Abstract
DRAIC is a 1.7 kb spliced long noncoding RNA downregulated in castration-resistant advanced prostate cancer. Decreased DRAIC expression predicts poor patient outcome in prostate and seven other cancers, while increased DRAIC represses growth of xenografted tumors. Here we show that cancers with decreased DRAIC expression have increased NF-κB target gene expression. DRAIC downregulation increased cell invasion and soft agar colony formation; this was dependent on NF-κB activation. DRAIC interacted with subunits of the IκB kinase (IKK) complex to inhibit their interaction with each other, the phosphorylation of IκBα and the activation of NF-κB. These functions of DRAIC mapped to the same fragment containing bases 701–905. Thus, DRAIC lncRNA inhibits prostate cancer progression through suppression of NF-κB activation by interfering with IKK activity.
Keywords: NF-κB, LncRNA, DRAIC, prostate cancer
Introduction
Prostate cancer is a very common malignancy occurring particularly in elderly males, with approximately 174,650 new cases per year and 31,620 deaths per year in the USA according to the 2019 estimates of the American Cancer Society. Androgen deprivation therapy is used to medically treat prostate cancers that cannot be cured by surgical resection. Although tumors often regress after androgen deprivation, most tumors eventually re-appear with androgen-independent (castration resistant, CR) cells and metastasize to other organs (1). Therefore, the emergence of castration-resistant prostate cancer is an important part of the problem for treating prostate cancer.
A number of non-conventional gene regulatory mechanisms have emerged in the form of long noncoding RNAs, miRNAs, tRFs etc (2–4). Long noncoding RNAs (lncRNAs) are RNA transcripts >200 nucleotides in length that do not encode any protein. LncRNAs regulate complex biological processes (5,6) and exhibit cell and tissue specific expression with specific subcellular localization (7–9). Cytoplasmic lncRNAs regulate the turnover of mRNA and proteins (10–12), act as decoys for RNA binding proteins and interact with miRNAs (13–15) to regulate different cellular signaling pathways (16) and serve as scaffolds that interact with multiple proteins to form a functional complex (17,18). Nuclear lncRNAs interact with different chromatin modifying proteins and transcription factors and help them to get recruited to different genomic loci to regulate gene expression (18,19). The expression of lncRNAs is altered in various cancers (20,21) and they play major roles in cellular development and differentiation (22). Recently, a number of lncRNAs, PCA3, PCAT-1, PCAT29, PCGEM1, PRNCR1, ARLNC1 and ANRIL have been shown to play a key role in the regulation of prostate cancer, though their mechanisms of action are still being deciphered (23–28). PCA3 has also shown promising results for urinary detection of prostate cancer with greater potential compared to PSA (28).
NF-κB is a major signaling pathway that regulates many genes involved in immune and inflammatory response. Aberrant NF-κB activation is associated with increased invasion and other cancer phenotypes (29–32). The activated NF-κB pathway is a key survival mechanism in a variety of cancer types (33). The classical NF-κB signaling pathway is stimulated by TNF-α, IL-1 and other cytokines leading to the activation of IκB kinase complex (IKK), consisting of the catalytic IKKα and IKKβ subunits and the regulatory IKKγ subunit (the latter also known as NEMO for NF-κB essential modulator) (34). The activated IKK complex phosphorylates IκBα protein, leading to the latter’s proteosomal degradation and release of NF-κB, which is then transported to the nucleus to bind to specific DNA sequences to activate the transcription of specific sets of genes (35,36). One lncRNA is known to regulate NF-κB: NF-κB interacting long noncoding RNA (NKILA) interacts with NF-κB/IκB complex and shields the phosphorylation site of IκBα, thereby inhibiting NF-κB activation and reducing breast cancer metastasis (36).
A previous study from our lab identified in prostate cancers an androgen-repressed cytoplasmic lncRNA, DRAIC, with low DRAIC expression being associated with poor disease-free survival in patients (37). DRAIC is a 1.7 kb long transcript consisting of five exons, located primarily in the cytoplasm and inhibiting invasion in prostate cancer cells (Sakurai et al., 2015). Interestingly, a low level of DRAIC was predictive of poor prognosis in seven other malignancies: bladder cancer, hepatocellular carcinoma, gliomas, lung adenocarcinoma, kidney renal clear cell carcinoma, stomach adenocarcinoma, and cutaneous melanoma. The activated androgen receptor binds to the DRAIC promoter to down-regulate its expression in prostate cancer cells. In castration resistant prostate cancer cells, the androgen driven pathways are activated at a very low concentration of androgen, and so DRAIC is constitutively down regulated as prostate cancers advance. The mechanism by which DRAIC exerts its protective effect on patients was unknown.
We now report that the tumor suppressive role of DRAIC in prostate cancer is through the repression of the NF-κB pathway and that DRAIC acts by interacting with and inhibiting the IKK complex. Given that there are attempts to inhibit the IKK complex pharmacologically, it is particularly interesting that a naturally occurring lncRNA acts as a natural inhibitor of IKK. Very few lncRNAs have till now been shown to directly interact with and thus regulate proteins in signal transduction pathways, and so our results anticipate that other lncRNAs will emerge with similar functions on other signal transduction pathways.
Materials and methods
Cell Culture and transfection
LNCaP, PC3M and C4–2B cells were cultured in RPMI medium supplemented with 10% FBS, 1% penicillin/streptomycin, 1 mM sodium pyruvate and 10 mM HEPES buffer. HEK293T and HeLa cells were cultured in DMEM medium supplemented with 10% FBS and 1% penicillin/streptomycin. All the cell lines were purchased from ATCC and maintained in humidified incubator at 37°C in the presence of 5% CO2. The cell lines were authenticated by Short Tandem Repeats (STR) analysis at 15 genomic loci and the amelogenin gene (Bio-Synthesis, Lewisville, Tx, USA). SiRNA against DRAIC, IKKα, IKKβ, NEMO, p65 and MISSION Universal Negative Control siRNA (NC) (50 nM) (Supplementary Table 1) were reverse transfected into LNCaP cells using RNAiMax (Life Technologies, Carlsbad, CA) reagent. 48 hours post transfection cells were harvested either for RNA isolation, cDNA synthesis and qPCR or for cell lysate preparation. For NF-κB reporter assay, 3x NF-κB firefly luciferase and CMV driven Renilla luciferase plasmids were transfected in LNCaP, C4–2B, HEK293T and PC3M cells using Lipofectamine 3000. 24 hours post transfection cells were lysed with 1X passive lysis buffer and luminescence signals were captured by the luminometer. Full length DRAIC was cloned into pcDNA3 vector as described earlier (37). All DRAIC fragments were amplified from pcDNA3-DRAIC backbone and cloned into BamH1/Xho1 site of pcDNA3 using in-fusion methods (Supplementary Table 1). Stable cell lines expressing full length DRAIC or DRAIC deletion constructs were generated in PC3M, HeLa and DRAIC KO LNCaP cells while DRAIC deletion constructs were generated in DRAIC KO background by transfecting either 1 μg of empty pcDNA3 vector or appropriate pcDNA3 plasmid with Lipofectamine 3000 (Life Technologies) reagent according to the manufacturer’s protocol. The stable cell lines were selected with 500 μg/ml G418 for two weeks and maintained under selection for the experiments.
Plasmids and reagents
The plasmids IKK-2 K44M ((item #1104), Flag-IKKβ (S177E, S181E) (item #64609) pX333 (item #64073) were purchased from Addgene. Antibodies to IKKα (Abcam,Cambridge, UK, ab109749), IKKβ (ab32135), NEMO (Abcam, ab178872), phospho IκBα (S32) (Abcam, ab92700) were purchased from Abcam. P65 (Catalog no. ABE347), Pan Ago (Catalog no. MABE56) antibodies were obtained from Sigma-Millipore (Burlington, MA) and antibodies to IκBα (#9242), STAT3 (#4904), TAK1 (#5206), TBK1 (#3504), β-Catenin (#8480), CENP-A (#2186), Chk2 (#6334) were from Cell Signaling Technology (Danvers, MA). Antibody against α-tubulin (sc-5286) and β-actin (sc-47778) were purchased from Santa Cruz Biotechnology (Dallas, Texas). The DRAIC siRNAs si226, si787 and siRNA against p65 were obtained from Invitrogen (Life Technologies) and siRNAs against IKKα, IKKβ and NEMO were procured from Dharmacon (Lafayette, CO). The universal negative control siRNA was purchased from Sigma. Bay11–7082 (Catalog no. B5556) was purchased from Sigma.
RNA immunoprecipitation, RNA pull down and Immunoblot analysis
RNA immunoprecipitation was carried out from LNCaP and PC3M cells using the Magna RNA Immunoprecipitation (RIP) kit (Millipore, catalog no. 17–700) according to the manufacturer’s protocol. 2×107 cells and 5 μg of each IKKα, IKKβ, NEMO, IκBα and p65 antibodies were used for each RIP assay. For RNA pull down experiments we used Ribo-Trap kit (MBL International Corporation) and followed the manufacturer’s protocol. The full-length DRAIC sense and antisense was prepared by PCR and used as a template for in vitro transcription reaction by T7 promoter using T7 polymerase and in vitro MEGAscript T7 transcription kit (Supplementary Table 1). The sense and antisense DRAIC RNA was labeled by random incorporation of 5-Bromo-UTP during the in vitro transcription. 5 μg of each sense and antisense in vitro transcribed DRAIC RNA was used per pull down experiment. The sense and antisense DRAIC RNA was heated at 85°C for 3 min in RNA structure buffer (20 mM Tris-HCl, pH 7.0, 100 mM and 10 mM MgCl2) and slowly allowed to cool to room temperature to promote proper RNA folding. Anti-BrdU antibody was incubated with the magnetic beads at 4°C for overnight and washed with beads wash buffer (supplied by kit) followed by incubation of the antibody-bead complex with in vitro transcribed sense and antisense RNA at 4°C for 2 hours. The unbound RNA was removed with beads washing buffer. The cytosolic cell lysates was prepared using the kit’s lysis buffer. The antibody-beads-RNA complex was incubated with cytosolic extract at 4°C for 1 hour followed by 6 times washing with kit wash buffer. The RNA bound protein was lysed with 1X Laemmli buffer and resolved on SDS-PAGE followed by immunoblotting with antibodies to IKKα (1:2000), IKKβ (1:2000), NEMO (1:3000), IκBα (1:2000) and p65 (1:3000), Chk2 (1:5000), CENP-A (1:5000) and β-Catenin (1:5000). For immunobloting of phospho IκBα, cells were pre-treated with 10 μM MG132 for 4 hours and lysed with lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, pH 8.0, 5 mM NaF, 5 mM MgCl2, 5 mM β-glycerophosphate, 0.5 mM sodium vanadate, 1 mM DTT, 0.5 mM PMSF and protease inhibitors) and centrifuged at 13000 rpm for 30 min. The supernatants containing equal amounts of proteins were resolved on SDS-PAGE and immunoblotted with anti-phospho IκBα antibody.
Cell proliferation and invasion assay
The MTT assay was performed with 5×104 cells plated in 24 well plates for WT and DRAIC KO LNCaP cells at different time points. The matrigel containing Boyden chamber was first rehydrated with serum-free RPMI medium at 37°C for 2 hours. 1×105 cells were seeded in serum-free medium in the top of the chamber, full growth medium containing 10% FBS (Thermo Fisher Scientific, catalog no. 10082147) was added to the bottom of the chamber as a chemo-attractant and the chamber incubated at 37°C in presence of 5% CO2 for 24 hours. After 24 hours, the invaded cells on the bottom surface of the membrane were gently washed with 1X PBS and fixed with 100% methanol for 5 min followed by 0.5% crystal violet staining at room temperature for 15 min. The non-invading cells from the upper surface of the chamber were removed by scrubbing. Randomly 10 fields were captured under microscope and the invaded cell number counted per field.
Chromatin Immunoprecipitation assay
Cells were washed twice with PBS pH 7.4 and crosslinked with 1% formaldehyde at room temperature with gentle shaking for 10 min. The reaction was quenched with 125 mM glycine for 5 min followed by washing the cells twice with PBS and lysing the cells with lysis buffer containing 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100, 1% SDS, 5 mM EDTA pH 8.0, protease inhibitor cocktails (Sigma) on ice for 20 min followed by sonication for (10s on/10s off with 20% amplitude for 10 min) with Sonic Dismembrator model 500 (Fisher Scientific) to get the chromatin fragmented to around 300–1000 bp. The supernatants were collected after centrifugation at 8000Xg for 10 min and incubated overnight with protein G Dynabeads (#10004D, Thermo Fisher Scientific) bound with the 2 μg p65 antibody (Millipore) The beads were sequentially washed with low salt buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, pH 8.0, 1%Triton X-100, 0.1% SDS), high salt wash buffer (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 2 mM EDTA, pH 8.0, 1%Triton X-100, 0.1% SDS) and LiCl wash buffer (10 mM Tris-HCl, pH 8.0, 1% NP-40, 250 mM LiCl, 2 mM EDTA, pH 8.0). DNA was eluted from the beads with the elution buffer (100 mM sodium bicarbonate and 1% SDS) at 30°C for 30 min. The DNA was de-crosslinked with 200 mM NaCl at 65°C for overnight followed by proteinase K treatment at 55°C for 1 hr. The DNA was then purified with Zymo column for PCR.
In vitro kinase assay
The IKKβ immunoprecipitates from WT and DRAIC KO clones of LNCaP cells were used for in vitro kinase assay in 1X kinase buffer (20 mM HEPES, pH 7.6, 5 mM MgCl2, 2 mM MnCl2, 10 mM PNPP, 0.5 mM PMSF, 1 mM benzamidine, 1 mM DTT, 10 μM ATP, 10 mM NaF, 5 mM β-glycerophosphate, 0.5 mM sodium vanadate) at 30°C for 60 min in presence of 10 μCi [γ−32P] ATP and 2 μg GST IκBα (1–54) as substrate. The reaction was terminated by addition of 5X SDS sample buffer and the sample was run on SDS-PAGE gel and developed by autoradiography.
In vitro DRAIC-IKK binding assay
To determine the direct association of DRAIC with IKK complex, the recombinant His taged IKKα was purchased from Thermo Fisher Scientific (Waltham, MA). Recombinant GST-NEMO was purified based on the previously published protocol (38) and when needed the GST released by thrombin cleavage. DRAIC and its different deletion constructs, APTR and luciferase RNAs were in vitro transcribed using MEGAscript T7 transcription kit (Supplementary Table 1). For direct binding assay, 50 ng His tag IKKα or GST-NEMO was immobilized with Ni2+ beads or glutathione beads in binding buffer (25 mM HEPES, pH 7.5, 150 mM NaCl, 0.05% NP-40, 10% glycerol, protease inhibitor and RNaseOUT) at 4°C for 1 hour followed by addition of 1 μg DRAIC or APTR or Luciferase RNA. All RNAs were pre-heated at 85°C in RNA structure buffer (20 mM Tris-HCl, pH 7.0, 100 mM KCl, 10 mM MgCl2) and cooled down to room temperature for 15 min. The RNA-protein-bead mix was incubated at 4°C for 1 hour. Bead-bound protein and RNA was washed 5 times with washing buffer (25 mM HEPES, pH 7.5, 150 mM NaCl, 25 mM Imidazole, 0.1% NP-40). The RNA was isolated using TRIzol reagent and qPCR was performed with DRAIC specific primers (Supplementary Table 1).
For IKK complex formation assay, 50 ng His6-IKKα was immobilized on nickel beads for 1 hour at 4°C and 1 μg DRAIC and control RNAs were added followed by 50 ng of (GST-cleaved) recombinant NEMO at 4°C for 2 hours. The beads were washed as previously mentioned, boiled in SDS sample buffer and loaded on SDS-PAGE and immunoblotted with the specific antibodies.
RNA isolation and cDNA synthesis and quantitative PCR
Total RNA was isolated from LNCaP and PC3M cells using RNeasy mini kit (Qiagen, catalog no. 74104). 1 μg of total RNA was reverse transcribed using SuperScript III First-Strand cDNA synthesis kit (Thermo Fisher Scientific, catalog no. 18080051). The qPCR was performed using Power SYBR green master mix reagent (Thermo Fisher Scientific, catalog no. 4367659) on a Step One plus qPCR machine. The qPCR (StepOne Plus Thermo Fisher Scientific, Waltham, MA) fold induction was calculated using the ΔΔCt method after normalizing with loading control 18S RNA or GAPDH (Supplementary Table 1).
Anchorage-independent growth assay
Soft agar colony formation assay was carried out in 6 well plates. Briefly, 1% and 0.6% noble agar (Sigma-Aldrich, catalog no. A5431) were prepared and sterilized and maintained at 40°C temperature throughout the experiment and the 2X medium was also maintained at the same temperature. The bottom layer of agar was prepared by mixing 0.75 ml of 1% agar with 0.75 ml of 2X RPMI medium consisting of 20% FBS supplemented with 2% penicillin/streptomycin, 2 mM sodium pyruvate and 20 mM HEPES buffer and added to each well and allowed 30 min to solidify. For the top layer, 1 × 104 cells were suspended in 0.75 ml 2X RPMI growth medium and mixed with 0.75 ml of 0.6% agar, added on the solid bottom layer and itself allowed to solidify for an additional 30 min. The plates were then incubated at 37°C in the presence of 5% CO2 for an additional 2–3 weeks. The medium was replaced every week. The visible colonies were checked under microscope and counted by taking pictures of 10 random fields from each set of experiments. For the Bay11–7082 experiment, the cells were pre-treated with 5 μM Bay11–7082 for 2 hours before plating in soft agar. Bay11–7082 was maintained at 1μM in the medium throughout the assay.
Mouse Xenograft
Six weeks old athymic nude mice were procured from Jackson laboratory and mice experiments were performed based on the University of Virginia institutional guidelines. 1×106 PC3M cells stably overexpressing either EV or full length DRAIC were harvested and re-suspended in matrigel and PBS at 1:1 ratio and injected in both flanks of mice. Tumor volume was monitored and measured weekly twice with slide caliper. Tumor volume was calculated as (π/6)ab2, where a is the maximum length of the tumor and b is the minimum length of the tumor.
DRAIC Exon 2–4 knockout by CRISPR/Cas9
To delete DRAIC exon 2–4, multiple sgRNAs were designed using the online tool http://crispr.mit.edu/. The sgRNAs were annealed and cloned into px333 plasmid backbone (Addgene #64073) using restriction enzyme site Bbs1 and Bsa1. The px333 plasmid was digested with Bbs1 at 37°C for 2 hours and ligated to the DRAIC exon 2 targeting sgRNA using quick ligation kit (NEB) at room temperature for 5 min followed by transformation, plasmid isolation and sgRNA insertion verification by Sanger sequencing. The 2nd sgRNA targeting exon 4 was cloned into the Bsa1 site of px333 plasmid in a similar way and the sgRNA insertion verified by Sanger sequencing. The LNCaP cells were transfected with px333 plasmid containing both the sgRNAs targeting exon 2 and 4 of DRAIC and pcDNA3 plasmids for G418 antibiotic selection. The cells were selected with 500 μg/ml G418 for around 2 weeks until the non-transfected cells were killed. The resistant cells were diluted and plated into 96 well plates for single cell expansion. The genomic DNA was isolated from each single clones using Quick extract DNA isolation kit (QE09050, Epicentre) and deletion was confirmed by conventional PCR. The PCR product was gel purified and analyzed by Sanger sequencing to validate the desired homozygous deletion. Quantitative RT-PCR was further performed to check the DRAIC RNA level in the KO clones (Supplementary Table 1).
Gene Set Enrichment and Guilt By Association Analysis
Gene set enrichment analysis (39) was performed to identify pathways upregulated or downregulated in patients from The Cancer Genome Atlas (TCGA) (40) in the bottom third of DRAIC expression relative to patients in the top third of DRAIC expression. The RNA-seq quantification results were downloaded from the TCGA data portal. The fold change of all genes expressed at greater than one fragment per kilobase per million mapped reads (FPKM) in each cancer was calculated in patients in the bottom third of DRAIC relative to patients in the top third of DRAIC expression. The genes were ranked by fold change and used as input for gene set enrichment analysis.
Guilt by association analysis was performed to identify pathways whose genes were negatively correlated with DRAIC expression. The Spearman correlation coefficients (rho) representing the association of DRAIC expression with each gene expressed greater than one FPKM in each cancer was calculated and used to rank genes for Guilt by Association analysis.
Statistical tests
All data are presented as mean ± SD from indicated numbers of measurements. The significance between the two groups is calculated by student’s t test (paired test, two sided). The differences are considered as statistically significant if the p value is <0.05.
Results
DRAIC decreased tumorigenic potential both in vitro and in vivo
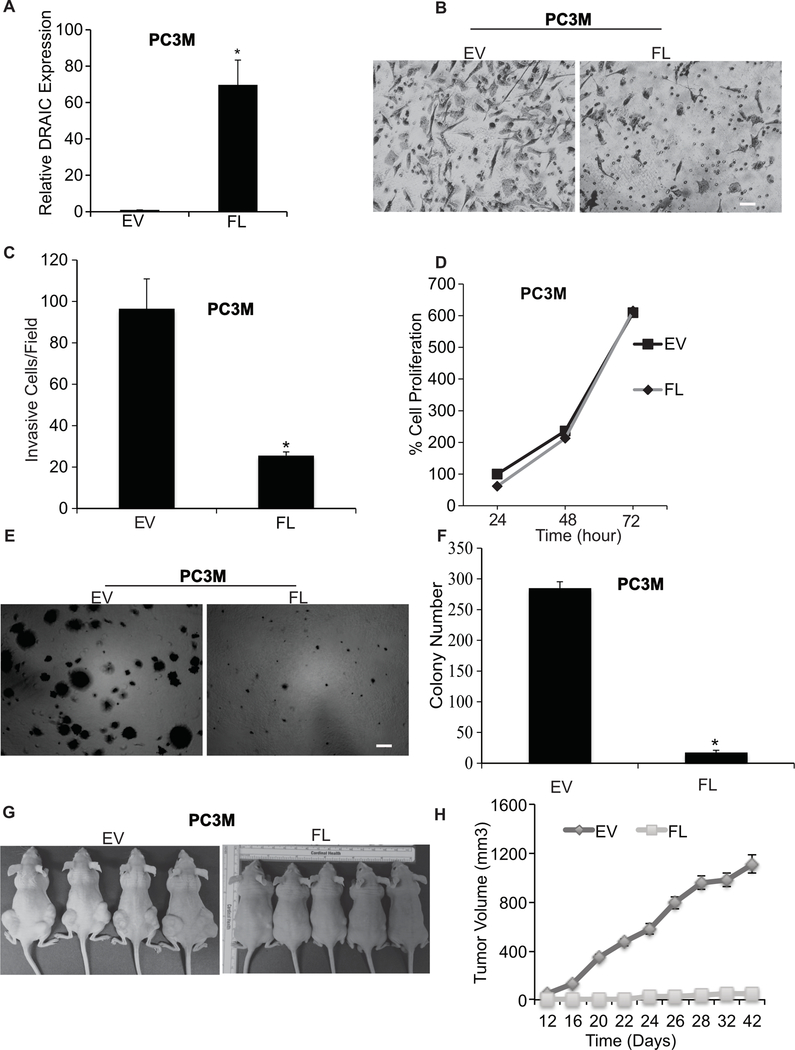
We first confirmed our previous result (37) that DRAIC suppresses invasion of prostate cancer cell lines through matrigel in Boyden Chamber assays (Fig. 1, A to C). The stable overexpression of DRAIC did not decrease the growth of the cells attached to plastic in a tissue culture dish (Fig. 1D), but decreased the growth of the cells in anchorage independent conditions, as evidenced by the decrease of colony formation in soft agar (Fig. 1E,F). To evaluate the effect of DRAIC overexpression on tumor growth in vivo, nude mice were injected with PC3M cells with or without overexpression of DRAIC (Fig. 1G,H). Three weeks after injection, the empty vector injection group started showing palpable tumors but no tumors were visible for DRAIC overexpressing PC3M cells. Six weeks after injection, the empty vector transfected PC3M cells reached a tumor volume of 1109±102 mm3 whereas the DRAIC overexpressing PC3M cells showed tumor volume of 48.43±19.77 mm3 (Fig. 1G,H). Therefore, the tumorigenic potential of PC3M cells is significantly inhibited by DRAIC overexpression.
Figure 1. DRAIC suppresses the tumorigenic property of prostate cancer.
(A) RT-qPCR analysis of DRAIC expression with and without overexpression in PC3M cells. We quantified DRAIC expression relative to GAPDH normalized this to the level of DRAIC in empty vector transfected cells. Results expressed as mean ± s.d, n = 3, *P<0.05. (B) Images showing matrigel invasion assay in PC3M cells with empty vector (EV) or overexpressing full length DRAIC (FL). Scale bar 20 μm. (C) The quantification of the invasion assay is done by counting 10 different random fields and plotted average cell number per field. Results expressed as mean ± s.d, n = 3, *P<0.05. (D) Cell proliferation of PC3M cells at indicated hr by MTT assay on cells attached to plastic cell culture dish. Results expressed as mean ± s.d, n = 3, *P<0.05. (E, F) Anchorage independent soft agar colony formation with 104 PC3M cells at 3 weeks. Bar graph represents average and S.D. of colony number per field from 10 fields. Scale bar 50 μm. Results expressed as mean ± s.d, n = 3, *P<0.05. (G) Nude mice (n = 10 per group) were injected with PC3M cells overexpressing EV and FL and Tumor volume is calculated twice in a week and plotted in (H). mean ± s.d, *P<0.05.
Prostate cancers with low level of DRAIC expressed more NF-κB targets
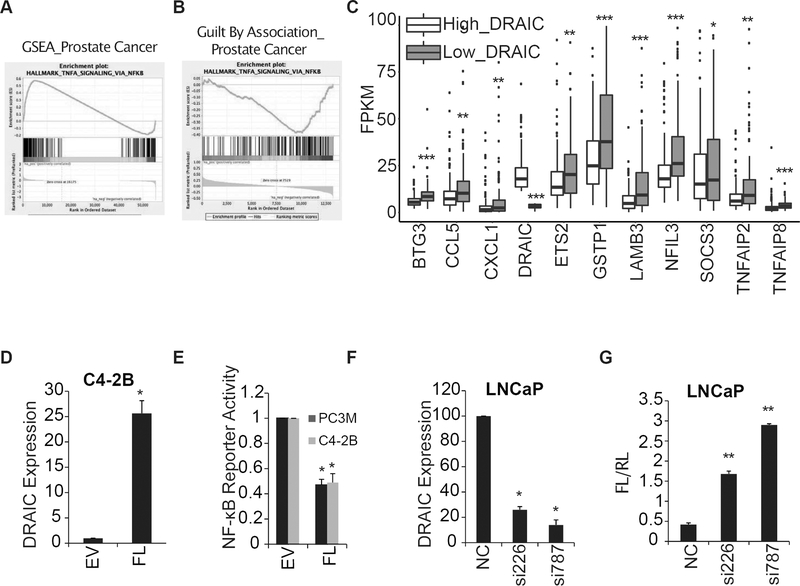
To determine what pathways are significantly altered in prostate cancers with high versus low levels of DRAIC, we analyzed the RNA sequencing data generated from 469 tumors from prostate cancer patients in TCGA and compared the RNA expression patterns in the tumors in the bottom third of DRAIC expression with those in the top third. Gene Set Enrichment Analysis (GSEA) (39) showed that genes activated by TNFα-NF-κB were significantly upregulated in the former (Fig. 2A). Guilt By Association analysis (41), Identified protein-coding genes and pathways significantly correlated with DRAIC expression in Prostate cancer TCGA dataset. Protein-coding genes that were negatively correlated with DRAIC expression showed enrichment of genes involved in TNFα signaling via NF-κB pathway (Fig. 2B). Indeed, many known NF-κB targets, particularly those activated by the canonical pathway, were upregulated in cancers with the lowest quartile of DRAIC expression (Fig. 2C).
Figure 2. Low DRAIC expression is associated with increased NF-κB activity.
(A) Gene set enrichment analysis revealed that NF-κB targets are upregulated in tumors in the lowest third of DRAIC expression compared to tumors in the highest third of DRAIC expression. (B) The expression of genes involved in TNFα signaling via NF-κB were found to be negatively correlated with DRAIC expression through Guilt by Association analysis. (C) Box-plot shows expression of indicated NF-κB target gene levels in prostate cancers in TCGA in the highest third of DRAIC expression (Red) versus those in the lowest third (Blue). ***P<0.001, **P <0.01, *P<0.05, Wilcoxon rank sum test. (D) DRAIC overexpression in C4–2B cells measured by RT-qPCR and normalized to 18S RNA. (E) NF-κB reporter assay (Firefly luciferase/Renilla luciferase) in PC3M and C4–2B cells with EV or overexpressing FL DRAIC. (F-G) DRAIC levels (F) and NF-κB luciferase reporter assay (G) in LNCaP cells transfected with negative control siRNA (NC) or two different siRNAs (50 nM) against DRAIC. For (D) to (G) results expressed as mean ± s.d, n = 3. The p values were calculated using Student’s t-test, **P<0.01, *P<0.05.
DRAIC represses NF-κB activity at or upstream from the step where IKK phosphorylates IκBα
Consistent with the results from human cancers, overexpression of DRAIC in androgen independent cell lines PC3M and C4–2B decreased NF-κB activity as measured by an NF-κB driven luciferase reporter (Fig. 2D, E). Conversely, depletion of DRAIC in androgen dependent LNCaP cells resulted in an increase of NF-κB activity (Fig. 2F, G).
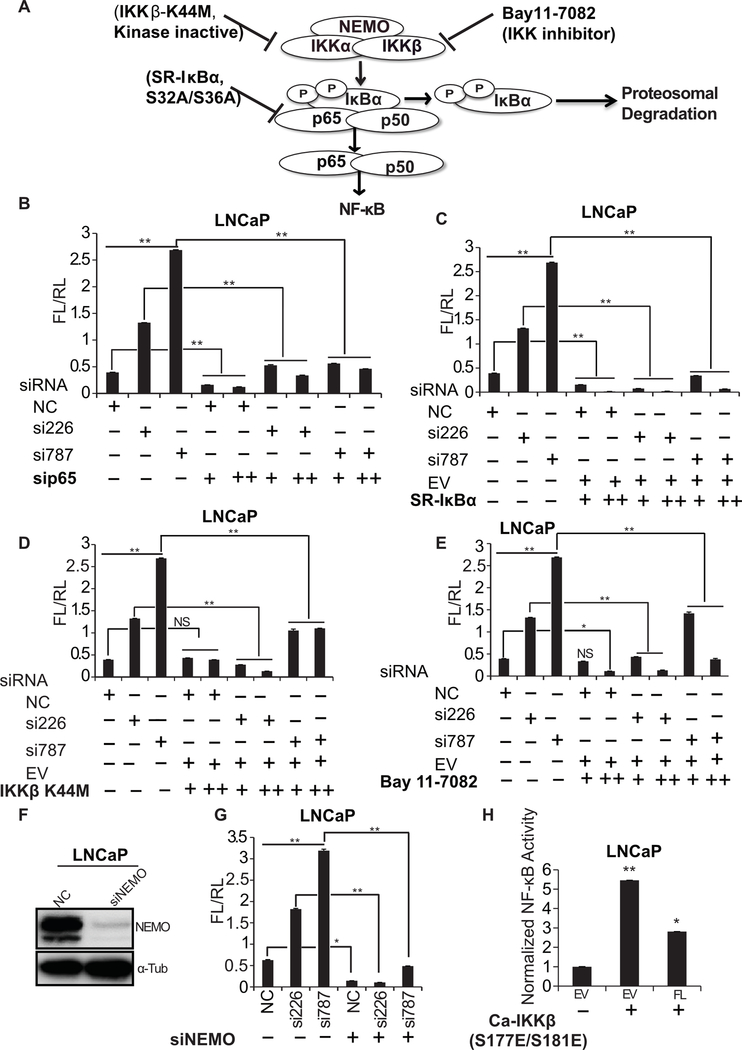
To identify the step at which the NF-κB pathway is activated in DRAIC-depleted cells, we interfered with the normal levels/activity of NF-κB, IκBα and IKK (Fig. 3A). The NF-κB activation seen after DRAIC knockdown was reversed by any of the following maneuvers: knockdown of p65 (Fig. 3B), overexpressing the constitutively active super repressor IκBα (S32A/S36A) (Fig. 3C), overexpressing the dominant negative IKKβ mutant (K44M) (Fig. 3D), inhibiting IKK by Bay11–7082 (Fig. 3E) or knockdown of NEMO from the IKK complex (Fig. 3F, G). These results strongly suggest that knockdown of DRAIC activates the NF-κB pathway at or upstream from the phosphorylation of IκBα by IKK. When we generated the constitutively active IKK by over-expressing the IKKβ (S177E/S181E) in androgen dependent LNCaP cells, the NF-κB activity was still repressed by DRAIC overexpression (Fig. 3H, compare with Fig. 2E), suggesting that DRAIC represses the NF-κB pathway downstream of active IKK. Intersecting these results leads to the conclusion that DRAIC is likely to repress the NF-κB pathway by interfering with the function of IKK.
Figure 3. DRAIC represses NF-κB pathway at the level of IKK.
(A) A Schematic representation of the canonical NF-κB pathway with different inhibitory molecules mentioned in this figure. (B-E) NF-κB luciferase reporter activity in LNCaP cells transfected with siRNAs (50 nM) against DRAIC followed by either co-transfection of si-p65 (25 nM and 50 nM) (B), overexpression of super repressive IκBα (C), overexpression of dominant negative IKKβ (K44M) (D), or, inhibition of IKK by Bay11–7082 (E). EV: empty vector. (F) Western blot showing knockdown of NEMO using siRNA in LNCaP cells. (G) NF-κB luciferase activity in LNCaP cells co-transfecting siRNAs against DRAIC and NEMO. (H) NF-κB luciferase reporter activity upon overexpression of constitutively active IKKβ (S177E/S181E) in LNCaP cells with transient overexpression of empty vector (EV) or a plasmid expressing full-length DRAIC (FL). Results expressed as mean ± s.d, n = 3, **P<0.01, *P<0.05.
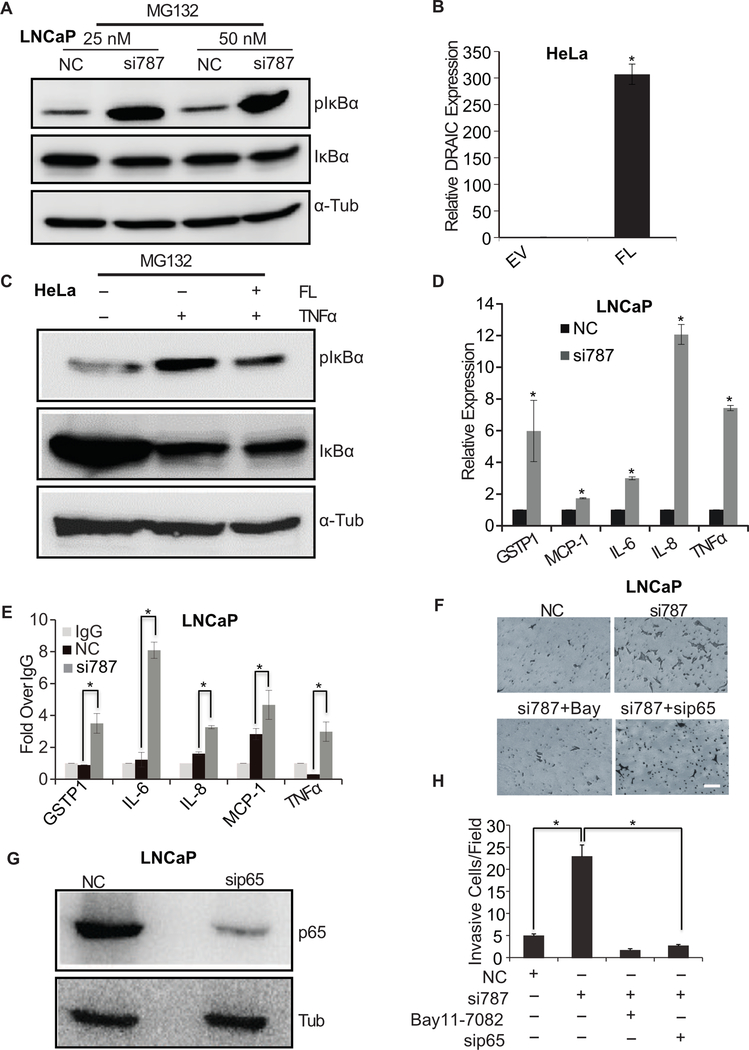
Knockdown of DRAIC in LNCaP cells increased the phosphorylation of IκBα (Fig. 4A, Supplementary Fig. S1A) consistent with the hypothesis that DRAIC interferes with IKK activity. The result was validated using another siRNA against DRAIC (Supplementary Fig. S1B). Conversely, overexpression of DRAIC in HeLa cells stimulated by TNFα suppressed the phosphorylation of IκBα that is normally stimulated by TNFα (Fig. 4B, C).
Figure 4. DRAIC knockdown increases NF-κB activity.
(A) LNCaP cells were transfected with two different concentrations of siRNA (25 and 50 nM) against DRAIC or negative control (NC), followed by 10 μM MG132 treatment for 4 hours. Cell lysates prepared using RIPA buffer and immunoblotted for phospho IκBα, total IκBα and α-Tubulin. (B, C) HeLa cells were stably transfected with EV (empty vector) or FL (expressing full length DRAIC) and RT-qPCR performed to measure DRAIC over-expression. Mean± s.d, n = 3, *P<0.05. (C) HeLa cells from (B) were treated with TNF-α followed by 10 μM MG132 for 4 hours and cell lysates immunoblotted for phospho IκBα, total IκBα and α-Tubulin. (D) LNCaP cells transfected with NC or siRNA against DRAIC. RT-qPCR of indicated NF-κB responsive genes, expressed after normalization to 18S RNA and then to level of expression in NC cells. Mean ± s.d, n = 3, *P<0.05. (E) ChIP-qPCR of p65 at sites known in the literature to bind NF-κB at promoters of NF-κB responsive genes. IgG ChIP is taken as 1. Mean± s.d, n = 3, *P<0.05. (F) Matrigel invasion assay performed in LNCaP cells treated with either siRNA NC or against DRAIC (si787). DRAIC depleted cells were treated with IKK inhibitor, Bay11–7082 (6 hr) or sip65 (48 hr). Scale bar 20 μm. (G) LNCaP cells transfected with siRNAs immunoblotted for p65 and Tubulin. (H) The invasive cells in (F) counted under microscope by taking 10 random fields. Results expressed as mean ± s.d, n = 3, *P<0.05. *P<0.05 relative to NC.
Finally, we determined that knockdown of DRAIC induced several endogenous NF-κB responsive genes, like GSTP1, MCP-1, IL-6, IL-8 and TNFα by qRT-PCR (Fig. 4D). ChIP-qPCR of p65 showed that DRAIC knockdown increased the binding of NF-κB to the promoters of above-mentioned genes (Fig. 4E).
Cell invasion induced by DRAIC knockdown is decreased upon inhibition of NF-κB activity in LNCaP cells
Increased NF-κB activity in different cancer cells is associated with more invasion (42,43). We therefore checked whether the increased invasion upon DRAIC depletion (37) is due to the increased NF-κB activity (Fig. 4F). Knockdown of p65 (Fig. 4G) or IKK inhibition with Bay11–7082 decreased the invasion seen upon knockdown of DRAIC (Fig. 4F, H). Therefore, the biological effect upon DRAIC depletion is likely mediated through the activation of NF-κB.
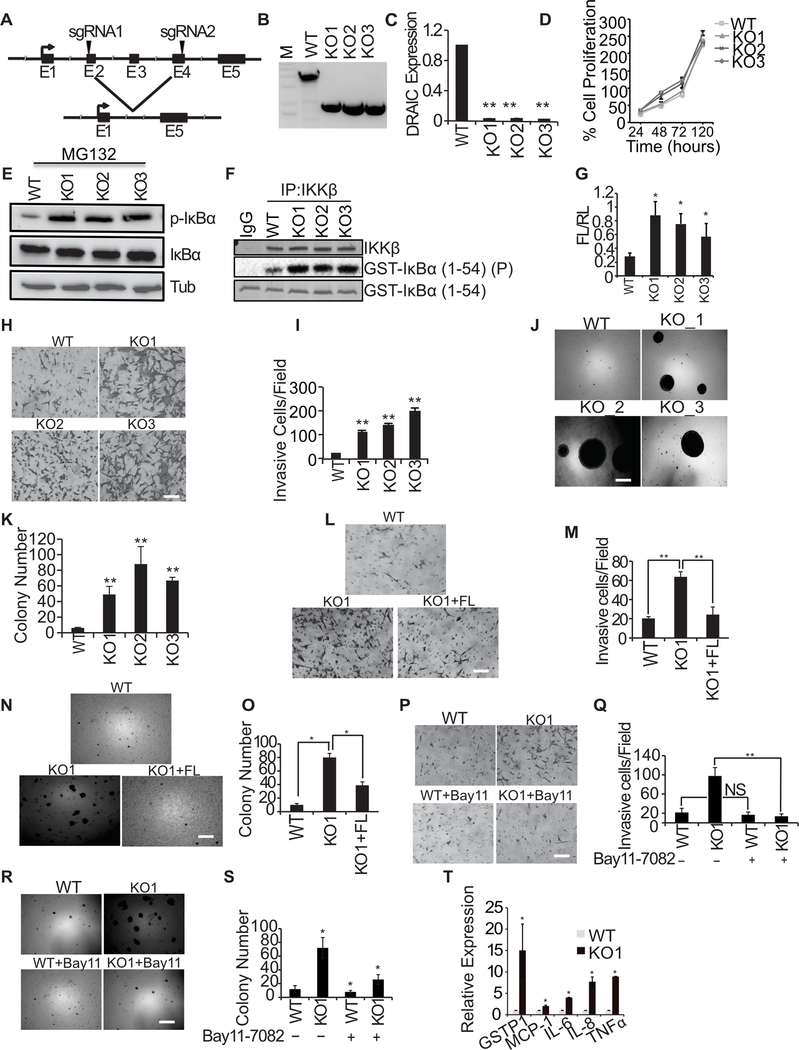
Deletion of DRAIC Exon 2–4 using CRISPR/Cas9 also increased invasion of LNCaP cells through the activation of NF-κB
To ensure that endogenous DRAIC indeed represses invasion and colony formation, we sought to eliminate DRAIC from LNCaP cells by a method that does not use siRNAs. We knocked out exons 2–4 of DRAIC in LNCaP cells by CRISPR/Cas9 (Fig. 5A, B and Supplementary Fig. S2), and this decreased the expression of even the residual exon 5 of DRAIC (Fig. 5C). Although proliferation of cells attached to plastic was unchanged (Fig. 5D), the knockout of DRAIC increased the phosphorylation of IκBα (Fig. 5E), consistent with the hypothesis that DRAIC interferes with IKK activity. IKK kinase activity, measured by an in vitro kinase assay of immunoprecipitated IKK on GST-IκBα, was increased after DRAIC knockout (Fig. 5F). The NF-κB reporter activity was stimulated in DRAIC knockout cells (Fig. 5G) consistent with the result that DRAIC represses the phosphorylation of the IκBα inhibitor of NF-κB. To eliminate the possibility that the siDRAIC mediated increase in invasion or colony formation was due to off-target activity of the siRNA, we also checked the phenotypes in the DRAIC knockout clones. DRAIC knockout increased invasion of cells through matrigel in a Boyden Chamber assay (Fig. 5H, I) and increased the ability of the LNCaP cells to form soft agar colonies (Fig. 5J, K). Conversely, the increased invasion and colony formation of the DRAIC knockout clones was suppressed by expressing DRAIC from an exogenous promoter (Fig. 5L–O). The increase in invasion and colony formation seen upon deletion of DRAIC was also reversed by inhibiting the NF-κB pathway by the IKK inhibitor BAY11–7082 (Fig. 5P–S), confirming by an independent assay that the biological effects of depletion of DRAIC are mediated by activation of NF-κB. Many of the NF-κB responsive genes were upregulated in DRAIC KO cells (Fig. 5T). Thus, the siDRAIC mediated increase in cell invasion, soft agar colony formation and stimulation of NF-κB responsive genes (Fig. 4D) was reproduced by knockout of DRAIC and these effects were not due to the off-target activity of the siRNAs.
Figure 5. DRAIC knockout increases tumorigenic property of prostate cancer cells through NF-κB pathway.
(A) A schematic illustration of DRAIC knockout strategy using CRISPR/Cas9 in LNCaP cells is shown. The sgRNAs were designed from DRAIC exon 2 and exon 4 as indicated by the arrowhead to knockout Exon 2–4. (B) Three representative single clones with KO of DRAIC identified by truncated PCR product from DRAIC genomic DNA. M: DNA molecular weight marker. (C) RT-qPCR of DRAIC RNA from WT and KO clones. Mean±s.d, n=4, **p<0.01. The sequence of the DRAIC knockout was validated from PCR product from DRAIC gene. (D) MTT assay on DRAIC WT and KO clones. (E, F) (E) WT and DRAIC KO clones were treated with 10 μM MG132 for 4 hours and cell lysates were prepared from WT LNCaP and DRAIC KO clones and immunoblotted for phospho-IκBα, total IκBα and α-Tubulin. (F) In vitro kinase assay with IKKβ immunoprecipitate from WT and DRAIC KO clones of LNCaP cells using GST-IκBα as substrate. Top: immunoblot of immunoprecipitated IKKβ. Middle: autoradiogram after kinase reaction. Bottom: substrate visualized by Coomasie staining.(G) NF-κB luciferase reporter activity with DRAIC KO clones. Mean±s.d, n=3, *p<0.05. (H, I) Matrigel invasion assay with DRAIC WT and KO clones. A representative image shown in H and results quantified in I. Mean±s.d, n=4, **p<0.01. Scale bar 20 μm. (J, K). The anchorage independent soft agar colony formation by DRAIC WT and KO clones. A representative image is shown in J and quantitation in K. The colony number was measured by counting 10 different fields. Mean±s.d, n=3, **p<0.01. Scale bar 50 μm. (L, M) FL DRAIC was overexpressed in DRAIC KO cells and invasion assay performed and quantified. Mean±s.d n=3,**p<0.01. Scale bar 20 μm. (N, O) The anchorage independent soft agar colony formation was carried out in DRAIC KO clones overexpressing FL DRAIC and quantified. Mean±s.d, n=4, *p<0.05. Scale bar 50 μm. (P, Q) DRAIC KO clones were treated with Bay11–7082 (for 6 hr) and invasion assay was performed. Mean±s.d, n=3, *p<0.05. Scale bar 20 μm. (R, S) DRAIC KO clones were treated with Bay11–7082 (for 6hr) and soft-agar colony assay was performed. Mean±s.d, n=3, *p<0.05. Scale bar 50 μm. (T) RT-qPCR of NF-κB responsive mRNAs from WT and DRAIC KO cells. Mean ± s.d, n = 3. *P<0.05.
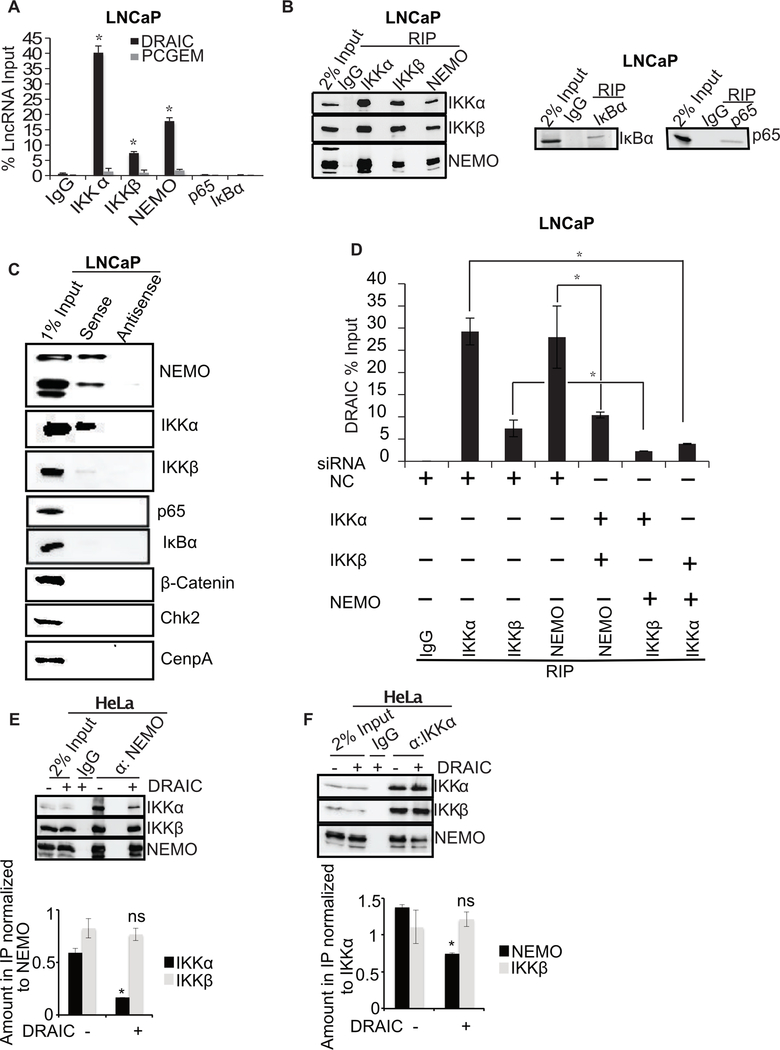
DRAIC interacts with the IKK complex and decreases the integrity of the complex
To understand how DRAIC inhibits IKK, we immunoprecipitated different proteins in the NF-κB pathway: IKKα, IKKβ, NEMO, NF-κB-p65 and IκBα and measured whether DRAIC was associated with any of them by RNA immunoprecipitation (RIP). DRAIC was co-immunoprecipitated with IKKα, IKKβ and NEMO but not with p65 or IκBα (Fig. 6A, B). DRAIC did not associate with several other negative control proteins, IgG, TAK1, TBK1, STAT3 or Ago (Supplementary Fig. S3A–E). LncRNAs like PCGEM or Linc00152, expressed at a level comparable to DRAIC, did not immunoprecitate with IKK complex (Fig. 6A, B and Supplementary Fig. S3E). Other cellular RNAs, MALAT1, PCA3, PCAT1, SchLp1, GAPDH, or GS1 did not associate with IKK complex (Supplementary Fig. S3F). These results collectively establish the specificity of the DRAIC-IKK interaction.
Figure 6. DRAIC interacts with IKK complex.
(A) RNA immunoprecipitation (RIP) assay from LNCaP using NF-κB pathway proteins with antibodies against IKKα, IKKβ, NEMO, p65 and IκBα. RT-qPCR for DRAIC and PCGEM (a prostate tissue specific androgen regulated gene) expressed as % of input lncRNA in precipitates. (B) The immunoprecipitated samples in RIP were immunoblotted to ensure pull down of indicated proteins. (C) In vitro transcribed BrU labeled DRAIC sense (S) and antisense (AS) RNA was incubated in LNCaP cell lysates and pulled down with anti-BrdU antibody. The precipitate was immunoblotted for indicated proteins. (D) LNCaP cells after knockdown of two subunits of the IKK complex (Supplementary Fig. S3G–J). RIP assay performed with the remaining subunit followed by q-PCR for DRAIC. (E, F) NEMO (E) or IKKα (F) was immunoprecipitated from HeLa cells in presence and absence of stably overexpressed DRAIC and immunoblotted for indicated proteins. The amount of IKKα or IKKβ associated with NEMO (E) and the amount of NEMO or IKKβ associated with IKKα (F) were quantitated and plotted below the immunoblots. Mean±s.d, n=3, *p<0.05.
Conversely, in vitro transcribed BrU labeled sense DRAIC RNA immobilized on beads associated with NEMO and IKKα but not with p65, IκBα, Chk2 or CenpA (Fig. 6C). IKKβ interaction with DRAIC was more variable (Fig. 6A, C), and was significantly less than the interaction with the other IKK subunits. The antisense DRAIC RNA does not pull down any of these proteins, demonstrating specificity of the interaction with sense DRAIC RNA.
In the pull-downs in either direction, NEMO and IKKα appear to bind DRAIC more than IKKβ (Fig. 6A, C). To confirm this, siRNA mediated knockdown of two of the three subunits of the IKK complex followed by immunoprecipitation of the remaining subunit showed that all three subunits associated with DRAIC independent of the other subunits, although IKKα and NEMO were better at associating with DRAIC than IKKβ (Fig. 6D and Supplementary Fig. S3G–J).
We next examined whether DRAIC disrupted the integrity of the IKK complex. Overexpression of DRAIC followed by immunoprecipitation of NEMO (Fig. 6E) or IKKα (Fig. 6F) revealed that DRAIC diminished the association of NEMO and IKKα with each other.
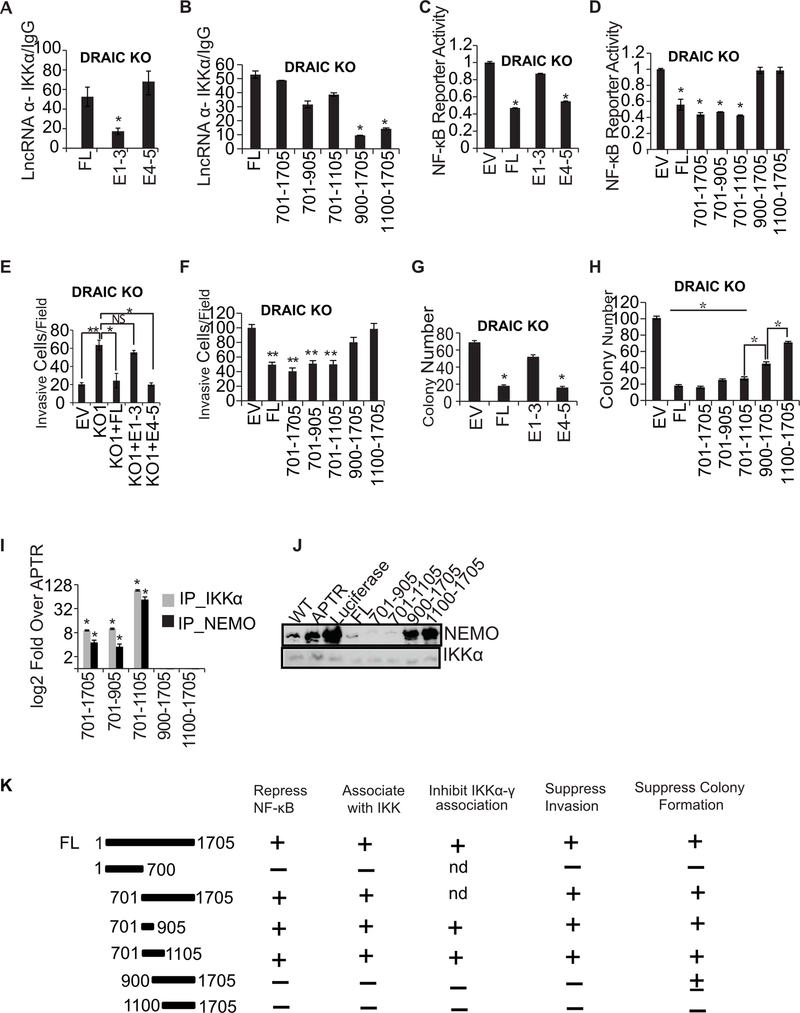
701–905 of exons 4–5 of DRAIC suppresses invasion and NF-κB activity
The deletion mutations in DRAIC showed that a fragment containing exons 4–5 (bases 701–1705), but not exons 1–3 (bases 1–700), was as good as full length DRAIC in co-immunprecipitating with IKKα from DRAIC KO cells expressing exogenous DRAIC fragments (Fig. 7A). When further deletions were made in steps of 200 nucleotides from exons 4–5, bases 701–905 described a minimal fragment that associated well with IKKα (Fig. 7B). Interestingly, exon 4–5 (relative to exon 1–3) and fragment 701–905 (relative to 900–1705), was best at repressing an NF-κB reporter when expressed in DRAIC KO cells (Fig. 7C, D). Exon 4–5 also repressed NF-κB in 293T cells that normally do not express DRAIC (Supplementary Fig. S4A) even when it was activated by constitutively active IKKβ (Supplementary Fig. S4B).
Figure 7. 701–905 bases of E4–5 associate with IKK and suppress invasion and NF-κB activity.
(A, B) RIP assay with IKKα in DRAIC KO cells after overexpressing DRAIC RNA and deletion constructs. RT-qPCR of DRAIC expressed as fold over signal from IgG IP. For A-H: Results expressed as mean ± s.d, n = 3, *P<0.05. (C, D) NF-κB luciferase reporter activity after expressing DRAIC or derivatives in DRAIC KO cells, normalized to activity in cells transfected with empty vector (EV). Mean ± s.d, n = 3, *P<0.05. (E, F) Invasion assay with DRAIC KO cells stably overexpressing DRAIC and derivatives. EV: empty vector. The number of invasive cells was quantified by taking 10 random microscopic fields. Mean± s.d, n = 3, **P<0.01, *P<0.05. (G, H) Soft-agar colony formation with DRAIC KO cells expressing DRAIC or derivatives. Mean ± s.d, n = 3, *P<0.05. (I) In vitro association of DRAIC or its derivatives with bacterially produced recombinant His-IKKα or GST-NEMO. RT-qPCR signal of DRAIC in the protein pull-downs normalized to the signal for APTR lncRNA (negative control). Mean±s.d, n=3, *p<0.05. (J) In vitro complex formation between recombinant NEMO and His6-IKKα in the presence of in vitro transcribed RNA indicated at top. His6-IKKα was immobilized on Nickel agarose beads and incubated with RNA followed by incubation with recombinant NEMO protein. The beads were then washed and boiled with Laemmli buffer and immunoblotted for the indicated proteins. WT: No RNA added. APTR and Luciferase were used as negative control RNA. (K) Summary of the results of the structure-function studies of DRAIC in prostate cancer cells. Nd: not done.
Most interestingly, the ability to suppress invasion also mapped to DRAIC 701–905 bases (Fig. 7E, F and Supplementary Fig. S4C–F), as did the ability to suppress colony formation in soft agar (Fig. 7G, H), though the DRAIC 900–1705 appeared to have residual colony suppression activity. Finally, bacterially produced recombinant IKKα or NEMO associated specifically with DRAIC in vitro, and this too was mediated by DRAIC (701–905), but not DRAIC (900–1705) (Fig. 7I). The in vitro association of NEMO with immobilized IKKα was diminished by pre-incubation with full length DRAIC, and this too mapped to DRAIC (701–905) (Fig. 7J). Luciferase RNA, APTR lncRNA, DRAIC (900–1705) or DRAIC (1100–1705) did not inhibit the IKKα-NEMO interaction, serving as negative controls. The summary of the results of the structure-function studies of DRAIC (Fig. 7K) shows that the minimal fragment of DRAIC that represses the NF-κB luciferase reporter by binding to IKK is also the minimal fragment that represses cell invasion and soft agar colony formation, supporting our hypothesis of the mechanism of action of DRAIC.
Discussion
Our lab previously identified a tumor suppressive long noncoding RNA, DRAIC which is expressed in the androgen dependent prostate cancer cell lines and whose expression is decreased in castration resistant aggressive prostate cancer (37). DRAIC inhibited cancer cell invasion and migration. In this study we show that DRAIC also suppresses soft agar colony formation and growth of xenograft tumors, without affecting cell proliferation when cells are adherent to tissue culture plates. Gene expression analysis in cancers with low levels of DRAIC alerted us to the possibility that DRAIC may suppress NF-κB regulated genes. Indeed, knockdown (or knockout) of DRAIC increased the NF-κB reporter activity and over-expression of DRAIC decreased the reporter activity. The increased invasion was seen upon knockdown/knockout of DRAIC is reversed by inhibiting the NF-κB pathway. DRAIC physically associates with the trimeric IKK complex, decreases the integrity of the complex and inhibits IKK kinase activity on IκBα, thus stabilizing this inhibitor of NF-κB. Finally, the minimal region of DRAIC responsible for suppressing NF-κB activity was also sufficient for inhibiting cancer cell invasion and soft agar colony formation. Altogether, our results suggest that DRAIC is acting as a tumor suppressor by inhibiting the NF-κB pathway by inhibiting cell invasion, migration, tumor growth and inflammatory gene expression.
NF-κB is a master transcription factor for regulating the inflammatory signaling pathway in different cancers including prostate (42) and it has been shown that the NF-κB pathway plays a critical role in regulating castration resistant and metastatic prostate cancer (44). The NF-κB pathway has been shown to stimulate soft agar colony formation in breast carcinomas, melanomas, colon carcinomas and osteocarcinomas (45) and invasion in breast and skin carcinomas (46,47). NF-κB is an attractive target for therapy in cancers (48,49). These reports support our conclusion that the increase in invasion and soft agar colony formation seen upon DRAIC down-regulation can be explained by the activation of the NF-κB pathway. Since NF-κB activation has also been associated with poor outcome in solid tumors (50), we hypothesize that the poor outcome associated with low DRAIC is also at least partly due to the activation of NF-κB.
The mechanism by which DRAIC inhibits IKK and thus inhibits NF-kB is also very interesting. There are reports in the literature that single agents like thalidomide block NF-κB activation via suppression of IKK kinase activity (51). A proteasomal inhibitor PS-341 can also block NF-κB activation and cause growth arrest and apoptosis in glioblastoma cell lines and tumor explants (52). The IKK inhibitor Bay11–7082 has also been effective at inhibiting NF-κB activity and thus increasing apoptosis (53). The IKK kinase activity is stimulated by several agents like TNFR, TLR or IL1R including intracellular DNA damage and reactive oxygen species (54). It will be interesting in the future if any of these agents mediate their effect on IKK through the induction or repression of DRAIC. Conversely, DRAIC may affect the modifications on IKK subunits that are associated with trimeric complex formation. Androgen activation has been shown to increase NF-κB activity (55), and to simultaneously repress DRAIC, so an interesting possibility is that this effect of androgen on NF-κB may be mediated, at least partially, through the reduction of DRAIC.
There is at least one other long noncoding RNA involved in regulating NF-κB pathway (36,56). Liu et al showed that the NF-κB inducing long noncoding RNA, NKILA interacts with the NF-κB: IκBα complex, shields IκBα from phosphorylation by IKK and thus inhibits NF-κB pathway and inhibits the migration and invasion in cancer cell (36). Other groups validated this finding independently in colon carcinomas and non-small lung cell carcinomas (57,58). Thus a future question will be if there are other lncRNAs that regulate this pathway at these stages and/or whether there is any co-regulation of DRAIC and NKILA to coordinately regulate NF-κB activity.
Long noncoding RNA might have different cellular function in different tissue lineages. DRAIC has been shown to be behaving as a tumor suppressor in eight different malignant tumors (37) but appears to be oncogenic in breast cancer (59). Tiessen et al. has shown that in breast cancer cells the knockdown of DRAIC increased the autophagic flux and suggested that this is mediated through mTORC1 activation (60). NF-κB pathway has been shown to be involved in autophagy but the relation between these two pathways is complex. NF-κB can repress autophagy by activating mTOR pathway but in the literature people have also found that autophagy is independent of NF-κB (61). We will examine in the future whether DRAIC reduces autophagy in prostate cancer cells, and if so, whether this process is dependent on the effects of DRAIC on NF-κB. It will be interesting to examine whether there are tissue-lineage specific differences in the ability of DRAIC to repress NF-κB.
There are two other examples of lncRNA specifically interacting with and regulating signal transduction proteins: lnc-DC interacts with STAT3 to decrease its interaction with SHP1 phosphatase and thus activates the STAT3 pathway (16) and AK023948 interacts with DHX9 which forms a complex with PI3K-p85 to activate the AKT pathway (62). We hypothesize that regulation of signal transduction proteins by physical interaction of lncRNAs may emerge as an important function of this enigmatic class of molecules.
Supplementary Material
Significance.
A cytoplasmic tumor-suppressive lncRNA interacts with and inhibits a major kinase that activates an oncogenic transcription factor in prostate cancer.
Acknowledgement
This work was supported by R01 AR067712 to AD, NCI R01 CA192399 to MWM, DOD award PC151085 to MK and T32 GM007267 to AC. We acknowledge the MAPS core facility at University of Virginia and Tiffany Melhuish-Baxter for help with the mouse xenograft experiment. M. Murat Koseoglu, Róża K. Przanowska for helpful discussions. Manikarna Dinda for critical reading and helpful comments.
Footnotes
Competing Interests: Authors declare there are no competing interests.
References
- 1.Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer 2001;1:34–45 [DOI] [PubMed] [Google Scholar]
- 2.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004;5:522–31 [DOI] [PubMed] [Google Scholar]
- 3.Kumar P, Kuscu C, Dutta A. Biogenesis and Function of Transfer RNA-Related Fragments (tRFs). Trends Biochem Sci 2016;41:679–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009;10:155–9 [DOI] [PubMed] [Google Scholar]
- 5.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011;147:358–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 2007;8:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci 2007;120:2498–506 [DOI] [PubMed] [Google Scholar]
- 8.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 2009;33:717–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A 2008;105:716–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature 2011;470:284–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang F, Zhang H, Mei Y, Wu M. Reciprocal regulation of HIF-1alpha and lincRNA-p21 modulates the Warburg effect. Mol Cell 2014;53:88–100 [DOI] [PubMed] [Google Scholar]
- 12.Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu M, et al. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis 2014;5:e1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell 2013;52:101–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J, Abdelmohsen K, Yang X, De S, Grammatikakis I, Noh JH, et al. LncRNA OIP5-AS1/cyrano sponges RNA-binding protein HuR. Nucleic Acids Res 2016;44:2378–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giovarelli M, Bucci G, Ramos A, Bordo D, Wilusz CJ, Chen CY, et al. H19 long noncoding RNA controls the mRNA decay promoting function of KSRP. Proc Natl Acad Sci U S A 2014;111:E5023–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 2014;344:310–3 [DOI] [PubMed] [Google Scholar]
- 17.Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun 2013;4:2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quinodoz S, Guttman M. Long noncoding RNAs: an emerging link between gene regulation and nuclear organization. Trends Cell Biol 2014;24:651–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016;29:452–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell 2015;28:529–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 2014;15:7–21 [DOI] [PubMed] [Google Scholar]
- 23.Prensner JR, Chen W, Han S, Iyer MK, Cao Q, Kothari V, et al. The long non-coding RNA PCAT-1 promotes prostate cancer cell proliferation through cMyc. Neoplasia 2014;16:900–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature 2013;500:598–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malik R, Patel L, Prensner JR, Shi Y, Iyer MK, Subramaniyan S, et al. The lncRNA PCAT29 inhibits oncogenic phenotypes in prostate cancer. Mol Cancer Res 2014;12:1081–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Pitchiaya S, Cieslik M, Niknafs YS, Tien JC, Hosono Y, et al. Analysis of the androgen receptor-regulated lncRNA landscape identifies a role for ARLNC1 in prostate cancer progression. Nat Genet 2018;50:814–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao B, Lu YL, Yang Y, Hu LB, Bai Y, Li RQ, et al. Overexpression of lncRNA ANRIL promoted the proliferation and migration of prostate cancer cells via regulating let-7a/TGF-beta1/ Smad signaling pathway. Cancer Biomark 2018;21:613–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang T, Qu X, Jiang J, Gao P, Zhao D, Lian X, et al. Diagnostic significance of urinary long non-coding PCA3 RNA in prostate cancer. Oncotarget 2017;8:58577–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto Y, Gaynor RB. Role of the NF-kappaB pathway in the pathogenesis of human disease states. Curr Mol Med 2001;1:287–96 [DOI] [PubMed] [Google Scholar]
- 30.Chen F, Castranova V, Shi X, Demers LM. New insights into the role of nuclear factor-kappaB, a ubiquitous transcription factor in the initiation of diseases. Clin Chem 1999;45:7–17 [PubMed] [Google Scholar]
- 31.Wang CY, Mayo MW, Baldwin AS Jr. TNF- and cancer therapy-induced apoptosis: potentiation by inhibition of NF-kappaB. Science 1996;274:784–7 [DOI] [PubMed] [Google Scholar]
- 32.Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol 2005;5:749–59 [DOI] [PubMed] [Google Scholar]
- 33.Ben-Neriah Y, Karin M. Inflammation meets cancer, with NF-kappaB as the matchmaker. Nat Immunol 2011;12:715–23 [DOI] [PubMed] [Google Scholar]
- 34.Hayden MS, Ghosh S. NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev 2012;26:203–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonizzi G, Karin M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol 2004;25:280–8 [DOI] [PubMed] [Google Scholar]
- 36.Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 2015;27:370–81 [DOI] [PubMed] [Google Scholar]
- 37.Sakurai K, Reon BJ, Anaya J, Dutta A. The lncRNA DRAIC/PCAT29 Locus Constitutes a Tumor-Suppressive Nexus. Molecular cancer research : MCR 2015;13:828–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hwang B, Phan FP, McCool K, Choi EY, You J, Johnson A, et al. Quantification of cellular NEMO content and its impact on NF-kappaB activation by genotoxic stress. PloS one 2015;10:e0116374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 2005;102:15545–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cancer Genome Atlas Research N. The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015;163:1011–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009;458:223–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia Y, Shen S, Verma IM. NF-kappaB, an active player in human cancers. Cancer Immunol Res 2014;2:823–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan M, Xu Q, Zhang P, Zhou XJ, Zhang ZY, Chen WT. Correlation of NF-kappaB signal pathway with tumor metastasis of human head and neck squamous cell carcinoma. BMC Cancer 2010;10:437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin R, Yi Y, Yull FE, Blackwell TS, Clark PE, Koyama T, et al. NF-kappaB gene signature predicts prostate cancer progression. Cancer Res 2014;74:2763–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Higgins KA, Perez JR, Coleman TA, Dorshkind K, McComas WA, Sarmiento UM, et al. Antisense inhibition of the p65 subunit of NF-kappa B blocks tumorigenicity and causes tumor regression. Proc Natl Acad Sci U S A 1993;90:9901–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu Y, Zhou BP. TNF-alpha/NF-kappaB/Snail pathway in cancer cell migration and invasion. Br J Cancer 2010;102:639–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith SM, Lyu YL, Cai L. NF-kappaB affects proliferation and invasiveness of breast cancer cells by regulating CD44 expression. PloS one 2014;9:e106966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bradford JW, Baldwin AS. IKK/nuclear factor-kappaB and oncogenesis: roles in tumor-initiating cells and in the tumor microenvironment. Adv Cancer Res 2014;121:125–45 [DOI] [PubMed] [Google Scholar]
- 49.Durand JK, Baldwin AS. Targeting IKK and NF-kappaB for Therapy. Adv Protein Chem Struct Biol 2017;107:77–115 [DOI] [PubMed] [Google Scholar]
- 50.Wu D, Wu P, Zhao L, Huang L, Zhang Z, Zhao S, et al. NF-kappaB Expression and Outcomes in Solid Tumors: A Systematic Review and Meta-Analysis. Medicine (Baltimore) 2015;94:e1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keifer JA, Guttridge DC, Ashburner BP, Baldwin AS Jr. Inhibition of NF-kappa B activity by thalidomide through suppression of IkappaB kinase activity. J Biol Chem 2001;276:22382–7 [DOI] [PubMed] [Google Scholar]
- 52.Yin D, Zhou H, Kumagai T, Liu G, Ong JM, Black KL, et al. Proteasome inhibitor PS-341 causes cell growth arrest and apoptosis in human glioblastoma multiforme (GBM). Oncogene 2005;24:344–54 [DOI] [PubMed] [Google Scholar]
- 53.Garcia MG, Alaniz L, Lopes EC, Blanco G, Hajos SE, Alvarez E. Inhibition of NF-kappaB activity by BAY 11–7082 increases apoptosis in multidrug resistant leukemic T-cell lines. Leuk Res 2005;29:1425–34 [DOI] [PubMed] [Google Scholar]
- 54.Hinz M, Scheidereit C. The IkappaB kinase complex in NF-kappaB regulation and beyond. EMBO Rep 2014;15:46–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang L, Altuwaijri S, Deng F, Chen L, Lal P, Bhanot UK, et al. NF-kappaB regulates androgen receptor expression and prostate cancer growth. Am J Pathol 2009;175:489–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu Y, Liu X, Xie M, Liu M, Ye M, Li M, et al. The NF-kappaB-Responsive Long Noncoding RNA FIRRE Regulates Posttranscriptional Regulation of Inflammatory Gene Expression through Interacting with hnRNPU. J Immunol 2017;199:3571–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang J, Zhao L, Chen W, Duan J, Shrestha D, Zhou R, et al. LncRNA NKILA suppresses colon cancer cell proliferation and migration by inactivating PI3K/Akt pathway. Translational Cancer Research 2018;7:1431–8 [Google Scholar]
- 58.Lu Z, Li Y, Wang J, Che Y, Sun S, Huang J, et al. Long non-coding RNA NKILA inhibits migration and invasion of non-small cell lung cancer via NF-kappaB/Snail pathway. J Exp Clin Cancer Res 2017;36:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao D, Dong JT. Upregulation of Long Non-Coding RNA DRAIC Correlates with Adverse Features of Breast Cancer. Noncoding RNA 2018;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tiessen I, Abildgaard MH, Lubas M, Gylling HM, Steinhauer C, Pietras EJ, et al. A high-throughput screen identifies the long non-coding RNA DRAIC as a regulator of autophagy. Oncogene 2019;38:5127–41 [DOI] [PubMed] [Google Scholar]
- 61.Comb WC, Cogswell P, Sitcheran R, Baldwin AS. IKK-dependent, NF-kappaB-independent control of autophagic gene expression. Oncogene 2011;30:1727–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koirala P, Huang J, Ho TT, Wu F, Ding X, Mo YY. LncRNA AK023948 is a positive regulator of AKT. Nat Commun 2017;8:14422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.