In the CRISPR system, a short RNA guide sequence and a variant of the Cas9 protein are introduced into cells. The RNA will bind to an identical sequence in the genome, as long as it is followed by the PAM sequence (usually -GG), and mark it for subsequent cutting or modification by the Cas9 protein. However, it is well known to most biologists from primers and siRNAs that nucleotide binding does not require a 100% identical sequence and single base pair mismatches are often tolerated. So unsurprisingly, CRISPR modifications were soon found at “off-target” positions in the genome, at chromosomal locations that were not intended but nevertheless cut in cells.
Off-target effects have evolved from a minor scientific detail to a topic picked up by television news. This is because off-targets could potentially be fatal for the whole CRISPR technique. If CRISPR cleave random locations in the genome, this could render the technique useless for most applications. The first studies [Hsu2013] using systematic mutations of a selected target sequence found that mutations of the target sequence are tolerated and that the nucleotides closer to the CRISPR recognition PAM site are less tolerant, similar to what was known from siRNA binding. Then, a series of studies from different labs using high-throughput sequencing of cell cultures found many more off-targets than expected, up to thousands for a single guide [Tsai2015] [Kim2018]. However, these studies also confirmed that most of the off-targets were sequences similar to the RNA guide sequence. In some cases, and unexpectedly, the guide even tolerated nucleotide insertions or deletions (indels) of the target, potentially increasing the range of possibly off-targets in the genome by one or two orders of magnitude. This seemed to cast doubt on the entire CRISPR technology.
However, indels in these studies were exceedingly rare, less than a handful of cases are known across thousands of validated off-targets. Also, the studies mostly confirmed what had been expected, that the majority of off-targets are sequences similar to the target. Depending on how one interprets the data [Haeussler2016], up to 80% of the off-targets have less than four mismatches and 97% have less than five mismatches to the guide. As such, they can be easily predicted with standard sequence alignment programs (though not seed-based ones like BLAST).
When thinking about the number of possible alignments in a genome, a rather theoretical property of most eukaryotic genomes becomes relevant: their biased sequence distribution. The nucleotides found in a genome are not random at all. While some sequences are extremely common (homopolymers like AAAAAAAA as an extreme example can be found tens of thousands of times), some 20mers can be found hundreds of times and most 20mers in protein-coding sequences are unique for a particular gene. But this is not true for all genes. As most biologists know, genes have complex similarity relationships, a few groups have many almost identical copies in the genome (like zinc fingers or olfactory receptors), many others appear in the genome only once. As a result, it is easy to see that anything written about off-targets depends very much on the gene and the 20mer target sequence within that gene. One can construct a guide sequence with millions of identical target sequences in the genome, which will shred the genome into thousands of small pieces, but one can often also find a guide that has no single similar sequence in the genome at all, even assuming that one would tolerate five mismatches. This is why researchers now use software when they select the target sequence. In the off-target high-throughput studies above, a few guides were very unspecific, which explains the high number of off-targets found. Most researchers who use design software would have never tried these guides in practice.
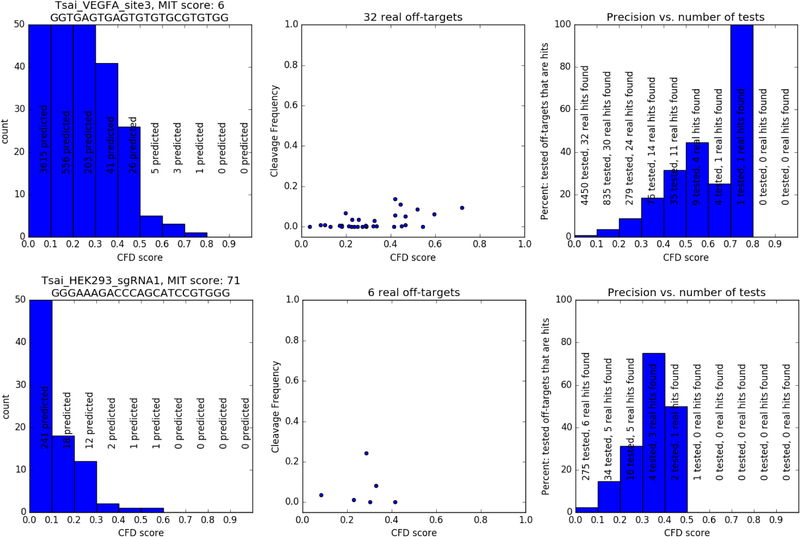
Another result from the high-throughput studies was that most similar sequences in the genome shown by the guide design software are not actually cut in cells. Even if they are bound by the RNA and the Cas9 protein as shown by Chip-Seq assays, this does not mean that the sites are cut [Wu2015]. The majority of sequences that align to the guide’s target are “false positives”, very similar to the target but not cut in cells at a frequency over the detection limit of the known assays. Out of hundreds of predicted off-targets shown by the software tools, only a few will typically be confirmed by PCR, the most sensitive assay. Figure 1 illustrates this on the two most extreme examples from the original Guide-seq study: the first guide has a low specificity score and thousands of predicted off-targets, the second guide is very specific and only has a few hundred predicted off-targets. Of the 4450 predicted off-targets of the first guide, only 32 were actually cleaved in the cell culture. It is hard to run 4450 PCRs. If one tests only the top-ranked 279 off-targets by PCR this would catch only 24 of the 32 off-targets. However, for the more specific guide, an exhaustive PCR means only 275 PCRs and screening only the top-ranked 34 predictions, a very manageable list, would yield 5 of the 6 off-targets.
Figure 1 -.
Comparison of two guides: the first row shows data for the guide VEGFA_site3, the second row shows data for HEK293_sgRNA1. The first column shows histograms of the number of predicted off-targets by CFD (Cutting-frequency determination) score, the second column shows the off-targets actually cleaved in cells, the third column shows for various CFD cutoffs how many off-targets would have to screened and how many of these would be real off-targets.
Also, the cleavage frequency, the concentration of Cas9 and the exposure time are important parameters: most off-targets are cut very rarely, so while they may appear in cell cultures and can be found there by PCR, they may be impossible to observe in transgenic lines, where the Cas9 is injected and only active very briefly. This lead some authors to dismiss the off-targets found through high-throughput assays as entirely irrelevant in practice [Iyver2015], as they were unable to find any off-targets in their transgenic mice, especially after inbreeding.
At this point, I hope it is clear to the reader that the topic of off-targets is impossible to discuss without taking into account the experimental context and the guide sequence. In bacteria, with their tiny genomes, off-targets will hardly play any role at all, as in most cases, it is hard to find similar sequences in the genome, as long as the guide has been checked. In fruit flies, with small genomes and followed by out-crossing that removes off-targets quickly, they seem to be not an issue either. In mice, with a Cas9 that is quickly removed, the topic probably has to be addressed, but with well-designed guides, screening with the existing assays, and possibly using modified Cas9s that trade efficiency for specificity (ref) they seem to be a manageable problem [Anderson2018]. There remains a lingering doubt if a mouse phenotype was caused by an off-target, possibly by large rearrangements [Kosicki2018], but as long as the guide has been selected well and the model has been screened with current assays, the risk seems very small.
So what should researchers do to reduce off-target effects? First and foremost, they should use design software to pick guides that have few off-targets. By citations, the two main software tools in this area are Chop-Chop [Labun 2019] the website I wrote, crispor.org [Concordet2018]. They differ in that Crispor tries to show as much data as possible on the guides and let the user make his/her choice, whereas Chop-chop shows less information but recommends an optimal guide based on rules. As often with bioinformatics, sequences should be checked on several websites, which provides some insurance against software problems. Do not hesitate to email the authors of these tools if their output is not clear. Both websites will predict a score for the guide and potential off-targets in the genome. Picking specific guides is the only advice that applies to all cellular contexts. Once the guide have been introduced into cells, researchers must either count on enough inbreeding to cross out any potential off-target modifications, validate the off-targets predicted by the software or use high-throughput cell culture assays to find any potential off-targets, irrespective of software predictions.
A specific guide will save a lot of work later, since the few predicted off-targets can be tested with a single plate of primers. If for some reason a very unspecific guide must be used and the context is human cells or a mouse model, the lab should plan to spend a lot of time or money on testing the resulting cells, as possibly thousands of PCRs may have to be run on the mouse model or human cells. Whole-genome sequencing, though easy thanks to Digenome-seq software [Park2017], is not necessarily cheap nor the most sensitive option, as it may miss many off-targets unless read coverage is very high and that across the whole genome, see [Anderson2018]. As an alternative, a strategy like in [Anderson2018] can be used: first, establish a list of potential off-targets by applying high-throughput assays on cell cultures, most likely Guide-seq [Tsai2015] (which can be bought as a service from commercial companies now) or possibly its more recent and reportedly more sensitive cousin Discover-Seq [Wiener2019]. Once off-targets that are cleaved in the cells have been identified, the lab can screen for these in the organism with targeted PCRs.
To remove the ones that are present even in the animal, one can try to change the Cas9 enzyme, e.g. by using one of the high-fidelity Cas9s, which increase off-target specificity but often unfortunately also lower on-target efficiency or sometimes can turn out to not cleave a given target sequence at all [Kulcsar2017]. One can also resort to alternate CRISPR enzymes with a very different off-target profile, like Cpf1 [Zetsche2015], though this may require a whole new round of high-throughput screening. Limiting the time Cas9 is active in the cells may be another way to reduce the frequency of off-target cleaveage. With either method, predicted or newly identified off-targets will probably require final quantitative validation with PCR followed by high-throughput sequencing.
The situation is special for medical applications in humans. Here, well-selected, specific guides need to undergo extensive testing with a combination of the most sensitive high-throughput assays available [DeWitt2016, Magis2018, Cai2018]. Once the list of all possible off-targets in the genome is known for a particular guide, it is addressed by a combination of measures e.g. through modifications to Cas9, by changing the guide or the delivery procedure, as described above and the testing restarts.
Despite these challenges, we should keep in mind that many off-targets will not be a problem if only a given cell type is modified. As Dana Carroll recently noted: “mutations in a muscle-specific gene, like dystrophin, which is not expressed or required in the hematopoietic lineages (as far as we know), are probably tolerable.” When it comes to human applications, the context that matters is even more important, from the specific guide, to the Cas9 used and its concentration, to the particular cell type and ultimately the medical gain from the genome modification relative to the potential risks and their likelihood. In tumors and in crop plants, we are already changing the genome with “broad random mutagenesis with radiation or chemicals”, leading to “a substantial load of background mutations that are never characterized or acknowledged” [Carroll2019]. At this stage, with the first CRISPR human trials just starting this year [Vertex2019], it is a matter of opinion, but at the moment, as described in this article, one can easily argue that off-target effects will not be the main obstacle for medical applications of CRISPR in humans.
Acknowledgements:
Many thanks to Jean-Paul Concordet for guiding the development of crispor.org, many useful discussions on CRISPR over the years and literature pointers for this editorial.
Funding: NHGRI 5U41HG002371, CZI Silicon Valley Community Foundation 2018-182809
Footnotes
Declaration of conflict of interest: Crispor.org is free and free to install for academics and nonprofits, but UCSC is selling licenses for Crispor.org to commercial companies and I may be getting a part of future licensing fees.
References:
- Anderson KR, Haeussler M, Watanabe C, Janakiraman V, Lund J, Modrusan Z, Stinson J, Bei Q, Buechler A, Yu C, Thamminana SR. CRISPR off-target analysis in genetically engineered rats and mice. Nature methods. 2018. July;15(7):512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concordet JP, Haeussler M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic acids research. 2018. May 14;46(W1):W242–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D Collateral damage: benchmarking off-target effects in genome editing. Genome biology. 2019. June 3;20(1):114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Bai H, Mahairaki V, Gao Y, He C, Wen Y, Jin YC, Wang Y, Pan RL, Qasba A, Ye Z. A universal approach to correct various HBB gene mutations in human stem cells for gene therapy of beta- thalassemia and sickle cell disease. Stem cells translational medicine. 2018. January;7(1):87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt MA, Magis W, Bray NL, Wang T, Berman JR, Urbinati F, Heo SJ, Mitros T, Muñoz DP, Boffelli D, Kohn DB. Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Science translational medicine. 2016. October 12;8(360):360ra134-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keep calm and edit on. Nat Biotechnol. 2018. August 6;36(8):667. doi: 10.1038/nbt.4221. [DOI] [PubMed] [Google Scholar]
- Haeussler M, Schönig K, Eckert H, Eschstruth A, Mianné J, Renaud JB, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, Joly JS, Concordet JP. Evaluation of off-target and on-target scoring algorithms and integration intothe guide RNA selection tool CRISPOR. Genome Biol. 2016. July 5;17(1):148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013. September;31(9):827–32. doi: 10.1038/nbt.2647. Epub 2013 Jul 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer V, Shen B, Zhang W, Hodgkins A, Keane T, Huang X, Skarnes WC. Off-target mutations are rare in Cas9-modified mice. Nature methods. 2015. June;12(6):479. [DOI] [PubMed] [Google Scholar]
- Park J, Childs L, Kim D, Hwang GH, Kim S, Kim ST, Kim JS, Bae S. Digenome-seq web tool for profiling CRISPR specificity. Nature methods. 2017. June;14(6):548. [DOI] [PubMed] [Google Scholar]
- Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nature biotechnology. 2018. August;36(8):765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Bae S, Park J, Kim E, Kim S, Yu HR, Hwang J, Kim JI, Kim JS. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015. March;12(3):237–43, 1 p following 243. doi: 10.1038/nmeth.3284. Epub 2015 Feb 9. [DOI] [PubMed] [Google Scholar]
- Kulcsár PI, Tálas A, Huszár K, Ligeti Z, Tóth E, Weinhardt N, Fodor E, Welker E. Crossing enhanced and high fidelity SpCas9 nucleases to optimize specificity and cleavage. Genome biology. 2017. December;18(1):190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magis W, DeWitt MA, Wyman SK, Vu JT, Heo SJ, Shao SJ, Hennig F, Romero ZG, Campo-Fernandez B, McNeill M, Rettig GR. In vivo selection for corrected β-globin alleles after CRISPR/Cas9 editing in human sickle hematopoietic stem cells enhances therapeutic potential. bioRxiv. 2018. January 1:432716. [Google Scholar]
- Labun K, Montague TG, Krause M, Torres Cleuren YN, Tjeldnes H, Valen E. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic acids research. 2019. May 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, Aryee MJ, Joung JK. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015. February;33(2):187–197. doi: 10.1038/nbt.3117. Epub 2014 Dec 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Scott DA, Kriz AJ, Chiu AC, Hsu PD, Dadon DB, Cheng AW, Trevino AE, Konermann S, Chen S, Jaenisch R, Zhang F, Sharp PA. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol. 2014. July;32(7):670–6. doi: 10.1038/nbt.2889. Epub 2014 Apr 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- http://ir.crisprtx.com/news-releases/news-release-details/crispr-therapeutics-and-vertex-announce-progress-clinical.
- Wienert B, Wyman SK, Richardson CD, Yeh CD, Akcakaya P, Porritt MJ, Morlock M, Vu JT, Kazane KR, Watry HL, Judge LM. Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science. 2019. April 19;364(6437):286–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, Van Der Oost J, Regev A, Koonin EV. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015. October 22;163(3):759–71. [DOI] [PMC free article] [PubMed] [Google Scholar]