Abstract
Ultrasound is the most commonly used clinical imaging modality. However, in applications requiring cell-labeling, the large size and short active lifetime of ultrasound contrast agents limit their longitudinal use. Here, 100 nm radius, clinically applicable, polymeric nanoparticles containing a liquid perfluorocarbon, which enhance ultrasound contrast during repeated ultrasound imaging over the course of at least 48 h, are described. The perfluorocarbon enables monitoring the nanoparticles with quantitative 19F magnetic resonance imaging, making these particles effective multimodal imaging agents. Unlike typical core–shell perfluorocarbon-based ultrasound contrast agents, these nanoparticles have an atypical fractal internal structure. The nonvaporizing highly hydrophobic perfluorocarbon forms multiple cores within the polymeric matrix and is, surprisingly, hydrated with water, as determined from small-angle neutron scattering and nuclear magnetic resonance spectroscopy. Finally, the nanoparticles are used to image therapeutic dendritic cells with ultrasound in vivo, as well as with 19F MRI and fluorescence imaging, demonstrating their potential for long-term in vivo multimodal imaging.
Keywords: 19F MRI, cell therapy, cell tracking, multimodal imaging, perfluorocarbons, ultrasound
1. Introduction
Imaging modalities, such as magnetic resonance imaging (MRI), single photon emission computed tomography (SPECT), and positron emission tomography (PET) are key players in personalized medicine, due to their noninvasive nature.[1] However, these imaging techniques are expensive, logistically difficult, and can involve ionizing radiation, all of which hinder their longitudinal and frequent use. Ultrasound does not suffer from these limitations and can be used at the bedside, with frequent time intervals, and provide real-time imaging with excellent resolution. Consequently, ultrasound has become the most widespread and cost-effective clinical imaging modality.
Ultrasound has, so far, been largely unsuitable for longitudinal applications, such as cell tracking, where labeling with a contrast agent is needed, mainly because of the absence of effective contrast agents with sufficient in vivo lifetime.[2] These agents are typically micrometer-sized, surfactant (mainly phospholipid)-coated microbubbles with a compressible gas core.[3] Microbubbles are rapidly cleared from the circulation, within minutes, by trapping in the lungs and liver, by acoustic destruction, or by deflation as a result of gas diffusion. Several modifications of microbubble formulations have been proposed to overcome the stability issues, which mainly focused on shell composition, in order to increase the circulation time and durability of the microbubble agents.[4] Examples of these modifications include the combination of different shell compositions, like albumins, phospholipids, or polymers[5] with different gaseous cores, such as high molecular weight gases, for example, perfluorocarbon (PFC, C3F8 or C4F10) or sulfur hexafluoride (SF6).[6] Nevertheless, none of these attempts resulted in an in vivo stability exceeding 1 h. Additionally, the large size of these agents (typically 2 μm) prevents extravasation and can effectively limit intracellular localization.
Sub-micrometer droplets[5] consisting of liquid PFCs are able to extravasate and were proposed as phase-change agents that can be vaporized with an acoustic trigger and subsequently generate contrast.[7–11] The droplet-to-bubble conversion upon vaporization, comes with a fivefold increase in diameter.[12] Both microbubbles and phase-change droplets have been used to temporarily disrupt cell membranes and form pores that allow the uptake of specific drugs (sonoporation).[4,13–15] However, the high acoustic pressures needed to reach the vaporization threshold of these phase-change droplets and the short-lived contrast generation capability of the transient microbubbles resulting from acoustic vaporization remain an issue for long-term monitoring with ultrasound, making cell labeling infeasible.
Liquid PFCs typically generate poor ultrasound contrast[16] unless very high concentrations are used.[17–20] PFCs are both extremely hydrophobic and lipophobic, and stabilization of these compounds in aqueous environments is challenging. Typically, PFCs used in vivo as a lipid-stabilized emulsion have limited stability.[21] All imaging agents described thus far that contain a liquid PFC have a core-shell-like structure, where the liquid PFC droplet is coated with a lipid or polymer shell. A significant improvement of stability can be achieved by the encapsulation of PFCs in inorganic shells, such as silica.[22,23] However, while well-suited for in vivo applications, these materials still have to pass approval for clinical use. Despite the stability improvements and excellent acoustic properties, current clinical ultrasound contrast agents are still one-shot contrast agents, ill-suited for long-term applications, such as cell tracking or long-term labeling and imaging of inflamed tissue. Shapiro et al. recently introduced a new class of reporters for ultrasound, based on genetically encoded gas nanostructures from microorganisms. These gas-filled protein nanostructures allow the possibility of imaging of targets outside the vasculature and monitoring of cellular signals such as gene expression.[24–26] A few other studies describe the use of PFC-free solid silica-based nanoparticles for in vivo monitoring of stem cells with ultrasound and MRI.[27–29] These nanoparticles form solid aggregates upon cellular uptake under physiological conditions and allow the imaging and tracking of human mesenchymal stem cells with ultrasound due to the large size of aggregates.[29] The application of these materials is likely limited in applications that require colloidally stable particles.
Here, we present nanoparticles that consist of a high-boiling-point liquid PFC, encapsulated in a poly(lactic-co-glycolic acid) (PLGA) matrix.[30] These nanoparticles are stable in ultrasound over a timeframe of days, including after exposure to extremely high-pressure ultrasound. With a radius of 100 nm, these particles are small enough for cell uptake, and to extravasate and leave the circulation. The nanoparticles are loaded with perfluoro-15-crown-5-ether (PFCE) to enable imaging with both ultrasound and 19F MRI. Additionally, these nanoparticles can be loaded with gadoteridol for 1H MRI imaging and with fluorescent dyes. PFC-loaded PLGA nanoparticles have been originally explored as imaging agents for 19F MRI.[31–33] 19F MRI has become a key player in cell tracking, owing to (1) the absence of radiotracers and ionizing radiation, (2) an imaging window not restricted by tracer decay, (3) the ability for in vivo quantification, and (4) the direct clinical translatability.[34–36] On the other hand, MRI and, in particular 19F MRI, can be slow and cumbersome, and therefore a combination with a bedside technique, such as ultrasound, is highly valuable, as it allows for an optimal combination of quick imaging with ultrasound for localization, followed by single time point 19F MRI for quantification.
We investigate these PFCE-PLGA nanoparticles for long-term ultrasound imaging, focusing on cell tracking for cell therapy applications. We first characterize the internal structure of the nanoparticles using nuclear magnetic resonance spectroscopy (NMR) and small angle neutron scattering (SANS). These analyses reveal that the nanoparticles have a fractal multicore structure. Next, we imaged free nanoparticles and nanoparticle-loaded cells, both in vitro using phantoms and in vivo in mice, using multimodal imaging (ultrasound, 19F MRI, and fluorescence). We show that the nanoparticles are stable upon ultrasound exposure, fully biocompatible, and applicable to in vivo cell tracking.
2. Results and Discussion
2.1. Nanoparticles Display Long-Term Stable Acoustic Contrast
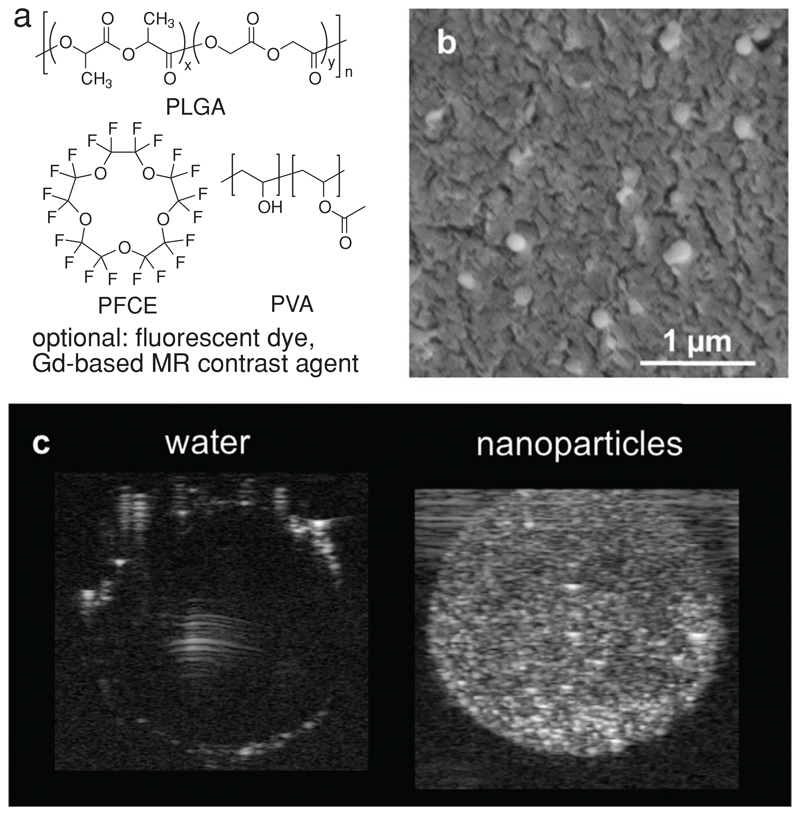
The nanoparticles are synthesized using a miniemulsion formulation technique.[31,33] They consist of a PLGA matrix loaded with PFCE and are stabilized with poly(vinyl alcohol) (PVA) (Figure 1a). Further modifications such as fluorescent dye or 1H MR imaging agent (typically gadoteridol) can be carried out, if needed. The nanoparticles have a mean radius of about 100 nm, as shown by cryogenic scanning electron microscopy (SEM, Figure 1b, radius 100 ± 20 nm (N = 20)). A PFCE encapsulation of 20–40 wt% (33 ± 11 wt% (number of batches N = 79)) was demonstrated by NMR spectroscopy (compare Table S2, Supporting Information, for some examples). We found that these nanoparticles, apart from being suitable for 19F MRI,[31,33] also generate acoustic contrast and can be imaged using standard ultrasound B-mode imaging (Figure 1c). Moreover, the acoustic contrast lasts at least a few hours with no signs of degradation, allowing for long-term imaging over a course of 48 h at least, as we show later in this study (Figure 2c).
Figure 1.
a) Chemical structures of the main components of PFCE-loaded PLGA nanoparticles: PLGA, PFCE, and PVA. Fluorescent dyes, or a Gd-based MR contrast agent, such as gadoteridol, can be further added for multimodal imaging. b) Cryo-SEM image of nanoparticles in water shows an average radius of 100 ± 20 nm. cNP = 10 mg mL−1. Scale bar: 1 μm. c) Ultrasound image of aqueous solution of nanoparticles (right) in a gelatin phantom compared to ultrapure water (left). cNP = 10 mg mL−1, 21 MHz, 50 dB.
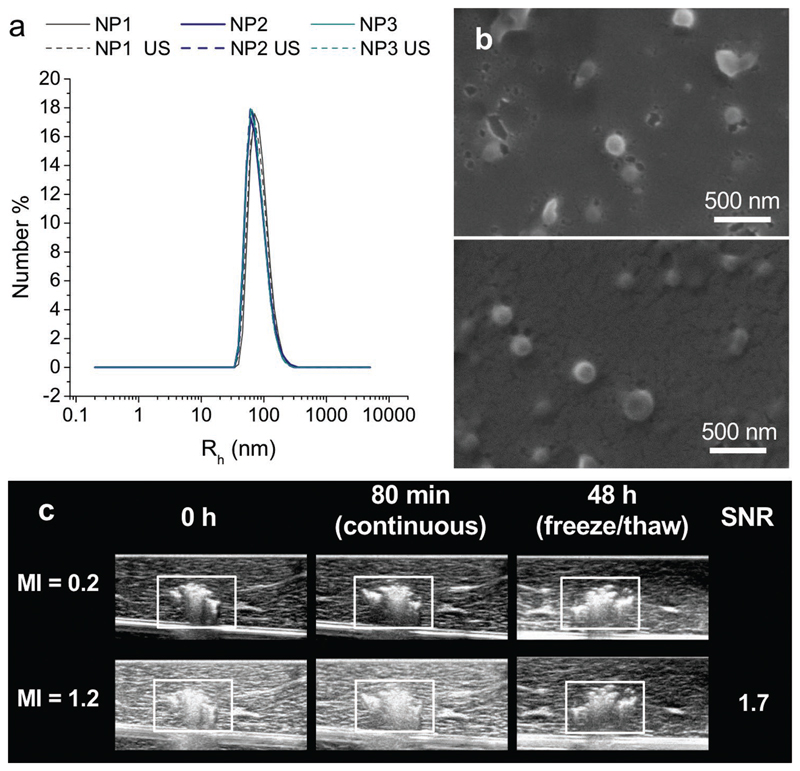
Figure 2.
PLGA-PFCE nanoparticles are stable in ultrasound. a) Number distributions of nanoparticles radius before (straight line) and after (dashed line) exposure to ultrasound measured at 173°. No changes were observed. Three different batches of nanoparticles were used (PFCE content NP1 24 wt%, NP2 and NP3 both 38 wt%; NP = nanoparticle). Each line represents an average of two independent measurements. b) SEM images were acquired on nanoparticles before (upper panel) and after (lower panel) probe sonication. No changes were observed in particle morphology. NPs with 39 wt% PFCE and hydrodynamic radius of 96 nm diameter (measured by DLS at 173°) were used. c) 0.5 mg of PFCE nanoparticles were injected in tissue (liver) and imaged at 0 and 80 min and 48 h postinjection at MI values of 0.2 and 1.2 (boxes indicate location of the particles). The transducer was left “on” continuously at MI = 1.2 in the same position for 80 min after injection. The sample underwent a freeze/thaw cycle before the 48 h time point. The SNR did not change for each MI over the period of 48 h.
To demonstrate that PFCE-loaded PLGA nanoparticles can be used for long-term ultrasound imaging, we carried out several in vitro experiments. First, we exposed the nanoparticles to ultrasound for 60 s and compared the diameter before and after ultrasound exposure using dynamic light scattering (DLS, Figure 2a). Next, we imaged the nanoparticles with SEM before and after probe sonication (Figure 2b). Both techniques demonstrated that the particles radius does not change indicating nanoparticle stability. The PFCE content also did not change after insonation. Thus, exposure to ultrasound did not significantly alter the properties of these nanoparticles.
To assess the stability of acoustic contrast in biological conditions, we injected nanoparticles in a tissue sample and imaged using a clinical ultrasound scanner at room temperature (Figure 2c) at high and low mechanical index (MI). After the first imaging session, the transducer was left “on” continuously for 80 min at high MI (MI = 1.2). This exposure was then followed by another imaging session at both MI values (Figure 2c). The sample was then frozen for 48 h and thawed to room temperature before imaging again. Even after this freezing and thawing of the sample, we observed no visible decrease in contrast, demonstrating that the imaging agents are extremely stable. Furthermore, the long stability of contrast strongly suggests the absence of gaseous components that could have come from gas diffusion or vaporization of the PFCE itself.
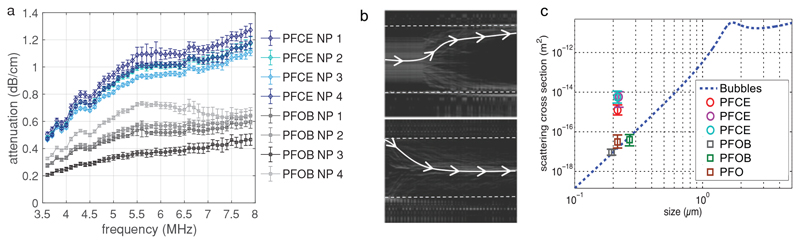
An ideal ultrasound contrast agent is characterized by a high scattering cross section and a low absorption of sound waves. The mechanical response of ultrasound contrast of microbubbles can be recorded with ultra-high-speed imaging.[37] However, the size of these PFCE-PLGA nanoparticles is well below the optical diffraction limit and necessitates alternative and more indirect methods to quantify their acoustic properties. For both microbubbles and solid particles, the scattering cross section is closely related to the attenuation cross section. Therefore, we measured acoustic attenuation of PFCE-loaded nanoparticles, using nanoparticles loaded with perfluorooctylbromide (PFOB) as control, both with and without gadoteridol. The characteristics of nanoparticles used for attenuation measurements are summarized in Table S1 (Supporting Information). Acoustic attenuation through nanoparticle solutions was measured at frequencies ranging from 3.5 to 8 MHz and pressures ranging from 50 kPa to 1 MPa (Figure 3a). These different pressures resulted in the error bars in Figure 3a. PFCE-PLGA nanoparticles showed the highest attenuation over the entire frequency range. This measurement, however, is not a direct measurement of the scattering properties of the particles, in particular because different mechanisms can affect the attenuation of dispersions.[38] To further assess the scattering properties of PFCE-PLGA nanoparticles, an additional measurement was performed. Here, we studied the displacement of the particles induced by acoustic radiation force, which can be more directly linked to the scattering cross section of the particles. Nanoparticle suspensions were injected in a microchannel subjected to an ultrasound standing wave (Figure 3b). Under the effect of primary acoustic radiation force, the particles move toward the node or the antinode of the standing wave.[39] The displacement of Sonovue(c) microbubbles was used to quantify the pressure field in the capillary. The amplitude of the acoustic radiation force (Figure 3c) was determined from a simple force balance, and the scattering cross section was subsequently calculated from radiation force as described in the Experimental Section. The theoretical scattering cross section[40] of microbubbles as a function of their size at a driving frequency of 2.0 MHz is depicted by the dashed blue line. The scattering coefficient of the perfluorooctane (PFO) and PFOB nanoparticles, used as controls, was similar to that expected for a coated bubble of comparable size. Interestingly, PFCE nanoparticles display a scattering coefficient that is two orders of magnitude higher than the predicted coefficient for a bubble of the same diameter. PFCE does not only improve echogenicity of nanoparticles but also changes the direction of the radiation force (Figure 3b): PFCE-loaded nanoparticles move toward the pressure antinode (upper panel), as expected for small bubbles. In contrast, nanoparticles loaded with other PFCs move toward the pressure node. This effect was detected at very low pressures (50 kPa), that is, in the absence of acoustic streaming and of spontaneous bubble formation from the PFCE. The latter is also excluded by the fact that the nanoparticles do not lose their echogenicity even after the application of high-intensity ultrasound, including tip sonication, or freeze/thaw procedures.
Figure 3.
Acoustic characterization of PFCE-PLGA nanoparticles. a) Attenuation of acoustic waves by nanoparticle solution (10 mg mL−1) at different frequencies. PFCE-loaded nanoparticles display higher attenuation than PFOB-loaded nanoparticles indicating that PFCE nanoparticles display higher scattering intensity. Each line represents an independent batch of nanoparticles; number of the batch corresponds to number in Table S1 (Supporting Information). b) Optical maximum intensity projection images for a sample of PFCE particles (upper) and PFOB particles (lower) flowing in a channel exposed to a standing acoustic wave. PFCE particles move toward high-pressure areas (channel edges indicated) whereas the PFOB particles move toward the pressure nodes located at the center of the channel. A sample path is highlighted in each panel. c) Scattering cross section of various particles compared to coated bubbles, as a function of the size of the particles. A square symbol denotes a positive sign for the radiation force, while circles denote a negative sign. Nanoparticles with PFCE show unexpectedly high scattering cross sections.
Overall, PFCE-loaded nanoparticles display sufficient acoustic contrast combined with long-term stability, up to several days in phantom, and thus are suitable for longitudinal imaging with ultrasound. The scattering cross section of PFCE-PLGA nanoparticles was lower compared to micrometer-sized microbubbles, as can be seen, for example, from the simulation of the scattering cross section of microbubbles in Figure 3c. However, their extraordinary high stability, small size, and the ease of loading with further compounds allow for different applications compared to microbubbles. Especially, applications that require extravasation or long-term monitoring of contrast agent, such as targeted drug delivery or cellular therapy, should be possible using the nanoparticles in the future.
2.2. PFCE-PLGA Nanoparticles Have a Fractal Multicore Structure
To explain the atypical stability and acoustic properties of PFCE-PLGA nanoparticles, we further investigated them by extensive physicochemical analyses. Particularly, we focused on potential differences in the structural configuration, which could result in the observed differences in contrast capability. As a reference, we synthesized PFCE-loaded core–shell PLGA nanocapsules (Figure 4a). To prepare these nanocapsules, we modified the procedure described previously by Tsapis et al. for the preparation of PFOB-loaded core–shell nanocapsules.[41] Thus, it could be expected that PFCE-loaded colloids will have a core–shell geometry, similar to PFOB-loaded colloids from the literature. In this method, sodium cholate, which is a small anionic surfactant, is used to stabilize the emulsion droplets during the emulsion formation, instead of nonionic poly(vinyl alcohol). After evaporation of the solvent, nanocapsules were coated with poly(vinyl alcohol) by adsorption.[41] In contrast to the nanoparticles, which are stable in solution for several days,[31] PFCE-loaded core–shell capsules displayed lower colloidal stability with the formation of visible aggregates after several hours, potentially as a consequence of coalescence or Ostwald ripening.[42] Similar to PFOB-loaded capsules from reference,[43] PFCE-loaded capsules showed ultrasound contrast in harmonic mode (Figure S10, Supporting Information).
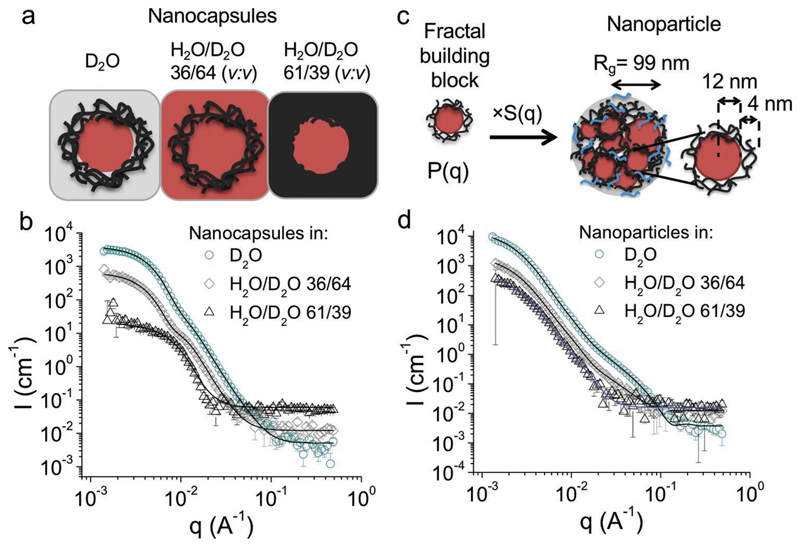
Figure 4.
Characterization of the nanoparticles and the core–shell capsules with SANS using contrast variation method. a) Schematic representation of core–shell capsules as they are “seen” by neutrons based on their scattering length density (SLD) in solvents used for SANS measurements. In D2O, both PLGA and PFCE contribute to scattering contrast, while in H2O/D2O 36/64 (v:v) or in H2O/D2O 61/39 (v:v) either PFCE (SLD = 3.86 × 10−6 Å−2) or PLGA (SLD = 2.11 × 10−6 Å−2) have the same scattering density as the solvent and are matched. b) SANS scattering patterns of PFCE-loaded core–shell capsules with fits of experimental data to core–shell structure. The form factor of nanocapsules can be fitted with a core–shell model (black lines). c) Schematic representation of the fractal core–shell model, which was used to fit the SANS data of nanoparticles. The scattering intensity of nanoparticles results from core–shell form factor, which is multiplied with a fractal structure factor to account for multiple domains within one particle. PFCE represented by red spheres, PLGA black lines, PVA blue lines. Rg is the radius of gyration. Note that the drawing of Rg is approximate; Rg is the root-mean-square distance of the parts of an object from its center of mass). d) SANS scattering patterns of PFCE-loaded nanoparticles with fits of the scattering intensity with a fractal model from (b) in different solvents. In D2O and H2O/D2O, 36/64 (v:v) (black lines) fractal core–shell model was used, while in H2O/D2O 61/39 (v:v) we applied the model for fractal aggregates with spherical building blocks (blue line).
To determine the internal structure of both types of PFCE-loaded colloids, we first measured transmission electron microscopy (TEM) and cryo-TEM. However, nanoparticles displayed only low contrast and were not stable in the electron beam. Therefore, we decided to use SANS, as it enables determination of internal structure of colloids in the solution. SANS experiments were performed with nanoparticles prepared with and without addition of gadoteridol (results in Figure 4 or the Supporting Information, respectively), as an additional 1H MR contrast agent, and core–shell nanocapsules. The summary of characterization of the nanocapsules and of the nanoparticles from Figure 4 with other techniques are in the Supporting Information (Table S3, Supporting Information; cryo-SEM images in Figures S2 and S3, Supporting Information; static light scattering (Guinier plots) in Figure S1, Supporting Information).
To determine the internal structure of nanoparticles by SANS, we applied a contrast variation method. In this technique, the neutron scattering contrast of the solvent is adjusted to highlight different compartments of nanoparticles.[44] A measure for the scattering contrast of the material is its scattering length density (SLD), which depends on the atomic composition and bulk density of the compound (Table S2, Supporting Information). As light water (H2O) and heavy water (D2O) have different scattering length densities, it is possible to match a specific compartment of the particles using a H2O/D2O mixture, which has the same SLD as the compartment. Thus, the matched compartments seem transparent for neutrons, as their scattering is the same as of the solvent background. Here, we used a H2O/D2O 36/64 (v:v) mixture to match PFCE (SLD = 3.87 × 10−6 Å−2) and H2O/D2O 61/39 (v:v) to match PLGA (SLD = 2.11 × 10−6 Å−2), while in D2O both components contributed to particle scattering. Figure 4a shows a schematic of how different components are “seen” by neutrons in different solvents.
The resulting SANS patterns of nanoparticles (with gadoteridol) and nanocapsules are shown in Figure 4b,d, nanoparticles without gadoteridol in Figure S6 (Supporting Information) and of non-loaded PLGA nanoparticles, which we used as an additional control in Figure S4 (Supporting Information) (compare Table S3, Supporting Information, and cryo-SEM images in Figures S2 and S3, Supporting Information, for further details on used samples). To determine the internal structure from SANS scattering patterns, we modeled the q-dependent absolute scattering intensities using theoretical models comprising particle form factors P(q) for the scattering of different geometrical shapes. The scattering profiles of the nanocapsules could be modeled with a core–shell structure, as expected (Figure 4b, Table S4, Supporting Information). By contrast, neither a core–shell model nor a homogeneous sphere model, which we used for nonloaded PLGA nanoparticles, matches well with the scattering pattern of PFCE-loaded nanoparticles. Instead, a model that describes a particle as a fractal aggregate of core–shell building blocks with a PFCE core and PLGA shell provided a good fit of the experimental SANS patterns.[45] A schematic representation of the model is shown in Figure 4b, the SANS patterns with the resulting model fit in Figure 4d, and the fit results are further summarized in Table S5 (Supporting Information). According to this fractal model, the small core–shell building blocks have a PFCE core radius of 9 nm and PLGA shell with a thickness of 4 nm. These building blocks are then assembled into a fractal multicore particle, which has a radius of gyration, Rg, of 98 nm (calculated from the fractal model as described by Teixeira).[45] Thus, the radius of gyration from model analysis is similar to the radius of 100 nm obtained from cryo-SEM further confirming the SANS model. Note, that Rg and radius from cryo-SEM measurement are not the same radius, as Rg is defined as the root-mean-square distance of the parts of an object from its center of mass. Further characterization of the samples from SANS with static and dynamic light scattering and cryo-SEM and the discussion of the results are presented in Section S1.2 (Supporting Information). SANS scattering patterns of PFCE nanoparticles prepared without addition of 1H MR contrast agent gadoteridol (Figure S6, Table S7, Supporting Information) showed similar results, suggesting that addition of gadoteridol did not affect the internal structure. This could be due to low encapsulation of gadoteridol. Additional SANS data comparing different batches of nanoparticles are available from the authors.
The fractal model agreed with experimental scattering patterns of nanoparticles only when the SLD of the PLGA core and PFCE shell were not fixed to the values of the pure compounds, but instead treated as free fitting parameters and calculated from the model. The resulting SLD values were between the SLD of pure compound and the solvent suggesting that both PLGA and PFCE are hydrated with the solvent used for SANS measurement (Table S5, Supporting Information). Similar results were obtained for several samples at two different SANS beamlines (see Figure S6, Table S6, Supporting Information, for results on a different sample).
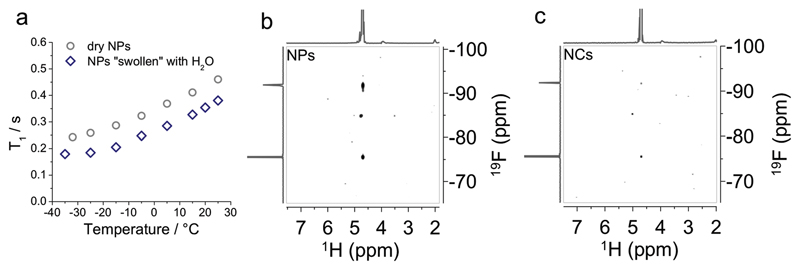
To prove that the highly hydrophobic PFCE is hydrated, we used NMR spectroscopy. First, we carried out relaxation time measurements at different temperatures with solid-state NMR spectroscopy (ssNMR) using the nanoparticles either as a freeze-dried powder or swollen with water. Here, the hydration of nanoparticles leads to a decrease of the spin-lattice relaxation time (T1) and an increase of the spin-spin relaxation time (T2) (Figure 5a and Figure S7, Supporting Information). These changes indicate that water is inside the polymer network making it more flexible, and thus, suggesting that water could be close to PFCE. Moreover, heteronuclear Overhauser enhancement spectroscopy (HOESY) measurements of fractal nanoparticles in solution also revealed that water is in close contact with PFCE cores (Figure 5b; tirifluoroacetic acid (TFA) as internal reference (δ − 76 ppm)). Nanoparticles show an HOE cross-peak between 19F nucleus in PFCE (−92 ppm) and 1H nucleus in water (4.79 ppm). The HOE effect is caused by crossspace dipole–dipole interaction and observed only when the internuclear distance is lower than a few Angstrom, and when the interaction is long-lived. Such interaction between water and PFCE is unexpected, as PFCE is highly hydrophobic. In contrast to nanoparticles, the HOE in PFCE core–shell nanocapsules, which were used as a control, is very low (Figure 5c). This result is also in accord with SANS measurements, where the SLD of PFCE–core in the capsules could be fitted with a value of pure PFCE. Also, the HOE of the internal TFA-reference appears lower in nanocapsules than in nanoparticles, indicating that TFA could also diffuse inside the nanoparticles, as discussed further in the Supporting Information (Figure S8, Table S8, Discussion S2, Supporting Information). HOESY measurements were reproduced several times with different batches of nanoparticles and nanocapsules showing the same result. The measurements shown in Figure 5b,c were done with different samples than those used for SANS measurements, as these were measured using exactly the same settings with TFA as internal reference. HOE was also observed in samples that were measured with SANS (data not shown, but available from the authors).
Figure 5.
Characterization of nanoparticles with solid-state NMR spectroscopy and heteronuclear Overhauser enhancement spectroscopy (HOESY). a) ssNMR spectroscopy: Spin-lattice relaxation time, T1, of freeze-dried nanoparticles, and of nanoparticles swollen with water reveals that water is present inside the nanoparticle (same batch of nanoparticles as used for SANS measurements in Figure 4d). b,c) HOESY NMR of PFCE-loaded nanoparticles (NPs prepared without gadoteridol, PFCE-content 26 wt%, Rh(173°) = 97 nm) and nanocapsules (PFCE-content 14 wt%, Rh(173°) = 82 nm) with TFA (δ = −76 ppm (CF3‒COOH)) as internal reference show that in nanoparticles 19F of PFCE is close to water.
Overall, SANS and NMR revealed that PFCE-PLGA nanoparticles have a multicore structure in which the highly hydrophobic PFCE is hydrated. The formation of multiple PFC domains has been reported previously in multicore micelles made of thermoresponsive polymers with covalently attached perfluorocarbon side chains,[46,47] where PFC side chains cluster due to the hydrophobic interaction. However, the noncovalent encapsulation of liquid perfluorocarbons in polymeric particles usually results in core–shell structures. Furthermore, the HOE, which results from dipole–dipole interactions between fluorocarbon and water, is completely unexpected for highly hydrophobic materials and has not reported previously. Thus, our results indicate that not only hydrophobic interactions but also other effects could play a role in the formation of multicore structure. Though it appears surprising that PFCE is close to water in the nanoparticles, it is known that PFCE can form complexes with some anions in the gas phase[48,49] and also weak complexes with cations.[50] The strong electron-withdrawing nature of fluorine could possibly be the reason for the occurrence of HOE effect. The fractal structure is obtained only with PVA as a surfactant, and the use of sodium cholate during the emulsification leads to core–shell structures. Though the PVA content in nanoparticles could not be determined exactly, the presence of PVA was shown by 1H NMR (Figure S9, Supporting Information). These observations suggest that the surfactant plays a pivotal role in the formation of the structure.
The relationship between the acoustic contrast and the structure of the particles remains unclear, especially considering small size of the PFCE nanoparticles. Our data, especially imaging in phantoms up to 48 h (Figure 2c), indicate that the generation of acoustic contrast does not involve vaporization of PFCE. In contrast, commercial microbubbles, such as Sonovue(c),[51] can be imaged only for a short time frame. Rapoport et al. previously observed cavitation in polymer-stabilized PFCE nanodroplets and suggested the evaporation of gases, which are dissolved in PFCE, as a possible reason for cavitation.[7] Similarly, the core–shell nanocapsules, which we used as a control in this study, displayed acoustic contrast in harmonic mode (compare Figure S10, Supporting Information, for images on nanocapsules in harmonic mode). In contrast, fractal PFCE-PLGA nanoparticles used in this study did not show harmonic signal but could be imaged using B mode. Additional ultra-highspeed imaging experiments with PFCE-PLGA nanoparticles using a Brandaris128 camera also did not show cavitation or bubble formation (data not shown due to insufficient resolution). Though nanoparticles were too small to be resolved with the Brandaris128 camera, we expect that in the case of cavitation, micrometer-sized bubbles would be visible, similar to cavitation of phase-change nanodroplets.[17,52] Thus, either no cavitation took place or the formed bubbles were very small and condensed back to nanoparticles quickly, and in a reversible manner. These observations suggest that the mechanism of acoustic contrast generation might be different from phase-change nanodroplets and microbubbles and does not involve cavitation.
Chen et al. recently reported the acoustic manipulation of silver nanocubes, demonstrating that the structure of nanocubes plays a more important role than the size.[53] Similarly, the multicore structure of PFCE-PLGA nanoparticles could be a key to the mechanism of acoustic contrast generation. However, a detailed systematic investigation of different parameters, affecting the structure of the nanoparticles and their behavior in the acoustic field, is needed to understand the mechanism of acoustic contrast generation, although many standard acoustic characterization assays are designed for much larger micrometer-sized agents.
2.3. Nanoparticles Are Suitable for In Vivo Cell Imaging
To demonstrate that PFCE-PLGA nanoparticles are suitable for long-term ultrasound imaging in vivo, we performed in vivo imaging in mice. With this experiment, we show that PFCE-loaded PLGA nanoparticles can be used for long-term tracking of therapeutic cells during cell therapy. We carried out in vivo imaging with a high-resolution preclinical ultrasound scanner, due to small size of the animals, and particularly murine lymph nodes (LNs) which cannot be resolved using clinical frequencies. The results were confirmed ex vivo with a clinical ultrasound scanner to demonstrate that nanoparticles can be imaged at frequencies that are usually used in the clinics. We used primary human therapeutic dendritic cells (DCs), as used in clinical trials[54] for cancer therapy of immunogenic tumors, such as melanoma. Here, we focus on nanoparticles rather than capsules, due to their superior stability. We have previously shown that labeling cells with nanoparticles does not result in the loss of cell viability, when compared to a nonlabeled control.[31–33] Furthermore, our previous MRI study has shown no effect of labeling on cells with respect to the expression of maturation markers by DCs, their ability to activate T cells or their migratory behavior.[31,55] Together, no effects on cell function were observed. Moreover, the cellular 19F loading, a key parameter for 19F MR-sensitivity, is the highest reported so far.[21]
Here, we first injected 2 × 106 cells labeled with nanoparticles in liver tissue and imaged the tissue with ultrasound, followed by fluorescence and 19F MR imaging to confirm the localization of the cells. We could detect the labeled cells using all three imaging modalities after the injection into excised liver tissue (Figure 6a). Note that the 19F MRI sensitivity matches the sensitivity reported previously,[21] as expected for liquid, rather than gaseous or solid, PFCE.
Figure 6.
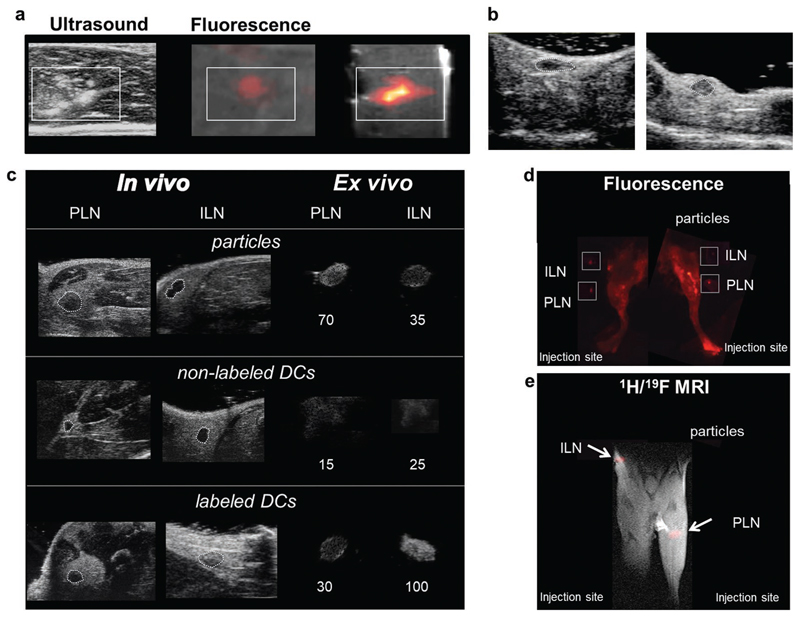
Nanoparticles are suitable for tracking the therapeutic cells with ultrasound in vivo. a) US, fluorescence, and 1H and 19F (false color) MR images of 2 million DCs labeled with particles containing PFCE, IC-Green, and Gd injected in a tissue sample (boxes indicate position of the cells). b) High-frequency in vivo US images of the inguinal lymph node (ILN) of a mouse before (left) and after (right) intranodal injection of 0.1 mg of PFCE nanoparticles show a tenfold increase in mean contrast in the node after injection (see Videos S1 and S2, Supporting Information). c) Mice were injected with 5 million labeled primary murine DCs in the footpad and imaged after 24 h using US on a clinical scanner (Figure S9, Supporting Information) and a high-resolution small animal scanner (48 MHz). Control mice received an equivalent number of nonlabeled cells or free particles. In vivo images of the draining lymph nodes (inguinal lymph node (ILN) and popliteal lymph node (PLN)) are shown at high resolution (48 MHz). Ex vivo images are also shown (circled) at high and low frequency (Figure S9, Supporting Information). Labeled DCs increased contrast in the node nearly fivefold compared to nonlabeled cells. d) A fluorescence image shows the same results, with labeled cells mainly in the INL and particles in the PLN and surrounding lymphatics. Excised lymph nodes were also imaged (boxes). e) The corresponding 1H/19F MRI, with 19F data in false color, also shows the same distribution. The injection site is just outside the field of view. Please see Figure S11 (Supporting Information) for further in vivo images, and Figure S12 (Supporting Information) for ex vivo images on a clinical scanner.
During clinical DC vaccinations, therapeutic cells can be injected directly in the lymph nodes of patients.[56] However, the total volume of the cells, which is needed for the injection in the murine lymph nodes of the mice, is too high for the intranodal injection (>10 μL). Therefore, we injected nanoparticle solution using the amount of nanoparticles, equivalent to 0.1 × 106 labeled DCs. In comparison, in human trials higher cell numbers are used, typically 3–15 × 106 cells injected intranodally.[56] High-frequency ultrasound imaging showed that the mean contrast of the lymph node changed over tenfold, owing to the presence of nanoparticles equivalent to 0.1 × 106 labeled cells (Figure 6b, and Videos S1 and S2, Supporting Information).
Finally, to see if migratory cells could be imaged using US, we injected labeled primary murine DCs in vivo in the footpad. Control mice were injected with the same number of untreated cells or free nanoparticles alone. Mice were imaged 24 h after injection using ultrasound, both at a clinical (Figure S12, Supporting Information, ex vivo) and at a high-frequency scanner (Figure 6c, and Figure S11, Supporting Information). Lower resolution clinical imaging at 7 MHz (Figure S12, Supporting Information) was not able to resolve mouse LNs in vivo; thus, the nodes were imaged in vivo at 25 MHz on a high-frequency preclinical US scanner. Additionally, we imaged the lymph nodes ex vivo with both scanners (Figure S12, Supporting Information, and Figure 6c) to corroborate the in vivo data. A sixfold increase in contrast was observed between nodes that contain nanoparticles or labeled cells, and those with nonlabeled cells (Figure 6c). The presence of cells in the lymph nodes was further confirmed in the fluorescence and 19F MRI images (Figure 6d,e). In contrast to labeled cells, nonlabeled cells were not detectable at any point. Typically, around 2% of injected DCs migrate from the injection site to draining lymph nodes,[56] which suggests that we can detect as little as 0.1 × 106 cells (2% of injected dose). A previous study showed that free nanoparticles are cleared from the site of injection, allowing for repeated injections.[31] Thus, the results presented here confirm that we imaged migratory labeled cells reaching a lymph node in vivo, and not free contrast nanoparticles or nonlabeled migratory cells. Finally, the sensitivity of imaging is well above that necessary for DC vaccination trials, and the labeling does not seem to hinder DC migration in vivo, demonstrating that these particles are suitable for the labeling and imaging of therapeutic cells.
Overall, our in vivo results demonstrate that these nanoparticles are suitable for long-term imaging with ultrasound, 19F MRI, and with addition of a dye to the formulation, also fluorescent and photoacoustic imaging.[17] Combining these complementary imaging techniques allows flexibility in clinical imaging. Thus, fast and easy screening with ultrasound at the bedside could be combined, for example, with quantitative 19F MRI, perhaps followed by fluorescence microscopy in biopsy samples. 19F MRI can provide quantitative information but requires more imaging time and transport of the patient to an MRI machine. Fluorescence provides better resolution, but it is hampered by the limited penetration of light through biological tissues.
Modern personalised medicine is complex, and there is no single imaging agent that can provide all the required information (e.g., cost-effective, accessible, penetration depth, quantitative, long-term).[57] Our multimodal imaging agent allows for the flexibility to combine the strengths of different imaging modalities as needed for a specific imaging need.
3. Conclusion and Outlook
Ultrasound imaging is ideally suited for the study of personalized therapeutics, but its potential is vastly unrealized due to the lack of stable, small, and biocompatible agents. Here, we present nano-particles (radius 100 nm), which are suitable for long-term in vivo ultrasound in combination with 19F MRI and fluorescence. The acoustic contrast generation does not involve vaporization of PFCE, evidenced by the lack of change in size, PFCE content, and the ability to perform 19F MRI after ultrasound. The nanoparticles in this study have a very atypical fractal structure, with multiple PFCE cores, each of which is surrounded by PLGA, as shown by SANS. Additionally, despite the hydrophobic nature of PFCE, NMR revealed that water is inside the particles and is in contact with the PFCE. This structure could be a key to the sustained acoustic contrast.
The physicochemical characteristics of fractal PFCE-PLGA nanoparticles can be adjusted for additional biomedical applications.[33] Possible modifications include changes in size or degradation rate, as well as surface modification with targeting ligands or a stealth PEG coating for additional applications.[33] Finally, these nanoparticles have recently been approved for a pilot clinical trial, where they will be used to label therapeutic dendritic cells in melanoma patients [NCT02574377]. Future clinical translation can be expected and these agents could potentially allow radiation-free and real-time ultrasound at bedside in clinical areas now dominated by other imaging modalities.
4. Experimental Section
Statistics
Standard two-tailed t-tests were carried out where indicated. In all cases, standard deviations and sample sizes are indicated.
Synthesis
Nanoparticles were made as described previously,[31] with or without the addition of gadoteridol from Prohance (Bracco Imaging Europe, Amsterdam). PLGA nanoparticles with entrapped perfluorocarbon alone or in combination with gadoteridol and a fluorescent dye, IC-Green (Akorn Inc., IL, USA) were prepared using a miniemulsion formulation method. Briefly, PLGA (100 mg, Resomer RG 502 H, lactide: glycolide molar ratio 48:52 to 52:48; Evonic Industry, Germany) were dissolved in dichloromethane (3 mL) and mixed rapidly with PFCE (900 μL, Exfluor Inc., TX, USA), or (230 μL, Perfluoron, Alcon Inc., TX, USA), or PFOB (275 μL, Fluorochem, UK). When needed, IC-Green (1 mg) in water and/or gadoteridol (1780 μL) were added. This mixture was premixed with a pipette and added rapidly to aqueous solution of poly(vinyl alcohol) (25.5 g, 1.96 wt%) and emulsified for 3 min under sonication at 40% amplitude using a digital sonicator from Branson Ultrasonics (Connecticut, USA). The solvent was evaporated overnight at 4 °C under stirring, and nanoparticles were collected by centrifugation at 21 000 g for 20 min, washed five times with distilled water, and lyophilized yielding ≈100 mg of nanoparticles as a colorless powder. 19F NMR: δ [ppm] −92 (C−F of PFCE); typical PFCE content 20–40 wt% (33 ± 11 wt% (N = 79)); corresponds to 4 × 1019 to 9 × 1018 19F atoms mg−1 determined with trifluoroacetic acid as internal reference). Gd content was measured using inductively coupled plasma mass spectrometry (ICP MS). The particles typically contain 0.5 μg mg−1. Depending on which batch of commercial PVA is used, the addition of some poly(propylene oxide) (PPO, Sigma-Aldrich, average Mn 2700) to the PLGA-PFCE mixture in dichloromethane (DCM) can improve the encapsulation of PFCE. Typically, PPO stock solution in DCM (15 mg of stock solution, prepared by adding 80 mg PPO to 150 μL DCM) was used. This addition is needed only with some batches of PVA, as PPO can be present in PVA as impurity, but the amount is different depending on which batch of PVA is used.
Nanocapsules were synthesized using sodium cholate as surfactant. PLGA (100 mg, resomer 502H) was dissolved in dichloromethane (3 mL) and mixed with perfluoro-15-crown-5 ether (900 μL) by pipetting it up and down with a glass pipette. The resulting primary emulsion was added to solution of sodium cholate (25 g, 1.5 wt% solution in water) and sonicated on ice for 3 min at an amplitude of 40% (Branson digital sonifier s250). After sonication, dichloromethane was evaporated overnight under stirring at room temperature. To exchange the surfactant, PVA solution (10 g of 1.96 wt% solution) was added to the suspension and the mixture was stirred at 4 °C for 5 d. The particles were washed twice with water at 16 000 g for 35 min. After washing, particles were resuspended in water (4 mL), frozen with liquid N2 and freeze-dried. Typical yield: ≈50–100 mg nanocapsules as a colorless powder. 19F NMR: δ [ppm] −92 (C−F of PFCE); typical PFCE content 10–20 wt% (determined with TFA as internal reference).
Acoustic Properties
The experimental setup used to measure the acoustic cross section consisted of a square cross-section capillary of about 380 μm in size attached to a piezoelectric crystal in order to generate an acoustic standing wave in the capillary at a frequency of 1.94 MHz. The motion of the particles was recorded at 125 frames s−1 using a high-speed camera (Photron, APX-RS) connected to an Olympus microscope. A 10× microscope objective was used for the visualization. The signals were generated with a Tabor AWG arbitrary waveform generator and amplified with an ENI 350L power amplifier. The experiments were realized at low pressures to limit the rapid build-up of acoustic streaming in the fluid. The data analysis was performed in Matlab, notably the PIV vector fields were obtained using OpenPIV, a freely available software. Two methods were used simultaneously to analyze the optical recording to obtain a better precision. The first is a PIV analysis based on the cross correlation between a subset of images. The relative displacement obtained from the PIV analysis can then be rescaled to the known geometry. The second method is based on averaging in the horizontal direction. The resulting pixel line was added to the previous ones in a single frame as presented in Figure 1f (panels). The measurement was done at least three times, with the average and standard deviation shown.
The direct measurement of the pressure scattered by the nanoparticles is delicate due to the low absolute pressure scattered, and the need to use the same transducer for the reception and the emission in a controlled environment with sufficient sensitivity. Theories indeed predict a high directivity of the signal scattered by solid particles, and the same could apply here. Therefore, the acoustic activity and echogenicity of the particles was further quantified using a method based on acoustic radiation force.[58] The signal backscattered by an US contrast agent and the radiation force acting on it have the same origin, and therefore a measure of the scattering coefficient can be achieved by measuring the particle displacement in a well-defined acoustic field. A piezoelectric single element transducer was used to generate a standing wave in a square glass capillary with homogeneously dispersed particles. The creation of an US standing wave induces migration toward the pressure node or antinode located in the middle and the sides of the channel, respectively. The motion of the particles was recorded with a high-speed camera and analyzed (Figure 1e). To do so, each frame was averaged in the direction of the channel to give a single pixel line in Matlab. The pixel line from the consecutive’s frames was added one after the other to create the time frame presented. The convergence observed in the time frame was then compared to the theoretical result of the force balance equation to extract the corresponding cross section.
Quantification of Scattering Cross Section
The forces acting on a random particle in an acoustic field include the drag force,[59] the added mass force that describes the acceleration of the surrounding fluid following the particle motion,[60] and the radiation force that drives the motion of the particle. The force balance equation can then be solved to determine the trajectory of the particle of any given size. In first approximation, the particles are assumed to obey the well-accepted theory describing the interaction of a rigid sphere with the acoustic field.[58] A field decomposition of the radiation force into spherical harmonics gives a frequency component f1 that only depends on the density mismatch and a frequency component f2 that depends on the relative compressibility. The radiation force, Fr, experienced by the particle in a standing wave is then given by
| (1) |
where ρ0 is the density of the fluid, PA is the driving acoustic pressure, Vp the volume of the particle, c0 the speed of sound in the fluid, and k the wave vector
| (2) |
Using the no-slip boundary condition at the wall of a solid sphere and Newton’s law comes
| (3) |
where the relative size of the particle with respect to the viscous boundary layer thickness and with v the dynamic viscosity of water, is the density ratio, and fus the frequency of the applied US wave.
Solving the wave equation on a spherical harmonic base leads to the following expression for the far-field backscattered scalar potential in a spherical referential having its origin at the center of the particle and the space variables r and θ
| (4) |
In a traveling ultrasound wave of the same frequency, the potential becomes
| (5) |
For a 100 nm particle in water in a 2 MHz frequency US wave,
| (6) |
For a solid polymer nanoparticle, the imaginary part of the coefficient f2 can then be neglected as compared to the real part. The backscattered pressure, ps, then relates to the radiation force in the direction of the transducer by
| (7) |
From this expression, the scattering cross section in intensity can be defined as the total scattered power over the incoming intensity
| (8) |
For these calculations, these following parameters were assumed:
cPFCE = 652 m s−1
cPFOB = 658 m s−1
cPFO = 598 m s−1 (from our pulse-echo data).
The specific gravity of the PFCE is stated by the manufacturer as 1.78.
Attenuation Measurements
For the attenuation measurement, a Tabor WW1281A arbitrary waveform generator was used to generate the acoustic signal, subsequently amplified by a Vectawave VBA100-200 amplifier, and sent to a Panametrics A308S transducer (5 MHz center frequency, 1.96 in. focal distance). The acoustic wave was allowed to propagate through a chamber built in-house (18 mm acoustical path length), sealed with two acoustically transparent membranes. The transducer was calibrated in emission using an optical hydrophone (Precision Acoustics, UK) in order to send a constant pressure across the whole frequency range investigated. The transmitted wave was then measured by a panametric A305S transducer (2.25 MHz center frequency, 1.88 in. focal length). 20 cycle waveforms were sent for each set of pressures and frequencies, and the measurement was repeated 200 times for each parameter set.
Physicochemical Characterization of Nanoparticles
DLS experiments were performed on a Malvern Zetasizer ZS Nano at a scattering angle θ = 173° using disposable cuvettes. The particle concentration was 0.01 mg mL−1. The typical hydrodynamic radius obtained at θ = 173° was around 100 nm (average over more than ten different batches).
Multiangle DLS and Static LS experiments were performed on ALV compact goniometer system equipped with ALV7004 correlator, ALV/LSE-5004 Goniometer, ALV/Dual High QE APD detector unit with a fiber splitting device setup of two off detection system, and a Uniphase Model 1145P He-Ne Laser. The laser wavelength and power were 632.8 nm and 22 mW, respectively. The temperature was controlled by a Julabo CF41 thermostatic bath. Water for dilutions was filtered with 0.45 μm hydrophilic filters. The concentration of samples was 0.01 mg mL−1
DLS data were collected at scattering angles θ = 30°–150° in 10° steps. Data analysis was done with HDRC software, which was kindly provided by Prof. Manfred Schmidt, University of Mainz, Germany. The apparent hydrodynamic radii at different angles were obtained either from the cumulant or the biexponential fitting of the autocorrelation function. Both provided a good approximation of the radius for particles with polydisperse, monomodal size distribution. The absolute inverse z-averages of hydrodynamic radii were obtained by extrapolating the apparent diffusion coefficients q→0.[61,62] The second cumulant values μ2 were obtained from cumulant fitting at θ = 90°.
Static light scattering (SLS) for determination of the radius of gyration (Rg) was measured at θ = 30°–60° in 3° steps. This measurement range was selected, as it is needed for Guinier analysis (q × Rg ≤ 1). The radius of gyration was determined from the angular dependence of the Rayleigh ratio according to the Guinier equation.[63] The Rayleigh ratio Rθ was obtained as follows
| (9) |
with a refractive index of solvent nwater = 1.333, refractive index of reference ntoluene = 1.494, and Rayleigh ratio of toluene Rtoluene = 1.02 × 10−3 m−1 calculated as described by Wu.[64]
Nanoparticle density was measured with an oscillating U tube using an Anton Paar DMA 5000. Aqueous solutions of nanoparticles with weight fractions of nanoparticles w = 0.01, 0.008, 0.005, 0.003, and 0.001 were used, and the density of solid particles was calculated from a linear extrapolation using
| (10) |
Typical density of PFCE and PFOB nanoparticles ranges between 1.4 and 1.5 mg cm−3.
SEM was done on an environmental scanning electron microscope FEI ESEM XL30 at 20 kV and magnifications up to 60 000×. The sample was fixed on a glass plate by evaporation of the suspension fluid. A layer of gold was then deposited on the sample before observation by SEM was done three times on different batches, and representative images are shown.
Cryo-SEM was done at JEOL 6330 Cryo Field Emission Scanning Electron Microscope (FESEM). For cryo-SEM analysis, the samples (8 μL, 10 mg mL−1) were pipetted in two rivets, which were then placed together. Next, the samples were frozen in liquid nitrogen slush and placed in an Oxford Alto 2500 cryo station with a cryo-transfer device. There the top rivet was broken, and the sample was heated to −95 °C for 5 min, followed by a coating of 60/40 Au/Pd and transfer to the cryo-SEM.
Nuclear magnetic resonance spectroscopy was measured on Brucker Avance III 400 MHz spectrometer equipped with BBFO probe at 298 K. Particles (typically 5–8 mg) were dissolved in 500 μL D2O and mixed with 1 μL TFA in 100 μL D2O.
2D HOESY was performed using a spectral width spanning from −110 to −60 ppm for 19F (377 MHz) and −0.5 to 10.5 ppm for 1H (400 MHz) using 2048 × 256 points. 16 scans per increment were acquired using an interscan relaxation delay of 2.0 s and a mixing time for dipole–dipole relaxation of 400 ms.
Quantitative 19F NMR was measured with an interscan relaxation delay of 20 s. The number of scans was between 8 and 32 depending on concentration of fluorinated compound.
Data evaluation was done with MestreNova 10.0 from Mestrelab. Internal projections of HOESY spectra were extracted using TopSpin 3.5 from Bruker.
19F solid-state NMR spectra and relaxation times were measured on a Varian VNMRS 850 MHz spectrometer. A 4 mm HXY probe was used, which was resonant for 19F at a frequency of 799.75 MHz; the samples were spun at an magic angle spinning (MAS) frequency of 10 kHz. A spin echo sequence was used for measuring the T2 relaxation time and inversion recovery experiments for T1. For measuring the particles in dry state, freeze-dried nanoparticle powder was used. To obtain nanoparticles that are swollen with water, freeze-dried powder was incubated with excess of water for 5 min and then centrifuged at 21 000 g. The pellet from centrifugation was then immediately filled into a rotor for solid-state NMR measurements. Data evaluation was performed with MatNMR.
SANS experiments were performed at the D11 small angle diffractometer at the ILL in Grenoble (France). The description of settings of D11 diffractometer is given in detail in the Supporting Information. SANS was also carried out on the SANS2D small-angle diffractometer at the ISIS Pulsed Neutron Source (STFC Rutherford Appleton Laboratory, Didcot, UK; http://www.isis.stfc.ac.uk).[65] A collimation length of 12 m and incident wavelength range of 1.75–12.5 Å were employed. Data were measured simultaneously on two 1 m2 detectors to give a q-range of 0.0015–0.85 Å−1. The small-angle detector was positioned 12 m from the sample and offset vertically 80 mm and sideways 100 mm. The wide-angle detector was position 5 m from the sample, offset sideways by 860 mm and rotated to face the sample. The magnitude of the scattering wave vector q is defined as
| (11) |
where θ is the scattered angle and λ is the incident neutron wavelength. The beam diameter was 8 mm. Each raw scattering data set was corrected for the detector efficiencies, sample transmission, and background scattering and converted to scattering cross-section data (∂Σ/∂Ω vs q) using the instrument-specific software (http://www.mantidproject.org). These data were placed on an absolute scale (cm−1) using the scattering from a standard sample (a solid blend of hydrogenous and perdeuterated polystyrene) in accordance with established procedures.[66]
Samples in D2O and H2O/D2O 36/64 (v:v) were measured in 2 mm quartz cuvettes, while for samples in H2O/D2O 61/39 (v:v) 1 mm cuvettes were used. In total, five different batches of nanoparticles with and without gadoteridol and two different batches were measured. Measurement data that were not included in this manuscript is available from the authors.
Data analysis was done using the NIST SANS[67] Macro for Igor Pro (Wavemetrics) or SasView 4.1.0.[68]
Imaging Techniques and Cell Labeling: In Vitro Imaging
In vitro US imaging was carried out on samples in a gel phantom or injected ex vivo in bovine liver tissue. A linear array transducer (L11-3) with center frequency 7 MHz was used for all the US scans (SONOS 7500, Philips Medical Systems, Best, The Netherlands). The MI was variable, from 0.1 to 1.2, as stated in the text. Gain was set to 90%. Gel phantoms consisted of 8% gelatin (Dr. Oetker, Ede, The Netherlands) and 2% agar (Agar Powder CMN, Boom, Meppel, The Netherlands) by weight. Analyses on the contrast (Figures 1b,c and 2c) were carried out by drawing a region of interest over the relevant area and measuring average pixel intensity using Image J (U.S. National Institutes of Health, Bethesda, MD).[69] The average intensity is shown (n = 3). Cells were labeled with the particles in at least three experiments (with different batches of particles and cells from different donors).
For Figure 2a, the particles were exposed to 60 s of US at an MI of 1.2 (8 MHz, focus at 0.5 cm; n = 3) before the measurement size parameters through DLS. In Figure 2b, the particles were exposed to probe sonication for 30 s at 40% power, as during synthesis (n = 3). SEM was performed before and after probe sonication. Injections of particles in tissue phantoms were done (n = 5) with different batches of particles and in different tissue phantoms. 1 mg of particles contains about 4 × 108 particles mg−1; Sonovue(c) contains about 2 × 108 bubbles mL−1 when reconstituted as per directions. For Figure 2c, 0.5 mg of particles were injected in a tissue phantom. Images were acquired immediately after injection at low (0.2) and high (1.2) mechanical index (0 h), and the transducer was left on and in place at MI = 1.2 for 80 min (80 min, continuous). After 80 min, the phantoms were imaged again at both MI. The phantoms were then placed in a freezer for 48 h and thawed at room temperature for 1 h, before further imaging (48 h).
MR imaging (Figure 6e) was performed on an 11.7 T MR system (Bruker Biospin, Ettlingen, Germany), equipped with a horizontal bore magnet, using a dual 1H/19F volume coil. Image settings were TR/TE of 800/10.5 ms, 2 × 2 × 2 mm voxels, 256 × 128 matrix, and 2 averages for 1H using a spin echo sequence; 960/46 ms, 4 × 4 × 4 mm voxels, 64 × 32 matrix, 512 averages using a RARE sequence with RARE factor 8. For fluorescence imaging, samples were placed in a FluorVivo 300 (INDEC BioSystems, Santa Clara, CA). Exposure times were between 0.05 and 0.15 s (Figure 6d). Imaging of labeled cells was done with three different batches of particles and cells from different donors.
Cell Isolation and Labeling
Primary human DCs were isolated from donor blood as previously described[55] and labeled with 5 mg of particles per million cells from days 3 to 8 of the in vitro culture period (n = 3). Cells were washed three times before use. Viable cells were counted using a cell counter with trypan blue exclusion.
Primary murine DCs were generated from bone marrow cells as previously described.[70] Cells were incubated with 1 mg of particles per million cells mL−1 overnight for labeling (n = 3); 1 μg mL−1 LPS was used to obtain mature DCs.
In Vivo Imaging
Mice were housed under specified pathogen-free conditions in the Central Animal Laboratory (Nijmegen, The Netherlands). All experiments were performed according to the guidelines for animal care of the Nijmegen Animal Experiments Committee. Untreated C57/BL/6J female mice (Harlan) at ten weeks of age were used for these experiments; 12 mice were used for imaging, with at least three mice per condition, with the experiment repeated three times. All mice were identical before experimentation, and thus no further randomization procedure was used.
High-resolution in vivo US imaging was carried out using a VisualSonics Vevo 2100 system (MS-700 probe, central frequency 50 MHz, 100% power, gain set to 37 dB). Mice were anesthetized using Isoflurane. Intranodal injections (Figure 6b) were performed with a NanoFil Microliter syringe (World Precision Instruments, Germany) under microscopic guidance (n = 3). For the cell tracking experiments (Figure 6c–e), 5 × 106 cells were injected in the footpad of a mouse, and the mouse was imaged 24 h later (popliteal and inguinal lymph nodes) with high-resolution in vivo US (n = 6) The nodes were also removed and imaged ex vivo in a gel at a high (MS-700 probe, VisualSonics Vevo 2100) and low frequency (7 MHz, Philips SONOS 7500; n = 2). MRI was carried out on a horizontal bore 7T system (Bruker, Clinscan) using a dual-tuned volume coil. Image settings were TR/TE of 1200/15 ms, 0.1 × 0.1 × 2 mm voxels, 512 × 512 matrix, and 2 averages for 1H using a TSE sequence with an acquisition time of 2.57 min; 100/2.8 ms, 1.6 × 0.8 × 4 mm voxels, 64 × 32 matrix, 256 averages using a GRE sequence with an acquisition time of 13.42 min (n = 2). Fluorescence imaging was done in a FluorVivo system, as with the ex vivo tissue samples (n = 6).
Supplementary Material
Supporting Information is available from the Wiley Online Library or from the author.
Acknowledgements
O.K. and G.L. contributed equally to this work. F.B.B., E.S., C.L.d.K., C.F., and I.J.M.d.V. contributed equally to this work. The authors would like to thank Jan van Hest, Daniela Wilson, Maaike van Hout-Kuijer, Tim Idzenga, Martin Kreutz, Anne Leferink, Tim Segers, Andor Veltien, Eric de Ronde, Paul Tinnemans, Neus Vilanova Garcia, Geert-Jan Janssen, Hans Janssen, Gerrit Janssen, Manfred Schmidt, Katharina Landfester, Andreas Riedinger, SasView developer team, particularly Miguel Gonzalez, Paul Butler, and Steve King, David Hess and PSCM labs for ALV-LS access at ILL; Philippe Trochet and Goran Rodsjo from VisualSonics Inc. M.S. was funded by a Netherlands Organization for Scientific Research (NWO) VENI 700.10.409 and an European Research Council (ERC) grant ERC-2014-StG-336454-CoNQUeST; A.H. by NWO investment Grants 91106021 and BIG (VISTA); C.F. by ERC-2010-AdG-269019-PATHFINDER and an NWO Spinoza award; J. M.d.V. by the Netherlands Institute of Regenerative Medicine (NIRM, FES0908) and the EU FP7 program ENCITE (HEALTH-F5-2008-201842). L.J.C. was supported by an NWO VIDI personal grant (Project No. 723.012.110). O.K. acknowledges Max Planck-University of Twente Center for Complex Fluid Dynamics and Erasmus+ staff travel grant. Part of this work was supported by NanoNextNL, a micro and nanotechnology consortium of the Government of The Netherlands and 130 partners. I.K.V. acknowledges the NWO ECHO-STIP Grant 717.013.005 and NWO VIDI Grant 723.014.006 for financial support. Support of NWO for the “Solid-State NMR facility for Advanced Materials Science” is gratefully acknowledged. SANS was supported by the Institut Laue-Langevin (ILL) [https://doi.org/10.5291/ILL-DATA.9-10-1478]. Experiments at the ISIS Pulsed Neutron and Muon Source were supported by a beamtime allocation from the Science and Technology Facilities Council. This work benefited from the use of the SasView application, originally developed under NSF Award DMR-0520547. SasView also contains code developed with funding from the EU Horizon 2020 programme under the SINE2020 project Grant No. 654000. The authors thankfully acknowledge the experimental help of Tim Segers in measuring the acoustic radiation force in a square capillary. All data are available from the corresponding author on a reasonable request.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Dr Olga Koshkina, Department of Tumor Immunology, Radboud Institute for Molecular Life Sciences (RIMLS), Geert Grooteplein Zuid 28, 6525 GA, Nijmegen, The Netherlands; Physical Chemistry of Polymers, Max Planck Institute for Polymer Research, Ackermannweg 10, 55128 Mainz, Germany.
Dr Guillaume Lajoinie, Physics of Fluids Group, Technical Medical (TechMed) Centre and MESA+ Institute for, Nanotechnology, University of Twente, Drienerlolaan 5, 7522 NB, Enschede, The Netherlands.
Prof Francesca Baldelli Bombelli, Laboratory of Supramolecular and BioNano Materials, (SupraBioNanoLab), Department of Chemistry, Materials, and Chemical Engineering, “Giulio Natta,”, Politecnico di Milano, Via Luigi Mancinelli 7, 20131 Milan, Italy.
Edyta Swider, Department of Tumor Immunology, Radboud Institute for Molecular Life Sciences (RIMLS), Geert Grooteplein Zuid 28, 6525 GA, Nijmegen, The Netherlands.
Dr Luis J. Cruz, Translational Nanobiomaterials and Imaging, Department of Radiology, Leiden University Medical Centre, Albinusdreef 2, 2333 ZA, Leiden, The Netherlands.
Dr Paul B. White, Institute for Molecules and Materials, Radboud University, Heyendaalseweg 135, 6525 AJ, Nijmegen, The Netherlands.
Dr Ralf Schweins, Institut Laue – Langevin, DS/LSS, 71 Avenue des Martyrs, CS 20 156, 38042 Grenoble CEDEX 9, France.
Dr Sarah E. Rogers, ISIS Pulsed Neutron and Muon Source, Science and Technology Facilities Council, Rutherford Appleton Laboratory, Harwell, Oxford OX11 0QX, UK.
Remco Fokkink, Department of Agrotechnology and Food Sciences, Physical Chemistry and Soft Matter, Wageningen University, 6708 WE, Wageningen, Netherlands.
Prof Ilja K. Voets, Laboratory of Self-Organizing Soft Matter, Laboratory of Macromolecular and Organic Chemistry, Department of Chemical Engineering and Chemistry and Institute for Complex Molecular Systems, Eindhoven University of Technology, De Rondom 70, 5612 AP, Eindhoven, The Netherlands.
Dr Ernst R. H. van Eck, Institute for Molecules and Materials, Radboud University, Heyendaalseweg 135, 6525 AJ, Nijmegen, The Netherlands.
Prof Arend Heerschap, Department of Radiology and Nuclear Medicine, Radboudumc, Geert Grooteplein Zuid 10, 6525 GA, Nijmegen, The Netherlands.
Prof Michel Versluis, Physics of Fluids Group, Technical Medical (TechMed) Centre and MESA+ Institute for, Nanotechnology, University of Twente, Drienerlolaan 5, 7522 NB, Enschede, The Netherlands.
Prof Chris L. de Korte, Physics of Fluids Group, Technical Medical (TechMed) Centre and MESA+ Institute for, Nanotechnology, University of Twente, Drienerlolaan 5, 7522 NB, Enschede, The Netherlands; Department of Radiology and Nuclear Medicine, Radboudumc, Geert Grooteplein Zuid 10, 6525 GA, Nijmegen, The Netherlands.
References
- [1].Srinivas M, Aarntzen EHJG, Bulte JWM, Oyen WJ, Heerschap A, de Vries IJM, Figdor CG. Adv Drug Delivery Rev. 2010;62:1080. doi: 10.1016/j.addr.2010.08.009. [DOI] [PubMed] [Google Scholar]
- [2].Qin S, Caskey CF, Ferrara KW. Phys Med Biol. 2009;54:R27. doi: 10.1088/0031-9155/54/6/R01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lee HJ, Yoon T-J, Yoon YI. Ultrasonography. 2017;36:378. doi: 10.14366/usg.17014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mehta KS, Lee JJ, Taha AA, Avgerinos E, Chaer RA. J Vasc Surg. 2017;66:266. doi: 10.1016/j.jvs.2016.12.133. [DOI] [PubMed] [Google Scholar]
- [5].Huang C, Zhang H, Bai R. Acta Pharm Sin B. 2017;7:447. doi: 10.1016/j.apsb.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Figueiredo M, Esenaliev R. J Drug Delivery. 2012;2012:1. doi: 10.1155/2012/767839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Rapoport NY, Kennedy AM, Shea JE, Scaife CL, Nam K-H. J Controlled Release. 2009;138:268. doi: 10.1016/j.jconrel.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kang ST, Yeh CK. Langmuir. 2011;27 doi: 10.1021/la203212p. 13183. [DOI] [PubMed] [Google Scholar]
- [9].Reznik N, Williams R, Burns PN. Ultrasound Med Biol. 2011;37:1271. doi: 10.1016/j.ultrasmedbio.2011.05.001. [DOI] [PubMed] [Google Scholar]
- [10].Shpak O, Verweij M, Vos HJ, de Jong N, Lohse D, Versluis M. Proc Natl Acad Sci USA. 2014;111:1697. doi: 10.1073/pnas.1312171111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Shpak O, Stricker L, Versluis M, Lohse D. Phys Med Biol. 2013;58:2523. doi: 10.1088/0031-9155/58/8/2523. [DOI] [PubMed] [Google Scholar]
- [12].Ishijima A, Tanaka J, Azuma T, Minamihata K, Yamaguchi S, Kobayashi E, Nagamune T, Sakuma I. Ultrasonics. 2016;69:97. doi: 10.1016/j.ultras.2016.04.002. [DOI] [PubMed] [Google Scholar]
- [13].Yu H, Xu L. J Controlled Release. 2014;174:151. doi: 10.1016/j.jconrel.2013.11.010. [DOI] [PubMed] [Google Scholar]
- [14].Liu W-W, Liu S-W, Liou Y-R, Wu Y-H, Yang Y-C, Wang C-RC, Li P-C. Sci Rep. 2016;6 doi: 10.1038/srep24753. 24753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Fix SM, Novell A, Yun Y, Dayton PA, Arena CB. J Ther Ultrasound. 2017;5:7. doi: 10.1186/s40349-017-0085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jafari S, Diou O, Mamou J, Renault G, Fattal E, Tsapis N, Bridal SL. IEEE Trans Ultrason, Ferroelectr, Freq Control. 2014;61:5. doi: 10.1109/TUFFC.2014.6689771. [DOI] [PubMed] [Google Scholar]
- [17].Reznik N, Shpak O, Gelderblom EC, Williams R, de Jong N, Versluis M, Burns PN. Ultrasonics. 2013;53:1368. doi: 10.1016/j.ultras.2013.04.005. [DOI] [PubMed] [Google Scholar]
- [18].Rui EMSM, Kolios MC, Gorelikov I, Matsuura N. IEEE Int Ultrason Symp Proc; IEEE; 2010. pp. 495–498. [Google Scholar]
- [19].Scheible FW, Gosink BB, Leopold GR, Long DM, Higgins CB. Radiology. 1982;145:759. doi: 10.1148/radiology.145.3.7146409. [DOI] [PubMed] [Google Scholar]
- [20].Díaz-López R, Tsapis N, Fattal E. Pharm Res. 2010;27:1. doi: 10.1007/s11095-009-0001-5. [DOI] [PubMed] [Google Scholar]
- [21].Srinivas M, Boehm-Sturm P, Figdor CG, de Vries IJ, Hoehn M. Biomaterials. 2012;33:8830. doi: 10.1016/j.biomaterials.2012.08.048. [DOI] [PubMed] [Google Scholar]
- [22].Nakamura T, Sugihara F, Matsushita H, Yoshioka Y, Mizukami S, Kikuchi K. Chem Sci. 2015;6:1986. doi: 10.1039/c4sc03549f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lee AL, Gee CT, Weegman BP, Einstein SA, Juelfs AR, Ring HL, Hurley KR, Egger SM, Swindlehurst G, Garwood M, Pomerantz WCK, et al. ACS Nano. 2017;11:5623. doi: 10.1021/acsnano.7b01006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Shapiro MG, Goodwill PW, Neogy A, Yin M, Foster FS, Schaffer DV, Conolly SM. Nat Nanotechnol. 2014;9:311. doi: 10.1038/nnano.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Maresca D, Lakshmanan A, Lee-Gosselin A, Melis JM, Ni Y-L, Bourdeau W, Kochmann DM, Shapiro MG. Appl Phys Lett. 2017;110 doi: 10.1063/1.4976105. 073704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bourdeau RW, Lee-Gosselin A, Lakshmanan A, Farhadi A, Kumar R, Nety SP, Shapiro MG. Nature. 2018;553:86. doi: 10.1038/nature25021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chen F, Ma M, Wang J, Wang F, Chern S-X, Zhao ER, Jhunjhunwala A, Darmadi S, Chen H, Jokerst JV. Nanoscale. 2017;9:402. doi: 10.1039/c6nr08177k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kempen PJ, Greasley S, Parker KA, Campbell JC, Chang H-Y, Jones JR, Sinclair R, Gambhir SS, Jokerst JV. Theranostics. 2015;5:631. doi: 10.7150/thno.11389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Jokerst JV, Khademi C, Gambhir SS. Sci Transl Med. 2013;5:177ra35. doi: 10.1126/scitranslmed.3005228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Figdor CG, Vries DIJM, Srinivas M, Cruz RLJ. 2014.
- [31].Srinivas M, Cruz LJ, Bonetto F, Heerschap A, Figdor CG, de Vries IJM. Biomaterials. 2010;31:7070. doi: 10.1016/j.biomaterials.2010.05.069. [DOI] [PubMed] [Google Scholar]
- [32].Srinivas M, Tel J, Schreibelt G, Bonetto F, Cruz L-J, Amiri H, Heerschap A, Figdor CG, de Vries IJM. Nanomedicine. 2015;10:2339. doi: 10.2217/NNM.15.76. [DOI] [PubMed] [Google Scholar]
- [33].Swider E, Staal AHJ, van Riessen NK, Jacobs L, White PB, Fokkink R, Janssen G-J, van Dinther E, Figdor CG, de Vries IJM, Koshkina O, et al. RSC Adv. 2018;8:6460. doi: 10.1039/c7ra13062g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Srinivas M, Morel PA, Ernst LA, Laidlaw DH, Ahrens ET. Magn Reson Med. 2007;58:725. doi: 10.1002/mrm.21352. [DOI] [PubMed] [Google Scholar]
- [35].Kislukhin AA, Xu H, Adams SA, Narsinh KH, Tsien RY, Ahrens ET. Nat Mater. 2016;15:662. doi: 10.1038/nmat4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Srinivas M, Heerschap A, Ahrens ET, Figdor CG, de Vries IJM. Trends Biotechnol. 2010;28:363. doi: 10.1016/j.tibtech.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Overvelde M, Garbin V, Sijl J, Dollet B, de Jong N, Lohse D, Versluis M. Ultrasound Med Biol. 2010;36:2080. doi: 10.1016/j.ultrasmedbio.2010.08.015. [DOI] [PubMed] [Google Scholar]
- [38].Bonacucina G, Perinelli DR, Cespi M, Casettari L, Cossi R, Blasi P, Palmieri GF. Int J Pharm. 2016;503:174. doi: 10.1016/j.ijpharm.2016.03.009. [DOI] [PubMed] [Google Scholar]
- [39].Bruus H, Dual J, Hawkes J, Hill M, Laurell T, Nilsson J, Radel S, Sadhal S, Wiklund M. Lab Chip. 2011;11:3579. doi: 10.1039/c1lc90058g. [DOI] [PubMed] [Google Scholar]
- [40].Leighton TG. The Acoustic Bubble. Academic; San Diego, CA: 1996. [Google Scholar]
- [41].Pisani E, Tsapis N, Paris J, Nicolas V, Cattel L, Fattal E. Langmuir. 2006;22:4397. doi: 10.1021/la0601455. [DOI] [PubMed] [Google Scholar]
- [42].Solans C, Izquierdo P, Nolla J, Azemar N, Garcia-Celma MJ. Curr Opin Colloid Interface Sci. 2005;10:102. [Google Scholar]
- [43].Pisani E, Tsapis N, Galaz B, Santin M, Berti R, Taulier N, Kurtisovski E, Lucidarme O, Ourevitch M, Doan BT, Beloeil JC, et al. Adv Funct Mater. 2008;18:2963. [Google Scholar]
- [44].Rübe A, Hause G, Mäder K, Kohlbrecher J. J Controlled Release. 2005;107:244. doi: 10.1016/j.jconrel.2005.06.005. [DOI] [PubMed] [Google Scholar]
- [45].Teixeira J. J Appl Crystallogr. 1988;21:781. [Google Scholar]
- [46].Kujawa P, Liu RCW, Winnik FM. J Phys Chem B. 2002;106:5578. [Google Scholar]
- [47].Kujawa P, Goh CCE, Calvet D, Winnik FM. Macromolecules. 2001;34:6387. [Google Scholar]
- [48].Brodbelt J, Maleknia S, Liou CC, Lagow R. J Am Chem Soc. 1991;113:5913. [Google Scholar]
- [49].Lin T-Y, Lin W-H, Clark WD, Lagow RJ, Larson SB, Simonsen SH, Lynch VM, Brodbelt JS, Maleknia SD, Liou C-C. J Am Chem Soc. 1994;116:5172. [Google Scholar]
- [50].Lai C-Z, Reardon ME, Boswell PG, Bühlmann P. J Fluorine Chem. 2010;131:42. doi: 10.1016/j.jfluchem.2009.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Greis C. Eur Radiol. 2004;14:P11. [PubMed] [Google Scholar]
- [52].Reznik N, Lajoinie G, Shpak O, Gelderblom EC, Williams R, de Jong N, Versluis M, Burns PN. Ultrasound Med Biol. 2014;40:1379. doi: 10.1016/j.ultrasmedbio.2013.11.025. [DOI] [PubMed] [Google Scholar]
- [53].Chen M, Cai F, Wang C, Wang Z, Meng L, Li F, Zhang P, Liu X, Zheng H. Adv Sci. 2017;4 doi: 10.1002/advs.201600447. 1600447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Aarntzen EHJG, Srinivas M, De Wilt JHW, Jacobs JFM, Lesterhuis WJ, Windhorst AD, Troost EG, Bonenkamp JJ, van Rossum MM, Blokx WAM, Mus RD et al. Proc Natl Acad Sci USA. 2011;108 doi: 10.1073/pnas.1113045108. 18396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Bonetto F, Srinivas M, Weigelin B, Cruz LJ, Heerschap A, Friedl P, Figdor CG, de Vries IJM. NMR Biomed. 2012;25:1095. doi: 10.1002/nbm.2774. [DOI] [PubMed] [Google Scholar]
- [56].Aarntzen EHJG, Srinivas M, Bonetto F, Cruz LJ, Verdijk P, Schreibelt G, Van De Rakt M, Lesterhuis WJ, Van Riel M, Punt CJA, Adema GJ, et al. Clin Cancer Res. 2013;19:1525. doi: 10.1158/1078-0432.CCR-12-1879. [DOI] [PubMed] [Google Scholar]
- [57].Zeelen C, Paus C, Draper D, Heskamp S, Signore A, Galli F, Griessinger CM, Aarntzen EH. Q J Nucl Med Mol Imaging. 2018;62:56. doi: 10.23736/S1824-4785.17.03052-7. [DOI] [PubMed] [Google Scholar]
- [58].Settnes M, Bruus H. Phys Rev E. 2012;85 doi: 10.1103/PhysRevE.85.016327. 016327. [DOI] [PubMed] [Google Scholar]
- [59].Mettin R, Doinikov AA. Appl Acoust. 2009;70:1330. [Google Scholar]
- [60].Magnaudet J, Eames I. Annu Rev Fluid Mech. 2000;32:659. [Google Scholar]
- [61].Fischer K, Schmidt M. Biomaterials. 2016;98:79. doi: 10.1016/j.biomaterials.2016.05.003. [DOI] [PubMed] [Google Scholar]
- [62].Schärtl W. Light Scattering from Polymer Solutions and Nanoparticle Dispersions. Springer; Berlin: 2007. [Google Scholar]
- [63].Guinier A, Fournet G. Small Angle Scattering of X-Rays. Wiley; New York: 1955. [Google Scholar]
- [64].Wu H. Chem Phys. 2010;367:44. [Google Scholar]
- [65].Heenan RK, Rogers SE, Turner D, Terry AE, Treadgold J, King SM. Neutron News. 2011;22:19. [Google Scholar]
- [66].Wignall GD, Bates FS. J Appl Crystallogr. 1987;20:28. [Google Scholar]
- [67].Kline SR. J Appl Crystallogr. 2006;39:895. [Google Scholar]
- [68].Doucet M. SasView Version 4.1.2 [Google Scholar]
- [69].Schneider CA, Rasband WS, Eliceiri KW. Nat Methods. 2012;9:671. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Den Brok MH, Nierkens S, Wagenaars JA, Ruers TJ, Schrier CC, Rijke EO, Adema GJ. Vaccine. 2012;30:737. doi: 10.1016/j.vaccine.2011.11.080. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.