Abstract
The immune system is designed to robustly respond to pathogenic stimuli but to be tolerant to endogenous ligands to not trigger autoimmunity. Here, we studied an endogenous damage-associated molecular pattern, mitochondrial DNA (mtDNA), during primary graft dysfunction (PGD) after lung transplantation. We hypothesized that cell-free mtDNA released during lung ischemia–reperfusion triggers neutrophil extracellular trap (NET) formation via TLR9 signaling. We found that mtDNA increases in the BAL fluid of experimental PGD (prolonged cold ischemia followed by orthotopic lung transplantation) and not in control transplants with minimal warm ischemia. The adoptive transfer of mtDNA into the minimal warm ischemia graft immediately before lung anastomosis induces NET formation and lung injury. TLR9 deficiency in neutrophils prevents mtDNA-induced NETs, and TLR9 deficiency in either the lung donor or recipient decreases NET formation and lung injury in the PGD model. Compared with human lung transplant recipients without PGD, severe PGD was associated with high levels of BAL mtDNA and NETs, with evidence of relative deficiency in DNaseI. We conclude that mtDNA released during lung ischemia–reperfusion triggers TLR9-dependent NET formation and drives lung injury. In PGD, DNaseI therapy has a potential dual benefit of neutralizing a major NET trigger (mtDNA) in addition to dismantling pathogenic NETs.
Keywords: primary graft dysfunction, mitochondrial DNA, neutrophil extracellular traps, TLR9
Clinical Relevance
In primary graft dysfunction (PGD) after lung transplantation, we now have a more complete understanding of the pathophysiologic events, which begin with the release of mitochondrial DNA (mtDNA) during early reperfusion triggering TLR9-dependent neutrophil extracellular trap (NET) formation. In clinical samples, increased BAL mtDNA and NETs are clearly associated with more severe PGD. There are now multiple potential targets for therapeutic intervention in PGD, including mtDNA, TLR9, and NETs.
Solid organ transplantation is a life-saving approach for thousands of patients with end-stage organ dysfunction. Major advances have been made in perioperative management and in the treatment of acute and chronic rejection; however, many challenges remain to improve clinical outcomes. In the United States, more than 2,300 lung transplant surgeries are performed each year for a variety of end-stage lung diseases (1). The major early complication after transplantation is primary graft dysfunction (PGD), which occurs within the first 72 hours after surgery, drives early morbidity and mortality, and is a risk factor for chronic lung allograft dysfunction (2–4). PGD is induced by ischemia-reperfusion lung injury and shares many similarities to the acute respiratory distress syndrome (ARDS) (5). Indeed, it is defined by similar clinical criteria, and the grading system is based on the severity of the oxygenation defect (6). The pathophysiology of PGD is complex, but innate immune activation, perhaps excessive, is an integral feature. Neutrophils prominently accumulate in the lungs during PGD, and their activation is likely responsible for the breakdown in the lung barrier and alveolar flooding (7).
We have previously reported that neutrophil extracellular traps (NETs) are produced in abundance in a mouse model of PGD on the basis of prolonged donor lung cold ischemia and in humans with severe PGD (8). Although they are potentially protective in infectious conditions through microbial containment, NETs have been implicated in a variety of noninfectious diseases, where they have generally been responsible for bystander tissue injury (9, 10). In our model of PGD in mice, for example, the application of intrabronchial DNaseI therapy at the time of transplantation significantly reduced lung inflammation and improved lung function (8). In a mouse model of bacterial pneumonia, an important cause of ARDS, DNaseI therapy also reduced lung injury and improved survival, while not impairing bacterial containment (11).
The trigger for NET production in PGD is unknown. Here, we hypothesized that NET formation could be induced by damage-associated molecular patterns (DAMPs) released from injured cells during ischemia-reperfusion. Potential DAMPs previously implicated in inflammatory states, and that could be involved in the pathogenesis of PGD, include release products from damaged mitochondria (DNA with CpG motifs, formylated peptides), HMGB1, and ATP among others (12). Disrupted mitochondria are implicated in trauma-induced organ dysfunction, including lung injury (13), and we therefore focused our studies on mitochondrial DNA (mtDNA) and the potential for NET induction via endosomal TLR9 activation.
Methods
Additional detail on the mouse surgical procedures, lung intravital microscopy, NET measurements, and human biospecimens are provided in the data supplement.
Mice
Eight- to 12-week-old male mice housed under specific pathogen-free conditions were used for all experiments. C57BL/6J and mTmG mice were purchased from The Jackson Laboratory. TLR9−/− mice were obtained from Charles Kim (University of California, San Francisco [UCSF]). PAD4−/− mice were obtained from Kerri Mowen (The Scripps Research Institute).
Orthotopic Lung Transplant Model of PGD
Orthotopic left lung transplants were performed using the specified mouse genotypes, as previously described (8). The prolonged cold ischemia (PCI) model of PGD was induced by storing the left donor lung in sterile gauze soaked in Perfadex at 4°C for 18 hours before transplantation into the recipient mouse with minimal warm ischemia (8). A second group of lung transplants with no cold ischemia and minimal warm ischemia (immediate transplantation) were included as surgical controls for the PCI transplants. In selected experiments, DNaseI (2,000 units in 15 μl diluent; Roche) versus diluent alone (20 mM TRIS-HCl, 1 mM MgCl2) were directly instilled into the donor lung bronchus before bronchial anastomosis. mtDNA was extracted from mouse liver and isolated using the XIT Mitochondrial DNA kit (G-Biosciences). In selected experiments, mtDNA (50 μg in 15 μl diluent) versus diluent alone were directly instilled into the donor lung bronchus before bronchial anastomosis. All animals were killed 8 hours after transplantation.
Lung Intravital Microscopy
A modified version (14) of the previously published method of stabilized lung intravital imaging (15) was used. mTmG donor and recipient mice were used in minimal warm ischemia and PCI transplant models. To stain extracellular DNA, SYTOX Green was instilled intratracheally (50 μl of 1.0 mM solution) and intravenously (50 μl of 0.5 mM solution).
mtDNA and TLR9 Quantitative PCR
mtDNA was quantified in cell-free BAL (1 μL) fluid samples (mouse and human) by amplification of the mitochondria-specific Cytochrome C Oxidase 1 gene using mouse- or human-specific primer/probe sets (Applied Biosystems). TLR9 receptor expression was measured in bone marrow neutrophils from wild-type (WT) or TLR9−/− mice using TLR9 primers and GAPDH primers (Taqman; Thermo Fisher Scientific).
In vitro NET Generation and Immunofluorescence
Neutrophils (5 × 105) were plated and stimulated for 4 hours at 37°C and 5% CO2 with 4 μM ionomycin (Adipogen) or 100 μg/ml of mtDNA or nuclear DNA. Nuclear DNA was isolated with the NucleoSpin DNA RapidLyse kit (Macherey-Nagel). In select experiments, mtDNA was pretreated with 70 U of DNaseI (Roche) for 30 minutes at 37°C and then 5 minutes at 75°C to deactivate DNaseI (16) before incubation with neutrophils.
NET and DNaseI ELISAs
NETs in cell culture supernatants and mouse or human BAL/plasma samples were quantified using ELISAs as previously described (11). DNaseI levels in human BAL/plasma were measured using a DNaseI ELISA (MBS2024612; MyBiosurce) following the manufacturer’s instructions.
Acute Lung Injury Measurements
BAL total protein and white bloods cells and differential were measured as previously described (8, 11).
Arterial Blood Gas Measurements
Blood was collected from the left ventricle into heparinized syringes for arterial blood gas measurements. Arterial blood gases were measured using a VetScan i-STAT 1 handheld analyzer and CG4+ cartridges (Abaxis).
Human Biospecimens
BAL and plasma samples were prospectively collected from two independent cohorts of subjects (UCSF and University of California, Los Angeles) on the morning of the first postoperative day after lung transplant surgery from April 2016 to August 2017. BAL was obtained as part of routine clinical bronchoscopy by the subjects’ lung transplant clinician. PGD was defined and scored per international guidelines on the basis of chest radiography and oxygenation (17).
Statistics
All in vivo and in vitro experiments were repeated a minimum of three independent times. Unless otherwise noted, all mouse data are presented as mean ± SEM, and human subject data are presented as minimum-to-maximum whiskers and box plots showing the median and interquartile ranges. A P value ≤0.05 was considered significant. All statistical analyses were performed using GraphPad Prism 7.0 (GraphPad Software, Inc.) and STATA v15.1 (StataCorp).
Study Approval
All experiments were approved by the Institutional Animal Care and Use Committee (UCSF) and the Committee on Human Research (UCSF and University of California, Los Angeles). Written informed consent was obtained from participants before inclusion in the study.
Results
mtDNA Is Released during Experimental PGD and Is Completely Degraded by DNaseI
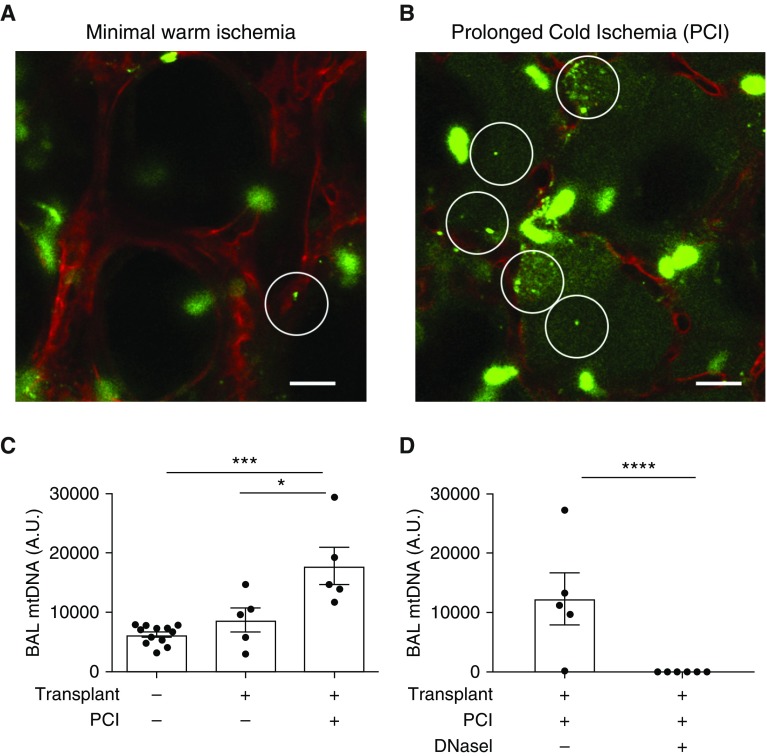
We used orthotopic lung transplantation in syngeneic mice and PCI of the donor lung (8) to test for the release of extracellular DNA (mtDNA or NETs) during organ reperfusion. Using intravital lung microscopy (18), we observed increased extracellular DNA in alveoli during graft reperfusion in mice transplanted with PCI compared with transplants with minimal warm ischemia (Figures 1A and 1B and Videos E1 and E2 in the data supplement). We reasoned that the intraalveolar DNA could be derived from NETs, as previously described (8), or released from injured lung cells during ischemia-reperfusion. We measured mtDNA accumulation in the BAL of transplanted mice by quantitative PCR using a mitochondrial-specific probe that binds to cytochrome C oxidase 1. Lungs transplanted with PCI produced higher mtDNA levels than mice transplanted with lungs with minimal warm ischemia or nontransplanted control BAL (Figure 1C). We instilled DNaseI intrabronchially immediately before lung anastomosis, which completely degraded mtDNA in the PCI model compared with the diluent control (Figure 1D).
Figure 1.
Mitochondrial DNA (mtDNA) is increased in experimental primary graft dysfunction (PGD) and is completely degraded by DNaseI treatment. (A and B) Lung intravital microscopy in orthotopic lung transplantation with donor lung minimal warm ischemia (A) or donor lung prolonged cold ischemia (PCI; B) (mTmG donor and recipient). Image is obtained within the first hour of reperfusion. SYTOX Green was administered (i.t. and i.v.) to highlight extracellular DNA (green). White circles indicate foci of intraalveolar extracellular DNA. Scale bars: 20 μm. (C and D) Cytochrome C Oxidase 1 (specific for mitochondrial DNA) was amplified by quantitative PCR (qPCR) from mouse BAL samples. All samples were obtained from wild-type (WT) mice in the following groups: nontransplanted mice (BAL only), lung transplants with no cold ischemia (immediate transplant), transplanted mice with donor lung PCI (18 h), transplanted mice with PCI treated with either diluent or DNaseI (2,000 units) intrabronchially at the time of transplantation. (C) Experimental PGD increased BAL mtDNA levels compared with controls. (D) mtDNA was undetectable in the PCI + DNaseI model. Diluent was added in the group without DNaseI. (C and D) Data were analyzed using one-way ANOVA (C) or Student’s t test (D) (n = 5–12). *P ≤ 0.05, ***P ≤ 0.001, and ****P ≤ 0.0001. A.U. = arbitrary unit; i.t. = intratracheal; i.v. = intravenous.
Intrabronchial Administration of mtDNA before Lung Anastomosis Induces NET Formation and Lung Injury
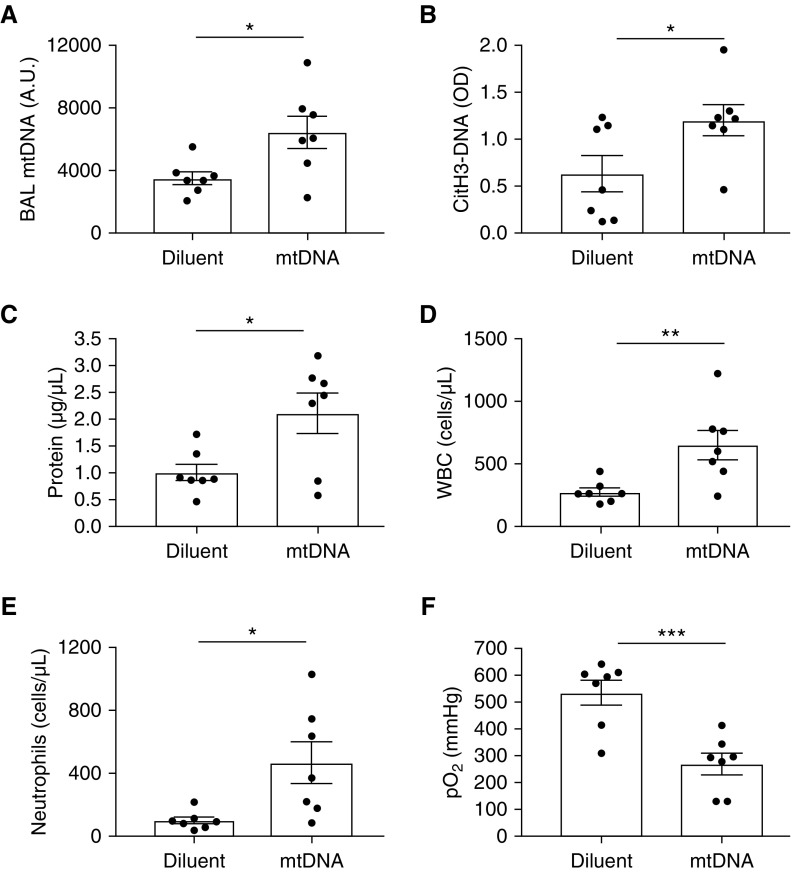
To determine if mtDNA is sufficient to cause lung injury in the setting of lung transplantation, we tested the adoptive transfer of mtDNA into the donor lung immediately before lung anastomosis in the minimal warm ischemia transplant model, which does not have elevated levels of BAL mtDNA during reperfusion (Figure 1C). As expected, mice instilled with intrabronchial mtDNA had higher values of BAL mtDNA at 8 hours after reperfusion than diluent controls (Figure 2A). The mtDNA group also had higher levels of BAL NETs (CitH3-DNA complexes; Figure 2B), increased protein permeability (Figure 2C), increased neutrophilic inflammation (Figures 2D and 2E), and decreased oxygenation (Figure 2F), all indicating that mtDNA is sufficient to induce NET formation and lung injury after lung transplantation.
Figure 2.
mtDNA is sufficient to induce neutrophil extracellular trap (NET) formation and lung injury after lung transplantation. Donor lungs with minimal warm ischemia were instilled intrabronchially with mtDNA (50 μg in 15 μl volume) or diluent (15 μl) immediately before bronchial anastomosis. (A) mtDNA measured by qPCR in mouse BAL samples. (B) Citrullinated NETs measured by citH3-DNA ELISA were higher in BAL samples of mice instilled with mtDNA than in diluent controls. (C–F) The mtDNA group had increased BAL protein (C), total white blood cells (WBC) (D), and neutrophils (E), and significantly reduced oxygenation capacity (F) versus diluent controls. Data were analyzed using Student’s t test (n = 7). *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001. citH3-DNA = citrullinated histone H3; OD = optical density; pO2 = arterial blood oxygen pressure.
mtDNA Induces TLR9-Dependent NET Formation
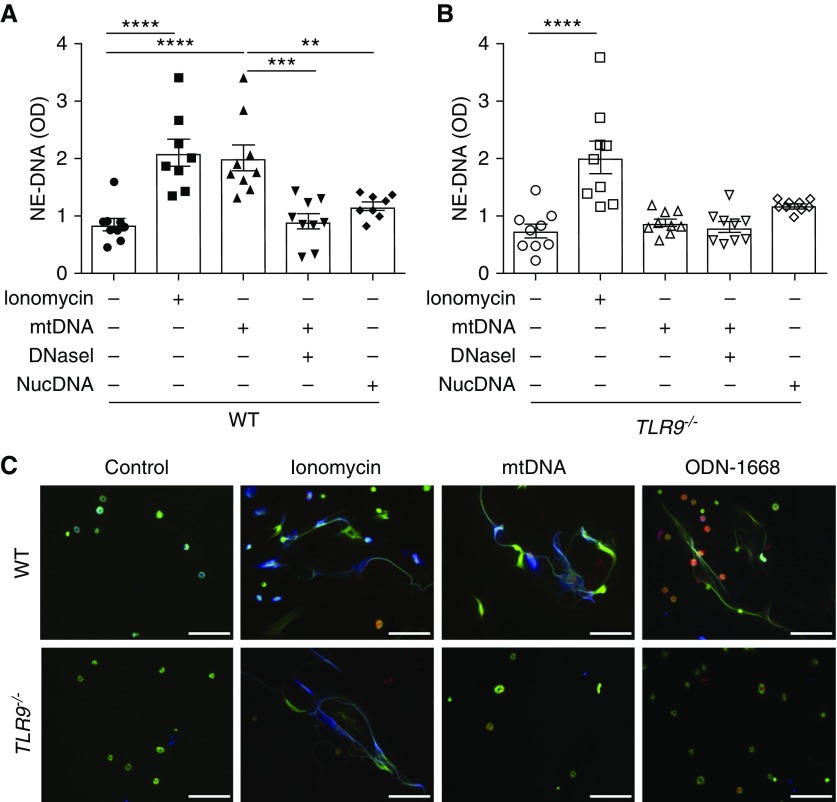
To further study the possible effect of cell-free mtDNA on NET induction, mouse neutrophils were isolated from bone marrow and stimulated with ionomycin (positive control) or mtDNA isolated from mouse liver. NETs were detected using a neutrophil elastase–DNA ELISA in cell-free supernatants and with immunofluorescence (8). Ionomycin and mtDNA-stimulated neutrophils both efficiently generated NETs (Figures 3A and 3C). Incubation of mtDNA with DNaseI before incubation with neutrophils failed to induce NETs, indicating that mtDNA was responsible for NET formation and not other products potentially contaminating the mtDNA during isolation and purification (Figure 3A). As an additional control, nuclear DNA was isolated from mouse liver and failed to induce NETs (Figure 3A).
Figure 3.
mtDNA stimulates TLR9-dependent NET formation. Bone marrow neutrophils from WT and TLR9−/− mice were isolated and stimulated with ionomycin (4 μM), mtDNA (100 μg/ml), DNaseI-degraded mtDNA (100 μg/ml), or nuclear DNA (nucDNA, 100 μg/ml) for 4 hours at 37°C. NETs were measured from the cell-free supernatants by ELISA. (A and B) Ionomycin stimulation increased NET production in WT and TLR9−/− neutrophils, but mtDNA only induced NETs in WT neutrophils. mtDNA digestion with DNaseI completely eliminated NET induction in WT neutrophils. Nuclear DNA did not stimulate NET production in either WT or TLR9−/− neutrophils. Data were analyzed using one-way ANOVA (n = 8–9). **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001. (C) Immunofluorescence staining of neutrophils from groups in A and B with antibodies against citrullinated histone 3 (blue) and neutrophil elastase (red), and DNA staining with SYTOX Green. The TLR9-specific agonist, ODN-1668 (10 μM), was added for this experiment. mtDNA and ODN-1668 stimulated NET formation in WT but not TLR9−/− neutrophils. Scale bars: 50 μm. NE-DNA = neutrophil elastase–DNA; ODN = oligodeoxynucleotides; TLR9 = Toll-like receptor 9.
We next tested the molecular pathway by which mtDNA induces NETs. Endosomal TLR9 recognizes CpG motifs in mtDNA, resulting in activation of NF-ĸB and downstream inflammatory pathways (13). Using quantitative PCR, we confirmed that TLR9 mRNA is present in isolated neutrophils (Figure E1). mtDNA failed to induce NETs in TLR9−/− neutrophils, whereas ionomycin stimulation efficiently induced NETs in these cells (Figures 3B and 3C). mtDNA incubated with DNaseI, or nuclear DNA, did not induce NET production in TLR9−/− neutrophils (Figure 3B). We also tested a TLR9-specific agonist (ODN-1668), which induced NETs in WT but not TLR9−/− neutrophils (Figure 3C).
TLR9 Deficiency Decreases NET Production and Lung Injury
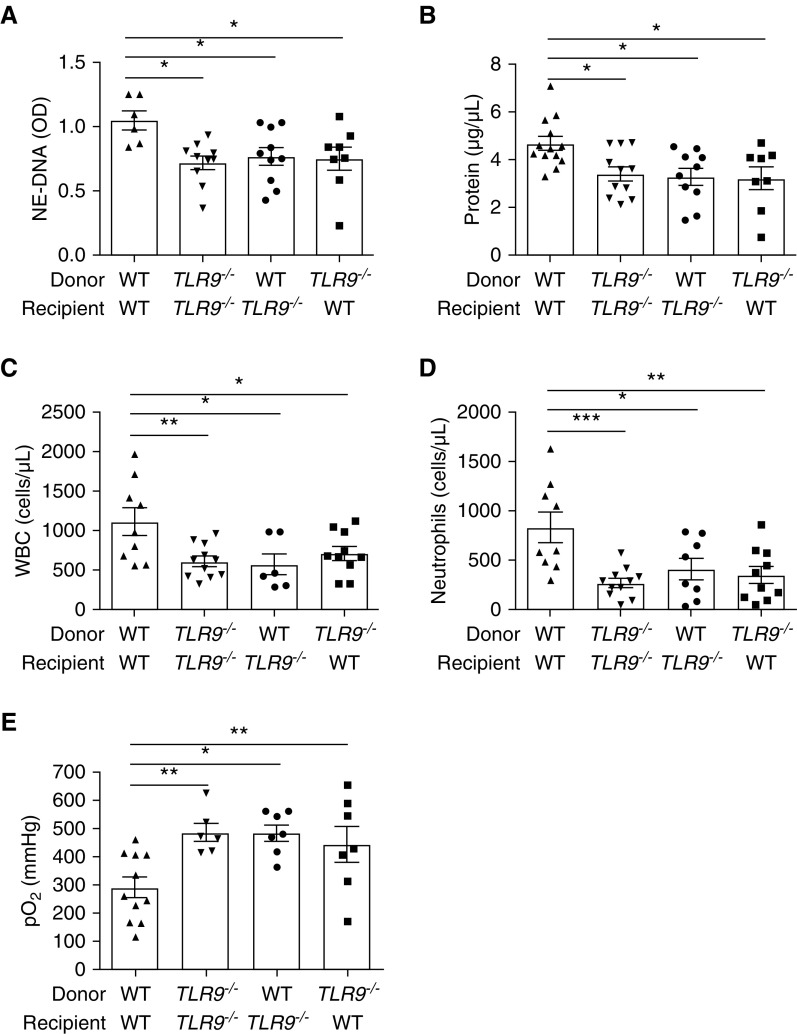
To test for the role of the mtDNA-TLR9 axis in the pathogenesis of PGD, we produced lung transplant chimeras in the PCI model in the following groups: WT donor lung transplanted into WT recipient, TLR9−/− donor lung transplanted into TLR9−/− recipient, WT donor lung transplanted into TLR9−/− recipient, or TLR9−/− donor lung transplanted into WT recipient. Mice were killed at 8 hours after transplantation and lung injury was measured. Compared with WT→WT transplants, any other combination of lung transplantation that incorporated TLR9 deficiency yielded decreased alveolar NETs (Figure 4A), decreased lung permeability (Figure 4B), decreased neutrophilic inflammation (Figures 4C and 4D), and improved lung function as measured by arterial oxygenation (Figure 4E). These data demonstrate that TLR9 is stimulated in both lung native cells and infiltrating immune cells in experimental PGD to generate NETs and produce lung injury.
Figure 4.
TLR9 deficiency decreases NET content and lung injury in experimental PGD. (A) In experimental PGD using PCI of the donor lung, NETs are present in BAL fluid of WT transplants and reduced in any combination of lung transplant that includes TLR9−/− donor or recipients. (B–E) The lack of TLR9 in either lung donor or recipient improves lung injury as measured by BAL total protein (B), BAL WBCs (C), BAL neutrophils (D), or pO2 (E). Data were analyzed using one-way ANOVA (n = 6–13). *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001.
Peptidyl Arginine Deiminase 4 Deficiency Reduces Citrullinated NETs and Limits the Extent of Lung Injury in PGD
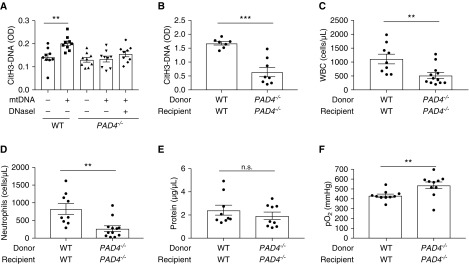
PAD4 (peptidyl arginine deiminase 4) is a critical mediator of NET formation by citrullinating histones and thus facilitating chromatin decondensation during NET formation (19). NET generation is decreased in PAD4-deficient mice (20) as well as with the use of PAD4-specific inhibitors (21). To further study the relevance of NET-induced lung injury in PGD, we analyzed NET generation and lung injury parameters in the absence of PAD4. We first confirmed the lack of citrullinated NETs (CitH3-DNA complexes) in PAD4−/− neutrophils after mtDNA stimulation relative to WT neutrophils (Figure 5A). Next, we tested PAD4-deficient lung transplants versus WT control transplants in the PGD model. PAD4-deficient transplants yielded decreased BAL citrullinated NETs (Figure 5B), neutrophilic inflammation (Figures 5C and 5D), and a trend toward decreased lung permeability (Figure 5E). Arterial oxygenation was significantly improved in PAD4-deficient transplants (Figure 5F).
Figure 5.
PAD4 deficiency reduces citrullinated NET generation and improves graft function. (A) Bone marrow neutrophils from WT or PAD4−/− mice were stimulated with mtDNA (100 μg/ml) or DNaseI-degraded mtDNA (100 μg/ml) for 4 hours at 37°C. WT neutrophils stimulated with mtDNA produced citrullinated NETs; however, no citH3-DNA complexes were produced in PAD4−/− neutrophils stimulated with mtDNA. (B) In experimental PGD, BAL NETs (citH3-DNA complexes) were reduced in the setting of donor and recipient PAD4 deficiency. (C, D, and F) The lack of PAD4 in both lung donor and recipient improves lung injury as measured by BAL WBCs (C), BAL neutrophils (D), or pO2 (F). (E) BAL total protein trended lower in the PAD4-deficient group but was not significant (n.s.). (A–F) Data were analyzed using one-way ANOVA (A) or Student’s t test (B–F) (n = 7–12). **P ≤ 0.01 and ***P ≤ 0.001. PAD4 = peptidyl arginine deiminase 4.
Severe PGD in Humans Is Associated with Increased BAL mtDNA and NET Levels and Increased NET/DNaseI Ratios
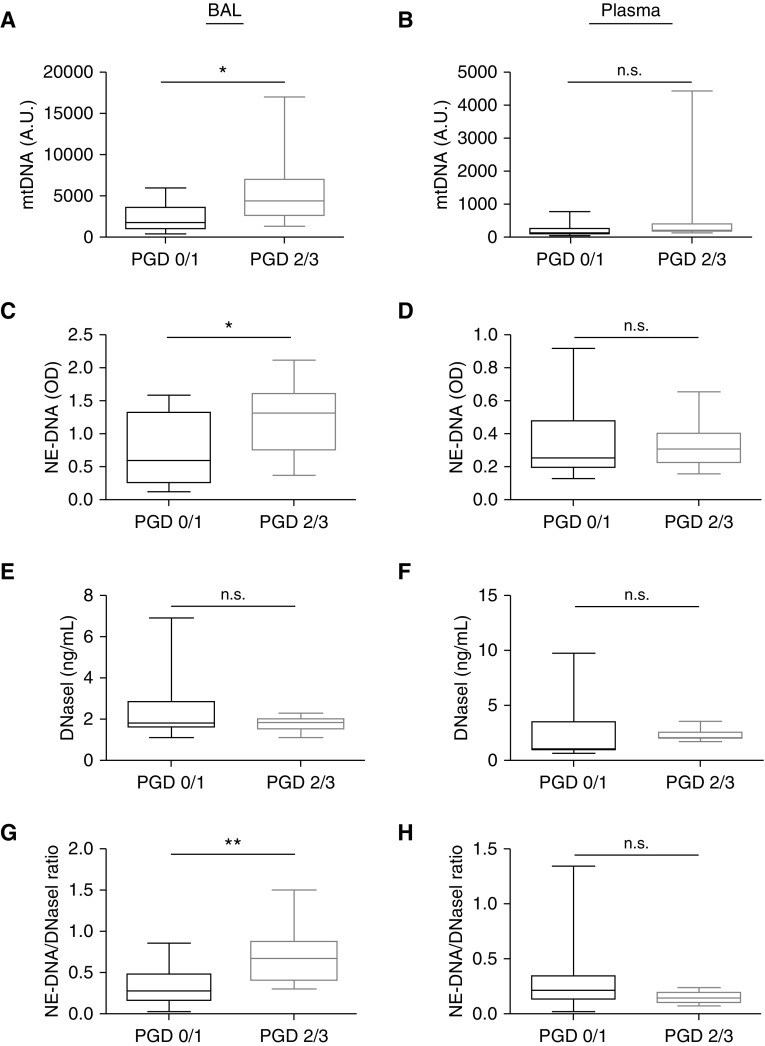
To verify our data in the experimental PGD model, we analyzed human BAL and plasma samples collected on the first postoperative day after lung transplantation at two medical centers. We grouped subjects into either no PGD at any time point (PGD 0/1 at 24, 48, and 72 h after transplant) or severe PGD (PGD 2/3) at each time point after transplant. There were no significant differences in the demographics and clinical characteristics between the severe PGD and no PGD groups (Table E1). We quantified total white blood cells, neutrophils, and monocytes in the BAL of these subjects, and although there was a trend toward higher neutrophil counts in patients with severe PGD, none of the three parameters were statistically different (Figures E2A–E2C). We detected higher mtDNA levels in BAL fluid, but not plasma, in severe PGD compared with no PGD (Figures 6A and 6B). NET levels were also increased in the BAL of severe PGD compared with no PGD (Figures 6C and 6D). Although DNaseI concentration was not significantly different between the two PGD groups (Figures 6E and 6F), there was a significant increase in the NETs to DNaseI ratio in the BAL of severe PGD compared with no PGD (Figures 6G–6H). Receiver operating characteristic curves generated for mtDNA, NETs, or NET/DNaseI measurements early after transplantation showed that BAL NETs/DNaseI was most associated with concurrent or subsequent severe PGD status (Figures E3A–E3D). We also correlated BAL mtDNA content with the arterial oxygen pressure/fraction of inspired oxygen in the human cohort and our mouse experiments and observed a negative correlation between BAL mtDNA and arterial oxygen pressure/fraction of inspired oxygen (human cohort correlation coefficient = −0.47, P = 0.0136; mouse model correlation coefficient = −0.53, P = 0.0333).
Figure 6.
Severe PGD in humans is associated with increased BAL mtDNA and NETs and relative deficiency of BAL DNaseI. (A and B) BAL and plasma mtDNA measurements (qPCR) in subjects with no PGD (PGD 0/1) or severe PGD (PGD 2/3). (C and D) BAL and plasma NETs (NE-DNA complexes) in no PGD or severe PGD. (E and F) BAL and plasma DNaseI levels in no PGD or severe PGD. (G and H) BAL and plasma NET/DNaseI levels in no PGD or severe PGD. Data were analyzed using the Mann-Whitney-Wilcoxon test. PGD 0/1 (n = 18); PGD 2/3 (n = 9). *P ≤ 0.05 and **P≤ 0.01.
Discussion
Ischemia-reperfusion injury is a complex process during which reduced ATP generation and increased reactive oxygen species can induce tissue injury and neutrophil accumulation/activation, leading to further injury (22). Neutrophil accumulation may lead to extracellular DNA trap formation that can perpetuate organ dysfunction by directly injuring endothelial and epithelial cells (9, 23). Previously, we described NET accumulation in experimental PGD and in humans with severe PGD and determined that dismantling extracellular chromatin with DNaseI in mice significantly improved graft function (8). However, the mechanism by which NETs were produced was not identified.
We have now discovered that mtDNA, released during lung ischemia-reperfusion injury, accumulates in the alveolar compartment, also the site of injury in PGD. Increased mtDNA levels have been reported in a variety of pathologic conditions, both sterile and pathogen induced, including sepsis, trauma, ARDS, and others (24–26). It is now clear that cell-free mtDNA is more than just a marker of cellular injury; it is a potent inducer of inflammatory responses. CpG motifs within mtDNA are recognized by TLR9 present in a variety of immune and nonimmune cells (27). mtDNA signals through TLR9 to induce NF-κB, MAPK, and other downstream inflammatory mediators (28, 29). TLR9, and other TLRs that recognize nucleic acids, initially traffic to the cell membrane, but, perhaps to restrict access to nucleic acids, ultimately reside on endosomal membranes. For example, cell-surface overexpression of TLR7, which is normally endosomal, leads to systemic autoimmunity through self-recognition of nucleic acids (30). Recent data indicate that red blood cells retain signaling-incompetent TLR9 on the cell surface that can serve to scavenge endogenous TLR9 agonists such as mtDNA to limit inflammation (31).
The administration of mtDNA to lung grafts immediately before transplantation was sufficient to induce lung injury and NET formation. mtDNA can trigger NET formation via TLR9 expressed in neutrophils, which concurs with a previous study in trauma, where mtDNA also induced NET production through TLR9 (32). mtDNA, but not nuclear DNA, robustly produces NETs in vitro. Preincubation of mtDNA with DNaseI (and subsequent DNaseI inactivation) completely blocks NET formation, as does the absence of TLR9. In chimeric lung transplants, the absence of TLR9 in either the lung donor or the recipient decreases NET formation and lung injury in experimental PGD.
We have studied the effects of targeting NETs to reduce lung injury using two independent approaches—the use of PAD4 deficiency and treatment with DNaseI. PAD4 is a critical enzyme that citrullinates histones to decondense chromatin during the process of NET formation. PAD4-deficient lung transplants have reduced citrullinated NETs and improved graft function. We have previously shown that intrabronchial DNaseI treatment at the time of syngeneic lung transplantation reduces NETs and lung injury and improves graft function (8). From our current study, it is now clear that DNaseI had a dual mode of protection in PGD. DNaseI efficiently degrades mtDNA, which is a major trigger of NETs in PGD, as well as efficiently degrading NET chromatin to limit tissue injury.
The treatment of ARDS is challenging, because the inflammatory/injury cascade is usually well established by the time ARDS is clinically recognized. However, with PGD, the at-risk period is known and injury can be anticipated and preventive strategies implemented, such as DNaseI delivered at the time of surgery or during ex vivo lung perfusion where NETs have been associated with worse recipient outcomes (33). These factors and the dual action of DNaseI in targeting NETs make it an appealing therapeutic option in PGD. Delivery of DNaseI at the time of transplantation may prevent mtDNA-induced NET formation entirely, which could be important if NET-cleavage fragments promote immune responses in the setting of alloimmunity (34).
Our human studies have enhanced our understanding of the relevance of these pathways in PGD. BAL and plasma samples were obtained during the early PGD at-risk period, and we confirmed previous results that elevation in BAL NETs, but not plasma, is associated with severe PGD status (8). In addition, we discovered that mtDNA is also elevated in severe PGD, similar to results obtained in our experimental PGD model. We hypothesized that DNaseI levels are reduced in severe PGD through consumption or underproduction. Although absolute DNaseI levels were not different in the PGD groups, we found that the NET/DNaseI ratio was most associated with severe PGD. In other words, NET overproduction and/or relative DNaseI deficiency correlates best with PGD. There are a variety of DNaseI polymorphisms that have been linked to reduced DNaseI levels, which should be studied in PGD susceptibility.
Our study is not without limitations. We did not determine the cellular origin of mtDNA in PGD, although we suspect that lung epithelial and endothelial cells are prominent sources, because these cells are the target of ischemia-reperfusion damage. These cells likely contribute the majority of mtDNA in experimental PGD (35). However, we previously identified that platelets prominently accumulate in the PGD model (8), and platelets can release intact mitochondria and mitochondria-containing microparticles (36). Therefore, it is possible that platelets significantly contribute mtDNA in PGD as well. Although we found that mtDNA was sufficient to induce PGD, there are likely other DAMPs released during lung ischemia-reperfusion injury that could trigger NET formation that we did not study (12). For example, formylated peptides released from damaged mitochondria could be involved (35), although these peptides (e.g., fMLP) do not induce NET formation (37–39). Finally, in our chimeric lung transplant experiments, we did not identify the critical cell(s) that mediate mtDNA-TLR9 signaling. Neutrophils are likely involved based on our in vitro experiments and the protection observed in transplant combinations that included a deficiency of TLR9 in the transplant recipient (hematopoietic cells). However, our data also clearly indicate that donor lung cells are responding to TLR9 agonists to facilitate lung inflammation and injury in PGD. TLR9 is expressed on a variety of lung stromal cells and alveolar macrophages (40), which could mediate these responses.
In summary, our work presents broad evidence that mtDNA is generated during ischemia-reperfusion injury after lung transplantation and is a potent trigger of pathogenic NET production via TLR9. It is likely that this pathophysiologic process is also important in other organ injuries and disease states that have reported dependency on the mtDNA-TLR9 axis. We also found a promising approach to treating NET-induced lung injury to improve lung transplant outcomes by targeting not only NET-associated chromatin but also mtDNA that is an important upstream driver of NET formation.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank the Biological Imaging Development Center at the University of California, San Francisco for assistance with the intravital imaging experiments.
Footnotes
Supported by National Institutes of Health grants R01 HL130324 and R01 AI125445 (M.R.L.), RO1 HL134851 (J.P.S.), and the Nina Ireland Program in Lung Health (M.R.L.).
Author Contributions: B.M. and M.R.L. designed the research studies. B.M., F.L., E.L., S.J.C., N.K., and M.M. conducted the experiments. B.M. and S.J.C. acquired the data. B.M., F.L., J.J.T., D.M.S., A.S., J.C., R.S., M.Y.S., D.J.R., A.D., J.P.L., A.A., S.S.W., J.A.B., S.R.H., J.A.G., L.E.L., R.J.S., M.E.K., A.V., J.K., J.P.S., and M.R.L. collected or processed the human biospecimens. B.M., E.L., S.J.C., N.K., M.M., J.P.S., and M.R.L. analyzed the data. B.M. and M.R.L. wrote the manuscript.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2019-0140OC on October 24, 2019
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Valapour M, Lehr CJ, Skeans MA, Smith JM, Carrico R, Uccellini K, et al. OPTN/SRTR 2016 annual data report: lung. Am J Transplant. 2018;18:363–433. doi: 10.1111/ajt.14562. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki Y, Cantu E, Christie JD. Primary graft dysfunction. Semin Respir Crit Care Med. 2013;34:305–319. doi: 10.1055/s-0033-1348474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chambers DC, Cherikh WS, Goldfarb SB, Hayes D, Jr, Kucheryavaya AY, Toll AE, et al. International Society for Heart and Lung Transplantation. The international thoracic organ transplant registry of the International Society for Heart and Lung Transplantation: thirty-fifth adult lung and heart-lung transplant report-2018. Focus theme: multiorgan transplantation. J Heart Lung Transplant. 2018;37:1169–1183. doi: 10.1016/j.healun.2018.07.020. [DOI] [PubMed] [Google Scholar]
- 4.Christie JD, Kotloff RM, Ahya VN, Tino G, Pochettino A, Gaughan C, et al. The effect of primary graft dysfunction on survival after lung transplantation. Am J Respir Crit Care Med. 2005;171:1312–1316. doi: 10.1164/rccm.200409-1243OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Perrot M, Liu M, Waddell TK, Keshavjee S. Ischemia-reperfusion-induced lung injury. Am J Respir Crit Care Med. 2003;167:490–511. doi: 10.1164/rccm.200207-670SO. [DOI] [PubMed] [Google Scholar]
- 6.Christie JD, Carby M, Bag R, Corris P, Hertz M, Weill D ISHLT Working Group on Primary Lung Graft Dysfunction. Report of the ISHLT working group on primary lung graft dysfunction part II: definition. A consensus statement of the International Society for Heart and Lung Transplantation. J Heart Lung Transplant. 2005;24:1454–1459. doi: 10.1016/j.healun.2004.11.049. [DOI] [PubMed] [Google Scholar]
- 7.Ross SD, Tribble CG, Gaughen JR, Jr, Shockey KS, Parrino PE, Kron IL. Reduced neutrophil infiltration protects against lung reperfusion injury after transplantation. Ann Thorac Surg. 1999;67:1428–1433. doi: 10.1016/s0003-4975(99)00248-9. [Discussion p. 1434.] [DOI] [PubMed] [Google Scholar]
- 8.Sayah DM, Mallavia B, Liu F, Ortiz-Muñoz G, Caudrillier A, DerHovanessian A, et al. Lung Transplant Outcomes Group Investigators. Neutrophil extracellular traps are pathogenic in primary graft dysfunction after lung transplantation. Am J Respir Crit Care Med. 2015;191:455–463. doi: 10.1164/rccm.201406-1086OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caudrillier A, Kessenbrock K, Gilliss BM, Nguyen JX, Marques MB, Monestier M, et al. Platelets induce neutrophil extracellular traps in transfusion-related acute lung injury. J Clin Invest. 2012;122:2661–2671. doi: 10.1172/JCI61303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jorch SK, Kubes P. An emerging role for neutrophil extracellular traps in noninfectious disease. Nat Med. 2017;23:279–287. doi: 10.1038/nm.4294. [DOI] [PubMed] [Google Scholar]
- 11.Lefrançais E, Mallavia B, Zhuo H, Calfee CS, Looney MR. Maladaptive role of neutrophil extracellular traps in pathogen-induced lung injury. JCI Insight. 2018;3:e98178. doi: 10.1172/jci.insight.98178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Todd JL, Palmer SM. Danger signals in regulating the immune response to solid organ transplantation. J Clin Invest. 2017;127:2464–2472. doi: 10.1172/JCI90594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Headley MB, Bins A, Nip A, Roberts EW, Looney MR, Gerard A, et al. Visualization of immediate immune responses to pioneer metastatic cells in the lung. Nature. 2016;531:513–517. doi: 10.1038/nature16985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Looney MR, Thornton EE, Sen D, Lamm WJ, Glenny RW, Krummel MF. Stabilized imaging of immune surveillance in the mouse lung. Nat Methods. 2011;8:91–96. doi: 10.1038/nmeth.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Z, Fasco MJ, Kaminsky LS. Optimization of Dnase I removal of contaminating DNA from RNA for use in quantitative RNA-PCR. Biotechniques. 1996;20:1012–1014, 1016, 1018–1020. doi: 10.2144/96206st02. [DOI] [PubMed] [Google Scholar]
- 17.Snell GI, Yusen RD, Weill D, Strueber M, Garrity E, Reed A, et al. Report of the ISHLT working group on primary lung graft dysfunction, part I: definition and grading-A 2016 consensus group statement of the International Society for Heart and Lung Transplantation. J Heart Lung Transplant. 2017;36:1097–1103. doi: 10.1016/j.healun.2017.07.021. [DOI] [PubMed] [Google Scholar]
- 18.Lefrançais E, Ortiz-Muñoz G, Caudrillier A, Mallavia B, Liu F, Sayah DM, et al. The lung is a site of platelet biogenesis and a reservoir for haematopoietic progenitors. Nature. 2017;544:105–109. doi: 10.1038/nature21706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hemmers S, Teijaro JR, Arandjelovic S, Mowen KA. PAD4-mediated neutrophil extracellular trap formation is not required for immunity against influenza infection. PLoS One. 2011;6:e22043. doi: 10.1371/journal.pone.0022043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li P, Li M, Lindberg MR, Kennett MJ, Xiong N, Wang Y. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J Exp Med. 2010;207:1853–1862. doi: 10.1084/jem.20100239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis HD, Liddle J, Coote JE, Atkinson SJ, Barker MD, Bax BD, et al. Inhibition of PAD4 activity is sufficient to disrupt mouse and human NET formation. Nat Chem Biol. 2015;11:189–191. doi: 10.1038/nchembio.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalogeris T, Baines CP, Krenz M, Korthuis RJ. Cell biology of ischemia/reperfusion injury. Int Rev Cell Mol Biol. 2012;298:229–317. doi: 10.1016/B978-0-12-394309-5.00006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saffarzadeh M, Juenemann C, Queisser MA, Lochnit G, Barreto G, Galuska SP, et al. Neutrophil extracellular traps directly induce epithelial and endothelial cell death: a predominant role of histones. PLoS One. 2012;7:e32366. doi: 10.1371/journal.pone.0032366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puskarich MA, Shapiro NI, Trzeciak S, Kline JA, Jones AE. Plasma levels of mitochondrial DNA in patients presenting to the emergency department with sepsis. Shock. 2012;38:337–340. doi: 10.1097/SHK.0b013e318266a169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Q, Itagaki K, Hauser CJ. Mitochondrial DNA is released by shock and activates neutrophils via p38 map kinase. Shock. 2010;34:55–59. doi: 10.1097/SHK.0b013e3181cd8c08. [DOI] [PubMed] [Google Scholar]
- 26.Nakahira K, Kyung SY, Rogers AJ, Gazourian L, Youn S, Massaro AF, et al. Circulating mitochondrial DNA in patients in the ICU as a marker of mortality: derivation and validation. PLoS Med. 2013;10:e1001577. doi: 10.1371/journal.pmed.1001577. [Discussion, p. e1001577.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 28.Gu X, Wu G, Yao Y, Zeng J, Shi D, Lv T, et al. Intratracheal administration of mitochondrial DNA directly provokes lung inflammation through the TLR9-p38 MAPK pathway. Free Radic Biol Med. 2015;83:149–158. doi: 10.1016/j.freeradbiomed.2015.02.034. [DOI] [PubMed] [Google Scholar]
- 29.Zhang JZ, Liu Z, Liu J, Ren JX, Sun TS. Mitochondrial DNA induces inflammation and increases TLR9/NF-κB expression in lung tissue. Int J Mol Med. 2014;33:817–824. doi: 10.3892/ijmm.2014.1650. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Pisitkun P, Deane JA, Difilippantonio MJ, Tarasenko T, Satterthwaite AB, Bolland S. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 2006;312:1669–1672. doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- 31.Hotz MJ, Qing D, Shashaty MGS, Zhang P, Faust H, Sondheimer N, et al. Red blood cells homeostatically bind mitochondrial DNA through TLR9 to maintain quiescence and to prevent lung injury. Am J Respir Crit Care Med. 2018;197:470–480. doi: 10.1164/rccm.201706-1161OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itagaki K, Kaczmarek E, Lee YT, Tang IT, Isal B, Adibnia Y, et al. Mitochondrial DNA released by trauma induces neutrophil extracellular traps. PLoS One. 2015;10:e0120549. doi: 10.1371/journal.pone.0120549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caldarone L, Mariscal A, Sage A, Khan M, Juvet S, Martinu T, et al. Neutrophil extracellular traps in ex vivo lung perfusion perfusate predict the clinical outcome of lung transplant recipients. Eur Respir J. 2019;53:e1801736. doi: 10.1183/13993003.01736-2018. [DOI] [PubMed] [Google Scholar]
- 34.Scozzi D, Wang X, Liao F, Liu Z, Zhu J, Pugh K, et al. Neutrophil extracellular trap fragments stimulate innate immune responses that prevent lung transplant tolerance. Am J Transplant. 2019;19:1011–1023. doi: 10.1111/ajt.15163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scozzi D, Ibrahim M, Liao F, Lin X, Hsiao HM, Hachem R, et al. Mitochondrial damage-associated molecular patterns released by lung transplants are associated with primary graft dysfunction. Am J Transplant. 2019;19:1464–1477. doi: 10.1111/ajt.15232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boudreau LH, Duchez AC, Cloutier N, Soulet D, Martin N, Bollinger J, et al. Platelets release mitochondria serving as substrate for bactericidal group IIA-secreted phospholipase A2 to promote inflammation. Blood. 2014;124:2173–2183. doi: 10.1182/blood-2014-05-573543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papayannopoulos V. Neutrophil extracellular traps in immunity and disease. Nat Rev Immunol. 2018;18:134–147. doi: 10.1038/nri.2017.105. [DOI] [PubMed] [Google Scholar]
- 38.Yuen J, Pluthero FG, Douda DN, Riedl M, Cherry A, Ulanova M, et al. NETosing neutrophils activate complement both on their own NETs and bacteria via alternative and non-alternative pathways. Front Immunol. 2016;7:137. doi: 10.3389/fimmu.2016.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Remijsen Q, Kuijpers TW, Wirawan E, Lippens S, Vandenabeele P, Vanden Berghe T. Dying for a cause: NETosis, mechanisms behind an antimicrobial cell death modality. Cell Death Differ. 2011;18:581–588. doi: 10.1038/cdd.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneberger D, Caldwell S, Kanthan R, Singh B. Expression of Toll-like receptor 9 in mouse and human lungs. J Anat. 2013;222:495–503. doi: 10.1111/joa.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.