ABSTRACT
During central nervous system development, spatiotemporal gene expression programs mediate specific lineage decisions to generate neuronal and glial cell types from neural stem cells (NSCs). However, little is known about the epigenetic landscape underlying these highly complex developmental events. Here, we perform ChIP-seq on distinct subtypes of Drosophila FACS-purified NSCs and their differentiated progeny to dissect the epigenetic changes accompanying the major lineage decisions in vivo. By analyzing active and repressive histone modifications, we show that stem cell identity genes are silenced during differentiation by loss of their activating marks and not via repressive histone modifications. Our analysis also uncovers a new set of genes specifically required for altering lineage patterns in type II neuroblasts (NBs), one of the two main Drosophila NSC identities. Finally, we demonstrate that this subtype specification in NBs, unlike NSC differentiation, requires Polycomb-group-mediated repression.
KEY WORDS: Drosophila, Neurogenesis, Histone modifications, Polycomb, Trithorax
Highlighted Article: A FACS-based method for sorting different cell types and their ChIP-seq have provided an in vivo dataset that reveals dynamic histone modifications during neuronal differentiation in Drosophila.
INTRODUCTION
During development of the central nervous system (CNS), neural stem cells (NSCs) divide asymmetrically to generate daughter cells with self-renewing capacity but also complex neurogenic and gliogenic lineages. Regulation of this process requires tight and highly dynamic control of multiple cell fate decisions. For cells to commit to their ultimate cell identity, spatiotemporal gene expression programs are required. It is assumed that activation of lineage-specific genes and silencing of stem cell genes is accompanied by changes in chromatin states. How histone modifications change over time during neurogenesis in vivo, however, is not very well described.
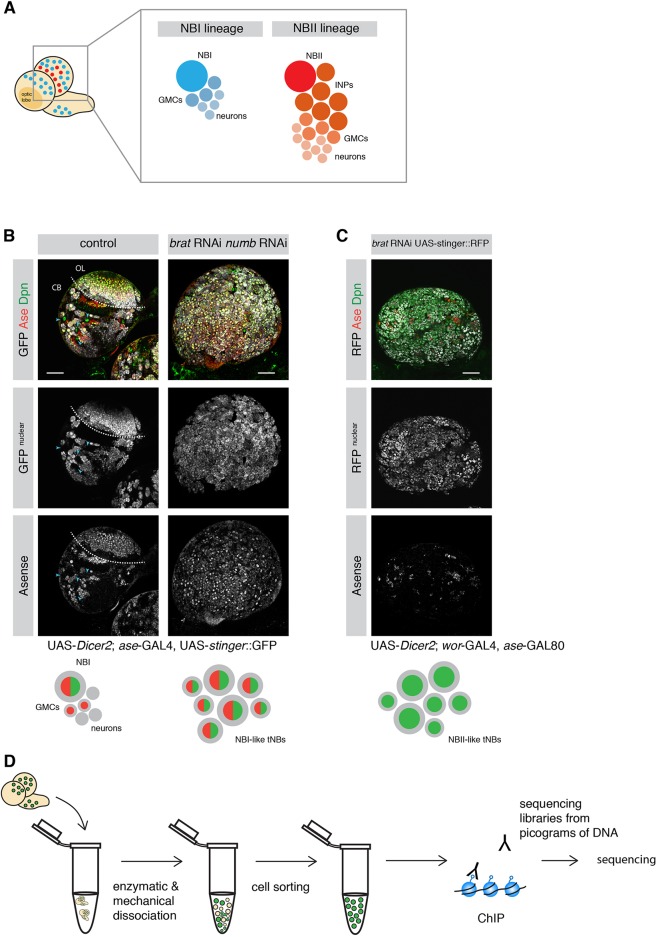
The Drosophila larval CNS has become a key model for the fundamental mechanisms underlying brain development and chromatin states (Homem and Knoblich, 2012). The larval CNS is populated by distinct types of NSCs, or neuroblasts (NBs), which vary in abundance, neuronal output and division mode. Together, these NBs give rise to the majority of the neurons of the adult brain (Truman and Bate, 1988). The majority of the central brain NBs are of type I (NBIs). Each NBI gives rise to another NBI and a ganglion mother cell (GMC), which divides once more to generate two differentiated neurons or glia. Type II NBs (NBIIs) are a rare subpopulation with only eight NBII per brain lobe (Fig. 1A) (Bello et al., 2008; Boone and Doe, 2008; Homem and Knoblich, 2012; Sousa-Nunes et al., 2010). Unlike NBIs, NBIIs divide into one NBII and one transit-amplifying cell called an intermediate neural progenitor (INP). NBIIs generate many more neurons, because INPs continue to divide asymmetrically five or six times, each time giving rise to a GMC that divides into two neurons or glia cells (Bello et al., 2008; Boone and Doe, 2008; Homem and Knoblich, 2012). Other than lineage structure and size, cell markers can also be used to differentiate NB subtypes. Whereas NBIs express both Asense (Ase) and Deadpan (Dpn) (Bowman et al., 2008), NBIIs only express Dpn (Bello et al., 2008). During neurogenesis, both NB subtypes divide asymmetrically to give rise to their respective progeny (Kang and Reichert, 2015). Brain tumors form if the asymmetric segregation of cell fate determinants during NB cell division is disrupted (Betschinger et al., 2006; Knoblich, 2010). Among these determinants are the TRIM-NHL protein Brain tumor (Brat) and the Notch inhibitor Numb (Arama et al., 2000; Betschinger et al., 2006; Bowman et al., 2008; Knoblich et al., 1995; Lee et al., 2006a,b). Although Brat depletion results in the generation of ectopic NBII-like tumor NBs (tNBs) at the expense of differentiated brain cells (Bowman et al., 2008), simultaneous loss of Brat and Numb causes the NBI-like tNBs to overproliferate (see Results).
Fig. 1.
Strategy to investigate histone marks in specific NB lineages. (A) Schematic depicting a larval brain with NBI (blue) and NBII (red) lineages. (B) The ase-GAL4 driver line marks NBI lineages with nuclear GFP but not NBII lineages (blue arrowheads). Combined knockdown of brat and numb results in ectopic Ase+ Dpn+ tNBs. Dashed line separates optic lobe (OL) and central brain (CB). (C) brat depletion with the NBII-specific driver line results in mainly Ase− Dpn+ tNBs. (D) Schematic showing an overview of the ChIP-seq strategy. Scale bars: 50 µm.
In many cell types, transitions in chromatin states are regulated by the evolutionarily conserved Polycomb group (PcG) and Trithorax group (TrxG) proteins. PcG and TrxG have emerged as antagonistic regulators that silence or activate gene expression, respectively (Kingston and Tamkun, 2014; Levine et al., 2002; Schuettengruber et al., 2007). These multimeric protein complexes regulate the transcriptional state of genes by post-translationally modifying amino acid residues of histone tails (Kingston and Tamkun, 2014; Levine et al., 2004). PcG proteins exert a repressive activity via two main complexes, the Polycomb repressive complexes 1 and 2 (PRC1 and PRC2). Although PRC1 and PRC2 can exist in various compositions and associate with context-specific accessory proteins, both have been shown to contain a specific core set of proteins including subunits with catalytic activity (Bracken et al., 2006; Simon and Kingston, 2009). Within PRC2, Enhancer of zeste [E(z) in Drosophila, EZH1/2 in mammals] catalyzes the trimethylation of lysine 27 on histone 3 (H3K27me3) (Cao and Zhang, 2004). H3K27me3 is recognized by PRC1, which in turn includes the histone H2A ubiquityltransferase Sce [RING1A (RING1) and RING1B (RNF2) in mammals] (de Napoles et al., 2004). Histone modifications associated with active transcription are deposited by TrxG proteins (Kassis et al., 2017), which counteract repressive marks by histone acetylation or methylation, in particular by trimethylation of lysine 4 on histone H3 at active promoters (Byrd and Shearn, 2003; Dou et al., 2005; Petruk et al., 2001; Kim et al., 2005).
Although well-known for their role in long-term transcriptional memory, PcG and TrxG complexes are highly dynamic during development and thus facilitate cellular plasticity (Kwong et al., 2008; Nègre et al., 2006). In the last decade, it has been shown that PcG and TrxG complexes are crucial for ensuring correct neurogenesis in mammals (Hirabayashi et al., 2009; Lim et al., 2009; Pereira et al., 2010) as well as in Drosophila (Bello et al., 2007; Touma et al., 2012). Despite the strength of genetic in vivo experiments, however, global analysis of the histone modifications underlying their function, and therefore target genes, has mainly been performed in vitro. This constitutes a real knowledge gap, as recent studies have demonstrated that the chromatin states may vary significantly between in vivo tissues and their related in vitro cell lines, mainly owing to culture conditions (Xie et al., 2013; Zhu et al., 2013). Given also that epigenetic changes are highly context- and developmental time-dependent, providing in vivo datasets to investigate chromatin states of different cell types in complex tissues will increase our understanding of how the epigenetic landscape dynamically defines cellular states.
In recent years, in vivo studies made use of Drosophila to shed light on the dynamics of chromatin state changes during embryonic neural differentiation (Ye et al., 2017) and during larval stages (Aughey et al., 2018; Marshall and Brand, 2017). Profiling the binding of chromatin remodelers has highlighted the plasticity of chromatin states during differentiation (Marshall and Brand, 2017). Although binding of chromatin factors is associated with active or repressive chromatin, binding does not necessarily reflect downstream histone modifications. For example, the histone marks can change drastically between parasegments of the Drosophila embryo, whereas the occupancy of PcG proteins remains unchanged (Bowman et al., 2014). Thus, investigating the dynamics of chromatin states based on chromatin marks is crucial for understanding the functional specialization of cells during development. Moreover, how PcG/TrxG complexes target genes on the chromatin level between different subtypes of progenitor cells during neuronal differentiation or tumorigenic transformation has remained elusive.
Here, we use the Drosophila larval CNS to track in vivo changes of histone modifications not only upon differentiation, but also between different populations of NSCs and their tumorigenic counterparts. We developed a fluorescence-activated cell sorting (FACS)-based method to sort different cell types and perform ChIP-seq for the active histone mark, H3K4me3, and the repressive mark, H3K27me3. Our FACS-based approach provides an in vivo dataset that reveals dynamic histone modifications during neuronal differentiation. In particular, we observed that self-renewal and cell-division genes are repressed independently of H3K27me3 levels. In contrast, we further show that H3K27me3-mediated repression is crucial for silencing lineage-specific stem cell factors, including known factors as well as a new set of genes that are specific to NBIIs. Finally, we present genetic evidence for the requirement of these new NBII-specific factors for self-renewal and demonstrate the role of PcG complexes in defining different subtypes of neural stem cells.
RESULTS
Profiling repressive and active histone modifications of NSCs and neurons
H3K4me3 and H3K27me3 are two major histone modifications associated with TrxG-activated and PcG-repressed states, respectively. However, these histone modifications have not yet been analyzed independently in distinct subtypes of NSCs in Drosophila. To analyze H3K4me3 and H3K27me3 histone marks in different brain cell types using ChIP-seq, we combined genetic labeling with a protocol for generating sequencing libraries from picogram quantities of DNA (Bowman et al., 2014). The NB subtype-specific GAL4 drivers ase-GAL4 (NBI lineage-specific) and wor-GAL4, ase-GAL80 (NBII lineage-specific), allowed us to preferentially label distinct NB lineages with nuclear-localized fluorophores (stinger::GFP or RFP). Indeed, GFP expressed by ase-GAL4 exclusively labeled NBI lineages (both Dpn+ and Ase+) and was not expressed in NBIIs (only Dpn+) (Fig. 1B). To amplify the production of rare NBIIs and at the same time generate tNBs, RNAi constructs against the cell fate determinants brat and numb were expressed using the mentioned driver lines. Depletion of brat in larval brains with an NBII-specific driver resulted in the overproliferation of NBII-like tNBs, evident by an increase in Dpn+, Ase− cells (Fig. 1C). In contrast, simultaneously depleting numb and brat using an NBI-specific driver resulted in overgrowth of Dpn+, Ase+ NBI-like tNBs (Fig. 1B). Therefore, this strategy allowed us to generate fluorescently labeled distinct NB cell types and neurons.
Besides central brain NB lineages, the larval brain consists of embryonic neurons, mushroom body neuroblasts and cells of the optic lobes. To avoid impurities from these structures NBI, neurons and tNBs were isolated according to fluorescence intensity and cell size by flow cytometry (Berger et al., 2012; Harzer et al., 2013). Purified cell populations were then analyzed by ChIP-seq for H3K27me3 and H3K4me3 histone modifications (Fig. 1D).
The H3K4me3 signal peaked around transcriptional start sites (TSSs), whereas the H3K27me3 signal occurred in broad domains covering gene bodies. For example, in all cell types the ubiquitously expressed gene RNA polymerase II 215 kD subunit contained a H3K4me3 peak at the TSS which was devoid of H3K27me3 signal (Fig. S1A). In contrast, the gene caudal showed no H3K4me3 peak, but instead high H3K27me3 levels over the gene body (Fig. S1B). This is in accordance with the fact that the function of Caudal is mostly restricted to the larval digestive system. Moreover, caudal is not expressed in the larval CNS (modENCODE data and Berger et al., 2012) and has been shown to inhibit NB specification upon mis-expression (Birkholz et al., 2013).
Self-renewal and cell cycle genes are repressed during differentiation in a H3K27me3-independent manner
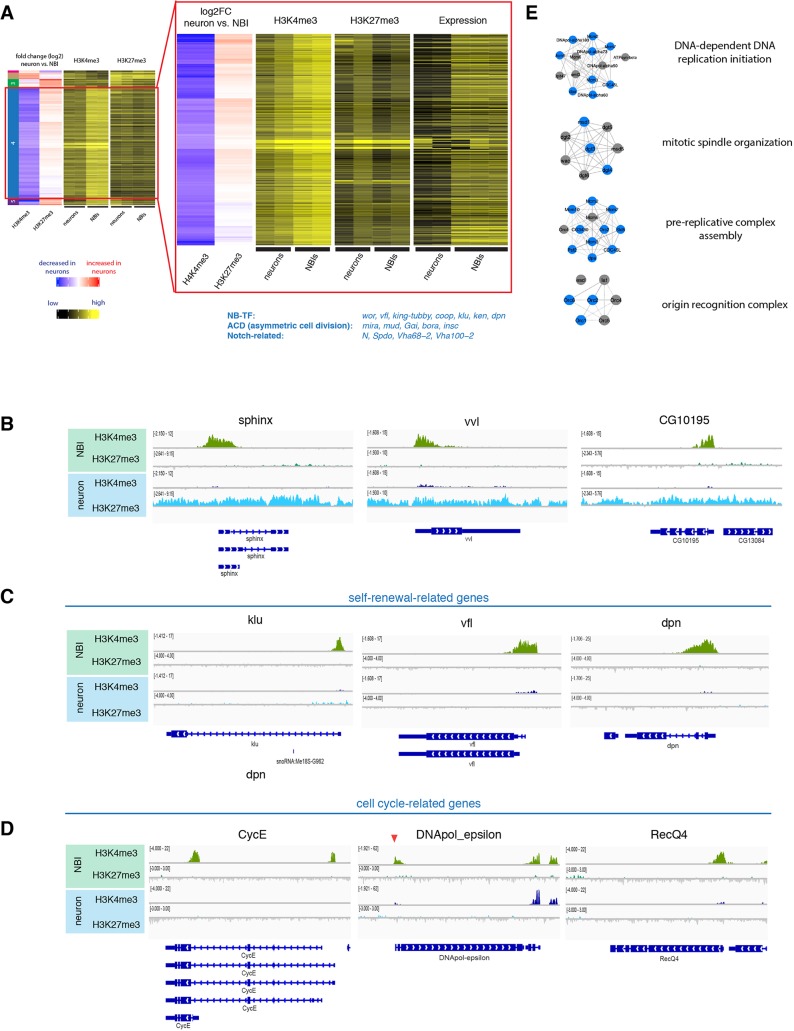
To investigate changes in the epigenetic landscape during neurogenesis, we collected NBIs and neurons as described above in duplicates. We subtracted the individual inputs from their respective samples to generate coverage tracks. The read counts for H3K27me3 localization were analyzed over the whole gene body, but the reads for H3K4me3 were counted from +1 to +500 bp downstream of the TSS. From this data, regions with differential signals were identified between different cell types. Finally, we performed unsupervised hierarchical clustering analysis on differentially marked genes in NBIs and neurons (Fig. 2A) identifying five distinct groups of genes. Three clusters were dependent on H3K27me3-mediated repression. Cluster 1 showed a decreased H3K27me3 signal upon differentiation, whereas cluster 3 showed an increased H3K27me3 signal in neurons. These clusters were not enriched for genes of a particular pathway or biological process when analyzed using Gene Ontology (GO) enrichment analysis. Another example of H3K27me3-mediated repression was cluster 5. Whereas clusters 1 and 3 showed changes in H3K27me3 signal and no or mild changes in H3K4me3, genes in cluster 5 (22 genes) showed a drastic switch from an H3K4me3+ H3K27me3− to an H3K4me3− H3K27me3+ chromatin state upon differentiation. Cluster 5 included the long non-coding RNAs cherub, pncR002:3R and sphinx, and transcription factors nab and vvl (Fig. 2B).
Fig. 2.
Changes of active and repressive histone modifications upon differentiation. (A) Unsupervised hierarchical clustering analysis of gene log2 fold change between NBIs and neurons. NB-related genes of cluster 4 according to literature are indicated blue. Close up to cluster 4 genes showing heatmap of TPM values, together with H3K4me3 and H3K27me3 changes between neurons versus NBIs. (B) ChIP-seq tracks of representative examples for genes of cluster 5. (C,D) ChIP-seq tracks of representative examples for cell-cycle-related genes (C) and self-renewal-related genes (D). (E) Examples of mitosis-related protein complexes. Blue indicates genes found in cluster 4.
The other two clusters (2 and 4) were mainly dependent on changes in the H3K4me3 signal. Cluster 2 showed an increase in H3K4me3 levels in neurons, whereas cluster 4 contained a large number of 318 genes and was characterized by a loss of H3K4me3 upon differentiation. These gene loci had either a small increase in H3K27me3 or were completely devoid of both marks in neurons (Fig. 2A,C,D). GO enrichment analysis (Table S1) showed that cluster 4 genes were enriched for genes involved in self-renewal (e.g. stem cell proliferation P=0.002) and mitosis-related processes (e.g. DNA replication P=3.04E-21), which are both processes that cease upon differentiation. To validate this, we took advantage of published transcriptome data of neurons and NBIs (Berger et al., 2012). Indeed, the cluster 4 genes showed a significant downregulation in neurons compared with NBIs (Fig. 2A). In support of this finding, protein complexes essential for cell division were enriched in cluster 4 (Fig. 2E), whereas bona fide NB self-renewal transcription factors such as vfl (also known as zld), klu and dpn as well as asymmetric cell division regulators (Gαi, mud, insc, bora) appeared in cluster 5. This suggests that during differentiation stem cell-promoting genes lack an H3K27me3 mark and that their repression could be mediated through mechanisms independent of PcG (Fig. 2A). This result is corroborated by previous data indicating that the genes mira, CycE, stg and dpn are enriched in HP1-associated chromatin in Drosophila neurons (Marshall and Brand, 2017).
Thus, these data suggest that a small group of genes is controlled by H3K27me3-mediated repression upon differentiation, whereas most stem cell-related genes are turned off via an additional mechanism, potentially involving HP1 enrichment. This result is surprising considering H3K27me3 datasets indicate spreading of PcG-repressed regions upon neural differentiation in mammals (Södersten et al., 2018; Zhu et al., 2013) and suggests that different strategies of epigenetic control of neurogenesis have been established across evolution.
In mammalian stem cells bivalent chromatin states, characterized by the simultaneous presence of H3K27me3 and H3K4me3, are suggested to poise genes, which upon differentiation are resolved into activating or repressive states. In our dataset, only eight of the 393 genes changing their H3K4me3 and/or H3K27me3 upon differentiation showed both histone marks. The dominance of monovalent loci suggests that neurogenesis in the Drosophila larval brain does not rely on bivalent domains. It is likely that the source of H3K4me3 and H3K27me3 co-occurrence is the differential expression of these genes within the NB population. This is well illustrated by the gene tll, which possesses both histone marks in our data and has been shown to be only expressed in a small subset of central brain NBs (Yang et al., 2015). Thus, our study in purified NBs and together with previous reports (Gan et al., 2010; Marshall and Brand, 2017; Schuettengruber et al., 2009; Ye et al., 2017) adds to the growing evidence that bivalent chromatin states are absent in Drosophila.
Subtype-specific NB genes are controlled by TrxG and PcG
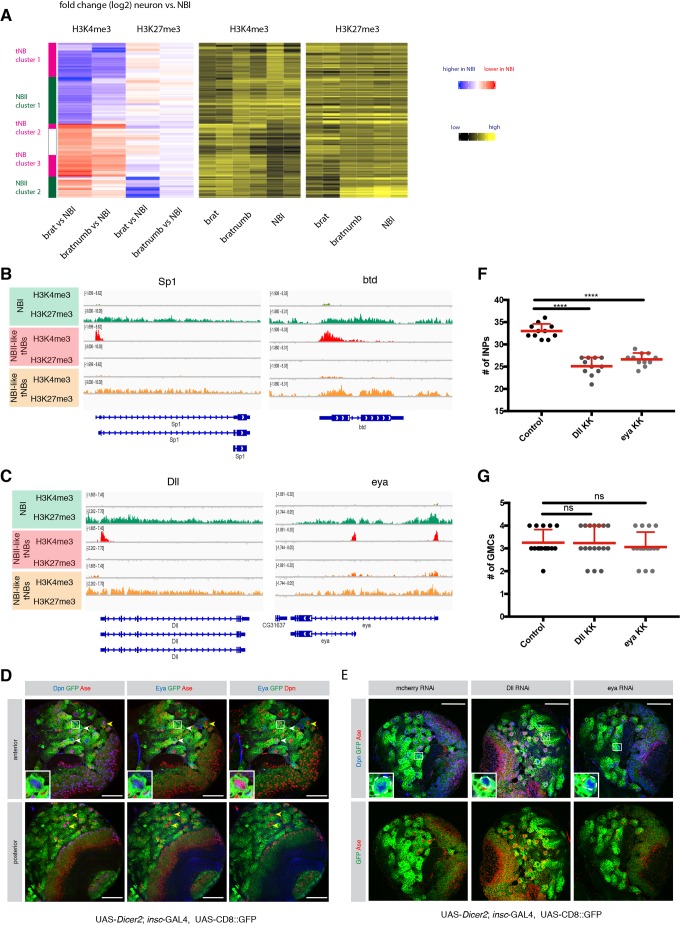
Next, we wanted to address whether alterations of histone modifications can be observed between different types of NB lineages. To this end, we made use of tNBs which are of a different origin. RNAi of both brat and numb induced tumors made of NBIs, whereas the depletion of brat alone initiates tumors consisting of NBIIs (Fig. 1B,C). We reasoned that features occurring in NBII-like tNBs but absent in NBI-like tNBs would likely be specific to NBII lineages rather than because of tumorigenesis. We performed hierarchical clustering analysis between NBIs, NBI-like tNBs and NBII-like tNBs as described above and identified two NBII-specific sets of genes (Fig. 3A and Fig. S2). The first cluster of genes showed a decrease in H3K4me3 in NBII-like tNBIIs compared with NBIs and no or only mild changes when compared with NBI-like tNBIs (NBII cluster 1). Moreover, these loci showed no or only modest increases in H3K27me3. As a key example, among this set of genes, we found the NBI-specific transcription factor ase, which showed clear H3K4me3 signals in NBI and brat numb-depleted NBI-like tNBs but no signal in NBII-like tNBs (Fig. S3A).
Fig. 3.
Comparison of different NB subtypes identifies PcG and TrxG-dependent NBII-specific factors. (A) Unsupervised hierarchical clustering analysis of gene log2 fold change between NBI, NBI-like tNB and NBII-like tNB (relevant section is shown, for full heatmap see Fig. S2). (B,C) ChIP-seq tracks of the known NBII factors Sp1 and btd (B) and novel NBII-specific factors Dll and eya (C). (D) Eya immunostaining in type I and type II neuroblasts. Driver line used was UAS-dicer2; insc-Gal4, UAS-CD8::GFP. White arrowheads show NBIIs, yellow arrowheads show NBIs. Inset shows magnification of boxed area, showing a close-up of NBIIs. (E) Immunostainings of larval brains expressing RNAi against Dll or eya show smaller NBIIs. Inset shows magnification of boxed area; NBIIs are circled with white dashed line. (F,G) Quantification of immediate progenies of NBIIs (F) and NBIs (G). The driver line used was UAS-dicer2; insc-Gal4, UAS-CD8::GFP. Data are mean+s.d. For INPs (F): control, 33±1.61; Dll, 25.09±1.97; eya, 26.64±1.43; n=11. For GMCs (G): control, 3.25±0.57 (n=16); Dll, 3.23±0.75 (n=17); eya, 3.05±0.65 (n=17). ****P<0.0001 (one-way ANOVA test). ns, not significant. n numbers are lineages quantified. Scale bars: 50 µm.
In the second gene cluster (NBII cluster 2), NBII-like tNBIIs showed increased H3K4me3 signal and lower H3K27me3 occupancy. Interestingly, previous genetic evidence has suggested a role of trx in maintaining different subtypes of NB lineages. In particular, the two loci buttonhead (btd) and Sp1 are required to specify NBII from neuroectoderm (Álvarez and Díaz-Benjumea, 2018) and to maintain NBII lineages (Komori, 2014; Xie et al., 2014). Indeed, both genes are H3K4me3+ in NBII-like tNBIIs and have reduced intensity of H3K27me3 signal in NBI-derived numb tumors as well as NBIs (Fig. 3B). Our clustering identified additional genes with a similar pattern (Fig. 3A). Two of these H3K4me3+ NBII-like NBII-specific genes were the homeodomain transcription factor Distal-less (Dll) and the transcriptional coactivator eyes absent (eya) (Fig. 3C). We decided to further focus on and characterize the contribution of these two genes in NBII specification, as previous work has shown that Dll enhancer was active in NBII lineages (Izergina et al., 2009) and eya was mainly expressed in NBIIs (which we confirmed using immunostaining; Fig. 3D). Dll- or eya-depleted brains resulted in smaller NBII cells (Fig. 3E and Fig. S3B) with a reduced number of INPs (Fig. 3F), which indicates reduced stemness (Song and Lu, 2011; Wissel et al., 2018). In contrast, NBI lineages showed normal NB growth and unaffected GMC numbers (Fig. 3G and Fig. S3C). Thus, our data indicate that these two genes are required to maintain NBII lineages. This would further suggest that NB subtype-specific genes are regulated by PcG and TrxG.
Finally, tNB-specific changes were mostly H3K4me3 changes (reduction in tNB cluster 1 and gain in tNB clusters 2 and 3) (Fig. 3A and Fig. S2), whereas only minor changes were observed for H3K27me3. This suggests that tumor-specific changes are mediated by TrxG proteins rather than PcG.
PRC1 and PRC2 are required for NB maintenance
Our data suggest that both TrxG and PcG complexes play an important role in maintaining NBI and NBII identities. TrxG-dependent maintenance of NBII identity was shown to rely on the target genes btd and Sp1 (Álvarez and Díaz-Benjumea, 2018; Komori et al., 2014). By contrast, the role of PcG in NB-subtype specification, besides Hox gene repression, remains largely unexplored. During brain development, loss of PcG repression leads to ectopic expression of Hox genes, which in turn induces apoptosis and depletion of both type I and type II NBs (Bello et al., 2007). In accordance with these previous findings, our ChIP-seq data revealed high levels of H3K27me3 at the two Hox gene clusters, the Antennapedia and Bithorax complexes, in both NBIs and NBIIs (Fig. S4A).
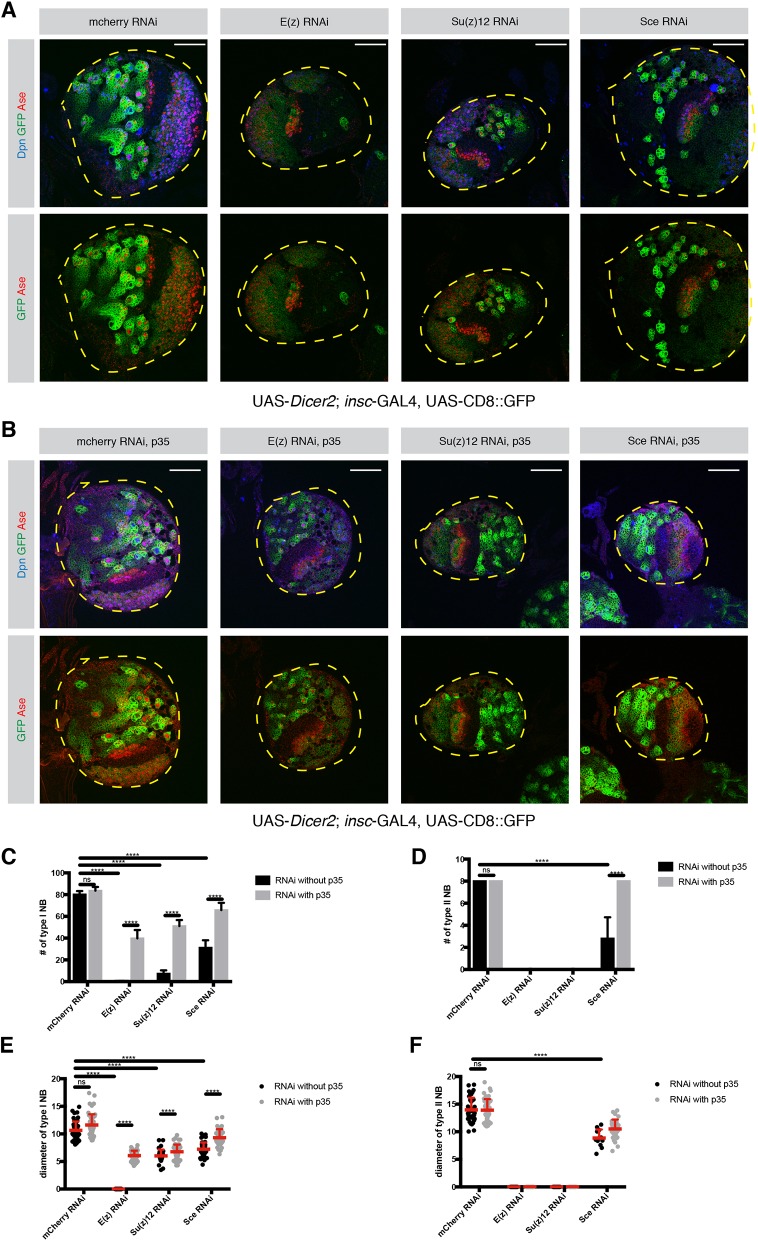
To investigate whether PRC-mediated repression is only required to prevent Hox gene-induced apoptosis or whether it plays a broader role in neurogenesis, we blocked apoptosis by expressing the baculovirus caspase inhibitor gene P35. To this end, RNAi constructs against components of PRC2 [E(z), Su(z)12] or PRC1 (Sce) were expressed in NBs using the general NB driver line insc-GAL4, which resulted in a great decrease in NBI and NBII cell numbers (Fig. 4A). Upon P35 expression in a PcG-depleted background, GFP+ NB lineages could be restored (Fig. 4B) as has been previously reported (Bello et al., 2007).
Fig. 4.
PRC1 and PRC2 are required for NB maintenance. (A,B) Loss of PRC1 and PRC2 causes a significant decrease in the number of NBs. (A) Larval brain lobes expressing RNAi against mCherry, E(z), Su(z)12 and Sce. The driver line used was UAS-dicer2; insc-Gal4, UAS-CD8::GFP. (B) Larval brains expressing apoptosis inhibitor P35 together with the PRC RNAi constructs in A. The driver line used was UAS-dicer2; insc-Gal4, UAS-CD8::GFP. In A and B, lobes are outlined with yellow dashed lines. (C) Quantification of NBI numbers in mCherry-, E(z)-, Su(z)12- and Sce-depleted larval brains with and without P35 expression. n=5 brain lobes. mCherry, 80.2±3.11; mCherry+P35, 83.6±3.46; E(z), 0.2±0.44; E(z)+P35, 39.8±7.66; Su(z)12, 7.4±2.96; Su(z)12+P35,51±5.61; Sce, 31.2±6.83; Sce+P35, 65.75±6.7. (D) Quantification of NBII numbers in mCherry-, E(z)-, Su(z)12- and Sce-depleted larval brains with and without P35 expression. n=5 brain lobes. mCherry, 8; mCherry+P35, 8; E(z), not applicable (NA); E(z)+P35, NA; Su(z)12, NA; Su(z)12+P35, NA; Sce, 2.8±1.92; Sce+P35, 8. (E) Quantification of NBI diameter in mCherry-, E(z)-, Su(z)12- and Sce-depleted larval brains with and without P35 expression. mCherry, 10.64±1.6 (n=50); mCherry+P35, 11.61±1.93 (n=50); E(z), NA; E(z)+P35, 6.05±0.9 (n=50); Su(z)12, 6.01±1.14 (n=34); Su(z)12+P35, 6.76±1.3 (n=50); Sce, 7.21±1.22 (n=50); Sce+P35, 9.31±1.54 (n=50). n=number of NBIs. (F) Quantification of NBII diameter in mCherry-, E(z)-, Su(z)12- and Sce-depleted larval brains with and without P35 expression. mCherry, 13.96±2.19 (n=40); mCherry+P35, 13.91±2 (n=40); E(z), NA; E(z)+P35, NA; Su(z)12, NA; Su(z)12+P35, NA; Sce, 8.85±1.56 (n=12); Sce+P35, 10.5±1.7 (n=32). n=number of NBIIs. Data are mean+s.d. ****P<0.0001 (two-way ANOVA). ns, not significant. Scale bars: 50 µm.
Although RNAi constructs were expressed with the same driver line in NBIs and NBIIs, blocking apoptosis in PRC2-depleted lineages restored NBI but not NBII cell numbers (Fig. 4C,D). In contrast, the number of NBs in PRC1-depleted brains was restored, suggesting that PRC1, unlike PRC2, appears to only target the Hox genes and therefore prevents apoptosis of NBs (Fig. 4D). However, these restored NBIIs still exhibited a smaller cell size (Fig. 4F) than their control NBIIs. These results indicate that in addition to its function as anti-apoptotic in both type I and type II NB, PRC2 is required specifically in NBIIs to maintain self-renewal potential. These data therefore suggest that PcG-dependent repression targets more than just the HOX genes to maintain NBII.
We have observed that, within the genome, there are loci that are heavily marked with H3K27me3 (Fig. S4B,C). Among these regions, we have found two transcription factors, opa and ham (Fig. S4D,E), which have previously been described as temporal switch genes in NBII lineages and their ectopic expression has been shown to limit NB self-renewal, resulting in the disappearance of NBIIs (Abdusselamoglu et al., 2019; Eroglu et al., 2014). Thus, given their role in NBII lineages, the de-repression of opa and ham might affect NBII lineages more than NBI lineages upon PcG-depletion. This might explain the increased sensitivity of NBII.
PcG proteins prevent premature NB differentiation
Although the number of NBIs was restored in apoptosis-inhibited PcG-depleted conditions, the NBI cell size was reduced (Fig. 4E). NBs must maintain a certain growth rate to maintain their self-renewal potential and prevent differentiation (Song and Lu, 2011). We therefore analyzed these apoptosis-inhibited NBIs and their self-renewal potential. RNAi constructs against PRC1 and PRC2 components were expressed together with P35, and NBs were analyzed at 6 h after pupal formation (APF), the timepoint at which NB start to exit proliferation (Fig. S5A). Although the number of NBI in both PRC1- and PRC2-depleted brains were restored in third instar larval brains upon P35 expression, NB numbers were significantly decreased at 6 h APF, and the diameter of the remaining NBs was significantly lower compared with the control (Fig. S5B,C). Altogether, these data show that, even though the number of NBs was restored in apoptosis-inhibited PRC-depleted lineages, these NBs fail to maintain their self-renewal potential, as reported by their smaller size and early differentiation compared with their wild-type counterparts. Together, these results indicate that PcG proteins are required to maintain stemness both in type I and type II NBs, but with different sensitivities.
To address the physiological consequences of premature NB differentiation, we analyzed the viability of PcG-depleted flies. RNAi-mediated knockdown of PcG proteins with and without P35 expression using insc-GAL4 led to lethality during development (Neumüller et al., 2011). This observation further confirms our previous results that neurogenesis of NBI lineages is not fully restored. However, this approach suffers from the caveat that the insc promotor is active in some cells of the larval gut and salivary glands. To exclude the possibility that lethality could originate from abnormal development of other tissues, we next used a brain-restricted NBI lineage-specific driver line ase-GAL4. Similar to insc-GAL4, ase-GAL4-mediated loss of PcG proteins led to a decline in NBI number and size, which could be rescued by blocking apoptosis (Fig. S6A,B). These phenotypes were nonetheless weaker, which we could explain by the strength of insc-GAL4, being more highly expressed than ase in NBIs (Berger et al., 2012). However, knockdown of both PcG and P35 using the ase-GAL4 driver line in NBI lineages showed a significant decrease in H3K27me3 signal (Fig. S6C). Therefore, these results further confirm that PcG promotes self-renewal as well as preventing apoptosis in NBI.
When PcG proteins were depleted with ase-GAL4-driven RNAi during NBI development, the majority of eggs failed to develop into adult flies (Fig. S6D). Between 3% and 18% of eggs that were laid hatched, but flies showed neurological abnormalities and became stuck in the fly food, which led to death. Similarly, preventing apoptosis in these PcG knockdown backgrounds did not rescue the number of viable flies (Fig. S6D). Therefore, NBIs and NBIIs depend on PcG proteins for proper neuron production, although at different sensitivities. In summary, these results suggest that PRC1 and PRC2 maintain NB neurogenesis by silencing the genes that induce apoptosis and the genes that instruct differentiation.
DISCUSSION
We provide a resource of histone modification datasets for different types of NSCs and their differentiated progeny. In combination with chromatin accessibility (Aughey et al., 2018) and binding maps of chromatin remodelers (Marshall and Brand, 2017) of Drosophila brain cells, we hope that our dataset will serve as a useful community resource. We show that, during differentiation, stem cell identity genes are silenced in a PcG-independent manner, which supports previous findings showing that these genes are silenced through HP1-enriched chromatin (Marshall and Brand, 2017). In addition, PcG-mediated silencing is unlikely to instruct the stepwise inactivation of stem cell genes during differentiation as loss of H3K27me3 did not induce ectopic NBs.
Here, we take advantage of in vivo genetic labeling to investigate chromatin dynamics of different NB subtypes. As the type II NBs are very lowly abundant, we used tumor NBs of type I and type II origins as a proxy in order to obtain enough material to be able to compare these two cell types. We further validated each change observed by comparing tumor with healthy type I NBs and excluded artifacts present because of the tumorigenic state of the cells. Our data show that both TrxG and PcG are required to establish NBII identity. We identify a set of NBII-specific genes, including previously identified btd (Komori et al., 2014) and Sp1 (Álvarez and Díaz-Benjumea, 2018). We further identify Dll and eya, which are specifically required for NBII maintenance. It has been previously described that btd acts as an activator of Dll in the development of the ventral imaginal discs (Estella et al., 2003). This suggests that in NBII-identity specification the Trithorax-target btd could act together with Dll and eya. Such a mechanism would explain why the loss of btd causes a distinct phenotype compared with the loss of Dll and eya. Interestingly, an NBI to NBII conversion is observed only in 18% of NBIs ectopically overexpressing btd, indicating that either co-factors are missing or that the chromatin of btd targets is inaccessible (Komori et al., 2014). Our data of NB subtype-specific genes being characterized by H3K27me3 repressive chromatin favor the latter. Therefore, as opposed to TrxG-activated stem cell and mitosis genes, the repression of NBII-specific genes is ensured by PcG-mediated H3K27me3 histone modifications, suggesting that Polycomb plays a role in defining the diversity of NSC lineages. Moreover, our data indicate that PcG repression is required not only for the silencing of HOX genes but also for the self-renewal capacity of NBs. Unlike TrxG (Komori et al., 2014), the loss of catalytic subunits of PcG complexes did not convert NBIIs to NBIs or vice versa. This suggests that NB subtype specification cannot be explained solely by an absence of repression but requires a further activation mechanism. Strikingly, loss of PcG complexes caused a significant decrease in the number of NBs. Interestingly, across all the cell types, developmental genes such as caudal, eve, peb, scr and slp1, as well as genes involved in embryonic NB temporal patterning [hb, kr, pdm (nub), cas and grh], are heavily marked with H3K27me3. It is therefore possible that PcG-mediated repression is required to silence these developmentally crucial genes in addition to the Hox genes. Thus, the observed reduction in NB stemness might be caused by the de-repression of these genes.
Besides an overall decrease in NB maintenance, we observed an increased sensitivity of NBII lineages to a reduction in PRC2 activity. Interestingly, opa and ham, two previously described temporal switch genes in NBII lineages (Abdusselamoglu et al., 2019), are also enriched in H3K27me3 marks in NBs. Opa and ham are expressed in the immediate NBII progeny, the INPs, and ectopic expression of these genes limits self-renewal of NBIIs and causes NBIIs to disappear (Abdusselamoglu et al., 2019; Eroglu et al., 2014). Even though these two genes are heavily marked with H3K27me3 across all NB samples, NBIIs might be specifically sensitive to PRC2 depletion because they could be more receptive to premature de-repression of genes, the expression of which is normally restricted NBII progeny only.
In the future, investigating the downstream targets of PcG in different NB subtypes could reveal the underlying mechanisms of subtype specification. In conclusion, our data provide a useful resource to investigate how chromatin state dynamics orchestrate the diversity and correct progression of NSC lineages.
MATERIALS AND METHODS
Fly strains
Fly strains used in this study were: UAS-Su(z)12 RNAi [31191, Bloomington Drosophila Stock Center (BDSC)], UAS-E(z) RNAi (36068, BDSC), UAS-Sce RNAi [106328, Vienna Drosophila Resource Center (VDRC)], UAS-p35 (5072, 5073, BDSC; both were tested for functionality), UAS-Dll RNAi (101750, VDRC), UAS-eya RNAi (108071, VDRC).
Immunofluorescence
Brains were dissected and fixed for 20 min in PBS with 5% paraformaldehyde and 0.1% Triton X-100. After three washes with 1× PBS with 0.1% Triton X-100 (PBST), brains were incubated for 1 h in blocking solution (PBST with 3% normal goat serum), incubated with blocking solution with primary antibodies and washed again three times with PBST. Secondary antibodies (1:500, goat Alexa Fluor®, A-31553, A-11077, A-21450, Invitrogen) were added for 1-2 h and then removed with three PBST washes. Brains were mounted in Vectashield Antifade Mounting Medium (Vector Laboratories). Primary antibodies used were: rat anti-Asense (1:500, Eroglu et al., 2014), guinea pig anti-Deadpan (1:1000, Eroglu et al., 2014), H3K27me3 (1:500, Active Motif, 39155).
Microscopy
Images were recorded using a Zeiss Confocal 780 microscope. Images of different conditions in one panel were recorded using the same settings.
Isolation of NBs using FACS
NB-sized cells were sorted from third instar larval brains according to GFP/RFP signal and cell size as previously described (Berger et al., 2012; Harzer et al., 2013). Briefly, brains were collected in 1× Rinaldini solution and then enzymatically and mechanically dissociated in Schneider's medium supplemented with fetal bovine serum (10%), penicillin-streptomycin (2%), insulin (20 µl/ml), L-glutamine (20 mM), L-glutathione (40 µg/ml), 20-hydroxyecdyson (5 µg/ml).
Statistics
Statistical analyses were performed with GraphPad Prism 7. Experiments were not randomized. Sample sizes were estimated depending on the previous experiences with similar setups and the investigator was not blinded. Two-way ANOVA was used to assess statistical significance between multiple samples, whereas unpaired two-tailed Student's t-test was used between two samples.
ChIP-seq
Preparation of soluble chromatin
We pelleted 50,000 sorted cells of interest by centrifuging at 3000 rpm (0.8 g) for 10 min. The cell pellet was resuspended in complete media. Fixation was performed with formaldehyde (final concentration 1%) for 5 min at room temperature. After quenching with glycine (final concentration 125 mM) for 3 min at room temperature, cells were centrifuged at 3000 rpm for 10 min and the supernatant was discarded. Cells were then resuspended in 100 µl 1× PBS with CaCl2 (final concentration: 1 mM) and Triton X-100 (final concentration: 0.1%) and incubated with 5 units of micrococcal nuclease (Worthington Biochemical, LS004798) at 37°C for 3 min. After incubation, the sample was immediately transferred on ice and 2.5 µl 0.5 M EDTA, 6.25 µl 0.2 M EGTA and 1.25 µl 1× PBS were added in order to stop the reaction. After adjusting the sample volume to 300 µl with 1× PBS, sonication was performed with a microtip sonicator (OmniZRuptor 250, Omni International; microtip, power output: 20) for 20 s in a prechilled metal holder. After sonication, the sample was snap frozen in liquid nitrogen. Fragment size was assessed using the Agilent High Sensitivity DNA Assay.
Chromatin immunoprecipitation
The volume of thawed chromatin samples was adjusted to 500 µl with 50 µl 10× lysis buffer [1 mM EDTA, 10 mM Tris-HCl (pH 8), 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine, 1% Triton X-100], 140 µl water and 10 µl of 50× complete protease inhibitor. After 5 min incubation on ice, samples were spun down at maximum speed (6.9 g) for 10 min at 4°C. While supernatant was transferred to fresh tubes, 5 µl was saved as input sample (1%) at 4°C. Samples were incubated with primary antibodies overnight at 4°C, and then incubated with 10 µl Dynabeads Protein A (Invitrogen, 10006D) for 1 h at 4°C. After six washes with lysis buffer containing 100 mM NaCl and 1% Triton X-100, ChIP DNA was eluted twice with 125 µl fresh elution buffer (0.2% SDS, 0.1 M NaHCO3, 5 mM DTT) at 65°C for 10 min. The input DNA volume was adjusted to 250 µl with elution buffer. To achieve reversal of crosslinking, 1 M Tris-HCl (10 mM final concentration) and 500 mM EDTA (2 mM final concentration) were added to samples. Antibodies used for ChIP were: H3K27me3 (Active Motif, 39155, 5 µg) and H3K4me3 (Millipore, 07-473, 1 µl).
Library construction
Library construction was performed as previously described (Bowman et al., 2013). In short, after the ChIP sample volume was adjusted to 37.5 µl, end polishing reaction (50 µl) was performed by incubating the sample with 1× T4 ligase buffer (New England Biolabs; NEB), 0.4 mM dNTPs, 7.5 U T4 polymerase (NEB), 2.5 U Klenow polymerase (NEB) and 25 U polynucleotide kinase for 30 min at 20°C in a thermocycler. To clean-up the samples, solid phase reversible immobilization (SPRI) beads (Agencourt AMPure XP, Beckman Coulter) were used at a 1.8× beads ratio. DNA was eluted with 16.5 µl water and an A-tailing reaction (25 µl) was performed. For this, a 16 µl sample with 1× NEB buffer 2, 0.2 mM dATP and 7.5 U Klenow 3′-5′ exo minus (NEB) were incubated for 30 min at 37°C. SPRI cleanup was performed with 1.8× beads ratio and DNA was eluted with 9.5 µl of water. An adapter ligation reaction (25 µl) was performed by incubating 9 µl of sample with 1× rapid T4 ligase buffer (Enzymatics), 0.01 µM annealed universal adapter and 150 U T4 rapid ligase (Enzymatics) for 15 min at room temperature. SPRI cleanup was performed once again with 1.6× beads ratio and DNA was eluted with 10.5 µl water.
Finally, library amplification was performed by setting up a PCR reaction (50 µl) with 1× Phusion HF master mix (NEB), 0.2 µM universal primer, 0.2 µM barcoded primer, 1× SYBR Green I (Invitrogen) and 0.5 µl Rox (USB). Then, the PCR reaction was performed using an Applied Biosystems 7500 Fast Real-Time PCR System. The program used was: an initial denaturing for 30 s at 98°C, followed by multiple cycles of 10 s denaturation at 98°C, 20 s annealing at 64°C and 45 s extension at 72°C. Reactions were terminated at the end of the extension phase, after SYBR green reported reaction kinetics in the log phase for several cycles.
Bioinformatics
Reads were aligned to dm3 with bowtie2 (v2.2.4) (Langmead and Salzberg, 2012). Coverage tracks were produced with deeptools2 (v2.5.0.1) (Ramírez et al., 2016) by subtracting the respective input (--ratio subtract --normalizeTo1x 121400000 -bs 1). Reads of ChIP alignments were counted with multiBamCov of bedtools (v2.25.0) (Quinlan and Hall, 2010). H3K4me3 reads were counted in a 500 bp region downstream of the first TSS. H3K27me3 reads were counted over the genebody. Flybase 5.44 was used as annotation. Differential regions were called with DESeq2 (v1.22.2) (Love et al., 2014). Heatmaps of differential regions were generated with ComplexHeatmaps (v2.1.0) (Gu et al., 2016). The hierarchical tree was based on log2FC (DEseq2) with method complete and Euclidean distance. In addition, log2TPMs are shown.
RNA-seq data were accessed from GEO (GSE38764). Adapters were clipped with trimgalore (v0.5.0; https://github.com/FelixKrueger/TrimGalore) (Martin, 2011). rRNA matching reads were removed with bowtie2 (v2.3.4.1) (Langmead and Salzberg, 2012). Abundances (TPM) were estimated with Salmon (v0.11.0) against the transcriptome (dmeI r5.44) (Patro et al., 2017).
Enrichment analysis
GO enrichment analysis was performed on www.flymine.org/ with Holm-Bonferroni correction with a maximum P-value of 0.05. For analysis of protein complexes the Compleat website (https://www.flyrnai.org/compleat/) was used (Vinayagam et al., 2013).
Supplementary Material
Acknowledgements
We thank all Knoblich lab members for support and discussions, Francois Bonnay and Joshua A. Bagley for comments on the manuscript, Elke Kleiner, the Research Institute of Molecular Pathology/IMBA Biooptics Facility for assistance and the Harvard TRiP collection, the BDSC and the VDRC for reagents.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: M.D.A., L.L., J.A.K.; Methodology: S.K.B., E.E.; Formal analysis: M.D.A., L.L., T.B.; Investigation: M.D.A., L.L.; Data curation: M.D.A., L.L.; Writing - original draft: M.D.A., L.L., J.A.K.; Writing - review & editing: S.K.B., J.A.K.; Visualization: M.D.A., L.L.; Supervision: R.E.K., J.A.K.; Funding acquisition: J.A.K.
Funding
Work in the Knoblich laboratory is supported by the Österreichischen Akademie der Wissenschaften, the Austrian Science Fund (Z_153_B09) and an advanced grant from the European Research Council.
Data availability
ChIP-seq data reported in this paper have been deposited in GEO under accession number GSE134509.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.183400.supplemental
Peer review history
The peer review history is available online at https://dev.biologists.org/lookup/doi/10.1242/dev.183400.reviewer-comments.pdf
References
- Abdusselamoglu M. D., Eroglu E., Burkard T. R. and Knoblich J. A. (2019). The transcription factor odd-paired regulates temporal identity in transit-amplifying neural progenitors via an incoherent feed-forward loop. eLife 8, 450 10.7554/eLife.46566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez J.-A. and Díaz-Benjumea F. J. (2018). Origin and specification of type II neuroblasts in the Drosophila embryo. Development 145, dev158394 10.1242/dev.158394 [DOI] [PubMed] [Google Scholar]
- Arama E., Dickman D., Kimchie Z., Shearn A. and Lev Z. (2000). Mutations in the β-propeller domain of the Drosophila brain tumor (brat) protein induce neoplasm in the larval brain. Oncogene 19, 3706-3716. 10.1038/sj.onc.1203706 [DOI] [PubMed] [Google Scholar]
- Aughey G. N., Estacio Gomez A., Thomson J., Yin H. and Southall T. D. (2018). CATaDa reveals global remodelling of chromatin accessibility during stem cell differentiation in vivo. eLife 7, 6061 10.7554/eLife.32341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello B., Holbro N. and Reichert H. (2007). Polycomb group genes are required for neural stem cell survival in postembryonic neurogenesis of Drosophila. Development 134, 1091-1099. 10.1242/dev.02793 [DOI] [PubMed] [Google Scholar]
- Bello B. C., Izergina N., Caussinus E. and Reichert H. (2008). Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 3, 5 10.1186/1749-8104-3-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger C., Harzer H., Burkard T. R., Steinmann J., van der Horst S., Laurenson A.-S., Novatchkova M., Reichert H. and Knoblich J. A. (2012). FACS purification and transcriptome analysis of Drosophila neural stem cells reveals a role for klumpfuss in self-renewal. Cell Rep. 2, 407-418. 10.1016/j.celrep.2012.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J., Mechtler K. and Knoblich J. A. (2006). Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell 124, 1241-1253. 10.1016/j.cell.2006.01.038 [DOI] [PubMed] [Google Scholar]
- Birkholz O., Vef O., Rogulja-Ortmann A., Berger C. and Technau G. M. (2013). Abdominal-B and caudal inhibit the formation of specific neuroblasts in the Drosophila tail region. Development 140, 3552-3564. 10.1242/dev.096099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone J. Q. and Doe C. Q. (2008). Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev. Neurobiol. 68, 1185-1195. 10.1002/dneu.20648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman S. K., Rolland V., Betschinger J., Kinsey K. A., Emery G. and Knoblich J. A. (2008). The tumor suppressors brat and numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev. Cell 14, 535-546. 10.1016/j.devcel.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman S. K., Simon M. D., Deaton A. M., Tolstorukov M., Borowsky M. L. and Kingston R. E. (2013). Multiplexed Illumina sequencing libraries from picogram quantities of DNA. BMC Genomics 14, 466 10.1186/1471-2164-14-466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman S. K., Deaton A. M., Domingues H., Wang P. I., Sadreyev R. I., Kingston R. E. and Bender W. (2014). H3K27 modifications define segmental regulatory domains in the Drosophila bithorax complex. eLife 3, e02833 10.7554/eLife.02833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken A. P., Dietrich N., Pasini D., Hansen K. H. and Helin K. (2006). Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 20, 1123-1136. 10.1101/gad.381706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd K. N. and Shearn A. (2003). ASH1, a Drosophila trithorax group protein, is required for methylation of lysine 4 residues on histone H3. Proc. Natl Acad. Sci. USA 100, 11535-11540. 10.1073/pnas.1933593100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R. and Zhang Y. (2004). The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 14, 155-164. 10.1016/j.gde.2004.02.001 [DOI] [PubMed] [Google Scholar]
- de Napoles M., Mermoud J. E., Wakao R., Tang Y. A., Endoh M., Appanah R., Nesterova T. B., Silva J., Otte A. P., Vidal M. et al. (2004). Polycomb group proteins ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell 7, 663-676. 10.1016/j.devcel.2004.10.005 [DOI] [PubMed] [Google Scholar]
- Dou Y., Milne T. A., Tackett A. J., Smith E. R., Fukuda A., Wysocka J., Allis C. D., Chait B. T., Hess J. L. and Roeder R. G. (2005). Physical association and coordinate function of the H3K4 methyltransferase MLL1 and the H4K16 acetyltransferase MOF. Cell 121, 873-885. 10.1016/j.cell.2005.04.031 [DOI] [PubMed] [Google Scholar]
- Eroglu E., Burkard T. R., Jiang Y., Saini N., Homem C. C. F., Reichert H. and Knoblich J. A. (2014). SWI/SNF complex prevents lineage reversion and induces temporal patterning in neural stem cells. Cell 156 1259-1273. 10.1016/j.cell.2014.01.053 [DOI] [PubMed] [Google Scholar]
- Estella C., Rieckhof G., Calleja M. and Morata G. (2003). The role of buttonhead and Sp1 in the development of the ventral imaginal discs of Drosophila. Development 130, 5929-5941. 10.1242/dev.00832 [DOI] [PubMed] [Google Scholar]
- Gan Q., Schones D. E., Eun S. H., Wei G., Cui K., Zhao K. and Chen X. (2010). Monovalent and unpoised status of most genes in undifferentiated cell-enriched Drosophilatestis. Genome Biol. 11, R42 10.1186/gb-2010-11-4-r42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Eils R. and Schlesner M. (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847-2849. 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- Harzer H., Berger C., Conder R., Schmauss G. and Knoblich J. A. (2013). FACS purification of Drosophila larval neuroblasts for next-generation sequencing. Nat. Protoc. 8, 1088-1099. 10.1038/nprot.2013.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi Y., Suzki N., Tsuboi M., Endo T. A., Toyoda T., Shinga J., Koseki H., Vidal M. and Gotoh Y. (2009). Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron 63, 600-613. 10.1016/j.neuron.2009.08.021 [DOI] [PubMed] [Google Scholar]
- Homem C. C. F. and Knoblich J. A. (2012). Drosophila neuroblasts: a model for stem cell biology. Development 139, 4297-4310. 10.1242/dev.080515 [DOI] [PubMed] [Google Scholar]
- Izergina N., Balmer J., Bello B. and Reichert H. (2009). Postembryonic development of transit amplifying neuroblast lineages in the Drosophila brain. Neural Dev. 4, 44 10.1186/1749-8104-4-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K. H. and Reichert H. (2015). Control of neural stem cell self-renewal and differentiation in Drosophila. Cell Tissue Res. 359, 33-45. 10.1007/s00441-014-1914-9 [DOI] [PubMed] [Google Scholar]
- Kassis J. A., Kennison J. A. and Tamkun J. W. (2017). Polycomb and trithorax group genes in Drosophila. Genetics 206, 1699-1725. 10.1534/genetics.115.185116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. H., Barrera L. O., Zheng M., Qu C., Singer M. A., Richmond T. A., Wu Y., Green R. D. and Ren B. (2005). A high-resolution map of active promoters in the human genome. Nature 436, 876-880. 10.1038/nature03877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston R. E. and Tamkun J. W. (2014). Transcriptional regulation by trithorax-group proteins. Cold Spring Harb. Perspect. Biol. 6, a019349 10.1101/cshperspect.a019349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich J. A. (2010). Asymmetric cell division: recent developments and their implications for tumour biology. Nat. Rev. Mol. Cell Biol. 11, 849-860. 10.1038/nrm3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich J. A., Jan, and Jan Y. N. (1995). Asymmetric segregation of numb and prospero during cell division. Nature 377, 624-627. 10.1038/377624a0 [DOI] [PubMed] [Google Scholar]
- Komori H., Xiao Q., Janssens D. H., Dou Y. and Lee C.-Y. (2014). Trithorax maintains the functional heterogeneity of neural stem cells through the transcription factor Buttonhead. eLife 3, e03502 10.7554/eLife.03502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong C., Adryan B., Bell I., Meadows L., Russell S., Manak J. R. and White R. (2008). Stability and dynamics of polycomb target sites in Drosophila development. PLoS Genet. 4, e1000178 10.1371/journal.pgen.1000178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. and Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357-359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-Y., Andersen R. O., Cabernard C., Manning L., Tran K. D., Lanskey M. J., Bashirullah A. and Doe C. Q. (2006a). Drosophila Aurora-A kinase inhibits neuroblast self-renewal by regulating aPKC/Numb cortical polarity and spindle orientation. Genes Dev. 20, 3464-3474. 10.1101/gad.1489406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-Y., Wilkinson B. D., Siegrist S. E., Wharton R. P. and Doe C. Q. (2006b). Brat is a miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev. Cell 10, 441-449. 10.1016/j.devcel.2006.01.017 [DOI] [PubMed] [Google Scholar]
- Levine S. S., Weiss A., Erdjument-Bromage H., Shao Z., Tempst P. and Kingston R. E. (2002). The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol. Cell. Biol. 22, 6070-6078. 10.1128/MCB.22.17.6070-6078.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine S. S., King I. F. G. and Kingston R. E. (2004). Division of labor in polycomb group repression. Trends Biochem. Sci. 29, 478-485. 10.1016/j.tibs.2004.07.007 [DOI] [PubMed] [Google Scholar]
- Lim D. A., Huang Y.-C., Swigut T., Mirick A. L., Garcia-Verdugo J. M., Wysocka J., Ernst P. and Alvarez-Buylla A. (2009). Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature 458, 529-533. 10.1038/nature07726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W. and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall O. J. and Brand A. H. (2017). Chromatin state changes during neural development revealed by in vivo cell-type specific profiling. Nat. Commun. 8, 2271 10.1038/s41467-017-02385-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J. 17, 10-12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Nègre N., Hennetin J., Sun L. V., Lavrov S., Bellis M., White K. P. and Cavalli G. (2006). Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol. 4, e170 10.1371/journal.pbio.0040170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumüller R. A., Richter C., Fischer A., Novatchkova M., Neumüller K. G., and Knoblich J. A. (2011). Genome-wide analysis of self-renewal in Drosophila neural stem cells by transgenic RNAi. Cell Stem Cell, 8, 580-593. 10.1016/j.stem.2011.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R., Duggal G., Love M. I., Irizarry R. A. and Kingsford C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417-419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira J. D., Sansom S. N., Smith J., Dobenecker M.-W., Tarakhovsky A. and Livesey F. J. (2010). Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc. Natl. Acad. Sci. USA 107, 15957-15962. 10.1073/pnas.1002530107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S., Sedkov Y., Smith S., Tillib S., Kraevski V., Nakamura T., Canaani E., Croce C. M. and Mazo A. (2001). Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene. Science 294, 1331-1334. 10.1126/science.1065683 [DOI] [PubMed] [Google Scholar]
- Quinlan A. R. and Hall I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841-842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F., Ryan D. P., Grüning B., Bhardwaj V., Kilpert F., Richter A. S., Heyne S., Dündar F. and Manke T. (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160-W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B., Chourrout D., Vervoort M., Leblanc B. and Cavalli G. (2007). Genome regulation by polycomb and trithorax proteins. Cell 128, 735-745. 10.1016/j.cell.2007.02.009 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B., Ganapathi M., Leblanc B., Portoso M., Jaschek R., Tolhuis B., van Lohuizen M., Tanay A. and Cavalli G. (2009). Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol. 7, e13 10.1371/journal.pbio.1000013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J. A. and Kingston R. E. (2009). Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 10, 697-708. 10.1038/nrm2763 [DOI] [PubMed] [Google Scholar]
- Södersten E., Toskas K., Rraklli V., Tiklova K., Björklund Å. K., Ringnér M., Perlmann T. and Holmberg J. (2018). A comprehensive map coupling histone modifications with gene regulation in adult dopaminergic and serotonergic neurons. Nat. Commun. 9, 1226 10.1038/s41467-018-03538-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y. and Lu B. (2011). Regulation of cell growth by Notch signaling and its differential requirement in normal vs. tumor-forming stem cells in Drosophila. Genes Dev. 25, 2644-2658. 10.1101/gad.171959.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa-Nunes R., Cheng L. Y. and Gould A. P. (2010). Regulating neural proliferation in the Drosophila CNS. Curr. Opin. Neurobiol. 20, 50-57. 10.1016/j.conb.2009.12.005 [DOI] [PubMed] [Google Scholar]
- Touma J. J., Weckerle F. F. and Cleary M. D. (2012). Drosophila Polycomb complexes restrict neuroblast competence to generate motoneurons. Development 139, 657-666. 10.1242/dev.071589 [DOI] [PubMed] [Google Scholar]
- Truman J. W. and Bate M. (1988). Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev. Biol. 125, 145-157. 10.1016/0012-1606(88)90067-X [DOI] [PubMed] [Google Scholar]
- Vinayagam A., Hu Y., Kulkarni M., Roesel C., Sopko R., Mohr S. E. and Perrimon N. (2013). Protein complex–based analysis framework for high-throughput data sets. Sci. Signal. 6, rs5 10.1126/scisignal.2003629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissel S., Harzer H., Bonnay F., Burkard T. R., Neumüller R. A. and Knoblich J. A. (2018). Time-resolved transcriptomics in neural stem cells identifies a v-ATPase/Notch regulatory loop. J. Cell Biol. 217, 3285-3300. 10.1083/jcb.201711167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie R., Everett L. J., Lim H.-W., Patel N. A., Schug J., Kroon E., Kelly O. G., Wang A., D'Amour K. A., Robins A. J. et al. (2013). Dynamic chromatin remodeling mediated by polycomb proteins orchestrates pancreatic differentiation of human embryonic stem cells. Cell Stem Cell 12, 224-237. 10.1016/j.stem.2012.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Li X., Zhang X., Mei S., Li H., Urso A. and Zhu S. (2014). The Drosophila Sp8 transcription factor buttonhead prevents premature differentiation of intermediate neural progenitors. eLife 3, e03596 10.7554/eLife.03596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.-P., Fu C.-C., Sugino K., Liu Z., Ren Q., Liu L.-Y., Yao X., Lee L. P. and Lee T. (2015). Transcriptomes of lineage-specific Drosophila neuroblasts profiled by genetic targeting and robotic sorting. Development 143, 411-421. 10.1242/dev.129163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Li M., Gu L., Chen X., Shi J., Zhang X. and Jiang C. (2017). Chromatin remodeling during in vivo neural stem cells differentiating to neurons in early Drosophila embryos. Cell Death Differ. 24, 409-420. 10.1038/cdd.2016.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Adli M., Zou J. Y., Verstappen G., Coyne M., Zhang X., Durham T., Miri M., Deshpande V., De Jager P. L. et al. (2013). Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell 152, 642-654. 10.1016/j.cell.2012.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.