ABSTRACT
The actin cytoskeleton and active membrane trafficking machinery are essential for polarized cell growth. To understand the interactions between myosin XI, vesicles and actin filaments in vivo, we performed fluorescence recovery after photobleaching and showed that the dynamics of myosin XIa at the tip of the spreading earthmoss Physcomitrella patens caulonemal cells are actin-dependent and that 50% of myosin XI is bound to vesicles. To obtain single-particle information, we used variable-angle epifluorescence microscopy in protoplasts to demonstrate that protein myosin XIa and VAMP72-labeled vesicles localize in time and space over periods lasting only a few seconds. By tracking data with Hidden Markov modeling, we showed that myosin XIa and VAMP72-labeled vesicles exhibit short runs of actin-dependent directed transport. We also found that the interaction of myosin XI with vesicles is short-lived. Together, this vesicle-bound fraction, fast off-rate and short average distance traveled seem be crucial for the dynamic oscillations observed at the tip, and might be vital for regulation and recycling of the exocytosis machinery, while simultaneously promoting vesicle focusing and vesicle secretion at the tip, necessary for cell wall expansion.
KEY WORDS: FRAP, Intracellular transport, Myosin V, Tip growth, Vesicle transport
Summary: Myosin XI rapidly associates and dissociates with vesicles and F-actin. This turnover is predicted to be essential for cell polarization and growth.
INTRODUCTION
Polarized cell growth, a mechanism by which plasma membrane and cell wall material are deposited to a defined region of the cell, is widespread across plants, fungi and oomycetes. As a result, plant cells exhibit a large variety of different shapes well-designed to achieve various functions (Geitmann and Ortega, 2009; Szymanski and Cosgrove, 2009). Tip growth is an extreme form of polarized cell growth where the cell expands only in one direction, generating elongated cells that can be hundreds of micrometers long (Hepler et al., 2001; Menand et al., 2007). This specialized form of growth is particularly well-suited for pollen tubes to reach the ovule during sexual reproduction (Cole and Fowler, 2006; Hepler et al., 2001), for root hairs to enhance uptake nutrients and water from the soil (Gilroy and Jones, 2000; Hepler et al., 2001), and for bryophytes seeking land colonization (Heckman et al., 2001; Kenrick and Crane, 1997). This process has been extensively studied in pollen tubes from lily, tobacco and Arabidopsis thaliana, as well as in root hairs from A. thaliana and protonemal cells from Physcomitrella patens. It is well known that tip growth requires a dynamic actin cytoskeleton network as well as an active membrane trafficking machinery (Cole and Fowler, 2006; Gilroy and Jones, 2000; Heckman et al., 2001; Hepler et al., 2001; Kenrick and Crane, 1997). Patterns of intracellular transport vary between tip-growing cells. In pollen tubes and root hairs, actin-myosin-dependent rapid transport of large organelles results in cytoplasmic streaming (Geitmann and Nebenführ, 2015; Hepler et al., 2001), whereas in moss protonemata, cytoplasmic streaming is absent and large organelles, with the exception of the chloroplasts, are not motile (Furt et al., 2012). Nevertheless, the growth of root hairs, pollen tubes and protonemata is strongly dependent on F-actin, most probably due to vesicle transport and organization of the apical cytosplasm (Park and Nebenführ, 2013; Vidali et al., 2001, 2009). In moss protonemata, modeling evidence suggests that endomembrane vesicles must be focused at the tip of the cell by F-actin to overcome diffusion limitations and sustain growth (Bibeau et al., 2018). A similar dependence for tip growth on F-actin but also on microtubules is present in fungal hyphae, where a vesicle-organizing center forms close to the cell apex (Riquelme et al., 2018). Because the wall material required for growth far exceeds the necessary membrane material, endocytosis occurs to recycle the excess plasma membrane (Grebnev et al., 2017; Ketelaar et al., 2008; Steer, 1988). However, how these different types of machinery are coordinated with the actin cytoskeleton is still unknown. To better understand how tip-growing cells self-organize their cytoplasmic components to achieve and maintain polarized growth, it is crucial to determine the function of each component separately as well as their interactions. Because of this limited information, quantitative modeling efforts must simplify the function of the cytoskeleton as fluxes of directed secretion (Campàs and Mahadevan, 2009; Dumais et al., 2006; Fayant et al., 2010; Kroeger et al., 2011; Luo et al., 2017; Rojas et al., 2011). Although these fluxes are a good first approximation, they provide little insight into the mechanisms by which the cytoskeleton functions. Modeling efforts that incorporate the true interactions of the cytoskeleton, its motors and cargos will help to answer questions regarding the establishment and maintenance of polarized secretion.
Several studies have shown that the actin-based molecular motor myosin XI is required for tip growth in plants. Silencing of the two myosin XI genes, myosin XIa and myosin XIb in the moss P. patens resulted in spherical cells where tip growth was completely abolished (Vidali et al., 2010). Dominant-negative inhibition and gene-knockout approaches suggested that among the 13 isoforms of myosin XI present in A. thaliana, myosin XIK is a primary contributor for root hair elongation by tip growth (Ojangu et al., 2007; Park and Nebenführ, 2013; Peremyslov et al., 2008; Prokhnevsky et al., 2008); whereas myosin XIC and myosin XIE are important for pollen tube growth (Madison et al., 2015). During the last decade, it was well-established that members of the myosin XI family are responsible for motility of large organelles in higher plants, even though their relative contribution and interdependency relationships are not fully understood (Avisar et al., 2012, 2009, 2008; Griffing et al., 2014; Henn and Sadot, 2014; Madison and Nebenführ, 2013; Peremyslov et al., 2008, 2010; Prokhnevsky et al., 2008; Sparkes et al., 2008; Ueda et al., 2010; Vick and Nebenführ, 2012). For example, organelle motility of Golgi stacks, peroxisomes and mitochondria in root hairs and leaf cells is reduced in myosin xik-knockout mutant plants, which also display reduced root hair elongation (Prokhnevsky et al., 2008). Nevertheless, a functional myosin XIK tagged with yellow fluorescent protein does not localize to these organelles, instead, the signal accumulates in the apical domain (Park and Nebenführ, 2013). In addition, the apical localization of myosin XIa in moss caulonemal cells does not correlate with the localization for large organelles observed in other plants (Furt et al., 2012; Vidali et al., 2010). Therefore, the function fulfilled by myosin XI, which is essential to achieve and/or maintain tip growth in plants, is still unknown.
Numerous studies have reported that members of the myosin V family, which are the animal and fungal homologues of those comprising the myosin XI family of plants, are involved not only in motility of large organelles but also impact on secretory, endocytic and recycling pathways through their ability to transport endomembrane vesicles (Fan et al., 2004; Hammer and Sellers, 2012; Lapierre et al., 2001; Li and Nebenführ, 2008; Lise et al., 2006; Nedvetsky et al., 2007; Ohyama et al., 2001; Pruyne et al., 1998; Rodriguez and Cheney, 2002; Roland et al., 2007; Schott et al., 1999; Volpicelli et al., 2002; Yan et al., 2005). Recent evidence suggests that myosin XI is involved in endomembrane vesicle transport in plants. Results from studies investigating colocalization of vesicle markers (i.e. RabA4b and SCAMP2), using fluorescence recovery after photobleaching (FRAP), and biochemical co-fractionation with vesicle (RabA4b) and exocytic (Sec6) markers inferred that myosin XIK in A. thaliana is associated with endomembrane vesicles in both leaf cells and root hairs (Park and Nebenführ, 2013; Peremyslov et al., 2012; Rybak et al., 2014). Using confocal microscopy combined with fluctuation cross-correlation analyses, we have previously demonstrated that myosin XIa and the SNARE protein VAMP72, an endomembrane vesicle marker, colocalize at the cell apex and synchronize during tip growth (Furt et al., 2013). Surprisingly, we have also shown that apical myosin XIa precedes F-actin during polarized growth of P. patens caulonemal cells (Furt et al., 2013). Pharmacological approaches using latrunculin B to depolymerize the actin filaments, further showed that myosin XIa stays associated with VAMP72-labeled vesicles at the apex of moss cells after disruption of the actin network (Furt et al., 2013). In addition, we observed the emergence of myosin XIa associated with VAMP72-labeled vesicles as ectopic clusters, which were then propelled in an actin-dependent manner through the cell when the actin network self-reorganizes (Furt et al., 2013).
Together, these results suggest the existence of a mechanism during which myosin XI-associated endomembrane structures organize the actin polymerization machinery. However, as the apex of tip-growing cells in plants is densely occupied by vesicles (Bove et al., 2008; de Graaf et al., 2005; Kroeger et al., 2009; Ovečka et al., 2005; Parton et al., 2001; Preuss et al., 2004; Szumlanski and Nielsen, 2009; Zonia and Munnik, 2008), and image resolution is diffraction limited when using confocal laser scanning microscopy, it is a challenge to visualize single vesicles (∼80 nm in diameter) in tip-growing cells of plants in order to uncover this mechanism. Therefore, the ability of myosin XIa to transport endomembrane vesicles and coordinate this transport with the actin polymerization machinery, and whether this is, indeed, the essential function fulfilled by myosin XIa to achieve polarized growth, remains to be demonstrated. Furthermore, regarding plant cells, there are no quantitative in vivo estimates of these binding interactions. Specifically, the average on- and off-rate between myosin XI and its cargo are unknown. In addition, myosin XI run lengths (=velocity/association time) and off-rates on actin filaments remain unknown. For these reasons, any quantitative insight concerning these interactions would shape the way the active transport machinery is viewed in plants, and would enable future modeling by constraining the large number of parameters regarding the cytoskeleton during polarized growth.
In this study, we used P. patens caulonemal cells and applied FRAP (Lorén et al., 2015; McNally, 2008) to demonstrate that myosin XIa recovers at the cell apex in an actin-dependent manner. After using the actin depolymerizing agent latrunculin B, we performed FRAP in the absence of actin filaments and determined a range of effective dissociation constants (Kd) for the binding of myosin XIa to VAMP72-labeled vesicles. To improve the precision of these measurements we used variable-angle epifluorescence microscopy (VAEM). We showed that myosin XIa and VAMP72-labeled vesicles colocalized at the cortex of moss cells to form highly dynamic punctate structures, whose persistent motility depends on actin filaments. With particle tracking and Hidden Markov modeling (HMM), we characterized the weak transient interactions between myosin XI, VAMP72-labeled vesicles and filamentous actin. Our results are consistent with the hypothesis that myosin XIa coordinates the traffic machinery and actin network dynamics to maintain polarized growth in moss cells.
RESULTS
After photobleaching, apical myosin XIa recovers faster than VAMP72-labeled vesicles at the cell apex in an actin-dependent manner
We have previously shown that the level of cytoplasmic myosin XIa and F-actin fluctuate at the apex of caulonemal cells, and that increases in myosin XIa levels anticipate those of F-actin, whereas cytoplasmic myosin XIa levels fluctuate in-phase with the endomembrane vesicle marker VAMP72 (Furt et al., 2013). To explain this, we proposed a model where myosin XIa, through its association with endomembrane vesicles, coordinates vesicular traffic and F-actin polymerization-driven motility at the cell apex to maintain polarized cell growth (Furt et al., 2013). This proposed model predicts that the dynamics of myosin XI at the cell tip exhibit some degree of coupling with filamentous actin. To verify this, we conducted FRAP experiments (Lorén et al., 2015; McNally, 2008) of myosin XIa at the growing cell tip and shank (Figs 1A, S1 and Movie 1). Compared to that at the shank, myosin XIa at the tip exhibited fluorescence recovery that is consistent with active transport. Specifically, during the same time span, fluorescence intensity of 3mEGFP-labeled myosin XIa at the tip recovered to a higher degree than that at the cell shank. Normalizing the recovered fluorescence intensity to the pre-bleach intensity of the shank highlights this effect (compare Fig. 1A with Fig. S1B). Given that the fluorescence recovery is reduced in the proximity of the cell boundary at the cell tip (Fig. 2A), this further indicates that myosin XI is actively transported to the cell tip. This suggests that F-actin that concentrates at the cell tip is coupled to an increased flow of myosin XIa. This result does not support a simple mechanism of F-actin statically capturing myosin XIa to increase accumulation at the cell tip, which should result in a slower myosin XIa recovery in FRAP.
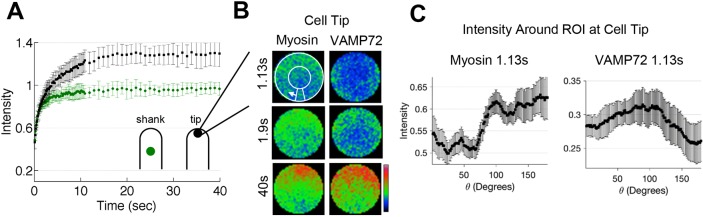
Fig. 1.
Dynamics of apical myosin XIa during polarized cell growth. (A) Fluorescence recovery of 3mEGFP-myosin XIa at the cell shank (green circles) and the cell tip (black circles). n=7 and 10 cells for shank and tip, respectively. Fluorescence intensities from tip and shank regions were normalized to pre-bleach fluorescence of the shank region. Error bars represent ±s.d. (B) Cropped and frame-averaged photobleached ROI at the cell tip of cells expressing 3mEGFP-myosin XIa (left) and 3mCherry-VAMP72 (right). White circles and arrow indicate how the ROI perimeter was measured during recovery. (C) Fluorescence intensity profile of 3mEGFP-myosin XIa (left) and 3mCherry-VAMP72 (right) along the perimeter of the ROI at 1.13 s after photobleaching.
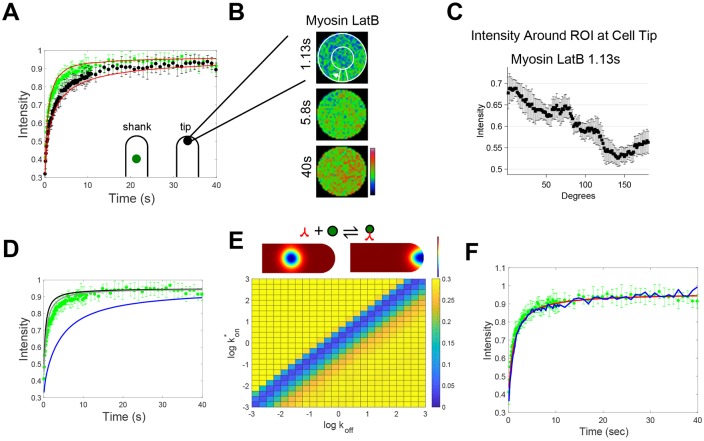
Fig. 2.
Treatment with latrunculin B allows to characterize binding between myosin XIa and vesicles. (A) Fluorescence recovery of 3mEGFP-myosin XIa at the shank (green circles) and the tip (black circles) in cells treated with latrunculin B. Red lines indicate the reaction diffusion FRAP model with best-fit dissociation constant (Kd), and a diffusion coefficient for myosin XI for its closed configuration (D=3.4 μm2/s). n=6 and 8 cells for the shank and tip, respectively. Fluorescence intensity from tip and shank regions was normalized to the pre-bleach fluorescence intensity of the shank region. Error bars ±s.d. (B) Cropped and frame-averaged photobleached ROI at the cell tip for cells expressing 3mEGFP-myosin XIa and treated with latrunculin B. White circles and arrow indicate how the perimeter of the ROI was measured during the recovery. (C) Fluorescence intensity profile of 3mEGFP-myosin XIa along the perimeter of the ROI at 1.13 s after photobleaching. (D) Fluorescence recovery of 3mEGFP-myosin XIa in cells treated with latrunculin B at the shank (green circles) compared to theoretical myosin XIa diffusion, where D=2.3 (gray line), 3.4 (black line) or 0.29 μm2/s (blue line). (E) Example reaction–diffusion model of FRAP at the cell shank and tip at 1.13 s after photobleaching (top). Red indicates concentrated pixels and blue indicates less concentrated pixels. kon and koff heat map of the sum of squared differences between the Comsol reaction–diffusion model at the cell shank and the experimental myosin XIa fluorescence recovery at the shank (bottom). Bright yellow indicates large differences and dark blue indicates small differences. (F) Fluorescence recovery of 3mEGFP-myosin XIa in cells treated with latrunculin B at the shank (green circles) and corresponding two-dimensional infinite-boundary reaction–diffusion model as described by Kang et al. (2010) (red line). Blue line indicates the fluorescence recovery from Digital Confocal Microscopy Suite. Both simulations were run by using: U/B = 0.5.
To better understand how F-actin coupling results in increased flow of myosin XIa to the cell tip, we analyzed its fluorescence recovery spatially (Fig. 1B and Movie 2). At early times during fluorescence recovery (1.13 s after photobleaching), we detected recovery of myosin XIa at the cell apex before that of VAMP72-labeled vesicles (Fig. 1B). Given that the bleach region of interest (ROI) is 4 μm in diameter, this fluorescence recovery is consistent with myosin motor speeds of a few microns per second. To quantify the directionality of this recovery, we measured the fluorescence of myosin XIa along the perimeter of the photobleached region and found that myosin recovered first at the cell apex (Fig. 1C). This spatial recovery profile is not consistent with diffusion – which we have previously characterized in detail – specifically when considering the effect of boundaries and cell shape (Bibeau et al., 2018; Kingsley et al., 2018). The pattern of recovery is inconsistent with simple diffusion, which should be from the shank towards the cell tip. The pattern of recovery is also inconsistent with a long-term association of myosin XIa with VAMP72-labeled vesicles, which recover along the actin cortex (Fig. 1C). Hence, fluorescence recovery of myosin XI is expected to be a combination of vesicle transport, diffusion and transport on actin that is independent of vesicles.
To determine whether the observed myosin XIa fluorescence recovery is actin dependent, we treated growing cells with latrunculin B. Depolymerization of F-actin reduced the flow of myosin XIa at the cell tip (Fig. 2A) and removed the myosin Xia-dense apical spot (Fig. 2B). Without F-actin, myosin XIa recovered from the cell shank towards the cell tip (Fig. 2B,C and Movie 3), in a pattern that is consistent with simple diffusion (Bibeau et al., 2018; Kingsley et al., 2018). This indicates that myosin XIa exhibits actin-dependent apical recovery in the absence of latrunculin B treatment.
FRAP reveals the fraction of myosin XI bound to VAMP72-labeled vesicles
To determine the degree to which myosin XIa binds to VAMP72-labeled vesicles, we conducted FRAP experiments in the presence of latrunculin B (Fig. 2A). By removing the actin cytoskeleton, we ensure that myosin XIa fluorescence recovery was a result of myosin XIa either bound or unbound to vesicles. Because myosin XI should diffuse more slowly when associated with vesicles, it is possible to infer the fraction of bound myosin XI by measuring the rate of the fluorescence recovery if the diffusion coefficients are known. Owing to the potential of vesicle binding, myosin XIa diffusion cannot be measured by using FRAP. We, therefore, estimated the potential range of diffusion coefficients on the basis of their similarity to the well-characterized myosin V (Krementsov et al., 2004; Li et al., 2004; Wang et al., 2004) and the measured viscosity of the moss cytoplasm (see Materials and Methods, and Kingsley et al., 2018). Since myosin V is found in the open or closed configuration, and the type of configuration modify the Stokes radius of the protein and, consequently, its diffusion coefficient, it is possible that myosin XI exhibits two unique diffusion coefficients. Given the known changes for myosin V hydrodynamic radius measured by analytical centrifugation (Krementsov et al., 2004; Li et al., 2004; Wang et al., 2004) and the known viscosity of the moss cytoplasm (Kingsley et al., 2018), we estimate the myosin XI diffusion coefficient to be between 2.3 and 3.4 μm2/s, for the open and closed configuration, respectively.
By using the diffusion coefficients for myosin XI and vesicle (Bibeau et al., 2018), we modeled the interaction as a reversible bimolecular reaction, i.e. U + S ⇌ B, in which ‘U’ is the concentration of unbound myosin XIa in the cytoplasm, ‘S’ the concentration of available vesicle binding sites for myosin XIa and ‘B’ the concentration of myosin XIa bound to a vesicle receptor. We further assumed that the number of myosin XIa-binding sites on a vesicle, i.e. S(x,y,t), is constant at equilibrium, i.e. S(x,y,t)=Seq (Kang and Kenworthy, 2008; Kang et al., 2010; Sprague and McNally, 2005). It then follows that we can calculate the effective on-rate kon* by using kon*=Seqkon (Kang and Kenworthy, 2008; Kang et al., 2010; Sprague and McNally, 2005). On the basis of these assumptions, we can now model the process as a reaction–diffusion system between two-species (Kang and Kenworthy, 2008; Kang et al., 2010) described by Eqns 1 and 2:
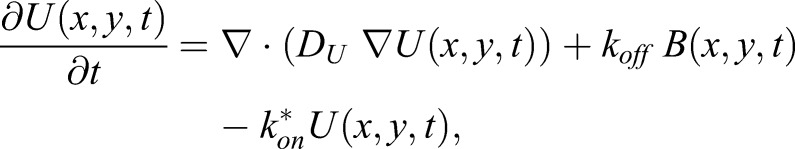 |
(1) |
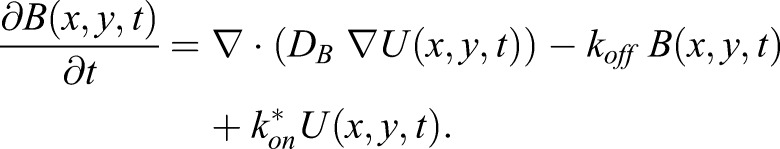 |
(2) |
Here, U(x,y,t) and B(x,y,t) are the concentrations of unbound and bound myosin XIa, DU and DB are the estimated diffusion coefficients for unbound myosin XIa and VAMP72-labeled vesicles, ∇ is the gradient operator and koff is the off-rate of myosin XIa from the vesicle.
Although Eqns 1 and 2 (Kang and Kenworthy, 2008; Kang et al., 2010; Sprague and McNally, 2005), can be solved analytically for simple boundary conditions, no analytical solution exists for our cell shape. We have previously shown, through extensive experimentation and simulations of FRAP, that cell shape is important to model FRAP (Bibeau et al., 2018; Kingsley et al., 2018). Here, we used the finite element modeling software Comsol (Comsol Inc, Stockholm, Sweden) to simulate FRAP in the presence of myosin vesicle binding (Fig. 2E) and cell boundaries, to solve this system of equations (see Materials and Methods for details). To highlight the sensitivity of our measurements, we demonstrate the extreme cases in which myosin XIa is completely bound to vesicles (D=0.29 µm2 s−1) (Bibeau et al., 2018) or completely free (Fig. 2D). A parameter sweep across potential Kon* and koff given by Eqns 1 and 2 (Fig. 2E) demonstrates that we only have the experimental sensitivity to determine the effective dissociation constant (Kd*) or, more intuitively, the ratio of unbound to bound myosin, Kd*=koff /kon*=U/B, and not the individual rate constants. Using the Matlab (MathWorks, Natick, MA) liveLink Comsol Multiphysics module, we were able to iteratively find an effective dissociation constant that matched experimental myosin XIa fluorescence recoveries, i.e. U/B=0.8−0.5, at both the tip and the shank (Fig. 2A, see Materials and Methods for details). Here, the variation in the U/B ratio reflects the possible open (0.8) and close (0.5) configurations of myosin XI. Through algebraic arrangement, the U/B ratio can be modified to be the fraction of myosin XI bound to a vesicle, which is ∼55% (open) and ∼66% (close). Similar recovery curves were found by applying these best-fit parameters to the two-dimensional infinite-boundary reaction–diffusion model from Kang et al. (Fig. 2F) (Kang et al., 2010) and by using the Digital Confocal Microscopy Suite (DCMS) with reaction-diffusion (Kingsley et al., 2018) (Fig. 2F).
Myosin XIa and VAMP72 puncta exhibit actin-dependent dynamics at the cortex of moss protonemal cells
To further assess the dynamics of myosin XIa and vesicles on the particle level, we used VAEM, which, by illuminating only a thin optical section, provides the signal-to-noise ratio necessary to image single endomembrane vesicles (Konopka and Bednarek, 2008). We found, as expected, that 3mEGFP-VAMP72 labeled highly dynamic small punctate structures at the cortex of apical caulonemal cells (Fig. S2A top, Movie 4). This is consistent with the localization to endomembrane vesicles and observations using the same probe, in which protoplasts of Arabidopsis cells cultured in suspension were analyzed (Uemura et al., 2004). We also showed that 3mEGFP-Myosin XIa localizes to similar dynamic punctate structures at the cortex of caulonemal cells (Fig. S2A, bottom panels; Movie 4). By using the detection tool of the TrackMate plugin from ImageJ (see Materials and Methods), we determined that the average density of myosin XIa-decorated structures is 0.49±0.04 per μm2 – which is similar to that of VAMP72-labeled endomembrane vesicles with 0.52±0.01 per μm2 (Fig. S2B). These densities are slightly higher but in the same range as those reported for class II formin, a nucleator of actin filaments present in P. patens (van Gisbergen et al., 2012).
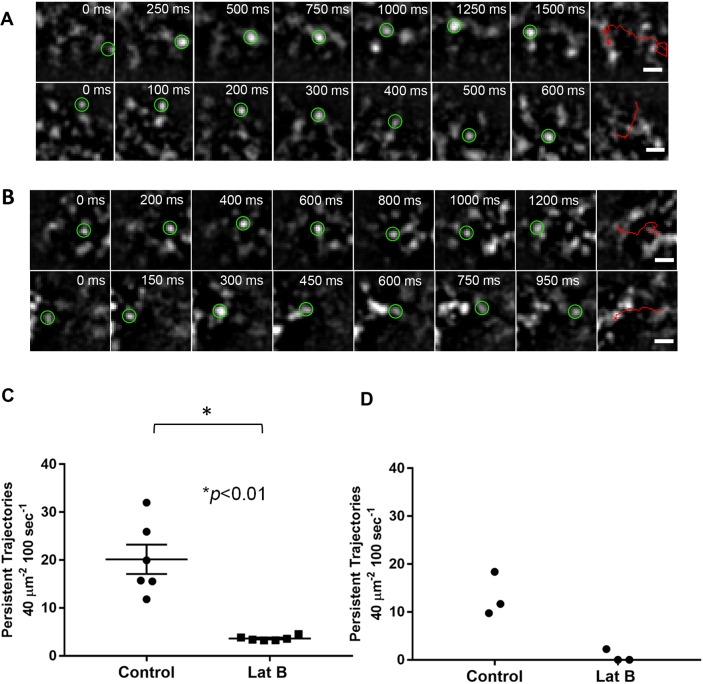
To evaluate whether the motility of both myosin XIa-puncta and VAMP72-labeled vesicles depends on actin, we imaged the apical caulonemal cells in the presence or absence of 25 μM latrunculin B that completely depolymerizes the actin filaments (Vidali et al., 2009). We tracked the movement of punctate structures labeled by myosin XIa and VAMP by using TrackMate in the presence or absence of the drug. At least three types of motion were observed: 1) some punctate structures that move rapidly in and out of the imaging field, 2) some that move in a persistent linear manner and, 3) some that stay confined to an area (displacement <0.5 μm). We only focused on punctate structures that display a directed movement for at least 12 consecutive frames (600 ms) and show a displacement >1.2 μm. Examples of such persistent linear trajectories recorded for myosin XIa and VAMP72 are shown in Fig. 3, panels A and B, respectively (Movie 4). Over a period of 100 s, we were able to detect 20.2±3.1 and 13.3±2.6 persistent linear trajectories for myosin XIa and VAMP72, respectively, per average imaged area per cell (40 μm2) (Fig. 3, panels C and D, respectively). However, after treatment with latrunculin B, we found only 3.6±0.2 and 0.8±0.8 persistent linear trajectories for myosin XIa and VAMP72, respectively, per average imaged area per cell (40 μm2) (Fig. 3C,D). Such a large decrease in the number of persistent linear trajectories in the absence of actin filaments supports the hypothesis that myosin XIa-puncta and VAMP72-labeled endomembrane vesicles move on actin filaments in apical caulonemal cells.
Fig. 3.
Myosin XIa and VAMP72-labeled vesicle directed motion at the cell cortex depends on actin. (A,B) Representative examples of persistent linear trajectories of 3mEGFP-Myosin XIa (A) and 3mEGFP-VAMP72-labeled vesicles (B) at the cortex of apical caulonema cells near the apex. Images were acquired by using VAEM at 50 ms intervals for the complete time series. Selected images of each trajectory are shown with the time from the series indicated at the top. Persistent linear trajectories are shown in the last image of each series. Tracked particles are indicated by green circles, trajectories are indicated by red lines. Scale bars: 1 μm. (C,D) Quantification of the number of persistent linear trajectories detected in control apical caulonema cells and those treated with latrunculin B for myosin Xia. n=6 cells, ±s.e.m., a Mann–Whitney statistical two-tailed test was used for comparison (C) and VAMP72-labeled vesicles (n=3 cells) (D).
Myosin XIa puncta and VAMP72-labeled vesicles are highly dynamic at the cortex of moss protoplasts
Although we were able to identify a portion of myosin XIa puncta and VAMP72-labeled vesicles that exhibited actin-dependent active motion, we needed to improve our imaging signal-to-noise ratio and tracking fidelity to track both myosin XIa and VAMP72-labeled vesicles together. To be able to do this, we imaged protoplasts with VAEM; we used protoplasts because, with this technique, it is possible to slightly flatten the cells and reliably get larger imaging areas, and more-consistent imaging data. Just as in caulonema cells, we found that both fluorescently tagged myosin XIa and VAMP72-labeled vesicles localize to dynamic punctate structures (Fig. 4A).
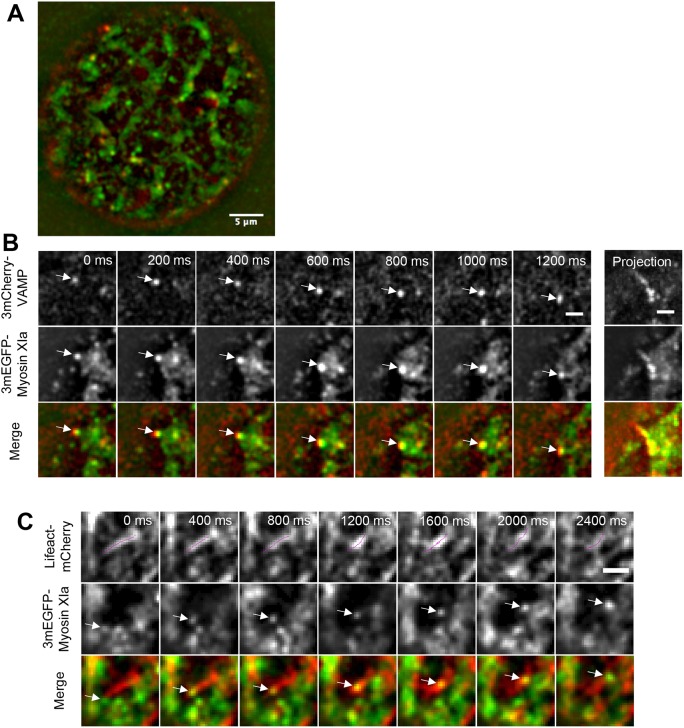
Fig. 4.
Myosin XIa puncta and VAMP72-labeled vesicles exhibit colocalized dynamic motion in protoplasts. (A) Representative VAEM image of a protoplast expressing 3mCherry-VAMP and 3mEGFP-myosin XIa. Scale bar: 5 µm (B) Selected images showing colocalized 3mCherry-VAMP and 3mEGFP-myosin XIa (top two rows); merged images in the third row show VAMP-labeled vesicles (red) and myosin XIa (green). Images from protoplasts were simultaneously acquired using VAEM at 100 ms intervals. The maximum projection of 26 frames from each corresponding time series is shown in the right panel. Scale bars: 2 µm. (C) Selected images showing Lifeact-mCherry-labeled F-actin and 3mEGFP-tagged myosin XIa (top two panels). Merged images are shown in the third row, with actin filaments in red and myosin XIa in green. Images from protoplasts were simultaneously acquired using VAEM at 100 ms intervals. Arrows indicate myosin XI-labeled punctate structures moving on actin filaments (indicated by the dotted lines). Scale bar: 2 μm.
In protoplasts, we observed that several punctate structures labeled with both 3mEGFP-myosin XIa and 3mCherry-VAMP72 move in a linear manner, as indicated by the maximum projections in the examples shown in Fig. 4B. To test whether these trajectories overlap with those of actin filaments, we simultaneously imaged 3mEGFP- tagged myosin XIa and Lifeact-mCherry-tagged F-actin (Vidali et al., 2009). We found that myosin XIa-labeled structures colocalize and move along cortical actin filaments in moss protoplasts (Fig. 4C). Together with the fact that myosin XIa is an actin-based motor, these results further support that the motion of the endomembrane vesicles labeled by both myosin XIa and VAMP72 occurs on actin filaments.
Myosin XIa-associated structures and VAMP72-labeled vesicles exhibit short persistent active motion in protoplasts
Because of the improved resolution in protoplasts, it was possible to quantitatively infer how myosin XIa and VAMP72-vesicles associate with F-actin by analyzing the persistent nature of their trajectories – without the need for tracking individual actin filaments. To this aim we used HMM (see Materials and Methods, and Fig. S2 for more details) (Rabiner, 1989). Briefly, an HMM is a statistical model used to determine the likelihood whether, at a given time, a system is in some unobserved state based on a known observable variable. As previously shown by Roding et al. (2014), an HMM can be applied to intracellular particle tracking data to determine the likelihood whether, at a specific point in a given trajectory, a particle is either in a Brownian state or an active transport state (Fig. S2A). Intuitively, a particle that moves in a random pattern is more likely to be in a Brownian state and a particle that moves in a ballistic fashion is more likely to be in an active transport state. Yet, without a quantitative model, it can be difficult to manually classify the state of the particle. The HMM makes the best statistical guess at the true state of the particle, on the basis of particle trajectories (observed sequences), known information about active and Brownian motion (emission matrix), and inferred unbinding and binding onto actin (transition matrix). Importantly, this model does not require a fluorescently labeled actin cytoskeleton or tracking of individual actin filaments within crowded environments. It only requires the tracking of particles from one channel of data (in our case, myosin XIa or VAMP72), which can reliably be carried out by using the Fiji plugin Trackmate (see Materials and Methods).
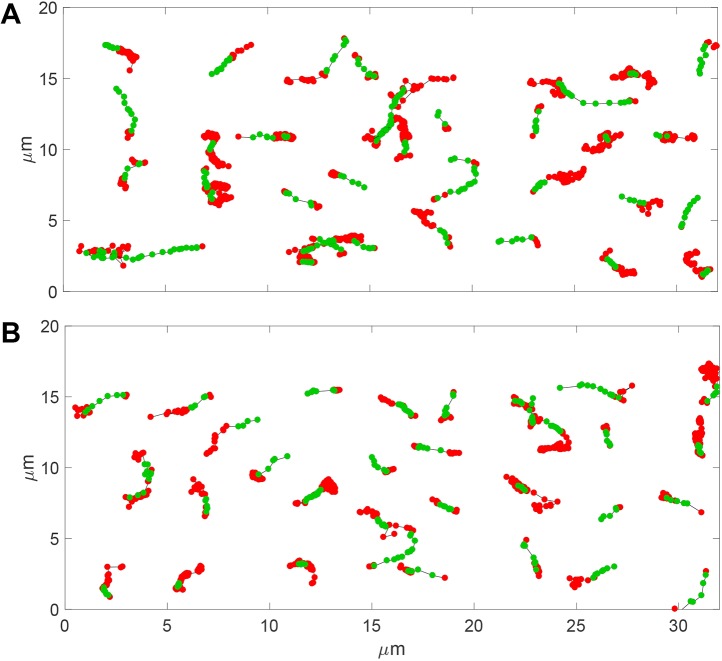
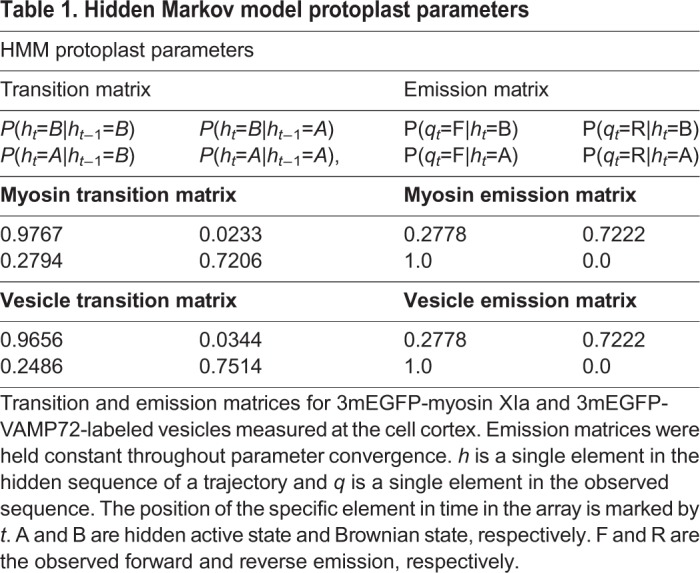
When the HMM was applied to tracking data for myosin XIa puncta or VAMP72-labeled vesicles in protoplasts (a total of four cells were analyzed and their trajectories combined), we, respectively, found 139 out of 307 (45%) or 126 out of 572 (22%) trajectories with an active component (Fig. 5A,B and Movies 5-7). The HMM also estimated the most likely state transition probabilities for myosin XIa puncta and VAMP72-labeled vesicles, Table 1 (see Materials and Methods). We found that both myosin XIa and VAMP72-labeled vesicles have an ∼75% chance of remaining on a filament [P(ht=A|ht−1=A)] during the exposure time of 50 ms. Consistent with this, we did not observe any continuously active sections of trajectories for more than 1.05 s. We also observed the actin-associated run lengths to have an average end-to-end distance of 1.24±0.06 and 1.30±0.07 μm for myosin XIa puncta and VAMP72-labeled vesicles, respectively; errors indicate the ±standard error of the mean (±s.e.m.), n=number of active trajectories. The instantaneous velocity for active trajectories of myosin XI puncta and VAMP72-labeled vesicles was 2.7±1.95 and 2.45±1.89 μm/s, respectively; errors indicate the ±standard deviation (±s.d.), n=number of active trajectories. Interestingly, the instantaneous velocity for Brownian trajectories was similar, i.e. 2.4±2.00 and 1.83±1.66 μm/s, for myosin XI puncta and VAMP72-labeled vesicles, respectively (errors indicate standard deviation, n=number of active trajectories).
Fig. 5.
Predicted trajectories in protoplasts using HMM. (A) Example of VAMP72-labeled vesicle trajectories containing a sequence in the active state, predicted by using the HMM. (B) Example of myosin XIa trajectories containing a sequence in the active state, predicted by using the HMM. The Brownian state is indicated by red circles, the active state is indicated by green circles.
Table 1.
Hidden Markov model protoplast parameters
Myosin XIa and VAMP72-labeled vesicles exhibit a weak and transient association
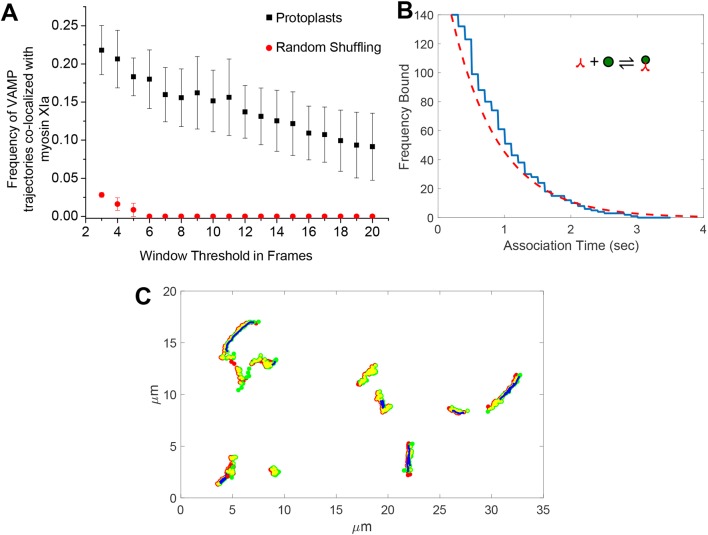
To determine whether myosin XIa and VAMP72 are associated with the same membranes, we simultaneously imaged myosin XIa-labeled structures and VAMP72-labeled vesicles in protoplasts. In an approach similar to that described by Deschout et al. (2013), we used automated analysis to determine whether the myosin XIa puncta and VAMP72-labeled vesicles exhibit spatiotemporal colocalization. For this, we used a minimal contact radius and a sliding correlation window to determine whether two particles from different channels showed correlated movement (see Materials and Methods, and Fig. S3). For each given window size, the frequency of VAMP72-labeled trajectories was calculated by dividing the number of VAMP72-labeled trajectories that colocalize with myosin XI, by the VAMP72-labeled trajectories in total (Fig. 6A, see also Materials and Methods). We found that a fraction of myosin XIa-decorated structures partially colocalize – in time and space – with VAMP72-labeled vesicles at the cell cortex in moss protoplasts. We observed that ≥20% of the VAMP72-labeled trajectories exhibited some colocalization over a period of 400 ms in total, and ≥15% over a period of 1 s in total (Fig. 6A and Movies 7-8). These colocalization frequencies are substantially higher than those obtained by randomly shuffling the observed trajectories (Fig. 6A, random shuffle). The reduction in localization frequency at increasing window sizes is the result of increased stringency for colocalization at bigger window sizes.
Fig. 6.
Colocalization of myosin XIa with VAMP72-labeled vesicles is specific and transient. (A) Frequency of 3mCherry-VAMP72 trajectories that colocalized with 3mEGFP-myosin XIa as detected in protoplasts (black squares) or randomly generated by a simulation (red circles). Each frame corresponds to 100 ms. The minimum number of frames (i.e. threshold of windows) that are required to classify two moving particles as colocalized is indicated at the x-axis (n=3 cells, ±s.e.m., P<0.0001, Fisher's exact test). (B) The number of myosin XIs observed on a vesicle in protoplasts for a given time (blue line) and the corresponding best fit to Eqn 7 (red dashed line). (C) Example of colocalized protoplast trajectories. VAMP72-labeled vesicle trajectories (red dots) and myosin XIa trajectories (green dots) colocalize (yellow dots) and exhibit active motion (blue lines), as predicted by HMM.
To examine the total colocalization time of both myosin XIa and VAMP72-labeled vesicles, we divided the total number of colocalization events by the total number of identified events for a window size of three frames. Of all identified and tracked myosin XIa, we found that ∼4.2% was localized to vesicles; we also found that 10% of identified VAMP72-labeled vesicles colocalized with a myosin XI. Although these percentages may seem low when compared with our FRAP results, they yield from a colocalization analysis that requires spatial colocalization specifically at the cortex of the cell. Also, some of the total events could be due to noise, which will increase the total number of events and reduces the proportion of colocalization events. Nevertheless, because of the stringency of this analysis, these measurements are well-above random colocalization events (Fig. 6A). This result is consistent with the hypothesis that myosin XIa associates with endomembrane vesicles.
The colocalization data also allow us to estimate the off-rate for interaction between myosin XI and VAMP72-labeled vesicles. To determine this rate, we examined the duration myosin XIa resides on a vesicle. Fitting the observed residence times to an exponential decay of the form:
| (3) |
yields an off-rate (koff) of ∼1.4 s−1 (Fig. 6B). Here, n is the number of molecules bound after the elapsed time t, and t0 is the minimum imaging time required to detect a colocalization event (i.e. 200 ms) by using the colocalization algorithm. This off-rate is fast, resulting in a half-life of the complex of ∼500 ms.
To determine whether VAMP72-labeled vesicle motion is dependent on myosin XIa, we examined the HMM results and any colocalized trajectories together (see Fig. 6C, Movies 5-8. We found that, of the 139 VAMP72-labeled vesicle-active trajectories, only 28 (20%) colocalized with a detectable myosin XIa, Movie 7. Since 80% of the VAMP72-labeled vesicles were able to move actively without a myosin XIa (Movie 5), this population of VAMP72-labeled vesicles seems to be propelled by formin-mediated actin polymerization (Furt et al., 2013; van Gisbergen et al., 2012). Moreover, because 19 out of the 126 (15%) myosin XIa active trajectories colocalized with a detectable VAMP72-labeled vesicle (Movie 7), myosin XIa might move on actin without any cargo or might transport cargo other than VAMP72-labeled vesicles (Movie 6).
DISCUSSION
We have shown here, that myosin XIa and VAMP72-labeled vesicles colocalize in vivo, and exhibit actin-dependent mobility observed as short but persistent trajectories. By using particle-tracking statistics, we found that myosin XIa interacts transiently with VAMP72-labeled vesicles and demonstrates a fast off-rate (∼1.4 s−1). Taken together, our results support a hypothesis in which myosin XIa transiently interacts with its binding constituents to coordinate vesicle motion and actin dynamics in a flexible manner.
To better understand how myosin XIa and VAMP72-labeled vesicles interact with the actin cytoskeleton at the tip of the caulonema cell during polarized growth, we first performed FRAP. We found that myosin XIa and VAMP72-labeled vesicles exhibited actin-dependent fluorescence recovery. During fluorescence recovery, we found that both myosin XIa and VAMP72-labeled vesicles restored apical localization at the cell tip in <2 s. Considering that we used a bleach radius of 2 μm and that the myosin XIa and VAMP72-labeled vesicles exhibit transport velocities of ∼2 μm/s, these recovery rates are consistent with active transport. This observation is similar to those described for pollen tubes (Bove et al., 2008), where the FRAP of vesicles shows a similar pattern. However, we did find that myosin XIa was able to achieve apical localization before that of VAMP72-labeled vesicles, indicating that myosin XIa displays an additional tip-localized recovery mode that is different to that of simple diffusion or VAMP72 vesicle association. This recovery mode might be the result of unloaded myosin XIa moving onto actin or of myosin XIa exchanging-off from bleached cargo at the tip, identifying the mechanism for this additional recovery is left for future work. A similar type of apical localization has been seen for myosin XIK in root hair of A. thaliana (Park and Nebenführ, 2013; Peremyslov et al., 2012), suggesting a conservation of function between species. However, additional research is required because an association of myosin XIK with VAMP72 positive vesicles has not been investigated in root hairs.
To demonstrate that myosin XIa associates with vesicles, we performed VAEM on protoplasts expressing fluorescently labeled myosin XIa and VAMP72-labeled vesicles. We found that myosin XIa colocalizes with VAMP72-labeled vesicles over periods of 0.5–2 s, which are consistent with an off-rate of 1.4 s−1. This off-rate is an upper limit estimate because we cannot be certain when each association occurred during our imaging time. Nevertheless, an off-rate like this suggests that myosin XIa and VAMP72-labeled vesicles do not reside together much longer than a few seconds. This is also supported by our FRAP analysis, where 55–66% of myosin is associated with vesicles. In contrast, by using single particle tracking, we found that only a small percentage of the tracked myosin XIa (4.3%) associates with VAMP72-labeled vesicles. This discrepancy could be because FRAP measures the average behavior of molecules, resulting in an apparent reduction on myosin XI diffusion. This, in turn, might be due to several factors, such as association with other vesicles and organelles, while tracking is done at the single-particle level at the cortex, and the use of VAEM that has a low signal-to-noise ratio and the potential to detect noise as particles. The important conclusion is that a substantial fraction of myosin XIa molecules remain free. Hence, our measurements suggest that, in vivo, a significant population of myosin XIa and vesicles exhibit brief transient interactions. The transient nature of the association of myosin XI to vesicles in the moss P. patens might be different to that necessary to transport larger organelles. In vascular plants, fast cytoplasmic streaming is driven by myosin XI (Shimmen and Yokota, 2004) but this type of streaming is not present in moss cells (Furt et al., 2012). The type of myosin XI-organelle association required for cytoplasmic streaming is still under investigation (Kurth et al., 2017; Madison et al., 2015; Park and Nebenführ, 2013; Peremyslov et al., 2013; Ueda et al., 2010) but we hypothesize that efficient vesicle transport and delivery also require a type of transient interaction in vascular plants.
We also observed several instances in which fluorescently labeled myosin XIa and VAMP72-labeled vesicles associated and moved along Lifeact-mCherry-decorated actin filaments. By using HMM on particle tracking data, we were able to characterize active motion. In protoplasts, we found that both myosin XIa and VAMP72-labeled vesicles actively moved not much more than a few microns over a period of <1 s. We did not successfully apply HMM to caulonemata as the tracking fidelity was reduced due to a reduced imaging area. Nevertheless, our tracking clearly shows that active trajectories were abolished in caulonema treated with latrunculin B, indicating that these active trajectories are actin dependent.
To better understand how the interaction between myosin XIa and VAMP72-labeled vesicles influences their active motion, we combined our colocalization and HMM analyses. We found that active motion of myosin XIa and VAMP72-labeled vesicles happens with or without concomitant colocalization. This indicates that myosin XIa either transports cargo other than VAMP72-labeled vesicles or that myosin XIa can move without any cargo onto filamentous actin. Results of our FRAP analysis of myosin XIa dynamics in growing cells is consistent with myosin without cargo being associated with F-actin at the tip. Unloaded myosin XIa on F-actin could be important for filament organization and maintenance. VAMP72-labeled vesicles might also move through the formin-mediated actin propulsion system, as previously suggested (Furt et al., 2013; Schuh, 2011), which is consistent with previously observed motility of class II formin (van Gisbergen et al., 2012). However, it is also possible that VAMP72-labeled vesicles are being transported by different myosins, such as myosin XIb (Vidali et al., 2010) or myosin VIII (Wu and Bezanilla, 2014). We do not expect any microtubule dependent motion of VAMP72 because latrunculin abolished active motion in caulonemata.
On the basis of the particle tracking evidence and the bulk FRAP measurements, we conclude that myosin XIa and VAMP72-labeled vesicles transiently interact with each other and exhibit short run lengths on actin filaments. Since P. patens does not exhibit large organelle cytoplasmic streaming (Furt et al., 2012), it is not surprising that myosin XIa only displays short-lived transport of vesicles. Additionally, the transport speeds of myosin XI (∼2.7 μm/s) found here are slightly less relative to other orthologs (Tominaga et al., 2013) and might be a result of decreased polarized growth observed in moss. We suggest this possibility because A. thaliana plants engineered to have decreased cytoplasmic streaming have smaller cells, whereas fast-streaming plants have larger cells (Tominaga et al., 2013). Nevertheless, the transport speed of vesicles is not a limiting factor because vesicles are present at high concentration at the apex of some tip-growing cells (Bove et al., 2008; de Win et al., 1999; Lee et al., 2008; Szumlanski and Nielsen, 2009). It is also possible that other mechanisms, such as vesicle retention and focusing of vesicles at a particular location are what is limiting (Bibeau et al., 2018). Future research into the intracellular transport of protonemata, pollen tubes and root hairs will help determine which pathways are unique or conserved (Orr et al., 2019).
Vascular plants might require additional levels to control organelle and vesicle transport in order to fulfill more-complex functions, which may have resulted in myosin XIs with a variety of activities (Haraguchi et al., 2018). It will be valuable to investigate how moss myosin XI compares with these molecules. Importantly, the weak interactions and short run lengths we observed here are consistent with a mechanism by which myosin XIa actively remodels and regulates oscillating apical F-actin during polarized growth, while still promoting vesicle focusing for exocytosis (see Fig. 7). Without the flexibility of these weak transient interactions, the apical actin spot may not be as dynamic or easily regulated. Future modeling and simulation strategies will help to evaluate some of these possibilities, in particular regarding the participation of myosin. The results provided here provide valuable quantitative information for this type of strategy, which can be complemented by mutations and in vivo studies. Furthermore, a quantitative exploration of these binding interactions will help to shape future modeling studies that incorporate the cytoskeleton in polarized cell growth.
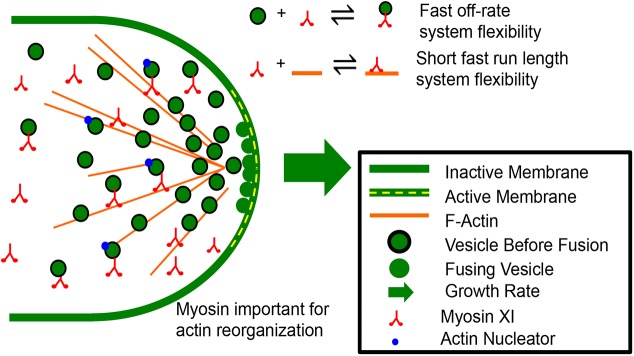
Fig. 7.
Transient interactions between myosin XIa, VAMP72-labeled vesicles and F-actin give rise to system flexibility in polarized cell growth of P. patens caulonemal cells. Here, myosin XIa and vesicles transiently interact with each other (fast off-rate) and the actin cytoskeleton (short fast run length=fast velocity and short association time), giving rise to short-lived persistent motion that is directed towards the secretion zone at the cell apex. These transient interactions allow for the dynamic actin oscillations observed during tip growth, while providing adequate vesicle focusing to sustain expansion of the cell wall.
MATERIALS AND METHODS
Constructs and cell lines
Two independent 3mEGFP-VAMP72 cell lines were obtained by transforming the Gransden strain of the spreading earthmoss P. patens with pTH-Ubi-3mEGFP-VAMP72, and by selecting for stable lines. The construct was obtained by a two-element LR Gateway reaction of entry clones pENT-L1-3mEGFP-R5 (three copies of the monomeric, enhanced, green fluorescent protein) and pENT-L5-VAMP72-L2 (coding sequence of VAMP72 from P. patens) (Furt et al., 2013), and the destination vector pTH-Ubi-Gateway (vector expressing hygromycin resistance for stable transformation in P. patens and driving constitutive expression of a gene of interest by the maize ubiquitin promoter) (Vidali et al., 2007). The 3mCherry-VAMP72 plus 3mEGFP-MyosinXIa and the Lifeact-mCherry plus 3mEGFP-MyosinXIa lines have been previously described (Furt et al., 2013).
Culture conditions
Physcomitrella cell lines were propagated using standard methods as previously described (Vidali et al., 2007). Isolation of protoplasts and DNA transformation were done according to Liu and Vidali (2011).
For VAEM imaging (Konopka and Bednarek, 2008), moss cell lines were grown on PpNO3 medium (Caisson Labs) (Vidali et al., 2007) for 7 days. One-week-old protonemal tissue was either directly mounted on glass microscope slides for imaging or used to prepare protoplasts, as previously described (Liu and Vidali, 2011) with some modifications. After counting, protoplasts were resuspended in 5 ml of 8% (w/v) mannitol and cultured in the growth chamber for 1 day. For imaging, moss protonemal tissue or protoplasts were deposited on a 1% (w/v) agar pad in PpNO3 medium on a glass microscope slide. 10 μl of liquid PpNO3 medium was applied before covering it with a glass coverslip (0.25 mm thick) and sealing it with Vaseline, lanolin, paraffin (1:1:1; VALAP). For treatment with latrunculin B, 10 μl latrunculin B (25 μM) resuspended in PpNO3 medium was added for 10 min; addition of PpNO3 medium alone was used as control. The concentration of latrunculin B was selected based on previous evidence that at least 10 μM of this drug is required to completely depolymerize actin and inhibit moss growth (Vidali et al., 2009), and 25 μM had been used recently to consistently depolymerize all the actin in caulonemal cells (Wu and Bezanilla, 2018). We verified, by using confocal laser scanning microscopy, that treatment with 25 μM latrunculin B completely depolymerizes all detectable apical and cortical F-actin within 10 min (Fig. S4). Apical caulonemal cells were imaged exactly 10 min after the treatment. Measurements on each cell were considered biological replicates because only one cell was analyzed from each prepared slide. Each preparation was repeated at least three times. For FRAP, moss cell lines were grown and cultured as described in Bibeau et al. (2018).
VAEM microscopy
Protoplasts were mounted on an inverted microscope (model Ti-E; Nikon Instruments) and imaged with an Apo TIRF, 60×, NA 1.49, oil immersion objective (Nikon Instruments). Apica caulonemal cells from protonemal tissue were imaged with an Apo TIRF, 100×, NA 1.49, oil immersion objective (Nikon Instruments). To increase magnification, the 1.5× optovar was used to collect all images. The 488 and 561 nm laser lines were used to excite mEGFP and mCherry, respectively. To provide the maximum signal-to-noise ratio, the laser illumination angle was finely tuned for each sample. Signals were simultaneously acquired for both channels using a beam splitter and a 512×512 electron-multiplying CCD camera iXON3 (Andor Technology).
Image processing and tracking
Images collected by VAEM, were first processed in ImageJ by using a background subtraction (rolling ball radius was tuned to 5 pixels for myosin XI and VAMP72), a FFT bandpass filter (a diameter comprising between 2 and 20 pixels was used) and an enhance contrast tool (enhancement without pixel saturation was normalized to all slices of each time series). Detection and tracking of the cortical punctate structures of myosin XIa and VAMP72 were performed using the TrackMate plugin in FIJI. We manually tracked several punctate structures to determine the best settings. Briefly, the DoG Detector tool was chosen because it is optimal for small spot sizes and it allows to differentiate between two spots that are close to each other. The diameter of the structures was ≤0.6 μm and the threshold was kept at 1000 for all molecules analyzed. For tracking, we used the simple LAP tracker tool with the following parameters: linking maximum distance of 0.5 μm; gap-closing maximum distance of 0.7 μm; gap-closing maximum frame gap of 0. Two other filters were applied to track the punctate structures that move in a linear persistent manner: spots per track >12 (equivalent to 600 ms) and a displacement >1.2 μm. Each trajectory recorded was verified manually using an in-lab developed Matlab routine (available upon request). To track the punctate structures that stay confined to an area, we adjusted the filters as follows: number of spots per track >90 (equivalent to 4.5 s) and displacement ≤0.5 μm. The number of trajectories was normalized to the average imaged area (40 μm2) for each cell. The instantaneous velocities of each tracked punctate structure were calculated using the coordinates obtained by the simple LAP tracker tool.
Colocalization frequency
To automate identification of colocalizing fluorescently labeled myosin XIa and VAMP72-labeled vesicles we used an approach similar to that described by Deschout et al. (2013). With a custom Matlab (MathWorks, Natick, MA) routine (available upon request), we compared the trajectories of myosin XIa and VAMP72-. An example analysis can be found in Fig. S5. For two trajectories to be considered colocalized they first needed to have a temporal overlap and be within a specified contact radius of 0.5 μm. Then a sliding correlation window was used to determine the correlations between the two channels in the x and y directions. If the correlation within the window was >0.7, the movement was classified as correlated for that direction. Window correlations were found using the Matlab function ‘corr’. If the trajectories exhibited correlated motion in both the x and y directions the time point within the selected window was classified as colocalized. Here we used a minimum correlation threshold of 0.7 and a maximum contact radius of 0.5 μm to account for tracking and alignment error.
To demonstrate that the observed colocalized trajectories were not a result of random Brownian motion, we randomly shuffled the existing trajectories in time and space with a custom written Matlab script (available upon request). The new trajectories were then analyzed for colocalization, as previously mentioned.
Hidden Markov model
To facilitate the use of a Hidden Markov model (HMM), trajectories were preprocessed such that they were converted from spatial coordinates into either reverse ‘R’ or forward ‘F’ moves (Roding et al., 2014). Specifically, we measured the angle between three consecutive points along the trajectory, namely, ti−1, ti and ti+1. This angle θi was then classified as a forward move when the angle was greater than a specific threshould or was otherwise classified as a reverse. Trajectories were also filtered by size such that, trajectories shorter than seven frames or longer than 150 frames were not used. We selected trajectories longer than seven frames to reduce tracking error, and trajectories shorter than 150 frames because longer trajectories correspond to a different population of particles that remain static during observation.
To classify the transport state of a particle for a given point on these modified trajectories, we employed a HMM (Roding et al., 2014) (Fig. S3) with two hidden states, namely, an active state ‘A’ and a Brownian state ‘B’ (available upon request). Essentially, the HMM determines the number of consecutive forward moves. Here, we represent each hidden sequence as H=h1,h2,…,hT and each observed sequence as Q=q1,q2,…,qT. In the active state ‘A’ we assumed that myosin XIa and VAMP72-labeled vesicles always move forward, whereas in the Brownian state ‘B’ an equal probability distribution over all angles ‘θ’ was assumed. Thus, the emission probabilities in the Brownian state are only a function of the angle threshold, where the probability of moving forward given the Brownian state and the probability of moving in the reverse direction given the Brownian state are P(qt=F|ht=B)=(180-130)/(180) and P(qt=R|ht=B)=1-P(qt=F|ht=B), respectively. This leaves the state-transition probabilities as the only open parameters in our model. The state-transition probabilities are the chances that the protein of interest will change its state on each successive portion of the sequence. The four possible state transitions are (i) active to active, (ii) active to Brownian, (iii) Brownian to Brownian and (iv) Brownian to active, which can be written formally as P(ht=A|ht−1=A), P(ht=A|ht−1=B), P(ht=B|ht−1=B) and P(ht=B|ht−1=A), respectively.
To find the most likely state transitions for each experimental condition, we used the forward and reverse trajectories as inputs to a modified version of the Matlab function hmmtrain for known emission matrices. hmmtrain uses the Baum–Welch algorithm (Rabiner, 1989), which recursively finds the transition and emission matrices that maximize the likelihood of the observed sequence. We modified this algorithm, such that the emission matrix did not change. This modified function reliably found the transition matrix that maximized the likelihood of the observed sequence Q, regardless of the initial guess for the transition matrix (Fig. S6A,B). With the most likely state transitions and the converted sequences, we then used the Matlab function hmmviterbi to determine the most likely state of a specific particle at a specific time.
Here, an angle threshold of 130° was chosen because it allowed for the detection of shorter active trajectories while minimally effecting the most likely transition probabilities (Fig. S6C,D). Larger angles placed a too stringent requirement on forward moves and began to influence the transition matrix (Fig. S6C,D).
Experimental FRAP processing
Experimental FRAP was processed as previously described by Bibeau et al. (2018). Briefly, FRAP confocal images were saved as tiff stacks and converted into intensity traces by averaging the mean fluorescence intensity within the 4 μm-wide ROI by using a custom written ImageJ macro. ROIs were selected manually; the tip ROI was at the most apical area of the cell, while the shank ROI was at ∼15 μm from the tip. To compare between experiments, both ROI signals were normalized by the shank signal. All replicate experiments were then averaged and further normalized by acquisition photobleaching controls with Matlab. To quantify the spatial fluorescence recovery FRAP tiff stacks had the respective ROIs cropped and averaged using a custom written ImageJ macro. Then the fluorescence intensities of the perimeters of the cropped ROIs were extracted, limited-volume corrected and plotted in Matlab.
Estimating the myosin XI diffusion coefficient
To estimate the myosin XI diffusion coefficient, we used the sedimentation coefficients of myosin V (Krementsov et al., 2004; Li et al., 2004; Wang et al., 2004). Sedimentation coefficients are used to quantify how quickly a protein of interests moves through gradients during ultracentrifugation. By using force balance analysis, it is possible to extract the hydrodynamic radius of a protein on the basis of its sedimentation coefficient (Erickson, 2009), i.e.:
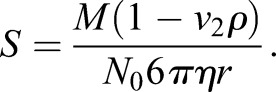 |
(4) |
There, S is the sedimentation coefficient, which is represented in 10−13 s, M is the molecular weight of the protein of interest, v2 is the inverse density of the protein, ρ is the density of the solvent, N0 is Avogadro's number, η is the viscosity of the solvent and r is the hydrodynamic radius of the protein. Based on Ca2+ concentrations, it has been shown that myosin V can either be in an open or closed configuration and its sedimentation coefficients were found to be 10.7S or 14S, respectively (Wang et al., 2004),values that are in agreement between laboratories (Krementsov et al., 2004; Li et al., 2004). On the basis of Eqn 4 and the parameters measured and described by Wang and colleagues, the corresponding hydrodynamic radii are 13.4 and 10.2 nm for the open and closed configurations, respectively. Application of the Stokes–Einstein equation for the hydrodynamic radii of the two configurations for myosin V to myosin XI in moss yields theoretical diffusion coefficients of 2.3 and 3.4 μm2/s for the open and closed configurations, respectively.
Modeling reaction diffusion FRAP
To solve the reaction diffusion equations Eqns 1 and 2 in COMSOL (Comsol Inc, Stockholm, Sweden), we used no-flux boundary conditions inside a two-dimensional approximation of the moss geometry that we have previously shown to be valid for our bleaching conditions (Bibeau et al., 2018; Kingsley et al., 2018). We also used initial conditions, in which U(x,t,0)=total/(1+Kd*) and B(x,y,0)=total-U(x,y,0) for everywhere outside the 4 μm-wide circular ROI (R). Here, the effective equilibrium constant is the quotient between the effective on-rate and off-rate Kd*=kon*/koff, and total is the arbitrary combined concentration of the unbound and bound myosin XIa. This concentration is arbitrary because we observe experimentally the normalized fluorescence intensity within the ROI (R), i.e.:
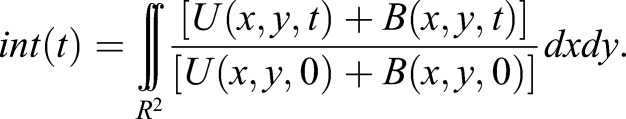 |
(5) |
Inside the bleach region the initial concentrations for U(x,y,0) and B(x,y,0) were multiplied by the bleach depth parameter ‘BD’ to simulate an instantaneous ideal bleach.
To solve these equations for arbitrary kon* and koff we used the finite element modeling software Comsol Multiphysics (Comsol Inc, Stockholm, Sweden). Simulations were created by using the ‘two-dimensional transport of dilute species’ interface with two species, a ‘time-dependent solution’ and a ‘normal physics controlled mesh’. We then performed a parameter sweep across potential values of kon* and koff by using the Matlab liveLink Comsol Multiphysics module, and calculated the sum of squares differences between the simulated recoveries and those measured experimentally. Based on these results, we concluded that we could only determine the effective dissociation constant Kd*=kon*:koff. To converge on a best-fit Kd* we imposed gradient descent on the following cost function,
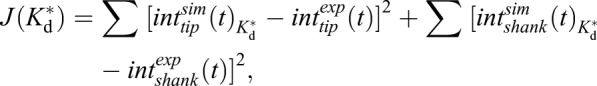 |
(6) |
by using a custom-written script for the Matlab liveLink Comsol Multiphysics module (available upon request). Here, superscript ‘sim’ and ‘exp’ indicate simulated and experimental FRAP recoveries, subscript ‘tip’ and ‘shank’ indicate the position of the photo-bleach, and J(Kd*) is the dissociation constant dependent cost function. We used the numerical forward approximation of a derivative to determine the local gradients during convergence.
Supplementary Material
Acknowledgements
We thank all members of the L.V. and E.T. labs for helpful discussions, and thank Prof. Arne Gericke (Worcester Polytechnic Institute, MA) for allowing us to use his TIRF microscope.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.P.B., F.F., L.V.; Methodology: J.P.B., F.F., J.L.K., L.V.; Software: J.P.B., S.I.M., J.L.K., M.F.L., E.T.; Validation: J.L.K., E.T., L.V.; Formal analysis: J.P.B., F.F., J.L.K., M.F.L., E.T.; Investigation: J.P.B., F.F., L.V.; Resources: E.T., L.V.; Data curation: J.P.B., F.F.; Writing - original draft: J.P.B., F.F.; Writing - review & editing: F.F., E.T., L.V.; Supervision: E.T., L.V.; Project administration: L.V.; Funding acquisition: L.V.
Funding
This work was supported by National Science Foundation (NSF) CBET 1309933 to E.T., and NSF-MCB 1253444 to L.V., and by the National Institutes of Health (NIH) R01GM121679 to E.T. E.T. and L.V. also acknowledge support from Worcester Polytechnic Institute Startup Funds. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.234682.supplemental
References
- Avisar D., Prokhnevsky A. I., Makarova K. S., Koonin E. V. and Dolja V. V. (2008). Myosin XI-K Is required for rapid trafficking of Golgi stacks, peroxisomes, and mitochondria in leaf cells of Nicotiana benthamiana. Plant Physiol. 146, 1098-1108. 10.1104/pp.107.113647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avisar D., Abu-Abied M., Belausov E., Sadot E., Hawes C. and Sparkes I. A. (2009). A comparative study of the involvement of 17 Arabidopsis myosin family members on the motility of Golgi and other organelles. Plant Physiol. 150, 700-709. 10.1104/pp.109.136853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avisar D., Abu-Abied M., Belausov E. and Sadot E. (2012). Myosin XIK is a major player in cytoplasm dynamics and is regulated by two amino acids in its tail. J. Exp. Bot. 63, 241-249. 10.1093/jxb/err265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibeau J. P., Kingsley J. L., Furt F., Tüzel E. and Vidali L. (2018). F-Actin mediated focusing of vesicles at the cell tip is essential for polarized growth. Plant Physiol. 176, 352-363. 10.1104/pp.17.00753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bove J., Vaillancourt B., Kroeger J., Hepler P. K., Wiseman P. W. and Geitmann A. (2008). Magnitude and direction of vesicle dynamics in growing pollen tubes using spatiotemporal image correlation spectroscopy and fluorescence recovery after photobleaching. Plant Physiol. 147, 1646-1658. 10.1104/pp.108.120212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campàs O. and Mahadevan L. (2009). Shape and dynamics of tip-growing cells. Curr. Biol. 19, 2102-2107. 10.1016/j.cub.2009.10.075 [DOI] [PubMed] [Google Scholar]
- Cole R. A. and Fowler J. E. (2006). Polarized growth: maintaining focus on the tip. Curr. Opin. Plant Biol. 9, 579-588. 10.1016/j.pbi.2006.09.014 [DOI] [PubMed] [Google Scholar]
- de Graaf B. H. J., Cheung A. Y., Andreyeva T., Levasseur K., Kieliszewski M. and Wu H.-M. (2005). Rab11 GTPase-regulated membrane trafficking is crucial for tip-focused pollen tube growth in tobacco. Plant Cell 17, 2564-2579. 10.1105/tpc.105.033183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschout H., Martens T., Vercauteren D., Remaut K., Demeester J., De Smedt S. C., Neyts K. and Braeckmans K. (2013). Correlation of dual colour single particle trajectories for improved detection and analysis of interactions in living cells. Int. J. Mol. Sci. 14, 16485-16514. 10.3390/ijms140816485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Win A. H. N., Pierson E. S. and Derksen J. (1999). Rational analyses of organelle trajectories in tobacco pollen tubes reveal characteristics of the actomyosin cytoskeleton. Biophys. J. 76, 1648-1658. 10.1016/S0006-3495(99)77324-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumais J., Shaw S. L., Steele C. R., Long S. R. and Ray P. M. (2006). An anisotropic-viscoplastic model of plant cell morphogenesis by tip growth. Int. J. Dev. Biol. 50, 209-222. 10.1387/ijdb.052066jd [DOI] [PubMed] [Google Scholar]
- Erickson H. P. (2009). Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol. Proced. Online 11, 32-51. 10.1007/s12575-009-9008-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G.-H., Lapierre L. A., Goldenring J. R., Sai J. and Richmond A. (2004). Rab11-family interacting protein 2 and myosin Vb are required for CXCR2 recycling and receptor-mediated chemotaxis. Mol. Biol. Cell 15, 2456-2469. 10.1091/mbc.e03-09-0706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayant P., Girlanda O., Chebli Y., Aubin C.-E., Villemure I. and Geitmann A. (2010). Finite element model of polar growth in pollen tubes. Plant Cell 22, 2579-2593. 10.1105/tpc.110.075754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furt F., Lemoi K., Tüzel E. and Vidali L. (2012). Quantitative analysis of organelle distribution and dynamics in Physcomitrella patens protonemal cells. BMC Plant Biol. 12, 70 10.1186/1471-2229-12-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furt F., Liu Y.-C., Bibeau J. P., Tüzel E. and Vidali L. (2013). Apical myosin XI anticipates F-actin during polarized growth of Physcomitrella patens cells. Plant J. 73, 417-428. 10.1111/tpj.12039 [DOI] [PubMed] [Google Scholar]
- Geitmann A. and Nebenführ A. (2015). Navigating the plant cell: intracellular transport logistics in the green kingdom. Mol. Biol. Cell 26, 3373-3378. 10.1091/mbc.E14-10-1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geitmann A. and Ortega J. K. (2009). Mechanics and modeling of plant cell growth. Trends Plant Sci. 14, 467-478. 10.1016/j.tplants.2009.07.006 [DOI] [PubMed] [Google Scholar]
- Gilroy S. and Jones D. L. (2000). Through form to function: root hair development and nutrient uptake. Trends Plant Sci. 5, 56-60. 10.1016/S1360-1385(99)01551-4 [DOI] [PubMed] [Google Scholar]
- Grebnev G., Ntefidou M. and Kost B. (2017). Secretion and endocytosis in pollen tubes: models of tip growth in the spot light. Front. Plant Sci. 8, 154 10.3389/fpls.2017.00154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffing L. R., Gao H. T. and Sparkes I. (2014). ER network dynamics are differentially controlled by myosins XI-K, XI-C, XI-E, XI-I, XI-1, and XI-2. Front. Plant Sci. 5, 218 10.3389/fpls.2014.00218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer J. A. and Sellers J. R. (2012). Walking to work: roles for class V myosins as cargo transporters. Nat. Rev. Mol. Cell Biol. 13, 13-26. 10.1038/nrm3248 [DOI] [PubMed] [Google Scholar]
- Haraguchi T., Ito K., Duan Z., Rula S., Takahashi K., Shibuya Y., Hagino N., Miyatake Y., Nakano A. and Tominaga M. (2018). Functional diversity of class XI myosins in arabidopsis thaliana. Plant Cell Physiol. 59, 2268-2277. 10.1093/pcp/pcy147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman D. S., Geiser D. M., Eidell B. R., Stauffer R. L., Kardos N. L. and Hedges S. B. (2001). Molecular evidence for the early colonization of land by fungi and plants. Science 293, 1129-1133. 10.1126/science.1061457 [DOI] [PubMed] [Google Scholar]
- Henn A. and Sadot E. (2014). The unique enzymatic and mechanistic properties of plant myosins. Curr. Opin. Plant Biol. 22, 65-70. 10.1016/j.pbi.2014.09.006 [DOI] [PubMed] [Google Scholar]
- Hepler P. K., Vidali L. and Cheung A. Y. (2001). Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 17, 159-187. 10.1146/annurev.cellbio.17.1.159 [DOI] [PubMed] [Google Scholar]
- Kang M. and Kenworthy A. K. (2008). A closed-form analytic expression for FRAP formula for the binding diffusion model. Biophys. J. 95, L13-L15. 10.1529/biophysj.108.135913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M. C., Day C. A., DiBenedetto E. and Kenworthy A. K. (2010). A quantitative approach to analyze binding diffusion kinetics by confocal FRAP. Biophys. J. 99, 2737-2747. 10.1016/j.bpj.2010.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenrick P. and Crane P. R. (1997). The origin and early evolution of plants on land. Nature 389, 33-39. 10.1038/37918 [DOI] [Google Scholar]
- Ketelaar T., Galway M. E., Mulder B. M. and Emons A. M. C. (2008). Rates of exocytosis and endocytosis in Arabidopsis root hairs and pollen tubes. J. Microsc. 231, 265-273. 10.1111/j.1365-2818.2008.02031.x [DOI] [PubMed] [Google Scholar]
- Kingsley J. L., Bibeau J. P., Mousavi S. I., Unsal C., Chen Z., Huang X., Vidali L. and Tüzel E. (2018). Characterization of cell boundary and confocal effects improves quantitative FRAP analysis. Biophys. J. 114, 1153-1164. 10.1016/j.bpj.2018.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka C. A. and Bednarek S. Y. (2008). Variable-angle epifluorescence microscopy: a new way to look at protein dynamics in the plant cell cortex. Plant J. 53, 186-196. 10.1111/j.1365-313X.2007.03306.x [DOI] [PubMed] [Google Scholar]
- Krementsov D. N., Krementsova E. B. and Trybus K. M. (2004). Myosin V: regulation by calcium, calmodulin, and the tail domain. J. Cell Biol. 164, 877-886. 10.1083/jcb.200310065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger J. H., Daher F. B., Grant M. and Geitmann A. (2009). Microfilament orientation constrains vesicle flow and spatial distribution in growing pollen tubes. Biophys. J. 97, 1822-1831. 10.1016/j.bpj.2009.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger J. H., Zerzour R. and Geitmann A. (2011). Regulator or driving force? the role of turgor pressure in oscillatory plant cell growth. PLoS ONE 6, e18549 10.1371/journal.pone.0018549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth E. G., Peremyslov V. V., Turner H. L., Makarova K. S., Iranzo J., Mekhedov S. L., Koonin E. V. and Dolja V. V. (2017). Myosin-driven transport network in plants. Proc. Natl. Acad. Sci. USA 114, E1385-E1394. 10.1073/pnas.1620577114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre L. A., Kumar R., Hales C. M., Navarre J., Bhartur S. G., Burnette J. O., Provance D. W., Mercer J. A., Bähler M. and Goldenring J. R. (2001). Myosin Vb is associated with plasma membrane recycling systems. Mol. Biol. Cell 12, 1843-1857. 10.1091/mbc.12.6.1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. J., Szumlanski A., Nielsen E. and Yang Z. B. (2008). Rho-GTPase-dependent filamentous actin dynamics coordinate vesicle targeting and exocytosis during tip growth. J. Cell Biol. 181, 1155-1168. 10.1083/jcb.200801086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.-F. and Nebenführ A. (2008). The tail that wags the dog: the globular tail domain defines the function of myosin V/XI. Traffic 9, 290-298. 10.1111/j.1600-0854.2007.00687.x [DOI] [PubMed] [Google Scholar]
- Li X.-D., Mabuchi K., Ikebe R. and Ikebe M. (2004). Ca2+-induced activation of ATPase activity of myosin Va is accompanied with a large conformational change. Biochem. Biophys. Res. Commun. 315, 538-545. 10.1016/j.bbrc.2004.01.084 [DOI] [PubMed] [Google Scholar]
- Lise M.-F., Wong T. P., Trinh A., Hines R. M., Liu L. D., Kang R. J., Hines D. J., Lu J., Goldenring J. R., Wang Y. T. et al. (2006). Involvement of myosin Vb in glutamate receptor trafficking. J. Biol. Chem. 281, 3669-3678. 10.1074/jbc.M511725200 [DOI] [PubMed] [Google Scholar]
- Liu Y. C. and Vidali L. (2011). Efficient polyethylene glycol (PEG) mediated transformation of the moss Physcomitrella patens. J. Vis. Exp. 50, e2560 10.3791/2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorén N., Hagman J., Jonasson J. K., Deschout H., Bernin D., Cella-Zanacchi F., Diaspro A., McNally J. G., Ameloot M., Smisdom N. et al. (2015). Fluorescence recovery after photobleaching in material and life sciences: putting theory into practice. Q. Rev. Biophys. 48, 323-387. 10.1017/S0033583515000013 [DOI] [PubMed] [Google Scholar]
- Luo N., Yan A., Liu G., Guo J., Rong D., Kanaoka M. M., Xiao Z., Xu G., Higashiyama T., Cui X. et al. (2017). Exocytosis-coordinated mechanisms for tip growth underlie pollen tube growth guidance. Nat. Commun. 8, 1687 10.1038/s41467-017-01452-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison S. L. and Nebenführ A. (2013). Understanding myosin functions in plants: are we there yet? Curr. Opin. Plant Biol. 16, 710-717. 10.1016/j.pbi.2013.10.004 [DOI] [PubMed] [Google Scholar]
- Madison S. L., Buchanan M. L., Glass J. D., McClain T. F., Park E. and Nebenführ A. (2015). Class XI myosins move specific organelles in pollen tubes and are required for normal fertility and pollen tube growth in Arabidopsis. Plant Physiol. 169, 1946-1960. 10.1104/pp.15.01161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally J. G. (2008). Quantitative FRAP in analysis of molecular binding dynamics in vivo. Methods Cell Biol. 85, 329-351. 10.1016/S0091-679X(08)85014-5 [DOI] [PubMed] [Google Scholar]
- Menand B., Calder G. and Dolan L. (2007). Both chloronemal and caulonemal cells expand by tip growth in the moss Physcomitrella patens. J. Exp. Bot. 58, 1843-1849. 10.1093/jxb/erm047 [DOI] [PubMed] [Google Scholar]
- Nedvetsky P. I., Stefan E., Frische S., Santamaria K., Wiesner B., Valenti G., Hammer J. A., Nielsen S., Goldenring J. R., Rosenthal W. et al. (2007). A role of myosin Vb and Rab11-FIP2 in the aquaporin-2 shuttle. Traffic 8, 110-123. 10.1111/j.1600-0854.2006.00508.x [DOI] [PubMed] [Google Scholar]
- Ohyama A., Komiya Y. and Igarashi M. (2001). Globular tail of myosin-V is bound to VAMP/synaptobrevin. Biochem. Biophys. Res. Commun. 280, 988-991. 10.1006/bbrc.2001.4236 [DOI] [PubMed] [Google Scholar]
- Ojangu E. L., Järve K., Paves H. and Truve E. (2007). Arabidopsis thaliana myosin XIK is involved in root hair as well as trichome morphogenesis on stems and leaves. Protoplasma 230, 193-202. 10.1007/s00709-006-0233-8 [DOI] [PubMed] [Google Scholar]
- Orr R. G., Cheng X., Vidali L. and Bezanilla M. (2019). Orchestrating cell morphology from the inside out - using polarized cell expansion in plants as a model. Curr. Opin. Cell Biol. 62, 46-53. 10.1016/j.ceb.2019.08.004 [DOI] [PubMed] [Google Scholar]
- Ovečka M., Lang I., Baluška F., Ismail A., Illeš P. and Lichtscheidl I. K. (2005). Endocytosis and vesicle trafficking during tip growth of root hairs. Protoplasma 226, 39-54. 10.1007/s00709-005-0103-9 [DOI] [PubMed] [Google Scholar]
- Park E. and Nebenführ A. (2013). Myosin XIK of Arabidopsis thaliana accumulates at the root hair tip and is required for fast root hair growth. PLoS ONE 8, e76745 10.1371/journal.pone.0076745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton R. M., Fischer-Parton S., Watahiki M. K. and Trewavas A. J. (2001). Dynamics of the apical vesicle accumulation and the rate of growth are related in individual pollen tubes. J. Cell Sci. 114, 2685-2695. [DOI] [PubMed] [Google Scholar]
- Peremyslov V. V., Prokhnevsky A. I., Avisar D. and Dolja V. V. (2008). Two class XI myosins function in organelle trafficking and root hair development in Arabidopsis. Plant Physiol. 146, 1109-1116. 10.1104/pp.107.113654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov V. V., Prokhnevsky A. I. and Dolja V. V. (2010). Class XI myosins are required for development, cell expansion, and F-Actin organization in Arabidopsis. Plant Cell 22, 1883-1897. 10.1105/tpc.110.076315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov V. V., Klocko A. L., Fowler J. E. and Dolja V. V. (2012). Arabidopsis myosin XI-K localizes to the motile endomembrane vesicles associated with F-actin. Front. Plant Sci. 3, 184 10.3389/fpls.2012.00184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov V. V., Morgun E. A., Kurth E. G., Makarova K. S., Koonin E. V. and Dolja V. V. (2013). Identification of myosin XI receptors in Arabidopsis defines a distinct class of transport vesicles. Plant Cell 25, 3022-3038. 10.1105/tpc.113.113704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss M. L., Serna J., Falbel T. G., Bednarek S. Y. and Nielsen E. (2004). The Arabidopsis Rab GTPase RabA4b localizes to the tips of growing root hair cells. Plant Cell 16, 1589-1603. 10.1105/tpc.021634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhnevsky A. I., Peremyslov V. V. and Dolja V. V. (2008). Overlapping functions of the four class XI myosins in Arabidopsis growth, root hair elongation, and organelle motility. Proc. Natl. Acad. Sci. USA 105, 19744-19749. 10.1073/pnas.0810730105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D. W., Schott D. H. and Bretscher A. (1998). Tropomyosin-containing actin cables direct the Myo2p-dependent polarized delivery of secretory vesicles in budding yeast. J. Cell Biol. 143, 1931-1945. 10.1083/jcb.143.7.1931 [DOI] [PubMed] [Google Scholar]
- Rabiner L. R. (1989). A tutorial on hidden markov-models and selected applications in speech recognition. Proc. IEEE 77, 257-286. 10.1109/5.18626 [DOI] [Google Scholar]
- Riquelme M., Aguirre J., Bartnicki-Garcia S., Braus G. H., Feldbrugge M., Fleig U., Hansberg W., Herrera-Estrella A., Kamper J., Kuck U. et al. (2018). Fungal morphogenesis, from the polarized growth of hyphae to complex reproduction and infection structures. Microbiol. Mol. Biol. Rev. 82, e00068-17 10.1128/MMBR.00068-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roding M., Guo M., Weitz D. A., Rudemo M. and Särkkä A. (2014). Identifying directional persistence in intracellular particle motion using hidden markov models. Math. Biosci. 248, 140-145. 10.1016/j.mbs.2013.12.008 [DOI] [PubMed] [Google Scholar]
- Rodriguez O. C. and Cheney R. E. (2002). Human myosin-Vc is a novel class V myosin expressed in epithelial cells. J. Cell Sci. 115, 991-1004. [DOI] [PubMed] [Google Scholar]
- Rojas E. R., Hotton S. and Dumais J. (2011). Chemically mediated mechanical expansion of the pollen tube cell wall. Biophys. J. 101, 1844-1853. 10.1016/j.bpj.2011.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roland J. T., Kenworthy A. K., Peranen J., Caplan S. and Goldenring J. R. (2007). Myosin Vb interacts with Rab8a on a tubular network containing EHD1 and EHD3. Mol. Biol. Cell 18, 2828-2837. 10.1091/mbc.e07-02-0169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak K., Steiner A., Synek L., Klaeger S., Kulich I., Facher E., Wanner G., Kuster B., Zarsky V., Persson S. et al. (2014). Plant cytokinesis is orchestrated by the sequential action of the TRAPPII and exocyst tethering complexes. Dev. Cell 29, 607-620. 10.1016/j.devcel.2014.04.029 [DOI] [PubMed] [Google Scholar]
- Schott D., Ho J., Pruyne D. and Bretscher A. (1999). The COOH-terminal domain of Myo2p, a yeast myosin V, has a direct role in secretory vesicle targeting. J. Cell Biol. 147, 791-808. 10.1083/jcb.147.4.791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M. (2011). An actin-dependent mechanism for long-range vesicle transport. Nat. Cell Biol. 13, 1431-1436. 10.1038/ncb2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimmen T. and Yokota E. (2004). Cytoplasmic streaming in plants. Curr. Opin. Cell Biol. 16, 68-72. 10.1016/j.ceb.2003.11.009 [DOI] [PubMed] [Google Scholar]
- Sparkes I. A., Teanby N. A. and Hawes C. (2008). Truncated myosin XI tail fusions inhibit peroxisome, Golgi, and mitochondrial movement in tobacco leaf epidermal cells: a genetic tool for the next generation. J. Exp. Bot. 59, 2499-2512. 10.1093/jxb/ern114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague B. L. and McNally J. G. (2005). FRAP analysis of binding: proper and fitting. Trends Cell Biol. 15, 84-91. 10.1016/j.tcb.2004.12.001 [DOI] [PubMed] [Google Scholar]
- Steer M. W. (1988). Plasma membrane turnover in plant cells. J. Exp. Bot. 39, 987-996. 10.1093/jxb/39.8.987 [DOI] [Google Scholar]
- Szumlanski A. L. and Nielsen E. (2009). The Rab GTPase RabA4d regulates pollen tube tip growth in Arabidopsis thaliana. Plant Cell 21, 526-544. 10.1105/tpc.108.060277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski D. B. and Cosgrove D. J. (2009). Dynamic coordination of cytoskeletal and cell wall systems during plant cell morphogenesis. Curr. Biol. 19, R800-R811. 10.1016/j.cub.2009.07.056 [DOI] [PubMed] [Google Scholar]
- Tominaga M., Kimura A., Yokota E., Haraguchi T., Shimmen T., Yamamoto K., Nakano A. and Ito K. (2013). Cytoplasmic streaming velocity as a plant size determinant. Dev. Cell 27, 345-352. 10.1016/j.devcel.2013.10.005 [DOI] [PubMed] [Google Scholar]
- Ueda H., Yokota E., Kutsuna N., Shimada T., Tamura K., Shimmen T., Hasezawa S., Dolja V. V. and Hara-Nishimura I. (2010). Myosin-dependent endoplasmic reticulum motility and F-actin organization in plant cells. Proc. Natl. Acad. Sci. USA 107, 6894-6899. 10.1073/pnas.0911482107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T., Ueda T., Ohniwa R. L., Nakano A., Takeyasu K. and Sato M. H. (2004). Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 29, 49-65. 10.1247/csf.29.49 [DOI] [PubMed] [Google Scholar]
- van Gisbergen P. A., Li M., Wu S. Z. and Bezanilla M. (2012). Class II formin targeting to the cell cortex by binding PI(3,5)P(2) is essential for polarized growth. J. Cell Biol. 198, 235-250. 10.1083/jcb.201112085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vick J. K. and Nebenführ A. (2012). Putting on the breaks: regulating organelle movements in plant cells(f). J. Integr. Plant Biol. 54, 868-874. 10.1111/j.1744-7909.2012.01180.x [DOI] [PubMed] [Google Scholar]
- Vidali L., McKenna S. T. and Hepler P. K. (2001). Actin polymerization is essential for pollen tube growth. Mol. Biol. Cell 12, 2534-2545. 10.1091/mbc.12.8.2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., Augustine R. C., Kleinman K. P. and Bezanilla M. (2007). Profilin is essential for tip growth in the moss Physcomitrella patens. Plant Cell 19, 3705-3722. 10.1105/tpc.107.053413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., Rounds C. M., Hepler P. K. and Bezanilla M. (2009). Lifeact-mEGFP reveals a dynamic apical F-actin network in tip growing plant cells. PLoS ONE 4, e5744 10.1371/journal.pone.0005744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., Burkart G. M., Augustine R. C., Kerdavid E., Tüzel E. and Bezanilla M. (2010). Myosin XI is essential for tip growth in Physcomitrella patens. Plant Cell 22, 1868-1882. 10.1105/tpc.109.073288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpicelli L. A., Lah J. J., Fang G., Goldenring J. R. and Levey A. I. (2002). Rab11a and myosin Vb regulate recycling of the M4 muscarinic acetylcholine receptor. J. Neurosci. 22, 9776-9784. 10.1523/JNEUROSCI.22-22-09776.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Thirumurugan K., Stafford W. F., Hammer J. A. III, Knight P. J. and Sellers J. R. (2004). Regulated conformation of myosin V. J. Biol. Chem. 279, 2333-2336. 10.1074/jbc.C300488200 [DOI] [PubMed] [Google Scholar]
- Wu S.-Z. and Bezanilla M. (2014). Myosin VIII associates with microtubule ends and together with actin plays a role in guiding plant cell division. Elife 3, e03498 10.7554/eLife.03498.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.-Z. and Bezanilla M. (2018). Actin and microtubule cross talk mediates persistent polarized growth. J. Cell Biol. 217, 3531-3544. 10.1083/jcb.201802039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q., Sun W., Kujala P., Lotfi Y., Vida T. A. and Bean A. J. (2005). CART: An hrs/actinin-4/BERP/myosin V protein complex required for efficient receptor recycling. Mol. Biol. Cell 16, 2470-2482. 10.1091/mbc.e04-11-1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonia L. and Munnik T. (2008). Vesicle trafficking dynamics and visualization of zones of exocytosis and endocytosis in tobacco pollen tubes. J. Exp. Bot. 59, 861-873. 10.1093/jxb/ern007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.