Abstract
Many microbial species have been recognized as enteropathogens for humans. Here, we predicted the causative agents of acute diarrhea using data from multiplex quantitative PCR (qPCR) assays targeting 19 enteropathogens. For this, a case-control study was conducted at eight hospitals in Thailand. Stool samples and clinical data were collected from 370 hospitalized patients with acute diarrhea and 370 non-diarrheal controls. Multiple enteropathogens were detected in 75.7% and 13.0% of diarrheal stool samples using multiplex qPCR and bacterial culture methods, respectively. Asymptomatic carriers of enteropathogens were found among 87.8% and 45.7% of individuals by qPCR and culture methods, respectively. These results suggested the complexity of identifying causative agents of diarrhea. An analysis using the quantification cut-off values for clinical relevance drastically reduced pathogen-positive stool samples in control subjects from 87.8% to 0.5%, whereas 48.9% of the diarrheal stool samples were positive for any of the 11 pathogens. Among others, rotavirus, norovirus GII, Shigella/EIEC, and Campylobacter were strongly associated with acute diarrhea (P-value < 0.001). Characteristic clinical symptoms, epidemic periods, and age-related susceptibility to infection were observed for some enteropathogens. Investigations based on qPCR approaches covering a broad array of enteropathogens might thus improve our understanding of diarrheal disease etiology and epidemiological trends.
Subject terms: Infectious-disease epidemiology, Clinical microbiology, Infectious-disease diagnostics, Viral epidemiology, Diarrhoea
Introduction
Diarrheal diseases are one of the major causes of mortality and morbidity worldwide, especially during the first 5 years of life for individuals subjected to malnutrition1–3. Diarrhea can be defined by increased stool frequency, liquidity, or volume4. A wide range of enteropathogens including bacteria, viruses, and protozoa have been recognized as the causative agents of infectious diarrhea5,6. Several enteropathogens act either directly by modulating epithelial ion transport systems and barrier functions or indirectly via neuropeptide secretion, induction of inflammation, or by compromising intestinal absorption7. Thus, the timely identification of causes of acute diarrhea can lead to appropriate treatment, prevention, and control measures. Molecular assays targeting nucleic acid markers facilitate the screening of a broad range of enteropathogens in stool samples, consuming much less time than conventional methods such as cell culture, microscopy, and antigen-based tests. However, due to the high sensitivity of these nucleic acid amplification assays, high rates of asymptomatic carriers and mixed infection cases are commonly reported, particularly in developing country settings8–12. Several studies have shown correlations between pathogen load and severity, or have accurately diagnosed individual pathogens using qPCR assays with optimal cut-off values13–21.
In this study, we predicted the etiological agents of acute diarrhea using stool samples from hospitalized patients in Thailand using our quantitative pathogen detection procedure22 with clinically relevant cut-off values. We further investigated characteristic clinical symptoms, epidemic periods, and age-related susceptibility to infection associated with the detected enteropathogens.
Results
Detection of diarrheagenic microbes in stool specimens using multiplex qPCR assays
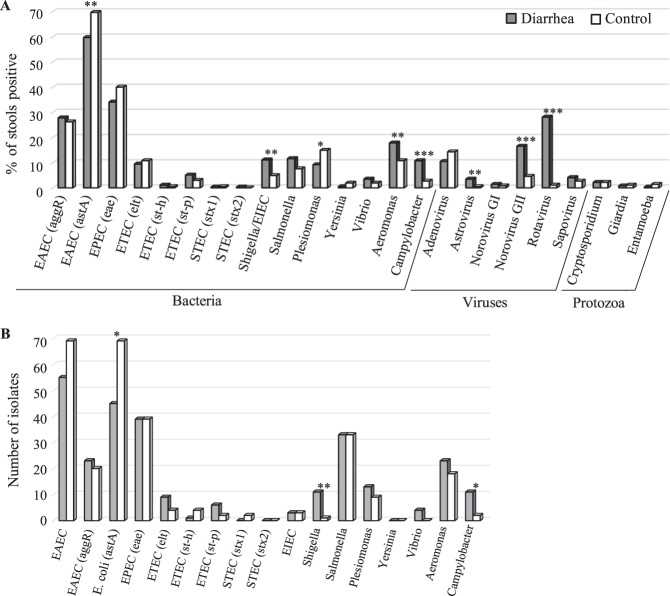
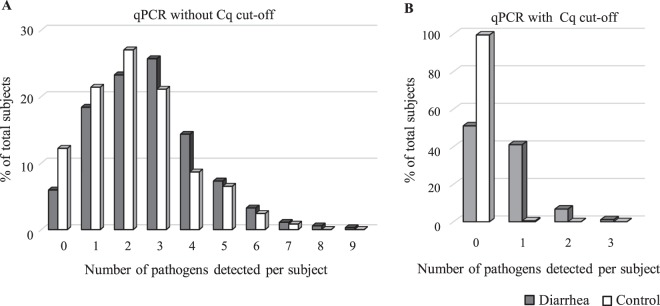
Stool specimens (370 each) from cases and controls were investigated using a multiplex qPCR panel assay to detect 24 target genes (Fig. 1A). The pathogen detection rates of bacteria, viruses, and parasites in diarrhea cases were 84.9%, 49.2%, and 3.2%, whereas the corresponding rates in the controls were 83.8%, 20.8%, and 4.9%, respectively. The numbers of Escherichia coli including enteroaggregative E. coli (EAEC) (aggR, astA) and enteropathogenic E. coli (EPEC) (eae) were relatively high in both cases and controls. Shigella/EIEC (odds ratio (OR) 2.40, P = 0.003), Aeromonas (OR 1.78, P = 0.008), Campylobacter (OR 4.21, P < 0.001), astrovirus (OR 5.57, P = 0.007), norovirus GII (OR 4.01, P < 0.001), and rotavirus (OR 31.94, P < 0.001) were detected more often in patients than in controls (Table 1). The detection rates of enteric adenovirus in the adenovirus-positive samples assessed using other qPCR assays23 were also not significant (OR 1.3, P = 0.393) (data not shown). EAEC (astA) (OR 0.64, P = 0.004) and Plesiomonas (OR 0.55, P = 0.013) were more common in non-diarrhea controls without an adjusted Cq cut-off value. An average of 2.7 ± 0.1 (standard error) pathogens was detected using qPCR in patients with diarrhea, whereas 2.3 ± 0.1 were detected in the controls (Fig. 2A). The percentages of “no pathogen detected” and mixed infection were 5.9% and 75.7% (IQR, 3; mean, 3.3) in cases, and 12.2% and 66.5% (IQR, 2; mean, 3.1) in controls, respectively.
Figure 1.
Detection of enteropathogens in 370 diarrheal and 370 non-diarrheal control subjects. (A) Proportions of patients and control subjects who tested positive by qPCR assays. (B) Number of isolates by bacterial culture methods. *P < 0.05; **P < 0.01; ***P < 0.001.
Figure 2.
Number of pathogens detected per subject using qPCR among 370 each of diarrheal (an average of 2.7 ± 0.1 (standard error) pathogens) and non-diarrheal control subjects (an average of 2.3 ± 0.1 (standard error) pathogens) (A) and the distribution based on the quantitative cycle (Cq) cut-off values mentioned in Table 1 (B).
Cultivation and identification of bacterial enteropathogens isolated from stool specimens
Two hundred and thirty-four and 230 bacterial pathogens consisting of Aeromonas caviae, A. hydrophila, A. sobria, A. veronii, Campylobacter coli, C. fetus, C. jejuni, E. coli, Plesiomonas shigelloides, Salmonella enterica, Shigella boydii, S. flexneri, S. sonnei, Vibrio fluvialis, and V. parahaemolyticus were isolated from 170 cases (45.9%) and 169 controls (45.7%), respectively (Fig. 1B, Table 2). Multiple bacterial pathogens were isolated from 13.0% cases and 12.7% controls (Table 3). The most common bacteria were E. coli (31.4% vs 37.0%), Salmonella (8.9% vs 8.9%), and Aeromonas (5.9% vs 4.3%) in both cases and controls (Table 2). The isolation rates of Shigella and Campylobacter species were statistically associated with diarrhea, and S. sonnei and C. jejuni were the dominant species. Further, 306 isolates of E. coli were classified into 44 genotypes by multiplex-PCR assays and ≥8 different O-serotypes were identified among 66 Salmonella isolates. Most bacterial pathogens were similarly distributed in both patients and healthy control subjects.
Table 2.
Bacterial pathogens isolated from cases and controls and their features.
| Pathogen | Genotype or Serogroup | Number of isolates in cases | Number of isolates in controls | |
|---|---|---|---|---|
| Aeromonas | 23 | 18 | ||
| A. caviae | 8 | 10 | ||
| A. hydrophila | 4 | 0 | ||
| A. sobria | 11 | 7 | ||
| A. veronii | 0 | 1 | ||
| Campylobacter | 11 | 2 | ||
| C. coli | 1 | 0 | ||
| C. fetus | 1 | 0 | ||
| C. jejuni | 9 | 2 | ||
| Escherichia | E. coli | 139 | 167 | |
| EAEC | 55 | 69 | ||
| uidA, aafII, astA | 0 | 1 | ||
| uidA, astA, pCVD432 | 3 | 8 | ||
| uidA, astA, pCVD432, pic | 3 | 0 | ||
| uidA, astA, pic | 2 | 2 | ||
| uidA, pic | 8 | 12 | ||
| uidA, pCVD432, pic | 4 | 2 | ||
| uidA, pCVD432 | 10 | 20 | ||
| astA, pCVD432 | 0 | 2 | ||
| pic | 2 | 2 | ||
| uidA, aggR | 3 | 3 | ||
| uidA, aggR, astA | 1 | 1 | ||
| uidA, aggR, astA, pCVD432 | 0 | 1 | ||
| uidA, aggR, astA, pCVD432, pic | 2 | 5 | ||
| uidA, aggR, pCVD432 | 7 | 2 | ||
| uidA, aggR, pCVD432, pic | 9 | 4 | ||
| uidA, aggR, pic | 1 | 2 | ||
| aggR, pic, pCVD432 | 0 | 1 | ||
| aggR, pCVD432 | 0 | 1 | ||
| EPEC | 39 | 42 | ||
| uidA, eae | 32 | 24 | ||
| uidA, eae, astA | 1 | 3 | ||
| uidA, eae, bfpA | 1 | 1 | ||
| uidA, eae, bfpB | 1 | 5 | ||
| uidA, bfpB | 0 | 2 | ||
| uidA, bfpB, astA | 0 | 1 | ||
| eae | 4 | 6 | ||
| ETEC | 14 | 9 | ||
| uidA, elt | 3 | 1 | ||
| uidA, elt, astA | 4 | 2 | ||
| uidA, elt, est-p | 1 | 0 | ||
| uidA, elt, est-p, astA | 0 | 1 | ||
| uidA, est-h | 1 | 1 | ||
| uidA, est-h, astA | 0 | 1 | ||
| uidA, est-h, astA, bfpA | 0 | 1 | ||
| uidA, est-p | 2 | 1 | ||
| uidA, est-p, astA | 2 | 0 | ||
| elt, est-p | 1 | 0 | ||
| est-h, astA, bfpA | 0 | 1 | ||
| STEC | uidA, stx1 | 0 | 2* | |
| EIEC | 3 | 3 | ||
| uidA, ipaH, virF | 3 | 1 | ||
| uidA, invE, astA | 0 | 1 | ||
| invE | 0 | 1 | ||
| DAEC | 1 | 5 | ||
| uidA, daaE | 1 | 4 | ||
| uidA, daaE, astA | 0 | 1 | ||
| Other | 27 | 37 | ||
| uidA, astA | 27 | 36 | ||
| astA | 0 | 1 | ||
| Shigella | 11 | 1 | ||
| S. boydii | 0 | 1 | ||
| S. flexneri | 3 | 0 | ||
| S. sonnei | 8 | 0 | ||
| Salmonella | S. enterica | 33 | 33 | |
| O4 | 20 | 9 | ||
| O7 | 4 | 10 | ||
| O8 | 1 | 4 | ||
| O9 | 3 | 2 | ||
| O3,10 | 3 | 2 | ||
| O13 | 2 | 1 | ||
| O35 | 0 | 1 | ||
| Other O-antigen groups | 0 | 4 | ||
| Plesiomonas | P. shigelloides | 13 | 9 | |
| Vibrio | 4 | 0 | ||
| V. fluvialis | 1 | 0 | ||
| V. parahaemolyticus | 3 | 0 | ||
*Production of verotoxin-1 was confirmed by reverse passive latex agglutination, using the VTEC-RPLA kit (Denka Seiken, Japan).
Table 3.
Number of bacterial pathogens isolated per subject.
| Number of pathogen | Diarrhea | Control |
|---|---|---|
| 0 | 200 (54.1%) | 201 (54.3%) |
| 1 |
122 (33.0%) |
122 (33.0%) |
| 2 |
33 (8.9%) |
35 (9.5%) |
| 3 |
14 (3.8%) |
11 (3.0%) |
| 4 |
1 (0.3%) |
0 (0%) |
| 5 |
0 (0%) |
1 (0.3%) |
| Total |
370 (100%) |
370 (100%) |
Statistical analysis of qPCR data with the clinically relevant cut-off values
Due to the issues associated with high rates of asymptomatic carriage and the detection of multiple pathogens in patients, we next compared the quantification cycle values (Cq) of qPCR for each target gene in the case and control samples (Fig. S1). Cq cut-off values for each pathogen were determined using receiver operating curve (ROC) analysis (see Methods). Results showed that 12 targets had statistically significant associations with diarrhea occurrence (Table 1). With the clinically relevant cut-off values, pathogen-positive stool samples in control subjects were drastically reduced from 87.8% to 0.5%, whereas 181 diarrheal stools (48.9%) were positive for any of the 11 enteropathogens - rotavirus, norovirus GII, Shigella/enteroinvasive E. coli (EIEC), Campylobacter, EAEC, Salmonella, Plesiomonas, Aeromonas, sapovirus, astrovirus, and enterotoxigenic E. coli (ETEC) – that were strongly associated with acute diarrhea (Fig. 2B, Table 1). Moreover, rotavirus, norovirus GII, Shigella/enteroinvasive E. coli (EIEC), Campylobacter, EAEC, Salmonella, Plesiomonas, Aeromonas, sapovirus, astrovirus, and enterotoxigenic E. coli (ETEC) were strongly associated with acute diarrhea. Among 181 patients, single and multiple infections with the causative agents accounted for 84.0% and 16.0%, cases, respectively (Table 4). Mixed infection cases including bacterial–viral (7.7%, 14/181), bacterial–bacterial (4.4%, 8/181), and viral–viral (3.9%, 7/181) infections were still recognized despite the use of the stringent cut-off values. These mixed infection cases were observed more frequently in younger age groups (≤15 years of age) than in older age groups (≥16 years old) (P = 0.037).
Table 1.
Detection of enteropathogen targets in stool samples from cases and control subjects, odds ratio, Cq cutoff value, and predicted causative agents by quantitative PCR analyses.
| Detection by quantitative PCR | Cq cutoff value | Detection based on optimal cut-off | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Diarrhea (%) | Control (%) | OR | P-value | Diarrhea (%) | Control (%) | OR | P-value | ||
| Rotavirus | 104 (28.1) | 4 (1.1) | 31.94 | <0.001 | 27.98 | 80 (21.6) | 0 (0) | 205.34 | <0.001 |
| Norovirus GII | 61 (16.5) | 17 (4.6) | 4.01 | <0.001 | 20.23 | 29 (7.8) | 0 (0) | 64.01 | <0.001 |
| Shigella/EIEC | 41 (11.1) | 18 (4.9) | 2.40 | 0.003 | 30.06 | 21 (5.7) | 0 (0) | 45.58 | <0.001 |
| Campylobacter | 40 (10.8) | 10 (2.7) | 4.21 | <0.001 | 28.97 | 18 (4.9) | 0 (0) | 38.89 | <0.001 |
| EAEC (aggR) | 103 (27.8) | 99 (26.8) | 1.06 | 0.805 | 22.54 | 11 (3.0) | 0 (0) | 23.70 | 0.001 |
| EAEC (astA) | 221 (59.7) | 259 (70.0) | 0.64 | 0.004 | 19.95 | 10 (2.7) | 1 (0.3) | 7.17 | 0.011 |
| Aeromonas | 66 (17.8) | 40 (10.8) | 1.78 | 0.008 | 27.42 | 9 (2.4) | 0 (0) | 19.47 | 0.004 |
| Sapovirus | 15 (4.1) | 10 (2.7) | 1.50 | 0.416 | 27.20 | 9 (2.4) | 0 (0) | 19.47 | 0.004 |
| Astrovirus | 13 (3.5) | 2 (0.5) | 5.57 | 0.007 | 26.21 | 8 (2.2) | 1 (0.3) | 5.78 | 0.038 |
| Salmonella | 43 (11.6) | 28 (7.6) | 1.60 | 0.080 | 25.56 | 7 (1.9) | 0 (0) | 15.29 | 0.015 |
| Plesiomonas | 33 (8.9) | 56 (15.1) | 0.55 | 0.013 | 26.17 | 6 (1.6) | 0 (0) | 13.21 | 0.031 |
| ETEC (elt) | 35 (9.5) | 40 (10.8) | 0.86 | 0.626 | 25.29 | 6 (1.6) | 0 (0) | 13.21 | 0.031 |
| Detection by quantitative PCR | Cq cutoff value | Detection based on optimal cut-off | |||||||
| Diarrhea (%) | Control (%) | OR | P-value | Diarrhea (%) | Control (%) | OR | P-value | ||
| EPEC | 125 (34.1) | 149 (40.3) | 0.77 | 0.094 | 21.08 | 5 (1.4) | 0 (0) | 11.15 | 0.062 |
| ETEC (st-p) | 19 (5.1) | 11 (3.0) | 1.73 | 0.191 | 26.24 | 4 (1.1) | 0 (0) | 9.10 | 0.124 |
| Adenovirus | 39 (10.5) | 53 (14.3) | 0.71 | 0.147 | 14.45 | 4 (1.1) | 0 (0) | 9.10 | 0.124 |
| Vibrio | 13 (3.5) | 8 (2.2) | 1.61 | 0.376 | 30.09 | 3 (0.8) | 0 (0) | 7.06 | 0.249 |
| Norovirus GI | 5 (1.4) | 3 (0.8) | 1.58 | 0.725 | 28.02 | 3 (0.8) | 0 (0) | 7.06 | 0.249 |
| ETEC (st-h) | 4 (1.1) | 2 (0.5) | 1.81 | 0.686 | — | 0 (0) | 0 (0) | 1 | 1 |
| Cryptosporidium | 8 (2.2) | 8 (2.2) | 1 | 1 | — | 0 (0) | 0 (0) | 1 | 1 |
| Giardia | 3 (0.8) | 4 (1.1) | 0.78 | 1 | 24.52 | 1 (0.3) | 0 (0) | 3.01 | 1 |
| Entamoeba | 1 (0.3) | 6 (1.6) | 0.23 | 0.123 | — | 0 (0) | 0 (0) | 1 | 1 |
| Yersinia | 2 (0.5) | 7 (1.9) | 0.33 | 0.177 | — | 0 (0) | 0 (0) | 1 | 1 |
| STEC (stx1) | 1 (0.3) | 2 (0.5) | 0.60 | 1 | — | 0 (0) | 0 (0) | 1 | 1 |
| STEC (stx2) | 1 (0.3) | 0 (0) | 3.01 | 1 | 37.74 | 1 (0.3) | 0 (0) | 3.01 | 1 |
The 12 targeted genes in 11 enteropathogens in the top half of the table were determined as the causative agents of acute diarrhea. Receiver operating curve (ROC) analyses were conducted to detect the optimal cut-off value. The cut-off value with the largest OR, which satisfied more than 95% specificity, was considered optimal. The relationships between case (control) and positive or negative control based on the calculated cut-off value were reported as ORs and P-values determined using the Fisher’s exact test.
Abbreviations: OR, odd ratio; Cq, quantification cycle; EIEC, Enteroinvasive E. coli; EAEC, Enteroaggregative E. coli; ETEC, Enterotoxigenic E. coli; EPEC, Enteropathogenic E. coli; STEC, Shiga toxin-producing E. coli.
Table 4.
Number of cases and combinations of causative agents estimated among 181 patients.
| Selected agents | Single infection (%) |
Mixed infection | Total | |
|---|---|---|---|---|
| with one pathogen (cases) | with two pathogens (cases) | |||
| Rotavirus | 70 | 8 | 2 | 80 |
| (87.5) | astA* (3), Camp (2), NorII (2), Astro (1) | aggR & astA (1), Camp & Sal (1) | ||
| Norovirus GII | 20 | 8 | 1 | 29 |
| (69.0) | Camp (3), Astro (2), Rota (2), Sapo (1) | aggR & astA (1) | ||
| Shigella/EIEC | 17 | 4 | 0 | 21 |
| (81.0) | Sapo (1), Ples (1), astA (1), elt (1) | |||
| Campylobacter | 10 | 7 | 1 | 18 |
| (55.6) | NorII (3), Rota (2), Sapo (1), aggR (1) | Rota & Sal (1) | ||
| EAEC (aggR) | 6 | 3 | 2 | 11 |
| (54.5) | astA (1), Camp (1), elt (1) | astA & Rota (1), astA & NorII (1) | ||
| EAEC (astA) | 2 | 6 | 2 | 10 |
| (20.0) | Rota (3), aggR (1), Ples (1), Shig (1) | Rota & aggR (1), NorII & aggR (1) | ||
| Aeromonas | 8 | 1 | 0 | 9 |
| (88.9) | Ples (1) | |||
| Sapovirus | 4 | 5 | 0 | 9 |
| (44.4) | Shig (2), Camp (1), Astro (1), NorII (1) | |||
| Astrovirus | 4 | 4 | 0 | 8 |
| (50.0) | NorII (2), Rota (1), Sapo (1) | |||
| Salmonella | 5 | 0 | 2 | 7 |
| (71.4) | Rota & Camp (1), Ples & elt (1) | |||
| Plesiomonas | 3 | 2 | 1 | 6 |
| (50.0) | Aero (1), astA (1) | elt & Sal (1) | ||
| ETEC (elt) | 3 | 2 | 1 | 6 |
| (50.0) | aggR (1), Shig (1) | Ples & Sal (1) | ||
*astA; EAEC (astA), Camp; Campylobacter, Astro; astrovirus, aggR; EAEC (aggR), NorII; norovirus GII, Rota; rotavirus, Sapo; sapovirus, Ples; Plesiomonas, Shig; Shigella/EIEC, Aero; Aeromonas, Sal; Salmonella, elt; ETEC (elt).
Patient features and clinical symptoms that might be caused by the detected pathogens
Patient features and clinical symptoms that might be caused by the detected pathogens were then investigated. Patients with a high frequency of defecation (more than 10 times per day) had significantly higher numbers of Aeromonas than all cases (P = 0.006) (Table S1). Rotavirus infection was significantly associated with fever (P = 0.034). Moreover, bacterial infections rather than viral infections tended to cause abdominal pain (P < 0.001). Shigella/EIEC (P = 0.020), Salmonella (P = 0.013), Aeromonas (P = 0.027), and Plesiomonas (P = 0.037) were more related to abdominal pain, whereas norovirus GII infection was rare in individuals with abdominal pain (P = 0.014). More than 20 WBCs/hpf (white blood cells per high power field) and red blood cells were present in the stool samples of patients with Shigella/EIEC infection (P < 0.001 and P = 0.012, respectively). Vomiting frequency was higher in rotavirus- (P < 0.001) and norovirus GII -infected patients (P = 0.009) than in all patients, but the incidence of nausea was significantly associated with Aeromonas- (P = 0.020) and sapovirus-infected patients (P = 0.037). Rotavirus (P = 0.001), norovirus GII (P = 0.004), and Campylobacter (P = 0.040) were more often detected in patients less than five years of age, whereas Shigella/EIEC (P = 0.001), Aeromonas (P = 0.010), and Plesiomonas (P = 0.021) were detected in patients older than five years of age (Table S2). The length of hospitalization for patients infected with each pathogen was not significantly different (P > 0.05). Pathogens that were associated with dry and cool seasons included rotavirus, followed by norovirus GII, whereas those associated with the other seasons (hot or rainy) included EAEC, Shigella/EIEC, and Plesiomonas (Table S3).
Discussion
In this study, we investigated the etiological agents of acute diarrhea in patients with severe symptoms by quantitatively detecting a broad range of known enteropathogens. Our highly sensitive molecular assays, as well as bacteriological culture methods, detected high rates of asymptomatic cases and mixed infections in the study population. As the pathogen quantities in stool specimens from case and control subjects estimated using qPCR assays were different (Fig. S1), we set cut-off values of Cq for the specific diagnosis of patients and then predicted the causative pathogens of acute diarrhea. This considerably improved the assay and allowed differentiation between symptomatic and asymptomatic carriage; it also assisted in specifying disease-associated pathogens in the patients. Most illnesses were caused by rotavirus, followed by norovirus GII, Shigella/EIEC, and Campylobacter.
The Cq values obtained from qPCR assays for case and control samples were compared in each age group or each month (season) to identify trends and patterns of enteropathogen infections. The detection rates of Shigella/EIEC, Aeromonas, and Plesiomonas were higher in the ≥5 years of age group than in the <5 years of age group. In contrast, in addition to rotavirus, norovirus GII, and Campylobacter, ETEC, and sapovirus were likely to be more abundant in younger age groups (Table S2 and Fig. S3). These results might be due to differences in susceptibility to infections and/or lifestyles such as eating habits. Notably, EAEC (aggR) infection was also associated with diarrhea under the cut-off condition and nine of 11 patients were 1 year of age or under. Persistent diarrhea with EAEC is most frequently reported in children aged ≤1 year24. Our results thus indicate that infants are more susceptible to EAEC infection than older individuals. Furthermore, regarding asymptomatic infections with norovirus GII, 16 of 17 cases were less than 5 years of age and the remaining one was 6 years old. Infected asymptomatic carriers in young children might be important for the transmission of norovirus infection. Moreover, the seasonal trends of viral infections were clearer than those of bacterial infections (Table S3 and Fig. S2). Rotavirus, norovirus, and astrovirus were more likely to be abundant in the cool and dry season (November to February), whereas Shigella/EIEC, EAEC (aggR), and Plesiomonas were likely to be abundant in other seasons (rainy and/or hot). This approach can be useful for tracking the distribution and emergence of subclinically-infected persons, as well as diarrheal patients.
Next, we focused on the clinical symptoms of patients who were infected with the detected causative pathogens (Table S1). The patients infected with rotavirus presented with fever and mild to severe diarrhea. Vomiting was more common in rotavirus- and norovirus-infected patients than in all diarrhea patients; however, abdominal pain was less frequent in norovirus-infected patients (P = 0.014) than in rotavirus-infected patients (P = 0.139). In addition, rates of nausea in rotavirus- and norovirus-infected patients were 18/77 (23.4%) and 3/29 (10.3%), respectively. This might indicate a characteristic symptom of norovirus infection, that is, the sudden onset of vomiting. Shigella/EIEC, Aeromonas, Salmonella, and Plesiomonas infections were associated with abdominal pain. In patients positive for Shigella/EIEC, the number of fecal leucocytes (WBCs) and the appearance of erythrocytes (RBCs) in stool were significantly higher, which is one of the known characteristics of Shigellosis25. Clinical symptoms deduced from the qPCR results and patient data thus appear to correspond with the generally known symptoms caused by rotavirus, norovirus, or Shigella.
Campylobacter was also detected at significantly higher levels in diarrhea cases using qPCR and culture methods. This genus was likely to cause fever and unlikely to cause high frequent defecation in children. The role of Aeromonas as an etiological agent of acute diarrhea has been controversial26,27. Previously, in a challenge study, diarrhea was demonstrated in only two of 57 human volunteers, with doses ranging from 104 to 1010 CFU26. In this study, many subclinical Aeromonas infections were recognized; however, such infections were associated with diarrhea in the cutoff condition and were accompanied by abdominal pain, nausea, and a high frequency of defecation. Thus, our analysis underscores the risk of illness caused by Aeromonas.
EAEC infection was not associated with any obvious characteristic clinical symptom. Investigations focusing on children (aged ≤1 year) who are probably highly susceptible to infection can be beneficial to understand this pathogen. Two genes, astA and aggR, were used in our qPCR panel assay to detect EAEC; however, astA was not always detected together with aggR. astA encodes EAST1 (EAEC heat-stable enterotoxin), which shares the functional properties of the enterotoxin (STa) secreted by ETEC24, whereas aggR is known as a transcriptional regulator (a key virulence regulator) and an important marker for virulent EAEC28. astA was detected not only in some isolates of EAEC, but also in isolates of EPEC, ETEC, EIEC, DAEC (Diffusely adherent E. coli), and non-categorized diarrheagenic E. coli (Table 2). Furthermore, single infection with astA-carrying agents was detected in only two of 10 cases (Table 4). These results imply that the presence of astA is not sufficient to conclusively identify the causative agents of diarrhea. Further, evaluation of the pathogenicity of these suspected pathogens will also be important.
This study has certain limitations. We could not determine the causative agents in approximately 50% of cases, although many enteropathogens were detected. Our estimates of causative agents were based on differences in pathogen loads between patient and control stool samples. In reality, the association between pathogen quantity and disease is host-pathogen-specific. In addition, pathogen quantities change in each stage of clinical manifestation8,29. These issues should be addressed in future studies to accurately predict etiological agents of disease. Further, the lower detection rates of certain enteropathogens such as Vibrio, norovirus GI, and STEC were not sufficient to interpret the qPCR results in this study. Further, the lack of consideration of matching pairs in the statistical analysis is also a limitation.
In a recent report, travelers’ diarrhea in foreign visitors to Thailand was investigated using a TaqMan array card assay30. One hundred and seventy-three cases from the in-patient or out-patient department in a private hospital and 165 non-diarrheal subjects were enrolled in that study. The results of that study were in agreement with our results regarding the high detection rates of Campylobacter, EAEC, EPEC, and norovirus GII, and the extremely low detection rates of protozoa including Cryptosporidium, Entamoeba histolytica, and Giardia. In contrast, rotavirus, adenovirus, and astrovirus were rarely detected in their study. The low detection rate of rotavirus might be due to the age of their enrolled patients, who were more than 18 years of age. Accordingly, massive surveillance using several approaches will assist in identifying a panel of microbial pathogens associated with acute diarrhea.
In conclusion, the causative agents of acute diarrhea were predicted using a multiplex qPCR panel assay with patient-specific cut-off values, and the characteristic clinical symptoms caused by each enteropathogen were partially revealed. This approach can provide new insights into infectious diarrheal diseases. Data accumulation, validation of the results, and the re-design/adjustment for known or unknown target enteropathogens in a multiplex quantitative assay could be critical to establish superior diagnostic procedures.
Methods
Specimen collection
Stool specimens were collected from patients hospitalized with acute diarrhea from eight Thai government hospitals nationwide during April 2016 to March 2018 (Table S4). Of all cases, 353 (95.4%) were patients with abnormally loose stools more than three times within 24 h and a diarrhea-free time of at least 7 days prior to the collection of stool specimens. The remaining 4.6% cases were associated with less-frequent defecation. Eligible cases were excluded from the study if they had known immunodeficiency or chronic causes of diarrheal symptoms, or if stool volume was too small (<3 g) for the experiments. Stool specimens were collected from patients prior to starting the antibiotic treatment in the hospitals or immediately after antibiotic treatment (<1 h). For each diarrhea case, one healthy volunteer who had no history of diarrhea for at least 30 days before enrolment and who was a match with the case regarding age (mostly within 3 years of the age of the case) and area of residence (living in at least the same province) was selected among the residents or visitors to the hospital by public health officers, coordinators, or nurses. Stool specimens collected in a sterile container were first stored at 4–10 °C at each site and shipped overnight on ice packs to the central reference laboratory of the National Institute of Health (NIH) of Thailand.
Culture methods
Stool samples collected from diarrheal and non-diarrheal control subjects were cultured as part of routine work—for example, direct plating/selective enrichment, isolation, and identification by biochemical tests according to the procedure of the NIH of Thailand. The viable enteric bacteria including Salmonella spp., Shigella spp., Vibrio spp., Aeromonas spp., P. shigelloides, E. coli, and Campylobacter spp. were isolated. Some isolates of enteropathogens were further confirmed or characterized by PCR and serotyping. Five colonies of E. coli on MacConkey agar, sorbitol MacConkey agar, Salmonella-Shigella agar, or xylose lysine deoxycholate agar were examined by multiplex-PCR assays31–34 for the detection and differentiation of pathogenic E. coli. For the isolation of Campylobacter spp., 1–2 g of stool was enriched in 2 ml Preston broth at 37 °C for 3–4 h before dropping stool suspensions on modified charcoal, cefoperazone, deoxycholate agar (mCCDA) and incubating them at 37 °C for 48 h in anaerobic jars under microaerophilic conditions (Anaero Pack-MicroAero, Mitsubishi Gas Chemical, Japan).
Quantitative PCR assay
We used TaqMan real-time PCR-based assays for the simultaneous detection of enteropathogens, namely astrovirus (ORF1a), sapovirus (polymerase/capsid junction), adenovirus (hexon), norovirus GI (RdRp/capsid junction), norovirus GII (ORF1-ORF2), rotavirus group A (NSP3), Cryptosporidium spp. (COWP), Giardia lamblia (ITS1), E. histolytica (18S rRNA), P. shigelloides (hugA), Campylobacter spp. (gyrB), Vibrio spp. (toxR), Salmonella spp. (invA), Aeromonas spp. (aerA), Yersinia spp. (lysP), Shigella/EIEC (ipaH), EPEC (eae), ETEC (elt, est-h, est-p), EAEC (aggR, astA), and Shiga toxin-producing E. coli (STEC) (stx1, stx2), following the detection procedure described previously22. Several of the defining markers of E. coli pathotypes are proven virulence determinants of the respective pathotype, but for EAEC, the essential virulence determinant has not been proven35.
Total RNA and DNA were extracted from individual stool samples using QIAamp viral RNA and QIAamp fast DNA stool mini kits (cat#52906 and #51604, Qiagen, USA), respectively, either manually or on a robotic workstation for the automated purification of nucleic acids. For viral RNA/DNA preparation, whole stool (200 mg) was suspended in 1.8 ml saline, which was centrifuged at 4,000 × g for 20 min and the supernatant containing viral particles was separated. For bacterial and parasitic DNA preparation, diarrheal stool samples were centrifuged at 15,000 × g for 1 min to obtain a stool pellet (wet weight, 200 mg). qPCR assays were performed using an Applied Biosystems 7500 Fast real-time PCR system (Foster City, CA, USA). Target genes were amplified using the QuantiFast Pathogen RT-PCR and PCR kits, respectively. Herein, pure water was used as an extraction control to monitor DNA cross-contamination and for environmental contaminants during extraction. For preventing laboratory contamination, preparation of the PCR master mix, extraction of DNA/RNA from stool specimens, addition of DNA/RNA templates to the master mix, and qPCR reactions were conducted in separate areas/rooms in our laboratory. Moreover, each batch of the master mix for real-time PCR reaction was always tested to monitor contamination, along with the test samples.
qPCR data were interpreted according to the following criteria: (i) quantification cycle (Cq) value, defined as the number of PCR cycles where the fluorescent signal exceeded the detection threshold, which was fixed at 0.2, 0.3, and 0.5 relative fluorescence units for viral, parasitic, and bacterial targets, respectively; (ii) greater than or equal to a Cq value of 41 was considered as negative for all pathogens in this study; (iii) signals from the internal control (IC) RNA and IC DNA had Cq values of 31 ± 3 and 32 ± 3, respectively; (iv) positive and no template controls (NTC) were used and verified for validity in every qPCR run22.
Statistical analysis
We reported the number of detections and their percentage for each pathogen by case and control groups. We also estimated the ORs and performed Fisher’s exact tests. The complex infections were counted and reported as proportions. For each pathogen, Cq values of qPCR-positive samples were presented as median, interquartile range (IQR), and total range. ROC analyses were conducted to detect the optimal cut-off value. The cut-off value with the largest OR, which satisfied more than 95% specificity, was considered optimal. In all procedures, we calculated the OR based on the contingency table by adding a 0.5 correction value to all cells (Haldane-Anscombe 1/2 correction) to address zero- cell in the two-by-two contingency table for case/control and positive/negative. The relationships between case (control) and positive or negative controls based on the calculated cut-off value were reported as ORs and P-values, as determined using the Fisher’s exact test. All statistical analyses were performed using R (The R Foundation for Statistical Computing, ver. 3.5.1) and all tests were two-tailed. P-values < 0.05 were considered statistically significant.
Ethical statements
The clinical protocol was approved by the Ethical Review Committee for Research in Human Subjects, Ministry of Public Health, Thailand (reference no. 44/2558), Institutional Review Board BIDI (S023q/58), Prapokklao Hospital (CTIREC044), Sunprasitthiprasong Hospital (063/2559), Chiang Rai Prachanukroh Hospital (CR 0032.102/9844), and Institutional Review Board of Maesot General Hospital and Samutsakhon Hospital. Stool specimens were collected from all the volunteers who provided written informed consent. The methods were carried out in accordance with the approved guidelines.
Supplementary information
Acknowledgements
The authors would like to thank all the volunteers and the health care professionals who participated in this study. This work was supported by the Japan Initiative for Global Research Network on Infectious Diseases (J-GRID) of the Ministry of Education, Culture, Sports, Science & Technology of Japan (MEXT), Japan Agency for Medical Research and Development (AMED) under grant number JP18fm0108003, and the Department of Medical Sciences (DMSc), the Ministry of Public Health, Thailand.
Author contributions
K.O., W.W. and S.H. conceived the study. K.O., W.W. and W.K. designed the protocol. P.K., N.C., W.M., N.A., P.K., P.U., C.T., S.K., L.J., C.J., S.N. and W.S. coordinated specimen collection with hospital staff and patients and clinical data analysis at the site. S.C., P.W., W.W., W.K., N.S., T.W. and P.A.O. performed bacterial culture and characterization of isolates. W.W., W.K. and N.S. performed real-time PCR and data analysis. S.K., W.W. and K.O. performed the statistical analysis. K.O. and S.H. were responsible for the overall design and writing. All authors reviewed the draft and approved the decision to submit for publication.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-60711-1.
References
- 1.Brown KH. Diarrhea and malnutrition. J. Nutr. 2003;133:328s–332s. doi: 10.1093/jn/133.1.328S. [DOI] [PubMed] [Google Scholar]
- 2.Petri WA, Jr., et al. Enteric infections, diarrhea, and their impact on function and development. J. Clin. Invest. 2008;118:1277–1290. doi: 10.1172/JCI34005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferdous F, et al. Severity of diarrhea and malnutrition among under five-year-old children in rural Bangladesh. Am. J. Trop. Med. Hyg. 2013;89:223–228. doi: 10.4269/ajtmh.12-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sweetser S. Evaluating the patient with diarrhea: a case-based approach. Mayo Clin. Proc. 2012;87:596–602. doi: 10.1016/j.mayocp.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischer Walker CL, Sack D, Black RE. Etiology of diarrhea in older children, adolescents and adults: a systematic review. PLoS Negl. Trop. Dis. 2010;4:e768. doi: 10.1371/journal.pntd.0000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Operario DJ, Houpt E. Defining the causes of diarrhea: novel approaches. Curr. Opin. Infect. Dis. 2011;24:464–471. doi: 10.1097/QCO.0b013e32834aa13a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodges K, Gill R. Infectious diarrhea: Cellular and molecular mechanisms. Gut Microbes. 2010;1:4–21. doi: 10.4161/gmic.1.1.11036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Platts-Mills JA, Liu J, Houpt ER. New concepts in diagnostics for infectious diarrhea. Mucosal Immunol. 2013;6:876–885. doi: 10.1038/mi.2013.50. [DOI] [PubMed] [Google Scholar]
- 9.Horwood PF, et al. A high burden of asymptomatic gastrointestinal infections in traditional communities in Papua New Guinea. Am. J. Trop. Med. Hyg. 2017;97:1872–1875. doi: 10.4269/ajtmh.17-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eibach D, et al. Application of a multiplex PCR assay for the detection of gastrointestinal pathogens in a rural African setting. BMC Infect. Dis. 2016;16:150. doi: 10.1186/s12879-016-1481-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, et al. Development and assessment of molecular diagnostic tests for 15 enteropathogens causing childhood diarrhoea: a multicentre study. Lancet Infect. Dis. 2014;14:716–724. doi: 10.1016/S1473-3099(14)70808-4. [DOI] [PubMed] [Google Scholar]
- 12.Taniuchi M, et al. Etiology of diarrhea in Bangladeshi infants in the first year of life analyzed using molecular methods. J. Infect. Dis. 2013;208:1794–1802. doi: 10.1093/infdis/jit507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phillips G, et al. Diagnosing norovirus-associated infectious intestinal disease using viral load. BMC Infect. Dis. 2009;9:63. doi: 10.1186/1471-2334-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phillips G, et al. Diagnosing rotavirus A associated IID: Using ELISA to identify a cut-off for real time RT-PCR. J. Clin. Virol. 2009;44:242–245. doi: 10.1016/j.jcv.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Bolotin S, et al. Correlation of real time PCR cycle threshold cut-off with Bordetella pertussis clinical severity. PloS One. 2015;10:e0133209. doi: 10.1371/journal.pone.0133209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jansen RR, et al. Frequent detection of respiratory viruses without symptoms: toward defining clinically relevant cutoff values. J. Clin. Microbio.l. 2011;49:2631–2636. doi: 10.1128/JCM.02094-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang G, et al. Quantitation of group A rotavirus by real-time reverse-transcription-polymerase chain reaction: correlation with clinical severity in children in South India. J. Med. Virol. 2004;73:118–122. doi: 10.1002/jmv.20053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barletta F, et al. Quantitative real-time polymerase chain reaction for enteropathogenic Escherichia coli: a tool for investigation of asymptomatic versus symptomatic infections. Clin. Infect. Dis. 2011;53:1223–1229. doi: 10.1093/cid/cir730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elfving K, et al. Real-time PCR threshold cycle cutoffs help to identify agents causing acute childhood diarrhea in Zanzibar. J. Clin. Microbiol. 2014;52:916–923. doi: 10.1128/JCM.02697-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li LL, et al. Aetiology of diarrhoeal disease and evaluation of viral-bacterial coinfection in children under 5 years old in China: a matched case-control study. Clin. Microbiol. Infect. 2016;22:381.e389–381.e316. doi: 10.1016/j.cmi.2015.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, et al. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: a reanalysis of the GEMS case-control study. Lancet (London, England) 2016;388:1291–1301. doi: 10.1016/S0140-6736(16)31529-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wongboot W, Okada K, Chantaroj S, Kamjumphol W, Hamada S. Simultaneous detection and quantification of 19 diarrhea-related pathogens with a quantitative real-time PCR panel assay. J. Microbiol. Methods. 2018;151:76–82. doi: 10.1016/j.mimet.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 23.van Maarseveen NM, Wessels E, de Brouwer CS, Vossen AC, Claas EC. Diagnosis of viral gastroenteritis by simultaneous detection of Adenovirus group F, Astrovirus, Rotavirus group A, Norovirus genogroups I and II, and Sapovirus in two internally controlled multiplex real-time PCR assays. J. Clin. Virol. 2010;49:205–210. doi: 10.1016/j.jcv.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 24.Hebbelstrup Jensen B, Olsen KE, Struve C, Krogfelt KA, Petersen AM. Epidemiology and clinical manifestations of enteroaggregative Escherichia coli. Clin. Microbiol. Rev. 2014;27:614–630. doi: 10.1128/CMR.00112-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hossain MA, Albert MJ. Effect of duration of diarrhoea and predictive values of stool leucocytes and red blood cells in the isolation of different serogroups or serotypes of Shigella. Trans. R. Soc. Trop. Med. Hyg. 1991;85:664–666. doi: 10.1016/0035-9203(91)90388-F. [DOI] [PubMed] [Google Scholar]
- 26.Morgan DR, Johnson PC, DuPont HL, Satterwhite TK, Wood LV. Lack of correlation between known virulence properties of Aeromonas hydrophila and enteropathogenicity for humans. Infect. Immun. 1985;50:62–65. doi: 10.1128/IAI.50.1.62-65.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teunis P, Figueras MJ. Reassessment of the enteropathogenicity of mesophilic Aeromonas species. Front. Microbiol. 2016;7:1395. doi: 10.3389/fmicb.2016.01395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarantuya J, et al. Typical enteroaggregative Escherichia coli is the most prevalent pathotype among E. coli strains causing diarrhea in Mongolian children. J. Clin. Microbiol. 2004;42:133–139. doi: 10.1128/JCM.42.1.133-139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fredricks DN, Relman DA. Sequence-based identification of microbial pathogens: a reconsideration of Koch’s postulates. Clin. Microbiol. Rev. 1996;9:18–33. doi: 10.1128/CMR.9.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lertsethtakarn P, et al. Travelers’ Diarrhea in Thailand: A quantitative analysis using TaqMan(R) array card. Clin. Infect. Dis. 2018;67:120–127. doi: 10.1093/cid/ciy040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller D, et al. Identification of unconventional intestinal pathogenic Escherichia coli isolates expressing intermediate virulence factor profiles by using a novel single-step multiplex PCR. Appl. Environ. Microbiol. 2007;73:3380–3390. doi: 10.1128/AEM.02855-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sjoling A, Sadeghipoorjahromi L, Novak D, Tobias J. Detection of major diarrheagenic bacterial pathogens by multiplex PCR panels. Microbiol. Res. 2015;172:34–40. doi: 10.1016/j.micres.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 33.Chandra M, Cheng P, Rondeau G, Porwollik S, McClelland M. A single step multiplex PCR for identification of six diarrheagenic E. coli pathotypes and Salmonella. Int. J. Med. Microbiol. 2013;303:210–216. doi: 10.1016/j.ijmm.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 34.Gunzburg ST, Tornieporth NG, Riley LW. Identification of enteropathogenic Escherichia coli by PCR-based detection of the bundle-forming pilus gene. J. Clin. Microbiol. 1995;33:1375–1377. doi: 10.1128/JCM.33.5.1375-1377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robins-Browne RM, et al. Are Escherichia coli pathotypes still relevant in the era of whole-genome sequencing? Front. Cell Infect Microbiol. 2016;18:141. doi: 10.3389/fcimb.2016.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.