CRISPR homing drive variants have different numbers of multiplexed gRNAs for optimal performance.
Abstract
The rapid evolution of resistance alleles poses a major obstacle for genetic manipulation of populations with CRISPR homing gene drives. One proposed solution is using multiple guide RNAs (gRNAs), allowing a drive to function even if some resistant target sites are present. Here, we develop a model of homing mechanisms parameterized by experimental studies. Our model incorporates several factors affecting drives with multiple gRNAs, including timing of cleavage, reduction in homology-directed repair efficiency due to imperfect homology, Cas9 activity saturation, gRNA activity level variance, and incomplete homology-directed repair. We find that homing drives have an optimal number of gRNAs, usually between two and eight, depending on the specific drive type and performance parameters. These results contradict the notion that resistance rates can be reduced to arbitrarily low levels by gRNA multiplexing and highlight the need for combined approaches to counter resistance evolution in CRISPR homing drives.
INTRODUCTION
An efficient gene drive could rapidly modify or suppress a target population (1–4). Such a mechanism could potentially be used to prevent transmission of vector-borne diseases such as malaria or dengue and could also have conservation applications (1–4). The best-studied form of an engineered gene drive mechanism is the homing drive, which uses the CRISPR-Cas9 system to cleave a wild-type allele. The drive allele is then copied into the wild-type site via homology-directed repair, increasing the frequency of the drive allele in the population. Thus far, CRISPR homing gene drives have been demonstrated in yeast (5–8), flies (9–16), mosquitoes (17–19), and mice (20).
However, homing drives typically suffer from high rates of resistance allele formation. These alleles can form when DNA is repaired by end joining, which often mutates the sequence. The consequence of a changed sequence is that the drive’s guide RNA (gRNA) can no longer target the allele for cleavage. Resistance alleles have been observed to arise both in germline cells as an alternative to homology-directed repair and in the early embryo due to deposition of Cas9 and gRNA into the egg by drive-carrying mothers (12). While formation of resistance alleles remains the primary obstacle to construction of efficient gene drives, substantial progress has been made toward overcoming this challenge. For example, a suppression-type drive in Anopheles gambiae (21) and a modification-type drive in Drosophila melanogaster (16) avoided issues with resistance alleles by targeting an essential gene. Because of this, resistance alleles that disrupted the function of the target gene had substantially reduced fitness. This allowed both drives to successfully spread through cage populations.
Multiplexing gRNAs has been proposed as a mechanism for increasing the efficiency of gene drives (1, 4). This would purportedly work by two mechanisms. First, having multiple cut sites would potentially allow drive conversion even if some of the sites have resistance sequences due to previous end joining repair at those sites. As long as at least one site remains wild type and, thus, cleavable, homology-directed repair can still occur. Second, the chance of forming a full resistance allele that preserves the function of the target gene would be substantially reduced due to the possibility of disruptive mutations forming at any of the gRNA target sites. Resistance alleles that disrupt the function of the target gene incur large fitness costs in several drive designs, which would make resistance substantially less likely to block the spread of the drive.
However, two studies using two gRNAs (13, 16) showed somewhat lower increases in efficiency than predicted by simple models of multiple gRNAs (22–24). This is partially because most models assume that cleavage and repair by either homology-directed repair or end joining occurs sequentially at each gRNA target site. However, it appears that some germline resistance alleles form before the narrow temporal window for homology-directed repair (10, 12, 13). Additional resistance sequences may also form as a direct alternative to successful homology-directed repair, while others could form after meiosis I when only end joining repair is possible. Furthermore, unless cleavage occurs in both of the outermost gRNA target sites during the window for homology-directed repair, the wild-type chromosome on either side of the cleavage would have imperfect homology to the drive allele because of nonhomologous DNA between the cut site and the homology arm (13). Imperfect homology likely reduces repair fidelity (i.e., less homology-directed repair and more end joining repair). This proposition is supported by the greatly reduced efficiency seen in a construct with four gRNA targets located far apart from one another (9). Last, it is unlikely that gRNAs are the limiting factor in Cas9/gRNA enzymatic activity (15). As the number of gRNAs increases, the total cleavage rate likely plateaus, thus reducing the cleavage rate at each individual site and thereby preventing further gains in drive efficiency.
Here, we systematically model these factors and show how they are expected to affect the performance of homing drives with multiple gRNAs. We verify and parameterize these models via experimental analysis of several homing drives in D. melanogaster. We additionally consider other factors that could reduce gene drive performance, such as partial homology-directed repair and uneven activity of gRNAs. We then apply our model to predict drive performance in Anopheles mosquitoes, assessing several types of homing drives for population modification or suppression. We find that the reduction in efficiency due to imperfect homology is synergistic, with the lower per-site cleavage rates from Cas9 activity saturation. Because of this, each type of drive has an optimal number of gRNAs that results in maximized overall performance, a finding that could inform future designs of homing gene drives.
METHODS
Drive variants
In our model, we consider five types of homing gene drive systems:
1) Standard drive. The standard homing drive is a population modification system. Its primary drive mechanism occurs in germline cells during early meiosis. When it operates successfully, the drive allele replaces wild-type alleles in the germline. However, resistance alleles can also form, preventing the spread of the drive.
2) Population suppression drive. This drive increases in frequency in the same manner as the standard homing drive, and resistance alleles develop under the same circumstances. However, the drive targets a recessive female fertility gene and disrupts the function of the gene with its presence. Resistance alleles can also disrupt the function of the target gene. Females with two disrupted copies of the gene are rendered sterile, while males are unaffected. Notably, unlike the standard homing drive, this drive does not carry any payload, as, it accomplishes its goal simply with its presence. Such a drive was successful in laboratory populations of the mosquito A. gambiae (21).
3) Haplolethal drive. This drive system is a modification of the standard homing drive system. It targets a gene that is critical to the viability of the individual. However, the drive contains a recoded portion of the gene that is immune to Cas9 cleavage, so the presence of the drive does not disrupt the function of the target. If an individual receives a resistance allele that disrupts the haplolethal target, then that individual will not be viable, preventing these resistance alleles from entering the population. A haplolethal homing drive was successful in a laboratory population of the fruit fly D. melanogaster (16).
4) Recessive lethal drive. This drive is similar to the haplolethal drive, but the target is recessive lethal. Only individuals carrying two resistance alleles that disrupt the target gene function are nonviable. Thus, resistance alleles are removed from the population more slowly. However, this drive may be easier to engineer because the drive can provide rescue even in the presence of a resistance allele. It is also more tolerant of a high rate of embryo resistance allele formation because this allows it to operate better as a toxin-antidote system (25, 26).
5) Gene disruption drive. The gene disruption homing drive is a population modification system that is similar to the suppression drive in that its presence disrupts the target gene, as do resistance alleles. However, individuals with two disrupted copies of this gene remain viable and fertile, although they suffer from a small additional fitness cost. The purpose of this drive is to remove the functionality of a particular gene from the population, which can provide benefits such as reduction in disease transmission (27, 28). An advantage of this drive is that there is no need for a recoded sequence. However, finding suitable targets for particular applications could potentially be difficult.
Computational model
We implemented each of the gene drive models using SLiM version 3.2.1 (29). SLiM is an individual-based, forward-time population genetic simulation framework. General parameters and ecology components are shared across all models.
Our model considers a single panmictic population of sexually reproducing diploid individuals with nonoverlapping generations. The model differs from a standard Wright-Fisher–type model in that population size is not regulated. Offspring are generated from random pairings throughout the population, with mate choice and female fecundity affected by genotype fitness. To determine mate choice, first, a random male is selected. This candidate is then accepted at a rate equal to his genotype fitness (e.g., a male with a fitness of 0.5 is accepted half of the time). If he is rejected, then another random candidate is selected, until either a mate has been chosen or the female fails to find an acceptable mate after a total of 10 attempts. Female fecundity is then multiplied by her genotype fitness, along with a factor representing the impact of population density in the system: 10/(1 + 9 N/K), where N is the total population and K is the carrying capacity. A number of offspring are then generated on the basis of a binomial distribution with a maximum of 50 and p = fecundity/25. This model produces logistic dynamics while allowing the population size to fluctuate around the expected capacity. After pairings and offspring have been determined, the genotypes of the offspring are modified according to the genetic component of the model.
In one set of simulations, a small number of drive/wild-type heterozygous flies were introduced into a wild-type population of 100,000 at an initial frequency of 1%. The simulation was then conducted for 100 generations. In another set of simulations, a wild-type female was crossed to a drive/wild-type heterozygote male, and a configurable number of offspring were generated from that single pairing. The genotype of each offspring was recorded to estimate drive performance parameters. Drive conversion was equal to the fraction of wild-type alleles in the germline converted to drive alleles, and resistance allele formation rates also represented rates of conversion from wild-type alleles. The genetic module of our model is described in the Supplementary Methods.
Experimental methods
Detailed descriptions of our plasmid construction techniques, the construction and sequencing primers used, the generation of transgenic lines, fly rearing, phenotyping and analysis techniques, and genotyping are available in the Supplementary Methods.
RESULTS
Simple model
To compare our results to previous work, we constructed a simple model of homing drive dynamics. This model considers each gRNA site independently, with parameters inspired by highly efficient homing drives in Anopheles mosquitoes (18, 19, 21, 30). At each gRNA target site, we assume a cut rate of 99%. If the site is cut, then there is a 7.8% chance that a resistance sequence will be formed. Otherwise, homology-directed repair occurs, and the entire allele (including all target sites, even if some have resistance sequences) is converted to a drive allele. In this model, increasing the number of gRNA target sites always increases the efficiency of the drive, and arbitrarily low resistance rates can be achieved by adding more target sites (Fig. 1). Since even relatively few gRNAs can reduce resistance allele formation to very low levels under this model, gRNA multiplexing has been considered as a highly promising and comparatively straightforward method to avert resistance in homing gene drives (22–24).
Fig. 1. Resistance allele formation.
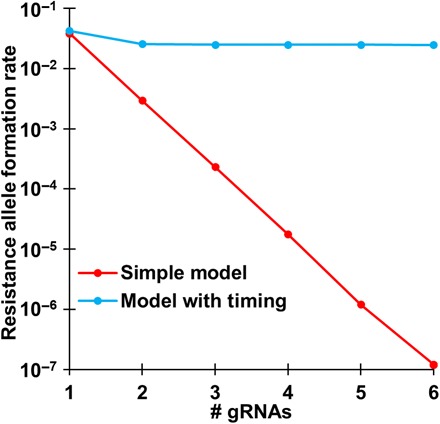
Five million offspring were generated from crosses between drive/wild-type heterozygotes and wild-type individuals for each model and number of gRNAs. The rate at which wild-type alleles are converted to resistance alleles in the germline of drive/wild-type heterozygous individuals is shown.
Model with timing
The simple model does not take timing of cleavage and repair into account. However, several lines of evidence indicate that cleavage timing can play a key role in drive conversion. Experiments indicated that wild-type alleles can only be converted to resistance alleles and not to drive alleles in the early embryo due to maternally deposited Cas9 (12, 13), where homology-directed repair for the purposes of drive conversion does not take place at appreciable rates. Furthermore, at least some resistance alleles form in pregonial germline cells that can affect the genotype of multiple offspring (10, 12, 13). After the chromosomes separate in late meiosis, homology-directed repair is no longer possible, and any cleavage at this time results in the formation of resistance alleles by end joining repair. It is thus likely that there is only a narrow temporal window in the germline, during which the drive can be successfully copied via homology-directed repair. This window presumably covers early meiosis when homologous chromosomes are close together, which would increase the chance that one chromosome could be used as a template for repair of a double-strand break in the other.
To explore the expected impact of these mechanisms on resistance rates, we constructed a model where cuts during a homology-directed repair phase occur simultaneously, and the DNA then has a single opportunity to undergo homology-directed repair. In this model, there are discrete temporal phases. In each phase, wild-type gRNA sites are cut before any repair takes place. In the first phase, end joining repair always occurs after cutting, so having more gRNAs allows more target sites to avoid being converted to resistance alleles. Only in the next phase is homology-directed repair possible, which takes place at a specific rate if any cutting occurs. If homology-directed repair (successful drive conversion) does not take place, then end joining is assumed to repair the cut, forming a resistance allele. Thus, as the probability of the DNA being cut approaches 100% due to many gRNAs, the overall rate of resistance formation does not decrease indefinitely. Instead, it reaches a minimum value equal to the chance that end joining takes place instead of successful homology-directed repair in the second phase (Fig. 1). This suggests that the simple model of multiple gRNAs is likely inadequate to accurately assess homing drive dynamics.
Synthetic target site experiments with one gRNA
Previous experiments with two gRNAs resulted in a lower-efficiency improvement than even that predicted by our model that included timing (13). This was shown even more starkly with a four-gRNA drive (9) that had a lower drive conversion efficiency than a one-gRNA drive. We hypothesize that two additional factors could account for this discrepancy. First, the rate at which homology-directed repair occurs after cleavage in the appropriate phase (which we refer to as repair fidelity) is likely reduced if the DNA on either side of the cut sites does not have immediate homology to the drive (meaning that end resection must proceed for several nucleotides before it reaches DNA with homology to the drive). This will often be the case because a drive allele is constructed to have DNA homologous only to that at the outermost cut sites (the leftmost and rightmost sites). Thus, drive efficiency is reduced except when both outer gRNAs are cleaved. Second, the amount of Cas9 enzyme is limited. Thus, as the number of gRNAs increases, Cas9 eventually becomes saturated with gRNAs and cleavage activity plateaus. This has the effect of decreasing the cleavage rate at individual gRNA sites as the total number of gRNAs increases. To test the impact of these two mechanisms on drive efficiency, we conducted a series of experiments.
We first constructed a drive system in D. melanogaster that targeted a synthetic enhanced green fluorescent protein (EGFP) site with one gRNA and Cas9 driven by the nanos promoter (fig. S1), similar to previous synthetic target site drives (15). Drive/wild-type heterozygotes displayed a drive conversion efficiency of 83% [95% confidence interval (CI), 79 to 86%] in females and 61% (95% CI, 57 to 65%) in males (data S1). These values were higher than previous synthetic target site drives (15), likely due to the different genomic location of the target site or the different gRNA target, which targeted further away from the 3xP3 promoter in EGFP. Drive conversion efficiency was significantly higher in females (P < 0.0001, Fisher’s exact test), which was consistent with previous studies (13, 15). It remains unclear why this may be the case, but it may be due to sex differences in levels of repair proteins in the germline (possibly related to the lack of male chromosomal recombination), resulting in a higher ratio of homology-directed repair to end joining in the appropriate temporal window. Expression of Cas9 by the nanos promoter could also be variable between the sexes. If expression started earlier in males, then an increased number of resistance alleles could form before the temporal window for homology-directed repair.
Since multiplexing of gRNAs can best be accomplished by expressing them from a single compact promoter, we modified our drive system to include a transfer RNA (tRNA) that must be spliced out of the gRNA gene to generate an active gRNA. By including additional tRNAs between gRNAs, several gRNAs can be expressed together with this system (31). We found that drive/wild-type heterozygote females had a drive conversion efficiency of 82% (95% CI, 78 to 86%) in females and 65% (95% CI, 62 to 70%) in males for the one-gRNA drive with the tRNA (data S2). This indicates that the tRNA system functions correctly in homing drives without apparent loss of efficiency, allowing its use in multiplexed gRNA experiments.
We next constructed a drive to determine the effects of reduced homology between the cleaved wild-type chromosome and the drive allele. We used a single gRNA as above with the tRNA system but with the right homology arm realigned to a hypothetical second gRNA cut site (Fig. 2). Thus, the first 114 nucleotides to the right of the cut site would not be homologous to any DNA around the drive allele. Drive conversion rates for females were only 84% (95% CI, 77 to 90%) of the rate of the one-gRNA drive that had full homology around the cut site, while the rate for males was 89% (95% CI, 83 to 96%) of the full homology drive (data S3). This indicates that a multiple-gRNA drive would indeed exhibit lower conversion efficiency when cleavage does not take place at both ends.
Fig. 2. Experimental performance of homing drives with different configurations.
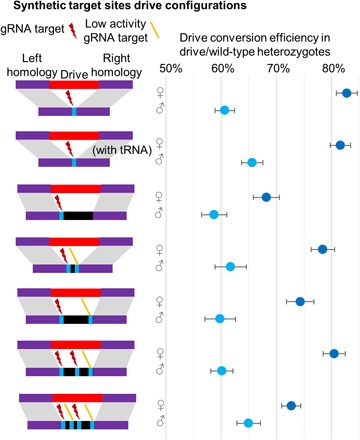
Blue shows gRNA target sites, and black shows regions of DNA that have no homology to the drive allele. Highly active gRNAs are shown by a red lightning bolt, and gRNAs with low activity are shown with an orange line icon.
Split drive experiments
To assess the effects of Cas9 activity saturation, we examined three constructs containing Cas9 with either zero, one, or four gRNAs targeting a genomic region located between two genes and downstream of both to minimize potential interference with native genes. Mutations resulting from the repair of cleavage events in this area are thereby unlikely to affect an individual’s fitness. These constructs were placed at the same genomic site as the synthetic target site constructs. Individuals with these constructs were crossed to those carrying the split drive targeting yellow developed previously (15) to generate individuals heterozygous for both a Cas9 element and a split-drive element. These individuals all had a single gRNA targeting yellow and a variable number of gRNAs that target a region where sequence changes produce no phenotype. The embryo resistance allele formation rates in individuals with zero, one, or four gRNAs that were not targeting yellow in the Cas9 element were 83% (95% CI, 80 to 87%), 72% (95% CI, 68 to 77%), and 65% (95% CI, 60 to 70%), respectively (data S4). The differences between the construct with zero additional gRNA elements and the others were statistically significant (P < 0.0001 in both cases, Fisher’s exact test), although the difference between the constructs with one and four additional gRNAs did not quite reach statistical significance (P = 0.06, Fisher’s exact test). The amount of the gRNA targeting yellow was constant in these drives. However, the rate at which yellow was cleaved decreased as the number of other gRNAs increased. This is consistent with the hypothesis that saturation of Cas9 activity reduces the cleavage rates at individual gRNA target sites when the total number of gRNAs is increased.
Nevertheless, Cas9 does not necessarily become fully saturated with a single gRNA. The total cleavage rate could potentially somewhat increase if additional gRNAs are provided, although it would likely plateau rapidly. When heterozygotes for the split drive targeting yellow (15) and the standard drive targeting yellow (12) (which had one copy of Cas9 and two copies of the gRNA gene) were crossed to w1118 males, the rate of embryo resistance allele formation and mosaicism was somewhat higher than for standard drive/resistance allele heterozygotes with one copy of Cas9 and only one gRNA gene (P = 0.0036, Fisher’s exact test) (data S5).
Experiments with multiple gRNAs
To assess the performance of drives with multiple gRNAs, we created several additional constructs targeting EGFP, but with two, three, or four gRNAs (Fig. 2). The left target site for each of these was the same as for the one-gRNA synthetic target site drives, and the homologous ends of all of these drives matched the left and right gRNA target sites. However, we found that of the four gRNAs used, only the first and the third had high cleavage activity, as indicated by sequencing of embryo resistance alleles (table S1). Although germline cleavage activity was likely somewhat higher than in the embryo for these gRNAs, their low activity undoubtedly reduced drive performance. Nevertheless, we found that the overall performance of these drives was consistent with the performance predicted by our model that included the effects of timing, repair fidelity, and Cas9 activity saturation (Fig. 2). The results show that adding additional gRNAs does not exponentially increase the efficiency of homing drives.
Specifically, we constructed two different two-gRNA drives. One of these had two closely spaced gRNA targets (36 nucleotides apart) and showed a drive conversion efficiency of 78% (95% CI, 74 to 83%) in females and 62% (95% CI, 56 to 67%) in males (data S6). This was slightly higher than the second drive where the two gRNAs were more widely spaced (114 nucleotides apart), which demonstrated a drive conversion efficiency of 74% (95% CI, 70 to 79%) in females and 60% (95% CI, 55 to 64%) in males (data S7). Because the second gRNA has low activity in each of these drives, the small difference in the performance between them could possibly be accounted for by the lower repair fidelity in the drive with more widely spaced gRNAs (Fig. 2) when only one target site is cut. The drive with three gRNAs was similar to the two-gRNA drive with widely spaced gRNAs, with the addition of a third active gRNA in between the two target sites of the two-gRNA drive. This likely increased the overall cleavage rate due to the higher proportion of active gRNAs and allowed for greater repair fidelity on the right end, since cleavage in this system usually takes place at the left and middle gRNA targets, instead of often only at the left gRNA target. Thus, this construct showed an improved drive conversion efficiency of 80% (95% CI, 77 to 84%) in females, although male drive conversion efficiency apparently remained at 60% (95% CI, 56 to 64%). A final construct added an additional gRNA between the left and middle gRNAs (the same gRNA that the closely spaced two-gRNA construct included). However, since this gRNA had low activity, overall drive performance may have been negatively affected by saturation of Cas9 by gRNAs, resulting in a reduced drive conversion efficiency of 73% (95% CI, 69 to 76%) in females, although male drive conversion efficiency appears to have improved to 65% (95% CI, 61 to 68%) (possibly due to an underestimation of conversion efficiency for males with the three-gRNA construct).
Refinement of the model
To further refine our model, we next incorporated distinct phases for homing drive dynamics in the germline (Fig. 3). In this model, we assume that first, early germline resistance alleles form, followed by a homology-directed repair phase, and then a late germline resistance allele formation phase. In the embryos of mothers with at least one drive allele, maternally deposited Cas9 and gRNA can result in the formation of additional resistance alleles. During this process, deletions can occur if cleavage occurs nearly simultaneously at different cut sites. If a second site is cleaved before a first cleavage has been repaired, then the section of DNA between the two sites is excised when the gap is closed by end joining repair.
Fig. 3. Steps in the model.
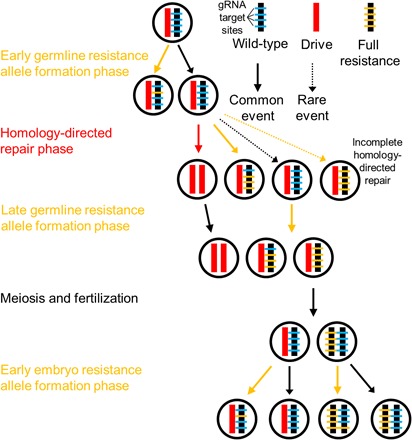
First, wild-type gRNA target sites can be cleaved in the early germline, forming resistance alleles. Next, cleavage occurs at a high rate in the homology-directed repair phase. Usually, this results in successful conversion to a drive allele. However, if homology-directed repair fails to occur, then end joining can form resistance alleles. Incomplete homology-directed repair can also convert the entire allele to a resistance allele, ignoring individual target sites. Next, another resistance allele formation phase converts most remaining wild-type sites into resistance sequences. Meiosis and fertilization take place, and then, if the female parent had at least one drive allele, a final phase of resistance allele formation takes place in the early embryo.
We additionally model reduced repair fidelity from imperfect homology around the cut sites, Cas9 activity saturation, and variance in the activity level of individual gRNAs. See the Supplementary Results for a detailed treatment of these model components and estimation of parameters based on our experiments. Models with repair fidelity or Cas9 activity saturation alone did not produce much deviance from our basic model with timing (Fig. 4). However, a model that includes both repair fidelity and Cas9 activity saturation demonstrated fundamentally changed dynamics. We found that there was a synergistic effect between the factor of the reduced cut rate per site caused by Cas9 activity saturation and the factor of reduced repair fidelity when the outermost target sites are not cut. Because of this, we find the emergence of an optimal number of gRNAs to maximize drive conversion efficiency (Fig. 4), which decreases rapidly when additional gRNAs are added. The additional modeling of gRNA activity variance had only a small negative effect on drive conversion performance (Fig. 4).
Fig. 4. Effects of model components on drive performance.
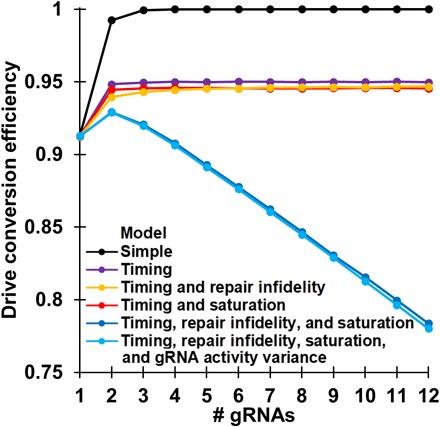
Five million offspring were generated from crosses between drive/wild-type heterozygotes and wild-type individuals for each model and number of gRNAs. The rate at which wild-type alleles are converted to drive alleles in the germline of drive/wild-type individuals is shown.
With parameters simulating an efficient A. gambiae construct, the optimal number of gRNAs in this model is two, although drives with three gRNAs have nearly as good conversion efficiency (Fig. 4). Thus, not only do further increases in the number of gRNAs fail to provide substantial benefits, they actually result in substantial reductions in drive efficiency. However, note that the optimal number of gRNAs for overall performance may be somewhat greater than the optimal number for drive conversion efficiency, as detailed below.
Types of resistance alleles
Resistance alleles can either preserve or disrupt the function of a target gene. The latter are expected to be more common due to frameshift mutations or other disruptions to the target sequence but should usually be less detrimental to drive performance. In our model, we assume that resistance sequences preserving the function of the target gene form in 10% of cases (12, 13), although this could be substantially reduced by targeting conserved sequences (13, 16, 21). We further assume that if even a single resistance sequence that disrupts the function of the target gene is present, the target gene is rendered nonfunctional. Any deletion due to simultaneous cleavage followed by end joining repair is also assumed to disrupt the target gene. One major advantage of multiple-gRNA drives is therefore that complete resistance alleles that preserve the function of the target gene should become exponentially less common as the number of gRNAs increases (Fig. 5, black line).
Fig. 5. Formation of resistance alleles that preserve the function of the target gene.
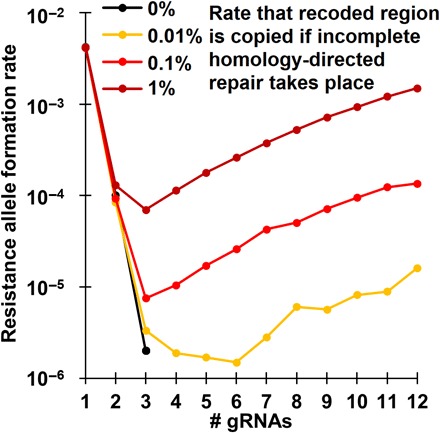
Five million offspring were generated from crosses between drive/wild-type heterozygotes and wild-type individuals for each number of gRNAs and each level of probability that incomplete homology-directed repair results in the formation of resistance alleles that preserve the function of the target gene. The formation rate of resistance alleles that preserve the function of the target gene is shown. No such resistance alleles were formed in systems with at least four gRNAs, except in drives where incomplete homology-directed repair was possible.
However, certain types of gene drives are vulnerable to incomplete homology-directed repair as another mechanism for forming resistance alleles that preserve the function of the target gene. These drives target a critical gene such that individuals are rendered nonviable if one (haplolethal) or both (recessive lethal) alleles are disrupted. The drives contain a recoded sequence of the targeted gene that is immune to cleavage by the drive’s gRNAs. If homology-directed repair copies the recoded portion of the drive, a complete resistance allele that preserves the function of the target gene is formed, regardless of the number of gRNA targets in the system. For modification drives, this is not an issue if the payload is also copied. These events are likely to be even more rare than copying of only the rescue element (because the rescue element is often located at the end of a drive). It is similarly unlikely for the rescue and drive elements to be copied without the payload. Thus, the only outcome of incomplete homology-directed repair that we model is full resistance formation, either disrupting or, more rarely, preserving the function of the target gene. A more detailed discussion of this mechanism is provided in the Supplementary Results covering incomplete homology-directed repair. With incomplete homology-directed repair as the last element in our “full model,” we find that there is an optimal number of gRNAs for this family of drives for minimizing resistance alleles that preserve the function of the target gene. This number is usually three, but it is somewhat higher when the rate of incomplete homology-directed repair copying the recoded region is very low (Fig. 5).
Results of the full model for multiplexed gRNAs
With our full model in place, we consider the performance of several types of drives. The first of these is the standard homing drive. This drive accomplishes its goal by carrying an engineered payload and targets a neutral locus. Consequently, there is no effect from disrupting the target, and all resistance alleles are treated the same. The next is a suppression drive targeting a recessive female fertility gene (21). In this drive, females are rendered sterile unless they have at least one wild-type allele or a resistance allele that preserves the function of the target gene. The dynamics of this drive result in complete population suppression when it is successful. We also consider approaches for population modification that target a haplolethal or recessive lethal gene, where the drive has a recoded sequence of the gene that is immune to gRNA cleavage (16). In the haplolethal approach, any individual with a resistance allele that disrupts the target gene is nonviable, removing these alleles from the population. In the recessive lethal approach, an individual is only nonviable if it has two such resistance alleles. Last, we consider a population modification drive that targets a gene of interest, such as a gene required for malaria transmission in Anopheles (27, 28). Rather than carrying a payload, this drive’s purpose is to disrupt its target in a manner similar to that of the suppression drive.
We found that the optimal number of gRNAs for the population modification drives to achieve a maximum drive frequency was three, although drives with two gRNAs were nearly as efficient (Fig. 6A). The haplolethal drive reached nearly 100% frequency when modeled with two or more gRNAs (Fig. 6A) due to rapid removal of resistance alleles. The recessive lethal drive is slower at removing resistance alleles when they form at low rates, so it reached a lower frequency (Fig. 6A). However, the haplolethal drive also removes drive alleles when they are present in the same individual as a resistance allele that disrupts the function of the target gene. Thus, this system spreads somewhat more slowly than other types of population modification drives, although not as slowly as the population suppression homing drive (Fig. 6B). Of particular interest, gRNAs beyond two reduce drive conversion efficiency, which results in a slower spread of the drive (Fig. 6B). However, having multiple gRNAs is essential for reducing the formation rate of resistance alleles that preserve the function of the target gene (Fig. 5), which would otherwise outcompete drive alleles over time (Fig. 6C).
Fig. 6. Comparison of performance parameters for different types of homing drive.
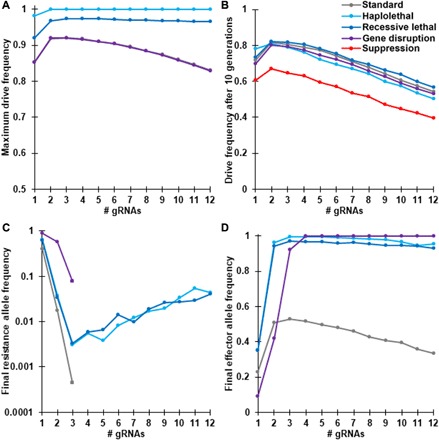
Drive/wild-type heterozygotes were released into a population of 100,000 individuals at an initial frequency of 1%. The simulation was then conducted for 100 generations using the full model. The displayed results are the average from 20 simulations for each type of drive and number of gRNAs. (A) The maximum drive allele frequency reached at any time in the simulations. Note that the standard drive and gene disruption drive have nearly identical values. (B) Number of generations needed for the drive to reach at least 50% total allele frequency. Note that the suppression drive is only shown in (B). (C) Final frequency of resistance alleles after 100 generations. The displayed values are only for resistance alleles that preserve the function of the target gene. No resistance alleles were present in the standard drive and gene disruption drive when at least four gRNAs were present. (D) Final effector allele frequency in the population after 100 generations. This was the drive allele for most drive types, but for the gene disruption drive, it includes resistance alleles that disrupt the function of the target gene as well.
Overall, having three gRNAs is usually optimal for population modification drives to attain maximum drive frequency after 100 generations (Fig. 6D). However, for the gene disruption homing drive, the optimal number of gRNAs for maximizing the frequency of “effector” alleles was four, five, or six (Fig. 6D). This is because effector alleles for this drive also include resistance alleles that disrupt the function of the target gene. In addition, this type of drive is not substantially impaired by incomplete homology-directed repair. This means that gene disruption drive can make efficient use of a higher number of gRNAs. Drives modeled with somewhat reduced performance based on our Drosophila experiments in this study (albeit with slightly lowered embryo resistance allele formation rates to represent an improved promoter) showed similar patterns, but with the optimal number of gRNAs increased by one for each drive (fig. S5 and see the Supplementary Results).
Multiplexing gRNAs for suppression drives
Suppression-type homing drives are particularly prone to failure if the resistance allele formation rate is high or if the drive conversion efficiency is too low. When examining the rate at which the drive was successful in completely suppressing the population (Fig. 7A), our high-performance drives with default parameters were usually successful, so long as there were sufficiently many gRNAs. However, drives with somewhat reduced performance (see the Supplementary Results) were less able to achieve successful suppression, regardless of the number of gRNAs. As with the default parameters, low numbers of gRNAs resulted in formation of resistance alleles that preserved the function of the target gene, which were able to quickly reach fixation in the population and prevent suppression (Fig. 7B). With an intermediate number of gRNAs, complete population suppression still usually occurred (Fig. 7C), but when the number of gRNAs was high, the rate of complete suppression declined. This is because with a high number of gRNAs, the drive suffered from reduced conversion efficiency and lacked the power to completely suppress the population (Fig. 7D). As the number of gRNAs is increased beyond two to three, the genetic load imposed by the drive at its final equilibrium (in the absence of resistance alleles that preserve the function of the target gene) is substantially reduced (fig. S6), preventing the drive from inducing complete suppression if the population growth rate at low densities is sufficiently high. With a choice of target sites with reduced formation of resistance sequences that preserve the function of the target gene, complete suppression becomes more likely, and the optimal number of gRNAs is reduced (fig. S7).
Fig. 7. Number of gRNAs needed for successful population suppression.
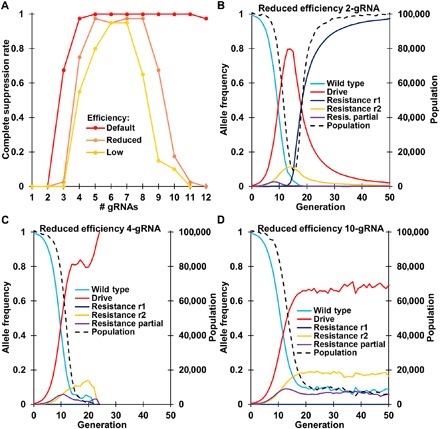
Drive/wild-type heterozygotes with a suppression drive were released into a population of 100,000 individuals at an initial frequency of 1%. The simulation was then conducted for 100 generations. (A) The displayed results are the average from 20 simulations for each type of drive and number of gRNAs. The fraction of simulations that resulted in complete suppression is shown. The full model was used. The default system based on the Anopheles parameters used an early germline cleavage rate of 2%, a homology-directed repair phase cleavage rate of 98%, and an embryo cleavage rate of 5%. For the reduced efficiency drive model, these parameters were changed to 5, 92, and 10%, respectively. The low efficiency drive model changed these parameters to 8, 90, and 15%, respectively. Allele frequency and population size trajectories are shown for individual simulations using the reduced efficiency model with (B) 2, (C) 4, and (D) 10 gRNAs. r1 refers to resistance alleles that preserve the function of the target gene, and r2 refers to resistance alleles that disrupt the function of the target gene.
DISCUSSION
Our study shows that homing drives likely have an optimal number of gRNAs that maximize drive efficiency while minimizing the formation of resistance alleles that preserve the function of the target gene. This result emerged naturally from a model that incorporated specific time steps for cleavage and repair, Cas9 activity saturation, and reduced repair fidelity when homology ends around the cut sites fail to line up perfectly. Even with a more basic model that differs from the model only by allowing a narrow timing window for homology-directed repair, we are able to reject the notion that homing gene drives can be made arbitrarily efficient by having a sufficiently high number of gRNAs. Overall, we showed that while multiple gRNAs are useful for improving drive efficiency and reducing resistance, these performance gains are far smaller than those predicted by simple models with sequential cutting and repair (22–24) or even models that include simultaneous cutting (22). This new model is consistent with our experimental results in this study, as well as previous work that observed smaller improvements from multiple gRNAs than predicted (13) or even marked declines in performance (9). Our model also takes germline cleavage timing into account, which is consistent with resistance allele sequencing in previous experimental studies (10, 12, 13).
While our model represents a step forward in our understanding of how multiplexed gRNAs affect homing drive efficiency, further improvements are needed to be able to more accurately predict homing drive performance. Earlier work indicated that the window for homology-directed repair is narrow, with only resistance alleles forming before and afterward (10, 12, 13). A better understanding of this window, the rate of successful homology-directed repair, and the proportion of resistance alleles formed before, during, and after this window would allow for improvements to our model. Homology of DNA on either side of a cut site is well known to be critical for the fidelity of homology-directed repair, and we showed that it indeed influences drive conversion efficiency. Last, Cas9 cleavage activity always reaches a maximum as more gRNAs are added, although details of this have not yet been thoroughly quantified. It is likely that for many gRNA promoters, a maximum cut rate would be reached quickly, thus reducing the cleavage rates at individual gRNA target sites as the total number is increased. Future studies could investigate how this saturation occurs and enable refinement of the quantitative model. In particular, the rate of resistance alleles formed due to incomplete homology-directed repair could be better quantified, with particular attention paid to the rate at which any recoded region is fully copied, thereby forming a resistance allele that preserves the function of the target gene. Last, variance in the activity level of gRNAs is well known, and we also observed this in our multiple gRNA homing drives in this study. These activity levels could potentially be predicted (32), but experimental assessment will likely remain necessary in the foreseeable future.
In our model, we also assumed that each gRNA cut site independently had the same chance of forming a resistance sequence that disrupts the function of the target gene. Thus, gRNA target sites would be best located close together to maximize repair fidelity. In practice, frameshifts between gRNA cut sites, but with restored frame after the last mutated site, may be insufficient to disrupt the function of the target gene. Thus, a good practice to minimize the formation of resistance alleles that preserve the function of the target gene would be to target conserved or important regions less tolerant of mutations, and perhaps to space gRNAs far enough apart, despite the cost to drive conversion efficiency, to ensure that a frameshift between any two gRNA sites disrupts the gene. At minimum, gRNAs should be placed far enough apart to prevent mutations at one site from converting an adjacent target site into a resistance allele.
Our models allowed us to gain insights about the relative strengths and weaknesses of the different types of homing gene drives. Standard drives lack any particular mechanism for removing resistance alleles (they need not even target a specific gene), which means that a successful drive of this nature requires a high drive efficiency, very low resistance allele formation rates, and low fitness costs to persist long enough to provide substantial benefits. The optimal number of gRNAs for these drives is likely low, perhaps two or three for a highly efficient system.
By contrast, drives that target haplolethal or recessive lethal genes can effectively remove resistance alleles that disrupt the function of the target gene and, thus, tolerate substantially higher overall rates of resistance allele formation. These drives are not expected to lose much efficiency with larger numbers of gRNAs, because although drive conversion efficiency is reduced, the drives also operate by toxin-antidote principles (25, 26), enabling removal of wild-type alleles and an accompanying relative increase in drive allele frequency even without drive conversion. However, we hypothesize that with reduced homology around the cut sites, incomplete homology-directed repair becomes more likely. This results in an optimal level of gRNAs that minimizes the formation of resistance alleles that preserve the function of the target gene due to incomplete homology-directed repair and end joining mechanisms. It is unclear how often incomplete homology-directed repair occurs, but it is likely that the optimal number of gRNAs for these drives is perhaps three or four. However, the rate of incomplete homology-directed repair could perhaps be minimized if the drive is located in an intron (possibly a synthetic intron), with essential recoded regions on either side of the intron. A system of this nature would only form a resistance allele that preserves the function of the target gene if incomplete homology-directed repair were to occur on both sides of the drive. This would allow for efficient use of a greater number of gRNAs. Improvements of this nature may not be necessary, however, if the rate of resistance allele formation that preserves the target function is substantially less than the rate at which payload genes are inactivated by mutations that occur during homology-directed repair (10−6 per nucleotide), which is approximately 1000-fold greater than the rate by DNA replication. If such a rate would preclude effective deployment of a homing drive, then toxin-antidote systems (25, 26) that rely only on DNA replication for copying of payload genes may be more suitable.
A gene disruption homing drive for population modification could potentially avoid both the need for a recoded region and inactivation of payload genes by targeting an endogenous gene. In this case, the end goal would be to disrupt this gene either by the presence of the drive or by formation of resistance alleles, rather than spreading a specific payload, and the formation of resistance alleles that disrupt the target gene may actually be beneficial due to their reduced fitness cost compared to the drive. For such a drive, the optimal number of gRNAs would be the minimum number necessary to prevent the formation of resistance alleles that preserve the function of the target gene, perhaps four to eight, depending on population size, target site, and drive performance.
A drive designed for population suppression has similar considerations, but with a narrower window for success. This is because any formation of complete resistance alleles that preserve the function of the target gene would likely result in rapid failure of the drive. In addition, if drive conversion efficiency is insufficient, then the drive may lack the power to completely suppress the population, at least within a reasonable timeframe. Thus, a narrower range of five to seven gRNAs would likely be optimal for such a drive. For all of these drive types, if the rate of resistance allele formation that preserves the function of the target gene is lower than in our models (such as by targeting a sequence that is highly intolerant of mutations (21)), then the optimal number of gRNAs is somewhat reduced.
Overall, we conclude that the total number of gRNA should be kept relatively low to achieve maximum effectiveness of multiple-gRNA drives: at least two, but well under a dozen, with the exact number depending on the type of drive and other performance characteristics. The gRNA target sites should also be placed as close together as possible while still far enough apart to prevent mutations at one target site from affecting adjacent sites. While our results suggest that multiplexing of gRNAs alone is unlikely to enable the development of highly effective homing drives, we expect that this approach will still be a critical component of any successful drive, especially when combined with additional strategies.
Supplementary Material
Acknowledgments
Funding: This study was supported by funding from New Zealand’s Predator Free 2050 program under award SS/05/01 to P.W.M. and the NIH awards R01GM127418 to P.W.M., R21AI130635 to J.C., A.G.C., and P.W.M., and F32AI138476 to J.C. Author contributions: S.E.C. performed and analyzed computational simulations. S.Y.O., C.L., and Z.W. performed and analyzed experiments. J.C. conceived and designed the study. S.E.C. and J.C. drafted the manuscript. All authors contributed to and revised the final version of the manuscript. Competing interests: All authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Data and models used in the paper are also available on github (https://github.com/MesserLab/Homing_Mechanisms_with_multiplexed_gRNAs). Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/10/eaaz0525/DC1
Supplementary Methods
Supplementary Results
Fig. S1. Synthetic target site drive schematic diagram.
Fig. S2. Effects of repair fidelity on drive performance.
Fig. S3. Effects of Cas9 activity saturation on drive performance.
Fig. S4. Effects of gRNA activity variance on drive performance.
Fig. S5. Comparison of performance parameters for different types of homing drives with lower cleavage efficiencies.
Fig. S6. Genetic load.
Fig. S7. Number of gRNAs needed for successful population suppression with a reduced functional resistance rate.
Table S1. gRNA cut site sequence analysis.
Data S1. Fly phenotype counts and analysis for the standard one-gRNA drive.
Data S2. Fly phenotype counts and analysis for the one-gRNA drive with a tRNA.
Data S3. Fly phenotype counts and analysis for the one-gRNA drive with poor right end homology.
Data S4. Fly phenotype counts and analysis for the split drive targeting yellow.
Data S5. Fly phenotype counts and analysis for combined drives targeting yellow.
Data S6. Fly phenotype counts and analysis for the two-gRNA drive with close target sites.
Data S7. Fly phenotype counts and analysis for the two-gRNA drive.
Data S8. Fly phenotype counts and analysis for the three-gRNA drive.
Data S9. Fly phenotype counts and analysis for the four-gRNA drive.
REFERENCES AND NOTES
- 1.Champer J., Buchman A., Akbari O. S., Cheating evolution: engineering gene drives to manipulate the fate of wild populations. Nat. Rev. Genet. 17, 146–159 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Burt A., Heritable strategies for controlling insect vectors of disease. Philos. Trans. R. Soc., B 369, 20130432 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alphey L., Genetic control of mosquitoes. Annu. Rev. Entomol. 59, 205–224 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Esvelt K. M., Smidler A. L., Catteruccia F., Church G. M., Concerning RNA-guided gene drives for the alteration of wild populations. eLife 3, e03401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DiCarlo J. E., Chavez A., Dietz S. L., Esvelt K. M., Church G. M., Safeguarding CRISPR-Cas9 gene drives in yeast. Nat. Biotechnol. 33, 1250–1255 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roggenkamp E., Giersch R. M., Schrock M. N., Turnquist E., Halloran M., Finnigan G. C., Tuning CRISPR-Cas9 gene drives inSaccharomyces cerevisiae. G3: Genes, Genomes, Genet. 8, 999–1018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basgall E. M., Goetting S. C., Goeckel M. E., Giersch R. M., Roggenkamp E., Schrock M. N., Halloran M., Finnigan G. C., Gene drive inhibition by the anti-CRISPR proteins AcrIIA2 and AcrIIA4 in Saccharomyces cerevisiae. Microbiology 164, 464–474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shapiro R. S., Chavez A., Porter C. B. M., Hamblin M., Kaas C. S., DiCarlo J. E., Zeng G., Xu X., Revtovich A. V., Kirienko N. V., Wang Y., Church G. M., Collins J. J., A CRISPR–Cas9-based gene drive platform for genetic interaction analysis in Candida albicans. Nat. Microbiol. 3, 73–82 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oberhofer G., Ivy T., Hay B. A., Behavior of homing endonuclease gene drives targeting genes required for viability or female fertility with multiplexed guide RNAs. Proc. Natl. Acad. Sci. 115, E9343–E9352 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.KaramiNejadRanjbar M., Eckermann K. N., Ahmed H. M. M., Sánchez C H. M., Dippel S., Marshall J. M., Wimmer E. A., Consequences of resistance evolution in a Cas9-based sex conversion-suppression gene drive for insect pest management. Proc. Natl. Acad. Sci. 115, 6189–6194 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gantz V. M., Bier E., Genome editing. The mutagenic chain reaction: a method for converting heterozygous to homozygous mutations. Science 348, 442–444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Champer J., Reeves R., Oh S. Y., Liu C., Liu J., Clark A. G., Messer P. W., Novel CRISPR/Cas9 gene drive constructs reveal insights into mechanisms of resistance allele formation and drive efficiency in genetically diverse populations. PLOS Genet. 13, e1006796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Champer J., Liu J., Oh S. Y., Reeves R., Luthra A., Oakes N., Clark A. G., Messer P. W., Reducing resistance allele formation in CRISPR gene drive. Proc. Natl. Acad. Sci. 115, 5522–5527 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Champer J., Wen Z., Luthra A., Reeves R., Chung J., Liu C., Lee Y. L., Liu J., Yang E., Messer P. W., Clark A. G., CRISPR gene drive efficiency and resistance rate is highly heritable with no common genetic loci of large effect. Genetics 212, 333–341 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Champer J., Chung J., Lee Y. L., Liu C., Yang E., Wen Z., Clark A. G., Messer P. W., Molecular safeguarding of CRISPR gene drive experiments. eLife 8, e41439 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Champer J., Yang E., Lee Y. L., Liu J., Clark A. G., Messer P. W., Resistance is futile: A CRISPR homing gene drive targeting a haplolethal gene. bioRxiv, 651737 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammond A. M., Kyrou K., Bruttini M., North A., Galizi R., Karlsson X., Kranjc N., Carpi F. M., D’Aurizio R., Crisanti A., Nolan T., The creation and selection of mutations resistant to a gene drive over multiple generations in the malaria mosquito. PLOS Genet. 13, e1007039 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammond A., Galizi R., Kyrou K., Simoni A., Siniscalchi C., Katsanos D., Gribble M., Baker D., Marois E., Russell S., Burt A., Windbichler N., Crisanti A., Nolan T., A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechnol. 34, 78–83 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gantz V. M., Jasinskiene N., Tatarenkova O., Fazekas A., Macias V. M., Bier E., James A. A., Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl. Acad. Sci. U.S.A. 112, E6736–E6743 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grunwald H. A., Gantz V. M., Poplawski G., Xu X.-R. S., Bier E., Cooper K. L., Super-Mendelian inheritance mediated by CRISPR–Cas9 in the female mouse germline. Nature 566, 105–109 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kyrou K., Hammond A. M., Galizi R., Kranjc N., Burt A., Beaghton A. K., Nolan T., Crisanti A., A CRISPR-Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat. Biotechnol. 36, 1062–1066 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prowse T. A. A., Cassey P., Ross J. V., Pfitzner C., Wittmann T. A., Thomas P., Dodging silver bullets: good CRISPR gene-drive design is critical for eradicating exotic vertebrates. Proc. R. Soc. B. 284, 20170799 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noble C., Olejarz J., Esvelt K., Church G., Nowak M., Evolutionary dynamics of CRISPR gene drives. Sci. Adv. 3, e1601964 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marshall J. M., Buchman A., Sanchez C. H. M., Akbari O. S., Overcoming evolved resistance to population-suppressing homing-based gene drives. Sci. Rep. 7, 3776 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Champer J., Kim I., Champer S. E., Clark A. G., Messer P. W., Performance analysis of novel toxin-antidote CRISPR gene drive systems. bioRxiv 10.1101/628362, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Champer J., Lee Y. L., Yang E., Liu C., Clark A. G., Messer P. W., A toxin-antidote CRISPR gene drive system for regional population modification. bioRxiv 10.1101/628354, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garver L. S., Dong Y., Dimopoulos G., Caspar controls resistance to Plasmodium falciparum in diverse Anopheline species. PLOS Pathog. 5, e1000335 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong Y., Simões M. L., Marois E., Dimopoulos G., CRISPR/Cas9 -mediated gene knockout of Anopheles gambiae FREP1 suppresses malaria parasite infection. PLOS Pathog. 14, e1006898 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haller B. C., Messer P. W., SLiM 3: Forward genetic simulations beyond the Wright–Fisher model. Mol. Biol. Evol. 36, 632–637 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hammond A. M., Kyrou K., Gribble M., Karlsson X., Morianou I., Galizi R., Beaghton A., Crisanti A., Nolan T., Improved CRISPR-based suppression gene drives mitigate resistance and impose a large reproductive load on laboratory-contained mosquito populations. bioRxiv, 360339 (2018). [Google Scholar]
- 31.Port F., Bullock S. L., Augmenting CRISPR applications in Drosophila with tRNA-flanked sgRNAs. Nat. Methods 13, 852–854 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doench J. G., Fusi N., Sullender M., Hegde M., Vaimberg E. W., Donovan K. F., Smith I., Tothova Z., Wilen C., Orchard R., Virgin H. W., Listgarten J., Root D. E., Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/10/eaaz0525/DC1
Supplementary Methods
Supplementary Results
Fig. S1. Synthetic target site drive schematic diagram.
Fig. S2. Effects of repair fidelity on drive performance.
Fig. S3. Effects of Cas9 activity saturation on drive performance.
Fig. S4. Effects of gRNA activity variance on drive performance.
Fig. S5. Comparison of performance parameters for different types of homing drives with lower cleavage efficiencies.
Fig. S6. Genetic load.
Fig. S7. Number of gRNAs needed for successful population suppression with a reduced functional resistance rate.
Table S1. gRNA cut site sequence analysis.
Data S1. Fly phenotype counts and analysis for the standard one-gRNA drive.
Data S2. Fly phenotype counts and analysis for the one-gRNA drive with a tRNA.
Data S3. Fly phenotype counts and analysis for the one-gRNA drive with poor right end homology.
Data S4. Fly phenotype counts and analysis for the split drive targeting yellow.
Data S5. Fly phenotype counts and analysis for combined drives targeting yellow.
Data S6. Fly phenotype counts and analysis for the two-gRNA drive with close target sites.
Data S7. Fly phenotype counts and analysis for the two-gRNA drive.
Data S8. Fly phenotype counts and analysis for the three-gRNA drive.
Data S9. Fly phenotype counts and analysis for the four-gRNA drive.


