Abstract
Time-resolved HIV-1 envelope (Env) interactions with CD4 and CCR5 or CXCR4 were visualized and quantified on the surface of cells by combining multicolour super-resolution localization microscopy (dSTORM) with fluorescence fluctuation spectroscopy imaging. Utilizing primary isolate JR-FL and laboratory HXB2 strains, we revealed the time-resolved stoichiometry of both CD4 and CCR5 or CXCR4 receptors in the prefusion complex upon arrival of HIV-1 Env. The HIV-1 Env pre-fusion dynamics for both R5 and X4 tropic strains consists of a three-step mechanism, which differs in stoichiometry. The action of the monoclonal neutralizing antibody (Nab) b12 was also tested, revealing that the mechanism of inhibition differs between JR-FL and HXB2 Env. These molecular insights into the precise Env-induced time-resolved stoichiometry of CD4 and CCR5 or CXCR4 reveal HIV-1 receptor and co-receptor assemblies as novel targets for inhibitor design.
Introduction
Entry of HIV-1 into a host cell requires an initial interaction between the viral-envelope displayed glycoprotein spike complex, Env, with cell surface displayed CD4 and co-receptors1. Although structural studies have revealed the intra-molecular basis for CD4 receptor and CXCR4/CCR5 co-receptor-induced conformational changes to the HIV-1 Env during host cell entry2, little is known about how the inter-molecular dynamics and stoichiometry of this process culminates in fusion with the host cell membrane in live cells3. This is due to the difficulty of working with live cells and the lack of temporal resolution of the techniques commonly employed (i.e. crystallography and cryo-EM). To overcome these difficulties and enable real-time observations of R5 and X4 tropic HIV-1 Env-induced CD4 and CCR5 or CXCR4 interactions between single HIV-1 virions and live cells, we multiplexed number and brightness (N&B)4–6 with real-time single virus tracking (SVT)7,8(Fig. 1). To corroborate our NandB data, we additionally employed total internal reflection microscopy (TIRFM) combined with super-resolution localization microscopy (dSTORM)9.
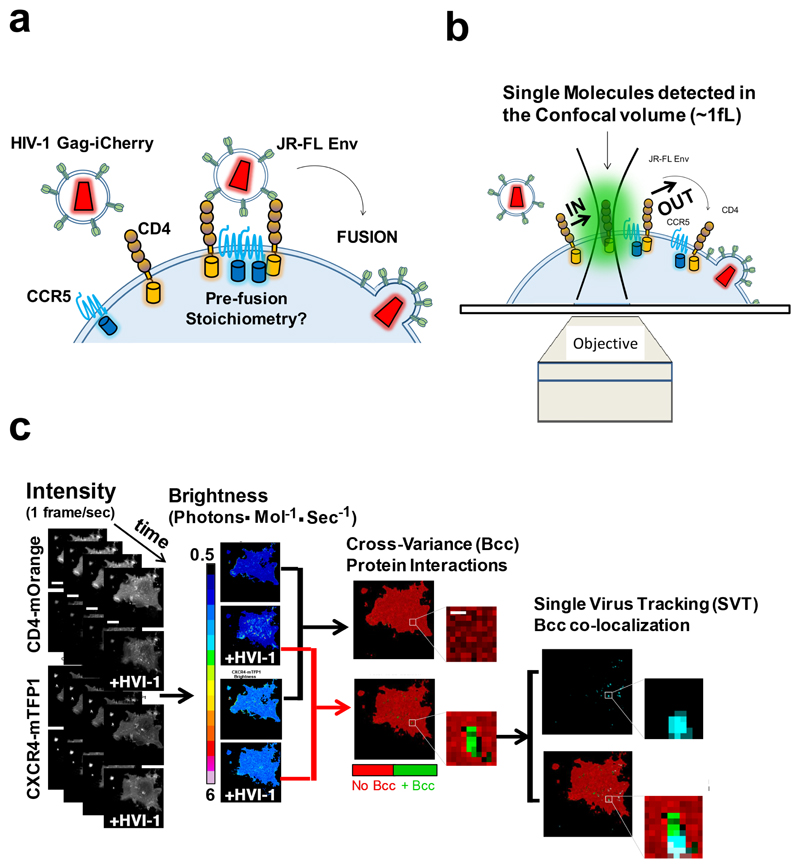
Figure 1. HIV-1 Env – CD4 and CCR5 or CXCR4 stoichiometry super-resolution imaging and fluorescence fluctuation spectroscopy in live cells.
a, The CD4 and CCR5 or CXCR4 receptors were labelled with mOrange and mTFP1. R5 and X4 tropic labelled HIV virions were produced using JR-FL and HXB2 Env and Gag-iCherry. The pre-fusion reaction of individual HIV-1 virions was assessed multiplexing real-time single virus tracking with two color number and brightness. b, Labelled CD4 receptors and co-receptors diffuse through a confocal volume generating fluorescence fluctuation traces that are informative of the oligomeric state of the labelled receptors as described in the NandB method. When cross-correlating both traces, the one coming from CD4-mOrange and the one coming from the co-receptor (e.g. CCR5-mTFP1) one can detect protein-protein interactions, as described in ccNandB theory. c, Flow diagram depicting the overall strategy from image acquisition until analysis. COS7 cells co-expressing CD4-mOrange and CCR5 or CXCR4-mTFP1 were exposed to HIVJR-FL/Gag-iCherry or HIVHXB2/GagiCherry virions and imaged using a confocal system equipped with two HyD detectors and one PMT (1, first column from the left). Number and Brightness analysis was performed on time-stacks of images for both CD4-mOrange Channel and CCR5-mTFP1 or CXCR4-mTFP1; in parallel single virus tracking (SVT) was performed on the third Gag-iCherry channel. Pixel-by-Pixel Brightness images containing information on the oligomeric state for CD4 and CXCR4 were produced using a software developed in house in R (2, second column from the left)). Finally, co-localization analysis was carried out between the x-y coordinates for the labelled virions (in this example HIVHXB2/GagiCherry) and the cross-variance (Bcc) (3, third column from the left) coming from CD4 and CXCR4 channels (4, fourth column from the left). The computation of Bcc was also carried out with a second R package developed in house.
Guided by the combined use of advanced quantitative light microscopy, we show with detail the mechanistic underpinnings of the inter-molecular dynamics of CD4 and co-Receptors during HIV-1 pre-fusion reaction for both: an X4 tropic HIV-1 Env lab strain (HXB210) and a R5 tropic HIV-1 primary isolate strain (JR-FL11). We also show how b12, a CD4-binding-neutralizing antibody, prevents exposure of the co-receptor binding by restricting V1/V2 loop recognition2, and blocks CD4 – CXCR4 interactions for HXB2 Env. JR-FL Env CD4 – CCR5 interactions were also disrupted at 100μm /mL but higher order oligomeric states of CD4 and co-receptors were not detected as opposed to HXB2 Env. Recent studies have stressed the importance of the role played by the host during HIV-1 entry and fusion12–15; the molecular insights presented here into the precise Env-induced time-resolved stoichiometry of CD4 and CCR5 or CXCR4 may contribute to the development of future therapeutic interventions directed to disrupt the earliest stages of the virus life cycle.
Results
HIV-1 X4 tropic virus interactions with CD4 and CXCR4: Fluorescence Fluctuation Spectroscopy
X4 tropic HIV-1 viruses carrying HXB2 Env and Gag-iCherry (HIVHXB2/GagiCherry)16 were exposed to COS7 cells co-transfected with CD4-mOrange and CXCR4-mTFP1 at 37 °C (Fig. 1). COS7 cells expressing CD4-mOrange and CXCR4-mTFP1 were imaged together with HIVHXB2/GagiCherry virions at a time resolution of one frame per second. Inspired by alternative laser excitation (ALEX)17, we implemented line interleaved excitation confocal microscopy; which also prevented bleed-through avoiding potential false positives in our cross-variance analysis. Time-resolved brightness and cross-variance brightness (Bcc)5 images were calculated from CXCR4-mTFP1 and CD4-mOrange intensity images, as described in (Fig. 1 and Supplementary Fig. 1).
Single particle tracking of HIVHXB2/GagiCherry performed in parallel with Bcc analysis using the red channel revealed that 12% virions induced positive Bcc regions of interest (i.e. CD4–CXCR4 interactions) when landing on the surface of COS7 cells (Fig. 1-2; Supplementary Fig. 1-4). By using the time-resolved coordinates of HIV-1 x-y virions that colocalized with positive cross-variance (Bcc) regions, we could recover the CD4–CXCR4 time-resolved stoichiometry for individual virions (Fig. 2e). The average time-course for the pre-fusion reaction for HXB2 Env (n = 10) shows that CXCR4 dimerization occurs at t = 1.7 mins, followed by CD4 hexamer formation at 3.4 min. This 6±1 CD4 – 2±0.3 CXCR4 configuration is transiently stable (3.4 – 5.1 mins) until an average 4.9±0.8 CD4 – 1.5±0.3 stoichiometry is adopted at t = 6.8 mins (Fig. 2e). Only 12% of HIVHXB2/GagiCherry virions induced positive Bcc (i.e. CD4 – CXCR4 interaction); similar percentages were found in other reports16,18. Time-resolved HIV-1 x-y virion coordinates that did not colocalize with positive cross-variance (Bcc) regions presented a reaction profile distinct from the ones that did (Supplementary Fig 5). Virions unable to induce positive Bcc regions were incapable of initiating the formation of a pre-fusion complex with CD4 and co-receptor and therefore unable to fuse with the host cell. We also produced HIVHXB2/GagiCherry virions in the presence of 320 nM of saquinavir19 (SQV), a protease inhibitor that binds to the active site of the viral protease, leading to immature viruses with uncleaved Gag-iCherry16. We reasoned that Env would be restricted and unable to cluster in these immature virions 20, so the second step of the HIVHXB2 pre-fusion reaction (i.e. CD4 hexamer formation) would not occur. None of the tracked HIVHXB2/GagiCherry immature virions (Supplementary Fig. 6) induced positive Bcc (i.e. CD4−CXCR4 interaction) or a significant increase in CD4 and CXCR4 oligomeric states (Supplementary Figure 6). This implies that HIVHXB2/GagiCherry virions that did induce positive Bcc (i.e. CD4 - CXCR4 interactions) were mature and potentially productive (Supplementary Figure 7). More negative controls using agents known to inhibit the HIV-1 fusion reaction were employed to further validate our approach. We employed b1221,22, CD4 antibodies like OKT423 or co-receptor ligands (CXCL1224) (Supplementary Fig.5-9) for HIV-1 virions and no positive Bcc was ever observed in all cases. The reaction profile for HIVHXB2/GagiCherry virions in the presence of 100 μl/mL of b12 is depicted in Fig.2e, rightpanel. In this case, the presence of b12 blocked the CD4 – CXCR4 interaction as no Bcc positive was observed in regions where HIVHXB2/GagiCherry virions were detected. Even if the absence of Bcc is indicative that the neutralizing antibody was functional, we did recover the oligomeric states of CD4 and co-recceptors in these locations (Fig. 2e, right panel). Strikingly, CD4 was able to oligomerize up to a tetramer, while CXCR4 formed a (the expected) dimeric state.
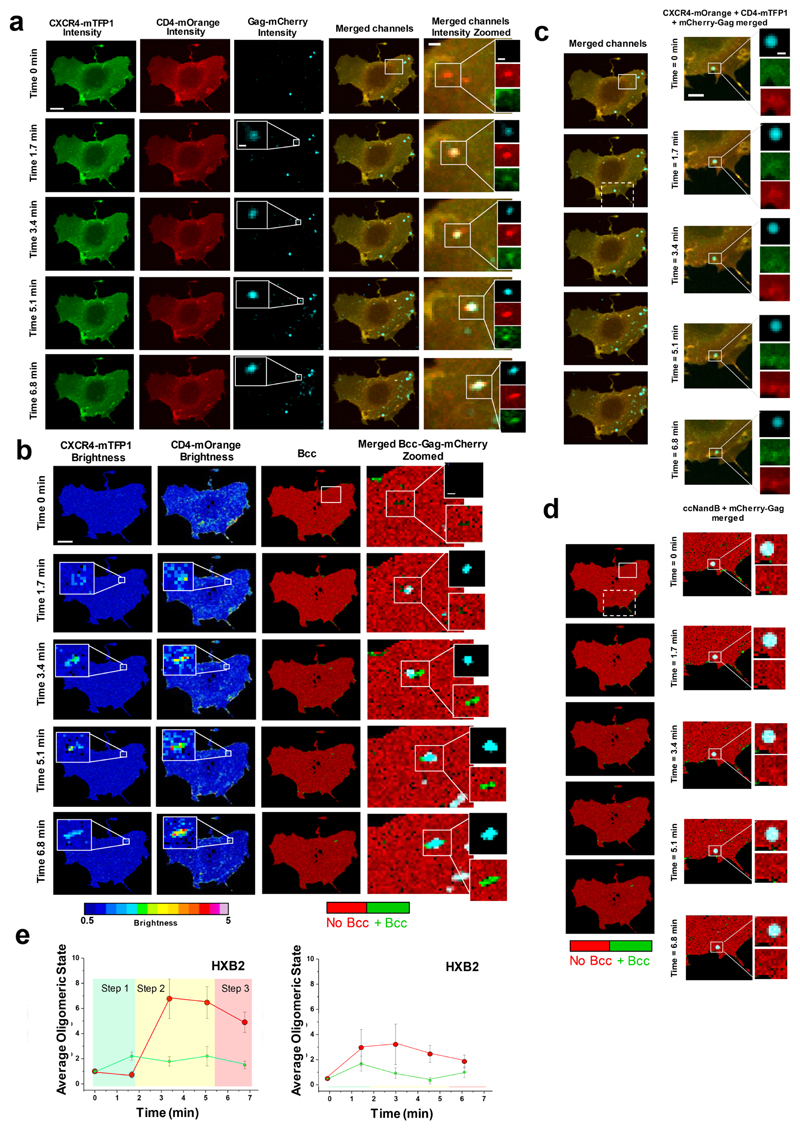
Figure 2. Visualization of HXB2 based HIV-1 virion receptor stoichiometry in live cells.
a, Fast time-resolved, three-color imaging was performed on a COS7 cell co-expressing CXCR4-mTFP1 (green micrographs) and CD4-mOrange (red micrographs) exposed to HIVHXB2 –Gag-iCherry virions (cyan micrographs), scale bar 1 μm. b, Time-resolved brightness analysis was performed for both CXCR4-mTFP1 (first column from the left) and CD4-mOrange (second column from the left). Time-resolved cross-variance (Bcc) analysis was also performed (third column from the left) rendering a small region of interest in which CD4-CXCR4 interactions occurred (green). Real-time single virus tracking was performed in parallel and the micrographs show co-localization of HIVHXB2 –Gag-iCherry (cyan pixels) and positive Bcc (green pixels). c, A second region of interest (dashed square) shows a HIVHXB2-Gag-iCherry virus that did not induce positive Bcc (d). These viruses most likely were immature and constitute robust built-in controls for our N&B and Bcc analysis. e, Time-resolved stoichiometry for CD4 (red dots) and CXCR4 (green dots) upon addition of HIVHXB2/GagiCherry (left panel). The circles indicate the average (n = 10) and the error bars indicate the standard error for each time point. The time-resolved homotypic interactions for CD4 (red dots) and CXCR4 (green dots) upon addition of HIVHXB2/GagiCherry in the presence of inhibitory concentrations of b12. CD4 and CXCR4 did not interact in this case. Eror bars indicate standard error for each time point wich is the average for n = 12.
HIV-1 X4 tropic virus interactions with CD4 and CXCR4: Super-resolution localization microscopy
Multicolor TIRF-dSTORM25 microscopy was also performed on COS7 cells co-expressing CD4-mRFP and CXCR4-eYFP exposed to HIVHXB2 during 10 min (Fig. 3). After this time, cells and viruses were fixed and treated with nanoboosters (anti-RFP and anti-eYFP nanobodies labelled with Atto dyes) and Env antibodies for HIV specifically designed for super-resolution localization microscopy (see Material and Methods). The pre-fusion reaction of individual HIV-1 virions was assessed by the interaction factor (IF) method26. As described in material and methods, both the average normalized sum of photons per interaction event (Supplementary Fig.9) and the real stoichiometry (normalized sum of photons per interaction event, Fig. 4c) were obtained. In the case of HIVHXB2, we found that the multi-colour dSTORM approach supported the three-step mechanism observed with Bcc (Fig. 3). For HXB2 Env (n = 103), the first step is also an asymmetric pre-hairpin intermediate2 consisting on 1-2 CD4 and 1 CXCR4 molecules; we found 69% of the events to correspond to this step with 1.6+/-0.03 CD4 : 0.7+/-0.02 CXCR4. Similar to the data acquired via N and B, step two followed right after step one and implies the formation of a CD4 hexamer and CXCR4 dimerization. Only 17% of the quantified events were found to belong to this step with 6.5+/0.2 CD4 : 2.1 +/-0.42 CXCR4 stoichiometry. Finally, according to the time-resolved data, the disassembly of the CD4 hexamer into a CD4 tetramer initiates the fusion reaction; where 14% of the events with stoichiometry 3.3+/-0.17 CD4: 1.2+/-0.13 CXCR4 were found to belong to this third step. We also calculated the number of HXB2 Envs engaged with CD4-CXCR4 interacting complexes belonging to the previously defined three steps of the pre-fusion mechanism. Only one or two HXB2 Env were found to be engaged with CD4-CXCR4 complexes (Supplementary Fig. 10). The presence of b12 also blocked the CD4 – CXCR4 interactions as measured with dSTORM (Supplementary Fig. 11) as no IF positive was observed in these cells.
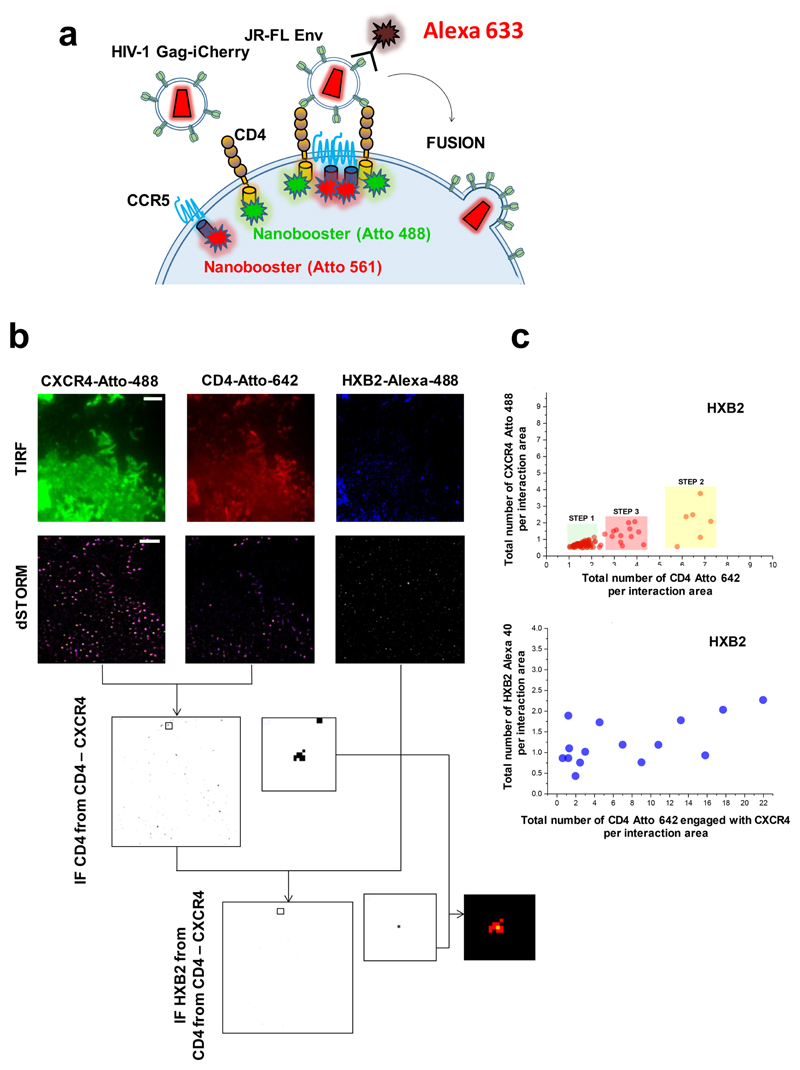
Figure 3. Visualization of HIV-1 virion receptor stoichiometry in cells with multi-colour dSTORM.
a, The labelled CD4 and CCR5 or CXCR4 receptors were labelled again with nanoboosters (Chromotek) specifically engineered for dSTORM imaging. b, HIV virions were exposed to COS7 cells and 10 minutes after fixed and imaged in a TIRF-dSTORM set up equipped with two EM-CCD cameras (Zeiss Elyra dual-cam, see Material and Methods section). The pre-fusion reaction of individual HIV-1 virions was assessed by co-localization analysis of single molecules with an average axial resolution of 30 nm per event. Colocalization positive images were produced to generate masks to recover their stoichiometry, defined as the normalized sum of photons per interaction event. CD4 and CCR5 or CXCR4 co-localization masks were used against the JR-FL or HXB2 labeled Env super-resolution image to recover the number of HIV-1 Env engaged with CD4 –CCR5 or CD4 –CXCR4 complexes. c, The total number of normalized events per interaction area of CXCR4 labeled with Atto 488 was plotted against the total number of events per interaction area of CD4 labeled with Atto 642 for COS7 cells exposed to HIVHSB2 also labelled with Alexa 405 against the Env (as described in material and methods) (top chart, n = 103). Three different regions corresponding to the three steeps of the pre-fusion reaction are populated only for cells exposed to HIVHXB2. In the bottom chart, the total number of HXB2 Env interacting with CD4 and CXCR4 labelled with Alexa 405 are plotted against the total number of CD4 – CXCR4 interacting complexes (n = 18).
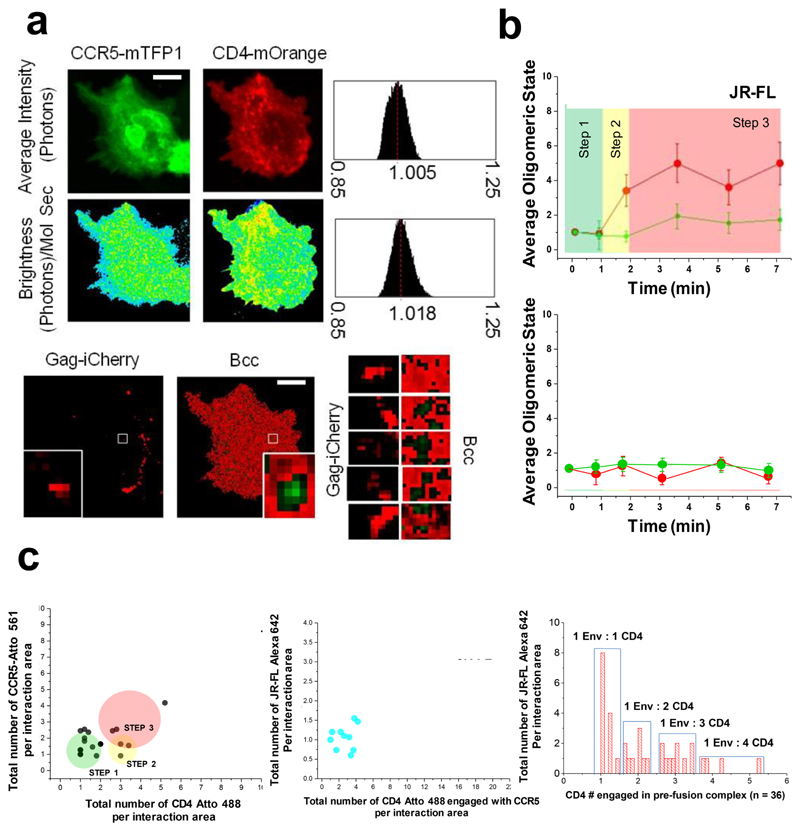
Figure 4. Visualization of JR-FL based HIV-1 virion receptor stoichiometry in live cells.
a, Fast time-resolved, three-color imaging was performed on a COS7 cell co-expressing CCR5-mTFP1 (green micrographs) and CD4-mOrange (red micrographs) exposed to HIVJR-FL –Gag-iCherry virions, scale bar 1 μm. The brightness histogram for each channel together with the pixel by pixel Brightnes maps are also presented (second row). In the last row, microsgraphs corresponding to the HIVHXB2-iCherry (in red) togher with the Bcc map is shown. Green regions represent positive Bcc. The time-resolved co-localization map coming from the white squares is also presented (third row, right panels) and correspond to regions of 0.5 X 0.5 μm. b, Time-resolved stoichiometry for CD4 (red dots) and CXCR4 (green dots) upon addition of HIVJR-FL/GagiCherry (right panel). The circles indicate the average value (n = 12) and the error bars indicate the standard error for each time point c, Time-resolved homotypic interactions for CD4 (red dots) and CCR5 (green dots) upon addition of HIVJR-FL/GagiCherry in the presence of inhibitory concentrations of b12. CD4 and CCR5 did not interact in this case. Eror bars indicate standard error for each time point which represents the average (n = 14). c, The total number of normalized events per interaction area of CCR5 labeled with Atto 561 was plotted against the total number of events per interaction area of CD4 labeled with Atto 488 for COS7 cells exposed to HIVJR-FL also labelled with Alexa 633 against the Env (as described in material and methods) (n = 23). The middle panel shows the total number of HXB2 Env interacting with CD4 and CXCR4 labelled with Alexa 405 are plotted against the total number of CD4 – CXCR4 interacting complexes (n = 12). The right panel shows the distribution of all CD4 – Env interactions and the relative frequency of their stoichiometry (n = 36).
HIV-1 R5 tropic virus interactions with CD4 and CCR5: Fluorescence Fluctuation Spectroscopy
R5 tropic HIV-1 viruses carrying JR-FL Env and Gag-iCherry (HIVJR-FL/GagiCherry)16 were exposed to COS7 cells co-transfected with CD4-mOrange and CCR5-mTFP1 at 37 °C (Fig. 4a-b). COS7 cells expressing CD4-mOrange and CCR5-mTFP1 were imaged together with HIVJR-FL/GagiCherry virions, respectively, at the same time resolution as before (one frame per second). Time-resolved brightness and cross-variance brightness (Bcc)5 images were also calculated for CCR5-mTFP1 and CD4-mOrange intensity images (Fig. 4a, left panel). For JR-FL Env (n = 12), during the initial attachment to the cognate host receptor, CD4-Env forms an asymmetric pre-hairpin intermediate (1.7 min) following oligomerisation of additional CD4 molecules on the trimer leading to the secondary intermediate (3.4 – 7 min). Concomitantly, dimerization of CCR5 co-receptors (3.7 min) results in the final fusion competent complex, with a total of 4 CD4±0.3 and 2±0.3 CCR5 molecules bound to a single HIV-1 Env trimer (Fig. 4b, top panel).
The addition of inhibitory concentrations of b12 (100 μg/mL) impeded both CD4 – CCR5 interactions and posterior CD4 trimer formation (Fig. 4b, bottom panel). Moreover, CCR5 dimerization was also not observed, indicating that homotypic interactions between receptors and co-receptors were also inhibited for JRFL Env decorated HIV-1 virions. This contrasts HXB2 Env, where the interaction between CD4 and CXCR4 was disrupted but not further homodimerization of CXCR4 or CD4 tetramerization (Fig. 2e).
HIV-1 R5 tropic virus interactions with CD4 and CCR5: Super-resolution localization microscopy
Multi-colour TIRF - dSTORM corroborated the stoichiometry found using time-resolved Bcc (Fig. 4c) for HIV-1 R5 tropic virions. Indeed, for JR-FL Env (n = 23), the first asymmetric pre-hairpin intermediate (Step 1, 1 min) was identified for 69% of the quantified interacting events (with an average stoichiometry of 1.3+/-0.1 CD4 : 1.6+/-0.1 CCR5). Events for the secondary intermediate (1-2 min) accounted for 17% of the total (2.9+/-0.1 CD4 : 1.9+/-0.3 CCR5). The final fusion competent complex, (2 – 7 min) accounted for 14 % of the events with an average stoichiometry of (3.4+/-0.1 CD4 : 1.5+/-0.2 CCR5). We also evaluated the number of JR-FL Env engaged with the different CD4 –CCR5 complexes (Fig. 4c middle and right panels). An average of 1.1+/-0.9 Env (n = 12) were found to be engaged with CD4-CCR5 interacting complexes, indicating that for R5 HIV-1 virus only one Env might be sufficient to trigger fusion (Supplementary Fig. 11). Overall, the combination of single molecule time-resolved Bcc with dSTORM shows a very good correlation with Bcc for the HIV-1 prefusion reaction in both Envs, JR-FL and HXB2. Low interaction factors (IF) from TIRF-dSTORM images were also recovered for these JR-FL virions (Supplementary Fig. 11) in the presence of inhibitory concentrations of b12, further corroborating our results with Bcc.
Discussion
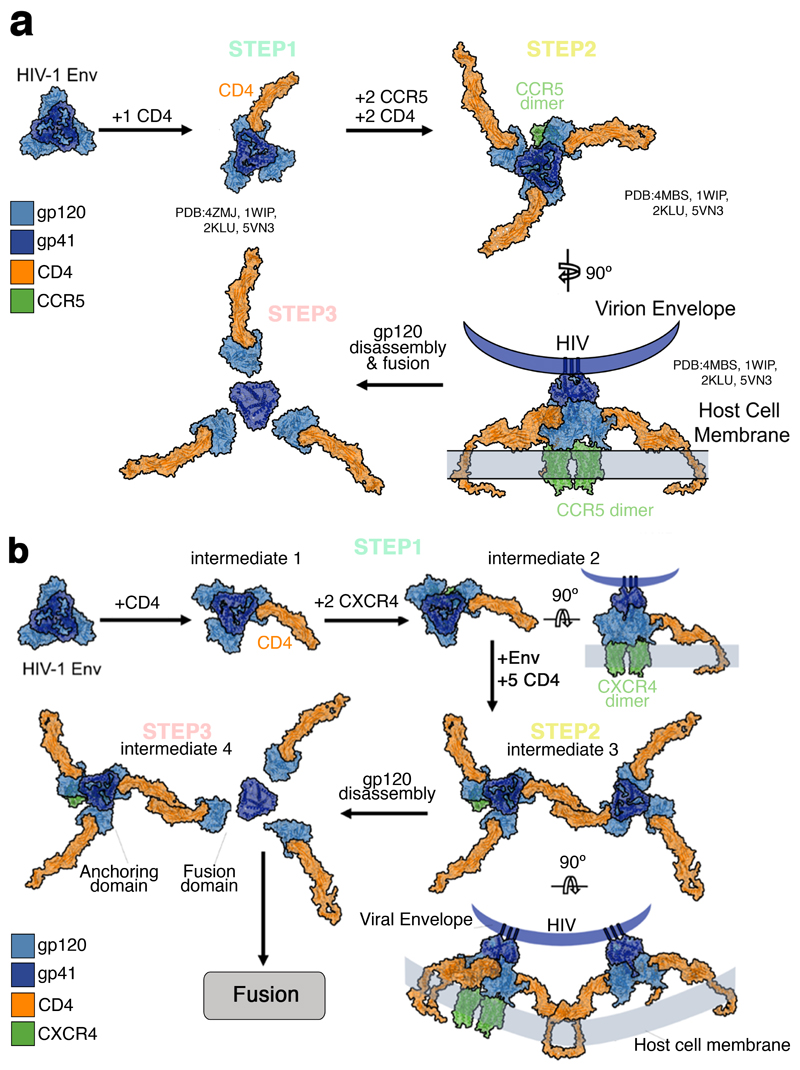
Altogether, our studies support a dynamic three step model for both HIVHXB2 and HIVJR-FL (Fig. 5). For X4 tropic virions, Env – CD4 interactions induce CXCR4 dimerization, CD4 then engages with two Env, - as shown by 3 color TIRF-dSTORM microscopy - to generate a hexamer that might serve as a scaffold to stabilise a final 4CD4 – 1/2CXCR4 conformation, with a single Env (Fig. 5b). We speculate that for HIVHXB2, step 2 is crucial to culminate the fusion reaction and there could be an anchoring domain and a fusion domain that undergoes gp120 disassembly leading to 6 helix bundle formation. For R5 tropic virions, Env – CD4 interactions form a the previously described asymmetric pre-hairpin intermediate 27–29; following binding and oligomerisation of 2 additional CD4 molecules with concomitant CCR5 dimerization. After this, the secondary intermediate leads to the final fusion competent complex with a total of 4±0.3 CD4, 2±0.3 CCR5 and 1 JR-FL Env, as seen with 3 colour TIRF-dSTORM microscopy. Interestingly, dSTORM experiments show an average stoichiometry for step 3 of 3.4+/-0.1 CD4 : 1.5+/-0.2 CCR5. In these single molecule experimetns the 4 CD4 molecules engaged with 1 Env spike was seen with a probability of 0.1 being 3 CD4 more likely to be detected (P = 0.36). The majority of CD4 molecules were found in a monomeric state before HIV-1 addition and this instance might favour the use of 3 CD4 moleuces engaged in the JR-FL complex. It is possible, however, that pre-existent CD4 dimers could also engage with Env in step 3 originating a complex with 4 CD4 molecules (Supplementary Figure 12).
Figure 5. A three-step stoichiometric model for HIV-1 Env-host receptor interactions.
a, The HIV-1 JR-FL Env glycoprotein (Blue) (PDB ID: 4ZMJ) is present as a trimer on the mature virion. b, Time-resolved stoichiometry pre-fusion reaction for CXCR4-CD4 (red dots) induced by HIVHXB2–Gag-iCherry virions.
(gp120: light blue, gp41: dark blue, CD4: orange, CXCR4: green, b12: yellow)
Our data indicate that both HXB2 Env and JR-FL Env start with an asymmetric intermediate bound to a single CD4, as previously suggested27. Our models also support the existence of important differences in the entry mechanisms of X4 and R5 strains. In the X4 strains, CXCR4 dimerization30,31 occurs prior to CD4 hexamer formation and following initial Env – CD4 recognition32 (as characterized by TIRF – dSTORM, Fig.3 and Fig.4c). For R5 tropic JR-FL, CCR5 dimerization31 occurs after Env-CD4 complexation and recruitment of 2 additional CD4 molecules33 around the complex (Fig. 5a). These data also suggests that the V3 loop of Env could be fully accessible with the formation of bridging sheet27,28 with 2 – 3 CD4 molecules 29. Interestingly, we demonstrate that the transition towards formation of a JR-FL Env-3CD4 complex occurs relatively rapidly (1.7 min, Fig. 4c) and prior to CCR5 dimerization, which contrasts the relatively slow transition suggested by Kwon and co-workers (2015)28. Recent in-vitro analyses by Ma and co-workers (2018)29, have further refined the initial conformations sampled by Env prior to fusion 29. Combined with our model of the higher order assembly of Env-receptor complexes, these data provide a holestic understanding for the Env dynamics on the virion surface. This, has potential implications about Env open configurations and how CD4 and co-receptors could be recruited around Env (either primary isolates or lab strains which cleary differ in their studies and ours). Here, we show in live cells, from the perspective of the host, the time-resolved stoichiometry for CD4 and co-receptors corroborating the model proposed in29 for both strains tested. Our 3 colour TIRF-dSTROM experiments shown that HIVHXB2 necessitates 2 Env spikes to complete the fusion reaction whilst HIVJR-FL only needs one. The different stoichiometry found for these two Env strains is in line with other reports3,34 and not surprising. Indeed, R5 tropic primary isolates HIVJR-FL, exhibits reduced dynamics as compared to X4 tropic lab strains HIVHXB2 and this have implicaitons on receptor stoichiometry29.
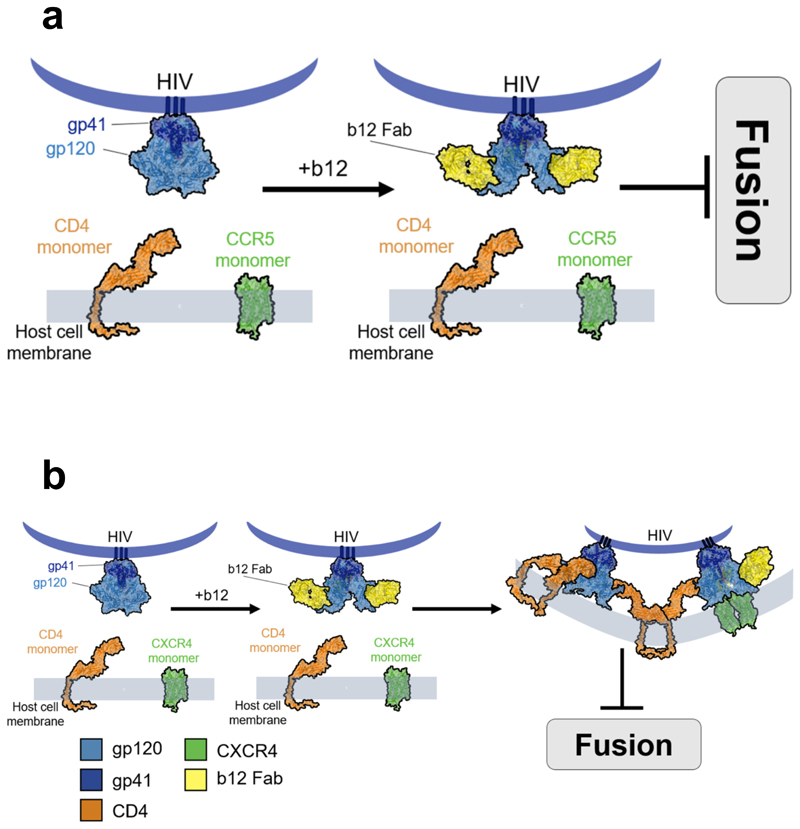
Ultimately, this study opens up novel avenues for testing putative therapeutic approaches for the treatment of HIV-1 infection, which prevent the formation of the required receptor and co-receptor oligomerization state necessary for viral entry. CD4-binding-neutralizing antibodies, like b12 (Fig. 6), that prevent exposure of the co-receptor binding by restricting V1/V2 loops2, blocked CD4 – CXCR4 interactions for HXB2 Env and CD4 – CCR5 interactions for JR-FL Env. It is possible that not all Env were engaged with b12 and therefore a small subpopulation would be able to interact with CD4, inducing CXCR4 oligomerization in the case of HXB2 Env. The b12, however, would hinder CD4 hexamer formation, impeeding the formation of the prebundle structure. In the case of JR-FL, this behaviour was not observed and disruption of CD4 – CCR5 interactions also prevented the formation of independent higher oligomeric states for both CD4 and co-receptor. These data also could indicate a higher affinity of b12 for JR-FL Env as compared to HXB2, as we found no JR-FL Env available able to interact with CD4 using the same b12 concentration (100 μg/mL); another explanation might be the differential number of spikes presented in both strains. The novel insights on the precise mechanism of broadly neutralizing antibodies (Fig. 6) presented in this study and most importantly, the molecular mechanisms of the HIV-1 prefusion reactions, may help to improve host-directed therapeutic interventions crucial to preclude the emergence of resistant viral variants. Recently, we have discussed on the importance of design alternative a novel therapeutic interventions for HIV-1 that focus on the host rather than the virus itself1. This study uses an integrative approach exploiting the combination of advanced imaging technologies to better understand the problem of heterogeneity in HIV-1 entry. Further inter-disciplinary studies focussing on primary cells and tissue are still needed to deepen our understanding of HIV-1 stoichiometry during fusion. This report, however, pioneers the use of single molecule approaches in live cells upon HIV-1 entry and paves the way for future studies aiming to quantify the response of host factors to HIV-1 infection with high spatial and temporal molecular resoltuions.
Figure 6. b12 disrupts CD4 co-receptors interactions.
a, Model showing how b12 totally impedes CD4 and CCR5 interactions inhibiting the fusion reaction from not allowing step 1 to occur. b, Model depicting one possibility for the targeting of mAb12 (yellow) to the CD4 binding region of HIV-1 Env, which prevents fusion into the host cell, but still allows CXCR4 oligomerisation, possibly due to incomplete coverage on the Env spike. Non-interacting time-resolved oligomeric states for CD4 and CXCR4 following HIVHXB2 –Gag-iCherry virions in the presence of 100 μg/mL b12.
Methods
Cloning and expression of CD4 and CXCR4
hCD4-YFP and hCXCR4-YFP constructs in pRluc-N1(Perkin-Elmer) and pEYFP-N1 (Clonetech) were kindly provided by Mario Mellado (CNB-CSIC, Madrid, Spain) and Thomas Hope (Northwestern University, U.S.A) and were transformed into E.coli DH5a. The plasmids were subsequently isolated and the cDNA of hCD4 and hCXCR4 were cloned into mOrange, mRFP and mTFP1 vectors by transferring the HindIII/BamHI and NheI/AgeI fragments into the corresponding sites of the vectors, to make CD4-mOrange, CD4-mRFP and CXCR4-mTFP1. COS-7 cells were transiently transfected 24 hours before the experiment using GeneJuice (Novagen) according to the recommendations from the manufacturer. COS-7 cells were imaged in DMEMcomp Fluobrite (Life Technologies).
Cell Culture
HEK293T cells and COS-7 cells were grown using DMEM (Life Technologies) supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, and 1% L-Glutamine to give DMEM complete (DMEMcomp). All cells were maintained in a 37°C incubator supplied with 5% CO2.
Virus production and expression of virus constructs
pR8DEnv (encoding the HIV-1 genome harbouring a deletion within Env), pcRev, Gag-iCherry, HXB2, JRFL, and VSV-G were kindly provided by James Binley (Torrey Pines Institute for Molecular Studies, San Diego, U.S.A) and Greg Melikyan (Emory University, Atlanta, U.S.A). Pseudotyped viral particles were produced by transfecting HEK293T cells plated at 60%– 70% confluence in T175 flasks. DNA components were transfected using GeneJuice (Novagen) in accordance with the manufacturer’s instructions. To produce particles harbouring the Gag-iCherry, cells were transfected with 2 μg pR8DEnv, 2 μg Gag-iCherry, 1 μg pcREV, and 3 μg of the appropriate viral envelope (either VSV-G or the CCR5-tropic HIV-1 strain JR-FL or the CXCR4- tropic HXB2). Transfection mixtures were then added to cells in DMEMcomp before returning flasks to the 37°C CO2 incubator. After 12 hours, the medium containing the transfection mixture was removed and cells were washed with 1 x PBS. Fresh DMEMcomp (lacking phenol red) was then added. Cells were subsequently incubated for a further 24 hours. At 48 hours post-transfection, viral supernatants were removed from cells and pushed through a 0.45 μm non-pyrogenic syringe filter (SARSTEDT). The virus was concentrated by incubation with the LENTI-X concentrator (Clonetch Laboratories) in accordance with the manufacturer’s instructions, aliquoted, and stored at -80°C. Pseudotyped immature viral particles were produced as described above, with the addition of 320nM of HIV protease inhibitor Saquinavir (European Pharmacopoeia, Sigma-Aldrich) one hour after transfection and re-added after 12 hours that the mixture (transfection) was removed and new medium was added onto the cells.
Viral infections
HIV-1 produced virions were tested for productive infectivity in the COS-7 reporter cell line by Gag-iCherry production. COS-7 cells were transiently transfected, as mentioned above, for CD4-mOrange and CXCR4-mTFP1 expression. 24 hours post transfection they were infected with either HXB2-, VSVG-, JRFL-, or Saquinavir treated HXB2-Gag-iCherry viruses. Cells and viruses were incubated for 4 hours at 37°C. Accutase solution (Sigma-Aldrich) was then used to remove viruses from the cell environment, cells were washed with 1x PBS, new DMEMcomp was added and cells were incubated at 37°C. Cells were imaged 24 and 48 hours post infection.
Antibody Neutralisation Assay
Gag-iCherry HXB2 virions were mixed with different amounts of neutralising antibody (nAb) b12 (POLYMUN) and incubated at 37°C for one hour. Cells expressing CD4-mOrange and CXCR4-mTFP1 were imaged before and after addition of the virus-nab mixture as described below. Cells and virion-nab were incubated at 37°C and imaged 24 and 48 hours post infection for Gag-iCherry production.
Light Microscopy
COS-7 cells co-expressing CD4-mOrange and CXCR4-mTFP1 were imaged before and after virion addition at 37°C using an SP8–X-SMD Leica microscope from Leica Microsystems (Mannheim, Germany). To rule out artefacts due to overexpression, only cells expressing low amounts of mTFP1 and mOrange were selected (1-10 photocounts per pixel). The selected cells mostly expressed CD4-mOrange as a monomer and CXCR4-mTFP1 as a mixture between monomers and dimers, indicating that these expression conditions did not cause over-expression of the proteins. Gag-iCherry virus aliquots were left on ice to thaw and were gently shaken before addition to the cells. During the experimental procedure a 63X/1.4 NA oil immersion objective was used. Fluorescent proteins were excited using a White light laser (WLL) tuned at 80 MHz. The WLL was set for two or three different pathways to avoid bleed-through between CXCR4-mTFP1, CD4-mOrange and Gag-iCherry: WLL tuned at 470, 514 and 594 nm to sequentially excite the fluorescent proteins by line scanning. The photon detection was performed by two gated hybrid internal detectors (HyD) on single photon counting mode tuned at 478-507 nm (mTFP1) and 520-586 nm (mOrange) and gated between 2.5-12.5 ns to avoid auto-fluorescence and refraction light from the coverslip, while the Gag-iCherry emission was detected by a Photomultiplier tube (PMT) tuned at 619-753 nm. The acquisition was continuous for 500 to 700 frames at 256x256 pixels, while the dwell time was 2.43μs and the frame rate was 1.02/s.
Number and Brightness Analysis
We used the established number and brightness method4–6. Briefly, first and second moment analysis based on fluorescence fluctuations of one channel (CXCR4-mTFP1 or CD4-mOrange) in a pixel over time provides a quantitative result of the average number of particles in one channel, based on their brightness in that given pixel over time (frames). Cross number and brightness analysis extends fluorescence cross correlation spectroscopy to two-dimensional cellular images by the co-variance analysis of fluorescence fluctuations in two channels5. The two-colour mode (from CXCR4-mTFP1 and CD4-mOrange) calculation of the co-variance between intensity fluctuations in ch1 and ch2 allows evaluation of the cross-Brightness or the co-variance of the intensities. Bcc is a quantitative measure of the overall number of molecules forming the stoichiometric ratio of the complex. The absence of FRET between the fluorescence proteins (mTFP1, mOrange, and iCherry) was checked prior to the accumulation of data. Fluorescence bleed-through in detection channels was prevented by line interleaved excitation. Photobleaching of fluorescent proteins during the acquisition was corrected using the detrending algorithm6,35. The data were analysed using an R package in R studio https://github.com/rorynolan/nandb).
Fluorescence Resonance Energy Transfer – Fluorescence Lifetime Imaging microscopy (FRET-FLIM) in live cells
COS-7 cells grown on glass-bottom 35 mm Petri dishes (Mattek) were transiently transfected with CD4-mOrange and CXCR4-mTFP1. Before imaging, Dulbecco’s modified eagle medium was replaced with PBS equilibrated at 37°C. Multicolour images were acquired two days post-transfection using a Leica SP8-X-SMD confocal microscope (Leica Microsystems) with a 63×/1.40 numerical aperture oil immersion objective. mTFP1 and mOrange were excited at 440 and 514 nm, respectively, and the fluorescence emission was detected using two hybrid detectors in photon counting mode at 470-520 and 560-595 nm, respectively. FRET detection was based on the time domain FLIM experiments which were performed using a Time-Correlated Single Photon Counting (TCSPC) system operated by a PicoHarp 300 module (PicoQuant) attached to the Leica SP8-X-SMD confocal microscope (Leica Microsystems) with a 63×/1.40 numerical aperture oil immersion objective at 37°C. A 440 nm picosecond pulsed diode laser PDL 800-B (PicoQuant) tuned at 40 MHz was used to excite the donor (CXCR4-mTFP1) and the emitted photons passing through the 460-500nm emission filter and were detected using an external hybrid detector in photon counting mode. At least 1000 photon events per pixel were collected in all cases and the lifetime analysis was carried out using a Symphotime (PicoQuant). The acquired fluorescent decays were fitted by mono- or bi-exponential model36.
Single Virus Tracking
The spot-enhancing filter 2D plugin from ImageJ37 was applied to background-subtracted images to improve the signal to noise ratio. Virus tracking was performed with the 64-bit software module from Imaris (BitPlane, Zurich, Switzerland), using an auto-regressive algorithm. Tracking provided quantitative information regarding the mean fluorescence intensities of the viral content and membrane markers, particle's instantaneous velocity, trajectory and the mean square displacements (MSD). Since N&B was computed every 100 frames, the x-y coordinates defined within these frames were taken as a reference when carrying out the clocalisation analysis between positive Bcc and HIV-1 virions.
dSTORM Immunostaining
COS-7 cells expressing CCR5-YFP or CXCR4-YFP and CD4-mRFP were incubated with non-labelled JRFL or HXB2 virions respectively for 10minutes at 37°C. After a single PBS wash to remove non-primed virions, cells were fixed with 3.7% PFA (Sigma) for 20 minutes at room temperature followed by 5 min permeabilisation with a 2% Saponin solution (Sigma), After 1h blocking with 4% BSA (Sigma), Nanoboosters (Chromotek, Germany) targeting the YFP (GFP_Booster_Atto488) or the RFP (RFP_Booster_Atto594) were added in 1:100 final concentration in blocking buffer and incubated for one additional hour. S Flag signal was introduced in HXB2, thus these virons were labelled with an mouse-anti-FLAG antibody and an anti-mouse Alexa 405 secondary. JRFL was detected with a 2G12 primary antibody and a anti-human- Alexa 633 secondary. These combination of colours was selected to avoid spectral overlap between the three channels (CD4, Co-Receptors and Env) and sequential acquisition was performed from the longest wavelength until the shortest to avoid collateral photobleaching during each dSTORM acquisition (see below).
Super Resolution Image Acquisition
dSTORM acquisitions were performed on a Zeiss Elyra TIRF system (Carl Zeiss Ltd, Jena) with a dual camera attachment and two Andor iXon Ultra 897 EMCCDs. For raw dSTORM data, 20000 images were acquired for each channel sequentially with an exposure time of 20 ms and a gain of 50 and using a 256 x256 region on the camera using a Plan-Apochromat 100x/1.46 Oil DIC Elyra objective with an additional 1.6x lens in front of the camera. The use of nanoboosters (see immunostaining details above; ChromoTek, Munich) allowed us to avoid significant photobleaching while still having proper fluorophore blinking for superresolution. The excitations were cleaned with either laser blocking filters (488/561; 561/642) for the Atto 488; 561 and 642 signals or a band pass filter (420-480) for the Alexa 405 signal.
dSTORM Image Analysis
The raw TIRF images were processed in Fiji/Imagej 1.51u with the QuickPALM v1.1 plugin to provide dSTORM images with 30 nm average axial resolution. The dSTORM multichannel images were further analysed for molecular interactions between CD4 and CXCR4; CD4 and Hxb2 and CXCR4 and Hxb2 using the Interaction Factor V1.1.0 plugin26. The interaction factor localisation maps were then used to create localisation masks to filter the localisation of positive molecular interactions in the dSTORM images. The dSTORM images where then normalised to the minimum number of photons per interaction detected. As the fluorescence value in the dSTORM images come from single molecular events, the lowest average integrated intensity measured corresponds to the monomer value. After this normalization, the Interaction factor filtered dSTORM images represent n-fold increase over monomers in the interacting regions. The interaction factor images coming from V1.1.0 plugin were employed to generate masks that were then applied to the different super-resolution images coming from the right channels (CD4-mOrange and CCR5-mTFP1 or CXCR4-mTFP1). Also, as depicted in New Figure 3 the Interaction Factor mask was also applied to the HIV-1 channel (for HXB2 Alexa 405 and for JRFL Alexa 633). The resulting images were integrated utilizing automatic particle recognition (ICY (http://www.bioimageanalysis.org/, institute Pasteur, Paris) and computed to recover the normalized number of events per pixel that gave how many HIV-1 Envs were engaged with CD4 (1-2 for HXB2 and 1 for JR-FL). Repetitive events coming from the multiple blinking in the same spot were discarded by applying a filter (Thunderstorm, ImageJ plugin and Zen software Elyra, Jena).
Structural Analysis and Model Building
A model for binding of the HXB2 and JR-FL HIV-1 glycoprotein spike to human CD4 and CXCR4/CCR5 was built using the following structures: ligand-free HIV-1 Env mimic (BG505 SOSIP.664) (PDB: 4ZMJ), human CD4 ectodomain (PDB: 1WIP), human CD4 transmembrane and cytoplasmic domains (PDB:2KLU), human CXCR4 chemokine receptor (PDB:3OE0), human CCR5 chemokine receptor (PDB: 4MBS), HIV-1 Env mimic (B41 SOSIP.664) in complex with the ectodomain of CD4 (PDB: 5VN3), HIV-1 Env mimic (B41 SOSIP.664) in complex with Fab variable domains of mAb b12 (PDB: 5VN8), and HIV-1 gp120 in complex with b12 (PDB: 2NY7).
Models were generated in Chimera38, Coot39, and Pymol (https://pymol.org). The HIV-host cell attachment model showing likely stoichiometries was generated by aligning the dimerised CD4 structure with the sCD4 chains of 5VN3. The transmembrane and cytoplasmic domains of CD4 were modelled onto the CD4 structure in Coot.
All data is available upon request
Supplementary Material
Methods Summary.
Details of molecular cloning, light microscopy, real-time single virus tracking, two color Number and Brigthness analysis are given in Methods.
Acknowledgments
This work has been supported by the Wellcome Trust to RN (105278) and G.M.J (203852), Medical Research Council (MR/L009528/1 to T.A.B.), the National Institutes of Health Oxford-Cambridge Fellowship to CAC. S.P-P acknowledges funding from the Nuffield Department of Medicine leadership fellowship and all authors from the Wellcome Trust Core Award (203141).
Footnotes
Author Contributions S.P-P conceived and supervised the study. M.I. and L.A performed experiments. R.N. produced computational algorithms for image processing. M.I., R.N., L.A., C.A.C., G.M.J. and S. P-P analyzed the data.. Y.W. and T.A.B. built atomic models. S.P-P wrote the manuscript with comments from all authors.
Author Information Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
Potential Conflict of Interests None declared
References
- 1.Jakobsdottir GM, et al. On the Whereabouts of HIV-1 Cellular Entry and Its Fusion Ports. Trends in molecular medicine. 2017;23:932–944. doi: 10.1016/j.molmed.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Ozorowski G, et al. Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike. Nature. 2017;547:360–363. doi: 10.1038/nature23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandenberg OF, Magnus C, Regoes RR, Trkola A. The HIV-1 Entry Process: A Stoichiometric View. Trends Microbiol. 2015;23:763–774. doi: 10.1016/j.tim.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Digman MA, Dalal R, Horwitz AF, Gratton E. Mapping the number of molecules and brightness in the laser scanning microscope. Biophysical Journal. 2008;94:2320–2332. doi: 10.1529/biophysj.107.114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Digman MA, Wiseman PW, Choi C, Horwitz AR, Gratton E. Stoichiometry of molecular complexes at adhesions in living cells. Proc Natl Acad Sci U S A. 2009;106:2170–2175. doi: 10.1073/pnas.0806036106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nolan R, et al. nandb-number and brightness in R with a novel automatic detrending algorithm. Bioinformatics (Oxford, England) 2017;33:3508–3510. doi: 10.1093/bioinformatics/btx434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Padilla-Parra S, Marin M, Kondo N, Melikyan GB. Synchronized Retrovirus Fusion in Cells Expressing Alternative Receptor Isoforms Releases the Viral Core into Distinct Sub-cellular Compartments. Plos Pathogens. 2012;8 doi: 10.1371/journal.ppat.1002694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones DM, Padilla-Parra S. Imaging real-time HIV-1 virion fusion with FRET-based biosensors. Scientific Reports. 2015;5 doi: 10.1038/srep13449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Healey EG, et al. Repulsive guidance molecule is a structural bridge between neogenin and bone morphogenetic protein. Nat Struct Mol Biol. 2015;22:458–465. doi: 10.1038/nsmb.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ratner L, et al. Complete nucleotide sequences of functional clones of the AIDS virus. AIDS Res Hum Retroviruses. 1987;3:57–69. doi: 10.1089/aid.1987.3.57. [DOI] [PubMed] [Google Scholar]
- 11.Schulke N, et al. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. Journal of virology. 2002;76:7760–7776. doi: 10.1128/JVI.76.15.7760-7776.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francis AC, Melikyan GB. Single HIV-1 Imaging Reveals Progression of Infection through CA-Dependent Steps of Docking at the Nuclear Pore, Uncoating, and Nuclear Transport. Cell host & microbe. 2018;23:536–548.e536. doi: 10.1016/j.chom.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaitseva E, et al. Fusion Stage of HIV-1 Entry Depends on Virus-Induced Cell Surface Exposure of Phosphatidylserine. Cell Host Microbe. 2017;22:99–110.e117. doi: 10.1016/j.chom.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones DM, et al. Dynamin-2 Stabilizes the HIV-1 Fusion Pore with a Low Oligomeric State. Cell Reports. 2017;18:443–453. doi: 10.1016/j.celrep.2016.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russell RA, et al. Astrocytes Resist HIV-1 Fusion but Engulf Infected Macrophage Material. Cell Reports. 2017;18:1473–1483. doi: 10.1016/j.celrep.2017.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Padilla-Parra S, et al. Fusion of Mature HIV-1 Particles Leads to Complete Release of a Gag-GFP-Based Content Marker and Raises the Intraviral pH. Plos One. 2013;8 doi: 10.1371/journal.pone.0071002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapanidis AN, et al. Alternating-laser excitation of single molecules. Accounts of chemical research. 2005;38:523–533. doi: 10.1021/ar0401348. [DOI] [PubMed] [Google Scholar]
- 18.Miyauchi K, Kozlov MM, Melikyan GB. Early steps of HIV-1 fusion define the sensitivity to inhibitory peptides that block 6-helix bundle formation. PLoS Pathog. 2009;5:e1000585. doi: 10.1371/journal.ppat.1000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deeks SG, Smith M, Holodniy M, Kahn JO. HIV-1 protease inhibitors - A review for clinicians. Jama-Journal of the American Medical Association. 1997;277:145–153. doi: 10.1001/jama.277.2.145. [DOI] [PubMed] [Google Scholar]
- 20.Chojnacki J, et al. Maturation-Dependent HIV-1 Surface Protein Redistribution Revealed by Fluorescence Nanoscopy. Science. 2012;338:524–528. doi: 10.1126/science.1226359. [DOI] [PubMed] [Google Scholar]
- 21.Binley JA, et al. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. Journal of Virology. 2004;78:13232–13252. doi: 10.1128/jvi.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clotet-Codina I, et al. HIV endocytosis after dendritic cell to T cell viral transfer leads to productive virus infection. Antiviral Research. 2009;83:94–98. doi: 10.1016/j.antiviral.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 23.Finnegan CM, et al. Sphingomyelinase restricts the lateral diffusion of CD4 and inhibits human immunodeficiency virus fusion. Journal of virology. 2007;81:5294–5304. doi: 10.1128/jvi.02553-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altenburg JD, Jin Q, Alkhatib B, Alkhatib G. The potent anti-HIV activity of CXCL12gamma correlates with efficient CXCR4 binding and internalization. Journal of virology. 2010;84:2563–2572. doi: 10.1128/jvi.00342-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nature methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bermudez-Hernandez K, et al. A Method for Quantifying Molecular Interactions Using Stochastic Modelling and Super-Resolution Microscopy. Sci Rep. 2017;7 doi: 10.1038/s41598-017-14922-8. 14882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munro JB, et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science. 2014;346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwon YD, et al. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol. 2015;22:522–531. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma X, et al. HIV-1 Env trimer opens through an asymmetric intermediate in which individual protomers adopt distinct conformations. Elife. 2018;7 doi: 10.7554/eLife.34271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan Q, et al. Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science (New York, NY) 2013;341:1387–1390. doi: 10.1126/science.1241475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin L, et al. Structural biology. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine. Science (New York, NY) 2015;347:1117–1122. doi: 10.1126/science.1261064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Q, et al. Quaternary contact in the initial interaction of CD4 with the HIV-1 envelope trimer. Nature structural & molecular biology. 2017;24:370–378. doi: 10.1038/nsmb.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu L, et al. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 34.Yang XZ, Kurteva S, Ren XP, Lee S, Sodroski J. Subunit stoichiometry of human immunodeficiency virus type 1 envelope glycoprotein trimers during virus entry into host cells. Journal of Virology. 2006;80:4388–4395. doi: 10.1128/jvi.80.9.4388-4395.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nolan R, Iliopoulou M, Alvarez L, Padilla-Parra S. Detecting protein aggregation and interaction in live cells: A guide to number and brightness. Methods. 2017 doi: 10.1016/j.ymeth.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 36.Padilla-Parra S, Auduge N, Coppey-Moisan M, Tramier M. Quantitative FRET analysis by fast acquisition time domain FLIM at high spatial resolution in living cells. Biophys J. 2008;95:2976–2988. doi: 10.1529/biophysj.108.131276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sage D, Neumann FR, Hediger F, Gasser SM, Unser M. Automatic tracking of individual fluorescence particles: application to the study of chromosome dynamics. IEEE Trans Image Process. 2005;14:1372–1383. doi: 10.1109/tip.2005.852787. [DOI] [PubMed] [Google Scholar]
- 38.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/s0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.