Abstract
Deregulated oncogenic signaling linked to PI3K/AKT and mTORC1 pathway activation is a hallmark of human T cell acute leukemia (T-ALL) pathogenesis and contributes to leukemic cell resistance and adverse prognosis. Notably, although the multi-agent chemotherapy of leukemia leads to a high rate of complete remission, options for salvage therapy for relapsed/refractory disease are limited due to the serious side effects of augmenting cytotoxic chemotherapy. We report that ablation of HSF1, a key transcriptional regulator of the chaperone response and cellular bioenergetics, from mouse T-ALL tumors driven by PTEN loss or human T-ALL cell lines, has significant therapeutic effects in reducing tumor burden and sensitizing malignant cell death. From a mechanistic perspective, the enhanced sensitivity of T-ALLs to HSF1 depletion resides in the reduced MAPK-ERK signaling and metabolic and ATP-producing capacity of malignant cells lacking HSF1 activity. Impaired mitochondrial ATP production and decreased intracellular amino acid (AA) content in HSF1-deficient T-ALL cells trigger an energy-saving adaptive response featured by attenuation of the mTORC1 activity, which is co-regulated by ATP, and its downstream target proteins (p70S6K and 4E-BP). This leads to protein translation attenuation that diminishes oncogenic signals and malignant cell growth. Collectively, these metabolic alterations in the absence of HSF1 activity reveal cancer cell liabilities and have a profound negative impact on T-ALL progression.
Introduction
T cell acute lymphocytic leukemia (T-ALL) is an aggressive hematological cancer arising from the malignant transformation of lymphoid progenitors primed for differentiation in the thymus represents approximately 15% of pediatric and 25% of adult ALL cases (1). Current multi-agent aggressive chemotherapy results in T-ALL cure rates close to 80% in pediatric patients and 60% in adults. Despite this therapeutic advantage, a significant fraction of T-ALL patients relapse and eventually experience refractory leukemia with poor outcome (2). Therefore, there is a need for greater understanding of the mechanisms of leukemic transformation and particularly the role of signaling networks enhancing cancer cell plasticity and adaptation, as well as of the targeted novel pathways to which tumor cells are specifically addicted.
Leukemic T cell transformation is commonly caused by ectopic expression of oncogenic signals involving chromosomal translocations of T cell receptor (TCR) genes with transcription factor genes such as MYC, TAL1/2, LMO1/2, TLX1/3, HOXA9/10, NKX2–1 or MEF2C (3, 4). Besides aberrant transcription factor expression, primary T-ALL harbors mutations that deregulate the NOTCH1, PTEN/PI3K or RAS/PI3K signaling pathways (4–6)). The vast majority of T-ALL patients (>50%) present activating mutations in the Notch1 receptor gene and inactivation of Cdkn2a (4, 5, 7). Silencing mutations in the Fbxw7 gene, which encodes E3-ubiquitin ligase, can extend activation of NOTCH signaling and contributes to T cell transformation (8). Notably, approximately 20% of T-ALL cases have two co-existing genetic lesions that activate NOTCH1 (7). T-ALL patients also display other genomic lesions including PTEN loss or RAS mutations that preferentially promote the outgrowth of clones escaping therapy with NOTCH1 or PI3K and MEK inhibitors (9–11). In particular, loss-of-function or deletion of the Pten gene, which occurs in 19% of pediatric and 11% of adult T-ALLs is a dominant oncogenic driver of PI3K-AKT-mTOR signaling that participates in the regulation of ribosome biogenesis and translation. Of note, targeting translation, for example, by mTORC1 inhibition is sufficient to extend survival in mouse models of Pten-loss-induced T-ALL, and inhibition of eukaryotic translation initiation factor 4E (eIF4E) or eIF2α-dependent protein translation have been demonstrated to have a broader anti-ALL effect (12). Finally, metabolic reprogramming caused by PTEN loss plays a key role in resistance to anti-NOTCH therapies in T-ALL (13), which provides the rationale for therapeutic targeting of cell metabolism in T-ALL.
The traditional cancer model posits that cancer initiation and progression are critically dependent on activation of adaptive and protective transcriptional networks that alleviate the immediate and chronic consequences of cell stress associated with cellular transformation. By facilitating the reprogramming of metabolic and bioenergetics and protein synthesis activity, these stress response networks support the high biosynthetic demand of cancer cells exerting a positive impact on cancer progression. Accordingly, the established role of the evolutionarily conserved transcriptional stress network regulator HSF1 in driving malignant transformation, as well as its downstream mediators of the heat shock response and cellular bioenergetics, makes the reduction/inhibition of HSF1, in level or activity, a potential therapeutic target for cancer. Indeed, we and others have shown that genetic ablation of HSF1 impacts tumor development in a broad range of cancer types (14–17). Previous reports by our group and others (18, 19) have also implicated HSF1 as a dominant orchestrator in acute leukemia and uncovered that HSF1 ablation interferes with growth of human T-ALL and delays or eradicates leukemia in mouse models of T-ALL driven by oncogenic NOTCH1 as well as TP53 deletion, while preserving normal hematopoiesis. Here, we investigated the potential dependency of T-ALLs induced by loss of PTEN on HSF1-dependent, non-oncogenic anabolic and energy-sensing adaptive pathways, and explored the therapeutic potential of targeting HSF1 in resistant T-ALL.
Materials and Methods
Animals.
The generation of mice containing loxp sites flanking exons 2 and 3 of the Hsf1 gene (Hsf1f/f) on a C56BL/6 genetic background has been described (20). Hsf1f/f mice were bred to mice containing loxp sites in exon 5 of the Pten (21), and then crossed with mice harboring a Cre recombinase transgene under control of the proximal Lck promoter B6.Cg-Tg (Lck-cre) (22) to yield Hsf1f/f-Ptenf/f-Lck-Cre+ (designated as Hsf1-PtenKO) mice, in which the floxed alleles are expected to be deleted exclusively in the T-cell lineage. Ptenf/f-Lck-cre+ mice with intact Hsf1 expression served as control. Ptenf/f and Lck-Cre+ strains were obtained from the Jackson Laboratory. We confirmed efficient inactivation of the Hsf1f/f and Ptenf/f alleles by Cre in T cells by PCR-based genotyping with DNA isolated from tails or thymus of the indicated genotypes.
In PCR analysis with thymic T-ALL DNA, the combination of primers p4: 5’-TGTCAGGCCTGGTT TTTAGG-3’ and p5: 5’ TGGCCACACTTCCATTTACA-3’ amplifies a fragment of 220 bp for the disrupted hsf1 allele and fragments of 1,130 bp and 979 bp for the hsf1f/f and WT alleles, respectively.
In the analysis performed for Pten on thymic T-ALL DNA, the primer combination of 5’-AATCTAGGGCCTCTTGTGCC-3’; and 5’-GCTTGATATCGAATTCCTGCAGC-3’ amplifies a fragment of 300 bp for the disrupted Pten alleles.
For detection of transgenic Cre, the combination of primers: 5’-GCGGTCTGGCAGTAAAAACTA TC-3’ and 5’-GTGAAACAGCATTGCTGTCACTT-3’ amplifies a fragment of 100 bp.
Animal care and experiments were performed in accordance with guidelines of the Institutional Animal Care and Use Committee and NIH. Unless otherwise indicated, all the analyses were performed on samples from tumor bearing-mice euthanized at a terminal leukemic stage.
Cell lines and drug treatment.
The following human T-ALL cell lines were used in this study: The γ-secretase inhibition (GSI)-sensitive CUTLL1 cells (generously provided by Dr. A Ferrando (Columbia University, NY) harbor a t(7;9)(q34;q34) translocation that induces the expression of a TCRβ-NOTCH1 fusion protein; display a hererozygous mutation that results in the substitution of arginine 248 for glutamine in the DNA-binding domain of the TP53 gene; and are positive for PTEN expression (23). CCRF-CEM (CCL119) cells are GSI-resistant, exhibit heterozygous mutations in Notch1 and TP53 alleles and harbor homozygous Pten mutations (9). The GSI-resistant Molt4 cell line with activating Notch1 that has been isolated from a relapsed T-ALL patient is deficient in Pten and displays heterozygous mutations in the TP53 and Nras genes (24, 25). The CCRF-HSB-2 T-ALL cell line expresses wild-type Notch1 (25). With the exception of CUTLL1, all cell lines were obtained from ATCC and were used in the first 3–5 passages. Cell lines were certified by the vendor.
To knock down Hsf1 in human T-ALL cell lines, we used the previously validated Doxycycline (Dox)-inducible short hairpin RNA (shRNA) retroviral system (16). T-ALL cells were infected with retroviruses expressing scramble (5’-TAAGGCTATGAAGAGATAC-3’) or Hsf1-specific (5’-GTGGACTCCAACCTGGATA-3’) shRNAs. For inducible shRNA expression transduced cells were cultured in medium containing Dox (500ng/ml), which was changed every other day.
Doxycycline was purchased from Sigma-Aldrich, and compounds KRIBB11 (Hy-100872) and Rocaglamide (Hy-19356) were obtained from MedChemExpress. For drug toxicity assays, cells were treated with different drug concentrations for 4 days, and cell viability was assessed by trypan blue dye exclusion.
Molecular T cell clonality analysis.
PCR-based T cell clonality assays detecting genomic TCRβ rearrangements were performed as described (26). Briefly, thymus genomic DNA (100 ng) prepared from T-ALL-bearing mice at different concentrations was used as template for PCR amplification. Primers were:
Dβ1-F, 5’-TTATCTGGTGGTTTCTTCCAGC-3’; Jβ1.6-R, 5’-GGTAGAAAGGTAGAGGGTTCCAGA-3’;
Dβ2ext-F, 5’-GCACCTGTGGGGAAGAAACT-3’; Jβ2.6ext-R, 5’-TGAGAGCTGTCTCCTACTATCGATT-3’; and Dβ2.1inter-F, 5’-GTATCACGATGTAACATTGTG; Jβ2.7int-R, 5’-GGAAGCGAGAGATGTAATC. Notably, for Dβ2 to Jβ2 rearrangement analysis, 0.5μl of the first amplification (Dβ2ext-β2.6ext) was subjected to a second PCR with primers Dβ2.1inter and Jβ2.7int.
Western blotting (WB) analysis:
Cells or tissues were homogenized in RIPA buffer with protease and phosphatase inhibitors (Roche Diagnostics, IN). Thirty micrograms of protein resolved by SDS-PAGE was immunoblotted as previously described (15). Membranes were probed with the following antibodies: α-tubulin (2144), β-actin (47778), AMPK (2603), p-AMPK (T172; 2535), HSF1 (4356), p-AKT (S473; 4060), p-AKT (T308; 2965), AKT (9272), p-mTOR (S2448; 2971), mTOR (2972), p-p70SK (T389; 9206), p70SK (9202), p-S6R (S240/244; 5364), S6R (2217), p-4E-BP1 (T37/46; 2855), p-CAD (S1859; 12662), p-GSK3 (S9/21; 9336), 4E-BP1(S65; 9451), Pten (9559), p-PDK1 (S241; 3438), p-ERK (4370), 4E-BP1(T70; 9455), and 4E-BP1 (9452) (Cell Signaling Technology), α-tubulin (5286), HSF1 (13516) and ERK (sc-94) (Santa Cruz Biotech) and c-Myc (790–4628) (Hoffmann La Roche).
Histology, immunohistochemistry (IHC) and immunofluorescence (IF) microscopy.
For these analyses, we followed the procedures described previously (20). Briefly, tissues were fixed in 4% paraformaldehyde (PFA) in PBS and embedded in paraffin. Hematoxylin and eosin (H&E) staining was performed on paraffin sections (7μm). For IHC, after antigen retrieval the sections were blocked in 3% BSA, incubated with a primary antibody against HSF1 (rabbit 4356; Cell Signaling Technology) or Ki67 (rabbit RM-9106; Thermo Scientific) overnight at 4°C and Alexa Fluor 555 or 594-conjugated secondary antibody for 1 hr at 25°C, and then examined under an Axio10 imager fluorescence microscope (Carl Zeiss, Germany). Nuclei were stained with DAPI (4’,6’-diamidino-2-phenylindole) reagent.
For IF staining, cells plated on a coated cover glass for 16 hr were fixed in 4% PFA for 10 min at 25°C, permeabilized with 0.1% Triton X-100 in PBS (PBST) for 5 min at 4°C, blocked in 5% BSA in PBST for 1 hr, and incubated overnight at 4°C with primary antibodies (anti-mTOR [rabbit 2983; Cell Signaling] or anti-LAMP2 [rat 81729; Santa Cruz]). The cover glasses were then rinsed with PBS and incubated with fluorescence-conjugated secondary antibodies (Alexa Fluor 488 [goat-anti-rabbit IgG; A11008] and 594 [goat-anti-rat; A11007] from Invitrogen, in PBS at 25°C for 1 hr. The slides were stained with DAPI in PBS for nuclear counterstaining and mounted with Vectashield H-1200 (Vectar). Image acquisition was performed on an AxioVision Imager fluorescent microscope (Zeiss) supported with an AxioCam MRC or HRC camera (Zeiss) using a 40x objective lens (numerical aperture [NA], 0.75). AxioVision REL 4.8 software was used for data analysis.
Apoptosis assays and flow cytometry (FACS) analysis:
Cell apoptosis was measured using Annexin V labelling and 7-AAD (420404; BioLegend) or TUNEL staining performed with ApoptoTag Red in situ apoptosis detection kit (S7165; Chemicon/Millipore). Nuclei were counterstained with DAPI reagent.
To determine the origin of T-ALL cells, we performed FACS-based immune-phenotypic analysis by staining of cells from thymus or peripheral lymphatic organs for markers characteristic for T cell development. Cells were stained with conjugated antibodies (CD4, CD8, CD3, TCRβ, CD25 and CD44; Bioledgend) for 30 min at 4 °C and washed twice with FACS buffer (PBS, 2 % FCS, 1 mM EDTA). Multicolor FACS analysis was performed with FACS Canto (Becton–Dickinson Pharmingen) and data analyzed with FlowJo software. Notably, early uncommitted thymocytes (CD4 and CD8-negative (DN) cells) can be subdivided into four stages of differentiation based on the expression of CD44 and CD25 (DN1, CD44+CD25−; DN2, CD44+CD25+; DN3, CD44–CD25+; and DN4, CD44–CD25–).
Measurement of ATP and protein synthesis:
Cellular ATP content in cells was measured using a bioluminescence ENLITENR ATP determination kit (FF2000; Promega, Madison WI). The amount was normalized to the protein content, which was determined using a protein assay (Bio-Rad Laboratories), and is presented as micromoles per gram protein. Protein synthesis was quantified using an 3H-leucine metabolic labeling assay, previously described (20).
Quantification of reactive oxygen species (ROS), mitochondrial membrane potential (ΔΨm) and glucose uptake assay:
For glucose uptake, 5×105 cells were incubated in glucose and serum-free medium for 2 hours. Cells were then incubated in medium containing 5% dialyzed FCS and 1μCi/ml of 3H-2-DG (MP Biomedicals-Radiochemical) for 10 min., rinsed three times with PBS and solubilized in 0.5 ml of 1% SDS. Radioactive counts were determined by a scintillation counter and scintillation reads were normalized to the protein concentration. For ΔΨm and cellular ROS measurements, cells were stained with 50mM TMRE (Molecular Probes) or 5μM dihydroethidium (DHE) for 30 min. at 37°C and analyzed by FACS.
Mitochondrial respiratory assays and metabolite determination:
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured using a XF96 Extracellular Flux Analyzer (Agilent Technology) as described previously (20). Detection of metabolites was performed using gas chromatography-mass spectrometry (GC-MS). Briefly, metabolites were extracted from T-ALL tumors or T-ALL cell lines and GC-MS analysis was carried out with a Waters GCT Premier mass spectrometer fitted with an Agilent 6890 gas chromatograph and a Gerstel MPS2 auto-sampler at the Metabolomics Core Facility of the University of Utah (Salt Lake City, UT).
Statistics:
For tumor survival studies, at least 25–30 female and male mice per group were used. Most other assays were performed with tissues from 4–5 male and female mice. Data are presented as means ± standard deviation (SD) of at least three independent experiments. Statistical significance between experimental groups was assessed using Student’s t test or a log-rank test for survival differences, and a p value of 0.05 was considered significant. For data sets with significant variant differences confirmed by Bartlett’s test for equality of variance, statistical significance was evaluated by Welch’s test.
Results
HSF1 is required for PTEN-loss-induced T-ALL development and maintenance.
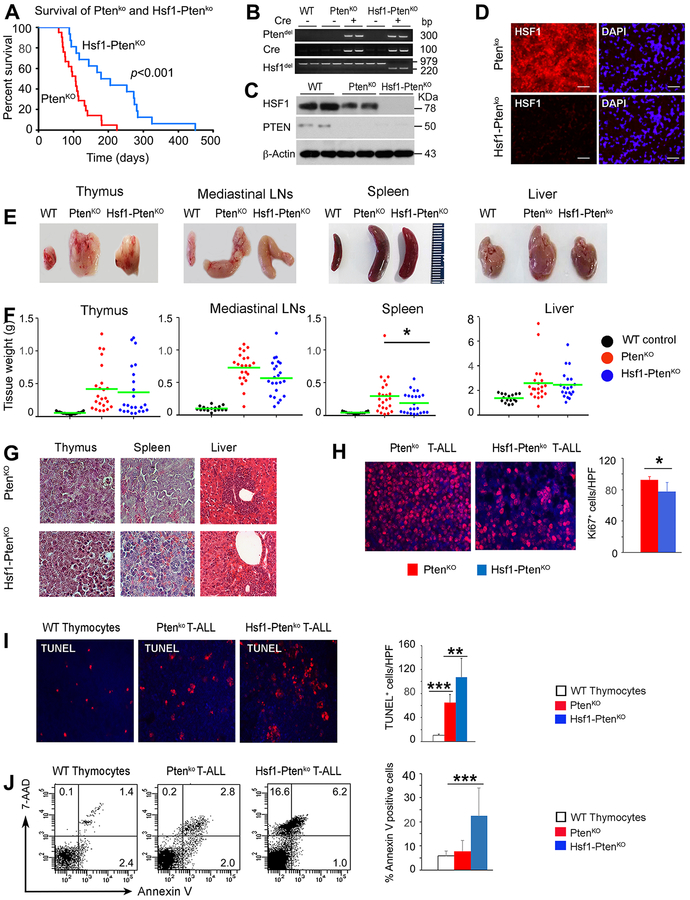
The significant upregulation and expression of HSF1 in neoplastic cells directs a non-oncogenic transcription program that is critically involved in promotion of diverse cancer types (14–16), including primary T-ALLs driven by deregulated NOTCH1 or TAL signaling (19). Additional genetic lesions, particularly associated with PTEN loss, seem to be a critical factor for refractory T-ALL development and leukemic disease progression. We thus evaluated the effects of Hsf1 inactivation on T-ALL development in a mouse model with T cell lineage-specific conditional loss of Pten or both Pten and Hsf1 via expression of the Cre recombinase under control of the lck proximal promoter. Notably, the lck promoter is active during T cell development in the thymus, starting from the double negative (DN) 2 stage. T-cell specific Pten loss induces an aggressive disseminating T-ALL disease, originating in the thymus, with 100% penetrance and lethality within 3–5 months (27). PtenKO or Hsf1-PtenKO mice before the onset of T-ALL development did not have abnormalities in distribution of various T cell populations in the thymus or peripheral lymphoid tissues (data not shown). However, particularly intriguing in our analysis was the markedly and statistically significant delay in T-ALL fatal disease progression in Hsf1-PtenKO mice compared to control PtenKO mice, indicating a potent anti-leukemic effect of HSF1 ablation (Fig. 1A). Efficient deletion of Pten and Hsf1 alleles in thymus and tumors of PtenKO or Hsf1-PtenKO mice was detected by genomic PCR, WB analysis and IHC staining (Fig. 1B, C and D). A survey of the induction and dissemination of leukemic cells in different organs performed at pre-terminal disease stage revealed that Hsf1-PtenKO mice had tumors with a significantly reduced prevalence in the thymus (17 of 19 PtenKO mice, 90% versus 12 of 22 Hsf1-PtenKO mice, 55%), lymph nodes (LNs) (17/19 PtenKO mice, 90% versus 13/22 Hsf1-PtenKO mice, 59%), and liver (10/19 PtenKO mice, 53% versus 4/22 Hsf1-PtenKO mice, 18%) but exhibited similar metastatic potential in the spleen (17/19 PtenKO mice, 90% versus 18/22 Hsf1-PtenKO mice, 82%). In addition, we observed a reduced size and weight of organs, including thymus, spleen, LNs and liver, in Hsf1-PtenKO mice, although this did not reach significance (Fig. 1E and F). As expected, tumor-bearing mice displayed a significant increase in lymphatic tissue (thymus, spleen and LNs) cellularity as compared to age-matched C57BL/6 wild-type control mice, but no significant difference was observed between the genotypes (data not shown). Histological analysis confirmed complete destruction of the cortical-medullary structure in the thymus and follicular structure in the spleen by lymphoblastic cells, found in both genotypes (Fig. 1G). Liver-infiltrating lymphoblastic cells were apparent to a similar degree in both mouse strains and formed large clusters in the portal areas and were localized as single cells scattered throughout the sinusoidal areas (Fig. 1G). Notably, while some PtenKO mice with metastatic lesions displayed large amounts of lipid deposition in the liver, Hsf1-PtenKO mice were free of such metabolic perturbations despite extensive lymphoblastic cell infiltration (data not shown). PTEN-loss-induced tumors in Hsf1-PtenKO mice exhibited a sustained slight decrease in cellular proliferation (Ki67 staining), but a marked increase in apoptosis (TUNEL staining) (Fig. 1H and I).
Figure 1. HSF1 ablation attenuates PTEN-loss-induced T-ALL development and progression.
(A) Kaplan-Meyer plot of tumor-free survival curves for PtenKO (n=30) and Hsf1-PtenKO (n=25) male and female mice. p value was derived using Log-Rank test.
(B) PCR-based verification of Hsf1 and/or Pten allele deletion or Cre allele expression performed with DNA isolated from thymus of WT control mice or T-ALLs of the indicated genotypes.
(C) Efficient inactivation of HSF1 and/or PTEN is demonstrated by Western blot analysis in T-ALL cell extracts from PtenKO or Hsf1-PtenKO mice. Samples from thymus of normal WT mice were used as a control. β-actin was used as loading control.
(D) Representative IHC of HSF1 in thymic T-ALL sections from PtenKO or Hsf1- PtenKO mice. DAPI (blue) represents nuclear staining. Bars=20μm (n=4 mice).
(E) Representative macroscopic images of thymus, spleen, MLN and liver from indicated genotypes.
(F) Quantification of the organ weight in T-ALL-bearing animals of the indicated genotype. Normal WT mice without lymphomas were included as control (for panel F, n=16–23 mice/group).
(G) Representative histological analysis of thymus, spleen and liver from PtenKO or Hsf1- PtenKO mice. Mag. 40x. (n=5 mice/group).
(H-I) Ki67 and TUNEL staining of thymic T-ALL sections from PtenKO or Hsf1- PtenKO mice. Thymus of normal WT mice was used as a control. DAPI (blue) represents nuclear staining. Bars=20μm. Quantification of Ki67+ proliferating cells or TUNEL+ apoptotic cells per high-power field is presented (right panel) (n=4–5 mice per group).
(J) Apoptosis or necrosis monitored by FACS of thymic T-ALL cells from PtenKO or Hsf1- PtenKO mice stained for annexin V and 7-AAD. Thymocytes from WT mice were used as control. The percent of apoptotic (annexinVpos-7-AADneg/pos) and necrotic (annexinVneg-7-AADpos) cells is indicated in the quadrants. Quantification of apoptotic and necrotic cells is presented in the right panel (n=4 mice).
Analyses were performed at an advanced (preterminal) stage of T-ALL disease. For all panels, scale bars represent mean ± SD. Statistical significance is indicated (* p<0.05, ** p< 0.01, *** p< 0.001).
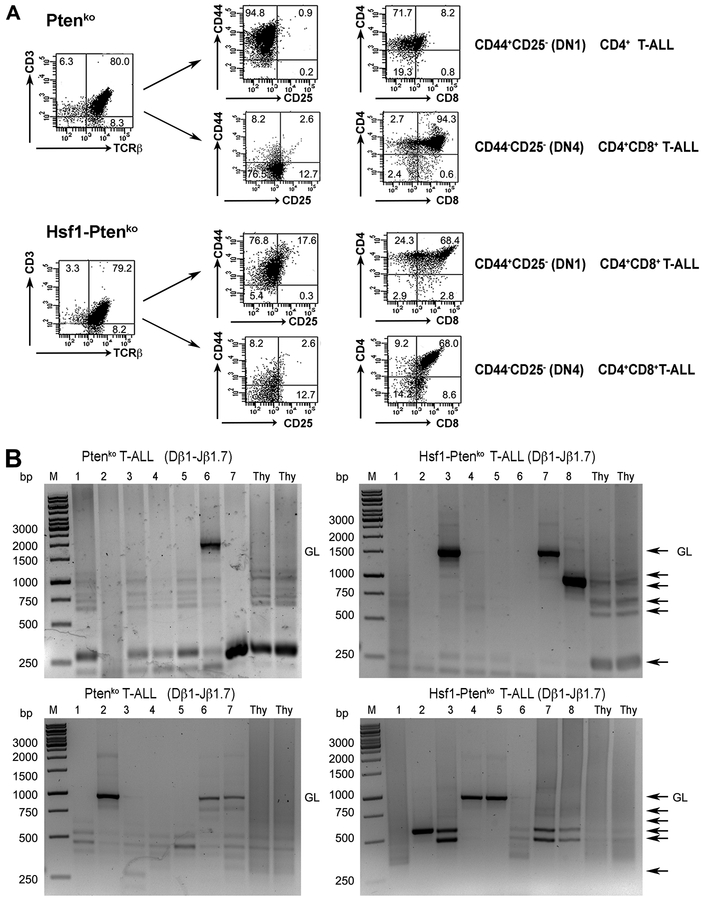
To determine the origin of neoplastic cells, we performed immuno-phenotypic analysis of lymphoma cells. Of the individual lymphomas analyzed in the thymus of PtenKO mice, all (21/21; 100%) were CD3+TCRβ+ and the vast majority (18/21; 85.7%) were CD4+CD8+ (Fig. 2A). Interestingly, a few of these lymphomas (3/21; 14.2%) exhibited aberrant expression of CD4 (CD4+CD8−), reminiscent of immature single positive (ISP) cells. Among these lymphomas, 12 of 21 (57.1%), were CD44+CD25−, 4 of 21 (19.1%) were CD25+CD44+ (corresponding to the DN2 stage); and 5 of 21 were CD25−CD44− (corresponding to the DN4 stage). A similar expression pattern was observed for lymphomas in the thymus of Hsf1-PtenKO mice with CD3+TCRβ+ (20/22; 90.9%), CD3+TCRβ− (2/22; 9.1%), CD4+CD8+ (20/22; 90.9%); and ISP CD4+CD8− (2/22; 9.1%) cells. The developmental stage was also comparable: 17 of 22 (77.3%) were CD44+CD25−; 2 of 22 (9.1%) were CD25+CD44+ (DN2 stage); and 3 of 22 (13.6%) were CD25−CD44− (DN4 stage). Representative expression profiles are displayed in Fig. 2A. We further determined the clonality of randomly selected thymus-derived TCRβ+ lymphomas for each genotype by examining genomic DNA for TCRβ rearrangement by PCR analysis. We detected a few predominant bands from individual T-ALLs from PtenKO or Hsf1-PtenKO mice (Fig. 2B), indicating that neoplastic cells in both genotypes could arise at least from a limited number of clones. Taken together, these results indicate an important contribution of HSF1 in promoting Pten-loss-induced T-ALL development and progression.
Figure 2. HSF1 is not required for T cell development and its genetic elimination does not affect the immune phenotype of Pten-loss-driven T-ALLs.
(A) Representative FACS-based immunophenotypic analysis of T-ALLs from PtenKO or Hsf1- PtenKO mice assayed at an advanced stage of disease. Initial gating was on T-ALL blasts based on their forward and side scatters. Surface markers analyzed on the leukemic blasts. DN, double negative. The percent of cell populations is indicated in the quadrants.
(B) Clonality assessment by genomic TCRβ rearrangement. T-ALL at an advanced stages of disease recovered from PtenKO or Hsf1- PtenKO mice were analyzed by semi-quantitative PCR analysis of the indicated Vβ1-to DJβ1.7 and Vβ2-to-DJβ2.7 rearrangements. DNA prepared from normal WT thymus (Thy) was included in the analysis. GL: germline, Thy: Thymus, M: molecular marker, black arrows: Dβ-Jβ rearrangements.
Suppression of mTORC1 and MAPK/ERK signaling by HSF1 ablation attenuates PTEN-loss-induced T-ALL progression.
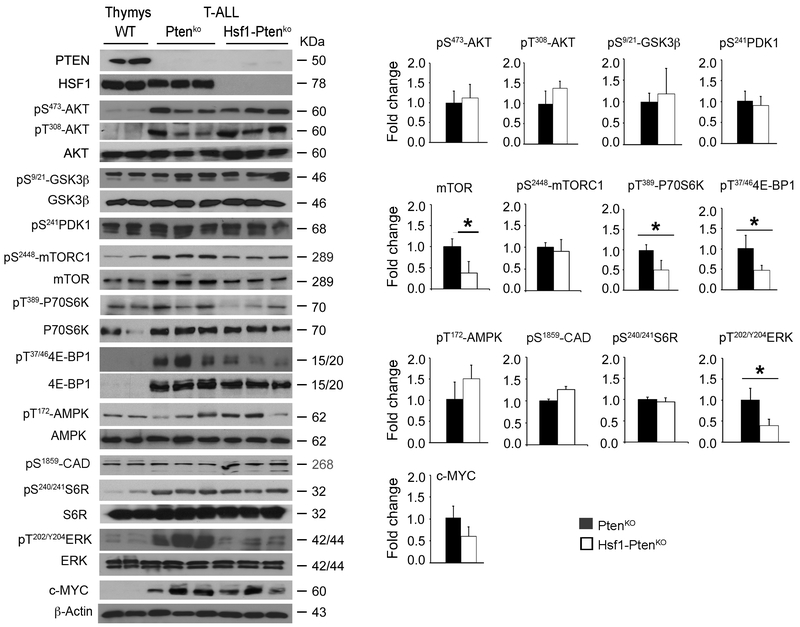
Key hallmarks of T-ALL adaptation and progression is the dysregulation and aberrant activation of PI3K-AKT-TORC1 signaling by genetic mutations associated with PTEN loss, which reprograms tumor cell mitochondrial biogenesis and anabolic metabolic and protein translation pathways (28, 29). Treatment of T-ALL with γ-secretase NOTCH1 signaling inhibitors or glucocorticoid dexamethasone often leads to drug resistance in patients, which can partly be attributed to PTEN loss and activation of PI3K-AKT-mTOR signaling (9, 30). Aberrant dysregulation of the RAS-MAPK-ERK signaling, occurring in about 50% of patients, is also a dominant oncogenic driver in aggressive drug resistance and poor prognosis of T-ALL (10). This prompted us to investigate the effects of HSF1 ablation on the PI3K-AKT and MAPK/ERK signaling pathways and its impact on cancer-anabolic-supportive pathways. In primary T-ALL, with intact or Hsf1 deletion, AKT and downstream target GSK3β activities, as assessed by their phosphorylation status (pS473-AKT, pT308-AKT, pS9/21-GSK3α/β), were markedly upregulated compared to normal control thymus; however, tumors of both genotypes showed similar amounts of increased activity levels (Fig. 3). Of note, PDK1, which is located directly downstream of PI3K, primes pT308-AKT, while mTORC2 primes pS473-AKT. Loss of the S6K1-mediated feedback loop resulting from mTORC1 inhibition also enhances PDK1-mediated pT308-AKT activation. HSF1 ablation did not cause significant reduction in PDK1 activity level assessed by its auto-phosphorylation at residue S241 in tumors. By a direct comparison of Hsf1-PtenKO and PtenKO tumors, however, we identified that the mTORC1 activity and its downstream signaling targets, including pT389-p70S6K and pT37/46-4E-BP that regulate mRNA translation and the rate of protein synthesis, were markedly decreased in Hsf1-PtenKO tumors. As expected, attenuation of p70S6K activity in Hsf1-PtenKO tumors induced an adaptive negative feedback loop that restricts mTORC1 phosphorylation (pS2448-mTORC1). No overt differences in activity of other p70S6K substrates, namely pS240/244-S6R, which stimulate CAP-dependent phosphorylation, and pS1859- CAD, which catalyzes the first step in pyrimidine synthesis (31), were observed between the genotypes. The activation state of mTORC1 is controlled directly by ATP levels (32) or indirectly by AMPK activation (33). We have reported that HSF1 loss in primary hepatocytes causes a marked cellular ATP level decline that sensitizes AMPK activation (15, 20). Although T-ALL displayed elevated AMPK activity (S172-AMPK), no significant differences were observed between the tumor genotypes. However, we found a significant reduction in the level of pT202/Y204-ERK1/2, which is a downstream target of receptor tyrosine kinase (RTK) signaling and activates mTORC1 via phosphorylation and functional inhibition of the tuberous sclerosis complex (TSC) (34). Hsf1-deficient T-ALL retained the ability to Pten-loss-evoked increased c-Myc level and activity, which synergizes with AKT signaling to promote glucose and glutamine metabolism and protein translation through activation of mTORC1 (35).
Figure 3. Suppression of MAPK/ERK signaling and mTORC1 activity and its downstream signaling molecules in Hsf1-PtenKO T-ALLs.
Representative WBs showing MAPK/ERK and PI3K/AKT/mTOR signaling protein expression in T-ALLs from PtenKO or Hsf1-PtenKO mice assayed at an advanced stage of disease. Thymocytes from WT were used as control. For quantification, the level of protein forms normalized to β-actin loading control was expressed as relative fold increase to the PtenKO level. Alternatively, the level of phosphorylated protein normalized to the total protein level is expressed as relative fold increase compared to the PtenKO level.
Scale bars represent mean ± SD. (n=4–6 mice per group). Statistical significance is indicated (* p<0.05).
Taken together, these data suggest that HSF1 promotes T-ALL cell growth via regulation of adaptive anabolism-supporting mTORC1 activity and its downstream signaling molecules p70S6K and 4E-BP, which are known to be involved in mitochondrial biogenesis and selective expression of mitochondrial proteins as well as other anabolic pathways required for cell survival (34, 36). A significant reduction in p-ERK1/2 level also suggests reduced RTK signaling, which is required for tumor cell proliferation.
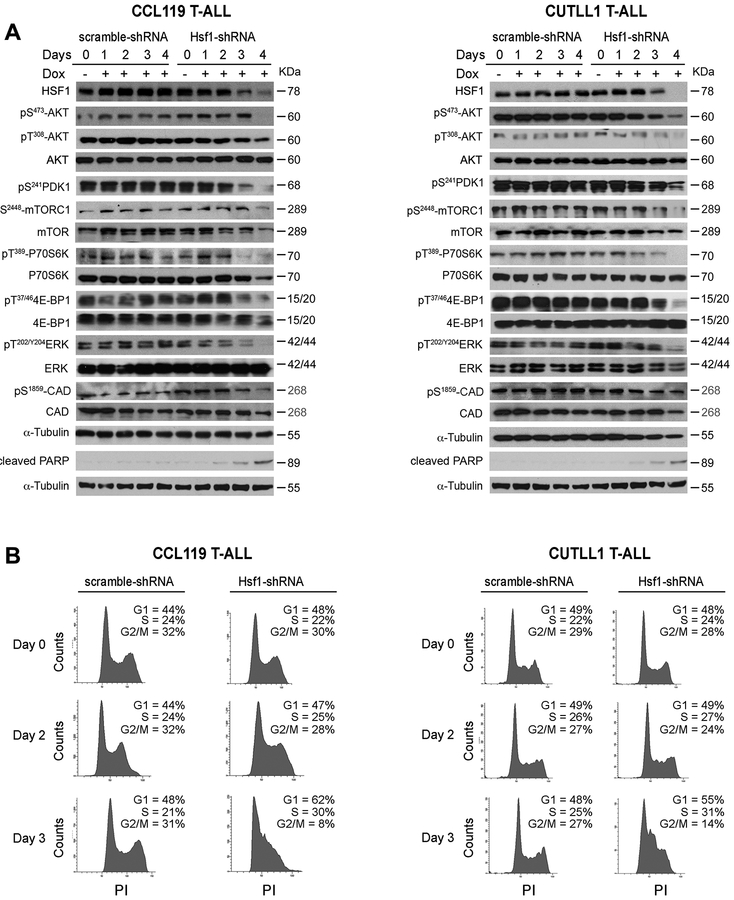
Hsf1 regulates the mTORC1 and MAPK/ERK signaling in human T-ALLs and its genetic or pharmacological inhibition impacts leukemic cell proliferation and survival.
To further investigate the role of HSF1 in human T-ALL survival, we monitored the levels of oncogenic PI3K-AKT-mTOR and MAPK-ERK signaling following knockdown of Hsf1 (using Dox-inducible stable shRNA) in CUTLL1 (intact PTEN expression) and CCL119 (harboring homozygous Pten mutations) cell lines. As a control for a potential Dox effect on metabolism and proliferation, experiments included control cells expressing Dox-inducible scramble shRNAs. Consistent with our mouse T-ALL analyses, we found that Hsf1 loss significantly reduced the mTORC1 and MAPK/ERK signaling and strongly affected leukemic cell survival (Fig. 4A and Fig. S1). Cell cycle analysis revealed a negative effect of Hsf1 knockdown on cell cycle progression (reduced fraction of leukemic cells in S and G2/M phase) and increased apoptotic response (Figs. 4B and Fig. S1). Consistent with our in vivo analyses and recent findings describing the effects of Hsf1 inhibition on T-ALL cell growth (19), we also confirmed that targeting HSF1 with pharmacological agents (KRIB11, Recoglamide) (37, 38) significantly affected human T-ALL cell viability, leading to increased apoptosis (Fig. S2).
Figure 4. Suppression of mTORC1 signaling following Hsf1 depletion in human T-ALL cell lines.
(A) Representative WBs showing MAPK/ERK and PI3K/AKT/mTOR signaling protein expression in human T-ALL cell lines (CCL119 and CUTLL1) following silencing of Hsf1 by doxycycline-inducible shRNA expression for the indicated times. Cells expressing scramble shRNA were used as control (n=3 experiments).
(B) Cell cycle analysis of CCL119 and CUTLL1 cell lines following silencing of Hsf1 as described in panel A. Cells were resuspended in propidium Iodide (PI) solution (100μg/ml ribonuclease A, 50μg/ml PI in PBS) and incubated for 30 min at room temperature and analyzed by FACS. The percentage of cells in G1, S, and G2/M cell-cycle phases are indicated (n=3 independent analyses).
Effects of Hsf1 loss on metabolic and bioenergetic pathways in T-ALLs.
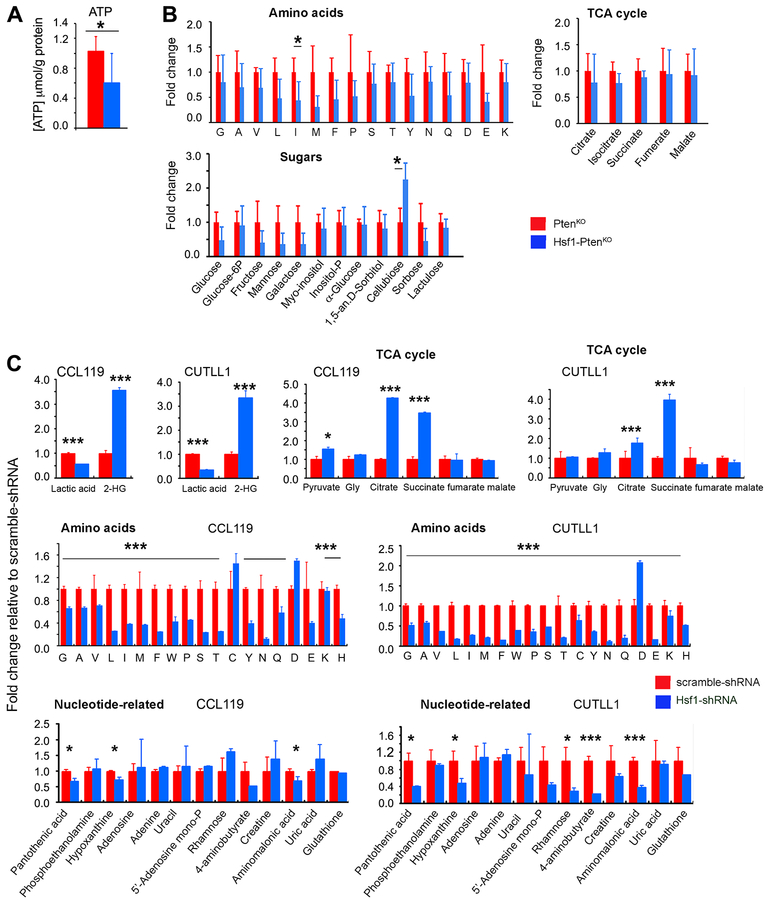
Based on previous reports, the upregulation and activation of the Hsf1 transcriptional program by a variety of stressors, including nutrient availability and malignant transformation, plays a critical role in monitoring the protein folding state by inducing heat shock proteins (HSPs) and optimizing the cellular metabolic bioenergetics and anabolic synthetic capacity (15, 20). In the context of physiological function and disease, Hsf1 loss triggers an energy-saving adaptive response associated with inhibition of energy consuming anabolic synthetic pathways (protein, lipid and glucose). To determine whether HSF1 loss can have a significant impact on metabolic homeostasis and bioenergetics pathways in T-ALL progression, we thus performed targeted metabolic and signaling analyses in tumors derived from Hsf1-PtenKO and control PtenKO mice. Loss of Hsf1 in T-ALL led to a significant reduction in ATP levels and affected the steady-state levels of several amino acids (AA) including alanine, leucine, isoleucine, tyrosine, and more significantly, methionine (Fig. 5, A–B). Notably, methionine is the substrate for the generation of cysteine and taurine, as well as for synthesis of the antioxidant glutathione and is required for the initiation of translation. We also found that HSF1 ablation caused a reduction in the intracellular level of glucose as well as other sugars, whereas levels of cellobiose, which can enzymatically convert to D-glucose, was significantly increased in Hsf1-PtenKO T-ALL tumors. This effect likely represents a compensatory response to reduced glucose levels. The potential impact of Hsf1 loss on lactic acid and metabolite levels associated with the tricarboxylic acid (TCA) cycle and fatty acids was not directly supported by our data.
Figure 5. Effects of HSF1 inactivation on metabolite levels in Pten-loss-induced T-ALL tumors or human T-ALL cell lines.
(A-B) ATP and steady-state levels of metabolites in T-ALLs from PtenKO or Hsf1-PtenKO mice assayed at an advanced stage of disease. For quantification, the values of each metabolite were expressed as relative fold changes to the mean value of PtenKO tumor samples that was arbitrarily set at 1 (n=4 mice per group).
(C) Levels of metabolites in CCL119 and CUTLL1 cell lines 3 days following silencing of Hsf1 by induced shRNA. The values of each metabolite were expressed as relative fold changes to the mean value of scramble shRNA-treated cells that was arbitrarily set at 1 (n=4 samples per group).
Scale bars represent mean ± SD. Statistical significance is indicated (* p<0.05, *** p< 0.001).
To validate these general findings, we then performed targeted metabolic analyses in human T-ALL cell lines (CCL119 and CUTLL1) stably expressing Dox-inducible Hsf1 shRNA. Analyses included control T-ALL cells expressing Dox-inducible scramble shRNA in the presence of Dox (Fig. 5C). The reduction of HSF1 protein levels in CCL119 and CUTLL1 cells led to reduced levels of lactate, whereas the levels of pyruvate either increased (CCL119 cells) or was not affected (CUTLL1). This effect, which was not evident in ex vivo analyzed tumors, resulted in a decrease in the lactate/pyruvate ratio in the Hsf1 knockdown cell lines, with potential metabolic consequences in the cellular redox status. Notably, lactate is generated from pyruvate reduction by the lactate dehydrogenase (LDH) system, and hsf1-deficient tumor cells have been shown to generate reduced levels of lactate (39). Inducible Hsf1 knockdown led also to a marked increase of intracellular levels of the TCA cycle metabolites, citrate and succinate, as well as 2-hydroxyglutarate (2-HG). Notably, 2-HG is a competitive inhibitor of α-ketoglutarate (α-KG)-dependent dioxygenases, and high levels of D-2-HG were detected in tumor cells harboring isocitrate dehydrogenase (IDH)1/2 mutations (40). On the other hand, production of the enantiomer L-2HG has been shown to be increased by hypoxia and other stress conditions in cells in the absence of IDH1/2 mutations (41). Furthermore, recent studies have suggested that perturbed mitochondrial and electron transport complex (ETC) function results in 2-HG accumulation along with an increase in succinate and citrate levels (42). The levels of other TCA cycle intermediates were not significantly affected. Consistent with our ex vivo tumor analyses, we detected a dramatic decrease in intracellular levels of several AA following Hsf1 knockdown.
Hsf1 ablation in human T-ALL reduces mitochondrial bioenergetic capacity and triggers an adaptive mTORC1 signaling attenuation that interferes with malignant cell progression.
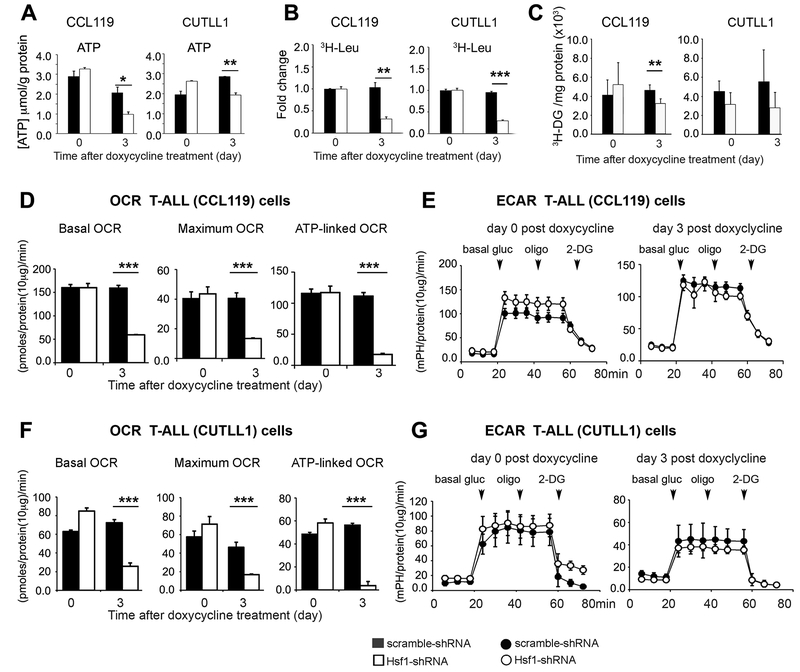
A decline in mTOR activity after Hsf1 knockdown in T-ALL may be expected, as HSF1 transcriptionally enhances mitochondrial ATP-producing capacity and therefore mTORC1 activity, which is strongly inhibited by depletion of ATP (20, 32). On the other hand, mTORC1 stimulates mitochondrial biogenesis through effects on mRNA translation and ribosome biogenesis, and therefore inhibition of mTORC1 signaling leads to mitochondrial impairment (29). We thus examined the mitochondrial metabolic function in human T-ALL cells at 3 days post Hsf1 knockdown induction, when a decline in HSF1 protein level is evident. Silencing Hsf1 substantially reduced ATP production and protein translation as measured by 3H-Leu labelling in CUTLL1 and CCL119 cells (Fig. 6, A and B). Remarkably, Hsf1 knockdown cells presented a low oxygen consumption rate (OCR), an indicator of mitochondrial oxidative phosphorylation (OXPHOS), while the extracellular acidification rate (ECAR) did not significantly differ compared to control cells expressing Dox-inducible scramble shRNA (Fig. 6, D–G). Notably, the knockdown of Hsf1 reduced glucose uptake in CCL119 cells, an effect that was not clearly evident in CUTLL1 cells (Fig. 6C). TMRE staining indicated that Hsf1 knockdown decreased the mitochondria membrane potential and this was associated by a moderately increase in the cytoplasmic reactive oxygen species (ROS) level, based on DHE staining (Fig. S3).
Figure 6. HSF1 knockdown in human T-ALL cell lines impairs mitochondrial respiratory capacity and reduces protein synthesis.
T-ALL cell lines (CCL119 and CUTLL1) without (day 0) or following silencing of Hsf1 by doxycycline-inducible shRNA expression for 3 days were assayed for (A) ATP content, (B) Protein synthesis rates, (C) Glucose uptake rates, and (D-G) OCR and ECAR profiles. Cells expressing scramble shRNA were used as control. Scale bars represent mean ± SD. (n=4–6 samples). For all panels, symbols or bars represent cells expressing Hsf1 shRNA (open) or scramble shRNA (filled). Statistical significance is indicated (* p<0.05, ** p< 0.01, *** p< 0.001).
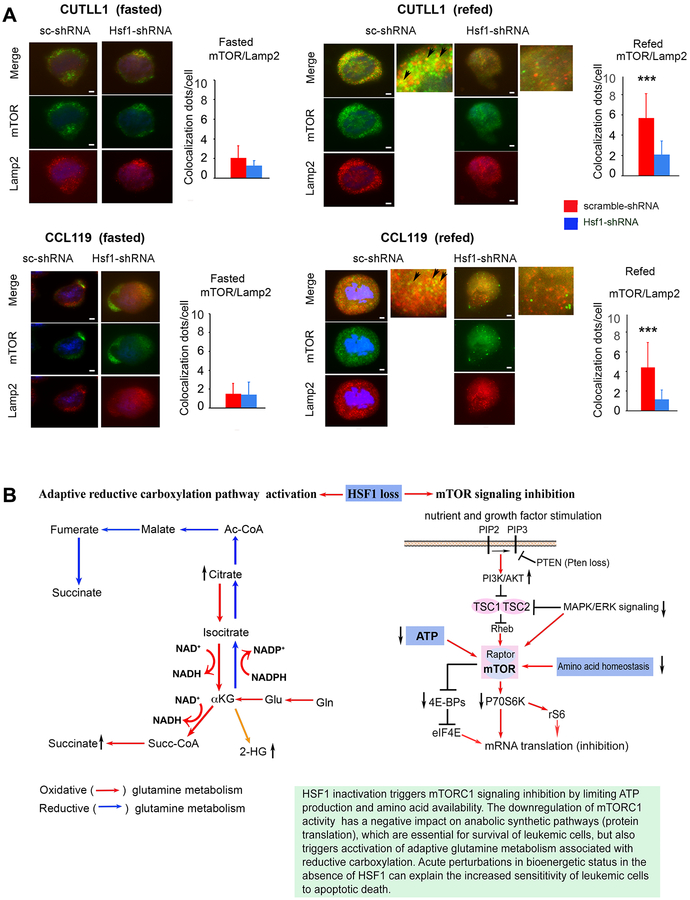
The findings that Hsf1 loss in T-ALL cells did not further augment the AMPK activity level (Fig. 3), suggest that the attenuation of mTORC1 signaling by Hsf1 loss is unlikely to be primarily mediated through an AMPK-dependent mechanism. Another possible explanation could be that Hsf1 inactivation in T-ALL suppresses levels of several intracellular AAs that signal mTORC1 activation (34, 36). Biochemical and structural analyses established that upon AA availability mTORC1 is active due to its lysosomal location mediated by Rag GTPases. We thus next sought to mechanistically link Hsf1 action to AA signals and mTORC1 activation in lysosomes. Consistent with our hypothesis, the intracellular location of endogenous mTORC1, as revealed in immunofluorescence assays, was strikingly different in T-ALL with intact versus knockdown Hsf1. In cells deprived of nutrients including AAs, mTORC1 remained diffusely cytosplasmic, in a pattern that is not concentrated on lysosomes/late endosomes (marked by LAMP2) (Fig. 7A). In starved tumor cells expressing HSF1 that are then briefly re-stimulated with AAs (complete medium), a large fraction of mTORC1 translocated to the lysosomal compartment (Fig. 7A). In contrast, Hsf1 knockdown markedly inhibited AA-induced mTORC1 lysosomal localization. Taken together, these results show that in spite of the increased nutrient-stimulated AKT-mediated signals in human T-ALL, a marked decrease in the activity of mTOR in the absence of Hsf1 was evident. This effect can be explained by the decline of mitochondrial capacity and intracellular ATP levels as well as reduced AA availability.
Figure 7. HSF1 knockdown attenuates mTORC1 activation by impairing nutrient-induced lysosomal mTORC1 translocation.
(A) Representative images of mTOR-Lamp2 colocalization in CUTLL1 or CCL119 cells following silencing of Hsf1 by doxycycline-inducible shRNA expression for 3 days. Cells expressing scramble shRNA were used as a control. Cells were cultured in PBS plus 0.5% dialyzed FCS for 50 min (Fasted) and either processed directly or after further stimulation with full medium (DMEM/10%FCS) for 10 minutes (refed). Quantification of mTORC-Lamp2 colocalization (yellow) dots (arrows) is displayed as a mean ± SD. (n=4–5 samples). 25 cells randomly selected and counted (*** p< 0.001).
(B) A schematic model for HSF1 action on mTORC1 activation and preservation of anabolic metabolic activity to remedy the oncogenic stress and promote drug resistant and relapsed malignant T cell progression. HSF1 inactivation suppresses mTORC1 activity by impacting ATP levels and AA availability, but triggers activation of adaptive glutamine metabolism associated with oxidative and/or reductive αKG carboxylation to antagonize the potential anti-leukemic effects of a HSF1-targeting approach. Oxidative and reductive carboxylation pathways (right panel) and mTOR signaling (right panel) are indicated.
Discussion
Progression model studies for many cancer types reveal that the transformation from a normal to malignant cell is driven by a vigorous clonal evolution and selection process, ultimately resulting in the acquisition of genetic aberrations that produce malignant cells with specific cellular features including drug therapy resistance. Undoubtedly, the survival and increased growth of well-adapted tumor-propagating cells is largely dependent on activation of regulatory programs that alleviate the immediate or chronic consequences of oncogenic and environmental stress and ensure a balanced proteome and optimal cellular metabolic bioenergetic capacity. The activation of this highly wired regulatory system that includes chaperone proteins and other quality control elements as well as several transcriptional and translational regulators has an essential role for survival of cancer cells (43). Our previous studies described a dynamic yet well-coordinated mechanistic model according to which HSF1, acting as a sensor and information hub, is capable of regulating transcriptional induction of HSPs and in parallel, re-adjusting the bioenergetic capacity of the cell to increase energetic efficiency and maximal mitochondrial ATP regeneration (20). Hence, in the context of physiological function and disease, HSF1 loss maintains an energy-saving adaptive response program that is coupled with inhibition of energy consuming anabolic synthetic pathways, particularly protein translation that are important drivers of leukemia progression. In the current study, we demonstrated that these energy-saving adaptive changes due to HSF1 loss may interfere with T-ALL development, raising the possibility of targeting HSF1 activity for leukemia treatment. Although the full spectrum of factors regulated by HSF1 that contribute to the development of resistant T-ALL in humans remain to be adequately elucidated, our data provide evidence for an unexplored functional crosstalk between two potent anabolic regulators, HSF1 and mTORC1, in T-ALL. Previous reports point out that HSF1 has a key role in NOTCH1-driven T-ALL through the control of chaperone/co-chaperone response (19). In this respect, our study further underlies the central role of HSF1 as a mediator of T-ALL development by potentiating metabolic and bioenergetic activity, thereby increasing overall anabolism-supportive pathways.
Our findings, which combine mouse and human T-ALL cell-based analyses, support a model (Fig. 7B), whereby HSF1 activity promotes the T-ALL malignant state through effects on energy-dependent mTOR signals and pro-growth anabolic processes. Notably, hyperactive PI3K-AKT-mTOR signaling stimulated by oncogenic lesions or reduced PTEN expression facilitate leukemic cell addiction to protein translational pathways, particularly eIF4E- or eIF2a-dependent mRNA translation (12, 44). Therefore, targeting this addiction is a therapeutic target for mTOR-dysregulated T-ALL hematological malignancy. Additionally, this model may explain how T cells finely tune oncogenic signals via HSF1 activity-specific reprogramming of the energetic state. Looking at this model from several perspectives yields the following insights:
A key aspect of this model is the elucidation of the mechanisms by which HSF1 depletion suppress mTORC1 activity. The mTORC1 kinase is known to localize in lysosomal compartments, where the kinase complex integrates a variety of environmental inputs, including nutrients (AAs), growth factors (insulin) and cellular energy (ATP) that stimulate mTORC1 signaling, whereas growth activates mTORC2 pathways (34, 36). The data here suggest that T-ALL, despite lacking HSF1 expression, displayed constitutively high AKT signaling, partially driven by hyperactive RTK signaling or PTEN loss-induced upregulated PI3K activity, but mTOR activity and its downstream target proteins (p70SK, S6R, and 4E-BP), which stimulate protein translation and mitochondrial biogenesis, was markedly repressed. It appears, thus, that this effect on mTOR signaling is independent of major AKT signaling and, as elaborated before, mTORC1 attenuation likely occurs without direct involvement of AMPK. This raised the possibility that HSF1 loss restricts mTORC1 activity in part by limiting cellular energy sources, thus impacting the sensing ATP-mTOR activity.
Another interesting finding is that the changes in bioenergetics caused by HSF1 loss appear to have a dramatic effect on the intracellular AA steady-state level sensed by mTORC1. It is well known that mTOR activity is transmitted by an array of substrate signals that orchestrate the balance between anabolic and catabolic metabolism (36, 45). The AA homeostasis, a critical aspect of protein translation, is determined by rates of extracellular AA import and export via AA transporters, protein synthesis and turnover, as well as AA oxidation (46). To maintain anabolic balance and alleviate stress, mTORC1 stimulates AA uptake and increases protein and nucleotide synthetic rates while suppressing autophagic catabolism of proteins. mTORC1 activity balances cellular AA supply and purine biosynthesis for protein translation in the presence of sufficient nutrients, in part through control of ATF4 translation and widespread induction of mRNAs encoding AA transporters (47). On the other hand, starvation of AAs increases ATF4 activity and AA transporter levels via a GCN2-eIF2α-regulated mechanism. This suggests diverse regulatory circuits couple translational demand to monomeric AA supply under nutrient availability or restriction states. The activation of an adaptive route of AA acquisition via extracellular protein uptake and catabolism can also be a major contributor to AA pools in cancer cells, especially under conditions of mTORC1 inhibition (48, 49). One consequence of the precipitous drop in ATP levels as a result of HSF1 depletion, therefore, is that T-ALL cells with constitutive growth factor signaling are unable to sustain minimal biosynthetic and bioenergetic activity required for cell adaptation and survival. By restricting translation through mTORC1 inhibition, the cell could minimize energy consumption and restore AA balance to a certain degree; however, we speculate that malignant T-ALL cells with HSF1 depletion are not well positioned to raise levels of mitochondrial ATP production and maintain AA homeostasis required to sustain minimal translational activity. Thus, the data suggest that HSF1-depleted cells are forced to harness the costly free energy of the intracellular AA catabolic oxidation to ameliorate the bioenergetic deficit and preserve cell survival.
Another central conclusion for this study is the powerful activation of adaptive glutamine metabolism associated with oxidative and/or reductive α-ketoglutarate (αKG) carboxylation in T-ALL cells with HSF1 depletion. Reductive carboxylation is known to support cancer cell proliferation with impaired mitochondrial metabolism and contributes to biosynthesis of lipids and citric acid cycle (CAC) metabolites and other macromolecular precursors as well as redox state (42, 50, 51). The conversion of glutamine-derived αKG to isocitrate through a reductive carboxylation reaction catalyzed by cytosolic and mitochondrial NADP+/NADPH isocitrate dehydrogenases (IDH) is stimulated by mitochondrial redox status changes and has been associated with mitochondrial dysfunction and decreased NAD+/NADH ratio (50, 51). In line with these and our previous findings indicating that the impact of silencing Hsf1 on mitochondrial bioenergetic capacity is associated with decreased NAD+ availability and NAD+/NADH ratio (20), we observed that Hsf1 loss in T-ALL cells significantly increased levels of citrate, succinate, 2-HG and reductive equivalents (NADH, NADPH), which are metabolic hallmarks of reductive and oxidative glutamine metabolism. The high succinate level could be explained in part by αKG oxidation driven by αKG dehydrogenase activity, an NADH-generating enzyme complex, and may also be the end product of AA catabolism and reduction of fumarate to succinate using the NADH-dependent fumarase reductase system (52). Elevated 2-HG synthesis, likely owing to a decrease in NAD+/NADH ratio, has been proposed to increase NADPH demand for its synthesis in competition with other NADPH-dependent pathways including the pentose phosphate pathway (PPP) that is essential for buffering ROS activity and promoting cell viability (53). As counterintuitive as it may seem, 2-HG synthesis, by creating a significant demand on NADPH, can limit the NADPH available for reductive carboxylation of αKG. Even though the rate of 2-HG synthesis increases in our study, the reductive carboxylation pathway operated to consume significant amounts of NADPH at a time when NADPH is critical to neutralize ROS. We propose, thus, that the increased demand on NADPH driven by 2-HG synthetic pathways and reductive glutamine metabolism can sensitize Hsf1-deficient leukemic cells to oxidative stress (ROS). However, we would like to emphasize that our work has not conclusively demonstrated and characterized the role of NAD+/NADH and NADP+/NADPH redox balance systems in our T-ALL human cell lines and that further biochemical analyses are required to establish the metabolic basis of the redox dysregulation in our experimental model. The increased aspartate levels that we observed in Hsf1-deficient T-ALL cells may be driven by a reverse αKG-citrate-oxaloacetate (OAA) pathway, although the precise mechanism remains to be explored. Since succinate and 2-HG are known antagonists of αKG, their elevated levels are also expected to inhibit the activity of αKG-dependent dioxygenases and alter the epigenetic landscape of malignant cells, possibly through changes in gene repressive DNA and histone hypermethylation marks (54). The impact of Hsf1 depletion on epigenetic regulation of T-ALL progression await further investigation.
Together, our results suggest a central role of HSF1 in preserving metabolic energy and mTORC1 anabolic activity and improving redox homeostatic capacity to remedy the oncogenic stress and promote drug resistance and relapsed malignant T cell progression. Targeting these compensatory adaptive changes through Hsf1 ablation offers a therapeutic approach.
Supplementary Material
Implications:
Targeting HSF1 and HSF1-dependent cancer-specific anabolic and protein homeostasis programs has a significant therapeutic potential for T-ALL and may prevent progression of relapsed/refractory disease.
Acknowledgments:
This work was supported by the grants from the National Cancer Institute to N. F. Mivechi and D. Moskophidis (R01 CA132640 and R01 CA062130).
Footnotes
Disclosure of potential conflicts of interest: Authors declare no potential conflict of interest.
References
- 1.Pui CH, Relling MV, Downing JR. 2004. Acute lymphoblastic leukemia. N Engl J Med 350:1535–48. [DOI] [PubMed] [Google Scholar]
- 2.Gokbuget N, Stanze D, Beck J, Diedrich H, Horst HA, Huttmann A, ET AL. German Multicenter Study Group for Adult Acute Lymphoblastic L. 2012. Outcome of relapsed adult lymphoblastic leukemia depends on response to salvage chemotherapy, prognostic factors, and performance of stem cell transplantation. Blood 120:2032–41. [DOI] [PubMed] [Google Scholar]
- 3.Van Vlierberghe P, Pieters R, Beverloo HB, Meijerink JP. 2008. Molecular-genetic insights in paediatric T-cell acute lymphoblastic leukaemia. Br J Haematol 143:153–68. [DOI] [PubMed] [Google Scholar]
- 4.Belver L, Ferrando A. 2016. The genetics and mechanisms of T cell acute lymphoblastic leukaemia. Nat Rev Cancer 16:494–507. [DOI] [PubMed] [Google Scholar]
- 5.Grabher C, von Boehmer H, Look AT. 2006. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat Rev Cancer 6:347–59. [DOI] [PubMed] [Google Scholar]
- 6.Girardi T, Vicente C, Cools J, De Keersmaecker K. 2017. The genetics and molecular biology of T-ALL. Blood 129:1113–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. 2004. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306:269–71. [DOI] [PubMed] [Google Scholar]
- 8.Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, Basso G, Ferrando A, Aifantis I. 2007. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med 204:1825–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M, et al. 2007. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med 13:1203–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dail M, Wong J, Lawrence J, O’Connor D, Nakitandwe J, Chen SC, Xu J, Lee LB, Akagi K, Li Q, Aster JC, Pear WS, Downing JR, Sampath D, Shannon K. 2014. Loss of oncogenic Notch1 with resistance to a PI3K inhibitor in T-cell leukaemia. Nature 513:512–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mendes RD, Cante-Barrett K, Pieters R, Meijerink JP. 2016. The relevance of PTEN-AKT in relation to NOTCH1-directed treatment strategies in T-cell acute lymphoblastic leukemia. Haematologica 101:1010–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanchez-Martin M, Ambesi-Impiombato A, Qin Y, Herranz D, Bansal M, Girardi T, Paietta E, et al. 2017. Synergistic antileukemic therapies in NOTCH1-induced T-ALL. Proc Natl Acad Sci U S A 114:2006–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herranz D, Ambesi-Impiombato A, Sudderth J, Sanchez-Martin M, Belver L, Tosello V, et al. 2015. Metabolic reprogramming induces resistance to anti-NOTCH1 therapies in T cell acute lymphoblastic leukemia. Nat Med 21:1182–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai C, Whitesell L, Rogers AB, Lindquist S. 2007. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell 130:1005–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin X, Moskophidis D, Mivechi NF. 2011. Heat shock transcription factor 1 is a key determinant of HCC development by regulating hepatic steatosis and metabolic syndrome. Cell Metab 14:91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xi C, Hu Y, Moskophidis D, Mivechi NF. 2012. Heat shock factor Hsf1 promotes Her2/Neu-induced mammary tumorigenesis and metastasis by modulating epithelial mesenchymal transition. Oncogene 287:35646–35657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai C, Santagata S, Tang Z, Shi J, Cao J, Kwon H, Bronson RT, Whitesell L, Lindquist S. 2012. Loss of tumor suppressor NF1 activates HSF1 to promote carcinogenesis. J Clin Invest 122:3742–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Min JN, Huang L, Zimonjic DB, Moskophidis D, Mivechi NF. 2007. Selective suppression of lymphomas by functional loss of Hsf1 in a p53-deficient mouse model for spontaneous tumors. Oncogene 26:5086–97. [DOI] [PubMed] [Google Scholar]
- 19.Kourtis N, Lazaris C, Hockemeyer K, Balandran JC, Jimenez AR, Mullenders J, et al. 2018. Oncogenic hijacking of the stress response machinery in T cell acute lymphoblastic leukemia. Nat Med 24:1157–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiao A, Jin X, Pang J, Moskophidis D, Mivechi NF. 2017. The transcriptional regulator of the chaperone response HSF1 controls hepatic bioenergetics and protein homeostasis. J Cell Biol 216:723–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lesche R, Groszer M, Gao J, Wang Y, Messing A, Sun H, Liu X, Wu H. 2002. Cre/loxP-mediated inactivation of the murine Pten tumor suppressor gene. Genesis 32:148–9. [DOI] [PubMed] [Google Scholar]
- 22.Hennet T, Hagen FK, Tabak LA, Marth JD. 1995. T-cell-specific deletion of a polypeptide N-acetylgalactosaminyl-transferase gene by site-directed recombination. Proc Natl Acad Sci U S A 92:12070–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palomero T, Barnes KC, Real PJ, Glade Bender JL, Sulis ML, Murty VV, Colovai AI, Balbin M, Ferrando AA. 2006. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to gamma-secretase inhibitors. Leukemia 20:1279–87. [DOI] [PubMed] [Google Scholar]
- 24.Ikediobi ON, Davies H, Bignell G, Edkins S, Stevens C, O’Meara S, et al. 2006. Mutation analysis of 24 known cancer genes in the NCI-60 cell line set. Mol Cancer Ther 5:2606–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao SS, O’Neil J, Liberator CD, Hardwick JS, Dai X, Zhang T, Tyminski E, Yuan J, Kohl NE, Richon VM, Van der Ploeg LH, Carroll PM, Draetta GF, Look AT, Strack PR, Winter CG. 2009. Inhibition of NOTCH signaling by gamma secretase inhibitor engages the RB pathway and elicits cell cycle exit in T-cell acute lymphoblastic leukemia cells. Cancer Res 69:3060–8. [DOI] [PubMed] [Google Scholar]
- 26.King AG, Kondo M, Scherer DC, Weissman IL. 2002. Lineage infidelity in myeloid cells with TCR gene rearrangement: a latent developmental potential of proT cells revealed by ectopic cytokine receptor signaling. Proc Natl Acad Sci U S A 99:4508–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hagenbeek TJ, Spits H. 2008. T-cell lymphomas in T-cell-specific Pten-deficient mice originate in the thymus. Leukemia 22:608–19. [DOI] [PubMed] [Google Scholar]
- 28.Tandon P, Gallo CA, Khatri S, Barger JF, Yepiskoposyan H, Plas DR. 2011. Requirement for ribosomal protein S6 kinase 1 to mediate glycolysis and apoptosis resistance induced by Pten deficiency. Proc Natl Acad Sci U S A 108:2361–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morita M, Gravel SP, Chenard V, Sikstrom K, Zheng L, Alain T, Gandin V, Avizonis D, Arguello M, Zakaria C, McLaughlan S, Nouet Y, Pause A, Pollak M, Gottlieb E, Larsson O, St-Pierre J, Topisirovic I, Sonenberg N. 2013. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab 18:698–711. [DOI] [PubMed] [Google Scholar]
- 30.Blackburn JS, Liu S, Wilder JL, Dobrinski KP, Lobbardi R, Moore FE, Martinez SA, Chen EY, Lee C, Langenau DM. 2014. Clonal evolution enhances leukemia-propagating cell frequency in T cell acute lymphoblastic leukemia through Akt/mTORC1 pathway activation. Cancer Cell 25:366–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robitaille AM, Christen S, Shimobayashi M, Cornu M, Fava LL, Moes S, Prescianotto-Baschong C, Sauer U, Jenoe P, Hall MN. 2013. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science 339:1320–3. [DOI] [PubMed] [Google Scholar]
- 32.Dennis PB, Jaeschke A, Saitoh M, Fowler B, Kozma SC, Thomas G. 2001. Mammalian TOR: a homeostatic ATP sensor. Science 294:1102–5. [DOI] [PubMed] [Google Scholar]
- 33.Hardie DG, Ross FA, Hawley SA. 2012. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol 13:251–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J, Guan KL. 2019. mTOR as a central hub of nutrient signalling and cell growth. Nat Cell Biol 21:63–71. [DOI] [PubMed] [Google Scholar]
- 35.Hsieh AL, Walton ZE, Altman BJ, Stine ZE, Dang CV. 2015. MYC and metabolism on the path to cancer. Semin Cell Dev Biol 43:11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saxton RA, Sabatini DM. 2017. mTOR Signaling in Growth, Metabolism, and Disease. Cell 168:960–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoon YJ, Kim JA, Shin KD, Shin DS, Han YM, Lee YJ, Lee JS, Kwon BM, Han DC. 2011. KRIBB11 inhibits HSP70 synthesis through inhibition of heat shock factor 1 function by impairing the recruitment of positive transcription elongation factor b to the hsp70 promoter. J Biol Chem 286:1737–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santagata S, Mendillo ML, Tang YC, Subramanian A, Perley CC, Roche SP, Wong B, Narayan R, Kwon H, Koeva M, Amon A, Golub TR, Porco JA, Whitesell L, Lindquist S 2013. Tight coordination of protein translation and HSF1 activation supports the anabolic malignant state. Science 341:1238303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao YH, Zhou M, Liu H, Ding Y, Khong HT, Yu D, Fodstad O, Tan M. 2009. Upregulation of lactate dehydrogenase A by ErbB2 through heat shock factor 1 promotes breast cancer cell glycolysis and growth. Oncogene 28:3689–701. [DOI] [PubMed] [Google Scholar]
- 40.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, et al. 2009. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462:739–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oldham WM, Clish CB, Yang Y, Loscalzo J. 2015. Hypoxia-Mediated Increases in L-2-hydroxyglutarate Coordinate the Metabolic Response to Reductive Stress. Cell Metab 22:291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mullen AR, Hu Z, Shi X, Jiang L, Boroughs LK, Kovacs Z, Boriack R, Rakheja D, Sullivan LB, Linehan WM, Chandel NS, DeBerardinis RJ. 2014. Oxidation of alpha-ketoglutarate is required for reductive carboxylation in cancer cells with mitochondrial defects. Cell Rep 7:1679–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klaips CL, Jayaraj GG, Hartl FU. 2018. Pathways of cellular proteostasis in aging and disease. J Cell Biol 217:51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwarzer A, Holtmann H, Brugman M, Meyer J, Schauerte C, Zuber J, Steinemann D, Schlegelberger B, Li Z, Baum C. 2015. Hyperactivation of mTORC1 and mTORC2 by multiple oncogenic events causes addiction to eIF4E-dependent mRNA translation in T-cell leukemia. Oncogene 34:3593–604. [DOI] [PubMed] [Google Scholar]
- 45.Valvezan AJM BD 2019. Molecular logic of mTORC1 signalling as a metabolic rheostat. Nature Metabolism 1:321–33. [DOI] [PubMed] [Google Scholar]
- 46.Broer S, Broer A. 2017. Amino acid homeostasis and signalling in mammalian cells and organisms. Biochem J 474:1935–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park Y, Reyna-Neyra A, Philippe L, Thoreen CC. 2017. mTORC1 Balances Cellular Amino Acid Supply with Demand for Protein Synthesis through Post-transcriptional Control of ATF4. Cell Rep 19:1083–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palm W, Park Y, Wright K, Pavlova NN, Tuveson DA, Thompson CB. 2015. The Utilization of Extracellular Proteins as Nutrients Is Suppressed by mTORC1. Cell 162:259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nofal M, Zhang K, Han S, Rabinowitz JD. 2017. mTOR Inhibition Restores Amino Acid Balance in Cells Dependent on Catabolism of Extracellular Protein. Mol Cell 67:936–946 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, DeBerardinis RJ. 2011. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 481:385–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang L, Shestov AA, Swain P, Yang C, Parker SJ, Wang QA, et al. 2016. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature 532:255–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomitsuka E, Kita K, Esumi H. 2010. The NADH-fumarate reductase system, a novel mitochondrial energy metabolism, is a new target for anticancer therapy in tumor microenvironments. Ann N Y Acad Sci 1201:44–9. [DOI] [PubMed] [Google Scholar]
- 53.Gelman SJ, Naser F, Mahieu NG, McKenzie LD, Dunn GP, Chheda MG, Patti GJ. 2018. Consumption of NADPH for 2-HG Synthesis Increases Pentose Phosphate Pathway Flux and Sensitizes Cells to Oxidative Stress. Cell Rep 22:512–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong CC, Qian Y, Yu J. 2017. Interplay between epigenetics and metabolism in oncogenesis: mechanisms and therapeutic approaches. Oncogene 36:3359–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.