Abstract
Comparisons of infectivity among the clinically important nontuberculous mycobacteria (NTM) species have not been explored in great depth. Rapid-growing mycobacteria, including Mycobacterium abscessus and M. porcinum, can cause indolent but progressive lung disease. Slow-growing members of the M. avium complex are the most common group of NTM to cause lung disease, and molecular approaches can now distinguish between several distinct species of M. avium complex including M. intracellulare, M. avium, M. marseillense, and M. chimaera. Differential infectivity among these NTM species may, in part, account for differences in clinical outcomes and response to treatment; thus, knowing the relative infectivity of particular isolates could increase prognostication accuracy and enhance personalized treatment. Using human macrophages, we investigated the infectivity and virulence of nine NTM species, as well as multiple isolates of the same species. We also assessed their capacity to evade killing by the antibacterial peptide cathelicidin (LL-37). We discovered that the ability of different NTM species to infect macrophages varied among the species and among isolates of the same species. Our biochemical assays implicate modified phospholipids, which may include a phosphatidylinositol or cardiolipin backbone, as candidate antagonists of LL-37 antibacterial activity. The high variation in infectivity and virulence of NTM strains suggests that more detailed microbiological and biochemical characterizations are necessary to increase our knowledge of NTM pathogenesis.
Keywords: nontuberculous mycobacteria, human macrophages, cathelicidin (LL-37) antibacterial peptide, phosphatidylinositol, cardiolipin
Clinical Relevance
Differences in virulence among the clinically relevant nontuberculous mycobacterial (NTM) species and the factors that promote host evasion are understudied. We show that macrophage infectivity varies among NTM species and even among isolates of the same species. Moreover, NTM-derived phospholipids may facilitate evasion from host innate antibacterial peptides such as cathelicidin.
More than 199 different species of nontuberculous mycobacteria (NTM) have been identified to date (1). NTM are common inhabitants of environmental water biofilms and soil, and can be aerosolized and inhaled into the lungs through daily activities such as showering and gardening (2, 3). Over the last 30 years, NTM lung disease (NTM-LD) has become increasingly prominent, even outnumbering tuberculosis cases in many regions of the world (4). Long-term therapy is often necessary to treat NTM-LD, and recurrence due to relapse or reinfection is common, resulting in substantial morbidity and mortality (5, 6).
Rapid-growing mycobacteria (RGM), such as Mycobacterium abscessus, are of great concern, especially with regard to patients with cystic fibrosis (CF). RGM require treatment with prolonged multidrug antibiotic regimes, and even then are often refractory to treatment, with a high likelihood of relapse (7). M. porcinum is another RGM that is increasingly recognized as a human pathogen (8). In patients without CF, the most common LD-causing NTM belong to the M. avium complex (MAC) and are slow-growing mycobacteria (SGM). In previous work, we applied rpoB gene sequencing to identify M. abscessus, M. porcinum, M. intracellulare, M. marseillense, and M. chimaera species from environmental and clinical samples (9). The differential ability of these NTM organisms to survive within host macrophages is an important observation that has furthered our understanding of NTM pathogenesis. Another barrier in the current field of NTM research is the lack of knowledge regarding the virulence mechanisms used by pathogenic NTM. The well-characterized virulence factors of M. tuberculosis include a variety of lipid molecules; however, much less is known about the NTM lipid components that contribute to host immune evasion (10). Previously, we reported a novel virulence mechanism of NTM that is mediated by lipids and results in resistance to and inactivation of the cathelicidin antibacterial peptide (LL-37) by NTM cell membrane (CM) and/or cell wall (CW) lipids (11).
In the current study, we assessed the ability of different NTM species and isolates of the same species to survive exposure to human macrophages and to evade LL-37–mediated bacterial killing. We discovered that 1) the number of cell-associated NTM varied markedly among species and even among isolates of the same species after infection of human macrophages, suggesting intrinsic differences in infectivity even among closely related NTM isolates; 2) increased virulence correlated with heightened numbers of macrophage-associated NTM and resistance to and neutralization of LL-37 antibacterial activity; and 3) NTM-modified phospholipid molecules likely function as virulence factors by neutralizing the antibacterial activity of LL-37.
Portions of this manuscript have been previously published in abstract form (12).
Methods
Please see the data supplement for additional details regarding the methods used in this work.
NTM Isolates Used in this Study
Table 1 lists all of the isolates used in this study. These isolates were chosen because they are either well-described type strains with available genomic information or were previously cultured and identified via partial gene sequencing from environmental or clinical samples by our laboratory (9).
Table 1.
Nontuberculous Mycobacteria Isolates Used in This Study
| Species | Identification | Source |
|---|---|---|
| M. abscessus subsp. abscessus | P2a | Clinical |
| 12-39-Sw-B-2 | Kitchen sink | |
| 12-45-Sw-A-1 | Kitchen sink | |
| 19977 | ATCC type strain | |
| M. porcinum | KAU12-9-Sw-B-2 | Kitchen sink |
| M. intracellulare | AH2 | Clinical, 90-yr-old woman |
| AH9 | Clinical, 70-yr-old woman | |
| AH24 | Clinical, 87-yr-old woman | |
| 12-56-S-1-2 | Garden soil | |
| 9141 | Clinical | |
| M. avium | Chester | ATCC type strain |
| subsp. hominissuis H87 | ATCC type strain | |
| M. interjectum | 12-26-S-1-1 | Garden soil |
| M. timonense | AH20 | Clinical, 50-yr-old woman |
| M. colombiense | 12-29-S-1-2 | Garden soil |
| M. marseillense | AH7 | Clinical, 78-yr-old man |
| AH17 | Clinical, 38-yr-old man | |
| 12-2-S-1-1 | Garden soil | |
| M. chimaera | AH6 | Clinical, 73-yr-old woman |
| AH13 | Clinical, 71-yr-old man | |
| AH25 | Clinical, 79-yr-old man | |
| 12-54-Sw-B-1 | Kitchen faucet | |
| 12-42-Sw-A-3 | Showerhead | |
| 12-40-Sw-A-1 | Showerhead | |
| 12-35-Sw-A-1 | Kitchen faucet | |
| 12-43-Sw-A-1 | Showerhead | |
| 12-29-Sw-A-1 | Kitchen faucet | |
| 12-44-Sw-A-1 | Showerhead | |
| 12-49-Sw-B-1 | Kitchen faucet | |
| 12-25-Sw-B-2 | Kitchen faucet | |
| 12-2-Sw-B-2 | Refrigerator spout | |
| 12-22-Sw-A-1 | Showerhead | |
| 12-7-S-1-3 | Fruit cannery soil |
Definition of abbreviations: M. = Mycobacterium; NTM = nontuberculous mycobacteria.
“P” or “AH” indicates isolates recovered from respiratory samples from patients suspected of having NTM lung disease. The number 12 indicates isolates recovered from environmental sources in Hawai’i. “Sw” indicates NTM that were recovered from swabs used to sample biofilms on the surface of showerheads, kitchen sinks, and refrigerator spouts. “S” indicates NTM that were recovered from environmental sources of soil.
NTM Infection of Human Immune Cells and LL-37 Immunoblotting
Human THP-1–derived macrophages were used because they are a standard cell line that is commonly used in mycobacterial experiments and because the isogenic nature of this cell line limits experimental variability (13). The infection protocol used was adapted from our published work of mycobacteria-infected THP-1 macrophages (14–18). Isolation and infection of primary human monocyte-derived macrophage (MDM) and alveolar macrophage (AM) cell cultures, and LL-37 immunoblotting are described in the data supplement.
Monitoring the Antibacterial Activity of LL-37 against NTM
The methods used to monitor the antibacterial activity of LL-37 against NTM were previously described (11) and are also detailed in the data supplement.
Thin-Layer Chromatography of NTM Lipids and LL-37 Binding Immunoblotting
To define the NTM lipids that bind LL-37, CM lipids, total lipids from the CW, and cytosolic fractions were prepared from NTM isolates. To determine whether lipids present in these fractions bind LL-37, fractions were spotted in triplicate onto identical silica plates and subjected to TLC. The first two plates were sprayed with CuSO4 and α-naphthol to visualize lipid species and glycolipids, respectively. To determine which NTM lipids LL-37 binds, the third plate was used for LL-37 binding IB as described in the data supplement.
Testing the LL-37 Inactivation Activity of Pure Phospholipids
Purified lipids were obtained from Avanti Polar Lipids and included soy L-α-lysophosphatidylinositol (PI; #840044), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (PC; #850457), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (PE; #850725), and 1,1′,2,2′ tetraoleoyl cardiolipin (CL; #710335). The lipids were stored dry at −20°C, resuspended in 2:1 chloroform:methanol, and subjected to TLC and LL-37 binding immunoblots. For LL-37 bioassays, lipids were dissolved in absolute ethanol and stored at −20°C.
We previously demonstrated that Escherichia coli is susceptible to 25 μg/ml of LL-37, and NTM-exposed LL-37 loses its potent activity against E. coli (11). The E. coli bioassay was used to determine whether these purified lipids and total lipid extracts (TLEs) from M. chimaera 12-54-Sw-B-1 and 12-7-S-1-3 neutralize the antibacterial activity of LL-37. LL-37 at 25 μg/ml was added to 1 μg of lipid in 250 μl of LL-37 media (defined in the data supplement) and preincubated at 37°C for 4 hours. Subsequently, 2 × 105 E. coli was inoculated into each of the samples, incubated for an additional 4 or 20 hours, and swabbed onto Luria-Bertani agar plates to assess E. coli growth. As an untreated control, E. coli was inoculated into 250 μl of LL-37 medium. As a positive control, E. coli was incubated with synthetic LL-37 peptide (25 μg/ml). The vehicle, 0.4% ethanol, was also incubated with and without LL-37 in the presence of E. coli.
Results
NTM Phylogenomic Tree
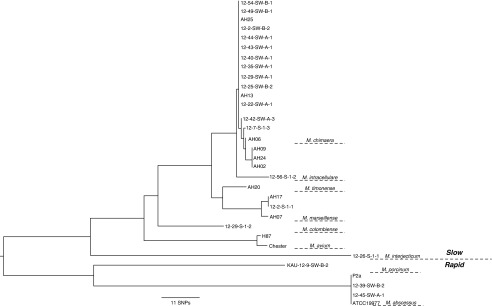
In our previously published work, we identified several NTM species using partial rpoB gene sequencing (9). Thirty-two NTM isolates from this earlier study were selected for the current experiments. Partial rpoB sequences were used to create an alignment matrix of 726 bp. Of the 726 sites in the alignment, 146 sites (∼20%) were variable and contained no missing data. The phylogenetic relationships between the 32 isolates in Figure 1 show the delineation between the growth rates (rapid vs. slow growing) and the included species.
Figure 1.
Phylogenetic analysis of the nontuberculous mycobacteria (NTM) isolates used in this study. A phylogeny based on the multiple sequence alignment of partial rpoB sequences inferred between isolate relationships and delineated as rapid- (RGM) and slow-growing NTM (SGM) isolates was obtained using the neighbor-joining method and observed SNPs. The scale bar represents an 11-SNP difference in nucleotide sequences. M. = Mycobacterium.
NTM Demonstrate a Range of Inter- and Intraspecies Infectivity in Human Macrophages
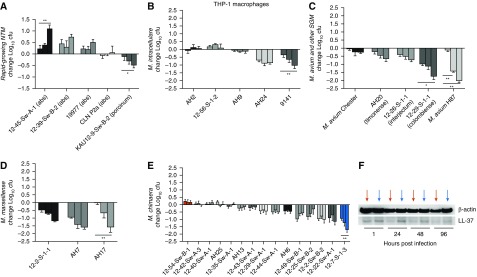
THP-1–derived macrophages were infected with a variety of RGM and MAC species. RGM isolates of M. abscessus and M. porcinum showed inter- and intraspecies variations in THP-1 cell–associated growth by 96 hours after infection (Figure 2A). Specifically, three different isolates of M. abscessus showed an increased cell-associated burden with time, and a fourth isolate (P2a) showed minimal change. In contrast, there was a marked temporal reduction in the macrophage-associated burden of M. porcinum.
Figure 2.
Differential infectivity of RGM and SGM species in human THP-1 macrophages. THP-1 macrophages were infected with environmental or clinical RGM or Mycobacterium avium complex species isolates. The change in log10 cfu is shown per species. Three bars are shown for each species corresponding to the change in cfu determined after 24 hours (first bar), 48 hours (second bar), and 96 hours after infection (third bar) compared with cfu quantified at 1 hour after infection of macrophages. n = 3–10 independent experiments. (A) M. abscessus (n = 4 isolates) and an isolate of M. porcinum; *P = 0.01 and **P = 0.002. (B) M. intracellulare (n = 5 isolates); **P = 0.003. (C) M. avium (n = 2 isolates), **P = 0.003; M. timonense (n = 1 isolate); M. interjectum (n = 1 isolate); M. colombiense (n = 1 isolate), *P = 0.02. (D) M. marseillense (n = 3 isolates), **P = 0.003. (E) M. chimaera (n = 15 isolates), **P = 0.004. (F) LL-37 immunoblot of cell lysates from THP-1 macrophages infected with the more-virulent M. chimaera isolate 12-54-Sw-B-1, as indicated by the red arrows, or the less-virulent M. chimaera isolate 12-7-S-1-3, as indicated by the blue arrows, at various time points after infection. β-actin was used as a loading control. cfu = colony-forming units; THP-1 = human leukemia monocytic cell line.
The MAC species tested also demonstrated a range of infectivity in THP-1 macrophages. Among the five M. intracellulare isolates examined, two (AH2 and 12-56-S-1-2) showed modest growth when cocultured with macrophages. In contrast, M. intracellulare AH9, AH24, and 9141 demonstrated decreases in viable bacteria over time (Figure 2B). Macrophages also showed effective control of two isolates of M. avium (Chester and H87), M. timonense, M. interjectum, and M. colombiense (Figure 2C). The three isolates of M. marseillense tested showed a progressive decline in bacterial colony-forming units (cfu) over time, with THP-1 macrophages significantly controlling the growth of isolate AH17 by 96 hours after infection (Figure 2D). The M. chimaera isolates tested showed a range of growth in macrophages, as demonstrated in Figure 2E, including significant differences between the cfu of M. chimaera isolate 12-54-Sw-B-1 (Figure 2E, red bars) and M. chimaera isolate 12-7-S-1-3 (Figure 2E, blue bars). Based on these findings, we considered M. chimaera 12-54-Sw-B-1 to be more virulent than M. chimaera 12-7-S-1-3.
To investigate the status of LL-37 expression in THP-1 macrophages before and after infection with the more- or less-virulent M. chimaera isolates, we performed LL-37 IB. Less LL-37 was detected in lysates from cells infected with the more-virulent 12-54-Sw-B-1 M. chimaera isolate (red arrows) compared with lysates from cells infected with the less-virulent 12-7-S-1-3 M. chimaera isolate (blue arrows) at the 24- and 48-hour time points (Figure 2F).
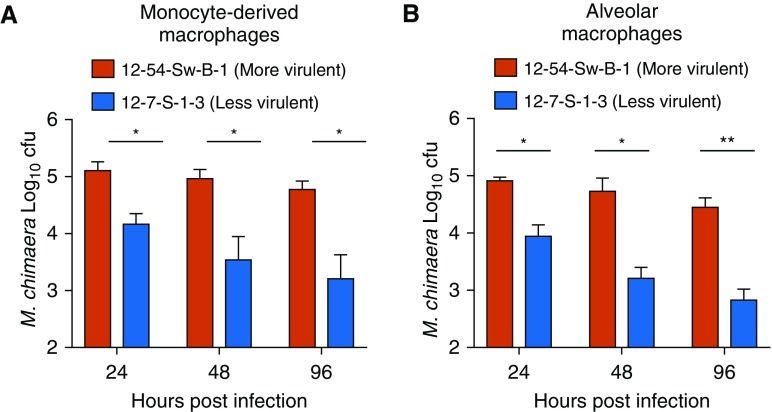
To determine whether the cfu trends observed in the THP-1 macrophages could also be seen in primary immune cells, we compared the temporal cell-associated burden of M. chimaera 12-54-Sw-B-1 with that of M. chimaera 12-7-S-1-3 in infected human MDMs and AMs. Similar to what was seen in the THP-1 cells, a significantly greater bacterial burden was observed with M. chimaera 12-54-Sw-B-1 than with M. chimaera 12-7-S-1-3 in both types of primary human macrophages (Figures 3A and 3B).
Figure 3.
Differential infectivity of more- and less-virulent M. chimaera in primary human monocyte-derived macrophages (MDMs) and alveolar macrophages (AMs). (A and B) MDMs (A) and AMs (B) were infected with the more-virulent M. chimaera 12-54-Sw-B-1 or less-virulent M. chimaera 12-7-S-1-3, and changes in cfu were recorded over time. n = 3 different donor samples. Red bars represent M. chimaera 12-54-Sw-B-1 and blue bars represent M. chimaera 12-7-S-1-3. *P < 0.05 and **P < 0.005.
More-Virulent NTM Also Resist LL-37–mediated Bacterial Killing
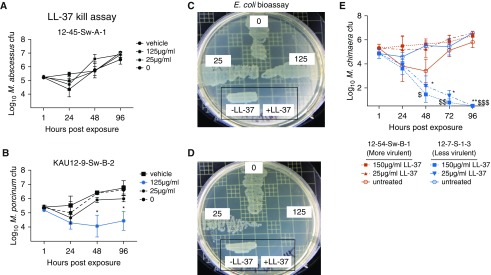
We previously reported resistance to and inactivation of the LL-37 antibacterial peptide as a virulence mechanism of NTM (11). Because M. abscessus 12-45-Sw-A-1 showed significantly higher cfu than M. porcinum in the THP-1 macrophage assay (Figure 2A), we tested these two isolates for their susceptibility to LL-37. M. abscessus 12-45-Sw-A-1 completely resisted exposure to varying concentrations of LL-37 (Figure 4A), whereas the M. porcinum isolate KAU12-9-Sw-B-2 exhibited significant sensitivity to 125 μg/ml of LL-37 (Figure 4B, blue line). Although 25 μg/ml of LL-37 effectively killed E. coli (Figures 4C and 4D, inset boxes), 25 μg/ml and 125 μg/ml of LL-37 incubated in the 96-hour conditioned media from M. abscessus lost its potent antibacterial activity against E. coli (Figure 4C), suggesting that a component in the conditioned media from this more virulent isolate inactivated LL-37. In contrast, 125 μg/ml of LL-37 incubated in 96-hour conditioned media from M. porcinum did not neutralize the antibacterial activity of LL-37 against E. coli (Figure 4D). Boiling the conditioned medium from M. abscessus before the addition of LL-37 did not abrogate the neutralizing activity of the medium (data not shown), indicating that a heat-stable, secreted NTM component was responsible for LL-37 inactivation.
Figure 4.
Differential susceptibility to LL-37 of more- and less-virulent NTM isolates. (A and B) The direct antibacterial activity of LL-37 against M. abscessus 12-45-Sw-A-1 (A) or M. porcinum KAU12-9-Sw-B-2 (*P < 0.05) (B) was assessed by monitoring changes in cfu over time in LL-37 bacterial-killing assays. (C and D) Next, the LL-37 neutralization capabilities of the RGM were determined by incubating fresh LL-37 (25 or 125 μg/ml) with 96-hour conditioned medium from a liquid culture of M. abscessus 12-45-Sw-A-1 (C) or M. porcinum KAU12-9-Sw-B-2 (D). The bioactivity of LL-37 against Escherichia coli was also assayed to assess whether NTM was capable of neutralizing LL-37. Also shown is the growth of E. coli incubated alone (no LL-37) or with neat LL-37 (i.e., LL-37 not exposed to NTM-conditioned medium [both surrounded by a black box]). (E) The direct effects of 25 or 150 μg/ml of LL-37 on the viability of M. chimaera 12-54-Sw-B-1 and 12-7-S-1-3 were assessed by cfu counts after the indicated times. n = 3 independent experiments. *P < 0.04, **P = 0.008, $P = 0.01, $$P = 0.001, and $$$P = 0.0004.
The more-virulent M. chimaera isolate, 12-54-Sw-B-1, and less-virulent isolate, 12-7-S-1-3, were also tested for resistance to LL-37. Although 12-54-Sw-B-1 showed a modest decrease in cfu in the absence of LL-37 at the 48-hour time point, bacterial cfu increased by the 96-hour time point (Figure 4E). Importantly, 12-54-Sw-B-1 was completely resistant to LL-37 (Figure 4E red lines). On the other hand, 25 and 150 μg/ml of LL-37 demonstrated significant antibacterial activity against the 12-7-S-1-3 isolate of M. chimaera (Figure 4E, blue lines).
Modified Phospholipids of NTM Bind LL-37
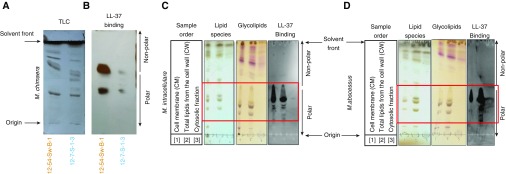
To investigate the potential of specific NTM lipids that may be responsible for LL-37 inactivation, TLEs were prepared from M. chimaera 12-54-Sw-B-1 and 12-7-S-1-3 and subjected to TLC (Figure 5A). To further dissect which NTM factor(s) bind LL-37, TLEs were also probed for lipids that bind to LL-37. The LL-37 binding immunoblot, in which a TLC plate was incubated with LL-37 peptide and then immunoblotted with anti-LL-37 antibody (19), revealed that LL-37 binds to M. chimaera polar lipids (Figure 5B).
Figure 5.
Phospholipids of NTM bind LL-37. (A) Total lipid extracts from M. chimaera 12-54-Sw-B-1 and 12-7-S-1-3 were separated by TLC. (B) LL-37 binding immunoblot of TLC-separated total lipids of M. chimaera 12-54-Sw-B-1 and 12-7-S-1-3 sequentially incubated with LL-37 peptide and immunoblotted with an anti-LL-37 antibody (LL-37 binding). (C and D) Next, the cell membrane (CM), cell wall (CW), and cytosolic fractions of M. intracellulare 9141 (C) and M. abscessus 19977 (D) were separated by TLC and sprayed with CuSO4 (to detect lipid species) or α-naphthol (to detect glycolipids), and TLC-separated total lipid fractions were sequentially incubated with LL-37 and immunoblotted with anti-LL-37 antibody.
To examine whether the binding of LL-37 to NTM polar lipids extends to another species of MAC and to RGM, as well as to further dissect which lipids of NTM bind LL-37, various lipid-containing fractions were separated from M. intracellulare and M. abscessus. These lipid fractions were subjected to TLC and stained with CuSO4 to detect lipid species, or with α-naphthol to detect glycolipids, and also probed for lipid–LL-37 complexes. LL-37 bound avidly to polar lipids in the CM and CW fractions (outlined by red box) of both species, but not to any of the nonpolar lipids (Figures 5C and 5D). Moreover, LL-37 did not bind to any component in the cytosolic fraction, indicating that the products responsible for this biological activity were present as surface molecules in M. intracellulare and M. abscessus. It is plausible that the cytosolic fraction contained less lipids than the CM and CW fractions; however, the bands in the upper portion of the lipid species and glycolipid TLC plates were similar in amount and intensity. Thus, we reasoned that the fractions contained comparable amounts of lipids, and thus the active component was primarily resident in the aforementioned fractions.
The current literature provides little information regarding detailed biochemical studies of NTM polar lipids (glycosylated or charged species decorated on smaller C8-C20 branched or single fatty acyl moieties) versus nonpolar lipids (wherein macromolecules include longer-chain branched or single fatty acyl backbones). Despite the scarcity of information, all mycobacteria are known to possess numerous phospholipids, including many PI derivatives, notably the PI mannosides (PIMs). To determine whether LL-37 binds these and/or other phospholipids of NTM, mass spectrometry (MS) was used to probe for NTM-specific phospholipids in BAL fluid samples from M. intracellulare–infected patients. A representative spectrum of the molecular ions detected shows a region of PI in which the most prominent species were PI(18:1/18:1), PI(16:0/18:1), and PI(18:0/20:4) (Figure E1 in the data supplement). Next, biotinylated LL-37 peptide was incubated with M. abscessus–conditioned medium for 24 hours in a pull-down assay. Results from the MS analyses support our conjecture that M. abscessus phospholipids interacted with LL-37 (the pulled-down phospholipids are identified in Figure E2 and Table E1).
Putative NTM Candidates as LL-37 Antagonists
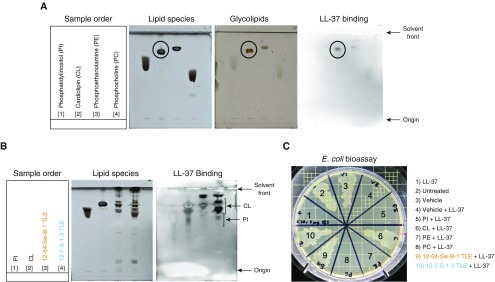
Because NTM-specific phospholipids are not commercially available, we used soy PI and synthetic CL, PE, and PC as substitutes to test their LL-37 neutralizing activity. TLC and LL-37 binding immunoblot analyses revealed that among the four phospholipids tested, LL-37 interacted most strongly to CL (Figure 6A). When PI and CL controls were subjected to TLC and LL-37 immunoblots alongside TLEs from M. chimaera 12-54-Sw-B-1 and M. chimaera 12-7-S-1-3, LL-37 bound more strongly to lipids that comigrated with CL and less strongly to lipid components that comigrated with PI (Figure 6B). Next, using the E. coli bioassay, we tested PI, CL, PE, and PC for their capacity to inactivate LL-37. As previously observed, E. coli was effectively killed when it was incubated with fresh LL-37 alone (inset boxes, Figures 4C and 4D). However, the bactericidal activity of 25 μg/ml of LL-37 against E. coli was completely lost after coincubation with CL (Figure 6C, no. 6). In stark contrast, LL-37 retained potent antibacterial activity against E. coli despite incubation with PI, PE, and PC (Figure 6C, nos. 5, 7, and 8). LL-37 antibacterial activity for E. coli was also abrogated when the peptide was incubated with TLEs of 12-54-Sw-B-1 or 12-7-S-1-3, but not with the vehicle control (Figure 6C, nos. 4, 9, and 10).
Figure 6.
Candidate antagonists of LL-37. (A) Phosphatidylinositol (PI), cardiolipin (CL), phosphoethanolamine (PE), and phosphocholine (PC) were subjected to the same TLC and LL-37 binding IB conditions as shown in Figure 5. (B) Comparison of the migration of M. chimaera 12-54-Sw-B-1 and 12-7-S-1-3 total lipid extracts (TLEs) with pure PI and CL by TLC and LL-37 binding IB. (C) E. coli was added to LL-37 (25 μg/ml) incubated with PI, CL, PE, or PC or M. chimaera 12-54-Sw-B-1 and 12-7-S-1-3 TLEs or vehicle controls. Loss of killing of E. coli was used as a readout for LL-37 inactivation.
Discussion
In this study, we used our collection of environmental and clinical isolates of RGM and SGM that we previously identified to the species and subspecies levels using gene sequencing to further explore NTM infectivity and virulence. Based on our data, we believe that in vitro assays of cell-associated NTM survival in the presence of macrophages and evasion of LL-37 bactericidal activity are useful for distinguishing more- and less-infective NTM isolates. Our data provide additional evidence supporting our hypothesis that more-virulent NTM isolates are more capable of neutralizing the antibacterial activity of LL-37 than less-virulent isolates. In this study, we took additional steps to identify the NTM-derived antagonist(s). First, TLC and LL-37 binding assays (Figure 5) demonstrated that LL-37 binds to polar NTM lipids. The MS data presented in Figures E1 and E2 support our TLC data, and further implicate NTM phospholipids as potential molecules that interact with LL-37. The data presented in Figure 6 provide additional evidence that the phospholipid CL is a viable candidate to be an antagonist of LL-37 antibacterial activity. However, because the TLEs from both the more- and less-virulent M. chimaera isolates inactivated LL-37 to similar degrees (Figure 6C), more studies are warranted to further dissect the nuances of this virulence mechanism.
CL is an important component of the inner mitochondrial membrane of eukaryotic cells, particularly in the heart muscle of mammals, as well as in the membranes of prokaryotes (20). Although as a phospholipid of mycobacteria, it often accounts for a large percentage (30–50%) of the total phospholipid content (21), the similarities between human and NTM-derived CL are unknown at this time. Higher amounts of CL are associated with more pathogenic mycobacteria, including M. tuberculosis (21–23), but little is known about its functional role in the pathogenesis of NTM lung infections. Mycobacterial PI is another phospholipid that is abundantly present in the membrane and plays an essential role in viability, growth, and infectivity (24, 25). PI is the basic subunit for the synthesis of PIM, lipomannan, and lipoarabinomannan—higher-order glycolipids and lipoglycans, and major constituents of the mycobacterial CW (26–28). The diversity and high amounts of phospholipids in the mycobacterial CM suggest the possibility that subtle and overt differences in the resistance to and inactivation of LL-37 among the different NTM species and strains may be due to differences in the strength of LL-37 antagonism as well as the relative abundance of these different phospholipids on the surface of these mycobacteria. Determining the differential amounts of CL, PI, and PIM in the CMs and CWs of more- and less-virulent NTM species, and their stochastic effects needed for LL-37 inactivation, was beyond the scope of this study and will be the focus of future studies.
In our experience, mycobacterial CL and PIM are notoriously difficult to separate and purify. Moreover, in the absence of robust, enriched fractions for NTM-derived CL, PIM, and other phospholipids, our MS analyses were unable to distinguish between CL and PIM (data not shown), or among the phospholipids phosphatidylethanolamine, phosphatidylserine, and phosphatidylcholine (Figure E2 and Table E1). For this purpose, high-quality fragmentation data generated via tandem MS will be needed. Purified phospholipid fractions from various NTM species should also be tested in vitro along with dose titration and kinetic assays to validate whether NTM-derived CL or PIM inactivate LL-37 to a higher degree than other phospholipids. Ideally, endogenous LL-37 peptide and NTM bioactive lipids should also be purified directly from patient samples and tested ex vivo for LL-37 antibacterial activity. Alternatively, specific neutralization of the bioactive phospholipid using enzymatic treatments in relevant biological material, such as BAL fluid from NTM-infected patients, would further support the notion that specific NTM-derived lipids neutralize LL-37. Further studies using purified or synthetic NTM-derived phospholipids have the potential to lead to the development of novel, targeted therapeutics to alleviate NTM infections.
We acknowledge that NTM species may possess unique virulence attributes and structural features, such that the aforementioned phospholipids may not be the only molecules capable of inactivating LL-37, and these lipid species may even act as molecular decoys. We do not discount the possibility that other neutral lipid species (e.g., methylmannose polysaccharides) coexist within this phospholipid-enriched polar lipid fraction and may also influence the antibacterial activity of host-protective innate immune peptides (29). Robust chemical classifications will be needed in future work to define and identify possible LL-37–interacting NTM bioactive products. Detailed lipidomic studies may also help to identify additional differences in lipids between more- and less-virulent NTM isolates.
In this study, we also show a wide range of viability and growth in THP-1 macrophages among the different isolates of M. abscessus subsp. abscessus tested (Figure 2A). The single isolate of M. porcinum tested demonstrated low infectivity in THP-1 macrophages, but we anticipate that examining a larger number of M. porcinum isolates will reveal a wide range of intraspecies infectivity similar to that seen with M. abscessus. Additionally, differences in inter-NTM-species infectivity were noted for MAC (Figures 2B–2D). Overall, we found that two of the five M. intracellulare isolates tested (40%) showed a numerical increase in macrophages in a temporal fashion. However, all of the M. avium, M. timonense, M. interjectum, and M. marseillense isolates tested showed decreased numbers of cell-associated NTM in the presence of macrophages. Regarding M. chimaera, three of 15 isolates tested (20%) (12-54-Sw-B-1, 12-42-Sw-A-3, and 12-40-Sw-A-1) showed positive growth in the number of cell-associated NTM in the presence of THP-1 macrophages, suggesting that these isolates are better equipped to survive in macrophages than other isolates of the same species. Importantly, these data indicate that although all were identified as M. chimaera, they showed an intraspecies variability in their ability to survive in human immune cells, including human MDMs and AMs (Figures 3A and 3B).
We did not uniformly observe that clinical isolates were more infective to macrophages than environmental isolates; rather, we found that infectivity varied among the different NTM species and among different strains within the same species. By comparison, in a study performed over 20 years ago, Pedrosa and colleagues reported that animal-derived MAC isolates were more virulent in mice, human MAC isolates were less virulent in mice, and environmental isolates were unable to infect mice (30). However, that study was limited in that it was published before the advent of high-throughput sequencing, and consequently MAC was treated as a single species. Thus, it is probable that the authors compared different MAC species rather than different isolates of the same species. After applying variable-number tandem-repeat typing, Tatano and colleagues identified M. avium isolates from patients with either nodular bronchiectasis or cavitary LD (n = 5 for each group), and compared their survival in THP-1 macrophages (31). They observed no significant difference in growth between the two groups, suggesting that M. avium isolates show similar intraspecies virulence. More recently, Rindi and colleagues demonstrated that infection with a M. avium subsp. hominissuis sequevar A (MAH-A) or B (MAH-B) resulted in similar bacterial burdens 2 hours after infection in peripheral blood mononuclear cells and THP-1 macrophages (32). However, MAH-A showed significantly higher cfu by day 5 after infection than MAH-B, which suggests that MAH also exhibits intraspecies differences in virulence.
In summary, we show that NTM infectivity correlates with resistance to and antagonism of LL-37, and that the leading candidate for this immune evasive mechanism is a modified phospholipid such as CL or PIM, both of which are major components of the NTM CM. We also provide evidence that macrophage infectivity varies among different species of related NTM and among different isolates of the same NTM species. Future work should also correlate in vitro NTM infectivity and LL-37 evasion with clinical and radiographic assessments of disease severity and prognosis in patients with NTM-LD, identification of point-of-care biomarkers of infectivity, and the design of novel therapeutics that target NTM-derived pathogenic determinants (e.g., CL and PIM) as an immunoenhancer by, for example, preventing the antagonism of LL-37.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank Pearlanne Zelarney and John Yang, Ph.D., for their assistance in downloading some of the data used in this study from the National Jewish Health (NJH) Research Database and providing access to stored samples in the NJH Biobank, respectively. They also thank Dr. Hong Wei Chu and Nicole Pavelka (NJH Human Cell Core) for providing alveolar macrophages and expertise and assistance with the ex vivo human cell experiments. They acknowledge Diagnostic Laboratory Services (Aiea, Hawai’i) for providing some of the clinical isolates used in this study.
Footnotes
Supported by the Division of Pulmonary Sciences and Critical Care Medicine, University of Colorado Anschutz Medical Campus, a National Jewish Health Challenge Grant, Shoot for the Cure, and the Padosi Foundation (J.R.H.). A 2017 Abstract Scholarship was awarded to J.R.H. from the Assembly on Microbiology, Tuberculosis, and Pulmonary Infections to present portions of this work at the 2017 American Thoracic Society International Conference, Washington, D.C.
Author Contributions: J.R.H. designed and performed experiments, and wrote, edited, and finalized the manuscript. T.H., R.C., P.K., L.M.N.R., G.J.N., R.V., M.N.I., C.M., and D.H. performed experiments, assisted with data presentation, and edited the manuscript. N.A.H., L.E.E., and M.S. assisted with NTM rpoB gene sequencing and phylogeny, and edited the manuscript. S.A., S.C.F., D.R.V., K.M.D., and E.D.C. edited and approved the final version of the manuscript for important intellectual content, supervised the study, and provided some funding.
This manuscript is dedicated to the memory of Tamara Hess, who contributed to the early NTM lipid studies presented herein.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2018-0278OC on September 23, 2019
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.LPSN. List of prokaryotic names with standing in nomenclature. 2018 doi: 10.1093/nar/gkt1111. [accessed 2019 Oct 21]. Available from: http://www.bacterio.net/mycobacterium.html. [DOI] [PMC free article] [PubMed]
- 2.Falkinham JO, III, Iseman MD, de Haas P, van Soolingen D. Mycobacterium avium in a shower linked to pulmonary disease. J Water Health. 2008;6:209–213. doi: 10.2166/wh.2008.032. [DOI] [PubMed] [Google Scholar]
- 3.De Groote MA, Pace NR, Fulton K, Falkinham JO., III Relationships between Mycobacterium isolates from patients with pulmonary mycobacterial infection and potting soils. Appl Environ Microbiol. 2006;72:7602–7606. doi: 10.1128/AEM.00930-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffith DE, Aksamit TR. Understanding nontuberculous mycobacterial lung disease: it’s been a long time coming. F1000 Res. 2016;5:2797. doi: 10.12688/f1000research.9272.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vinnard C, Longworth S, Mezochow A, Patrawalla A, Kreiswirth BN, Hamilton K. Deaths related to nontuberculous mycobacterial infections in the United States, 1999-2014. Ann Am Thorac Soc. 2016;13:1951–1955. doi: 10.1513/AnnalsATS.201606-474BC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fleshner M, Olivier KN, Shaw PA, Adjemian J, Strollo S, Claypool RJ, et al. Mortality among patients with pulmonary non-tuberculous mycobacteria disease. Int J Tuberc Lung Dis. 2016;20:582–587. doi: 10.5588/ijtld.15.0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park IK, Olivier KN. Nontuberculous mycobacteria in cystic fibrosis and non-cystic fibrosis bronchiectasis. Semin Respir Crit Care Med. 2015;36:217–224. doi: 10.1055/s-0035-1546751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wallace RJ, Jr, Brown-Elliott BA, Wilson RW, Mann L, Hall L, Zhang Y, et al. Clinical and laboratory features of Mycobacterium porcinum. J Clin Microbiol. 2004;42:5689–5697. doi: 10.1128/JCM.42.12.5689-5697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honda JR, Hasan NA, Davidson RM, Williams MD, Epperson LE, Reynolds PR, et al. Environmental nontuberculous mycobacteria in the Hawaiian Islands. PLoS Negl Trop Dis. 2016;10:e0005068. doi: 10.1371/journal.pntd.0005068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tran T, Bonham AJ, Chan ED, Honda JR. A paucity of knowledge regarding nontuberculous mycobacterial lipids compared to the tubercle bacillus. Tuberculosis (Edinb) 2019;115:96–107. doi: 10.1016/j.tube.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 11.Honda JR, Hess T, Malcolm KC, Ovrutsky AR, Bai X, Irani VR, et al. Pathogenic nontuberculous mycobacteria resist and inactivate cathelicidin: implication of a novel role for polar mycobacterial lipids. PLoS One. 2015;10:e0126994. doi: 10.1371/journal.pone.0126994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honda JR, Nieto L, Zhou Y, Hesser D, Hasan NA, Mehaffy C, et al. Mycobacterium avium complex species virulence and lipid profiles Am J Respir Crit Care Med 2017195A5060. [Google Scholar]
- 13.Theus SA, Cave MD, Eisenach KD. Activated THP-1 cells: an attractive model for the assessment of intracellular growth rates of Mycobacterium tuberculosis isolates. Infect Immun. 2004;72:1169–1173. doi: 10.1128/IAI.72.2.1169-1173.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bai X, Oberley-Deegan RE, Bai A, Ovrutsky AR, Kinney WH, Weaver M, et al. Curcumin enhances human macrophage control of Mycobacterium tuberculosis infection. Respirology. 2016;21:951–957. doi: 10.1111/resp.12762. [DOI] [PubMed] [Google Scholar]
- 15.Bai X, Kinney WH, Su WL, Bai A, Ovrutsky AR, Honda JR, et al. Caspase-3-independent apoptotic pathways contribute to interleukin-32γ-mediated control of Mycobacterium tuberculosis infection in THP-1 cells. BMC Microbiol. 2015;15:39. doi: 10.1186/s12866-015-0366-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai X, Feldman NE, Chmura K, Ovrutsky AR, Su WL, Griffin L, et al. Inhibition of nuclear factor-κ B activation decreases survival of Mycobacterium tuberculosis in human macrophages. PLoS One. 2013;8:e61925. doi: 10.1371/journal.pone.0061925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bai X, Kim SH, Azam T, McGibney MT, Huang H, Dinarello CA, et al. IL-32 is a host protective cytokine against Mycobacterium tuberculosis in differentiated THP-1 human macrophages J Immunol 201018438303840 [DOI] [PubMed] [Google Scholar]
- 18.Oberley-Deegan RE, Lee YM, Morey GE, Cook DM, Chan ED, Crapo JD. The antioxidant mimetic, MnTE-2-PyP, reduces intracellular growth of Mycobacterium abscessus. Am J Respir Cell Mol Biol. 2009;41:170–178. doi: 10.1165/rcmb.2008-0138OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taki T, Gonzalez TV, Goto-Inoue N, Hayasaka T, Setou M. TLC blot (far-eastern blot) and its applications. Methods Mol Biol. 2009;536:545–556. doi: 10.1007/978-1-59745-542-8_55. [DOI] [PubMed] [Google Scholar]
- 20.Schlame M, Hostetler KY. Cardiolipin synthase from mammalian mitochondria. Biochim Biophys Acta. 1997;1348:207–213. doi: 10.1016/s0005-2760(97)00119-7. [DOI] [PubMed] [Google Scholar]
- 21.Sarma PV, Srikanth L, Venkatesh K, Murthy PS, Sarma PU. Isolation, purification and characterization of cardiolipin synthase from Mycobacterium phlei {PRIVATE} Bioinformation. 2013;9:690–695. doi: 10.6026/97320630009690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akamatsu Y, Nojima S. Separation and analyses of the individual phospholipids of mycobacteria. J Biochem. 1965;57:430–439. doi: 10.1093/oxfordjournals.jbchem.a128097. [DOI] [PubMed] [Google Scholar]
- 23.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 24.Brennan PJ, Nikaido H. The envelope of mycobacteria. Annu Rev Biochem. 1995;64:29–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- 25.Guerin ME, Kordulakova J, Schaeffer F, Svetlikova Z, Buschiazzo A, Giganti D, et al. Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria. J Biol Chem. 2007;282:20705–20714. doi: 10.1074/jbc.M702087200. [DOI] [PubMed] [Google Scholar]
- 26.Khoo KH, Dell A, Morris HR, Brennan PJ, Chatterjee D. Structural definition of acylated phosphatidylinositol mannosides from Mycobacterium tuberculosis: definition of a common anchor for lipomannan and lipoarabinomannan. Glycobiology. 1995;5:117–127. doi: 10.1093/glycob/5.1.117. [DOI] [PubMed] [Google Scholar]
- 27.Nigou J, Gilleron M, Puzo G. Lipoarabinomannans: from structure to biosynthesis. Biochimie. 2003;85:153–166. doi: 10.1016/s0300-9084(03)00048-8. [DOI] [PubMed] [Google Scholar]
- 28.Briken V, Porcelli SA, Besra GS, Kremer L. Mycobacterial lipoarabinomannan and related lipoglycans: from biogenesis to modulation of the immune response. Mol Microbiol. 2004;53:391–403. doi: 10.1111/j.1365-2958.2004.04183.x. [DOI] [PubMed] [Google Scholar]
- 29.Mendes V, Maranha A, Alarico S, Empadinhas N. Biosynthesis of mycobacterial methylglucose lipopolysaccharides. Nat Prod Rep. 2012;29:834–844. doi: 10.1039/c2np20014g. [DOI] [PubMed] [Google Scholar]
- 30.Pedrosa J, Flórido M, Kunze ZM, Castro AG, Portaels F, McFadden J, et al. Characterization of the virulence of Mycobacterium avium complex (MAC) isolates in mice. Clin Exp Immunol. 1994;98:210–216. doi: 10.1111/j.1365-2249.1994.tb06127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tatano Y, Yasumoto K, Shimizu T, Sano C, Sato K, Yano S, et al. Comparative study for the virulence of Mycobacterium avium isolates from patients with nodular-bronchiectasis- and cavitary-type diseases. Eur J Clin Microbiol Infect Dis. 2010;29:801–806. doi: 10.1007/s10096-010-0930-2. [DOI] [PubMed] [Google Scholar]
- 32.Rindi L, Lari N, Garzelli C. Virulence of Mycobacterium avium subsp: hominissuis human isolates in an in vitro macrophage infection model. Int J Mycobacteriol. 2018;7:48–52. doi: 10.4103/ijmy.ijmy_11_18. [DOI] [PubMed] [Google Scholar]
- 33.Ovrutsky AR, Chan ED, Kartalija M, Bai X, Jackson M, Gibbs S, et al. Cooccurrence of free-living amoebae and nontuberculous mycobacteria in hospital water networks, and preferential growth of Mycobacterium avium in Acanthamoeba lenticulata Appl Environ Microbiol 20137931853192 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.