Abstract
Neuroblastoma is a deadly pediatric solid tumor with infrequent recurrent somatic mutations. Particularly, the pathophysiology of tumors without MYCN amplification remains poorly defined. Utilizing an unbiased approach, we performed gene set enrichment analysis of RNA-seq data from 498 neuroblastoma patients and revealed a differentially overexpressed gene signature in MYCN non-amplified neuroblastomas with telomerase reverse transcriptase (TERT) gene overexpression and coordinated activation of oncogenic signaling pathways, including E2Fs, Wnt, Myc, and the DNA repair pathway. Promoter rearrangement of the TERT gene juxtaposes the coding sequence to strong enhancer elements, leading to TERT overexpression and poor prognosis in neuroblastoma, but TERT-associated oncogenic signaling remains unclear. ChIP-seq analysis of the human CLB-GA neuroblastoma cells harboring TERT rearrangement uncovered genome-wide chromatin co-occupancy of Brd4 and H3K27Ac and robust enrichment of H3K36me3 in TERT and multiple TERT-associated genes. Brd4 and cyclin-dependent kinases (CDKs) had critical regulatory roles in the expression and chromatin activation of TERT and multiple TERT-associated genes. Epigenetically targeting Brd4 or CDKs with their respective inhibitors suppressed the expression of TERT and multiple TERT-associated genes in neuroblastoma with TERT overexpression or MYCN amplification. ChIP-seq and ChIP-qPCR provided evidence that the CDK inhibitor directly inhibited Brd4 recruitment to activate chromatin globally. Therefore, inhibiting Brd4 and CDK concurrently with AZD5153 and dinaciclib would be most effective in tumor growth suppression, which we demonstrated in neuroblastoma cell lines, primary human cells, and xenografts. In summary, we describe a unique mechanism in neuroblastoma with TERT overexpression and an epigenetically targeted novel therapeutic strategy.
Introduction
Neuroblastoma (NB) is the most common pediatric extracranial solid tumor and accounts for approximately 15% of childhood cancer mortality (1, 2). Despite recent treatment advances, the prognosis of high-risk NB remains poor. Activating intragenic rearrangements involving the upstream promoter region of the TERT gene were found to define a subgroup of approximately 20% high-risk NB mutually exclusive from MYCN amplifications or ATRX mutations (3, 4). The rearrangements led to placement of a super-enhancer (SE) sequence proximal to the TERT promoter, chromatin remodeling and robust enrichment of active histone marks H3K27Ac in the promoter or enhancer and H3K36me3 in the gene body. This resulted in increased TERT transcription and productive transcriptional elongation (3, 4).
Bromodomain-containing protein 4 (Brd4) modulates SE-associated transcriptional initiation and elongation through recruitment of the Mediator complex and positive transcriptional elongation factor b (p-TEFb) complex consisting of Cdk9 and cyclin T1 (5). Cdk9 phosphorylates the serine-2 residues on the C-terminal repeat domain (CTD) of RNA Pol II and leads to productive transcriptional elongation (5). Brd4 preferentially bind to acetylated histones at promoters and enhancers or SE and recruit Cdk9 (6, 7). BET proteins also function as master transcriptional elongation factors, activating global transcription elongation and enhancer-directed gene transcription through a mechanism independent of CDK9 recruitment (7). Decreased binding of Brd4 to chromatin through Brd4 inhibitors (JQ1) or small-molecular degrader of BET family proteins (dBET1 or dBET6) results in diminished transcription and profound anti-proliferative effects (6, 7).
No comprehensive study has yet explored the molecular pathways and gene signatures associated with TERT overexpression in NB. We hypothesized that the elucidation of transcriptional networks and epigenetic alteration associated with TERT overexpression can lead to identification of druggable targets to improve therapeutic efficacy. In this study, we identified a coordinated activation of the gene networks driven by a variety of transcriptional factors in a NB subgroup with TERT overexpression. We uncovered a direct regulatory role of Brd4 and Cdks in the transcription of TERT and TERT-associated genes and a novel mechanism of action wherein dinaciclib (Cdk inhibitor) inhibits Brd4 recruitment to active chromatin. Brd4 inhibitors and dinaciclib could each inactivate the expression of TERT and multiple TERT-associated genes and profoundly suppress the NB tumor growth. BET inhibitors JQ1 or AZD5153 and dinaciclib in combination act synergistically or additively to inhibit the growth of NB cells with TERT overexpression or MYCN amplification in vitro and NB xenograft growth in vivo. We found overlapping molecular phenotypes between NB with MYCN amplification and TERT overexpression and demonstrated an epigenetically co-targeting strategy to treat NB with TERT overexpression.
Materials and Methods
NB cell lines, patient tumor and bone marrow samples
NB cell lines were purchased from the American Type Culture Collection (ATCC; Rockville, MD). All cell lines were authenticated by ATCC and free from mycoplasma contamination. Kelly and CLB-GA cells were grown in RPMI 1640 w/ HEPES, 5% FBS, 2mM glutamine, and 1X Insulin-Transferrin-Selenium (ITS), CHLA-90 cells in IMDM with 10% FBS, and 1X ITS, SK-N-AS cells in DMEM with 10% FBS, and 0.1mM NEAA. All media contained 100Uml−1 penicillin and 100μg ml −1 streptomycin.
Fresh resected NB patient tumor samples and bone marrow (BM) were collected and utilized via Stanford University Institutional Review Board (IRB)-approved protocols (#45458, #46426). Mononuclear cells from BM used were Ficoll-purified.
Gene Set Enrichment Analysis (GSEA)
We performed Gene Set Enrichment Analysis (GSEA) (http://www.broadinstitute.org/gsea) to interpret gene expression data between two biological groups using the Molecular Signatures Database, MSigDB (http://www.broad.mit.edu/gsea/msigdb/index.jsp) (8). Default parameters and T-test were used. Gene sets with a false discovery rate (FDR) <25% were considered significant.
R2 genomics analysis and visualization
We analyzed the Tumor Neuroblastoma-SEQC-498-seqcnb1 dataset, Tumor Neuroblastoma (TERT)-Fischer-394-custom (9), and a third Tumor Neuroblastoma Gene-TARGET-161 dataset using the R2 platform [Koster J. R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl) (2011)].
ChIP–qPCR and ChIP-seq
ChIP–qPCR and ChIP-seq were performed as described previously (10, 11). Briefly, approximately 20–50 million cells per IP were collected and crosslinked with formaldehyde. Antibodies used: anti-Brd4 (A301–985A50, BETHYL Laboratories), Anti-H3K27ac (ab4729, Abcam), Anti-H3K36me3 (ab9050, Abcam). The specific primers used are listed in the Supplementary Table S1. ChIP-seq libraries were prepared according to the Illumina protocol and sequenced using a HiSeq (Illumina) at the Stanford Functional Genomics Facility. Single-end reads of 75-base were obtained, yielding a minimum of 20 million reads/sample. Sequencing data were uploaded and analyzed using the Galaxy web platform (12). ChIP-seq data have been deposited at Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo), accession number GSE133453.
Real time RT-PCR analysis
The RNA extraction and cDNA synthesis were previously described (11). The specific primers used are listed in the Supplementary Table S1.
Orthotopic NB PDX and CLB-GA xenograft mouse model and in vivo treatment
Establishment of orthotopic NB patient derived xenograft (PDX) and CLB-GA xenograft mouse models were previously described (13) and approved by Stanford University Institutional Animal Care and Use Committee (#32942). PDXs-NB-ST16 were generated using collagenase-digested patient adrenal NB tumor (MYCN-amplified). When the tumor reached >50mm3, the CLB-GA injected mice (n=10 per group) were treated with dinaciclib IP (40mg/kg/day) (14) alone, by oral gavage with AZD5153 (5mg/kg/day) alone (15), dinaciclib and AZD5153 combined, or vehicle control daily daily. Tumor volume was measured via ultrasound as previously described (13).
Immunoblot
Cell lysis and western blot were performed as described previously (11). Antibodies used: anti-actin (A5441, Sigma), anti-Phospho-Rb #9308 (Ser807/811) and anti-Rb (4H1) Mouse mAb #9309 (Cell Signaling), anti-β-catenin (sc-7963), anti-Dvl2 (sc-8026, Santa Cruz Biotechnology), and anti-Myc (ab32072, abcam).
MTS cell viability assay
Cell viability was assessed using CellTiter-AQueous MTS assay kit (Promega) and measured by microplate reader with a 96-well format.
Statistical analysis
Results were expressed as mean value ± SD (four independent experiments) and analysed using Student’s t-test. Survival functions were plotted within Prism using Kaplan-Meier analysis and compared using the log-rank test.
Results
High TERT mRNA expression is associated with poor prognosis and enrichment of oncogenic pathways in neuroblastoma
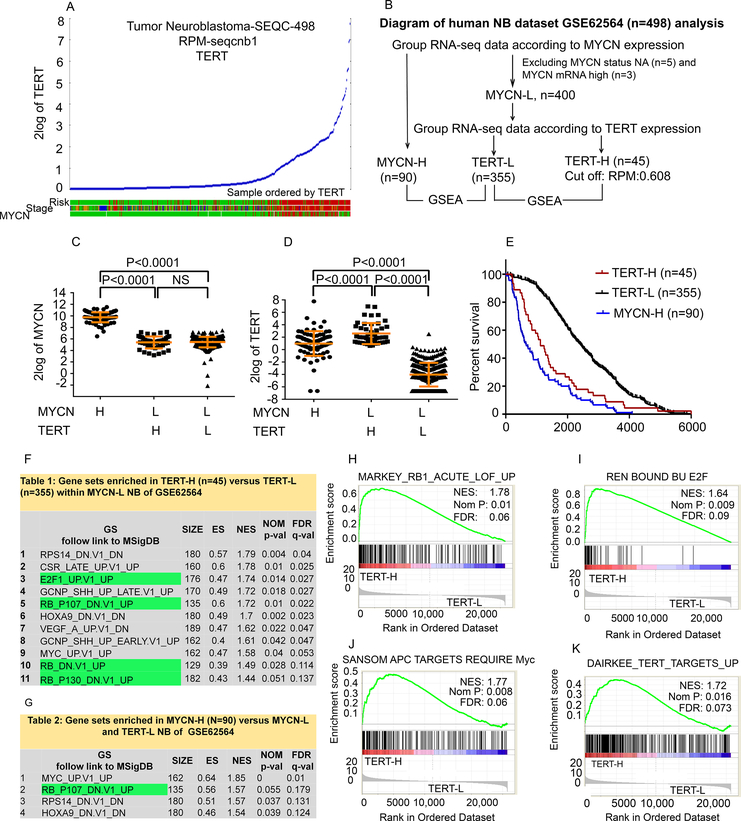
Using R2 genomics analysis and visualization application, we validated the direct link between the level of TERT expression and tumor risk, stage, and MYCN status in an RNA-seq dataset GSE62654 from a cohort of 498 NB patients (16). We identified a distinct subpopulation of NB which overexpressed TERT and was associated with high-risk, high-stage, and absence of MYCN amplification (Fig. 1A). We categorized this dataset into MYCN-High (MYCN-H, n=90) and MYCN-Low (MYCN-L). MYCN-L was further subdivided into TERT-high (TERT-H, n=45) and TERT-Low (TERT-L, n=355) (Fig. 1B). The levels of MYCN and TERT mRNA expression in these categories are shown in Fig. 1C and 1D. TERT mRNA was increased in MYCN-H tumors compared to that in the MYCN-L TERT-L subgroup (Fig. 1D). Strikingly, the TERT-H subgroup expressed a significantly higher level of TERT than both the MYCN-H and TERT-L subgroups (Fig. 1D) and had a similar decreased overall survival as the MYCN-H subgroup (Fig. 1E). Among the high risk patients in this dataset, 52% had amplified MYCN (MYCN-H) and 23% had TERT overexpression (TERT-H) without MYCN amplification (cut off Reads Per Million Reads (RPM)=0.608).
Fig. 1. TERT mRNA expression and identification of top enriched pathways in TERT-H NB.
(A) High TERT mRNA expression was associated with high-risk and high-stage NB as analyzed with R2 using GSE62564 dataset consisting of 498 human NB patient samples. High-Risk and stage 4 (red), Stage 4 (red), MYCN amplified (red), MYCN non-amplified (green). (B) RNA-seq results from 490 NB patients within the GSE62564 dataset were assigned into three subgroups: MYCN-H, TERT-H (MYCN-L), and TERT-L (MYCN-L). (C, D) Dot plots showing mRNA expression levels of MYCN and TERT in the three subgroups. Each dot represents a value from an individual patient. (E) Comparison of the overall survival among the three subgroups. (F, G) The top enriched pathways in TERT-H and MYCN-H subgroups; (H-K) Representative enrichment plots for selected genes sets differentially expressed in the TERT-H (MYCN-L) compared to TERT-L (MYCN-L) are presented. On the x-axis are genes ranked according to their expression, starting with the up-regulated genes on the left and all the way down to the downregulated genes on the far right. NES: normalized enrichment score. Nom P: normalized p value; FDR: false discovery rate. (H) Genes up-regulated in adult fibroblasts with inactivated RB1 by Cre-lox: acute loss of function (LOF) of RB1; (I) Genes whose promoters were bound by E2F1 and E2F4 in the primary fibroblasts WI-38, by ChIP on chip assay; (J) Genes up-regulated after Cre-lox knockout of APC in the small intestine that require functional MYC; (K) Genes up-regulated in non-spontaneously immortalizing (NSI) primary breast cancer tumor cultures upon expression of TERT off a retroviral vector.
We next applied GSEA transcriptome analysis to identify the differentially upregulated gene sets which are enriched in the TERT-H (MYCN-L) or MYCN-H subgroups as compared with the TERT-L (Fig. 1F and 1G). Using MSigDB curated C6 Oncogenic signatures consisting of 189 gene sets often deregulated in cancer, we found that more oncogenic gene sets were significantly enriched in the TERT-H than the MYCN-H subgroup as compared with that in the TERT-L subgroup. Several gene sets were similarly enriched in both TERT-H and MYCN-H subgroups, corresponding to loss of RB1 function and over-expression of Myc, RPS14- or HoxA9-modulated set of genes (Fig. 1F and 1G). Similarly, using C2 CGP (chemical and genetic perturbations) which contained 3433 gene sets, we found genes upregulated upon acute loss of RB1 (Fig. 1H) and genes whose promoters were directly bound by E2F1 and E2F4 (Fig. 1I) as the top enriched gene sets. The APC dependent gene set was also enriched in the TERT-H subgroup, indicative of Wnt/β-catenin pathway activation (Fig. 1J). Additionally, genes induced by ectopic TERT overexpression were among the top enriched gene sets in the TERT-H subgroup (Fig. 1K), demonstrating a causal relationship between TERT overexpression and upregulation of this set of genes. Similarly, TERT-overexpression and significant enrichment of oncogenic or molecular gene sets were found in NB with TERT rearrangement by analyzing a second Fischer-394 dataset of 310 MYCN non-amplified NB patients including patients with (n=32) and without (n=278) TERT-gene rearrangement (9) and a third Tumor Neuroblastoma Gene-TARGET–161 dataset within R2 platform (Supplementary Fig. S1, A–R and Table S2–S3).
Brd4 is essential to activation of TERT-associated gene expression signature
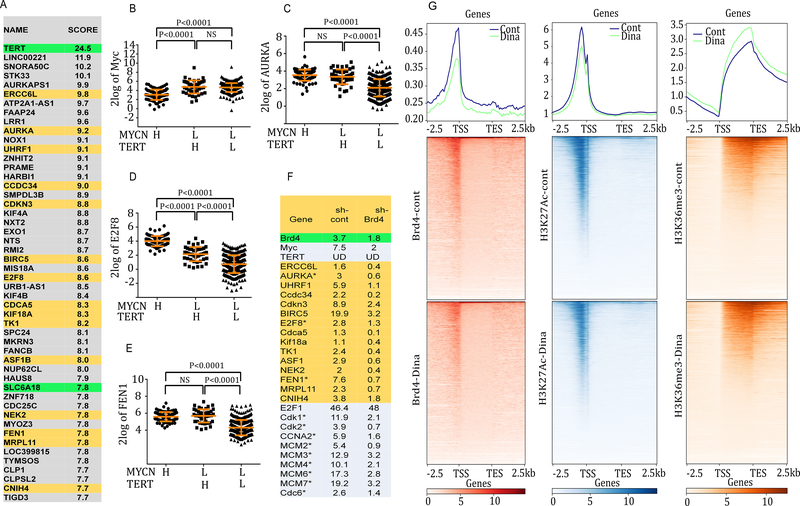
We examined the differentially up-regulated genes between the TERT-H and TERT-L subgroups of NB without MYCN amplification to further explore the potential mechanisms involved in the activation of TERT-associated gene signature. GSEA revealed the top 50 overexpressed genes in the TERT-H subgroup versus TERT-L, ranking from the largest to the smallest fold change in Fig. 2A. Both c-Myc and MYCN are known TERT transcriptional regulators that directly activate its expression via promoter binding and association with other transcription factors (17–19). However, the TERT-H and TERT-L subgroups which are MYCN-L expressed equivalent levels of MYCN and c-Myc mRNA (Fig. 1C and 2B; Supplementary Fig. S1, A–C), suggesting that TERT overexpression in the TERT-H subgroup was not due to an increase in c-Myc or MYCN mRNA expression. In addition to TERT itself, Aurora kinase A (AURKA), E2F8, and flap endonuclease-1(FEN1) are also among the top 50 up-regulated genes in the TERT-H subgroup, and the expression of AURKA, E2F8, and FEN1 are all significantly higher in the TERT-H than the TERT-L subgroup (Fig. 2, C–E; Supplementary Fig. S1,D–F).
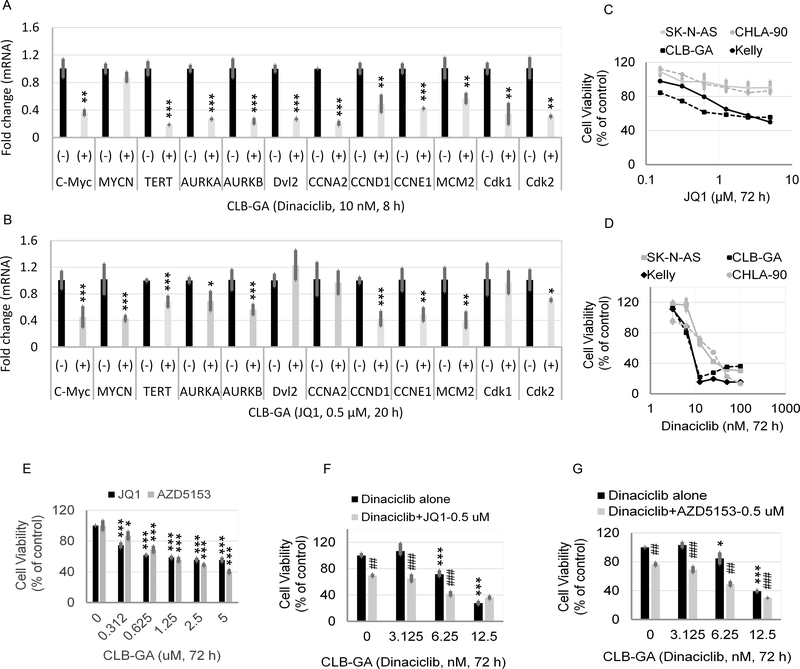
Fig. 2. Identification of potential Brd4 target genes within the top 50 up-regulated genes in TERT-H subgroup of NB and effects of dinaciclib on genome-wide chromatin occupancy of epigenetic modulators in CLB-GA cells with known TERT gene rearrangement.
(A) Top 50 differentially up-regulated genes in TERT-H (n=45) versus TERT-L subgroup (n=355) within MYCN-L of GSE62564 dataset by GSEA. (B, C, D, E) Comparison of gene expression profiles of Myc, AURKA, E2F8, and FEN1 among MYCN-H, TERT-H (MYCN-L), and TERT-L (MYCN-L) subgroup of GSE62564 dataset. Each dot represents a value from an individual patient. (F) Gene expression changes analyzed by RNA-seq. Effects of Brd4 shRNA on the expression of TERT-associated genes and E2F target genes in Brd4 active MLL-AF9 AML cells. Known E2F target genes are labelled with an asterisk (*). Data were extracted from microarray analysis of murine leukemia cells expressing MLL-AF9 fusion with or without Brd4 shRNA (15). (G) Effects of dinaciclib on genome-wide chromatin occupancy of Brd4, H3K27Ac, and H3K36me3 in CLB-GA cells. Cells were treated in the absence or presence of 10 nM dinaciclib for 8 h, followed by Brd4-, H3K27Ac-, and H3K36me3-ChIP-seq. The binding profiles were plotted using computeMatrix in scale-region mode, and each gene body was shrunken to the same length as 2500 bp. Sequences up to 2.5 kb upstream of the TSS and 2.5 kb downstream of the 3’ gene end were shown. Average occupation for each 100 bin region is indicated. The top panel histograms show the average genome-wide occupancies in the presence or absence of dinaciclib for Brd4, H3K27Ac, and H3K36me3. The lower panels of density heat maps show genome-wide chromatin occupancy by Brd4, H3K27Ac, and H3K36me3 at the TSS, the transcriptional end site (TES), and gene scaled gene body region and effects of dinaciclib.
TERT promoter rearrangement juxtaposes the TERT coding sequence to stronger SE elements (3), resulting in an increased expression of TERT (3). Brd4 plays a critical role in co-activating diverse key transcriptional regulators by positively regulating SE function (5). We hypothesized that SE-driven transcriptional programs are required for the active transcription of TERT and TERT-associated pathways in the TERT-H subgroup. In support of this hypothesis, we found that 15 of the top 50 differentially up-regulated genes in the TERT-H subgroup are either proven or potential Brd4 target genes (Fig. 2A). The specific dependency of these genes on Brd4 was further validated by reanalyzing the previously published microarray data in which Brd4 was depleted with shRNA in a leukemia cell model with known activated Brd4 pathways driven by MLL-fusion protein (20). The mRNA expression of these genes was largely abolished by Brd4 depletion (Fig. 2F). Other known E2F target genes and cell cycle related genes (marked with asterisk in Fig. 2F) were among the top differentially up-regulated genes in the TERT-H subgroup, and the expression of these genes was also sensitive to Brd4 depletion (Fig. 2F), indicative of an essential role of Brd4 in the regulation of E2F target genes.
Genome-wide chromatin occupancy of epigenetic modulators and effects of dinaciclib
To further understand the chromatin status globally and locally at the loci of TERT or TERT-associated genes we performed ChIP-seq analysis to examine the chromatin binding profiles of Brd4 along with active chromatin marks H3K27Ac and H3K36me3 in CLB-GA, a NB cell line that harbors TERT gene rearrangement (3). ChIP-seq results were analyzed and plotted with computeMatrix in either scale-region mode (Fig. 2G) or select reference-point mode (Supplementary Fig. S2). We found that Brd4 and H3K27Ac preferentially bind to the gene promoter region around the transcription start site (TSS) while H3K36me3 is associated with the gene body (Fig. 2G and Supplementary Fig. S2). We then hypothesized that targeting both Cdk9, the core component of the p-TEFb elongation complex, and Cdk2 could impact the chromatin occupancies of these epigenetic modulators. We treated CLB-GA cells for 8 h with 10 nM dinaciclib, a Cdk inhibitor (21) and examined the genome-wide binding profiles of Brd4, H3K27Ac, and H3K36me3. This treatment resulted in a marked global decrease of the Brd4 occupancy, a mild reduction of the H3K27Ac occupancy, and a slight increase in the H3K36me3 binding occupancy (Fig. 2; Supplementary Fig. S2).
Dinaciclib inhibits TERT mRNA expression and chromatin occupancy of epigenetic modulators
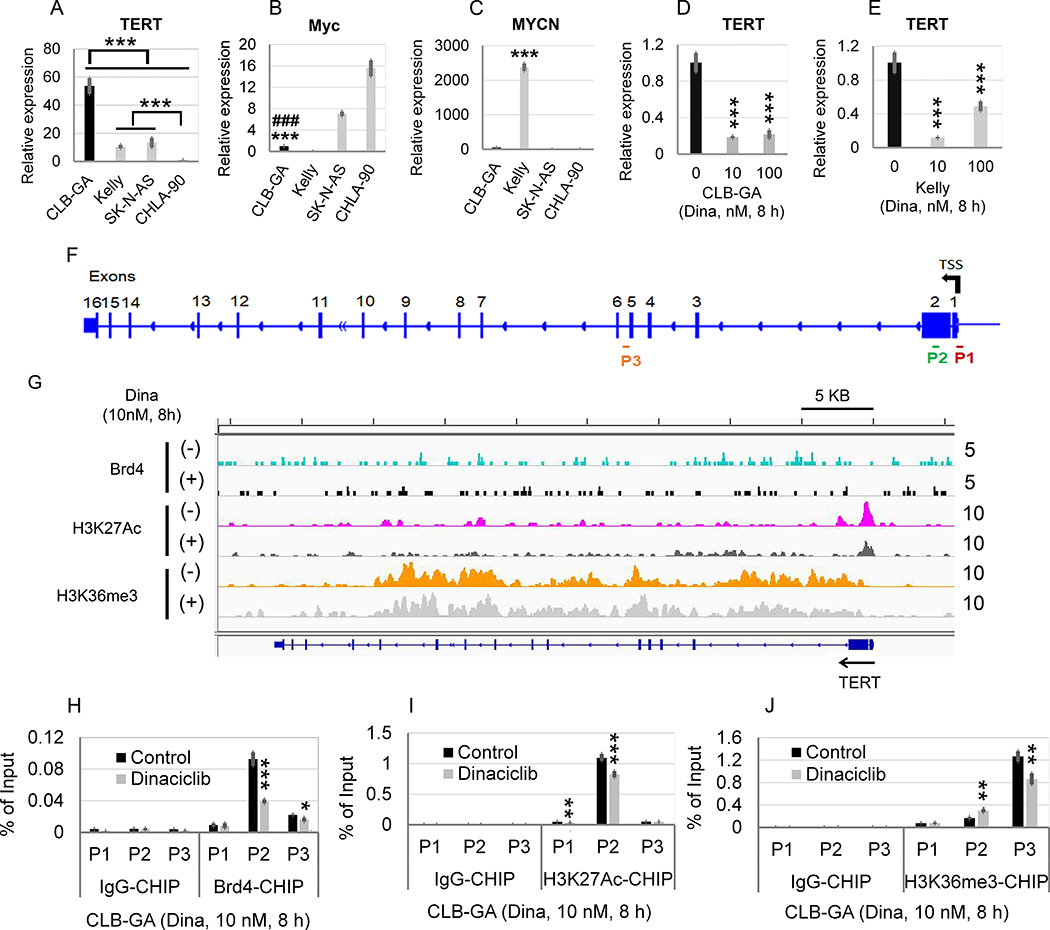
We hypothesized that dinaciclib could decrease TERT expression and affect the chromatin occupancy of Brd4, H3K27Ac, and H3K36me3 at the TERT promoter or gene body. Consistent with what was observed in the large NB patient cohort (Fig. 1D), the CLB-GA cell line bearing a known TERT gene rearrangement (3) expressed a significantly higher level of TERT than the MYCN-amplified Kelly cell line, and two other MYCN non-amplified NB cell lines: CHLA-90 and SK-N-AS (Fig. 3A). Moreover, there was no direct correlation between TERT overexpression and the mRNA or protein expression of c-Myc or MYCN among four NB cell lines (Fig. 3, B and C; supplementary Fig. S3). Strikingly, treatment of CLB-GA or Kelly cells with dinaciclib 10 nM for 8 h resulted in a profound inhibition of TERT expression in both CLB-GA (Fig. 3D) and Kelly cells (Fig. 3E), suggesting that dinaciclib effectively abrogates the TERT expression driven by TERT or MYCN overexpression. We then hypothesized that dinaciclib directly disrupts the recruitment of Brd4, H3K27Ac, and H3K36me3 to TERT promoter or gene body. We designed three ChIP-qPCR primers (Fig. 3F) that probe regions of the TERT promoter (P1 and P2) or gene body (P3). Consistent with the general pattern seen in Fig. 2G and Supplementary Fig. S1, H3K27Ac binds primarily to the TERT promoter region whereas the H3K36me3 is enriched in the TERT gene body shown as multiple peaks in the H3K36me3 track (Fig. 3G). Brd4 was shown to bind to the TERT promoter and gene body region, shown as multiple small peaks. Treatment of CLB-GA cells with 10 nM dinaciclib decreased the binding peaks of Brd4 and H3K27Ac at TERT promoter or gene body but had less effect on the enrichment of H3K36me3 at TERT promoter or gene body (Fig. 3G). We next quantified the occupancy of Brd4, H3K27Ac, and H3K36me3 at the TERT promoter (P1/P2) or gene body (P3 primer set) in control- or dinaciclib-treated CLB-GA cells with ChIP-qPCR. Similarly, the binding of Brd4 or H3K27Ac to the TERT promoter was significantly decreased after dinaciclib treatment (Fig. 3, H and I). By contrast, the binding of H3K36me3 was significantly increased at TERT promoter (P2) but decreased at TERT gene body region (P3) (Fig. 3J).
Fig. 3. Effects of dinaciclib on the TERT mRNA expression and chromatin occupancy of epigenetic modulators in CLB-GA.
(A, B, C) Comparison of mRNA expression of TERT, Myc, and MYCN in human NB cell lines CLB-GA, Kelly, SK-N-AS, and CHLA-90. (D, E) Effects of dinaciclib on the mRNA expression of TERT in CLB-GA and Kelly cells. Cells were untreated or treated with dinaciclib at 10 nM and 100 nM for 8 h, followed by RT-qPCR analysis. The relative levels of TERT mRNA expression were normalized to the GAPDH level in D and E and expressed as fold changes relative to control (set at 1). The mean ± SD of four replicates is shown. (F) Diagram of TERT gene body and promoter. The locations of CHIP-qPCR primer sets for TERT are shown. P1: TERT promoter (−13 to +121 bp relative to TSS); P2: TERT promoter (389 to 531 bp relative to TSS, on exon 2; P3: 53 bp relative to the last amino acid of TERT exon 5 (TERT gene body). (G) Effects of dinaciclib on binding of Brd4, H3K27Ac, and H3K36me3 to the promoters or enhancers of TERT gene. CLB-GA cells were treated in the absence or presence of 10 nM dinaciclib for 8 h, followed by Brd4-, H3K27Ac, and H3K36me3-ChIP-seq, as described in the Materials and Methods. IGV visualization of ChIP-seq peaks at the TERT loci. The selected region spans the whole gene body of TERT as shown at the bottom and the calculated ChIP-seq enrichment values are indicated on the right. (H, I, J) Effect of dinaciclib on TERT promoter binding by Brd4 (H), H3K27Ac (I), and HK36me3 (J), as assessed by ChIP-qPCR.
Dinaciclib inhibits the expression and chromatin occupancy of epigenetic modulators at genes co-targeted by E2F, Wnt-β catenin, and Brd4
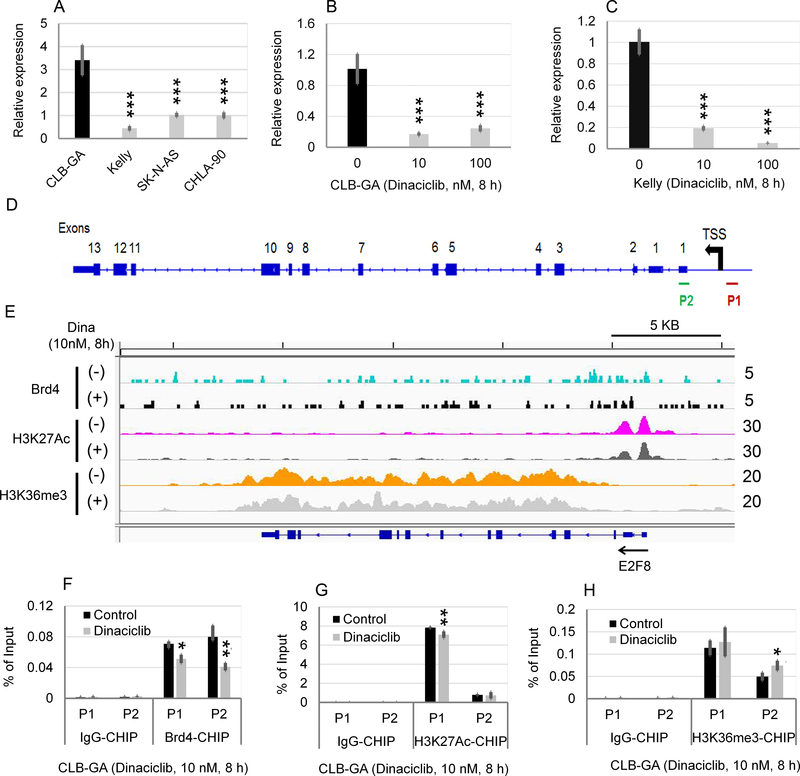
As shown in Fig. 1F and Supplementary Table S1, E2F-dependent gene sets were identified as the top enriched gene sets associated with high TERT in NB. E2F8, a recently identified atypical member of the E2F transcription factor family (22), was identified as one of the top 50 differentially overexpressed genes associated with TERT overexpression (Fig. 2A). We thus examined the effects of dinaciclib on E2F8 mRNA expression and the chromatin occupancy of Brd4, H3K27Ac, and H3K36me3 at E2F8 promoter or gene body. Similar to what was observed with TERT mRNA expression (Fig. 3A), CLB-GA cell line expressed the highest level of E2F8 mRNA in four NB cell lines (Fig. 4A). Dinaciclib treatment at 10 nM for 8 h significantly inhibited the E2F8 mRNA expression in CLB-GA (Fig. 4B) and Kelly cells (Fig. 4C) and markedly decreased the binding of Brd4 and H3K27Ac to E2F8 promoter (Fig. 4, D to G) while increasing the binding of H3K36me3 to the E2F8 promoter (Fig. 4H).
Fig. 4. Effects of dinaciclib on the E2F8 mRNA expression and chromatin occupancy of epigenetic modulators in CLB-GA with known TERT chromosomal translocation.
(A) Comparison of E2F8 expression in four human NB cell lines. Fold changes in the TERT expression was shown in CLB-GA, Kelly, and SK-N-AS cell line in relation to that in CHLA-90. (B, C) Effects of Dinaciclib on the mRNA expression of E2F8 in CLB-GA and Kelly cells. Cells were untreated or treated with Dinaciclib at 10 nM and 100 nM for 8 h, followed by q-PCR analysis. The relative levels of E2F8 mRNA expression in A, B, and C were normalized to the GAPDH, and expressed as fold changes relative to (A) CHLA-90 and (B, C) control (set at 1). The mean ± SD of four replicates is shown. (D) Diagram of E2F8 gene body and promoter. The locations of ChIP-qPCR primer sets for TERT were shown. P1: E2F8 promoter (−158 to 42 bp relative to TSS of E2F8); P2: E2F8 promoter (573 to 822 bp relative to TSS of E2F8). (E) Effects of Dinaciclib on binding of Brd4, H3K27Ac, and H3K36me3 to the promoters or enhancers of E2F8 gene. CLB-GA cells were treated in the absence or presence of 10 nM Dinaciclib for 8 h, followed by Brd4-, H3K27Ac, and H3K36me3-ChIP-seq. Genome browser (hg38) views of ChIP-seq peaks at the following loci. The location of each gene is shown at the bottom of the panels and the calculated ChIP-seq enrichment values are indicated on the right. (F, G, H) Effect of Dinaciclib on E2F8 promoter binding by Brd4 (F), H3K27Ac (G), and HK36me3 (H), as assessed by ChIP-qPCR.
We next examined the effects of dinaciclib on the mRNA expression and chromatin occupancies of several Wnt-β-catenin signaling or target genes. Our results showed that CLB-GA expressed significantly higher levels of Dishevelled protein, Dvl2, a known activator of Wnt-β-catenin signaling pathway (23), AURKA, and AURKB (Supplementary Fig. S4, A–C). The mRNA expression of Dvl2, AURKA and AURKB was also potently suppressed by dinaciclib (Fig. 5A). Moreover, Brd4, H3K27Ac, and H3K36me3 directly bind to the promoters of Dvl2, AURKA, and AURKB, all of which were reduced with dinaciclib treatment. By contrast, dinaciclib had little or no effect on the chromatin occupancy of H3K36me3 on all three genes (Supplementary Fig. S4, D–F).
Fig. 5. JQ1 sensitive Brd4 binding profiles and effects of BET inhibitor or/and dinaciclib on CLB-GA cells.
(A, B) Effects of dinaciclib and JQ1 on the mRNA expression of genes shown in CLB-GA cells. Cells were untreated or treated with 10 nM dinaciclib or 0.5 μM JQ1 for 8 or 20 h, respectively, followed by RT-qPCR analysis. The relative levels of mRNA expression were normalized to the GAPDH level and were expressed as fold changes relative to control (set at 1). (C, D, E) Effects of JQ1, dinaciclib, or AZD5153 on the cytotoxicity of human NB cell lines. Cells were treated with JQ1 (C, E), dinaciclib (D), or AZD5153 (E) at the indicated concentrations for 72 h, followed by MTS assay. (F, G) Additive or synergistic effects of JQ1/AZD5153 in combination with dinaciclib. CLB-GA cells were treated with JQ1, AZD5153, or dinaciclib alone or combination at the indicated concentrations for 72 h, followed by MTS assay. Asterisk or hash is in relation to control or dinaciclib treatment alone, respectively.
Comparing single-gene expression and survival, we showed that overexpression of several TERT-associated genes such as FEN1, BIRC5 (survivin), and Ubiquitin-like PHD and RING domain-containing 1 (UHRF1) was independently associated with high risk, advanced stage, and poor overall survival of NB, and to a lesser extent with MYCN amplification. Epigenetically, we observed an enrichment of Brd4 and H3K27Ac at the promoters of FEN1, BIRC5, and UHRF1, which was sensitive to dinaciclib inhibition. There were also multiple binding peaks of H3K36me3 on the gene body regions of all three genes, which were largely unchanged upon dinaciclib treatment (Supplementary Fig. S5, A–L).
Targeting TERT and TERT-associated genes with BET inhibitors and dinaciclib alone or in combination
To further verify the direct link between Brd4 activation and the TERT-associated gene signature, we validated the direct binding of Brd4 on the chromatin of gene interested by reanalyzing a previously published ChIP-seq dataset GSE104745 deposited at GEO (11). As shown in supplementary Fig. S6, Brd4 directly marked the promoters of TERT, E2F8, AURKA, AURKB, FEN1, and c-Myc (the putative Brd4 target gene) in Brd4 active OCI-AML3 leukemia cells, and such binding to each of these gene promoters was sensitive to Brd4 inhibitor JQ1. To further understand the inhibition of Brd4 or Cdk9 by their corresponding inhibitors on the overexpression of TERT and TERT-associated genes, we compared the effects of dinaciclib and JQ1 on the mRNA expression of the genes activated by E2F- or β-catenin pathways in CLB-GA cells. Unlike JQ1, dinaciclib markedly inhibited the expression of c-Myc but not MYCN (Fig. 5, A and B). JQ1 was less or ineffective in suppressing the expression of TERT, Dvl2, AURKA, AURKB, CCNA2, Cdk1, and Cdk2. By contrast, Dinaciclib was more potent in blocking the expression of those genes (Fig. 5, A and B). Next, we examined the effects of JQ1 and dinaciclib on the growth of CLB-GA cells. As shown in Fig. 5, C and D, both JQ1 and dinaciclib showed more potent cytotoxicity for CLB-GA cells (TERT gene rearrangement) and Kelly cells (MYCN amplification) than to that of CHLA-90 and SK-N-AS cells, both lacking TERT gene rearrangement and MYCN amplification. AZD5153, a novel bivalent inhibitor of Brd4, showed comparable cytotoxicity to JQ1, a conventional monovalent Brd4 inhibitor in CLB-GA cells (Fig. 5E). Combined treatment of JQ1 or AZD5153 with dinaciclib showed additive or synergistic effects on the viability of CLB-GA, Kelly and CHLA-90 (Fig. 5, F and G; Supplementary Fig. S7, A–B and S8), but had little effect on SK-N-AS (Supplementary Fig. S7C and S8).
BET inhibitors and dinaciclib alone or in combination inhibit cell growth and mRNA expression of multiple genes in NB patient primary cells
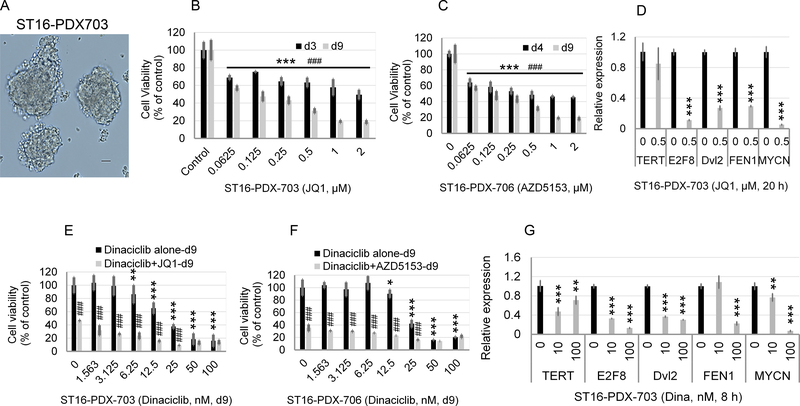
To validate the cell growth inhibition by BET inhibitors, ST16, primary cells isolated from a NB patient tumor carrying MYCN amplification were treated with JQ1 or AZD5153 for 3, 4, or 9 days. The isolated NB tumor cells grew in spheres, as shown in Fig. 6A. Both JQ1 and AZD5153 potently inhibited the tumor cell growth at both day 4 and day 9 with AZD5153 being slightly more effective than JQ1 (Fig. 6, B and C). Treatment of ST16 cells with JQ1 for 20 h drastically suppressed the expression of E2F8, Dvl2, FEN1, and MYCN (Fig. 6D), indicative of Brd4 dependency. Accordingly, AZD5153 or JQ1 alone showed potent inhibitory effects on the growth of ST16 cells (Fig. 6, E and F) while showing less effect on the TERT expression (Fig. 6D). Unlike BET inhibitors, dinaciclib at 10 nM for 8 h exhibited significant inhibition on the expression of TERT, E2F8, and Dvl2 but had little to no effect on the expression of FEN1 and MYCN (Fig. 6G). Higher concentration of dinaciclib (100 nM) was required to achieve marked inhibition on FEN1 and MYCN expression (Fig. 6G). Accordingly, dinaciclib alone at concentrations greater than 12.5 nM showed potent cytotoxicity to ST16 cells (Fig. 6, E and F). Combined treatment with low nanomolar concentrations of dinaciclib and JQ1 or AZD5153 showed additive cytotoxicity (Fig. 6, E and F). Collectively, our results suggest that Brd4 inhibitors and dinaciclib may functionally compensate each other to simultaneously inhibit the expression of multiple growth promoting genes in NB cells with amplified MYCN.
Fig. 6. Effects of BET inhibitors or dinaciclib treatment alone or in combination on the growth of human primary NB cells.
(A) Sphere growth of in vitro cultured NB cells isolated from patient-derived xenograft (PDX) generated using un-cultured human NB tumor cells. (B, C) Primary NB cells were treated with JQ1 or AZD5153 at the indicated concentrations and times, followed by MTS assay. Bar graphs represent mean values ± SD of 4 biological replicates. (D, G) Effects of JQ1 on the mRNA expression of genes shown. Primary NB cells were untreated or treated with JQ1 at 0.5 μM for 20 h, followed by RT-qPCR analysis. The relative levels of mRNA expression were normalized to the GAPDH level and were expressed as fold changes relative to control (set at 1). (E, F) Effects of JQ1, AZD5153, or dinaciclib alone or in combination on the cytotoxicity of human primary NB cells. Cells were treated with either dinaciclib alone at indicated concentrations or JQ1 or AZD5153 alone at 0.5 μM or in combination for 9 days, followed by MTS colorimetric assay of cell viability.
Coordinated expression of TERT and TERT-associated genes in primary human NB cells and inhibition of CLB-GA xenograft growth in vivo by either dinaciclib or AZD5153 alone or in combination
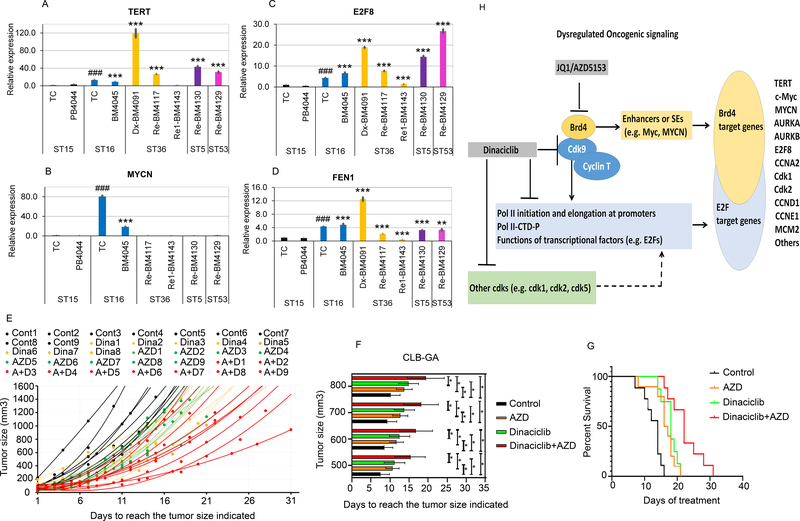
We validated by RT-qPCR the association between TERT overexpression and upregulation of E2F8 and FEN1 using patient primary NB cells. The NB patient and sample information were listed in Supplementary Table S4. The MYCN non-amplified ST15 patient tumor cells (ST15-TC) from adrenal tumor or peripheral blood mononuclear cells (PBMC, ST15-PB) that expressed low levels of TERT were used as baseline control (Fig. 7A). ST16 showed an 80.4-fold and 18.5-fold increase in MYCN expression (Fig. 7B) as compared to that of ST15-TC and ST15-PB (PB4044). Consistent with what was found in the cohort of 498 NB patients (GSE62654) (Fig. 1D), we observed 13-fold and 3-fold increase in TERT mRNA expression in the MYCN amplified ST16-TC (isolated from adrenal tumor) and ST16-BM (BM4045 cells, isolated from hypercellular bone marrow infiltrated with NB cells) compared to that of the ST15 counterparts, respectively (Fig. 7A; Supplementary Table S4). Strikingly, the expression of TERT mRNA was much higher in three hypercellular and metastatic human bone marrow NB samples lacking MYCN amplification, including one at diagnosis (ST36-BM4091) and two at relapse (ST5-BM4130 and ST53-BM4129) (Fig. 7, A and B). Consistent with the expression pattern observed in the large NB patient cohort, the expression of E2F8 and FEN1 correlated well with that of TERT (Fig. 7, A–D). In particular, we were able to follow one NB patient (ST36) whose bone marrow was infiltrated with tumor cells that overexpressed TERT at diagnosis over the course of therapy: the expression of all three genes (TERT, E2F8, and FEN1) was significantly elevated at diagnosis (ST36-BM4091), decreased at d20 Cycle 4-induction therapy (ST16-BM4771), and further decreased to a level that was nearly equivalent to the respective control ST15 at post Cycle 6-induction therapy (ST16-BM4143). This corresponded to a reduced hypercellularity and infiltration of tumor cells in this patient’s bone marrow (Fig. 7, A–D; Supplementary Table S4), raising the possibility that the expression of these genes might serve as biomarkers for therapy response or bone marrow NB tumor cell infiltration.
Fig. 7. Comparison of TERT, E2F8, FEN1, and MYCN expression in human primary NB cells and effects of either dinaciclib or AZD5153 alone or in combination on the CLB-GA orthotopic xenograft growth in vivo.
(A, B, C, D) The mRNA expression of TERT, E2F8, Dvl2, FEN1, and MYCN in human primary NB cells was compared after isolation from primary tumor tissue (TC) or bone marrow (BM), followed by q-PCR analysis. The relative levels of mRNA expression were calculated using the 2−ΔΔCt method after normalization to the GAPDH level and were expressed as fold changes relative to a NB tumor without MYCN amplification (ST15) (set at 1). Asterisk or Hash is in relation to isolated ST15-TC or ST15-BM, respectively. MYCN (+): ST16; MYCN (−): ST15, ST36, and ST5. ST15, ST16, and ST36 Dx-BM4091 were collected at diagnosis. ST36 Re-BM4117 and ST5 BM4130 were collected at refractory disease. ST36 Re1-BM4143 was collected after the refractory tumor was treated. (E) Tumor volume scatter plots of orthotopic CLB-GA xenograft in athymic nude mice. X-axis shows the days after injection of CLB-GA cells. Y-axis shows the tumor size. The dots or curves of best fit with the associated equation were plotted to show individual tumor size of each animal in each group. (F) The bar graph presents the average tumor size. X-axis shows the days after injection of CLB-GA cells. Y-axis shows the average tumor size ± SD of each of the four groups. (G) Survival curve analysis shows the probability of mouse overall survival. Two-sided log-rank test was used to generate the p-value. P <0.001: AZD or dinaciclib versus the vehicle control; AZD+dinaciclib versus with either alone. (H) Proposed working model of regulatory pathways and mechanism of action for BET inhibitors and dinaciclib in human NB with TERT overexpression.
Finally, we hypothesized that AZD5153 and dinaciclib would each be effective alone and more effective in combination than either alone in inhibiting tumor growth in vivo using mice with CLB-GA-derived orthotopic xenograft. Strikingly, the tumor growth from either dinaciclib-(yellow dots/lines, Fig. 7E) or AZD5153-treated mice (green dots/lines, Fig. 7E) was markedly attenuated as compared to that in the mice treated with control vehicle (black dots/lines, Fig.7E). Dinaciclib and AZD5153 in combination exhibited more dramatic suppression of tumor growth (red dots/lines, Fig. 7E). Similarly, the days to reach the tumor sizes indicated and the probability of survival were significantly increased compared to controls in the mouse groups treated with either dinaciclib or AZD5153 alone, and the two drugs combined were more potent than either alone (Fig. 7, F–G). Altogether, NB tumors with TERT overexpression and without MYCN amplification were more sensitive to the concurrent epigenetic targeting with BET inhibitors and dinaciclib in vivo.
Discussion
In this study, using a combination of approaches including GSEA, genome wide epigenetic analysis, and functional validation experiments, we have gained insights into the epigenetic regulation of TERT expression and TERT-associated molecular pathways in the subgroup of high-risk NB with TERT overexpression and without MYCN amplification. We found that TERT overexpression in this subgroup of NB is associated with a coordinated activation of the gene networks driven by epigenetic regulators and diverse transcriptional factors or oncogenic signaling pathways such as E2Fs, Myc, and Wnt pathways.
Compelling evidence suggests a strong link between E2Fs-, Wnt/β-catenin-, or MYCN-pathway and elevated expression of several of the top 50 upregulated genes associated with TERT overexpression, including BIRC5, AURKA, UHRF1, and FEN1(24–28). Overexpression of these genes was found to be significantly associated with poor prognosis of NB. BIRC5 and AURKA are direct target genes of E2Fs (24, 25, 29, 30), Wnt/β-catenin (26, 31), MYCN (25, 32), and Myc (33). In MYCN amplified NB, BIRC5 is induced via a functional cooperation between MYCN and E2F1 (25). UHRF1 was recently identified as one of the E2F8 target genes (27, 28). FEN1 was previously reported to be a direct E2F target gene whose promoter was bound by both E2F1 and E2F4 (34).
BET proteins such as Brd4 co-activate diverse oncogenic transcription and elongation via direct binding of BDs to acetylated histones at enhancers or SEs (7, 35, 36). SE-dependent NB-specific core transcriptional programs driven by MYCN and Myc can be repressed transcriptionally by inhibition of Brd4 (37–39). Recently, E2F-dependent transcriptional program and transcription of cell cycle-related genes were identified as the primary downstream targets of BET proteins in both glioblastoma (GBM) cell lines and patient-derived GBM spheres with integrative ChIP-seq and RNA-sequencing analysis (35).
In this study, we hypothesized that BET protein Brd4 cooperates with oncogenic transcriptional factors in the E2Fs and Wnt pathways and upregulates TERT and TERT-associated genes in the subgroup of NB with TERT overexpression (proposed model in Fig. 7H). Profiling the chromatin binding profiles of epigenetic modulators Brd4, H3K27Ac, and H3K36me3 enabled us to verify the direct chromatin activation of TERT and TERT-associated genes in CLB-GA, a NB cell line harboring TERT gene rearrangement and TERT overexpression. The transcriptional dependence on Brd4 by these upregulated genes was further validated by reanalyzing the published datasets using Brd4-shRNA (20) or BET inhibitor (11). Moreover, inhibition of Brd4 with BET inhibitor JQ1 or AZD5153 simultaneously targeted and inhibited the expression of multiple TERT-associated genes and attenuated the tumor growth of NB cell lines harboring TERT gene rearrangement or MYCN amplification.
Dinaciclib was previously shown to inhibit both Cdk2 and Cdk9 activity in MYCN-amplified NB cells, leading to inhibition of Rb phosphorylation at Serine 807/811 (21). However, our results indicate that the inhibitory effects of dinaciclib on the transcription of E2Fs-, Myc-, Wnt-β-catenin, and Brd4 target genes and tumor growth are mediated through a mechanism independent of p-Rb inhibition at a lower nanomolar concentration (Supplementary Fig. S9, A–C). We showed that dinaciclib mediates a marked global reduction in the chromatin occupancy by Brd4 in CLB-GA cells, suggesting a previously undefined mechanism of dinaciclib directly affecting Brd4 chromatin recruitment. This is supported by a crystal structure study showing dinaciclib interacting with the acetyl-lysine recognition site of BRDT (BD testis-specific protein, a member of the BET family of bromodomains) through hydrogen bonding interactions within the KAc site (37, 38, 40, 41). Our ChIP-seq and ChIP-qPCR results also provide evidence that dinaciclib directly inhibited the recruitment of Brd4 to activate chromatin globally through either directly interfering Brd4 binding or inhibiting Cdk9 in p-TEFb-mediated elongation.
Our results enable us to postulate that a significant portion of NB patients with TERT overexpression and without MYCN amplification in the 498 NB GSE62654 dataset may carry TERT gene rearrangement, a genetically distinct subtype of high-risk NB. This was supported by: 1) Similar to what was observed in the TERT-Fischer-394-custom dataset (Supplemental Fig. S1A), there was a clear “on” and “off” TERT mRNA expression pattern with a distinct RPM cutoff (Fig. 1, A and D); 2) TERT was overexpressed in 22.5% of high-risk NB, similar to the percentage of NB with TERT gene rearrangement reported in recent studies (3, 4); 3) TERT promoter mutation is less likely to be involved, since none to date has been identified (18); 4) No direct correlation between the expression of c-Myc or MYCN and TERT mRNA or protein in NB cell lines and the large cohort of NB patient dataset.
Our GSEA analysis results revealed that TERT is also overexpressed in the subgroup of NB with MYCN amplification as compared to that without overexpressed MYCN and TERT (Fig. 1D). We found that the molecular pathways in the TERT overexpression subgroup overlapped significantly with those in the MYCN amplification subgroup (Fig. 1, F and G). We therefore speculated that the TERT overexpression subgroup and the MYCN amplification subgroup could be therapeutically targeted with similar approaches such as BET inhibition (37, 38, 41). However, the clinical efficacy of BET inhibitors could be limited by innate or acquired resistance through an aberrant Wnt signaling pathway (42). Our GSEA transcriptome analysis also revealed upregulation of the Wnt/β-catenin signaling pathway in the TERT overexpression subgroup, potentially limiting the effect of BET inhibition (43, 44). We addressed this by combining AZD5153 with dinaciclib and found the combination treatment to be more effective than either alone in inhibiting growth of primary tumor cells with TERT gene rearrangement or MYCN amplification and growth of xenograft tumor established from NB cell lines. We postulated that the additive to synergistic effects of BET inhibitors and dinaciclib were driven by non-overlapping ability to suppress the transcriptome of various genes in TERT overexpressing NB cells. BET inhibitors preferentially target SE-driven transcription whereas dinaciclib targets the promoter proximal transcription of the aberrantly upregulated genes in the E2Fs or β-catenin pathways. Concomitant inhibition of the transcription or activities of other Cdks such as Cdk1, Cdk2, and Cdk5 (14) by dinaciclib may contribute to dinaciclib-mediated transcription inhibition and the observed synergy with BET inhibitors (proposed model in Fig. 7H).
Currently, Next Generation Sequencing has not been used to define the subtype of NB with TERT rearrangement in a clinical setting. Although feasible to integrate whole genome sequencing and whole exome sequencing of TERT into clinical decision-making, limitations such as high cost, time consuming workflow, and the need for advanced bioinformatics exist. There is a lack of biomarkers for the diagnosis, prognosis, or treatment evaluation of NB patients with TERT gene rearrangement. Considering the distinct TERT mRNA expression observed from analyzing the large NB patient cohort and our RT-qPCR validation with primary NB patient samples, we propose that quantifying the level of TERT and MYCN overexpression by RT-qPCR could serve as screening biomarkers. Further studies with a larger sample size are warranted to validate this potential application.
Supplementary Material
Statement of Significance.
Epigenetically co-targeting Brd4 and Cdks suppresses human neuroblastoma with TERT overexpression by inhibiting the TERT-associated gene expression networks.
Acknowledgments
The authors thank the Stanford Division of Hematology/Oncology faculty and fellows, the Stanford Cancer Institute pediatric clinical research staff, and Lucile Packard Children’s Hospital Stanford for their support in banking patient tissues used in this study. We thank Prof. Julien Sage for his insightful comments for the study and manuscript. This work was supported by the National Institutes of Health grant R01 NS094218 (Chiu).
Footnotes
The authors declare no potential conflicts of interest.
References
- 1.Matthay KK, Maris JM, Schleiermacher G, Nakagawara A, Mackall CL, Diller L, et al. Neuroblastoma. Nat Rev Dis Primers 2016, 2:16078. [DOI] [PubMed] [Google Scholar]
- 2.Trigg RM, Turner SD. ALK in Neuroblastoma: Biological and Therapeutic Implications. Cancers (Basel) 2018;10(4): E113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peifer M, Hertwig F, Roels F, Dreidax D, Gartlgruber M, Menon R, et al. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature 2015, 526(7575):700–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valentijn LJ, Koster J, Zwijnenburg DA, Hasselt NE, van Sluis P, Volckmann R, et al. TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat Genet 2015, 47(12):1411–1414. [DOI] [PubMed] [Google Scholar]
- 5.Sengupta S, George RE. Super-Enhancer-Driven Transcriptional Dependencies in Cancer. Trends Cancer 2017;3(4):269–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013, 153(2):320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winter GE, Mayer A, Buckley DL, Erb MA, Roderick JE, Vittori S, et al. BET Bromodomain Proteins Function as Master Transcription Elongation Factors Independent of CDK9 Recruitment. Mol Cell 2017, 67(1):5–18 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. : Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005, 102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ackermann S, Cartolano M, Hero B, Welte A, Kahlert Y, Roderwieser A, et al. A mechanistic classification of clinical phenotypes in neuroblastoma. Science 2018, 362:1165–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc 2006;1(2):729–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang M, Zhu L, Garcia JS, Li MX, Gentles AJ, Mitchell BS. 2018. Brd4 regulates the expression of essential autophagy genes and Keap1 in AML cells. Oncotarget. 2018;9(14):11665–11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Afgan E1, Baker D1, Batut B2, van den Beek M3, Bouvier D4, Cech M4, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 2018, 46(W1):W537–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeki J, Taylor JS, Yavuz B, Coburn J, Ikegaki N, Kaplan DL, et al. Disseminated injection of vincristine-loaded silk gel improves the suppression of neuroblastoma tumor growth. Surgery 2018, 164(4):909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parry D, Guzi T, Shanahan F, Davis N, Prabhavalkar D, Wiswell D, et al. : Dinaciclib (SCH 727965), a novel and potent cyclin-dependent kinase inhibitor. Mol Cancer Ther 2010, 9(8):2344–2353. [DOI] [PubMed] [Google Scholar]
- 15.Rhyasen GW, Hattersley MM, Yao Y, Dulak A, Wang W, Petteruti P, et al. : AZD5153: A Novel Bivalent BET Bromodomain Inhibitor Highly Active against Hematologic Malignancies. Mol Cancer Ther 2016, 15(11):2563–2574. [DOI] [PubMed] [Google Scholar]
- 16.Su Z, Fang H, Hong H, Shi L, Zhang W, Zhang W, et al. : An investigation of biomarkers derived from legacy microarray data for their utility in the RNA-seq era. Genome Biol 2014, 15(12):523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu KJ, Grandori C, Amacker M, Simon-Vermot N, Polack A, Lingner J, et al. Direct activation of TERT transcription by c-MYC. Nat Genet 1999, 21(2):220–224. [DOI] [PubMed] [Google Scholar]
- 18.Lindner S, Bachmann HS, Odersky A, Schaefers S, Klein-Hitpass L, Hero B, et al. Absence of telomerase reverse transcriptase promoter mutations in neuroblastoma. Biomed Rep 2015, 3(4):443–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rickman DS, Schulte JH, Eilers M. The Expanding World of N-MYC-Driven Tumors. Cancer Discov 2018, 8(2):150–163. [DOI] [PubMed] [Google Scholar]
- 20.Roe JS, Mercan F, Rivera K, Pappin DJ, Vakoc CR. BET Bromodomain Inhibition Suppresses the Function of Hematopoietic Transcription Factors in Acute Myeloid Leukemia. Mol Cell 2015;58(6):1028–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Z, Wang Z, Pang JC, Yu Y, Bieerkehazhi S, Lu J, et al. Multiple CDK inhibitor dinaciclib suppresses neuroblastoma growth via inhibiting CDK2 and CDK9 activity. Sci Rep 2016, 6:29090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lv Y, Xiao J, Liu J, Xing F. E2F8 is a Potential Therapeutic Target for Hepatocellular Carcinoma. J Cancer 2017;8(7):1205–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smalley MJ, Signoret N, Robertson D, Tilley A, Hann A, Ewan K, et al. Dishevelled (Dvl-2) activates canonical Wnt signalling in the absence of cytoplasmic puncta. J Cell Sci 2005, 118(Pt 22):5279–5289. [DOI] [PubMed] [Google Scholar]
- 24.Jiang Y, Saavedra HI, Holloway MP, Leone G, Altura RA. Aberrant regulation of survivin by the RB/E2F family of proteins. J Biol Chem 2004;279(39):40511–40520. [DOI] [PubMed] [Google Scholar]
- 25.Eckerle I, Muth D, Batzler J, Henrich KO, Lutz W, Fischer M, et al. Regulation of BIRC5 and its isoform BIRC5–2B in neuroblastoma. Cancer Lett 2009, 285(1):99–107. [DOI] [PubMed] [Google Scholar]
- 26.Ma H, Nguyen C, Lee KS, Kahn M. Differential roles for the coactivators CBP and p300 on TCF/beta-catenin-mediated survivin gene expression. Oncogene 2005;24(22):3619–3631. [DOI] [PubMed] [Google Scholar]
- 27.Park SA, Platt J, Lee JW, Lopez-Giraldez F, Herbst RS, Koo JS. E2F8 as a Novel Therapeutic Target for Lung Cancer. J Natl Cancer Inst 2015;107(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sidhu H, Capalash N. UHRF1: The key regulator of epigenetics and molecular target for cancer therapeutics. Tumour Biol 2017;39(2):1010428317692205. [DOI] [PubMed] [Google Scholar]
- 29.Ohtani K, DeGregori J, Nevins JR. Regulation of the cyclin E gene by transcription factor E2F1. Proc Natl Acad Sci U S A 1995;92(26):12146–12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka M, Ueda A, Kanamori H, Ideguchi H, Yang J, Kitajima S, et al. Cell-cycle-dependent regulation of human aurora A transcription is mediated by periodic repression of E4TF1. J Biol Chem 2002, 277(12):10719–10726. [DOI] [PubMed] [Google Scholar]
- 31.Dutta-Simmons J, Zhang Y, Gorgun G, Gatt M, Mani M, Hideshima T, et al. : Aurora kinase A is a target of Wnt/beta-catenin involved in multiple myeloma disease progression. Blood 2009, 114(13):2699–2708. [DOI] [PubMed] [Google Scholar]
- 32.Otto T, Horn S, Brockmann M, Eilers U, Schuttrumpf L, Popov N, et al. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer cell 2009, 15(1):67–78. [DOI] [PubMed] [Google Scholar]
- 33.den Hollander J, Rimpi S, Doherty JR, Rudelius M, Buck A, Hoellein A, et al. : Aurora kinases A and B are up-regulated by Myc and are essential for maintenance of the malignant state. Blood 2010, 116(9):1498–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, et al. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev 2002, 16(2):245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu L, Chen Y, Mayakonda A, Koh L, Chong YK, Buckley DL, et al. : Targetable BET proteins- and E2F1-dependent transcriptional program maintains the malignancy of glioblastoma. Proc Natl Acad Sci U S A 2018, 115(22):E5086–E5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin HY. Targeting Super-Enhancers for Disease Treatment and Diagnosis. Mol Cells 2018;41(6):506–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puissant A, Frumm SM, Alexe G, Bassil CF, Qi J, Chanthery YH, et al. : Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov 2013, 3(3):308–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henssen A, Althoff K, Odersky A, Beckers A, Koche R, Speleman F, et al. : Targeting MYCN-Driven Transcription By BET-Bromodomain Inhibition. Clin Cancer Res 2016, 22(10):2470–2481. [DOI] [PubMed] [Google Scholar]
- 39.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. : BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011, 146(6):904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin MP, Olesen SH, Georg GI, and Schonbrunn E. Cyclin-dependent kinase inhibitor dinaciclib interacts with the acetyl-lysine recognition site of bromodomains. ACS Chem Biol 2013, 8:2360–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Felgenhauer J, Tomino L, Selich-Anderson J, Bopp E, Shah N. Dual BRD4 and AURKA Inhibition Is Synergistic against MYCN-Amplified and Nonamplified Neuroblastoma. Neoplasia 2018;20(10):965–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fong CY, Gilan O, Lam EY, Rubin AF, Ftouni S, Tyler D, et al. BET inhibitor resistance emerges from leukaemia stem cells. Nature 2015, 525(7570):538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature 2014, 510(7504):278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fu LL, Tian M, Li X, Li JJ, Huang J, Ouyang L, et al. Inhibition of BET bromodomains as a therapeutic strategy for cancer drug discovery. Oncotarget 2015, 6(8):5501–5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.