Abstract
Tissue survival responses to ionizing radiation are nonlinear with dose, rather yielding tissue-specific descending curves that impede straightforward analysis of biologic effects. Apoptotic cell death often occurs at low doses while at clinically relevant intermediate doses double-strand break misrepair yields mitotic death that determines outcome. As researchers frequently use a single low dose for experimentation, such strategies may inaccurately depict inherent tissue responses. Cutting edge radiobiology has adopted full dose survival profiling and devised mathematical algorithms to fit curves to observed data to generate highly reproducible numerical data that accurately define clinically relevant inherent radiosensitivities. Here we established a protocol for irradiating organoids that delivers radiation profiles simulating the organ of origin. This technique yielded highly similar dose survival curves of small and large intestinal crypts in vivo and their cognate organoids analyzed by the single hit multi-target (SHMT) algorithm, outcomes reflecting the inherent radiation profile of their respective Lgr5+ stem cell populations. As this technological advance is quantitative, it will be useful for accurate evaluation of intestinal (patho)physiology and drug screening.
Keywords: stem cells, organoids, irradiation, intestines, Lgr5
Introduction
An extensive literature shows that organoids derived from small and large intestines recapitulate structural and functional characteristics of the tissue of origin (1,2). The purpose of the current set of investigations was to define conditions for delivery of radiation to intestinal organoids that would accurately reflect impact of ionizing radiation on the organ of origin. In contrast to the many biologic events that can be defined by straight lines that relate input to outcome, for instance ligand-receptor binding, Michaelis-Menten enzyme kinetic analysis, and stimulus-secretion coupling of hormone, mitogen and cytokine release to list a few, ionizing radiation-induced dose survival data in eukaryotic cells are best fit by non-linear descending curves, analyzed by complex mathematical algorithms. Whereas radiation is increasingly employed as a tool in biological and translational research, frequently conducted by scientists who may not have had formal radiobiologic research training, we highlight here straightforward methods and data analytic techniques to facilitate high quality radiobiologic data management using organoid culture. The specific approach taken in the present studies was to exploit recently-detailed in vivo small and large intestinal crypt radiation dose-survival data to establish organoid growth settings that yield comparable radiation dose survival curves ex vivo (3). We provide a set of growth conditions for radiation dose survival colony assays and a simple analytic tool that yields a single quantitative readout that defines the inherent radiosensitivity of the organ of origin from which organoids were derived. Using these protocols, we provide evidence in organoid culture that the established radiation sensitivities of the small and large intestine reflect the inherent sensitivity of their Lgr5+ stem cell populations.
Materials and Methods
Mice:
Lgr5-lacZ and Lgr5-EGFP-ires-CreERT mice were used as described (4). C57/Bl6 mice were purchased from Jackson Laboratories (Bar Harbor, ME). Mouse protocols were approved by Memorial Sloan Kettering Cancer Center Institutional Animal Care and Use Committee.
Crypt Microcolony Assay:
Whole-body radiation was delivered with a Shepherd Mark-I unit (Model 68, SN643, J. L. Shepherd & Associates, San Fernando, CA) operating a 137Cs source. The Crypt Microcolony Assay was performed as described (5). Briefly, at 3.5 days after irradiation small intestines were excised, fixed in 10% formalin overnight, embedded in paraffin, cut into transverse cross-sections, and circumferences were stained with hematoxylin and eosin. Surviving (regenerating) crypts are defined as containing 10 or more violaceous, thick-walled non-Paneth cells that are enclosed with a lumen. Number of surviving crypts was counted in each circumference. At least 4 or 5 circumferences were scored per mouse, and 4 mice were used to generate each data point.
Crypt isolation:
Crypt isolation and the dissociation of cells for flow cytometry were performed as described (6,7) modified as follows. Small intestine (approx. 10 cm) and distal colon were removed and flushed with cold phosphate buffer saline (PBS) containing 100 U/ml penicillin and 100 U/ml streptomycin. Colonic fragments were sliced into 1–3 mm pieces, then suspended in 10 ml of Dulbecco’s Modified Eagle Medium (DMEM high glucose) containing 1% FBS, 500 U/ml collagenase IV. The mixture was incubated for 30 min at 37°C in a shaking water bath. Thereafter tissue fragments were vigorously shaken using a 10-ml pipette to isolate crypts, allowed to settle under gravity for 1 min, and the supernatant was removed for inspection by inverted microscopy. The resuspension/sedimentation procedure was repeated three times, liberated crypts in suspension were combined, passed through 100 μm-cell strainer to remove muscle material, and then centrifuged at 4°C at 300g for 5 min. Isolated crypts were pelleted, washed with cold 1% FBS/DMEM, and centrifuged three times at 60g for 3 min to separate crypts from single cells. For small intestine, tissue was chopped into 3–5 mm pieces and incubated in 5 mM EDTA/PBS for 20–30 min on ice. After removal of EDTA buffer, tissue was washed once with cold PBS and vigorously resuspended in cold buffer using a 10-mL pipette to isolate intestinal crypts. This step was repeated twice. Supernatant was collected through a 100 μm cell strainer and centrifuged at 300g for 5 min. SI crypts were washed in cold PBS three times centrifuging at 60g for 3 min.
Single stem cell isolation:
Single epithelial cell preparation and flow cytometry analysis were performed as described (8) with the modifications below. For the large intestine we used what we called the “sausage protocol”. The whole large intestine was tied on one end with surgical thread, filled with 30 mM EDTA/PBS with 1 mM DTT and tied on the opposite end afterwards. Five colons were pooled and incubated in the same solution (30 mM EDTA in PBS with 1 mM DTT) for 90 min in a rocking platform at 4°C. After this incubation, knots were cut out, large intestines were washed once, fat was discarded, tissues were opened longitudinally, cut into 1–2 mm pieces and shaken in PBS for 5 min to get single crypts. For small intestine, tissue fragments (3–5 mm) were incubated with 30 mM EDTA/PBS with 1 mM DTT for 20 min on ice, followed by washing with PBS and shaken for 3 min to get single crypts. For both tissues, after shaking, crypt suspension was passed through a 100 μm cell strainer and spin down twice at 300 g for 5 min. Pelleted crypts were dissociated in 2–3 ml of TryplE containing 200 U/ml DNAase, 0.5 mM N-acetylcysteine (NaC) and 10 μmol/L Y-27632 for 3 min at 37°C in a water bath, shaking every minute to make a single cell suspension. Dissociated cells were washed with 1% FBS/DMEM and passed sequentially through 70, 40 and 20 μm-cell strainer. Single stem cells were either obtained from Lgr5-GFP mice, sorting for GFP+ cells or from B6 mice in which case cells were stained with the following antibodies: Brilliant Violet 421 anti-mouse CD45, anti-mouse CD117 (c-Kit) Alexa Fluor 700 and anti-mouse PE/Cy7-conjugated anti-CD44 for 30 min on ice, washed once with 1% FBS/DMEM and analyzed in a BD FACSaria, as described (8,9). DAPI (1μg/ml) was used to exclude dead cells in all sortings. Flow cytometry data was analyzed using FCS Express Flow Cytometry software (De Novo Software, California). The working concentrations of the antibodies were determined by titration of each one of them (Table S1).
Organoid and colony culture:
After centrifugation, pure crypts were counted and resuspended in Matrigel covered with Advanced DMEM/F12 media containing 1mM Hepes, 1 mM Glutamax and 100 U/ml antibiotics (ADF), 10% R-spondin conditioned media and 50% Wnt Conditioned Media (when indicated); supplemented with 1X B27, 1X N2 and 1mM N-acetylcystein containing 50 ng/mL murine recombinant EGF and 100 ng/mL mouse recombinant Noggin. Wnt3a conditioned media was produced using a cell line kindly provided by Robert Vries (L-Wnt3a cell line, Hubrecht Institute) and R-spondin conditioned media was prepared as we described previously (10). Intestinal stem cell colonies were cultured from sorted single Lgr5-GFP cells based upon the method reported by Yin et al (11). Approximately 3,000 intestinal stem cells were plated in 30 μl Matrigel and cultured in ENR-VC media with 1.5 mM Valproic Acid (V) and CHIR99021 (C), containing 10 μM Rho-kinase/ROCK inhibitor Y-27632 and 1 μM Jagged-1 only for the first 48h. Under these culture conditions intestinal stem cells divide symmetrically without differentiating, growing into homogeneous stem cell colonies. Intestinal stem cell colonies were passaged once per week as single cells following a 3-min incubation with TrypLE containing 2 kU ml−1 DNase1, N-acetylcysteine, and Y-27632.
Organoid irradiation:
Irradiation was delivered with a Shepherd Mark-I unit (Model 68, SN643, J. L. Shepherd & Associates, San Fernando, CA) operating a 137Cs source. Organoids were passaged one day prior to the experiment, plated at a density of 100–150 organoids/well in at least 3 wells for each corresponding dose. Organoids were exposed to single fraction radiation (range 0–16Gy). Manual counting under an inverted brightfield microscope of surviving organoids at day 4 to 10 post-radiation was used to generate classic dose response curves. Brightfield imaging of organoids after radiation was performed with Citation 5 Cell Imaging Multi-mode reader (Biotek). Survival fraction was calculated as number of surviving organoids/number of organoids in the unirradiated sample.
Radiation dose survival curves:
Radiation dose survival curves are characterized on a log-linear plot by an initial shoulder, or the quasi-threshold region, and an exponential region, where the curve approximates a straight line (12–17). In the present study, we adapted the Single Hit Multi-Target (SHMT) model to characterize radiosensitivity profiles of small and large intestinal organoids. The SHMT model assumes that each cell contains [n] identical targets for pro-lethal radiation damage, and while inactivation of a single target is sublethal, creating the shoulder region of the radiation dose-survival curve, inactivation of [n] targets is required for cell lethality, expressed in the exponential portion of the curve (12,18,19). Whereas Intestinal organoids are multicell “mini-guts” dependent on a subset of Lgr5+ stem cells that drive organoid growth and radiation dose-survival in vitro, we modified the baseline SHMT paradigm, introducing the concept that each organoid contains [n] identical stem cell targets. Inactivation of one stem cell is sublethal, creating a shoulder region of the organoid radiation dose-survival curve, and dose-dependent inactivation of [n] stem cells is required for organoid lethality, expressed in the exponential portion of the dose-survival curve. GraphPad Prism 6 software (GraphPad, La Jolla, CA) was used to generate the dose survival curves applying the following equation:
where S is a survival rate of the cell population, D is the dose, D0 is the reciprocal slope value of the exponential portion of the curve, n is the extrapolation number and Dq is the quasi-threshold dose range of the shoulder region. D0 that defines inherent radiosensitivity of the irradiated organoids is the dose required to reduce the fraction of surviving organoids to 37%, and systematically predicts organoid-type specific 10% survival (D10) as the product of 2.3xD0, where 2.3 is the natural log of 10. Dq defines the low dose range where organoid reparable sublethal radiation damage prevails, resulting in disproportionate organoid survival per dose unit relative to the inherent D0. The algorithm fits the dose survival curve to the measured end point at each tested dose by iterative weighted least square regression analysis of all data points, estimating covariance of survival curve parameters and corresponding confidence limits, and reports the computed D0, Dq and the n number. D0 values are used in the current study to compare inherent radiosensitivities between tissues, organoids and stem cell colonies.
Whole mount staining of organoids:
β-galactosidase (LacZ) staining protocol was described by Barker et al. (4) for tissue staining and here we adapted that protocol for whole mount organoids. Matrigel-embedded SI organoids grown for 1 week in a 24-well plate were washed with PBS 1x and incubated for 30 min in ice-cold fixative (1% formaldehyde, 0.2% gluteraldehyde and 0.02% NP40 in PBS 1X) at 4°C on a rolling platform. The fixative was removed and the organoids washed twice in PBS for 20 min at room temperature (RT) on a rolling platform. The β-galactosidase substrate (5 mM K3FE(CN)6, 5 mM K4Fe(CN)6 •3H2O, 2 mM MgCl2, 0.02% NP40, 0.1% sodium deoxycholate and 1mg/ml X-gal (5-Bromo-4-chloro-3-indolyl β-D-galactopyranoside, Sigma) in PBS was then added and incubated in the dark overnight at RT. The substrate was removed and the organoids washed twice in PBS for 20 min at RT on a rolling platform. The tissues were then fixed for 30 min with 4% PFA/PBS at RT in the dark on a rolling platform. PFA was removed and the organoids washed twice in PBS for 20 min at RT on a rolling platform. Originally, we used the LacZ Tissue Staining Kit from Invivogen, but the company stopped producing it, and now all the reagents can be obtained separately (Table S1). Imaging of LacZ stained organoids was performed under a Zeiss Lumar v.12 Stereoscope at Molecular Cytology Core Facility (MCCF) from MSKCC. To confirm organoid death, Matrigel-embedded intestinal organoids were stained with 1 mg/ml Hoechst 33342 and 1.5 μM propidium iodide one hour before taking images. Hoechst 33342 was also used to determine the number of cells at day 3. For these experiments imaging was performed with inverted confocal Zeiss LSM 5 Live microscope at MCCF.
DNA double-strand break (DSB) repair focus staining and quantitation:
Single Lgr5-GFP+ cells obtained from SI and LI colonies were seeded onto 8-well Lab-Tek chamber (NUNC) pre-coated with 20 μl of 100% Matrigel (~150–200 cells per well) and overlaid with growth medium. Lgr5-GFP+ cells were irradiated with 8Gy either on day 1 or day 4 post-plating. At the indicated times, immunostaining was carried out using a standard protocol. Briefly, colonies were washed twice with PBS, fixed with 4% paraformaldehyde for 30 min, washed, and quenched with 50 mM NH4Cl for 15 min. After blocking for 2 h (PBS containing 0.5% BSA, 0.2 mg/ml Na-azide, 0.3 μM DAPI and 0.25% Triton X-100), colonies were incubated with primary antibodies in blocking buffer overnight at 4°C. After three washes with PBS, colonies were incubated with secondary antibodies in blocking solution for 2 h at room temperature. γH2AX mouse monoclonal antibody against γH2AX Ser139 (Millipore [clone JBW 301], #05–636) was used at a dilution 1:1000 v/v, and secondary antibody Alexa Fluor 488 F(ab’)2 fragment of goat anti-mouse IgG (H+L) (2 mg/ml) was used at a dilution of 1:400. Samples were mounted with ProLong Diamond Antifade Reagent (Life Technologies). Fluorescent images were acquired with a 63x/1.4NA objective oil lens on a Zeiss widefield microscope (Axio Observer.Z1) equipped with an AxioCam HRc camera. Ten consecutive 0.2μm-thin sections were imaged, deconvoluted with a theoretical PSF using AutoQuant X software, and max-projected into a single image using Image J (FIJI).
On average 100 nuclei/experiment were randomly selected from each sample for quantitation of focus numbers. Fiji software was used for image analysis (20). Briefly, foci were scored within a nucleus whose boundary was defined automatically from a DAPI image. Focus threshold was determined manually as a mean = 8.28 pixels, based on discreet γH2AX foci generated at a low radiation dose (2 Gy at 30 min), consistent with published results (21). Due to extensive overlap of foci at 30 min after high-dose irradiation, focus number was estimated at this time by measuring total γH2AX fluorescence per nucleus normalized to 8.28 mean pixels/focus.
Results
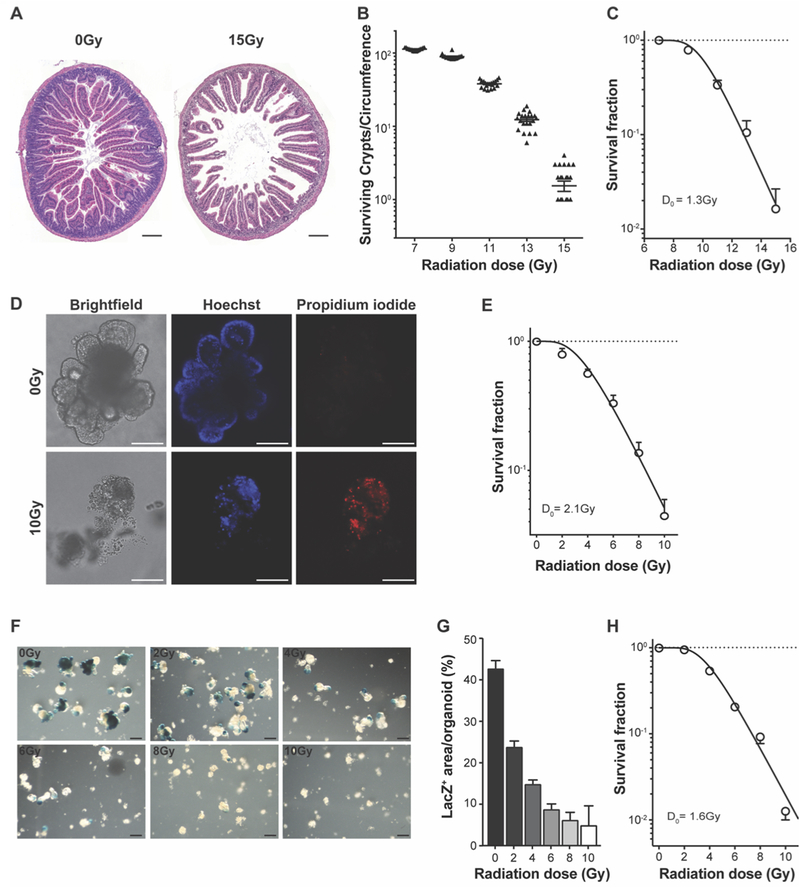
The Microcolony Assay (also termed Clonogenic Assay) developed by Withers and Elkind (5) directly quantifies radiation dose-dependent lethality of the crypt compartment, is predictive of eventual animal death from Radiation Gastrointestinal (GI) Syndrome, and is considered a “gold standard” in the radiation field for evaluating normal tissue response to ionizing radiation (22,23). This assay, which measures number of regenerating crypts per small intestinal (SI) circumference at 3.5 days post irradiation (typical histologic sections are shown in Fig. 1A), is regarded as a surrogate for stem cell survival, as a single surviving stem cell is considered sufficient for regeneration of an entire crypt. Numerous studies performed in mice reveal that 10–15 regenerating crypts per circumference, or approximately 10% crypt survival, is the minimum required to fully recover the GI mucosa post irradiation (24). Figure 1B shows that crypt survival in C57BL/6 mice generates a descending exponential curve typical of radiation responses in mammalian tissues (25) with 113±1 crypts/SI circumference after 7Gy (a value not different from the 114±3 crypts/circumference in unirradiated mice), reduced to 12±1 crypts after 13Gy (P≤0.001) and further to 1 crypt/circumference at 15Gy (P≤0.001 vs. 13Gy). These data are consistent with a large body of literature on this topic (26,27). Whereas cell death at low radiation doses often occurs by mechanisms that are independent of DNA misrepair (28–30), the clinically-relevant mechanism of radiation killing, algorithms have been developed to quantify inherent radiosensitivity in the intermediate radiation dose range (12–17). Hence, to further characterize inherent radiosensitivity from our data, we transformed crypt survival data by non-linear regression fitting according to the well-established SHMT algorithm, yielding a single D0 value that serves as a numerical estimate of the efficiency of DSB repair (31). Note, the higher the D0 value the greater the radioresistance. Figure 1C shows that SHMT transformation of our crypt survival data yields a D0=1.3 ± 0.1Gy, virtually identical to D0 values previously calculated by ourselves and others in this strain (22,32) (Fig. 1C). In this context, we previously reported highly similar D0 values for SI crypt survival in vivo in multiple mouse strains, as follows: 1.3 ± 0.2Gy for B6.129X1/J mice; 1.5 ± 0.1Gy for C57BL/6 mice; 2.0 ± 0.2Gy for sv129/BL6 mice and 1.9 ± 0.1Gy for C3HeB/FeJ mice (32).
Figure 1. SI organoids recapitulate in vivo response to ionizing radiation.
(A) Representative full-transverse section of proximal jejunum from C57BL/6 mice at 3.5 days post treatment with 15Gy whole body radiation compared with unirradiated mice. (B) GI damage assessed by the Crypt Microcolony Assay of Withers and Elkind (5). Triangles represent number of crypts in individual circumferences. (C) Transformation of data in (B) using the SHMT algorithm. (D) Representative live images of SI organoids grown for 6 days from isolated crypts that had been irradiated 24h after plating with 10Gy compared with unirradiated organoids. Shown are bright field (left), Hoechst (middle), and propidium iodide (right) images. (E) Dose survival curve of SI organoids derived from irradiated SI crypts handled as in (D). (F) Representative images of SI organoids grown from crypts isolated from Lgr5-lacZ mice stained for lacZ expression 6 days post-irradiation. (G) Quantification of LacZ+ area (blue) per organoid area (white+blue). (H) Dose survival curve of organoids derived from single Lgr5-GFP+ SI stem cells that had been irradiated at 24h after plating, and counted at 6 days post-irradiation. Data (mean ± standard deviation) are from four mice/dose analyzing four circumferences/mouse (B and C), or from triplicate determinations collated from 7 independent experiments in (E), 3 independent experiments in (H), or at least 2 experiments in (G). Scale bars = 200 μm in all images (A, D and F).
Using organoid technology established by Clevers and colleagues (1) that allows growth of intestinal organoids from crypts and from single intestinal stem cells in a defined media containing EGF, Noggin and R-spondin (ENR), we adapted principles of the in vivo Clonogenic Assay to profile radiation dose survival of SI organoids ex vivo. For initial studies, SI crypts were isolated from Lgr5-lacZ mice. Crypts contain Lgr5+ stem cells at their base intermingled with Paneth cells overlaid by progenitor cells (4). Under these media conditions, once the upper opening seals, crypts undergo continuous budding events, creating organoids that consist of a lumen filled with apoptotic cells lined by a villus-like epithelium surrounded by multiple crypt domains (1). For our studies, isolated SI crypts were irradiated 24 h after plating to allow for recovery from isolation stress (which interferes with reliable radiation dose profiling). While unirradiated organoids grow logarithmically and exclude propidium iodide (PI), an indication of membrane integrity, Fig. 1D shows that a dose of 10Gy is lethal for SI crypts (shown at day 7) as the structure completely disintegrates, manifesting as a cloud of cell debris that readily incorporates PI. Radiation dose survival analysis of SI organoids irradiated with escalating doses (2–10Gy) yields a curve with a D0=2.1 ± 0.1Gy at 6 days post-radiation (Fig. 1E), well within the range of our D0 values found for the in vivo Microcolony Assay of Withers and Elkind as described above. These results indicate that SI organoids maintain the radiosensitivity profile of the intact organ.
Further, organoids derived from Lgr5-lacZ mice when stained for β-galactosidase (LacZ) to evaluate stem cell response at day 6 post-radiation (2–10Gy) show dose-dependent reduction in the stem cell compartment (LacZ+ area/organoid) with escalating radiation doses (whole mount staining shown in Fig. 1F, quantified in Fig. 1G). To provide evidence supporting intestinal stem cells as determinant of organoid and organ radiosensitivity, Lgr5-GFP+ stem cells isolated from Lgr5-EGFP-ires-CreERT mice by fluorescence-activated cell sorting (FACS) were irradiated with the same dose range as for intact organoids at 24h after plating and allowed to grow into intact organoids (Fig. 1H). At day 6 post-radiation, number of organoids was counted and SHMT transformation of dose survival data generated a D0=1.6 ± 0.1Gy, indicating that indeed, for the small intestine, intrinsic radiosensitivity of Lgr5+ stem cells is highly similar to that of crypts in the intact organ in vivo and to organoids derived thereof.
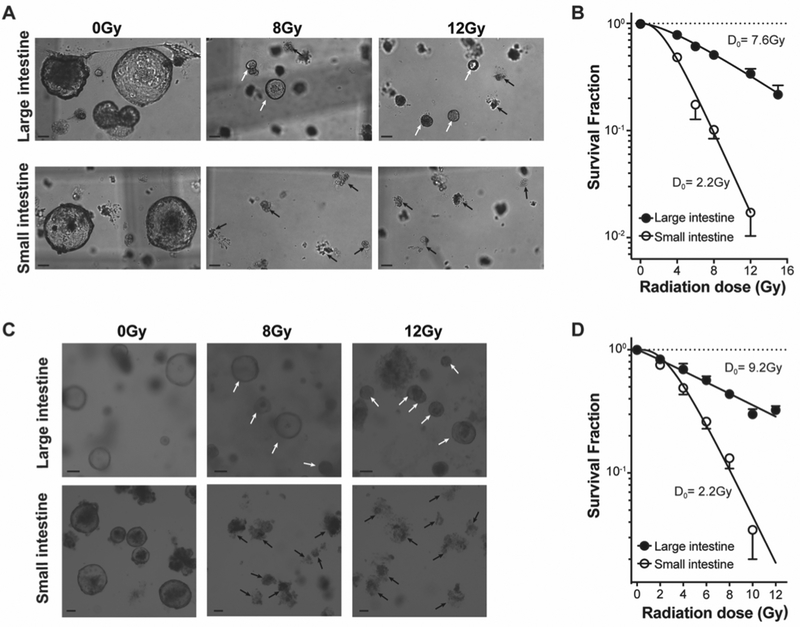
Employing Lgr5-lacZ transgenic mice, we recently published that Lgr5+ stem cells in the distal large intestine are more radiation resistant than their counterpart Lgr5+ stem cells in the small intestine in vivo (3). To extend these findings to the in vitro setting, initial studies generated organoids from crypts isolated from small and distal large intestines of Lgr5-lacZ mice. In contrast to SI organoids which contain Paneth cells that provide Wnt large intestinal (LI) organoids require exogenous Wnt, since in vivo Wnt is provided by stromal cells (33) not included in our SI culture conditions. Hence SI and LI organoids were grown in WENR (Wnt, EGF, Noggin, R-spondin) media for these studies. Fig. 2A displays representative bright-field images of organoids derived from LI and SI crypts irradiated at 24h after plating and shows that LI organoids are more radioresistant than SI organoids. While LI organoids are alive at 6 days post 8Gy and 12Gy, near complete lethality is detected in SI organoids, manifested as a cloud of cell debris in place of organoids. Radiation dose survival analysis of SI and LI organoids using 4–16Gy (Fig. 2B) confirms that LI organoids are markedly radioresistant with a D0=7.6 ± 1.3Gy as compared with SI organoids that exhibit a D0=2.2 ± 0.3Gy (p<0.001, Fig. 2B). Furthermore, consistent with the well-described dose-dependent cellular growth delay post-radiation, all LI organoids treated with 4–16Gy are smaller than unirradiated organoids on day 7 post-irradiation, with growth recovery displaying a clear radiation dose dependence (Fig. S1 A–B). In addition, mechanically passaged LI and SI organoids retain the radiosensitivities observed with organoids from fresh crypts, studied up to passage 10. Fig. 2C shows that passage 10 LI organoids retain radioresistance compared with SI organoids, displaying live organoids at 6 days post 8Gy and 12Gy, which appear capable of faster post-radiation growth than freshly-isolated organoids (compare Fig. S1 A–B with Fig. S1 C–D). SHMT transformation of radiation dose survival curves of SI and LI organoids grown from passaged organoids confirm LI organoids retain radioresistance with a higher D0=9.2 ± 1.5Gy than SI organoids D0=2.2 ± 0.2Gy (Fig. 2D, p<0.001). These results validate ex vivo organoids as legitimate surrogates for in vivo radiation responses, a phenotype that is stable over time.
Figure 2. LI organoids are significantly more resistant to ionizing radiation than SI organoids.
(A) Representative brightfield images of LI and SI organoids grown for 7 days from freshly-isolated crypts irradiated 24h after plating with 0Gy (control, left panels), 8Gy (middle panels) and 12Gy (right panels). LI organoids are more resistant to ionizing radiation than SI organoids as live organoids can still be detected after 8Gy and 12Gy (white arrows), while SI organoids are dead, displaying only a debris cloud, at 8Gy (black arrows). (B) Dose survival curves of murine LI and SI organoids grown from isolated crypts irradiated 24h after plating. Organoids were counted 10 days post-irradiation. (C, D) Similar data are derived using passaged LI and SI organoids, handled as in (A,B). Data (mean ± standard error) are collated from 3 independent experiments each performed in triplicate in (B and D). Scale bars = 100 μm (A and C).
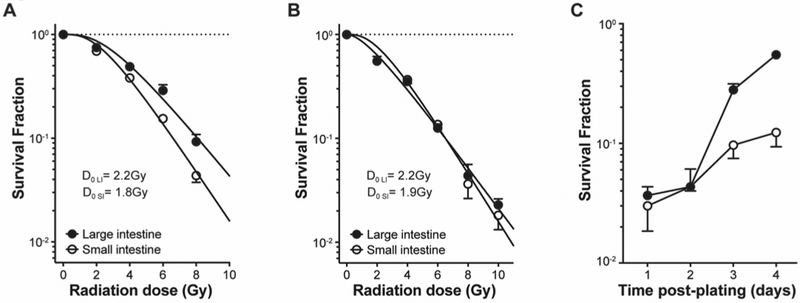
Unexpectedly, organoids derived from single LI stem cells sorted from Lgr5-GFP mice and irradiated 24h after plating display loss of radioresistance yielding a D0=2.2 ± 0.2Gy as compared to organoids derived from SI stem cells which retain their original radiosensitivity with D0=1.8 ± 0.1Gy (Fig. 3A). This selective loss of LI stem cell radioresistance was confirmed using an alternative sorting strategy that allows obtaining a larger number of cells by FACS using CD44 and cKit surface markers (8,9) (see Fig. S2 for sorting strategy). For these studies, stem cell populations marked by CD44highcKit− were isolated from large and small intestines of C57BL/6 mice and irradiated 24h after plating as above. Radiation dose survival curves of these cell populations generated D0=2.2 ± 0.1Gy for LI stem cells and D0=1.9 ± 0.2Gy for SI stem cells (Fig. 3B). However, by delaying delivery of radiation beyond 24 h post plating, we found that LI stem cells recover their inherent radioresistance by day 3 of culture (Fig. 3C), at a moment when single cells have developed into mini-organoids containing an average of 30 (with lower 95% CI= 12 and upper 95% CI= 40) cells per organoid, consistent with a doubling time of approximately 12h (1). The mechanism by which development of cell-cell contact interactions in LI organoids restores radioresistance remains unknown, although preliminary data (see below) suggest these radiation phenotypes reflect differences in DNA repair, which will require further studies for elucidation.
Figure 3. Transient loss of LI stem cell radiation resistance upon plating of single cells.
(A) Dose survival curves of organoids derived from single Lgr5-GFP+ intestinal stem cells that were obtained from symmetrically-dividing stem cell colonies, irradiated at 24h after plating and counted at 6 days post-irradiation. (B) Dose survival curve of organoids derived from single CD44highcKit− intestinal stem cells that had been sorted from C57BL/6 mice irradiated at 24h after plating and counted at 6 days post-irradiation. (C) LI stem cells recover resistance to radiation at 3 days after plating. Single CD44highcKit- intestinal cells were sorted from C57BL/6 mice, irradiated with 8Gy on successive days after plating, and organoids were counted at 6–7 days post-irradiation. Data (mean±standard error) are collated from at least 2 independent experiments (A-C).
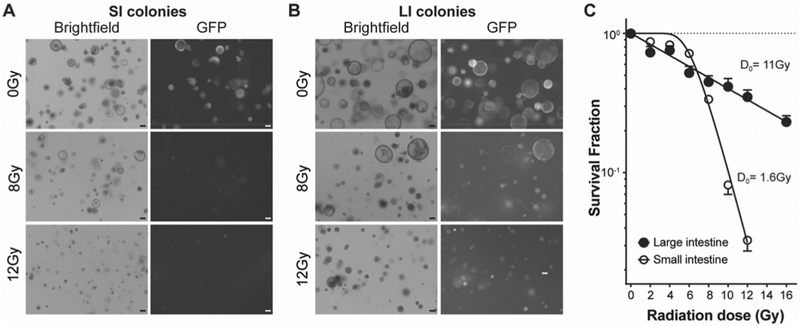
To further evaluate the inherent radiosensitivity of SI and LI stem cells, we used a niche-independent high-purity Lgr5+ intestinal stem cell culture system. In this system, combination of a GSK3β inhibitor and histone deacetylase inhibitor enables culture of homogenous symmetrically-dividing Lgr5+ stem cell colonies from single purified stem cells. The potent activation of the Wnt and Notch pathways in this culture system maintains the high frequency of Lgr5+ cells (60%−100%, see Fig. S3), in contrast to SI and LI organoids that display 14±11% and 4±3% Lgr5+ stem cells, respectively. For these studies, LI and SI colonies were grown from sorted Lgr5-GFP+ cells [isolated from 5 (for large intestines) or 3 (for small intestines) mice], which allows for availability of an unlimited number of stem cells. It is worth pointing out that although this culture system has been reported (11), it has never been used before to our knowledge to study biologic processes. For radiation dose survival experiments, these pure stem cell colonies were passaged as single cells by trypsinization and irradiated at day 3 after plating, at the time when developing LI organoids regain their radioresistance. Note that single LI stem cells lose their radioresistance irrespective of the media in which they are resuspended, as this radiosensitive phenotype is observed in single LI stem cells grown in either WENR (where they grow as organoids) or ENR-VC (where they grow as stem cell colonies), associated with reduced resolution of γH2AX foci, indicative of diminished DNA DSB repair capacity (Fig. S4 A–B). Fig. 4A and 4B show that LI stem cell colonies are more radioresistant than SI colonies, displaying live colonies at 7 days post 8Gy and 12Gy. In contrast, a dose of 8Gy eliminates most SI stem cell colonies and a dose of 12Gy is almost completely lethal, manifested as near absence of colonies under brightfield and GFP imaging. Radiation dose survival SHMT analysis of SI and LI colonies, grown for 7 to 10 days post 2–16Gy shows that LI stem cell colonies, like LI organoids, LI crypts and stem cells in the intact organ in vivo, are markedly radioresistant (3) with a D0=10.9 ± 2.0Gy compared with SI colonies that manifest a D0=1.6 ± 0.1Gy (p<0.001, Fig. 4C).
Figure 4. Pure stem cell colonies recapitulate the radiosensitivity of small and large intestines.
(A) Representative brightfield (left) and GFP (right) images of LI colonies grown for 7 days from single Lgr5-GFP+ cells irradiated at 3 days after plating with 8Gy and 12Gy (lower panels) compared with unirradiated colonies (control, upper panels). (B) Representative brightfield (left) and GFP (right) images of SI colonies grown for 7 days from single Lgr5-GFP+ cells and irradiated at 3 days after plating with 8Gy and 12Gy (lower panels) compared with unirradiated colonies (control, upper panels). (C) Dose survival curve of SI and LI stem cell colonies irradiated at 3 days after plating and counted at 6–10 days post-irradiation. Data (mean ± standard error) are collated from triplicate determinations from 3 independent experiments. Scale bars = 100 μm in all images (A and B).
Discussion
Altogether, our results indicate that we have generated an in vitro radiation sensitivity assay validated against in vivo published data using classic radiobiology concepts. Using this technology, we provide strong evidence that in vivo responses reflect inherent radiosensitivity of the Lgr5+ stem cell compartment of these organs. Whether the Bmi-1 population of resting small intestinal stem cells at position +4 from the crypt base, or an analogous population in the large intestine, might also play a role in this radiation response is currently unknown (34).
A large body of literature reports data on mammalian normal and tumor stem cell radiosensitivity, but correlation with organ/tumor radiosensitivity remains inconclusive (35–42). A potentially confounding factor in this analysis is that SI and LI stem cell radiosensitivity is often measured by employing methodologies that did not comply with the rigorous criteria developed and validated in the field of radiobiology over the past four decades (43–45). For example, while a recent study argues that a dose of 12Gy renders colonic Lgr5+ stem cells more radiosensitive than colonic KRT19+ stem cells based on lineage tracing experiments (43), we have shown that at this dose level colonic Lgr5+ cells are totally resistant to radiation (3), and hence there should be no stress-induced incentive for colonic stem cell lineage tracing. We thus posit that the reason lineage tracing is observed in KRT19+ cells is that unlike colonic Lgr5+ CBCs, KRT19 cells undergo significant damage at 12Gy, leading to KRT19 lineage tracing during the rapid division phase of population recovery. One recent study utilized ionizing radiation to study whether Dll1high precursor cells of the small intestinal secretory lineage might revert to stem cells upon radiation damage using a dose of 6Gy, repeatedly referring to this dose as one that induces “extensive” radiation damage (45). In fact, radiation dose survival curves in C57BL/6 mice uniformly show 6Gy to be within the Dq region of the dose survival curve, which reflects potentially lethal damage that is mostly reversible. Of note, the observation that 6Gy is a relatively low dose in no way contradicts the interpretation of the data that Dll1high precursor cells can revert to stem cells upon genotoxic stress. These and other studies (46–48) point to a need for standardized assays to achieve the goal of translating in vitro data successfully into animal models and humans.
We believe that the current study will help to establish conditions for organoid irradiation that accurately reflect in vivo responses, and provide a simplified analytic tool, the D0, to allow for quantitative comparison of impact of pharmacologic and genetic regulation of genotoxic stress on the small and large intestine. In this context, our preliminary data indicate that this assay can be translated into a clinical setting in colorectal cancer. Treatment with neo-adjuvant chemoradiation (nCR) followed by total meso-rectal excision has been the standard of care for locally-advanced rectal cancers for more than two decades. At present, only ~30% of neo-adjuvant patients will achieve clinical or pathological complete responses to nCR therapy and are candidates for organ preservation. Moreover, these patients have better outcomes in terms of disease free and overall survival (49). However, success of this approach is hindered by lack of molecular and/or functional assays that predict response to therapy, emphasizing the need for development of personalized models that can predict treatment response. Based on results established in the current manuscript, we have applied the assays described here to test radiation responses of patient-derived organoids (PDOs) and early evidence indicates that PDO radiation responses predict treatment response to nCR (Adileh, Paty and Kolesnick, manuscript in preparation).
Finally, we propose these data will be of benefit to the scientific community interested in using organoids to investigate organ biology other than radiation biology, as validation against a high-quality in vivo assay, the Clonogenic Assay of Withers and Elkind, indicates that growth conditions identified here closely resemble those of intact organs in live animals.
Supplementary Material
Significance:
Findings establish standards for irradiating organoids that deliver radiation profiles that phenocopy the organ of origin.
Acknowledgements
This work was supported by the generous donations of Corinne Berezuk, Mike Stieber, Patrick Gerschel and Edouard Gerschel. This research was funded in part through the NIH/NCI Cancer Center Support Core Grant P30 CA008748. We also thank Jim Russell for his insightful comments about the manuscript.
Financial support: This work was supported by the generous donations of Corinne Berezuk, Mike Stieber, Patrick Gerschel and Edouard Gerschel (P.P.). This research was funded in part through the NIH/NCI Cancer Center Support Core Grant P30 CA008748.
Footnotes
Conflict of Interest Statement:
Patents unrelated to this work: RK (US7195775B1, US7850984B2, and US10052387B2), RK and JAR (US8562993B2, US9592238B2, US20150216971A1, and US20170335014A1), RK and ZF (US20170333413A1 and US20180015183A1) RK and ZF are co-founders of Ceramedix Holding L.L.C. The remaining authors have declared that no conflicts of interest exist.
References
- 1.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009;459:262–5 [DOI] [PubMed] [Google Scholar]
- 2.Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 2011;141:1762–72 [DOI] [PubMed] [Google Scholar]
- 3.Hua G, Wang C, Pan Y, Zeng Z, Lee SG, Martin ML, et al. Distinct Levels of Radioresistance in Lgr5(+) Colonic Epithelial Stem Cells versus Lgr5(+) Small Intestinal Stem Cells. Cancer Res 2017;77:2124–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007;449:1003–U1 [DOI] [PubMed] [Google Scholar]
- 5.Withers HR, Elkind MM. Microcolony survival assay for cells of mouse intestinal mucosa exposed to radiation. International Journal of Radiation Biology and Related Studies in Physics, Chemistry, and Medicine 1970;17:261–7 [DOI] [PubMed] [Google Scholar]
- 6.Yui S, Nakamura T, Sato T, Nemoto Y, Mizutani T, Zheng X, et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5(+) stem cell. Nature Medicine 2012;18:618–23 [DOI] [PubMed] [Google Scholar]
- 7.Sato T, Clevers H. Primary mouse small intestinal epithelial cell cultures. Methods Mol Biol 2013;945:319–28 [DOI] [PubMed] [Google Scholar]
- 8.Wang F, Scoville D, He XC, Mahe MM, Box A, Perry JM, et al. Isolation and characterization of intestinal stem cells based on surface marker combinations and colony-formation assay. Gastroenterology 2013;145:383–95 e1-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothenberg ME, Nusse Y, Kalisky T, Lee JJ, Dalerba P, Scheeren F, et al. Identification of a cKit(+) colonic crypt base secretory cell that supports Lgr5(+) stem cells in mice. Gastroenterology 2012;142:1195–205 e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L, Adileh M, Martin ML, Klingler S, White J, Ma X, et al. Establishing estrogen-responsive mouse mammary organoids from single Lgr5+ cells. Cellular Signalling 2016;29:41–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin X, Farin HF, van Es JH, Clevers H, Langer R, Karp JM. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat Methods 2014;11:106–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elkind MM, Sutton H. Radiation response of mammalian cells grown in culture. 1. Repair of X-ray damage in surviving Chinese hamster cells. Radiat Res 1960;13:556–93 [PubMed] [Google Scholar]
- 13.Powers EL. Considerations of survival curves and target theory. Phys Med Biol 1962;7:3–28 [DOI] [PubMed] [Google Scholar]
- 14.Fowler JF. The first James Kirk memorial lecture. What next in fractionated radiotherapy? Br J Cancer Suppl 1984;6:285–300 [PMC free article] [PubMed] [Google Scholar]
- 15.Withers HR. Failla memorial lecture. Contrarian concepts in the progress of radiotherapy. Radiat Res 1989;119:395–412 [PubMed] [Google Scholar]
- 16.Fuks Z, Vlodavsky I, Andreeff M, McLoughlin M, Haimovitz-Friedman A. Effects of extracellular matrix on the response of endothelial cells to radiation in vitro. Eur J Cancer 1992;28A:725–31 [DOI] [PubMed] [Google Scholar]
- 17.Chapman JD. Can the two mechanisms of tumor cell killing by radiation be exploited for therapeutic gain? J Radiat Res 2014;55:2–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nomiya T Discussions on target theory: past and present. J Radiat Res 2013;54:1161–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman JD. Single-hit mechanism of tumour cell killing by radiation. Int J Radiat Biol 2003;79:71–81 [PubMed] [Google Scholar]
- 20.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012;9:676–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bocker W, Iliakis G. Computational Methods for analysis of foci: validation for radiation-induced gamma-H2AX foci in human cells. Radiat Res 2006;165:113–24 [DOI] [PubMed] [Google Scholar]
- 22.Potten CS. A comprehensive study of the radiobiological response of the murine (BDF1) small intestine. Int J Radiat Biol 1990;58:925–73 [DOI] [PubMed] [Google Scholar]
- 23.Withers HR. Radiation biology and treatment options in radiation oncology. Cancer Res 1999;59:1676s–84s [PubMed] [Google Scholar]
- 24.Hendry JH, Potten CS, Roberts NP. The gastrointestinal syndrome and mucosal clonogenic cells: relationships between target cell sensitivities, LD50 and cell survival, and their modification by antibiotics. Radiat Res 1983;96:100–12 [PubMed] [Google Scholar]
- 25.Thames HD, Hendry DH. Fractination in Radiotherapy. London, New York, Philadelphia: Taylor & Francis; 1987. [Google Scholar]
- 26.Maj JG, Paris F, Haimovitz-Friedman A, Venkatraman E, Kolesnick R, Fuks Z. Microvascular function regulates intestinal crypt response to radiation. Cancer Res 2003;63:4338–41 [PubMed] [Google Scholar]
- 27.Booth D, Potten CS. Protection against mucosal injury by growth factors and cytokines. Journal of the National Cancer Institute Monographs 2001:16–20 [DOI] [PubMed] [Google Scholar]
- 28.Ling CC, Chen CH, Fuks Z. An equation for the dose response of radiation-induced apoptosis: possible incorporation with the LQ model. Radiother Oncol 1994;33:17–22 [DOI] [PubMed] [Google Scholar]
- 29.Fuks Z, Persaud RS, Alfieri A, McLoughlin M, Ehleiter D, Schwartz JL, et al. Basic fibroblast growth factor protects endothelial cells against radiation-induced programmed cell death in vitro and in vivo. Cancer Res 1994;54:2582–90 [PubMed] [Google Scholar]
- 30.Sato Y, Ebara T, Sunaga N, Takahashi T, Nakano T. Interaction of radiation and gefitinib on a human lung cancer cell line with mutant EGFR gene in vitro. Anticancer Res 2012;32:4877–81 [PubMed] [Google Scholar]
- 31.Hall EJ, Giaccia AJ. Radiobiology for the radiologist. 8th ed. Philadelphia: Lippincott Williams & Wilkins; 2018. [Google Scholar]
- 32.Rotolo JA, Maj JG, Feldman R, Ren D, Haimovitz-Friedman A, Cordon-Cardo C, et al. Bax and Bak do not exhibit functional redundancy in mediating radiation-induced endothelial apoptosis in the intestinal mucosa. International Journal of Radiation Oncology, Biology, Physics 2008;70:804–15 [DOI] [PubMed] [Google Scholar]
- 33.Farin HF, Van Es JH, Clevers H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology 2012;143:1518–29 e7 [DOI] [PubMed] [Google Scholar]
- 34.Yan KS, Chia LA, Li X, Ootani A, Su J, Lee JY, et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A 2012;109:466–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kummermehr JC. Tumour stem cells--the evidence and the ambiguity. Acta Oncol 2001;40:981–8 [DOI] [PubMed] [Google Scholar]
- 36.Milas L, Hittelman WN. Cancer stem cells and tumor response to therapy: current problems and future prospects. Semin Radiat Oncol 2009;19:96–105 [DOI] [PubMed] [Google Scholar]
- 37.Gerweck LE, Wakimoto H. At the Crossroads of Cancer Stem Cells, Radiation Biology, and Radiation Oncology. Cancer Res 2016;76:994–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greenberger JS, Epperly M. Bone marrow-derived stem cells and radiation response. Semin Radiat Oncol 2009;19:133–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pajonk F, Vlashi E, McBride WH. Radiation resistance of cancer stem cells: the 4 R’s of radiobiology revisited. Stem Cells 2010;28:639–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koch U, Krause M, Baumann M. Cancer stem cells at the crossroads of current cancer therapy failures--radiation oncology perspective. Semin Cancer Biol 2010;20:116–24 [DOI] [PubMed] [Google Scholar]
- 41.Rycaj K, Tang DG. Cancer stem cells and radioresistance. Int J Radiat Biol 2014;90:615–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fabbrizi MR, Warshowsky KE, Zobel CL, Hallahan DE, Sharma GG. Molecular and epigenetic regulatory mechanisms of normal stem cell radiosensitivity. Cell Death Discov 2018;4:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Asfaha S, Hayakawa Y, Muley A, Stokes S, Graham TA, Ericksen RE, et al. Krt19(+)/Lgr5(−) Cells Are Radioresistant Cancer-Initiating Stem Cells in the Colon and Intestine. Cell Stem Cell 2015;16:627–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yin T, Wei H, Leng Z, Yang Z, Gou S, Wu H, et al. Bmi-1 promotes the chemoresistance, invasion and tumorigenesis of pancreatic cancer cells. Chemotherapy 2011;57:488–96 [DOI] [PubMed] [Google Scholar]
- 45.van Es JH, Sato T, van de Wetering M, Lyubimova A, Yee Nee AN, Gregorieff A, et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol 2012;14:1099–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu W, Chen Q, Wu S, Xia X, Wu A, Cui F, et al. Radioprotector WR-2721 and mitigating peptidoglycan synergistically promote mouse survival through the amelioration of intestinal and bone marrow damage. J Radiat Res 2015;56:278–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ciorba MA, Riehl TE, Rao MS, Moon C, Ee X, Nava GM, et al. Lactobacillus probiotic protects intestinal epithelium from radiation injury in a TLR-2/cyclo-oxygenase-2-dependent manner. Gut 2012;61:829–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun ZJ, Zhang YZ, Liu F, Chen JJ, Chen DX, Liu HB, et al. A fusion protein composed of the DSL domain of Dll1 and RGD motif protects cryptic stem cells in irradiation injury. Biosci Rep 2018;38(2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith JJ, Strombom P, Chow OS, Roxburgh CS, Lynn P, Eaton A, et al. Assessment of a Watch-and-Wait Strategy for Rectal Cancer in Patients With a Complete Response After Neoadjuvant Therapy. JAMA Oncol 2019:e185896. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.