To the Editor:
Tuberculosis (TB), the disease caused by Mycobacterium tuberculosis (Mtb), remains a leading cause of morbidity and mortality. Current tools to identify Mtb-infected individuals, specifically interferon-γ release assays (IGRAs) and the tuberculin skin test (TST), cannot distinguish between asymptomatic Mtb-infected individuals (latent Mtb infection (LTBI)) and those with TB [1]. Advancement of TB diagnostics and their application in TB-endemic settings requires an assay that distinguishes between individuals with LTBI and TB. In this pilot study, we compared the ability of three CD8+ T-cell-based assays to distinguish Ugandan adults with: confirmed pulmonary TB (HIV-uninfected and HIV-infected), LTBI (HIV-uninfected only), and those who are TST-negative (HIV-uninfected only).
Participants were recruited from the National TB Treatment Center at Mulago Hospital, hospital staff and the community surrounding Kampala, Uganda, between 2001 and 2014, into four cohorts: culture-confirmed pulmonary TB/HIV-uninfected (TB cohort); culture-confirmed pulmonary TB/HIV-infected (TB/HIV cohort; CD4 count >150 cells·μL−1); LTBI/HIV-uninfected (LTBI cohort; TST ≽10 mm); and TST negative/HIV-uninfected (TST-negative cohort; TST<10 mm). Assignment to the LTBI and TST-negative cohorts was based on TST result and absence of signs or symptoms of TB (prolonged cough, haemoptysis, fever, weight loss and night sweats). This study received Institutional Review Board approval from all sponsoring institutions and written, informed consent was obtained from all participants.
A total of 170 participants were enrolled: 43 TB cohort, 42 TB/HIV cohort, 46 LTBI cohort, and 39 TST-negative cohort. Whole blood was drawn and peripheral blood mononuclear cells (PBMC) isolated and cryopreserved at the time of cohort assignment, and prior to TB treatment (if indicated). Three assays were used to quantify Mtb-specific CD8+ T-cells: intracellular cytokine staining (ICS) assay to measure CD8+ T-cell interferon-γ (IFN-γ), tumour necrosis factor-α (TNF-α), and interleukin-2 (IL-2) production in response to ESAT6 and CFP10 peptide pools (ESAT6/CFP10 ICS assay); ICS assay to measure CD8+ T-cell IFN-γ, TNF-α, and IL-2 production in response to a pool of Mtb-specific epitopes (epitope-pool ICS assay); and a flow cytometry assay to measure CD8+ T-cell binding to a tetramer-pool (tetramer-pool assay).
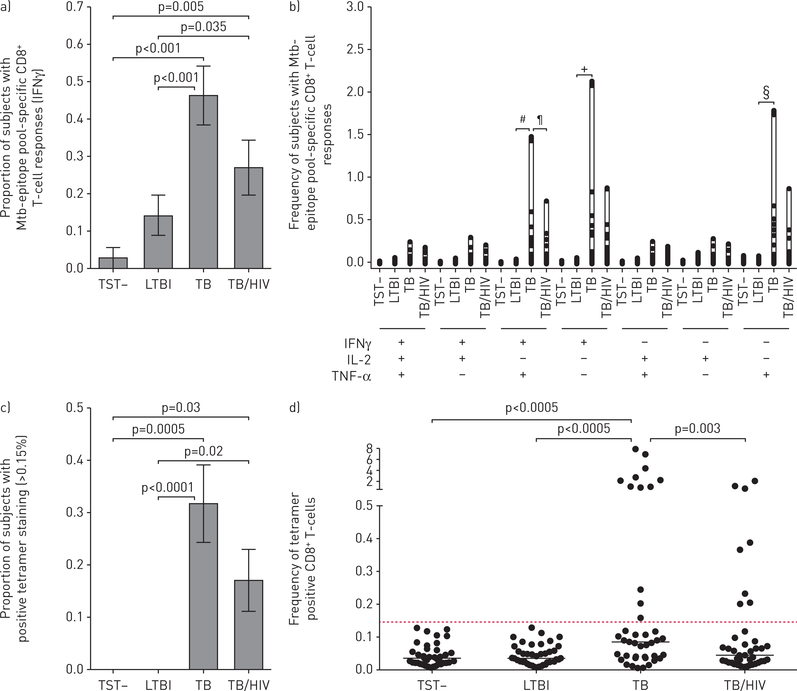
We hypothesised that eight minimal epitopes from four Mtb-specific genes, previously identified and validated by our group to be recognised by CD8+ T-cells from Mtb-infected individuals, would elicit strong CD8+ T-cell responses from individuals with confirmed TB regardless of their HIV status, when pooled for use in an immune-based assay [2–4]. The gene, location and restriction alleles are: CFP102–9/HLAB45, CFP103–11/HLAB08, CFP102–11/HLAB44, CFP1049–58/HLAB35, CFP1075–83/HLAB15, Mtb8.433–41/HLAB15, EsxG3–11/HLAA02 and EsxJ24–34/HLAB57. To test this hypothesis, we used this pool of eight epitopes in our ICS epitope-pool assay. We observed significant differences in the proportion of individuals with a positive IFN-γ response to the epitope-pool among cohorts (figure 1a). When comparing the magnitude and pattern of cytokine production, we also observed significant differences in the frequency of CD8+ T-cells productive of either IFN-γ or TNF-α in the TB as compared with the LTBI cohort (figure 1b). Individuals in the TB cohort were also more likely to have CD8+ T-cells productive of both IFN-γ and TNF-α than individuals with LTBI, and those with TB/HIV (figure 1b). These findings contrast to those obtained in our ESAT6/CFP10 ICS assay, where there were no significant differences in the proportion of individuals with positive cytokine responses among the TB, TB/HIV and LTBI cohorts (68.3%, 61% and 64.3%, respectively).
FIGURE 1.
CD8+ T-cell cytokine and tetramer responses to Mycobacterium tuberculosis (Mtb)-epitope pool distinguishes latent tuberculosis infection (LTBI) from tuberculosis (TB). a) The proportion of subjects with detectable CD8+ T-cell responses to a pool of eight Mtb-specific epitopes, as measured by interferon (IFN)-γ production in an intracellular cytokine staining (ICS) assay is shown (error bars indicate SEM). A positive response is defined as having at least 0.05% of CD8+ T-cells expressing IFN-γ after subtraction of background (no stimulation) responses. The proportion of positive responses between groups were compared using Chi-squared tests. b) The frequency of Mtb-specific CD8+ T-cells based on their production of either IFN-γ, tumour necrosis factor (TNF)-α and/or interleukin (IL)-2 is shown. The combinations of different cytokine functions are shown on the x-axis, with the frequency of distinct cytokine-producing cells within the Mtb-epitope pool responsive CD8+ T-cell compartment on the y-axis. The frequency of Mtb-specific CD8+ T-cells productive of each cytokine were compared using Kruskal–Wallis test, following adjustment for multiple comparisons using the Dwass, Steel, and Critchlow-Fligner method. c) The proportion of subjects with detectable CD8+ T-cell binding to a pool of eight Mtb-specific tetramers, as measured by flow cytometry, is shown (error bars indicate SEM). A positive response is defined as having at least 0.15% of CD8+ T-cells staining positive for tetramer. Statistical analysis with Chi-squared followed by pairwise comparison (Bonferroni adjustment). d) The frequencies of CD8+tetramer+ T-cells in different cohorts are shown. Statistical analysis with Kruskal–Wallis test followed by pairwise comparison (Bonferroni adjustment). Statistical Analysis System v9.4 (SAS Institute Inc., Cary, NC, USA) was used for analysis. #: p=0.02; ¶: p=0.0009; +: p=0.012; §: p=0.006.
We sought to determine if a tetramer-based assay could distinguish individuals with confirmed TB (TB and TB/HIV cohorts) from individuals without TB disease (LTBI and TST-negative cohorts). To investigate, we used the identical pool of eight epitopes in a tetramer-pool assay. Significant differences in the proportion of CD8+ T-cells that bound the tetramer-pool between cohorts were observed (figure 1c). The frequencies of CD8+ T-cell tetramer-pool binding were also significantly different between TB and LTBI, and TB and TB/HIV cohorts (figure 1d). Spearman’s correlation assessed associations between donor responses to the tetramer-pool and epitope-pool ICS assays; however, correlation between production of either cytokine and tetramer-pool binding was low (rs=0.349 for IFN-γ and rs=0.368 for TNF-α; p<0.001 both comparisons). We identified two TB/HIV co-infected individuals who bound the tetramer-pool, but did not produce IFN-γ or TNF-α in the epitope-pool ICS assay, and upon further investigation noted that CD8+tetramer+ T-cells from these discordant individuals demonstrated high expression (11.4% and 6.6%) of the exhaustion marker PD-1 (median CD8+tetramer+PD-1+ T-cells from TB/HIV donors was 1.99%; interquartile range 1.04–10.2%). Conversely, 11 individuals (five TB cohort; four HIV/TB cohort; and two LTBI cohort) who produced IFN-γ in response to the epitope-pool failed to bind the tetramer-pool. We suspect that human leukocyte antigen (HLA)-mismatching between these donors and the HLA-restricting alleles used to generate the tetramers limited the strength of the relationship between epitope-pool ICS and tetramer-pool assays.
Several groups have used Mtb-specific CD8+ T-cell functional or phenotypic responses to distinguish individuals with LTBI from those with TB [5–11]. Unlike some prior reports, we could not detect significant differences in the proportion of CD8+ T-cells that produced pro-inflammatory cytokines in response to ESAT6/CFP10 peptide pools between individuals with LTBI and TB [6, 9, 10]. However, when using an epitope-pool as a stimulus in our ICS assay, CD8+ T-cell cytokine responses were significantly more common among adults with TB compared to those with LTBI. Activation of Mtb-specific CD4+ T-cells can lead to indirect activation of CD8+ T-cells, as has been shown in a murine model of TB [12]. We hypothesise that the use of minimal epitopes limited amplification of Mtb-specific CD4+ T-cell responses in donors with LTBI, and only elicited cytokine production from Mtb-specific CD8+ T-cells in individuals with TB who had a high bacillary burden. Thus, the use of a pool of minimal epitopes in an ICS assay provided a gain in specificity for individuals with TB.
Our CD8+ T-cell tetramer-pool assay was also highly specific for individuals with TB. Tetramer-based assays have several technical advantages compared to ICS-based assays, and can be analysed using a flow cytometer with limited laser capacity. Tetramers detect low affinity T-cell receptor/HLA interactions and identify antigen-specific T-cells with limited capacity to mount a cytokine response, as we observed among two TB/HIV participants who exhibited tetramer pool binding but no cytokine production in response to the epitope pool. A major biological restriction of tetramer-based assays is the requirement for HLA-compatibility for tetramer-binding to occur. Nonetheless, in this pilot study, we have shown the feasibility of such an approach.
Our study was limited by the lack of a LTBI cohort with HIV-infection (LTBI/HIV). Individuals with LTBI/HIV are a heterogeneous population, with advanced imaging revealing evidence for subclinical, active disease among asymptomatic individuals with normal chest radiographs [13]. Thus, as some individuals with LTBI/HIV harbour subclinical, active disease, they may have a bacillary-load sufficient to elicit a Mtb-specific CD8+ T-cell response in our assays. We hypothesise that CD8+ T-cell responses to both our assays would be quite heterogeneous among individuals with LTBI/HIV, and that this population should be included in future studies.
In summary, CD8+ T-cell recognition of a pool of eight immunodominant Mtb-specific epitopes was highly selective for subjects with pulmonary TB, including individuals with TB/HIV co-infection. Once validated, a CD8+ T-cell epitope-pool assay may show promise to distinguish Mtb-infected individuals without TB disease, from those individuals suffering from active TB.
Acknowledgements
We would like to thank the Ugandan participants who gave their time and dedication to this health research; Francis Adatu Engwau and Alphonse Okwera and their staff at the National Tuberculosis Treatment Centre, Mulago Hospital; the Ugandan National Tuberculosis and Leprosy Program; Sam Ogwang and Moses Joloba, JCRC TB laboratory; Brenda Okwera and the nurses and home visitors of Ugandan field team; and Erin Merrifield, Dept of Pediatrics, OHSU, for their contributions to this study.
Support statement: This project has been funded in whole or in part with Federal funds from the National Institutes of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract no. HHSN272200900053C and by the Tuberculosis Research Unit established with Federal funds from the United States National Institutes of Allergy and Infectious Diseases and the United States National Institutes of Health and Human Services, under contract nos. NO1-AI-95383 and HHSN266200700022C/NO1-AI-70022 and with Papé Family Research Institute funds. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Funding information for this article has been deposited with the Crossref Funder Registry.
Conflict of interest: C. Lancioni has nothing to disclose. G.M. Swarbrick is an employee of ViTi, Inc, a company that may have commercial interest in the results of this research. This potential conflict has been reviewed and managed by OHSU. B. Park has nothing to disclose. M. Nyendak reports other from ViTi, Inc., outside the submitted work; Required language from OHSU: OHSU and M. Nyendak have a financial interest in ViTi, a company that may have a commercial interest in the results of this research and technology. These potential individual and institutional conflicts of interest have been reviewed and managed by OHSU. M. Nsereko has nothing to disclose. H. Mayanja-Kizza has nothing to disclose. M.D. Null is an employee of ViTi, Inc, a company that may have commercial interest in the results of this research. This potential conflict has been reviewed and managed by OHSU. M.E. Cansler is an employee of ViTi, Inc, a company that may have commercial interest in the results of this research. This potential conflict has been reviewed and managed by OHSU. R.B. Duncan has nothing to disclose. J. Baseke has nothing to disclose. K. Chervenak has nothing to disclose. L. Malone has nothing to disclose. E.G. Heaphy has nothing to disclose. W.H. Boom reports grants from NIH/NIAID, during the conduct of the study. D.M. Lewinsohn reports grants from NIH (HHSN272200900053C), grants from NIH (NO1-AI-95383 and HHSN266200700022C/NO1-AI-70022), other from Papé Family Research Institute, during the conduct of the study; other from ViTi Inc., outside the submitted work; OHSU and D.M. Lewinsohn have a financial interest in ViTi, a company that may have a commercial interest in the results of this research and technology. These potential individual and institutional conflicts of interest have been reviewed and managed by OHSU. D.A. Lewinsohn reports grants from NIH (HHSN272200900053C), grants from NIH (NO1-AI-95383 and HHSN266200700022C/NO1-AI-70022), other from Papé Family Research Institute (OHSU institutional funds), during the conduct of the study; other from ViTi Inc., outside the submitted work; OHSU and D.A Lewinsohn have a financial interest in ViTi, a company that may have a commercial interest in the results of this research and technology. These potential individual and institutional conflicts of interest have been reviewed and managed by OHSU.
References
- 1.Metcalfe JZ, Everett CK, Steingart KR, et al. Interferon-gamma release assays for active pulmonary tuberculosis diagnosis in adults in low- and middle-income countries: systematic review and meta-analysis. J Infect Dis 2011; 204: Suppl. 4, S1120–S1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lewinsohn DM, Swarbrick GM, Cansler ME, et al. Human Mycobacterium tuberculosis CD8 T cell antigens/epitopes identified by a proteomic peptide library. PLoS One 2013; 8: e67016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewinsohn DA, Winata E, Swarbrick GM, et al. Immunodominant tuberculosis CD8 antigens preferentially restricted by HLA-B. PLoS Pathog 2007; 3: 1240–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewinsohn DA, Swarbrick GM, Park B, et al. Comprehensive definition of human immunodominant CD8 antigens in tuberculosis. NPJ Vaccines 2017; 2: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caccamo N, Guggino G, Meraviglia S, et al. Analysis of Mycobacterium tuberculosis-specific CD8 T-cells in patients with active tuberculosis and in individuals with latent infection. PLoS One 2009; 4: e5528. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Day CL, Abrahams DA, Lerumo L, et al. Functional capacity of Mycobacterium tuberculosis-specific T cell responses in humans is associated with mycobacterial load. J Immunol 2011; 187: 2222–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Day CL, Moshi ND, Abrahams DA, et al. Patients with tuberculosis disease have Mycobacterium tuberculosis-specific CD8 T cells with a pro-apoptotic phenotype and impaired proliferative capacity, which is not restored following treatment. PLoS One 2014; 9: e94949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pollock KM, Whitworth HS, Montamat-Sicotte DJ, et al. T-cell immunophenotyping distinguishes active from latent tuberculosis. J Infect Dis 2013; 208: 952–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rozot V, Patrizia A, Vigano S, et al. Combined use of Mycobacterium tuberculosis-specific CD4 and CD8 T-cell responses is a powerful diagnostic tool of active tuberculosis. Clin Infect Dis 2015; 60: 432–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rozot V, Vigano S, Mazza-Stalder J, et al. Mycobacterium tuberculosis-specific CD8+ T cells are functionally and phenotypically different between latent infection and active disease. Eur J Immunol 2013; 43: 1568–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silva BD, Trentini MM, da Costa AC, et al. Different phenotypes of CD8+ T cells associated with bacterial load in active tuberculosis. Immunol Lett 2014; 160: 23–32. [DOI] [PubMed] [Google Scholar]
- 12.Bold TD, Ernst JD. CD4+ T cell-dependent IFN-gamma production by CD8+ effector T cells in Mycobacterium tuberculosis infection. J Immunol 2012; 189(5): 2530–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esmail H, Lai RP, Lesosky M, et al. Characterization of progressive HIV-associated tuberculosis using 2-deoxy-2-[(18)F]fluoro-D-glucose positron emission and computed tomography. Nat Med 2016; 22: 1090–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]