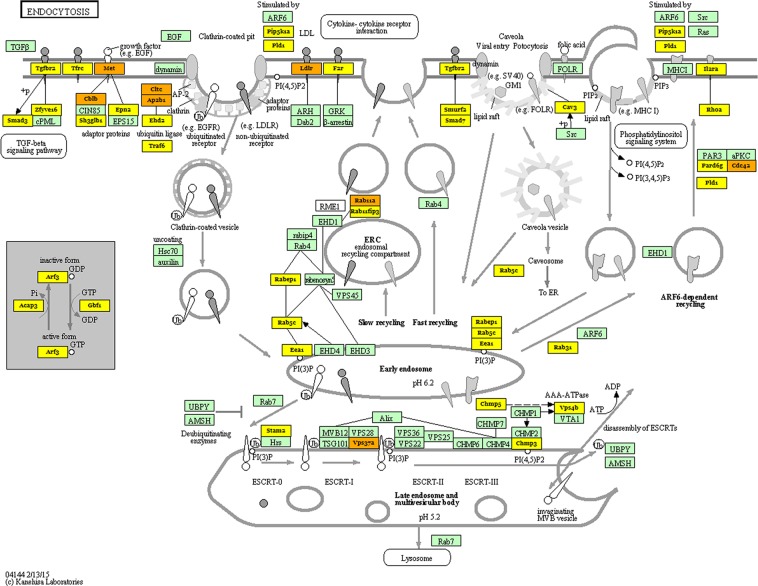
All three different algorithms predicted that miR-101-3p, miR-137, miR-186-5p, miR-326, miR-382-5p and miR-876-5p can modulate cellular endocytosis (Fig. 2), consistently with dysbindin main functions32–34.
Figure 2.
Genes belonging to the “endocytosis pathway” are targets of miRNAs up-regulated in the Dys−/− retina. Yellow genes are targets of one miRNA, while orange genes are targets of more than one miRNA. This picture is the output of Diana miRPath software and recalls the KEGG pathway deposited in the database (KEGG permission n°190309)80: https://www.genome.jp/kegg/pathway.html.
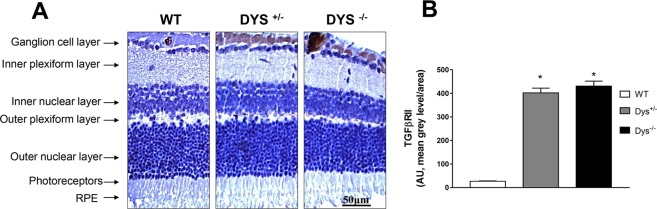
Both Tarbase and microT-CDS predictions evidenced that TGF-β signaling pathway can be dysregulated by miRNAs up-regulated in the Dys−/− retina. The microT-CDS algorithm predicted that miR-101-3p, miR-137, miR-186-5p, miR-326 and miR-382 can control the TGF-β signaling pathway, which is known to be involved in collagen 1 synthesis and induction of fibrosis35. This result may be relevant for pulmonary fibrosis, often occurring in HPS-7. Specifically, miR-101-3p (TGFBRI, ACVR2) and miR-186-5p experimentally target genes of the TGFβ signaling pathway (ACVR1, SMAD4, SMAD5, ACVR2A). Therefore, we tested the hypothesis that also the retina of Dys−/− mice could be affected by a dysfunctional TGFβ signaling. To this aim, we stained the retina for TGFβ1, TGFβRI and TGFβRII. We found a significant (p < 0.05) increased staining for TGFβRII, but not for TGFβ1 and TGFβRI, in the retinal ganglion cell layer of Dys+/− and Dys−/− mice, compared to WT mice (Fig. 3).
Figure 3.
TGFβRII staining in the retinal ganglion cell layer. Representative IHC images (A) and densitometric analysis (B) for TGFβRII staining in the retinal ganglion cell layer of WT (white bars), Dys+/− (gray bars) and Dys−/− (black bars). Each column represents the average ± SD (N = 12). *p < 0.05 vs. WT.
Furthermore, targetscan and microT-CDS predictions suggested the interaction of dysregulated miRNAs in Dys−/− mice with genes belonging to the pathways named “amphetamine addiction”, “morphine addiction” and “glutamatergic synapse”. These genes are listed in Table 5, and these results suggest that retinal dysregulation of these miRNAs may affect dopaminergic, GABAergic signaling and glutamatergic signaling, which were found to be involved in retinal function and retinal light adaptation36,37.
Table 5.
Genes that are target of miRNAs dysregulated in retina of Dys−/− mice (microT-CDS algorithm).
| Amphetamine addiction |
| SLC6A3, GRIN3A, CAMK2A, STX1A, PP1CA, and PDYN |
| Morphine addiction |
| OPRM1, GABRG3, GNB3, KGNJ5, and PDE7B |
| Glutamatergic synapse |
| PPP3CC, GRIA2, PPP3R2, SLC1A1, PPP3CB, ADCY1, SHANK2, HOMER2, GNB4, ADCY3, GRM5, PPP3R1, DLGAP1, PPP3CA, PLCB1, GRIK3, GNB3, SLC17A8, HOMER1, and SLC17A6 |
Scotopic ERG analysis
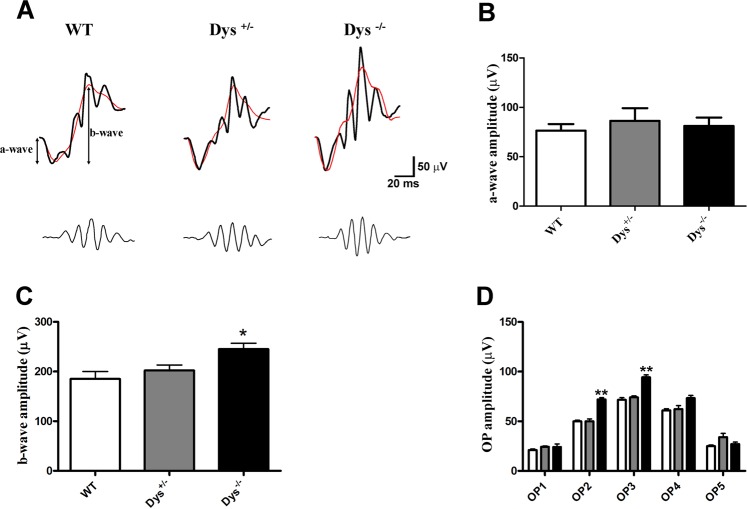
ERG was recorded in mice belonging to the three experimental groups: WT, Dys+/− and Dys−/−. The representative recordings in Fig. 4 (panel A) show scotopic ERG waveforms (a-wave, the b-wave and oscillatory potentials (OPs) on the rising part of the b-wave) recorded at light intensities of 1 log cd-s/m2. It can be noticed that the ERG amplitude was altered in Dys−/− dark-adapted mice. Comparing the average amplitude of the a-wave (Fig. 4B) and b-waves (Fig. 4C) in controls WT, Dys+/− and Dys−/− mice, we found a statistically significant difference between b-wave amplitude records in WT vs. Dys−/− mice (p < 0.05). Oscillatory potentials (OPs) represent high-frequency oscillations on the leading edge of the ERG b-wave. It should be noted that OP2 and OP3 in the Dys−/− mice showed amplitudes larger than control animals, which may indicate alterations in the activity of outer and inner retina, respectively (Fig. 4D). Furthermore, the lack of dysbindin was associated with an increase in b-wave amplitude in mice, consistently with a previous report of increased b-wave amplitude in dark-adapted albinos13. Moreover, a-wave and b-wave abnormalities have been previously found in schizophrenic patients38 or individuals bearing high genetic risk of schizophrenia39.
Figure 4.
ERG records in wild type (WT), Dys+/− and Dys −/− mice. Representative ERG waveforms (top) w/o OPs (black and red lines, respectively) and representative OP waveforms (bottom) (A), a-wave amplitudes (B), b-wave amplitudes (C), and OPs amplitude (D) in WT (white bars), Dys+/− (gray bars) and Dys−/− (black bars) mice recorded at light intensity of 1 log cd-s/m2. *p < 0.05 and **p < 0.01 vs. WT. Each column represents the average ± SD (N = 12).
Discussion
Dysbindin-1 is widely expressed in the brain and plays a role in protein trafficking and synaptic transmission. Dysbindin alterations have been linked to altered neuronal plasticity attributed to its influence on neurodevelopment during the embryonal stage.
Dysbindin mutations have been associated to a rare disease, named Hermansky-Pudlak syndrome 7 (HPS-7), characterized by pulmonary fibrosis, abnormalities in platelet aggregation and oculocutaneous albinism, which alters retinal functions13.
We analyzed miRNA expression profiles and function in the retina of Dys−/− mice, to shed light on pathological mechanisms and retinal defects upon deletion of dysbindin, which may have translational value for the Hermansky-Pudlak syndrome 7 (HPS-7). We first predicted with bioinformatic tools a small set of miRNAs, putatively involved in oculocutaneous albinism, and then the expression profile of these miRNAs was analyzed in the retina and serum of Dys−/− mice. No significative differences were found in serum of Dys−/− mice, compared to either Dys+/− or WT mice (data not shown). On the contrary, 6 out of 13 miRNAs were found to be significantly up-regulated in Dys−/− mouse retina, compared to WT or Dys+/− groups. These data suggest that DTNBP1 depletion can perturb a yet unknown molecular signaling pathway, which increases expression of a specific set of miRNAs potentially related to albinism and/or retinal neurodevelopment. It was previously found that FMRN gene deletion in mice led to the up-regulation of a series of miRNAs, likely influencing the degradation of pre-miRNAs instead of promoting their expression24. Moreover, BDNF was found to influence miRNA maturation and stability by modulation of DICER and lin28b expression40. Furthermore, gene deletion led to miRNAs dysregulation in several experimental paradigms25–27. Dysbindin is a component of BLOC-1, which mediates the sorting of lysosome-related organelles (LROs) and the lack of dysbindin leads to an altered sorting of lysosomes and melanosome maturation7. Additionally, Argonaute 2 (Ago2)-miRNA lysosomal sorting was found to be modulated by BLOC3 in Drosofila41. Thus, we hypothesize that the lack of dysbindin could also influence the sorting of miRNAs, leading to their up-regulation in retinal tissues.
Diana miRPath post-hoc analysis revealed that miRNAs that are up-regulated in the Dys−/− mice retina are linked to “endocytosis” and “protein processing in the endoplasmatic reticulum”, which are pathways consistent with the functions of dysbindin mentioned above. Additionally, the bioinformatic analysis with Diana miRPath revealed that up-regulated miRNAs in Dys−/− mice can dysregulate the TGFβ signaling pathway. This bioinformatic result might partially explain a feature of HPS-7, i.e. pulmonary fibrosis. In fact, the TGF-β signaling pathway, when over activated, leads to pro-fibrotic events and its involvement in pulmonary fibrosis has been widely reported42–44. Interestingly, the role of regulation of fibrosis by miR-101-3p and miR-326 was also reported in a recent review35. Moreover, the induced expression of miR-101-3p decreased signs of pulmonary fibrosis in an animal model45; while, miR-326 was found to be down-regulated in lung samples of patients with idiopathic pulmonary fibrosis46. Based on the above data, we hypothesize that the up-regulation of miRNAs upon dysbindin deletion could be a negative feed-back process, aimed at attenuating TGF-β signaling linked to lung fibrosis46. In this perspective, miRNAs found up-regulated in the Dys−/− retina deserve to be analyzed in pulmonary exudates of HPS-7 patients and/or animal models of pulmonary fibrosis. Moreover, these miRNAs could be further developed as pharmacological targets for treatment of pulmonary fibrosis in HPS-7. Additionally, we analyzed retinal staining of upstream effectors of TGFβ signaling pathway (TGFβ1, TGFβRI and TGFβRII), and we found a significant increase of TGFβRII staining in retinal ganglion cell layer of Dys+/− and Dys−/− mice, compared to control wild type mice. In physiological conditions retinal TGFβRII is localized in the retinal ganglion cell layer. TGFβRII depletion was found to be detrimental on retina development and function, leading to a low b-wave amplitude and to a degeneration of proximal retinal neurons47 (amacrine and ganglion cells48). On the basis of previous reports, TGFβRII expression levels can modulate the intensity of TGFβ1 signaling (smad or smad-independent signaling)49, in accordance with comorbidity linked to fibrosis in Hermansky Pudlak 7 patients7–9,35.
Furthermore, bioinformatic analysis, with the Tarbase algorithm, evidenced that miR-101-3p and miR-186-5p could target genes involved in LTP and LTD, the basic processes of neuronal plasticity that are affected in neurodevelopmental disorders50, consistently with the link between dysbindin dysfunctions and inefficient neuronal plasticity2,51. Dysbindin was found to regulate the expression of NMDA receptors, regulating hippocampal LTP in mice31. The DISC1 “disrupted in the schizophrenia 1 gene” when mutated increases the risk of schizophrenia in humans52,53, and DISC1 mutations impair LTP and LTD in mice54. DISC1 interacts with dysbindin influencing its own stability; moreover, DISC1 regulates neurite outgrowth induced by dysbindin55. Interestingly, some reports support the evidence that LTP occurs also at excitatory retinal synapses, formed by bipolar cells and retinal ganglion cells, in the developing retina56. Besides regulation of synaptic plasticity, dysbindin has been associated with various mechanisms underlying synaptic function57–59 and the regulation of dopamine and glutamate signaling in the brain5,38,60. Moreover, in vitro experiments suggest that dysbindin suppresses DA release61 and cultured neurons from Dys−/− mice show increased cell surface expression of the dopamine D2 receptor, due to an increase of receptor membrane insertion16.
It is noteworthy that some dysbindin gene variants have been linked to visual dysfunctions6,14, which in turn were linked to a-wave and b-wave ERG aberrations38,39. Dopaminergic signaling and GABAergic transmission regulate adaptive mechanisms in the retina, in response to light brightness modifications. These two systems controls the light adaptation process in the retina, relying on rod bipolar cells sensitization and desensitization in dim-light or bright light conditions, respectively36,37. Moreover, glutamatergic and dopaminergic systems were found to be linked in retinal light adaptation62,63. Therefore, similar to effects in the brain, dysbindin mutations could affect the retina by altering the dopaminergic signaling and retinal synaptic plasticity. This hypothesis was supported by bioinformatic analysis, which predicted that dysregulated miRNAs in the retina of Dys−/− mice could modulate dopaminergic, GABAergic and glutamatergic signaling, by targeting genes belonging to “amphetamine addiction”, “morphine addiction” and “glutamatergic synapse” pathways. Furthermore, it has been demonstrated that decreased retinal DA levels contributed to early visual and retinal dysfunction64. Furthermore, in vitro studies show that antipsychotics increase light sensitivity of retinal ganglion cells (RGCs) and this effect can be explained by D2 receptor antagonism65. Previous data clearly showed that the block of dopamine D1 receptors increases the b-wave amplitude66. This indicates that endogenous DA decreases the amplitudes of the ERG waves by acting through D1 receptors66. Moreover, it was found that the dopaminergic system could bi-directionally modulate the ERG response; i.e. the excitatory action on the b-wave could be mediated mainly by D2 receptors, while the suppressive effect upon the d-wave is mediated mostly by D1 receptors, which is counteracted by the dopamine excitatory action mediated through D2 receptors67. Furthermore, oscillatory potentials are regulated mainly by dopaminergic neurons, but also by GABAergic and glutamatergic system68. We analyzed the retinal function of Dys−/− mice and found a significant increase in the b-wave, OP2 and OP3, compared to WT mice. This result is consistent with the hypothesis that dysbindin deletion could affect the dopaminergic signaling in the retina of Dys−/− mice, due to an imbalance of DA retinal neurotransmission, similarly to what was found in neurons, where dysbindin siRNA treatment increased D2 receptor signaling along with decreased D2 receptor internalization69.
In conclusion, our findings on retinal miRNAs dysregulation in Dys−/− mice, along with retinal dysfunction data, suggest that the Dys−/− mouse represents a good paradigm for HSP-7, beyond a schizophrenia model as previously demonstrated11,17,19. Our data suggest that retinal function in ocular albinism could be modulated by miRNAs and also by dopaminergic tone. Further studies need to be carried out to directly assess dopaminergic signaling in the retina of Dys−/− mice (e.g. ERG measurements on Dys−/− mice treated with dopaminergic agonists or antagonists). Finally, our findings support the hypothesis that pharmacological modulation of dysregulated miRNAs could be potentially relevant in clinical practice to counteract visual acuity reduction, and possibly pulmonary fibrosis in HSP-7 patients.
Methods
Bioinformatic miRNA prediction
High-throughput expression analysis of miRNAs is expensive and may requires a large number of samples, and post-hoc validation through quantitative real-time PCR28; for this reason, we first carried out an integrated bioinformatic approach to select miRNAs potentially dysregulated in Dys−/−mice. The first step included the prediction of newly disrupted or created miRNA-binding sites upon mutation of the dysbindin gene, through access to the miRSNPs database70. Predictions were carried out by filtering for European Caucasian ancestry. In order to decrease the false discovery rate of predictions, we mined at microRNA.org71 miRNAs that can regulate genes associated with albinism, such as: TYR, TYRP1, OCA2, SLC45A2, DTNBP1, PAX3, MITF, SRY, SOX10, EDNRB, and EDN3. Another bioinformatic analysis was carried out with the Diana miRPath v 3.0 tool29 to predict a focused miRNA set, to be further analyzed in WT, Dys+/− and Dys−/− mice. Targets were predicted applying the Tarbase algorithm. Pathways putatively dysregulated by miRNAs, listed in Tables 1 and 2, were rescored on the basis of their regulatory capability on the following signaling pathways and according to literature search data (pathway AND albinism; pathway AND neurodevelopment): “TGF-β1”72, “Protein Processing”1,2,32, “Fatty acid elongation”73,74, “endocytosis”1,2, “ubiquitin”75, “TNF signaling”76, “RNA transport”1,2,77, and “actin cytoskeleton”78,79. A post-hoc bioinformatic analysis was carried out with the Diana miRPath v 3.0 tool on the six out of 13 miRNAs, that were found to be dysregulated in retinal samples of Dys +/− and Dys−/−, compared to WT mice. In this case we iteratively used the algorithms Tarbase, microT-CDS and Targetscan30. If the Tarbase algorithm is applied, Diana miRPath predictions recall experimentally validated microRNA:RNA interactions, while microT-CDS and Targetscan rely on prediction and scoring of miRNA:RNA interactions30. Diana miRpath provided enriched depiction of KEGG pathways targeted by dysregulated miRNAs (KEGG permission n° 190309)80.
Animals
All experimental procedures were approved by the IACUC of the University of Catania (approval # 640/2017-PR). Procedures were carried out in accordance to the Association for Research in Vision and Ophthalmology (ARVO) Statement for the Use of Animals in Ophthalmic and Vision Research. Dys−/−, Dys+/− and WT mice used in this study, were produced using a heterozygous (Dys+/−) breeding strategy as previously described11,19. Wild-type C57BL/6 J mice were purchased from Envigo (San Pietro al Natisone, Udine, Italy). Six-month-old male Dys mutant mice (Dys+/− and Dys−/−) and their wild-type littermates (Dys+/+) were used (N = 18 per genotype). N = 6 mice per genotype were used for miRNA expression analysis, while N = 12 per genotype animals were used for ERG and immunohistochemical assessment. All mice were genotyped using a duplex polymerase chain reaction (PCR) as previously described10,14. Primers for the WT gene, yielding a PCR product of 472 base pairs, were 5′-AGCTCCACCTGCTGAACATT-30 and 50-TGAGCCATTAGGAGATAAGAGCA-3′. Primers for the Dys mutant gene, yielding a product of 274 base pairs, were 5′-TCCTTGCTTCGTTCTCTGCT-3′ and 5′-CTTGCCAGCCTTCGTATTGT-3′. The 472-base-pair product was detected only in Dys+/+ and Dys+/− mice, while the 274-base-pair product was detected only in the Dys+/− and Dys−/− mice. One-week prior ERG analysis, mice were moved from the animal colony room to a climate-controlled holding room (21 ± 1 °C), weighed, singly housed, and maintained on a 12-hour reverse light/dark cycle with free access to food and water. After sacrifice by means of cervical dislocation, eye globes were excised, and retina collected. Plasma samples were obtained, through intracardiac puncture, immediately after sacrifice of the mice.
Extraction and qPCR of miRNAs
Total RNA was isolated from retina samples with TRIzol reagent (Thermo Fisher Scientific, Boston, MA, USA), according to the manufacturer’s instructions. Serum samples were centrifuged at 300 × g for 15 minutes at 4 °C to remove circulating cells and/or debris. Total RNA was isolated from 200 μl serum by using the Qiagen miRNeasy mini kit (Qiagen, Hilden, Germany), according to the Qiagen supplementary protocol for purification of small RNAs from serum and plasma; RNA was finally eluted in 45 μl of RNAse-free water and quantified by GenQuant pro spectrophotometer (Biochrom, Cambridge, UK). MiRNA expression analysis was performed through TaqMan assays. Reverse transcription of miRNAs was performed by TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific, Boston, MA, USA); the resulting miRNA-specific cDNA was amplified with the TaqMan microRNA Assays (Thermo Fisher Scientific, Boston, MA, USA) and the TaqMan Universal Master Mix II, no UNG (Thermo Fisher Scientific, Boston, MA, USA), according to the manufacturer’s instructions. MiR-16 was used as endogenous control for both retina and serum81,82. Real Time PCR reactions were performed on a 7900 HT Fast Real Time PCR System (Applied Biosystems, Monza, Italy). Differentially expressed miRNAs were identified by applying an unpaired T test (p-value ≤ 0.05); expression fold changes were calculated by the 2−ΔΔCt method. PCR experiments followed MIQE guidelines.
Tissue preparation for immunohistochemical staining
Eyes were enucleated and processed as previously described by Castorina A. et al.83. For each eye different sections where used for TGFβ1 or TGFβ Receptor I or TGFβ Receptor II staining. Immunohistochemical analysis was performed in accordance with the standard avidin–biotin complex (ABC) method. Briefly, sections were incubated with TGFβ1 (Abcam, #ab92486; 1:100) or TGFβ Receptor I (Abcam, #ab31013, 1:100) or TGFβ Receptor II (Cell Signaling, #3713; 1:100). Therefore, sections were incubated with a 1:200 diluted biotinylated goat anti–rabbit IgG for 1 h at room temperature. The sections were then rinsed and treated with reagents from an ABC Kit and counterstained with hematoxylin. All sections were examined, and images were taken with a light microscope (Axiovert, Carl Zeiss Inc). Densitometric analysis was carried with ImageJ84. TGFβ Receptor II staining in the retinal ganglion cell layer was quantified as follows: i. images were converted in black and white; ii. blue channel colour was switched off; iii. grey scale measurements were then normalized for the corresponding area of retinal ganglion cell layer.
Scotopic ERG analysis
Before ERG testing, mice were dark adapted overnight. Scotopic ERG was carried out accordingly to previous reports about scotopic b-wave amplitude increase in albinos13. In anesthetized mice, pupils were dilated with 0.5% atropine, the cornea was intermittently irrigated with saline solution to prevent clouding of the ocular media, and a heating pad was used to keep the body temperature at 38 °C. The ERG responses were recorded through silver/silver chloride corneal electrodes and a forehead reference electrode. A ground electrode was placed on the tail. Scotopic ERG responses, which primarily measures rod function, were evoked by a 1 log cd-s/m2 flash generated through a Ganzfeld stimulator (Biomedica Mangoni, Pisa, Italy). The electrodes were connected to a two-channel amplifier. Signals were amplified at 1,000 gain and bandpass filtered between 0.2 and 500 Hz before being digitized at 5 kHz rate with a data acquisition device (Biomedica Mangoni, Pisa, Italy). The amplitude of the a-wave was measured at a fixed time of 8 ms after stimulus onset to minimize contamination from contributions of non-photoreceptor cells85. The b-wave amplitude was measured from the trough of the a-wave to the peak of the b-wave. Mean amplitudes of a- and b-wave ERG responses were plotted. For each experimental condition, ERG analysis was performed on 12 eyes per group (2 eyes for each animal), 6 mice per experimental group. Data was analyzed with respect to several parameters: a-, b-wave amplitudes and amplitudes of oscillatory potentials. Peak a-wave amplitude was measured from baseline to the initial negative going voltage, whereas peak b-wave amplitude was measured from the trough of the a-wave to the peak of the positive b-wave. In order to determine the amplitude of the oscillatory potentials (OP1-OP5), the ERG was low pass filtered at 16.5 Hz (red trace in Fig. 3) and subtracted from the original ERG wave. To evaluate the amplitude of OPs, ERG responses recorded at light intensity of 1 log cd-s/m2 were digitally filtered with a bandpass of 65–300 Hz to eliminate the a- and b-wave interference and to avoid the 60 Hz line noise.
Statistical analysis
Experimental groups were masked to investigators during sample analysis for miRNA expression, immunohistochemistry, and ERG assessment. Labels were unveiled after analysis by investigators that carried out genotype and tissue collection. For the ERG and immunohistochemistry analysis in vivo experiments, all values are expressed as mean ± SD (N = 12 eyes per group, 2 eyes per animal, 6 mice per group). The quantitative PCR for miRNAs expression analysis was carried out on a total of 12 retinas for each experimental group, two retinas from each animal were pooled (N = 6 samples; each run in triplicate). Circulating miRNAs were evaluated in 18 serum samples from 6 mice per each genotype (N = 6 per group; each run in triplicate). Three independent experimental animal sets were used in this study for miRNA expression analysis, immunohistochemistry and ERG assessment, respectively. Statistical significance was assessed by one-way ANOVA with Tukey-Kramer post-hoc test for multiple comparisons. Differences were considered statistically significant for p-values < 0.05. GraphPad Prism v.7 (GraphPad Software, La Jolla, CA, USA) was used for statistical analysis and graph-figure design.
Supplementary information
Acknowledgements
We would like to thank Dr. Vittorio Porciatti (Bascom Palmer Eye Institute, University of Miami Miller School of Medicine, Miami, USA) for fruitful scientific discussions on ERG issue and for reading the manuscript. This study was supported in part by a grant from University of Catania “Piano triennale per la Ricerca – Linea Intervento 2, University of Catania, Italy”. We also wish to thank the Scientific Bureau of the University of Catania for language support.
Author contributions
C.Bucolo, F.P., G.M.L. conceived the research. G.L.R., C.B.M.P., G.M.L., S.A.T., CBarbagallo, S.G., carried out the experiments. G.L.R., C.B.M.P., CBarbagallo, R.M., F.M., M.C., carried out formal analysis. C.Bucolo, M.C., S.S., G.M.L., F.P. provided animals, developed and designed methodology along with creation of models. Development or design of methodology; creation of models. G.L.R., C.B.M.P., C.Bucolo. wrote the initial draft of the manuscript. C.Bucolo, C.B.M.P., S.S., M.R., M.P., F.P., F.D., F.J.G. revised and edited the final version of the manuscript.
Data availability
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Giovanni Luca Romano, Chiara Bianca Maria Platania and Gian Marco Leggio.
Supplementary information
is available for this paper at 10.1038/s41598-020-60931-5.
References
- 1.Ghiani CA, et al. The dysbindin-containing complex (BLOC-1) in brain: developmental regulation, interaction with SNARE proteins and role in neurite outgrowth. Molecular psychiatry. 2010;15(115):204–215. doi: 10.1038/mp.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mullin AP, et al. Gene dosage in the dysbindin schizophrenia susceptibility network differentially affect synaptic function and plasticity. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2015;35:325–338. doi: 10.1523/JNEUROSCI.3542-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson MA, Newey SE, Martin-Rendon E, Hawkes R, Blake DJ. Dysbindin, a novel coiled-coil-containing protein that interacts with the dystrobrevins in muscle and brain. The Journal of biological chemistry. 2001;276:24232–24241. doi: 10.1074/jbc.M010418200. [DOI] [PubMed] [Google Scholar]
- 4.Iijima S, et al. Immunohistochemical detection of dysbindin at the astroglial endfeet around the capillaries of mouse brain. Journal of molecular histology. 2009;40:117–121. doi: 10.1007/s10735-009-9221-6. [DOI] [PubMed] [Google Scholar]
- 5.Shao L, et al. Schizophrenia susceptibility gene dysbindin regulates glutamatergic and dopaminergic functions via distinctive mechanisms in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18831–18836. doi: 10.1073/pnas.1114569108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donohoe G, et al. Early visual processing deficits in dysbindin-associated schizophrenia. Biological psychiatry. 2008;63:484–489. doi: 10.1016/j.biopsych.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 7.Li W, et al. Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1) Nature genetics. 2003;35:84–89. doi: 10.1038/ng1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryan MM, et al. Clinical and molecular phenotyping of a child with Hermansky-Pudlak syndrome-7, an uncommon genetic type of HPS. Molecular genetics and metabolism. 2017;120:378–383. doi: 10.1016/j.ymgme.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orphanet. Hermansky-Pudlak syndrome. available at: https://www.orpha.net/consor/cgi-bin/OC_Exp.php?lng=EN&Expert=79430.
- 10.Bahadori Ronja, Rinner Oliver, Schonthaler Helia Berrit, Biehlmaier Oliver, Makhankov Yuri V., Rao Prashanth, Jagadeeswaran Pudur, Neuhauss Stephan C. F. The Zebrafishfade outMutant: A Novel Genetic Model for Hermansky–Pudlak Syndrome. Investigative Opthalmology & Visual Science. 2006;47(10):4523. doi: 10.1167/iovs.05-1596. [DOI] [PubMed] [Google Scholar]
- 11.Papaleo F, Yang F, Garcia S, Chen J, Lu B, Crawley J N, Weinberger D R. Dysbindin-1 modulates prefrontal cortical activity and schizophrenia-like behaviors via dopamine/D2 pathways. Molecular Psychiatry. 2010;17(1):85–98. doi: 10.1038/mp.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fagadau Warren R., Heinemann Murk H., Cotlier Edward. Hermansky-Pudlak syndrome: albinism with lipofuscin storage. International Ophthalmology. 1981;4(1-2):113–122. doi: 10.1007/BF00139585. [DOI] [PubMed] [Google Scholar]
- 13.Krill AE. The electroretinogram and electro-oculogram: clinical applications. Investigative ophthalmology. 1970;9:600–617. [PubMed] [Google Scholar]
- 14.Mechelli A, et al. Dysbindin modulates brain function during visual processing in children. NeuroImage. 2010;49:817–822. doi: 10.1016/j.neuroimage.2009.07.030. [DOI] [PubMed] [Google Scholar]
- 15.Straub RE, et al. Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. American journal of human genetics. 2002;71:337–348. doi: 10.1086/341750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji Y, et al. Role of dysbindin in dopamine receptor trafficking and cortical GABA function. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19593–19598. doi: 10.1073/pnas.0904289106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papaleo F, Lipska BK, Weinberger DR. Mouse models of genetic effects on cognition: relevance to schizophrenia. Neuropharmacology. 2012;62:1204–1220. doi: 10.1016/j.neuropharm.2011.04.025. [DOI] [PubMed] [Google Scholar]
- 18.Savage JE, et al. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nature genetics. 2018;50:912–919. doi: 10.1038/s41588-018-0152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheggia D, et al. Variations in Dysbindin-1 are associated with cognitive response to antipsychotic drug treatment. Nature communications. 2018;9:2265. doi: 10.1038/s41467-018-04711-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leggio, G. M. et al. The epistatic interaction between the dopamine D3 receptor and dysbindin-1 modulates higher-order cognitive functions in mice and humans. Molecular Psychiatry, 10.1038/s41380-019-0511-4 (2019). [DOI] [PubMed]
- 21.De Groef L, Cordeiro MF. Is the Eye an Extension of the Brain in Central Nervous System Disease? Journal of ocular pharmacology and therapeutics: the official journal of the Association for Ocular Pharmacology and Therapeutics. 2018;34:129–133. doi: 10.1089/jop.2016.0180. [DOI] [PubMed] [Google Scholar]
- 22.Kutzbach B, Summers CG, Holleschau AM, King RA, MacDonald JT. The prevalence of attention-deficit/hyperactivity disorder among persons with albinism. Journal of child neurology. 2007;22:1342–1347. doi: 10.1177/0883073807307078. [DOI] [PubMed] [Google Scholar]
- 23.Saadeh R, Lisi EC, Batista DAS, McIntosh I, Hoover-Fong JE. Albinism and developmental delay: the need to test for 15q11-q13 deletion. Pediatric neurology. 2007;37:299–302. doi: 10.1016/j.pediatrneurol.2007.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu T, et al. A MicroRNA Profile in Fmr1 Knockout Mice Reveals MicroRNA Expression Alterations with Possible Roles in Fragile X Syndrome. Molecular neurobiology. 2015;51:1053–1063. doi: 10.1007/s12035-014-8770-1. [DOI] [PubMed] [Google Scholar]
- 25.Ma, A. J. et al. Associations of CXCL16, miR-146a and miR-146b in atherosclerotic apolipoprotein E-knockout mice. Molecular Medicine Reports, 10.3892/mmr.2018.9270 (2018). [DOI] [PubMed]
- 26.Ham Suji, Kim Tae Kyoo, Lee Sangjoon, Tang Ya-Ping, Im Heh-In. MicroRNA Profiling in Aging Brain of PSEN1/PSEN2 Double Knockout Mice. Molecular Neurobiology. 2017;55(6):5232–5242. doi: 10.1007/s12035-017-0753-6. [DOI] [PubMed] [Google Scholar]
- 27.Takao, A. et al. Generation of PTEN-knockout (−/−) murine prostate cancer cells using the CRISPR/Cas9 system and comprehensive gene expression profiling. Oncology Reports, 10.3892/or.2018.6683 (2018). [DOI] [PubMed]
- 28.de Ronde Maurice W J, Ruijter Jan M, Moerland Perry D, Creemers Esther E, Pinto-Sietsma Sara-Joan. Study Design and qPCR Data Analysis Guidelines for Reliable Circulating miRNA Biomarker Experiments: A Review. Clinical Chemistry. 2018;64(9):1308–1318. doi: 10.1373/clinchem.2017.285288. [DOI] [PubMed] [Google Scholar]
- 29.Vlachos IS, et al. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic acids research. 2015;43:W460–6. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riffo-Campos Ángela, Riquelme Ismael, Brebi-Mieville Priscilla. Tools for Sequence-Based miRNA Target Prediction: What to Choose? International Journal of Molecular Sciences. 2016;17(12):1987. doi: 10.3390/ijms17121987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang Tina Tze-Tsang, Yang Feng, Chen Bo-Shiun, Lu Yuan, Ji Yuanyuan, Roche Katherine W., Lu Bai. Dysbindin regulates hippocampal LTP by controlling NMDA receptor surface expression. Proceedings of the National Academy of Sciences. 2009;106(50):21395–21400. doi: 10.1073/pnas.0910499106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marley A, von Zastrow M. Dysbindin promotes the post-endocytic sorting of G protein-coupled receptors to lysosomes. PloS one. 2010;5:e9325. doi: 10.1371/journal.pone.0009325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tropea D, Hardingham N, Millar K, Fox K. Mechanisms underlying the role of DISC1 in synaptic plasticity. The Journal of physiology. 2018;596:2747–2771. doi: 10.1113/JP274330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang W, Zhu C, Shen Y, Xu Q. The pathogenic mechanism of dysbindin-1B toxic aggregation: BLOC-1 and intercellular vesicle trafficking. Neuroscience. 2016;333:78–91. doi: 10.1016/j.neuroscience.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 35.Kang Hara. Role of MicroRNAs in TGF-β Signaling Pathway-Mediated Pulmonary Fibrosis. International Journal of Molecular Sciences. 2017;18(12):2527. doi: 10.3390/ijms18122527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herrmann R, et al. Rod vision is controlled by dopamine-dependent sensitization of rod bipolar cells by GABA. Neuron. 2011;72:101–110. doi: 10.1016/j.neuron.2011.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Travis AM, Heflin SJ, Hirano AA, Brecha NC, Arshavsky VY. Dopamine-Dependent Sensitization of Rod Bipolar Cells by GABA Is Conveyed through Wide-Field Amacrine Cells. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2018;38:723–732. doi: 10.1523/JNEUROSCI.1994-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balogh Z, Benedek G, Keri S. Retinal dysfunctions in schizophrenia. Progress in neuro-psychopharmacology & biological psychiatry. 2008;32:297–300. doi: 10.1016/j.pnpbp.2007.08.024. [DOI] [PubMed] [Google Scholar]
- 39.Hebert M, et al. Retinal response to light in young nonaffected offspring at high genetic risk of neuropsychiatric brain disorders. Biological psychiatry. 2010;67:270–274. doi: 10.1016/j.biopsych.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 40.Huang Y-WA, Ruiz CR, Eyler ECH, Lin K, Meffert MK. Dual regulation of miRNA biogenesis generates target specificity in neurotrophin-induced protein synthesis. Cell. 2012;148:933–946. doi: 10.1016/j.cell.2012.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris DA, Kim K, Nakahara K, Vasquez-Doorman C, Carthew RW. Cargo sorting to lysosome-related organelles regulates siRNA-mediated gene silencing. The Journal of cell biology. 2011;194:77–87. doi: 10.1083/jcb.201102021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneider DJ, et al. Cadherin-11 contributes to pulmonary fibrosis: potential role in TGF-beta production and epithelial to mesenchymal transition. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2012;26:503–512. doi: 10.1096/fj.11-186098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Agassandian M, et al. VCAM-1 is a TGF-beta1 inducible gene upregulated in idiopathic pulmonary fibrosis. Cellular signalling. 2015;27:2467–2473. doi: 10.1016/j.cellsig.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Margadant C, Sonnenberg A. Integrin-TGF-beta crosstalk in fibrosis, cancer and wound healing. EMBO reports. 2010;11:97–105. doi: 10.1038/embor.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang C, et al. MicroRNA-101 attenuates pulmonary fibrosis by inhibiting fibroblast proliferation and activation. The Journal of biological chemistry. 2017;292:16420–16439. doi: 10.1074/jbc.M117.805747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Das S, et al. MicroRNA-326 regulates profibrotic functions of transforming growth factor-beta in pulmonary fibrosis. American journal of respiratory cell and molecular biology. 2014;50:882–892. doi: 10.1165/rcmb.2013-0195OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Braunger B. M., Pielmeier S., Demmer C., Landstorfer V., Kawall D., Abramov N., Leibinger M., Kleiter I., Fischer D., Jagle H., Tamm E. R. TGF- Signaling Protects Retinal Neurons from Programmed Cell Death during the Development of the Mammalian Eye. Journal of Neuroscience. 2013;33(35):14246–14258. doi: 10.1523/JNEUROSCI.0991-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ren Jason Q., Li Lei. A circadian clock regulates the process of ERG b- and d-wave dominance transition in dark-adapted zebrafish. Vision Research. 2004;44(18):2147–2152. doi: 10.1016/j.visres.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 49.Rojas A, Padidam M, Cress D, Grady WM. TGF-beta receptor levels regulate the specificity of signaling pathway activation and biological effects of TGF-beta. Biochimica et biophysica acta. 2009;1793:1165–1173. doi: 10.1016/j.bbamcr.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moretto E, Murru L, Martano G, Sassone J, Passafaro M. Glutamatergic synapses in neurodevelopmental disorders. Progress in neuro-psychopharmacology & biological psychiatry. 2018;84:328–342. doi: 10.1016/j.pnpbp.2017.09.014. [DOI] [PubMed] [Google Scholar]
- 51.Gokhale A, et al. The N-ethylmaleimide-sensitive factor and dysbindin interact to modulate synaptic plasticity. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2015;35:7643–7653. doi: 10.1523/JNEUROSCI.4724-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Millar JK, et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Human molecular genetics. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- 53.Bradshaw NJ, Porteous DJ. DISC1-binding proteins in neural development, signalling and schizophrenia. Neuropharmacology. 2012;62:1230–1241. doi: 10.1016/j.neuropharm.2010.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greenhill SD, et al. NEURODEVELOPMENT. Adult cortical plasticity depends on an early postnatal critical period. Science (New York, N.Y.) 2015;349:424–427. doi: 10.1126/science.aaa8481. [DOI] [PubMed] [Google Scholar]
- 55.Lee S-A, et al. Disrupted-in-schizophrenia 1 (DISC1) regulates dysbindin function by enhancing its stability. The Journal of biological chemistry. 2015;290:7087–7096. doi: 10.1074/jbc.M114.614750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei Hong-ping, Yao Yuan-yuan, Zhang Rong-wei, Zhao Xiao-feng, Du Jiu-lin. Activity-Induced Long-Term Potentiation of Excitatory Synapses in Developing Zebrafish Retina In Vivo. Neuron. 2012;75(3):479–489. doi: 10.1016/j.neuron.2012.05.031. [DOI] [PubMed] [Google Scholar]
- 57.Carr GV, Jenkins KA, Weinberger DR, Papaleo F. Loss of dysbindin-1 in mice impairs reward-based operant learning by increasing impulsive and compulsive behavior. Behavioural brain research. 2013;241:173–184. doi: 10.1016/j.bbr.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cox MM, et al. Neurobehavioral abnormalities in the dysbindin-1 mutant, sandy, on a C57BL/6J genetic background. Genes, brain, and behavior. 2009;8:390–397. doi: 10.1111/j.1601-183X.2009.00477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dickman DK, Davis GW. The schizophrenia susceptibility gene dysbindin controls synaptic homeostasis. Science (New York, N.Y.) 2009;326:1127–1130. doi: 10.1126/science.1179685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leggio, G. M., Bucolo, C., Platania, C. B. M., Salomone, S. & Drago, F. Current drug treatments targeting dopamine D3 receptor. Pharmacology and Therapeutics, 165 (2016). [DOI] [PubMed]
- 61.Kumamoto N, et al. Hyperactivation of midbrain dopaminergic system in schizophrenia could be attributed to the down-regulation of dysbindin. Biochemical and biophysical research communications. 2006;345:904–909. doi: 10.1016/j.bbrc.2006.04.163. [DOI] [PubMed] [Google Scholar]
- 62.Pflug Renate, Nelson Ralph, Huber Sonja, Reitsamer Herbert. Modulation of horizontal cell function by dopaminergic ligands in mammalian retina. Vision Research. 2008;48(12):1383–1390. doi: 10.1016/j.visres.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang D.-Q., Zhou T.-R., McMahon D. G. Functional Heterogeneity of Retinal Dopaminergic Neurons Underlying Their Multiple Roles in Vision. Journal of Neuroscience. 2007;27(3):692–699. doi: 10.1523/JNEUROSCI.4478-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim MK, et al. Dopamine Deficiency Mediates Early Rod-Driven Inner Retinal Dysfunction in Diabetic Mice. Investigative ophthalmology & visual science. 2018;59:572–581. doi: 10.1167/iovs.17-22692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jensen RJ. Effects of Antipsychotic Drugs Haloperidol and Clozapine on Visual Responses of Retinal Ganglion Cells in a Rat Model of Retinitis Pigmentosa. Journal of ocular pharmacology and therapeutics: the official journal of the Association for Ocular Pharmacology and Therapeutics. 2016;32:685–690. doi: 10.1089/jop.2016.0102. [DOI] [PubMed] [Google Scholar]
- 66.Popova E, Kostov M, Kupenova P. Effects of dopamine D1 receptor blockade on the ERG b- and d-waves during blockade of ionotropic GABA receptors. Eye and vision (London, England) 2016;3:32. doi: 10.1186/s40662-016-0064-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Popova E., Kupenova P. Effects of dopamine receptor blockade on the intensity-response function of ERG b- and d-waves in dark adapted eyes. Vision Research. 2013;88:22–29. doi: 10.1016/j.visres.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 68.Wachtmeister Lillemor. Oscillatory potentials in the retina: what do they reveal. Progress in Retinal and Eye Research. 1998;17(4):485–521. doi: 10.1016/S1350-9462(98)00006-8. [DOI] [PubMed] [Google Scholar]
- 69.Iizuka Y., Sei Y., Weinberger D. R., Straub R. E. Evidence That the BLOC-1 Protein Dysbindin Modulates Dopamine D2 Receptor Internalization and Signaling But Not D1 Internalization. Journal of Neuroscience. 2007;27(45):12390–12395. doi: 10.1523/JNEUROSCI.1689-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu C, et al. MirSNP, a database of polymorphisms altering miRNA target sites, identifies miRNA-related SNPs in GWAS SNPs and eQTLs. BMC genomics. 2012;13:661. doi: 10.1186/1471-2164-13-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic acids research. 2008;36:D149–53. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang L, Lyerla T. Histochemical and cellular changes accompanying the appearance of lung fibrosis in an experimental mouse model for Hermansky Pudlak syndrome. Histochemistry and cell biology. 2010;134:205–213. doi: 10.1007/s00418-010-0724-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tech K, Gershon TR. Energy metabolism in neurodevelopment and medulloblastoma. Translational pediatrics. 2015;4:12–19. doi: 10.3978/j.issn.2224-4336.2015.01.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mita T, et al. Docosahexaenoic Acid Promotes Axon Outgrowth by Translational Regulation of Tau and Collapsin Response Mediator Protein 2 Expression. The Journal of biological chemistry. 2016;291:4955–4965. doi: 10.1074/jbc.M115.693499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ando H, Ichihashi M, Hearing VJ. Role of the ubiquitin proteasome system in regulating skin pigmentation. International journal of molecular sciences. 2009;10:4428–4434. doi: 10.3390/ijms10104428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stenina MA, Krivov LI, Voevodin DA, Yarygin VN. Phenotypic differences between mdx black mice and mdx albino mice. Comparison of cytokine levels in the blood. Bulletin of experimental biology and medicine. 2013;155:376–379. doi: 10.1007/s10517-013-2158-5. [DOI] [PubMed] [Google Scholar]
- 77.Fei E, et al. Nucleocytoplasmic shuttling of dysbindin-1, a schizophrenia-related protein, regulates synapsin I expression. The Journal of biological chemistry. 2010;285:38630–38640. doi: 10.1074/jbc.M110.107912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ryder PV, et al. The WASH complex, an endosomal Arp2/3 activator, interacts with the Hermansky-Pudlak syndrome complex BLOC-1 and its cargo phosphatidylinositol-4-kinase type IIalpha. Molecular biology of the cell. 2013;24:2269–2284. doi: 10.1091/mbc.e13-02-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Palmisano I, et al. The ocular albinism type 1 protein, an intracellular G protein-coupled receptor, regulates melanosome transport in pigment cells. Human molecular genetics. 2008;17:3487–3501. doi: 10.1093/hmg/ddn241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kanehisa Minoru, Sato Yoko, Furumichi Miho, Morishima Kanae, Tanabe Mao. New approach for understanding genome variations in KEGG. Nucleic Acids Research. 2018;47(D1):D590–D595. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tea M, Michael MZ, Brereton HM, Williams KA. Stability of small non-coding RNA reference gene expression in the rat retina during exposure to cyclic hyperoxia. Molecular vision. 2013;19:501–508. [PMC free article] [PubMed] [Google Scholar]
- 82.Mi Q-S, et al. Identification of mouse serum miRNA endogenous references by global gene expression profiles. PloS one. 2012;7:e31278. doi: 10.1371/journal.pone.0031278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Castorina Alessandro, Leggio Gian Marco, Giunta Salvatore, Magro Gaetano, Scapagnini Giovanni, Drago Filippo, D’Agata Velia. Neurofibromin and Amyloid Precursor Protein Expression in Dopamine D3 Receptor Knock-Out Mice Brains. Neurochemical Research. 2010;36(3):426–434. doi: 10.1007/s11064-010-0359-0. [DOI] [PubMed] [Google Scholar]
- 84.Rueden, C. T. et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics, 10.1186/s12859-017-1934-z (2017). [DOI] [PMC free article] [PubMed]
- 85.Robson John G., Saszik Shannon M., Ahmed Jameel, Frishman Laura J. Rod and cone contributions to thea-wave of the electroretinogram of the macaque. The Journal of Physiology. 2003;547(2):509–530. doi: 10.1113/jphysiol.2002.030304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.