Optimized protocols for protein production and purification as well as cryoEM grid preparation allow the reconstruction of a 3D map of yeast fatty acid synthase (FAS) at ∼3 Å resolution from a comparatively small number of particle images within a short time. The first complete model of yeast FAS is presented.
Keywords: purification of protein complexes, 3D reconstruction and image processing, single-particle cryoEM, cryo-electron microscopy, macromolecular machines, protein structure, yeast fatty acid synthase
Abstract
Single-particle electron cryo-microscopy (cryoEM) has undergone a ‘resolution revolution’ that makes it possible to characterize megadalton (MDa) complexes at atomic resolution without crystals. To fully exploit the new opportunities in molecular microscopy, new procedures for the cloning, expression and purification of macromolecular complexes need to be explored. Macromolecular assemblies are often unstable, and invasive construct design or inadequate purification conditions and sample-preparation methods can result in disassembly or denaturation. The structure of the 2.6 MDa yeast fatty acid synthase (FAS) has been studied by electron microscopy since the 1960s. Here, a new, streamlined protocol for the rapid production of purified yeast FAS for structure determination by high-resolution cryoEM is reported. Together with a companion protocol for preparing cryoEM specimens on a hydrophilized graphene layer, the new protocol yielded a 3.1 Å resolution map of yeast FAS from 15 000 automatically picked particles within a day. The high map quality enabled a complete atomic model of an intact fungal FAS to be built.
1. Introduction
Recent developments in single-particle cryoEM make it possible to determine the structures of large macromolecular complexes that are not available in sufficiently large amounts or that resist crystallization. In cryoEM, individual complexes are imaged in a thin layer of vitrified buffer (McDowall et al., 1983 ▸). With the recently developed direct electron detectors (McMullan et al., 2009 ▸) and image-processing packages (Cheng et al., 2015 ▸), cryoEM has become increasingly powerful, and it is now the method of choice for determining the structures of large macromolecular assemblies at high resolution. New image-processing algorithms can deal with sample heterogeneity, and analyzing this heterogeneity often provides direct insights into molecular mechanisms (Zivanov et al., 2018 ▸; Punjani et al., 2017 ▸; Grant et al., 2018 ▸; Murphy et al., 2019 ▸). It is no longer uncommon for cryoEM to achieve resolutions of 3 Å or better. To date, more than 200 cryoEM structures in this high resolution range have been deposited in the Electron Microscopy Data Bank (EMDB; http://emdatabank.org/). In the same way as X-ray structures, the new high-resolution cryoEM structures serve as a base for designing inhibitors or mutants and for analyzing biomolecular interfaces.
The yeast fatty acid synthase (FAS) was one of the first protein complexes to be analyzed in structural biology. Since the mid 1960s, dozens of studies have described the overall structure of the 2.6 MDa complex and its individual domains (Lynen et al., 1980 ▸; Maier et al., 2010 ▸). Although today the mechanism of modular fatty-acid synthesis is well understood, FAS remains an important target for structural and functional studies. Yeast FAS is a prime example for revealing co-translational subunit association as a mechanism in the assembly of eukaryotic proteins (Shiber et al., 2018 ▸; Fischer et al., 2020 ▸). It is also critical for the production of fatty acids in microbes as a platform for chemical synthesis (Gajewski, Pavlovic et al., 2017 ▸; Zhu et al., 2017 ▸). So far, FAS has been purified from natural sources (Lynen, 1969 ▸), but it is now becoming increasingly important to develop mutants, which are expressed in recombinant systems (Maier, 2017 ▸; Heil et al., 2019 ▸). At the same time, requirements for high-quality protein preparations for structural studies are becoming more stringent.
To meet these requirements, we developed a new protocol for the rapid preparation of recombinantly expressed yeast FAS. Our protocol includes vector-based expression under the native promoter, non-invasive affinity chromatography and strict monitoring of protein integrity. Taking advantage of a companion protocol that prevents protein denaturation at the air–water interface (D’Imprima et al., 2019 ▸), we show that we can reconstruct a 3D map of yeast FAS at ∼3 Å resolution from a comparatively small number of particle images within a short time. The same approach can now be used for other macromolecular assemblies.
2. Materials and methods
2.1. Strain cultivation and protein purification
Yeast cultures were grown and FAS was purified as reported previously (Gajewski, Pavlovic et al., 2017 ▸; D’Imprima et al., 2019 ▸). Haploid FAS-deficient Saccharomyces cerevisiae cells were transfected with plasmids carrying FAS-encoding genes and grown in YPD medium. After bead disruption and differential centrifugation, the soluble components were purified by Strep-Tactin affinity chromatography followed by size-exclusion chromatography. The main peak was collected. During purification, FAS was kept in buffer P1 (100 mM sodium phosphate pH 6.5). Purification was monitored by SDS–PAGE.
2.2. Thermal shift assay (TSA) and activity assay
Buffers P1, P2 (100 mM sodium phosphate pH 7.4), P3 (100 mM sodium phosphate pH 8), P4 (100 mM sodium phosphate, 100 mM NaCl pH 7.4), P5 (100 mM Tris–HCl pH 7.4) and distilled water were used in thermal shift assays (see also Fig. 1 ▸). Briefly, 2 µl protein solution (0.9 mg ml−1) was mixed with 21 µl buffer and 2 µl 62.5× SYPRO Orange protein gel stain, and fluorescence was then measured from 5 to 95°C with a step of 0.5°C min−1 with the excitation wavelength set to 450–490 nm and the emission wavelength set to 560–580 nm. FAS activity was determined by tracing NADPH consumption at 334 nm as reported in Gajewski, Buelens et al. (2017 ▸) and adapted for plate-reader readout (120 µl scale containing 200 mM NaH2PO4/Na2HPO4 pH 7.3, 1.75 mM 1,4-dithiothreitol, 0.03 mg ml−1 BSA, 0.7 µg FAS, 500 µM malonyl-CoA, 417 µM acetyl-CoA and 250 µM NADPH).
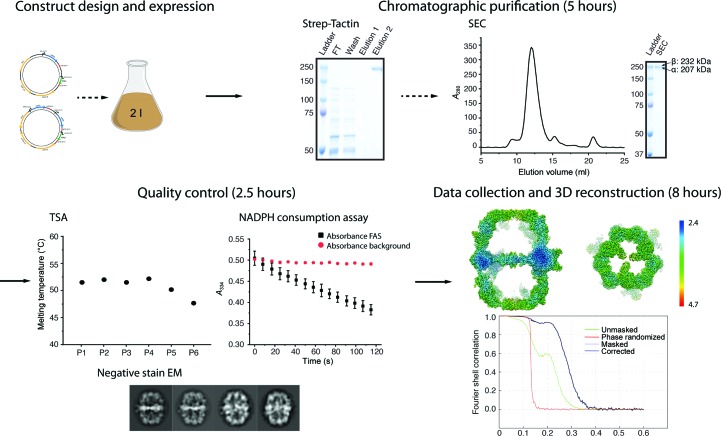
Figure 1.
Structural analysis of yeast FAS. Yeast FAS was expressed overnight from pRS vector-encoded FAS1 and FAS2 genes. Gravity flow of the cleared lysate over a Strep-Tactin column and subsequent size-exclusion chromatography (SEC) delivered pure protein within 5 h. Protein quality was monitored by NADPH consumption, thermal shift assays (TSA) and negative-stain transmission EM within 2.5 h. Thermal stability was tested for a set of conditions (P1, 100 mM sodium phosphate pH 6.5; P2, 100 mM sodium phosphate pH 7.4; P3, 100 mM sodium phosphate pH 8; P4, 100 mM sodium phosphate, 100 mM NaCl pH 7.4; P5, 100 mM Tris–HCl pH 7.4; P6, distilled water). The activity of the preparation was 2310 ± 48 mU mg−1 and the error in the melting point varied by less than 0.5°C; both values were within technical replication. Protein integrity was assessed further by negative-stain EM and 2D single-particle image analysis (within 1.5 h). CryoEM images were collected in movie mode in 4.5 h. 20 000 particles were picked automatically, of which 15 000 were selected by 2D and 3D classification, to yield a map at 3.1 Å resolution in 3.5 h of image processing.
2.3. Negative-stain electron microscopy
FAS was diluted to 0.05 mg ml−1 in purification buffer P1 and was negatively stained with 2%(w/v) sodium silicotungstate (Agar Scientific, Stansted, England). Specimens were prepared by applying a 3 µl droplet of protein solution to 300 mesh carbon-coated copper grids freshly glow-discharged at 15 mA for 45 s (Structure Probe Inc., West Chester, Pennsylvania, USA). The sample was incubated for 1 min before blotting with Whatman No. 1 filter paper (Sigma–Aldrich, Munich, Germany). Subsequently, two changes of 3 µl of stain were applied to the specimens for 15 s before blotting. Finally, the grids were left at room temperature to dry. Micrographs were recorded with an FEI Tecnai G2 Spirit (FEI Company, Hillsboro, Oregon, USA) operated at 120 kV on a Gatan Ultrascan 4000 CCD camera at a pixel size of 2.68 Å.
2.4. CryoEM grid preparation
Specimen preparation was carried out as described by D’Imprima et al. (2019 ▸). Briefly, Quantifoil R1.2/1.3 grids (Quantifoil Micro Tools, Jena, Germany) were washed overnight in chloroform (Sigma–Aldrich, Munich, Germany). The grids were coated with a single layer of graphene (Graphenea, Cambridge, Massachusetts, USA) stored in a sandwich of polyethylene terephthalate (PET) support and a protective layer of poly(methyl methacrylate) (PMMA). Graphene pads (1 cm2) were floated onto Quantifoil grids in a water bath where they were released from the PET support. Subsequently, the water was drained and graphene was layered carefully onto the grids. To ensure good adherence of the graphene, the grids were annealed at 150°C for 30 min. The graphene-coated grids were then washed in pure acetone and 2-propanol for 1 h each to remove the PMMA film and the grids were dried under a nitrogen stream. Other than during annealing, the graphene-coated grids were kept under a nitrogen stream in order to minimize air contamination of the graphene. Finally, the grids were dipped into 5 µM 1-pyrenecarboxylic acid (Sigma–Aldrich, Munich, Germany) dissolved in DMSO (Sigma–Aldrich, Munich, Germany) for 1 min, rinsed in one change of 2-propanol and ethanol, and dried under a nitrogen stream. For all grids, the graphene layer was deposited on the carbon side of the Quantifoils, whereas the protein sample was subsequently applied to the copper side.
2.5. Single-particle cryoEM
3 µl FAS solution (0.3 mg ml−1), incubated with 1 mM NADPH and 1 mM malonyl-CoA for 5 min at room temperature, was applied to the graphene-coated Quantifoil grids. The grids were vitrified in a Vitrobot Mark IV plunge-freezer at 100% humidity and 10°C after blotting for 6–8 s. CryoEM images were collected with a Titan Krios (FEI Company, Hillsboro, Oregon, USA) electron microscope operating at 300 kV. Images were recorded automatically with EPU at a pixel size of 0.833 Å on a Falcon III EC direct electron detector (FEI Company, Hillsboro, Oregon, USA) operating in counting mode. A total of 792 dose-fractionated movies were recorded with a cumulative dose of ∼32 e− Å−2. Image drift correction and dose weighting were performed using MotionCor2 (Zheng et al., 2017 ▸) within the RELION-3 pipeline (Zivanov et al., 2018 ▸). CTF determination was performed with CTFFIND 4.1.13 (Rohou & Grigorieff, 2015 ▸). From a data set of 19 981 particles picked automatically with crYOLO (Wagner et al., 2019 ▸), 15 320 remained after 2D and 3D classification in cryoSPARC (Punjani et al., 2017 ▸). The particles contributing to the best 3D class were subjected to homogeneous and non-uniform refinement in cryoSPARC, yielding a map at 3.1 Å resolution, as determined by the post-processing procedure in RELION (Chen et al., 2013 ▸).
2.6. Model building
The X-ray model of yeast FAS (PDB entry 3hmj; Johansson et al., 2009 ▸) was docked into the cryoEM map with USCF Chimera (Pettersen et al., 2004 ▸) and manually rebuilt and completed in Coot (Emsley et al., 2010 ▸). The model was refined using phenix.real_space_refinement (Liebschner et al., 2019 ▸) with geometry and secondary-structure restraints, followed by manual inspection and adjustments in Coot. The geometry of the model was validated by MolProbity (Chen et al., 2010 ▸).
3. Results
3.1. Developing a protocol for FAS purification
Previous procedures for the preparation of yeast FAS from baker’s yeast followed a sequence of ammonium sulfate fractionation, chromatography on calcium phosphate gels, ultracentrifugation and hydroxyapatite chromatography (Lynen, 1969 ▸). An improved variant that included additional chromatographic steps was used for the 3.1 Å resolution X-ray structure of baker’s yeast FAS (Leibundgut et al., 2007 ▸; Lomakin et al., 2007 ▸). A significantly shorter protocol was based on the modification of yeast FAS with a His tag integrated into the FAS1 gene by homologous recombination, which enabled nickel-chelating chromatography as the first purification step (Johansson et al., 2008 ▸).
We recently established a plasmid-based expression system suitable for expressing FAS-encoding genes in baker’s yeast deletion strains (D’Imprima et al., 2019 ▸). Here, the FAS1 gene was tagged with a Strep-Tag at the C-terminus of subunit β (Schmidt & Skerra, 2007 ▸). Strep-Tactin affinity chromatography followed by size-exclusion chromatography (SEC) delivered pure protein within 5 h. The protein is pure as judged by SDS–PAGE and has a specific activity of 2100 ± 300 mU mg−1, which is in the range reported for the best previous preparations of fungal FASs (Kolodziej et al., 1996 ▸; Fichtlscherer et al., 2000 ▸; Wieland et al., 1979 ▸; Fischer et al., 2015 ▸). The standard deviation of the specific enzymatic activities of FAS from nine independent preparations indicates that the protocol delivers protein of a significantly better, more reproducible quality than previous protocols. The normalized standard deviation of specific enzymatic activities in our study was 0.14, whereas previously it was 0.52 (Lynen, 1969 ▸).
The C-terminus of the β subunit was selected for affinity tagging, because it is stably anchored in the MPT domain, which is itself stably integrated into the main protein body (Johansson et al., 2009 ▸; Gipson et al., 2010 ▸). The suitability of the C-terminus of the β subunit for modifications with peptides and proteins has also recently been demonstrated by others: a 3×FLAG-tag fusion aided in the purification of FAS for studying ACP-mediated substrate shuttling (Lou et al., 2019 ▸), and the FAS co-translational assembly pathway protein (Shiber et al., 2018 ▸) as well as the autophagic degradation of FAS (Shpilka et al., 2015 ▸) were monitored using a GFP fusion construct. To keep as closely as possible to physiological conditions, we put the encoding sequence on single copy number centromeric pRS shuttle vectors of types pRS313 and pRS315 (Sikorski & Hieter, 1989 ▸; Gajewski, Pavlovic et al., 2017 ▸). Expression yielded 1.4 ± 0.4 mg yeast FAS from a 2 l culture within 5 h. The plasmid-encoded expression system enables rapid and economical mutagenesis and tolerates lethal phenotypes induced by FAS mutations when external fatty acids are supplied (Fig. 1 ▸).
3.2. Quality measures for protocol development
Large macromolecular complexes tend to be structurally unstable and often assume several different, simultaneously present conformations. Unsuitable purification methods can induce disassembly and aggregation or small structural changes that may be misinterpreted as conformational variability. It is therefore essential to use appropriate protein-purification methods to prevent disassembly and denaturation during purification and cryoEM sample preparation (Chari et al., 2015 ▸). The small percentages of picked particles in many cryoEM reconstructions suggest that the majority are damaged. In many instances the proportion of intact particles is below 20% [19% for human synaptic GABAA receptor (Zhu et al., 2018 ▸), 15% for human P-glycoprotein (Kim & Chen, 2018 ▸), 11.8% for nucleosome (Takizawa et al., 2018 ▸), 8.9% for human γ-secretase (Bai et al., 2015 ▸) and 5.7% for sodium channel from electric eel (Yan et al., 2017 ▸)]. Frequently, it is not clear whether the macromolecular complex suffered during protein production or sample preparation for cryoEM.
Each step in our protocol for the rapid preparation of yeast FAS for high-resolution structural studies was examined rigorously. Quality criteria included oligomeric state and thermal stability, monitored by size-exclusion chromatography (SEC), and thermal unfolding, monitored by sparse-matrix screening (TSA) (Ericsson et al., 2006 ▸). Both methods are sensitive tools for screening protein preparation conditions. Further, the catalytic activity of FAS served as a measure of overall protein integrity. Specific catalytic activity, determined as the catalytic activity of the probe related to the FAS concentration as judged by SDS–PAGE, proved to be ideal for optimizing the vector-based expression system and assessing progress in the purification protocol. Amongst other things, we found that the C-terminus of subunit β tolerated tagging, while tagging at the C-terminus of subunit α [in the phosphopantetheine transferase (PPT) domain] prevented complex assembly (data not shown). As outlined in Fig. 1 ▸, SEC, TSA and activity assays were used routinely to check the protein quality of each preparation.
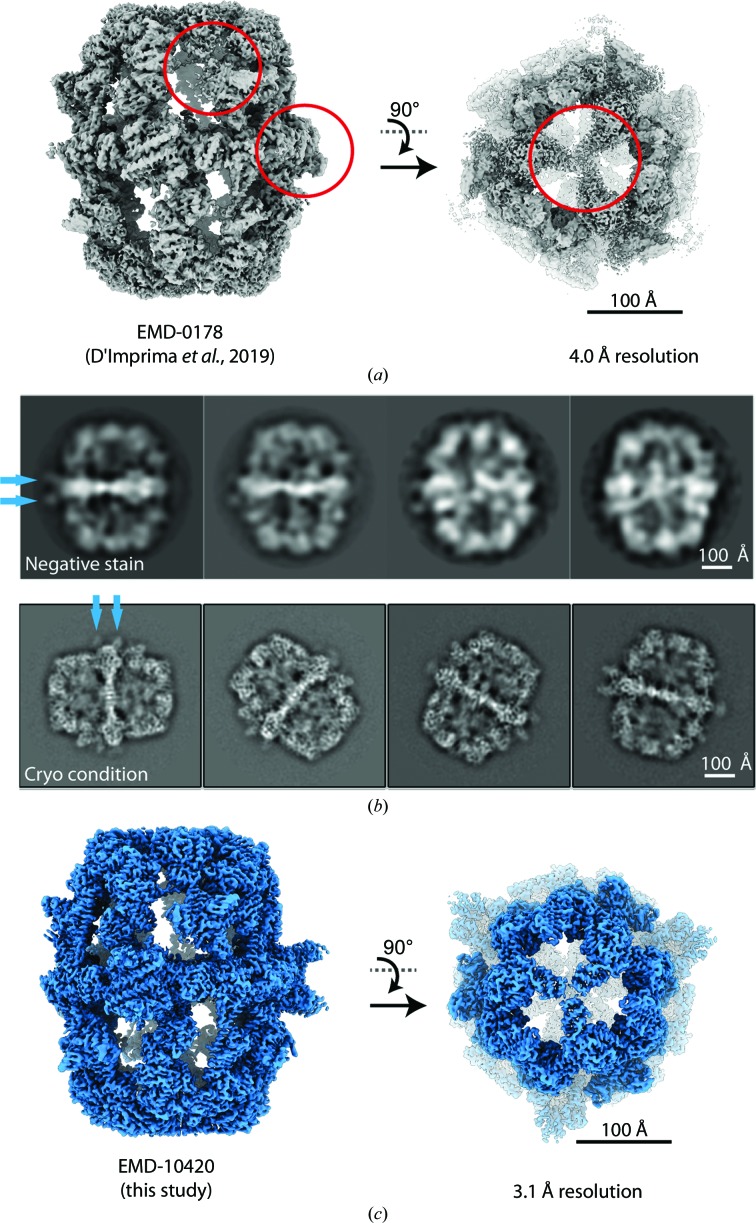
As another valuable diagnostic of protein stability (Gao et al., 2016 ▸; Thompson et al., 2016 ▸), negative-stain EM identified the FAS PPT domain as a major source of structural heterogeneity. When the FAS complex was purified by SEC and concentrated by centrifugation through a semipermeable membrane, the PPT domain was absent in 2D class averages and 3D classes (D’Imprima et al., 2019 ▸) [Fig. 2 ▸(a)]. When the concentration step was omitted, 2D class averages of negatively stained particles consistently showed the PPT domain on the outside of the FAS central wheel. The concentration step proved to be unnecessary when we used a continuous support layer on the EM grids, which reduces the sample concentration required for specimen preparation by at least one order of magnitude (D’Imprima et al., 2019 ▸). The partial unfolding of the PPT domain was only observable by EM, as it escapes quality control by enzymatic activity and protein-stability measurements. The PPT domain is only required for the initial step of post-translational modification of the carrier protein (ACP) domain, without being directly involved in the fatty-acid synthesis cycle, and poor PPT domain quality is therefore not visible in the NADPH consumption assay. Furthermore, the PPT domain is not integrated into the FAS barrel and does not contribute to its thermal and oligomeric stability (Johansson et al., 2009 ▸). CryoEM was performed with the same FAS batch as used for negative-stain EM [Fig. 2 ▸(b)]. CryoEM data indicated that avoiding the concentration step not only preserves the PPT domain density, but also those of other poorly resolved domains [Fig. 2 ▸(c)], including the trimerization domain and the acetyltransferase (AT) domain, in particular its interface with the enoyl reductase (ER) domain, which are now equally as well defined as the other FAS domains.
Figure 2.
Comparison of FAS preparations. (a) The published map (D’Imprima et al., 2019 ▸) lacks the PPT domain and parts of the β-domes are poorly resolved (red circles). (b) Data collected using protein prepared by the optimized protocol described here. The 2D class averages show structured PPT domains (blue arrows) and resolved secondary-structure features at the β-domes. (c) CryoEM map from 15 000 particles at 3.1 Å resolution.
3.3. CryoEM of stable, intact FAS
For cryoEM, the FAS sample purified as above was incubated with NADPH and malonyl-CoA prior to plunge-freezing. Although this treatment results in a slight decrease in the thermal protein stability as determined by TSA (Supplementary Fig. S1), it reduces sample heterogeneity by driving the synthesis of bound fatty acids to completion. Protein denaturation at the air–water interface was avoided by applying the sample to a film of graphene on the carbon side of the Quantifoil EM grids. The graphene support was rendered hydrophilic by using 1-pyrenecarboxylic acid as a noncovalent chemical doping agent (D’Imprima et al., 2019 ▸; Section 2). 2D unsupervised class averages revealed that the complex was very stable [Fig. 2 ▸(b) and Supplementary Fig. S2]. Three-dimensional reconstruction yielded a map at a global resolution of 3.1 Å [Fig. 3 ▸(a)]. In distinction from our previous cryoEM map (EMD-0178; D’Imprima et al., 2019 ▸), the resolution is isotropic (Supplementary Fig. S3) and we were able to build a complete model of yeast FAS (Table 1 ▸).
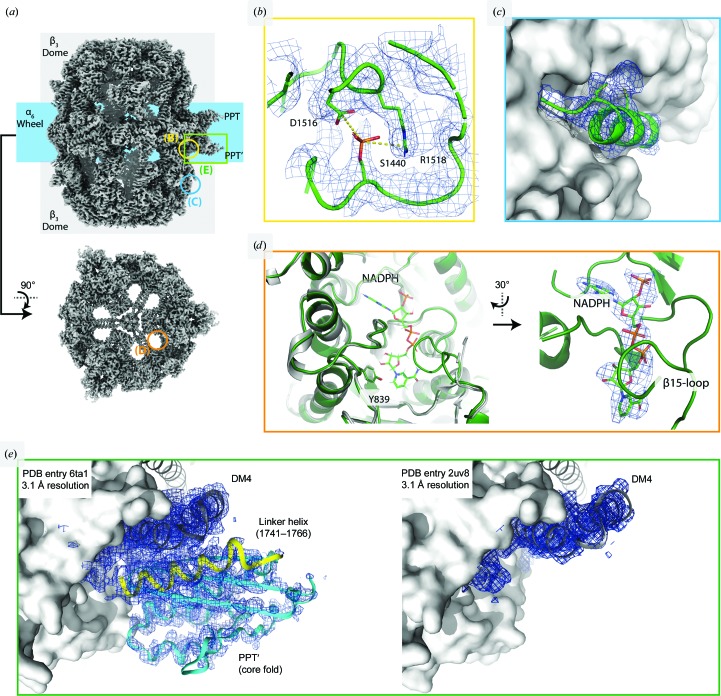
Figure 3.
3.1 Å resolution map of FAS. (a) Overview of the EM map. The square and circles labelled (B), (C), (D) and (E) indicate the map regions that are enlarged in (b), (c), (d) and (e), respectively. (b) Density at Ser1440 suggesting phosphorylation. (c) Density at residues Cys820 and Cys824 (subunit β) not accounted for by the atomic model. (d) NADPH cofactor density in mesh representation, bound to the active site of the KR domain. Left: the KR active site in the apo form as in the X-ray structure (PDB entry 2uv8; gray) superimposed on our cryoEM structure (green). NADPH and the catalytically active Tyr839 are shown in stick representation. (e) The PPT domain and the dimerization module DM4, which acts as an adaptor to anchor the PPT domain at the perimeter of the FAS barrel (PPT domain in cyan, DM4 in gray and linker helix in yellow; both densities are shown at 1.0σ). Left: the PPT domain traced in the 3.1 Å resolution cryoEM density. Right: the 3.1 Å resolution X-ray map (data from PDB entry 2uv8; Leibundgut et al., 2007 ▸) shows that DM4 is well resolved, whereas there is no density for the PPT domain or linker helix.
Table 1. Statistics of 3D reconstruction and model refinement.
| Data collection | |
| Electron microscope | Titan Krios |
| Electron detector | Falcon III |
| Voltage (kV) | 300 |
| Defocus range (µm) | 0.5–2.1 |
| Pixel size (Å) | 0.833 |
| Electron dose (e− Å−2) | 32 |
| Images | 792 |
| 3D reconstruction | |
| Final particles | 15320 |
| Applied symmetry | D3 |
| Resolution (Å) | 3.1 |
| B factor (Å2) | −72 |
| Model composition | |
| Peptide chains | 2 |
| Residues | 3780 |
| Cofactors | FMN, NADPH |
| Ramachandran plot | |
| Favored (%) | 94.27 |
| Outliers (%) | 0.13 |
| Validation | |
| MolProbity score | 1.96 |
| Rotamer favored (%) | 94.01 |
| Rotamer outliers (%) | 1.49 |
The new cryoEM map revealed additional density at Ser1440, suggesting that this serine is phosphorylated, as was previously observed in a large-scale phosphorylation analysis in S. cerevisiae (Li et al., 2007 ▸) [Fig. 3 ▸(b)]. Ser1440 is located in the dimerization module DM4 that holds the PPT domain at the perimeter of the barrel. The phosphate group is embedded in a pocket near Asp1516 and Arg1518. Sequence comparisons revealed high conservation of the Ser1440–Asp1516–Arg1518 motif (Grininger, 2014 ▸). In addition, we found density at Cys820 and Cys824 that is not accounted for by the atomic model [Fig. 3 ▸(c)]. The two cysteines are not conserved in fungal FASs, and the density possibly originates from the malonyl group, which binds to cysteine(s) owing to the high malonyl-CoA concentration in solution. In the structure, the NADPH cofactor is bound to the active site of the KR domain [Fig. 3 ▸(d)], but not to the ER domain. The active nicotinamide unit is exposed at the inner surface, which contains the acyl-ACP docking sites. Tyr839 sits at the entrance to the binding pocket and is responsible for the protein transfer that neutralizes the hydroxyl anion in the reduction of the carbonyl group by NADPH. This residue was recently mutated to a phenylalanine, turning FAS into a nonreducing, lactone-producing enzyme (Zha et al., 2004 ▸; Gajewski, Buelens et al., 2017 ▸). A comparison with the cofactor-free X-ray structure of baker’s yeast FAS shows the structuring of the β15 loop upon NADPH binding, as observed in the homologous Thermomyces lanuginosus type I FAS and type II KR domain (Jenni et al., 2007 ▸) [Fig. 3 ▸(e)].
4. Discussion
Within the past five years, cryoEM has developed into a powerful technique for biological structure determination. This is documented by a sharp increase in the number of maps released by the EMDB (from eight in 2002 to 417 in 2012 and 1771 in 2018). Fast and easy access to purified samples is a prerequisite for fully exploiting the technical developments in cryoEM for molecular biology. We have revisited the process of resolving the structure of yeast FAS, a major milestone in early cryoEM and crystallographic studies, and have derived a rapid protocol for determining its complete structure at high resolution.
A number of challenges and pitfalls were revealed during the development of our protocol. In the case of yeast FAS, neither the vector-based expression strategy nor affinity tagging at the C-terminus of subunit β affected the protein quality. However, the PPT domain turned out to be particularly sensitive to partial denaturation. The PPT domain may be prone to denaturation because it is monomeric in the yeast FAS complex (Lomakin et al., 2007 ▸), while it forms trimers as a separate protein (Johansson et al., 2009 ▸). Earlier structures of yeast FAS confirm that the PPT domain is unstable. The PPT domain was not traced in electron densities in the landmark X-ray structures at 3.1–4 Å resolution (Jenni et al., 2007 ▸; Leibundgut et al., 2007 ▸; Lomakin et al., 2007 ▸; Johansson et al., 2008 ▸) [Fig. 3 ▸(e)] or in cryoEM maps at 3–4 Å resolution (Lou et al., 2019 ▸; D’Imprima et al., 2019 ▸). We conclude that the PPT domain denatures easily during protein purification, crystallization or cryoEM grid preparation. It is likely that the PPT domain partly unfolds when the protein is concentrated at the solid–liquid interface of the semipermeable membrane (Rabe et al., 2011 ▸). Changes in protein structure resulting from adsorption to solid surfaces are well documented (Tunc et al., 2005 ▸; Norde, 1986 ▸; Höök et al., 1998 ▸; Maste et al., 1997 ▸), ranging from protein denaturation at membranes for water purification (Lee et al., 2016 ▸) to modified behavior of key drug candidates such as amyloid peptides (Zhou et al., 2013 ▸). Strikingly, yeast FAS does not denature upon adsorption to a graphene support film on EM grids, whereas it does denature by interaction with semipermeable membranes or at the air–water interface. Whether and how adsorption to solid surfaces induces protein damage and impairs structure determination at atomic resolution of conformationally weak or unstable proteins will require further investigation.
In conclusion, we present a rapid pipeline for the preparation of the 2.6 MDa yeast FAS with high quality. Together with a companion protocol (D’Imprima et al., 2019 ▸), structural analysis of yeast FAS at ∼3 Å resolution by cryoEM is achievable within a day. While the presented pipeline is unlikely to be directly applicable to other protein complexes, the approach of monitoring and optimizing the individual steps of a purification procedure may serve as a blueprint for other macromolecular assemblies.
Supplementary Material
EMDB reference: yeast FAS, EMD-10420
PDB reference: yeast FAS, 6ta1
Supplementary Figures. DOI: 10.1107/S2052252519017366/eh5006sup1.pdf
Acknowledgments
We thank Deryck J. Mills, Simone Prinz and Mark Linder for EM support. We are grateful to Dr David Wöhlert, Martin Centola and Dr Emanuele Rossini for discussions. Author contributions are as follows. Mirko Joppe: protein purification, protein quality control, protocol development, data validation, and review and editing of the final draft. Edoardo D’Imprima: design of the project, supervision, EM sample preparation, EM data collection and validation, writing of the original draft, and review and editing of the final draft. Nina Salustros: EM sample preparation, EM data collection and validation. Karthik S. Paithankar: data analysis and review and editing of the final draft. Janet Vonck: supervision, model building, and review and editing of the final draft. Martin Grininger: design of research, data validation, writing of the final draft, resources, supervision and funding acquisition. Werner Kühlbrandt: design of research, review and editing of the final draft, resources, supervision and funding acquisition. Werner Kühlbrandt is an editor of IUCrJ. The other authors declare no competing interests.
Funding Statement
This work was funded by Hessisches Ministerium für Wissenschaft und Kunst grant . Volkswagen Foundation grant 85701 to Martin Grininger. Max-Planck-Gesellschaft grant .
References
- Bai, X.-C., Yan, C., Yang, G., Lu, P., Ma, D., Sun, L., Zhou, R., Scheres, S. H. W. & Shi, Y. (2015). Nature, 525, 212–217. [DOI] [PMC free article] [PubMed]
- Chari, A., Haselbach, D., Kirves, J.-M., Ohmer, J., Paknia, E., Fischer, N., Ganichkin, O., Möller, V., Frye, J. J., Petzold, G., Jarvis, M., Tietzel, M., Grimm, C., Peters, J.-M., Schulman, B. A., Tittmann, K., Markl, J., Fischer, U. & Stark, H. (2015). Nat. Methods, 12, 859–865. [DOI] [PMC free article] [PubMed]
- Chen, S., McMullan, G., Faruqi, A. R., Murshudov, G. N., Short, J. M., Scheres, S. H. W. & Henderson, R. (2013). Ultramicroscopy, 135, 24–35. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Cheng, Y., Grigorieff, N., Penczek, P. A. & Walz, T. (2015). Cell, 161, 438–449. [DOI] [PMC free article] [PubMed]
- D’Imprima, E., Floris, D., Joppe, M., Sánchez, R., Grininger, M. & Kühlbrandt, W. (2019). eLife, 8, e42747. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Ericsson, U. B., Hallberg, B. M., DeTitta, G. T., Dekker, N. & Nordlund, P. (2006). Anal. Biochem. 357, 289–298. [DOI] [PubMed]
- Fichtlscherer, F., Wellein, C., Mittag, M. & Schweizer, E. (2000). Eur. J. Biochem. 267, 2666–2671. [DOI] [PubMed]
- Fischer, M., Joppe, M., Mulinacci, B., Vollrath, R., Konstantinidis, K., Kötter, P., Ciccarelli, L., Vonck, J., Oesterhelt, D. & Grininger, M. (2020). Sci. Rep. 10, 895. [DOI] [PMC free article] [PubMed]
- Fischer, M., Rhinow, D., Zhu, Z., Mills, D. J., Zhao, Z. K., Vonck, J. & Grininger, M. (2015). Protein Sci. 24, 987–995. [DOI] [PMC free article] [PubMed]
- Gajewski, J., Buelens, F., Serdjukow, S., Janssen, M., Cortina, N., Grubmüller, H. & Grininger, M. (2017). Nat. Chem. Biol. 13, 363–365. [DOI] [PubMed]
- Gajewski, J., Pavlovic, R., Fischer, M., Boles, E. & Grininger, M. (2017). Nat. Commun. 8, 14650. [DOI] [PMC free article] [PubMed]
- Gao, Y., Cao, E., Julius, D. & Cheng, Y. (2016). Nature, 534, 347–351. [DOI] [PMC free article] [PubMed]
- Gipson, P., Mills, D. J., Wouts, R., Grininger, M., Vonck, J. & Kühlbrandt, W. (2010). Proc. Natl Acad. Sci. USA, 107, 9164–9169. [DOI] [PMC free article] [PubMed]
- Grant, T., Rohou, A. & Grigorieff, N. (2018). eLife, 7, e35383. [DOI] [PMC free article] [PubMed]
- Grininger, M. (2014). Curr. Opin. Struct. Biol. 25, 49–56. [DOI] [PubMed]
- Heil, C. S., Wehrheim, S. S., Paithankar, K. S. & Grininger, M. (2019). Chembiochem, 20, 2298–2321. [DOI] [PubMed]
- Höök, F., Rodahl, M., Kasemo, B. & Brzezinski, P. (1998). Proc. Natl Acad. Sci. USA, 95, 12271–12276. [DOI] [PMC free article] [PubMed]
- Jenni, S., Leibundgut, M., Boehringer, D., Frick, C., Mikolásek, B. & Ban, N. (2007). Science, 316, 254–261. [DOI] [PubMed]
- Johansson, P., Mulinacci, B., Koestler, C., Vollrath, R., Oesterhelt, D. & Grininger, M. (2009). Structure, 17, 1063–1074. [DOI] [PubMed]
- Johansson, P., Wiltschi, B., Kumari, P., Kessler, B., Vonrhein, C., Vonck, J., Oesterhelt, D. & Grininger, M. (2008). Proc. Natl Acad. Sci. USA, 105, 12803–12808. [DOI] [PMC free article] [PubMed]
- Kim, Y. & Chen, J. (2018). Science, 359, 915–919. [DOI] [PubMed]
- Kolodziej, S. J., Penczek, P. A., Schroeter, J. P. & Stoops, J. K. (1996). J. Biol. Chem. 271, 28422–28429. [DOI] [PubMed]
- Lee, A., Elam, J. W. & Darling, S. B. (2016). Environ. Sci. (Camb.), 2, 17–42.
- Leibundgut, M., Jenni, S., Frick, C. & Ban, N. (2007). Science, 316, 288–290. [DOI] [PubMed]
- Li, X., Gerber, S. A., Rudner, A. D., Beausoleil, S. A., Haas, W., Villén, J., Elias, J. E. & Gygi, S. P. (2007). J. Proteome Res. 6, 1190–1197. [DOI] [PubMed]
- Liebschner, D., Afonine, P. V., Baker, M. L., Bunkóczi, G., Chen, V. B., Croll, T. I., Hintze, B., Hung, L.-W., Jain, S., McCoy, A. J., Moriarty, N. W., Oeffner, R. D., Poon, B. K., Prisant, M. G., Read, R. J., Richardson, J. S., Richardson, D. C., Sammito, M. D., Sobolev, O. V., Stockwell, D. H., Terwilliger, T. C., Urzhumtsev, A. G., Videau, L. L., Williams, C. J. & Adams, P. D. (2019). Acta Cryst. D75, 861–877.
- Lomakin, I. B., Xiong, Y. & Steitz, T. A. (2007). Cell, 129, 319–332. [DOI] [PubMed]
- Lou, J. W., Iyer, K. R., Hasan, S. M. N., Cowen, L. E. & Mazhab-Jafari, M. T. (2019). Sci. Rep. 9, 12987. [DOI] [PMC free article] [PubMed]
- Lynen, F. (1969). Methods Enzymol. 14, 17–33.
- Lynen, F., Engeser, H., Foerster, E., Fox, J. L., Hess, S., Kresze, G., Schmitt, T., Schreckenbach, T., Siess, E., Wieland, F. & Winnewisser, W. (1980). Eur. J. Biochem. 112, 431–442.
- Maier, T. (2017). Nat. Chem. Biol. 13, 344–345. [DOI] [PubMed]
- Maier, T., Leibundgut, M., Boehringer, D. & Ban, N. (2010). Q. Rev. Biophys. 43, 373–422. [DOI] [PubMed]
- Maste, M. C. L., Norde, W. & Visser, A. (1997). J. Colloid Interface Sci. 196, 224–230. [DOI] [PubMed]
- McDowall, A. W., Chang, J.-J., Freeman, R., Lepault, J., Walter, C. A. & Dubochet, J. (1983). J. Microsc. 131, 1–9. [DOI] [PubMed]
- McMullan, G., Faruqi, A. R., Henderson, R., Guerrini, N., Turchetta, R., Jacobs, A. & van Hoften, G. (2009). Ultramicroscopy, 109, 1144–1147. [DOI] [PMC free article] [PubMed]
- Murphy, B. J., Klusch, N., Langer, J., Mills, D. J., Yildiz, Ö. & Kühlbrandt, W. (2019). Science, 364, eaaw9128. [DOI] [PubMed]
- Norde, W. (1986). Adv. Colloid Interface Sci. 25, 267–340. [DOI] [PubMed]
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C. & Ferrin, T. E. (2004). J. Comput. Chem. 25, 1605–1612. [DOI] [PubMed]
- Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. (2017). Nat. Methods, 14, 290–296. [DOI] [PubMed]
- Rabe, M., Verdes, D. & Seeger, S. (2011). Adv. Colloid Interface Sci. 162, 87–106. [DOI] [PubMed]
- Rohou, A. & Grigorieff, N. (2015). J. Struct. Biol. 192, 216–221. [DOI] [PMC free article] [PubMed]
- Schmidt, T. & Skerra, A. (2007). Nat. Protoc. 2, 1528–1535. [DOI] [PubMed]
- Shiber, A., Döring, K., Friedrich, U., Klann, K., Merker, D., Zedan, M., Tippmann, F., Kramer, G. & Bukau, B. (2018). Nature, 561, 268–272. [DOI] [PMC free article] [PubMed]
- Shpilka, T., Welter, E., Borovsky, N., Amar, N., Shimron, F., Peleg, Y. & Elazar, Z. (2015). Proc. Natl Acad. Sci. USA, 112, 1434–1439. [DOI] [PMC free article] [PubMed]
- Sikorski, R. S. & Hieter, P. (1989). Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed]
- Takizawa, Y., Tanaka, H., Machida, S., Koyama, M., Maehara, K., Ohkawa, Y., Wade, P. A., Wolf, M. & Kurumizaka, H. (2018). Open Biol. 8, 170255. [DOI] [PMC free article] [PubMed]
- Thompson, R. F., Walker, M., Siebert, C. A., Muench, S. P. & Ranson, N. A. (2016). Methods, 100, 3–15. [DOI] [PMC free article] [PubMed]
- Tunc, S., Maitz, M. F., Steiner, G., Vázquez, L., Pham, M. T. & Salzer, R. (2005). Colloids Surf. B Biointerfaces, 42, 219–225. [DOI] [PubMed]
- Wagner, T., Merino, F., Stabrin, M., Moriya, T., Antoni, C., Apelbaum, A., Hagel, P., Sitsel, O., Raisch, T., Prumbaum, D., Quentin, D., Roderer, D., Tacke, S., Siebolds, B., Schubert, E., Shaikh, T. R., Lill, P., Gatsogiannis, C. & Raunser, S. (2019). Commun. Biol. 2, 218. [DOI] [PMC free article] [PubMed]
- Wieland, F., Renner, L., Verfürth, C. & Lynen, F. (1979). Eur. J. Biochem. 94, 189–197. [DOI] [PubMed]
- Yan, Z., Zhou, Q., Wang, L., Wu, J., Zhao, Y., Huang, G., Peng, W., Shen, H., Lei, J. & Yan, N. (2017). Cell, 170, 470–482. [DOI] [PubMed]
- Zha, W., Shao, Z., Frost, J. & Zhao, H. (2004). J. Am. Chem. Soc. 126, 4534–4535. [DOI] [PubMed]
- Zheng, S. Q., Palovcak, E., Armache, J.-P., Verba, K. A., Cheng, Y. & Agard, D. A. (2017). Nat. Methods, 14, 331–332. [DOI] [PMC free article] [PubMed]
- Zhou, X., Zhang, Y., Zhang, F., Pillai, S., Liu, J., Li, R., Dai, B., Li, B. & Zhang, Y. (2013). Nanoscale, 5, 4816–4822. [DOI] [PubMed]
- Zhu, S., Noviello, C. M., Teng, J., Walsh, R. M. Jr, Kim, J. J. & Hibbs, R. E. (2018). Nature, 559, 67–72. [DOI] [PMC free article] [PubMed]
- Zhu, Z., Zhou, Y. J., Krivoruchko, A., Grininger, M., Zhao, Z. K. & Nielsen, J. (2017). Nat. Chem. Biol. 13, 360–362. [DOI] [PubMed]
- Zivanov, J., Nakane, T., Forsberg, B. O., Kimanius, D., Hagen, W. J., Lindahl, E. & Scheres, S. H. W. (2018). eLife, 7, e42166. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
EMDB reference: yeast FAS, EMD-10420
PDB reference: yeast FAS, 6ta1
Supplementary Figures. DOI: 10.1107/S2052252519017366/eh5006sup1.pdf