Abstract.
Resistance to anti-leprosy drugs is on the rise. Several studies have documented resistance to rifampicin, dapsone, and ofloxacin in patients with leprosy. We looked for point mutations within the folP1, rpoB, and gyrA gene regions of the Mycobacterium leprae genome predominantly in the neural form of leprosy. DNA samples from 77 nerve tissue samples were polymerase chain reaction (PCR)-amplified for M leprae DNA and sequenced for drug resistance–determining regions of genes rpoB, folP1, and gyrA. The mean age at presentation and onset was 38.2 ± 13.4 (range 14–71) years and 34.9 ± 12.6 years (range 10–63) years, respectively. The majority had borderline tuberculoid leprosy (53 [68.8%]). Mutations associated with resistance were identified in 6/77 (7.8%) specimens. Mutations seen were those associated with resistance to rifampicin, ofloxacin, and dapsone. All the six patients were drug-naive. The clinical and pathological manifestations in this group did not differ from the drug-sensitive group. This study highlights the occurrence of resistance to the standard multidrug therapy and ofloxacin in leprosy. Among the entire cohort, 1/77 (1.3%) showed resistance to rifampicin, 2/77 (2.6%) to dapsone, and 5/77 (6.4%) to ofloxacin. Six new patients showing infection by mutant strains indicated the emergence of primary resistance. Resistance to ofloxacin could be due to frequent use of quinolones for many bacterial infections. The results of the study indicate the need for development of a robust and strict surveillance system for detecting drug resistance in leprosy in India.
INTRODUCTION
Leprosy is one of the leading causes of peripheral neuropathy, particularly in low-resource or developing countries. Leprosy affects the sensory, motor, and autonomic nerve functions, leading to prominent sensory loss and muscle weakness.1–3 Despite three decades of effective treatment with multidrug therapy (MDT), leprosy remains a public health problem. Recently, there has been an increase in relapsing cases, failures in MDT treatment, and the emergence of drug-resistant strains of Mycobacterium leprae, which could undermine existing leprosy control measures.
Standard treatment for leprosy is an MDT with dapsone, rifampicin, and clofazimine.4 Resistance to dapsone and rifampicin is reported from many leprosy-endemic regions of the world.5–7 Although prevalence of leprosy is declining it remains a global problem with potential for resurgence in many areas as borders open and worldwide travel is fast.8
The emergence of MDR organisms for any infectious disease is a public health concern. Currently, recommended control measures for treating leprosy with MDT are designed to prevent the spread of dapsone-resistant M. leprae strains. In the current study, patients with leprous neuropathy were studied for resistance to rifampicin, dapsone, and ofloxacin by PCR sequencing of the drug resistance–determining regions (DRDR) of the genes rpoB, folP1, and gyrA. Clinical, histopathological, and treatment status in the drug-resistant and drug-sensitive groups has been reported.
MATERIALS AND METHODS
Study subjects.
This ambispective cohort study was conducted at the National Institute of Mental Health and Neuro Sciences (NIMHANS), Bangalore, located in the southern part of India. It is a national quaternary care center for all neurological disorders. National Institute of Mental Health and Neuro Sciences is not a center identified specifically for leprosy patients, but patients with predominantly pure neuritic form of leprosy or leprous neuropathy with unidentified skin lesions attend the outpatient clinics. In this study, spanning over a period of 12 years (2007 to 2018), a total of 83 nerve samples of leprosy patients were obtained. A final number of 77 samples were selected for reporting. We were able to procure 44 preserved specimens of nerve tissue from histopathologically characterized patients of leprosy, after careful scrutiny of all neuropathology records with diagnosis of leprosy. The remaining 33 patients were prospectively recruited from 2015, and fresh nerve tissue was taken up for PCR study.
Ethics statement.
All patients included in the study were adults. As part of the hospital requirement, written informed consent was obtained from all the patients for diagnostic biochemical tests and biopsy procedure. Ethics approval from the “NIMHANS Ethics Committee” (Institutional Review Board) was obtained to collect all clinical, biochemical, and histopathological characteristics from the hospital medical records and process all stored nerve tissue samples of leprosy patients for PCR study and sequencing for DRDR in rpoB, folP1, gyrA—the genes responsible for rifampicin, dapsone, and ofloxacin drug resistance, respectively.
Detection of mutations by PCR targeting rpoB, folP, and gyrA.
All DNA samples isolated from preserved nerve tissue and fresh nerve tissue stored in RNAlater solution were transferred to Stanley Browne Laboratory, The Leprosy Mission Trust India in Delhi. Nerve tissue from paraffin blocks were analyzed with FFPE DNA (M/s. Analytik Jena AG, Jena, Germany) kit method for DNA extraction. Mycobacterium leprae–specific DNA was amplified by PCR and sequenced for the aforementioned genes. Of the 83 specimens tested, the M. leprae DNA from 77 samples was PCR-amplified (18/77 were fresh nerve tissue samples preserved in RNAlater solution).
PCR methodology.
PCR-based gene amplification was performed using primers according to the guidelines of WHO “Global Surveillance of Drug Resistance in Leprosy 2008” for detection of mutation in rpoB, gyrA, and folP1 genes in M. leprae genome.9 The PCR mixture contained 12.5 µL Hot Start Taq polymerase PCR Master Mix (2 ×) (Qiagen, Hilden, Germany), 1.25 µL forward primer and reverse primer at a final concentration of 0.5 µM and 5 µL of template DNA from the processed sample. The final volume of the reaction mix was made up to 25 µL with nuclease-free water. Primer sequences used in this study were as follows: folP primers: folPF CTTGATCCTGACGATGCTGT; folPR CCACCAGACACATCGTTGAC, rpoB primers: rpoBF GTCGAGGCGATCACGCCGCA; rpoBR CGACAATGAACCGATCAGAC, and gyrA primers: gyrAF ATGGTCTCAAACCGGTACATC; gyrAR TACCCGGCGAACCGAAATTG. The reaction was cycled 40 times at 94°C, 60°C, and 70°C for 1 minute each; preceded by initial denaturation at 95°C for 15 minutes; and ended by final extension at 72°C for 10 minutes. Each reaction setup contained one negative and one positive control. After detection of the PCR product on a 2% agarose gel, amplicons were excised from the gel and later purified using the Qiagen Gel Extraction Kit. The PCR products were sent for commercial sequencing (at Eurofin Technologies Pvt. Limited, Bangalore, India). Sequence data were analyzed using MEGA version 5.1 (http://megasoftware.net/).
RESULTS
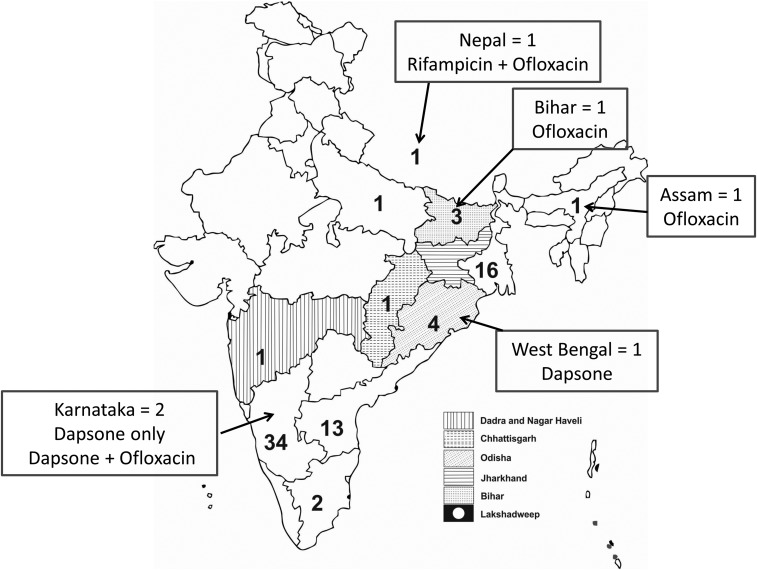
Patients came from different states of India and also Nepal (Figure 1). Based on the clinical and nerve histopathology findings, 83 patients of leprosy with neuropathy were selected for the study. DNA could be isolated from all the 83 nerve tissue specimens. Mycobacterium leprae–specific DNA was amplified by PCR from 77 of 83 (93%) specimens, and these were included for the final analysis. The biopsied nerves were superficial radial nerve = 33 (42.9%); dorsal cutaneous branch of ulnar nerve = 18 (23.4%); sural nerve = 17 (22.1%); and superficial peroneal nerve = 9 (11.7%).
Figure 1.
Schematic diagram of map of India and Nepal. Embedded numbers represent number of leprosy patients from each state in India and from Nepal. Data in boxes indicate the number of resistant patients and the drug. The shaded regions represent the areas endemic for leprosy.
The six patients’ DNA samples not amplified by PCR were also reported to have leprosy by histopathology (healed leprosy = 2, lepra reaction = 1, and borderline tuberculoid [BT] = 3 [one showed acid fast bacilli]). Among the 77 patients, the clinical diagnoses were leprous neuropathy in 70/77 (91%), mononeuritis multiplex in four (6.3%), vasculitic neuropathy in two (3.2%), and distal symmetric sensory motor neuropathy in one (1.6%). Patients were from different states of the country and neighboring nation of Nepal. Family history of leprosy was reported in 5/77 (6.5%) patients. For the entire group, the mean age at the onset of disease, age at presentation, and duration of illness were 34.9 ± 12.6 (range 10–63) years, 38.2 ± 13.4 (range 14–71) years, and 3.4 years (range 1 month to 55 years), respectively. The M: F ratio was 70:7 (10:1). The patient with 55 years duration is a 71-year-old man with recurrence of neuropathic symptoms. This duration was purely based on the patient’s version of his prior chronic neuropathic illness at 16 years of age, when he was prescribed anti-leprosy drugs for 6 months. His past medical records were not available for verification. The majority of patients (43/77, 56.0%) presented between 20 and 40 years of age (60.0%), and 46/77 (60.0%) also had onset between 20 and 40 years. Among the 77 specimens, 72 had a definitive diagnosis by histopathology. One patient demonstrated mild axonopathy in the thickened but unaffected superficial radial nerve. However, fine needle aspiration of the ulnar nerve abscess revealed M. leprae DNA. The six patients with resistance had a mean age at onset of 30.0 ± 16.8 (range 13–61) years, mean age at presentation of 33.0 ± 17.0 (range 14–61) years, and mean duration of illness of 3.1 ± 4.4 (range 1–12) years. Five of these six patients had BT and one had borderline lepromatous (BL) leprosy by histopathology (Table 1). Among the 71 remaining patients with no evidence of resistance, the mean age at onset of symptoms was 35.1 ± 13.0 (range 10–63) years, age at presentation was 38.3 ± 13.8 (range 14–71) years, and duration of illness was 4.7 ± 7.4 years (1 month–2 years). The histopathological diagnosis was as follows: BT = 48, tuberculoid (TT) = 8, BL = 7, mid borderline (BB) = 3, healed/fibrosed = 4, and axonopathy = 1. The place of origin and salient features of the six patients with resistance are summarized in Table 1. All except one patient from Bihar hailed from nonendemic regions for leprosy in India. The demographics and clinical and histopathological features of the group with resistance did not differ from those of the remaining larger drug-sensitive group. No unusual features were encountered in the drug-resistant group.
Table 1.
Salient demographics, and clinical and histopathological characteristics in the six leprosy patients with resistant mutations
| Variables | Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 |
|---|---|---|---|---|---|---|
| Place of origin | Darjeeling, West Bengal | Nepal | Kadapa, Andhra Pradesh | Dharbhanga, Bihar | Dhubri, Assam | Chikkaballapur, Karnataka |
| Year of evaluation | 2008 | 2013 | 2014 | 2015 | 2017 | 2018 |
| Endemic region | No | No | No | Yes | No | No |
| Follow-up in December 2018 | Lost | Lost | Deficits stable. MDT for 12 months 5 years ago. No active disease | Lost | Deficits stable. On MDT for 12 months | Deficits stable. Stopped MDT after the 12-month course |
| Gender | M | M | M | F | M | M |
| Age at presentation (years) | 45 | 22 | 61 | 14 | 32 | 24 |
| Age at onset (years) | 33 | 20 | 60 | 13 | 31 | 22 |
| Duration of illness (months) | 144 | 24 | 18 | 12 | 24 | 48 |
| Affected limb(s) | Lower limbs | Lower limbs | Upper limbs | Right upper limb | Lower limbs | Right upper limb |
| Histopathology diagnosis | BT | BT | BL | BT | BT | BT |
| Biopsied nerve | DCUN | Superficial peroneal nerve | DCUN | DCUN | SRN | SRN |
| Histopathology findings | ||||||
| Plasma cells | – | – | Yes | – | – | – |
| Histiocytes | Yes | Yes | Yes | – | – | – |
| Lymphocytes | Yes | Yes | Yes | Yes | Yes | Yes |
| Perineurial thickening | Yes | Yes | Yes | Yes | Yes | Yes |
| Endoneurial thickening | Yes | Yes | Yes | Yes | Yes | Yes |
| Perivascular inflammation | Yes | Yes | Yes | Yes | Yes | Yes |
| Giant cells | – | – | – | – | Yes | – |
| Foam cells | – | Yes | – | – | – | – |
| Caseous necrosis | – | Yes | – | – | – | – |
| AFB | No | No | Yes | No | No | No |
| Skin lesions | Yes | Yes | No | No | Yes | Yes |
| Hypopigmented patches | Yes | – | – | – | Yes | Yes |
| Nonhealing ulcers | Yes | Yes | – | – | – | – |
| Mutations in drug resistance–determining region | ||||||
| Dapsone | Pro55Ser | – | – | – | – | Pro55Lue |
| Rifampicin | – | Lue436Gln | – | – | – | – |
| Ofloxacin | – | Ala91Pro | Ala91Pro | Gly89Ala | Gly89Cys | Ala91Val |
| Prior drug therapy | None | None | None | None | None | None |
BT = borderline tuberculoid; DCUN = dorsal cutaneous branch of ulnar nerve; SRN = superficial radial nerve.
Resistance mutations were identified in 6/77 (7.8%) specimens: rifampicin (Leu436Gln) combined with ofloxacin (Ala91Pro) in one, dapsone (Pro55Leu) combined with ofloxacin (Ala91Val) in one, only dapsone (Pro55Ser) in one, and only ofloxacin in three patients (Gly89Ala, Ala91Pro, and Gly89Cys) (Table 2).
Table 2.
Mutation pattern of drug resistance for multidrug therapy
| Sl.No | Drugs | Leprosy type | Mutation | Position | Change in nucleotide |
|---|---|---|---|---|---|
| 1 | Rifampicin and ofloxacin | BT | Leu436Gln and Ala91Pro | 279 bp (2275228–2275506) 2275524, 225 bp (7499–7682) | CTG → CAG (T is replaced by A) GCA → CCA (G is replaced by C) |
| 2 | Dapsone and ofloxacin | BT | Pro55Leu and Ala91Val | 254 bp (296800–297025) 296859, 225 bp (7499–7682) | CCC → CTC (C is replaced by T) GCA → GTA (C is replaced by T) |
| 3 | Dapsone | BT | Pro55Ser | 254 bp (296800–297025) 296859 | CCC → TCC (C is replaced by T) |
| 4 | Ofloxacin | BT | Gly89Ala | 225 bp (7499–7682) | GGC → GCC (G is replaced by C) |
| 5 | Ofloxacin | BT | Gly89Cys | 225 bp (7499–7682) | GGC → TGC (G is replaced by T) |
| 6 | Ofloxacin | BL | Ala91Pro | 225 bp (7499–7682) | GCA → GTA (C is replaced by T) |
BT = borderline tuberculoid. Bold indicates position of mutation and change in nucleotide.
The clinically affected nerve was biopsied in 75 (97.4%) patients. The nerve selection, histopathological findings in the biopsied nerve, and its classification are represented in Table 1. AFB was demonstrated in 11 (14.3%) specimens. Skin lesions were reported in 27 patients (35.0%), but examination revealed lesions in 42 patients (54.5%). In addition to nerve biopsy, skin biopsy from the affected area was performed in 15 patients, and this showed the following: BT = 4, BB = 1, TT = 3, nonspecific inflammation = 5, and no changes = 2. Nerve conduction studies carried out in the 77 patients showed sensory motor demyelinating polyradiculoneuropathy in 76 (98.7%) patients and motor sensory axonopathy in 1 (1.3%) patient.
DISCUSSION
This is a 12-year ambispective cross-sectional observational study conducted at a quaternary center for neurological disorders. The retrospective group had 44 patients, from 2007 to 2015, and the prospective group had 33 patients, from 2016 to 2018. The main aim was to look for mutations in the folP1, rpoB, and gyrA genes in M. leprae DNA isolated from 77 biopsied peripheral nerve tissue characterized to have leprosy by histopathology. Both drug-naive and MDT-treated patients were studied, and findings with regard to the resistance pattern for M. leprae along with the demographics and histopathological features have been presented. There were six patients identified to have resistance. Importantly, all patients were drug-naive, indicating primary infection with resistant strains. Among these, one patient came from Bihar which is reported to be an endemic region for leprosy.
Although the prevalence of leprosy in India is less than 1/10,000 since 2005,10 it still accounts for 62% of the total new patients reported worldwide. Emergence of multidrug-resistant M. leprae strains is reported in several studies from Southeast Asia and other parts of the developing world. This indicates a serious threat to effective control of leprosy, particularly in South Asia. It is proposed that development of drug-resistant strains of M. leprae is particularly due to premature discontinuation of treatment and monotherapy.11,12 In India, dapsone monotherapy has been discontinued since 1985. Mycobacterium leprae resistance to dapsone was first reported in 1964,13 for rifampicin in 1976,12 and for ofloxacin in 1996 in nude mice with established M. leprae infection,14 and in the year 2000 in a Japanese patient for dapsone, rifampicin, and sparfloxacin.15
The program for Global Sentinel Surveillance for drug resistance in leprosy was established by the WHO in 2009 to monitor drug resistance among patients of relapse.9 Nine cases of dapsone and one case of rifampicin resistance were detected among 72 leprosy relapse patients.16 In contrast, in our cohort, six (7.8%) new drug-naive leprous neuropathy patients demonstrated resistance to either one or more anti-leprosy drugs. Five of the six patients had BT and one had BL leprosy. Two of our BT patients had resistance to dapsone: one to dapsone alone and the other to dapsone and ofloxacin. The other three BT patients had resistance to rifampicin and ofloxacin and one each to ofloxacin only, and the BL patient to ofloxacin. Thus, the findings suggest that the infection was caused by drug-resistant M. leprae, an indication of emergence of primary resistance. The clinical features, histopathological characteristics, type of leprosy, and treatment regime did not differ between the drug-resistant and drug-sensitive groups.
Point mutations in M. leprae genome responsible for dapsone and rifampicin resistance have been identified in several studies, and this has facilitated the development of new tools for monitoring drug resistance.17,18 In a large study from Tamil Nadu, India, 214 clinically suspected patients of relapse/drug resistance were investigated for MDT susceptibility by mouse foot pad inoculation. Authors identified 15 (16%) secondary dapsone-resistant strains.19 In comparison, two patients with primary resistance to dapsone were identified.
Mycobacterial resistance to rifampicin correlates with changes in the structure of the subunit of the DNA-dependent RNA polymerase, primarily because of missense mutations of the rpoB gene.20 Our single patient with resistance to rifampicin also had a missense mutation but with dual resistance to ofloxacin also.
In a study from Kolkata, India, skin biopsies from 50 patients with a relapse of leprosy were studied for rifampicin resistance. Two patients who had completed 2 years of MB-MDT showed a missense mutation in the CAC codon.21 Our case with resistance to rifampicin also had the same mutation. In a report from Brazil, M. leprae DNA from nerve tissue was tested for mutations in rpoB, folP1, gyrA, and gyrB genes. Mutations in the rpoB and folP1 genes were detected, suggesting multidrug resistance. Seventy percent of the M. leprae mutations were isolated from relapsed patients. No resistance to ofloxacin was detected.22 One of the mutations (Pro55Leu) for dapsone resistance was the same as seen in this study. Thus, the mutations identified for dapsone resistance seem to be common.
In a study carried out in Vietnam, skin smears/skin biopsy samples from 423 leprosy patients showed 19 mutations in 187 folP1 samples, but none related to drug resistance in 163 rpoB and 147 gyrA gene samples was noted. Only two of the 19 were new cases. In comparison, all the six (7.8%) patients with resistance were new cases in the present study group.
Patients with relapse and those who had received dapsone monotherapy in the past have shown high mutation rates (78%), compared with patients with relapse who had previously received MDT (33%).11 In contrast to that report, the findings in this study indicate that resistant strains are causing infection in our group. The first case in the present study had undergone nerve biopsy in 2007, and it means that resistant strains have caused leprosy several years ago. No follow-up information on this patient is available.
In a study from Louisiana, among 39 patients, one case each with dapsone alone and dapsone- and rifampin-resistant M. leprae was detected,23,24 and both were drug naive. Dapsone-resistant patients in this study also had distinct mutations with primary resistance.
In the 1980s, the WHO recommended dapsone, rifampicin, and clofazimine as the primary agents for MDT.4 In this series, none of the patients exhibiting drug resistance had received any anti-leprosy drug(s). There were 28 (36.4%) patients who had received MDT in the past for varying periods before nerve biopsy, and 49 (64.6%) were drug-naive at the time of biopsy.
In a previous study carried out on slit skin smears of 250 samples, from India, MDR M. leprae was detected from relapsed leprosy patients. Seven had showed mutations for rifampicin and dapsone, seven for dapsone and ofloxacin, and one for rifampicin and ofloxacin resistance.25 By contrast, the present cohort was studied for M. leprae DNA in nerve tissue and showed dual resistance in two patients similar to the aforementioned report and single drug resistance in the remaining four. However, in our comparatively smaller sample size, a case with dual rifampicin and ofloxacin resistance was identified, which otherwise is a rare occurrence.
In a worldwide study conducted in 19 countries, including India, 1932 patients were studied for mutations in rpoB, folP1, and gyrA genes. Dual resistance to rifampicin and dapsone and to dapsone and ofloxacin was identified, but none to a combination of rifampicin and ofloxacin. In our cohort, a single case of rifampicin resistance (1/77, 1.3%), who also had resistance to ofloxacin, was identified. A similar case was reported in only one earlier study.25
In the current cohort, the overall resistance rate among 77 patients was 7.8%, similar to the report in the large multinational study. However, individually, it was less for rifampicin (1/77, 1.3%) and dapsone (2/77, 2.6%), except for ofloxacin (4/77, 5.2%) which was similar. The factor(s) responsible for primary resistance despite MDT practice is of immense concern and, thus, an intense surveillance mechanism should be established under the leprosy control program. More studies on Indian leprosy patients should be carried out to identify the burden of resistance. There are a limited number of studies assessing the importance of identification of M. leprae DNA predominantly in the neural form and pure neural leprosy (PNL).26–30 This form of leprosy presents as peripheral neuropathy without skin lesions, and in India and Nepal, it accounts for 3.9–18.0% of all diagnosed patients.31–35 In this study, there were 40 (52.0%) patients qualified for PNL, which is much higher when compared with previous reports, and this could be due to a referral bias. PCR identified M. leprae DNA in all these patients.
With the findings of resistance in the cohort, it may be valuable to subject tissue samples for PCR testing from all suspected leprosy patients and also to look for the presence of drug resistance. In patients with TT leprosy, histopathological confirmation is not certain as AFB is uncommonly seen and nonspecific inflammation is more frequent. In such situations, subjecting the nerve tissue for PCR for Mycobacterium lepra identification may be considered. Furthermore, in all pure neuritic leprosy patients identification of M. lepra DNA by PCR will confirm the diagnosis as mislabeling leprosy as chronic inflammatory demyelinating neuropathy, neurosarcoidosis, and vasculitis could unnecessarily lead to long-term incorrect therapy and, thus, adverse effects of immunomodulation.
In conclusion, the current series presents the pattern of drug resistance identified in a large cohort of leprosy patients with primarily neural form of leprosy. For the first time, sequencing for the DRDR for MDT drugs was performed on DNA isolated from nerve tissue in India. In this study, 6/77 patients with drug resistance and two patients with dual resistance were identified. PCR testing for identification of M. leprae DNA is a simple, fast, and affordable method of confirming leprosy and should be taken up based on the type of leprosy. Furthermore, the isolated M. leprae DNA should be subjected to determination of drug resistance so that treatment strategies can be modified accordingly and also the spread of resistant strains can be curtailed.
Acknowledgments:
We immensely thank all the patients for permitting us to publish their medical data. The American Society of Tropical Medicine and Hygiene (ASTMH) assisted with publication expenses.
REFERENCES
- 1.Croft RP, Nicholls PG, Richardus JH, Smith WC, 2000. Incidence rates of acute nerve function impairment in leprosy: a prospective cohort analysis after 24 months (The Bangladesh Acute Nerve Damage Study). Lepr Rev 71: 18–33. [DOI] [PubMed] [Google Scholar]
- 2.Saunderson P, Gebre S, Desta K, Byass P, Lockwood DN, 2000. The pattern of leprosy-related neuropathy in the AMFES patients in Ethiopia: definitions, incidence, risk factors and outcome. Lepr Rev 71: 285–308. [DOI] [PubMed] [Google Scholar]
- 3.van Brakel WH, Nicholls PG, Das L, Barkataki P, Suneetha SK, Jadhav RS, Maddali P, Lockwood DNJ, Wilder-Smith E, Desikan KV, 2005. The INFIR cohort study: investigating prediction, detection and pathogenesis of neuropathy and reactions in leprosy. Methods and baseline results of a cohort of multibacillary leprosy patients in north India. Lepr Rev 76: 14–34. [PubMed] [Google Scholar]
- 4.WHO Study Group on Chemotherapy of Leprosy for Control Programmes, WHO , 1982. Chemotherapy of Leprosy for Control Programmes: Report of a WHO Study Group. Meeting, October12–16, 1981. Geneva, Switzerland: World Health Organization, Available at: http://apps.who.int/iris/handle/10665/38984. Accessed January 6, 2019. [Google Scholar]
- 5.Cambau E, Perani E, Guillemin I, Jamet P, Ji B, 1997. Multidrug-resistance to dapsone, rifampicin, and ofloxacin in Mycobacterium leprae. Lancet Lond Engl 349: 103–104. [DOI] [PubMed] [Google Scholar]
- 6.Williams DL, Gillis TP, 2012. Drug-resistant leprosy: monitoring and current status. Lepr Rev 83: 269–281. [PubMed] [Google Scholar]
- 7.Benjak A, et al. 2018. Phylogenomics and antimicrobial resistance of the leprosy bacillus Mycobacterium leprae. Nat Commun 9: 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson R, 2014. Neglected tropical diseases take hold in the USA. Lancet Infect Dis 14: 1050–1051. [DOI] [PubMed] [Google Scholar]
- 9.WHO, Regional Office for South-East Asia , 2009. Guidelines for Global Surveillance of Drug Resistance in Leprosy. WHO Regional Office for South-East Asia, Available at: http://apps.who.int/iris/handle/10665/205158. Accessed January 6, 2019. [Google Scholar]
- 10.Desikan KV, 2012. Elimination of leprosy & possibility of eradication - the Indian scenario. Indian J Med Res 135: 3–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kai M, et al. 2011. Analysis of drug-resistant strains of Mycobacterium leprae in an endemic area of Vietnam. Clin Infect Dis 52: e127–e132. [DOI] [PubMed] [Google Scholar]
- 12.Jacobson RR, Hastings RC, 1976. Rifampin-resistant leprosy. Lancet Lond Engl 2: 1304–1305. [DOI] [PubMed] [Google Scholar]
- 13.Pettit JH, Rees RJ, 1964. Sulphone resistance IN leprosy. AN experimental and clinical study. Lancet Lond Engl 2: 673–674. [DOI] [PubMed] [Google Scholar]
- 14.Ji B, Perani EG, Petinom C, Grosset JH, 1996. Bactericidal activities of combinations of new drugs against Mycobacterium leprae in nude mice. Antimicrob Agents Chemother 40: 393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuoka M, Kashiwabara Y, Namisato M, 2000. A Mycobacterium leprae isolate resistant to dapsone, rifampin, ofloxacin and sparfloxacin. Int J Lepr Other Mycobact Dis 68: 452–455. [PubMed] [Google Scholar]
- 16.Surveillance of Drug Resistance in Leprosy , 2010. Weekly Epidemiological Record (WER). Vol. 86, 233–240. WHO; Available at: https://www.who.int/wer/2011/wer8623/en/. Accessed January 6, 2019. [Google Scholar]
- 17.Williams DL, Spring L, Harris E, Roche P, Gillis TP, 2000. Dihydropteroate synthase of Mycobacterium leprae and dapsone resistance. Antimicrob Agents Chemother 44: 1530–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cambau E, Carthagena L, Chauffour A, Ji B, Jarlier V, 2006. Dihydropteroate synthase mutations in the folP1 gene predict dapsone resistance in relapsed cases of leprosy. Clin Infect Dis 42: 238–241. [DOI] [PubMed] [Google Scholar]
- 19.Sekar B, Elangeswaran N, Jayarama E, Rajendran M, Kumar SS, Vijayaraghavan R, Anandan D, Arunagiri K, 2002. Drug susceptibility of Mycobacterium leprae: a retrospective analysis of mouse footpad inoculation results from 1983 to 1997. Lepr Rev 73: 239–244. [PubMed] [Google Scholar]
- 20.Honore N, Cole ST, 1993. Molecular basis of rifampin resistance in Mycobacterium leprae. Antimicrob Agents Chemother 37: 414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasanoor Reja AH, et al. 2015. Report of rpoB mutation in clinically suspected cases of drug resistant leprosy: a study from Eastern India. Indian J Dermatol Venereol Leprol 81: 155–161. [DOI] [PubMed] [Google Scholar]
- 22.Jardim MR, et al. 2003. Criteria for diagnosis of pure neural leprosy. J Neurol 250: 806–809. [DOI] [PubMed] [Google Scholar]
- 23.Williams DL, Hagino T, Sharma R, Scollard D, 2013. Primary multidrug-resistant leprosy, United States. Emerg Infect Dis 19: 179–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams DL, Lewis C, Sandoval FG, Robbins N, Keas S, Gillis TP, Scollard DM, 2014. Drug resistance in patients with leprosy in the United States. Clin Infect Dis 58: 72–73. [DOI] [PubMed] [Google Scholar]
- 25.Lavania M, et al. 2018. Molecular detection of multidrug-resistant Mycobacterium leprae from Indian leprosy patients. J Glob Antimicrob Resist 12: 214–219. [DOI] [PubMed] [Google Scholar]
- 26.Dharmendra, Ramanujam K, 1966. Pure polyneuritic leprosy of tuberculoid type. Lepr India 38: 152–158. [Google Scholar]
- 27.Noordeen SK, 1972. Epidemiology of (poly) neuritic type of leprosy. Lepr India 44: 90–96. [Google Scholar]
- 28.Nations SP, Katz JS, Lyde CB, Barohn RJ, 1998. Leprous neuropathy: an American perspective. Semin Neurol 18: 113–124. [DOI] [PubMed] [Google Scholar]
- 29.Pannikar VK, Arunthathi S, Chacko CJ, Fritschi EP, 1983. A clinico-pathological study of primary neuritic leprosy. Lepr India 55: 212–221. [PubMed] [Google Scholar]
- 30.Suneetha S, Arunthathi S, Chandi S, Kurian N, Chacko CJ, 1998. Histological studies in primary neuritic leprosy: changes in the apparently normal skin. Lepr Rev 69: 351–357. [DOI] [PubMed] [Google Scholar]
- 31.Kaur G, Girdhar BK, Girdhar A, Malaviya GN, Mukherjee A, Sengupta U, Desikan KV, 1991. A clinical, immunological, and histological study of neuritic leprosy patients. Int J Lepr Mycobact Dis 59: 385–391. [PubMed] [Google Scholar]
- 32.Rao PN, Suneetha S, 2016. Pure neuritic leprosy: current status and relevance. Indian J Dermatol Venereol Leprol 82: 252–261. [DOI] [PubMed] [Google Scholar]
- 33.Wilder-Smith E, 2002. Diagnosis of pure neuritic leprosy. Neurol J Southeast Asia 7: 61–63. [Google Scholar]
- 34.Hui M, Uppin MS, Challa S, Meena AK, Kaul S, 2015. Pure neuritic leprosy: resolving diagnostic issues in acid fast bacilli (AFB)-negative nerve biopsies: a single centre experience from South India. Ann Indian Acad Neurol 18: 292–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ooi WW, Srinivasan J, 2004. Leprosy and the peripheral nervous system: basic and clinical aspects. Muscle Nerve 30: 393–409. [DOI] [PubMed] [Google Scholar]