Abstract.
Arboviral diagnosis has been complicated throughout the tropical and subtropical Americas by the recent co-circulation of Zika virus (ZIKV), chikungunya virus (CHIKV), and dengue virus (DENV). The aim of this study was to implement a multiplex real-time RT-PCR (rRT-PCR) for ZIKV, CHIKV, and DENV in Paraguay to test patients who were clinically suspected of having dengue. We tested 110 sera from patients who presented to the Hospital de Clínicas in 2016 and had testing for DENV nonstructural protein 1 (NS1; 40 positive and 70 negative). Using a composite reference standard, we confirmed 51 dengue cases (46.4%): 38/40 NS1 positive and 13/70 NS1 negative. Chikungunya virus and ZIKV were detected in one sample each, both were DENV NS1 negative. The NS1 test demonstrated good agreement with rRT-PCR for DENV. However, multiplex rRT-PCR identified a subset of dengue cases and additional arboviral infections that would not be detected if NS1 assays are relied upon for diagnosis.
Arboviruses are important agents of human disease worldwide and represent a significant public health problem in Paraguay. According to data from the Dirección General de Vigilancia de la Salud (DGVS), the first cases of ZIKV in the country were detected in November 2015, amidst ongoing transmission of dengue virus (DENV, serotypes 1, 2, and 4) and CHIKV.1 Confirmed cases of DENV (serotypes 1, 3, and 4) as well as CHIKV and ZIKV were also documented in 2016, the year in which samples were obtained for the current study.1 Accurate diagnosis of these arboviruses is complicated by nonspecific clinical manifestations and limitations to available laboratory tests.2 Serological assays, in particular, may be inaccurate as antibodies do not reliably develop until day 5 after the onset of symptoms, and there is significant antibody cross-reactivity between DENV and ZIKV.3 The objective of this study was to implement a sensitive and specific, multiplex real-time RT-PCR (rRT-PCR) for ZIKV, CHIKV, and DENV (the “ZCD assay”) to test patients who were clinically suspected of having dengue. Dengue virus detection in this assay was evaluated in the setting of an existing diagnostic algorithm based on rapid diagnostic testing for the DENV nonstructural protein 1 (NS1) antigen. The multiplex assay was also used to screen for acute cases of chikungunya, Zika, and viral coinfections.
Acute-phase sera, collected ≤ 5 days post-symptom onset, were included from 110 patients who were clinically suspected of having dengue and presented from March–April 2016. Samples were obtained at the Laboratorio Central of the Hospital de Clínicas (Facultad de Ciencias Médicas, Universidad Nacional de Asunción, Paraguay) and tested immediately for DENV NS1 antigen using the immunochromatographic Dengue NS1 Ag Test (“the NS1 test,” SD Standard Diagnostics, Inc., Korea) according to the manufacturer’s instructions. Serum aliquots were stored at −80°C until processing for RNA and IgM detection. For the current study, we selected 40 NS1-positive samples to confirm DENV detection and 70 NS1-negative samples to screen for Zika, chikungunya, and undetected DENV infections. The study was reviewed and approved by the Scientific and Ethical Committees of the Instituto de Investigaciones en Ciencias de la Salud, Universidad Nacional de Asunción (IICS-UNA; IRB00011984, code M05/2018).
Viral RNA was extracted from 140 μL of patient serum and eluted into 60 μL of buffer using the QIAamp viral RNA mini kit (Qiagen, Germantown, MD) according to the manufacturer’s recommendations. The samples were screened for ZIKV, CHIKV, and DENV RNA using the ZCD assay, as previously described and validated.4 RNase-P detection was used as internal control.5 For all samples that tested positive for DENV RNA in the ZCD assay, DENV serotype and quantitated viral load (expressed in log10 copies/mL of serum) were determined using a serotype-specific rRT-PCR as described.6 Sera were tested for anti-DENV IgM antibodies using a commercial MAC-ELISA (Dengue ELISA IgM Capture, Vircell Microbiologists, Granada, Spain) according to the manufacturer’s recommendations; optical density was measured on an Inc. Stat Fax 2100 ELISA reader (Awareness Technology, Palm City, FL). Qualitative results were recorded and used in this study.
Dengue cases were considered confirmed if the patients tested positive for 1) DENV RNA in both the ZCD and DENV serotyping assays or 2) NS1 and anti-DENV IgM. This definition was applied to limit potential false-positive diagnoses for patients who tested positive in only one assay. Assay performance for individual tests was calculated in reference to this set of confirmed cases. Statistical analysis was performed using GraphPad software version 8.0.1 (San Diego, CA). Categorical variables were compared using Fisher’s exact test. Continuous variables were compared by t test or the Mann–Whitney test (viral load).
In total, 51 patients had a confirmed dengue case (51/110, 46.4%). Of the dengue cases, 47 were positive by rRT-PCR (92.2%) and four additional patients were positive for NS1 and anti-DENV IgM (7.8%). General characteristics and DENV test results are shown in Table 1. The mean age was 27.4 years (SD 19.9), and 38 children (< 18 years of age; 34.5%) were included. Dengue virus serotypes were confirmed in all 47 cases positive by rRT-PCR: DENV-1 (n = 40) and DENV-4 (n = 7). Chikungunya virus and ZIKV were detected in one sample each (1/110 [0.9%]). The viral load for CHIKV and ZIKV were 8.0 and 4.2 log10 copies/mL of serum, respectively. The detection of ZIKV RNA was confirmed using a second published rRT-PCR, as described, which targets a separate region of the ZIKV genome.7 All samples that tested negative for a viral target in the ZCD assay were positive for RNase-P.
Table 1.
Patient characteristics, dengue virus (DENV) test results, and routine laboratory findings among dengue cases and non-dengue cases
| Clinical factors, n (%) | All patients | Dengue cases* | Non-dengue cases | Odds ratio (95% CI) |
|---|---|---|---|---|
| Patients | 110 (100) | 51 (100) | 59 (100) | – |
| Female | 62 (56.4) | 27 (52.9) | 35 (59.3) | – |
| Age (years), mean (SD) | 27.4 (19.9) | 25.5 (20.4) | 29.0 (19.4) | – |
| DENV results, positive | ||||
| Real-time RT-PCR | 47 (42.7) | 47 (92.2) | 0 | – |
| Nonstructural protein 1 | 40 (36.4) | 38 (34.5) | 2 (3.4) | – |
| IgM | 8 (7.3) | 5 (9.8) | 3 (5.1) | – |
| Laboratory findings | ||||
| Anemia† | 33 (30.0) | 15 (29.4) | 18 (30.5) | – |
| Elevated hemoglobin | 4 (3.6) | 3 (5.9) | 1 (1.7) | – |
| Thrombocytopenia | 30 (27.3) | 16 (31.4) | 14 (23.7) | 1.5 (0.6–3.5) |
| 100,000–150,000 | 18 (16.4) | 10 (19.6) | 8 (13.6) | – |
| 50,000–100,000 | 9 (8.2) | 5 (9.8) | 4 (6.8) | – |
| < 50,000 | 3 (2.7) | 1 (2.0) | 2 (3.4) | – |
| White blood cell counts† | ||||
| Leukopenia | 27 (24.5) | 13 (25.5) | 14 (23.7) | 1.1 (0.5–2.7) |
| Leukocytosis | 14 (12.7) | 1 (2.0) | 13 (22.0) | 0.07 (0.01–0.42) |
| Neutropenia | 15 (13.6) | 4 (7.8) | 11 (18.6) | 0.4 (0.1–1.2) |
| Lymphopenia | 65 (59.1) | 38 (74.5) | 27 (45.8) | 3.5 (1.5–8.1) |
Bold text indicates a statistical significance with a P-value less than 0.05.
* Cases confirmed by rRT-PCR or NS1 and anti-DENV IgM detection.
† Abnormal results were defined as the following: anemia, hemoglobin concentration < 12 g/dL; leukopenia, leukocytes < 4,000 cells/mm3; leukocytosis, leukocytes > 10,000 cells/mm3; neutropenia, neutrophils < 2,000 cells/mm3; and lymphopenia, lymphocytes < 3,000 cells/mm3.20
Of the 40 patients who originally tested positive for NS1, 34 (85.0%) were positive for DENV by rRT-PCR (Table 2) and four were positive for anti-DENV IgM. Good agreement between the NS1 test and rRT-PCR for DENV was observed (κ = 0.64, 95% CI 0.49–0.79; Supplemental Table 1). With reference to the study definition of a confirmed dengue case, the NS1 assay demonstrated 74.5% sensitivity (38/51 cases) and 96.7% specificity (2/59 negative cases).
Table 2.
Patient characteristics and laboratory findings among NS1-positive and NS1-negative dengue cases.
| Dengue cases | P-value | ||
|---|---|---|---|
| NS1 positive | NS1 negative | ||
| Patients | 38 (100) | 13 (100) | |
| Female | 22 (57.9) | 5 (38.5) | 0.33 |
| Age (years), mean (SD) | 25.6 (20.6) | 25.42 (20.4) | – |
| Dengue results | |||
| Serotype | 34 (100) | 13 (100) | – |
| DENV-1 | 31 (91.2) | 9 (69.2) | 0.439 |
| DENV-4 | 3 (8.8) | 4 (30.8) | 0.060 |
| Viral load, mean (SD)* | 7.09 (1.02) | 5.91 (1.09) | 0.001 |
| DENV-1, mean (SD) | 7.21 (0.81) | 6.04 (0.69) | < 0.001 |
| DENV-4, mean (SD) | 5.80 (2.17) | 5.63 (1.82) | 0.914 |
Bold text indicates a statistical significance with a P-value less than 0.05.
DENV = dengue virus; NS1 = nonstructural protein 1.
* In log10 copies/mL of serum.
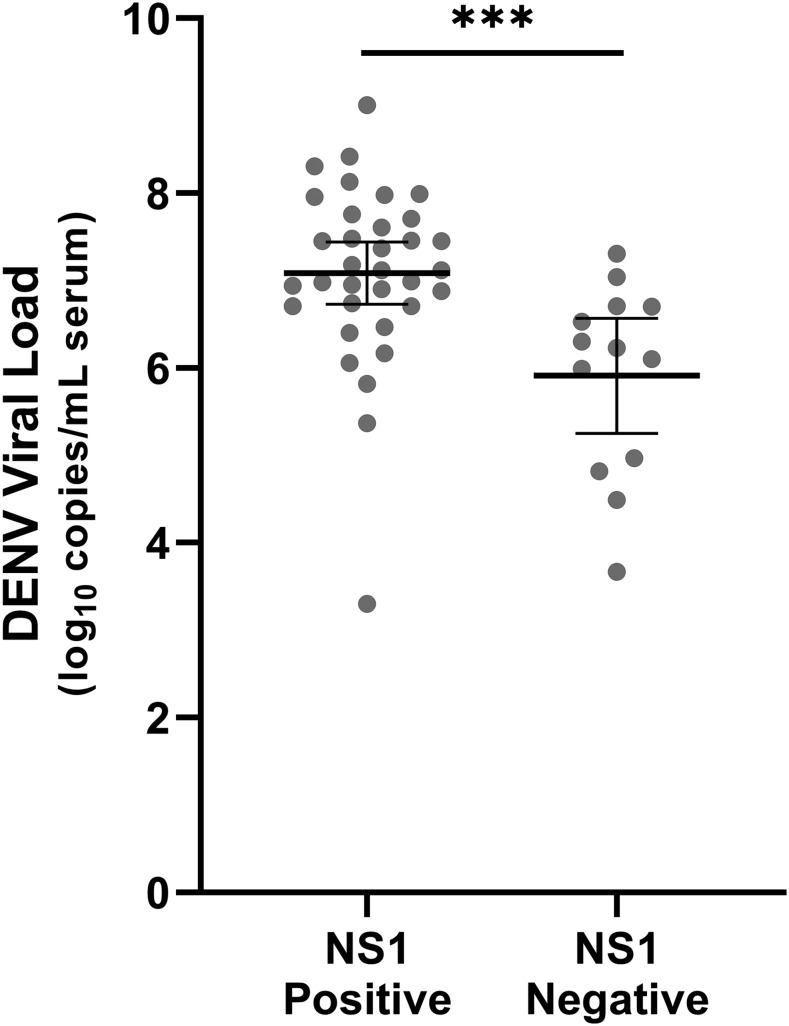
Thirteen dengue cases tested positive by rRT-PCR but negative in the NS1 test (13/70 NS1-negative samples, 18.6%). Mean viral load was significantly higher in NS1-positive samples (7.09 log10 copies/mL of serum, SD 1.02) NS1-negative samples (5.91 log10 copies/mL of serum, SD 1.09; P < 0.001; Table 2, Figure 1). There was a trend toward the decreased detection of NS1 in DENV-4 cases (3/7, 42.9%) compared with DENV-1 (31/40, 77.5%), but this did not reach significance (P = 0.080). The distribution of viral load of DENV-1 and DENV-4 was not significantly different (P = 0.170).
Figure 1.
Dengue virus load at presentation was higher in patients with detectable NS1. Bars represent mean viral load and 95% CIs. P < 0.001 (***).
Hemograms were ordered as part of standard clinical care, and results were obtained by chart review. Routine hematologic parameters were analyzed as categorical variables to identify differences between dengue cases and non-dengue cases. Lymphopenia was associated with dengue cases (odds ratio [OR] 3.5, 95% CI 1.5–8.1) and leukocytosis was associated with a non-dengue cause (OR for dengue 0.07, 95% CI 0.01–0.42). Thrombocytopenia was not associated with dengue in these patients (OR 1.5, 95% CI 0.6–3.5).
The judicious use of diagnostic tests is an important consideration in regions of DENV endemicity, and in the current study, we implemented a sensitive and specific, multiplex rRT-PCR for ZIKV, CHIKV, and DENV to test patients who were clinically suspected of having dengue but had previously only been tested for NS1. Our findings generally agree with arboviral cases reported by DGVS in 2016.8 Circulating DENV serotypes had a marked predominance of DENV-1 accompanied by co-circulation, in a smaller proportion, of DENV-4. Few cases of CHIKV and ZIKV were reported from Paraguay in 2016. For the cases of CHIKV and ZIKV detected here, viral loads were consistent with published values,9–11 and notably, the ZIKV case was negative for NS1.
In our patient population, the NS1 test had a specificity of 96.7%, indicating that this can be used to rule-in dengue cases in the acute phase. However, 13 dengue cases (13/70, 18.6%) were identified among patients who tested negative in the NS1 test. Leukocytosis was strongly associated with a non-dengue case, and the only dengue case with this finding was also NS1 positive. Based on these data, patients with a negative NS1 result and leukocytosis should be worked-up for non-DENV causes for their illness. In the remaining NS1-negative patients, multiplex molecular testing may have a particularly important role in the diagnostic work-up, as 15 arboviral infections were detected among these cases, including cases of CHIKV and ZIKV.
Dengue virus viral load was significantly lower among NS1-negative dengue cases than the NS1-positive cases. NS1 test sensitivity is significantly reduced among secondary dengue cases, likely resulting from preexisting anti-NS1 antibodies that interfere with detection.12,13 This would be consistent with our population, as viral load also tends to be lower among secondary cases at presentation, although peak viral loads may be higher.12–15 In addition, the Paraguayan population has been exposed to several epidemics of dengue, and it is expected that a high proportion of our cases resulted from secondary infections. The significance of NS1 detection for dengue prognosis has varied between studies.16,17 Given that the sensitivity of NS1 detection differs between serotypes,18,19 consistent with the trend observed in our study for DENV-4, some of these differences between studies may be related to the predominant serotype. The clinical prognostic value of NS1 detection, therefore, warrants further study in our population.
Detailed clinical information was not available for these patients, which limits our ability to associate findings with the day of illness. However, only 5/51 (9.8%) dengue cases had detectable anti-DENV IgM, indicating that most patients presented within the first 5 days of symptom onset. Although only DENV-1 and DENV-4 were detected in our patients, this distribution of serotypes is consistent with reported dengue cases in Paraguay during that time.
In conclusion, the ZCD assay provides a useful tool for screening suspected dengue cases in endemic areas. Its use may be targeted to certain patient populations to maximize the added benefit to arboviral detection and identify cases that would be missed if rapid NS1 assays are relied upon in the acute setting.
Supplemental table
Acknowledgments:
We thank the staff at the Laboratorio Central of the Hospital de Clínicas (Facultad de Ciencias Médicas, UNA, Paraguay) for their involvement in patient sample collection and processing as part of routine care.
Note: Supplemental table appears at www.ajtmh.org.
REFERENCES
- 1.MSPyBS , 2016. Boletín Epidemiológico República del Paraguay, Vol. 48 Asunción, Paraguay: DGVS, 1–34. [Google Scholar]
- 2.WHO , 2017. World Health Organization, Pan American Health Organization. Tool for the Diagnosis and Care of Patients with Suspected Arboviral Diseases. Washington, DC: Pan American Health Organization. [Google Scholar]
- 3.Stettler K, et al. 2016. Specificity, cross-reactivity and function of antibodies elicited by Zika virus infection. Science 353: 823–826. [DOI] [PubMed] [Google Scholar]
- 4.Waggoner JJ, Gresh L, Mohamed-Hadley A, Ballesteros G, Vargas Davila MJ, Tellez Y, Sahoo MK, Balmaseda A, Harris E, Pinsky BA, 2016. Single-reaction multiplex reverse transcription PCR for detection of Zika, chikungunya, and dengue viruses. Emerg Infect Dis 22: 1295–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waggoner JJ, Abeynayake J, Sahoo MK, Gresh L, Tellez Y, Gonzalez K, Ballesteros G, 2013. Development of an internally controlled real-time reverse transcriptase PCR assay for pan-dengue virus detection and comparison of four molecular dengue virus detection assays. J Clin Microbiol 51: 2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waggoner JJ, et al. 2013. Single-reaction, multiplex, real-time RT-PCR for the detection, quantitation, and serotyping of dengue viruses. Negl Trop Dis 7: e2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, Stanfield SM, Duffy MR, 2008. Genetic and serologic properties of Zika virus associated with an epidemic, Yap state, Micronesia, 2007. Emerg Infect Dis 14: 1232–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MSPyBS , 2016. Boletín Epidemiológico República del Paraguay, Vol. 52 Asunción, Paraguay: DGVS, 1–15. [Google Scholar]
- 9.Staikowsky F, Grivard P, Souab A, Schuffenecker I, Roux K. Le, Lecuit M, Michault A, 2009. Prospective study of chikungunya virus acute infection in the island of La réunion during the 2005–2006 outbreak. PLoS One 4: e7603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waggoner JJ, et al. 2016. Viremia and clinical presentation in Nicaraguan patients infected with Zika virus, chikungunya virus, and dengue virus. J Clin Infect Dis 63: 1584–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Musso D, Rouault E, Teissier A, Lanteri MC, Zisou K, Broult J, Grange E, Maite TN, 2017. Molecular detection of Zika virus in blood and RNA load determination during the French polynesian outbreak. J Med Virol 89: 1505–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tricou V, Vu HTT, Quynh NVN, Nguyen CVV, Tran HT, Farrar J, Wills B, Simmons CP, 2010. Comparison of two dengue NS1 rapid tests for sensitivity, specificity and relationship to viraemia and antibody responses. BMC Infect Dis 10: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teoh B, Sam S-S, Tan K-K, Johari J, Abd-Jamil J, Hooi P-S, 2016. The use of NS1 rapid diagnostic test and QRT-PCR to complement IgM ELISA for improved dengue diagnosis from single specimen. Sci Rep 6: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duyen HTL, Ngoc TV, Ha DT, Hang VTT, Kieu NTT, Young PR, Farrar JJ, Simmons CP, Wolbers M, Wills BA, 2011. Kinetics of plasma viremia and soluble nonstructural protein 1 concentrations in dengue: differential effects according to serotype and immune status. J Infect Dis 203: 1292–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pok K-Y, Lai Y-L, Sng J, Ng L-C, 2010. Evaluation of nonstructural 1 antigen assays for the diagnosis and surveillance of dengue in Singapore. Vector Zoonotic Dis 10: 1009–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuan NM, et al. 2017. An evidence-based algorithm for early prognosis of severe dengue in the outpatient setting. Clin Infect Dis 64: 656–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duong V, et al. 2011. Clinical and virological factors influencing the performance of a Ns1 antigen-capture assay and potential use as a marker of dengue disease severity. PLoS Negl Trop Dis 5: e1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guzman MG, et al. 2010. Multi-country evaluation of the sensitivity and specificity of two commercially-available NS1 ELISA assays for dengue diagnosis. PLoS Negl Trop Dis 4: e811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sea VRF, Cruz ACR, Gurgel RQ, Nunes BTD, Silva EVP, Dolabella SS, Santos RLCdos, 2013. Underreporting of dengue-4 in Brazil due to low sensitivity of the NS1 Ag test in routine control programs. PLoS One 8: e64056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wallach JB, 2013. Interpretação de Exames de Laboratóriais, 9a edição Williamson MA, Snyder LM, Eds. Rio de Janeiro, Brazil: Editora Guanabara Koogan Ltda. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.