Abstract
Malaria parasites adopt a remarkable variety of morphological life stages as they transition through multiple mammalian host and mosquito vector environments. We profiled the single-cell transcriptomes of thousands of individual parasites, deriving the first high-resolution transcriptional atlas of the entire Plasmodium berghei life cycle. We then used our atlas to precisely define developmental stages of single cells from three different human malaria parasite species, including parasites isolated directly from infected individuals. The Malaria Cell Atlas provides both a comprehensive view of gene usage in a eukaryotic parasite and an open-access reference dataset for the study of malaria parasites.
Single-cell RNA sequencing (scRNA-seq) is revolutionizing our understanding of heterogeneous cell populations, revealing rare cell types, unraveling developmental processes, and enabling greater resolution of gene expression patterns than has previously been possible (1). The ambition of cataloging the complete cellular composition of an animal is already becoming reality (2, 3), but thus far, atlasing efforts have focused on multicellular organisms. Here, we present the first comprehensive cell atlas of a unicellular eukaryote, the malaria parasite, across the entirety of its life cycle.
Although malaria parasites are unicellular, they display remarkable cellular plasticity during their complex life cycle, with stages ranging from 1.2 to 50 μm and spanning vastly different human and mosquito environments. Clinical symptoms of malaria result from asexual replication within red blood cells, whereas transmission to new hosts relies on replication in the mosquito. Both disease development and transmission are therefore underpinned by the parasite’s ability to serially differentiate into morphologically distinct forms, including invasive, replicative, and sexual stages (Fig. 1A). This versatility is orchestrated by tight regulation of a compact genome, where the function of ~40% of genes remains unknown (4). Better understanding of gene use and gene function throughout the parasite’s life cycle is needed to inform the development of much-needed new drugs, vaccines, and transmission-blocking strategies.
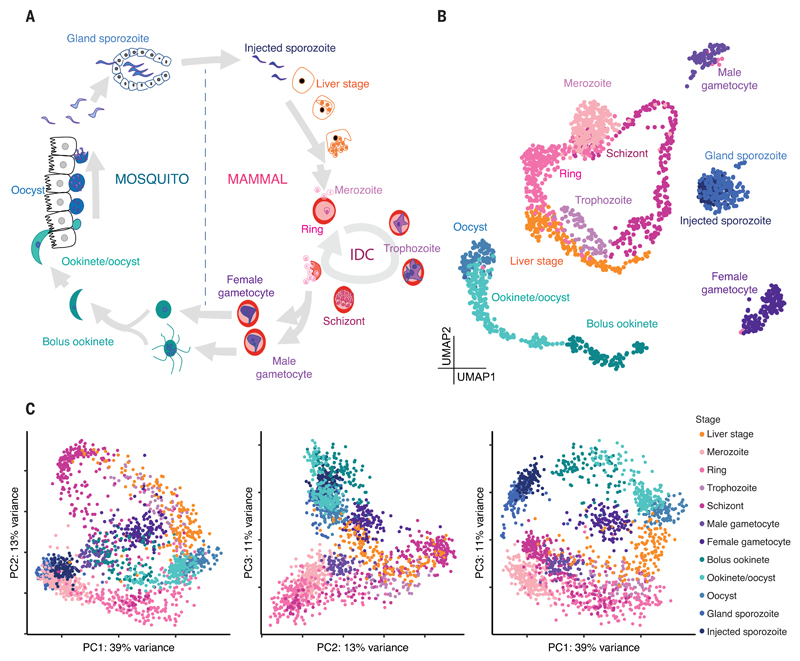
Fig. 1. A single-cell atlas of the P. berghei life cycle.
(A) The life cycle begins when an infected mosquito injects sporozoites into the mammalian host. From here, parasites enter the liver, where they develop, replicate, and then egress to enter the IDC. During the IDC, parasites invade erythrocytes, where they develop, replicate asexually, burst, and re-invade erythrocytes cyclically. Sexual forms are taken up by the mosquito, and if fertilization is successful, parasites invade the midgut and subsequently the salivary glands of the mosquito. In these different environments, parasites adopt different cellular strategies: replicative stages (liver stage, schizont, oocyst), invasive stages (merozoite, ookinete, and sporozoite), and sexual stages (male and female gametocytes). (B) UMAP of single-cell transcriptomes sampled from all stages of the life cycle, with cells colored according to their stage from (A). (C) The first three principal components from transcriptomes of all stages in the life cycle.
Single-cell resolution transcriptional variation provides insights into gene usage
To begin to build the Malaria Cell Atlas, we profiled 1787 single-cell transcriptomes across the entire life cycle of Plasmodium berghei using a modified Smart-seq2 approach (5). Purification methods were adapted to isolate each stage of the life cycle, including challenging samples such as rings, which have low levels of RNA, and ookinetes, which are difficult to sort (fig. S1). Ninety percent of sequenced cells passed quality control (1787/1982 cells) and poor-quality cells were identified in each stage according to the distribution of the number of genes per cell (fig. S2). After quality control, we detected a mean of 1527 genes per cell across the entire dataset; however, the number of genes detected was highly dependent on parasite stage (P < 0.001; fig. S2). Transcriptomes were normalized with TMM (trimmed mean of M-values) in groups of related stages for further analysis. For samples expected to be overlapping or heterogeneous (e.g., the blood stages), we used k-means clustering to delineate stages and confirm their classification based on known marker genes and correlations with bulk reference datasets (figs. S3 and S4). This allowed for differentiation of male, female, and asexual stages in the blood, as well as between ookinetes and oocysts in the heterogeneous population of parasites taken from the mosquito midgut during ookinete invasion.
All cell transcriptomes were visualized using uniform manifold approximation and projection (UMAP) (6) (Fig. 1B) and the first three components of a principal components analysis (PCA) (Fig. 1C). The results showed orientation of cells along a developmental path and also, to some extent, the formation of groups according to cellular strategy and host environment [e.g., actively replicative stages such as trophozoites and oocysts are near each other in UMAP and the first two principal components, whereas the third principal component separates the cells by host; Fig. 1,B and C]. All stages displayed marker genes concordant with known expression patterns (fig. S5). Merozoites, rings, trophozoites, and schizonts formed a circle, capturing the cyclical nature of the asexual intraerythrocytic developmental cycle (IDC) (Fig. 1B and fig. S3). A portion of this developmental trajectory was closely paralleled by the 44-hour liver schizonts. For these cells, we also captured the host’s transcriptome, confirming at a single-cell level that the parasite’s developmental progression is independent of the host cell’s cell cycle state (7) (fig. S6). In the mosquito stages, we observed a clear and abrupt transition from ookinetes to oocysts 48 hours after an infectious blood meal as the parasite crossed the mosquito midgut (fig. S4), and we were able to identify genes that are differentially expressed along this understudied developmental trajectory (data S1). In the two sporozoite collections (direct from salivary glands versus injected by infected mosquitoes into a mock host), we detected that upon release from the glands into the host, sporozoites express twice as many genes and up-regulate genes necessary for host invasion (data S1). This increase in the number of detected genes might indicate an activation of sporozoites in the mammalian host prior to liver cell invasion.
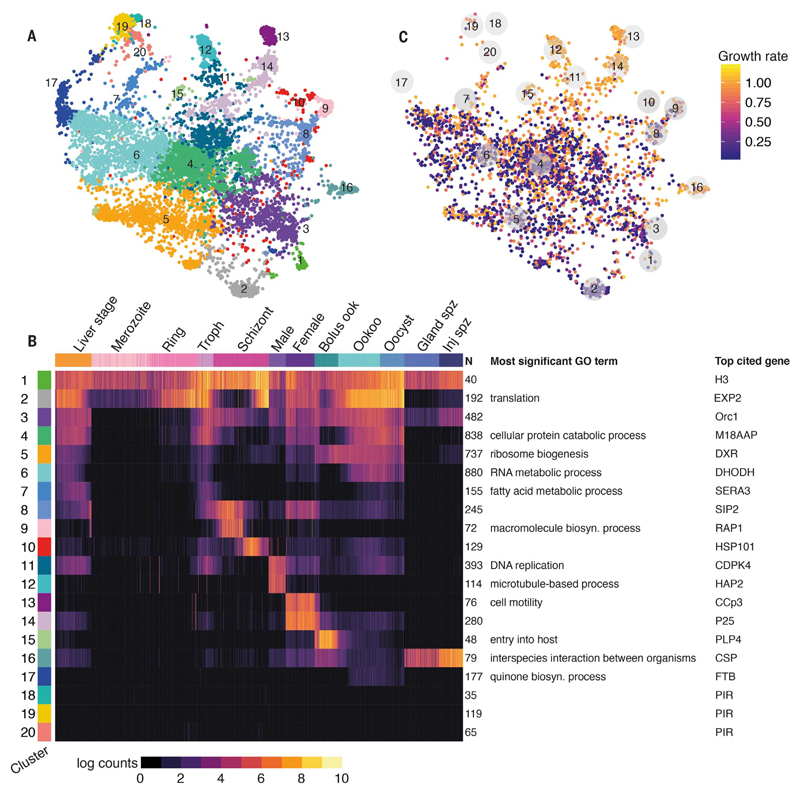
Our survey of the P. berghei life cycle enables a global view of gene expression and “guilt-by-association” prediction of function based on coexpression patterns. We constructed a force-directed k–nearest neighbor (k-NN) graph, where each node represents one of 5156 genes detected in the dataset (data S1). Graph spectral clustering (8) was used to assign each gene to one of 20 modules based on the graph distance matrix (Fig. 2A and data S1). Clusters were enriched for genes involved in specific biological processes and displayed distinct patterns of expression throughout the life cycle (Fig. 2B, fig. S7, and data S1). Some gene clusters (1 and 2) consisted mainly of housekeeping genes and ribosomal RNA components highly expressed across the full life cycle. At the other extreme, several clusters (clusters 18 to 20) showed low mean expression across cells and were primarily composed of genes from rapidly evolving multigene families (pirs and fams), which have no 1:1 orthologs with P. falciparum (Fig. 2B and fig. S8). Ten gene clusters (clusters 7 to 16) were highly expressed in a single stage. We corroborated these stage-specific gene modules using two additional methods. First, we identified marker genes based on level of expression relative to all other stages (data S1). Additionally, for each stage we defined a core transcriptome of genes where transcripts were detected in >50% of cells (data S1 and S2). The number of genes unique to the core transcriptome for each canonical stage ranged from 0 in merozoites to 237 in oocysts (data S1). Core genes for each stage were over-represented in clusters that coincided with expression at that stage and contained genes involved in the cellular strategy of that stage (e.g., DNA replication, invasion, sexual development; fig. S9), confirming our module assignment to specific stages.
Fig. 2. Graph-based clustering of genes reveals gene usage throughout the life cycle.
(A) A k-NN force-directed graph of all 5156 detected genes. Each node represents a gene. Nodes are colored according to their graph-based spectral clustering assignment, and each cluster is labeled by cluster number. (B) A heat map of mean expression for each cluster across all cells in the dataset. Cells are ordered by their developmental progression. Shown at the right are the number of genes in each cluster (N), the most significant Gene Ontology term (biological process) associated with the cluster (from data S1, Benjamini P < 0.05), and the top cited gene in the cluster (based on PlasmoDB). In the clusters where there is a tie for lowest P value, the GO term with the greatest percentage of genes in the cluster relative to the background is shown. If terms had identical representation, the term with the lowest GO number is shown (see complete table in data S1). (C) The same graph as in (A) colored according to relative growth rate of knockout mutants in asexual blood-stage parasites (11).
The majority of gene clusters showed pre-dominant expression in specific stages (Fig. 2B and fig. S7), offering new guidance as to where and how these genes might function. For example, genes encoding CelTOS and circumsporozoite protein (CSP), both important for invasion (9, 10), were found in cluster 16, which contains 79 genes most highly expressed in the invasive ookinete and sporozoite stages. Among these 79 genes are 34 annotated only as “conserved Plasmodium protein with no known function.” Their highly correlated expression with known invasion genes and their high expression in invasive stages will help to inform future functional studies. We also overlaid asexual growth rate data from a genome-scale knockout screen (11). Genes expressed primarily in transmission stages (clusters 11 to 16) tended to show normal growth rates in asexual blood stages (Fig. 2C), offering further support for the idea that genes in these clusters are primarily important in transmission stages. For genes in each cluster, we also identified motifs enriched in their upstream regulatory region [–1000 base pairs (bp)], which could be binding sites for apetala-2 (AP2) transcription factors that play critical roles in parasite progression through the life cycle (fig. S10 and data S1). Several of these motifs are conserved with P. falciparum in asexual stages (12) and transmission stages (13) (fig. S10). This categorization will help to functionally annotate genes with no known function, thereby enabling informed studies on gene regulation and supporting efforts to identify good candidates for transmission-blocking drug and vaccine development.
Development is the primary driver of differences in gene expression across the life cycle. However, variation between individual parasites within developmental stages is important for adaptation to the host environment (14). The principal mechanism for intrastage variation is thought to be driven by variation in expression among members of large multigene families whose functions are poorly defined (15). Nearly all life cycle stages showed highly variable expression in at least one of such multigene families (fig. S11). The largest of these families, pir, has a role in establishing chronic blood-stage infections (16). Subsets of pir genes showed variable expression in different stages, coupled with distinctive upstream sequences (fig. S12). Such putative promoter architectures could define stage-specific expression, with epigenetic control defining which subset is expressed. Interestingly, we found five coexpressed pairs of pir genes in merozoites and rings (fig. S12). Pir genes within each pair were split across different chromosomes but shared similar promoter architectures, with different pairs having different promoters (fig. S12). Although the function of these coexpression patterns is as yet unknown, such coexpression in a single cell can only be detected using scRNA-seq, highlighting another use of scRNA-seq toward identifying novel expression patterns.
Expanding the Malaria Cell Atlas across technologies and species
Droplet-based approaches to generate single-cell transcriptomes are less than 10% as costly as Smart-seq2 per cell, enabling the exploration of many more cells. To more deeply sample parasites along the entire IDC, we used 10x Genomics’ droplet-based Chromium platform to simultaneously capture P. berghei and another parasite species, P. knowlesi, in a single inlet (17). We found that 6.34% of cells were dual-species doublets, confirming a doublet rate as expected for mammalian cells (fig. S13). After removal of doublets and additional quality control, we captured 4884 P. berghei cells and 4237 P. knowlesi cells.
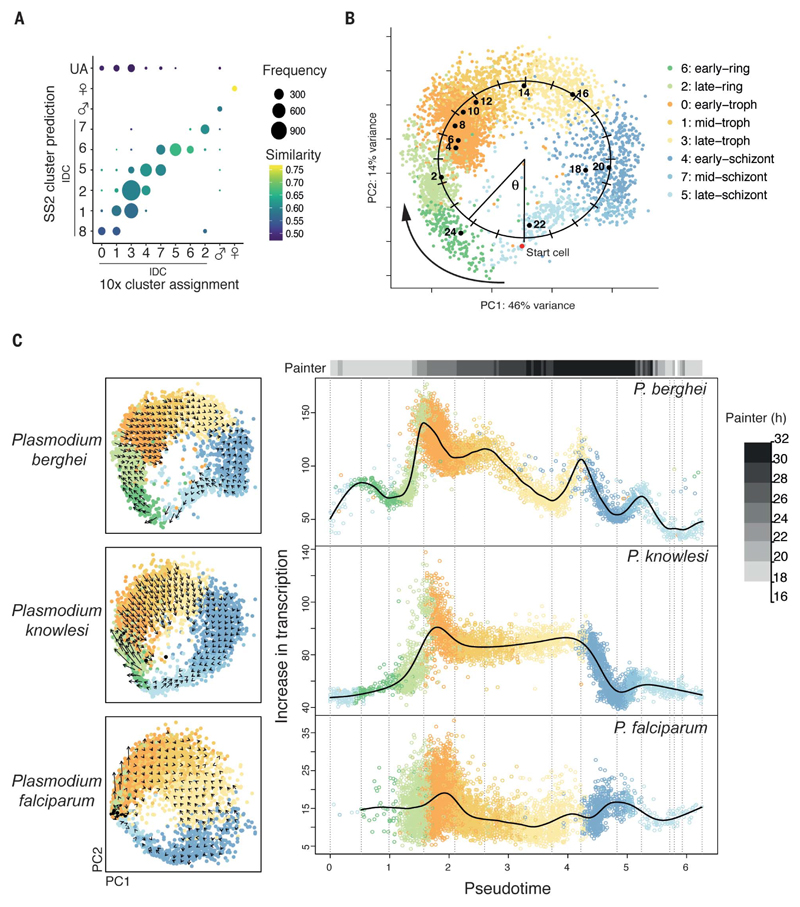
We used canonical correlation analysis (CCA) and scmap (Fig. 3A and fig. S13) (18, 19) to map P. berghei life stages between the 10x and Smart-seq2 technologies. CCA clustering showed good representation of all stages in both Smart-seq2 and 10x datasets within the IDC (fig. S13). Using scmap-cell, 94% of cells in the 10x data were assigned to a Smart-seq2 cell with high confidence, allowing us to align datasets (Fig. 3A). The additional P. berghei 10x data increased the coverage of cells in our atlas and confirmed our ability to evaluate single cells characterized by different methodologies. To account for the continuous cyclical nature of the data, we ordered the 10x cells in pseudotime by fitting an ellipse to the first two principal components and calculating the angle around the center of this ellipse for each cell relative to a start cell (Fig. 3B and methods). To annotate the orientation of the cycle with real time, we correlated each single-cell transcriptome with published bulk reference datasets and observed a high correspondence between bulk time point and pseudotime order (Fig. 3B and fig. S14).
Fig. 3. Alignment of datasets reveals transcriptional rates in the IDC.
(A) P. berghei 10x data mapped to Smart-seq2 data using scmap-cell. Cells are grouped according to their 10x cluster assignment (figs. S13 and S15) and the cluster of the Smart-seq2 cell it mapped to (fig. S15). A cosine similarity threshold of 0.5 led to classification of 283 cells (<6% of cells) as unassigned (UA). (B) PCA of P. berghei IDC cells from 10x. Pseudotime of each cell was measured by fitting an ellipse to the data and calculating the angle (radians) around the center of this ellipse for each cell relative to the start cell (red point). Black points represent the mean PCA coordinates of the bulk prediction for each cell (22) (fig. S14). (C) Left: PCAs of three Plasmodium species colored by their P. berghei cell assignment based on scmap. Arrows represent the relative change in transcriptional state based on RNA velocity. Right: Scaled increase in expression over the IDC. Cells are ordered according to the pseudotime of their scmap-assigned cell in the IDC P. berghei index. The top bar represents the matched time point between the P. berghei RNA velocity–derived transcription rates and P. falciparum transcription rates reported by Painter et al. (21) using Pearson correlations. Vertical gray lines mark peaks and troughs determined from the P. berghei data, as described in the methods.
Additionally, we generated a 10x dataset comprising 6737 cells from the IDC stages of the human parasite P. falciparum. We used scmap-cell to assign each P. falciparum and P. knowlesi cell to the P. berghei 10x reference index built with 1:1 orthologs (data S3), thus enabling us to align the developmental trajectories of these three species (Fig. 3C and fig. S15). We used this alignment to examine patterns of transcription in all three species in a time-resolved manner across the IDC. We calculated RNA velocity (20), which measures the rate of change of mRNA molecule abundances, across this deeply sampled set of Plasmodium parasites. We found that the rate of change varies markedly over the IDC in P. berghei, with peak velocity occurring in late rings, consistent with bulk studies of nascent RNA transcription (Fig. 3C and fig. S16) (21). We compared patterns of transcription as measured by RNA velocity at key transitions across species (Fig. 3C and fig. S16). Overall, the alignment of developmental trajectories across species revealed generally similar patterns of RNA velocity in the IDC of different malaria species; however, some life cycle stages and genes showed more similarity than others (fig. S17, table S2, and data S4). These parasite species naturally infect vastly different hosts and have different IDC lengths (24 hours for P. berghei, 27 hours for P. knowlesi, and 48 hours for P. falciparum). RNA velocity analyses highlight that the pace of transcription varies to support development to the next life stage in the IDC, with some similarity across these three species (Fig. 3C and fig. S16).
Transcriptomic studies of both in vivo and in vitro malaria parasites are often confounded by multiple life stages within a single sample. The Malaria Cell Atlas can be used to deconvolve bulk transcriptomic data and identify the specific life stages that were present in a bulk RNA-seq sample. We demonstrated this with the use of published bulk RNA-seq datasets from P. berghei (22, 23) and P. falciparum (24) (fig. S18). Future bulk RNA-seq studies can use the atlas to identify differences in cell type composition, potentially regressing these out to calculate more accurate differential expression between conditions or samples. Furthermore, scRNA-seq data generated by other groups (25) using a different droplet-based technology (Drop-seq) are also easily classified using scmap and the atlas data provided here (fig. S19). Together these findings confirm our ability to evaluate single cells characterized by different methodologies across parasite species that diverged more than 12.5 million years ago (26).
The Malaria Cell Atlas as a reference for clinical samples
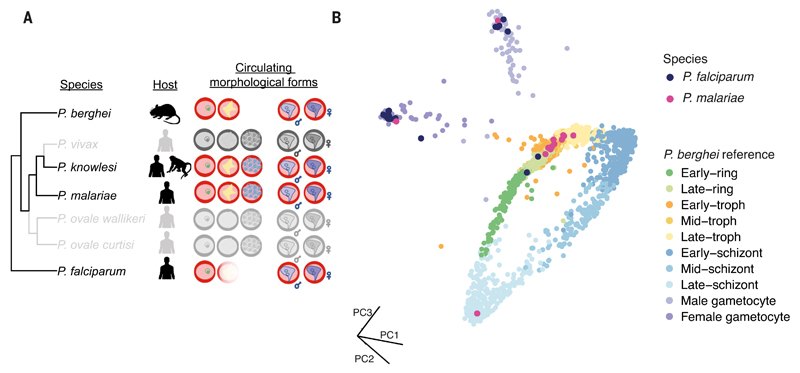
In vitro systems, although critical for experimental studies of Plasmodium parasites, are unlikely to fully capture the breadth of expression variation of parasites circulating in naturally infected carriers. Moreover, there are six phylogenetically distinct human-infecting species (Fig. 4A), several of which do not have any existing expression data and cannot be cultured in vitro. We therefore explored whether scRNA-seq data from wild parasites taken straight from infected people could be used to place wild parasites in developmental time using our atlas. We developed a methanol-based preservation protocol that produced Smart-seq2 transcriptomes with equivalent quality to unpreserved cells in the lab (fig. S20). Next, we used the protocol to preserve samples from three naturally infected asymptomatic carriers in Mbita, Kenya, which we then sorted and sequenced in the UK. We recovered single-cell transcriptomes from all three volunteers, and these field-collected samples displayed quality similar to that of laboratory samples (fig. S20). P. falciparum cells mapped to our atlas revealed male, female, and early asexual parasites (Fig. 4B), which are the expected circulating stages for this species (Fig. 4A). Cells clustered by stage and not by donor, indicating that comparisons both within and between hosts are possible; this indicates that scRNA-seq on field parasites will enable transcriptional characterization of natural infections. One of the volunteers was also infected with P. malariae, leading to the acquisition of transcriptomic data for this underexplored species. Notably, we observed late developmental stages; this was expected because, unlike P. falciparum, P. malariae late stages do not sequester in deep tissue (Fig. 4, A and B). As a proof of concept, we have shown that parasite species that have previously been inaccessible for expression analysis can now be characterized by combining scRNA-seq with the atlas.
Fig. 4. The Malaria Cell Atlas enables high-resolution mapping of field-derived single-cell transcriptomes of P. falciparum and P. malariae.
(A) Phylogeny of Plasmodium showing the mammalian host and the stages found in circulation for each species. P. falciparum and P. berghei sequester their late stages in deep tissue, whereas other species have all morphological forms in circulation. Species in color were profiled in the atlas. (B) P. falciparum and P. malariae field-derived cells mapped onto the P. berghei 10x reference index using scmap-cell. The field-derived samples mapped to developmental stages that were expected in circulation for each species.
The Malaria Cell Atlas reference dataset comprises 15,858 parasite transcriptomes covering every life stage along the parasite’s life cycle at single-cell resolution, and spans different technologies and different parasite species. The data are freely accessible as a processed dataset and through a user-friendly web interface (27, 28). As such, this will be a key resource for the malaria community in the study of transcriptional regulation and control of developmental progression at the highest resolution. Because the Malaria Cell Atlas provides a foundation for studying the biology of individual parasites directly from their natural environment, it represents an important endeavor toward characterizing phenotypes critical for malaria control, including those related to pathogenicity, drug resistance, and transmission biology.
Methods
Parasite culturing in vivo and in vitro
P. berghei parasites came from drug selection marker-free reporter line RMgm-928 that expressed mCherry, under the control of the hsp70 promoter, throughout the life cycle (29). Parasites were propagated in female 6- to 8-week-old Theiler’s Original outbred mice supplied by Envigo UK. Mosquito infections were performed in 2- to 5-day-old Anopheles stephensi mosquitoes.
P. falciparum (3D7) was maintained in O+ blood using RPMI 1640 culture medium (Gibco) supplemented with 25 mM HEPES (Sigma), 10 mM d-glucose (Sigma), hypoxanthine (50 mg/liter, Sigma), and 10% human serum (obtained locally in accordance with ethically approved protocols) in a mix containing 5% O2, 5% CO2, and 90% N2.
P. knowlesi (strain A1-H.1) was maintained in continuous culture (30) in O+ blood, using RPMI 1640 culture medium (Gibco) supplemented with 25 mM HEPES (Sigma), 22.2 mM d-glucose (Sigma), hypoxanthine (50 mg/liter, Sigma), 0.5% (wt/vol) Albumax II, and 10% horse serum, in a mix containing 5% O2, 5% CO2, and 90% N2. Cultures were maintained for >6 weeks without synchronization to ensure good representation of all stages in the IDC.
Human O+ erythrocytes were supplied by NHS Blood and Transplant, Cambridge, UK. All samples were anonymized. Use of erythrocytes from human donors for Plasmodium culture was approved by the NHS Cambridgeshire 4 Research Ethics Committee (REC reference 15/EE/0253) and the Wellcome Sanger Institute Human Materials and Data Management Committee.
Parasite isolation, cell sorting, and library preparation for Smart-seq2 scRNA-seq
Isolation of extraerythrocytic forms from HeLa cells
HeLa cells were cultured in DMEM supplemented with 10% FCS. P. berghei sporozoites were produced by homogenization of 50 dissected sets of salivary glands from female An. stephensi mosquitoes 22 days after an infectious blood meal. Sporozoites were counted on a hemocytometer, resuspended in DMEM, and added to an 80% confluent monolayer of HeLa cells at multiplicity of infection of 1. The plate was spun at 300g for 3 min and incubated at 37°C for 2 hours; cells were then washed twice with PBS and placed back in complete medium. After 24 hours, cells were split back at 70% confluency. Cells were harvested by trypsinization 44 hours after infection, washed once in PBS, and sorted immediately.
Isolation of blood-stage merozoites
P. berghei parasites were purified from an overnight (24 hours) 50-ml culture with 1 ml of infected blood using a 55% Histodenz cushion (Sigma) as detailed in (31). Purified schizont stages were stained with Hoechst 33342 (ThermoFisher) at a final concentration of 2.5 μg/ml for 10 min on ice, pelleted at 450g for 3 min, resuspended in 1 ml of medium, and passed through a 1.2-μm filter (Pall Life Sciences). Merozoites in the filtered fraction were sorted immediately.
Isolation of ring-stage parasites
A mouse infected with RMgm-928 was terminally bled by cardiac puncture using a syringe containing heparin. The ~1-ml blood sample was immediately transferred onto ice and stained with Hoechst 33342 (2.5 μg/ml) in PBS for 15 min (along with unstained controls for cell sorting). Cells were washed in RPMI and spun at 800g for 3 min and washed once more with PBS. Parasites were then incubated in 0.02% saponin in PBS for 3 min and then spun down at 1100g at 4°C. Parasites were washed once in PBS, pelleted at 1100g for 3 min, and then resuspended in 1 ml of PBS prior to sorting.
Isolation of ookinetes from the blood bolus
Ookinetes were isolated from the blood bolus of An. stephensi midguts at 18 and 24 hours after blood feeding from an RMgm-928–infected mouse at approximately 5% parasitemia. A lateral incision was made along the dissected mosquito midgut tissue to release the blood bolus and remaining blood was rinsed out using a syringe with PBS. Boluses from five midguts were pooled, diluted in 500 μl of PBS, and stained with SYBR green. To discriminate ookinetes from other stages in the blood bolus, a control feed was performed using a HAP2–-mCherry–infected mouse. HAP2 is essential for fertilization, so the bolus contained parasites but no ookinetes (32). This allowed us to enrich our sample for ookinetes by gating on the level of mCherry and SYBR green fluorescence (fig. S1, A to C).
Isolation of invading ookinetes and oocysts from the midgut
At 48 hours and 4 days post–blood meal, invading ookinetes and oocysts were isolated from 10 pooled infected midguts. Dissected midguts were disassociated in 200 μl of an enzymatic cocktail of collagenase IV (1 mg/ml) and elastase (1 mg/ml). The dissociation mixture was incubated at 30°C for 30 min with shaking at 300 rpm. Every 15 min, tissue was mechanically disrupted by pipetting up and down 40 times. To capture only invading ookinetes at 48 hours, the remaining blood bolus was removed as described above. As a control, midguts from mosquitoes that had fed on a HAP2–-mCherry mouse were disassociated to confirm that no remnants of the blood meal and noninvading parasites remained in the gut.
Isolation of salivary gland and injected sporozoites
Salivary glands from 20 An. stephensi infected with RMgm-928 were dissected on day 26 post–blood meal. Sporozoites were released from the glands by homogenizing the samples manually with a pestle in PBS. Samples were filtered with a 20-μm filter prior to sorting to remove large fragments of mosquito tissue. Simultaneously, female An. stephensi mosquitoes from the same infectious feed were fed using a membrane feeding assay containing approximately 600 μl of fructose solution (80 g/liter) with 10% human serum (filter-sterilized and heat-inactivated). Mosquitoes were exposed to the feeder for 12 min. After this, the remaining fructose/serum solution was removed from the feeder, and the presence of sporozoites in this solution was microscopically confirmed. Samples were then taken directly to cell sorting.
Preservation and isolation of cells from fresh peripheral blood samples
Samples were procured in the district of Mbita (Kenya) from asymptomatic volunteers in accordance with a study protocol reviewed and approved by the KEMRI Scientific and Ethics Review unit (KEMRI/RES/7/3/1). After screening with a rapid diagnostic test [SD BIOLINE Malaria Ag P.f/Pan (HRP-II/pLDH)], venous blood samples from infected volunteers were collected in EDTA-vacutainers. For each sample, two different purification methods were applied on 1 ml of the sample each to recover the later IDC and sexual stages on the one hand and the early IDC stages on the other. For the former, 1 ml of blood was resuspended in 5 ml of suspended animation buffer (10 mM Tris, 150 mM NaCl, 10 mM glucose, pH 7.37) (33) and placed on a magnetic column (MACs, Miltenyi). Late-stage parasites were eluted, washed once in suspended animation buffer, and resuspended in 200 μl of PBS. These were then fixed with 800 μl of methanol (Sigma) and preserved at –20°C. For the early IDC sample, another 1 ml was leucodepleted with a Plasmodipur filter (EuroProxima), washed twice in PBS, lysed twice with 0.15% saponin in PBS (Sigma), washed twice in PBS, and resuspended in 200 μl of PBS. Samples were then also fixed with 800 μl of methanol (Sigma) and preserved at –20°C. Both sample types were rehydrated with PBS, stained with Hoechst (2.5 μg/ml) in PBS for 15 min, and washed once in PBS prior to sorting.
Cell sorting
All parasite cell sorting was conducted on an Influx cell sorter (BD Biosciences) with a 70-μm nozzle. The HeLa samples were sorted on a Sony SH800 with a 100-μm nozzle chip. Parasites were sorted by gating on single-cell events and mCherry fluorescence (all stages) or Hoechst fluorescence (merozoites, field parasites). All parasites were sorted into nuclease-free 96- or 384-well plates (ThermoFisher) containing lysis buffer as described (5). Sorted plates were spun at 1000g for 10 s and immediately placed on dry ice.
Library preparation and sequencing
Reverse transcription, PCR, and library preparation were performed as detailed (5). All libraries were prepared in 96-well plates except a single 384-well plate of late blood stages. In the latter case, the lysis buffer volume was reduced to 2 μl, and the elongation temperature of the PCR was reduced to 68°C. Cells were multiplexed up to 384 and sequenced on a single lane of HiSeq 2500 v4 with 75-bp paired-end reads.
Parasite preparation and loading of 10x scRNA-seq
Parasite preparation
For P. berghei samples, blood was obtained by terminal bleed and passed through a prewetted Plasmodipur syringe filter (Europroxima) to filter out white blood cells prior to culturing. Three cultures were generated: cultured for 30 min, 10 hours, and 20 hours at 36.5°C with shaking at 65 RPM. Cultures were smeared prior to harvesting in order to ascertain parasitemia. After harvesting, the total number of red blood cells in each sample was counted using a disposable hemocytometer. This count was corroborated using a Countess cell counter. The number of infected red blood cells in each culture was used as a cell count, and cells were pooled 1:1:1 from the three time points and kept on ice. For P. knowlesi, the parasitemia of the cultured desynchronized parasites was measured and then cells were harvested by centrifugation at 450g for 3 min at 4°C. Supernatant was removed and parasites were washed twice in PBS before resuspension in PBS. The concentration of red blood cells was then calculated by manual hemocytometer, before calculating the final infected red blood cell concentration using the parasitemia. Cells were then pooled 1:1 with the P. berghei cell mixture described above in order to run a dual-species 10x analysis to evaluate doublet rates. P. falciparum parasites were prepared in the same manner as P. knowlesi but were run on their own 10x inlet.
10x loading
Cells were loaded according to manufacturer’s instructions to recover 10,000 cells per inlet. 10x Chromium Single Cell 3′ Library v2 chemistry was used and libraries were prepared according to manufacturer’s instructions. Each 10x input library was sequenced across two Hiseq 2500 Rapid Run lanes using 75-bp paired-end sequencing.
Bulk transcriptomics
Three P. berghei samples were prepared for bulk RNA-seq including early asexuals, late asexuals, and ookinetes. Mice infected with hsp70p:mCherry P. berghei were terminally bled by cardiac puncture using a syringe containing heparin. For the two asexual samples, the blood was treated with ammonium chloride to remove uninfected erythrocytes (34) either straight after the bleed (early) or after 24 hours of ex vivo culture (late). For the ookinete sample, the blood was cultured for 24 hours as described (35). RNA was extracted with TriZol according to the manufacturer’s recommendations and assayed with an Agilent RNA 6000 Nano assay, and transcriptomes were generated as described. A modified RNA-seq protocol was used. PolyA+ RNA (mRNA) was selected using magnetic oligo-d(T) beads. Reverse transcription using Superscript III (Life) was primed using oligo d(T) primers; second-strand cDNA synthesis included dUTP. The resulting cDNA was fragmented using a Covaris AFA sonicator. A “with-bead” protocol was used for dA-tailing, end repair, and adapter ligation using “PCR-free” barcoded sequencing adaptors (NEB) (36). After two rounds of SPRI cleanup (Agencourt), the libraries were eluted in EB buffer and USER enzyme mix (NEB) was used to digest the second-strand cDNA, generating directional libraries. The libraries were quantified by qPCR and sequenced on an Illumina HiSeq 2500.
Mapping and generation of expression matrices for scRNA-seq transcriptomes
Smart-seq2 mapping
Single-cell Plasmodium transcriptomes were mapped as reported previously (5). Briefly, trimmed reads were mapped using HISAT2 (v 2.0.0-beta) (37) to the P. berghei v3 genome (October 2016), and using STAR (v 2.5.0a) to the P. falciparum v3 (January 2016) and P. malariae v1 (March 2018) genomes using default parameters (38). Reads were summed against genes using HTseq (v 0.6.0) (39). For the coexpression of HeLa cells and liver-stage parasite analysis, both HeLa cells and parasites were mapped to respective genomes with STAR (v 2.5.1b) using default parameters (38).
10x data alignment, cell barcode assignment, and UMI counting
Cell Ranger single-cell software (version 2.1.0) was used to process sequencing reads, assigning each read to a cell barcode and UMI using standard parameters (17). After barcode assignment, the cDNA insert read was aligned using Cell Ranger (v 2.1.0) to a combined reference genome of P. knowlesi (March 2014) and P. berghei (July 2015), and the P. falciparum run was aligned to the 3D7 genome v3 (January 2016). These reference genomes were all obtained from www.sanger.ac.uk/resources/downloads/protozoa/.
Filtering and normalization of scRNA-seq data
Smart-seq2 filtering and normalization
Poor-quality cells were identified on a per-stage basis according to the distribution of the number of genes per cell, given the high variability of genes detected between stages (fig. S2, A and B). Cells with fewer than 1000 genes per cell and 2500 reads per cell were removed from the liver-stage parasites, trophozoites, male and female gametocytes, ookinetes, ookinetes/oocysts, and oocyst stages. Cells with fewer than 500 genes per cell and 2500 reads per cell were removed from schizonts and injected sporozoites. Cells with fewer than 40 genes per cell and 1000 reads per cell were removed from merozoites, rings, and gland sporozoites (fig. S2 and table S1). Additionally, we removed genes from further analysis that were detected in fewer than two cells across the entire dataset. The final dataset contained 1787 high-quality single cells from 1982 sequenced cells and 5156 genes out of 5245 genes with annotated transcripts. Transcriptomes were normalized with the weighted TMM method (40). Cells were normalized either all together or in five groups containing biologically similar stages; groups included IDC, liver-stage, gametocytes, ookinetes/oocysts, and sporozoites. Visual inspection of the relative expression plot (fig. S2D) showed little difference between normalization by biological group versus all together. Unless otherwise specified, further analysis was done on cells normalized by biological group.
10x filtering and normalization
For P. berghei, the output filtered matrix from Cell Ranger was read into Seurat (v 2.3.4) (18). Low-quality P. berghei cells with fewer than 230 detected genes were removed from further analysis. Initial inspection of filtered cells in the P. knowlesi and P. falciparum datasets showed that early-stage and late-stage IDC parasites were missing. These stages express fewer genes per cell relative to later stages based on our Smart-seq2 data, and we have previously observed a lower detection of genes per cell in P. falciparum (5), suggesting that these cells may have been removed by Cell Ranger’s default thresholding. Using the raw output matrices for these species, we adjusted thresholds to retain cells with >100 genes per cell for P. falciparum and >150 genes per cell for P. knowlesi. Intraspecies doublets were identified and removed from all three species using doubletFinder (v 1.0.0) (41). For the P. berghei/P. knowlesi run, we identified interspecies doublets as cells that contained >50 UMIs that mapped to each species (fig. S13A). The expected intraspecies doublet rate was calculated on the basis of this interspecies doublet rate, the relative proportion of each species, and the additional quality control thresholding. For P. falciparum, the intraspecies doublet rate was calculated from the expected doublet rate table provided by 10x Genomics. Thus, the numbers of intraspecies doublet cells removed were as follows: P. berghei = 200, P. knowlesi = 287, P. falciparum = 530 (fig. S13B). Doublets do not show a stage-specific bias (fig. S13B).
Single-cell transcriptome analysis of Smart-seq2 data
Cell clustering and projection
For timepoints where a heterogeneous population of stages was collected, we used k-means clustering using SC3 (version 1.7.7) to delineate stages and confirmed their classification based on known marker genes (42). This method was used for classification of males, females, trophozoites, and schizonts, as well as ookinetes and oocysts (figs. S3 and S4). For visualization in two dimensions, we performed UMAP (6) with the python package umap version 0.1.1 using the correlation distance metric, k–nearest neighbors of 10, min_dist of 1, spread of 2, and bandwidth of 1.
HeLa cell quality control and cell-cycle analysis
We performed initial filtering to identify the most robustly expressed genes across single cells. Genes were required to be expressed in >30 cells (of 164 cells) and cells needed to express >500 genes in both parasite and matched HeLa cells to be retained. This resulted in 163 matched cells with 4480 parasite genes and 8059 HeLa cell genes. We performed clustering of single HeLa and parasite cells independently using either all highly variable genes or subsets of annotated cell cycle genes. The highly variable genes were identified by plotting the averaged gene expression against gene dispersion [similar to Seurat (18)]. Louvain clustering was performed on single HeLa cells using only cell cycle genes, resulting in four louvain groups (fig. S6D). These groups are highly indicative of cell cycle progression starting from group 0 (G0/G1) to group 1 (G1S) to group 3 (G2) to group 4 (G2M).
Pseudotime
To order cells in a developmental trajectory, we reconstructed pseudotime using SLICER (43). Variable genes were identified within SLICER and then selected to build the trajectory based on a neighborhood variance that identifies genes that vary smoothly across the cell sets. SLICER was run independently on three groups of cells: (i) the liver-stage parasites, (ii) the entire IDC (merozoites, rings, trophozoites, and schizonts), and (iii) the ookinete-to-oocyst transition (bolus ookinetes, ookinete/oocyst, and oocyst). We assessed the performance of the algorithm by confirming that the pseudotime order matched the ground truth time point collections and expression of known marker genes over development (e.g., fig. S4E). To order all cells across the life cycle, we compiled these pseudotime orders with known timing of other stages that did not show a developmental signature (mature gametocytes and sporozoites).
Differential expression
Differential expression over the ookinete-to-oocyst transition and between gland and injected sporozoites was performed in monocle using the differentialGeneTest function (44).
Gene clustering and visualization of Smart-seq2 data
The gene count matrix was normalized by dividing by the mean counts for each gene and log scaling. This was done to reduce the amount to which gene clusters were driven by total gene expression and instead focus on the pattern of expression across cells. A k-NN graph was formed on the gene-normalized expression matrix with the Nearest Neighbors subpackage of python’s scikitlearn version 0.19.2 with parameters of k = 5 and a manhattan distance metric (45). We chose a k value of 5 because it was smaller than the smallest cluster we were interested in detecting, and the graph appeared robust from k =3 to k = 20. We then performed spectral graph clustering on this k-NN graph using the SpectralClustering subpackage of python’s scikitlearn version 0.19.2 (8, 46). The graph was visualized in Gephi with the forceatlas 2 graph layout algorithm in linlog mode to better show the clustering structure of the data (47, 48). Gene Ontology analysis was conducted with the dedicated PlasmoDB tool (49). The top cited gene in each cluster was identified using the literature tool on PlasmoDB (49).
Marker genes
Marker genes for each stage were identified in two ways. First, differentially expressed genes were calculated using the findMarkers function in scran (50). This function performs a Welch t test between pairs of stages and then identifies genes that are uniquely expressed in that cluster (pval.type = “all,” direction = “up”). This method was used to identify markers for each canonical stage, as well as marker genes within each host (mouse versus mosquito) and each cellular strategy (invasive, replicative, and sexual forms) (data S1). Second, marker genes were identified by defining a core set of genes for each stage as all genes that are expressed in more than 50% of cells. To avoid bias from the number of cells sampled within a stage, 60 cells were randomly selected per stage (data S2). We defined the unique core transcriptome as genes from each stage’s core that were unique to that stage’s core (i.e., not also found in more than 50% of cells from any other stage) (data S1).
Motif discovery
Motif discovery was performed using DREME, which searches for short (8 bp) motifs expressed as regular expressions (consensus sequences allowing for wildcards but not variable length gaps) in a given set of sequences (51). The 1000 bp upstream of the start codon for each gene detected in the Smart-seq2 dataset was used in the analysis. For each cluster, the input dataset was the upstream regions of each gene within that cluster and the negative set was the upstream region of genes that were not in that cluster. Clusters 18, 19, and 20 were not included in this analysis because they contain many paralogous genes from gene families with large duplicated upstream regions. The top motif of each cluster was compared to motifs from (12, 13, 52, 53) using Tomtom (54).
Analysis of development-independent gene expression variability
Almost all genes in Plasmodium genomes vary in expression over the life cycle. This is mainly thought to be related to the development of the parasite as it transitions between different life stages. We first identified highly variable genes in each stage independently. In the Smart-seq2 data, we used a general linear model to regress out the effect of pseudotime within developing stages [liver-stage exo-erythrocytic forms (EEFs), merozoites, rings, trophozoites, schizonts, ookinetes, and oocysts]. We preserved the mean expression of each gene by adding the predicted value of the mean to the residuals of the general linear model; in addition, we set any negative corrected values to zero in order to preserve the non-negativity of gene expression values. Finally, because the correction often shifted zeros to values only slightly above zero, we rounded these values down in order to meet the assumptions of the M3Drop model. We then used M3Drop (55) to identify genes with remaining heterogeneity [false discovery rate (FDR) ≤ 0.05], adjusted for mean expression level. Enrichment of each gene family within each stage was determined using the hypergeometric test with correction by FDR and a cutoff of 0.05.
We examined pir gene promoter architectures to determine whether particular gene expression patterns might be driven by transcription factors. First, we identified the 5′ UTR and upstream intergenic (putative promoter) regions of the pir genes shown in fig. S12. This was done manually by browsing the genome and referring to three P. berghei bulk RNA-seq samples of mixed early and late asexual stages as well as ookinete stages. Illumina reads from these libraries were mapped to the P. berghei v3 genome sequence using HISAT2 v2.0.0 (37), with–rna-strandness RF–max-intronlen 5000. The data were viewed using Artemis v18.0.0 (56). 5′ UTRs were defined as the region between the start codon and where RNA-seq coverage dropped to zero in at least two of the three samples. Upstream intergenic regions were defined from the start of the 5′ UTR to the next, upstream increase in coverage from one or more RNA-seq libraries. The upstream intergenic regions were BLASTed against each other (blastall 2.2.25, -p blastn -e 1e-20). The sequences involved in each hit were extracted, excluding those overlapping others with lower E-values. These sequences were then BLASTed against each other (blastall 2.2.25, -p blastn -e 0.01) and the resulting similarity matrix was used to cluster them with MCL v12-068 (57) with the inflation parameter set to 1.4. Sequences were collected together based on the clustering and aligned using MUSCLE v3.8.31 (58). The alignments were then trimmed by identifying highly conserved regions. Alignments in nonoverlapping windows of 10 nucleotides were evaluated, counting the proportion of sequences that were ungapped. An alignment position was called as good if ≥70% of sequences were ungapped at that position. A window of 10 nucleotides was called as a block if it contained no more than three bad positions. If there was more than one bad block in a row, a conserved region was ended. Only the longest conserved region from an alignment was kept. Sequences that began or ended within the conserved region were then removed. These alignments were used to build nucleotide profile hidden Markov models (HMMs) using HMMer i1.1rc3 (59). The models were then searched against the P. berghei v3 genome sequence, also using HMMer, to identify further members of the sequence families. Each hit was associated with the nearest downstream protein-coding gene. We identified eight upstream intergenic (promoter) sequence families associated with pir genes that we called A, C, D, F, G, H, I, and J (fig. S12).
RNA velocity
For each IDC (ring, trophozoite, schizont) Smart-seq2 cell that passed quality control, the exonic, intronic, and mixed reads were counted using RNA velocity (20). Intronic and mixed reads were combined to estimate the total unspliced reads in the data, whereas purely exonic reads were assumed to represent spliced transcripts. Cells were split by life cycle stage as for the pseudotime analysis, the expected ratio of spliced to unspliced reads for each gene was fit using RNA velocity, and residuals for each cell were estimated for each group independently. To ensure that we only considered genes that were fit well by the RNA velocity model, we required a minimum slope of 0.1 (increased from the default setting of 0.05) and a minimum correlation between spliced and unspliced reads of 0.5 (increased from the default setting of 0.05). To improve fits, we used the cells with the top and bottom 7.5% of expression levels for the fitting. Genes where more than 90% of residuals were either positive or negative were excluded as poorly fit genes. This resulted in 1345 genes × 548 cells for the IDC.
10x single-cell transcriptome analysis
Cell clustering
To identify male and female gametocytes in the P. berghei data, data were log-normalized, and clusters were identified using the shared nearest neighbor modularity optimization-based clustering algorithm in the FindClusters() function in Seurat (18). Two clusters corresponded to gametocytes based on expression of marker genes. These clusters were removed when comparing the data to the Smart-seq2 via CCA, as well as for the pseudotime assignment, and alignment of the three datasets in scmap (Fig. 3). The three species IDC PCAs were generated on TMM normalized data in scater (version 1.6.3) (60). Additionally, we identified clusters using the CCA in Seurat to compare the two methods of scRNA-seq (Smart-seq2 and 10x) (18). We identified nine clusters of cells that had good representation in both datasets (fig. S13C). One cluster, 8, contained only 15 cells across the two datasets and was removed from further analyses.
SCmap
We used scmap (version 1.1.5) (19) to compare datasets. We built three sets of cell indices that could be queried with the scmapCell() function that would allow each individual cell in the query dataset to be mapped to a reference index (19). To compare the Smart-seq2 and 10x data, we built an index of the blood-stage Smart-seq2 data (including gametocytes) and mapped the full P. berghei 10x dataset (including gametocytes) onto it. Because the IDC consists of a continuous set of cell stages and not discrete clusters, we modified the cell assignment method in scmap: Cells were assigned based on the top nearest neighbor. If the top cell had a cosine similarity of greater than 0.5, the query cell was assigned to that indexed cell along with its supporting metadata (cluster assignment, bulk prediction, pseudotime value). Using this cosine similarity threshold, 94% of 10x P. berghei cells were assigned to a cell in the SS2 P. berghei reference dataset.
To align the IDC trajectories across the three 10x datasets, we first compiled a set of one-to-one orthologs among 10 Plasmodium species (P. berghei, P. knowlesi, P. falciparum, P. malariae, P. ovale, P. vivax, P. gallinaceum, P. yoelii, P. chabaudi, P. cynomolgi) from OrthoMCL (61) (data S3). Using these orthologs, we built a scmap reference index that contained all P. berghei 10x IDC cells (gametocytes removed). We mapped both the P. falciparum and P. knowlesi data to this ortholog reference index. In addition to identifying the top nearest neighbor cell, we were able to incorporate information from the top three nearest neighbors to assign each cell based on the principal component space. To do this, we took a mean of the first two principal components of the top three nearest neighbors. Given this coordinate assignment, we located the nearest cell on the PCA and assigned the query cell to this index cell. If all three of the nearest neighbors had a cosine similarity of 0.3, then the query cell was given an assignment. With this lower cosine similarity threshold to account for cross-species differences, we were able to assign more than 96% of P. falciparum and 99% of P. knowlesi cells to a P. berghei index cell.
Finally, to map single-cell samples from the field, we built a 1:1 ortholog index of the complete 10x P. berghei dataset, including the gametocytes that were excluded for the IDC evaluations. We used this reference because (i) it fully represents the IDC and mature gametocytes, and (ii) it originates from an in vivo system like the volunteer cells. Because the gametocyte data were more sparse, the IDC cell assignment was based on the top nearest neighbor alone along with a cosine similarity threshold of 0.4. Using this method, we were able to map 13 P. malariae cells and 22 P. falciparum cells, assigning each cell to a developmental time.
“Clock” pseudotime
For the three 10x datasets, pseudotime around the IDC was calculated by fitting an ellipse to the data projected into the first two principal components using direct least squares (Fig. 3C and fig. S15). Angles around the center of this ellipse were calculated for each cell and oriented to a starting cell, which was defined using known markers. To align the three species in pseudo-time, 10x data from P. knowlesi and P. falciparum were projected directly onto the P. berghei reference using scmap (19) and cells were given the pseudotime of their P. berghei assigned cell. This “clock” pseudotime was aligned to real-time progression through the IDC using two methods. First, bulk RNA-seq data (22) from synchronized P. berghei parasites across 12 equally spaced time points around the IDC were projected onto our single-cell reference and their position in the first two principal components’ space was estimated from the average of their three nearest neighbors. These principal components’ locations were used to calculate a respective pseudotime for each bulk sample. Second, we mapped our single-cell RNA-seq transcriptomes onto the densely sampled P. falciparum bulk RNA-seq time course generated by Painter et al. (21). Genes were mapped across species using 1:1 orthologs (see above) and log-normalized RNA velocity-derived transcription rates were matched to the log-normalized transcription rates reported in (21) using Pearson correlations.
RNA velocity
We also ran RNA velocity on the 10x single-cell RNA-seq data from each species independently. Cells were filtered as described above; genes were filtered to exclude those that did not have at least one unspliced transcript in at least 10 cells and one spliced transcript in at least 20 cells. To account for the high number of zeros present in 10x data, we increased the k for the cell and gene k-NN smoothing included in RNA velocity to 50 and 5, respectively. To ensure good fits to the genes, we required a minimum slope of 0.2 and minimum correlation of 0.2 and used the top and bottom 20% of cells for the fitting. In addition, we excluded poorly fit genes as above. After this filtering, P. knowlesi data contained 1235 genes × 4237 cells, P. berghei data contained 1368 genes × 4763 cells, and P. falciparum data contained 645 genes × 6737 cells.
Comparing transcriptional waves
In the transcriptional waves through the IDC in P. berghei, we called 12 peaks and troughs by fitting a smoothed curve to the transcriptional rate through pseudotime using smooth.spline in R (Fig. 3C), with smoothing parameter (spar) equal to 0.9. We then identified all inversion points in the slope of consecutive points in the smoothed curve. For each of the 13 segments defined by these peaks and troughs, we used piecewise linear regression to test for significant increases or decreases in RNA velocity in each of the three species (Bonferroni multiple testing correction). In addition, we used piecewise linear regression on the individual genes for significant increases/decreases in RNA velocity across each segment in each species (5% FDR). To examine the conservation of the genes involved in the transcriptional waves through the IDC, we matched one-to-one orthologs of the genes used in the RNA velocity analyses across all three species. We plotted a heat map of the slopes for each of the 306 genes that were orthologous across all three species and passed RNA velocity quality control filters, and counted the number of genes that had a consistent direction with the pattern observed in P. berghei. Time points were matched across species by determining which P. berghei time point had the highest proportion of genes with slopes in the same direction as each P. knowlesi or P. falciparum time point. Any time point where fewer than 10 genes had consistent slopes with any P. berghei time point was considered unmatched. Significance of the matches was evaluated using a Binomial test of whether significantly more genes had a slope agreeing with the P. berghei reference than disagreeing. We used this test because is was robust to the fact that the numbers of cells per time point were not consistent across species, particularly for the early time points, which may cause so many of the DE genes to lose significance in the other species because of low numbers of cells (low power for the statistical test).
Deconvolution of bulk transcriptomic samples using scRNA-seq
The P. berghei 10x data were used as a reference and marker genes were called for each cluster in Seurat (v 2.3.4) using the Standard AUC classifier method. Genes that were not detected in >40% of cells and negative markers were excluded. The top 10 marker genes in each cluster, by power, were identified and used for deconvolution (n = 107). BSeq-sc (v 1.0) (62) was then used to estimate the proportion of cell types in each bulk sample using the default analysis pipeline.
Supplementary Material
Introduction
Plasmodium parasites, the causative agent of malaria, are single-celled organisms with distinct morphological developmental stages each specialized to inhabit vastly different environments and host cell types. Underlying this morphological diversity is tight regulation of a compact genome, where the functions of ~40% of genes remain unknown, hampering the rate of effective drug and vaccine development. Single-cell RNA sequencing (scRNA-seq) has allowed high-resolution mapping of developmental processes, cellular diversity, and cell-to-cell variation, and its application to unicellular organisms reveals individual-level variation between parasites across their full life cycle.
Rationale
We have assembled a Malaria Cell Atlas that presents the transcriptomic profiles of individual Plasmodium parasites across all morphological life cycle stages. The ambition of such an atlas is to (i) inform gene function and usage throughout the life cycle, (ii) understand the gene regulatory mechanisms underlying developmental transitions, (iii) discover parasite bet-hedging patterns, and (iv) provide a reference dataset that can be used to understand parasite biology by the malaria community in both lab and natural infections for multiple Plasmodium species.
Results
We isolated 1787 parasites using cell sorting and profiled full-length transcriptomes at 10 time points covering all life cycle stages across both the vector mosquito and the mammalian host. From these data, we could understand fine-scale transcriptional patterns of development and identify marker genes associated with parasite stage, cellular strategy (replicative, growth, and sexual phases), and host environment. Comparing single-cell gene expression patterns across the life cycle revealed groups of genes expressed in similar patterns during development. The resulting clusters of genes that behave similarly enables inference of possible function for the ~40% of genes that remain uncharacterized. Using droplet sequencing, we sequenced a further 15,858 cells from the intraerythrocytic developmental cycle for three different species, including two human pathogens. We aligned developmental trajectories across species during the pathogenic phase of the life cycle, establishing a cross-species comparison method. Finally, we developed a protocol for preserving wild parasites collected from naturally infected carriers and used scRNA-seq, together with the Malaria Cell Atlas as a reference, to identify wild parasite developmental stages and characterize a natural mixed-species infection at single-cell resolution.
Conclusion
We generated transcriptomes for all life cycle stages of Plasmodium and released these via the interactive Malaria Cell Atlas website, www.sanger.ac.uk/science/tools/mca/mca/. The Malaria Cell Atlas provides new insights into gene function and parasite developmental progression. We have demonstrated that it can serve as a transcriptomic reference, facilitating the interpretation of data from multiple species and multiple technologies. The characterization of wild Plasmodium parasites with immense genetic diversity will advance the study of the pathology and transmission of malaria directly from infected carriers. We envision that the Malaria Cell Atlas will support the development of much-needed new drugs, vaccines, and transmission-blocking strategies.
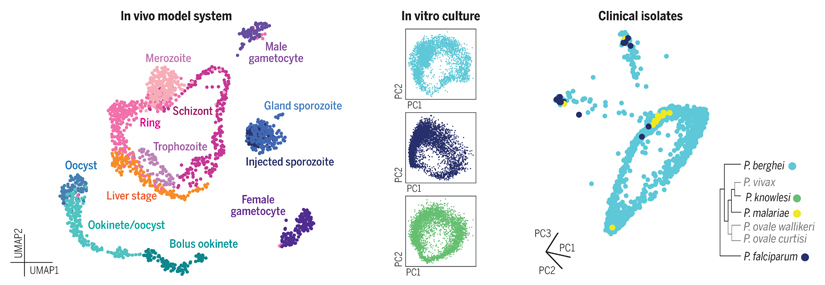
Single-cell RNA-seq references for the Plasmodium genus.
Left: Single-cell transcriptomes from across the life cycle of Plasmodium berghei were profiled (including liver, blood, and mosquito life stages). Center: Deep exploration of blood-stage parasites captured transcriptomic diversity at single-cell resolution across three different Plasmodium species by droplet sequencing. Right: Such datasets can serve as references to understand wild parasites isolated from clinical samples.
Acknowledgments
We thank the staff of the Illumina Bespoke Sequencing and Core Cytometry teams at the Wellcome Sanger Institute for their contribution, as well as J. Lee (QIMR Berghofer Medical Research Institute) and A. Cunnington (Imperial College London) for their contribution to the deconvolution of bulk RNA-seq data.
Funding
The Wellcome Sanger Institute is funded by the Wellcome Trust (grant 206194/Z/17/Z), which supports M.K.N.L., M.H., O.B., M.B., and J.C.R. Also supported by MRC Career Development Award G1100339 (M.K.N.L.); the Danish Institute of Advanced Study (D-IAS) and the University of Southern Denmark (SDU) (K.N.); Wellcome Trust grant 107372, the UK Department for International Development (DFID), the Swedish International Development Cooperation Agency (Sida), the Swiss Agency for Development and Cooperation (SDC), the German Academic Exchange Service (DAAD), and the Kenyan government (J.K.H. and H.B.); NIH/NIAID grant R01AI137154 (J.R. and L.V.); and Chan Zuckerberg Initiative grant reference 183501 (T.A.).
Footnotes
Author contributions: M.K.N.L. and A.M.T. conceived of and designed the study. Experiments were designed by V.M.H., A.J.C.R., M.K.N.L., and A.M.T. Parasite isolation optimization was performed by V.M.H. (mosquito stages), A.C.J.R. (blood stages, field parasites), and A.M.T. (all stages). V.M.H. and A.C.J.R. performed cell sorting and library preparation for Smart-seq2 and 10x, respectively. V.M.H. performed and oversaw the Smart-seq2 data analysis. V.M.H., A.C.J.R., and T.A. performed the 10x data analysis. H.H. performed the k-NN gene graph and spectral clustering analyses. T.A. performed the RNA velocity analyses. K.N. performed analyses on HeLa cells. A.J.R. performed analyses on bulk transcriptomes and pirs. L.H.V. and T.M. assisted with the lab work to produce P. knowlesi and P. berghei, respectively. O.B., J.C.R., M.B., and M.H. provided supervisory support to T.M., L.H.V., A.J.R., and T.A., respectively. M.H. also assisted with data analysis. H.B. and J.K.H. assisted with field work to capture P. falciparum and P. malariae from naturally infected donors. V.M.H., A.J.C.R., A.M.T., and M.K.N.L. wrote the manuscript, with contributions from other authors.
Competing interests: H.H. was previously an employee of 10x Genomics and holds shares in that company.
Data and materials availability: All raw sequencing data have been deposited in the European Nucleotide Archive at European Molecular Biology Laboratory European Bioinformatics Institute (www.ebi.ac.uk/ena/) under accession number ERP110344, and processed data are accessible via Github, and the Malaria Cell Atlas website (28). Expression matrices and analysis code are available on Github and archived on Zenodo (27) (DOI: 10.5281/zenodo.2843883).
References
- 1.Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA. The technology and biology of single-cell RNA sequencing. Mol Cell. 2015;58:610–620. doi: 10.1016/j.molcel.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Han X, et al. Mapping the Mouse Cell Atlas by Microwell-Seq. Cell. 2018;172:1091–1107.e17. doi: 10.1016/j.cell.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Fincher CT, Wurtzel O, de Hoog T, Kravarik KM, Reddien PW. Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science. 2018;360 doi: 10.1126/science.aaq1736. eaaq1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otto TD, et al. A comprehensive evaluation of rodent malaria parasite genomes and gene expression. BMC Biol. 2014;12:86. doi: 10.1186/s12915-014-0086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reid AJ, et al. Single-cell RNA-seq reveals hidden transcriptional variation in malaria parasites. eLife. 2018;7:e33105. doi: 10.7554/eLife.33105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McInnes L, Healy J, Melville J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. [6 December 2018]; arXiv1802.03426 [stat.ML]
- 7.Hanson KK, March S, Ng S, Bhatia SN, Mota MM. In vitro alterations do not reflect a requirement for host cell cycle progression during Plasmodium liver stage infection. Eukaryot Cell. 2015;14:96–103. doi: 10.1128/EC.00166-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi J, Malik J. Normalized cuts and image segmentation. IEEE Trans Pattern Anal Mach Intell. 2000;22:888–905. doi: 10.1109/34.868688. [DOI] [Google Scholar]
- 9.Kariu T, Ishino T, Yano K, Chinzei Y, Yuda M. CelTOS, a novel malarial protein that mediates transmission to mosquito and vertebrate hosts. Mol Microbiol. 2006;59:1369–1379. doi: 10.1111/j.1365-2958.2005.05024.x. [DOI] [PubMed] [Google Scholar]
- 10.Stewart MJ, Nawrot RJ, Schulman S, Vanderberg JP. Plasmodium berghei sporozoite invasion is blocked in vitro by sporozoite-immobilizing antibodies. Infect Immun. 1986;51:859–864. doi: 10.1128/iai.51.3.859-864.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bushell E, et al. Functional Profiling of a Plasmodium Genome Reveals an Abundance of Essential Genes. Cell. 2017;170:260–272. e8. doi: 10.1016/j.cell.2017.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Modrzynska K, et al. A Knockout Screen of ApiAP2 Genes Reveals Networks of Interacting Transcriptional Regulators Controlling the Plasmodium Life Cycle. Cell Host Microbe. 2017;21:11–22. doi: 10.1016/j.chom.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young JA, et al. In silico discovery of transcription regulatory elements in Plasmodium falciparum. BMC Genomics. 2008;9:70. doi: 10.1186/1471-2164-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guizetti J, Scherf A. Silence, activate, poise and switch! Mechanisms of antigenic variation in Plasmodium falciparum. Cell Microbiol. 2013;15:718–726. doi: 10.1111/cmi.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rovira-Graells N, et al. Transcriptional variation in the malaria parasite Plasmodium falciparum. Genome Res. 2012;22:925–938. doi: 10.1101/gr.129692.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brugat T, et al. Antibody-independent mechanisms regulate the establishment of chronic Plasmodium infection. Nat Microbiol. 2017;2:16276. doi: 10.1038/nmicrobiol.2016.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng GXY, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8 doi: 10.1038/ncomms14049. 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiselev VY, Yiu A, Hemberg M. scmap: Projection of single-cell RNA-seq data across data sets. Nat Methods. 2018;15:359–362. doi: 10.1038/nmeth.4644. [DOI] [PubMed] [Google Scholar]
- 20.La Manno G, et al. RNA velocity of single cells. Nature. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Painter HJ, et al. Genome-wide real-time in vivo transcriptional dynamics during Plasmodium falciparum blood-stage development. Nat Commun. 2018;9 doi: 10.1038/s41467-018-04966-3. 2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoo R, et al. Integrated analysis of the Plasmodium species transcriptome. EBioMedicine. 2016;7:255–266. doi: 10.1016/j.ebiom.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeoh LM, Goodman CD, Mollard V, McFadden GI, Ralph SA. Comparative transcriptomics of female and male gametocytes in Plasmodium berghei and the evolution of sex in alveolates. BMC Genomics. 2017;18:734. doi: 10.1186/s12864-017-4100-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee HJ, et al. Integrated pathogen load and dual transcriptome analysis of systemic host-pathogen interactions in severe malaria. Sci Transl Med. 2018;10 doi: 10.1126/scitranslmed.aar3619. eaar3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poran A, et al. Single-cell RNA sequencing reveals a signature of sexual commitment in malaria parasites. Nature. 2017;551:95–99. doi: 10.1038/nature24280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Böhme U, et al. Complete avian malaria parasite genomes reveal features associated with lineage-specific evolution in birds and mammals. Genome Res. 2018;28:547–560. doi: 10.1101/gr.218123.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howick VM, Reid AJ. vhowick/MalariaCellAtlas: Malaria Cell Atlas. 2019 doi: 10.5281/zenodo.2843883. [DOI] [Google Scholar]
- 28.Malaria Cell Atlas. www.sanger.ac.uk/science/tools/mca/mca/
- 29.Burda P-C, et al. A Plasmodium phospholipase is involved in disruption of the liver stage parasitophorous vacuole membrane. PLOS Pathog. 2015;11:e1004760. doi: 10.1371/journal.ppat.1004760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moon RW, et al. Adaptation of the genetically tractable malaria pathogen Plasmodium knowlesi to continuous culture in human erythrocytes. Proc Natl Acad Sci USA. 2013;110:531–536. doi: 10.1073/pnas.1216457110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomes AR, et al. A genome-scale vector resource enables high-throughput reverse genetic screening in a malaria parasite. Cell Host Microbe. 2015;17:404–413. doi: 10.1016/j.chom.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, et al. The conserved plant sterility gene HAP2 functions after attachment of fusogenic membranes in Chlamydomonas and Plasmodium gametes. Genes Dev. 2008;22:1051–1068. doi: 10.1101/gad.1656508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carter R, Chen DH. Malaria transmission blocked by immunisation with gametes of the malaria parasite. Nature. 1976;263:57–60. doi: 10.1038/263057a0. [DOI] [PubMed] [Google Scholar]
- 34.Martin WJ, Finerty J, Rosenthal A. Isolation of Plasmodium berghei (malaria) parasites by ammonium chloride lysis of infected erythrocytes. Nat New Biol. 1971;233:260–261. doi: 10.1038/newbiO233260a0. [DOI] [PubMed] [Google Scholar]
- 35.Siden-Kiamos I, et al. Plasmodium bergheicalcium-dependent protein kinase 3 is required for ookinete gliding motility and mosquito midgut invasion. Mol Microbiol. 2006;60:1355–1363. doi: 10.1111/j.1365-2958.2006.05189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozarewa I, et al. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nat Methods. 2009;6:291–295. doi: 10.1038/nmeth.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim D, Langmead B, Salzberg SL. HISAT: A fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anders S, Pyl PT, Huber W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11 doi: 10.1186/gb-2010-11-3-r25. R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst. 2019;8:329–337.e4. doi: 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kiselev VY, et al. SC3: Consensus clustering of single-cell RNA-seq data. Nat Med. 2017;14:483–486. doi: 10.1038/nmeth.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Welch JD, Hartemink AJ, Prins JF. SLICER: Inferring branched, nonlinear cellular trajectories from single cell RNA-seq data. Genome Biol. 2016;17:106. doi: 10.1186/s13059-016-0975-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pedregosa F, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 46.Saelens W, Cannoodt R, Saeys Y. A comprehensive evaluation of module detection methods for gene expression data. Nat Commun. 2018;9 doi: 10.1038/s41467-018-03424-4. 1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bastian M, Heymann S, Jacomy M. Third International AAAI Conference on Weblogs and Social Media. 2009 www.aaai.org/ocs/index.php/ICWSM/09/paper/viewPaper/154.
- 48.Jacomy M, Venturini T, Heymann S, Bastian M. ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PLOS ONE. 2014;9:e98679. doi: 10.1371/journal.pone.0098679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aurrecoechea C, et al. PlasmoDB: A functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:D539–D543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lun ATL, McCarthy DJ, Marioni JC. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000 Res. 2016;5:2122. doi: 10.12688/f1000research.9501.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bailey TL. DREME: Motif discovery in transcription factor ChIP-seq data. Bioinformatics. 2011;27:1653–1659. doi: 10.1093/bioinformatics/btr261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Campbell TL, De Silva EK, Olszewski KL, Elemento O, Llinás M. Identification and genome-wide prediction of DNA binding specificities for the ApiAP2 family of regulators from the malaria parasite. PLOS Pathog. 2010;6:e1001165. doi: 10.1371/journal.ppat.1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaneko I, Iwanaga S, Kato T, Kobayashi I, Yuda M. Genome-Wide Identification of the Target Genes of AP2-O, a Plasmodium AP2-Family Transcription Factor. PLOS Pathog. 2015;11:e1004905. doi: 10.1371/journal.ppat.1004905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8 doi: 10.1186/gb-2007-8-2-r24. R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andrews TS, Hemberg M. M3Drop: Dropout-based feature selection for scRNASeq. Bioinformatics. 2018 doi: 10.1093/bioinformatics/bty1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. Artemis: An integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 2012;28:464–469. doi: 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edgar RC. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eddy SR. Accelerated Profile HMM Searches. PLOS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCarthy DJ, Campbell KR, Lun ATL, Wills QF. Scater: Pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017;33:1179–1186. doi: 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baron M, et al. A Single-Cell Transcriptomic Map of the Human and Mouse Pancreas Reveals Inter- and Intra-cell Population Structure. Cell Syst. 2016;3:346–360.e4. doi: 10.1016/j.cels.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.