Abstract
Due to its dismal outcome, pancreatic cancer (PDAC) remains a therapeutic challenge making the testing of new pharmacological tools a goal of paramount importance. Here, we developed a rational approach for inhibiting PDAC growth based on leveraging cell cycle arrest of malignant cells at a phase that shows increased sensitivity to distinct epigenomic inhibitors. Specifically, we simultaneously inhibited Checkpoint kinase 1 (Chk1) by prexasertib and the G9a histone methyltransferase with BRD4770, thereby targeting two key pathways for replication fork stability. Methodologically, the antitumor effects and molecular mechanisms of the combination were assessed by an extensive battery of assays, utilizing cell lines and patient-derived cells as well as 3D spheroids and xenografts. We find that the prexasertib-BRD4770 combination displays a synergistic effect on replication-associated phenomena, including cell growth, DNA synthesis, cell cycle progression at S phase, and DNA-damage signaling, ultimately leading to a highly efficient induction of cell death. Moreover, cellular and molecular data reveal that the synergistic effect of these pathways can be explained, at least in large part, by the convergence of both Chk1 and G9a functions at the level of the ATR-RPA-checkpoint pathway, which is operational during replication stress. Thus, targeting the epigenetic regulator G9a, which is necessary for replication fork stability, combined with inhibition of the DNA damage checkpoint, offers a novel approach for controlling PDAC growth through replication catastrophe.
Implications
This study offers an improved, context-dependent, paradigm for the use of epigenomic inhibitors and provides mechanistic insight into their potential therapeutic use against PDAC.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) ranks third as a leading cause of cancer-related deaths in the U.S., with a median survival of 6 months and a devastating 5-year survival of 3–5%(1). This rate continues to rise with predictions that PDAC will hold the second position for cancer-related deaths by 2030(2). The aggressive biology, rapid dissemination, and late diagnosis advance this malignancy to an incurable stage, making therapy a challenge. Surgery, which offers the best chance for survival, is applicable to fewer than 20% of patients(3). Even with surgery, the disease recurs in approximately 80 percent of these patients, who die within five years of recurrence.
Unfortunately, PDAC is also highly resistant to chemotherapy and radiation. In fact, during the last 4 decades, only four drugs have been approved by the FDA to treat PDAC, which include gemcitabine (1996), erlotinib (2005), albumin-bound paclitaxel (2013) and irinotecan liposome injection (2015)(4,5). While FOLFIRINOX and gemcitabine plus nab-paclitaxel have been shown to improve survival(6,7), the improvement is incremental with the majority of patients still rapidly succumbing to their disease. Thus, there remains an urgent need of novel therapies for PDAC, in particular, targeting pathways highly relevant to its pathobiology.
PDAC, like many other malignancies, is a disease that involves the accumulation of both, genetic and epigenetic aberrations, and an interplay between them(8–11). In fact, gene expression networks that support tumorigenesis are modulated by epigenetic regulators and ultimately fixed by altered signaling from mutated oncogenes and tumor suppressors to define the PDAC phenotype. As a result, the development of small molecules that reversibly modify the cancer-associated epigenome is rapidly growing, and their most promising use, in particular in the context of solid tumors, is thought to be in combination therapies. However, most of these agents are being studied within the framework of their gene regulatory activity without taking into consideration their effects during the distinct cell cycle phases, which we believe to be critical for better understanding cancer. In fact, we have recently shown that arresting cells in G2/M with an Aurora kinase A inhibitor while combining them with an inhibitor of the epigenetic H3K9 methylation pathway is an effective approach for altering chromatin structure in a manner that gives rise to an aberrant mitotic checkpoint response leading to rapid death(12). This approach suggested that the capacity of cell-cycle inhibitors could be harnessed to enhance the use of epigenetic inhibitors. Here, we sought to combine targeting of Checkpoint kinase 1 (Chk1), a key regulator of cell cycle transition through its checkpoint function in response to DNA damage and G9a, a histone methyltransferase (HMT) for histone H3 lysine 9 mono- and di-methylation (H3K9me1 and H3K9me2), which remodels chromatin during DNA replication. Notably, we report that prexasertib (LY2606368), a Chk1 inhibitor, and BRD4770, a G9a inhibitor, together reduce the growth of PDAC cells, in both cell monolayer and 3D cultures as well as in vivo xenografts, achieving a synergistic effect. This dual inhibition causes cells to arrest in S-phase and results in cell death. Moreover, while cell death coincided with increased levels of cleaved caspase 3, pan-caspase inhibition did not rescue the effect, indicating that the main mechanism involved in this process is not caspase-dependent, a feature that characterizes several, recently described, types of death that differ from the canonical apoptotic pathway of cells dying in interphase(13). We found that combination-treated cells undergo extensive DNA damage with activation of the ATR-RPA replication stress response, an effect that is also observed in tumors from xenografts treated with both inhibitors. In summary, these data establish that targeting the epigenetic regulator G9a in the context of cell cycle inhibition holds promise as a novel combinatorial therapy for PDAC and provides mechanistic insights on how this process occurs. Therefore, this new knowledge extends our mechanistic understanding in the field of experimental therapeutics, which bears significant biomedical relevance.
Materials and methods
Cell lines and reagents
Human PDAC cell lines were obtained from ATCC and maintained according to recommendations. L3.6 (14) was cultured in appropriate media(15). Primary cell cultures were obtained from patient-derived xenografts and maintained as described(8). All cells were cultured at 37ºC in a humidified incubator with 5% CO2. BxPC3, L3.6 and Mia-Paca2 were used a maximum of 30 passages, while primary cultures were utilized a maximum of 15 passages. All cell lines tested negative for mycoplasma with last testing performed on March 8, 2019 (IDEXX BioAnalytics). Authentication of established PDAC lines was confirmed by STR testing at the same time. For in vitro studies, compounds were dissolved in DMSO and stored as 10mM (BRD4770, Cayman Chemicals; Z-VAD-FMK, ApexBio) and 10μM (prexasertib/LY2606368, ApexBio) aliquots at −20ºC for a maximum of 3 months. Antibody information for all relevant experiments is provided in Supplementary table S1.
Cell growth, synergy calculations, live cell viability, apoptosis analysis, clonogenic survival and spheroid viability
Relative cell viability was determined by MTS (CellTiter 96 Aqueous One Solution Cell Proliferation Assay, Promega). For synergy evaluation, cells were treated with seven increasing concentrations (prexasertib) or three independent concentrations (BRD4770). The fraction affected (Fa) was calculated normalized to controls. Pharmacological interaction was calculated using two different methods: 1) Combenefit software(16) applying Highest Single Agent (HSA) and Loewe models, and 2) combination index (CI) utilizing the Chou and Talalay method(17) with CompuSyn software (ComboSyn, Inc), where CI <1, 1 or >1 indicates synergistic, additive and antagonistic effect, respectively. For real-time live cell imaging, 5e3 cells/well were plated and treated. Cell confluency and apoptosis were monitored by the IncuCyte® S3 Live-Cell Analysis System coupled with Caspase-3/7 Apoptosis Green reagent (Essen Bioscience). For clonogenic assays, 3e4 of L3.6 or 1e4 of BxPC3 or Mia-Paca2 cells were plated on 60mm dishes and treated for 7d. Cells were fixed for 5min with 1:7 acetic acid/methanol solution and stained with 0.5% crystal violet (Sigma-Aldrich). Cell density was quantified by Image-J (NIH)(18). For spheroid viability, L3.6 (5e2 for 3d or 1e2 for 7d) were allowed to form spheroids in a three-dimensional (3D) Matrigel matrix. After 4d, media was replaced, and inhibitors added. After 72h or 7d, viability was assessed using the CellTiter-Glo 3D assay (Promega) on Tecan Infinite M200 plate reader. Bright field images were captured by a Canon EOS Rebel Xsi camera mounted on a Nikon Eclipse TS100 microscope at 10X magnification.
Subcutaneous and orthotopic xenografts
Animal experiments were approved by the Medical College of Wisconsin Institutional Animal Care and Use Committee (AUA00006130) and followed the guidelines outlined in the Guide for Care and Use of Laboratory animals (NIH). Right flanks of athymic nude NU/J mice (Foxn1nu, 5–6-week old females; The Jackson Laboratory) were injected with 1e6 L3.6 cells in 1:1 PBS/Matrigel (Corning). When tumors reached an average volume of 100mm3, mice were randomized by Studylog® (Studylog Systems, Inc) into groups (n=17/group) based on tumor volume and body weight. Animals were given vehicle (20% Captisol®, Ligand Pharmaceuticals Inc., in water + 10% DMSO in corn oil), prexasertib (2mg/kg in 20% Captisol®, subcutaneously for 3d followed by 4d off), BRD4770 (10mg/kg in corn oil, subcutaneously 3X/week on days 1, 3 and 5) or combination. Tumors were measured twice weekly, and animals euthanized after 14d. Tumor tissues were formalin-fixed and paraffin-embedded. For orthoptic xenografts, NU/J mice were anesthetized, received a small incision, and injected in the tail of the pancreas with 2.5e5 L3.6+luciferase cells in 1% Matrigel. Mice were observed daily and recovered 5d before treatment initiation. Mice were randomly divided by Studylog® into the 4 treatment groups (n=5/group) based on initial total flux (p/s). For tumor monitoring twice weekly, mice were anesthetized with 0.5% inhaled isoflourane, injected with d-luciferin (112.5mg/kg), and total flux (p/s) was quantified by the PerkinElmer IVIS Spectrum CT In Vivo imaging system. Animals were euthanized after clinical criteria indicated a body condition score of 2(19) or after 20% weight loss.
Cell-cycle and BrdU incorporation assays
For fluorescence-activated cell cycle sorting (FACS), cells were prepared as described(20). DNA content was analyzed on a FACSAria™ II (BD Biosciences). Events (1–5e4) were analyzed with appropriate gating in ModFit LT™ software (v5.0; Verity Software House). For BrdU proliferation assay, L3.6 grown on coverslips were treated for 48h. Cells were pulsed with 10μM BrdU (Sigma-Aldrich) for 15min and fixed with pre-chilled methanol for 5min. DNA was denatured sequentially by 1 and 2N HCl for 10min each and washed 3X with PBS. Cells were blocked with 1% BSA in PBS-Tween 0.1% for 30min, incubated with anti-BrdU overnight at 4ºC, followed by anti-mouse secondary (1:250; Alexa Fluor-488) for 1h at RT and mounted using ProLong™ Gold Antifade with DAPI (Thermo Fisher Scientific). For ssDNA detection by native BrdU assay, cells were grown on coverslips in 10μM BrdU during 24h before treatments. Inhibitors were added in the presence of 10μM BrdU for an additional 24h. Cells were fixed using 4% formaldehyde for 10min at RT, blocked in 1% BSA PBS-T and incubated overnight at 4ºC with anti-BrdU. Slides were incubated with anti-mouse secondary (1:250; Alexa Fluor-488) for 1h at RT. Coverslips were mounted using ProLong™ Gold Antifade with DAPI.
Electron and Immunofluorescence microscopy
For EM, L3.6 were treated, fixed in 2.5% glutaraldehyde in 0.1M sodium cacodylate buffer pH 7.4 for 1h at RT, and washed 3X 5min in cacodylate buffer. Post-fixation was performed in 1% osmium tetroxide solution containing 1.25% potassium hexacyanoferrate for 2h on ice(21). Cells were washed 3X with distilled water, dehydrated through graded methanol and processed into Embed-812 epoxy resin. Ultrathin sections (70nm) were stained with uranyl acetate + lead citrate for examination with a Hitachi H600 transmission electron microscope (TEM). Images were acquired on a Hamamatsu digital camera using AmtV602 image processing software. Immunofluorescence and confocal microscopy were performed as previously described(12). Primary antibodies are indicated in Supplementary table S1, followed by anti-mouse or anti-rabbit secondary antibody, as appropriate. DNA replication sites were visualized by incorporation of IdU and CldU into DNA as described(22). Briefly, L3.6 cells were labeled with 100μM IdU for 1h, washed, then treated with indicated drugs for 24h before incubation with 100μM CldU (Sigma-Aldrich) for 1h. Primary antibodies CldU (rat anti-BrdU) and IdU (mouse anti-BrdU) were diluted in 1% BSA buffer and incubated on slides overnight at 4°C. Fluorescence density of IdU or CldU was quantified by Image-J.
Comet assays
The Trevigen CometAssay® Kit was used to evaluate combined single-stranded (ss) and double-stranded (ds) DNA breaks (alkaline assay) or dsDNA breaks alone (neutral assay)(23). Individual cells at 1e5/ml were suspended in cold PBS and mixed with agarose at a 1:10 ratio, and then 50ul/well was pipetted onto slides. For combined ssDNA and dsDNA evaluation, slides were treated with lysis buffer for 30min and alkaline unwinding solution for 1h in the dark to denature DNA. Slides were then subjected to alkaline electrophoresis at 21V for 30min. For dsDNA breaks alone, slides were incubated 1h with lysis solution and washed in 1X TBE buffer for 15min. Electrophoresis was conducted in neutral conditions using 1X TBE buffer at 21V for 40min. Slides were dried and stained with SYBR safe Green DNA dye (ApexBio) for 30min and viewed by epifluorescence microscopy using 488nm filter. Analysis of DNA damage was conducted using Image-J with Open Comet plug-in (v1.3.1)(24).
Western blot analysis
Western blots were performed as described (25). Membranes were blocked in 5% Milk or 5% BSA (for phosphorylated proteins) in Tris-buffered saline Tween-20 (TBS-T) and incubated overnight at 4ºC with primary antibodies (Supplementary table S1). Blots were developed by Clarity Western ECL Substrate (BioRad).
DNA fiber assay
L3.6 were grown with indicated treatments for 24h. Cells were pulsed with 25μM IdU for 15min, washed 2X with ice-cold PBS, and pulsed with 250μM CldU for an additional 15min. Cells were harvested and resuspended in PBS at a concentration of 5e5/ml, and 2μL of cell suspensions were placed on glass slides and mixed with 7μL of lysis buffer (200mM Tris-HCl, pH 7.5, 50mM EDTA and 0.5% SDS) for 2min. Slides were tilted at 15° to allow fiber spreading and air dried. The slides were fixed in 70% ethanol overnight at 4°C and treated with 2.5N HCL for 30min followed by washes with PBS before blocking in 5% BSA for 30min at 37°C. DNA fibers were incubated with mouse anti-BrdU and rat anti-BrdU to detect incorporated IdU and CldU, respectively, overnight at 4°C, followed by 3 PBS washes, and incubation with anti-mouse (1:400; Alexa Fluor-546; Thermo Fisher Scientific) and anti-rat (1:300; Alexa Fluor-488) secondary for 1h at RT. Coverslips were mounted using ProLong™ Gold Antifade with DAPI.
Immunohistochemistry (IHC)
IHC was performed on xenograft tissue as described(26). Primary antibodies (Supplementary table S1) were incubated overnight at 4ºC. For TUNEL, tissues were processed using the ApopTag® Peroxidase in Situ Apoptosis Detection Kit (Millipore, S7100). Slides were developed with Nova Red (Vector Laboratories) and counterstained with Mayer’s hematoxylin. Five random fields (20X objective) per tumor, containing at least 1000 cells per field, were imaged and counted.
Results
Combined pharmacological inhibition of the G9a and CHK1 pathways inhibits pancreatic cancer cell growth in a synergistic manner
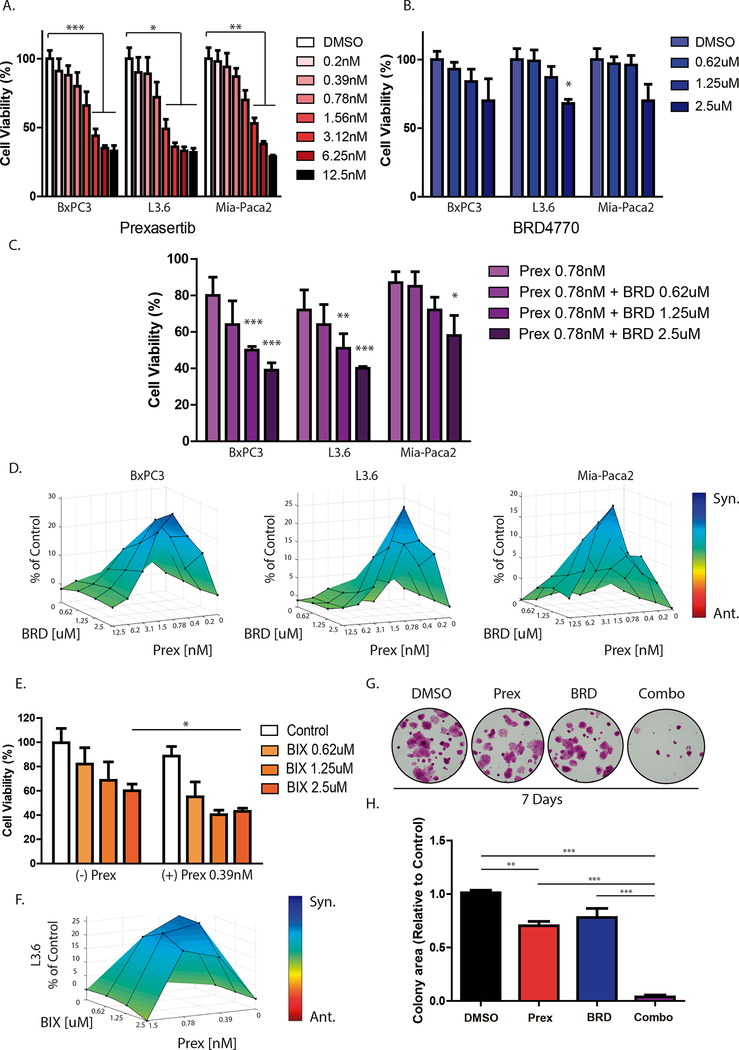
To evaluate the antitumor effects of inhibiting both, the cell cycle checkpoint protein CHK1 and the epigenetic regulator G9a, we first tested the effect of the CHK1 inhibitor, prexasertib (LY2606368), and the G9a HMT inhibitor, BRD4770, on cell proliferation, individually, in PDAC cell lines. We found that prexasertib (0.39 to 12.5nM) reduced cell viability in a dose-dependent manner in 3 well-characterized, aggressive human PDAC cell lines (BxPC3, L3.6 and Mia-Paca2) after 72 hours (Fig. 1A). Cell viability was significantly reduced on BxPC3 and Mia-Paca2 cells above 3.12nM (BxPC3: 44±5% vs. control at 3.12nM, 35±2% at 6.25nM, and 33±4% at 12.5nM; P< 0.01; Mia-Paca2: 53±4% vs. control at 3.12nM, 38±2% at 6.25nM, and 29±1% at 12.5nM; P< 0.01). For L3.6 cells, this effect was significant above 1.56nM (49±7% vs. control at 1.56nM, 36±3% at 3.12nM, 33±3% at 6.25nM, and 32±3% at 12.5nM; P<0.05). Treatment with BRD4770 alone (0.63–2.5μM) showed only a partial inhibition of growth in these cells (Fig. 1B). For BxPC3 and Mia-Paca2, this effect was not significant at 72 hours (BxPC3: 93±5% vs. control at 0.62μM; 84±9% vs. control at 1.25μM; and 70±16% vs. control at 2.5μM; P>0.05; Mia-Paca2: 97±5% vs. control at 0.62μM; 96±7% vs. control at 1.25μM; 70±12% vs. control at 2.5μM; P>0.05). For L3.6, 2.5μM of BRD4770 demonstrated a significant reduction in viability (68±3% vs control; P<0.05). Therefore, to evaluate if the combination of these inhibitors improves their individual efficacy, we treated PDAC cells with prexasertib at a dose that did not significantly impact cell viability (0.78nM) and three different concentrations of BRD4770 (0.62, 1.25 and 2.5μM). After 72 hours, the combination of prexasertib with BRD4770 significantly reduced cell viability in a dose-dependent manner more than either agent individually (Fig. 1C), starting at 1.25μM on BxPC3 and L3.6 cells (BxPC3: 80±10% with 0.78nM prexasertib alone vs. 50±2% combined with 1.25μM BRD4770 and vs. 39±4% combined with 2.5μM BRD4770; P<0.001; L3.6: 72±11% with 0.78nM prexasertib alone vs. 51±8% combined with 1.25μM BRD4770; P<0.01; and vs. 40±1% combined with 2.5μM BRD4770; P<0.001). For Mia-Paca2 cells, cell viability was 87±6% with 0.78nM prexasertib without BRD4770 compared to 58±11% in combination with 2.5μM BRD4770 (Fig. 1C; P<0.05). Thus, the dual inhibition of CHK1 and G9a reduces cell viability in PDAC cells, cultured in monolayer, more effectively than either treatment alone.
Figure 1. Combined CHK1 and G9a inhibition has a synergistic effect on PDAC cells.
PDAC cell lines (BxPC3, L3.6 and MiaPaca2) were treated with increasing concentrations of (A) the CHK1 inhibitor, prexasertib (Prex) or (B) the G9a inhibitor BRD4770 (BRD). Cell viability was evaluated after 72 hours of treatment using CellTiter MTS assay, and effect of treatments was compared against vehicle (DMSO) only treatment. Mean ± SEM are shown. (*, P<0.05; ***, P<0.001; t-test) C, PDAC cell lines were treated with prexasertib (0.78nM) in combination with BRD4770 at increasing concentrations (0.62–2.5μM) and viability evaluated after 72 hours. (*, P<0.05; **, P<0.01; ***, P<0.001; t-test) D, 3D representations of calculated pharmacological interactions using Combenefit are shown for combined treatment of prexasertib with BRD4770 after 72h at the indicated doses. Synergistic interactions are shown in blue. E, L3.6 treated with another G9a inhibitor, BIX01294 (BIX), in absence or presence of Prex further validate this effect. F, Combenefit analysis confirmed synergistic interactions. G, Clonogenic cell survival assays were performed on L3.6 with sustained inhibition of CHK1 and G9a for 7d using Prex (0.78nM) and BRD (1.25μM), alone or in combination. Representative images are shown. H, Graph depicts quantification of colony density by area relative to control (DMSO) condition. (**, P<0.01; ***, P<0.001; t-test).
Subsequently, we quantified the drug combination effects, determining the combination index (CI) of prexasertib and BRD4770. An 8×3 matrix format with MTS assay was used to measure the type of interaction between these two inhibitors. For this purpose, treatments were combined at a non-fixed concentration ratio, using dose ranges of 0.19–12.5nM for prexasertib and 0.62–2.5μM for BRD4770. Using Combenefit software(16), the synergy distribution demonstrated the highest HSA and Loewe scores when BRD4770 (2.5μM) was combined with low doses of prexasertib (0.78nM for BxPC3 and L3.6 cells, 1.56 or 3.12nM for MiaPaca2 cells) (Fig. 1D and Supplementary Fig 1A). Similar results were obtained using Compusyn, which determines an additive effect (CI=1), synergism (CI<1), or antagonism (CI>1) based on the theory of Chou-Talalay(17). The majority of combination treatments at 72 hours had CIs less than 1, indicating synergism between these two inhibitors, even at low concentrations of prexasertib (0.78nM), with CI values of 0.27, 0.35 and 0.27 in BxPC3, L3.6 and Mia-Paca2, respectively (Supplementary Fig 1B). This synergistic effect was confirmed using two primary patient-derived PDAC cell lines, 03.096 and LIPC, to better represent the complexity of human tumor biology. For this purpose, we evaluated cell viability in response to treatments, using prexasertib (6.25nM) combined with two different doses of BRD4470 (1.25 and 2.5μM) and calculated the synergy distribution (Supplementary Fig. 2). We found that the combinations synergistically reduced cell viability over individual treatments. For 03.096, cell viability was 63.6±2.9% with 6.25nM prexasertib and 1.25μM BRD4770 (P<0.01) for a CI of 0.76 and 54.6±4.8% with 6.25nM prexasertib and 2.5μM BRD4770 (P<0.05) for a CI of 0.62. For LIPC cells, cell viability was 50.3±15.9% (CI= 0.85) and 24.6±7.2% (P<0.01; CI= 0.58) with the same dose combinations, respectively.
To further evaluate the specificity of this effect, we tested the combination of prexasertib with a different, yet well-known, G9a inhibitor, BIX01294, which has a mechanism of action different from that of BRD4770 at the molecular level. BRD4770 is a S-adenosylmethionine (SAM) cofactor-competitive inhibitor of G9a, whereas BIX01294 behaves as a H3 peptide substrate-competitive inhibitor(27). Similar to BRD4770, BIX01294 showed a synergistic effect when combined with 0.39 and 0.78nM prexasertib, in particular at a concentration of 1.25μM, with CIs of 0.56 for 0.39nM prexasertib and 0.58 for 0.78nM prexasertib (Fig. 1E and F; Supplementary Fig. 2I). The conserved synergy observed between prexasertib and two different G9a inhibitors further supports the specificity of attributing this effect to targeting of the G9a complex in combination with CHK1 inhibition.
To determine the long-term effect of prexasertib and BRD4770 treatment on cell growth, we performed clonogenic survival assays. The clonogenic assay provides insight into the ability of a cell to undergo several rounds of cell division, thus assessing long-term effects on cell survival and the potential to form colonies of cancerous cells. Toward this end, cells were exposed to treatments for 7 days. The dose of BRD4770 was reduced for this long-term assay, since sustained inhibition of G9a with 2.5μM BRD4770 alone essentially eliminated L3.6 colony formation (Supplementary Fig. 3A–B). Individually, the treatment with 0.78nM prexasertib or 1.25μM BRD4770 partially reduced surviving colonies (Fig. 1G and H; 70.3±4.0% of control for prexasertib; P<0.01; and 78.1±7.2% of control for BRD4770; P>0.05). However, the combination of both inhibitors nearly abolished clonogenic survival with 97.0% of inhibition (3.0±1.5% of control; P<0.001). Similar effects on clonogenic survival after 7 days were demonstrated for BxPC3 (with 3.12nM prexasertib and 2.5μM BRD4770; Supplementary Fig. 3C–D) and Mia-Paca2 (with 6.25nM prexasertib and 2.5μM BRD4770; Supplementary Fig. 3E–F). Thus, the combination of prexasertib with BRD4770 demonstrated a significant reduction of survival and the ability of these cells to form colonies.
Combined inactivation of CHK1 and G9a is effective to reduce pancreatic cancer cell growth in 3D Culture Models and xenografts in vivo
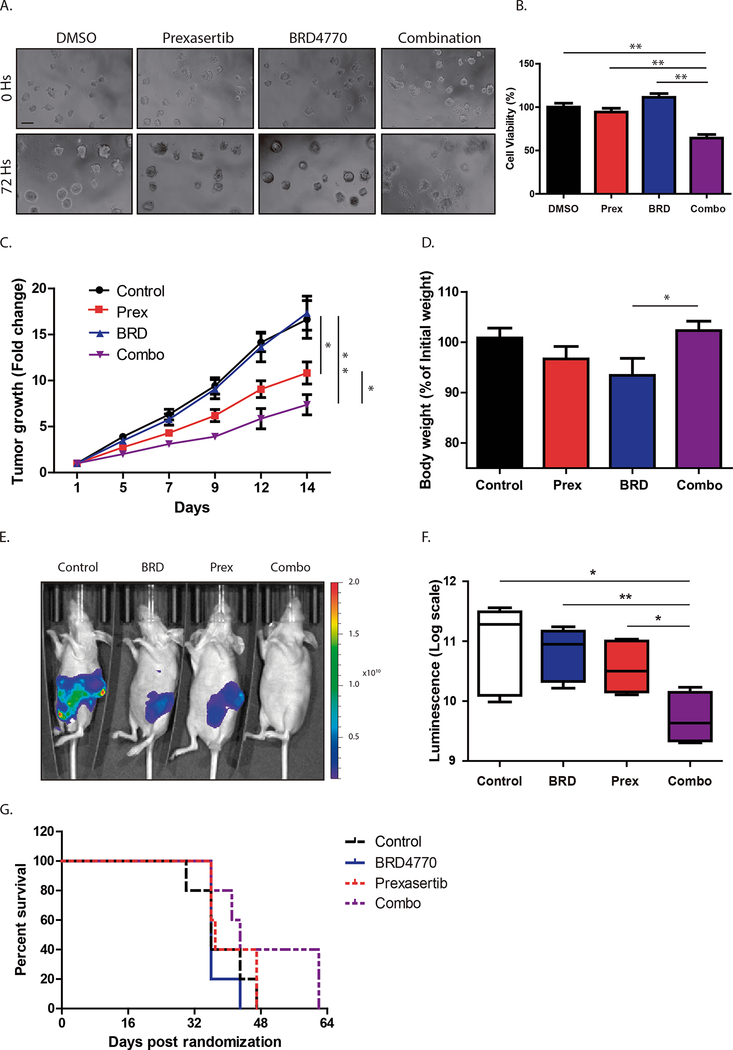
Since 3D cultures more closely simulate in vivo conditions than monolayer conditions, we treated L3.6 spheroids to evaluate the effect of the combination. Spheroids, which represent multilayer aggregates of cells, often demonstrate increased resistance to treatments(28). Cells were seeded in Matrigel and grown for 4 days to allow spheroid formation before adding prexasertib, BRD4770, or their combination. For this assay, we used a slightly higher concentration of prexasertib at 1.2nM and compared spheroid viability after 72 hours (Fig. 2A and B) or 7 days (Supplementary Fig. 4A–B) of treatment. After 72 hours of exposure, individual treatments with prexasertib (94.3±4.3%) or BRD4770 (111.3±4.4%) did not show any significant difference in comparison with control conditions (Fig. 2B; P>0.05). However, combination treatment significantly reduced spheroid viability (65±6.2%; P<0.01) when compared to control or individual treatments. In addition, there was a notable disruption of spheroid structure following combination treatment (Fig. 2A). Similarly, after 7 days of treatment, while prexasertib (109.7±7.8%) and BRD4770 (93.3±3.3%) individually did not show any significant difference compared to vehicle control (Supplementary Fig. 4A–B; P>0.05), combination treatment significantly reduced spheroid viability (32.3±5.9%; P<0.001). Thus, the combined inhibition of both targets antagonizes PDAC cell growth, even in 3D culture models that more closely simulate in vivo conditions.
Figure 2. Dual targeting of CHK1 and G9a inhibits growth of PDAC 3D spheroids and in vivo xenografts.
L3.6 spheroids treated with control (DMSO), prexasertib (Prex; 1.2nM), BRD4770 (BRD; 2.5μM) or their combination. A, Representative pictures after 72h are shown. (Scale bar, 200μm) B, ATP production was measured using CellTiter Glo 3D assay, and mean cell viability was normalized relative to vehicle-treated control (Mean ± SEM, n=3). (***, P<0.001; t-test) C, L3.6 cells were injected subcutaneously into mice. Once tumor volumes reached approximately 100mm3, randomized animals (n=17/group) were treated with control, prexasertib (2mg/kg), BRD4770 (10mg/kg) or their combination. Tumor volume after 14d of treatment is expressed relative to initial tumor volume. (*, P<0.05; **, P<0.01; t-test) D, Graph depicts body weights of mice at the end of treatment expressed as percentage of initial weights (Mean ± SEM, n=17). (*, P<0.05; t-test) E, Representative images of L3.6 luciferase-expressing orthotopic xenografts after 4wks of treatments. Luciferase activity was measured as total flux (photons/second). F, Boxplot graph depicts total luciferase activity expressed as log scale. Values are expressed as mean±SEM (n=5/group; *, P< 0.05; **, P< 0.01; t-test). G, Kaplan-Meier survival analysis showing prolonged survival of combination-treated mice compared to animals receiving control or individual treatments.
Subsequently, we evaluated the effect of the CHK1 and G9a inhibitors in vivo on the growth of subcutaneous L3.6 xenografts. Once tumors reached an average volume of 100mm3, mice were randomized to receive vehicle control, prexasertib alone, BRD4770 alone or combination treatments (n=17/group), and tumor growth was evaluated as fold-change (FC) in volume from initial tumor volume (Fig. 2C). With individual treatments, we found that prexasertib alone significantly diminished tumor growth (FC= 10.83±1.21) when compared with the control group (FC= 16.64±2.05; P<0.05) while BRD4770 alone did not produce any significant change in tumor growth (FC= 17.33±1.85) compared to controls (P>0.05). The group receiving combination treatment displayed significantly reduced tumor growth (FC= 7.37±1.10) when compared to control (P<0.01), prexasertib alone (P<0.05) or BRD4770 alone (P<0.001). By the end of treatment, animals that received individual treatments showed a slight reduction in body weight when compared to controls (Fig. 2D; 100.8±1.9% of starting weight for controls vs. 96.7±0.5% with prexasertib or vs. 93.4±3.3% with BRD4770). However, combination-treated animals maintained or slightly improved body weights to that of controls (102.3±1.8%), which was significant compared to BRD4770 alone treated animals (Fig. 2D; P<0.05). Xenograft tumors were then evaluated by distribution of volume FC in quartiles. The majority of combination-treated tumors distributed in the two lower quartiles (94.1%), with the majority (70.6%) falling exclusively in the lowest quartile (Supplementary Fig. 4C–E), representing the smallest tumor volumes. Finally, to evaluate the long-term effect of the combination on a model more representative of the complex biology of PDAC, we established orthotopic xenograft tumors. L3.6 cells stably expressing luciferase were injected into the pancreas of athymic nude mice to allow accurate monitoring of time-dependent in vivo tumor growth. After 5 days, mice were randomized by baseline bioluminescence measurements and treated with vehicle, prexasertib alone, BRD4770 alone, or their combination (n = 5/group). While individual treatments did not demonstrate a significant reduction in tumor growth (Fig. 2E–F; bioluminescence emission values expressed in logarithmic scale of 10.89±0.32 for control animals vs. 10.79±0.19 with BRD4770 alone; P>0.05 or vs. 10.56±0.19 with prexasertib alone; P>0.05), combination-treated animals displayed a significant reduction in tumor growth when compared with all groups after 4 weeks of treatment (Fig. 2E–F; 9.72±0.18; P< 0.05 vs. control and prexasertib alone; P>0.01 vs. BRD4770 alone). Furthermore, combination treatment prolonged overall survival as shown by Kaplan-Meier curve (Fig. 2G; median survival of 36 days for control and BRD4770 alone, 37 days for prexasertib alone and 43 days for combination). Therefore, the prexasertib-BRD4770 combination is more effective than each agent alone for inhibiting tumor cell growth both, in vitro and in vivo, and demonstrates clinical signs of improved health and survival in vivo.
Dual inhibition of CHK1 and G9a arrests cells in S-phase with a concomitant increase in cell death
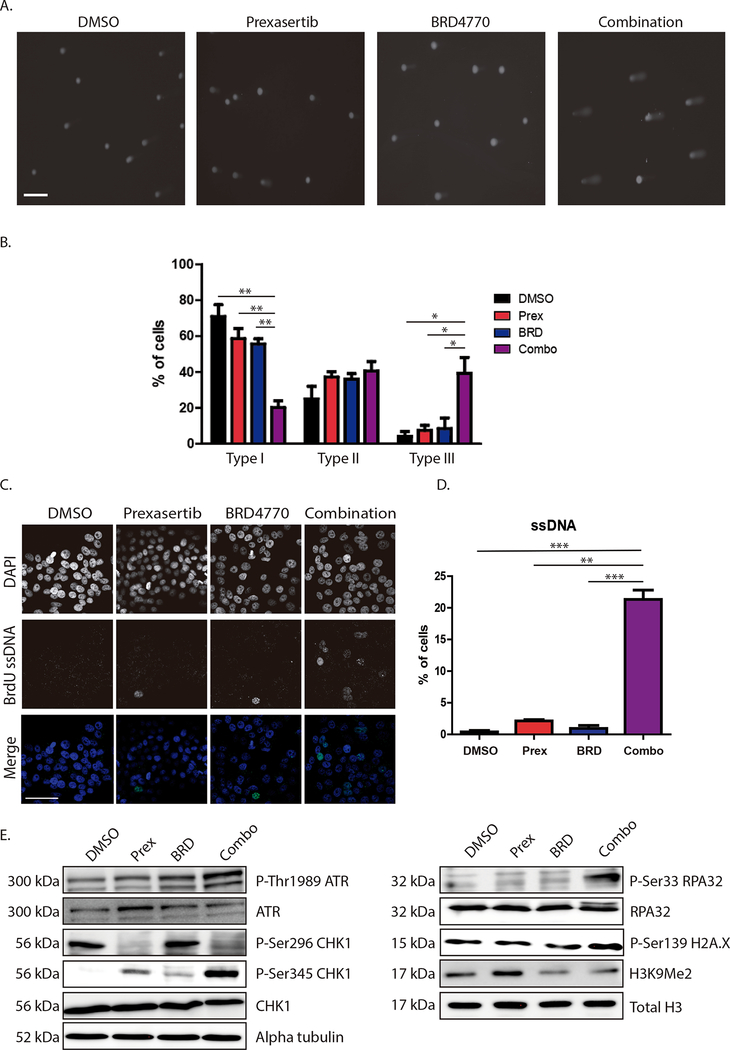
Since CHK1 is part of a checkpoint pathway that regulates cell cycle in response to DNA damage and G9a is an epigenetic regulator necessary for remodeling chromatin during DNA replication, we examined the mechanisms by which antagonizing these pathways together leads to PDAC growth inhibition. We first measured how these inhibitors impact cell cycle distribution using FACS analysis. We found that BRD4770 treatment alone (2.5μM) caused a slight increase in the number of cells in G2-M, while the combination resulted in a slight decrease of cells in this phase (Fig. 3A and 3B; 23.3±6.1% with BRD4770 vs. 16.0±2.3% in vehicle control, vs. 17.7±2.7% with prexasertib and vs. 10.7± 3.9% for combination-treated cells). Prexasertib alone at 0.78nM did not significantly alter cell cycle distribution compared to the vehicle control. However, the combination of prexasertib with BRD4770 had a significantly higher number of cells in S-phase compared with individual treatments or control (56.4±4.5% with combination vs. 37.3±4.6% in control; P<0.05; vs. 31.7±6.3% for prexasertib alone; P<0.05; and vs. 35.8±6.1% for BRD4770 alone; P=0.05). In addition, combination treatment increased the sub-G1 population compared to the other conditions (15.2±6.2% with combination vs. 2.3±0.6% in control, 1.6±0.3% with prexasertib alone and 4.1±0.8% for BRD4470 alone), demonstrating that the combination triggers cell death along with a higher proportion of cells in S-phase. Consequently, to evaluate whether the increased percentage of cells in S-phase was due to enhanced proliferation or cell cycle arrest during DNA replication, we measured DNA synthesis by BrdU incorporation in L3.6 cells (Fig. 3C). After 48 hours, prexasertib treatment alone (0.78nM) did not change the percentage of cells incorporating BrdU compared to control (Fig. 3D; 52.5±2.2% with prexasertib vs. 52.6±1.4% in control cells, P>0.05), while BRD4770 alone (2.5μM) significantly reduced the percentage of BrdU-positive cells (37.3±4.8 vs. control; P<0.05). Evaluation of combination-treated cells revealed an even greater reduction (Fig. 3D; 22.0±5.6%; P<0.01) when compared against control and prexasertib, indicating that, in the context of CHK1 and G9a inhibition, cells accumulate in the S-phase of the cell cycle, and their DNA replication capacity is significantly hindered.
Figure 3. Combined prexasertib-BRD4770 treatment synergizes to trigger DNA replication arrest and cell death.
A, FACS of L3.6 treated for 48h with control (DMSO), prexasertib (prex; 0.78nM), BRD4470 (BRD; 2.5μM) or their combination (Combo) was performed to determine cell cycle distribution. Combination treatment resulted in S-phase arrest, as indicated by the augmented blue hatch mark area, and cell death, as shown by an increase in Sub-G1 fraction. B, Graph depicts quantification of cell cycle distribution. Percentage of cells in each phase is expressed as Mean±SEM, n=3. (*, P<0.05; t-test) C, BrdU incorporation was evaluated to measure DNA replication. L3.6 were treated with DMSO, prexasertib (0.78nM), BRD4470 (2.5μM) or their combination for 48h, followed by pulsing with BrdU. Representative images are shown. (Scale bar, 50μm). D, Percentage of BrdU-positive cells for each condition was quantified and expressed as Mean±SEM, n=3. (*, P<0.05; **, P<0.01; t-test)
Subsequently, we tested whether cells arrested in S-phase were undergoing cell death, causing the sub-G1 population. We performed time-lapse microscopy using the Incucyte© live imaging system on L3.6 treated with vehicle, prexasertib (0.78nM), BRD4770 (2.5μM) or their combination. As previously observed by MTS assay (Fig. 1C), after 72 hours of treatment, L3.6 cells treated with the combination showed a significant reduction of cell confluence when compared to control and single treatments (Supplementary Fig. 5A). To evaluate apoptosis as a potential mechanism, we performed fluorescent detection of caspase 3/7 activation. While control-treated cells demonstrated a baseline level of caspase 3/7 activation (1±0.12, calculated as fold-change over control), prexasertib and BRD4770 each moderately increased the amount of active caspase 3/7 (1.94±0.29 and 3.66±1.74, respectively). Notably, the combination significantly increased the activation of caspase 3/7 over control and individual treatments (Supplementary Fig. 5B and C; 10.01±1.41; P<0.01 vs. control; P<0.01 vs. prexasertib alone; and P<0.05 vs. BRD4770 alone). Western blot confirmed an increase in cleaved capsase-3 from lysates of combination-treated cells (Supplementary Fig. 5D), and the presence of cell blebbing characteristic of apoptotic cell death was observed by TEM (Supplementary Fig. 5E). However, addition of the pan-caspase inhibitor Z-VAD-FMK (10μM) did not rescue cell death induced by combination treatment (57.2±4.6% for combination vs. 57.9±8.5% for combination + Z-VAD; P>0.05), despite a marked reduction of caspase 3/7 activation (Supplementary Fig. 5A and B; 10.01±1.41 for combination vs. 3.47±0.63 for combination + Z-VAD; P<0.01). Thus, while cell death caused by the prexasertib-BRD4770 combination involves activation of caspases, their inhibition is not able to rescue this process, suggesting that an additional mechanism makes this death irreversible.
The synergistic combination of CHK1 and G9a inhibition results in a failure to compensate the replication stress response
In an attempt to identify the mechanisms underlying the caspase-positive, yet irreversible cell death response, we evaluated the contribution that decreased replication processivity, evidenced by reduced BrdU incorporation with concurrent S-phase arrest, has on this phenomenon. Arrest in S-phase could indicate activation of replication checkpoints, which constitute a specialized branch of DNA damage-associated pathways(29). To investigate whether the combination induced DNA damage-associated pathways, we treated L3.6 cells for 48 hours and performed alkaline comet assay, which evaluates the presence of single- and double-stranded DNA breaks. The presence of a comet tail is indicative of DNA damage, and the length of the tail moment is used to quantify the degree of DNA damage. Tail moments were classified in three groups according to their length, each representing a different level of DNA damage (Type I: 0 to 5, no or low damage; Type II: 5 to 20, mild damage and Type III: more than 20, high damage). In the Type I group with no to low damage, we found that cells with control and individual treatments distributed in this group at similar rates (Fig. 4A and B; 71.0±6.5% for control, 58.6±5.5% for prexasertib and 55.7±2.8% for BRD4770 alone; P>0.05) while combination-treated cells falling within this group were remarkably decreased (20.2±3.7%; P<0.01 compared to each condition). In the type II group, we found similar distribution between four groups (25.0±7.0% for control, 37.2±3.1% with prexasertib, 35.2±3% with BRD4770 and 40.5±5.3% with the combination; P>0.05). Concurrently, we found a significant increase in the percentage of combination-treated cells in the Type III group compared to control or individual treatments (Fig. 4A and B; 39.2±8.9% for the combination vs. 4.0±2.8% for Control; 7.5±2.8% with prexasertib alone and 8.4±6.0% with BRD4770 alone; P<0.05), reflecting a higher level of DNA damage in response to combination treatment. To determine whether the DNA damage was attributed more to single- or double-stranded DNA breaks, we also employed the comet assay under neutral conditions, which specifically measures double-stranded DNA breaks. Tail moments were classified with the same criteria as above. We found no significant differences among the treatments (Supplementary Fig. 6) classified as Type I (56.9±8.9% for control, 64.0±9.3% with prexasertib, 40.6±18.7% with BRD4770 and 61.2±11.7% for the combination; P>0.05), Type II (23.7±4.8% for control, 17.5±4.2% with prexasertib, 30.3±8.4% with BRD4770 and 26.4±7.9% for the combination; P>0.05) or Type III (19.4±4.0% for control, 18.5±5.1% with prexasertib, 29±10.6% with BRD4770 and 12.3±3.8% for the combination; P>0.05), indicating that the differences observed with the alkaline comet assay were mainly due to the presence of single-stranded DNA breaks.
Figure 4. Combined CHK1 and G9a inhibition triggers DNA damage and RS.
A, DNA damage was evaluated using alkaline comet assay in L3.6 treated with control (DMSO), prexasertib (Prex; 0.78nM), BRD4770 (BRD; 2.5 μM) or combination (Combo), for 48h. Representative pictures are shown. (Scale bar, 200μm). B, Extent of DNA damage was classified according to comet tail length: Type I: No or low damage, Type II: Mild damage and Type III: High damage. Percentage of cells in each category was quantified as Mean±SEM, n=3. (*, P<0.05; **, P<0.01; t-test). C, Native BrdU assay was used to detect ssDNA in response to treatments with Prex (0.78nM), BRD (2.5μM) or their combination for 24h in L3.6. Representative images are shown. Nuclei positive for ssDNA (green) were quantified as percentage of total nuclei (DAPI; blue). (Scale bar, 50μm) D, Graph shows quantification expressed as Mean±SEM, n=3. (**, P<0.01; ***, P<0.001; t-test) E, Immunoblot of L3.6 cells after 24h of treatment (DMSO, Prex 0.78nM, BRD 2.5μM, or combo) indicates that RS markers (P-Thr1989-ATR, P-Ser345-CHK1, P-Ser33-RPA32) are increased with combination treatment. Total ATR, CHK1 and RPA are relevant controls. Increase in P-Ser139-H2A.X corroborates enhanced DNA damage. Decrease of P-Ser296-CHK1 (Prex and Combo) verifies on-target inhibition of CHK1, while decrease of H3K9Me2 (BRD and Combo) confirms G9a inhibition. Total H3 and α-tubulin are loading controls.
DNA damage during S-phase can arise from many different circumstances that trigger a slow down or halting of DNA replication, a phenomenon known as replication stress (RS). Typically, RS generates stretches of single-stranded DNA (ssDNA), which results from the replicative helicase continuing to unwind the parental DNA after the polymerase has stalled(30). If not resolved, RS leads to further DNA replication defects, replication fork collapse, and ultimately, DNA damage, which if not appropriately compensated, results in cell death. To determine whether RS was occurring in response to the combination, we investigated the generation of ssDNA. For this purpose, we performed BrdU incorporation assay under native conditions after 24 hours of treatment followed by immunofluorescence. We found that from individual treatments, prexasertib increased amount of ssDNA when compared to control (Fig. 4D; 2.1±0.2% for prexasertib alone vs. 0.4±0.2% in control; P<0.05), while BRD4470 alone did not show any significant difference (0.9±0.5% for BRD4770 alone; P>0.05 vs. control). When cells were treated with the combination, the percentage of BrdU-positive cells was significantly increased (21.3±1.5%; P<0.001 vs. control; P<0.01 vs. prexasertib alone; and P<0.001 vs. BRD4770 alone). Notably, BrdU signal in the combination-treated cells revealed a punctuate pattern (Fig. 4C), indicating the presence of ssDNA likely due to stalled replication forks.
The response of cells to RS also includes the activation of a series of key regulatory proteins, in particular the ATR-CHK1 DNA damage response pathway, which functions as a critical effector of the cellular response during RS and DNA damage(30). Hence, we evaluated different molecular markers of this pathway by western blot. First, we found that phosphorylation of CHK1 at Ser296, a marker of the catalytic activity of this enzyme, was abolished in the prexasertib and combination treated cells, while it remained unmodified in control and BRD4770 alone-treated cells, serving as a valuable “on target” effect for the CHK1 inhibitor at the dose used in these assays (Fig. 4E). In a similar manner, H3K9me2 was reduced in BRD4470 alone and combination treated cells, showing effective inhibition of G9a enzymatic activity under these treatments (Fig. 4D). Activation of the ATR-RPA pathway in response to the generation of ssDNA and RS was confirmed in the combination of prexasertib with BRD4770 by increased phosphorylation of RPA32 at Ser33 and CHK1 at Ser345, both direct targets of ATR activity (Fig. 4E) (31,32). In addition, we evaluated phosphorylation of ATR at Thr1989 in response to the different treatments. While levels of phosphorylation for this threonine residue were similar between control, prexasertib and BRD4470 conditions, increased phosphorylation was seen in combination-treated cells (Fig. 4E). H2A.X, a well-known marker of DNA damage, is also phosphorylated by ATR at serine 139 (Ser139) upon generation of ssDNA breaks and during RS conditions, such as replication fork arrest(33). This marker was increased in the combination treatment (Fig. 4E), further corroborating activation of the ATR-RPA32-CHK1 axis. However, inhibition of CHK1 results in failure of this checkpoint, ultimately leading to severe DNA damage.
Congruently, we obtained similar results by immunofluorescence. Immunostaining for P-Ser33-RPA32 showed that its nuclear activation occurs at similar rates under individual treatments and control conditions (Fig. 5A and B; 7.2±4.0% for control; 8.2±1.6% for prexasertib alone and 10.9±0.6% for BRD4770 alone; P>0.05). However, combination treatment induced a significant increase in P-Ser33-RPA32 (60.1±7.2%; P<0.01 vs. control; P<0.01 vs. prexasertib alone; and P<0.05 vs. BRD4770 alone). Hyperphosphorylation of RPA32, marked by the presence of phosphorylation at Ser4/Ser8, occurred at similar rates between control and individual treatments (Fig. 5C and D; 1.8±1.4% for control vs. 5.0±1.4% for prexasertib alone and vs. 5.7±1.9% for BRD4770 alone; P>0.05). Conversely, hyperphosphorylated RPA32 was significantly increased in combination-treated cells (Fig. 5D; 28.6±5.2%; P<0.05 for all conditions). Immunostaining for P-Ser139-H2A.X confirmed a significant increase in the overall amount of DNA damage in cell nuclei with combination treatment compared to control or individual treatments (Fig. 5E and F; 36.5±5.1% with combination vs. 2.8±1.1% for control; P<0.01; vs. 1.3±0.6% from prexasertib alone; P<0.05; and vs. 3.2±1.8% from BRD4770 alone; P<0.01). Notably, P-Ser33-RPA32, P-S4/S8-RPA32 and P-Ser139-H2A.X not only increased under combination treatment, but appeared simultaneously in a higher percentage of cells that received combination treatment for both, P-Ser33-RPA32 (Supplementary Fig. 7A and B; 30.0±5.4% for combination treatment vs. 0.9±0.5% for control; P<0.01; 0.3±0.1% for prexasertib alone and 1.2±0.3% for BRD4770 alone; P<0.05) and P-S4/S8-RPA32 (Supplementary Fig. 7C and D; 22.±2.8% for combination vs. 2.5±1.6% for control, 2.6±0.5% with prexasertib, and 3.3±0.5% with BRD4770; P<0.01). Furthermore, based on calculations taken from the Combenefit software, we did not observe such a response in P-Ser33-RPA32, P-S4/S8-RPA32 or P-Ser139-H2A.X when using a predicted non-synergistic combination of 0.39nM prexasertib with 1.25μM BRD4770 (Supplementary Fig. 8), further supporting the specificity of the synergistic effect. Taken together, these results suggest a mechanistic model whereby the combined inhibition of CHK1 and G9a induce cell death through genome-wide damage caused by uncompensated failure in DNA replication during S-phase. This result is in agreement with the notion that both proteins function as guardians against RS, through checkpoint signaling and chromatin remodeling at the replication fork(34,35).
Figure 5. Dual targeting of CHK1 and G9a triggers the RS response.
Immunofluorescence of P-Ser33-RPA32, P-S4/S8-RPA32, and P-Ser139-H2A.X on L3.6 after 24h of treatment with vehicle (DMSO), prexasertib (Prex; 0.78 nM), BRD4770 (BRD; 2.5 μM) or their combination (Combo). Representative images are shown for (A) P-Ser33-RPA32, (C) P-S4/S8-RPA32, and (E) P-Ser139-H2A.X. Cells positive for (B) P-Ser33-RPA32, (D) P-S4/S8-RPA32 or (F) P-Ser139-H2A.X were quantified as Mean ± SEM, demonstrating increases in these markers of RS after combination treatment. (n=3; scale bar, 50μm; *, P<0.05; **, P<0.01; ***, P<0.001; t-test)
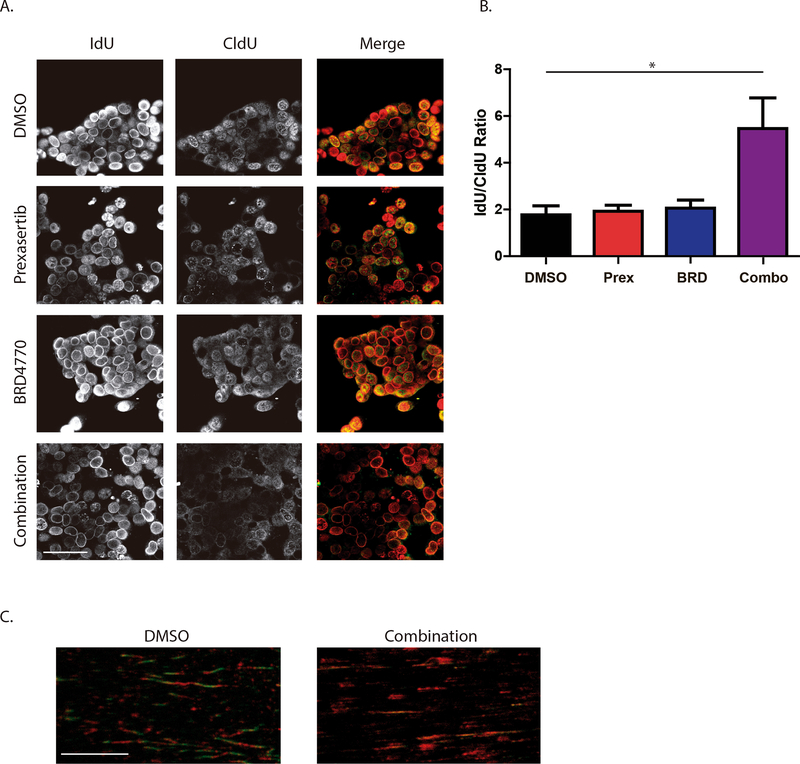
We further investigated the effect of combination treatment on activity of the replication fork. Cells were sequentially labelled with the halogenated nucleotide IdU, followed by 24 hours of treatments and CldU labeling to stain DNA replication foci (Fig. 6A and B). While the IdU/CldU ratio did not differ between control, prexasertib or BRD4770 conditions (1.77±0.38 for DMSO, 1.92±0.26 for prexasertib and 2.06±0.34 for BRD4770; P>0.05), the combination resulted in an increased IdU/CldU ratio (5.46±1.31; P<0.05), indicating a significant reduction in global fork activity 24 hours after treatment, as evidenced by lower incorporation of the second label, CldU. To visualize the effect of the combination at the level of the replication fork, we performed DNA fiber assay on L3.6 cells after 24 hours of treatment with vehicle or combination. We found that the combination treatment increased the overall number of IdU only-labelled tracks when compared to vehicle-control treated cells, substantiating a reduction in DNA replication and fork stalling (Fig. 6C).
Figure 6. CHK1 and G9a dual targeting inhibits DNA replication.
A, Replication foci labelling with IdU and CldU. Replication foci were labelled in L3.6 by IdU (Red) pulse, treatment for 24h with vehicle (DMSO), prexasertib (Prex; 0.78nM), BRD4770 (BRD; 2.5μM) or their combination (Combo), followed by second pulse with CldU (Green). B, Ratio of IdU to CldU was measured as mean fluorescence intensity for 40 cells each per experiment. (Mean±SEM, n=3; scale bar, 50μm; *, P<0.05; One-way ANOVA). C, DNA fiber assay was performed after 24h of control or combination treatment, followed by sequential IdU (Red) and CldU (Green) incorporation. Increase in IdU-Red tracks indicates stalled forks. (n=2, scale bar, 10μm).
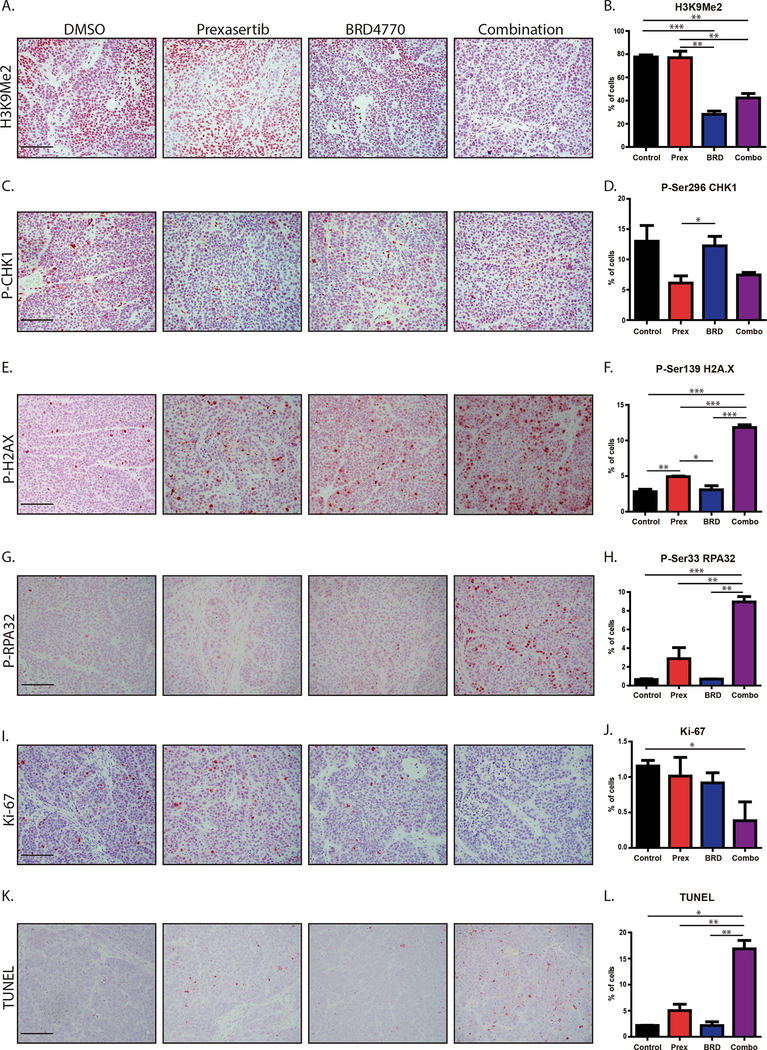
To confirm our observations in vivo, we checked tissues from treatment of the subcutaneous xenografts by IHC. We evaluated H3K9me2 staining (Fig. 7A and B) and found no difference between control and prexasertib-treated animals (77.6±1.7% vs. 76.9±5.7%). BRD4770 and combination-treated animals showed reduced H3K9me2 staining when compared to controls (28.1±2.7% for BRD4770 and 42.3±3.8% for combination vs. 77.6±1.7% for control; P<0.01), confirming the on-target effect of this inhibitor in vivo. To verify in vivo targeting of CHK1, we evaluated staining for P-Ser296-CHK1 (Fig. 7C and D). We found no difference between controls and animals given BRD4770 alone (13.0±2.6% for control vs. 12.2±1.5% with BRD4770). However, animals with prexasertib or the combination showed a reduction in P-Ser296-CHK1 staining (6.1±1.2% with prexasertib and 7.5±0.4% with the combination). We also checked the tissues for P-Ser139-H2A.X (Fig. 7E and F) and P-Ser33-RPA32 (Fig. 7G and H). Consistently, we found that xenografts from combination-treated mice displayed a higher percentage of P-Ser139-H2A.X positive cells (Fig. 7E and F; 11.8±0.4%) compared to control (2.8±0.3%; P<0.001), prexasertib (4.9±0.1%; P<0.001), and BRD4770 (3.1±0.6%; P<0.001). Similarly, we found a significant increase in P-Ser33-RPA32 positive cells in tumors from combination-treated animals (Fig. 7G and H; 8.9±0.6%) compared to control (0.7±0.1%; P<0.001), prexasertib (2.9±1.2%; P<0.01), and BRD4770 (0.7±0.01%; P<0.01). Further evaluation of xenograft tissues demonstrated that combined targeting of these pathways reduced tumor growth through reduced cell proliferation (Fig. 7I and J) with increased cell death (Fig. 7K and L). The proliferation marker Ki-67 was decreased in the combination (Fig. 7I and J; 0.4±0.3%) compared to control (1.2±0.1%; P<0.05), while individual treatments did not significantly affect Ki-67 levels (1.0±0.3% and 0.9±0.1% for prexasertib and BRD4770, respectively). Conversely, TUNEL staining, indicative of cell death, was increased in combination-treated tumors (Fig. 7K and L; 16.9±1.7%) compared with control (2.2±0.03%; P<0.05), prexasertib (5.0±1.2%; P<0.01), and BRD4770 (2.2±0.7%; P<0.01). Combined, these results validate our experiments with in vitro cultures that the accumulation of DNA damage and activation of the replication stress response also occur in vivo, thereby resulting in antitumor activity through critically affecting the processes of DNA replication, DNA damage signaling, cell proliferation and death.
Figure 7. Dual targeting of CHK1 and G9a induces cell death through RS and DNA damage in vivo.
Immunohistochemistry on L3.6 xenograft tissues. Representative images are shown for (A) H3K9Me2, (C) P-Ser296 CHK1, (E) P-Ser139 H2A.X, (G) P-Ser33 RPA32, (I) Ki-67 and (K) TUNEL. Percentage of cells positive for (B) H3K9Me2, (D) P-Ser296-CHK1, (F) P-Ser139-H2A.X, (H) P-Ser33-RPA32, (J) Ki-67 and (L) TUNEL were quantified as Mean±SEM, demonstrating an increase in markers of RS after combination treatment with decreased proliferation as measured by Ki-67 and increased cell death reflected by TUNEL. H3K9Me2 and P-Ser296-CHK1 served as on-target controls for BRD4770 and prexasertib, respectively. (n=3; scale bar, 100μm; *, P<0.05; **, P< 0.01; ***, P< 0.001; t-test).
Discussion
PDAC has a bleak prognosis resulting from its aggressive biology, rapid dissemination, and late diagnosis. Unfortunately, this recalcitrant cancer is also often highly resistant to chemotherapy and radiation with gemcitabine/nab-paclitaxel and FOLFIRINOX the only two standard chemotherapies(36,37). Numerous trials have aimed to improve these regimens through drug combinations or alternatives, but with limited success(38–40). Thus, there is an urgent need to identify effective novel therapies for this dismal disease. The current study reports data to support the innovative concept that small molecules against cell cycle checkpoints create a window of mechanistic opportunity to inhibit PDAC cell growth by appropriately choosing to antagonize the enzymatic activity of epigenomic regulators, which support processes that are critical for a particular phase of the cell cycle. Specifically, results from experiments performed here not only support the validity of this paradigm, but also extend our knowledge of experimental therapeutics, through key findings from inhibition of the cell cycle checkpoint protein, CHK1, and the epigenomic regulator G9a.
Our findings reveal that combined inhibition of CHK1 and G9a antagonizes PDAC cell growth, which is observed in various models, including cells grown as monolayers, clonogenic assays, 3D cultures, and in vivo xenografts. The synergistic effect of this combination is predominantly mediated, at the cellular level, by arrest of cells during S-phase with reduced DNA synthesis, increased DNA damage, and ultimately caspase-positive, irreversible cell death. These results suggest that inhibition of this histone methyltransferase impairs DNA synthesis and the functional stability of replication forks. Indeed, the combination treatment triggers a replication stress response evidenced by generation of ssDNA, slowing of replication forks, ssDNA breaks, and activation of the ATR-RPA-CHK1 signaling pathway, which is deficient due to the targeted inhibition of CHK1. Experimentally, these results have been gathered using stringent models and assays, including live-cell monitoring, caspase 3/7 activation assay along with caspase inhibition, electron microscopy, FACS-based cell cycle analysis, BrdU incorporation to assess DNA replication, alkaline and neutral comet assays to distinguish dsDNA and ssDNA breaks, native BrdU incorporation to visualize ssDNA, dual-pulse thymidine analog incorporation and DNA fiber assays to examine replication forks, along with western blot and immunofluorescence microscopy to evaluate activation of the ATR-RPA pathway and simultaneous presence of P-Ser139 H2A.X. Furthermore, we confirmed activation of RPA and H2A.X in combination-treated xenograft tissues. Thus, we are confident that the results reported here reflect alterations in the processes that support the model. Together, these observations support a mechanistic model in which targeting G9a would inhibit PDAC cell growth through its effect at the replication fork.
The functional impact that G9a inhibition has at the replication fork in this mechanistic model in combination with CHK1 inhibition could stem from recent findings that implicate G9a as a partner with RPA to stabilize chromatin-mediated events at the replication fork(35,41). G9a inhibition is also expected to alter the remodeling of chromatin that is necessary to complete the process of DNA replication(42,43). Other contributing effects may involve the function that this protein exerts on gene expression(44). This function has been previously characterized in detail by a large amount of publications, and, in the context of its inhibition with CHK1, did not justify being examined here. Inhibition of CHK1 appears to cause a similar effect as G9a on stability of the replication fork, though likely more so directly at the level of cell cycle checkpoints through its participation in the ATR-RPA signaling pathway(31,32). The disturbance at the level of the replication fork to activate replication stress, followed by a failure in appropriate checkpoints to allow restoration of replication dynamics likely leads to a catastrophic state to ultimately trigger cell death. Thus, these considerations explain, at least in part, the highly synergistic effect of both inhibitors since the pathways seem to converge.
In light of these results, it becomes important to discuss both the novelty and the biomedical importance of this study. For instance, our work proposes a new approach to utilize epigenomic inhibitors for the treatment of cancer. In this regard, it is important to underscore that most epigenomic agents being applied to the treatment of cancer are utilized without taking into consideration the synchronization of cells at a moment in which they can achieve their maximum effect. In fact, most of the small molecules targeting epigenomic regulators have been used within the context of the effects that they display on gene expression or to reach cytotoxic levels as a monotherapy(45). However, it is also known that many epigenomic regulators play significant roles in DNA replication, chromosome condensation, the structure and function of telomeres and centromeres, and coupled events such as spindle pole formation, stability, chromosome separation, and reconstitution of the nuclear membrane(46). Therefore, by focusing on the role that G9a plays in one of these processes rather than gene expression, our studies harness a window of mechanistic opportunity to offer an innovative therapeutic approach.
In terms of the pathobiological importance, this approach is particularly important for PDAC, a painful and devastating disease, for which new targeted therapies are urgently needed. Predisposition to PDAC is known to occur in carriers of alterations in the molecular machinery that mediates DNA damage and replication, such as BRCA proteins, ATM, PALB, among others. In fact, germline and somatic mutations in members of this pathway are present in almost 20% of PDAC patients(47). In addition, this disease initiates through the activation of known oncogenes (e.g. KRAS), which trigger an increased firing of replication origins, leading to replication stress that must be compensated to sustain cancer growth(48–50). For instance, oncogenes such as KRAS are not sufficient to cause cancer growth, unless additional alterations in tumor suppressor pathways that are guardians of genomic stability and cell cycle control (P16 and p53) occur subsequent to the initiation step. Therefore, our targeted strategy creates vulnerabilities in pathways, directly or indirectly, that thwart compensatory mechanisms of replication stress.
In summary, our study presents a novel combination treatment for PDAC based on inhibition of the cell cycle checkpoint protein, CHK1, and the epigenomic regulator G9a. The synergistic effect achieved from this combination occurs through the disruption of coordinated events at the replication fork, triggering replication stress and ultimately replication catastrophe from a deficiency in vital checkpoints. This two-tiered approach based on amplifying replication stress and impeding a compensatory mechanism also offers a unique consideration for the utility of epigenomic inhibitors within the context of the cell cycle to provide a mechanistic window of opportunity. Therefore, we believe strategies, such as the combination presented here, represent a promising therapeutic opportunity for PDAC, expanding the potential arsenal to fight this dismal disease.
Supplementary Material
Acknowledgements
Research reported in this manuscript was supported by funding from the NIH (grant R01 CA178627, to G. Lomberk and R01 DK052913, to R. Urrutia), the Advancing a Healthier Wisconsin Endowment (to each G. Lomberk and R. Urrutia) and The Linda T. and John A. Mellowes Endowed Innovation and Discovery Fund (to R. Urrutia).
Footnotes
Potential Conflicts of Interest: J.L. Iovanna is a consultant/advisory board member for Dynabio SA. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Iovanna J, Mallmann M, Goncalves A, Turrini O, Dagorn J. Current knowledge on pancreatic cancer. Front Oncol 2012;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting Cancer Incidence and Deaths to 2030: The Unexpected Burden of Thyroid, Liver, and Pancreas Cancers in the United States. Cancer Research 2014;74:2913. [DOI] [PubMed] [Google Scholar]
- 3.Kleeff J, Korc M, Apte M, La Vecchia C, Johnson CD, Biankin AV, et al. Pancreatic cancer. Nature Reviews Disease Primers 2016;2:16022. [DOI] [PubMed] [Google Scholar]
- 4.Cinar P, Ko AH. Best practices for the treatment of metastatic pancreatic adenocarcinoma: the therapeutic landscape in 2017. Chinese Clinical Oncology 2017;6 [DOI] [PubMed] [Google Scholar]
- 5.Ojima I, Lichtenthal B, Lee S, Wang C, Wang X. Taxane anticancer agents: a patent perspective. Expert opinion on therapeutic patents 2016;26:1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conroy T, Desseigne F, Ychou M, Bouché O, Guimbaud R, Bécouarn Y, et al. FOLFIRINOX versus Gemcitabine for Metastatic Pancreatic Cancer. New England Journal of Medicine 2011;364:1817–25 [DOI] [PubMed] [Google Scholar]
- 7.Goldstein D, El-Maraghi RH, Hammel P, Heinemann V, Kunzmann V, Sastre J, et al. nab-Paclitaxel Plus Gemcitabine for Metastatic Pancreatic Cancer: Long-Term Survival From a Phase III Trial. JNCI: Journal of the National Cancer Institute 2015;107:dju413–dju [DOI] [PubMed] [Google Scholar]
- 8.Bian B, Bigonnet M, Gayet O, Loncle C, Maignan A, Gilabert M, et al. Gene expression profiling of patient‐derived pancreatic cancer xenografts predicts sensitivity to the BET bromodomain inhibitor JQ1: implications for individualized medicine efforts. EMBO Molecular Medicine 2017;9:482–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lomberk G, Blum Y, Nicolle R, Nair A, Gaonkar KS, Marisa L, et al. Distinct epigenetic landscapes underlie the pathobiology of pancreatic cancer subtypes. Nature Communications 2018;9:1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lomberk GA, Urrutia R. The Triple Code Model for Pancreatic Cancer: Crosstalk Among Genetics, Epigenetics, and Nuclear Structure. The Surgical clinics of North America 2015;95:935–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer cell 2012;22:9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathison A, Salmonson A, Missfeldt M, Bintz J, Williams M, Kossak S, et al. Combined AURKA and H3K9 Methyltransferase Targeting Inhibits Cell Growth By Inducing Mitotic Catastrophe. Mol Cancer Res 2017;15:984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell death and differentiation 2018;25:486–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruns CJ, Harbison MT, Kuniyasu H, Eue I, Fidler IJ. In vivo selection and characterization of metastatic variants from human pancreatic adenocarcinoma by using orthotopic implantation in nude mice. Neoplasia (New York, NY) 1999;1:50–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura T, Fidler IJ, Coombes KR. Gene Expression Profile of Metastatic Human Pancreatic Cancer Cells Depends on the Organ Microenvironment. Cancer Research 2007;67:139. [DOI] [PubMed] [Google Scholar]
- 16.Di Veroli GY, Fornari C, Wang D, Mollard S, Bramhall JL, Richards FM, et al. Combenefit: an interactive platform for the analysis and visualization of drug combinations. Bioinformatics 2016;32:2866–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul 1984;22:27–55. [DOI] [PubMed] [Google Scholar]
- 18.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods 2012;9:671–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ullman-Cullere MH, Foltz CJ. Body condition scoring: a rapid and accurate method for assessing health status in mice. Lab Anim Sci 1999;49:319–23 [PubMed] [Google Scholar]
- 20.Fernandez-Zapico ME, Lomberk GA, Tsuji S, DeMars CJ, Bardsley MR, Lin Y-H, et al. A functional family-wide screening of SP/KLF proteins identifies a subset of suppressors of KRAS-mediated cell growth. Biochem J 2011;435:529–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russel L, Burguet S. Ultrastructure of leydig cells as revealed by secondary tissue treatment with a ferrocyanide-osmium mixture. Tissue Cell 1977;9:751–66. [DOI] [PubMed] [Google Scholar]
- 22.Bulgar AD, Weeks LD, Miao Y, Yang S, Xu Y, Guo C, et al. Removal of uracil by uracil DNA glycosylase limits pemetrexed cytotoxicity: overriding the limit with methoxyamine to inhibit base excision repair. Cell death & disease 2012;3:e252–e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumaravel TS, Vilhar B, Faux SP, Jha AN. Comet Assay measurements: a perspective. Cell Biology and Toxicology 2009;25:53–64 [DOI] [PubMed] [Google Scholar]
- 24.Gyori BM, Venkatachalam G, Thiagarajan PS, Hsu D, Clement M-V. OpenComet: An automated tool for comet assay image analysis. Redox Biology 2014;2:457–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grzenda A, Leonard P, Seo S, Mathison AJ, Urrutia G, Calvo E, et al. Functional impact of Aurora A-mediated phosphorylation of HP1γ at serine 83 during cell cycle progression. Epigenetics & chromatin 2013;6:21- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathison A, Grzenda A, Lomberk G, Velez G, Buttar N, Tietz P, et al. Role for Krüppel-like transcription factor 11 in mesenchymal cell function and fibrosis. PloS one 2013;8:e75311–e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan Y, Wang Q, Paulk J, Kubicek S, Kemp M, Adams D, et al. A Small-Molecule Probe of the Histone Methyltransferase G9a Induces Cellular Senescence in Pancreatic Adenocarcinoma. ACS Chem Biol 2012;7:1152–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Longati P, Jia X, Eimer J, Wagman A, Witt M-R, Rehnmark S, et al. 3D pancreatic carcinoma spheroids induce a matrix-rich, chemoresistant phenotype offering a better model for drug testing. BMC cancer 2013;13:95- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hustedt N, Gasser SM, Shimada K. Replication checkpoint: tuning and coordination of replication forks in s phase. Genes 2013;4:388–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nature cell biology 2014;16:2–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu S, Opiyo SO, Manthey K, Glanzer JG, Ashley AK, Amerin C, et al. Distinct roles for DNA-PK, ATM and ATR in RPA phosphorylation and checkpoint activation in response to replication stress. Nucleic acids research 2012;40:10780–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Q, Guntuku S, Cui X-S, Matsuoka S, Cortez D, Tamai K, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes & Development 2000;14:1448–59 [PMC free article] [PubMed] [Google Scholar]
- 33.Podhorecka M, Skladanowski A, Bozko P. H2AX Phosphorylation: Its Role in DNA Damage Response and Cancer Therapy. Journal of nucleic acids 2010;2010:920161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saldivar JC, Cortez D, Cimprich KA. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nature reviews Molecular cell biology 2017;18:622–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dungrawala H, Rose KL, Bhat KP, Mohni KN, Glick GG, Couch FB, et al. The Replication Checkpoint Prevents Two Types of Fork Collapse without Regulating Replisome Stability. Molecular cell 2015;59:998–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghosn M, Ibrahim T, Assi T, El Rassy E, Kourie HR, Kattan J. Dilemma of first line regimens in metastatic pancreatic adenocarcinoma. World J Gastroenterol 2016;22:10124–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu X, Zhang Y, Chen C, Yao Q, Li M. Targeted drug delivery in pancreatic cancer. Biochim Biophys Acta 2010;1805:97–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Louvet C, Labianca R, Hammel P, Lledo G, Zampino M, Andre T, et al. Gemcitabine in Combination With Oxaliplatin Compared With Gemcitabine Alone in Locally Advanced or Metastatic Pancreatic Cancer: Results of a GERCOR and GISCAD Phase III Trial. J Clin Oncol 2005;23:3509–16 [DOI] [PubMed] [Google Scholar]
- 39.Rocha Lima C, Green M, Rotche R, Miller W, Jeffrey G, Cisar L, et al. Irinotecan Plus Gemcitabine Results in No Survival Advantage Compared With Gemcitabine Monotherapy in Patients With Locally Advanced or Metastatic Pancreatic Cancer Despite Increased Tumor Response Rate. J Clin Oncol 2004;22:3776–83 [DOI] [PubMed] [Google Scholar]
- 40.Scheithauer W, Schull B, Ulrich-Pur H, Schmid K, Raderer M, Haider K, et al. Biweekly high-dose gemcitabine alone or in combination with capecitabine in patients with metastatic pancreatic adenocarcinoma: a randomized phase II trial. Ann Oncol 2003;14:97–104 [DOI] [PubMed] [Google Scholar]
- 41.Yang Q, Zhu Q, Lu X, Du Y, Cao L, Shen C, et al. G9a coordinates with the RPA complex to promote DNA damage repair and cell survival. Proc Natl Acad Sci U S A 2017;114:E6054–E63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olsen JB, Tropepe V. The expanding role of the Ehmt2/G9a complex in neurodevelopment AU - Deimling, Steven J. Neurogenesis 2017;4:e1316888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu Y, Song C, Zhang Q, DiMaggio PA, Garcia BA, York A, et al. Histone H3 lysine 56 methylation regulates DNA replication through its interaction with PCNA. Molecular cell 2012;46:7–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shankar SR, Bahirvani AG, Rao VK, Bharathy N, Ow JR, Taneja R. G9a, a multipotent regulator of gene expression. Epigenetics 2013;8:16–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahuja N, Sharma AR, Baylin SB. Epigenetic Therapeutics: A New Weapon in the War Against Cancer. Annu Rev Med 2016;67:73–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allis D, Caparros M, Jenuwein T, Reinberg D. Epigenetics, 2nd Edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2015. [Google Scholar]
- 47.Bailey P, Chang DK, Nones K, Johns AL, Patch A-M, Gingras M-C, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47. [DOI] [PubMed] [Google Scholar]
- 48.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 2006;444:633. [DOI] [PubMed] [Google Scholar]
- 49.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 2006;444:638. [DOI] [PubMed] [Google Scholar]
- 50.Halazonetis TD, Gorgoulis VG, Bartek J. An Oncogene-Induced DNA Damage Model for Cancer Development. Science 2008;319:1352. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.