Abstract
Antibiotic-resistant bacteria (ARB) and antibiotic resistance genes (ARGs) have been recognized as one of the biggest public health issues of the 21st century. Both ARB and ARGs have been determined in water after treatment with conventional disinfectants. Ultraviolet (UV) technology has been seen growth in application to disinfect the water. However, UV method alone is not adequate to degrade ARGs in water. Researchers are investigating the combination of UV with other oxidants (chlorine, hydrogen peroxide (H2O2), peroxymonosulfate (PMS), and photocatalysts) to harness the high reactivity of produced reactive species (Cl·, ClO·, Cl2·−, ·OH, and SO4·−) in such processes with constituents of cell (e.g., deoxyribonucleic acid (DNA) and its components) in order to increase the degradation efficiency of ARGs. This paper briefly reviews the current status of different UV-based treatments (UV/chlorination, UV/H2O2, UV/PMS, and UV-photocatalysis) to degrade ARGs and to control horizontal gene transfer (HGT) in water. The review also provides discussion on the mechanism of degradation of ARGs and application of q-PCR and gel electrophoresis to obtain insights of the fate of ARGs during UV-based treatment processes.
Keywords: Antibiotic resistance bacteria, Advanced oxidation processes, Disinfection, Reactive chlorine species, Sulfate radicals, Reactive oxygen species
1. Introduction
One of the most significant discoveries in the field of medicine is the development of antibiotics to treat human and animal diseases (Travis et al., 2018). The consumption of antibiotics is rising because of increase in human population and its demands (e.g., use of antibiotics increased up to 35% between 2000 and 2010) (Van Boeckel et al., 2014, 2015). Consumption of antibiotics in livestock and aquaculture is also on the rise (Kim et al., 2013; Sharma et al., 2016; Blaskovich, 2018; Hu et al., 2019a). Antibiotics undergo incomplete metabolic transformation that results in significant excretion of antibiotics and their residues into the environment (Khetan and Collins, 2007; Lee et al., 2017). In addition, the excessive consumption of antibiotics has led to significant increase of antibiotic residues in different compartments of the environment such as sources of drinking water supply, wastewater effluent, sludge, and soil (Rodríguez-Chueca et al., 2019; Zhang et al., 2019b). In the past few years, studies have shown that applications of antibiotics in human and animal health care could accelerate the occurrence of antibiotic-resistant bacteria (ARB) and antibiotic resistance genes (ARGs) in different water bodies, which include hospital waste effluents, water treatment plant effluents, and tap water (Michael et al., 2013; Shao et al., 2018; Yang et al., 2018). Excessive antibiotics have caused the proliferation of ARB and subsequently horizontal gene transfer (HGT) results in widespread ARGs in the environment (Sharma et al., 2016; Garner et al., 2018). In different water bodies of the world, diverse ARGs correspond to macrolides, tetracyclines, quinolones, and sulfonamides (erm, tet, qnr, and sul, respectively) (Lee et al., 2017). The appearance of ARB and ARGs in water resources has caused a great concern due to the possibility of millions of deaths by 2050 (Vindenes et al., 2016). According to the Center for Disease Control (CDC) and the World Health Organization (WHO), ARGs in the environment are one of the most critical public health issues of this century (World Health Organization, 2018). Consequently, control of the release of ARB and ARGs to the aquatic environment is of utmost importance.
Most of the wastewater treatment plants (WWTPs) are not adequate to be fully designed to remove antibiotics (Cizmas et al., 2015; Sousa et al., 2017, 2018). Unfortunately, when high levels of activated sludge are applied in biological treatments, the microbes caused proliferation of ARB and ARGs (Li et al., 2017; Ezzariai et al., 2018; Krzeminski et al., 2019). In treatment of drinking water, chlorination, chloramination, and ultraviolet (UV) radiation are employed to inactivate pathogenic microorganisms (Wang et al., 2017a; Yoon et al., 2017; Ren et al., 2018; Chang et al., 2019; Rodríguez-Chueca et al., 2019; Zhang et al., 2019c). The use of chlorine to inactivate ARB and ARGs has given conflicting effectiveness (Zhang et al., 2009; Munir et al., 2011; Li et al., 2016a). Sodium hypochlorite was able to inactivate tetracycline-resistant bacteria containing tetA and terC (Zhang et al., 2009). In another study, chlorination was found to have no effectiveness in treating ARB and ARGs (Munir et al., 2011). However, one of the studies showed that the residual chlorine after treatment behaved like antibiotics that prompted development of ARGs (Li et al., 2016b). Additionally, low dosages of free chlorine and chloramine caused facilitation of the HGT of ARGs through altering the cell permeability of bacteria (Sinha and Häder, 2002). Direct UV photolysis may be suitable to control ARGs because it can transform nitrogen bases of DNA (Cutler and Zimmerman, 2011). However, the damage to DNA varies with cell structure and environmental conditions (Zhang et al., 2019c). To completely inactivate ARGs, high dose of UV may be needed (Chen and Zhang, 2013; Lee et al., 2017; Mauter et al., 2018). The effectiveness of UV technology to control ARGs remains a challenge. In recent years, researchers have been investigating a combination of UV technology with other oxidation processes to achieve efficient control of ARGs and HGT (Lin et al., 2016; Pang et al., 2016; Sharma et al., 2016; Wang et al., 2017c; Michael-Kordatou et al., 2018; He et al., 2019; Hu et al., 2019b; Yang et al., 2019). This review focuses on the recent investigations on the combination of UV with oxidants, which include chlorine, hydrogen peroxide, and photocatalysts (i.e., to create advanced oxidation processes (AOPs)). Examples of each combined process are briefly summarized.
2. UV/chlorination
Studies have shown that the combination of UV and chlorination (UV/chlorine) is more effective in degrading micropollutants than UV or chlorination alone (Cheng et al., 2018; Zhang et al., 2018). Studies on micropollutants included hormones, pharmaceuticals, taste and odor compounds (Ting and Praveena, 2017; Fang et al., 2018; Chang et al., 2019). In the UV/chlorine process, hydroxyl radical (•OH/O• – ) and reactive chlorine species (Cl•, ClO•, and Cl2• – ) are generated (reactions 1–5) (Wu et al., 2016, 2017; Kong et al., 2018; Hua et al., 2019; Zhao et al., 2019).
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
The advantages of UV/chlorine process through involvement of reactive oxidizing species have been investigated in removing sul1, tetX, tetG, intI1, and 16S rRNA genes in municipal wastewater treatment plant (MWTP) effluent by chlorination, UV, and sequential UV/chlorine process (Zhang et al., 2015b, 2019c). Higher N-content in samples decreased the removal efficiency by chlorination process. This study showed that the removal efficiency of ARGs was higher by the UV irradiation followed by chlorination than UV or chlorination alone (Zhang et al., 2015b).
In a recent study, the removal of ARGs was studied by isolating ARB (HLS-6) with sul1 and sul2 from a drinking water sample (Zhang et al., 2019c). UV, chlorination, and UV/chlorine treatments of drinking water were sought to remove ARGs and to control HGT. The order of degradation of both sul1-qPCR and intI1-qPCR in the less than 20 min using different processes was UV/ chlorine>chlorination>UV. The UV/chlorine was able to obtain log reductions of 3.50 and 4.00 for sul1-qPCR and intI1-qPCR, respectively. Increase in pH of the water from 5.0 to 9.0 decreased the removal efficiency of ARGs. In presence of sulfamethoxazole antibiotic, removal efficiency of ARGs by UV/chlorine was more than either UV or chlorination alone (Zhang et al., 2019c). Significantly, damage of DNA was possible in the UV/chlorine process, confirmed by electrophoresis technique.
3. UV/hydrogen peroxide
UV/hydrogen peroxide (UV/H2O2) process has been studied extensively to degrade contaminants in water (Wols and Hofman-Caris, 2012; Wols et al., 2013; Miklos et al., 2018; Nihemaiti et al., 2018). This method is based on the production of •OH, which has a high redox potential (1.8–2.7 V) (Buxton et al., 1988). The nature of •OH is electrophilic and can attack electron-rich organic contaminants at high rate constant (~108 –109 L/mol/s (Buxton et al., 1988; Sharma, 2008; von Gunten, 2018) that ultimately leads to their transformation to CO2 and H2O. The interest in the role of •OH in treatment of ARGs is because of damage of DNA, induced by •OH attack to sugar backbone oxidized products (Naumov and von Sonntag, 2008). Investigation has been performed on UV/H2O2 treatment of plasmid-encoded ARGs (Chang et al., 2017; Guo et al., 2017; Yoon et al., 2017; Hu et al., 2018; Yoon et al., 2018; Chen et al., 2019; Rodríguez-Chueca et al., 2019). Both ARGs of Escherichia coli (E. coli), i.e., extracellular (e-ARG) and intercellular (i-ARG), were determined (Yoon et al., 2017, 2018). Concentrations of ARGs were evaluated by qPCR and structure changes were determined by gel electrophoreses (Buxton, 2008; Yoon et al., 2017).
A recent study investigated UV254 nm and UV254 nm/H2O2 treatments of E. coli and ARGs (Yoon et al., 2017). The damages to ampR (850 bp) and kanR (806 bp) amplicons, situated in the pUC4K of ARGs in host E. coli, were observed. The inactivation of E. coli was faster than the damages to ARGs. The dose of UV254 nm to obtain 4 log reduction of ARGs was 50–130 mJ/cm. The increase in pH did not have any influence on damage to ARGs by UV254 nm/H2O2. In contrast, in the case of chlorination, performance decreased with increasing pH (Yoon et al., 2017). In the UV254 nm/H2O2 treatment, the damage of eARGs was at a much faster rate than the damage to iARGs. This was expected because the reactive •OH species in the case of i-ARGs could not penetrate the cell and the oxidizing species were easily consumed by the cellular components.
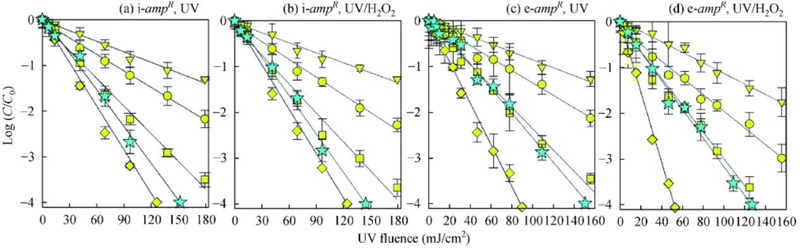
In a recent study, deactivation of transforming activity of an ARG was investigated to elucidate the mechanism of elimination of ARGs in E. coli (host) (Yoon et al., 2018). The log10 reduction was ~1 for the plasmid-encoded ARGs under UV254 nm of 40 mJ/cm2, which is usually applied in UV disinfection processes (Yoon et al., 2018). However, UV254 nm of 150 mJ/cm2 achieved more than 4 log reduction. When UV254 nm/H2O2 method was applied, no enhanced degradation efficiency was seen (i.e., compared with UV254 nm irradiation). Figure 1 presents the logarithmic relative concentrations of ampR amplicons that had variable length after UV254 nm and UV254 nm/H2O2 treatments of i-ARGs (Yoon et al., 2018). The degradation followed first-order kinetics (Fig. 1). Significantly, the rate of destruction of ampR increased with increase in size of ampR in both treatment processes. The obtained first-order rate constants for ampR (pUC19) were 3.4 × 10–2, 7.0 x 10–2, 1.1 × 10–1, and 1.5 × 10−1 cm2/mJ for 192 bps, 400 bps, 603 bps, and 851 bps qPCR amplicons for both UV254 nm and UV254 nm/H2O2 treatments, respectively. The suggested mechanisms of the elimination of transforming activity of plasmids (both extra- and intracellular) in both treatments include damage to DNA. The rate of the transformation of pUC19 was lower than the rate of the formation of cyclobutene-pyrimidine dimer (CPD); suggesting the repair of CPD in the cell (host).
Fig. 1.
Logarithmic relative concentration of the transforming activity (green color star) and ampR qPCR amplicons (192 bps (trigonal), 400 bps (circle), 603 bps (square) and 851 bps (diamond)) as a function of UV fluence during treatment of ((a) and (b)) intracellular and ((c) and (d)) extracellular pUC19 with ((a) and (c)) UV and ((b) and (d)) UV/H2O2 ([H2O2]0 = 10 mg/L) at pH 7.0. The symbols represent the measured data and the error bars represent one standard deviation from triplicate experiments. The lines are linear regressions of the data. (Adapted from Yoon et al. (2018) with the permission of the Royal Society of Chemistry).
4. UV/peroxymonosulfate
The UV/peroxymonosulfate (PMS) process has shown increasing attention in the last few years due to high redox potential of PMS (+1.82 V) and generation of sulfate radical (SO4•– ) (Rodríguez-Chueca et al., 2019; Umar et al., 2019). The generated SO4•– has comparable redox potential to •OH (2.5–3.1 V versus 1.8–2.7 V) (Neta and Huie, 1985; Sharma, 2013). Many investigations showed the effective degradation of organic contaminants by SO4•– (Zhang et al., 2015a; Ghanbari and Moradi, 2017; Liu et al., 2018; Wojnárovits and Takács, 2019). An attempt has been made on studying the role of UV/PMS in deactivation of ARGs (Rodríguez-Chueca et al., 2019). This study tested the UV-C, UV-C/H2O2 (0.05–0.5 mmol/L), and UV-C/PMS (0.05–0.5 mmol/L) processes at 4–18 s contact time of UV-C. The combined methods of UV-C/ PMS and UV-C/H2O2 using 0.5 mmol/L PMS were more effective to remove antibiotics than UV-C alone at a contact time of 0.7 s. Interestingly, the removal of ARGs had the quite opposite trend, i.e., UV-C was more effective in removing ARGs than combined UV-C/oxidant methods. It suggests that the oxidants in the combined methods (H2O2 and PMS) could also absorb UV-C irradiation that decreased the direct photolysis of DNA compared with UV-C alone. Furthermore, in the removal of ARGs by UV/H2O2 and UV/PMS methods, both photolysis of DNA and oxidation by generated •OH and SO4•– were operational, but cumulative effects were lower than the direct removal of ARGs by UV-C alone. In removing antibiotics, generated radicals were efficient because of high reactivity with target contaminants in water, while UV-C alone had no or low effect.
5. UV-photocatalyst
Heterogeneous titanium dioxide (TiO2) photocatalytic process has numerous applications, including the inactivation of microorganisms (Wang et al., 2017b; Uyguner Demirel et al., 2018). TiO2 under UV irradiation generates reactive oxygen species (ROS), which could inactivate ARB and ARGs (Rizzo et al., 2014; Dunlop et al., 2015; Krzeminski et al., 2019; Özkal et al., 2019; Zhang et al., 2019a). In an earlier work, photocatalytic disinfection of three strains of E. coli (J-53R and HT-99, rifampicin and chloramphenicol resistant strains, respectively, and K12, antibiotic sensitive strain) resulted in decrease of viable cell numbers of ARB from 3 log10 to 0.5 log10 after 180 min of treatment (Dunlop et al., 2015). Interestingly, after post-treatment incubation for 180 min for both ARB (37°C and 24 h), a bacterial recovery of 3 log10 was observed. Comparatively, at 150 min post-treatment, no E. coli K12 was recovered. This study recommended that the photocatalytic disinfection must be carried out for a long period of time to avoid post-treatment recovery, which will minimize the highly unwanted transfer of ARGs among bacteria (Dunlop et al., 2015).
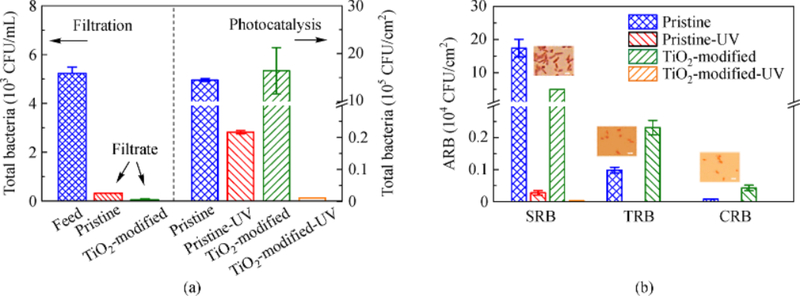
An attempt has been made to remove ARB and ARGs from wastewater effluent using photocatalytic reactive ultrafiltration membrane (Ren et al., 2018). In this study, TiO2 modified polyvinylidene fluoride (PVDF) membrane was used in the wastewater effluent, which could retain ARB. The removal efficiency was tested for ARGs (plasmid-mediated floR, sul1, and sul2). Almost complete removal and inactivation of total ARB were observed by both filtration and UV treatment (Fig. 2(a)). Significantly, retention of ARB by using pristine PVDF membrane was 98.9%, which was lower than the retention by TiO2-modified PVDF (>99.9%) (Fig. 2(a), left). As shown in Fig. 2(a) (right), removal of total bacteria by TiO2-modified PVDF membrane was much higher than that by pristine PVDF membrane. This finding was again confirmed by determining abundance of eRNA in the permeate (Ren et al., 2018).
Fig. 2.
((a), left) Total bacteria abundance in the feed and filtrate obtained from UF experiments. The secondary wastewater effluent was filtered by the pristine PVDF and TiO2-modified PVDF membranes until the permeate volume reached 250 mL, at a pressure of 1.4 bar (20 psi) and temperature of 25.0°C ± 0.5°C. ((a), right) Photocatalytic degradation of total bacteria on the surface of the pristine PVDF and TiO2-modified PVDF membranes before and after exposure to UV for 1 h. (b) CFU of antibiotic resistant bacteria (ARB) on the surfaces of the pristine PVDF membrane and TiO2-modified PVDF membrane, respectively, before and after exposure to UV irradiation, measured via spread plate method. Microscopic images of ARB are shown in the inset with a 2-μm scale bar. (Adapted from Ren et al. (2018) with the permission of the American Chemical Society).
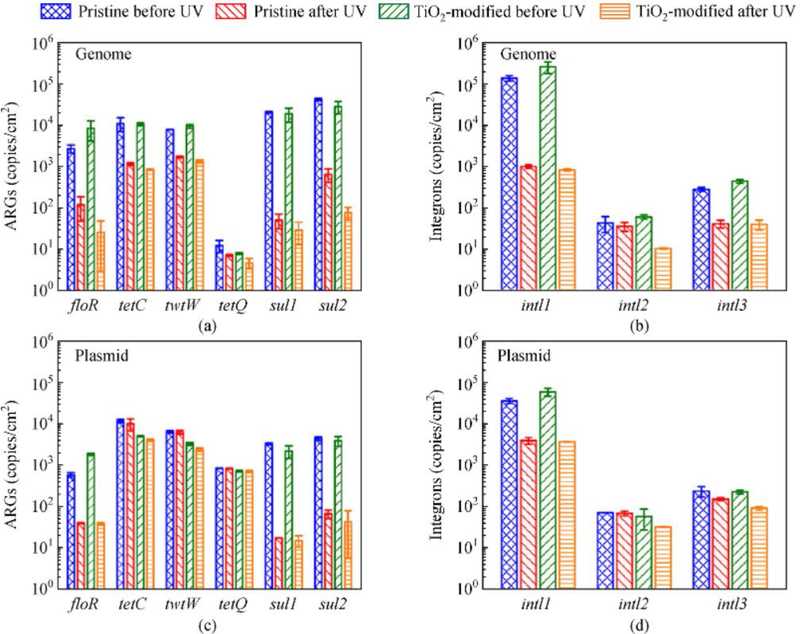
Figure 3 shows the results of photocatalytic experiments performed to study the degradation of ARGs and integrons, retained by pristine PVDF and TiO2-modified PVDF membranes. The membrane was exposed to UV irradiation for 1 h. Higher degradation efficiency of ARGs and integrons present in genome was seen than those located in plasmid (Figs. 3(a) and 3(b) versus Figs. 3(c) and 3(d)). Results clearly demonstrated the role of location of ARGs and integrons in the bacterial cell. It appears that the large size genome could allow the attack by ROS easily compared with the feasibility of the reactions of ROS with ARGs and integrons of relatively small size plasmid. Generally, the UV-treatment using the TiO2-modified PVDF membrane had higher efficiency of degradation of ARG and integrons than that of applying PVDF membrane alone (Fig. 3). Results of Fig. 3 agree with the results seen on inactivation of ARB (see Fig. 2). The efficiencies of degradation of floR, tetC, sul1, and intI1 in the plasmid in the UV-TiO2-modified PVDF membrane were somewhat higher than those by UV-pristine PVDF membrane. The located sulfonamides resistance genes (sul1 and sul2) in genome and plasmid had higher degradation efficiency than that of tetracycline resistance genes (tetC, tetW, and tetQ). The sulfonamides resistance genes tend to be degraded more easily than tetracycline resistance genes (Auerbach et al., 2007). Overall, results of Fig. 3 indicated the enhanced activity of UV-TiO2 to degrade ARG and integrons.
Fig. 3.
Photocatalytic degradation of ARGs and integrons on the surface of pristine PVDF and TiO2-modified PVDF membranes after UV treatment. ARGs and integrons in genome ((a), (b)) and plasmid ((c), (d)) were extracted using bacteria DNA kit and plasmid kit, respectively, and analyzed via quantitative PCR method (Adapted from Ren et al. (2018) with the permission of the American Chemical Society).
Research is also in progress on using TiO2-reduced graphene oxide (TiO2-rGO) composite photocatalysts under solar radiation to remove selected ARGs (namely sul1, ampC, ermB, mecA) and species-specific sequences (ecfX for Pseudomonas aeruginosa and enterococcispecific 23S rRNA) in real urban wastewaters (Karaolia et al., 2018). Results showed complete inactivation in 180 min of photocatalytic treatment using the composite. Significantly, post-treatment of 24 h had no regrowth of E. coli bacteria (less than limit of detection). The composite photocatalysts removed ampC. Also, abundance of ecfX decreased. However, sul1, ermB and 23S rRNA remained persistent throughout photocatalytic treatment. Importantly, the total concentration of DNA did not change significantly in the photocatalytic process, which suggested high stability of genomic DNA in treated wastewater (Karaolia et al., 2018).
6. Conclusions
Presence of ARB and ARGs in water has become a major issue of public health and studies are forthcoming to determine their removal efficiency during disinfection. In the past decade, the use of UV disinfection has increased and recent efforts are exploring increase in UV effectiveness for degrading ARB and ARGs in water by combining UV radiation with other oxidants/catalysts (chlorine, H2O2, PMS, and photocatalysts) to generate reactive species that can more effectively destroy ARB and ARGs. A few studies using UV/chlorine and UV/H2O2 to destroy ARB and ARGs have been performed. The UV/chlorine had shown better efficiency to remove ARGs than the individual UV or chlorination process. The trend of UV/ H2O2 is not clear compared with UV alone to degrade ARGs. A few studies on UV/PMS method have been conducted and SO4•– generated in the UV/PMS has advantages under certain conditions over •OH, generated in UV/H2O2 to degrade ARGs. The qPCR and gel electrophoresis techniques are being applied to understand mechanism of damages to ARGs. Mechanistic studies showed that e-ARGs are easier to be degraded than iARGs. The photocatalytic treatment has effectiveness to decrease ARGs, but constituents in the matrix of the treated water greatly influence the efficiency of removal of ARGs. The inorganic and organic constituents of the water scavenge ROS to reduce the efficiency of photocatalytic treatment method. The photocatalytic degradation of ARGs using TiO2-modified PVDF membrane was about 98%; more efficient in the genome than in plasmid. The UV-TiO2-modified PVDF membrane treatment process was effective in controlling HTG. Future studies should emphasize to elucidate mechanism of degradation of ARGs and controlling HGT. Future work may include removal of ARGs in real water samples by UV-based disinfection/treatment methods.
Acknowledgements
V. K. Sharma thanks the Program of Environmental and Sustainability, SPH, TAMU for the support. C. Jinadatha was supported by the Central Texas Veterans Health Care System (Temple, Texas). The views expressed in this article are those of the author(s) and do not necessarily represent the views of the Department of Veterans Affairs.
References
- Auerbach E A, Seyfried E E, McMahon K D (2007). Tetracycline resistance genes in activated sludge wastewater treatment plants. Water Research, 41(5): 1143–1151 [DOI] [PubMed] [Google Scholar]
- Blaskovich M A T (2018). The fight against antimicrobial resistance is confounded by a global increase in antibiotic usage. ACS Infectious Diseases, 4(6): 868–870 [DOI] [PubMed] [Google Scholar]
- Buxton G V (2008). An overview of the radiation chemistry of liquids In: Spotheim-Maurizot M, Mostafavi M, Jacquline TD, eds. Radiation Chemistry: From Basics to Applications in Material and Life Science. Paris, France: L’Editeur, 3–16 [Google Scholar]
- Buxton G V, Greenstock C L, Helman W P, Ross A B (1988). Critical review of rate constants for reactions of hydrated electrons, hydrogen atoms and hydroxyl radicals in aqueous solution. Journal of Physical and Chemical Reference Data, 17(2): 513–886 [Google Scholar]
- Chang F, Shen S, Shi P, Zhang H, Ye L, Zhou Q, Pan Y, Li A (2019). Antimicrobial resins with quaternary ammonium salts as a supplement to combat the antibiotic resistome in drinking water treatment plants. Chemosphere, 221: 132–140 [DOI] [PubMed] [Google Scholar]
- Chang P H, Juhrend B, Olson T M, Marrs C F, Wigginton K R (2017). Degradation of extracellular antibiotic resistance genes with UV254 treatment. Environmental Science & Technology, 51(11): 6185–6192 [DOI] [PubMed] [Google Scholar]
- Chen H, Zhang M (2013). Effects of advanced treatment systems on the removal of antibiotic resistance genes in wastewater treatment plants from Hangzhou, China. Environmental Science & Technology, 47 (15): 8157–8163 [DOI] [PubMed] [Google Scholar]
- Chen X, Yin H, Li G, Wang W, Wong P K, Zhao H, An T (2019). Antibiotic-resistance gene transfer in antibiotic-resistance bacteria under different light irradiation: Implications from oxidative stress and gene expression. Water Research, 149: 282–291 [DOI] [PubMed] [Google Scholar]
- Cheng S, Zhang X, Yang X, Shang C, Song W, Fang J, Pan Y (2018). The multiple role of bromide ion in PPCPs degradation under UV/ chlorine treatment. Environmental Science & Technology, 52(4): 1806–1816 [DOI] [PubMed] [Google Scholar]
- Cizmas L, Sharma V K, Gray C M, McDonald T J (2015). Pharmaceuticals and personal care products in waters: Occurrence, toxicity, and risk. Environmental Chemistry Letters, 13(4): 381–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler T D, Zimmerman J J (2011). Ultraviolet irradiation and the mechanisms underlying its inactivation of infectious agents. Animal Health Research Reviews, 12(1): 15–23 [DOI] [PubMed] [Google Scholar]
- Dunlop P S M, Ciavola M, Rizzo L, McDowell D A, Byrne J A (2015). Effect of photocatalysis on the transfer of antibiotic resistance genes in urban wastewater. Catalysis Today, 240: 55–60 [Google Scholar]
- Ezzariai A, Hafidi M, Khadra A, Aemig Q, El Fels L, Barret M, Merlina G, Patureau D, Pinelli E (2018). Human and veterinary antibiotics during composting of sludge or manure: Global perspectives on persistence, degradation, and resistance genes. Journal of Hazardous Materials, 359: 465–481 [DOI] [PubMed] [Google Scholar]
- Fang J, Liu J, Shang C, Fan C (2018). Degradation investigation of selected taste and odor compounds by a UV/chlorine advanced oxidation process. International Journal of Environmental Research and Public Health, 15(2): 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner E, Chen C, Xia K, Bowers J, Engelthaler D M, McLain J, Edwards M A, Pruden A (2018). Metagenomic characterization of antibiotic resistance genes in full-scale reclaimed water distribution systems and corresponding potable systems. Environmental Science & Technology, 52(11): 6113–6125 [DOI] [PubMed] [Google Scholar]
- Ghanbari F, Moradi M (2017). Application of peroxymonosulfate and its activation methods for degradation of environmental organic pollutants. Chemical Engineering Journal, 310: 41–62 [Google Scholar]
- Guo C, Wang K, Hou S, Wan L, Lv J, Zhang Y, Qu X, Chen S, Xu J (2017). H2O2 and/or TiO2 photocatalysis under UV irradiation for the removal of antibiotic resistant bacteria and their antibiotic resistance genes. Journal of Hazardous Materials, 323(Pt B): 710–718 [DOI] [PubMed] [Google Scholar]
- He H, Zhou P, Shimabuku K K, Fang X, Li S, Lee Y, Dodd M C (2019). Degradation and deactivation of bacterial antibiotic resistance genes during exposure to free chlorine, monochloramine, chlorine dioxide, ozone, ultraviolet light, and hydroxyl radical. Environmental Science & Technology, 53(4): 2013–2026 [DOI] [PubMed] [Google Scholar]
- Hu Y, Jiang L, Zhang T, Jin L, Han Q, Zhang D, Lin K, Cui C (2018). Occurrence and removal of sulfonamide antibiotics and antibiotic resistance genes in conventional and advanced drinking water treatment processes. Journal of Hazardous Materials, 360: 364–372 [DOI] [PubMed] [Google Scholar]
- Hu Y, Zhang T, Jiang L, Luo Y, Yao S, Zhang D, Lin K, Cui C (2019a). Occurrence and reduction of antibiotic resistance genes in conventional and advanced drinking water treatment processes. Science of the Total Environment, 669: 777–784 [DOI] [PubMed] [Google Scholar]
- Hu Y, Zhang T, Jiang L, Yao S, Ye H, Lin K, Cui C (2019b). Removal of sulfonamide antibiotic resistant bacterial and intracellular antibiotic resistance genes by UVC-activated peroxymonosulfate. Chemical Engineering Journal, 368: 888–895 [Google Scholar]
- Hua Z, Guo K, Kong X, Lin S, Wu Z, Wang L, Huang H, Fang J (2019). PPCP degradation and DBP formation in the solar/free chlorine system: Effects of pH and dissolved oxygen. Water Research, 150: 77–85 [DOI] [PubMed] [Google Scholar]
- Karaolia P, Michael-Kordatou I, Hapeshi E, Drosou C, Bertakis Y, Christofilos D, Armatas G S, Sygellou L, Schwartz T, Xekoukoulotakis N P, Fatta-Kassinos D (2018). Removal of antibiotics, antibiotic-resistant bacteria and their associated genes by graphenebased TiO2 composite photocatalysts under solar radiation in urban wastewaters. Applied Catalysis B: Environmental, 224: 810–824 [Google Scholar]
- Khetan S K, Collins T J (2007). Human pharmaceuticals in the aquatic environment: A challenge to Green Chemistry. Chemical Reviews, 107(6): 2319–2364 [DOI] [PubMed] [Google Scholar]
- Kim H, Hong Y, Park J E, Sharma V K, Cho S I (2013). Sulfonamides and tetracyclines in livestock wastewater. Chemosphere, 91(7): 888–894 [DOI] [PubMed] [Google Scholar]
- Kong X, Wu Z, Ren Z, Guo K, Hou S, Hua Z, Li X, Fang J (2018). Degradation of lipid regulators by the UV/chlorine process: Radical mechanisms, chlorine oxide radical (ClO•)-mediated transformation pathways and toxicity changes. Water Research, 137: 242–250 [DOI] [PubMed] [Google Scholar]
- Krzeminski P, Tomei M C, Karaolia P, Langenhoff A, Almeida C M R, Felis E, Gritten F, Andersen H R, Fernandes T, Manaia C M, Rizzo L, Fatta-Kassinos D (2019). Performance of secondary wastewater treatment methods for the removal of contaminants of emerging concern implicated in crop uptake and antibiotic resistance spread: A review. Science of the Total Environment, 648: 1052–1081 [DOI] [PubMed] [Google Scholar]
- Lee J, Jeon J H, Shin J, Jang H M, Kim S, Song M S, Kim Y M (2017). Quantitative and qualitative changes in antibiotic resistance genes after passing through treatment processes in municipal wastewater treatment plants. Science of the Total Environment, 605–606: 906–914 [DOI] [PubMed] [Google Scholar]
- Li D, Chen D, Yao Y, Lin J, Gong F, Wang L, Luo L, Huang Z, Zhang L (2016a). Strong enhancement of dye removal through addition of sulfite to persulfate activated by a supported ferric citrate catalyst. Chemical Engineering Journal, 288: 806–812 [Google Scholar]
- Li D, Zeng S, He M, Gu A Z (2016b). Water disinfection byproducts induce antibiotic resistance–Role of environmental pollutants in resistance phenomena. Environmental Science & Technology, 50(6): 3193–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Sheng G P, Lu Y Z, Zeng R J, Yu H Q (2017). Removal of antibiotic resistance genes from wastewater treatment plant effluent by coagulation. Water Research, 111: 204–212 [DOI] [PubMed] [Google Scholar]
- Lin W, Li S, Zhang S, Yu X (2016). Reduction in horizontal transfer of conjugative plasmid by UV irradiation and low-level chlorination. Water Research, 91: 331–338 [DOI] [PubMed] [Google Scholar]
- Liu C, Wu B, Chen X (2018). Sulfate radical-based oxidation for sludge treatment: A review. Chemical Engineering Journal, 335: 865–875 [Google Scholar]
- Mauter M S, Zucker I, Perreault F, Werber J R, Kim J, Elimelech M (2018). The role of nanotechnology in tackling global water challenges. Nature Sustainability, 1(4): 166–175 [Google Scholar]
- Michael I, Rizzo L, McArdell C S, Manaia C M, Merlin C, Schwartz T, Dagot C, Fatta-Kassinos D (2013). Urban wastewater treatment plants as hotspots for the release of antibiotics in the environment: A review. Water Research, 47(3): 957–995 [DOI] [PubMed] [Google Scholar]
- Michael-Kordatou I, Karaolia P, Fatta-Kassinos D (2018). The role of operating parameters and oxidative damage mechanisms of advanced chemical oxidation processes in the combat against antibiotic-resistant bacteria and resistance genes present in urban wastewater. Water Research, 129: 208–230 [DOI] [PubMed] [Google Scholar]
- Miklos D B, Remy C, Jekel M, Linden K G, Drewes J E, Hübner U (2018). Evaluation of advanced oxidation processes for water and wastewater treatment: A critical review. Water Research, 139: 118–131 [DOI] [PubMed] [Google Scholar]
- Munir M, Wong K, Xagoraraki I (2011). Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in Michigan. Water Research, 45(2): 681–693 [DOI] [PubMed] [Google Scholar]
- Naumov S, von Sonntag C (2008). The energetics of rearrangement and water elimination reactions in the radiolysis of the DNA bases in aqueous solution (eaq ‒ and OH attack): DFT calculations. Radiation Research, 169(3): 355–363 [DOI] [PubMed] [Google Scholar]
- Neta P, Huie R E (1985). Free-radical chemistry of sulfite. Environmental Health Perspectives, 64: 209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihemaiti M, Miklos D B, Hübner U, Linden K G, Drewes J E, Croué J P (2018). Removal of trace organic chemicals in wastewater effluent by UV/H2O2 and UV/PDS. Water Research, 145: 487–497 [DOI] [PubMed] [Google Scholar]
- Özkal C B, Venieri D, Gounaki I, Meric S (2019). Assessment of thin-film photocatalysis inactivation of different bacterial indicators and effect on their antibiotic resistance profile. Applied Catalysis B: Environmental, 244: 612–619 [Google Scholar]
- Pang Y, Huang J, Xi J, Hu H, Zhu Y (2016). Effect of ultraviolet irradiation and chlorination on ampicillin-resistant Escherichia coli and its ampicillin resistance gene. Frontiers of Environmental Science & Engineering, 10(3): 522–530 [Google Scholar]
- Ren S, Boo C, Guo N, Wang S, Elimelech M, Wang Y (2018). Photocatalytic reactive ultrafiltration membrane for removal of antibiotic resistant bacteria and antibiotic resistance genes from wastewater effluent. Environmental Science & Technology, 52(15): 8666–8673 [DOI] [PubMed] [Google Scholar]
- Rizzo L, Sannino D, Vaiano V, Sacco O, Scarpa A, Pietrogiacomi D (2014). Effect of solar simulated N-doped TiO2 photocatalysis on the inactivation and antibiotic resistance of an E. coli strain in biologically treated urban wastewater. Applied Catalysis B: Environmental, 144: 369–378 [Google Scholar]
- Rodríguez-Chueca J, Varella Della Giustina S, Rocha J, Fernandes T, Pablos C, Encinas Á, Barceló D, Rodríguez-Mozaz S, Manaia C M, Marugán J (2019). Assessment of full-scale tertiary wastewater treatment by UV-C based-AOPs: Removal or persistence of antibiotics and antibiotic resistance genes? Science of the Total Environment, 652: 1051–1061 [DOI] [PubMed] [Google Scholar]
- Shao S, Hu Y, Cheng J, Chen Y (2018). Research progress on distribution, migration, transformation of antibiotics and antibiotic resistance genes (ARGs) in aquatic environment. Critical Reviews in Biotechnology, 38(8): 1195–1208 [DOI] [PubMed] [Google Scholar]
- Sharma V K (2008). Oxidative transformations of environmental pharmaceuticals by Cl2, ClO2, O3, and Fe(VI): kinetics assessment. Chemosphere, 73(9): 1379–1386 [DOI] [PubMed] [Google Scholar]
- Sharma V K (2013). Oxidation of Amino Acids, Peptides, and Proteins. New Jersey, USA: Wiley, Inc. [Google Scholar]
- Sharma V K, Johnson N, Cizmas L, McDonald T J, Kim H (2016). A review of the influence of treatment strategies on antibiotic resistant bacteria and antibiotic resistance genes. Chemosphere, 150: 702–714 [DOI] [PubMed] [Google Scholar]
- Sinha R P, Häder D P (2002). UV-induced DNA damage and repair: A review. Photochemical & Photobiological Sciences, 1(4): 225–236 [DOI] [PubMed] [Google Scholar]
- Sousa J C G, Ribeiro A R, Barbosa M O, Pereira M F R, Silva A M T (2018). A review on environmental monitoring of water organic pollutants identified by EU guidelines. Journal of Hazardous Materials, 344: 146–162 [DOI] [PubMed] [Google Scholar]
- Sousa J M, Macedo G, Pedrosa M, Becerra-Castro C, Castro-Silva S, Pereira M F R, Silva A M T, Nunes O C, Manaia C M (2017). Ozonation and UV254 nm radiation for the removal of microorganisms and antibiotic resistance genes from urban wastewater. Journal of Hazardous Materials, 323(Pt A): 434–441 [DOI] [PubMed] [Google Scholar]
- Ting Y F, Praveena S M (2017). Sources, mechanisms, and fate of steroid estrogens in wastewater treatment plants: A mini review. Environmental Monitoring and Assessment, 189(4): 178. [DOI] [PubMed] [Google Scholar]
- Travis A, Chernova O, Chernov V, Aminov R (2018). Antimicrobial drug discovery: lessons of history and future strategies. Expert Opinion on Drug Discovery, 13(11): 983–985 [DOI] [PubMed] [Google Scholar]
- Umar M, Roddick F, Fan L (2019). Moving from the traditional paradigm of pathogen inactivation to controlling antibiotic resistance in water-Role of ultraviolet irradiation. Science of the Total Environment, 662: 923–939 [DOI] [PubMed] [Google Scholar]
- Uyguner Demirel C S, Birben N C, Bekbolet M (2018). A comprehensive review on the use of second generation TiO2 photocatalysts: Microorganism inactivation. Chemosphere, 211: 420–448 [DOI] [PubMed] [Google Scholar]
- Van Boeckel T P, Brower C, Gilbert M, Grenfell B T, Levin S A, Robinson T P, Teillant A, Laxminarayan R (2015). Global trends in antimicrobial use in food animals. Proceedings of the National Academy of Sciences of the United States of America, 112(18): 5649–5654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Boeckel T P, Gandra S, Ashok A, Caudron Q, Grenfell B T, Levin S A, Laxminarayan R (2014). Global antibiotic consumption 2000 to 2010: An analysis of national pharmaceutical sales data. The Lancet Infectious Diseases, 14(8): 742–750 [DOI] [PubMed] [Google Scholar]
- Vindenes T, Beaulac K R, Doron S (2016). The legislative momentum of antimicrobial stewardship: An international perspective. Current Treatment Options in Infectious Diseases, 8(2): 72–83 [Google Scholar]
- von Gunten U (2018). Oxidation processes in water treatment: Are we on track? Environmental Science & Technology, 52(9): 5062–5075 [DOI] [PubMed] [Google Scholar]
- Wang M, Shen W, Yan L, Wang X H, Xu H (2017a). Stepwise impact of urban wastewater treatment on the bacterial community structure, antibiotic contents, and prevalence of antimicrobial resistance. Environmental Pollution, 231(Pt 2): 1578–1585 [DOI] [PubMed] [Google Scholar]
- Wang W, Li G, Xia D, An T, Zhao H, Wong P K (2017b). Photocatalytic nanomaterials for solar-driven bacterial inactivation: Recent progress and challenges. Environmental Science. Nano, 4(4): 782–799 [Google Scholar]
- Wang Y, Zhan S, Zhou Q (2017c). The progress on removal techniques of antibiotic resistant genes from water environment. Shengtaixue Zazhi, 36(12): 3610–3616 [Google Scholar]
- Wojnárovits L, Takács E (2019). Rate constants of sulfate radical anion reactions with organic molecules: A review. Chemosphere, 220: 1014–1032 [DOI] [PubMed] [Google Scholar]
- Wols B A, Hofman-Caris C H M (2012). Review of photochemical reaction constants of organic micropollutants required for UV advanced oxidation processes in water. Water Research, 46(9): 2815–2827 [DOI] [PubMed] [Google Scholar]
- Wols B A, Hofman-Caris C H M, Harmsen D J H, Beerendonk E F (2013). Degradation of 40 selected pharmaceuticals by UV/H2O2. Water Research, 47(15): 5876–5888 [DOI] [PubMed] [Google Scholar]
- World Health Organization (2018). High levels of antibiotic resistance found worldwide, new data shows, 2018. Geneva: World Health Organization [Google Scholar]
- Wu Z, Fang J, Xiang Y, Shang C, Li X, Meng F, Yang X (2016). Roles of reactive chlorine species in trimethoprim degradation in the UV/ chlorine process: Kinetics and transformation pathways. Water Research, 104: 272–282 [DOI] [PubMed] [Google Scholar]
- Wu Z, Guo K, Fang J, Yang X, Xiao H, Hou S, Kong X, Shang C, Yang X, Meng F, Chen L (2017). Factors affecting the roles of reactive species in the degradation of micropollutants by the UV/chlorine process. Water Research, 126: 351–360 [DOI] [PubMed] [Google Scholar]
- Yang L, Wen Q, Zhao Y, Chen Z, Wang Q, Bürgmann H (2019). New insight into effect of antibiotics concentration and process configuration on the removal of antibiotics and relevant antibiotic resistance genes. Journal of Hazardous Materials, 373: 60–66 [DOI] [PubMed] [Google Scholar]
- Yang Y, Song W, Lin H, Wang W, Du L, Xing W (2018). Antibiotics and antibiotic resistance genes in global lakes: A review and meta-analysis. Environment International, 116: 60–73 [DOI] [PubMed] [Google Scholar]
- Yoon Y, Chung H J, Wen Di D Y, Dodd M C, Hur H G, Lee Y (2017). Inactivation efficiency of plasmid-encoded antibiotic resistance genes during water treatment with chlorine, UV, and UV/H2O2. Water Research, 123: 783–793 [DOI] [PubMed] [Google Scholar]
- Yoon Y, Dodd M C, Lee Y (2018). Elimination of transforming activity and gene degradation during UV and UV/H2O2 treatment of plasmid-encoded antibiotic resistance genes. Environmental Science. Water Research & Technology, 4(9): 1239–1251 [Google Scholar]
- Zhang B, Zhang Y, Teng Y, Fan M (2015a). Sulfate radical and its application in decontamination technologies. Critical Reviews in Environmental Science and Technology, 45(16): 1756–1800 [Google Scholar]
- Zhang C, Li Y, Shuai D, Shen Y, Wang D (2019a). Progress and challenges in photocatalytic disinfection of waterborne Viruses: A review to fill current knowledge gaps. Chemical Engineering Journal, 355: 399–415 [Google Scholar]
- Zhang M, Wang L, Xu M, Zhou H, Wang S, Wang Y, Bai M, Zhang C (2019b). Selective antibiotic resistance genes in multiphase samples during biofilm growth in a simulated drinking water distribution system: Occurrence, correlation and low-pressure ultraviolet removal. Science of the Total Environment, 649: 146–155 [DOI] [PubMed] [Google Scholar]
- Zhang R, Meng T, Huang C H, Ben W, Yao H, Liu R, Sun P (2018). PPCP degradation by chlorine-UV processes in ammoniacal water: New reaction insights, kinetic modeling, and DBP formation. Environmental Science & Technology, 52(14): 7833–7841 [DOI] [PubMed] [Google Scholar]
- Zhang T, Hu Y, Jiang L, Yao S, Lin K, Zhou Y, Cui C (2019c). Removal of antibiotic resistance genes and control of horizontal transfer risk by UV, chlorination and UV/chlorination treatments of drinking water. Chemical Engineering Journal, 358: 589–597 [Google Scholar]
- Zhang T, Zhang M, Zhang X, Fang H H (2009). Tetracycline resistance genes and tetracycline resistant lactose-fermenting Enterobacteriaceae in activated sludge of sewage treatment plants. Environmental Science & Technology, 43(10): 3455–3460 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhuang Y, Geng J, Ren H, Zhang Y, Ding L, Xu K (2015b). Inactivation of antibiotic resistance genes in municipal wastewater effluent by chlorination and sequential UV/chlorination disinfection. Science of the Total Environment, 512–513: 125–132 [DOI] [PubMed] [Google Scholar]
- Zhao X, Jiang J, Pang S, Guan C, Li J, Wang Z, Ma J, Luo C (2019). Degradation of iopamidol by three UV-based oxidation processes: Kinetics, pathways, and formation of iodinated disinfection byproducts. Chemosphere, 221: 270–277 [DOI] [PubMed] [Google Scholar]