Abstract
Purpose
Glioblastoma is the most frequent and lethal primary brain tumor. Development of novel therapies relies on the availability of relevant preclinical models. We have established a panel of 96 glioblastoma patient derived xenografts (PDX) and undertaken its genomic and phenotypic characterization.
Experimental Design
PDX were established from glioblastoma, IDH-wildtype (n=93), glioblastoma, IDH-mutant (n=2), diffuse midline glioma, H3 K27M-mutant (n=1), and both primary (n=60) and recurrent (n=34) tumors. Tumor growth rates, histopathology, and treatment response were characterized. Integrated molecular profiling was performed by whole exome sequencing (WES, n=83), RNA-sequencing (n=68), and genome-wide methylation profiling (n=76). WES data from 24 patient tumors was compared with derivative models.
Results
PDXs recapitulate many key phenotypic and molecular features of patient tumors. Orthotopic PDXs show characteristic tumor morphology and invasion patterns, but largely lack microvascular proliferation and necrosis. PDXs capture common and rare molecular drivers, including alterations of TERT, EGFR, PTEN, TP53, BRAF, and IDH1, most at frequencies comparable to human glioblastoma. However, PDGFRA amplification was absent. RNA-sequencing and genome-wide methylation profiling demonstrated broad representation of glioblastoma molecular subtypes. MGMT promoter methylation correlated with increased survival in response to temozolomide. WES of 24 matched patient tumors showed preservation of most genetic driver alterations, including EGFR amplification. However, in four patient-PDX pairs, driver alterations were gained or lost on engraftment, consistent with clonal selection.
Conclusions
Our PDX panel captures the molecular heterogeneity of glioblastoma and recapitulates many salient genetic and phenotypic features. All models and genomic data are openly available to investigators.
Keywords: patient derived xenograft, glioblastoma, glioma, tumor models, neuro-oncology
INTRODUCTION
Glioblastoma is the most common and aggressive primary brain tumor. Despite significant advances in understanding the underlying genetic drivers, limited progress has been made in developing new and effective therapies over the past decades, and glioblastoma remains uniformly lethal. Multiple factors contribute to the treatment refractory nature of glioblastoma including infiltration of normal brain, limited drug delivery into regions of tumor with an intact blood-brain barrier, inter- and intra-tumoral molecular heterogeneity, and inherent chemo- and radio-resistance of tumor-initiating cells. Additionally, many pre-clinical studies have relied heavily on a relatively narrow repertoire of glioma cell lines maintained in conventional cell culture for long periods of time. These cell lines have diverged genetically from human tumors or have uncertain provenance (1,2), which limits their translational utility. As a result, many promising studies in vitro have led to disappointing outcomes in clinical trials. Thus, a concerted effort was undertaken over the past decade to develop new, more biologically relevant model systems.
Most modern glioma models fall into one of two types, genetically engineered mouse models (GEMM) or patient-derived models established from surgical tumor tissue. Each approach has inherent advantages and limitations. GEMMs are established through introduction of defined genetic alterations into the mouse germline, resulting in de novo tumor formation (3). GEMMs have the advantage of an intact immune system and matched mouse tumor/stromal microenvironment, features particularly important for studies of immunotherapy. However, GEMMs are highly time intensive to develop and have only a limited repertoire of genetic driver alterations. Patient-derived models can be more readily established using several methods, the most common ones being brain tumor initiating cell (BTIC) culture and patient-derived xenografts (PDXs). BTICs are generated through culture of patient tumor tissue in serum-free media supplemented with exogenous growth factors including EGF and bFGF. BTICs genetically more closely mirror their corresponding primary tumors than conventional cell lines (4). However, BTICs lack the stromal microenvironment, and high levels of exogenous EGF can lead to loss of EGFR amplification, which is a key molecular driver in glioblastoma (5). PDX models are generated through implantation of patient tumor tissue into immunocompromised mice and can be established as heterotopic tumors, typically in the flank subcutaneous tissue, or as intracranial orthotopic tumors. PDX models maintain EGFR amplification (6) and the typical copy-number profile of glioblastoma (7). However, PDXs are limited by the lack of an intact immune system, a differing tumor microenvironment, particularly for heterotopic xenografts, as well as being more costly and labor-intensive to maintain. Despite the growing use of both BTIC and PDX models, the extent to which they recapitulate human glioblastoma remains incompletely understood.
Glioblastoma is a molecularly heterogeneous disease. A set of core genetic pathways drive glioblastoma with the majority of tumors showing dysregulation of the receptor tyrosine kinase, PI3-kinase, p53, and retinoblastoma pathways (8). However, combinations of multiple distinct genetic alterations may affect a particular pathway, resulting in tumors with differing therapeutic vulnerabilities. Genetic heterogeneity has presented a challenge in clinical trials as targeted therapies often benefit only a small subset of patients, and predictive biomarkers are needed to identify patients likely to respond to a particular therapy. Thus, large panels of molecularly heterogeneous patient-derived models can be particularly useful for these types of pre-clinical analyses.
In an effort to expedite basic and translational research, we have established a panel of 96 glioblastoma PDXs. This manuscript provides the first comprehensive description of the Mayo glioma PDX collection. Our PDX models capture the molecular heterogeneity of glioblastoma and recapitulate salient features of the patient tumors from which they derive. Models have already been widely distributed and Mayo Clinic continues to be committed to openly sharing these important research tools and accompanying genetic data with the broader research community.
METHODS
Establishment of PDX lines
PDX were established through injection of surgical tissue into the flank of athymic nude mice. Detailed procedures for establishment and maintenance of flank xenografts, orthotopic xenografts, and short-term explant cultures have been previously published (9) and are summarized in the Supplementary Methods. All experiments were conducted in accordance with the Belmont Report and U.S. Common Rule with approval from the Mayo Clinic Institutional Review Board and written consent from participating patients. All animal experiments were approved by the Mayo Clinic Institutional Animal Care and Use Committee.
Histopathologic Assessment
Orthotopic tumors have been established from 78 PDX lines and 58 utilized for detailed histologic analyses. At moribund, whole brains were formalin fixed and paraffin embedded and tissue blocks used to create a 360-core tissue microarray. Tumor histology was examined and invasion assessed using a human specific Lamin A/C antibody (Abcam, Cambridge, UK). Detailed methods are provided in Supplementary Methods.
Statistical Analysis
Overall survival was computed using the Kaplan-Meier method. The log rank test was used to compare survival across groups. The chi-square test (exact) or the two-sample t-test (rank sum) were used to compare characteristics across groups.
Genomic Analyses
DNA was extracted from frozen flank PDX tissue. Hotspot mutations in IDH1/2 and TERT promoter were assessed by Sanger sequencing and MGMT promoter methylation by methylation specific PCR (10,11). WES was performed at Mayo Clinic or Translational Genomics Institute (TGen) using the SureSelect (Agilent, Santa Clara, CA) or TGen Strexome V2 capture kits. When available, WES of patient germline (n=55) and patient tumor tissue (n=24) was performed. Mouse sequencing reads were removed using Xenome (12) prior to mutation calling. CNVs were detected using PatternCNV (13). Genome wide methylation profiling was performed at MD Anderson Cancer Center using the MethylationEPIC BeadChip Array (Illumina, San Diego, CA). RNA was extracted from orthotopic PDX tissue and RNAseq performed at Columbia University, Mayo Clinic, or TGen using TrueSeq libraries (Illumina). PDXs were assigned to glioblastoma molecular subgroups based on RNAseq (14) or methylation data (15). Further details are available in Supplementary Methods.
Therapy Response
Mice with established orthotopic tumors were randomized in groups of 5–10 mice and treated with placebo, radiation therapy (RT), temozolomide (TMZ), concomitant RT and TMZ, or bevacizumab. TMZ was administered by oral gavage (66 or 50 mg/kg daily × 5 days). RT [20 Gy total] was delivered at 2 Gy in 10 fractions. Bevacizumab was administered by intraperitoneal injection at a dose of 5 mg/kg twice weekly until moribund. For comparison of patient and PDX survival, patients receiving additional experimental therapies are grouped with the standard therapy they received. PDX survival was matched with the treatment received by the corresponding patient (RT or RT/TMZ) and association assessed using Spearman’s correlation.
RESULTS
Patient population and PDX establishment
Ninety-four PDX lines were established from 261 glioma patients undergoing surgery at Mayo Clinic between May 2000 and May 2017. An additional two PDX (GBM15 and GBM16) were obtained as established lines. PDX were generated through direct implantation of fresh tumor tissue into the flank of athymic nude mice (9). The overall rate of successful engraftment was 36% (Table 1A). However, this included 59 lower-grade (WHO grades II-III) glioma samples from which no viable PDX were established (Table S1). The engraftment rate for WHO grade IV gliomas was 47% and viable PDXs were generated from IDH-wildtype glioblastoma (n=91), IDH-mutant glioblastoma (n=2), and diffuse midline glioma, H3 K27M-mutant (n=1). Successful PDX were established from both newly diagnosed (n=60) and recurrent (n=34) tumors, with engraftment rates of 50% and 42%, respectively (Table 1B). Two PDX lines were derived from recurrent tumors from the same patient (Table S2). Seven recurrent tumors arose in progression from previously diagnosed lower-grade gliomas, but all PDX were IDH-wildtype. All patients with recurrent tumors received therapy prior to PDX engraftment, most receiving standard radiation therapy and temozolomide with or without other therapies (Table 1B, S2).
Table 1.
| A: Characteristics of all gliomas (WHO grades II-IV) implanted | ||
| Viable (N=94) | Non-viable (N=167) | |
| WHO Histologic Grade (at Implant) | ||
| II | 0 | 9 (5) |
| III | 0 | 50 (30) |
| IV | 94 (100) | 107 (64) |
| Indeterminate | 0 | 1 (1) |
| Diagnosis† | ||
| Glioblastoma, IDH-mutant (WHO IV) | 2 (2) | 11 (7) |
| Glioblastoma, NOS* (WHO IV) | 0 | 37 (22) |
| Glioblastoma, IDH-wildtype (WHO IV) | 91 (97) | 59 (35) |
| Gliosarcoma‡ | 4 | 3 |
| Glioblastoma with primitive neuronal component‡ | 1 | 0 |
| Diffuse midline glioma, H3 K27M-mutant (WHO IV) | 1 (1) | 0 |
| Other diagnoses** | 0 | 60 (36) |
| B: Patient characteristics of WHO grade IV gliomas implanted | ||
| Viable N=94 | Non-Viable N=107 | |
| Mean age, years (range) | 58.3 (21 – 84) | 56.5 (16 – 85) |
| Male, n (%) | 57 (61) | 65 (61) |
| Tumor Type Implanted | ||
| Primary | 60 (64) | 60 (56) |
| Recurrent | 27 (29) | 40 (37) |
| Recurrent in progression from lower-grade | 7 (7) | 7 (6) |
| Therapy prior to engraftment | ||
| None | 60 (64) | 60 (56) |
| RT, TMZ | 12 (13) | 20 (19) |
| RT, TMZ, other therapy | 19 (20) | 25 (23) |
| RT | 0 | 0 (0) |
| Other therapy combination | 3 (3) | 2 (2) |
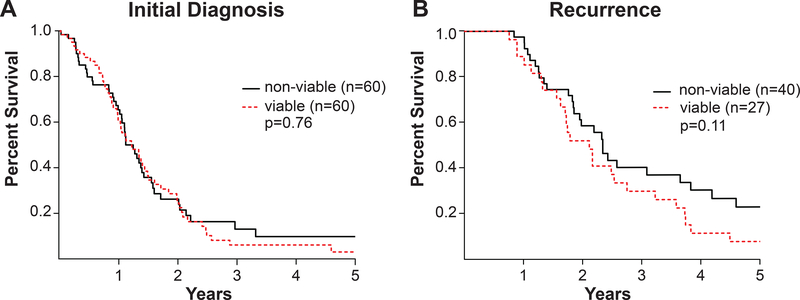
As no PDX engrafted from WHO grade II-III gliomas, analysis of clinical parameters was limited to WHO grade IV tumors. Clinical characteristics were similar for patients with grade IV astrocytomas forming viable or non-viable xenografts. There was no association of patient age, gender, or other clinical parameters with successful PDX engraftment (Table 1B). No overall survival difference was observed for patients with newly-diagnosed glioblastoma forming viable vs. non-viable xenografts (p=0.76; Fig. 1A). For recurrent tumors, associations were less clear as patient overall survival was slightly shorter for viable tumors (p=0.11; Fig. 1B) and engraftment rates were lower than for primary tumors (42% vs. 50%, p=0.26), although not statistically significant. As recurrent tumors frequently show therapy-related necrosis, this could reflect mixed tumor viability in the post-treatment resection specimens. Taken together, these findings suggest that while PDX engraftment is limited to WHO grade IV gliomas, the established PDXs are representative of this patient population and engraftment is not associated with clinically more aggressive primary tumors within this group.
Figure 1: Patient overall survival.
(A-B) Overall survival of patients with grade IV astrocytomas forming viable vs. non-viable xenografts. Kaplan-Meier plots for (A) newly diagnosed and (B) recurrent grade IV astrocytomas. p-value, log-rank test.
Phenotypic characterization of PDX lines
The PDXs have undergone extensive phenotypic characterization. For all viable PDXs, tissue provenance was established using short tandem repeat (STR) genotyping of PDX tissue and compared with patient blood and patient tumor tissue when available (Table S3A). Flank tumor growth rates were assessed and showed considerable variability between different PDX lines, with the median time to exceed 2 cm3 ranging from 20 to 189 days (Table S3B). Explanted flank tumor tissue could be used to establish short-term cultures for all PDX lines, either in standard cell culture media containing fetal bovine serum or in BTIC neural stem cell media supplemented with EGF/FGF (Table S3B). Cultures were utilized within one to two passages to generate orthotopic PDX. Intracranial injection of short-term explant cultured tumor cells generated viable orthotopic tumors in all PDX lines tested (Table S3B). Time to moribund for orthotopic tumors varied between lines from 20 to 180 days. The cataloging of PDX growth characteristics is useful for planning in vitro and in vivo studies.
Histopathologic examination demonstrated that orthotopic PDX recapitulate characteristic morphologic features of high-grade glioma (Figure S1). Tumors were cellular with typical astrocytic morphology and varying degrees of nuclear atypia, pleomorphism, and high mitotic activity. However, necrosis and microvascular proliferation, diagnostic features of glioblastoma, were identified in less than 20% of orthotopic PDX and were typically present as focal or early changes. This was in keeping with a detailed morphologic study of four early PDX lines, which found no evidence of microvascular proliferation or necrosis in orthotopic PDX (6). However, more than 80% of flank PDX demonstrated palisading necrosis, which might suggest that limited tumor size within the mouse brain may lessen appearance of this feature. Five PDX lines were established from glioblastoma histologic variants, including one glioblastoma with primitive neuronal component and four gliosarcoma and the PDX closely recapitulated the patient tumor morphology (Figure S2). Expression of GFAP was assessed in 12 patient tumors and derivative PDX (Figure S3) and was concordant in 11 pairs. Notably, tumors derived from gliosarcoma or glioblastoma with primitive neuronal component lacked GFAP expression or showed partial loss (Figure S3), in keeping with the typical histologic features of these glioblastoma variants.
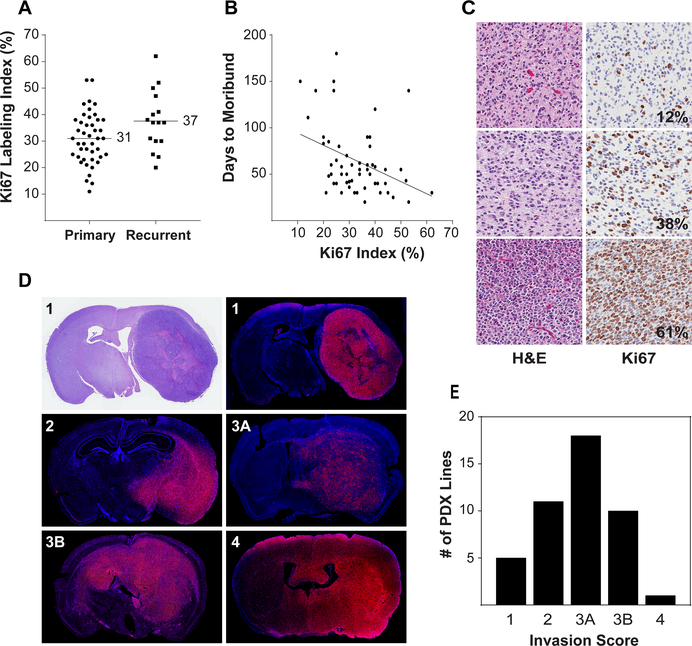
Proliferation was assessed by Ki67 immunohistochemistry using a tissue microarray (TMA) generated from 58 orthotopic PDXs. The Ki67 labeling index ranged from 11 to 62% overall (mean 33%) (Figure 2A, 2C). Ki67 was marginally higher in recurrent (37%) than primary tumors (31%, p=0.07) and was inversely correlated with time to moribund (Figure 2B). To assess invasiveness, 45 orthotopic PDXs were examined by immunofluorescence (Figure 2D). The majority of (n=40) demonstrated infiltration of the brain parenchyma, with most (n=29) showing contralateral hemisphere involvement (Figure 2E). This range of invasion parallels imaging findings in glioblastoma patient populations (16). Blood-brain barrier integrity is a key aspect of CNS drug delivery. To date, the blood-brain barrier has been assessed in a limited number of PDX models and demonstrated varying degrees of integrity (Figure S4), from intact (GBM6) to partial (GBM108) or more complete (GBM39) disruption. A detailed assessment of blood-brain barrier integrity in additional PDX models is planned.
Figure 2: Proliferation and invasiveness of orthotopic PDX.
(A) Distribution of Ki67 labeling in PDX derived from primary and recurrent tumors. Ki67 labeling index was determined from tissue microarray cores from 58 orthotopic PDX lines. A mean of 4.5 cores was analyzed from one (n=4) or two (n=54) independent orthotopic tumors for each line. (B) Inverse correlation of Ki67 labeling with time to moribund following intracranial injection of tumor cells. (C) Representative H&E and Ki67 staining for PDX with low, moderate, and high Ki67 labeling indexes. (D) Tumor cell invasion was assessed by immunofluorescence using human specific antibodies to Lamin A/C. Representative sections showing invasion pattern scoring: 1 = unilateral, well-demarcated tumor; 2 = unilateral with infiltrating border; 3A = bilateral, restricted to contralateral midline structures; 3B = bilateral with clear involvement of both hemispheres; 4 = diffuse infiltration bilaterally. (E) Distribution of invasion patterns for 55 orthotopic PDX.
Genomic Profiling of PDX tumors
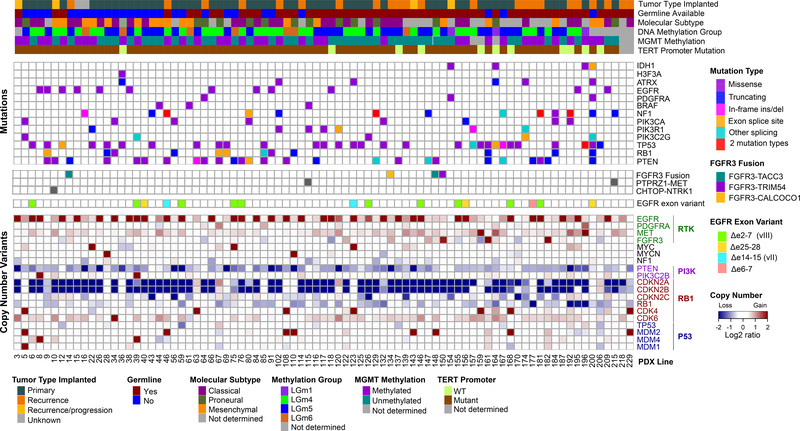
The PDX lines have undergone extensive genomic characterization, including whole exome sequencing (WES) (n=83), RNA sequencing (n=68), and genome-wide methylation profiling (n=76). To a high degree, the PDXs capture the genetic heterogeneity of IDH-wildtype glioblastoma, with the majority of known glioblastoma driver alterations represented (Figure 3) at a frequency roughly similar to the TCGA glioblastoma dataset (8) (Table S4). The majority of PDX showed gain of chromosome 7, as whole chromosome (n=60) or partial (n=15) gain, and loss of chromosome 10, as whole (n=62) or partial (n=14) loss. Alterations of EGFR were frequent and diverse, with one or more alterations identified in 37 PDX lines (45%), most commonly high-level amplification (n=35). A total of 14 EGFR single nucleotide variants (SNVs) were identified in 12 PDX lines, with concurrent EGFR amplification in 11 of the 12 lines. EGFR exon deletion variants were present in 16 PDX, including EGFRvIII (n=11, Δexon2–7), EGFRvII (n=2, Δexon14–15), c-terminal deletions (n=2, Δexon25–27), and a previously uncharacterized Δexon6–7 variant (n=1). All exon deletion variants were identified in EGFR amplified PDXs, with the exception of GBM170, which showed EGFRvIII in the setting of trisomy 7. By immunohistochemistry, EGFR expression was strongly correlated with copy number (Figure S5). In contrast, although present in approximately 15% of human glioblastoma, no PDX lines showed PDGFRA amplification. PDGFRA SNVs were identified in three PDX, including the oncogenic variant D842V. Despite the lack of PDGFRA CNVs, a high proportion of PDX demonstrated strong (29%) or moderate (24%) expression of PDGFRA (Figure S5). Less common RTK pathway alterations were also represented, including amplification of MET (n=2) and FGFR3 (n=1). Overall, with the exception of PDGFRA amplification, the PDX panel provides a broad representation of RTK alterations typical of glioblastoma.
Figure 3: Integrated genomic characterization of 83 PDX lines.
Mutations and copy number variants (CNVs) from WES are shown for core glioma genetic drivers. When available (n=55), patient germline variants were subtracted. Molecular gene expression subtype was determined from RNAseq (14). DNA methylation group was determined from genome wide methylation profiling according to TCGA pan-glioma classification (15). MGMT promoter methylation was assessed by quantitative methylation-specific PCR performed at Mayo Clinic. TERT promoter mutations (C228T and C250T) were detected by Sanger sequencing.
Similarly, other driver alterations paralleled human glioblastoma (Figure 3). TERT promoter mutations were the most frequent overall (86%) followed by homozygous deletion of CDKN2A (n=59, 70%). Multiple alterations were identified in the p53 pathway, including TP53 mutations (n=30, 36%) and MDM2 (n=8, 10%) or MDM4 (n=2, 2%) amplification. PI3-kinase pathway alterations involved PTEN (n=40, 48%) or other PI3K genes (Table S4). The retinoblastoma pathway was inactivated by RB1 loss/mutation (n=13, 16%) or amplification of CDK4 (n=7, 8%) or CDK6 (n=1, 1%). MAP kinase pathway alterations were present in a subset of PDX, including alterations of NF1 (n=14, 17%) and BRAF (n=3, 4%). Several PDX harbored amplification of MYC (n=3) or MYCN (n=2). The frequencies of these alterations are comparable to human glioblastoma.
Three PDX lines were derived from tumor types other than IDH-wildtype glioblastoma. GBM36 was engrafted from a cerebellar diffuse midline glioma harboring H3F3A p.K27M and mutations of ATRX and TP53. Two PDX lines, GBM164 and GBM196, were established from IDH-mutant glioblastoma. Both demonstrated SNVs of TP53 as well as homozygous deletion of CKDN2A, a marker of poor prognosis in IDH-mutant gliomas (17). Both lines also showed copy number gains involving MET, representing a potentially targetable alteration described in a high proportion of secondary glioblastoma (18). Taken together, these data suggest that PDX capture both common and rare genetic alterations characteristic of glioblastoma.
Genomic analysis included 28 PDXs established at recurrence. Overall, PDX derived from primary and recurrent tumors showed similar mutation burden (Figure S6). However, four PDXs derived from recurrent tumors harbored multiple mutations of the mismatch repair gene MSH6 (Table S5A) and a high mutation burden, consistent with a hypermutation phenotype (19). In addition to MSH6, mutations of LTBP4, IGF1R, and PRDM2 have been reported to be limited to recurrent glioblastoma (20). We identified variants of LTBP4 (4/28, 14%), IGF1R (2/28, 7%), and PRDM2 (2/28, 7%) in a similar proportion of our recurrent PDX models (Table S5B) as previously described (20). However, alterations of these genes were also identified in PDXs derived from primary tumors, although they may represent rare germline rather than somatic variants. GBM154 was a particularly unusual recurrent tumor, which had arisen 10 years after the initial diagnosis and treatment of a low-grade oligdendroglioma. Targeted next-generation sequencing of the initially diagnosed oligodendroglioma showed IDH1 p.R132H, TERT promoter, CIC, FUBP1, and CHEK2 mutations (Table S5C) and 1p/19q co-deletion. The GBM154 PDX, which was established at tumor recurrence, lacked IDH1 p.R132H but harbored a new IDH1 p.F397L mutation and retained only the 1p/19q co-deletion, CHEK2, and TERT promoter mutations. IDH1 loss has been recently described on recurrence and high-grade transformation of IDH-mutant gliomas (21). Thus, PDX capture unique genetic changes associated with tumor recurrence and prior therapy.
Gene expression was assessed by RNAseq in 68 orthotopic PDX (Figure 3). PDX were assigned to one of three glioblastoma gene expression subgroups based on an updated classification scheme, which excludes the neural subtype (14). PDX showed classical (n=31, 46%), mesenchymal (n=21, 31%), or proneural (n=16, 23%) expression subtypes (Table S6A). In 59 tumors with both WES and RNAseq, the classical expression subgroup was enriched for tumors with EGFR amplification (n=21, 78%) relative to the mesenchymal (n=5, 29%) and proneural groups (n=1, 7%). Principal component analysis showed grouping of tumors relative to expression subtype (Figure S7). These patterns of gene expression are comparable to what is observed in human glioblastoma. Analysis of expressed fusion transcripts identified the recurrent FGFR3-TACC3 fusion in three PDX lines (GBM14, GBM148, GBM232) as well as two non-canonical FGFR3 fusion transcripts, FGFR3-TRIM54 and FGFR3-CALCOCO1 (Table S6B, Figure 3). Additionally, previously reported fusions of PTPRZ1-MET and CHTOP-NTRK1 were identified in three PDX and represent potentially targetable alterations with specific inhibitors in development or clinical trials (18,22).
Epigenetic characterization included genome-wide DNA methylation profiling (n=76) and assessment of MGMT promoter methylation by methylation-specific PCR (n=78). Based on genome-wide methylation data, PDX were assigned to one of six TCGA pan-glioma methylation subclasses, LGm1-LGm6 (Figure 3 and Table S6A) (15). Three PDX were assigned to the IDH-mutant LGm1 group, the two IDH-mutant PDXs (GBM164 and GBM196) and GBM154, which arose from an oligodendroglioma. All others were assigned to one of three IDH-wildtype methylation classes: LGm4 (n=34, 46%), LGm5 (n=32, 44%), or LGm6 (n=7, 10%). However, principal component analysis did not clearly delineate tumors relative to methylation class (Figure S7). This may reflect the predominance of IDH-wildtype tumors, which are not fully separated by methylation profiling. Additionally, the inability to remove artifacts from contaminating mouse stroma and analysis at higher PDX passage number may more adversely affect the methylation data. MGMT promoter methylation was assessed in our laboratory and the CLIA laboratory at MD Anderson Cancer Center (Figure 3, Table S6A). Excluding indeterminate results, MGMT methylation status was concordant between laboratories for 82% (56 of 68) of PDXs. MGMT methylation was present in 45% (35 of 78) of PDX analyzed at Mayo and 55% (42 of 77) analyzed at MD Anderson. For 22 patients, MGMT methylation was assessed during clinical care; the majority of testing was performed at LabCorp (Burlington, NC). In 20 cases with MGMT assessed for both patient and PDX (Mayo analysis), 19 showed concordant methylation (Table S6A). Thus, PDXs reflect the epigenetic spectrum of glioblastoma.
Response to Standard Therapy
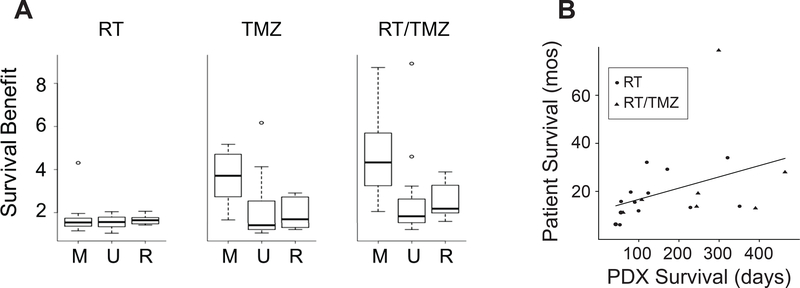
PDX models have particular utility for pre-clinical studies addressing response to therapy. To assess if PDXs recapitulate human treatment response, mice with established orthothotopic tumors from 37 PDX lines were treated with radiation therapy (RT), temozolomide (TMZ), combination RT/TMZ, or bevacizumab (Figure 4A, Table S7). Detailed response data from 20 PDX lines to RT and TMZ have been previously published (23). Across the 37 PDXs, RT and TMZ extended survival, with a median survival ratio relative to placebo of 1.6 for RT, 2.5 for TMZ monotherapy, and 2.8 for concurrent RT/TMZ. MGMT promoter methylation was strongly associated with response to RT/TMZ or TMZ alone, but did not correlate with response to RT alone. Single agent Bevacizumab showed a survival ratio greater than 1.3 in 23% of PDX (7 of 30) (Table S7). A positive correlation (ρ=0.53) was observed between patient and PDX survival following treatment with standard therapies (Figure 4B). Overall, these findings suggest that PDX reflect patient treatment response.
Figure 4: Response to standard therapies.
(A) Survival benefit across PDX models treated with RT, TMZ and RT/TMZ. Mice with established orthotopic tumors from 37 PDX lines were randomized to treatment with RT, TMZ, or RT/TMZ. Survival benefit was calculated as a ratio of survival of the treated mice to placebo treated mice. PDXs are grouped by MGMT status: M=MGMT methylated (N=14), U=MGMT unmethylated (N=14) and R=recurrent (N=9). (B) Comparison of patient and PDX survival following standard therapies for 20 matched pairs with xenografts established at initial diagnosis. Patient survival is shown in months and PDX survival in days.
Genomic Comparison of Patient and Derivative PDX
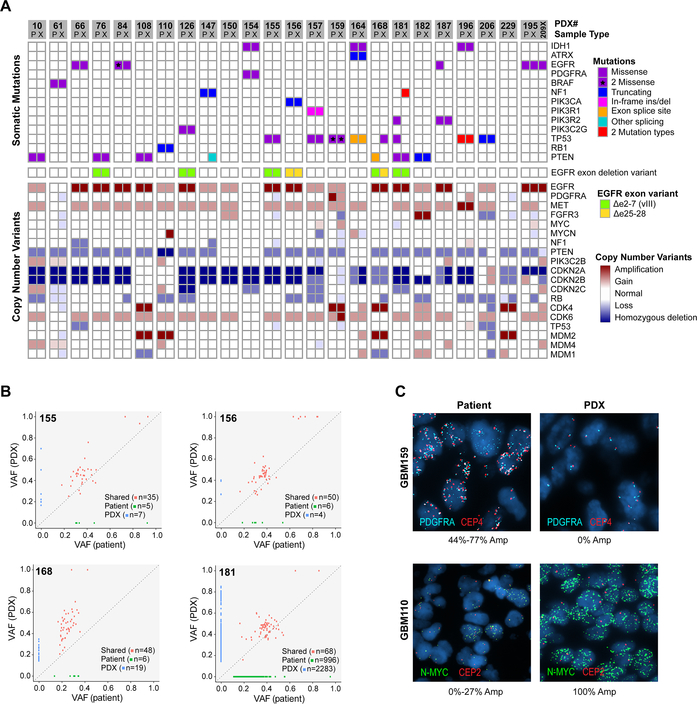
WES was performed for 24 matched PDX and patient tumors with available frozen tumor tissue. Overall, PDX showed a high degree of similarity to their parent tumors, preserving the vast majority of driver alterations (Figure 5). In all pairs, EGFR amplification was concordant between patient and PDX. This was confirmed by fluorescence in situ hybridization (FISH) of four matched patient-PDX pairs, which demonstrated high-level EGFR amplification in all tumor cells from both patient and matched PDX (Figure S8). MDM2 amplification was also identified in 100% of GBM108 patient and PDX cells (Figure S8). However, several tumor pairs showed genetic differences between patient and PDX. In two EGFR amplified patient-PDX pairs (GBM 84 and GBM187), an EGFR SNV was detected in the patient tumor that was absent from the derivative PDX. Two PDX lines (GBM164 and GBM187) showed CDKN2A homozygous loss compared to single copy loss in the patient sample (Figure 5). Four PDXs showed more significant divergence from the primary patient tumors. The GBM168 patient and PDX shared amplifications of EGFR, CKD4, and MDM2 (Figure 5), however, different EGFR exon deletion variants were identified. The patient sample also harbored a private PTEN alteration and the PDX a private TP53 variant. In the GBM110 patient-PDX pair, MYCN amplification was detected only in the PDX sample, while other alterations, including MDM2 amplification and PTEN homozygous deletion, were identified in both patient and PDX. FISH of matched patient and PDX tumors demonstrated amplification of N-MYC in a subset of patient tumor cells (0 to 27%) and in 100% of PDX tumor cells (Figure 5C). In contrast, MDM2 was amplified in 100% of both patient and PDX cells (Figure S8). In GBM159, the patient and PDX shared multiple alterations, including CDK4 amplification and two TP53 SNVs. However, PDGFRA amplification was detected by WES only in the patient sample and CDK6 amplification only in the PDX. FISH of the patient tumor demonstrated PDGFRA amplification in approximately 45–75% of tumor cells. Surprisingly, two distinct PDGFRA amplified subclones could be distinguished by co-amplification of the centromeric probe (Figure 5C). Neither amplified subclone was detected in the PDX tumor, although a subset of PDX cells showed additional copies of PDGFRA. In contrast, GBM181 patient and PDX showed many private SNVs, a pattern consistent with MSH6 mutation and an MMR-deficient hypermutation phenotype (Table S5). In summary, 4 of 24 PDX demonstrated notable differences with the original patient specimen, while other pairs were largely concordant.
Figure 5: Comparison of somatic alterations in 24 matched patient tumors and derivative PDX.
(A) Somatic mutations and CNVs involving core glioma associated genes. Matched patient (P) and derivative PDX (X) models are adjacent for each pair. The GBM195 patient-PDX pair is also shown with GBM209 (209X), a PDX line established from the patient’s subsequent tumor recurrence. (B) Representative scatterplots comparing variant allele frequency (VAF) of four patient-PDX pairs. The four selected pairs reflect the spectrum of SNV conservation across the matched patient-PDX pairs. (C) FISH of matched patient and PDX tissue showing subclonal amplifications of N-MYC (GBM110) and PDGFRA (GBM159). Two distinct PDGFRA amplified subclones could be distinguished in the patient tumor by involvement of the centromeric probe (CEP4). %Amp denotes the percentage of tumor cells with >8 copies of the target probe with range given across 3 patient tumor blocks.
The concordance of SNVs for all coding regions varied across the 24 patient-PDX pairs. Representative pairs are shown in Figure 5B. Private mutations were identified in both patient and PDX samples, but were predominantly present at low VAF. There were no recurrent SNVs private to patient or PDX samples. CNVs were also gained and lost during PDX engraftment, often at subclonal frequencies (Figure 5A). However, there were no consistent patterns of CNV change. In the setting of preserved genetic driver alterations, these changes likely represent passenger alterations and are of unclear biological significance. Overall, PDX show a high degree of similarity to the patient tumor from which they derive or show changes suggestive of clonal selection.
DISCUSSION
Patient-derived tumor models are increasingly utilized in neuro-oncology research for pre-clinical and translational studies. However, the extent to which these models faithfully recapitulate human gliomas has remained an open question. We have undertaken extensive genomic and phenotypic characterization of a large panel of 96 glioblastoma PDX models. Unsurprisingly, successful PDX establishment was limited to grade IV astrocytomas with the vast majority (93 of 96) being IDH-wildtype glioblastoma. Exactly 50% of newly diagnosed glioblastoma formed viable xenografts (Table 1 & S2). However, the factors determining successful PDX engraftment were enigmatic. Although PDX engraftment was limited to grade IV astrocytomas, within this group, there was no difference in overall survival, age or other characteristics between patients with primary tumors forming viable vs. non-viable xenografts (Figure 1, Table 1), suggesting that successful engraftment does not correlate with more aggressive tumors. As other groups have reported highly variable engraftment rates (24,25), technical factors are likely a strong contributor to PDX viability. A variety of factors may affect engraftment rates, including mouse strain and injection site (intracranial vs. flank) as well as time from tissue acquisition to injection, tissue size, and extent of necrosis. Our approach of serial heterotopic tumor passage was established based on early experience and has many practical advantages, including the ease of PDX establishment and maintenance, and transferability to less experienced laboratories. Although initially established in flank, all of our PDX models tested to date have generated successful orthotopic tumors that closely resemble human astrocytomas histologically (Figure S1–S2). While some early studies suggested that flank xenografts form non-infiltrative tumors (26), the majority of our PDXs formed highly infiltrative tumors when injected intracranially (Figure 2). However, necrosis and microvascular proliferation were infrequent in orthotopic PDX, in keeping with previous studies. Thus, establishment of flank xenografts is a practical and versatile approach, generating PDX models that largely recapitulate human gliomas.
The 96 glioblastoma patient-derived models described herein is the largest and most thoroughly characterized collection of glioma PDXs published to date. Previously, a panel of 53 glioblastoma PDXs established at Samsung Medical Center were shown to recapitulate histopathologic features of the corresponding patient tumors (24). However, genetic characterization of the PDXs was limited to a subset of glioma-associated genes (24). Recent studies have also described smaller, more extensively annotated libraries of glioblastoma PDX (27). In the present study, genomic analyses of 83 PDX lines demonstrate that, to a remarkable degree, our PDX library captures the diverse genetic alterations typically associated with glioblastoma (Figure 3). Notably, a range of EGFR alterations were identified in our models, including high-level amplifications, oncogenic missense mutations, and four distinct exon deletion variants. Consistent with human glioblastoma (8), the PI3-kinase, p53 and Rb pathways were dysregulated by a variety of genetic mechanisms in the majority of PDXs. Additionally, several uncommon genetic alterations were captured by our panel, including BRAF mutations, amplification of MYC and NMYC, as well as several recurrent glioblastoma with a MMR-deficient hypermutation phenotype. Although the engraftment rate was low (15%) for IDH-mutant glioblastoma, two IDH-mutant PDX were established. However, a striking exception was an absence of PDGFRA amplified PDX, which is an alteration with an expected frequency of approximately 15% in IDH-wildtype glioblastoma. This may reflect an inability of PDGFRA amplified tumors to form viable xenografts or, alternatively, selection pressure may be insufficient to maintain PDGFRA amplification during serial PDX passage. In keeping with this latter possibility, early passage GBM5 PDX was described as harboring PDGFRA amplification (6), however, the alteration was no longer detected by WES at passage eight. Nonetheless, a high proportion of PDX demonstrated moderate or strong PDGFRA expression (Figure S5), suggesting other mechanisms are still operative that can modulate expression. RNAseq and methylation profiling showed broad representation of glioblastoma molecular subgroups and both MGMT methylated and unmethylated tumors (Figure 3). Taken together, these data demonstrate that our panel generally captures the diversity of glioblastoma at the population level.
At the level of individual models, WES of 24 matched patient-PDX pairs showed high concordance between patient tumors and derivative models. EGFR amplification was preserved in all models, assessed by both WES and FISH (Figure 5). This finding is significant as EGFR amplification is often lost during glioma cell culture as was observed in two recent studies comparing the genomic features of matched patient and BTIC models (28,29). When changes in core genetic pathways were observed in our PDXs, the patterns were suggestive of clonal selection events rather than acquisition of de novo alterations (Figure 5). EGFR exon deletion variants (30) and RTK amplifications, particularly PDGFRA (31), are known to show intra-tumoral heterogeneity. In keeping with this, by FISH, N-MYC amplification (GBM110) and PDGFRA amplification (GBM159) were subclonal alterations, which underwent positive and negative selection, respectively, upon PDX engraftment. These findings parallel a recent study comparing 12 glioblastoma patient tumors with derivative models, which showed that extrachromosomal oncogene amplifications, present at variable levels in patient tumors, could undergo rapid selection during PDX engraftment (32). Clonal selection in PDX may occur either through a genetic bottleneck during engraftment or due to differing selection pressure in the mouse tumor microenvironment. Studies of breast cancer xenografts suggest the latter, showing initial preservation of intratumoral heterogeneity during PDX engraftment (33).
While core pathways were largely preserved in our models, a variable number of private alterations were identified in patient or PDX. The significance of these alterations in unclear as the private alterations were non-recurrent and largely subclonal (Figure 5). Additionally, it is likely that PDX undergo further evolution and clonal selection across serial passage, a concern which our study does not directly address. To date, few studies have examined ex vivo evolution in glioblastoma PDX or in other tumor types. However, one recent study compiled copy number data, predominantly extrapolated from gene expression profiling, across PDX from multiple tumor types and showed rapid acquisition of CNV changes across serial passage (34). Of note, glioblastoma PDX showed the lowest rate of acquired CNVs of the 13 tumors types examined in the study. CNV changes are not unique to PDX and have been observed over extended passage of BTIC models (35). However, in the setting of preserved core genetic alterations, the biological impact of these acquired alterations is uncertain. PDX growth kinetics have also been shown to vary over extended PDX passage, typically with increased growth rate over time (9). Further studies are needed to address fidelity across extended passage of both PDX and cell culture models. As such, the approach taken by our laboratory is to minimize genetic and phenotypic changes by restoring early passage PDX from frozen stocks (9). Nonetheless, we would recommend verifying the alterations of interest prior to subsequent studies.
Diverse patient-derived model libraries are particularly valuable for pre-clinical studies defining therapeutic benefit in individuals and the overall population. In the current and previous work (23), we have characterized PDX response to standard glioblastoma therapy, including RT and TMZ (Figure 4A and Table S7). The largest survival benefit for TMZ and RT/TMZ was seen in tumors with MGMT promoter methylation, consistent with results from human clinical trials (36). A positive correlation was observed between patient and PDX response to therapy (Figure 4B). Testing targeted therapies, at the individual level, models have been used as “avatars” to tailor treatment for patients based on response in a derivative model (37,38), with good correlation between patient and model (37). At the population level, panels of patient-derived models are increasingly utilized in preclinical studies to predict therapeutic response across a heterogeneous population. For testing novel therapies, a new framework for preclinical trials has been developed, which more closely mimics a phase II clinical trial (39). Therapies are tested across a diverse panel of PDX models with a single or small number of animals per treatment per PDX line (1×1×1 design) (40). This approach does not require that the models perfectly match the patient tumors, but rather that the panel reflects the disease population as a whole. A comprehensive study evaluated this design across 62 therapies in 6 tumor types and demonstrated a strong correlation with established molecular biomarkers and development of known therapy resistance mechanisms (41). In glioblastoma, this approach was used to test the efficacy of the MDM2 inhibitor RG7112 across a panel of 36 patient-derived BTICs and a subset of derivative xenograft models (42). MDM2-amplified tumors were highly sensitive to RG7112 while TP53-mutant tumors were inherently resistant. A similar study in our laboratory investigated combined temozolomide and veliparib in a panel of 28 orthotopic PDX models (43). Efficacy was limited to tumors with MGMT promoter methylation, a finding that has directed the biomarker enrichment strategy in the Alliance Phase II/III clinical trial testing the benefit of adding veliparib to adjuvant temozolomide (). These studies exemplify how large panels of glioblastoma patient-derived models can be used to develop predictive biomarkers a priori to identify and enroll on clinical trials only those patients most likely to benefit from targeted therapies.
The goal of the Mayo PDX National Resource is to provide an extensively annotated panel of glioblastoma PDX models as a freely available resource to the neuro-oncology research community. Similar efforts have been undertaken across a range of tumor types, (33,40,44,45), including a recently described panel of pediatric brain tumor models (46). Our panel is the largest and most robustly characterized glioblastoma collection described to date. Annotated genomic data are accessible through cBioPortal (47) with raw sequencing data available through the NCBI Sequence Read Archive (WES Accession PRJNA543854; RNAseq Accession PRJNA548556) and methylation data through Gene Expression Omnibus (GSE132650). Updated information and instructions for obtaining PDX samples are available at the Mayo PDX National Resource website: https://www.mayo.edu/research/labs/translational-neuro-oncology/mayo-clinic-brain-tumor-patient-derived-xenograft-national-resource. PDX models are provided at minimal cost to investigators and have been shared extensively, being utilized in over 150 peer-reviewed papers (Supplemental Bibliography) and numerous grant applications. Like any model system, PDX are not perfect. They are limited by the use of immunocompromised mice, potential differences within the human/mouse tumor microenvironment, and variable genetic drift over extended passage. Regardless, patient-derived models remain the most practical and clinically relevant model system for many preclinical and translational studies, particularly those investigating novel therapeutics and therapy combinations. Large, genetically heterogeneous glioma PDX panels are uniquely suited to address two important factors contributing to the lack of effective new drugs for glioblastoma—the inter-tumoral genetic heterogeneity of glioblastoma and issues of drug delivery to the CNS. The Mayo PDX National Resource has the potential to greatly accelerate studies addressing these questions and is a powerful tool for neuro-oncology research.
Supplementary Material
Translational Relevance.
Development of new and molecularly targeted therapies for glioblastoma has lagged behind other tumor types. Glioblastoma presents unique challenges due to its inherent molecular heterogeneity, infiltration of normal brain, and presence of the blood brain barrier. Clinically relevant glioma models are essential for pre-clinical and translational studies aimed at testing novel therapeutics and identifying biomarkers predictive of response. To this end, we have established a broad panel of 96 glioblastoma patient derived xenograft (PDX) models and characterized its genomic and phenotypic features. While PDX engraftment was limited to WHO grade IV astrocytomas, the established PDX lines reflect patient characteristics and capture the heterogeneity of glioblastoma, with representation of most major genetic driver alterations and molecular subgroups. All PDX models and accompanying genomic data are openly available to investigators and been shared extensively. Our PDX panel has the power to greatly expedite neuro-oncology research.
Acknowledgements
We are grateful for the services of the Mayo Clinic Cytogenetics Core Laboratory which contributed to this manuscript. The Core is supported, in part, by the Mayo Clinic Comprehensive Cancer Center Grant, funded by National Cancer Institute (P30CA15083). We are also grateful for the Mayo Clinic Medical Genome Facility Genome Analysis Core for contribution to this manuscript.
Financial Support: This work was supported by the Mayo SPORE in Brain Tumors (CA108961), RO1 CA184320, U01 CA227954, R24 NS092940, Accelerate Brain Cancer Cure, and the Mayo Clinic. DAR was supported by R25 HL92621 and Alpha Omega Alpha Carolyn L. Kuckein Student Research Fellowship. AC was supported by R35 CA197745 and U01 CA217858.
Conflicts of Interest: JNS has research funding from Novartis, Sanofi-Aventis, Merck, Genentech, BeiGene, GlaxoSmithKline, Eli Lilly and Company, Curtana Pharmaceuticals, Basilea Pharmaceutica, and Peleton Therapeutics. No authors have a personal financial interest in the PDX models. Mayo Clinic does have a financial interest in the PDX models and may stand to gain from the licensing of these models.
Footnotes
Dr. Califano is founder, equity holder, consultant, and director of DarwinHealth Inc., a company that has licensed some of the algorithms used in this manuscript from Columbia University. Columbia University is also an equity holder in DarwinHealth Inc.
References
- 1.Allen M, Bjerke M, Edlund H, Nelander S, Westermark B. Origin of the U87MG glioma cell line: Good news and bad news. Sci Transl Med 2016;8(354):354re3 doi 10.1126/scitranslmed.aaf6853. [DOI] [PubMed] [Google Scholar]
- 2.Torsvik A, Stieber D, Enger PO, Golebiewska A, Molven A, Svendsen A, et al. U-251 revisited: genetic drift and phenotypic consequences of long-term cultures of glioblastoma cells. Cancer Med 2014;3(4):812–24 doi 10.1002/cam4.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lenting K, Verhaak R, Ter Laan M, Wesseling P, Leenders W. Glioma: experimental models and reality. Acta Neuropathol 2017;133(2):263–82 doi 10.1007/s00401-017-1671-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 2006;9(5):391–403 doi 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 5.Schulte A, Gunther HS, Martens T, Zapf S, Riethdorf S, Wulfing C, et al. Glioblastoma stem-like cell lines with either maintenance or loss of high-level EGFR amplification, generated via modulation of ligand concentration. Clin Cancer Res 2012;18(7):1901–13 doi 10.1158/1078-0432.CCR-11-3084. [DOI] [PubMed] [Google Scholar]
- 6.Giannini C, Sarkaria JN, Saito A, Uhm JH, Galanis E, Carlson BL, et al. Patient tumor EGFR and PDGFRA gene amplifications retained in an invasive intracranial xenograft model of glioblastoma multiforme. Neuro Oncol 2005;7(2):164–76 doi 10.1215/S1152851704000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodgson JG, Yeh RF, Ray A, Wang NJ, Smirnov I, Yu M, et al. Comparative analyses of gene copy number and mRNA expression in glioblastoma multiforme tumors and xenografts. Neuro Oncol 2009;11(5):477–87 doi 10.1215/15228517-2008-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell 2013;155(2):462–77 doi 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlson BL, Pokorny JL, Schroeder MA, Sarkaria JN. Establishment, maintenance and in vitro and in vivo applications of primary human glioblastoma multiforme (GBM) xenograft models for translational biology studies and drug discovery. Curr Protoc Pharmacol 2011;Chapter 14:Unit 14 6 doi 10.1002/0471141755.ph1416s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kitange GJ, Carlson BL, Mladek AC, Decker PA, Schroeder MA, Wu W, et al. Evaluation of MGMT promoter methylation status and correlation with temozolomide response in orthotopic glioblastoma xenograft model. J Neurooncol 2009;92(1):23–31 doi 10.1007/s11060-008-9737-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vlassenbroeck I, Califice S, Diserens AC, Migliavacca E, Straub J, Di Stefano I, et al. Validation of real-time methylation-specific PCR to determine O6-methylguanine-DNA methyltransferase gene promoter methylation in glioma. J Mol Diagn 2008;10(4):332–7 doi 10.2353/jmoldx.2008.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conway T, Wazny J, Bromage A, Tymms M, Sooraj D, Williams ED, et al. Xenome--a tool for classifying reads from xenograft samples. Bioinformatics 2012;28(12):i172–8 doi 10.1093/bioinformatics/bts236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang C, Evans JM, Bhagwate AV, Prodduturi N, Sarangi V, Middha M, et al. PatternCNV: a versatile tool for detecting copy number changes from exome sequencing data. Bioinformatics 2014;30(18):2678–80 doi 10.1093/bioinformatics/btu363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q, Hu B, Hu X, Kim H, Squatrito M, Scarpace L, et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 2017;32(1):42–56 e6 doi 10.1016/j.ccell.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell 2016;164(3):550–63 doi 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baldock AL, Ahn S, Rockne R, Johnston S, Neal M, Corwin D, et al. Patient-specific metrics of invasiveness reveal significant prognostic benefit of resection in a predictable subset of gliomas. PLoS One 2014;9(10):e99057 doi 10.1371/journal.pone.0099057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shirahata M, Ono T, Stichel D, Schrimpf D, Reuss DE, Sahm F, et al. Novel, improved grading system(s) for IDH-mutant astrocytic gliomas. Acta Neuropathol 2018;136(1):153–66 doi 10.1007/s00401-018-1849-4. [DOI] [PubMed] [Google Scholar]
- 18.Hu H, Mu Q, Bao Z, Chen Y, Liu Y, Chen J, et al. Mutational Landscape of Secondary Glioblastoma Guides MET-Targeted Trial in Brain Tumor. Cell 2018;175(6):1665–78 e18 doi 10.1016/j.cell.2018.09.038. [DOI] [PubMed] [Google Scholar]
- 19.Hunter C, Smith R, Cahill DP, Stephens P, Stevens C, Teague J, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res 2006;66(8):3987–91 doi 10.1158/0008-5472.CAN-06-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Cazzato E, Ladewig E, Frattini V, Rosenbloom DI, Zairis S, et al. Clonal evolution of glioblastoma under therapy. Nat Genet 2016;48(7):768–76 doi 10.1038/ng.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mazor T, Chesnelong C, Pankov A, Jalbert LE, Hong C, Hayes J, et al. Clonal expansion and epigenetic reprogramming following deletion or amplification of mutant IDH1. Proc Natl Acad Sci U S A 2017;114(40):10743–8 doi 10.1073/pnas.1708914114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cocco E, Scaltriti M, Drilon A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol 2018;15(12):731–47 doi 10.1038/s41571-018-0113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carlson BL, Grogan PT, Mladek AC, Schroeder MA, Kitange GJ, Decker PA, et al. Radiosensitizing effects of temozolomide observed in vivo only in a subset of O6-methylguanine-DNA methyltransferase methylated glioblastoma multiforme xenografts. Int J Radiat Oncol Biol Phys 2009;75(1):212–9 doi 10.1016/j.ijrobp.2009.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joo KM, Kim J, Jin J, Kim M, Seol HJ, Muradov J, et al. Patient-specific orthotopic glioblastoma xenograft models recapitulate the histopathology and biology of human glioblastomas in situ. Cell Rep 2013;3(1):260–73 doi 10.1016/j.celrep.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 25.Kim KM, Shim JK, Chang JH, Lee JH, Kim SH, Choi J, et al. Failure of a patient-derived xenograft for brain tumor model prepared by implantation of tissue fragments. Cancer Cell Int 2016;16:43 doi 10.1186/s12935-016-0319-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antunes L, Angioi-Duprez KS, Bracard SR, Klein-Monhoven NA, Le Faou AE, Duprez AM, et al. Analysis of tissue chimerism in nude mouse brain and abdominal xenograft models of human glioblastoma multiforme: what does it tell us about the models and about glioblastoma biology and therapy? J Histochem Cytochem 2000;48(6):847–58 doi 10.1177/002215540004800613. [DOI] [PubMed] [Google Scholar]
- 27.Stringer BW, Day BW, D’Souza RCJ, Jamieson PR, Ensbey KS, Bruce ZC, et al. A reference collection of patient-derived cell line and xenograft models of proneural, classical and mesenchymal glioblastoma. Sci Rep 2019;9(1):4902 doi 10.1038/s41598-019-41277-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis B, Shen Y, Poon CC, Luchman HA, Stechishin OD, Pontifex CS, et al. Comparative genomic and genetic analysis of glioblastoma-derived brain tumor-initiating cells and their parent tumors. Neuro Oncol 2016;18(3):350–60 doi 10.1093/neuonc/nov143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenberg S, Verreault M, Schmitt C, Guegan J, Guehennec J, Levasseur C, et al. Multi-omics analysis of primary glioblastoma cell lines shows recapitulation of pivotal molecular features of parental tumors. Neuro Oncol 2017;19(2):219–28 doi 10.1093/neuonc/now160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Francis JM, Zhang CZ, Maire CL, Jung J, Manzo VE, Adalsteinsson VA, et al. EGFR variant heterogeneity in glioblastoma resolved through single-nucleus sequencing. Cancer Discov 2014;4(8):956–71 doi 10.1158/2159-8290.CD-13-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell 2011;20(6):810–7 doi 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 32.deCarvalho AC, Kim H, Poisson LM, Winn ME, Mueller C, Cherba D, et al. Discordant inheritance of chromosomal and extrachromosomal DNA elements contributes to dynamic disease evolution in glioblastoma. Nat Genet 2018;50(5):708–17 doi 10.1038/s41588-018-0105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bruna A, Rueda OM, Greenwood W, Batra AS, Callari M, Batra RN, et al. A Biobank of Breast Cancer Explants with Preserved Intra-tumor Heterogeneity to Screen Anticancer Compounds. Cell 2016;167(1):260–74 e22 doi 10.1016/j.cell.2016.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ben-David U, Ha G, Tseng YY, Greenwald NF, Oh C, Shih J, et al. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat Genet 2017;49(11):1567–75 doi 10.1038/ng.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baskaran S, Mayrhofer M, Kultima HG, Bergstrom T, Elfineh L, Cavelier L, et al. Primary glioblastoma cells for precision medicine: a quantitative portrait of genomic (in)stability during the first 30 passages. Neuro Oncol 2018;20(8):1080–91 doi 10.1093/neuonc/noy024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005;352(10):987–96 doi 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 37.Izumchenko E, Paz K, Ciznadija D, Sloma I, Katz A, Vasquez-Dunddel D, et al. Patient-derived xenografts effectively capture responses to oncology therapy in a heterogeneous cohort of patients with solid tumors. Ann Oncol 2017;28(10):2595–605 doi 10.1093/annonc/mdx416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weroha SJ, Becker MA, Enderica-Gonzalez S, Harrington SC, Oberg AL, Maurer MJ, et al. Tumorgrafts as in vivo surrogates for women with ovarian cancer. Clin Cancer Res 2014;20(5):1288–97 doi 10.1158/1078-0432.CCR-13-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Migliardi G, Sassi F, Torti D, Galimi F, Zanella ER, Buscarino M, et al. Inhibition of MEK and PI3K/mTOR suppresses tumor growth but does not cause tumor regression in patient-derived xenografts of RAS-mutant colorectal carcinomas. Clin Cancer Res 2012;18(9):2515–25 doi 10.1158/1078-0432.CCR-11-2683. [DOI] [PubMed] [Google Scholar]
- 40.Townsend EC, Murakami MA, Christodoulou A, Christie AL, Koster J, DeSouza TA, et al. The Public Repository of Xenografts Enables Discovery and Randomized Phase II-like Trials in Mice. Cancer Cell 2016;30(1):183 doi 10.1016/j.ccell.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 41.Gao H, Korn JM, Ferretti S, Monahan JE, Wang Y, Singh M, et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med 2015;21(11):1318–25 doi 10.1038/nm.3954. [DOI] [PubMed] [Google Scholar]
- 42.Verreault M, Schmitt C, Goldwirt L, Pelton K, Haidar S, Levasseur C, et al. Preclinical Efficacy of the MDM2 Inhibitor RG7112 in MDM2-Amplified and TP53 Wild-type Glioblastomas. Clin Cancer Res 2016;22(5):1185–96 doi 10.1158/1078-0432.CCR-15-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gupta SK, Kizilbash SH, Carlson BL, Mladek AC, Boakye-Agyeman F, Bakken KK, et al. Delineation of MGMT Hypermethylation as a Biomarker for Veliparib-Mediated Temozolomide-Sensitizing Therapy of Glioblastoma. J Natl Cancer Inst 2016;108(5) doi 10.1093/jnci/djv369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krepler C, Sproesser K, Brafford P, Beqiri M, Garman B, Xiao M, et al. A Comprehensive Patient-Derived Xenograft Collection Representing the Heterogeneity of Melanoma. Cell Rep 2017;21(7):1953–67 doi 10.1016/j.celrep.2017.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stewart E, Federico SM, Chen X, Shelat AA, Bradley C, Gordon B, et al. Orthotopic patient-derived xenografts of paediatric solid tumours. Nature 2017;549(7670):96–100 doi 10.1038/nature23647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brabetz S, Leary SES, Grobner SN, Nakamoto MW, Seker-Cin H, Girard EJ, et al. A biobank of patient-derived pediatric brain tumor models. Nat Med 2018;24(11):1752–61 doi 10.1038/s41591-018-0207-3. [DOI] [PubMed] [Google Scholar]
- 47.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2(5):401–4 doi 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Ellison DW, Figarella-Branger D, et al. WHO Classification of Tumours of the Central Nervous System. Lyon: International Agency for Research on Cancer; 2016. 408 p. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.