Abstract
Inversion of chromosome 16 (inv(16)) generates a fusion gene CBFB-MYH11, which is a driver mutation for acute myeloid leukemia (AML). Gene expression profiling suggests that Gata2, a hematopoietic transcription factor, is a top upregulated genes in preleukemic Cbfb-MYH11 knockin mice and is expressed in human inv(16) AML. On the other hand, we have also identified recurrent monoallelic deletions of GATA2 in relapsed human CBF-AML patients. To clarify the role of Gata2 in leukemogenesis by Cbfb-MYH11, we generated conditional Cbfb-MYH11 knockin mice with Gata2 heterozygous knockout. Gata2 heterozygous knockout reduced abnormal myeloid progenitors, which are capable of inducing leukemia in the Cbfb-MYH11 mice. Consequently, Cbfb-MYH11 mice with Gata2 heterozygous knockout developed leukemia with longer latencies than those with intact Gata2. Interestingly, leukemic cells with Gata2 heterozygous knockout gained higher number of mutations and showed more aggressive phenotype in both primary and transplanted mice. Moreover, leukemic cells with Gata2 heterozygous knockout showed higher repopulating capacity in competitive transplantation experiments. In summary, reduction of Gata2 activity affects mutational dynamics of leukemia with delayed leukemia onset in Cbfb-MYH11 knockin mice, but paradoxically results in a more aggressive leukemia phenotype, which may be correlated with leukemia relapse or poor prognosis in human patients.
Introduction
Inversion of chromosome 16 (inv(16)) generates a CBFB-MYH11 fusion gene, which causes acute myeloid leukemia (AML) subtype M4Eo.1 The encoded fusion protein, CBFβ-SMMHC, contributes to the pathogenesis of CBFB-MYH11 AML. Accumulating evidence has shown that CBFB-MYH11 AML follows a ‘two-hit’ model for leukemogenesis,2, 3 which states that AML is the consequence of collaborations between at least two broad classes of mutations; class I mutations that confer proliferative and/or survival advantage to cells (e.g. RAS signaling) and class II mutations that impair hematopoietic differentiation (e.g. CBFB-MYH11). We have shown that Cbfb-MYH11 (the mouse-human hybrid version of CBFB-MYH11 expressed in a mouse knockin model4) is necessary but not sufficient for leukemia and initiates leukemogenesis by blocking normal hematopoietic differentiation through inhibition of RUNX1, consistent with two-hit hypothesis.4–6 In vitro studies have shown that CBFβ-SMMHC may serve as a transcriptional repressor and it may sequester RUNX1 in the cytoplasm.7–9 Additionally, recent studies suggest CBFβ-SMMHC has roles not only for RUNX1 suppression but also for transcriptional activation of various genes in the process of leukemogenesis.10, 11 Thus, although recent studies have gradually unraveled the pathogenesis of CBFB-MYH11 AML, further investigation is still needed to better understand leukemogenesis and disease progression.
To further unveil the mechanism of pathogenesis for this type of leukemia, we performed gene expression profiling on Cbfb-MYH11 knockin mouse embryos.11 As shown below, we have also analyzed microarray and RNA-seq data to identify the most important dysregulated genes in Cbfb-MYH11 pre-leukemic mice.11, 12 Using these two datasets, we have identified Gata2, a critical hematopoietic transcription factor,13 as one of six commonly dysregulated genes shared by these two datasets. These findings suggest Gata2 may play an important role in the development of this type of leukemia.
Recently we also performed comprehensive somatic mutational analysis of CBF-AML patient samples, and identified deletions of a region on chromosome 3 in relapse samples from three patients.14 Interestingly, the minimal overlapping region of these deletions includes the GATA2 gene. This observation suggests that GATA2 haploinsufficiency may contribute to relapse of CBF-AML. Recent studies have also shown that patients with advanced myeloid malignancy associated with GATA2 deficiency have poor prognosis.15
From these findings, we proposed two hypotheses; 1) up-regulation of GATA2 contributes to CBFB-MYH11 leukemogenesis in the initiation phase; and 2) GATA2 deficiency contributes to the relapse/evolution of CBF-AML. To test these hypotheses, we generated conditional Cbfb-MYH11 knockin mice with heterozygous Gata2 knockout. Our findings suggest that Gata2 plays important but distinct roles in two different stages of Cbfb-MYH11 leukemia: sufficient Gata2 activity is important for Cbfb-MYH11 induced leukemogenesis, while Gata2 deficiency may contribute to the relapse of the disease.
Materials and Methods
Animals
All animal studies were approved by the National Human Genome Research Institute Animal Care and Use Committee, and all the procedures performed followed relevant National Institutes of Health guidelines and regulations. Cbfb-MYH11 conditional knockin (Cbfb+/56MMx1-Cre)16, 17, mice harboring a Gata2 exon 5 flanked by loxP sites (Gata2+/f)18 (obtained through the Mutant Mouse Resource and Research Center, mmrrc.org), mice harboring a Runx1 exon 4 flanked by loxP sites (Runx1f/f)19, and R26R-Brainbow2.1 (BB+/f) mice20 harboring fluorescent protein have been previously described. The mice were backcrossed to C57BL/6 for >10 generations, then crossed to each other. All mice were genotyped by polymerase chain reaction (PCR) with gene-specific primers (Supplemental Table1) using tail-snip DNA. Eight- to 12-week-old mice were injected intraperitoneally with 250 mg of poly(I:C) (pIpC; InvivoGen) every other day for 3 doses to activate Cre-recombinase under the control of the Mx1 promotor, which induces lox-recombination in bone marrow (BM) cells. All mice were observed for leukemia development for 12 months. Bulk spleen cells (1×106) from Gata2+/+Cbfb+/56MMx1-cre or Gata2+/fCbfb+/56MMx1-cre (CD45.1−CD45.2+) mice were transplanted into sublethally irradiated (650 rads) wildtype C57BL/6×129Sv (CD45.1+/CD45.2+) mice. For competitive transplantation, various numbers of c-Kit+ leukemic cells from Gata2+/fCbfb+/56MMx1-cre (CD45.1−CD45.2+YFP−) were mixed with those of BB+/fCbfb+/56MMx1-cre (CD45.1−CD45.2+YFP+; 3×105) and injected intravenously into sublethally irradiated wildtype mice (CD45.1+/CD45.2+).
FACS
Peripheral blood cells, spleen and BM cells from mice were isolated and stained as previously described for flow cytometry assay.21 Flow cytometry was performed using BD LSRII, and sorting was performed using BD ARIA-II. See Supplemental Methods for details regarding the antibodies used in this study.
Quantitative PCR
Quantitative PCR was performed by using Power SYBR Green PCR Master Mix (Applied Biosystems) according to the manufacturer’s instructions. RNA and DNA were extracted from BM cells using RNA MiniPrep Plus kit (Zymo Reserch) or DNA mini kit (Qiagen). Exon5/6 primers were used to detect unexcised Cbfb-MYH11 flox allele; Gata2 unexcised primers were used to detect unexcised Gata2 flox allele. Genomic control primers were used to as internal control for genomic DNA. Primers are listed in Supplemental Table 1.
Colony forming assay
Colony forming assay was performed by using BM cells 12 days after pIpC injection and MethoCult GF M3434 (STEMCELL Technologies) according to the manufacturer’s instructions.
Apoptosis and cell cycle analysis
For apoptotic analysis, cells were stained for Annexin V and 7AAD according to the manufacturer’s protocol (eBioscience). For cell cycle analysis, after staining surface markers, cells were fixed and stained for DAPI to evaluate DNA content. Then flow cytometry was performed and data was analyzed by FlowJo.
RNA-seq and whole exome sequencing
RNA-seq and whole exome sequencing (WES) were performed on messenger RNA (mRNA) and genomic DNA isolated from c-Kit+ BM or spleen cells of end-stage leukemic mice. The RNA-seq data and WES data have been deposited at Gene Expression Omnibus and Sequence Read Archive, respectively (accession numbers GSE 130343 and SRP193773). Details on all of the above procedures and data analysis are provided in Supplemental Methods.
Statistical analysis
For all in vitro experiments, at least three samples were tested per condition or genotype in order to perform statistical analysis. For animal studies, the numbers of animals used in each condition and/or genotype were determined based on the estimations of effect sizes and desired power, which was set at 1. No randomization was used, since we compared animals based on their genotype. No samples were excluded from data analysis. The researchers were not blinded for the animal studies. Data were analyzed with Prism software (GraphPad Software, La Jolla, CA, USA). Results are expressed as mean ± standard error of the mean. Differences between 2 groups were tested with a Student t test. The survival times of mice were analyzed with the Kaplan-Meier method and log-rank test. A two-sided t-test value of P < 0.05 was considered statistically significant (in figure legends: *P < 0.05, **P < 0.01, ***P < 0.001).
Results
Gata2 is upregulated in preleukemic hematopoietic cells in Cbfb-MYH11 knockin mice
Cbfb-MYH11 is thought to contribute to leukemogenesis by affecting the expression of various genes.10, 11 To identify candidate genes affected by Cbfb-MYH11, we screened the genes that differentially expressed in Cbfb-MYH11 knockin mouse model using two different cohorts. The first cohort was composed of a microarray analysis of the peripheral blood cells in the Cbfb-MYH11 conventional knockin (Cbfb+/MYH11) embryos compared with those in wild-type (WT) littermates (GSE19194).11 Top genes (24) were selected if they had fold change greater than 5 or less than −5 between Cbfb-MYH11 and WT mice and had adjusted p-value < 0.001 (Supplemental Figure 1). The second cohort is an RNA-seq analysis of the c-Kit+ BM cells from Cbfb-MYH11 conditional knockin (Cbfb+/56M) mice, collected at the preleukemic stage (2 weeks after treatment with pIpC) compared with c-Kit+ BM cells from wild-type littermates (GSE102388).12 Top genes (66) were selected if they had fold change greater than 3 or less than −3 and had adjusted p-value < 0.001 (Supplemental Figure 2). For both cohorts, most of the selected genes were upregulated in the cells from the Cbfb+/56M mice compared with those from WT mice. Interestingly, Gata2 was among 6 genes which were shared by these two cohorts as differentially expressed genes in Cbfb-MYH11 knockin mice (Figure 1A and Supplemental Table 2). We also evaluated Gata2 gene expression by quantitative reverse transcription polymerase chain reaction (qRT-PCR) and western blot in the conditional Cbfb-MYH11 knockin mice (Cbfb+/56MMx1-Cre), which were injected with pIpC to induce the expression of Cbfb-MYH11 through Cre-recombinase activation. At 12 days after pIpC treatment, before the development of leukemia, BM cells were collected from the mice. Gata2 was expressed higher in Cbfb+/56MMx1-Cre preleukemic BM cells compared to WT BM cells at both mRNA and protein levels (Supplemental Figure 3A and 3B). CBFβ-SMMHC, the fusion protein encoded by CBFB-MYH11, is a dominant negative repressor of RUNX1.1 Therefore, we evaluated Gata2 gene expression in BM cells in the Runx1f/fMx1-Cre mice. Gata2 was also expressed higher in Runx1f/fMx1-Cre BM cells compared to WT BM cells, 2 weeks after pIpC treatments (Supplemental Figure 4A). We also analyzed a published ChIP-seq study of ME-1 (a human inv(16) leukemia cell line that expresses CBFB-MYH11) using anti- Runx1, CBFβ and SMMHC antibodies (GSE46044)22, and the results suggested that GATA2 was likely a transcriptional target of CBFB-MYH11 (Supplemental Figure 4B). These findings suggest that both Runx1 suppression and transcriptional activation by CBFβ-SMMHC can lead to higher Gata2 expression in the Cbfb-MYH11 knockin mice.
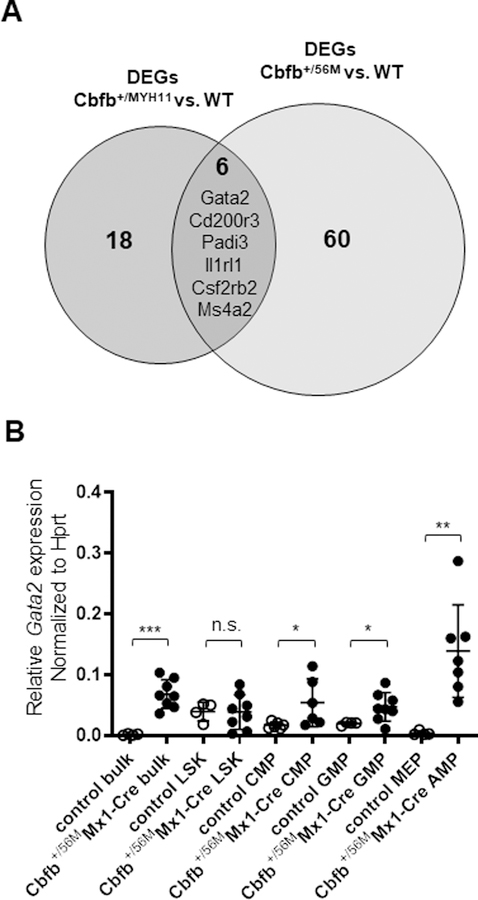
Figure 1. Analysis of Gata2 expression in preleukemic Cbfb+/56MMx1-Cre mice.
(A) The Venn diagram of common upregulated genes in the Cbfb+/MYH11 embryos and c-kit+ cells in the Cbfb+/56MMx1-Cre mice. (B) Relative levels of Gata2 mRNA in sorted bulk, LSK, CMP, GMP, MEP and AMP populations in BM cells at 12 days after pIpC injection. Each dot represents data derived from one sample and all measurements were made in triplicates. Values are presented relative to those of Hprt mRNA (n=4–8 for each group). *p<0.05; **p<0.01; ***p<0.001.
Gata2 is highly expressed in hematopoietic stem/progenitor cells.13 We therefore compared the expression levels of Gata2 in the hematopoietic stem/progenitor compartments of wildtype and Cbfb-MYH11 preleukemic BM cells. At 12 days after pIpC treatment, lineage negative BM cells from the wild type mice could be further divided into LSK (Lin−Sca-1−c-Kit+), granulocyte-monocyte progenitors (GMPs) (Lin−CD34+CD16/32−), common myeloid progenitors (CMPs) (Lin−CD34+CD16/32+), and megakaryocyte-erythroid progenitors (MEPs) (Lin−CD34−CD16/32−), as described previously (Supplemental Figure 5A).12, 23, 24 Cbfb+/56MMx1-Cre preleukemic BM cells contained LSK, CMPs, and GMPs as well, and an abnormal myeloid progenitor population (AMP) (Lin−CD34−CD16/32dim-+) instead of the MEPs in WT control (Supplemental Figure 5A). This AMP population is specific to Cbfb+/56MMx1-Cre mice and a putative leukemia initiating population in this leukemic mouse model.12, 16 To confirm which populations are responsible for the Gata2 high expression, LSKs, GMPs, CMPs, MEPs, and AMPs were sorted by FACS (Supplemental Figure 5B). qRT-PCR showed that LSKs had the highest Gata2 gene expression in the control mice (Figure 1B), but Gata2 was expressed higher in Cbfb+/56MMx1-Cre than WT control for every population examined except LSK (Figure 1B). Moreover, the highest level of Gata2 expression was detected in the AMPs of the Cbfb-MYH11 knockin mice. Considering that AMPs are specific to Cbfb-MYH11 mice and have been reported as responsible for leukemia initiation, these data suggest Gata2 has important roles in leukemogenesis during preleukemic phase of this mouse model.
Establishment of Gata2+/fCbfb+/56MMx1-Cre mice
To determine the functional significance of Gata2 in Cbfb-MYH11 induced leukemia, we generated Gata2+/+Cbfb+/56MMx1-Cre and Gata2+/fCbfb+/56MMx1-Cre mice.16, 18 The Gata2 knockout mice were injected with pIpC to induce the expression of Cbfb-MYH11 and the knockout of Gata2. Quantitative and conventional PCR reactions were performed to determine the excision efficiency of Cbfb-MYH11 and Gata2 flox alleles in BM cells 12 days after pIpC injection. The data confirmed that the excisions were almost complete for both genes (Supplemental Figures 6A, B and 7A, B). Moreover, qRT-PCR showed that the expression of the intact Gata2 mRNA was reduced in Gata2+/fCbfb+/56MMx1-Cre mice at 12 days after pIpC injection (Supplemental Figure 8A). These results revealed that appropriate induction of Cbfb-MYH11 and knockout of Gata2 in the hematopoietic cells occurred in the pIpC-treated Gata2+/fCbfb+/56MMx1-Cre mice.
Heterozygous knockout of Gata2 leads to reduced colony forming ability and smaller AMP in Cbfb-MYH11 preleukemic BM cells
As we showed previously, activation of Cbfb-MYH11 expression in the knockin mice causes increase of colony forming ability and emergence of the AMP population in the BM.16 We were therefore interested in the effect of Gata2 knockout on these phenotypes in the Cbfb-MYH11 knockin mice. At 2 weeks after pIpC treatment (before leukemia initiation), in vitro colony formation assay demonstrated that Gata2 knockout reduced colony forming ability of the Cfb-MYH11 - expressing cells (Figure 2A). However, Gata2 status had no effect on colony types formed by Cbfb-MYH11 - expressing cells (Supplemental Figure 8B). The AMP population accumulated in both Gata2+/+Cbfb+/56MMx1-Cre and Gata2+/fCbfb+/56MMx1-Cre mice (Figure 2B), but the AMP population was smaller in the Gata2+/fCbfb+/56MMx1-Cre mice than in the Gata2+/+Cbfb+/56MMx1-Cre mice (Supplemental Figure 8C). The LK population was also decreased in Gata2+/fCbfb+/56MMx1-Cre mice but the decrease did not reach statistical significance. Conversely, the LSK population, known to contain hematopoietic stem cells, was not different between these 2 groups of mice. These results suggest that Gata2 deficiency reduced the colony forming ability and the size of the AMP population in Cbfb-MYH11 knockin mice during early preleukemic phase. One the other hand, Gata2+/fMx1-Cre mice did not show any hematopoietic phenotype (Supplemental Figures 9A–D).
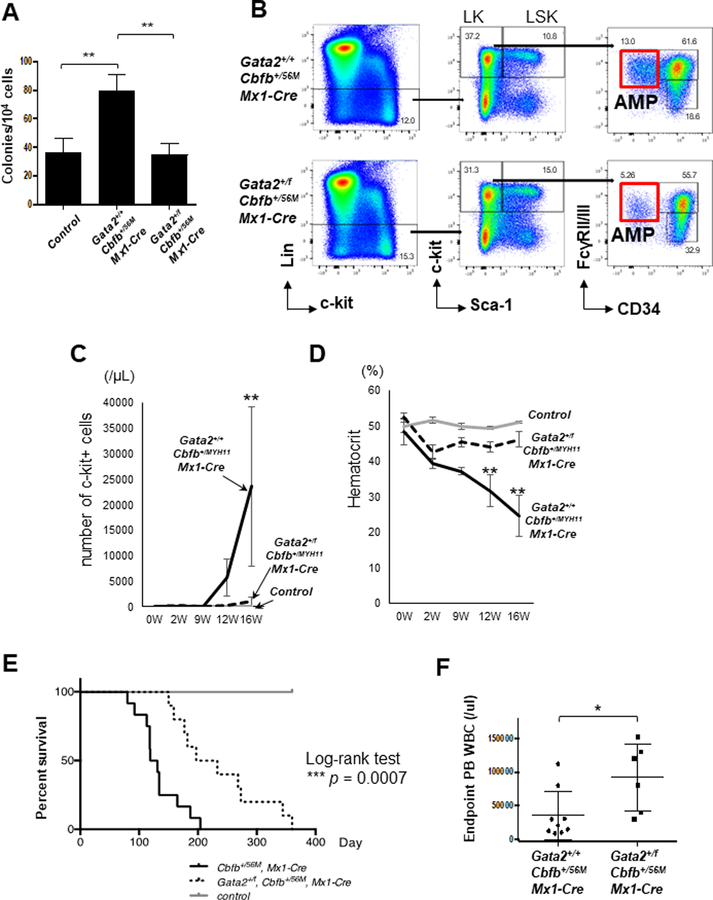
Figure 2. Gata2 heterozygous knockout delays leukemogenesis induced by Cbfb-MYH11.
(A) Total colonies produced from in vitro erythroid and myeloid differentiation assays with BM cells from the indicated genotypes (n=5 for each group). (B) Representative FACS plots of bone marrow cells gated on total cells (left plots), Lin- cells (middle plots), LK cells and AMP cells (right plots). (C) Numbers of c-Kit+ cells in the peripheral blood at the indicated time points after pIpC injection (n=5–6 for each group). (D) Hematocrit levels in the peripheral blood at the indicated time points after pIpC injection (n=5–6 for each group). (E) Kaplan-Meier curves of mice with the indicated genotypes during 12-month observation of leukemia development. Black line: Gata2+/+Cbfb+/56MMx1-Cre, n=12; dotted line: Gata2+/fCbfb+/56MMx1-Cre, n=10; grey line: control mice, n=17. The P-values were calculated with log-rank test. The difference between Gata2+/+Cbfb+/56MMx1-Cre mice and Gata2+/fCbfb+/56MMx1-Cre mice was highly significant (P=0.0007). (F) Peripheral blood white blood cell count at the end points for the mice with the indicated genotypes. Each dot represents data derived from one mouse (n=6–9 for each group). *p<0.05; **p<0.01.
Heterozygous knockout of Gata2 delays leukemia development in Cbfb-MYH11 knockin mice
To test our hypotheses that increased expression of Gata2 contributes to leukemogenesis by CBFb-MYH11, we observed leukemia development in Gata2+/fCbfb+/56MMx1-Cre and Gata2+/+ Cbfb+/56MMx1-Cre mice. Consistent with Gata2 playing an important role during leukemogenesis, the number of c-Kit+ leukemic cells in the peripheral blood was significantly lower in Gata2+/fCbfb+/56MMx1-Cre mice when compared to Gata2+/+Cbfb+/56MMx1-Cre mice at the same time points (Figure 2C). Hematocrit levels were also higher in Gata2+/fCbfb+/56MMx1-Cre mice than in Gata2+/+Cbfb+/56MMx1-Cre mice (Figure 2D), which indicated that progression of anemia due to leukemia development was slower in the Cbfb-MYH11 knockin mice with Gata2 knockout. Eventually, both groups developed leukemia with similar phenotype. BM cells from each group showed two types of leukemic cells, immature cells with predominantly monocytic features and blast-like progenitors, as shown in a previous study (Supplemental Figure 10A and B).16 Analysis of cell surface markers also showed similar phenotype between these two genotypes (Supplemental Figure 11). Various organs such as BM, spleen and liver were infiltrated with leukemic cells (Supplemental Figure 10A). Importantly, even though all the mice with the genotype of Gata2+/+Cbfb+/56MMx1-Cre or Gata2+/fCbfb+/56MMx1-Cre developed leukemia, Gata2+/fCbfb+/56MMx1-Cre leukemic mice had a longer survival than Gata2+/+Cbfb+/56MMx1-Cre mice (median survival, 215 vs 125 days; P=.0007; Figure 2E). As expected, Gata2 gene expression in c-Kit+ leukemic cells was reduced in Gata2+/fCbfb+/56MMx1-Cre mice than in Gata2+/+Cbfb+/56MMx1-Cre mice, but the expression of Cbfb-MYH11 and Runx1 was not different between these two groups of mice (Supplemental Figure 12A–C). Taken together, these results suggest that Gata2 heterozygous knockout delays leukemogenesis by Cbfb-MYH11 but does not change the morphology of the developed leukemia cells. Interestingly, despite the slower development of leukemia, there were more circulating white blood cells (WBCs) in the Gata2+/fCbfb+/56MMx1-Cre mice at the end-stage than in the Gata2+/+Cbfb+/56MMx1-Cre mice (Figure 2G). Conversely, % of c-Kit+ cells, hematocrit and spleen weight were not different between the two groups (Supplemental Figure 13A–C). Since hyperleukocytosis is a well-known poor prognostic marker in human leukemia,25 these findings led us to further characterizations of the leukemia cells of each genotype in the context of transplantation.
Leukemia developed faster in mice transplanted with Gata2+/fCbfb+/56MMx1-Cre leukemia cells
To evaluate the characteristics of leukemic cells in both groups, we performed transplantations with spleen cells (1.0×106 per recipient) isolated from end-stage leukemic mice (Figure 3A). Four different donors from each group showed similar characteristics including % of c-Kit+ cells in spleen (Supplemental Table 3). Surprisingly, despite slower development of leukemia, transplanted leukemic cells from Gata2+/fCbfb+/56MMx1-Cre mice led to much faster leukemia development in the recipient mice than those from Gata2+/+Cbfb+/56MMx1-Cre mice. WBC count, % and number of c-Kit+ positive cells were higher in recipient mice transplanted with Gata2+/fCbfb+/56MMx1-Cre leukemia cells than those transplanted with Gata2+/+Cbfb+/56MMx1-Cre leukemia cells at 4 weeks after transplantation (Supplemental Figure 14A–C). Hematocrit was lower in mice transplanted with Gata2+/fCbfb+/56MMx1-Cre leukemia cells than those transplanted with Gata2+/+Cbfb+/56MMx1-Cre leukemia cells at the same time point (Supplemental Figure 14D). The above findings were reproducible among the 4 donors for each genotype (Supplemental Figure 14E–H). Furthermore, mice transplanted with Gata2+/fCbfb+/56MMx1-Cre leukemia cells had a shorter survival than mice transplanted with Gata2+/+Cbfb+/56MMx1-Cre leukemia cells (median survival, 35.5 vs 83.5 days; P<.0001; Figure 3B). These findings revealed that leukemic cells from Gata2+/fCbfb+/56MMx1-Cre mice showed more aggressive phenotype than those from Gata2+/+Cbfb+/56MMx1-Cre mice.
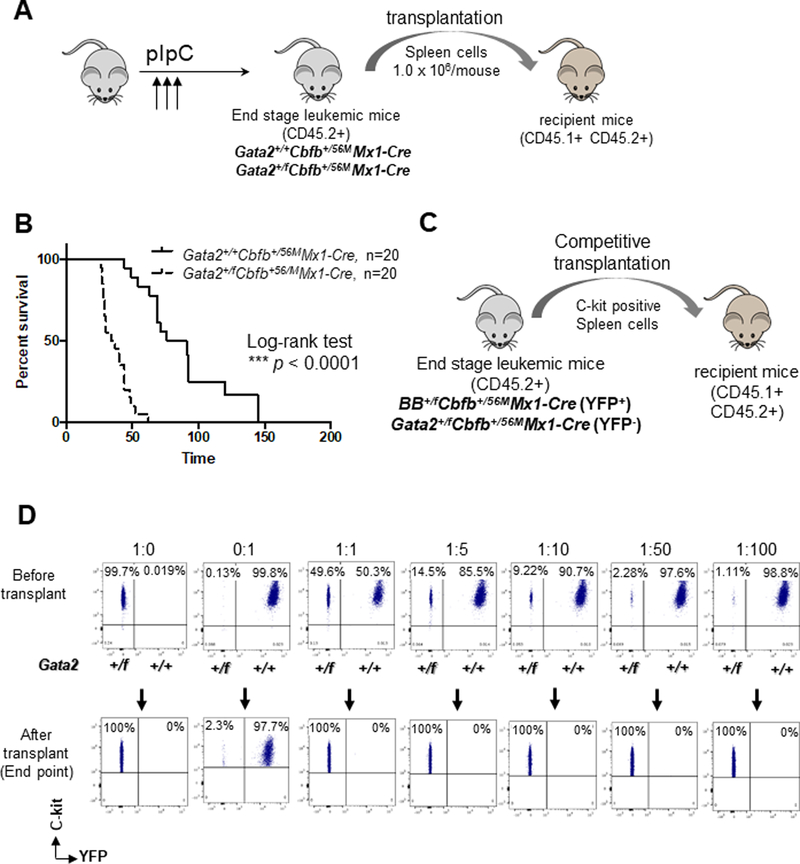
Figure 3. Accelerated leukemia development in recipient mice transplanted with Gata2+/fCbfb+/56MMx1-Cre leukemia cells.
(A) Experimental design. Equal numbers (1 million) of donor spleen cells obtained from end point mice (CD45.2+) were transplanted into sublethally irradiated WT recipient mice (CD45.1+CD45.2+). (B) Kaplan-Meier survival curves of transplanted mice. Black line: Gata2+/+Cbfb+/56MMx1-Cre, n=20; dotted line: Gata2+/fCbfb+/56MMx1-Cre, n=20. The p-values were calculated with log-rank test. The difference between Gata2+/+Cbfb+/56MMx1-Cre mice and Gata2+/fCbfb+/56MMx1-Cre mice was highly significant (P<0.0001).(C) For competitive transplantation, spleen cells from donor mice (CD45.2+; Gata2+/fCbfb+/56MMx1-Cre) and competitor mice (CD45.2+; BB+/+Cbfb+/56MMx1-Cre) were mixed in various ratios and injected through the tail vein of irradiated recipient mice (CD45.1+ CD45.2+). The efficacy of BM reconstitution was determined in peripheral blood (PB) at 4, 8, 12, and 16 weeks post-transplantation and BM at endpoint by FACS. (D) Representative FACS plots of cells injected samples immediately after sorting and recipients’ peripheral blood samples at endpoints. Leukemic cells from BB+/+Cbfb+/56MMx1-Cre (Gata2 wildtype) mice were c-Kit+YFP+; those from Gata2+/fCbfb+/56MMx1-Cre mice were c-Kit+YFP-.
Gata2+/fCbfb+/56MMx1-Cre leukemia cells had higher leukemia repopulating potential than Gata2+/+Cbfb+/56MMx1-Cre leukemia cells
To further characterize the advantage of leukemia repopulating capacity attributed to Gata2 deficiency, we compared BB+/fCbfb+/56MMx1-Cre (Gata2 WT) and Gata2+/fCbfb+/56MMx1-Cre (Gata2 knockout) leukemia cells in competitive transplantation assays (Figure 3C).26 Brainbow or BB+/f is a genetic cell-labeling technique by stochastic expression of distinct fluorescent proteins induced by Cre-recombinase activation.20 BB+/fCbfb+/56MMx1-Cre mice developed leukemia with fluorescent protein expression (Supplemental Figure 15A), which had similar latency and pathology as Gata2+/+Cbfb+/56MMx1-Cre mice (Supplemental Figure 15B). We sorted c-Kit+, yellow fluorescent protein (YFP) positive spleen cells from an end-stage BB+/fCbfb+/56MMx1-Cre mouse and c-Kit+, YFP− spleen cells from an end-stage Gata2+/fCbfb+/56MMx1-Cre mouse (Supplemental Figure 16A), which were then transplanted at various ratios (Supplemental Figure 16B) into CD45.1+CD45.2+ recipient mice. As shown in Figure 3D, the YFP-, Gata2+/fCbfb+/56MMx1-Cre leukemia cells were responsible for leukemia development in all recipients, even for those that received Gata2+/fCbfb+/56MMx1-Cre and Gata2+/+Cbfb+/56MMx1-Cre cells at 1:100 ratio. Even though both YFP+ and YFP- populations were present in the recipient mice in the early weeks after transplantation, leukemia cells from the Gata2+/fCbfb+/56MMx1-Cre (YFP-) mouse overwhelmed those from the BB+/fCbfb+/56MMx1-Cre mouse by the end stage (Supplemental Figure 17). These findings suggest that Cbfb-MYH11 leukemic cells with Gata2 deficiency have higher leukemia repopulating capacity than those with WT Gata2.
Pathway analysis revealed difference in cell cycle status and apoptosis between Gata2+/+ and Gata2+/f leukemic cells
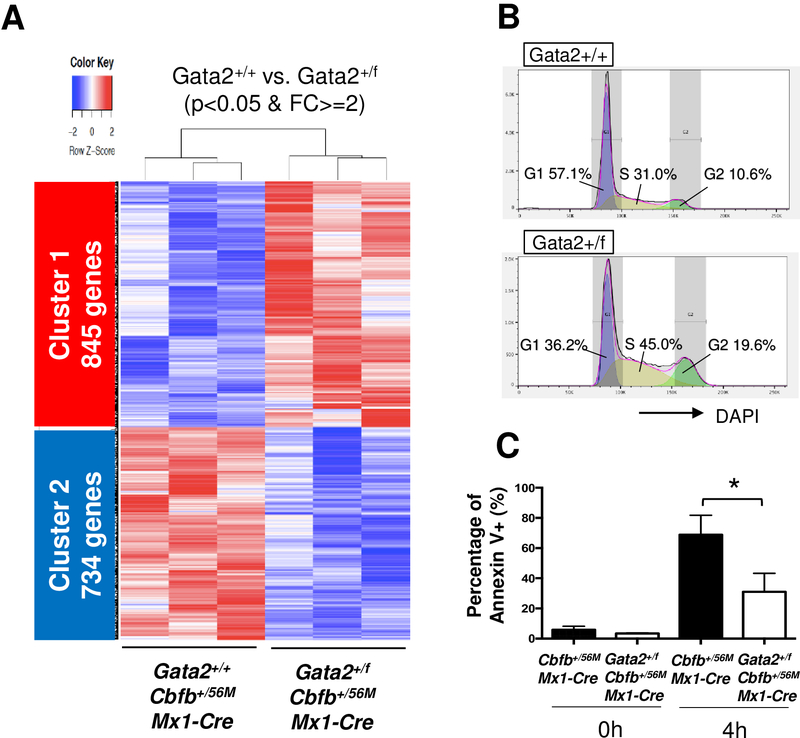
To further investigate the difference between leukemic cells from Gata2+/+Cbfb+/56MMx1-Cre mice and those from Gata2+/fCbfb+/56MMx1-Cre mice, we performed global gene expression profiling with RNA-seq (Supplemental Figure 18A and B). PCA plot and sample distance matrix showed a clear separation between Gata2+/+Cbfb+/56MMx1-Cre and Gata2+/fCbfb+/56MMx1-Cre leukemia cells (Supplemental Figure 18C and D). Compared with Gata2+/+Cbfb+/56MMx1-Cre cells, 1579 genes in Gata2+/fCbfb+/56MMx1-Cre cells were differentially expressed with Padj<.05 and absolute fold changes ≥2; 845 genes were upregulated (cluster 1) and 734 genes were downregulated (cluster 2) in the Gata2+/fCbfb+/56MMx1-Cre mice (Figure 4A). IPA upstream analysis showed Gata2 was the second most important putative upstream transcription factors that affect these differentially expressed genes (DEGs) (Supplemental Figure 19). Gene ontology (GO) analysis showed that genes associated with cell cycle and mitosis were significantly enriched in cluster 1 (Supplemental Figure 20A). For cluster 2 the enriched GO terms are related to cell differentiation and regulation of apoptotic process. To validate these observations we performed cell cycle analysis, which revealed that G1 phase was reduced while S and G2 phases were increased in Gata2+/fCbfb+/56MMx1-Cre leukemia cells (Figure 4B and Supplemental Figure 20B). Apoptosis assay using Annexin V revealed that apoptosis was reduced in Gata2+/fCbfb+/56MMx1-Cre leukemia cells after short term incubation (Figure 4C). These observations suggest that Gata2 deficiency results in cell cycle acceleration and reduction of apoptosis in Cbfb-MYH11 induced leukemia.
Figure 4. Gene expression profile of Gata2+/fCbfb+/56MMx1-Cre leukemia cells.
(A) Heatmap shows differentially expressed genes (DEGs) between c-kit+ BM cells from Gata2+/+Cbfb+/56MMx1-Cre mice and Gata2+/fCbfb+/56MMx1-Cre mice at the end points. Cluster 1 is up-regulated genes in Gata2+/fCbfb+/56MMx1-Cre, and cluster 2 is down-regulated genes in Gata2+/fCbfb+/56MMx1-Cre. Only DEGs with p<0.05 and FC>=2 are shown. (B) Representative data of cell cycle analysis using DAPI staining of c-Kit+ BM cells from endpoint leukemic mice. Data was analyzed by FlowJo. (C) Bar graph showing percentage of Annexin V positive c-Kit+ leukemic cells of the indicated genotypes after 4 hours incubation (n=4 for each group). *p<0.05.
Difference in cooperating mutations between Gata2+/+Cbfb+/56MMx1-Cre and Gata2+/fCbfb+/56MMx1-Cre mice
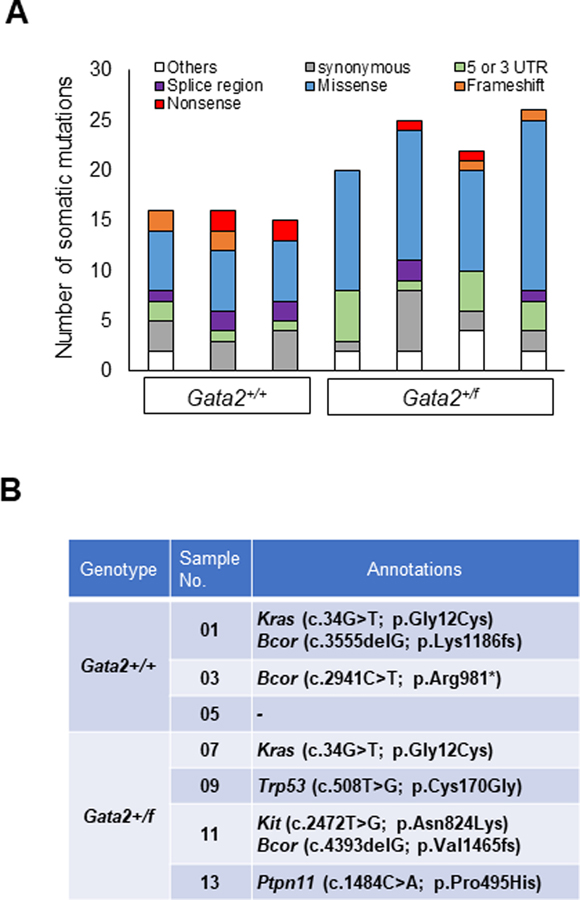
The leukemic cells likely have additional somatic mutations, which may affect their phenotype. Therefore, we performed whole exome sequencing (WES) of c-Kit+ leukemic cells from the spleens of primary leukemic mice (Gata2+/+Cbfb+/56MMx1-Cre and Gata2+/fCbfb+/56MMx1-Cre). DNA samples from CD3+ or CD19+ lymphocytes were used as germline controls. The average depth of sequencing coverage was 73X (Supplemental Figure 21), and the identified somatic mutations are listed in Supplemental Table 4. The types of mutations in each mouse with variant allele frequency (VAF) above 0.1 are shown in Figure 5A. On average, 19.71 ± 4.78 (s.d.) somatic mutations were identified per mouse, with more mutations found in Gata2+/fCbfb+/56MMx1-Cre mice than in Gata2+/+Cbfb+/56MMx1-Cre mice (23.25 ± 3.03 versus 15.0 ± 1.41, p= 0.0141) (Supplemental Figure 22A). Supplemental Table 5 lists nonsense, missense, and frameshift mutations in each mouse. On average, 11.6 ± 4.10 (s.d.) such mutations were identified per mouse, again with more mutations found in the Gata2+/fCbfb+/56MMx1-Cre mice than in the Gata2+/+Cbfb+/56MMx1-Cre mice (14.5 ± 2.87 versus 7.67± 1.25, p= 0.0225) (Supplemental Figure 22B). By reviewing the variants for genes likely involved in leukemogenesis, we identified mutations in Kras, Bcor, Trp53, Kit and Ptpn11 (Figure 5B), which are well known mutated genes in human leukemia.27 The expression of these variants in the leukemia cells was confirmed by RNA-seq. For example, the Bcor (c.2941C>T; p.R981X, c.4393delG; p.V1465fs), Kras (c.34G>T; p.G12C), Trp53 (c.508T>G; p.C170G), and Kit (c.2472T>G, p.N824K) mutations were observed in the corresponding leukemia RNA samples by RNA-seq (Supplemental Figure 22C–G). Importantly, these mutations (Kras p.G12C and Trp53 p.C170G) correspond to the mutations previously detected in human neoplastic patients (KRAS p.G12C and TP53 p.C176G, respectively) (Supplemental Figure 23A and B). In addition, the detected Kit mutation in Gata2+/fCbfb+/56MMx1-Cre (c.2472T>G, p.N824K; in sample 11) corresponds to the KIT p.N822K mutation that is frequently observed in human leukemia patients and is also known as a poor prognostic marker in human CBFB-MYH11 leukemia (Supplemental Figure 23C and D).28 These observations suggest that our Cbfb-MYH11 leukemic mouse model recapitulate the process of human leukemogenesis. Furthermore, preleukemic Gata2 status may affect the mutational landscape in leukemic cells and also their disease phenotype in the mouse model.
Figure 5. Profiling somatic mutations in leukemic cells from Gata2+/+Cbfb+/56MMx1-Cre and Gata2+/fCbfb+/56MMx1-Cre mice.
(A) Bar graphs showing the numbers and types of somatic mutations (with VAF>0.1) in c-Kit+ leukemic cells in 3 Gata2+/+Cbfb+/56MMx1-Cre mice and 4 Gata2+/fCbfb+/56MMx1-Cre mice. Others: upstream, downstream, and non-coding exons. (B) Case-specific mutations that are likely to contribute to leukemia pathogenesis.
Discussion
The understanding of the functional consequences of genetic events at early stages of transformation is critical for the understanding of leukemogenesis. Development of CBFB-MYH11 AML is classically explained by a clonal evolution model which is featured with multiple stepwise cooperating mutations.29, 30 The cooperating mutations in CBFB-MYH11 leukemia are supposed to be class I mutations, e.g., mutations in receptor tyrosine kinases that provide proliferation or survival signal to hematopoietic cells, since CBFB-MYH11 is a class II mutation 2. Using pre-leukemic hematopoietic cells from Cbfb-MYH11 knockin mice, gene expression profiling allowed us to identify 6 most upregulated genes at pre-leukemic stage. Among the human homologues of these 6 genes, GATA2 and CSF2RB2 are frequently expressed in CBFB-MYH11 leukemia patients (Supplemental Table 2). Of these two genes, Gata2 encodes a master regulator of hematopoiesis,13 and a recent study demonstrated increased Gata2 expression in preleukemic cells in the Cbfb-MYH11 knockin mice.31 To clarify the roles of Gata2 in the Cbfb-MYH11 leukemic mouse model, we established Gata2+/fCbfb+/56MMx1-Cre (conditional CBFB-MYH11 expression and Gata2 heterozygous knockout) mice and evaluated them in both ‘preleukemic’ and ‘leukemic’ phases. From the perspective of class I and class II mutations for leukemogensis, this mouse model is unique and interesting as it combined mutations of two transcription factors (both class II mutations) in a mouse model.
In the analysis of the ‘preleukemic’ stage in the Cbfb-MYH11 mice, we found that Gata2 was upregulated, especially in a putative leukemia initiating population16 in the bone marrow. Runx1 deletion in murine hematopoietic cells also led to higher Gata2 expression (Supplemental Figure 4A). Moreover, GATA2 may be a direct transcriptional target of CBFβ-SMMHC (Supplemental Figure 4B). Heterozygous knockout of Gata2 in the Cbfb-MYH11 mice delayed leukemia onset, suggesting that Gata2 has a significant role in the initiation of Cbfb-MYH11 induced leukemia. Interestingly, despite slower development of leukemia, the Cbfb-MYH11 induced leukemia cells in the Gata2 deficient mice showed more aggressive phenotype at the ‘leukemic’ stage (higher WBC counts in the primary mice, higher leukemia repopulating potential through transplantation, accelerated cell cycle and reduced apoptosis) than leukemia cells with intact Gata2 gene. These findings are consistent with the previous report of recurrent deletions of GATA2 in relapsed human CBF-AML patients.14 Thus, Gata2 deficiency in preleukemic phase seems to have contrasting roles in Cbfb-MYH11 leukemia: it represses or delays leukemogenesis from normal hematopoietic progenitors, but increases the repopulating potential of the leukemia cells (Supplemental Figure 24).
In order to understand the molecular mechanisms underlying this functional dichotomy we performed whole exome sequencing and RNA-seq of the leukemia cells. Interestingly, we found that Gata2+/fCbfb+/56MMx1-Cre leukemic cells had more somatic mutations than Gata2+/+Cbfb+/56MMx1-Cre leukemia cells, suggesting that preleukemic cells with Gata2 deficiency may needed more mutations for leukemia transformation, which explains why leukemia development was slower in Gata2+/fCbfb+/56MMx1-Cre mice. Although we could not see a clear difference in the number of somatic mutations between human CBF-AML samples with and without GATA2 deficiency (Supplemental Figure 25A) in our previous study,14 in another dataset32 CBFB-MYH11 AML samples with lower GATA2 expression had relatively higher number of somatic mutations, although the increase did not reach statistical significance (Supplemental Figure 25B and C). On the other hand, RNA-seq analysis revealed a large number of genes whose expression levels were altered in the Gata2+/fCbfb+/56MMx1-Cre leukemia cells as compared to Gata2+/+Cbfb+/56MMx1-Cre leukemia cells. The upregulated pathways included cell cycle, cell division, DNA replication, and cell proliferation. These gene expression changes were consistent with phenotypic observations of cell cycle changes and reduced apoptosis of the Gata2+/fCbfb+/56MMx1-Cre leukemia cells, which could be associated with more aggressive leukemia in the Gata2+/fCbfb+/56MMx1-Cre mice. It should be pointed out that these observed phenotype in the Gata2+/fCbfb+/56MMx1-Cre leukemic cells may be due to additional mutations in these cells rather than Gata2 knockout itself.
Although we have not seen any hematopoietic phenotype in heterozygous Gata2 knockout (Gata2f/+Mx1-cre) mice, which is consistent with previous studies (Supplemental Figure 9),33, 34 Gata2 may affect multiple aspects of hematopoietic cells in transplantation studies, especially in the presence of the Cbfb-MYH11 fusion gene. Previous studies suggested Gata2 knockout contribute to reduced hematopoietic stem cell activity.35, 36 Nevertheless, higher leukemia repopulating potential and aggressive phenotype in the Gata2+/fCbfb+/56MMx1-Cre leukemic cells were observed in our experiment, likely due to modifying interaction between Gata2 and Cbfb-MYH11. In addition, the aggressive phenotype of the Gata2+/fCbfb+/56MMx1-Cre leukemia cells may be related to the additional mutations observed in the Gata2+/fCbfb+/56MMx1-Cre leukemia cells rather than Gata2 deficiency itself. Our findings also raised a possibility that dosage of Gata2 was not that critical for leukemic cells to proliferate once the disease was initiated. Moreover, drug resistant cancer stem cells responsible for disease relapse are thought to be dormant in the previous studies.37 Therefore, the observed aggressive phenotype of the Gata2+/fCbfb+/56MMx1-Cre leukemia cells may not necessarily be related to CBF leukemia relapse in human patients.
Recent genomic sequencing studies in AML patients revealed the presence of preleukemic stem cells, harboring only a few founding mutations and capable of persisting during remission, which thus serve as a potential reservoir for subsequent leukemia relapse.38, 39 In fact, relapsed leukemia often arises from such preleukemic clones.40, 41 Our observation may reflect the process of leukemogenesis in human leukemic patients, in which such delayed leukemic transformation could contribute to relapse or poor prognosis.
GATA2 deficiency, a hereditary leukemia predisposition syndrome, is caused by germline heterozygous mutations in the GATA2 gene and is characterized by bone marrow failure and immunodeficiency with systemic features.42 The Gata2 deficiency in our leukemic mouse model is more like a ‘somatic’ mutation (conditional heterozygous knockout), therefore we have to interpret the results of this study carefully in comparison with the human patients bearing germline GATA2 deficiency. Interestingly, we could find no CBF-AML in human germline GATA2 deficiency patients reported in the literature, even though CBF-AML can be found in 10–15% of leukemia patients without GATA2 deficiency.43, 44 Although our mouse model might not be a precise model for leukemia with germline GATA2 deficiency, the fact that leukemic cells with Gata2 deficiency have more aggressive phenotype than those without it may be consistent with the clinical observation that patients with advanced hematological malignancy and germline GATA2 deficiency have poor prognosis.15
Currently, approximately half of the CBF-AML patients relapse after initial treatment, and survival post-relapse is poor.45, 46 Better understanding of how leukemia develop and relapse will lead to better treatments of the leukemia patients. Our findings further highlight the complex dynamic clonal evolution and the contribution from multiple gene mutation events, including from those that are unexpected.
Supplementary Material
Acknowledgments
The authors thank Stacy Anderson, Martha Kirby, Irene C. Ginty, Wendy Pridgen, NHGRI Transgenic Mouse Core and the National Institutes of Health (NIH) Intramural Sequencing Center for their technical help. Special thanks to Erica Bresciani and Hirotsugu Oda for scientific comments. This work used the computational resources of the NIH High Performance Computing Biowulf cluster (http://hpc.nih.gov).
This work was supported by the Intramural Research Programs of the National Human Genome Research Institute and National Cancer Institute, as well as a JSPS Oversea Research Fellowship (to S.S.).
Footnotes
Conflict-of-interest disclosure
The authors declare no competing financial interests.
References
- 1.Liu P, Tarle SA, Hajra A, Claxton DF, Marlton P, Freedman M, et al. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science (New York, NY) 1993. August 20; 261(5124): 1041–1044. [DOI] [PubMed] [Google Scholar]
- 2.Gilliland DG. Molecular genetics of human leukemias: new insights into therapy. Semin Hematol 2002. October; 39(4 Suppl 3): 6–11. [DOI] [PubMed] [Google Scholar]
- 3.Shigesada K, van de Sluis B, Liu PP. Mechanism of leukemogenesis by the inv(16) chimeric gene CBFB/PEBP2B-MHY11. Oncogene 2004. May 24; 23(24): 4297–4307. [DOI] [PubMed] [Google Scholar]
- 4.Castilla LH, Garrett L, Adya N, Orlic D, Dutra A, Anderson S, et al. The fusion gene Cbfb-MYH11 blocks myeloid differentiation and predisposes mice to acute myelomonocytic leukaemia. Nature genetics 1999. October; 23(2): 144–146. [DOI] [PubMed] [Google Scholar]
- 5.Castilla LH, Perrat P, Martinez NJ, Landrette SF, Keys R, Oikemus S, et al. Identification of genes that synergize with Cbfb-MYH11 in the pathogenesis of acute myeloid leukemia. Proceedings of the National Academy of Sciences of the United States of America 2004. April 6; 101(14): 4924–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castilla LH, Wijmenga C, Wang Q, Stacy T, Speck NA, Eckhaus M, et al. Failure of embryonic hematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MYH11. Cell 1996. November 15; 87(4): 687–696. [DOI] [PubMed] [Google Scholar]
- 7.Adya N, Stacy T, Speck NA, Liu PP. The leukemic protein core binding factor beta (CBFbeta)-smooth-muscle myosin heavy chain sequesters CBFalpha2 into cytoskeletal filaments and aggregates. Molecular and cellular biology 1998. December; 18(12): 7432–7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lutterbach B, Hou Y, Durst KL, Hiebert SW. The inv(16) encodes an acute myeloid leukemia 1 transcriptional corepressor. Proceedings of the National Academy of Sciences of the United States of America 1999. October 26; 96(22): 12822–12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang YY, Shi J, Zhang L, Davis A, Bravo J, Warren AJ, et al. Energetic and functional contribution of residues in the core binding factor beta (CBFbeta ) subunit to heterodimerization with CBFalpha. The Journal of biological chemistry 2000. December 15; 275(50): 39579–39588. [DOI] [PubMed] [Google Scholar]
- 10.Mandoli A, Singh AA, Jansen PWTC, Wierenga ATJ, Riahi H, Franci G, et al. CBFB-MYH11/RUNX1 together with a compendium of hematopoietic regulators, chromatin modifiers and basal transcription factors occupies self-renewal genes in inv(16) acute myeloid leukemia. Leukemia 2014. 04//print; 28(4): 770–778. [DOI] [PubMed] [Google Scholar]
- 11.Hyde RK, Kamikubo Y, Anderson S, Kirby M, Alemu L, Zhao L, et al. Cbfb/Runx1 repression-independent blockage of differentiation and accumulation of Csf2rb-expressing cells by Cbfb-MYH11. Blood 2010. February 18; 115(7): 1433–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhen T, Kwon E, Zhao L, Hsu J, Hyde RK, Lu Y, et al. CHD7 deficiency delays leukemogenesis in mice induced by CBFB-MYH11. Blood 2017. October 10. [DOI] [PMC free article] [PubMed]
- 13.Vicente C, Conchillo A, Garcia-Sanchez MA, Odero MD. The role of the GATA2 transcription factor in normal and malignant hematopoiesis. Critical reviews in oncology/hematology 2012. April; 82(1): 1–17. [DOI] [PubMed] [Google Scholar]
- 14.Sood R, Hansen NF, Donovan FX, Carrington B, Bucci D, Maskeri B, et al. Somatic mutational landscape of AML with inv(16) or t(8;21) identifies patterns of clonal evolution in relapse leukemia. Leukemia 2016. February; 30(2): 501–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donadieu J, Lamant M, Fieschi C, de Fontbrune FS, Caye A, Ouachee M, et al. Natural history of GATA2 deficiency in a survey of 79 French and Belgian patients. Haematologica 2018. August; 103(8): 1278–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuo YH, Landrette SF, Heilman SA, Perrat PN, Garrett L, Liu PP, et al. Cbf beta-SMMHC induces distinct abnormal myeloid progenitors able to develop acute myeloid leukemia. Cancer cell 2006. January; 9(1): 57–68. [DOI] [PubMed] [Google Scholar]
- 17.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science (New York, NY) 1995. September 08; 269(5229): 1427–1429. [DOI] [PubMed] [Google Scholar]
- 18.Charles MA, Saunders TL, Wood WM, Owens K, Parlow AF, Camper SA, et al. Pituitary-specific Gata2 knockout: effects on gonadotrope and thyrotrope function. Molecular endocrinology (Baltimore, Md) 2006. June; 20(6): 1366–1377. [DOI] [PubMed] [Google Scholar]
- 19.Growney JD, Shigematsu H, Li Z, Lee BH, Adelsperger J, Rowan R, et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood 2005. July 15; 106(2): 494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 2010. October 1; 143(1): 134–144. [DOI] [PubMed] [Google Scholar]
- 21.Hyde RK, Zhao L, Alemu L, Liu PP. Runx1 is required for hematopoietic defects and leukemogenesis in Cbfb-MYH11 knock-in mice. Leukemia 2015. August; 29(8): 1771–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mandoli A, Singh AA, Jansen PW, Wierenga AT, Riahi H, Franci G, et al. CBFB-MYH11/RUNX1 together with a compendium of hematopoietic regulators, chromatin modifiers and basal transcription factors occupies self-renewal genes in inv(16) acute myeloid leukemia. Leukemia 2014. April; 28(4): 770–778. [DOI] [PubMed] [Google Scholar]
- 23.Okada S, Nakauchi H, Nagayoshi K, Nishikawa S, Miura Y, Suda T. In vivo and in vitro stem cell function of c-kit- and Sca-1-positive murine hematopoietic cells. Blood 1992. December 15; 80(12): 3044–3050. [PubMed] [Google Scholar]
- 24.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 2000. March 9; 404(6774): 193–197. [DOI] [PubMed] [Google Scholar]
- 25.Dutcher JP, Schiffer CA, Wiernik PH. Hyperleukocytosis in adult acute nonlymphocytic leukemia: impact on remission rate and duration, and survival. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 1987. September; 5(9): 1364–1372. [DOI] [PubMed] [Google Scholar]
- 26.Harrison DE. Competitive repopulation: a new assay for long-term stem cell functional capacity. Blood 1980. January; 55(1): 77–81. [PubMed] [Google Scholar]
- 27.Faber ZJ, Chen X, Gedman AL, Boggs K, Cheng J, Ma J, et al. The genomic landscape of core-binding factor acute myeloid leukemias. Nature genetics 2016. 12//print; 48(12): 1551–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimada A, Taki T, Kubota C, Itou T, Tawa A, Horibe K, et al. N822 mutation of KIT gene was frequent in pediatric acute myeloid leukemia patients with t(8;21) in Japan: a study of the Japanese childhood AML cooperative study group. Leukemia 2007. October; 21(10): 2218–2219. [DOI] [PubMed] [Google Scholar]
- 29.Gilliland DG. Hematologic malignancies. Current opinion in hematology 2001. July; 8(4): 189–191. [DOI] [PubMed] [Google Scholar]
- 30.Pulikkan JA, Castilla LH. Preleukemia and Leukemia-Initiating Cell Activity in inv(16) Acute Myeloid Leukemia. Frontiers in oncology 2018; 8: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai Q, Jeannet R, Hua WK, Cook GJ, Zhang B, Qi J, et al. CBFbeta-SMMHC creates aberrant megakaryocyte-erythroid progenitors prone to leukemia initiation in mice. Blood 2016. September 15; 128(11): 1503–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018. October; 562(7728): 526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai FY, Keller G, Kuo FC, Weiss M, Chen J, Rosenblatt M, et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 1994. September 15; 371(6494): 221–226. [DOI] [PubMed] [Google Scholar]
- 34.Onodera K, Fujiwara T, Onishi Y, Itoh-Nakadai A, Okitsu Y, Fukuhara N, et al. GATA2 regulates dendritic cell differentiation. Blood 2016. July 28; 128(4): 508–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ling KW, Ottersbach K, van Hamburg JP, Oziemlak A, Tsai FY, Orkin SH, et al. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. The Journal of experimental medicine 2004. October 04; 200(7): 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim KC, Hosoya T, Brandt W, Ku CJ, Hosoya-Ohmura S, Camper SA, et al. Conditional Gata2 inactivation results in HSC loss and lymphatic mispatterning. The Journal of clinical investigation 2012. October; 122(10): 3705–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ebinger S, Ozdemir EZ, Ziegenhain C, Tiedt S, Castro Alves C, Grunert M, et al. Characterization of Rare, Dormant, and Therapy-Resistant Cells in Acute Lymphoblastic Leukemia. Cancer cell 2016. December 12; 30(6): 849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 2014. February 20; 506(7488): 328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proceedings of the National Academy of Sciences of the United States of America 2014. February 18; 111(7): 2548–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science (New York, NY) 2008. November 28; 322(5906): 1377–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 2012. January 11; 481(7382): 506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hyde RK, Liu PP. GATA2 mutations lead to MDS and AML. Nature genetics 2011. September 28; 43(10): 926–927. [DOI] [PubMed] [Google Scholar]
- 43.Grimwade D, Ivey A, Huntly BJ. Molecular landscape of acute myeloid leukemia in younger adults and its clinical relevance. Blood 2016. January 07; 127(1): 29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med 2016. June 9; 374(23): 2209–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Appelbaum FR, Kopecky KJ, Tallman MS, Slovak ML, Gundacker HM, Kim HT, et al. The clinical spectrum of adult acute myeloid leukaemia associated with core binding factor translocations. British journal of haematology 2006. October; 135(2): 165–173. [DOI] [PubMed] [Google Scholar]
- 46.Bhatt VR, Kantarjian H, Cortes JE, Ravandi F, Borthakur G. Therapy of core binding factor acute myeloid leukemia: incremental improvements toward better long-term results. Clinical lymphoma, myeloma & leukemia 2013. April; 13(2): 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.