Abstract
Digestive cancers-including gastric cancer (GC), colorectal cancer, hepatocellular carcinoma, esophageal cancer, and pancreatic cancer-accounted for 26% of cancer cases and 35% of cancer deaths worldwide in 2018. It is crucial and urgent to develop biomarkers for the diagnosis, prognosis, and therapeutic benefits of digestive cancers, especially for GC, since the incidence of GC is lower only than lung cancer in China, is hard to detect at an early stage, and is associated with poor prognosis. Mucins, glycoproteins encoded by MUC family genes, act as a part of a physical barrier in the digestive tract and participate in various signaling pathways. Some mucins have been used or proposed as biomarkers for carcinomas, such as MUC16 (CA125) and MUC4. However, there are no systematic investigations on the association of MUC family members with diagnoses and clinical outcomes even though relevant data have been largely accumulated in the past decade. By analyzing transcriptomic and clinical data of digestive cancer samples from TCGA involving colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), liver hepatocellular carcinoma (LIHC), stomach adenocarcinoma (STAD), and pancreatic adenocarcinoma (PAAD), it was found that expressions levels of MUC15, MUC13, and MUC21 were individually associated with survival for digestive cancers, and high expressions of EMCN (MUC14) and MUC15 were correlated with poor survival for STAD. Cox regression analysis indicated the predictive power of an EMCN/MUC15 combination for overall survival (OS) of GC patients, which was validated on an independent dataset from GEO. EMCN/MUC15 correlated genes were identified to be enriched in cancer-related processes, such as vasculature development, mitosis, and immunity. Therefore, we propose that an EMCN/MUC15 combination could be a potential prognostic signature for gastric cancer.
Keywords: MUC family, EMCN, MUC15, prognostic, gastric cancer
Introduction
Digestive cancers are a group of cancers that occur in the digestive tract, and include gastric cancer (GC), colorectal cancer, hepatocellular carcinoma, esophageal cancer, and pancreatic cancer. Digestive cancers accounted for around 26% of cancer cases and 35% of cancer deaths in the world in 2018 (Bray et al., 2018). Among them, the morbidity and mortality of GC in Eastern Asia is much higher than the worldwide average level. In China, the incidence of GC is only lower than lung cancer, and the mortality is third to lung cancer and liver cancer (Chen et al., 2014). Most patients suffering from early stage GC are asymptomatic and always develop distant metastasis at the time of diagnosis (Van Cutsem et al., 2016; Bray et al., 2018). Surgery is the main treatment for GC. Adjuvant or neoadjuvant therapy combined with surgery is commonly used to treat advanced GC, while targeted drugs for advanced GC, such as the HER2 (also known as ERBB2) antibody trastuzumab, and the VEGFR-2 antibody ramucirumab, are still in clinical trials (Van Cutsem et al., 2016). Therefore, developing biomarkers for the diagnosis, prognosis, and therapeutic response of digestive cancers, especially of GC, is necessary and urgent for reducing the mortality rate.
Mucins represent a group of glycoproteins encoded by MUC family genes. These high-molecular weight and filamentous glycoproteins could be classified into secreted mucins and membrane-bound mucins. In the digestive tract, secreted mucins form a mucus layer and act as part of a physical defensive barrier against external aggressive forces (Dekker et al., 2002; Dhanisha et al., 2018); membrane-bound mucins possess membrane specific domains which enable their diverse roles in signaling pathways (Dekker et al., 2002; Dhanisha et al., 2018). Not surprisingly, dysfunction of mucins in their fundamental roles is implicated in disease development at mucosal surfaces (Corfield, 2015; Dhanisha et al., 2018), and some mucins have been reported to display diagnostic or prognostic significance in different types of cancer. For example, MUC16, also known as CA125, is a widely used biomarker for the diagnosis of ovarian cancer (Yonezawa et al., 2011; Jonckheere and Van Seuningen, 2018) and was also found to be over-expressed in several other human malignancies, including pancreas, breast, and lung (Aithal et al., 2018). MUC4 promotes carcinogenetic progression and has been proposed as a promising biomarker for pancreatic, ovarian, esophagus, and lung cancers (Kaur et al., 2013; Jonckheere and Van Seuningen, 2018). MUC15 overexpression is significantly correlated with several types of cancers, including colon cancer, hepatocellular carcinoma, and thyroid cancer (Huang et al., 2009; Nam et al., 2011; Wang et al., 2013; Choi et al., 2018). Moreover, MUC4/MUC16/MUC20 high-expression signature was very recently reported to be correlated with poor overall survival (OS) in several types of digestive cancers including pancreatic, colon, and GCs (Jonckheere and Van Seuningen, 2018). However, there are no systematic investigations, so far, on the association of MUC family members with diagnosis, prognosis, and/or therapeutic benefits, even though the Cancer Genome Atlas (TCGA) project is producing massive genomic, transcriptomic, proteomic, and clinical data involving more than 11,000 patients of 33 different types of tumors (Weinstein et al., 2013), and meanwhile, a number of web tools, such as GEPIA (Tang et al., 2017) and cBioPortal for Cancer Genomics (Cerami et al., 2012; Gao et al., 2013), have been developed that enable users to easily and effectively mine TCGA data.
In the present study, by analyzing digestive cancer samples from TCGA involving colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), liver hepatocellular carcinoma (LIHC), stomach adenocarcinoma (STAD), and pancreatic adenocarcinoma (PAAD), we found that expression levels of MUC15, MUC13, and MUC21 were individually associated with survival for all these digestive cancers, and high expressions of EMCN (MUC14) and MUC15 were correlated with poor survival for STAD. Cox regression analysis showed that EMCN/MUC15 combination still exhibited a significant correlation with the OS of GC patients. The prognostic prediction power of signature EMCN/MUC15 was further validated on an independent GC dataset, GSE84437. EMCN/MUC15 top 50 correlated genes were identified to be enriched in cancer-related processes, including vasculature development, mitosis, immunity, and so on. Taken together, we propose EMCN/MUC15 combination as a potential prognostic signature for GC.
Materials and Methods
Datasets
Datasets were collected from TCGA1 and GEO2 (Barrett et al., 2012). Specifically, gene expression data (TPM, Transcripts Per Kilobase Million) and clinical data for digestive cancers including COAD, ESCA, LIHC, STAD, and PAAD, were analyzed with the online webserver GEPIA 1.0 (Tang et al., 2017). Among them, MUC family mRNA expression data (mRNA expression z-scores, which is based on RNASeqV2 processed and normalized using RSEM) and clinical profiles involving 407 STAD samples were extracted by using an online web tool cBioPortal for Cancer Genomics (Cerami et al., 2012; Gao et al., 2013). Additionally, GSE84437 were extracted from the GEO database, which involves mRNA microarray data and clinical profiles of 433 GC samples.
Survival Analysis
Kaplan–Meier (KM) survival analysis for digestive cancer samples as a whole was carried out by using the webserver GEPIA 1.0 (Tang et al., 2017), and for GC samples (TCGA-STAD from cBioPortal and GSE84437 R package survival3 was used. KM analysis was based on individual gene expression value and survival data. By using the median expression value of a query gene in a certain sample group as a cutoff, the samples were split into high and low expression groups with the expression level of the query gene not less than and less than the cutoff. The Cox proportional hazard model was built by using R package survival, fitted with two genes’ expression values for OS or disease free survival (DFS). Similar to the individual gene analysis, the median value of weighted expression value (shortened as WEV) of a gene combination in a certain cohort were used as a group cutoff, where WEV was calculated as the sum of cox-regression coefficient weighted expression value of each gene involved in the combination. Log rank p-values, cox proportional hazard ratios (HRs), and HR p-values were calculated to compare the survival between two groups split by the median value of gene expression or WEV. A p-value of less than 0.05 and HR greater than 1.05 or less than 0.95 suggest statistical significance of the survival difference between high and low groups, which indicates the corresponding gene or gene combination has a prognostic potential.
Gene Co-expression Analysis and Enrichment Analysis
Gene co-expression analysis was carried out using webserver cBioPortal, and the top 25 positively correlated and top 25 negatively correlated genes were selected according to Spearman correlation coefficients, which were taken together and simplified as “top 50 correlated genes” in our results. Here, correlated genes met two criteria: the absolute value of Spearman correlation coefficient is greater than 0.25, and the p-value is less than 0.01. Gene set enrichment analysis (GSEA) was performed by using R package clusterProfiler (Yu et al., 2012). The pathways enriched for GO (Gene Ontology) (Ashburner et al., 2000; The Gene Ontology Consortium, 2019) were plotted based on the negative logarithm of p-value.
Results
MUC15, 13, and 21 Display Prognostic Potential for Digestive Cancer on TCGA
Aiming to assess the prognostic potentials of every MUC gene, KM survival analysis was applied to TCGA digestive cancer samples as a whole involving COAD, ESCA, LIHC, STAD, and PAAD by using the webserver GEPIA 1.0 (Tang et al., 2017). Among the 14 MUC family members with expression data available, the expression levels of MUC1, MUC5AC, MUC6, OVGP1 (MUC9), MUC13, EMCN (MUC14), MUC15, MUC16, MUC17, and MUC21 individually exhibited significant correlations with OS, with HR p-values less than 0.05 and HR greater than 1.05 or less than 0.95; similarly, MUC2, MUC3A, MUC12, MUC13, MUC15, MUC17, MUC20, and MUC21 were significantly correlated with DFS (Table 1 and Supplementary Figure S1). MUC13, MUC15, MUC17, and MUC21 were significant for both OS and DFS, among which MUC15 performed best for OS correlation and the second best for DFS correlation. In comparison, MUC13 displayed the best performance in DFS analysis, while ranked relatively lower (9th) in OS analysis; MUC21 ranked 3rd for OS, and 8th for DFS (Table 1 and Supplementary Figure S1). These indicate that MUC15 represents a promising candidate for developing strategies for prognosis prediction for digestive cancers.
TABLE 1.
Survival analysis of TCGA digestive cancer samples for prognostic potentials of MUC family genes.
| Gene | HR p-value for OS | OS p-value rank | HR p-value for DFS | DFS p-value rank |
| MUC1 | 1.3E−05 | 6 | 0.23 | 11 |
| MUC2 | 0.69 | 14 | 5.2E−08 | 3 |
| MUC3A | 0.49 | 12 | 1.7E−06 | 4 |
| MUC5AC | 7.9E−06 | 5 | 0.74 | 14 |
| MUC6 | 2.7E−07 | 3 | 0.41 | 12 |
| OVGP1 (MUC9) | 0.0021 | 7 | 0.094 | 10 |
| MUC12 | 0.58 | 13 | 0.00012 | 5 |
| MUC13 | 0.032 | 9 | 2.1E−08 | 1 |
| EMCN (MUC14) | 0.044 | 10 | 0.71 | 13 |
| MUC15 | 1.7E−09 | 1 | 3.6E−08 | 2 |
| MUC16 | 6.4E−09 | 2 | 0.059 | 9 |
| MUC17 | 0.0053 | 8 | 0.00051 | 6 |
| MUC20 | 0.36 | 11 | 0.01 | 7 |
| MUC21 | 9.8E−07 | 4 | 0.01 | 8 |
OS stands for overall survival and DFS stands for disease free survival (DFS). The p-values less than 0.05 are displayed in bold.
MUC14 (EMCN) and 15 Display Prognostic Potential for Gastric Cancer on TCGA-STAD
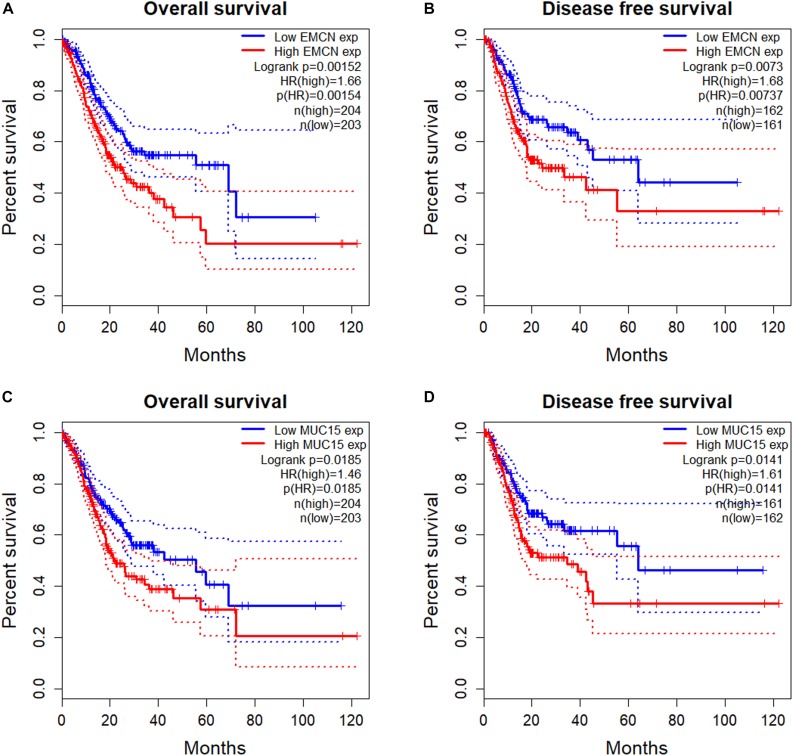
To investigate the prognostic potentials of MUC family genes for STAD, we performed KM survival analysis exclusively on STAD samples from TCGA with R package survival. It was found that the expression levels of EMCN (MUC14) and MUC15 individually showed significant correlations with both OS and DFS, and MCAM (MUC18) was significant only with OS (Table 2). KM survival plots, together with log rank p-values, cox proportional HRs, and HR p-values summarized in Figure 1 indicated that EMCN performed better than MUC15 in both OS and DFS analyses. Overall, EMCN and MUC15 could be potential biomarkers for STAD prognosis.
TABLE 2.
Survival analysis of TCGA STAD samples for prognostic potentials of MUC family genes.
| Gene | HR p-value for OS | HR p-value for DFS |
| MUC1 | 0.654 | 0.591 |
| MUC2 | 0.129 | 0.364 |
| MUC4 | 0.9 | 0.203 |
| MUC5B | 0.441 | 0.753 |
| MUC6 | 0.67 | 0.0854 |
| OVGP1 (MUC9) | 0.662 | 0.925 |
| MUC12 | 0.957 | 0.637 |
| MUC13 | 0.0511 | 0.234 |
| EMCN (MUC14) | 0.00154 | 0.00737 |
| MUC15 | 0.0185 | 0.0141 |
| MUC16 | 0.825 | 0.0975 |
| MUC17 | 0.145 | 0.406 |
| MCAM (MUC18) | 0.0167 | 0.323 |
| MUC20 | 0.891 | 0.62 |
| MUC21 | 0.224 | 0.745 |
OS stands for overall survival and DFS stands for disease free survival. The p-values less than 0.05 are displayed in bold.
FIGURE 1.
Survival analysis of TCGA STAD samples for prognostic potentials of EMCN (MUC14) and MUC15. (A) Overall Survival (OS) of EMCN. (B) Disease Free Survival (DFS) of EMCN. (C) Overall Survival of MUC15. (D) Disease Free Survival of MUC15. Log rank p-values, hazard ratios (HRs) and hazard ratio p-values were calculated. The 95% confidence intervals for survival time were shown in as dotted lines in the Kaplan–Meier (KM) survival plot.
EMCN/MUC15 Combination Could Serve as Prognostic Signature for Gastric Cancer
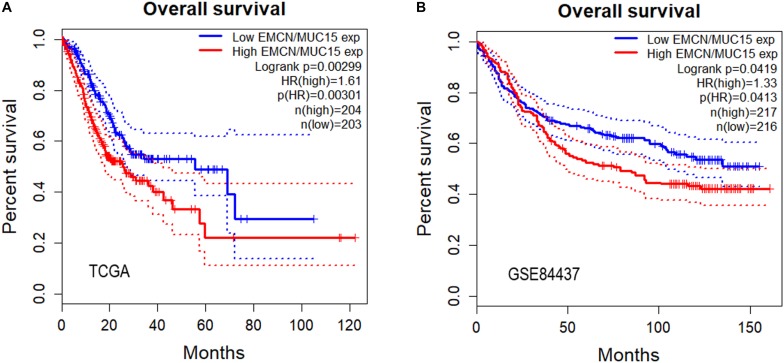
So far we have observed that high expressions of both EMCN and MUC15 were associated with poor prognosis in GC, and that EMCN and MUC15 displayed the strongest correlation to survival for GC and digestive cancers, respectively (Table 2 and Figure 1). Thus, we set out to investigate whether EMCN/MUC15 combination could be a prognostic signature for GC. Cox proportional hazards regression analysis was performed based on the two genes’ expression values and OS data derived from TCGA STAD dataset. As expected, the expression of EMCN/MUC15 combination exhibited significant correlation with OS, with log rank p-value of 0.00299 and HR p-value of 0.00301 (Figure 2A).
FIGURE 2.
Overall survival analyses of gastric cancer (GC) samples from TCGA STAD (A) and GSE84437 (B) for predictive power of EMCN/MUC15 signature. Log rank p-values, hazard ratios (HRs) and hazard ratio p-values were calculated. The 95% confidence intervals for survival time were shown as dotted lines in the Kaplan–Meier survival plot.
We then separately tested the prognostic prediction power of EMCN, MUC15 and their combination on an independent dataset, GSE84437, which involved 433 GC samples. Again, significant results of EMCN/MUC15 combination (HR = 1.33) were obtained with log rank p-value being 0.0419 and HR p-value being 0.0413 (Figure 2B); while one single gene, EMCN (HR p-value of 0.0807, HR = 1.27) or MUC15 (HR p-value of 0.156, HR = 0.82), had no significant prognostic prediction power, as shown in Supplementary Figure S2. We therefore proposed that EMCN/MUC15 combination could be a potential prognostic signature for GC.
EMCN/MUC15 Correlated Genes Are Functionally Enriched in Cancer Related Processes
By using webserver cBioPortal, the top 50 EMCN- (Table 3) or MUC15- (Table 4) correlated genes were identified based on mRNA expression data of TCGA STAD samples, including the top 25 positively correlated genes and top 25 negatively correlated genes. It is noticeable that there is no intersection between the two top 50 gene lists at all and no co-expression between EMCN and MUC15 (Spearman’s Correlation of 0.0264 with p-value of 0.592) either, implying the functional complementarity between EMCN and MUC15 and thus the rationality of the combination of the two genes in predicting prognosis for GC.
TABLE 3.
Top 50 genes correlated with EMCN based on TCGA STAD dataset.
| Correlated gene | Cytoband | Spearman correlation | p-value |
| CYYR1 | 21q21.3 | 0.931414 | 2.19E−183 |
| MYCT1 | 6q25.2 | 0.929044 | 1.90E−180 |
| ERG | 21q22.2 | 0.894179 | 3.19E−146 |
| DIPK2B | Xp11.3 | 0.887525 | 4.57E−141 |
| ADGRL4 | 1p31.1 | 0.886757 | 1.71E−140 |
| CD34 | 1q32.2 | 0.880383 | 6.99E−136 |
| TEK | 9p21.2 | 0.873397 | 4.03E−131 |
| PECAM1 | 17q23.3 | 0.871639 | 5.73E−130 |
| S1PR1 | 1p21.2 | 0.870224 | 4.72E−129 |
| LDB2 | 4p15.32 | 0.860092 | 8.59E−123 |
| RHOJ | 14q23.2 | 0.859913 | 1.10E−122 |
| CLEC14A | 14q21.1 | 0.854201 | 2.25E−119 |
| GNG11 | 7q21.3 | 0.853027 | 1.03E−118 |
| EBF1 | 5q33.3 | 0.846286 | 5.16E−115 |
| MMRN2 | 10q23.2 | 0.846005 | 7.29E−115 |
| CLEC1A | 12p13.2 | 0.843416 | 1.71E−113 |
| CALCRL | 2q32.1 | 0.841594 | 1.53E−112 |
| LRRC70 | 5q12.1 | 0.84015 | 8.47E−112 |
| MEF2C | 5q14.3 | 0.839354 | 2.16E−111 |
| ARHGEF15 | 17p13.1 | 0.836065 | 9.86E−110 |
| CDH5 | 16q21 | 0.828483 | 4.80E−106 |
| PALMD | 1p21.2 | 0.828283 | 5.97E−106 |
| SHE | 1q21.3 | 0.826792 | 3.01E−105 |
| SPARCL1 | 4q22.1 | 0.823121 | 1.52E−103 |
| JAM2 | 21q21.3 | 0.821442 | 8.85E−103 |
| RAD54L | 1p34.1 | –0.53926 | 1.10E−32 |
| CDCA5 | 11q13.1 | –0.53612 | 2.96E−32 |
| PKP3 | 11p15.5 | –0.53108 | 1.41E−31 |
| CDCA8 | 1p34.3 | –0.5303 | 1.79E−31 |
| ZWINT | 10q21.1 | –0.52817 | 3.44E−31 |
| KIF2C | 1p34.1 | –0.52339 | 1.46E−30 |
| HJURP | 2q37.1 | –0.51982 | 4.21E−30 |
| MCM2 | 3q21.3 | –0.51829 | 6.63E−30 |
| CDT1 | 16q24.3 | –0.51369 | 2.54E−29 |
| MYO19 | 17q12 | –0.51058 | 6.24E−29 |
| TONSL | 8q24.3 | –0.50684 | 1.82E−28 |
| CCNA2 | 4q27 | –0.5056 | 2.58E−28 |
| NCAPH | 2q11.2 | –0.5018 | 7.48E−28 |
| POC1A | 3p21.2 | –0.50165 | 7.81E−28 |
| NELFA | 4p16.3 | –0.50116 | 8.95E−28 |
| UBE2T | 1q32.1 | –0.50026 | 1.15E−27 |
| POLD2 | 7p13 | –0.49997 | 1.25E−27 |
| DTL | 1q32.3 | –0.49967 | 1.35E−27 |
| PTBP1 | 19p13.3 | –0.49959 | 1.38E−27 |
| CNOT11 | 2q11.2 | –0.49871 | 1.76E−27 |
| STIP1 | 11q13.1 | –0.49718 | 2.69E−27 |
| MAP7 | 6q23.3 | –0.49631 | 3.41E−27 |
| ESPL1 | 12q13.13 | –0.49591 | 3.81E−27 |
| TBRG4 | 7p13 | –0.49548 | 4.29E−27 |
| CDC25A | 3p21.31 | –0.49474 | 5.24E−27 |
Genes mentioned in Discussion section are highlighted in bold and italic.
TABLE 4.
Top 50 genes correlated with MUC15 based on TCGA STAD dataset.
| Correlated gene | Cytoband | Spearman correlation | p-value |
| ANO3 | 11p14.3-p14.2 | 0.558879 | 1.82E−35 |
| FSTL4 | 5q31.1 | 0.4959 | 3.82E−27 |
| TMPRSS13 | 11q23.3 | 0.469609 | 3.76E−24 |
| ZNF750 | 17q25.3 | 0.464898 | 1.21E−23 |
| LGALS7 | 19q13.2 | 0.454428 | 1.54E−22 |
| NCCRP1 | 19q13.2 | 0.452369 | 2.52E−22 |
| PCLO | 7q21.11 | 0.449054 | 5.50E−22 |
| GABRA3 | Xq28 | 0.446711 | 9.51E−22 |
| DLX3 | 17q21.33 | 0.443637 | 1.94E−21 |
| LIN28B | 6q16.3-q21 | 0.440243 | 4.21E−21 |
| ADGRV1 | 5q14.3 | 0.439028 | 5.55E−21 |
| USH1G | 17q25.1 | 0.436641 | 9.52E−21 |
| C12ORF56 | 12q14.2 | 0.429849 | 4.32E−20 |
| RSPO4 | 20p13 | 0.428819 | 5.41E−20 |
| SPAG17 | 1p12 | 0.425992 | 1.00E−19 |
| MARK1 | 1q41 | 0.424353 | 1.43E−19 |
| HTR2C | Xq23 | 0.423044 | 1.90E−19 |
| CT45A5 | Xq26.3 | 0.420712 | 3.13E−19 |
| PRPF40B | 12q13.12 | 0.419994 | 3.64E−19 |
| C3ORF67 | 3p14.2 | 0.419376 | 4.16E−19 |
| RIPPLY3 | 21q22.13 | 0.417437 | 6.27E−19 |
| CNGB3 | 8q21.3 | 0.417398 | 6.32E−19 |
| ATP6V0A4 | 7q34 | 0.413452 | 1.45E−18 |
| LINC00964 | 8q24.13 | 0.412548 | 1.74E−18 |
| VGLL1 | Xq26.3 | 0.409463 | 3.30E−18 |
| MCUB | 4q25 | –0.35985 | 3.92E−14 |
| FAS | 10q23.31 | –0.32779 | 7.51E−12 |
| IRF1 | 5q31.1 | –0.32732 | 8.08E−12 |
| ZIC2 | 13q32.3 | –0.31402 | 5.99E−11 |
| CDC42SE2 | 5q31.1 | –0.31243 | 7.55E−11 |
| HK3 | 5q35.2 | –0.30198 | 3.37E−10 |
| NUB1 | 7q36.1 | –0.30007 | 4.41E−10 |
| GBP4 | 1p22.2 | –0.29733 | 6.45E−10 |
| BBC3 | 19q13.32 | –0.29722 | 6.55E−10 |
| AIM2 | 1q23.1-q23.2 | –0.29707 | 6.68E−10 |
| NLRC5 | 16q13 | –0.29669 | 7.04E−10 |
| MAX | 14q23.3 | –0.29642 | 7.30E−10 |
| MTHFD1 | 14q23.3 | –0.29437 | 9.67E−10 |
| AGAP2 | 12q14.1 | –0.29096 | 1.54E−09 |
| IFNG | 12q15 | –0.29068 | 1.59E−09 |
| RASSF1 | 3p21.31 | –0.28787 | 2.32E−09 |
| GZMA | 5q11.2 | –0.28696 | 2.62E−09 |
| CCL4 | 17q12 | –0.28515 | 3.32E−09 |
| MAT2B | 5q34 | –0.28231 | 4.82E−09 |
| FCGR3A | 1q23.3 | –0.28226 | 4.85E−09 |
| THG1L | 5q33.3 | –0.28207 | 4.97E−09 |
| TK2 | 16q21 | –0.28202 | 5.01E−09 |
| PRKX | Xp22.33 | –0.27772 | 8.71E−09 |
| JAK2 | 9p24.1 | –0.27752 | 8.94E−09 |
| EEF2 | 19p13.3 | –0.2774 | 9.07E−09 |
Genes mentioned in Discussion section are highlighted in bold and italic.
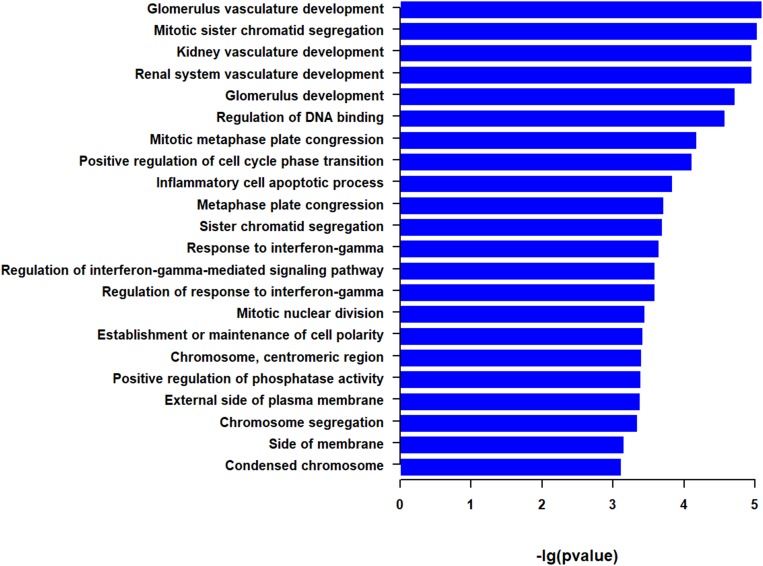
We then performed functional enrichment analysis with the two top 50 correlated genes as a whole. GSEA identified a total of 22 GO terms (Figure 3 and Supplementary Table S1). Among them, the most significant pathways were associated with vasculature development, such as glomerulus vasculature development and renal system vasculature development. Some enriched pathways are associated with mitosis, such as mitotic sister chromatid segregation and mitotic metaphase plate congression. Some pathways were associated with immunity, such as inflammatory cell apoptotic process and response to interferon-gamma. The other enriched pathways were involved in DNA binding, cell cycle phase transition, cell polarity, phosphatase activity, and side of plasma membrane (Figure 3 and Supplementary Table S1). These indicate that genes correlated with EMCN and MUC15 in GC tend to be enriched in cancer related processes, such as vasculature development, mitosis, and immunity.
FIGURE 3.
Go terms derived from gene set enrichment analysis (GSEA) for top 50 EMCN- and MUC15-correlated genes. The pathways are ranked by -log p-value. The 95% confidence intervals for survival time were shown as dotted lines in the Kaplan–Meier survival plot.
Discussion
In the present study, by systematically analyzing mRNA expression and clinical data of TCGA digestive cancer samples and GEO GC samples, we propose MUC15 as a promising candidate for prognosis prediction of digestive cancers, and EMCN/MUC15 combination as a potential prognostic signature for GC.
Gene signature identification is essentially a process of dimension reduction of high dimensional data. On one hand, a signature involving less features or genes obviously has more practicality; on the other hand, a signature is also expected to have sufficient interpretability, although it is far from achieved. In this sense, a good signature is supposed to consist of orthogonal or mutually exclusive features which are able to hold a testable hypothesis from a systematic viewpoint while also sustaining the robustness and reliability of the signature. However, most current efforts in this field focus on reducing dimension over enhancing explanatory power of the signature. In our work, although EMCN and MUC15 coding genes belong to the same gene family, it is noted that there is no expression correlation between the two genes and no intersection between their top 50 correlated genes, implying the orthogonality and functional complementarity between EMCN and MUC15. As we expected, the combination of EMCN/MUC15 shows more robust prognostic power than the individual genes in GC according to the testing result implemented on an independent dataset GSE84437. These observations not only support the rationality of the combination of the two genes in predicting prognosis, but also indicate the explanatory power of EMCN/MUC15 signature, which is supposed to play an important role in the robustness improvement.
EMCN, i.e. MUC14, encodes a membrane-bound protein, endothelial sialomucin or mucin-like sialo glycoprotein, which was reported to inhibit cell and extracellular matrix interaction, interfere with leukocyte-endothelial cell adhesion, and even promote the peritoneal metastasis process of GC cells (Liu et al., 2001; Zahr et al., 2016; Dhanisha et al., 2018; Bao et al., 2019). Among the 22 enriched functions for top 50 EMCN-correlated genes and top 50 MUC15-correlated genes, the most significant one is glomerulus vasculature development that is associated with four EMCN/MUC15 correlated genes including CD34, TEK, PECAM1, and IFNG (Tables 3, 4 and Supplementary Table S1). After carefully checking functional annotations of the four genes, we focused on two cancer relevant genes, CD34 and PECAM1. Both genes are significantly coexpressed with EMCN with correlation coefficients of 0.880 and 0.871, respectively (Table 3). CD34, a marker of vascular endothelial cells, is capable of supporting cell adhesion by increasing surface expression (Nielsen and McNagny, 2008). PECAM1, also known as CD31, encodes platelet endothelial cell adhesion molecule 1 that is necessary for leukocyte transendothelial migration (TEM) (Dasgupta et al., 2009). It is noteworthy that EMCN/COL4A5/CCL11 combination was very recently reported as a prognostic signature for diffuse type GC (Bao et al., 2019). In our study, among MUC family members, EMCN exhibits the strongest correlation with survival for GC. Taken together, EMCN may play crucial roles in tumorigenesis and progression of GC via cell adhesion and TEM of lymphocytes.
MUC15 also encodes a membrane-bound protein, which could promote cell proliferation, cell-extracellular matrix adhesion, colony forming ability, and invasion in colon cancer cells (Huang et al., 2009). Its overexpression is significantly correlated with diverse cancers (Pallesen et al., 2002; Shyu et al., 2007; Huang et al., 2009; Nam et al., 2011; Wang et al., 2013; Choi et al., 2018). However, it was also found that the expression of MUC15 decreased in hepatocellular carcinoma cells and negatively regulated metastasis of hepatocellular carcinoma (Wang et al., 2013). This suggests that MUC15 may perform diverse functions in tumorigenesis and progression. In our study, MUC15 displays the strongest correlation among the MUC family with survival for digestive cancers and MUC15 overexpression seems to be a promising candidate for a prognosis biomarker of digestive cancers. Combined with EMCN, the two genes provide a potential prognostic signature for GC and show more robustness in the prognostic prediction power than individual genes. As far as we know, the association of MUC15 with GC is rarely reported.
In summary, we propose EMCN/MUC15 combination as a prognostic signature with mechanistic interpretability. It not only possesses prognostic capability for GC, but also offers clues for further exploring systematic mechanisms of carcinogenesis of GC and other digestive cancers.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: TCGA-STAD, TCGA-COAD, TCGA-ESCA, TCGA-LIHC, TCGA-PAAD, and GSE84437.
Author Contributions
Y-YL and WD designed the study. WD and JL implemented the data analysis. BL, QL, and QS provided the valuable suggestions. JL and WD drafted the manuscript. Y-YL revised the manuscript and coordinated the study. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The results shown here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Funding. This work was supported by the grants from the National Key R&D Program of China (2018YFC0910500), the National Natural Science Foundation of China (81672736 and 31600750), the Shanghai Municipal Science and Technology Major Project (2017SHZDZX01 and 18DZ2294200), and the NIH CPTAC (Cancer Proteomic Tumor Analysis Consortium) program.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2020.00019/full#supplementary-material
References
- Aithal A., Rauth S., Kshirsagar P., Shah A., Lakshmanan I., Junker W. M., et al. (2018). MUC16 as a novel target for cancer therapy. Expert Opin. Ther. Targets 22 675–686. 10.1080/14728222.2018.1498845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., et al. (2000). Gene Ontology: tool for the unification of biology. Nat. Genet. 25 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao B., Zheng C., Yang B., Jin Y., Hou K., Li Z., et al. (2019). Identification of subtype-specific three-gene signature for prognostic prediction in diffuse type gastric cancer. Front. Oncol. 9:1243. 10.3389/fonc.2019.01243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T., Wilhite S. E., Ledoux P., Evangelista C., Kim I. F., Tomashevsky M., et al. (2012). NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Res. 41 D991–D995. 10.1093/nar/gks1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68 394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data: figure 1. Cancer Discov. 2 401–404. 10.1158/2159-8290.cd-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Zheng R., Zhang S., Zhao P., Zeng H., Zou X. (2014). Report of cancer incidence and mortality in China, 2010. Ann. Transl. Med. 2:61. 10.3978/j.issn.2305-5839.2014.04.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C., Thi Thao Tran N., Van Ngu T., Park S. W., Song M. S., Kim S. H., et al. (2018). Promotion of tumor progression and cancer stemness by MUC15 in thyroid cancer via the GPCR/ERK and integrin-FAK signaling pathways. Oncogenesis 7:85. 10.1038/s41389-018-0094-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corfield A. P. (2015). Mucins: a biologically relevant glycan barrier in mucosal protection. Biochim. Biophys. Acta 1850 236–252. 10.1016/j.bbagen.2014.05.003 [DOI] [PubMed] [Google Scholar]
- Dasgupta B., Dufour E., Mamdouh Z., Muller W. A. (2009). A novel and critical role for tyrosine 663 in platelet endothelial cell adhesion molecule-1 trafficking and transendothelial migration. J. Immunol. 182 5041–5051. 10.4049/jimmunol.0803192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J., Rossen J. W. A., Büller H. A., Einerhand A. W. C. (2002). The MUC family: an obituary. Trends Biochem. Sci. 27 126–131. 10.1016/s0968-0004(01)02052-2057 [DOI] [PubMed] [Google Scholar]
- Dhanisha S. S., Guruvayoorappan C., Drishya S., Abeesh P. (2018). Mucins: structural diversity, biosynthesis, its role in pathogenesis and as possible therapeutic targets. Crit. Rev. Oncol. 122 98–122. 10.1016/j.critrevonc.2017.12.006 [DOI] [PubMed] [Google Scholar]
- Gao J., Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., Sumer S. O., et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6:l1. 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Che M. I., Huang Y. T., Shyu M. K., Huang Y. M., Wu Y. M., et al. (2009). Overexpression of MUC15 activates extracellular signal-regulated kinase 1/2 and promotes the oncogenic potential of human colon cancer cells. Carcinogenesis 30 1452–1458. 10.1093/carcin/bgp137 [DOI] [PubMed] [Google Scholar]
- Jonckheere N., Van Seuningen I. (2018). Integrative analysis of the cancer genome atlas and cancer cell lines encyclopedia large-scale genomic databases: MUC4/MUC16/MUC20 signature is associated with poor survival in human carcinomas. J. Transl. Med. 16:259. 10.1186/s12967-018-1632-1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S., Kumar S., Momi N., Sasson A. R., Batra S. K. (2013). Mucins in pancreatic cancer and its microenvironment. Nat. Rev. Gastroenterol. Hepatol. 10 607–620. 10.1038/nrgastro.2013.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Shao Z.-M., Zhang L., Beatty P., Sartippour M., Lane T., et al. (2001). Human endomucin is an endothelial marker. Biochem. Biophys. Res. Commun. 288 129–136. 10.1006/bbrc.2001.5737 [DOI] [PubMed] [Google Scholar]
- Nam K.-H., Noh T.-W., Chung S.-H., Lee S. H., Lee M. K., Won Hong S., et al. (2011). Expression of the membrane mucins MUC4 and MUC15, potential markers of malignancy and prognosis, in papillary thyroid carcinoma. Thyroid 21 745–750. 10.1089/thy.2010.0339 [DOI] [PubMed] [Google Scholar]
- Nielsen J. S., McNagny K. M. (2008). Novel functions of the CD34 family. J. Cell Sci. 121 4145–4145. 10.1242/jcs.03504 [DOI] [PubMed] [Google Scholar]
- Pallesen L. T., Berglund L., Rasmussen L. K., Petersen T. E., Rasmussen J. T. (2002). Isolation and characterization of MUC15, a novel cell membrane-associated mucin. Eur. J. Biochem. 269 2755–2763. 10.1046/j.1432-1033.2002.02949.x [DOI] [PubMed] [Google Scholar]
- Shyu M. K., Lin M. C., Shih J. C., Lee C. N., Huang J., Liao C. H., et al. (2007). Mucin 15 is expressed in human placenta and suppresses invasion of trophoblast-like cells in vitro. Hum. Reprod. 22 2723–2732. 10.1093/humrep/dem249 [DOI] [PubMed] [Google Scholar]
- Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. (2017). GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45 W98–W102. 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium (2019). The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 47 D330–D338. 10.1093/nar/gky1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cutsem E., Sagaert X., Topal B., Haustermans K., Prenen H. (2016). Gastric cancer. Lancet 388 2654–2664. 10.1016/s0140-6736(16)30354-30353 [DOI] [PubMed] [Google Scholar]
- Wang R. Y., Chen L., Chen H. Y., Hu L., Li L., Sun H. Y., et al. (2013). MUC15 inhibits dimerization of EGFR and PI3K–AKT signaling and is associated with aggressive hepatocellular carcinomas in patients. Gastroenterology 145 1436–1448.e1-12. 10.1053/j.gastro.2013.08.009 [DOI] [PubMed] [Google Scholar]
- Weinstein J. N., Collisson E. A., Mills G. B., Shaw K. R. M., Ozenberger B. A., Ellrott K., et al. (2013). The cancer genome atlas pan-cancer analysis project. Nat. Genet. 45 1113–1120. 10.1038/ng.2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa S., Higashi M., Yamada N., Yokoyama S., Kitamoto S., Kitajima S., et al. (2011). Mucins in human neoplasms: clinical pathology, gene expression and diagnostic application. Pathol. Intern. 61 697–716. 10.1111/j.1440-1827.2011.02734.x [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L.-G., Han Y., He Q.-Y. (2012). clusterProfiler: an r package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 16 284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahr A., Alcaide P., Yang J., Jones A., Gregory M., dela Paz N. G., et al. (2016). Endomucin prevents leukocyte–endothelial cell adhesion and has a critical role under resting and inflammatory conditions. Nat. Commun. 7:10363. 10.1038/ncomms10363 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: TCGA-STAD, TCGA-COAD, TCGA-ESCA, TCGA-LIHC, TCGA-PAAD, and GSE84437.