Abstract
Two isolates of a Gram-negative, non-spore-forming coccobacillus cultured from the blood and cerebrospinal fluid of immunocompromised patients in the United States were described previously. Biochemical and phylogenetic analyses revealed that they belong to a novel species within the Francisella genus. Here we describe a third isolate of this species, recovered from blood of a febrile patient with renal failure, and formally name the Francisella species. Whole genome comparisons indicated the three isolates display greater than 99.9% average nucleotide identity (ANI) to each other and are most closely related to the tick endosymbiont F. persica, with only 88.6–88.8% ANI to the type strain of F. persica. Based on biochemical, metabolic and genomic comparisons, we propose that these three isolates should be recognized as Francisella opportunistica sp. nov, with the type strain of the species, PA05–1188T, available through the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSM 107100) and the American Type Culture Collection (ATCC BAA-2974).
The genus Francisella currently comprises seven named species, notably including F. tularensis, the causative agent of tularemia. It also includes the rare or opportunistic human pathogens F. novicida (also validly published as F. tularensis subsp. novicida [1–3]), F. philomiragia, and F. hispaniensis, as well as pathogens of aquatic organisms (F. noatunensis and F. halioticida) and tick endosymbionts (F. persica and other unclassified Francisella) [1, 2, 4–7].
We previously described two isolates of a novel Francisella species cultured from blood and cerebrospinal fluid of two US patients with immune compromising conditions [8]. One of these isolates, MA06–7296, was fully sequenced and provisionally named F. opportunistica [9]. The 16S rRNA and sdhA genes from the first isolate, PA05–1188, have also been sequenced and shown to be identical to those of MA06–7296 [8]. A third isolate, 14–2155, not previously described in the literature, was cultured in 2014 from blood of a febrile patient in Arizona with type II diabetes, renal failure, and cardiopulmonary failure.
As previously described, the F. opportunistica isolates are indole and catalase negative, oxidase positive, and urease negative (Table 1). All three are beta-lactamase negative, which is a unique characteristic among Francisella species described to date. Although MA06–7296 was previously reported as urease weak, this was not confirmed in subsequent testing [8]. F. opportunistica isolates do not grow in 6.5% NaCl and require cysteine supplementation for optimal growth [8, 9].
Table 1. Phenotypic characterization.
W, weak. V, variable among tested strains. See text for reactions identical among all Francisella tested.
| F. opportunistica | F. tularensis | F. novicida | F. philomiragia | |
|---|---|---|---|---|
| Oxidase* | + | − | − | + |
| Catalase* | − | W | W | W |
| Indole* | − | − | − | + |
| Urease* | − | − | − | − |
| β-lactamase* | − | + | + | + |
| α-cyclodextrin | − | − | V | − |
| Dextrin | + | V | V | V |
| Glycogen | + | V | V | V |
| Tween-40 | + | − | V | − |
| N-acetyl-D-glucosamine | + | V | + | + |
| Adonitol | − | − | V | − |
| L-arabinose | − | V | V | V |
| D-cellobiose | − | V | V | V |
| i-erythritol | − | − | V | V |
| D-galactose | + | V | + | V |
| lactulose | − | − | V | − |
| maltose | − | − | V | V |
| D-mannose | + | V | + | + |
| β-methyl-D-glucoside | − | − | − | V |
| D-psicose | − | V | V | V |
| sucrose | − | − | + | + |
| D-trehalose | − | − | + | V |
| turanose | − | − | V | − |
| mono-methyl-succinate | + | V | + | + |
| acetic acid | + | V | + | V |
| cis-aconitic acid | − | − | V | V |
| formic acid | − | − | V | − |
| D-galacturonic acid | − | − | − | V |
| α-hydroxybutyric acid | − | V | + | V |
| β-hydroxybutyric acid | + | − | + | + |
| itaconic acid | − | − | V | V |
| α-keto glutaric acid | + | V | V | V |
| α-keto valeric acid | V | − | − | − |
| D,L-lactic acid | + | V | + | + |
| succinic acid | + | V | + | V |
| bromosuccinic acid | + | V | V | V |
| succinamic acid | − | V | − | V |
| L-alaninamide | + | V | + | + |
| D-alanine | + | V | + | + |
| L-alanine | + | V | + | + |
| L-alanyl-glycine | + | V | + | + |
| L-asparagine | + | V | + | + |
| L-aspartic acid | + | V | + | + |
| L-glutamic acid | + | V | + | + |
| glycyl-L-aspartic acid | − | V | V | V |
| glycyl-L-glutamic acid | − | V | + | V |
| hydroxy-L-proline | + | − | V | − |
| L-ornithine | + | − | + | V |
| L-proline | + | V | + | + |
| L-pyroglutamic acid | + | − | + | + |
| D-serine | − | V | − | − |
| L-threonine | + | V | + | + |
| D,L-carnitine | − | − | − | V |
| urocanic acid | − | V | − | − |
| inosine | + | − | + | V |
| uridine | − | V | + | + |
| thymidine | − | V | V | V |
| 2,3-butanediol | − | − | V | − |
| glycerol | + | V | + | V |
| D,L-α-glycerol phosphate | + | + | + | V |
| glucose-1-phosphate | − | − | V | V |
| glucose-6-phosphate | + | V | V | V |
Reactions marked with an asterisk are not on the Biolog MicroLog GN2 plates, and were run separately for F. opportunistica isolates only. Results of these reactions for other Francisella species were compiled from the literature.
The PA05–1188 and MA06–7296 isolates were further tested for metabolic characteristics using the Biolog MicroLog system (Gram-negative GN2 MicroPlates), and compared to a selection of other Francisella isolates (Table 1). These included F. novicida (n = 11), F. philomiragia (n = 5), F. tularensis subsp. tularensis (n = 6, including five human and animal isolates and the attenuated strain ATCC 6223), and F. tularensis subsp. holarctica (n = 9, including eight human and animal isolates and the attenuated strain LVS). Results for the two subspecies of F. tularensis are combined in Table 1, as they cannot be consistently distinguished by any metabolic characteristic except the ability to metabolize glycerol. All tested Francisella strains were negative for tween-80, N-acetyl-D-galactosamine, D-arabitol, L-fucose, gentiobiose, m-inositol, α-D-lactose, D-mannitol, D-melibiose, D-raffinose, L-rhamnose, D-sorbitol, xylitol, citric acid, D-galactonic acid lactone, D-gluconic acid, D-glucosaminic acid, D-glucuronic acid, γ-hydroxybutyric acid, p-hydroxyphenylacetic acid, malonic acid, propionic acid, quinic acid, D-saccharic acid, sebacic acid, glucuronamide, L-histidine, L-leucine, L-phenylalanine, γ-aminobutyric acid, phenylethylamine, putrescine, and 2-aminoethanol. All tested strains were positive for D-fructose, α-D-glucose, methyl pyruvate, α-ketobutyric acid, and L-serine. PA05–1188 was positive for α-ketovaleric acid and L-aspartic acid, while MA06–7296 was negative. Results that vary among Francisella species are shown in Table 1. No single metabolic reaction on the MicroLog GN2 plate is unique to F. opportunistica; however, the overall pattern of reactivity is characteristic.
To further clarify the relationships among the three isolates and their position in the Francisella genus, we sequenced their full genomes using both PacBio and Illumina technology. For PacBio sequencing, 5 μg of input DNA was used for 10-kb fragment library preparation, and sequencing was performed on the PacBio RSII platform and assembled using the Hierarchical Genome Assembly Process (HGAP). For Illumina sequencing, libraries were prepared using the Nextera XT DNA Library Prep kit, and sequencing was performed on the MiSeq platform with a 300-cycle V2 reagent kit. Sequencing statistics and GenBank accession numbers are listed in Table 2. De novo assembly of PacBio reads yielded a single complete circular contig encompassing the chromosome for each isolate. Illumina sequences were assembled both using PacBio sequences as a reference and de novo using SPAdes 3.10.1 [10]. No plasmids were identified by either technology. Six errors in the PacBio consensus, all in 14–2155, were corrected by assembly of Illumina sequences to the PacBio consensus. Final sequences used for analysis and submitted to GenBank represent the corrected sequence obtained by comparison of both methods. All three genomes are approximately 1.8 Mb in length, with 32.5% GC content.
Table 2.
Genome sequencing
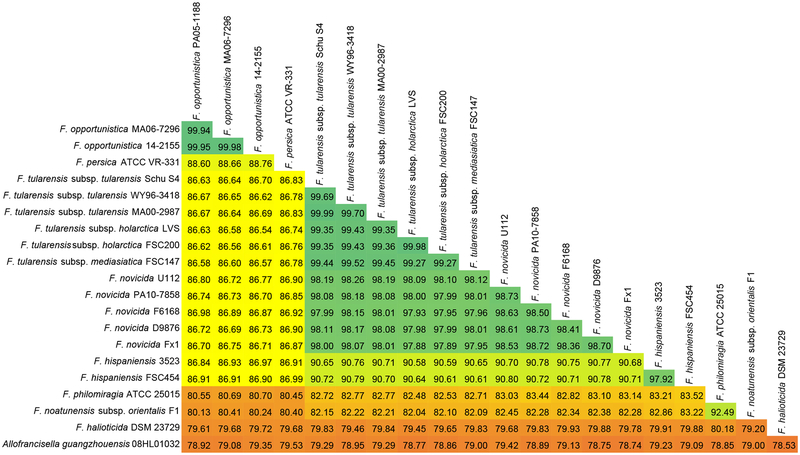
Average nucleotide identity (ANI) was calculated using the ANI calculator tool (http://enve-omics.ce.gatech.edu/ani/) with default settings of 1000 bp window size, 200 bp step size, and 70% identity over 700 bp [11]. The three genomes displayed >99% ANI to each other and <90% ANI to any other Francisella species, consistent with their designation as three isolates of a distinct Francisella species (Figure 1) [11]. The most closely related species is F. persica (formerly Wolbachia persica), a tick endosymbiont, with approximately 89% ANI to each of the three isolates [7] (Figure 1). The three F. opportunistica genomes displayed approximately 87% ANI to F. tularensis, F. novicida, and F. hispaniensis genomes. These analyses are consistent with the previously described clustering of F. opportunistica MA06–7296 with F. persica, based on phylogenetic analysis of 36 conserved housekeeping proteins [9, 12].
Fig. 1.
Distance matrix showing ANI among Francisellaceae family members. Matrix is color-coded according to ANI value, with the highest values shown in green and the lowest in orange.
Genomes were annotated using the NCBI Prokaryotic Genome Annotation Pipeline [13]. As previously described for PA05–1188T and MA06–7296, the three isolates show 100% identity in their 16S rRNA and sdhA gene sequences [8]. The F. opportunistica 16S rRNA and sdhA genes display 97.6% and 84.0% pairwise nucleotide identity, respectively, to their homologs in F. tularensis Schu. Sporadic polymorphisms vary between the three genomes. Single nucleotide polymorphisms are largely concentrated in a few genomic regions, including the Francisella pathogenicity island (FPI; see below), a type I restriction-modification system, and a region that contains multiple genes annotated as being involved in sulfur metabolism. The MA06–7296 and 14–2155 nucleotide sequences are identical in all three of these regions, while PA05–1188T contains multiple polymorphisms.
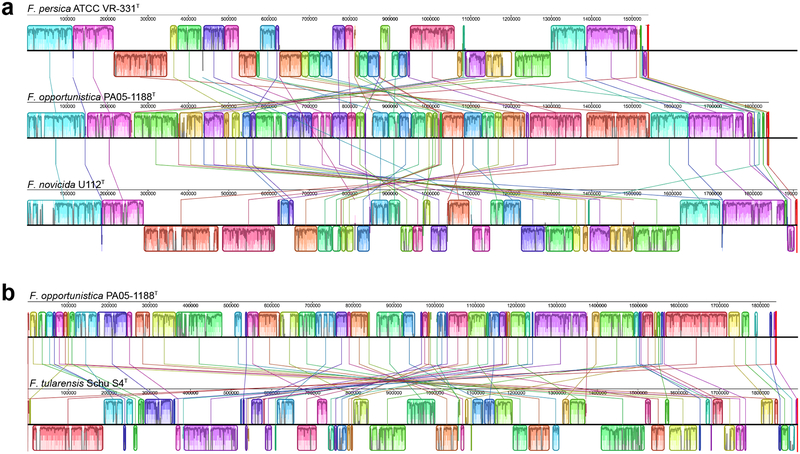
The three F. opportunistica genomes were aligned using the progressiveMauve algorithm within Geneious 11.0.4 software, and found to be collinear and extensively rearranged compared to other sequenced Francisella species (Figure 2) [14, 15]. IS elements were analyzed using ISFinder, and the sequences were deposited in the ISFinder database [16]. Six IS elements are polymorphic in F. opportunistica, with PA05–1188T containing the largest number of unique IS element insertions (Table 3). The most frequent of these elements is 99.2% identical in its consensus sequence to ISFw2, an element previously described in the German environmental Francisella isolate W12–1067. Three additional elements are homologous to ISFtu elements named in F. tularensis. ISFop2 is a homolog of ISFtu2, with 69.5% amino acid similarity (52.2% identity). Interestingly, a single copy of a more closely related ISFtu2 element (93.5% similarity/87.1% identity) is present and conserved in all three genomes. A truncated element found only in PA05–1188 is homologous to ISFtu4, with 71.9% amino acid similarity (66.1% identity). ISFop1 shows some limited similarity to ISFtu1 (39.2% similarity/19.4% identity).
Fig. 2.
Genome structure comparisons showing extensive rearrangement. (A) F. persica, F. opportunistica, and F. novicida. (B) F. opportunistica and F. tularensis (shown separately to highlight more extensive rearrangement). Locally collinear blocks are color coded, and local average nucleotide conservation is shown in graphs within the blocks. Homologous blocks are connected by colored lines. Blocks above the black line are in forward orientation compared to F. opportunistica, and those below the line are in reverse orientation. Figure was generated using progressiveMauve and Geneious 11.0.4 [14, 15, 19].
Table 3.
IS elements polymorphic in F. opportunistica
| Name | Family | Group | # conserved | # unique to PA05–1188 | # unique to MA06–7296 | # unique to 14–2155 | Closest Francisellaceae homologa |
|---|---|---|---|---|---|---|---|
| ISFw2 | IS5 | IS5 | 9 | 5 | 0 | 0 | Francisella sp. W12–1067 element (99% nt/100% aa identity) |
| ISFop1 | IS630 | N/A | 1 | 1 | 0 | 0 | Unnamed Allofrancisella element |
| ISFop2 | IS5 | IS427 | 11 | 1 | 0 | 1 | ISFtu2 (69.5% aa similarity) |
| ISFop3 | IS110 | N/A | 5 | 2 | 0 | 0 | Unnamed element in F. novicida (some strains), and F. uliginis |
| ISFop4 | IS3 | IS150 | 2 | 2 | 3 | 0 | None |
| Truncated elementb | IS982 | N/A | 0 | 1 | 0 | 0 | ISFtu4 (71.9% aa similarity) |
Homologs were found by BLAST searches limited to Francisellaceae. Amino acid similarity is given for defined, named elements listed in ISFinder.
This element is truncated and was not able to be annotated within ISFinder.
The FPI in each of the F. opportunistica genomes is present in a single copy and lacks the pdpC and pdpE genes, as described previously [9]. Sequences corresponding to these genes were found elsewhere in the genomes, outside the FPI, by BLAST search. The pdpE gene is intact in all three genomes, but pdpC is pseudogenized. Notably, in F. persica and other Francisella-like endosymbionts, pdpC and pdpE are present in the FPI region, but flanked by IS elements not present in either copy of the F. tularensis FPI, suggesting that these two genes have been subject to transposition [17].
The absence of beta-lactamase activity detected by biochemical testing is unusual among Francisella species. F. tularensis genomes include two annotated class A beta-lactamase genes, one of which is disrupted by insertion sequences in F. opportunistica. Interestingly, the same gene is also disrupted in F. persica, suggesting that this species may also lack beta-lactamase activity.
Taken together, these phenotypic and whole genome analyses reveal a novel Francisella species, most closely related to the endosymbiont F. persica. The genomes of the three isolates display >99% ANI to each other and are collinear, but contain differences in IS element content and nucleotide sequence in certain genomic regions, including the FPI. The isolation of F. opportunistica from three ill patients with immune compromising conditions in different regions of the United States (Pennsylvania, Massachusetts, and Arizona) suggests that the species is widespread. Correct identification of rare Francisella species is critical in order to avoid biosafety and biosecurity concerns associated with F. tularensis. Clinical laboratories should be aware of the possibility of infection with non-tularensis Francisella spp., especially in patients with immune compromising conditions [8, 18]. F. opportunistica can be distinguished from F. tularensis by beta-lactamase, antigen detection (DFA and slide agglutination) and by molecular methods including PCR and sequencing [8].
Description of Francisella opportunistica sp. nov.
F. opportunistica (op.por.tu.nis’ti.ca N.L. fem. adj. opportunistica, opportunistic, referring to the organism’s ability to opportunistically infect immunocompromised humans). The type strain, PA05–1188, was isolated in 2005 from a patient with hemophagocytic syndrome and juvenile rheumatoid arthritis; the MA06–7296 and 14–2155 strains were isolated in 2006 and 2014, respectively, from patients with end-stage renal disease. Cells are small Gram-negative coccobacilli. They are catalase negative, oxidase positive, and beta-lactamase negative. Other biochemical and metabolic characteristics are as shown in Table 1. Slide agglutination and direct fluorescent antibody assay for F. tularensis are both negative. [8, 9]. Whole genome sequence information for all three strains is available from GenBank with accession numbers CP022375 (14–2155), CP022376 (MA06–7296), and CP022377 (PA05–1188T). The genome length is approximately 1.8 Mb, with a GC content of 32.5%. The three isolates display >99% ANI to each other and <89% ANI to any other species, with the most closely related species being F. persica. They are 100% identical across the 16S rRNA and sdhA genes, with 97.6% identity to F. tularensis 16S rRNA and 84.0% identity to F. tularensis sdhA. The type strain, PA05–1188T, is available through the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSM 107100) and the American Type Culture Collection (ATCC BAA-2974).
Acknowledgments
We thank Elaine McCaffrey for initial phenotypic characterization of 14-2155, and Christopher Sexton and Brook Yockey for assistance with Biolog data. PacBio sequencing was performed by the CDC Genome Sequencing Lab, with special thanks to Dhwani Batra and Lori Rowe.
Funding Information
This work received no specific grant from any funding agency.
Footnotes
Ethical Approval
Patient follow-up was conducted as part of routine public health surveillance. Characterization of bacterial isolates derived from human specimens was approved by the CDC Institutional Review Board.
Conflict of Interest
The authors declare that there are no conflicts of interest. The findings and conclusions in this manuscript are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
References
- 1.Huber B, Escudero R, Busse HJ, Seibold E, Scholz HC et al. Description of Francisella hispaniensis sp. nov., isolated from human blood, reclassification of Francisella novicida (Larson et al. 1955) Olsufiev et al. 1959 as Francisella tularensis subsp. novicida comb. nov. and emended description of the genus Francisella. Int J Syst Evol Microbiol 2010;60(Pt 8):1887–1896. [DOI] [PubMed] [Google Scholar]
- 2.Johansson A, Celli J, Conlan W, Elkins KL, Forsman M et al. Objections to the transfer of Francisella novicida to the subspecies rank of Francisella tularensis. Int J Syst Evol Microbiol 2010;60:1717–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kingry LC, Petersen JM. Comparative review of Francisella tularensis and Francisella novicida. Frontiers in cellular and infection microbiology 2014;4(March):35–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hollis DG, Weaver RE, Steigerwalt AG, Wenger JD, Moss CW et al. Francisella philomiragia comb. nov. (formerly Yersinia philomiragia) and Francisella tularensis biogroup novicida (formerly Francisella novicida) associated with human disease. J Clin Microbiol 1989;27(7):1601–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brevik OJ, Ottem KF, Kamaishi T, Watanabe K, Nylund A. Francisella halioticida sp. nov., a pathogen of farmed giant abalone (Haliotis gigantea) in Japan. J Appl Microbiol 2011;111:1044–1056. [DOI] [PubMed] [Google Scholar]
- 6.Ottem KF, Nylund A, Karlsbakk E, Friis-Møller A, Kamaishi T. Elevation of Francisella philomiragia subsp. noatunensis Mikalsen et al. (2007) to Francisella noatunensis comb. nov. [syn. Francisella piscicida Ottem et al. (2008) syn. nov.] and characterization of Francisella noatunensis subsp. orientalis subsp. nov., two important fish pathogens. J Appl Microbiol 2009;106:1231–1243. [DOI] [PubMed] [Google Scholar]
- 7.Larson MA, Nalbantoglu U, Sayood K, Zentz EB, Cer RZ et al. Reclassification of Wolbachia persica as Francisella persica comb. nov. and emended description of the family Francisellaceae. Int J Syst Evol Microbiol 2016;66(3):1200–1205. [DOI] [PubMed] [Google Scholar]
- 8.Kugeler KJ, Mead PS, McGowan KL, Burnham JM, Hogarty MD et al. Isolation and characterization of a novel Francisella sp. from human cerebrospinal fluid and blood. J Clin Microbiol 2008;46(7):2428–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Challacombe JF, Petersen JM, Gallegos-Graves V, Hodge D, Pillai S et al. Whole-Genome Relationships among Francisella Bacteria of Diverse Origins Define New Species and Provide Specific Regions for Detection. Appl Environ Microbiol 2017;83(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012;19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P et al. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 2007;57(Pt 1):81–91. [DOI] [PubMed] [Google Scholar]
- 12.Ahlinder J, Ohrman C, Svensson K, Lindgren P, Johansson A et al. Increased knowledge of Francisella genus diversity highlights the benefits of optimised DNA-based assays. BMC Microbiol 2012;12:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 2016;44(14):6614–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 2010;5(6):e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012;28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 2006;34(Database issue):D32–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerhart JG, Dutcher HA, Brenner AE, Moses AS, Grubhoffer L et al. Multiple acquisitions of pathogen-derived Francisella endosymbionts in soft ticks. Genome Biol Evol 2018;10(2):607–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brett ME, Respicio-Kingry LB, Yendell S, Ratard R, Hand J et al. Outbreak of Francisella novicida bacteremia among inmates at a Louisiana correctional facility. Clinical Infectious Diseases 2014;59(6):826–833. [DOI] [PubMed] [Google Scholar]
- 19.Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: Multiple Alignment of Conserved Genomic Sequence With Rearrangements. Genome Res 2004;14(7):1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]